Abstract
In the previous study, it was found that long intergenic noncoding RNA-p21 (lincRNA-p21) was downregulated in hepatocellular carcinoma (HCC) and lincRNA-p21 overexpression inhibited tumor invasion through inducing epithelial–mesenchymal transition. However, the underlying mechanism was not fully elaborated. In this study, lincRNA-p21 expression was measured in 12 paired HCC and nontumor adjacent normal tissues by quantitative real-time polymerase chain reaction. The effects of lincRNA-p21 on HCC cells were studied using lentivirus expressing lincRNA-p21 vector in vitro. The association between lincRNA-p21 level and miR-9 level was tested with the Spearman rank correlation. The effects of miR-9 on HCC cells were studied by using miR-9 inhibitor in vitro. Luciferase assay was used to validate the target of miR-9. The results showed that lincRNA-p21 was downregulated in human HCC tissues and cell lines. LincRNA-p21 overexpression significantly inhibited HCC cell migration and invasion in vitro. Besides, lincRNA-p21 negatively regulated miR-9 expression level, and miR-9 was upregulated in human HCC tissues and cells. MiR-9 knockdown inhibited HCC cell migration and invasion in vitro. Finally, the luciferase assay results showed that E-cadherin was a direct target of miR-9. The expression level of E-cadherin was found to be regulated by lincRNA-p21 and miR-9. Altogether, the results suggested that lincRNA-p21 inhibits migration and invasion of HCC cells through regulating miR-9-mediated E-cadherin cascade signaling pathway.
Keywords: hepatocellular carcinoma, lincRNA-p21, miR-9, E-cadherin, epithelial–mesenchymal transition
Introduction
Hepatocellular carcinoma (HCC) is a highly aggressive solid malignancy and is the second leading cause of cancer-related death worldwide.1,2 Despite the significant advances in diagnosis and management of HCC, the 5-year overall survival rate remains very poor. Thus, understanding the molecular mechanisms of HCC is of great importance for the development of therapeutic strategies or applicable diagnostics for HCC.
The long noncoding RNAs (lncRNAs) are messenger RNA (mRNA)-like transcripts longer than 200 bp that have no protein-coding potential.3 It has been confirmed that lncRNAs play important regulatory roles in HCC carcinogenesis and cancer progression including cancer cell growth, cell differentiation, cell metastasis and fate decision.4,5 The most widely investigated lncRNAs in HCC include MALAT1, HOX antisense intergenic RNA H19, MEG3, MVIH and HULC.6 Besides, in the previous study, lincRNA-p21, a long intergenic noncoding RNA (lincRNA), was identified as an lincRNA that inhibits invasion and metastasis of HCC, and the underlying mechanism was found to be associated with the enhancement of epithelial–mesenchymal transition (EMT).7 The aim of this study was to explore the further mechanism underlying the effects of lincRNA-p21 on EMT in HCC cells.
Some evidence demonstrated that lincRNAs serve as miRNA sponges/decoys, have the capacity to decrease the amount of available miRNAs and thus contribute to the enhanced translations of their target mRNAs.8 For example, linc-MD1, a muscle-specific lincRNA, could enhance the mRNA expression of MAML1 and MEF2C by sponging miR-135 and miR-133 away from target mRNAs.9 Besides, lincRNA HOTAIR was reported to regulate HER2 expression by sponging miR-331-3p in gastric cancer, functioning as a competing endogenous RNA.10 Based on these investigations and the previous study, it was thus assumed that lincRNA-p21 could regulate its downstream miRNA to induce EMT, which finally affects the growth of HCC cells.
Tan et al indicated that miR-9 was highly expressed in HCC cell line and downregulation of miR-9 significantly reduced HCC cell invasion.11 Mechanism study suggested that EMT-related protein E-cadherin, a tumor invasion suppressor in HCC, was identified as a putative gene target of miR-9.11 Besides, the previous study by the authors of this current paper showed that the expression of E-cadherin was upregulated by overexpression of lincRNA-p21.7 This has led to the speculation that lincRNA-p21 may regulate the expression of E-cadherin via mediating miR-9 expression, finally leading to the inhibition of HCC progression and development. The aim of this study is to validate this speculation.
