Fibroblast-adapted rhesus CMV–vectored vaccines protect macaques from SIV challenge and elicit unconventional CD8 T cell responses. In contrast, Murray et al. show that humans vaccinated with fibroblast-adapted human CMV vaccines generate conventional CD8 T cell responses.
Abstract
Cytomegalovirus (CMV)-based vaccines have shown remarkable efficacy in the rhesus macaque model of acquired immune deficiency syndrome, enabling 50% of vaccinated monkeys to clear a subsequent virulent simian immunodeficiency virus challenge. The protective vaccine elicited unconventional CD8 T cell responses that were entirely restricted by MHC II or the nonclassical MHC I molecule, MHC-E. These unconventional responses were only elicited by a fibroblast-adapted rhesus CMV vector with limited tissue tropism; a repaired vector with normal tropism elicited conventional responses. Testing whether these unusual protective CD8 T responses could be elicited in humans requires vaccinating human subjects with a fibroblast-adapted mutant of human CMV (HCMV). In this study, we describe the CD8 T cell responses of human subjects vaccinated with two fibroblast-adapted HCMV vaccines. Most responses were identified as conventional classically MHC I restricted, and we found no evidence for MHC II or HLA-E restriction. These results indicate that fibroblast adaptation alone is unlikely to explain the unconventional responses observed in macaques.
Introduction
CMV is a common human herpesvirus that establishes lifelong infection in most people worldwide. Although infection is usually asymptomatic, CMV causes severe disease in the context of congenital infection and in transplant patients. This has prompted a long, and so far unsuccessful, search for a vaccine against CMV. In asymptomatic healthy carriers, CMV elicits an extremely large, sustained humoral and cell-mediated immune response. Despite this, CMV has the unusual capacity to superinfect CMV-seropositive individuals. For these reasons, CMV is being ardently pursued as a vaccine vector, in particular for vaccination against HIV.
In a series of groundbreaking studies, Louis Picker’s group vaccinated rhesus macaques with a rhesus CMV (RhCMV) vector encoding simian immunodeficiency virus (SIV) antigens (Hansen et al., 2011, 2013a,b, 2016). These macaques were then challenged repeatedly with SIV until they were unequivocally SIV infected. Of these vaccinated SIV-infected monkeys, 50% completely cleared the SIV infection, a result not previously observed with any vaccine or in rare elite controllers (Hansen et al., 2011, 2013b). Moreover, the protection afforded by RhCMV-based vaccines was associated with very unusual CD8 T cell responses to both the vector and inserted SIV antigens (Hansen et al., 2013a, 2016). Normal immunodominance hierarchies were abolished; instead, vaccine-elicited CD8 T cells recognized a broad array of peptides covering about two thirds of the antigenic proteins. Many responses were promiscuous, recognizing peptide presented by allogeneic cells, and some supertopes were recognized by all animals regardless of MHC haplotype. Most strikingly, two thirds of CD8 T cells recognized peptide in the context of MHC II, and the remainder recognized peptide in the context of the nonclassical MHC Ib molecule MHC-E. This vaccine completely failed to elicit classically MHC I–restricted CD8 T cells in any macaque. The authors suggested that these unconventional CD8 T cell responses were responsible for the RhCMV-SIV vaccine efficacy (Hansen et al., 2013a,b, 2016).
Such unconventional CD8 T cell responses are not a normal feature of the response to natural CMV infection in either monkeys or humans. In fact, these unconventional CD8 T cells were only elicited by fibroblast-adapted RhCMV vectors, i.e., viruses that had lost the pentameric glycoprotein complex that confers ability to infect most nonfibroblast cell types. Hansen et al. (2013a) propose that the altered tropism is a probable mechanism for induction of these atypical CD8 T cell responses. How loss of the pentameric complex so profoundly impacts CD8 T cell responses is not yet understood. Nevertheless, if unconventional T cells are a key feature of the RhCMV vaccine efficacy, it is important to know whether similar responses are elicited by fibroblast-adapted, pentameric complex–deficient CMV vaccines in humans.
