Abstract
Signal transducer and activator of transcription 3 (STAT3) is associated with various physiological and pathological functions, mainly as a transcription factor that translocates to the nucleus upon tyrosine phosphorylation induced by cytokine stimulation. In addition, a small pool of STAT3 resides in the mitochondria, where it serves as a sensor for various metabolic stressors including reactive oxygen species (ROS). Mitochondrially localized STAT3 largely exerts its effects through direct or indirect regulation of the activity of the electron transport chain (ETC). It has been assumed that the amounts of STAT3 in the mitochondria are static. We showed that various stimuli, including oxidative stress and cytokines, triggered a signaling cascade that resulted in a rapid loss of mitochondrially localized STAT3. Recovery of the mitochondrial pool of STAT3 over time depended on phosphorylation of Ser727 in STAT3 and new protein synthesis. Under these conditions, mitochondrially localized STAT3 also became competent to bind to cyclophilin D (CypD). Binding of STAT3 to CypD was mediated by the amino terminus of STAT3, which was also important for reducing mitochondrial ROS production after oxidative stress. These results outline a role for mitochondrially localized STAT3 in sensing and responding to external stimuli.
INTRODUCTION
Signal transducer and activator of transcription 3 (STAT3), a member of the STAT family of nuclear transcription factors, plays a key role in the regulation of a diverse set of physiological processes. Ideally situated as a molecular link between cellular inputs and nuclear gene regulation, STAT3 is aberrantly regulated under various pathological conditions, including cancer, cardiovascular disease, and disorders in immune responses. The intricacies of this signaling network have been teased out to better appreciate the mechanisms behind STAT3’s regulation of nuclear gene expression. The discovery of a distinct pool of STAT3 that resides in the mitochondria (1, 2) has added a new layer to the importance of STAT3 in controlling cellular homeostasis.
The functional importance of mitochondrial STAT3 (mitoSTAT3) has been extensively explored. mitoSTAT3 has been linked to the control of the electron transport chain (ETC) and adenosine 5′-triphosphate (ATP) production, mitochondrially encoded RNA regulation, modulation of reactive oxygen species (ROS) generation, Ras transformation, cellular growth, and protection from ischemia-reperfusion injuries through regulation of the mitochondrial permeability transition pore (MPTP) (3). Most reports attribute the phosphorylation of STAT3 at Ser727 to being key for its mitochondrial role, with the mitogen-activated protein kinase kinase (MEK)–extracellular signal–regulated kinase (ERK) signaling axis potentially being an important player (4, 5). However, the signaling pathways that control mitoSTAT3 largely still remain to be elucidated. Understanding such a pathway is especially important, considering that most studies looking to target STAT3 for therapeutic purposes have neglected its mitochondrial role, which could partly explain the absence of a clinically usable STAT3 inhibitor (6, 7). To more effectively study and target mitoSTAT3, we sought to examine its regulation at the signaling level.
We report that mitoSTAT3 was dynamically regulated by various cellular inputs, including oxidative stress and cytokines. Under these conditions, mitoSTAT3 abundance initially decreased. This was followed by a reequilibration of STAT3 into the mitochondria that depended on Ser727 phosphorylation. During this recovery phase, mitoSTAT3 bound to cyclophilin D (CypD), which could play a role in stabilizing the mitoSTAT3 pool. The N terminus of mitoSTAT3 appeared to be necessary for its association with CypD, thereby pointing to an additional site besides Ser727 in STAT3 that is important for its mitochondrial function. In this system, the N terminus of STAT3 was also important for regulating the mitochondrial superoxide production after an oxidative insult, likely as a consequence of the CypD association. These results provide insight into a signaling pathway that controls the amount of mitoSTAT3, manipulation of which may provide new therapeutic modalities to treat diseases associated with the dysfunctional actions of STAT3.
RESULTS
Rapid loss of mitoSTAT3 after an oxidative stress insult
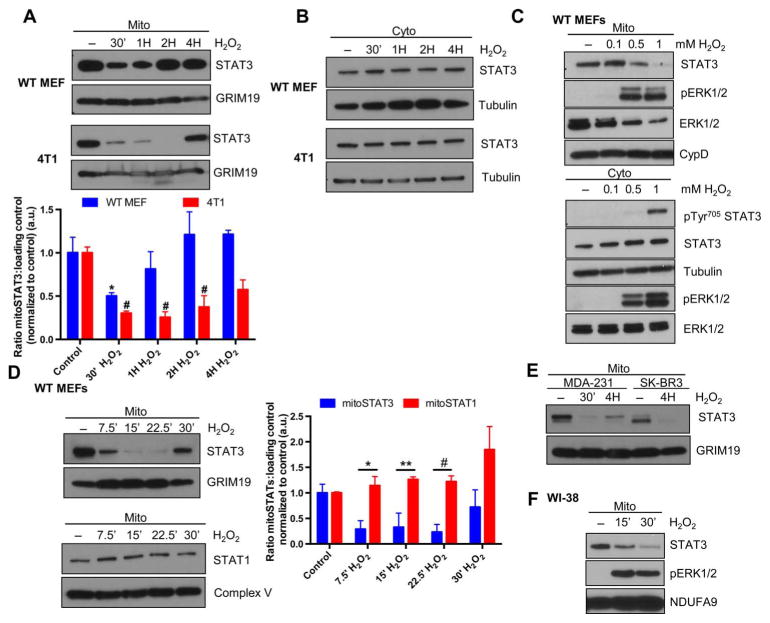
mitoSTAT3 limits mitochondrial and cellular ROS production (8–12). STAT3 is also posttranslationally modified by oxidation that in turn regulates its function (13, 14), and ROS may be important for controlling mitoSTAT3 amounts (15). Therefore, we examined whether mitoSTAT3 was regulated after an oxidative insult. When wild-type mouse embryonic fibroblasts (MEFs) or 4T1 breast cancer cells were treated with H2O2, we observed an initial loss of STAT3 from the mitochondria that recovered with time (Fig. 1A). No changes were noted in STAT3 amounts in cytosolic fractions from H2O2-treated wild-type MEFs or 4T1 cells (Fig. 1B), suggesting against nonspecific degradation of mitoSTAT3 as being responsible for the loss of the protein. H2O2 also induced phosphorylation of STAT3 at both Tyr705 and Ser727 (fig. S1, A and B), which was accompanied by an increase in the nuclear amounts of STAT3 (fig. S1C). The loss of mitoSTAT3 upon treatment with H2O2 was dependent on concentration and also coincided with the activation of mitochondrial ERK1/2 (Fig. 1C). We noted potent Ser727 phosphorylation of mitoSTAT3 after H2O2 treatment (fig. S1D), but mutation of either Ser727 or Tyr705 did not abolish the decrease in mitoSTAT3 amounts (fig. S1E).
Fig. 1. Oxidative stress triggers a loss of mitoSTAT3.
Wild-type (WT) MEFs or 4T1 cells were treated with 1 mM H2O2 for the indicated times, and mitochondrial (mito) (A) or cytosolic (cyto) (B) extracts were probed for STAT3. Quantification of mitoSTAT3 as compared to a loading control is depicted in (A). *P < 0.05 (compared to control); #P < 0.05 (compared to control). Blots are representative of three independent experiments. (C) WT MEFs treated with the indicated concentrations of H2O2 and extracts probed for mitoSTAT3 (top) and phosphorylated (p) Tyr705 and total STAT3 in the cytosol (bottom) (tubulin, cytosolic loading control; CypD, mitochondrial loading control). Blots are representative of two independent experiments. (D) Isolated mitochondria from WT MEFs treated for the indicated times with H2O2 were immunoblotted for mitoSTAT3 or mitoSTAT1 and quantified. *P < 0.01; **P < 0.04; #P < 0.005. Blots are representative of three independent experiments. a.u., arbitrary units. (E) MDA-231 and SK-BR3 human breast cancer cells were treated for the indicated times with H2O2 and immunoblotted for STAT3 and GRIM19 (mitochondrial loading control). Blots are representative of two independent experiments. (F) WI-38 human lung fibroblasts were treated with H2O2, and purified mitochondria were immediately lysed in sample buffer. Blots were probed for mitoSTAT3, pERK1/2, and NDUFA9 (mitochondrial loading control). Blots are representative of two independent experiments.
The kinetics of this pathway were extremely rapid because treatment with H2O2 for only 7.5 min triggered a loss of mitoSTAT3, with recovery starting to occur by 30 min (Fig. 1D). The loss of mitoSTAT3 appeared to be specific because mitoSTAT1 amounts were unchanged after H2O2 treatment (Fig. 1D). Probing mitochondrial extracts with a diverse set of STAT3 antibodies targeted to both the N and C termini all showed the same loss of mitoSTAT3 upon H2O2 treatment, making it unlikely that STAT3 was modified such that it is no longer immunoreactive (fig. S1F). Treatment of MDA-231 (ER−/PR−/HER2−) or SK-BR3 (ER−/PR−/HER2+) human breast cancer cell lines with H2O2 led to a similar loss of mitoSTAT3 (Fig. 1E), indicating that this pathway is active in receptor-negative and receptor-positive cancer lines alike. Mitochondria isolated from WI-38 human lung fibroblasts that were immediately lysed after treatment with H2O2 also displayed a decrease in mitoSTAT3 (Fig. 1F). This result suggested that it was unlikely that mitoSTAT3 was going to an insoluble fraction in the mitochondria and pointed to a true loss of the protein from this organelle. It appeared from these experiments and others (fig. S1G) that the exact timing of the loss and recovery was cell-dependent.
Loss and recovery of mitoSTAT3 with cytokine administration
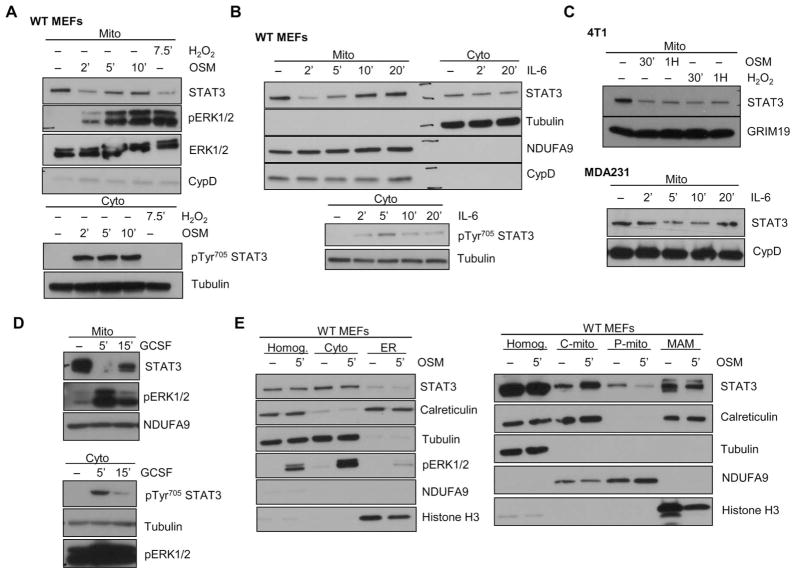
Because STAT3 is activated by interleukin-6 (IL-6) family cytokines, which bind to their respective receptor and transduce signals through glycoprotein 130 (gp130), we investigated whether typical STAT3 stimuli produce similar effects on mitoSTAT3. Treatment of MEFs with oncostatin M (OSM) triggered a loss of mitoSTAT3 within minutes that recovered again with time (Fig. 2A). Similar results were obtained when cells were treated with IL-6 with no apparent decrease in cytosolic STAT3 amounts (Fig. 2B). We observed the loss and recovery of mitoSTAT3 with cytokine treatment in several mouse and human cell lines (Fig. 2C and fig. S2A). Longer-term treatment of cells with cytokines (16) or growth factors (5, 17, 18) drives an increase in mitoSTAT3 amounts. This effect was also recapitulated in MEFs stimulated with OSM for 2 hours, suggesting that acute and long-term exposure differentially regulated mitoSTAT3 (fig. S2B). Likewise, extended treatment of cells with buthionine sulfoximine, an inhibitor of γ-glutamylcysteine synthetase that with time triggers oxidative stress, also led to an increase in mitoSTAT3 (fig. S2C).
Fig. 2. Cytokine treatment of cells induces mitoSTAT3 loss and recovery.
(A and B) WT MEFs were treated with either OSM (A) and IL-6 (B) for the indicated times, and mitochondrial extracts were immunoblotted for STAT3, pERK, total ERK (ERK1/2), and a loading control (NDUFA9 and/or CypD) (top). Cytosolic extracts probed for pTyr705 STAT3 are shown to confirm cell activation (bottom). Blots are representative of two independent experiments. (C) Mitochondria from 4T1 cells treated with OSM (top) or MDA-231 cells treated with IL-6 (bottom) were probed for STAT3 and loading control (GRIM19 or CypD). Blots are representative of two independent experiments. (D) Rat hepatoma cell lines expressing the chimeric G-CSF–gp130 receptor were stimulated with recombinant human G-CSF and immunoblotted for STAT3 in the mitochondrial fraction or pTyr705 STAT3 in the cytosolic fraction. Blots are representative of two independent experiments. (E) Purified mitochondria (P-mito) from control or OSM-stimulated WT MEFs were fractionated into cytosolic (cyto), ER (endoplasmic reticulum), crude mitochondria (C-mito), pure mitochondria (P-mito), and a mitochondria-associated membrane (MAM) fraction and blotted against a total homogenate fraction (Homog.). The abundance of calreticulin (ER and MAM marker), tubulin (cytosolic marker), NDUFA9 (mitochondrial marker), histone H3 (nuclear marker), and STAT3 is shown. Blots are representative of two independent experiments.