Materials and methods
Human samples
This study enrolled 12 patients with HCC, who had underwent surgical removal of tumors at the Department of Infectious Diseases of People’s Hospital of Zhengzhou University, Henan Provincial People’s Hospital (Zhengzhou, Henan, China). Partial tumors and nontumor adjacent normal tissues were obtained during surgery, frozen immediately with liquid nitrogen and stored in a freezer at −70°C. All HCC patients had no history of other malignancies. Written informed consent was obtained from all the patients. The study protocol was approved by the Ethics Committee of People’s Hospital of Zhengzhou University (Zhengzhou, Henan, China).
Cell culture, cell transfection and retroviral transduction
A normal liver cell line L02 and human HCC cell lines HepG2, SMMC7721, Hep3B, MHCC97H and SK-Hep1 were obtained from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China). Cells were cultured in a 5% CO2 humidified incubator at 37°C, and were maintained in Dulbecco’s Modified Eagle’s Medium supplemented with 10% heat-inactivated fetal bovine serum (FBS), 100 μg/mL streptomycin and 100 units/mL penicillin. MiR-9 mimics and miR-9 inhibitor were synthesized by GenePharma Corporation (Shanghai, China). Lipofectamine 2000 reagents (Thermo Fisher Scientific, Waltham, MA, USA) were used for oligonucleotide transfection. Anti-lincRNA-p21 and control lentivirus particles were purchased from GenePharma Corporation. Lentivirus expressing human lincRNA-p21 was constructed as previously described.12 For transduction, cells were incubated with virus-containing supernatant in the presence of 8 mg/mL polybrene for 48 hours. Then, the infected cells were incubated for 72 hours with hygromycin (200 mg/mL) or puromycin (2 mg/mL).
Cell migration and invasion assays
For the detection of in vitro cell migration and invasion ability, the 24-well Transwell chambers (Corning Incorporated, Corning, NY, USA) were used. The cells (5×104 per well) were cultured in the top chamber with serum-free media. Five hundred microliters of media with 10% FBS were added as a chemoattractant in the lower chamber. The media were removed from the chamber and the Transwell after cultivation for 24 hours. A cotton swab was used to gently wipe the chamber. The migrated cells were fixed in 4% paraformaldehyde and were then stained with crystal violet solution and finally counted under a microscope. For cell invasion assay, the Transwell membranes were precoated with Matrigel (BD Biosciences, San Jose, CA, USA), and the other procedures were similar to the cell migration assay.
Quantitative real-time polymerase chain reaction (qRT-PCR) analysis
Total RNA was extracted using TRIzol reagent according to the manufacturer’s protocol (Thermo Fisher Scientific). RNA was reverse-transcribed into cDNA by Quantscript RT Kit (Tiangen, Beijing, China). For detection of miR-9 expression level, TaqMan® 2× Universal PCR Master Mix (Thermo Fisher Scientific) was used, and qRT-PCR was performed on CFX96™ Real-Time PCR Detection System (Bio-Rad Laboratories, Hercules, CA, USA). The primers for miR-9 were as follows: forward, 5′-GTGCAGGGTCCGAGGT-3′; reverse, 5′-GCGCTCTTTGGTTATCTAGC-3′. U6-F was used as a reference gene, and the primers were as follows: forward, 5′-CTCGCTTCGGCAGCACA-3′; reverse, 5′-AACGCTTCACGAATTTGCGT-3′. For detection of the abundance of lincRNA-p21 transcript, total RNA was extracted 48 hours later and transcribed into cDNAs using Rever Tra Ace qPCR RT Kit (Toyobo, Osaka, Japan); the primers for lincRNA-p21 and reference gene and procedures were described in the previous study.7
Luciferase activity assay
By using the Dual-Luciferase Reporter Assay System (Promega, Fitchburg, WI, USA), the luciferase reporter gene assay was performed. Cells were seeded in 96-well plates, and wild-type or mutant reporter constructs (termed wild or mut) were co-transfected into HepG2 and SK-Hep1 cells with miR-9 (100 nmol/L) or miR-NC (100 nmol/L) and Renilla plasmid by using lipofectamine 2000 (Thermo Fisher Scientific). Dual-luciferase assay (Promega) was performed 48 hours after transfection.