Development of CMV vaccines is complicated by the limited host range of the virus. CMVs are found in most mammalian species and are highly species specific. Thus, the only way to test whether these unusual, protective CD8 T cell responses to fibroblast-adapted CMV occur in humans is to study the immune response in human subjects vaccinated with a fibroblast-adapted strain of human CMV (HCMV). We recently reported a phase I clinical trial to test the safety and immunogenicity of four live fibroblast-adapted HCMV vaccines that are chimeras of Towne and Toledo strains (Adler et al., 2016). As with the RhCMV vaccines, these viruses lack the pentameric complex and have cellular tropism essentially limited to fibroblasts, although the specific defect in the pentameric complex genes is different (Hansen et al., 2013a; Adler et al., 2016). Here, we report that the human CD8 T cell response to fibroblast-adapted Towne/Toledo HCMV does not mirror the CD8 T cell response observed in rhesus macaques. On the contrary, humans vaccinated with Towne/Toledo HCMV mounted CD8 T cell responses that were predominantly conventional in terms of immunodominance, breadth, core epitope length, and MHC restriction. These discrepant results may reflect differences between rhesus and human immune systems or differences, other than tropism, between RhCMV and HCMV vaccines.
Results and discussion
Towne/Toledo HCMV chimeras are primarily fibroblast adapted
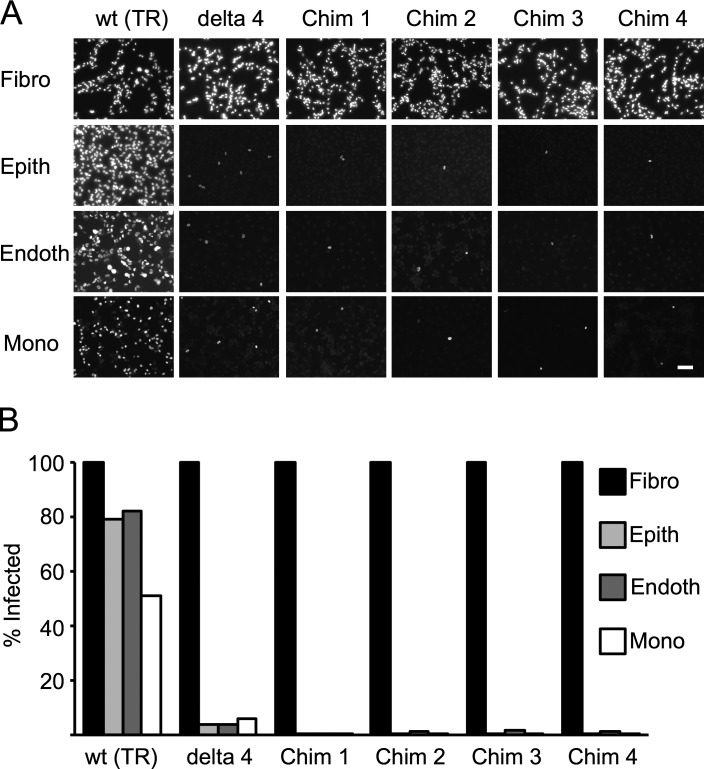
Sequence analysis showed that, in all four Towne/Toledo HCMV vaccines, the UL128-131 region derives from Toledo, which harbors a nonfunctional mutation in UL128 (GenBank accession nos. KX101021, KX101022, KX101023, and KX101024; Adler et al., 2016; Suárez et al., 2017). Therefore, these viruses should lack formation of the pentameric complex, which is required for entry into epithelial, endothelial, and other nonfibroblast cell types and is believed to be important to elicit robust epithelial cell entry–specific neutralizing antibody responses (Cui et al., 2008). Consistent with the genetic predictions that all four vaccines lack pentameric complex expression, all subjects who seroconverted after vaccination had low epithelial-specific neutralizing antibody responses (Adler et al., 2016). Because tissue tropism is likely involved in the unconventional CD8 T cell responses observed in rhesus macaques, we tested the tropism of the Towne/Toledo chimeras. We infected fibroblast, epithelial, endothelial, and monocytic cell lines with wild-type HCMV strain TR; TRΔ4, which is derived from TR and lacks UL128-150; or each of the four Towne/Toledo HCMV vaccines (Fig. 1). All virus strains infected 100% of fibroblasts. In contrast, only wild-type TR infected epithelial and endothelial cells efficiently. Wild-type TR infected monocytic cells more poorly than other cell types, so a multiplicity of infection (MOI) of 100 was needed to infect ∼35% of differentiated THP-1 cells. At this higher MOI, TRΔ4 infected 6% and the four chimeras each infected <1% of THP-1 cells. Thus, Towne/Toledo HCMV vaccines demonstrate a cell tropism that is consistent with disruption of the pentameric complex and is similar to the RhCMV68-1 strain used as an SIV vector in macaques.
