Abstract
Mutations in growth factor receptor signaling pathways are common in cancer cells, including the highly lethal brain tumor glioblastoma (GBM) where they drive tumor growth through mechanisms including altering the uptake and utilization of nutrients. However, the impact of changes in micro-environmental nutrient levels on oncogenic signaling, tumor growth and drug resistance is not well understood. We recently tested the hypothesis that external nutrients promote GBM growth and treatment resistance by maintaining the activity of mechanistic target of rapamycin complex 2 (mTORC2), a critical intermediate of growth factor receptor signaling, suggesting that altered cellular metabolism is not only a consequence of oncogenic signaling, but also potentially an important determinant of its activity. Here, we describe the studies that corroborate the hypothesis and propose others that derive from them. Notably, this line of reasoning raises the possibility that systemic metabolism may contribute to responsiveness to targeted cancer therapies.
Keywords: genetics, genetic-environment interaction, glioblastoma, metabolic reprogramming, mTOR
Introduction
Normal cells require growth factors to instruct them to take up and utilize nutrients. In contrast, mutations in the growth factor receptor signaling apparatus can render tumor cells independent of such cues, enabling them to metabolize nutrients in a cell-autonomous fashion [1]. The enhanced flux of nutrients into metabolic intermediates enables tumor cells to meet their energetic and anabolic demands, and may also globally alter gene transcription and the epigenetic landscape through generation of acetyl coenzyme A (acetyl-CoA) [2,3]. However, the effect of altered external nutrient levels on oncogenic signaling and therapeutic response is not well understood. Here, we propose three main and testable hypotheses. First, we hypothesize that altered cellular metabolism is not only a consequence of oncogenic signaling, but also potentially an important determinant of its activity. Second, we hypothesize that mTORC2, a component of the growth factor receptor signaling system, is a critical node that integrates growth factor receptor signaling with nutrient availability, influencing tumor growth and response to treatment. Third, we hypothesize that glucose or acetate-derived acetyl-CoA directly contributes to tumor growth and drug resistance by acetylation of Rictor, a core component of mTORC2 to promote its activity. The evidence for these three hypotheses is based on a recent study of the highly lethal brain cancer glioblastoma (GBM) [4].
GBM is characterized by frequent growth factor receptor pathway mutations and by altered cellular metabolism [5]. GBM was one of the first cancers sequenced by The Cancer Genome Atlas (TCGA) project and is now one of the most genomically well-characterized forms of human cancer [6,7]. Mutations in protein coding genes occurring at frequencies ≥5% above background are likely to have been identified [8], revealing a mutational landscape dominated by genetic alterations that potently activate growth factor receptor signaling pathways including the epidermal growth factor receptor (EGFR) [7], converging on the phosphoinositide 3-kinase (PI3K)-Akt-mechanistic target of rapamycin (mTOR) signaling network [3,5,7,9,10]. PI3K can also be hyper-activated in tumor cells by non-mutational events including hypoxia, partly through activation of the insulin and insulin-like growth factor (IGF) receptors [11–13].
We recently identified a new mechanism by which altered growth factor receptor signaling through the PI3K/Akt pathway in GBM cells is “tuned” in response to glucose and acetate levels via acetylation of Rictor, a core component of mTORC2. Thus in GBM cells, glucose or acetate are sufficient to maintain mTORC2 activity, rendering GBMs resistant to EGFR, PI3K and Akt- targeted treatments when extracellular nutrient levels are sufficiently high.
mTORC2, a critical component of altered growth factor receptor signaling
mTOR is a serine/threonine kinase that exists in two distinct complexes, mTORC1 and mTORC2, which differ in their regulation, function and responsiveness to the allosteric inhibitor, rapamycin [14,15]. mTORC1 contains mTOR kinase in association with Raptor, PRAS40, mLST8, Deptor and Tti1/Tel2. mTORC1 is a validated cancer target, integrating growth factor receptor signaling, energy status and amino acid sensing, to regulate downstream activity, including protein translation, cellular metabolism and nucleotide synthesis [16]. In contrast, mTORC2 is less well-understood. mTORC2 is composed of mTOR kinase in association with Rictor, mSIN1, Protor1/2, mLST8, Deptor and Tti1/Tel2 [14,15]. Upon growth factor receptor activation [10,17], the plasma membrane-embedded phospholipid, phosphatidylinositol (3,4,5)-trisphosphate (PIP3), physically interacts with mSIN1 to activate mTORC2 kinase activity, which consequently phosphorylates Akt at Ser473 [18] and two other members of the protein kinase A/G/C (AGC) subfamilies, PKCα, and serum and glucocorticoid-inducible kinase 1 (SGK1) [19,20]. Mutations in growth factor receptor signaling components including EGFR alterations, PIK3CA mutations and PTEN deletions highlight potential therapeutic pathways [5,7], which converge on mTORC2 to promote cellular proliferation and tumor growth [10,21,22].