Western blotting analysis
Total protein was isolated, and bicinchoninic acid protein assay (Beyotime, Haimen, China) was used to determine the protein concentration. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis was used to separate the proteins. The protein was then transferred onto a nitrocellulose membrane (EMD Millipore, Billerica, MA, USA). The membrane was then blocked at room temperature for 30 minutes and incubated with primary antibody anti-E-cadherin (Santa Cruz Biotechnology, Inc, Dallas, TX, USA) at 4°C overnight. By incubation with horseradish peroxidase-conjugated secondary antibody at room temperature for 1 hour, the protein bands were detected. The chemiluminescence reagent (Pierce, Waltham, MA, USA) was used for visualization.
Statistical analysis
All graphical data were presented as mean ± standard deviation. Data and statistics were analyzed using GraphPad Prism version 5 software (GraphPad Software, Inc., La Jolla, CA, USA). The Student’s t-test or one-way analysis of variance test was used to determine the significant difference between groups. The association between lincRNA-p21 level and miR-9 level was tested with the Spearman rank correlation. A statistically significant difference was indicated as P<0.05.
Results
LincRNA-p21 is downregulated in human HCC and inhibits HCC cell migration and invasion
To investigate the functions of lincRNA-p21 in HCC, its expression level in HCC tissues and nontumor adjacent normal tissues was detected (n=12 in each group). The results showed that lincRNA-p21 level was significantly downregulated in HCC tissues (tumor) than nontumor adjacent normal tissues (normal) (Figure 1A). It was also found that lincRNA-p21 level was significantly downregulated in HCC cell lines (HepG2, SMMC7721, Hep3B, MHCC97H and SK-Hep1) as compared to the normal cell line L02 (Figure 1B). Especially, HepG2 and SK-Hep1 cells were selected for the following investigation in the present work.
Figure 1.
LincRNA-p21 is downregulated in human HCC and inhibits HCC cell migration and invasion.
Notes: (A) LincRNA-p21 level was significantly reduced in tissues from HCC (tumor) than that from nontumor adjacent normal tissues (normal) (n=12 in each group). (B) The low level of lincRNA-p21 was observed in HCC cell lines (HepG2, SMMC7721, Hep3B, MHCC97H and SK-Hep1). (C) Real-time quantitative polymerase chain reaction analysis was used to detect the transduction efficiency of lentivirus expressing lincRNA-p21 vector in HepG2 and SK-Hep1 cells. (D) LincRNA-p21 overexpression significantly inhibited the migration of HepG2 and SK-Hep1 cells. (E) LincRNA-p21 overexpression markedly reduced invasion of HepG2 and SK-Hep1 cells. A statistically significant difference was indicated as P<0.05. *P<0.05 and **P<0.01 vs the corresponding controls. The magnification in D and E = ×100. Black dots and squares represent human samples.
Abbreviations: lincRNA, long intergenic noncoding RNA; HCC, hepatocellular carcinoma; qRT-PCR, quantitative real-time polymerase chain reaction.
LincRNA-p21 was overexpressed in HepG2 and SK-Hep1 cell lines (Figure 1C). It was found that lincRNA-p21 overexpression significantly inhibited the migration of HepG2 and SK-Hep1 cells (Figure 1D). LincRNA-p21 overexpression also markedly inhibited invasion of HepG2 and SK-Hep1 cells (Figure 1E).
