Abstract
Advances in human genetics in recent years have largely been driven by next-generation sequencing (NGS); however, the discovery of disease-related gene mutations has been biased toward the exome because the large and very repetitive regions that characterize the non-coding genome remain difficult to reach by that technology. For autosomal-dominant spinocerebellar ataxias (SCAs), 28 genes have been identified, but only five SCAs originate from non-coding mutations. Over half of SCA-affected families, however, remain without a genetic diagnosis. We used genome-wide linkage analysis, NGS, and repeat analysis to identify an (ATTTC)n insertion in a polymorphic ATTTT repeat in DAB1 in chromosomal region 1p32.2 as the cause of autosomal-dominant SCA; this region has been previously linked to SCA37. The non-pathogenic and pathogenic alleles have the configurations [(ATTTT)7–400] and [(ATTTT)60–79(ATTTC)31–75(ATTTT)58–90], respectively. (ATTTC)n insertions are present on a distinct haplotype and show an inverse correlation between size and age of onset. In the DAB1-oriented strand, (ATTTC)n is located in 5′ UTR introns of cerebellar-specific transcripts arising mostly during human fetal brain development from the usage of alternative promoters, but it is maintained in the adult cerebellum. Overexpression of the transfected (ATTTC)58 insertion, but not (ATTTT)n, leads to abnormal nuclear RNA accumulation. Zebrafish embryos injected with RNA of the (AUUUC)58 insertion, but not (AUUUU)n, showed lethal developmental malformations. Together, these results establish an unstable repeat insertion in DAB1 as a cause of cerebellar degeneration; on the basis of the genetic and phenotypic evidence, we propose this mutation as the molecular basis for SCA37.
Keywords: DAB1 reelin adaptor protein, unstable repeat insertion, RNA-mediated toxicity, SCA37, repeat expansion, repeat instability, neurodegeneration, neurodegenerative disease, neurodevelopmental gene, large Alu pentanucleotide repeat
Introduction
Spinocerebellar ataxias (SCAs) are a group of clinically and genetically very heterogeneous diseases, usually characterized by adult onset of progressive gait, limb, and speech ataxia caused by loss of cerebellar neurons.1, 2 The estimated prevalence of autosomal-dominant SCAs varies considerably in different world regions from 1.6 to 5.6 out of 100,000 inhabitants;1, 3 Machado-Joseph disease (MJD; also known as SCA3 [MIM: 109150]) is the most frequent in Portugal and worldwide, but a definite genetic diagnosis is still lacking for more than half of the families affected by these neurodegenerative diseases.1, 4
Currently 28 genes are known to harbor mutations causing SCA pathology. Seven of these are CAG repeat expansions that encode aberrant polyglutamine tracts in the protein products, rendering them toxic. Point mutations, deletions, or duplications in genes functioning in a wide range of neuronal processes explain 16 additional SCA types.5, 6, 7 Unstable repeat expansions in non-coding regions of protein-encoding genes have been reported only in five SCAs.8
The disease mechanism(s) associated with non-coding repeat expansions remained obscure until the concept emerged that RNA transcripts harboring expanded repeats were toxic in myotonic dystrophy type 1 (DM1 [MIM: 160900]).9, 10 RNA-associated pathogenesis was subsequently identified in other neurodegenerative diseases also caused by non-coding expanded repeats, such as myotonic dystrophy 2 (DM2 [MIM: 602668]), fragile X tremor/ataxia syndrome (FXTAS [MIM: 300623]), HD-like disease type 2 (HDL2 [MIM: 606438]), frontotemporal dementia and/or amyotrophic lateral sclerosis (FTDALS [MIM: 105550]), and several SCAs caused by non-coding repeat mutations.8, 10, 11
In SCAs caused by non-coding mutations, such as SCA8 (MIM: 608768), SCA10 (MIM: 603516), SCA36 (MIM: 614153), and SCA31 (MIM: 117210), the untranslated RNA repeat accumulates in affected brain tissue and correlates with the pathogenesis.1, 10, 12, 13, 14 SCA8 is associated with bidirectional transcription of a CTG/CAG repeat located in the UTR of ATXN8OS (MIM: 603680) and in a short open reading frame in the overlapping ATXN8 (MIM: 613289).15 In SCA10, SCA31, and SCA36, very large intronic penta- or hexanucleotide repeats are found in ATXN10 (MIM: 611550), TK2 (MIM: 188250) and BEAN1 (MIM: 612051), and NOP56 (MIM: 614154), respectively.12, 13, 14, 16 Studies on RNA-mediated pathogenesis point toward a complex combination of different effects, including aberrant transcript expression and processing and the formation of RNA aggregates in affected cells and tissues, which are also detected in cell and animal models that overexpress the repeat.8, 11, 17, 18, 19
Here, we describe a SCA in three large Portuguese families identified during a population-based survey of hereditary ataxias in Portugal.3 The affected individuals from these families share a pure cerebellar ataxia phenotype and, distinctively, onset of dysarthria in late adolescence to adulthood. In this study, we mapped this SCA by genome-wide linkage analysis of the families and identified a pathological (ATTTC)n repeat insertion in the non-coding region of DAB1 (DAB1, reelin adaptor protein [MIM: 603448]) in chromosomal region 1p32.2, which overlaps the SCA37 linkage region (MIM: 615945).20 Given the significant death caused by injection of the RNA repeat insertion in zebrafish, we suggest that RNA-mediated toxicity could be one pathogenic mechanism implicated in this SCA.
Subjects and Methods
Subjects
Families were ascertained during a nationwide, population-based, systematic survey of hereditary ataxias and spastic paraplegias performed in Portugal from 1994 to 2004.3 Affected individuals were referred for diagnostic purposes to the authorized Center for Predictive and Preventive Genetics (CGPP) at the Institute for Molecular and Cell Biology; they were tested for mutations in genes associated with SCA1, DRPLA, SCA2, MJD (SCA3), SCA6, SCA7, SCA8, SCA10, SCA12, SCA14, and SCA17. This study used the de-identified, previously collected DNA samples that were stored at the CGPP biobank, as well as anonymized samples from the Fundación Pública Galega de Medicina Xenómica and from the Montréal Neurological Institute at McGill University. For the study of relevant polymorphisms in the Portuguese population, anonymized DNA samples were obtained from both dried-blood spots from the public health laboratory newborn screening program and internal lab control samples. For skin biopsies, informed written consent was given for participation in this study, which was approved by the health ethics board of the Hospital Prof. Doutor Fernando Fonseca.
Genotyping and Linkage Analysis
DNA samples from 65 individuals from three Portuguese families were hybridized to Affymetrix GeneChip Human Mapping 10K Array Xba 142 2.0 according to the manufacturer’s recommendations. Each individual genotype was obtained with GeneChip DNA Analysis Software 3.0. After batch import of pedigree and sample information, data were checked for Mendelian errors. The file obtained was imported into ALOHOMORA21 and analyzed with PEDSTATS22 for a check of the consistency of the pedigree and identification of uninformative and/or mistyped SNPs for removal from the dataset. SNPs were reformatted as individual chromosomes for linkage in Merlin.23 Multipoint linkage analysis was performed with a parametric model assuming an autosomal-dominant mode of inheritance, a disease-allele frequency of 0.0001, and 95% penetrance.
Short tandem repeat (STR) markers from chr1: 51,245,557–59,958,568 (UCSC Genome Browser build hg19) were analyzed in families by PCR. Allele sizes were assessed by an ABI3730xl DNA Analyzer with GeneMapper software v.4.0 (Applied Biosystems).
Mutation Screening
Mutation screening was performed in affected individuals and unaffected relatives by Sanger sequencing of exon and exon-intron boundaries and by NGS. NGS was carried out in six affected individuals (II.9 and III.1 from family M, II.1 and III.6 from family G, and II.7 and III.5 from family R) and four unaffected relatives (II.10 from family M, II.6 and III.5 from family G, and II.3 from family R) at the sequencing facility of the Fundación Pública Galega de Medicina Xenomica. Enrichment was performed with SureSelect Target Enrichment (Agilent Technologies) targeting 3.4 Mb of genomic sequence at chr1: 55,800,000–59,200,000 (hg19). Each genomic library was sequenced with a SOLiD 5500xl system (Life Technologies) as paired-end reads (75 + 35 bp). Reads were aligned to the reference genome (hg19) with SOLiD BioScope software v.1.3.0, and then variants were called with Genome Analysis Toolkit v.2.1.24 In the ten individuals analyzed by NGS, the number of reads matching the target region ranged from 2,366,593 to 3,444,594. 77.7%–84.12% of the target region was covered at least 20×, whereas 92.2%–93.4% was covered at least 5×; the average depth of coverage ranged from 35.11× to 51.29×. Variant calling required at least 6× minimum depth of coverage. The next variant-filtering steps required that the variant be (1) heterozygous and (2) absent (or with a frequency below 0.01) from public databases (dbSNP135 and 1000 Genomes Project; February 2012).
Screening for repeat expansions was performed by standard PCR analysis in a family branch from each of the M, G and R pedigrees. Approximate allele size was assessed in 2% agarose gel. Non-polymorphic repeats were confirmed by PCR amplification with a 6-FAM labeled reverse primer and detected on an ABI3730xl DNA Analyzer.
Southern Blot Analysis
Genomic DNA (10 μg) was digested with XbaI (Fermentas Life Sciences). Digestion products were separated by electrophoresis on a 0.8% agarose gel, denatured with 0.4 M NaOH, and transferred in 10× saline sodium citrate (SSC) onto a Hybond-N+ nylon membrane (GE Healthcare) with a vacuum blotter (Bio-Rad). DNA was immobilized onto the membrane by UV cross-linking. The hybridization probe was synthesized from genomic DNA by PCR with HotStarTaq Master Mix Kit (QIAGEN) and primers SB24F and SB24R (Table S1 and Figure 2D). The probe was labeled with [α-32P]-dCTP with a specific activity of 3000Ci/mmol with the Prime It II Random Primer Labeling Kit (Agilent Technologies). Hybridization was carried out overnight at 65°C, and the signal was detected by X-ray film exposure or by phosphor storage screen and digital image acquisition on a Typhoon imaging system (GE Healthcare).