mTORC2 signaling is elevated in many cancers, including GBM
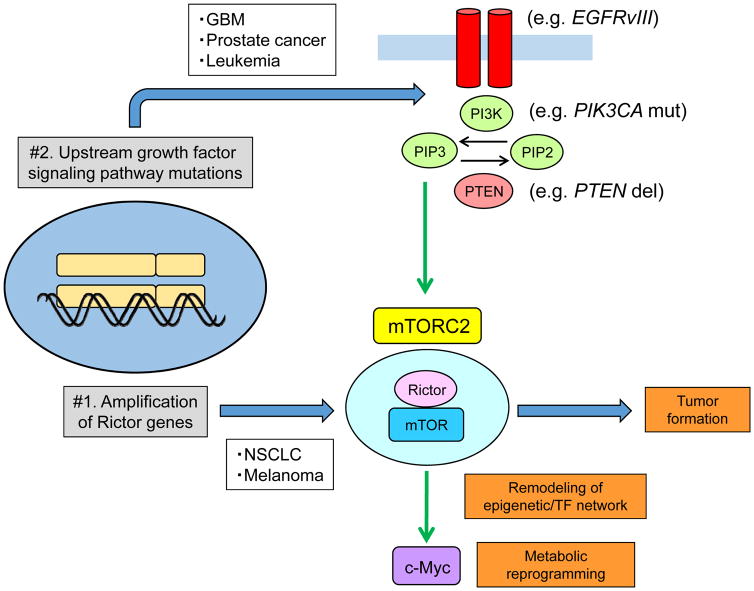
mTORC2 signaling can be aberrantly activated in cancer in two ways (Fig. 1). In non-small cell lung cancer and melanoma, Rictor amplification drives persistent mTORC2 signaling [23,24]. In prostate cancer, leukemia and GBM, mTORC2 signaling is hyper-activated by upstream growth factor signaling pathway mutations [9,10,22,25].
Figure 1.
mTORC2 signaling is elevated in many cancers via two distinct mechanisms.
In non-small cell lung cancer and melanoma, Rictor amplification drives mTORC2 signaling while mTORC2 signaling is hyper-activated by upstream growth factor signaling pathway mutations in prostate cancer, leukemia and GBM. Hyper-activated mTORC2 can drive tumor formation, remodel an epigenome and transcription factor network, and facilitate metabolic reprogramming through an oncogenic transcription factor c-Myc. del, deletion; EGFRvIII, epidermal growth factor receptor variant III; GBM, glioblastoma multiforme; mut, mutation; NSCLC, non-small cell lung cancer; PI3K, phosphoinositide 3-kinase; PIK3CA, phosphatidylinositol (4,5)-bisphosphate 3-kinase catalytic subunit alpha; PIP2, phosphatidylinositol (4,5)-bisphosphate; PIP3, phosphatidylinositol (3,4,5)-trisphosphate; PTEN, phosphatase and tensin homolog deleted on chromosome 10; TF, transcriptional factor.
In a Drosophila glioma model driven by persistently activated EGFR, RAS and PI3K signaling, loss of function Rictor and SIN1 alleles prevented glioma formation, demonstrating that mTORC2 is required for glioma formation [21]. Consistent with these data, conditional Rictor overexpression in astrocytes was sufficient to cause gliomas in mice [26]. These results indicate that mTORC2 may play an essential role in GBM pathogenesis, but its underlying molecular mechanisms are not understood.
We recently showed that mTORC2 promotes GBM growth and drug resistance, at least in part, through metabolic reprogramming. Many types of cancer cells, including GBM cells, convert the majority of the glucose they take up into lactate, providing a supply of glycolytic intermediates as carbon-containing precursors for macromolecular biosynthesis (i.e. the Warburg effect), even in the presence of sufficient oxygen to support oxidative phosphorylation [27–29]. This biochemical adaptation enables cancer cells to meet the coordinately elevated anabolic and energetic demands imposed by rapid tumor growth. We found that mTORC2, which is potently activated by EGFRvIII, the most common gain of function EGFR mutation in GBM, promotes the Warburg effect by controlling c-Myc through a novel acetylation dependent cascade to drive tumor growth [3,9]. We also showed that EGFRvIII remodels an epigenome and transcription factor network that regulates c-Myc, including via mTORC2, to control GBM metabolism [30]. Thus, mTORC2 plays a central role in GBM pathogenesis downstream of altered growth factor receptor signaling, at least in part, by reprogramming cellular metabolism.
mTORC2 signaling is regulated by glucose and acetate through acetyl-CoA
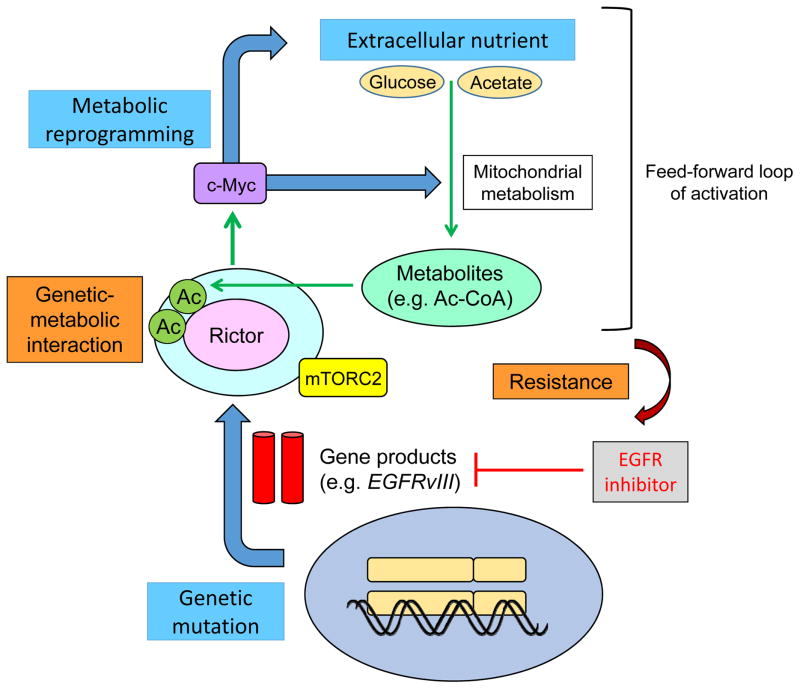
Given the importance of mTORC2-dependent metabolic reprogramming in GBM pathogenesis [3,9], we asked whether mTORC2 signaling itself, could be responsive to the nutrient microenvironment, and made the surprising discovery that glucose or acetate, two “fuel sources” that are widely available in the brain and readily taken up by tumor cells [31,32] are required to activate oncogenic EGFR-mTOR signaling and promote tumor growth [4] (Fig. 2).