The above results implied that signals originating from the plasma membrane could be transduced to the mitochondria. To further explore this possibility, we took advantage of cell lines containing chimeric receptors expressing the extracellular domain of granulocyte colony-stimulating factor (G-CSF) coupled intracellularly to gp130 (19). Treatment with G-CSF produced the same loss of mitoSTAT3 and subsequent recovery (Fig. 2D). This mitochondrial signaling pathway was not exclusively driven by gp130-coupled stimuli because interferon β (IFN-β), a type I IFN that activates STAT3, also caused a loss of mitoSTAT3 (fig. S2D). Not all stimuli activate this cascade because IFN-γ (fig. S2E) and epidermal growth factor (fig. S2F) failed to substantially decrease mitoSTAT3. To minimize contamination from the cytosol and other organelles, crude mitochondria used in these studies were always trypsinized to remove external proteins. We further purified mitochondria over a Percoll gradient (20) to separate the mitochondria from the tightly linked MAMs. Under these conditions, mitoSTAT3 still decreased with OSM treatment (Fig. 2E, P-mito), confirming a loss of STAT3 selectively from the mitochondrial fraction. This pathway could also be functionally relevant in vivo but may be difficult to fully explore because mice injected with saline alone display a robust decrease in mitoSTAT3 amounts, likely due to stress pathways induced by injecting the animal (fig. S2G). Together, these results show that mitoSTAT3 is dynamically regulated by various stressors.
JAK inhibition and mitoSTAT3 loss after cytokine treatment
Because the Janus kinase (JAK) receptor–associated tyrosine kinases drive downstream signaling from the IL-6 family cytokines and IFNs, we tested whether inhibition of the JAK activity prevented the loss of mitoSTAT3. Pretreatment of MEFs with the pan-JAK inhibitor ruxolitinib potently suppressed the loss of mitoSTAT3 induced with either OSM (Fig. 3A) or IL-6 (fig. S3A). Ruxolitinib did not inhibit H2O2-induced loss of mitoSTAT3 (Fig. 3B), which can likely bypass receptor-mediated activation and act on other downstream signaling cascades. These results were mirrored in MDA-231 cells preincubated with ruxolitinib and then subjected to either OSM or H2O2 treatment (Fig. 3C). Inhibition of STAT3 directly through its Src homology 2 (SH2) domain with the small molecule cryptotanshinone also did not substantially suppress OSM-induced mitoSTAT3 loss (fig. S3B). This finding is consistent with previous reports that demonstrated that the SH2 domain is dispensable for mitoSTAT3’s actions (1, 2, 11). Cryptotanshinone also blocks Tyr705 phosphorylation of STAT3, and consistent with our results using Y705F STAT3 mutants (fig. S1E), it would appear that Tyr705 is not involved in the mitochondrial localization of STAT3 after stimulation.
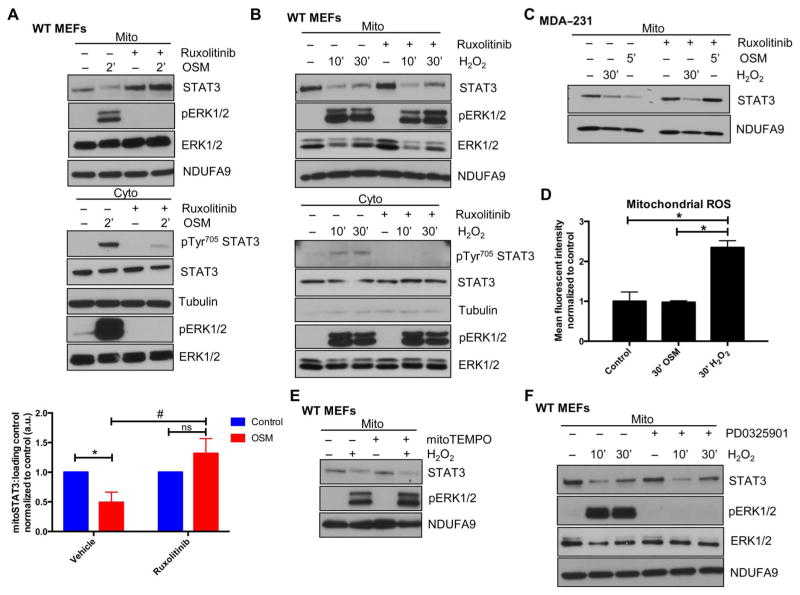
Fig. 3. Inhibition of JAKs prevents mitoSTAT3 loss upon cytokine treatment.
(A and B) MEFs were incubated with or without the JAK inhibitor ruxolitinib for 1 hour before the addition of OSM (A) or H2O2 (B), and mitochondrial and cytosolic extracts were immunoblotted for STAT3 or pTyr705 STAT3, respectively. Blots are representative of three independent experiments. Densitometric quantification of mitochondrial results from (A) is depicted. *P < 0.01; #P < 0.001; ns, not significant. (C) Mitochondria were isolated from MDA-231 breast cancer cells treated with ruxolitinib for 1 hour before the addition of OSM or H2O2 for the indicated times and immunoblotted for STAT3 and NDUFA9 (loading control). Blots are representative of two independent experiments. (D) Mitochondrial superoxide production was assayed by flow cytometry analysis using the MitoSOX dye in WT MEFs treated for 30 min with either OSM or H2O2. *P = 0.0002. n = 3 independent experiments. (E) WT MEFs were pretreated for 1 hour with the mitochondrial restricted antioxidant mitoTEMPO and then stimulated for 30 min with H2O2 with mitochondrial extracts blotted for STAT3, pERK1/2, or NDUFA9 (loading control). Blots are representative of two independent experiments. (F) WT MEFs were pretreated with the MEK inhibitor PD0325901 for 1 hour, followed by H2O2 treatment for the indicated times, and isolated mitochondria were subjected to SDS–polyacrylamide gel electrophoresis (PAGE) and immunoblotted for STAT3, pERK1/2, tERK1/2, and NDUFA9 (loading control). Blots are representative of two independent experiments.
We explored possible downstream mediators that transduce the signal(s) to the mitochondria from OSM and H2O2. Notably, H2O2 produced a robust increase in mitochondrial ROS production, which was not seen with OSM (Fig. 3D). Considering that STAT3 can be regulated by oxidative modifications (21) and that various signaling pathways can be activated independently of upstream events in the presence of ROS (22), we tested whether antioxidants could block the loss of mitoSTAT3 with H2O2 treatment. The mitochondrially targeted antioxidant mitoTEMPO (23) did not suppress the H2O2-induced decrease in mitoSTAT3 (Fig. 3E), likely ruling out an oxidative-driven program.
Further evaluation of the signaling pathway driving mitoSTAT3 loss revealed that ruxolitinib inhibited ERK activation induced by OSM but not by H2O2. STAT3 and, in particular, mitoSTAT3 have been linked to regulation by the MEK-ERK pathway (4), and mitoSTAT3 amounts may be controlled by this cascade (5). The MEK inhibitor PD0325901 completely blocked the H2O2-mediated activation of ERK but did not prevent mitoSTAT3 loss (10-min H2O2 treatment) or its initial recovery (30-min H2O2 treatment) (Fig. 3F). This result suggested that the MEK-ERK pathway was dispensable for these actions on mitoSTAT3 or, alternatively, that there was compensation from another signaling cascade. Treatment with the broad-spectrum kinase inhibitor staurosporine or the protein kinase inhibitor H7 dihydrochloride also did not block mitoSTAT3 loss (fig. S3C). H7 dihydrochloride inhibits protein kinase A (PKA) and PKC, both of which have been implicated in mitoSTAT3 biology (17, 24). Other signaling cascades converge on STAT3, including p38, phosphatidylinositol 3-kinase (PI3K)/Akt, c-Jun N-terminal kinase (JNK), and mechanistic target of rapamycin (mTOR) (25). Targeting each of these pathways individually did not prevent mitoSTAT3 loss (fig. S3, D to G). These results imply that multiple signaling events, which likely complement each other, may affect mitoSTAT3, making identification of the downstream regulation of this pathway challenging. Although the pharmacological inhibitors used in these studies have other cellular effects, the lack of a response in terms of blockade of the mitoSTAT3 loss lends greater support to the complexity of this signaling cascade.
Although we observed H2O2- and cytokine-induced loss of mitoSTAT3 in most cell lines, those with low basal amounts of mitoSTAT3, as in the case of the human breast cancer cell line MCF-7, often failed to show any further reduction in the mitochondrial levels of STAT3 (fig. S3H). This is consistent with tight regulation of mitoSTAT3 levels and cell line and context-dependent stimulation of the pathway.
Role of Ser727 phosphorylation and new protein synthesis on mitoSTAT3 recovery
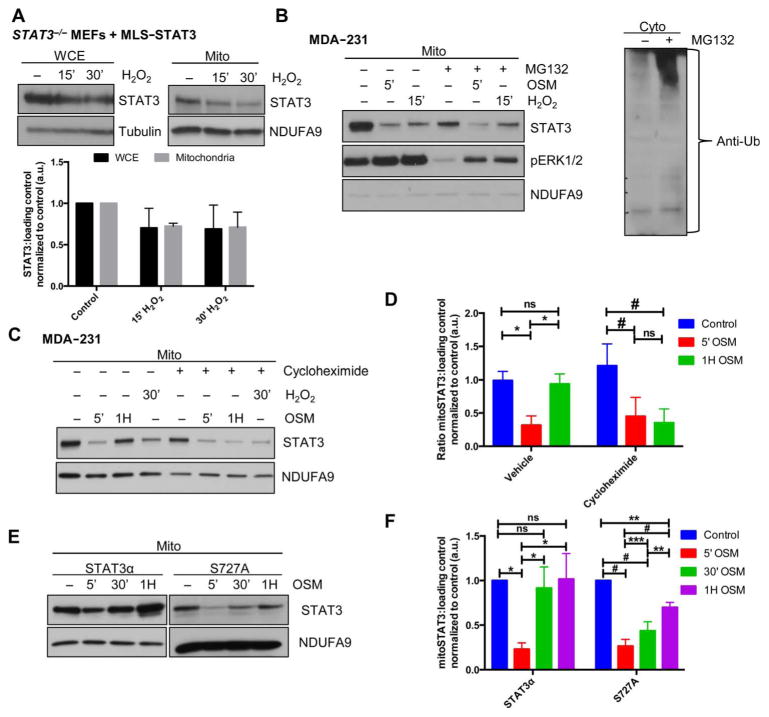
Because we could not definitively link a kinase pathway with the decreases in mitoSTAT3 after application of a stimulus, we investigated the potential contribution of signaling-driven proteolysis as a mechanism behind mitoSTAT3 regulation. Consistent with this idea, STAT3−/− MEFs expressing a mitochondrially targeted STAT3 and treated with H2O2 showed the same loss of STAT3 in whole-cell extract samples as from mitochondrial fractions (Fig. 4A). Although not statistically significant, the trend suggested that the mitoSTAT3 loss could be due to protein degradation rather than the export of STAT3 from the mitochondria and trafficking to another compartment, but we cannot exclude this latter possibility. STAT3 is cleaved by proteases such as caspases (26) and calpains (27), the latter of which is active in the mitochondria (28). However, treatment with the calpain inhibitor MDL-28170 did not block H2O2-induced mitoSTAT3 loss (fig. S4A).
Fig. 4. Recovery of mitoSTAT3 is dependent on new protein synthesis and Ser727 phosphorylation of STAT3.
(A) STAT3−/− MEFs were engineered to stably express mitochondrially targeted STAT3 (MLS-STAT3) and subjected to H2O2 treatment. Whole-cell extract (WCE) and mitochondria were Western blotted for STAT3 or the respective loading control. Blots are representative of three independent experiments and densitometric quantification is shown. (B) Mitochondria from MDA-231 cells were pretreated for 4 hours with vehicle or MG132 and treated with OSM or H2O2 and extracts probed for STAT3. Cytosolic extracts from vehicle- or MG132-treated cells were probed for ubiquitin (Ub) (right). Blots are representative of two independent experiments. (C and D) Mitochondrial extracts from MDA-231 cells treated with or without cycloheximide and stimulated with OSM or H2O2 were subjected to Western blotting (C). Results from OSM experiments were quantified in (D) by densitometry analysis. *P = 0.009; #P = 0.023. n = 3 independent experiments. (E and F) STAT3−/− MEFs expressing WT STAT3 (STAT3α) or the nonphosphorylatable mutant (STAT3 S727A) were treated for the indicated times with OSM, and mitochondrial lysates were immunoblotted for STAT3 (E). Quantification of these results is presented in (F). *P < 0.005; **P < 0.002; ***P < 0.02; #P ≤ 0.0002. n = 3 independent experiments.