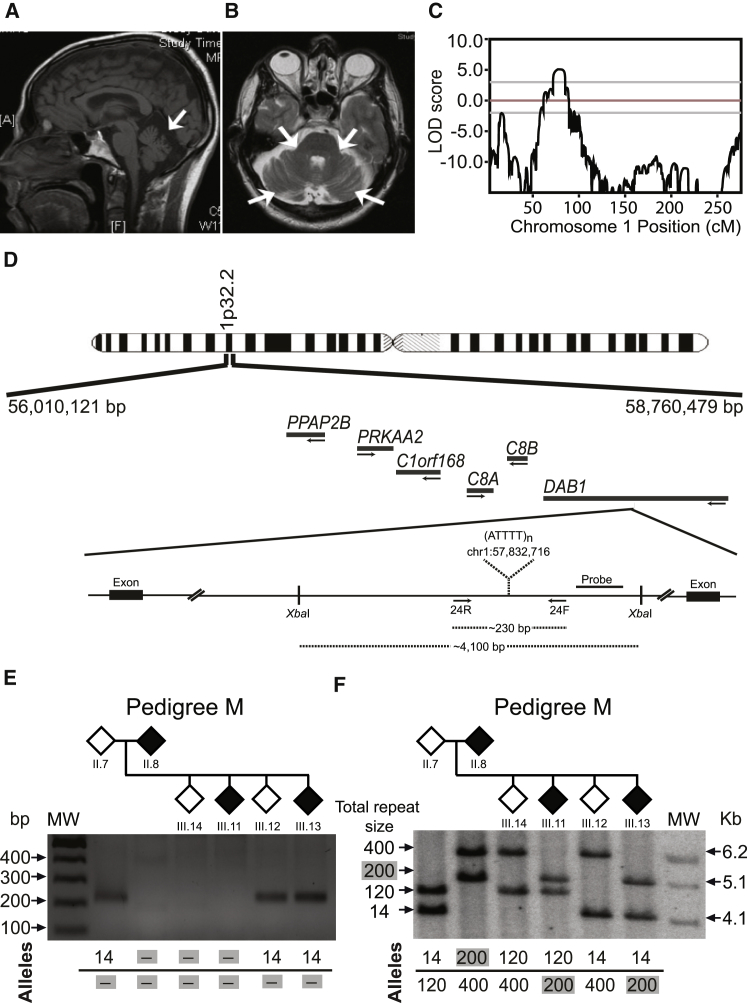
Figure 2.
Chromosome Mapping and Mutation Screening
(A and B) Brain MRI of affected individual M8 (Table 1) shows atrophy of (A) the cerebellar cortex on sagittal T1 and (B) the middle cerebellar peduncles on an axial T2-weighted image.
(C) Linkage analysis using genotypes from 18 individuals from family M shows a maximum multipoint LOD score of 5.1 obtained in chromosomal region 1p32.
(D) Schematic physical map of the SCA candidate region; also depicted are the intronic repeat region with the XbaI restriction sites, the location of the Southern blot probe, and primers used for standard PCR. The (ATTTT)n position according to hg19 is shown.
(E) Standard PCR analysis of the pentanucleotide repeat ATTTT/AAAAT in the intronic 5′ UTR of DAB1 shows a lack of PCR amplification of large repeat tracts that were not amplified by standard PCR.
(F) Using a probe hybridizing near the pentanucleotide repeat ATTTT/AAAAT in DAB1, corresponding Southern blot analysis for the same family branch in (E) shows that an ∼5.1-kb fragment corresponding to ∼200 pentanucleotide repeats was transmitted from the affected mother to affected offspring. Large non-pathogenic (ATTTT)n alleles are unstable, and the estimated allele size of 120 on Southern blot was shown by sequencing analysis to vary from 127 to 139 units. Individual ID numbers are the same as those in each pedigree in Figure 1.
PCR Amplification and Sequencing of Pentanucleotide Repeat Alleles
Short pentanucleotide repeats were analyzed by standard PCR with primers 24F and 24R (Table S1 and Figure 2D) with HotStarTaq Master Mix Kit (QIAGEN). PCR products were purified, and repeat size was assessed by Sanger sequencing. Large pentanucleotide repeat alleles were analyzed by long-range PCR with 200 ng DNA, 0.3 μM primers ALU24F and ALU24R (Table S1), 200 μM dNTPs, 1.4 mM MgCl2, 60 mM Tris-SO4 (pH 9.1), 18 mM (NH4)2SO4, 2 mM MgSO4, and 1 μL of Elongase Enzyme Mix (Invitrogen) in 50 μL. After 3 min at 94°C, DNA samples underwent ten cycles of amplification (94°C for 30 s and 62°C for 7 min) followed by an additional 30 cycles (94°C for 30 s and 62°C for 7 min with 20 s increments per cycle). PCR products were separated by electrophoresis in 1% agarose gels, DNA was extracted from the gel, and the number of repeat units and insertion composition were determined by Sanger sequencing with internal primers 24F and 24R4 (Table S1).
Repeat-Primed PCR for (ATTTC)n Insertion Alleles
The repeat insertion sequence was amplified by repeat-primed PCR (RP-PCR) with primers 24R, FLAG, and RP-TTCAT (Table S1). Reverse primer 24R was a 6-FAM-labeled locus-specific primer, RP-TTCAT was a repeat insertion-specific primer with a DNA tail sequence at the 5′ end, and reverse primer FLAG contained the same 5′ tail sequence as RP-TTCAT. PCR was performed with 100 ng genomic DNA, 0.4 μM primer 24F and primer FLAG, and 0.04 μM primer RP-TTCAT with HotStarTaq Master Mix (QIAGEN). The initial RP-PCR step was at 95°C for 15 min and was followed by 35 cycles (95°C for 45 s, 58°C for 30 s, and 72°C for 2 min with 20 s increments per cycle). Products of RP-PCR were detected on an ABI3730xl DNA Analyzer.
Pentanucleotide Repeat Cloning
For cloning, PCR amplification of normal alleles with 7–139 ATTTTs and the pathological (ATTTT)57(ATTTC)58(ATTTT)73 insertion (abbreviated ins(ATTTC)58) allele was carried out with primers flanking the Alu sequence, containing restriction sites for EcoRI and NotI (Table S1). PCR products were separated by agarose gel electrophoresis, and DNA was extracted with the QIAquick Gel Extraction Kit (QIAGEN), ligated into pCDH-CMV-MCS-EF1-GFP-T2A-Puro (System Biosciences), and transformed into the DH5α E. coli strain. Plasmid DNA was isolated with the QIAGEN Plasmid Midi Kit, and the insert sequence was confirmed by Sanger sequencing.
Cell Culture
Primary cultures of fibroblasts, established from skin biopsies, and HEK293T cells were cultured in DMEM with 10% fetal bovine serum at 37°C with 5% CO2.
Analysis of Gene Expression
For RT-PCR, RNA first-strand cDNA was synthesized with SuperScript III First-Strand Synthesis SuperMix (Invitrogen) from total human cerebellum RNA (Clontech).
Cell Transfection and RNA Fluorescence In Situ Hybridization Analysis
For RNA in situ hybridization, a digoxigenin 3′- and 5′-labeled locked nucleic acid (LNA) probe (Exiqon) was synthesized to contain the sequence (TGAAA)5TGA, predicted to hybridize to (AUUUC)n. HEK293T cells were transfected with the plasmid N(ATTTT)7, N(ATTTT)139, or ins(ATTTC)58 with jetPRIME transfection reagent (Polyplus-transfection). 48 hr after transfection, cells were fixed in 4% paraformaldehyde. Permeabilization was performed in 0.2% Triton in PBS for 10 min. After this, the coverslips were incubated in prehybridization solution (50% formamide, 10% dextran sulfate, 2× SSC, 50 mM sodium phosphate buffer, 10 mM ribonucleosyde vanadyl complex, and 100 μg/mL yeast tRNA) for 20 min at 60°C. The LNA probe was used at a working solution of 40 nM after dilution in prehybridization buffer and incubated for 2 hr at 60°C. This was followed by several washes at room temperature (5 min of 2× SSC and 0.1% Tween 20) and at 60°C (10 min of 0.2× SSC). Coverslips were blocked with 5% BSA in PBS with Tween 20 for 30 min and incubated overnight with anti-digoxigenin-rhodamine antibody (Roche Diagnostics). They were counterstained with DAPI. Fluorescence in situ hybridization (FISH) signals were scored with a Leica Microsystems SP5 II confocal microscope with a 63× glycerol objective. For DNase or RNase treatment, before prehybridization, coverslips were incubated for 1 hr at 37°C with 200 U/mL DNaseI or 100 μg/mL RNase A (Thermo Fisher Scientific). After acquisition, images were analyzed with Fiji.25
Protein Analysis
Total protein extracts were prepared from cultured cells or frozen human cerebellum in radioimmunoprecipitation assay solution containing a cocktail of protease and phosphatase inhibitors. For cells, direct lysis was performed on ice by scrapping. Tissue was homogenized on ice, sonicated, incubated on ice for 30 min, and centrifuged. Protein lysates were resolved by standard PAGE and transferred onto a nitrocellulose membrane with a rapid transfer system (Bio-Rad). Membranes were incubated overnight with a rabbit polyclonal anti-DAB1 antibody (sc-13981, Santa Cruz Biotechnology, at 1:500 dilution) at 4°C, incubated for 1 hr with a horseradish peroxidase (HRP)-conjugated antibody, and analyzed for signal detection with an enhanced chemilumescent HRP substrate (SuperSignal West Pico). Signal acquisition was performed digitally with a ChemiDoc XRS+ (Bio-Rad).
In Vitro Synthesis of RNA
Two complementary oligonucleotides were used for cloning a T7 promoter in the pCDH-CMV-MCS-EF1-GFP-T2A-Puro vector harboring the normal N(ATTTT)7 and N(ATTTT)139 alleles and the pathological ins(ATTTC)58 allele. Oligonucleotides had overhanging sequences compatible with XbaI and EcoRI, restriction sites used for cloning (Table S1). A mix of T7A and T7B oligonucleotides were denatured at 95°C for 5 min, annealed at room temperature, and used in the pCDH-CMV-MCS-EF1-GFP-T2A-Puro ligation. After cloning, T7-pCDH-CMV-MCS-EF1-GFP-T2A-Puro vectors were linearized with NotI and purified by phenol-chloroform, and RNA was transcribed in vitro with T7 RNA polymerase. Cas9 mRNA was used as control RNA for the injections and was transcribed in vitro as previously described.26
Zebrafish RNA Microinjection
All zebrafish (Danio rerio) experiments complied with standard animal care guidelines and national legislation for animal experimentation. Zebrafish were crossed for the generation of embryos for RNA injections; 5 nL in-vitro-synthesized RNA for each of the normal N(ATTTT)7 and N(ATTTT)139 alleles and the pathological ins(ATTTC)58 allele and control RNA was microinjected in the yolk of 1- to 2-cell stage zebrafish embryos27 at a concentration of 100 ng/μL. Each RNA sequence was microinjected three times in at least 200 embryos per injection. Embryos were raised at 28°C in E3 medium and observed at 6 and 24 hr postfertilization (hpf). Embryos were anesthetized by the addition of tricaine (ethyl 3-aminobenzoate; Sigma) to the E3 medium, and phenotypes were documented at 24 hpf with a stereomicroscope Leica M205FA (Leica Microsystems) on the imaging acquisition system Orca Flash 4.0LT (Hamamatsu Photonics). For analysis of lethality rate and normal developmental, the statistical significance was assessed with the χ2 test.