Figure 2.
Extracellular nutrients are required to maintain growth factor receptor signaling through mTORC2.
mTORC2 forms an auto-activation loop (i) by promoting glucose/acetate uptake and acetyl-CoA production through its downstream pathways of c-Myc [11] and (ii) by an activation of mTORC2 through acetyl-CoA-dependent acetylation of Rictor [36]. By these mechanisms, GBM cells with activated mTORC2 are resistant to molecularly targeted therapies toward their upstream stimulators, including EGFR. This study suggests that altered cellular metabolism is not only a consequence of oncogenic signaling, but also an important determinant of its activity, raising the possibility that systemic metabolism may contribute to resistance to targeted cancer therapies [59]. Ac, acetyl-group; Ac-CoA, acetyl-CoA.
In GBM cells, glucose and acetate-derived carbons can be used to produce the intermediary metabolite acetyl-CoA, the production of which is facilitated by oncogene-driven metabolic reprogramming [3]. Specifically, we found that glucose is metabolized into acetyl-CoA via pyruvate dehydrogenase (PDH) and acetate is similarly metabolized via acetyl-CoA synthetase 2 (ACSS2) to activate mTORC2 signaling in GBM cells. Importantly, a major part of carbon metabolism is regulated by acetyl-CoA which plays a key role in bioenergetics metabolism, cellular proliferation, and the regulation of gene expression [31]. Moreover, acetyl-CoA producing enzymes including PDH, ACSS2 and ATP citrate lyase (ACLY) are required by tumor cells for acetate utilization [31–35]. These results nominate mTORC2 as a critical integrator of nutrient status and growth factor receptor signaling and suggest that this activity is dependent upon levels of acetyl-CoA.
A variety of human tumors overexpress ACSS2, and its activity is responsible for the majority of cellular acetate uptake into both lipids and histones, qualifying ACSS2 as a targetable metabolic “Achilles heel” of a wide array of tumors [31–33,36]. PDH complex (PDC) has also been shown to be present and functional in the nucleus of mammalian cells in a cell-cycle-dependent manner, and also in response to serum, epidermal growth factor or mitochondrial stress [34]. Dynamic, spatial and temporal regulation of acetyl-CoA producing enzymes by oncogenic signaling provides a pathway for nuclear acetyl-CoA synthesis required for histone acetylation and epigenetic regulation [34,35,37]. Further studies will be necessary to determine how altered growth factor receptor signaling regulates these enzymes in order to facilitate post-translational modifications of oncogenic proteins in the cytosol, as well as the epigenetic landscape including histone modifications which will promote the growth of many types of human cancers.
Glucose-dependent Rictor acetylation promotes mTORC2 signaling
Next we assessed how intermediate metabolites, specifically acetyl-CoA, promote mTORC2 oncogenic signaling. Recent mass spectrometry and metabolomics analyses indicate that lysine acetylation is a critical post-translational modification in regulating essential cellular functions [38,39], including the activity of many metabolic enzymes [40–42]. Rictor contains several lysine residues that when acetylated increase mTORC2 activity, thus providing a critical link between nutrient-sensitive apparatus and mTORC2 signaling [43]. Further, the acetyl-CoA produced from glucose and acetate could potentially provide the acetyl group [44] used for Rictor acetylation to regulate mTORC2 signaling.
Biochemical analyses demonstrated that glucose and acetate promote the acetylation of Rictor protein through acetyl-CoA to activate mTORC2 [4]. PDH and ACSS2, which are the critical enzymes for acetyl-CoA production from glucose and acetate, are involved in the Rictor acetylation process, linking nutrient availability and metabolism to oncogenic mTORC2 signaling. Additionally, bioinfomatic analyses suggested that specific, evolutionarily conserved lysine residues within the Rictor protein may be the targets of acetylation that regulate the signaling of mTORC2 [4,43,45,46]. We therefore constructed a mutant of Rictor that substitutes arginine for these specific lysine residues (K1116, K1119, and K1125) which confers resistance to acetylation, and found that the presence of glucose significantly increased the acetylation of the wild-type Rictor construct, but could not promote that of the mutant, indicating that these lysines are essential in the glucose-dependent acetylation of Rictor [4]. Importantly, the substitution of the lysine residues by arginine in Rictor indeed prevents glucose-dependent increase of mTORC2 signaling. The results indicate that glucose promotes Rictor acetylation to activate mTORC2 signaling.
mTORC2 forms an auto-activation loop through HDAC-mediated Rictor acetylation
Protein acetylation, including the acetylation of Rictor, can be controlled through the balance between histone acetyltransferase (HAT) and histone deacetylase (HDAC) activities [43,47,48]. We recently demonstrated that mTORC2 suppresses the activity of class IIa HDACs in EGFR-mutant GBMs through a signal cascade that results in their inactivating phosphorylation [9]. Thus, if class IIa HDACs negatively regulate mTORC2 in response to extracellular nutrient via deacetylation of Rictor, mTORC2 can establish a feed-forward auto-activation loop through inactivation of class IIa HDACs to keep Rictor in an acetylated state, maintaining downstream signaling.