The association between lincRNA-p21 expression and miR-9 level
As shown in Figure 2A, it was found that lincRNA-p21 overexpression significantly inhibited the expression level of miR-9 in HepG2 and SK-Hep1 cells. However, lincRNA-p21 knockdown (anti-lincRNA-p21) significantly enhanced the expression level of miR-9 in HepG2 and SK-Hep1 cells (Figure 2B). Then, the expression level of miR-9 was detected in HCC tissues, and the results revealed that miR-9 level was significantly upregulated in HCC tissues than normal tissues (Figure 2C). Besides, the Spearman rank correlation analysis showed the negative relationship between lincRNA-p21 expression and miR-9 level (Figure 2D).
Figure 2.
The association between lincRNA-p21 expression and miR-9 level.
Notes: (A) LincRNA-p21 overexpression significantly inhibited the expression level of miR-9 in HepG2 and SK-Hep1 cells. (B) LincRNA-p21 knockdown (anti-lincRNA-p21) significantly enhanced the expression level of miR-9 in HepG2 and SK-Hep1 cells. (C) MiR-9 level was significantly induced in tissues from HCC (tumor) than that from nontumor adjacent normal tissues (normal). (D) The Spearman rank correlation analysis showed the negative relationship between lincRNA-p21 expression and miR-9 level. *P<0.05 and **P<0.01 vs the corresponding controls. Black dots and squares represent human samples.
Abbreviations: lincRNA, long intergenic noncoding RNA; HCC, hepatocellular carcinoma.
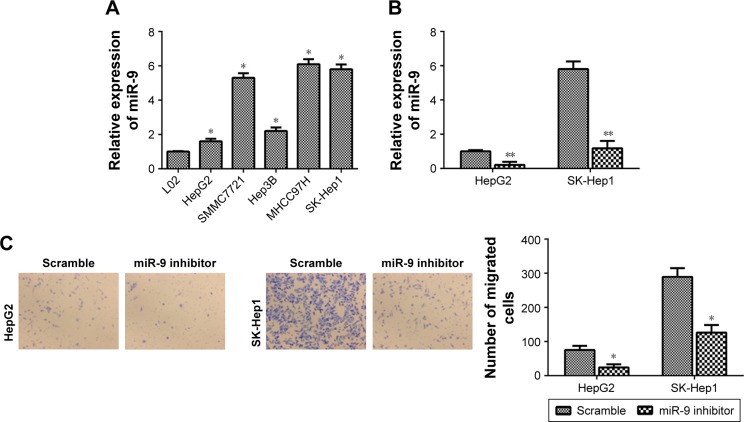
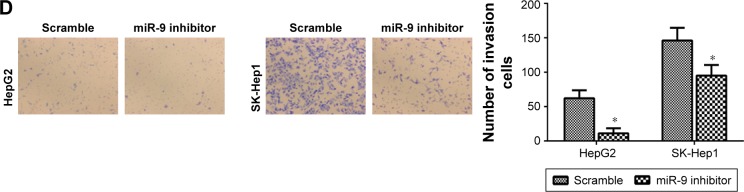
MiR-9 knockdown inhibits HCC cell migration and invasion
The findings of this study strongly implicated the participation of miR-9 in human HCC, which prompted us to study its functions in HCC. First, the expression level of miR-9 was detected in HepG2, SMMC7721, Hep3B, MHCC97H and SK-Hep1 cells. Interestingly, miR-9 was relatively higher in HepG2 and SK-Hep1 cells than other HCC cell lines (Figure 3A). Then, miR-9 was knocked down in HepG2 and SK-Hep1 cells (Figure 3B). It was found that miR-9 knockdown inhibited the migration of HepG2 and SK-Hep1 cells (Figure 3C). MiR-9 knockdown also reduced the invasion of HepG2 and SK-Hep1 cells (Figure 3D).