Results
Mapping SCA to Chromosomal Region 1p32.2
We studied three large Portuguese families affected by an unknown genetic type of autosomal-dominant SCA with adult onset and characterized by dysarthria as the first symptom in most affected individuals (Figure 1). All affected subjects had a similar clinical presentation consisting of progressive pure cerebellar ataxia starting in their late teens to early 60s (Table 1). Brain MRI showed cerebellar atrophy in all affected individuals for whom it was available (Figures 2A and 2B). Analysis of genes previously associated with SCA failed to detect mutations (data not shown), and therefore we performed a whole-genome linkage analysis on 65 individuals with the Affymetrix GeneChip Human Mapping 10K Array Xba 142 2.0. Linkage analysis with MERLIN showed maximum multipoint LOD scores of 5.1, 4.4, and 2.2 in chromosomal region 1p32 for families M, R, and G, respectively (Figure 2C). Haplotype analysis in these families established an ∼8 Mb candidate region containing the disease mutation. Fine-mapping with 14 STRs spanning this critical region positioned the mutation between markers D1S200 and D1S2869, comprising approximately 2.8 Mb on chr1: 56,010,121–58,760,479 (hg19) (Figure 1). This candidate region contained the genes PPAP2B, PRKAA2, C1orf168, C8A, C8B, and DAB1 (Figure 2D) and overlapped the SCA37 locus mapped in a Spanish family affected by pure SCA, which began in most affected individuals as falls, dysarthria, or clumsiness followed by a complete cerebellar syndrome.20
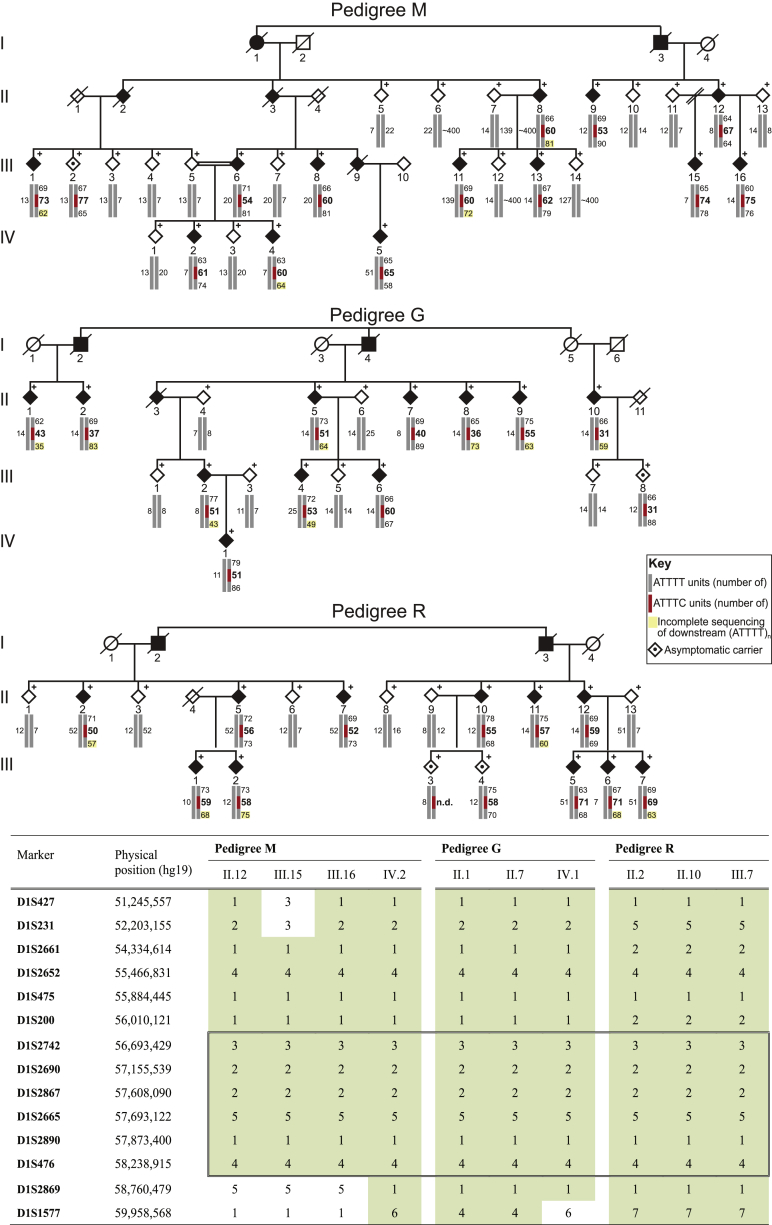
Figure 1.
Pedigrees of SCA-Affected Families and STR Haplotypes
Pedigree structures of the three families used for genome-wide linkage analysis and the corresponding individual genotypes and pentanucleotide repeat configuration. Below are the disease haplotypes from selected affected individuals, identified by pedigree number, for 14 markers from locus D1S427 (51,245,557 bp) to D1S1577 (59,958,568). The disease haplotypes found in the families established an initial candidate region spanning 2.8 Mb between markers D1S200 and D1S2869 in chromosomal region 1p32.2 (hg19) (shown in a box). Symbols in pedigrees were modified for privacy protection. Abbreviations are as follows: n.d., not determined; and +, individual for whom DNA was available.
Table 1.
Clinical and Genetic Features of Selected Affected Individuals
| Family | Affected Individual | Gender | Onset Age (Years) | Disease Progression (Years) | Onset Symptoms | Ataxia | (ATTTC)na |
|---|---|---|---|---|---|---|---|
| M | M1 | F | 30 | 34 | DA | +++ | 60 |
| M2 | M | 30 | 40 | DA | NA | 67 | |
| M3 | F | 18 | 24 | DA | NA | 75 | |
| M4 | F | 32 | 3 | DA | NA | 74 | |
| M5 | F | 30 | 27 | DA | ++ | 53 | |
| M6 | F | 34 | 6 | DA | ++ | 62 | |
| M7 | F | 40 | 6 | DA | ++ | 60 | |
| M8 | M | 27 | 30 | GI | +++ | 73 | |
| M9 | F | 23 | 38 | DA, GI | ++ | 54 | |
| M10 | F | 46 | 6 | DA, GI | + | 60 | |
| M11 | M | 30 | 8 | DA, GI | + | 61 | |
| M12 | F | 26 | 15 | DA, GI | ++ | 60 | |
| M13 | F | 29 | 2 | DA | + | 65 | |
| G | G2 | F | 58 | 10 | DA, GI | ++ | 43 |
| G6 | F | 55 | 20 | GI | ++ | 31 | |
| G8 | F | 40 | 14 | DA | ++ | 51 | |
| G9 | M | 45 | 13 | DA | ++ | 53 | |
| G11 | F | 35 | 2 | DA | + | 50 | |
| R | R1 | F | 30 | 47 | DA | ++ | 56 |
| R2 | F | 40 | 29 | DA | ++ | 52 | |
| R3 | F | 31 | 47 | DA | ++ | 57 | |
| R4 | M | 55 | 14 | DA | ++ | 59 | |
| R5 | F | 28 | 12 | DA | + | 71 | |
| R6 | F | 30 | 5 | DA | + | 71 | |
| R7 | M | 32 | 5 | DA | + | 69 | |
| R8 | F | 23 | 54 | DA | +++ | 55 | |
| MS | MS1 | M | 40 | 38 | DA, GI | +++ | 46 |
| MS2 | M | 47 | 31 | GI | +++ | 52 | |
| MS3 | M | 57 | 14 | DA, GI | +++ | 46 | |
| MS4 | F | 31 | 6 | DA | ++ | 61 |
Abbreviations are as follows: DA, dysarthria; GI, gait imbalance; and NA, not available.
The ATTTC repeat insertion is always flanked on both sides by (ATTTT)>58.
A Rare Haplotype in 1p32.2 Is Shared by Affected Individuals
We carried out mutation screening of candidate genes first by Sanger sequencing of known exons and corresponding exon-intron boundaries and then by NGS of the entire candidate region in affected individuals and unaffected relatives. In affected subjects, we detected 20 heterozygous intergenic and intronic variants not found in healthy relatives and absent from or with a frequency below 1% in the 1000 Genomes Project and dbSNP (Table S3). None of the identified variants was located in exonic or consensus splicing regions. We then carried out genotype analysis of variants with a frequency lower than 0.1% in 150 families affected by autosomal-dominant SCA of unknown genetic type (34 of whom were from southern Portugal) and in the general Portuguese population. We found three additional pedigrees sharing a core disease haplotype with the three linked families and confirmed that these variants were very rare in the control population. All six families with the disease haplotype were from southern Portugal. We identified four different haplotypes around the core haplotype in families affected by this SCA (Table 2). Haplotype analysis in the additional affected individuals narrowed the candidate region down to 1.8 Mb from rs537634498 to marker D1S2869. We then performed multiplex ligation-dependent probe amplification on genes that are within this region and are expressed in brain tissue, namely PPAP2B (MIM: 607125), PRKAA2 (MIM: 600497), and DAB1, but we detected no copy-number variations (data not shown).
Table 2.
Refinement of the Candidate Region
| Gene or Intergenic Region | Marker | Position on Chr1 (hg19) |
Haplotypes |
|||
|---|---|---|---|---|---|---|
| A | B | C | D | |||
| Intergenic | rs565332393 | 56,090,535 | C | C | T | T |
| rs762335464 | 56,223,397 | C | C | T | T | |
| rs142969184 | 56,251,658 | A | A | C | C | |
| rs761751006 | 56,305,070 | G | G | A | A | |
| ss2137493855 | 56,453,113 | G | G | A | A | |
| D1S2742 | 56,693,429 | 3 | 3 | 4 | 2 | |
| rs777060331 | 56,517,710 | C | C | T | T | |
| ss2137493856 | 56,545,567 | G | G | A | A | |
| rs138928773 | 56,753,932 | C | C | T | T | |
| rs528859858 | 56,810,200 | A | A | A | G | |
| PPAP2B | rs537634498 | 56,964,113 | CCCAGC | CCCAGC | CCCAGC | delCCCAGC |
| PRKAA2 | D1S2690 | 57,155,539 | 2 | 2 | 2 | 2 |
| C1orf168 | rs555296478 | 57,250,815 | delT | delT | delT | delT |
| C8A | rs572272180 | 57,367,559 | delTTG | delTTG | delTTG | delTTG |
| Intergenic | rs115293800 | 57,438,757 | C | C | C | C |
| rs866411539 | 57,444,772 | T | T | T | T | |
| DAB1 | rs145962085 | 57,481,145 | C | C | C | C |
| ss2137493861 | 57,491,966 | G | G | G | G | |
| D1S2867 | 57,608,090 | 2 | 2 | 2 | 2 | |
| D1S2665 | 57,693,122 | 5 | 5 | 5 | 5 | |
| (ATTTC)n | 57,832,716 | (ATTTC)n | (ATTTC)n | (ATTTC)n | (ATTTC)n | |
| D1S2890 | 57,873,400 | 1 | 1 | 1 | 1 | |
| ss2137493857 | 57,551,605 | A | A | A | A | |
| rs192485043 | 57,926,567 | A | A | A | A | |
| rs145097803 | 58,201,704 | A | A | A | A | |
| ss2137493862 | 58,215,160 | G | G | G | G | |
| D1S476 | 58,238,915 | 4 | 4 | 4 | 4 | |
| Intergenic | D1S2869 | 58,760,479 | 5 | 1 | ND | ND |
The following abbreviation is used: ND, not determined.