We demonstrated that among AGC subfamily kinases, PKCα phosphorylates and inactivates class IIa HDACs downstream of mTORC2 signaling, and Rictor is in turn physically associated with class IIa HDACs and deacetylated by them [4]. This signaling cascade forms an auto-activation loop of mTORC2 and promotes the activity of mTORC2. Importantly, the circuitry of mTORC2 signaling, inactivating phosphorylation of class IIa HDACs, and Rictor acetylation is coordinately up-regulated in human GBM patients and may be involved in the GBM pathogenesis in the clinic. Together, these results indicate that mTORC2 forms an auto-activation loop through acetyl-CoA- and HDAC-mediated Rictor acetylation, which underlies the mechanism of mTORC2’s response to nutrient availability and metabolic reprogramming in EGFR-mutant GBMs (Fig. 2).
Therapeutic implications: Glucose-dependent Rictor acetylation renders GBM cells resistant to EGFR, PI3K and Akt targeted therapies
We hypothesize that an acetylation-dependent auto-activation loop maintains mTORC2 signaling independent of upstream stimulation. Therefore, GBM cells with activated mTORC2 in the presence of glucose would be expected to be resistant to therapies that target its upstream elements including EGFR and PI3K [49]. Using genetic and pharmacological approaches, we showed that glucose-dependent acetylation of Rictor makes tumor cells resistant to therapies targeting key components of a growth factor receptor signaling pathway commonly hyper-activated in cancer by maintaining persistent downstream signaling (Fig. 2) [4,50]. The observations make the surprising prediction that GBM cells may use nutrients to escape targeted therapies.
These results have a number of potentially important and unanticipated implications. First, the therapeutic efficacy including chemotherapeutics [51] and molecularly targeted treatments [4] could be affected by the local and systemic metabolism, including hyperglycemia, which is common in GBM patients, particularly if they are treated with dexamethasone to reduce brain swelling [52]. Second, these results raise the possibility that diet could potentially influence response to treatment [53]. Third, it is likely that the glucose-dependent mTORC2 regulation is one component of a set of interlacing mechanisms by which tumor cells adjust their activity in response to metabolic changes. Recent work suggests that in addition to modulating Akt signaling in response to high glucose levels, cells can also maintain Akt signaling when glucose levels are low, through heightened activity of protein kinase RNA-like endoplasmic reticulum kinase (PERK) as part of an unfolded protein response [54]. Therefore, tumor cells appear to have evolved multiple complementary mechanisms to control the central nodes of core signaling pathways in response to changes in the nutrient environment, enabling tumor cells to tune their responses to maximize survival [55].
Future Research
In general, many of the epidemiological studies linking cancer propensities with obesity, diabetes, western life style and diet are relatively weak, in part, because they have not been designed to take tumor genotype into account. Thus, moving forward, it will be necessary to design rigorous mechanistic studies that will permit molecular dissection of the interaction between nutrients, metabolism and cancer genotype, and their impact both on tumor pathogenesis, aggressiveness and response to treatment. Further, our studies are based on one tumor type, although the molecular players that drive this behavior are common to many forms of cancer. We have proposed three hypotheses, each of which can be directly tested. First, to determine whether altered cellular metabolism is not only a consequence of oncogenic signaling, but also potentially an important determinant of its activity, it will be necessary to rigorously control diet and nutrient levels and to assess their impact on signal transduction, tumor growth, and response to treatment in genotyped patient-derived orthotopic mouse models and/or mouse genetic models. Critically, it is important to recognize that the micro-environmental levels of specific nutrients are not merely a consequence of the diet, but rather they are regulated by the interplay between dietary uptake, de novo synthesis and cellular utilization [1,56–58]. Thus, directly measuring nutrient levels and tracing their uptake and utilization in tumor tissue will be needed. Importantly, it is essential to recognize that mTORC2 is only one of the critical nodes integrating nutrient status and altered growth factor receptor signaling. Understanding how it interacts with other nutrient sensing pathways will also be important.
Second, to test our hypothesis that mTORC2 integrates growth factor receptor signaling with nutrient availability, influencing tumor growth and response to treatment, it will be necessary to study the role of mTORC2 and its modulation by nutrients in other cancer types including in patient-derived orthotopic or genetic mouse models.
Third, to test the hypothesis that glucose or acetate-derived acetyl-CoA directly contributes to tumor growth and drug resistance by acetylation of Rictor, a core component of mTORC2 to promote its activity, genetic studies will be needed in other cancer models, particularly to confirm the hypothesized importance of Rictor acetylation, and to determine if persistent downstream mTORC2 signaling caused by glucose or acetate is sufficient to maintain tumor growth and cause drug resistance. Further, it will be critical to elucidate the mechanisms by which mTORC2 promotes tumor growth and drug resistance, including by examining the impact of elevated acetyl-CoA levels on enhancer activation and transcriptional reprogramming.