Within the mitochondria, protein degradation is driven by two key serine proteases, Lon protease and ClpP protease, which are essential for mitochondrial protein quality control (29). We used the synthetic triterpenoid CDDO-Me to target the Lon protease, which inhibits its proteolytic activity in cells (30, 31). Pretreatment with CDDO-Me also did not block the loss of mitoSTAT3 after an oxidative stress insult (fig. S4B). The general proteasome inhibitor MG132, which also inhibits the Lon protease and attenuates proteasome-mediated outer mitochondrial membrane (OMM) degradation (32), failed to block the decrease in mitoSTAT3 seen with either OSM or H2O2 (Fig. 4B). Similarly, primary ClpP−/− MEFs treated with OSM and H2O2 still demonstrated the loss of mitoSTAT3 (fig. S4C).
Because degradation may play a role in mitoSTAT3 regulation, we tested whether protein synthesis was needed to drive full recovery of mitoSTAT3 amounts. Inhibition of protein synthesis with cycloheximide reduced the return of mitoSTAT3 after either OSM or H2O2 treatment in MDA-231 cells (Fig. 4, C and D). Similar results were obtained from H2O2-treated wild-type MEFs incubated with cycloheximide (fig. S4D). However, cycloheximide pretreatment had little effect on the cytoplasmic pool of STAT3 (fig. S4E), making it more likely that cycloheximide may be affecting the chaperone protein that targets STAT3 to the mitochondria. We also cannot exclude the possibility that there is a specific mitochondrial pool of STAT3 that is selectively affected by cycloheximide under these conditions or, alternatively, that cycloheximide affects some component of the signaling pathway through an off-target effect. Ser727 phosphorylation of STAT3 is important for STAT3’s trafficking to the mitochondria and may at least partially regulate its import (33). Hence, we speculated that mutation of Ser727 to alanine (S727A) would also negatively affect the reequilibration of mitoSTAT3 after its loss from the mitochondria. STAT3−/− MEFs expressing STAT3 S727A did not recover to the same extent as STAT3−/− MEFs expressing STAT3α after 30 min of OSM treatment, and the amounts of mitoSTAT3 were only starting to normalize after 1 hour of treatment in S727A mutant–expressing STAT3−/− MEFs (Fig. 4, E and F). These results provide insight into the regulation of mitoSTAT3 and produced a system through which the signaling pathways controlling STAT3 and its mitochondrial localization can be more effectively explored.
Interaction of mitoSTAT3 with CypD
mitoSTAT3 has been previously linked to the regulation of the MPTP, potentially through an interaction with CypD (34). Like other members of the cyclophilin family, CypD is a peptidyl-prolyl isomerase that triggers the opening of the permeability transition pore (35). It is also an important therapeutic target for ischemia-reperfusion injuries. Prolyl isomerase proteins mediate proper folding of target proteins, thus affecting protein stability, localization, and activity (36). Inhibition of CypD with cyclosporin A (CsA) drives an unfolded protein response pointing to a crucial role of CypD as a chaperone protein (37). Likewise, deletion of CypD impairs the function of several mitochondrial proteins (38, 39).
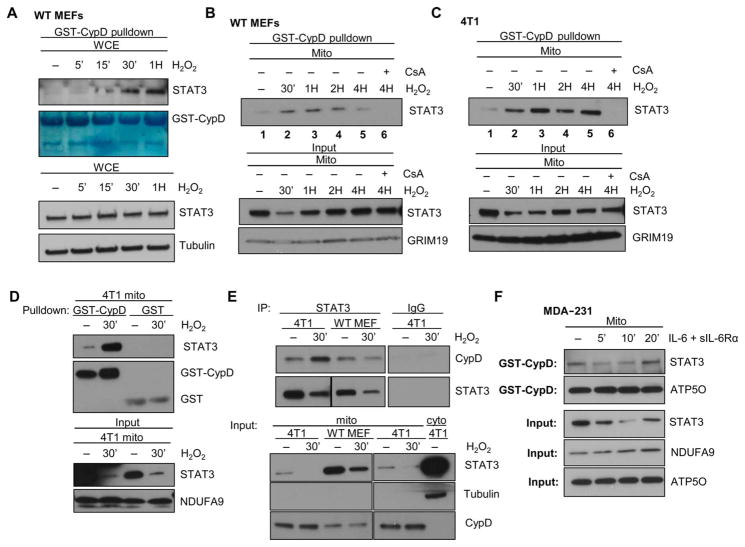
We noted that STAT3 became competent to bind to CypD after 15 to 30 min of H2O2 treatment (Fig. 5A), which corresponds with re-recruitment of STAT3 to the mitochondria after mitoSTAT3 loss (Fig. 1D). This result was confirmed in purified mitochondrial extracts incubated with recombinant GST-CypD from H2O2-treated wild-type MEFs (Fig. 5B) and 4T1 cells (Fig. 5C). In wild-type MEFs, mitoSTAT3 associated with CypD after 30 min of H2O2 treatment, which was maintained until 2 to 4 hours after stimulation when mitoSTAT3 amounts had returned to baseline [Fig. 5B, input (bottom)]. Notably, binding of mitoSTAT3 to GST-CypD was maintained up to 4 hours in 4T1 mitochondrial lysates, likely reflecting the longer time it takes for mitoSTAT3 to reequilibrate after its loss after H2O2 treatment (Fig. 1A). Incubation of cells with the CypD inhibitor CsA blocked the association of mitoSTAT3 with CypD, making the nonspecific interaction between these proteins less likely (Fig. 5, B and C). Similar results were seen in the mitochondria from STAT3+/+ or STAT3−/− MEFs treated with H2O2 (fig. S5A). Incubation with GST alone did not pull down any detectable mitoSTAT3, further supporting a specific interaction (Fig. 5D). Immunoprecipitation of STAT3 from mitochondrial lysates from wild-type MEFs, 4T1 cells, or MDA-231 breast cancer cells treated for 30 min with H2O2 also showed an increased association between mitoSTAT3 and CypD compared to untreated controls (Fig. 5E and fig. S5, B and C). Binding of mitoSTAT3 to CypD in IL-6–treated MDA-231 cells and various OSM-treated cell lines coincided with the recovery phase of mitoSTAT3 after stimulation (Fig. 5F and fig. S5, D to F). These results suggest that upon STAT3’s reentry to the mitochondria, it binds to CypD, potentially as part of its import and proper folding mechanism.
Fig. 5. mitoSTAT3 interacts with CypD.
(A) Lysates from WT MEF whole-cell extracts treated with H2O2 were subjected to pull-down with bead-bound glutathione S-transferase (GST)–CypD (Coomassie stain) and probed for STAT3 (input; bottom). Blots are representative of two independent experiments. (B and C) WT MEFs (B) and 4T1 cells (C) were incubated with H2O2 for various times before isolation of the mitochondria. Extracts were incubated with GST-CypD in the presence (lane 6) or absence of CsA, and STAT3 was assayed by immunoblotting (top). Bottom panels show the input (10% of that used for the pull downs). Blots are representative of two independent experiments. (D) Mitochondria from 4T1 cells either untreated or treated with H2O2 were subjected to GST-CypD or GST pull-down with immunoblotting for STAT3 or GST. Input is shown in the bottom panels. Blots are representative of two independent experiments. (E) mitoSTAT3 immunoprecipitations (IPs) from MEFs and 4T1 cells treated 30 min with H2O2. Immunoblots were probed for STAT3 (bottom) or CypD (top). Input (10% of that used for immunoprecipitation) is shown. Blots are representative of two independent experiments. (F) GST-CypD pull-down of mitochondrial extracts from MDA-231 cells treated with IL-6 and the soluble IL-6Rα with input is shown below. Blots were probed for STAT3. ATP5O is shown as a loading control for the pull-down and input samples. Blots are representative of two biological replicates.
The N terminus of STAT3 as key for CypD binding
Aside from being critical for the actions of mitoSTAT3, Ser727 in STAT3 may also be necessary to facilitate binding to CypD (34). We evaluated whether mutation of Ser727 affected STAT3’s interaction with CypD. GST-CypD pull-down of either 293T cells or STAT3−/− MEFs extracts expressing STAT3 S727A or 4T1 cells expressing STAT3 Y705F/S727A or Y705F/S727D variants revealed that Ser727 did not affect STAT3-CypD binding (fig. S6, A to C). Mutation of Ser727 to the phosphomimetic aspartic acid (S727D) seemed to increase basal association of STAT3 with CypD, potentially reflecting the fact that increased Ser727 phosphorylation of STAT3 may promote this protein-protein interaction. However, STAT3β, a naturally occurring splice variant that lacks the Ser727 site, bound to CypD after H2O2 treatment (fig. S6D), suggesting that Ser727 was dispensable for the binding of STAT3 to CypD.
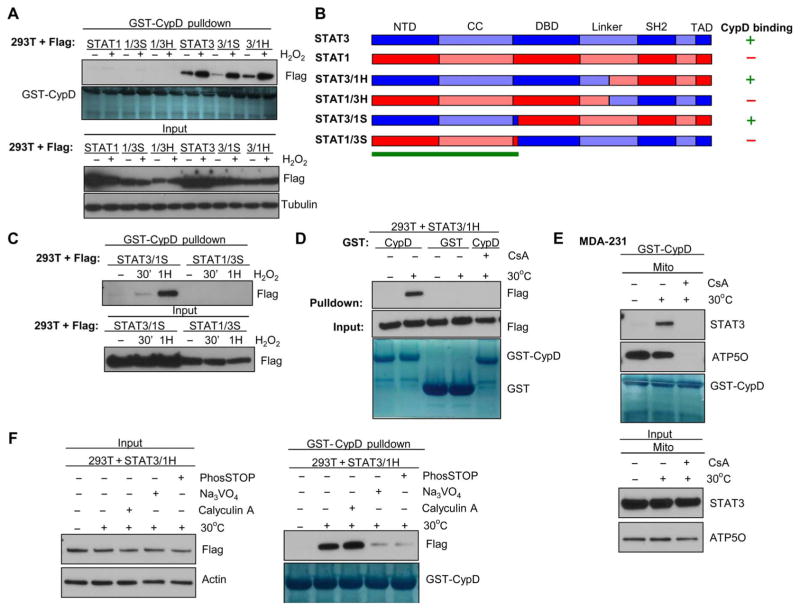
To further evaluate how STAT3 interacts with CypD, we used mitoSTAT1, which did not bind to CypD (fig. S6E). Using chimeric STAT3:STAT1 proteins (13, 40) expressed in 293T cells, we mapped the region of STAT3 that was necessary for the H2O2-induced STAT3-CypD interaction to the N terminus of STAT3 (amino acids 1–330 of STAT3) (Fig. 6, A and B). This result was validated with constructs that contain the N terminus of STAT3 (STAT3/1S) and those with the N terminus of STAT1 and the remaining STAT3 C-terminal portion (STAT1/3S) (Fig. 6C). To further examine the mechanism behind this association, we developed a cell-free system that used solubilized cell extracts warmed to 30°C. STAT3 bound to CypD in extracts from both 293T cells overexpressing STAT3 and from purified mitochondria from MDA-231 cells with endogenous STAT3 (Fig. 6, D and E). In both cases, binding of STAT3 to CypD was inhibited in the presence of CsA, and STAT3 was not detectable in lysates incubated with GST alone (Fig. 6, D and E). This cell-free system provides a useful tool to interrogate the regulation of the interaction of mitoSTAT3 and CypD in future studies.
Fig. 6. Binding of mitoSTAT3 to CypD is mediated through STAT3’s N terminus and can be recapitulated in a cell-free system.
(A) 293T cells expressing various chimeric versions of Flag-tagged STAT3/STAT1 were treated with H2O2, and whole-cell extracts were incubated with GST-CypD and probed for Flag (top). Bottom panels show the input (10% of that used for the pull-downs) with Coomassie stain of the GST-CypD input. Blots are representative of three independent experiments. (B) Diagram of the chimeric constructs used in (A), indicating the critical region (green bar) in STAT3 that mediates its binding to CypD (13). NTD, N-terminal domain; CC, coiled-coil domain; DBD, DNA binding domain; TAD, transcriptional activation domain. (C) 293T cells expressing STAT3/1S or STAT1/3S were treated with H2O2, subjected to GST-CypD pull-down and probed for Flag [pull-down (top) and input (bottom)]. Blots are representative of two independent experiments. (D) 293T cell lysates expressing STAT3/1H were warmed to 30°C and incubated with either GST-CypD or GST alone and immunoblotted for Flag. CsA was incubated with warmed lysates to also show the specificity of the interaction (lane 5). Bottom panel depicts Coomassie stain for GST-CypD and GST. Blots are representative of two independent experiments. (E) Mitochondrial lysates from MDA-231 cells tested in the cell-free system were incubated at 30°C, and a GST-CypD pull-down was performed in the presence or absence of CsA with immunoblotting for STAT3 and ATP5O. Blots are representative of two independent experiments. (F) Extracts from 293T cells expressing the chimeric protein STAT3/1H were warmed to 30°C in the established cell-free system in the presence of vanadate (Na3VO4), calyculin A, or the pan-phosphatase inhibitor cocktail PhosSTOP (Roche). Lysates (left; input) were subjected to GST-CypD pull-down (right) and probed for Flag (STAT3). Coomassie stain of GST-CypD is shown (right, bottom) to confirm equal loading of recombinant CypD. Blots are representative of two independent experiments.