An (ATTTC)n Insertion in DAB1 Segregates with SCA
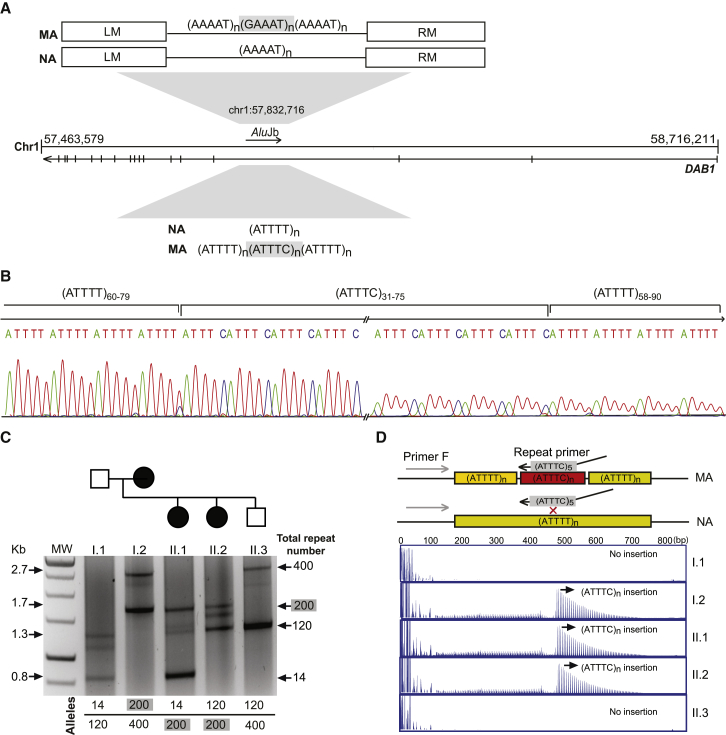
Next, we used standard PCR to screen 278 repetitive sequences within the candidate region containing tri-, tetra-, penta-, hexa-, and heptanucleotide and other complex repeats (Table S2 and Figure 2E). PCR analysis of a pentanucleotide repeat (ATTTT/AAAAT) in the 5′ UTR intron 3 of DAB1 (GenBank: NM_021080) showed no apparent transmission of this pentanucleotide repeat (Figure 2E and Figure S1A). We consistently observed a missing PCR product in all affected parent-to-affected-offspring transmissions analyzed (Figure S1A) and hypothesized that this most likely resulted from the presence of large alleles that were not amplifiable under standard PCR conditions. To investigate this further, we performed Southern blot analysis and identified an approximately 5.1 kb band that was transmitted only from affected individuals to their affected offspring (Figure 2F and Figure S1B). The amplification of these large alleles by long-range PCR of the repetitive region, followed by direct sequencing, enabled us to find a heterozygous (ATTTC)n insertion within the simple ATTTT/AAAAT repeat at nucleotide 57,832,716 on chromosome 1 (hg19). We identified complex pentanucleotide repeats with the structure [(ATTTT)60–79(ATTTC)31–75(ATTTT)58–90] in 3′ fully sequenced alleles (Figures 1 and 3A–3C and Table S4). We confirmed the presence of this (ATTTC)n insertion with its flanking (ATTTT)n by Sanger sequencing analysis in all 35 affected individuals from the initial three large kindreds and in the six available affected subjects from the three additional pedigrees with the shared core haplotype (Figure 1 and Table S4). The heterozygous (ATTTC)n insertion, ranging from 31 to 75 repeats (Figure 4A), was always flanked by (ATTTT)n tracts larger than 58 repeats and could be detected by repeat-primed PCR (Figure 3D). This (ATTTC)n insertion segregated with the disease in all families and was not detected by sequencing analysis of 520 chromosomes from the normal Portuguese population. In every disease allele, the insertion site was identical and placed in the middle of the normal ATTTT repeat, thus maintaining the pentanucleotide repeat structure. This genetic evidence establishes the (ATTTC)n insertion in DAB1 as causative for this type of autosomal-dominant SCA.
Figure 3.
Identification of an Unstable ATTTC Repeat Insertion in DAB1
(A) Schematic representation of the ATTTT/AAAAT simple repeat flanked by left (LM) and right (RM) monomers in the polymorphic middle A-rich region of an AluJb sequence in an intron of the DAB1 5′ UTR; also depicted is the structure of normal alleles (NAs) with pure ATTTT/AAAAT repeats and mutant alleles (MAs) with the (ATTTC)n insertion.
(B) Sequencing analysis of the DAB1 repeat shows the (ATTTC)n insertion in the middle of the ATTTT simple repeat in affected individuals.
(C) Long-range PCR across the Alu sequence was used for assaying alleles up to 2.7 kb.
(D) In this family branch shown for long-range PCR, the ATTTC repeat insertion was readily detected by RP-PCR analysis in affected individuals (mother I.2 and offspring II.1 and II.2) but not in the unaffected individuals I.1 and II.3.
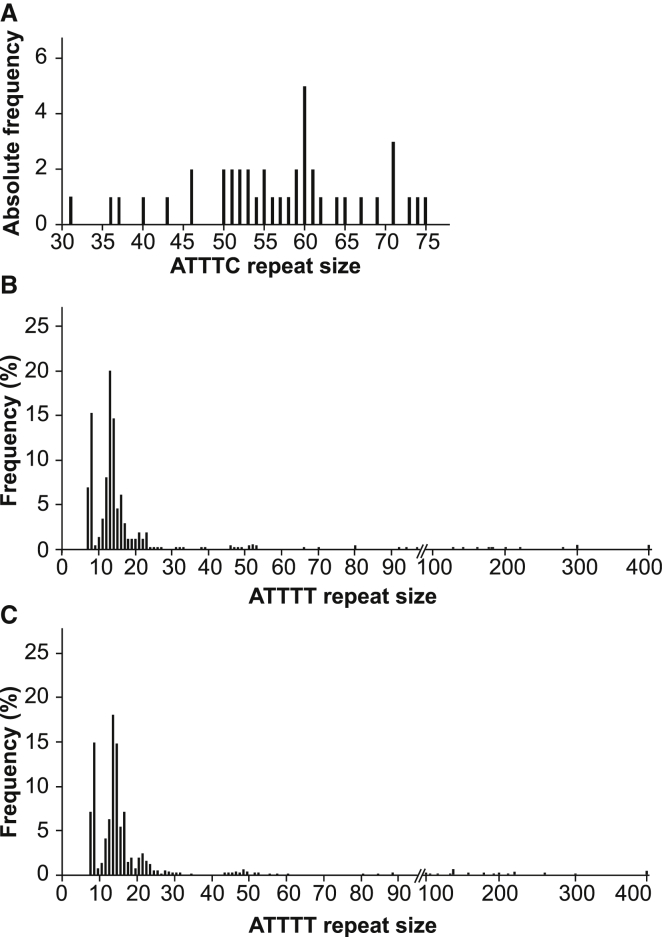
Figure 4.
Distribution of Pathological and Normal Alleles
(A) The absolute frequency of pathological (ATTTC)n allele sizes in SCA subjects (n = 41).
(B and C) Distribution of (ATTTT)n allele sizes in chromosomes from (B) the normal population (n = 520) and (C) affected individuals with other neurodegenerative diseases (n = 904).
Normal Polymorphic ATTTT Alleles Are Rarely Very Large
The ATTTT/AAAAT repeat localizes to the polymorphic middle A-rich region of an AluJb sequence (Figure 3A). After sequencing analysis in 260 subjects from the normal population, we found normal alleles of 7–400 ATTTT/AAAAT repeat tracts that never contained the pathological ATTTC repeat insertion (Figure 4B). We also performed sequencing analysis of a cohort of 452 subjects with neurodegenerative diseases of European and North American origin, including 101 unrelated subjects presenting with ataxia, tremor, or cognitive decline with onset after the age of 50 years and no family history; we did not find the (ATTTC)n insertion in these subjects, and the distribution of the ATTTT/AAAAT repeat allele was similar to that in the normal population (Figure 4C). Overall, the distribution of normal ATTTT/AAAAT repeats comprised mostly alleles shorter than 30 repeats and a rare group of larger alleles ranging from 30 to 400 repeats (with a frequency of 7.3% in the normal population and 6.7% in subjects with neurodegenerative diseases).
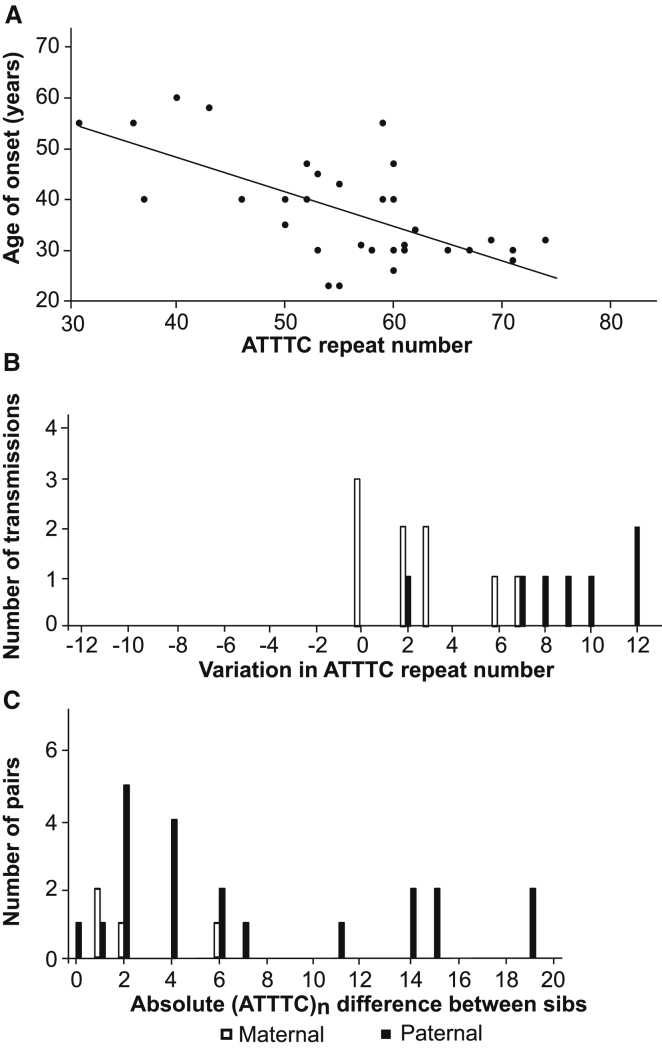
Length of the Unstable (ATTTC)n Insertion Correlates Inversely with Age of Onset
We observed a significant inverse correlation between ATTTC insertion size and age of onset (Figure 5A) whereby affected individuals with larger insertion sizes presented with earlier onset (r = −0.68, p < 0.001, n = 33). Thus, the ATTTC insertion size explains approximately 50% of variation in the age of disease onset (R2 = 0.46). Analysis of the ATTTC repeat insertion in families showed instability upon transmission in 81% (13/16) of parent-offspring transmissions (Figure 5B). The repeat insertion increased in length in all paternal transmissions by 2–12 ATTTCs but in only 67% (6/9) of maternal transmissions by two to seven ATTTC repeats. Figure 5C shows the difference in (ATTTC)n insertion size inherited by pairs of siblings from paternal and maternal transmissions. In sibships, paternal transmissions led to inherited repeat insertions varying by 0–19 units, whereas the difference was of 1–6 ATTTCs upon maternal transmissions. These results suggest that the (ATTTC)n insertion is highly unstable, even more so when the father is the transmitting parent.