Figure 1.
Towne/Toledo chimeras are fibroblast adapted. (A and B) Cell lines were infected with the indicated Towne/Toledo HCMV vaccines or control viruses at the following MOIs: fibroblasts (Fibro; NHDF), 3; epithelial (Epith; ARPE-19) and endothelial (Endoth; HUVEC) cells, 10; and monocytes (Mono; THP-1), 100. Delta 4 is a deletion mutant for UL128-150 and, thus, cannot form the pentameric complex. 24 h after infection, cells were stained for IE-1 and analyzed by immunofluorescence microscopy. (A) Immunofluorescence photomicrographs of IE-1 expression in infected cells. Bar, 100 µm. (B) Quantitation of proportion of infected cells. Data depict results from one representative of three independent experiments. Chim, chimera.
Vaccination with Towne/Toledo chimeras elicits narrowly focused CD8 T cell responses to IE-1
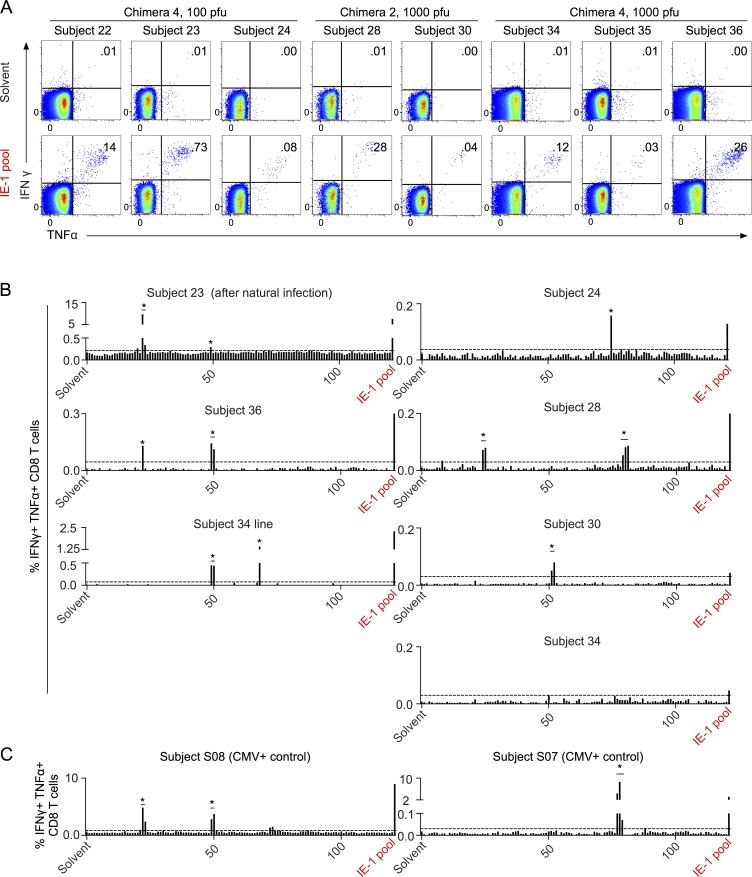
As previously reported, 8 out of 11 subjects who seroconverted after vaccination with low-dose (100 or 1,000 PFU) Towne/Toledo HCMV vaccines also mounted CD8 T cell responses. T cell responses were observed only in recipients of chimeras 2 and 4 (Table S1). All of these subjects responded to a peptide pool covering the immediate early 1 (IE-1) protein (Fig. 2 A). We previously reported that three subjects responded to one or more additional antigens (Adler et al., 2016), but as IE-1 was the only antigen that elicited a measurable CD8 T cell response from more than two subjects, we focused our detailed analysis on this antigen. Furthermore, assessment of the rhesus macaque CD8 T cell response to RhCMV antigens was restricted to IE-1 (Hansen et al., 2013a). In that study, RhCMV-vaccinated macaques mounted CD8 T cell responses that showed broad epitope specificity, in contrast to the narrow focus on a few epitopes that is characteristic of most CD8 T cell responses (Hansen et al., 2013a). We assessed the breadth of IE-1 peptide responses in all eight subjects who had measurable HCMV-specific CD8 T cell responses, and we found that each subject responded to only one to two peptides or peptide hot spots (two adjacent overlapping peptides; Fig. 2 B). This was similar to the breadth of responses in naturally HCMV-infected controls (Fig. 2 C). This contrasts with the extremely broad epitope specificity observed in RhCMV-vaccinated macaques, which responded to a mean of 36 IE-1 peptides (25% of all the overlapping peptides; Hansen et al., 2013a).