Role of a tyrosine phosphatase in mitoSTAT3-CypD interactions
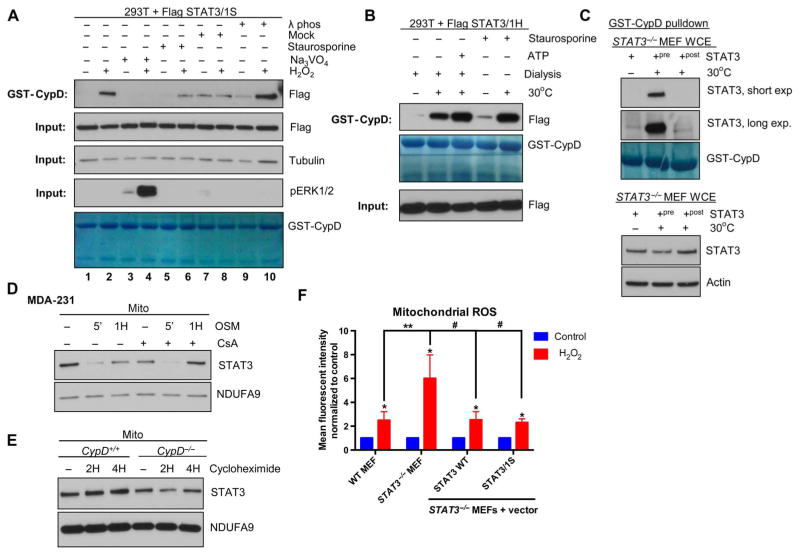
Using the cell-free system for studying the binding of mitoSTAT3 to CypD, we sought to further explore the mechanism driving this protein-protein interaction. Because warming of the extracts was sufficient to drive the association of these two proteins, we hypothesized that some enzymatic activity was required. Incubation of extracts with the tyrosine phosphatase inhibitor vanadate (Na3VO4) prevented the inducible binding of STAT3 to CypD to the same extent as a pan-phosphatase inhibitor (PhosSTOP) (Fig. 6F), which also contains serine/threonine phosphatase inhibitors. A role for a tyrosine phosphatase in this pathway seemed likely because the serine/threonine phosphatase inhibitor calyculin A did not affect this interaction (Fig. 6F). These results were confirmed in cells pretreated with vanadate, which blocked the H2O2-induced binding of STAT3 to CypD (Fig. 7A). In cell extracts subjected to λ phosphatase treatment (Fig. 7A), STAT3 bound to CypD to a greater extent, further supporting a role for dephosphorylation. Dialysis of cell extracts to remove small molecules, such as ATP, did not affect mitoSTAT3’s interaction with CypD, pointing to an ATP-independent mechanism for this interaction (Fig. 7B). Similarly, incubation of the kinase inhibitor staurosporine in extracts did not prevent the association of mitoSTAT3 with CypD (Fig. 7, A and B). Together, these results point to a role of the tyrosine phosphatase in mediating mitoSTAT3 binding to CypD.
Fig. 7. A tyrosine phosphatase–mediated event drives mitoSTAT3-CypD interactions.
(A) 293T cells expressing STAT3/1S were pretreated with either vanadate (Na3VO4) or staurosporine, followed by treatment with H2O2 for 1 hour. Untreated lysates were also subjected to λ phosphatase (λ phos) treatment or mock treatment. GST-CypD pull-down samples (top) and the corresponding input (bottom) were probed for Flag (STAT3), with the input also immunoblotted for tubulin (loading control) and pERK1/2. Coomassie stain of GST-CypD is shown (bottom) to confirm equal loading of recombinant CypD. Blots are representative of two independent experiments. (B) Extracts from 293T cells expressing STAT3/1H were dialyzed before mock treatment (30°C) with or without the addition of ATP (20 mM). Staurosporine was included during the mock treatment period for lysates in lanes 4 and 5. Binding of STAT3 and CypD was assessed by GST-CypD pull-down. Blots are representative of three independent experiments. (C) Whole-cell extracts from STAT3−/− MEFs were incubated with recombinant STAT3 on ice or either during (+pre) or after (+post) warming of the extract at 30°C for 30 min. Input (bottom) and GST-CypD pull-downs were probed for STAT3. Blots are representative of three independent experiments. (D) MDA-231 human cancer cells were pretreated with CsA and then treated with OSM, and mitochondrial lysates were probed for STAT3. Blots are representative of two independent experiments. (E) CypD+/+ and CypD−/− MEFs were incubated with cycloheximide, and mitoSTAT3 abundance was assessed from purified mitochondrial extracts. Blots are representative of two independent experiments. (F) WT, STAT3−/− MEFs, or STAT3−/− MEFs reconstituted with STAT3 WT or STAT3/1S were treated with H2O2, stained with MitoSOX and then analyzed by flow cytometry. Results are normalized to each respective untreated sample. *P < 0.01; **P < 0.001; #P = 0.0005. n = 5 independent experiments.
We examined whether STAT3 was the target of the tyrosine phosphatase and was needed to be modified to interact with CypD. STAT3 added to whole-cell extract STAT3−/− MEFs that were then incubated at 30°C bound to CypD (Fig. 7C). However, STAT3 added after extracts were warmed and then placed on ice did not bind to CypD (Fig. 7C). This result suggested that STAT3 needs to be present during the stimulation and that STAT3 itself is likely modified to interact with CypD.
Importance of the STAT3-CypD association
The absence of either STAT3 or CypD compromises mitochondrial metabolism and overall mitochondrial health (41–43). Because STAT3 associates with CypD upon its return to the mitochondria, we hypothesized that the chaperone function of CypD might be important for the stability of mitoSTAT3. mitoSTAT3 amounts were similar in CypD+/+ and CypD−/− MEFs after cytokine treatment (fig. S6F), with the latter, at times, showing a slight delay in full restoration of mitoSTAT3. Pre-treatment with CsA did not affect mitoSTAT3’s signaling regulation, suggesting that CypD is not involved in the loss or initial recovery of mitoSTAT3 after H2O2 or cytokine treatment (Fig. 7D and fig. S6G). Because CsA also prevents permeability transition, opening of the MPTP and its sequelae is unlikely to be an important driving force in the observed loss of mitoSTAT3 with external stimuli. However, these results do not completely address whether CypD is ultimately required for the stability of mitoSTAT3 because changes in the acute setting may only reflect whether STAT3 is appropriately targeted and imported into the mitochondria. In some instances, there appeared to be decreased basal amounts of mitoSTAT3 in the absence of CypD or after CsA treatment (Fig. 7D and fig. S6, F and G). On the basis of the sensitivity of the full recovery of mitoSTAT3 to cycloheximide (Fig. 4C), we examined whether cells lacking CypD relied more heavily on protein synthesis to maintain mitoSTAT3 amounts. mitoSTAT3 amounts decreased in CypD−/− MEFs treated with cycloheximide more readily than CypD+/+ MEFs (Fig. 7E). These results suggest that CypD may be an important binding partner and regulator of mitoSTAT3 stability, most likely through facilitating STAT3’s proper folding once it reaches the mitochondria.
To examine the functional role of the STAT3-CypD association, we took advantage of the link between mitoSTAT3 and ROS production. mitoSTAT3 limits mitochondrial and cellular ROS generation (8–12). We speculated that the N terminus of STAT3 would be essential for contributing to this protective effect because of its association with CypD. STAT3−/− MEFs generated more mitochondrial superoxide production than their wild-type counterparts after an oxidative stress (Fig. 7F). Reconstitution of STAT3−/− MEFs with either wild-type STAT3 or the chimeric STAT3/1S construct that contains the N terminus of STAT3 restored superoxide amounts to wild-type amounts (Fig. 7F). These results implicate the N terminus of STAT3, which mediates STAT3’s binding to CypD, as being important in limiting stress-induced mitochondrial ROS generation. Because we have not evaluated the role of wild-type STAT1 in this system, we cannot definitively exclude the possibility that the C terminus of STAT1 is not contributing at least in part to the ROS mitigating effect observed with the STAT3/1S construct. However, considering the role of STAT3 in mitochondrial ROS regulation (8–12), we anticipate that majority of the observed decrease in superoxide production with reexpression of full-length STAT3 is driven by its N terminus based on the comparable results seen with the STAT3/1S chimera.
DISCUSSION
mitoSTAT3 affects the ETC, ATP production, mitochondrial ROS generation, mitochondrial DNA regulation, mitochondrial Ca2+ content, and MPTP susceptibility. Together, this regulation drives both normal biological processes and pathological states. We now add to and extend those findings by describing a signaling pathway that acutely regulates mitoSTAT3 and drives its association with CypD (Fig. 8).
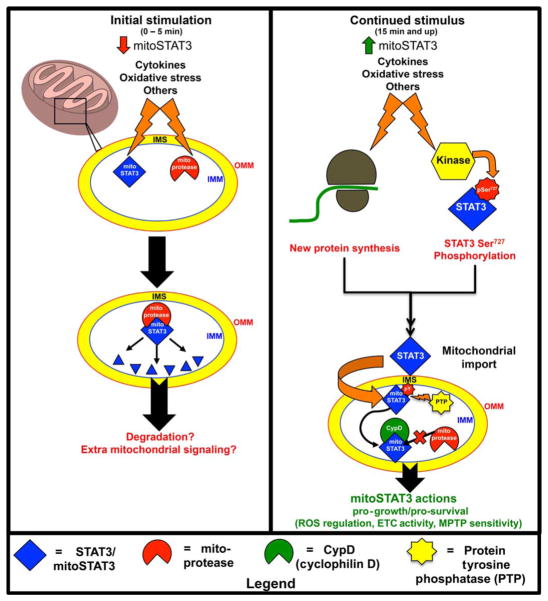
Fig. 8. Model of mitoSTAT3 regulation.
Upon initial stimulation (left), mitoSTAT3 and/or mitochondrial proteases (mitoproteases) are posttranslationally modified, which induces their association and leads to the proteolytic cleavage of mitoSTAT3. Presumably, proteolytic fragments of mitoSTAT3 may be further degraded or may contribute to extramitochondrial signaling. With continued stimulation (right), new protein synthesis (either of STAT3 or a chaperone protein required for STAT3 mitochondrial targeting) couples with Ser727 phosphorylation of STAT3 to mediate targeting of STAT3 to the mitochondria. Likely, mitoSTAT3 is also tyrosine-phosphorylated (pY; non-Tyr705 site). After its import into the mitochondria, mitoSTAT3 is dephosphorylated by a protein tyrosine phosphatase (PTP). This facilitates its interaction with CypD, which is likely important for mitoSTAT3’s proper folding and mitochondrial stability. This association determines ROS abundance and is also likely important for other downstream effects of mitoSTAT3, including regulation of the ETC and maintenance of the permeability transition pore (mitoSTAT3’s actions). IMS, intermembrane space.
mitoSTAT3 is implicated in both cardioprotection in ischemia-reperfusion injuries (8, 9, 34, 44, 45) and transformation and growth of cancer cells (2, 11, 46–48). Understanding how mitoSTAT3 is regulated will be a valuable tool to more effectively target the full STAT3 program driving these pathological states. This is especially important because most STAT3 inhibitors designed to date target STAT3’s SH2 domain, which likely would not affect mitoSTAT3’s activity because the SH2 domain is dispensable for its mitochondrial function (1, 2). Certain pharmacological treatments affect mitoSTAT3 abundance, although their widespread use in manipulating the mitoSTAT3 pathway remains to be determined (48, 49). For instance, phospho–valproic acid limits the translocation of mitoSTAT3 in a pancreatic cancer cell model, which leads to increased cancer cell death (48), pointing to the value in targeting mitoSTAT3.
STAT3’s mitochondrial localization can be altered during cell differentiation (15, 16), with ROS being a mechanism to decrease mitoSTAT3 (15). Increased mitoSTAT3 amounts drive T cell activation and axonal growth in cells under chronic stimulation with cytokines (16) and growth factors (5, 18). Together with the results presented here, these reports would suggest that mitoSTAT3 is differentially regulated in the acute compared to the chronic setting. mitoSTAT3 is important in the proliferation of embryonic stem cells, where it supports oxidative phosphorylation to couple STAT3’s transcriptional effects with maintaining pluripotency (50). Fine-tuning mitoSTAT3 amounts is then likely important in controlling mitochondrial adaptation to the real-time cellular demands, suggesting that this pathway is a crucial regulatory step in normal biological processes as well.