Figure 3.
MiR-9 knockdown inhibits HCC cell migration and invasion.
Notes: (A) The expression level of miR-9 in HepG2, SMMC7721, Hep3B, MHCC97H and SK-Hep1 cells. (B) qRT-PCR analysis was used to detect the transfection efficiency of miR-9 inhibitor in HepG2 and SK-Hep1 cells. (C) MiR-9 knockdown inhibited the migration of HepG2 and SK-Hep1 cells. (D) MiR-9 knockdown also reduced the invasion of HepG2 and SK-Hep1 cells. A statistically significant difference was indicated as P<0.05. *P<0.05 and **P<0.01 vs the corresponding controls. The magnification in C and D = ×100.
Abbreviations: HCC, hepatocellular carcinoma; qRT-PCR, quantitative real-time polymerase chain reaction.
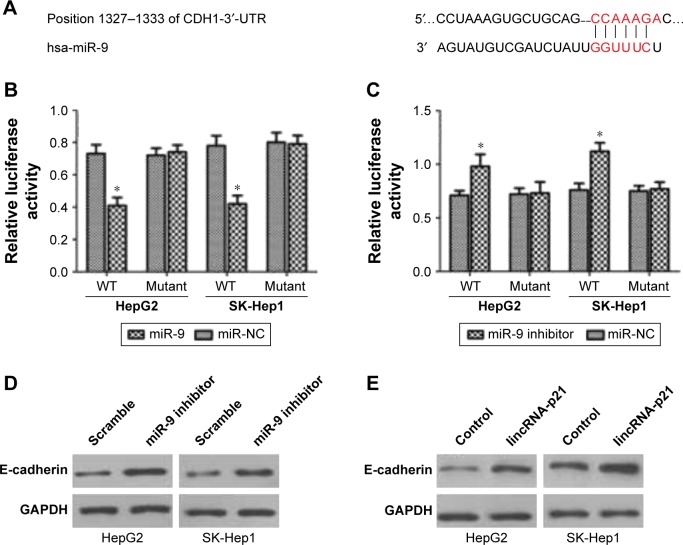
E-cadherin is a direct target of miR-9
A previous study indicated that E-cadherin (CDH1) was a putative gene target of miR-9 by TargetScan and PicTar programs.11 These programs showed that miR-9 may target sequence at nt1327–1333 of the CDH1-3′-UTR (Figure 4A). The direct effect of miR-9 on the translation of CDH1 mRNA into protein was measured by luciferase reporter assay in that study. Fluorescence activity was significantly decreased after cells were co-transfected with the miR-9 mimic and 3′-UTR of CDH1 mRNA luciferase reporter vector as compared to cells transfected with 3′-UTR of CDH1 luciferase reporter vector and miR-NC (Figure 4B). However, fluorescence activity was significantly increased after cells were co-transfected with the miR-9 inhibitor and 3′-UTR of CDH1 mRNA luciferase reporter vector as compared to cells transfected with 3′-UTR of CDH1 luciferase reporter vector and miR-NC (Figure 4C). Besides, it was found that miR-9 knockdown increased CDH1 expression in HepG2 and SK-Hep1 cells (Figure 4D), and lincRNA-p21 overexpression also increased CDH1 expression in HepG2 and SK-Hep1 cells (Figure 4E).
Figure 4.
E-cadherin is a target of miR-9.
Notes: (A) MiR-9 target sequence was at nt1327–1333 of the CDH1-3′-UTR. (B) Fluorescence activity was significantly decreased after cells were co-transfected with the miR-9 mimic and 3′-UTR of CDH1 mRNA luciferase reporter vector as compared to cells transfected with 3′-UTR of CDH1 luciferase reporter vector and miR-NC. (C) Fluorescence activity was significantly increased after cells were co-transfected with the miR-9 inhibitor and 3′-UTR of CDH1 mRNA luciferase reporter vector as compared to cells transfected with 3′-UTR of CDH1 luciferase reporter vector and miR-NC. (D) MiR-9 knockdown increased CDH1 expression in HepG2 and SK-Hep1 cells. (E) LincRNA-p21 overexpression also increased CDH1 expression in HepG2 and SK-Hep1 cells. A statistically significant difference was indicated as P<0.05. *P<0.05 vs the corresponding controls.