Figure 2.
Towne/Toledo chimera vaccination elicits IE-1–specific CD8 T cell responses with narrow epitope breadth. (A) PBMCs were stimulated with solvent control (top) or an overlapping peptide pool covering the entire AD169 IE-1 protein (bottom), and antigen-specific responses were assessed by ICS. Plots are gated on live CD3+CD8+CD4− lymphocytes. Numbers on plots indicate the proportion of gated cells that are IFN-γ+TNF+. Subject 24 did not respond to this commercially available peptide pool (not depicted) but did respond to an overlapping peptide pool covering the entire IE-1 protein sequence of the immunizing vaccine (shown). (B) PBMCs were epitope screened by stimulating triplicate wells with the individual peptides comprising the overlapping peptide pool. Peptides are numbered from the N terminus and are indicated on the x axis. Responses were considered positive if the proportion of CD8 T cells that was IFN-γ+TNF+ was >0.03% and >3 SD above background. Dotted lines represent cutoffs for positive responses for each subject. No individual peptide responses reached this threshold for subject 34, but upon growing a T cell line from subject 34 PBMCs, we identified two responses. The large response in subject 23 was subsequently determined to have arisen after natural HCMV infection between 1 and 2 yr after vaccination, but this response was present, albeit much lower, immediately after vaccine administration (Figs. 2 A and 5). Insufficient PBMCs were available to screen subjects 22 and 35 for responses to individual 15mers. Asterisks represent responses studied in detail for core epitope identity and HLA restriction, summarized in Table S1. (C) PBMCs from naturally infected HCMV-seropositive controls were epitope screened as in panel B. Data show FACS plots from every subject analyzed and are representative of two or more experiments (A) or one to two screening experiments (B and C).