Currently, the fate of mitoSTAT3 after stimulation is unknown. Proteolysis of mitoSTAT3 is consistent with the kinetics of its loss from the mitochondria. We detected proteolytic fragments of STAT3 in the mitochondria but not in cytoplasmic extracts, suggesting, but not proving, that the observed loss of mitoSTAT3 was due to proteolysis. Various inhibitors of mitochondrial proteases did not prevent the loss of mitoSTAT3. It is possible that these inhibitors do not efficiently enter the mitochondria or that the protease that cleaves mitoSTAT3 is insensitive to the inhibitors used. Intriguingly, both p50 and IκBα are lost from the mitochondria after stimulation with tumor necrosis factor–α, with IκBα likely being degraded after phosphorylation in part by a calpain-dependent mechanism (51). STAT3’s degradation may be linked with its phosphorylation status (52), and calpains target and cleave STAT3 (27). However, calpain inhibition did not affect treatment-induced mitoSTAT3 loss. Pharmacological blockade of the Lon protease and deletion of the ClpP protease, which are serine proteases in the mitochondria, did not attenuate mitoSTAT3’s loss. Other mitochondrial proteases, including inner membrane–associated mitochondrial AAA proteases, which are emerging as important targets in mitochondrial protein quality control and dynamics (29), may be the primary driver of mitoSTAT3 loss. Simultaneous blockade of multiple protease pathways may be necessary to prevent STAT3 cleavage because inhibition of a single cascade may lead to altered enzymatic activity and compensation by another protein. Consistent with this possibility, inhibition of only the Lon protease in HeLa cells does not grossly change mitochondrial function or protein content due to increased activity from another ATP-dependent protease (53). Another explanation for the loss of mitoSTAT3 is that it is removed from the mitochondria after stimulation through the PINK1 (PTEN-induced putative kinase 1)–Parkin–dependent mitochondrial-derived vesicle (MDV) pathway (54, 55). These vesicles remove damaged mitochondrial cargo for transport to the lysosome and are critical to mitochondrial quality control (56). The mitochondrial protein Tom20 is trafficked through these MDVs, and STAT3 is reported to interact with Tom20 (34).
Our results demonstrate that aside from H2O2, which has broad effects on intracellular signaling cascades, cytokines also affected mitoSTAT3, suggesting that signals emanating from the plasma membrane were rapidly transduced to the mitochondria. We do not know what signaling cascade is responsible for H2O2- and cytokine-induced loss of STAT3 from the mitochondria. Although various kinases (including Akt, ERK, and JNK) have been reported to reside in the mitochondria, where they regulate mitochondrial dynamics through phosphorylation events (57–60), inhibition of these mitochondrially localized signaling proteins in isolation did not affect stimulus-dependent decreases in mitoSTAT3.
The dependence of the recovery of mitoSTAT3 after stimulation on Ser727 phosphorylation agrees with a previous report that demonstrated that the Ser727 site is important for its mitochondrial import (33). However, the absence of an effect on basal mitoSTAT3 amounts when Ser727 is mutated to alanine (S727A) suggests that there are likely other means of bypassing this import requirement that are compromised under stress conditions. Like other proteins that are imported into the mitochondria, the mitochondrial import of STAT3 requires an intact mitochondrial membrane potential (61). STAT3 is ultimately being targeted to the inner mitochondrial membrane (IMM) or matrix (33, 34). Under stress conditions, a reduction in mitochondrial membrane potential disables the mitochondrial import machinery, which affects the import efficiency and mitochondrial abundance of various proteins (62, 63). However, continuous import of STAT3 to maintain its mitochondrial abundance is unlikely because mitoSTAT3 was markedly stable in wild-type cells hours after cycloheximide treatment (Fig. 7E). Although the MEK-ERK pathway phosphorylates STAT3 at Ser727 and has been linked to regulating mitoSTAT3 (4, 5), MEK and ERK inhibition did not affect mitoSTAT3 abundance. This is not surprising because various other kinases can phosphorylate STAT3 at this residue, and there is likely a large degree of cross-talk between multiple signaling pathways. The full recovery of mitoSTAT3 was blunted when protein synthesis was inhibited, despite the large cytoplasmic pool of STAT3 that was still present under these conditions. These results suggest that either the chaperone protein that is responsible for the translocation of STAT3 to the mitochondria has a short half-life or there is a limited fraction of STAT3 that can go to the mitochondria. The latter idea is interesting and could be analogous to p53, of which there is a distinct mitochondrial isoform (64). Other isoforms of STAT3 have been reported, but to date, there is no information on whether they, or others, are mitochondrially localized (65).
On the basis of the above results (Fig. 4), we anticipate that mitoSTAT3’s loss is likely due to proteolysis. However, a fraction of mitoSTAT3 could traffic to another cellular compartment. Signaling to the nucleus would be an attractive target, considering STAT3’s role as a nuclear transcription factor. Under the conditions that induced the loss of mitoSTAT3, STAT3 was potently recruited to the nucleus, making it at least plausible that the mitochondrial pool of STAT3 might contribute to the nuclear actions of STAT3. However, this possibility is difficult to assess because of the high cytosolic abundance of STAT3. Other mitochondrial proteins, including the pyruvate dehydrogenase complex and SSBP1, can translocate from the mitochondria to the nucleus to regulate transcription under stress conditions (66, 67). This mitochondrial retrograde signaling event occurs in Caenorhabditis elegans, where the protein ATFS-1 links coordinate mitochondrial and nuclear programs (62, 68, 69). Intriguingly, ATFS-1 is also proteolyzed in the mitochondria and accumulates in the nucleus under stress conditions. A similar pathway has not been detected in mammalian cells, but it is thought that signaling molecules such as ROS or Ca2+ released from the mitochondria might activate pathways (such as JNK) to drive mitonuclear cross-talk (70). STAT3 not only modulates mitochondrial ROS production (8, 9, 11, 12, 15), but it also directly regulates mitochondrial calcium content (16). It is tempting to speculate that mitoSTAT3, as well as other transcription factors that localize to the mitochondria such as nuclear factor κB, might be part of this communication network, especially when one considers the dynamic regulation we have observed in mitoSTAT3. Whether this is through direct trafficking to the nucleus, or indirectly through regulation of intracellular ROS and Ca2+ content, remains to be determined.
CypD plays a role in retrograde signaling through its effects on STAT3 activation (71). It is thus interesting that mitoSTAT3 binds to CypD in an inducible manner. The interaction of STAT3 and CypD coincides with mitoSTAT3’s return to the mitochondria. Reentry of proteins into the mitochondria requires their proper folding after mitochondrial import, and the chaperone function of CypD may play a critical role in this process. The decreased mitoSTAT3 amounts in cells lacking CypD after cycloheximide treatment suggest that the stability of the mitoSTAT3 pool at least partly depends on CypD. mitoSTAT3, which is serine-phosphorylated, associates with CypD in cardiomyocyte mitochondria, where it plays a role in regulation of MPTP sensitivity (34). We observed that Ser727 in STAT3, which has been implicated in its mitochondrial action, was not required for the binding of mitoSTAT3 to CypD. Rather, the N terminus of STAT3 mediates the association, suggesting that other domains in mitoSTAT3 are relevant for its non-canonical mitochondrial role. Selective targeting of the N terminus of STAT3 with peptide-based inhibitors affects mitochondrial membrane potential and leads to cancer cell death (72).
The action of a tyrosine phosphatase on STAT3 was necessary for its interaction with CypD. This is interesting because the binding of STAT3 to CypD can be potently induced with H2O2, a stimulus that normally inhibits tyrosine phosphatases through oxidation of key cysteines in their catalytic pocket (73). A major regulator of STAT3 signaling is the SH2 domain containing protein tyrosine phosphatase 2 (SHP2) (74). Coincidentally, SHP2 is also paradoxically activated by oxidative stress (75), and it can localize to the mitochondria, where it regulates mitoSTAT3 (76). Future work will clarify the identity and role of this phosphatase in this signaling cascade.
Consistent with previous reports (8–12), we have demonstrated a role for STAT3 in the modulation of cellular ROS. We extend that work by demonstrating that the N terminus of STAT3 may be key for limiting mitochondrial ROS production after a stress insult. Because the N terminus of STAT3 was also the critical region for binding to CypD, we anticipate that the STAT3-CypD association is important for regulating ROS generation at the mitochondrial level. This could be through stabilization of the mitoSTAT3 pool, allowing STAT3 to more effectively alter ETC activity under stress conditions as previously proposed (8). Increased amounts of mitoSTAT3 could also serve as a greater redox reserve, effectively scavenging mitochondrial ROS as they are generated (14). It is also conceivable that the association of mitoSTAT3 with CypD directly controls the opening of the MPTP (34) that is closely linked to cellular redox status. Work is ongoing to better understand how this protein-protein interaction affects this functional end point. Tight regulation of cellular redox state is paramount during cellular transformation and progression of cancer. CypD is necessary for Ras transformation and tumorigenesis (77). Because mitoSTAT3 is necessary for Ras transformation (2), it is tempting to speculate that CypD’s requirement in this process is through its binding and stabilization of the mitoSTAT3 pool. Further characterization of this cascade and how mitoSTAT3 is controlled are necessary to provide insight into how to effectively target the mitochondrial pool of STAT3 for the purposes of promoting health and combating disease.
MATERIALS AND METHODS
Cell culture
The following cell lines were used in this study: wild-type MEFs, STAT3−/− MEFs, CypD−/− MEFs, ClpP+/+ MEFs, ClpP−/− MEFs, 4T1 murine breast adenocarcinoma cells, MDA-MB-231 triple-negative human breast cancer cells, WI-38 human lung fibroblasts, SKBR3 HER2+ human breast cancer cells, BT474 HER2+ human breast cancer cells, MDA-MB-453 HER2+ human breast cancer cells, 293T human embryonic kidney cells, HeLa human cervical cancer cells, and rat hepatoma cell lines expressing the G-CSF–gp130 chimeric receptor. The human breast cancer cell lines were provided by G. Ginder, C. Clevenger, and F. Fang (Virginia Commonwealth University). WI-38 human lung fibroblasts were provided by S. Deb (Virginia Commonwealth University). HeLa cells were provided by T. Harris (University of Virginia). CypD−/− MEFs were provided by U. Moll (Stony Brook University). ClpP+/+ and ClpP−/− MEFs were provided by G. Auburger (Goethe-Universität). Rat hepatoma cell lines were provided by H. Baumann (Roswell Park Cancer Institute). Cells were routinely grown in an appropriate growth culture medium [Dulbecco’s modified Eagle’s medium (DMEM), DMEM/F-12, or RPMI] supplemented with 10% heat-inactivated fetal bovine serum (Serum Source International) and penicillin (50 U/ml) and streptomycin (50 μg/ml) (Life Technologies). Before experimental use, cell lines were determined to be free of mycoplasma contamination.
Reagents
The following chemicals and inhibitors were used: ruxolitinib (JAK inhibitor; 5 μM, Selleck Chemicals), PD0325901 (MEK inhibitor; 10 μM, Selleck Chemicals), LY294002 (PI3K inhibitor; 25 μM, Cell Signaling), SB203580 (p38 MAPK inhibitor; 10 μM, Calbiochem), SP600125 (JNK inhibitor; 50 μM, Cell Signaling), Torin 1 (mTOR inhibitor; 10 μM, Tocris Bioscience), CsA (CypD inhibitor; 5 μM or 10 μg/ml, Sigma-Aldrich), cycloheximide (protein synthesis inhibitor; 50 μg/ml, MP Biomedicals), staurosporine (tyrosine kinase inhibitor; 100 nM, Sigma-Aldrich), H7 dihydrochloride (PKA/PKC/PKG inhibitor; 20 μM, Sigma-Aldrich), cryptotanshinone (STAT3 inhibitor; 10 μM, Selleck Chemicals), mitoTEMPO (mitochondrial antioxidant; 100 μM, Sigma-Aldrich), MG132 (proteasome inhibitor; 25 μM, Tocris Bioscience), MDL-28170 [calpain inhibitor; 10 μM, gift from E. Lesnefsky (Virginia Commonwealth University, Richmond, VA)], CDDO-Me (Lon protease inhibitor; 50 μM, Sigma-Aldrich), recombinant murine IL-6 (50 ng/ml, PeproTech), recombinant human IL-6 (50 ng/ml, Pepro-Tech), soluble human IL-6Rα (50 ng/ml, PeproTech), human OSM with carrier (12.5 ng/ml, Cell Signaling), mouse OSM with carrier (12.5 ng/ml, Cell Signaling), H2O2 30% solution [1 mM (unless otherwise indicated), Sigma-Aldrich], cOmplete protease inhibitor cocktail (per the manufacturer’s instructions, Roche), PhosSTOP phosphatase inhibitor cocktail (per the manufacturer’s instructions, Roche). All other chemicals used for buffer formulations were purchased from Sigma-Aldrich or Thermo Fisher Scientific, unless otherwise indicated.
Treatments
All stimulations were carried out in cells cultured in full serum medium (10%) to avoid any effect of serum starvation on mitoSTAT3 amounts. Aside from protease inhibitor studies (4-hour pretreatment), all other inhibitors were preincubated on cells for no more than 1 hour before H2O2 or cytokine treatment. Vehicle or dimethyl sulfoxide treatments were used when appropriate as controls.