Abbreviations: lincRNA, long intergenic noncoding RNA; mRNA, messenger RNA; WT, wild type.
Discussion
In the previous study, it was found that the lincRNA-p21 expression was downregulated in HCC tissues and cells, and lincRNA-p21 overexpression contributed to the inhibition of invasion and metastasis in HCC.7 This study also demonstrated that lincRNA-p21 expression was down-regulated in human HCC and lincRNA-p21 overexpression inhibited HCC cell migration and invasion, suggesting that lincRNA-p21 acts as a tumor suppressor in HCC. The expression of lincRNA-p21 is also found to be downregulated in human colorectal cancer and prostate cancer.13,14 Besides, a previous study indicated that lincRNA-p21 impacts prognosis in resected non-small-cell lung cancer patients through angiogenesis regulation.15
Recent studies have suggested that lincRNAs can serve as “sponges” or “decoys” for miRNAs, decreasing the amount of available miRNAs and leading to enhanced translations of their target mRNA.8 In this study, lincRNA-p21 overexpression significantly inhibited the expression level of miR-9. However, lincRNA-p21 knockdown significantly enhanced the expression level of miR-9. Besides, the Spearman rank correlation analysis showed the negative relationship between lincRNA-p21 expression and miR-9 level. This study indicates that lincRNA-p21 can decrease the expression of miR-9 by serving as “sponges” or “decoys” for miR-9.
The effects of miR-9 were also explored in this study. It was revealed that miR-9 was upregulated in human HCC tissues and cells. Besides, miR-9 knockdown markedly inhibited HCC cell migration and invasion. Cai et al suggested that upregulation of miR-9 expression predicate advanced clinicopathological features and poor prognosis in HCC patients.16 Sun et al indicated that miR-9 could act as a HCC tumor activator and might be a potentially valuable biomarker for the prognosis in HCC patients.17 These research indicated the important role of miR-9 in HCC progression and development.
A previous study suggested that E-cadherin, a tumor invasion suppressor in HCC, was a putative gene target of miR-9.11 By luciferase assay, E-cadherin was identified as a direct target of miR-9. Besides, it was found that miR-9 knockdown increased CDH1 expression, and lincRNA-p21 overexpression also increased CDH1 expression. The previous study by the authors of this current paper also showed that the expression of E-cadherin was upregulated by overexpression of lincRNA-p21.7 To some extent, this study suggests that lincRNA-p21 regulates the expression of miR-9 and leads to the altered expression of E-cadherin, which finally contributes to the inhibition of HCC cell migration and invasion.
Conclusion
To sum up, this study revealed that lincRNA-p21 was downregulated in human HCC tissues and its overexpression significantly inhibited HCC cell migration and invasion in vitro. Besides, lincRNA-p21 negatively regulated miR-9 expression level. MiR-9 was upregulated in human HCC tissues and cell lines. MiR-9 knockdown inhibited HCC cell migration and invasion. Finally, the results suggested that E-cadherin is a direct target of miR-9. Altogether, this study indicated that lincRNA-p21 inhibits migration and invasion of HCC through miR-9/E-cadherin cascade signaling pathway.
Acknowledgments
This research was in part supported by the National Key Clinical Specialty Construction Projects of China.