Most IE-1–specific CD8 T cells from Towne/Toledo-vaccinated subjects recognize peptides presented by MHC I
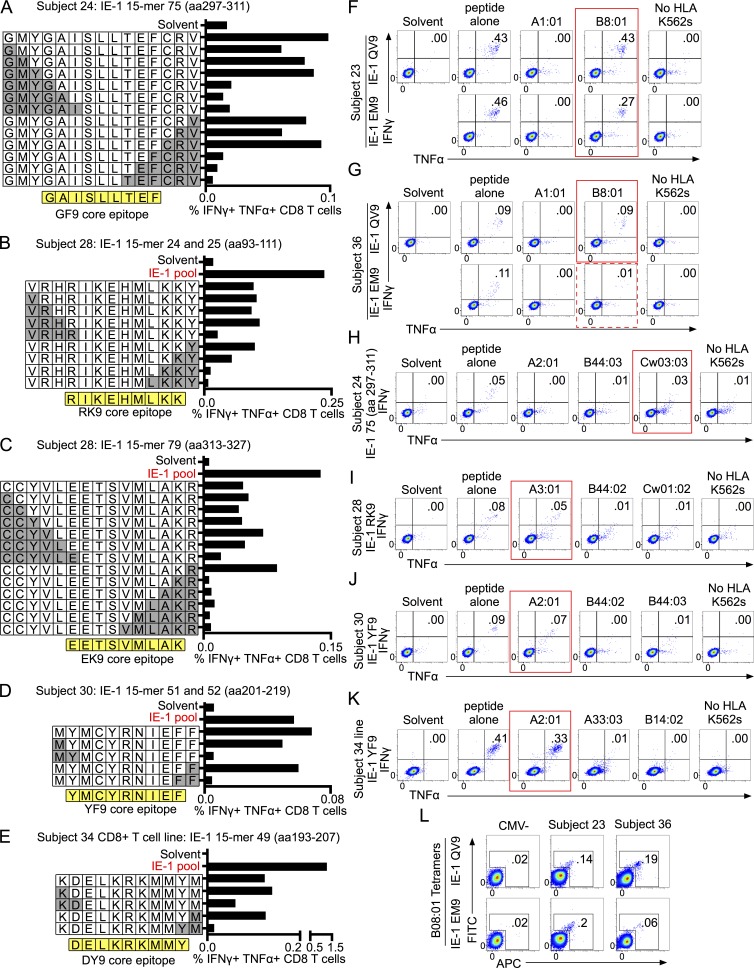
The second unusual feature of the CD8 T cell responses to RhCMV fibroblast-adapted vectors is the complete lack of responses that are restricted by classical MHC I molecules; two thirds are MHC II restricted, and the remaining are MHC-E restricted (Hansen et al., 2013a, 2016). We used a combination of peptide truncations (Fig. 3, A–E), single HLA transfectant antigen-presenting cell lines (Fig. 3, F–K), and tetramer staining (Fig. 3 L) to assess the core epitope identity and HLA restriction of CD8 T cell responses in Towne/Toledo vaccine recipients. The 10 response hotspots that were sufficiently robust to study are indicated with asterisks on the peptide maps (Fig. 2 B).
Figure 3.
Most CD8 T cells respond to epitopes presented by MHC I. (A–E) Peptide truncations were generated and used to stimulate PBMCs from the indicated subjects. Truncations were generated from either the single 15mer to which a subject responded or the 11mer-overlapping region when a subject responded to two consecutive 15mers. White boxes show the single letter abbreviation for amino acids of the truncated peptides. Gray boxes show the amino acids that were removed in each truncation. The core epitope identified through truncations is shown in yellow below each graph. CD8 T cell responses were defined as in Fig. 2. (C) The 7mer peptide that overlaps peptides 79–81 to which subject 28 responded did not elicit a response, which indicates that this region contains two separate peptides. We identified the N-terminal peptide as EK9. Truncations of peptide 81 did not yield interpretable results. (F–K) Single HLA transfectants were pulsed with the indicated peptides for 1.5 h, washed extensively, and then used to stimulate PBMCs from subjects positive for that HLA allele. Peptide alone denotes that PBMCs were incubated directly with the indicated peptide rather than incubated with peptide-pulsed/washed single HLA transfectants. No HLA K562s denotes that PBMCs were cultured with peptide-pulsed, but untransfected, APC. Red boxes highlight HLA restriction of peptide responses. The dashed red box indicates HLA restriction that accounts for only a minority of the peptide response. Unless otherwise noted, the peptides used are those shown in Table S1. Transfectants were not available for every MHC I isoform of every subject, but all available transfectants were tested, and all transfectants strongly expressed HLA as assessed by flow cytometry (Fig. 4 and not depicted). Plots are gated on live CD3+CD8+CD4− cells. (F) Subject 23’s responses were assessed on PBMC samples obtained before the putative natural infection event. (L) PBMCs were stained with HLA-B08 tetramers folded with the indicated peptide. Plots are gated on live CD3+CD4−CD8+ lymphocytes. Data show responses from every subject analyzed and depict results of one experiment (A, B, D–F, H, and K) or are representative of two independent experiments (C, G, I, J, and L).