Plasmids
pGEX-CypD was a gift from U. Moll (Stony Brook University) (78). pRC-CMV STAT3, pRC-CMV STAT1, pRC-CMV STAT3/1H, pRC-CMV STAT3/1S, pRC-CMV STAT1/3H, and pRC-CMV STAT1/3S have been described previously (13). MSCV (murine stem cell virus)–IRES (internal ribosomal entry site)–GFP (green fluorescent protein) expression vectors containing STAT3α, STAT3β, STAT3 Y705F, STAT3 S727A, STAT3 S727D, MLS-STAT3, and the Phoenix helper construct have been described previously (1).
Transfections
Transfections were carried out with the FuGENE reagent (Promega). Briefly, plasmid DNA (1 to 3 μg) was combined with the FuGENE reagent at a 1:3 ratio (μg/ml) and incubated in Opti-MEM medium for 5 min before adding to the cells. For short hairpin RNA (shRNA) viral transductions, 293T cells were transfected via FuGENE with the indicated shRNA vector (1.5 μg), pCMV-VSV-G envelope protein vector (1 μg), and pCMV-dR8.2 dvpr packaging plasmid vector (1.5 μg). Viral supernatants were collected after 48 hours, spun down, and filtered through a 0.45-μm filter before adding to the cells in the presence of polybrene (10 μg/ml) (Millipore). MSCV retroviruses were similarly packaged in 293T cells after transfection with the appropriate MSCV viral vector (4 μg) and the Phoenix helper construct (1 μg). Cell transductions were carried out for 24 hours in the presence of the virus and after 48 hours of puromycin selection (2 μg/ml, shRNA experiments), or GFP fluorescence-activated cell sorting was performed to generate a pure, stable pool of cells expressing the indicated viral vector.
Recombinant proteins
pGEX-CypD and pGEX vectors were transformed into BL-21(DE3) competent bacteria. Bacterial clones containing the plasmid of interest were grown overnight in 25 ml of LB medium and, on the following morning, diluted (1:15) to a 500-ml culture. Bacteria were grown until the optical density at 600 nm reached 0.6, at which point protein expression was induced with 1 mM isopropyl-β-D-thiogalactopyranoside for 6 hours. The bacteria were pelleted and lysed in bacterial lysis buffer, followed by sonication. The bacterial cell extract was then incubated with glutathione Sepharose 4B beads (GE Healthcare, #17-0756-01) for 1 hour at 4°C. Bead-bound GST proteins were pelleted and washed three times in bacterial lysis buffer before storage at −80°C. A portion of the purified proteins were resolved by SDS-PAGE, and gels were fixed and Coomassie-stained for the quantification of GST recombinant proteins as compared to a bovine serum albumin (BSA) standard curve. Recombinant Flag-STAT3 protein was a gift from C. Mertens (Darnell Laboratory, Rockefeller University) (79).
SDS-PAGE and immunoblotting
Cell samples were lysed in 20 mM Hepes (pH 7.4), 300 mM NaCl, 10 mM KCl, 1 mM MgCl2, 20% glycerol, and 1% Triton X-100. Equal protein was loaded on tris-glycine gels and subjected to SDS-PAGE. Gels were transferred to polyvinylidene difluoride membranes (Millipore, IPVH00010) using a semidry transfer apparatus. The following antibodies were used overnight at 4°C with shaking after blocking for 1 hour in 5% milk or 5% BSA in 1× tris-buffered saline (TBS) + 0.1% Tween 20: STAT3 (Cell Signaling #9139, mouse monoclonal, clone 124H6), STAT3 (N-terminal, BD Biosciences, mouse monoclonal), STAT3 (Cell Signaling, rabbit polyclonal), pTyr705 STAT3 (Cell Signaling, mouse monoclonal), pSer727 STAT3 (Cell Signaling, rabbit polyclonal), Complex V subunit I (Abcam, mouse monoclonal), STAT1 (BD Biosciences, mouse monoclonal), pTyr701 STAT1 (Cell Signaling, rabbit polyclonal), CypD (PPIF, Abcam, mouse monoclonal), calreticulin (Cell Signaling, rabbit monoclonal), histone H3 (Cell Signaling, mouse monoclonal), lamin A/C (Cell Signaling, mouse monoclonal), ATP5O (Abcam, mouse monoclonal), GRIM19 (Abcam, mouse monoclonal), pAkt (Cell Signaling, rabbit polyclonal), pc-Jun (Cell Signaling, rabbit polyclonal), NDUFA9 (Abcam #ab14713, mouse monoclonal), pERK1/2 (Cell Signaling #9101, rabbit polyclonal), tubulin (Sigma-Aldrich #T8203, mouse monoclonal, α-tubulin), and actin (Cell Signaling, mouse monoclonal). Secondary antibodies were used at a concentration of 1:5000 for 1 hour at room temperature in 5% milk in 1× TBS + 0.1% Tween 20.
After washing, blots were incubated with Amersham ECL (GE Healthcare, RPN2106) or enhanced chemiluminescence (ECL2) Western blotting (Thermo Fisher Scientific, 80196) detection reagents and then developed. Densitometry quantification was performed using ImageJ software [NIH, Bethesda, MA (http://imagej.nih.gov/ij/)].
Mitochondria isolation
Pure mitochondria and MAM fractions were isolated as previously described (20). For the crude mitochondria, adherent cells were treated and then washed with ice-cold 1× phosphate-buffered saline (PBS), trypsinized, and collected with an ice-cold full serum medium. Cells were spun down for 5 min at 1000 rpm, and the pellets were washed once with 3 ml of ice-cold 1× PBS. Cells were spun down again for 5 min at 1000 rpm, and the supernatant was aspirated. The cell pellets were resuspended in an appropriate volume (1 to 2 ml) of sucrose buffer [10 mM Hepes (pH 7.4), 250 mM sucrose, 1 mM EDTA, and protease and phosphatase inhibitors) and incubated on ice for 10 min. Cells were then added to a douncer (all steps are on ice), and cells were homogenized with manual strokes until roughly 90% of the cells were broken (verified with Trypan blue during douncing). Cells were collected and spun down at 800g for 5 min at 4°C to pellet the unbroken cells and nuclei. The supernatant was collected and spun down at 8800g for 10 min at 4°C to pellet the crude mitochondria. The supernatant was collected again and transferred to a new tube labeled as cytosolic fraction, which was spun down at 10,000g for 10 min at 4°C to remove any contaminants from the cytosolic sample. After this spin, the cytosolic supernatant (700 μl) was transferred to a new tube and frozen at −80°C until further analysis. The crude mitochondrial pellet from above was resuspended in 490 μl of sucrose buffer, and 10 μl of a stock solution of trypsin (5 mg/ml) was added to each tube (for a final concentration of 100 μg/ml). Samples were then rotated for 10 min at 4°C, after which 500 μl of 5% BSA solution was added to each tube to inactivate the trypsin. Samples were rotated again for 1 min at 4°C and then spun down at 10,000g for 10 min at 4°C. The supernatant was aspirated off (including the trypsin-digested material surrounding the mitochondrial pellet), and the mitochondrial pellets were washed in 500 μl of 2× sucrose buffer (spins were done at 10,000g for 10 min at 4°C). After the final wash, the mitochondrial pellets were resuspended in an appropriate volume of sucrose buffer and stored at −80°C until further analysis. For protein analysis, mitochondrial supernatants were lysed in an equal volume of sucrose buffer plus 1× PBS with 2% Triton X-100 plus protease and phosphatase inhibitors.
Pull-down assays
Pull-down assays were performed as previously described (78). Briefly, recombinant bead-bound GST-CypD or GST alone was blocked in H buffer [20 mM Hepes (pH 7.7), BSA (1 mg/ml), 75 mM KCl, 0.1 mM EDTA, 2.5 mM MgCl2, 0.05% NP-40, and 1 mM dithiothreitol (DTT)]. Two hundred fifty micrograms of mitochondrial or whole-cell extracts (lysed as described above) were incubated with 20 μg of bead-bound GST-CypD or GST alone and incubated overnight at 4°C. Beads were pelleted and washed in H buffer, and bound proteins were analyzed by immunoblotting.
Immunoprecipitation
Immunoprecipitation was carried out as previously described (80). Mitochondrial or whole-cell extracts were incubated overnight at 4°C with an antibody against STAT3 (Santa Cruz Biotechnology, sc-482, C-20, 1:100 dilution) plus agarose beads (Protein G Sepharose Fast Flow, GE Healthcare, 17-0618-01) or control immunoglobulin G plus agarose beads in immunoprecipitation buffer (pH 7.4, 150 mM NaCl, 50 mM tris-HCl, 1% Triton X-100, and 1 mM EDTA with protease and phosphatase inhibitor cocktails; Roche). Immunoprecipitates were washed three times with wash buffer (150 mM NaCl, 50 mM tris-HCl, 1% Triton X-100, and protease and phosphatase inhibitor cocktails) and separated by SDS-PAGE with blotting against the indicated antibodies.
Cell-free system for studying mitoSTAT3-CypD interactions
Mitochondria or whole-cell extracts were prepared as described but in the absence of protease or phosphatase inhibitors. Lysates were incubated on ice or at 30°C for 30 min in 1× NEBuffer Pack for Protein MetalloPhosphatases [50 mM Hepes (pH 7.5), 10 mM NaCl, 1 mM MnCl2, 2 mM DTT, and 0.01% Brij 35]. After incubation, extracts (100 μg) were subjected to GST-CypD pull-down as described. When applicable, cell lysates were dialyzed for 3 hours using a 10,000 MWCO dialyzer cassette according to the manufacturer’s protocol (Slide-A-Lyzer, Thermo Fisher Scientific).
Animals
Animals were treated in compliance with the Guide for the Care and Use of Laboratory Animals under the protocols approved by the Virginia Commonwealth University’s Institutional Animal Care and Use Committee.
Liver mitochondrial assays
Eight- to 12-week-old male CD-1 mice (Charles River Laboratories) were tail vein–injected with recombinant murine IL-6 (500 μg/kg) (PeproTech) and sacrificed at the indicated times. Livers were harvested from mice and washed with ice-cold PBS. The livers were minced and homogenized in 10-ml MSM buffer (220 mM mannitol, 70 mM sucrose, and 5 mM Mops) on ice. A homogenate fraction was collected for each sample and solubilized as described above. Homogenized tissues were then centrifuged at 500g for 10 min at 4°C to remove the cell debris. The supernatants were centrifuged at 3700g for 10 min at 4°C to pellet the mitochondria. Mitochondria were resuspended in 5 ml of ice-cold MSM-EDTA (MSM buffer plus 2 mM EDTA) and spun at 3700g for 10 min at 4°C. Supernatants were discarded, and the mitochondria were resuspended in 2.5 ml of MSM-EDTA buffer and spun down at 3700g for 10 min at 4°C (81). Mitochondria in MSM-EDTA buffer were lysed via the addition of an equal volume of 2% Triton X-100 in 1× PBS with protease and phosphatase inhibitor cocktails and analyzed via immunoblotting.
Statistical analysis
Results are presented as means ± SEM. The statistical analysis was carried out as follows. For Fig. 1A and fig. S1D, differences between the control and the groups under comparison were assessed using a two-sided two-sample t test at 5% level of significance. For Fig. 1D, differences between mitoSTAT3 and mitoSTAT1 at various times of H2O2 treatment were assessed using two-sided two-sample t test at 5% level of significance. For all others, the differences between groups were initially assessed using a one-way analysis of variance (ANOVA), followed by false discovery rate–adjusted (82) multiple (pairwise) comparisons at a 5% threshold. If P values were <0.05, we considered the group difference to be significant. n values are indicated in the figure legends for each panel and always represent independent experiments. Representative blots are shown.
Supplementary Material
Acknowledgments
We would like to thank T. Kordula (Virginia Commonwealth University), E. Lesnefsky (Virginia Commonwealth University), G. Shadel (Yale University), J. E. Darnell (The Rockefeller University), G. Auburger (Goethe University Medical School), D. Kashatus (University of Virginia), T. Harris (University of Virginia), and D. Brautigan (University of Virginia) for their helpful advice and invaluable discussions regarding this work. We would also like to thank G. Ginder, C. Clevenger, and F. Fang (Virginia Commonwealth University); S. Deb (Virginia Commonwealth University); U. Moll (Stony Brook University); G. Auburger (Goethe-Universität); H. Baumann (Roswell Park Cancer Institute); E. Lesnefsky (Virginia Commonwealth University); and C. Mertens (Rockefeller University) for providing cell lines, plasmids, and reagents. We would also like to thank J. Farnsworth and Q. Zhang in the Flow Cytometry Core for their continuous help in the generation of stable cell lines used for these studies.
Funding: This research was supported by NIH-NIGMS R01 GM101677 (to A.C.L.), Massey Cancer Center 2013-MIP-03 (to A.C.L. and K.T.), and NIH-NCI F30 CA183175 (to J.A.M.). K.T. is also supported by NIH-NCI (R01CA160688) and the Susan G. Komen Foundation (Investigator-Initiated Research grant, IIR12222224). P.E.S. is supported by the British Heart Foundation (grant no. FS/11/58/28937 and FS/15/43/31565). Additional support was provided by the Clinical and Translational Science Awards (award no. UL1TR000058) from the National Center for Advancing Translational Sciences. Services and products in support of the research project were generated by the Virginia Commonwealth University (VCU) Massey Cancer Center Flow Cytometry Shared Resource, supported in part with funding from NIH-NCI Cancer Center Support grant P30 CA016059. Microscopy was performed at the VCU Microscopy Facility, supported in part by funding from NIH-NCI Cancer Center Support grant P30 CA016059.