Conclusion and outlook
Cancer is a disease of endogenous somatic mutations. However, tumor development, progression and response to therapy are also profoundly influenced by the exogenous tumor environment. Therefore, in order to truly “personalize” cancer therapy, at least for patients with certain cancers where the epidemiological and molecular links between life style factors and cancer have been suggested [59–61], it may be necessary to broaden the concept of personalization to include an analysis of the ways in which each person’s diet, exercise and medications influence their tumor’s behavior and response to treatment. Many epidemiological studies have suggested an important role for nutrition in cancer pathogenesis [62], including by showing associations between blood glucose and cancer. Diabetes, hyperglycemia and hyper-insulinemia all increase the risk of cancer occurrence and reduce the efficacy of anti-cancer treatment [63–66], and pre-diabetic hyperglycemia and diabetes are significant risk factors for cancer death [52,67]. Several studies also suggest that anti-diabetic drugs, including metformin, may reduce cancer risk via several pathways related to cellular metabolism [68,69]. Further, the ketogenic diet is in clinical trials for some cancer patients [70,71], although the ability of GBM cells to maintain Rictor acetylation in response to acetate when glucose is not available, raises the possibility that a ketogenic diet may not be sufficient to prevent nutrient-dependent maintenance of mTORC2 signaling [4,72].
Rigorous studies that are molecularly based are needed to complement epidemiological analysis, in order to dissect the mechanisms by which local or systemic nutrients regulate the activity of oncogenes and the relative fitness of tumor cells bearing specific mutations. For example, KRAS-mutant colon cancer cells are preferentially selected in low glucose environments because of their enhanced ability to take up and utilize this nutrient [73]. It is anticipated, that the biochemical environment can shape the behavior of tumor cells in a genotype-specific fashion, potentially by altering the relative fitness of cells bearing a mutation to grow within that metabolic niche and also potentially by directly regulating downstream signaling. Future studies are need to mechanistically determine precisely how extracellular nutrients modulate oncogenic signaling and to translate these insights into more effective treatments for cancer patients.
Supplementary Material
Acknowledgments
This work is supported by National Institute for Neurological Diseases and Stroke Grant NS73831; the Defeat GBM Research Collaborative, a subsidiary of National Brain Tumor Society, the National Cancer Institute Grant CA119347; The Ben and Catherine Ivy Foundation; generous donations from the Ziering Family Foundation in memory of Sigi Ziering; and a grant provided by Uehara Memorial Foundation, The Novartis Foundation (Japan) for the Promotion of Science, The Cell Science Research Foundation, Grant-in-Aid from the Tokyo Biochemical Research Foundation and JSPS KAKENHI Grant Number 15K19067. The authors declare no conflicts of interest.
Abbreviations
- acetyl-CoA
acetyl coenzyme A
- EGFR
epidermal growth factor receptor
- GBM
glioblastoma multiforme
- HDAC
histone deacetylase
- mTORC2
mechanistic target of rapamycin complex 2
References
- 1.Pavlova NN, Thompson CB. The Emerging Hallmarks of Cancer Metabolism. Cell Metab. 2016;23:27–47. doi: 10.1016/j.cmet.2015.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kaelin WG, McKnight SL. Influence of metabolism on epigenetics and disease. Cell. 2013;153:56–69. doi: 10.1016/j.cell.2013.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Masui K, Cavenee WK, Mischel PS. mTORC2 in the center of cancer metabolic reprogramming. Trends Endocrinol Metab. 2014;25:364–73. doi: 10.1016/j.tem.2014.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Masui K, Tanaka K, Ikegami S, Villa GR, Yang H, et al. Glucose-dependent acetylation of Rictor promotes targeted cancer therapy resistance. Proc Natl Acad Sci USA. 2015;112:9406–11. doi: 10.1073/pnas.1511759112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Furnari FB, Cloughesy TF, Cavenee WK, Mischel PS. Heterogeneity of epidermal growth factor receptor signalling networks in glioblastoma. Nat Rev Cancer. 2015;15:302–10. doi: 10.1038/nrc3918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Network CGAR. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–8. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brennan CW, Verhaak RG, McKenna A, Campos B, Noushmehr H, et al. The somatic genomic landscape of glioblastoma. Cell. 2013;155:462–77. doi: 10.1016/j.cell.2013.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lawrence MS, Stojanov P, Mermel CH, Robinson JT, Garraway LA, et al. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature. 