Figure 5.
Inverse Correlation between (ATTTC)n Size and Age of Onset in SCA Subjects and Instability
(A) A scatterplot showing an inverse correlation between the length of the (ATTTC)n insertion and age of onset. Affected individuals with larger insertion sizes present with earlier onset (r = −0.68, p < 0.001, n = 33).
(B) Parent-offspring transmissions with varying ATTTC repeat numbers with paternal or maternal origin.
(C) Difference in ATTTC insertion size in pairs of siblings upon paternal and maternal transmissions.
Cerebellar Expression of DAB1 Transcripts with the (ATTTC)n Insertion Intron
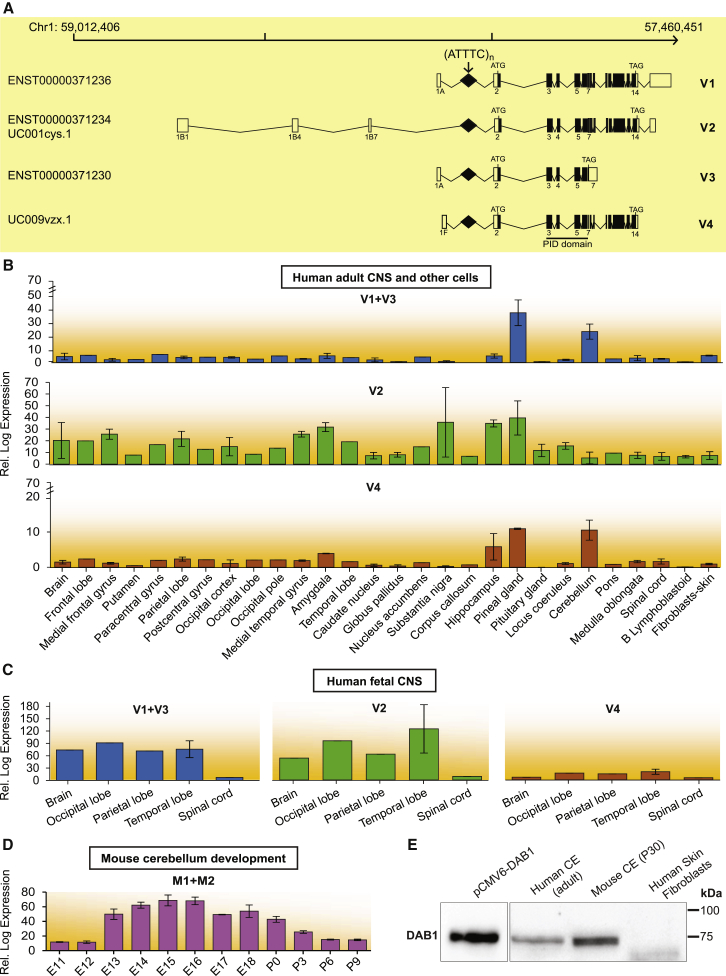
In the DAB1-oriented strand, the ATTTC repeat insertion is located in a 5′ UTR intron. DAB1 encompasses a region of 1.25 Mb genomic DNA. The 5′ UTR is composed of very large introns and spreads over 961 kb.28 At the expression level, the gene is very complex in that it contains several alternative first exons, which can result in transcripts with variable 5′ UTRs. The largest known coding transcript (Ensembl: ENST00000371236) comprises 15 exons and encodes a 5,298-bp mRNA and a 555-aa protein. Two validated DAB1 transcripts have exons flanking the repeat insertion site, and both encode a known 555-aa protein, represented in Figure 6A as V1 and V2. Additionally, two other transcripts found in the UCSC Genome Browser (hg19) and/or Ensembl position the (ATTTC)n insertion region in 5′ UTR introns. These are variants V3, encoding a predicted 213-aa protein, and V4 encoding a known protein of 555 residues (Figure 6A). We confirmed the expression of transcripts encompassing the intronic repeat insertion region by full-length RT-PCR of total human cerebellar RNA (Figure S2). We also analyzed the data obtained by cap analysis of gene expression (CAGE) from the FANTOM5 promoterome project29 and compared the expression of these transcripts in multiple human CNS regions, skin fibroblasts, and lymphoblastoid cell lines (Figure 6B). In human adults, the transcript variants DAB1 V1 and V3 were highly expressed in the cerebellum and modestly expressed in the hippocampus, the reverse of which was true for variant V2. The transcript variants V1, V3, and V4 showed low levels of expression in CNS regions except for the cerebellum and pineal gland, where they showed high levels. The expression of DAB1 was mainly brain specific, and lymphoblastoid cells and fibroblasts showed very low levels of each transcript. All transcript variants showed much higher expression in human fetal brain tissue than in adult brains (Figure 6C). During development of the mouse cerebellum, Dab1 expression is tightly regulated from embryonic day 11 to postnatal day 9, as shown by analysis of the data from FANTOM5 (Figure 6D),30 which is in line with its known essential function in the formation of this brain structure. DAB1 is a reelin signal transducer responsible for the accurate neuronal positioning of cerebellar, hippocampal, and cortical neurons during development.31 DAB1 function relies on an N-terminal PID/PTB (protein interaction/phosphotyrosine binding) domain involved in reelin receptor binding. Importantly, all four transcript variants spanning the repeat insertion region, including those abundantly expressed in the adult cerebellum, encode the functional PID/PTB domain (Figure 6A). Concomitant with the transcriptional data analysis, we found that DAB1 was present in human adult and mouse cerebellum, whereas no protein could be detected in primary skin fibroblasts in these experimental conditions or with an antibody against the C terminus of DAB1 (Figure 6E).
Figure 6.
Expression of DAB1 Transcripts Spanning the Region Containing the Repeat Insertion and DAB1
(A) Schematics of DAB1 genomic position on chromosome 1 show transcripts identified by CAGE (FAMTOM5) to result from usage of alternative promoters. These transcripts are also annotated in Ensembl or the UCSC Genome Browser (hg19), and all have the repeat insertion region in the 5′ UTR. Coding and non-coding exons in transcripts are represented by closed and open boxes, respectively. The location of the (ATTTC)n insertion region in transcript variants is represented by a diamond and indicated with an arrow, and the locations of the ATG start codon and TAG stop codon are shown. The following abbreviation is used: PID, phosphotyrosine interaction domain.
(B–D) Mean expression levels of DAB1 transcripts in different CNS regions, skin fibroblasts, and B lymphoblastoid cells from human adults, as analyzed from CAGE data (B);29 mean expression levels of DAB1 transcript variants in CNS regions of 20- to 29-week-old human fetuses (C); and mean developmental expression levels of Dab1 transcripts in mouse cerebellum from embryonic day 11 (E11) to postnatal day 9 (P9), as analyzed from CAGE data (D).30 Samples available for expression analysis of each CNS region or cell type ranged from one to three in human adults and from one to two in human fetuses, whereas in mouse cerebellum, three samples were available for all stages. Data represent the mean ± SD.
(E) Western blot analysis of DAB1 localization in adult human and mouse cerebellum and in primary skin fibroblasts. DAB1 from HEK293T cells overexpressing human DAB1 cDNA plasmid (pCMV6-hDAB1, OriGene Technologies) was used as a control. Boxes indicate cropped areas from digital images of the same membrane obtained at different exposure times (30 s for the first lane and 5 min for the other three lanes).
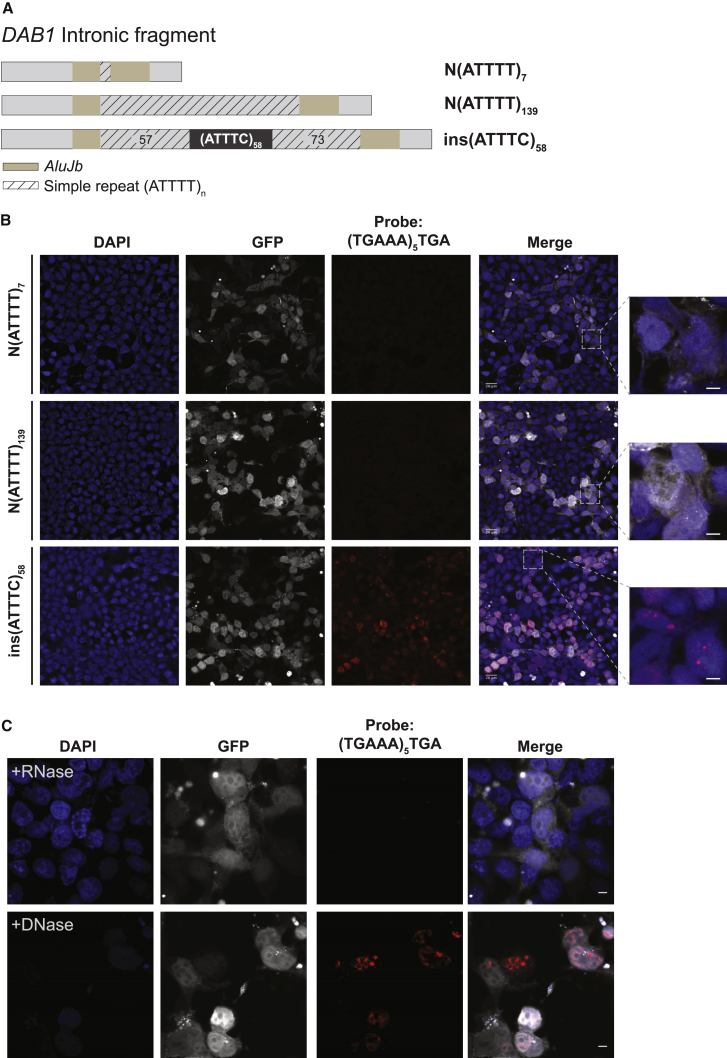
RNA Aggregates in Human Cells Overexpressing the (ATTTC)n Insertion
A plethora of different pathogenic mechanisms have been associated with neurodegeneration caused by repeat expansion in the non-coding region of genes. Formation of abnormal secondary structures by transcripts harboring repeat expansions leading to aberrant nuclear RNA aggregation is a common feature of these neurodegenerative diseases.32 RNA aggregates have been extensively described in affected tissues and can be observed upon overexpression of repeat-containing RNAs in cell models or organisms, even if they are removed from their normal gene context.9 To assess whether the ATTTC repeat insertion in DAB1 causes the formation of RNA aggregates, we overexpressed it in the human embryonic cell line HEK293T. Using a (TGAAA)5TGA probe predicted to hybridize to (AUUUC)n, we detected widespread formation of RNA aggregates by FISH 48 hr after transfection of a plasmid containing the repeat insertion with its flanking ATTTTs and AluJb monomers, ins(ATTTC)58, whereas none were detected in cells transfected with the corresponding normal N(ATTTT)7 or N(ATTTT)139 sequence (Figure 7A). These RNA aggregates generally had a nuclear localization (Figure 7B). Enzymatic treatment of the cells with RNase, but not with DNase, led to the disappearance of the FISH signal, suggesting that the labeled probe was hybridizing specifically to RNA (Figure 7C).