First, we noticed that several responses were identical to previously published HLA-B08–restricted responses to natural HCMV infection. Subjects 23 and 36 responded to IE-1 15mers 22 and 49–50 (Fig. 2 B). These peptides contain the 9mer sequences QIKVRVDMV (QV9) and ELKRKMMYM (EM9), which are previously described HLA-B08–restricted HCMV epitopes (Kern, 1999; Wills et al., 2002; Elkington et al., 2003). In both subjects, the QV9 responses were identified as HLA-B08 restricted using HLA-B08 transfectants and tetramers (Fig. 3, F, G, and L), and in subject 23, the EM9 response was likewise HLA-B08 restricted (Fig. 3, F and L). It is important to note that we discovered that subject 23 acquired a natural HCMV infection between 1 and 2 yr after vaccination (Adler et al., 2016). Leukapheresis occurred at 2 yr after vaccination, and that was the only time point at which there were sufficient PBMCs to perform the peptide-spanning screen of IE-1 (Fig. 2). However, we assessed MHC class I restriction of the two IE-1 peptide responses from subject 23 on PBMC samples taken before the natural infection. Therefore, this assessment is measuring the response to the vaccination only (Fig. 3 F). In subject 36, only ∼10% of the EM9 response could be accounted for by HLA-B08 restriction (Fig. 3 G). We conjecture that the remainder of the EM9 response may be HLA-B18 restricted in subject 36 (Table S1); however, HLA-B18 single transfectants were not available for this study.
We definitively identified four additional classical MHC I–restricted responses using peptide truncations and peptide-pulsed single HLA transfectants. These were Cw3-, A3-, A2-, and A2-restricted responses from subjects 24, 28, 30, and 34, respectively (Fig. 3, A, B, D, E, and H–K; and Table S1). In all cases, the proportion of CD8 T cells responding to peptide-pulsed single HLA transfectants was similar to the proportion responding when peptide was added directly to the PBMCs (Fig. 3, H–K). For subject 34, no individual IE-1 peptides elicited a response that rose consistently above background levels (Fig. 2 B), but we were able to grow a T cell line from this subject’s PBMCs. This line responded to two IE-1 peptides (Fig. 2 B), and one of these is HLA-A2 restricted (Fig. 3 K).
Of the three remaining responses (two in subject 28 and one in subject 34), there is reason to believe that two are also MHC I restricted. Single HLA transfectants were not available for all MHC I alleles for each of these subjects; however, peptide truncations revealed a pattern consistent with MHC I–restricted epitopes. First, responses were maintained until a critical single amino acid was lost from either the N or C terminus, at which point the response declined precipitously (Fig. 3, C and E). This pattern is typical of MHC I–restricted epitopes because the N and C termini of the peptide bind to the peptide-binding groove of the MHC I molecule and contribute important binding affinity, in contrast to the long MHC II–restricted peptides, which overhang the groove. Second, the core epitopes required for the responses were eight to nine amino acids long, which is typical for MHC I–restricted responses. Subject 28 responded to a 9mer from the N terminus of the region spanning peptides 79–81 (Fig. 3 C and Table S1), and subject 34 responded to a 9mer shared between peptides 49 and 50 (Fig. 3 E and Table S1). Overall, we identified the classical MHC I allele that restricted 8 out of 12 detectable responses. Of the remaining four responses, three are consistent with MHC I restriction, as assessed by peptide truncations.