2014;505:495–501. doi: 10.1038/nature12912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Masui K, Tanaka K, Akhavan D, Babic I, Gini B, et al. mTOR complex 2 controls glycolytic metabolism in glioblastoma through FoxO acetylation and upregulation of c-Myc. Cell Metab. 2013;18:726–39. doi: 10.1016/j.cmet.2013.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tanaka K, Babic I, Nathanson D, Akhavan D, Guo D, et al. Oncogenic EGFR signaling activates an mTORC2-NF-κB pathway that promotes chemotherapy resistance. Cancer Discov. 2011;1:524–38. doi: 10.1158/2159-8290.CD-11-0124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Treins C, Giorgetti-Peraldi S, Murdaca J, Semenza GL, Van Obberghen E. Insulin stimulates hypoxia-inducible factor 1 through a phosphatidylinositol 3-kinase/target of rapamycin-dependent signaling pathway. J Biol Chem. 2002;277:27975–81. doi: 10.1074/jbc.M204152200. [DOI] [PubMed] [Google Scholar]
- 12.Yu J, Li J, Zhang S, Xu X, Zheng M, et al. IGF-1 induces hypoxia-inducible factor 1α-mediated GLUT3 expression through PI3K/Akt/mTOR dependent pathways in PC12 cells. Brain Res. 2012;1430:18–24. doi: 10.1016/j.brainres.2011.10.046. [DOI] [PubMed] [Google Scholar]
- 13.Mohlin S, Hamidian A, von Stedingk K, Bridges E, Wigerup C, et al. PI3K-mTORC2 but not PI3K-mTORC1 regulates transcription of HIF2A/EPAS1 and vascularization in neuroblastoma. Cancer Res. 2015;75:4617–28. doi: 10.1158/0008-5472.CAN-15-0708. [DOI] [PubMed] [Google Scholar]
- 14.Laplante M, Sabatini DM. mTOR signaling in growth control and disease. Cell. 2012;149:274–93. doi: 10.1016/j.cell.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu SH, Bi JF, Cloughesy T, Cavenee WK, Mischel PS. Emerging function of mTORC2 as a core regulator in glioblastoma: metabolic reprogramming and drug resistance. Cancer Biol Med. 2014;11:255–63. doi: 10.7497/j.issn.2095-3941.2014.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hung CM, Garcia-Haro L, Sparks CA, Guertin DA. mTOR-dependent cell survival mechanisms. Cold Spring Harbor perspectives in biology. 2012:4. doi: 10.1101/cshperspect.a008771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zinzalla V, Stracka D, Oppliger W, Hall MN. Activation of mTORC2 by association with the ribosome. Cell. 2011;144:757–68. doi: 10.1016/j.cell.2011.02.014. [DOI] [PubMed] [Google Scholar]
- 18.Sarbassov DD, Guertin DA, Ali SM, Sabatini DM. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science. 2005;307:1098–101. doi: 10.1126/science.1106148. [DOI] [PubMed] [Google Scholar]
- 19.Oh WJ, Jacinto E. mTOR complex 2 signaling and functions. Cell Cycle. 2011;10:2305–16. doi: 10.4161/cc.10.14.16586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pearce LR, Sommer EM, Sakamoto K, Wullschleger S, Alessi DR. Protor-1 is required for efficient mTORC2-mediated activation of SGK1 in the kidney. Biochem J. 2011;436:169–79. doi: 10.1042/BJ20102103. [DOI] [PubMed] [Google Scholar]
- 21.Read RD, Cavenee WK, Furnari FB, Thomas JB. A drosophila model for EGFR-Ras and PI3K-dependent human glioma. PLoS Genet. 2009;5:e1000374. doi: 10.1371/journal.pgen.1000374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guertin DA, Stevens DM, Saitoh M, Kinkel S, Crosby K, et al. mTOR complex 2 is required for the development of prostate cancer induced by Pten loss in mice. Cancer Cell. 2009;15:148–59. doi: 10.1016/j.ccr.2008.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cheng H, Zou Y, Ross JS, Wang K, Liu X, et al. RICTOR Amplification Defines a Novel Subset of Patients with Lung Cancer Who May Benefit from Treatment with mTORC1/2 Inhibitors. Cancer Discov. 2015;5:1262–70. doi: 10.1158/2159-8290.CD-14-0971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Laugier F, Finet-Benyair A, André J, Rachakonda PS, Kumar R, et al. RICTOR involvement in the PI3K/AKT pathway regulation in melanocytes and melanoma. Oncotarget. 2015;6:28120–31. doi: 10.18632/oncotarget.4866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Magee JA, Ikenoue T, Nakada D, Lee JY, Guan KL, et al. Temporal changes in PTEN and mTORC2 regulation of hematopoietic stem cell self-renewal and leukemia suppression. Cell Stem Cell. 2012;11:415–28. doi: 10.1016/j.stem.2012.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bashir T, Cloninger C, Artinian N, Anderson L, Bernath A, et al. Conditional astroglial Rictor overexpression induces malignant glioma in mice. PLoS One. 2012;7:e47741. doi: 10.1371/journal.pone.0047741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–33. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ward PS, Thompson CB. Metabolic reprogramming: a cancer hallmark even warburg did not anticipate. Cancer Cell. 2012;21:297–308. doi: 10.1016/j.ccr.2012.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Koppenol WH, Bounds PL, Dang CV. Otto Warburg’s contributions to current concepts of cancer metabolism. Nat Rev Cancer. 2011;11:325–37. doi: 10.1038/nrc3038. [DOI] [PubMed] [Google Scholar]