Figure 7.
Formation of (AUUUC)n RNA Aggregates in a Human Cell Line
(A) Two normal Alu fragments with a simple ATTTT repeat of different size and the pathogenic ATTTC repeat insert (with the configuration (ATTTT)57(ATTTC)58(ATTTT)73) were directly amplified from genomic DNA and cloned into the pCDH-CMV-EF1-GFP-Puro vector.
(B) Transient overexpression of the pathogenic ATTTC repeat insertion, but not the normal ATTTT repeat of 7 or 139 units, in HEK293T cells leads to widespread formation of nuclear RNA aggregates visible after FISH staining with a probe, (TGAAA)5TGA, predicted to hybridize to (AUUUC)n. GFP expression was used as a marker for transfection. Represented are single-plane confocal images. Scale bars, 5 μm.
(C) Aggregates, variable in size and number in individual cells, were sensitive to enzymatic treatment with RNase but not DNase. Representative images are projections of z stacked images collected at 0.21 μm intervals with a confocal microscope. Scale bars, 5 μm.
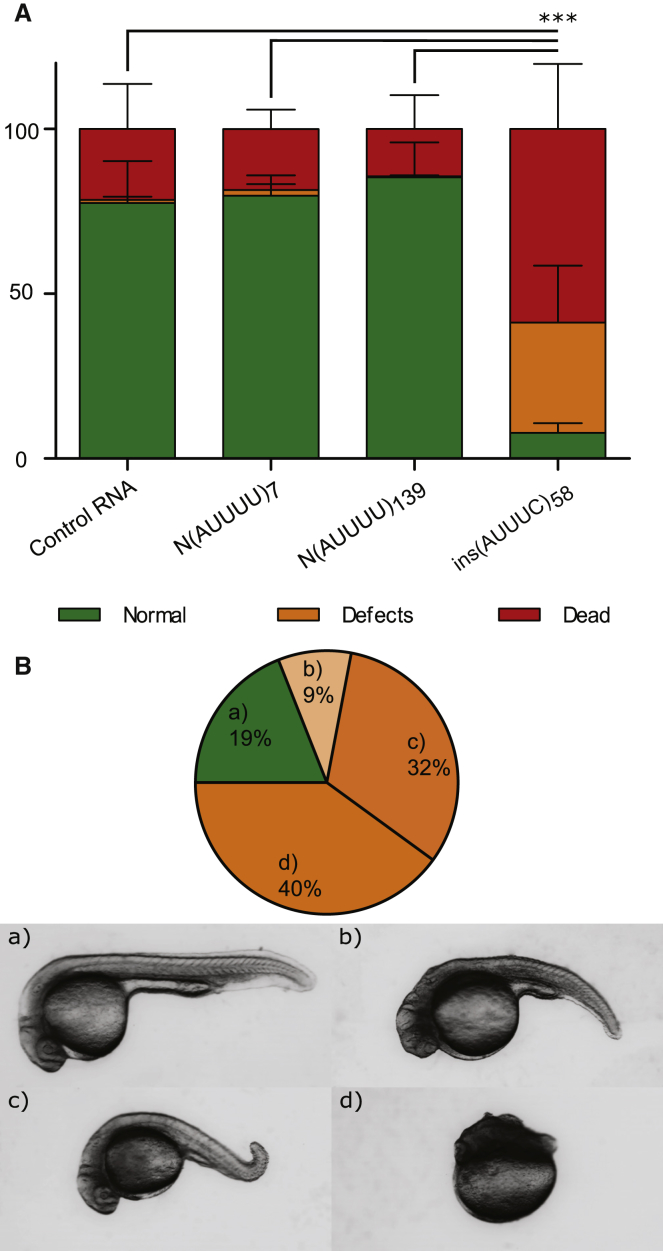
(AUUUC)n-Containing RNA Impairs Early Embryonic Development
To investigate whether the (ATTTC)n repeat insertion is deleterious in vivo, we injected 1- to 2-cell-stage zebrafish embryos with RNA containing the pathological DAB1 repeat insertion with its flanking AUUUUs and AluJb monomers and assessed animal viability, morphology, and survival. We injected RNA obtained from the normal alleles N(ATTTT)7 and N(ATTTT)139 and from the pathological ins(ATTTC)58. At 24 hpf, the lethality rate detected in the embryos injected with ins(AUUUC)58 was significantly higher than in the other conditions (21.51% for control RNA, 18.51% for N(AUUUU)7, 14.40% for N(AUUUU)139, and 58.79% for ins(AUUUC)58; average of three replicas with at least 200 embryos per replica; χ2 test p ≤ 0.001; Figure 8A). This difference was even higher among phenotypes in the surviving embryos, showing that the number of embryos that developed normally in the ins(AUUUC)58 injection was significantly lower than in the other conditions (77.52% for control RNA, 79.79% for N(AUUUU)7, 85.37% for N(AUUUU)139, and 7.76% for ins(AUUUC)58; χ2 test p < 0.0001; Figure 8A). Figure 8B shows the distribution of phenotypic classes observed in embryos injected with ins(AUUUC)58. These results further suggest that the DAB1 repeat insertion identified in this SCA is potentially deleterious in vivo.
Figure 8.
In Vivo Deleterious Effects of the (ATTTC)n Insertion
(A) Percentage of embryos that presented with lethality (dead), developmental defects (defects), or a wild-type phenotype (normal) at 24 hpf after RNA injection of control Cas9RNA, N(AUUUU)7, N(AUUUU)139, or ins(AUUUC)58 (average of three replicas with at least 200 embryos per replica; ∗∗∗p ≤ 0.001; χ2 test for the lethality rate). Data represent the mean ± SD.
(B) Distribution of phenotypic classes (a–d) observed in ins(AUUUC)58-injected embryos at 24 hpf and representative images of the observed phenotypic classes: (a) wild-type, (b) severe defects in the tail and head, (c) mild defect in the tail, and (d) severe defects in the anterior-posterior axis.
Discussion
We report the identification of an autosomal-dominant SCA characterized by pure cerebellar ataxia, as well as dysarthria as the first clinical manifestation, and caused by an insertion of an unstable ATTTC repeat in the non-coding region of the neurodevelopmental DAB1, which locates to the mapped SCA37 locus. The mutation consists of an insertion of an ATTTC repeat in the middle of an (ATTTT)n tract. The (ATTTC)n insertion (ranging from 31 to 75 pure ATTTC units) was found in affected individuals and absent from 520 control chromosomes. It completely segregates with the disease in the six families studied. The DAB1 mutation leads to an aberrant behavior of RNAs containing the AUUUC repeat insertion, rendering them prone to aggregation in a human cell line. In vivo assessment of the pathogenicity of this DAB1 insertion suggests that this abnormal RNA is toxic to zebrafish embryos by causing developmental defects and increased lethality.
The systematic epidemiological survey of hereditary ataxias performed in Portugal has allowed the genetic characterization of a large number of families.3 All affected subjects with this genetic type of SCA are originally from southern Portugal. It is interesting to notice that whereas MJD (SCA3) is the most frequent in the archipelago of Azores and in northern and central mainland Portugal, it is rarely found in the south.3 In the southern provinces of the country, the SCA described here is the most common by representing 14% of autosomal-dominant forms.
It is remarkable that an (ATTTC)n of 31–75 units is disease causing, whereas an (ATTTT)n of much larger size that can reach 400 repeats is not pathogenic. The (ATTTT)n length in healthy individuals ranged from 7–400 pentanucleotide units, whereas the ATTTT repeat tracts that flank the (ATTTC)n insertion in affected individuals from this study were larger than 58 units. The total pentanucleotide repeat size for the complex (ATTTT)n(ATTTC)n(ATTTT)n assessed in this cohort varied from approximately 190 to 220 units. For these sizes, RP-PCR can efficiently detect the repeat insertion in the middle of this complex repeat, but larger flanking (ATTTT)n sizes would not allow RP-PCR insertion detection; sequencing is required for confirmation of its presence.
We observed significant intergenerational instability of the (ATTTC)n insertion, a feature common to most diseases involving repeat expansions.8, 33 This repeat insertion is more unstable when the father is the transmitting parent, when its size tends to be larger than in maternal transmissions. This suggests that meiotic instability could play a relevant role in this dynamic feature, but we cannot exclude the possibility that mitotic instability during development and aging causes somatic mosaicism in the affected brain regions.
Notably, this cohort shows an inverse correlation between age of disease onset and size of the (ATTTC)n insertion, such that larger repeats result in earlier clinical manifestation, a feature of many disorders of repeat expansions. In fact, the size of the repeat insertion accounts for approximately 50% of the variability in age of onset (from late adolescence to the early 60s). Interestingly, recent evidence from a mouse model of SCA1 points to the existence of neuronal dysfunction, from development to adulthood, preceding the appearance of the neurological symptoms and neuronal cell death.34 Here, we identified a SCA-causing mutation in a neurodevelopmental gene, raising the possibility that neuronal dysfunction starting early in brain development could contribute to disease pathogenesis.
The expression of DAB1 is tightly regulated during development and across various brain regions. In the human CNS, expression analysis showed alternative promoter usage for DAB1 and region-specific expression of different transcripts. The (ATTTC)n insertion region reported here is included in all four transcripts identified in the human brain. Three of them encode the same protein of 555 residues (V1, V2, and V4) but differ in their 5′ UTRs. The promoter of transcript V2 is the most widely used in the CNS, whereas the promoters for V1, V3, and V4 are less used in general, except in the cerebellum. Altogether, this suggests that transcriptional regulation most likely plays an important role in tuning DAB1 function, especially in the cerebellum, the brain structure most affected in this SCA. Future studies will address to what extent the ATTTC repeat insertion in the non-coding DAB1 could alter its expression and normal function.
Given that injection of AUUUC repeat RNA into 1-cell-stage zebrafish embryos led to lethal developmental malformations causing significant death, we hypothesize that an RNA-mediated mechanism could contribute to the pathogenesis of this SCA. This RNA-mediated lethality seen in zebrafish embryos might be initiated by different, non-mutually exclusive mechanisms. Considering also that overexpression of the ATTTC repeat insertion leads to the formation of nuclear RNA aggregates in human cell lines, and in light of what we learned from other diseases involving non-coding repeats, these aberrant RNAs can be pathogenic by (1) sequestering critical RNA-binding proteins and preventing them from performing their normal function,32 (2) initiating non-AUG-dependent translation and producing toxic peptides,19, 35 and (3) disrupting RNA metabolism processes such as splicing or nuclear export.8 A possible pathological contribution of bidirectional transcription across the repeat insertion region, described in several repeat diseases,8, 15 remains to be investigated.