Footnotes
Author contributions: J.A.M., M.H., M.C., A.R., C.M., V.R., J.S., E.T., S.T.-O., S.G., and D.B. performed the experiments and collected and analyzed the data. J.A.M., M.H., P.E.S., H.B., K.T., and A.C.L. conceived and planned the experiments. J.A.M. and A.C.L. wrote the manuscript.
Competing interests: The authors declare that they have no competing interests.
REFERENCES AND NOTES
- 1.Wegrzyn J, Potla R, Chwae YJ, Sepuri NB, Zhang Q, Koeck T, Derecka M, Szczepanek K, Szelag M, Gornicka A, Moh A, Moghaddas S, Chen Q, Bobbili S, Cichy J, Dulak J, Baker DP, Wolfman A, Stuehr D, Hassan MO, Fu XY, Avadhani N, Drake JI, Fawcett P, Lesnefsky EJ, Larner AC. Function of mitochondrial Stat3 in cellular respiration. Science. 2009;323:793–797. doi: 10.1126/science.1164551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gough DJ, Corlett A, Schlessinger K, Wegrzyn J, Larner AC, Levy DE. Mitochondrial STAT3 supports Ras-dependent oncogenic transformation. Science. 2009;324:1713–1716. doi: 10.1126/science.1171721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Meier JA, Larner AC. Toward a new STATe: The role of STATs in mitochondrial function. Semin Immunol. 2014;26:20–28. doi: 10.1016/j.smim.2013.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gough DJ, Koetz L, Levy DE. The MEK-ERK pathway is necessary for serine phosphorylation of mitochondrial STAT3 and Ras-mediated transformation. PLOS ONE. 2013;8:e83395. doi: 10.1371/journal.pone.0083395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Luo X, Ribeiro M, Bray ER, Lee DH, Yungher BJ, Mehta ST, Thakor KA, Diaz F, Lee JK, Moraes CT, Bixby JL, Lemmon VP, Park KK. Enhanced transcriptional activity and mitochondrial localization of STAT3 co-induce axon regrowth in the adult central nervous system. Cell Rep. 2016;15:398–410. doi: 10.1016/j.celrep.2016.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wake MS, Watson CJ. STAT3 the oncogene—Still eluding therapy? FEBS J. 2015;282:2600–2611. doi: 10.1111/febs.13285. [DOI] [PubMed] [Google Scholar]
- 7.Yang R, Rincon M. Mitochondrial Stat3, the need for design thinking. Int J Biol Sci. 2016;12:532–544. doi: 10.7150/ijbs.15153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Szczepanek K, Chen Q, Derecka M, Salloum FN, Zhang Q, Szelag M, Cichy J, Kukreja RC, Dulak J, Lesnefsky EJ, Larner AC. Mitochondrial-targeted signal transducer and activator of transcription 3 (STAT3) protects against ischemia-induced changes in the electron transport chain and the generation of reactive oxygen species. J Biol Chem. 2011;286:29610–29620. doi: 10.1074/jbc.M111.226209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boengler K, Ungefug E, Heusch G, Schulz R. The STAT3 inhibitor stattic impairs cardiomyocyte mitochondrial function through increased reactive oxygen species formation. Curr Pharm Des. 2013;19:6890–6895. doi: 10.2174/138161281939131127115940. [DOI] [PubMed] [Google Scholar]
- 10.Lachance C, Goupil S, Leclerc P. Stattic V, a STAT3 inhibitor, affects human spermatozoa through regulation of mitochondrial activity. J Cell Physiol. 2013;228:704–713. doi: 10.1002/jcp.24215. [DOI] [PubMed] [Google Scholar]
- 11.Zhang Q, Raje V, Yakovlev VA, Yacoub A, Szczepanek K, Meier J, Derecka M, Chen Q, Hu Y, Sisler J, Hamed H, Lesnefsky EJ, Jr, Valerie K, Dent P, Larner AC. Mitochondrial-localized Stat3 promotes breast cancer growth via phosphorylation of serine 727. J Biol Chem. 2013;288:31280–31288. doi: 10.1074/jbc.M113.505057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garama DJ, Harris TJ, White CL, Rossello FJ, Abdul-Hay M, Gough DJ, Levy DE. A synthetic lethal interaction between glutathione synthesis and mitochondrial reactive oxygen species provides a tumor-specific vulnerability dependent on STAT3. Mol Cell Biol. 2015;35:3646–3656. doi: 10.1128/MCB.00541-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li L, Cheung S-h, Evans EL, Shaw PE. Modulation of gene expression and tumor cell growth by redox modification of STAT3. Cancer Res. 2010;70:8222–8232. doi: 10.1158/0008-5472.CAN-10-0894. [DOI] [PubMed] [Google Scholar]
- 14.Sobotta MC, Liou W, Stöcker S, Talwar D, Oehler M, Ruppert T, Scharf AND, Dick TP. Peroxiredoxin-2 and STAT3 form a redox relay for H2O2 signaling. Nat Chem Biol. 2015;11:64–70. doi: 10.1038/nchembio.1695. [DOI] [PubMed] [Google Scholar]
- 15.Kramer AH, Edkins AL, Hoppe HC, Prinsloo E. Dynamic mitochondrial localisation of STAT3 in the cellular adipogenesis model 3T3-L1. J Cell Biochem. 2015;116:1232–1240. doi: 10.1002/jcb.25076. [DOI] [PubMed] [Google Scholar]
- 16.Yang R, Lirussi D, Thornton TM, Jelley-Gibbs DM, Diehl SA, Case LK, Madesh M, Taatjes DJ, Teuscher C, Haynes L, Rincón M. Mitochondrial Ca2+ and membrane potential, an alternative pathway for Interleukin 6 to regulate CD4 cell effector function. eLife. 2015;4:e06376. doi: 10.7554/eLife.06376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Macias E, Rao D, Carbajal S, Kiguchi K, DiGiovanni J. Stat3 binds to mtDNA and regulates mitochondrial gene expression in keratinocytes. J Invest Dermatol. 2014;134:1971–1980. doi: 10.1038/jid.2014.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou L, Too HP. Mitochondrial localized STAT3 is involved in NGF induced neurite outgrowth. PLOS ONE. 2011;6:e21680. doi: 10.1371/journal.pone.0021680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim H, Hawley TS, Hawley RG, Baumann H. Protein tyrosine phosphatase 2 (SHP-2) moderates signaling by gp130 but is not required for the induction of acute-phase plasma protein genes in hepatic cells. Mol Cell Biol. 1998;18:1525–1533. doi: 10.1128/mcb.18.3.1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wieckowski MR, Giorgi C, Lebiedzinska M, Duszynski J, Pinton P. Isolation of mitochondria-associated membranes and mitochondria from animal tissues and cells. Nat Protoc. 2009;4:1582–1590. doi: 10.1038/nprot.2009.151. [DOI] [PubMed] [Google Scholar]
- 21.Shaw PE. Could STAT3 provide a link between respiration and cell cycle progression? Cell Cycle. 2010;9:4294–4296. doi: 10.4161/cc.9.21.13677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schieber M, Chandel NS. ROS function in redox signaling and oxidative stress. Curr Biol. 2014;24:R453–R462. doi: 10.1016/j.cub.2014.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dikalova AE, Bikineyeva AT, Budzyn K, Nazarewicz RR, McCann L, Lewis W, Harrison DG, Dikalov SI. Therapeutic targeting of mitochondrial superoxide in hypertension. Circ Res. 2010;107:106–116. doi: 10.1161/CIRCRESAHA.109.214601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dorsch M, Behmenburg F, Raible M, Blase D, Grievink H, Hollmann MW, Heinen A, Huhn R. Morphine-induced preconditioning: Involvement of protein kinase A and mitochondrial permeability transition pore. PLOS ONE. 2016;11:e0151025. doi: 10.1371/journal.pone.0151025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rawlings JS, Rosler KM, Harrison DA. The JAK/STAT signaling pathway. J Cell Sci. 2004;117:1281–1283. doi: 10.1242/jcs.00963. [DOI] [PubMed] [Google Scholar]
- 26.Darnowski JW, Goulette FA, Guan Y-j, Chatterjee D, Yang Z-F, Cousens LP, Chin YE. Stat3 cleavage by caspases: Impact on full-length Stat3 expression, fragment formation, and transcriptional activity. J Biol Chem. 2006;281:17707–17717. doi: 10.1074/jbc.M600088200. [DOI] [PubMed] [Google Scholar]
- 27.Oda A, Wakao H, Fujita H. Calpain is a signal transducer and activator of transcription (STAT) 3 and STAT5 protease. Blood. 2002;99:1850–1852. doi: 10.1182/blood.v99.5.1850. [DOI] [PubMed] [Google Scholar]
- 28.Thompson J, Hu Y, Lesnefsky EJ, Chen Q. Activation of mitochondrial calpain and increased cardiac injury: Beyond AIF release. Am J Physiol Heart Circ Physiol. 2016;310:H376–H384. doi: 10.1152/ajpheart.00748.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Quirós PM, Langer T, Lopez-Otín C. New roles for mitochondrial proteases in health, ageing and disease. Nat Rev Mol Cell Biol. 2015;16:345–359. doi: 10.1038/nrm3984. [DOI] [PubMed] [Google Scholar]
- 30.Gibellini L, Pinti M, Bartolomeo R, De Biasi S, Cormio A, Musicco C, Carnevale G, Pecorini S, Nasi M, De Pol A, Cossarizza A. Inhibition of Lon protease by triterpenoids alters mitochondria and is associated to cell death in human cancer cells. Oncotarget. 2015;6:25466–25483. doi: 10.18632/oncotarget.4510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bernstein SH, Venkatesh S, Li M, Lee J, Lu B, Hilchey SP, Morse KM, Metcalfe HM, Skalska J, Andreeff M, Brookes PS, Suzuki CK. The mitochondrial ATP-dependent Lon protease: A novel target in lymphoma death mediated by the synthetic triterpenoid CDDO and its derivatives. Blood. 2012;119:3321–3329. doi: 10.1182/blood-2011-02-340075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bragoszewski P, Gornicka A, Sztolsztener ME, Chacinska A. The ubiquitin-proteasome system regulates mitochondrial intermembrane space proteins. Mol Cell Biol. 2013;33:2136–2148. doi: 10.1128/MCB.01579-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tammineni P, Anugula C, Mohammed F, Anjaneyulu M, Larner AC, Sepuri NBV. The import of the transcription factor STAT3 into mitochondria depends on GRIM-19, a component of the electron transport chain. J Biol Chem. 2013;288:4723–4732. doi: 10.1074/jbc.M112.378984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Boengler K, Hilfiker-Kleiner D, Heusch G, Schulz R. Inhibition of permeability transition pore opening by mitochondrial STAT3 and its role in myocardial ischemia/reperfusion. Basic Res Cardiol. 2010;105:771–785. doi: 10.1007/s00395-010-0124-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Elrod JW, Molkentin JD. Physiologic functions of cyclophilin D and the mitochondrial permeability transition pore. Circ J. 2013;77:1111–1122. doi: 10.1253/circj.cj-13-0321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lu KP, Zhou XZ. The prolyl isomerase PIN1: A pivotal new twist in phosphorylation signalling and disease. Nat Rev Mol Cell Biol. 2007;8:904–916. doi: 10.1038/nrm2261. [DOI] [PubMed] [Google Scholar]
- 37.Hibino M, Sugiura K, Muro Y, Shimoyama Y, Tomita Y. Cyclosporin A induces the unfolded protein response in keratinocytes. Arch Dermatol Res. 2011;303:481–489. doi: 10.1007/s00403-010-1099-3. [DOI] [PubMed] [Google Scholar]
- 38.Nguyen TT, Wong R, Menazza S, Sun J, Chen Y, Wang G, Gucek M, Steenbergen C, Sack MN, Murphy E. Cyclophilin D modulates mitochondrial acetylome. Circ Res. 2013;113:1308–1319. doi: 10.1161/CIRCRESAHA.113.301867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Menazza S, Wong R, Nguyen T, Wang G, Gucek M, Murphy E. CypD−/− hearts have altered levels of proteins involved in Krebs cycle, branch chain amino acid degradation and pyruvate metabolism. J Mol Cell Cardiol. 2013;56:81–90. doi: 10.1016/j.yjmcc.2012.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li L, Shaw PE. A STAT3 dimer formed by inter-chain disulphide bridging during oxidative stress. Biochem Biophys Res Commun. 2004;322:1005–1011. doi: 10.1016/j.bbrc.2004.08.014. [DOI] [PubMed] [Google Scholar]