- 30.Liu F, Hon GC, Villa GR, Turner KM, Ikegami S, et al. EGFR Mutation Promotes Glioblastoma through Epigenome and Transcription Factor Network Remodeling. Mol Cell. 2015;60:307–18. doi: 10.1016/j.molcel.2015.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lyssiotis CA, Cantley LC. Acetate fuels the cancer engine. Cell. 2014;159:1492–4. doi: 10.1016/j.cell.2014.12.009. [DOI] [PubMed] [Google Scholar]
- 32.Mashimo T, Pichumani K, Vemireddy V, Hatanpaa KJ, Singh DK, et al. Acetate is a bioenergetic substrate for human glioblastoma and brain metastases. Cell. 2014;159:1603–14. doi: 10.1016/j.cell.2014.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schug ZT, Peck B, Jones DT, Zhang Q, Grosskurth S, et al. Acetyl-CoA synthetase 2 promotes acetate utilization and maintains cancer cell growth under metabolic stress. Cancer Cell. 2015;27:57–71. doi: 10.1016/j.ccell.2014.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sutendra G, Kinnaird A, Dromparis P, Paulin R, Stenson TH, et al. A nuclear pyruvate dehydrogenase complex is important for the generation of acetyl-CoA and histone acetylation. Cell. 2014;158:84–97. doi: 10.1016/j.cell.2014.04.046. [DOI] [PubMed] [Google Scholar]
- 35.Lee JV, Carrer A, Shah S, Snyder NW, Wei S, et al. Akt-dependent metabolic reprogramming regulates tumor cell histone acetylation. Cell Metab. 2014;20:306–19. doi: 10.1016/j.cmet.2014.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Comerford SA, Huang Z, Du X, Wang Y, Cai L, et al. Acetate dependence of tumors. Cell. 2014;159:1591–602. doi: 10.1016/j.cell.2014.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wellen KE, Hatzivassiliou G, Sachdeva UM, Bui TV, Cross JR, et al. ATP-citrate lyase links cellular metabolism to histone acetylation. Science. 2009;324:1076–80. doi: 10.1126/science.1164097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Choudhary C, Kumar C, Gnad F, Nielsen ML, Rehman M, et al. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009;325:834–40. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- 39.Zhao S, Xu W, Jiang W, Yu W, Lin Y, et al. Regulation of cellular metabolism by protein lysine acetylation. Science. 2010;327:1000–4. doi: 10.1126/science.1179689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xiong Y, Guan KL. Mechanistic insights into the regulation of metabolic enzymes by acetylation. J Cell Biol. 2012;198:155–64. doi: 10.1083/jcb.201202056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lin R, Zhou X, Huang W, Zhao D, Lv L, et al. Acetylation control of cancer cell metabolism. Curr Pharm Des. 2014;20:2627–33. doi: 10.2174/13816128113199990487. [DOI] [PubMed] [Google Scholar]
- 42.Xu W, Li Y, Liu C, Zhao S. Protein lysine acetylation guards metabolic homeostasis to fight against cancer. Oncogene. 2014;33:2279–85. doi: 10.1038/onc.2013.163. [DOI] [PubMed] [Google Scholar]
- 43.Glidden EJ, Gray LG, Vemuru S, Li D, Harris TE, et al. Multiple site acetylation of Rictor stimulates mammalian target of rapamycin complex 2 (mTORC2)-dependent phosphorylation of Akt protein. J Biol Chem. 2012;287:581–8. doi: 10.1074/jbc.M111.304337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cai L, Tu BP. On acetyl-CoA as a gauge of cellular metabolic state. Cold Spring Harb Symp Quant Biol. 2011;76:195–202. doi: 10.1101/sqb.2011.76.010769. [DOI] [PubMed] [Google Scholar]
- 45.Wang L, Du Y, Lu M, Li T. ASEB: a web server for KAT-specific acetylation site prediction. Nucleic Acids Res. 2012;40:W376–9. doi: 10.1093/nar/gks437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li T, Du Y, Wang L, Huang L, Li W, et al. Characterization and prediction of lysine (K)-acetyl-transferase specific acetylation sites. Mol Cell Proteomics. 2012;11:M111011080. doi: 10.1074/mcp.M111.011080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mihaylova MM, Vasquez DS, Ravnskjaer K, Denechaud PD, Yu RT, et al. Class IIa histone deacetylases are hormone-activated regulators of FOXO and mammalian glucose homeostasis. Cell. 2011;145:607–21. doi: 10.1016/j.cell.2011.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang B, Moya N, Niessen S, Hoover H, Mihaylova MM, et al. A hormone-dependent module regulating energy balance. Cell. 2011;145:596–606. doi: 10.1016/j.cell.2011.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Masui K, Cloughesy TF, Mischel PS. Review: molecular pathology in adult high-grade gliomas: from molecular diagnostics to target therapies. Neuropathol Appl Neurobiol. 2012;38:271–91. doi: 10.1111/j.1365-2990.2011.01238.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Masui K, Cavenee WK, Mischel PS. mTORC2 and Metabolic Reprogramming in GBM: at the Interface of Genetics and Environment. Brain Pathol. 2015;25:755–9. doi: 10.1111/bpa.12307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Weiler M, Blaes J, Pusch S, Sahm F, Czabanka M, et al. mTOR target NDRG1 confers MGMT-dependent resistance to alkylating chemotherapy. Proc Natl Acad Sci USA. 2014;111:409–14. doi: 10.1073/pnas.1314469111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Derr RL, Ye X, Islas MU, Desideri S, Saudek CD, et al. Association between hyperglycemia and survival in patients with newly diagnosed glioblastoma. J Clin Oncol. 