This form of cerebellum-specific degeneration is caused by a repeat insertion in DAB1, which encodes an adaptor protein of the reelin signaling pathway that controls neuronal migration and maturation of synaptic connections in the brain during development.36 Homozygous mutations in mouse Dab1 cause the scrambler and yotari phenotypes, characterized by aberrant splicing forms of Dab1 transcripts and little or no DAB1, leading to widespread misplacement of neurons in the cerebellum, hippocampus, and cortex and the associated ataxia.31, 36, 37 In young-adult mouse hippocampus, the reelin-DAB1 pathway regulates the maturation of dendritic spines, synaptogenesis, and glial ensheathment of newborn granule cells.38 In humans, a homozygous deletion of VLDLR (MIM: 192977), which encodes a receptor for reelin, originates an autosomal-recessive nonprogressive cerebellar ataxia and mental impairment (MIM: 224050).39 The presence of cerebellar symptoms is a common feature when the reelin-DAB1 pathway is impaired; however, the cerebellar atrophy exhibited by the affected individuals in this study is very mild in comparison with the recessive phenotypes referred above.
It is interesting that the (ATTTC)31–75 insertion in the intronic 5′ UTR of DAB1 is structurally similar to the SCA10-causing (ATTCT)800–4,500 expansion in intron 9 of ATXN10.16 The size ranges of the pathological pentanucleotide repeats for fully penetrant alleles differ greatly between these two diseases. One can speculate that these different size requirements for pathology are related to the specific gene context and localization in genomic elements.40 Nevertheless, given the bias toward increased (ATTTC)n size upon transmission, larger alleles might be expected to appear in affected individuals from other families with this repeat insertion. Notably, alleles of 280–850 units show reduced penetrance in SCA10, and alleles of 33–280 repeats have not been observed.41, 42, 43
DAB1 lies within the mapped SCA37 locus in chromosomal region 1p32 in a family originating from Spain.20 The occurrence of multiple SCA gene mutations clustered in small genomic regions is rare and has been seen only in PDYN (MIM: 131340; associated with SCA23 [MIM: 610245]), NOP56 (associated with SCA36), and TGM6 (MIM: 613900; associated with SCA35 [MIM: 613908]), which are contained in a 750 kb region on human chromosomal arm 20p.12, 44, 45 DAB1 encompasses 1.25 Mb within the SCA37 locus, and there is a possibility that the same ATTTC repeat insertion described here is the mutation in the SCA37-affected family, especially given that this kindred is from Spain, a country that borders Portugal. Thus, it is possible that the high incidence of DAB1 repeat insertions found in Portuguese SCA subjects could extend to Spain and probably other populations of European ancestry.
The highly polymorphic (ATTTT)n where the ATTTC insertion occurred is located in the middle A-rich stretch of an AluJb element. Other repeat diseases, such as Friedrich ataxia (MIM: 229300), DM2, SCA10, and SCA31, originate from the expansion or insertion of repeats in poly-A tracts in the middle or 3′ end of Alu elements.14, 46, 47, 48 These A-rich tracts can be the basis for nucleotide substitution leading to microsatellite birth either through errors introduced during Alu reverse transcription or through the accumulation of mutations in Alu poly-A stretches after insertion.40
This is the second neurodegenerative disease (in addition to SCA31) caused by an insertion in a polymorphic repeat located in an A-rich region of an Alu element.14 In both cases, the insertion of the pathogenic repeat is flanked by ATTTT/AAAAT tracts, creating a large unstable repeat that shares similarities with expanded repeats. These unstable repeat insertions might thus constitute a new class of dynamic mutations, difficult to identify with the common strategies currently in use. Furthermore, its occurrence in two genetic types of SCA suggests that they could be implicated in other brain diseases of unknown molecular etiology. The discovery of a pathogenic (ATTTC)n insertion in DAB1 could contribute toward a better understanding of neurodegenerative diseases.
Acknowledgments
We thank the families who participated in this study. We are grateful to Gonçalo Abecasis, Miguel Costa, Tito Vieira, and André Torres for help with MERLIN analysis; Beatriz Sobrino, Jorge Amigo, and Pilar Cacheiro for next-generation sequencing analysis, performed at the Santiago de Compostela node of the Spanish National Genotyping Center; Nuno Santarém and Anabela Cordeiro-da-Silva for assistance with cloning; António Amorim, Laura Vilarinho, and Paula Jorge for samples from the Portuguese population; and Paula Magalhães from the Institute for Molecular and Cell Biology Cell Culture and Genotyping Core for DNA extraction. This work was financed by Fundo Europeu de Desenvolvimento Regional (FEDER) funds through the COMPETE 2020 Operational Program for Competitiveness and Internationalization (POCI) of Portugal 2020 and by Portuguese funds through the Fundação para a Ciência e a Tecnologia (FCT) and Ministério da Ciência, Tecnologia, e Inovação in the framework of the project “Institute for Research and Innovation in Health Sciences” (POCI-01-0145-FEDER-007274); and by FCT grant PTDC/SAU-GMG/098305/2008 to I.S. A.I.S. was the recipient of an FCT scholarship (SFRH/BD/30702/2006). J.R.L. was supported by scholarships from PEst-C/SAU/LA0002/2013 and the European Molecular Biology Organization (ASTF494-2015). C.L.O. was supported by a scholarship from PEst-C/SAU/LA0002/2013. This work was also financed by the Porto Neurosciences and Neurologic Disease Research Initiative at the Instituto de Investigação e Inovação em Saúde (Norte-01-0145-FEDER-000008), supported by Norte Portugal Regional Operational Programme (NORTE 2020) under the PORTUGAL 2020 Partnership Agreement through FEDER, and by the Fondo de Investigación Sanitaria of the Instituto de Salud Carlos III (grant PI12/00742).
Published: July 6, 2017
Footnotes
Supplemental Data include two figures and four tables and can be found with this article online at http://dx.doi.org/10.1016/j.ajhg.2017.06.007.
Accession Numbers
The accession numbers for the data reported in this paper are ClinVar: SCV000579455 and dbSNP: ss2137493862, ss2137493857, ss2137493861, ss2137493856, and ss2137493855.
Web Resources
1000 Genomes, http://www.1000genomes.org/
Ensemble Genome Browser, http://www.ensembl.org/index.html
GenBank, http://www.ncbi.nlm.nih.gov/genbank/
Genome Analysis Toolkit, http://www.broadinstitute.org/genome-analysis-toolkit
OMIM, http://www.omim.org
UCSC Genome Browser, http://genome.ucsc.edu/
Supplemental Data
References
- 1.Sequeiros J., Martins S., Silveira I. Epidemiology and population genetics of degenerative ataxias. Handb. Clin. Neurol. 2012;103:227–251. doi: 10.1016/B978-0-444-51892-7.00014-0. [DOI] [PubMed] [Google Scholar]
- 2.Durr A. Autosomal dominant cerebellar ataxias: polyglutamine expansions and beyond. Lancet Neurol. 2010;9:885–894. doi: 10.1016/S1474-4422(10)70183-6. [DOI] [PubMed] [Google Scholar]
- 3.Coutinho P., Ruano L., Loureiro J.L., Cruz V.T., Barros J., Tuna A., Barbot C., Guimarães J., Alonso I., Silveira I. Hereditary ataxia and spastic paraplegia in Portugal: a population-based prevalence study. JAMA Neurol. 2013;70:746–755. doi: 10.1001/jamaneurol.2013.1707. [DOI] [PubMed] [Google Scholar]
- 4.Silveira I., Miranda C., Guimarães L., Moreira M.C., Alonso I., Mendonça P., Ferro A., Pinto-Basto J., Coelho J., Ferreirinha F. Trinucleotide repeats in 202 families with ataxia: a small expanded (CAG)n allele at the SCA17 locus. Arch. Neurol. 2002;59:623–629. doi: 10.1001/archneur.59.4.623. [DOI] [PubMed] [Google Scholar]
- 5.Carlson K.M., Andresen J.M., Orr H.T. Emerging pathogenic pathways in the spinocerebellar ataxias. Curr. Opin. Genet. Dev. 2009;19:247–253. doi: 10.1016/j.gde.2009.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coutelier M., Blesneac I., Monteil A., Monin M.L., Ando K., Mundwiller E., Brusco A., Le Ber I., Anheim M., Castrioto A. A Recurrent Mutation in CACNA1G Alters Cav3.1 T-Type Calcium-Channel Conduction and Causes Autosomal-Dominant Cerebellar Ataxia. Am. J. Hum. Genet. 2015;97:726–737. doi: 10.1016/j.ajhg.2015.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Di Gregorio E., Borroni B., Giorgio E., Lacerenza D., Ferrero M., Lo Buono N., Ragusa N., Mancini C., Gaussen M., Calcia A. ELOVL5 mutations cause spinocerebellar ataxia 38. Am. J. Hum. Genet. 2014;95:209–217. doi: 10.1016/j.ajhg.2014.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Loureiro J.R., Oliveira C.L., Silveira I. Unstable repeat expansions in neurodegenerative diseases: nucleocytoplasmic transport emerges on the scene. Neurobiol. Aging. 2016;39:174–183. doi: 10.1016/j.neurobiolaging.2015.12.007. [DOI] [PubMed] [Google Scholar]
- 9.Mankodi A., Logigian E., Callahan L., McClain C., White R., Henderson D., Krym M., Thornton C.A. Myotonic dystrophy in transgenic mice expressing an expanded CUG repeat. Science. 2000;289:1769–1773. doi: 10.1126/science.289.5485.1769. [DOI] [PubMed] [Google Scholar]