- 41.Carreira RS, Lee Y, Ghochani M, Gustafsson ÅB, Gottlieb RA. Cyclophilin D is required for mitochondrial removal by autophagy in cardiac cells. Autophagy. 2010;6:462–472. doi: 10.4161/auto.6.4.11553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tavecchio M, Lisanti S, Bennett MJ, Languino LR, Altieri DC. Deletion of cyclophilin D impairs β-oxidation and promotes glucose metabolism. Sci Rep. 2015;5:15981. doi: 10.1038/srep15981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Elschami M, Scherr M, Philippens B, Gerardy-Schahn R. Reduction of STAT3 expression induces mitochondrial dysfunction and autophagy in cardiac HL-1 cells. Eur J Cell Biol. 2013;92:21–29. doi: 10.1016/j.ejcb.2012.09.002. [DOI] [PubMed] [Google Scholar]
- 44.Szczepanek K, Xu A, Hu Y, Thompson J, He J, Larner AC, Salloum FN, Chen Q, Lesnefsky EJ. Cardioprotective function of mitochondrial-targeted and transcriptionally inactive STAT3 against ischemia and reperfusion injury. Basic Res Cardiol. 2015;110:53. doi: 10.1007/s00395-015-0509-2. [DOI] [PubMed] [Google Scholar]
- 45.Boengler K, Buechert A, Heinen Y, Roeskes C, Hilfiker-Kleiner D, Heusch G, Schulz R. Cardioprotection by ischemic postconditioning is lost in aged and STAT3-deficient mice. Circ Res. 2008;102:131–135. doi: 10.1161/CIRCRESAHA.107.164699. [DOI] [PubMed] [Google Scholar]
- 46.Capron C, Jondeau K, Casetti L, Jalbert V, Costa C, Verhoeyen E, Masse JM, Coppo P, Bene MC, Bourdoncle P, Cramer-Borde E, Dusanter-Fourt I. Viability and stress protection of chronic lymphoid leukemia cells involves overactivation of mitochondrial phosphoSTAT3Ser727. Cell Death Dis. 2014;5:e1451. doi: 10.1038/cddis.2014.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gough DJ, Marie IJ, Lobry C, Aifantis I, Levy DE. STAT3 supports experimental K-RasG12D-induced murine myeloproliferative neoplasms dependent on serine phosphorylation. Blood. 2014;124:2252–2261. doi: 10.1182/blood-2013-02-484196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mackenzie GG, Huang L, Alston N, Ouyang N, Vrankova K, Mattheolabakis G, Constantinides PP, Rigas B. Targeting mitochondrial STAT3 with the novel phosphovalproic acid (MDC-1112) inhibits pancreatic cancer growth in mice. PLOS ONE. 2013;8:e61532. doi: 10.1371/journal.pone.0061532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Qureshi R, Yildirim O, Gasser A, Basmadjian C, Zhao Q, Wilmet JP, Désaubry L, Nebigil CG. FL3, a synthetic flavagline and ligand of prohibitins, protects cardiomyocytes via STAT3 from doxorubicin toxicity. PLOS ONE. 2015;10:e0141826. doi: 10.1371/journal.pone.0141826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Carbognin E, Betto RM, Soriano ME, Smith AG, Martello G. Stat3 promotes mitochondrial transcription and oxidative respiration during maintenance and induction of naive pluripotency. EMBO J. 2016;35:618–634. doi: 10.15252/embj.201592629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cogswell PC, Kashatus DF, Keifer JA, Guttridge DC, Reuther JY, Bristow C, Roy S, Nicholson DW, Baldwin AS., Jr NF-κB and IκBα are found in the mitochondria. Evidence for regulation of mitochondrial gene expression by NF-κ B. J Biol Chem. 2003;278:2963–2968. doi: 10.1074/jbc.M209995200. [DOI] [PubMed] [Google Scholar]
- 52.Murase S. Signal transducer and activator of transcription 3 (STAT3) degradation by proteasome controls a developmental switch in neurotrophin dependence. J Biol Chem. 2013;288:20151–20161. doi: 10.1074/jbc.M113.470583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bayot A, Gareil M, Chavatte L, Hamon M-P, L’Hermitte-Stead C, Beaumatin F, Priault M, Rustin P, Lombes A, Friguet B, Bulteau AL. Effect of Lon protease knockdown on mitochondrial function in HeLa cells. Biochimie. 2014;100:38–47. doi: 10.1016/j.biochi.2013.12.005. [DOI] [PubMed] [Google Scholar]
- 54.Soubannier V, McLelland GL, Zunino R, Braschi E, Rippstein P, Fon EA, McBride HM. A vesicular transport pathway shuttles cargo from mitochondria to lysosomes. Curr Biol. 2012;22:135–141. doi: 10.1016/j.cub.2011.11.057. [DOI] [PubMed] [Google Scholar]
- 55.McLelland GL, Soubannier V, Chen CX, McBride HM, Fon EA. Parkin and PINK1 function in a vesicular trafficking pathway regulating mitochondrial quality control. EMBO J. 2014;33:282–295. doi: 10.1002/embj.201385902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sugiura A, McLelland GL, Fon EA, McBride HM. A new pathway for mitochondrial quality control: Mitochondrial-derived vesicles. EMBO J. 2014;33:2142–2156. doi: 10.15252/embj.201488104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Su CC, Yang JY, Leu HB, Chen Y, Wang PH. Mitochondrial Akt-regulated mitochondrial apoptosis signaling in cardiac muscle cells. Am J Physiol Heart Circ Physiol. 2012;302:H716–H723. doi: 10.1152/ajpheart.00455.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rasola A, Sciacovelli M, Chiara F, Pantic B, Brusilow WS, Bernardi P. Activation of mitochondrial ERK protects cancer cells from death through inhibition of the permeability transition. Proc Natl Acad Sci USA. 2010;107:726–731. doi: 10.1073/pnas.0912742107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hanawa N, Shinohara M, Saberi B, Gaarde WA, Han D, Kaplowitz N. Role of JNK translocation to mitochondria leading to inhibition of mitochondria bioenergetics in acetaminophen-induced liver injury. J Biol Chem. 2008;283:13565–13577. doi: 10.1074/jbc.M708916200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bijur GN, Jope RS. Rapid accumulation of Akt in mitochondria following phosphatidylinositol 3-kinase activation. J Neurochem. 2003;87:1427–1435. doi: 10.1046/j.1471-4159.2003.02113.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tammineni P, Anugula C, Mohammed F, Anjaneyulu M, Larner AC, Sepuri NB. The import of the transcription factor STAT3 into mitochondria depends on GRIM-19, a component of the electron transport chain. J Biol Chem. 2012;288:4723–4732. doi: 10.1074/jbc.M112.378984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nargund AM, Pellegrino MW, Fiorese CJ, Baker BM, Haynes CM. Mitochondrial import efficiency of ATFS-1 regulates mitochondrial UPR activation. Science. 2012;337:587–590. doi: 10.1126/science.1223560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang X, Chen XJ. A cytosolic network suppressing mitochondria-mediated proteostatic stress and cell death. Nature. 2015;524:481–484. doi: 10.1038/nature14859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Senturk S, Yao Z, Camiolo M, Stiles B, Rathod T, Walsh AM, Nemajerova A, Lazzara MJ, Altorki NK, Krainer A, Moll UM, Lowe SW, Cartegni L, Sordella R. p53Ψ is a transcriptionally inactive p53 isoform able to reprogram cells toward a metastatic-like state. Proc Natl Acad Sci USA. 2014;111:E3287–96. doi: 10.1073/pnas.1321640111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hevehan DL, Miller WM, Papoutsakis ET. Differential expression and phosphorylation of distinct STAT3 proteins during granulocytic differentiation. Blood. 2002;99:1627–1637. doi: 10.1182/blood.v99.5.1627. [DOI] [PubMed] [Google Scholar]
- 66.Sutendra G, Kinnaird A, Dromparis P, Paulin R, Stenson TH, Haromy A, Hashimoto K, Zhang N, Flaim E, Michelakis ED. A nuclear pyruvate dehydrogenase complex is important for the generation of acetyl-CoA and histone acetylation. Cell. 2014;158:84–97. doi: 10.1016/j.cell.2014.04.046. [DOI] [PubMed] [Google Scholar]
- 67.Tan K, Fujimoto M, Takii R, Takaki E, Hayashida N, Nakai A. Mitochondrial SSBP1 protects cells from proteotoxic stresses by potentiating stress-induced HSF1 transcriptional activity. Nat Commun. 2015;6:6580. doi: 10.1038/ncomms7580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nargund AM, Fiorese CJ, Pellegrino MW, Deng P, Haynes CM. Mitochondrial and nuclear accumulation of the transcription factor ATFS-1 promotes OXPHOS recovery during the UPRmt. Mol Cell. 2015;58:123–133. doi: 10.1016/j.molcel.2015.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pellegrino MW, Nargund AM, Haynes CM. Signaling the mitochondrial unfolded protein response. Biochim Biophys Acta. 2013;1833:410–416. doi: 10.1016/j.bbamcr.2012.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Quirós PM, Mottis A, Auwerx J. Mitonuclear communication in homeostasis and stress. Nat Rev Mol Cell Biol. 2016;17:213–226. doi: 10.1038/nrm.2016.23. [DOI] [PubMed] [Google Scholar]
- 71.Tavecchio M, Lisanti S, Lam A, Ghosh JC, Martin NM, O’Connell M, Weeraratna AT, Kossenkov AV, Showe LC, Altieri DC. Cyclophilin D extramitochondrial signaling controls cell cycle progression and chemokine-directed cell motility. J Biol Chem. 2013;288:5553–5561. doi: 10.1074/jbc.M112.433045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Timofeeva OA, Gaponenko V, Lockett SJ, Tarasov SG, Jiang S, Michejda CJ, Perantoni AO, Tarasova NI. Rationally designed inhibitors identify STAT3 N-domain as a promising anticancer drug target. ACS Chem Biol. 2007;2:799–809. doi: 10.1021/cb700186x. [DOI] [PubMed] [Google Scholar]
- 73.Östman A, Frijhoff J, Sandin Å, Böhmer FD. Regulation of protein tyrosine phosphatases by reversible oxidation. J Biochem. 2011;150:345–356. doi: 10.1093/jb/mvr104. [DOI] [PubMed] [Google Scholar]
- 74.Salmond RJ, Alexander DR. SHP2 forecast for the immune system: Fog gradually clearing. Trends Immunol. 2006;27:154–160. doi: 10.1016/j.it.2006.01.007. [DOI] [PubMed] [Google Scholar]
- 75.Park SJ, Kim HY, Kim H, Park SM, Joe E-h, Jou I, Choi YH. Oxidative stress induces lipid-raft-mediated activation of Src homology 2 domain-containing protein-tyrosine phosphatase 2 in astrocytes. Free Radic Biol Med. 2009;46:1694–1702. doi: 10.1016/j.freeradbiomed.2009.03.026. [DOI] [PubMed] [Google Scholar]
- 76.Zheng H, Li S, Hsu P, Qu CK. Induction of a tumor-associated activating mutation in protein tyrosine phosphatase Ptpn11 (Shp2) enhances mitochondrial metabolism, leading to oxidative stress and senescence. J Biol Chem. 2013;288:25727–25738. doi: 10.1074/jbc.M113.462291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bigi A, Beltrami E, Trinei M, Stendardo M, Pelicci PG, Giorgio M. Cyclophilin D counteracts P53-mediated growth arrest and promotes Ras tumorigenesis. Oncogene. 2016;35:5132–5143. doi: 10.1038/onc.2016.42. [DOI] [PubMed] [Google Scholar]
- 78.Vaseva AV, Marchenko ND, Ji K, Tsirka SE, Holzmann S, Moll UM. P53 opens the mitochondrial permeability transition pore to trigger necrosis. Cell. 2012;149:1536–1548. doi: 10.1016/j.cell.2012.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mertens C, Haripal B, Klinge S, Darnell JE. Mutations in the linker domain affect phospho-STAT3 function and suggest targets for interrupting STAT3 activity. Proc Natl Acad Sci USA. 2015;112:14811–14816. doi: 10.1073/pnas.1515876112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Derecka M, Gornicka A, Koralov SB, Szczepanek K, Morgan M, Raje V, Sisler J, Zhang Q, Otero D, Cichy J, Rajewsky K, Shimoda K, Poli V, Strobl B, Pellegrini S, Harris TE, Seale P, Russell AP, McAinch AJ, O’Brien PE, Keller SR, Croniger CM, Kordula T, Larner AC. Tyk2 and Stat3 regulate brown adipose tissue differentiation and obesity. Cell Metab. 2012;16:814–824. doi: 10.1016/j.cmet.2012.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sisler JD, Morgan M, Raje V, Grande RC, Derecka M, Meier J, Cantwell M, Szczepanek K, Korzun WJ, Lesnefsky EJ, Harris TE, Croniger CM, Larner AC. The Signal transducer and activator of transcription 1 (STAT1) inhibits mitochondrial biogenesis in liver and fatty acid oxidation in adipocytes. PLOS ONE. 2015;10:e0144444. doi: 10.1371/journal.pone.0144444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J R Stat Soc B Methodol. 1995;57:289–300. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.