2009;27:1082–6. doi: 10.1200/JCO.2008.19.1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mihaylova MM, Sabatini DM, Yilmaz Ö. Dietary and metabolic control of stem cell function in physiology and cancer. Cell Stem Cell. 2014;14:292–305. doi: 10.1016/j.stem.2014.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shin S, Buel GR, Wolgamott L, Plas DR, Asara JM, et al. ERK2 Mediates Metabolic Stress Response to Regulate Cell Fate. Mol Cell. 2015;59:382–98. doi: 10.1016/j.molcel.2015.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hensley CT, Faubert B, Yuan Q, Lev-Cohain N, Jin E, et al. Metabolic Heterogeneity in Human Lung Tumors. Cell. 2016;164:681–94. doi: 10.1016/j.cell.2015.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Romero IL, Mukherjee A, Kenny HA, Litchfield LM, Lengyel E. Molecular pathways: trafficking of metabolic resources in the tumor microenvironment. Clin Cancer Res. 2015;21:680–6. doi: 10.1158/1078-0432.CCR-14-2198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Martinez-Outschoorn UE, Peiris-Pagés M, Pestell RG, Sotgia F, Lisanti MP. Cancer metabolism: a therapeutic perspective. Nat Rev Clin Oncol. 2016 doi: 10.1038/nrclinonc.2016.60. [DOI] [PubMed] [Google Scholar]
- 58.Seyfried TN, Flores RE, Poff AM, D’Agostino DP. Cancer as a metabolic disease: implications for novel therapeutics. Carcinogenesis. 2014;35:515–27. doi: 10.1093/carcin/bgt480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Uzunlulu M, Telci Caklili O, Oguz A. Association between Metabolic Syndrome and Cancer. Ann Nutr Metab. 2016;68:173–9. doi: 10.1159/000443743. [DOI] [PubMed] [Google Scholar]
- 60.Micucci C, Valli D, Matacchione G, Catalano A. Current perspectives between metabolic syndrome and cancer. Oncotarget. 2016 doi: 10.18632/oncotarget.8341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hua F, Yu JJ, Hu ZW. Diabetes and cancer, common threads and missing links. Cancer Lett. 2016;374:54–61. doi: 10.1016/j.canlet.2016.02.006. [DOI] [PubMed] [Google Scholar]
- 62.Calle EE, Rodriguez C, Walker-Thurmond K, Thun MJ. Overweight, obesity, and mortality from cancer in a prospectively studied cohort of U.S. adults. N Engl J Med. 2003;348:1625–38. doi: 10.1056/NEJMoa021423. [DOI] [PubMed] [Google Scholar]
- 63.Giovannucci E, Harlan DM, Archer MC, Bergenstal RM, Gapstur SM, et al. Diabetes and cancer: a consensus report. Diabetes Care. 2010;33:1674–85. doi: 10.2337/dc10-0666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Perseghin G, Calori G, Lattuada G, Ragogna F, Dugnani E, et al. Insulin resistance/hyperinsulinemia and cancer mortality: the Cremona study at the 15th year of follow-up. Acta Diabetol. 2012;49:421–8. doi: 10.1007/s00592-011-0361-2. [DOI] [PubMed] [Google Scholar]
- 65.Tieu MT, Lovblom LE, McNamara MG, Mason W, Laperriere N, et al. Impact of glycemia on survival of glioblastoma patients treated with radiation and temozolomide. J Neurooncol. 2015;124:119–26. doi: 10.1007/s11060-015-1815-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Adeberg S, Bernhardt D, Foerster R, Bostel T, Koerber SA, et al. The influence of hyperglycemia during radiotherapy on survival in patients with primary glioblastoma. Acta Oncol. 2016;55:201–7. doi: 10.3109/0284186X.2015.1043397. [DOI] [PubMed] [Google Scholar]
- 67.Hirakawa Y, Ninomiya T, Mukai N, Doi Y, Hata J, et al. Association between glucose tolerance level and cancer death in a general Japanese population: the Hisayama Study. Am J Epidemiol. 2012;176:856–64. doi: 10.1093/aje/kws178. [DOI] [PubMed] [Google Scholar]
- 68.Hardie DG. The LKB1-AMPK pathway-friend or foe in cancer? Cancer Cell. 2013;23:131–2. doi: 10.1016/j.ccr.2013.01.009. [DOI] [PubMed] [Google Scholar]
- 69.Quinn BJ, Kitagawa H, Memmott RM, Gills JJ, Dennis PA. Repositioning metformin for cancer prevention and treatment. Trends Endocrinol Metab. 2013;24:469–80. doi: 10.1016/j.tem.2013.05.004. [DOI] [PubMed] [Google Scholar]
- 70.Schwartz K, Chang HT, Nikolai M, Pernicone J, Rhee S, et al. Treatment of glioma patients with ketogenic diets: report of two cases treated with an IRB-approved energy-restricted ketogenic diet protocol and review of the literature. Cancer Metab. 2015;3:3. doi: 10.1186/s40170-015-0129-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Allen BG, Bhatia SK, Anderson CM, Eichenberger-Gilmore JM, Sibenaller ZA, et al. Ketogenic diets as an adjuvant cancer therapy: History and potential mechanism. Redox Biol. 2014;2C:963–70. doi: 10.1016/j.redox.2014.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rieger J, Bähr O, Maurer GD, Hattingen E, Franz K, et al. ERGO: a pilot study of ketogenic diet in recurrent glioblastoma. Int J Oncol. 2014;44:1843–52. doi: 10.3892/ijo.2014.2382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yun J, Rago C, Cheong I, Pagliarini R, Angenendt P, et al. Glucose deprivation contributes to the development of KRAS pathway mutations in tumor cells. Science. 2009;325:1555–9. doi: 10.1126/science.1174229. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.