- 10.La Spada A.R., Taylor J.P. Repeat expansion disease: progress and puzzles in disease pathogenesis. Nat. Rev. Genet. 2010;11:247–258. doi: 10.1038/nrg2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wojciechowska M., Krzyzosiak W.J. Cellular toxicity of expanded RNA repeats: focus on RNA foci. Hum. Mol. Genet. 2011;20:3811–3821. doi: 10.1093/hmg/ddr299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kobayashi H., Abe K., Matsuura T., Ikeda Y., Hitomi T., Akechi Y., Habu T., Liu W., Okuda H., Koizumi A. Expansion of intronic GGCCTG hexanucleotide repeat in NOP56 causes SCA36, a type of spinocerebellar ataxia accompanied by motor neuron involvement. Am. J. Hum. Genet. 2011;89:121–130. doi: 10.1016/j.ajhg.2011.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.García-Murias M., Quintáns B., Arias M., Seixas A.I., Cacheiro P., Tarrío R., Pardo J., Millán M.J., Arias-Rivas S., Blanco-Arias P. ‘Costa da Morte’ ataxia is spinocerebellar ataxia 36: clinical and genetic characterization. Brain. 2012;135:1423–1435. doi: 10.1093/brain/aws069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sato N., Amino T., Kobayashi K., Asakawa S., Ishiguro T., Tsunemi T., Takahashi M., Matsuura T., Flanigan K.M., Iwasaki S. Spinocerebellar ataxia type 31 is associated with “inserted” penta-nucleotide repeats containing (TGGAA)n. Am. J. Hum. Genet. 2009;85:544–557. doi: 10.1016/j.ajhg.2009.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moseley M.L., Zu T., Ikeda Y., Gao W., Mosemiller A.K., Daughters R.S., Chen G., Weatherspoon M.R., Clark H.B., Ebner T.J. Bidirectional expression of CUG and CAG expansion transcripts and intranuclear polyglutamine inclusions in spinocerebellar ataxia type 8. Nat. Genet. 2006;38:758–769. doi: 10.1038/ng1827. [DOI] [PubMed] [Google Scholar]
- 16.Matsuura T., Yamagata T., Burgess D.L., Rasmussen A., Grewal R.P., Watase K., Khajavi M., McCall A.E., Davis C.F., Zu L. Large expansion of the ATTCT pentanucleotide repeat in spinocerebellar ataxia type 10. Nat. Genet. 2000;26:191–194. doi: 10.1038/79911. [DOI] [PubMed] [Google Scholar]
- 17.Cleary J.D., Ranum L.P. New developments in RAN translation: insights from multiple diseases. Curr. Opin. Genet. Dev. 2017;44:125–134. doi: 10.1016/j.gde.2017.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pearson C.E. Repeat associated non-ATG translation initiation: one DNA, two transcripts, seven reading frames, potentially nine toxic entities! PLoS Genet. 2011;7:e1002018. doi: 10.1371/journal.pgen.1002018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ishiguro T., Sato N., Ueyama M., Fujikake N., Sellier C., Kanegami A., Tokuda E., Zamiri B., Gall-Duncan T., Mirceta M. Regulatory Role of RNA Chaperone TDP-43 for RNA Misfolding and Repeat-Associated Translation in SCA31. Neuron. 2017;94:108–124.e7. doi: 10.1016/j.neuron.2017.02.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Serrano-Munuera C., Corral-Juan M., Stevanin G., San Nicolás H., Roig C., Corral J., Campos B., de Jorge L., Morcillo-Suárez C., Navarro A. New subtype of spinocerebellar ataxia with altered vertical eye movements mapping to chromosome 1p32. JAMA Neurol. 2013;70:764–771. doi: 10.1001/jamaneurol.2013.2311. [DOI] [PubMed] [Google Scholar]
- 21.Rüschendorf F., Nürnberg P. ALOHOMORA: a tool for linkage analysis using 10K SNP array data. Bioinformatics. 2005;21:2123–2125. doi: 10.1093/bioinformatics/bti264. [DOI] [PubMed] [Google Scholar]
- 22.Wigginton J.E., Abecasis G.R. PEDSTATS: descriptive statistics, graphics and quality assessment for gene mapping data. Bioinformatics. 2005;21:3445–3447. doi: 10.1093/bioinformatics/bti529. [DOI] [PubMed] [Google Scholar]
- 23.Abecasis G.R., Cherny S.S., Cookson W.O., Cardon L.R. Merlin--rapid analysis of dense genetic maps using sparse gene flow trees. Nat. Genet. 2002;30:97–101. doi: 10.1038/ng786. [DOI] [PubMed] [Google Scholar]
- 24.McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., Garimella K., Altshuler D., Gabriel S., Daly M., DePristo M.A. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hwang W.Y., Fu Y., Reyon D., Maeder M.L., Tsai S.Q., Sander J.D., Peterson R.T., Yeh J.R., Joung J.K. Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat. Biotechnol. 2013;31:227–229. doi: 10.1038/nbt.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mitsuhashi H., Mitsuhashi S., Lynn-Jones T., Kawahara G., Kunkel L.M. Expression of DUX4 in zebrafish development recapitulates facioscapulohumeral muscular dystrophy. Hum. Mol. Genet. 2013;22:568–577. doi: 10.1093/hmg/dds467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bar I., Tissir F., Lambert de Rouvroit C., De Backer O., Goffinet A.M. The gene encoding disabled-1 (DAB1), the intracellular adaptor of the Reelin pathway, reveals unusual complexity in human and mouse. J. Biol. Chem. 2003;278:5802–5812. doi: 10.1074/jbc.M207178200. [DOI] [PubMed] [Google Scholar]
- 29.Forrest A.R., Kawaji H., Rehli M., Baillie J.K., de Hoon M.J., Haberle V., Lassmann T., Kulakovskiy I.V., Lizio M., Itoh M., FANTOM Consortium and the RIKEN PMI and CLST (DGT) A promoter-level mammalian expression atlas. Nature. 2014;507:462–470. doi: 10.1038/nature13182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Arner E., Daub C.O., Vitting-Seerup K., Andersson R., Lilje B., Drabløs F., Lennartsson A., Rönnerblad M., Hrydziuszko O., Vitezic M., FANTOM Consortium Transcribed enhancers lead waves of coordinated transcription in transitioning mammalian cells. Science. 2015;347:1010–1014. doi: 10.1126/science.1259418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Howell B.W., Gertler F.B., Cooper J.A. Mouse disabled (mDab1): a Src binding protein implicated in neuronal development. EMBO J. 1997;16:121–132. doi: 10.1093/emboj/16.1.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Todd P.K., Paulson H.L. RNA-mediated neurodegeneration in repeat expansion disorders. Ann. Neurol. 2010;67:291–300. doi: 10.1002/ana.21948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Silveira I., Alonso I., Guimarães L., Mendonça P., Santos C., Maciel P., Fidalgo De Matos J.M., Costa M., Barbot C., Tuna A. High germinal instability of the (CTG)n at the SCA8 locus of both expanded and normal alleles. Am. J. Hum. Genet. 2000;66:830–840. doi: 10.1086/302827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hatanaka Y., Watase K., Wada K., Nagai Y. Abnormalities in synaptic dynamics during development in a mouse model of spinocerebellar ataxia type 1. Sci. Rep. 2015;5:16102. doi: 10.1038/srep16102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zu T., Gibbens B., Doty N.S., Gomes-Pereira M., Huguet A., Stone M.D., Margolis J., Peterson M., Markowski T.W., Ingram M.A. Non-ATG-initiated translation directed by microsatellite expansions. Proc. Natl. Acad. Sci. USA. 2011;108:260–265. doi: 10.1073/pnas.1013343108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sheldon M., Rice D.S., D’Arcangelo G., Yoneshima H., Nakajima K., Mikoshiba K., Howell B.W., Cooper J.A., Goldowitz D., Curran T. Scrambler and yotari disrupt the disabled gene and produce a reeler-like phenotype in mice. Nature. 1997;389:730–733. doi: 10.1038/39601. [DOI] [PubMed] [Google Scholar]
- 37.Goldowitz D., Cushing R.C., Laywell E., D’Arcangelo G., Sheldon M., Sweet H.O., Davisson M., Steindler D., Curran T. Cerebellar disorganization characteristic of reeler in scrambler mutant mice despite presence of reelin. J. Neurosci. 1997;17:8767–8777. doi: 10.1523/JNEUROSCI.17-22-08767.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bosch C., Masachs N., Exposito-Alonso D., Martínez A., Teixeira C.M., Fernaud I., Pujadas L., Ulloa F., Comella J.X., DeFelipe J. Reelin Regulates the Maturation of Dendritic Spines, Synaptogenesis and Glial Ensheathment of Newborn Granule Cells. Cereb. Cortex. 2016;26:4282–4298. doi: 10.1093/cercor/bhw216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Boycott K.M., Flavelle S., Bureau A., Glass H.C., Fujiwara T.M., Wirrell E., Davey K., Chudley A.E., Scott J.N., McLeod D.R., Parboosingh J.S. Homozygous deletion of the very low density lipoprotein receptor gene causes autosomal recessive cerebellar hypoplasia with cerebral gyral simplification. Am. J. Hum. Genet. 2005;77:477–483. doi: 10.1086/444400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kelkar Y.D., Eckert K.A., Chiaromonte F., Makova K.D. A matter of life or death: how microsatellites emerge in and vanish from the human genome. Genome Res. 2011;21:2038–2048. doi: 10.1101/gr.122937.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Alonso I., Jardim L.B., Artigalas O., Saraiva-Pereira M.L., Matsuura T., Ashizawa T., Sequeiros J., Silveira I. Reduced penetrance of intermediate size alleles in spinocerebellar ataxia type 10. Neurology. 2006;66:1602–1604. doi: 10.1212/01.wnl.0000216266.30177.bb. [DOI] [PubMed] [Google Scholar]
- 42.Seixas A.I., Maurer M.H., Lin M., Callahan C., Ahuja A., Matsuura T., Ross C.A., Hisama F.M., Silveira I., Margolis R.L. FXTAS, SCA10, and SCA17 in American patients with movement disorders. Am. J. Med. Genet. A. 2005;136:87–89. doi: 10.1002/ajmg.a.30761. [DOI] [PubMed] [Google Scholar]
- 43.Matsuura T., Fang P., Pearson C.E., Jayakar P., Ashizawa T., Roa B.B., Nelson D.L. Interruptions in the expanded ATTCT repeat of spinocerebellar ataxia type 10: repeat purity as a disease modifier? Am. J. Hum. Genet. 2006;78:125–129. doi: 10.1086/498654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bakalkin G., Watanabe H., Jezierska J., Depoorter C., Verschuuren-Bemelmans C., Bazov I., Artemenko K.A., Yakovleva T., Dooijes D., Van de Warrenburg B.P. Prodynorphin mutations cause the neurodegenerative disorder spinocerebellar ataxia type 23. Am. J. Hum. Genet. 2010;87:593–603. doi: 10.1016/j.ajhg.2010.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang J.L., Yang X., Xia K., Hu Z.M., Weng L., Jin X., Jiang H., Zhang P., Shen L., Guo J.F. TGM6 identified as a novel causative gene of spinocerebellar ataxias using exome sequencing. Brain. 2010;133:3510–3518. doi: 10.1093/brain/awq323. [DOI] [PubMed] [Google Scholar]
- 46.Clark R.M., Dalgliesh G.L., Endres D., Gomez M., Taylor J., Bidichandani S.I. Expansion of GAA triplet repeats in the human genome: unique origin of the FRDA mutation at the center of an Alu. Genomics. 2004;83:373–383. doi: 10.1016/j.ygeno.2003.09.001. [DOI] [PubMed] [Google Scholar]
- 47.Kurosaki T., Matsuura T., Ohno K., Ueda S. Alu-mediated acquisition of unstable ATTCT pentanucleotide repeats in the human ATXN10 gene. Mol. Biol. Evol. 2009;26:2573–2579. doi: 10.1093/molbev/msp172. [DOI] [PubMed] [Google Scholar]
- 48.Kurosaki T., Ueda S., Ishida T., Abe K., Ohno K., Matsuura T. The unstable CCTG repeat responsible for myotonic dystrophy type 2 originates from an AluSx element insertion into an early primate genome. PLoS ONE. 2012;7:e38379. doi: 10.1371/journal.pone.0038379. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.