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Yang X, Xie X, Xiao YF, et al. The emergence of long non-coding RNAs in the tumorigenesis of hepatocellular carcinoma. Cancer Lett. 2015;360(2):119–124. doi: 10.1016/j.canlet.2015.02.035. [DOI] [PubMed] [Google Scholar]
- 2.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. CA Cancer J Clin. 2012;62(1):10–29. doi: 10.3322/caac.20138. [DOI] [PubMed] [Google Scholar]
- 3.Tay Y, Rinn J, Pandolfi PP. The multilayered complexity of ceRNA crosstalk and competition. Nature. 2014;505(7483):344–352. doi: 10.1038/nature12986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Geisler S, Coller J. RNA in unexpected places: long non-coding RNA functions in diverse cellular contexts. Nat Rev Mol Cell Biol. 2013;14(11):699–712. doi: 10.1038/nrm3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ulitsky I, Bartel DP. LincRNAs: genomics, evolution, and mechanisms. Cell. 2013;154(1):26–46. doi: 10.1016/j.cell.2013.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yu Z, Dong F. Long non-coding RNAs and hepatocellular carcinoma (Review) Mol Clin Oncol. 2015;3(1):13–17. doi: 10.3892/mco.2014.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jia M, Jiang L, Wang YD, Huang JZ, Yu M, Xue HZ. LincRNA-p21 inhibits invasion and metastasis of hepatocellular carcinoma through Notch signaling-induced epithelial-mesenchymal transition. Hepatol Res. 2016;46(11):1137–1144. doi: 10.1111/hepr.12659. [DOI] [PubMed] [Google Scholar]
- 8.Deng K, Wang H, Guo X, Xia J. The cross talk between long, non-coding RNAs and microRNAs in gastric cancer. Acta Biochim Biophys Sin. 2016;48(2):111–116. doi: 10.1093/abbs/gmv120. [DOI] [PubMed] [Google Scholar]
- 9.Cesana M, Cacchiarelli D, Legnini I, et al. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell. 2011;147(2):358–369. doi: 10.1016/j.cell.2011.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu XH, Sun M, Nie FQ, et al. Lnc RNA HOTAIR functions as a competing endogenous RNA to regulate HER2 expression by sponging miR-331-3p in gastric cancer. Mol Cancer. 2014;13(1):2739–2748. doi: 10.1186/1476-4598-13-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tan HX, Wang Q, Chen LZ, et al. MicroRNA-9 reduces cell invasion and E-cadherin secretion in SK-Hep-1 cell. Med Oncol. 2010;27(3):654–660. doi: 10.1007/s12032-009-9264-2. [DOI] [PubMed] [Google Scholar]
- 12.Yang N, Fu Y, Zhang H, Hui S, Zhu N, Yang G. LincRNA-p21 activates endoplasmic reticulum stress and inhibits hepatocellular carcinoma. Oncotarget. 2015;6(29):28151–28163. doi: 10.18632/oncotarget.4661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhai H, Fesler A, Schee K, Fodstad Ø, Flatmark K, Ju J. Clinical significance of long intergenic noncoding RNA-p21 in colorectal cancer. Clin Colorectal Cancer. 2013;12(4):261–266. doi: 10.1016/j.clcc.2013.06.003. [DOI] [PubMed] [Google Scholar]
- 14.Wang X, Ruan Y, Wang X, et al. Long intragenic non-coding RNA lincRNA-p21 suppresses development of human prostate cancer. Cell Prolif. 2017;50(2) doi: 10.1111/cpr.12318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yoon JH, Abdelmohsen K, Srikantan S, et al. LincRNA-p21 suppresses target mRNA translation. Mol Cell. 2012;47(4):648–655. doi: 10.1016/j.molcel.2012.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cai L, Xi C. Up-regulation of miR-9 expression predicate advanced clinicopathological features and poor prognosis in patients with hepatocellular carcinoma. Diagn Pathol. 2014;9(1):1000. doi: 10.1186/s13000-014-0228-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sun J, Fang K, Shen H, Qian Y. MicroRNA-9 is a ponderable index for the prognosis of human hepatocellular carcinoma. Int J Clin Exp Med. 2015;8(10):17748. [PMC free article] [PubMed] [Google Scholar]