No evidence for MHC II or HLA-E restriction in CD8 T cell response to Towne/Toledo HCMV vaccines
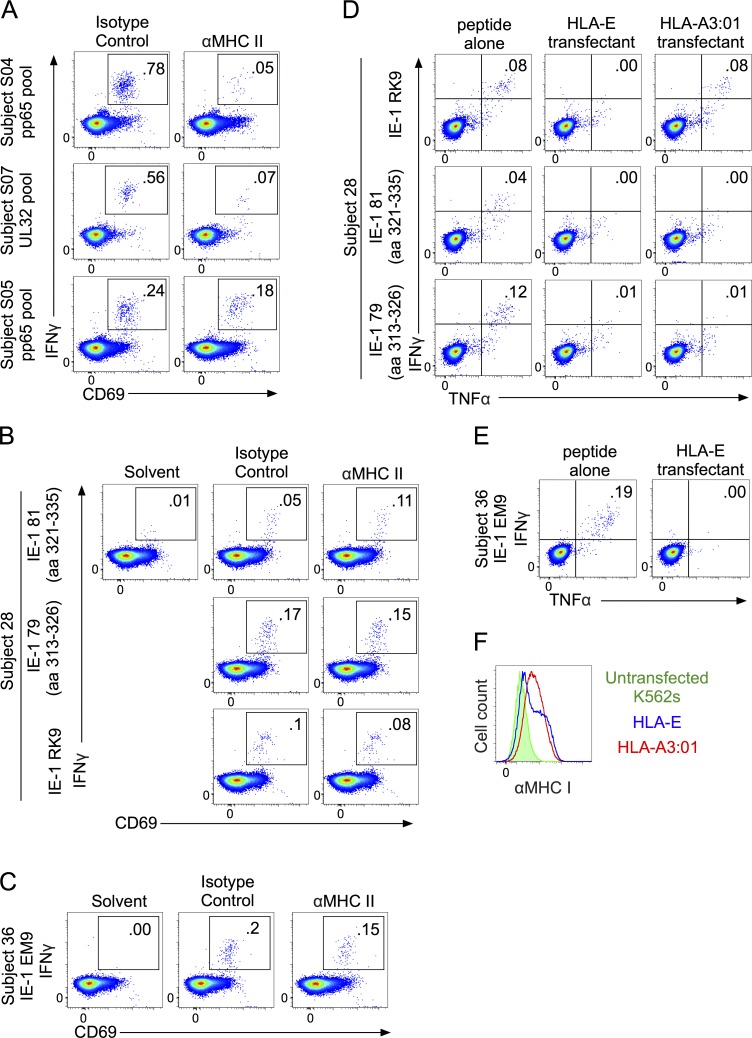
Although we found most CD8 T cell responses to the HCMV vaccine were MHC Ia restricted, the restriction element for four responses from subjects 36, 28, and 34 remained unidentified. Responses in subject 34 could not be studied further because the T cell line failed to survive. We tested whether the other three could be either MHC II or HLA-E restricted. To test for MHC II restriction, we tried to block responses using anti–MHC II antibodies or the class II–associated invariant chain peptide (CLIP), as described in rhesus macaque studies (Hansen et al., 2013a, 2016). We used CMV-specific CD4 T cell responses from three naturally infected subjects as positive controls for MHC II–restricted responses. Preincubating PBMCs with CLIP had no impact on the CD4 T cell–positive control responses or on any of the vaccine subjects’ responses (not depicted). For antibody blockade, we used the same antibodies that had blocked the monkey responses and also an antibody recently reported to block an MHC II–restricted HIV-specific human CD8 T cell response (Ranasinghe et al., 2016). As reported in the Ranasinghe study, we found that MHC II–restricted responses in humans could only be blocked by a 20-fold higher concentration of antibody than was used in the Rhesus study (Hansen et al., 2013a). Using 100 µg/ml of each of two anti–MHC II antibodies, we were able to block the majority of a CD4 T cell response in two of three HCMV-seropositive control subjects (Fig. 4 A). In the third subject, the same antibody combination blocked approximately one third of the CD4 T cell response. In contrast, anti–MHC II antibodies did not block any portion of the CD8 T cell responses to EK9 or peptides 79–81 in subject 28 or to the EM9 peptide in subject 36 (Fig. 4, B and C). We observed even more inconsistent blockade of CD8 T cell responses in naturally infected CMV-seropositive controls with the anti–MHC I antibody W6/32; thus, in the absence of appropriate positive controls, we could not directly test whether the three orphan CD8 T cell responses in vaccinated subjects were MHC I restricted. We assessed potential HLA-E restriction of these responses by peptide pulsing HLA-E–expressing single transfectants (Fig. 4 F) but found no evidence for HLA-E restriction (Fig. 4, D and E). These results cannot definitively exclude the possibility that these responses are restricted by either MHC II or HLA-E, particularly because antibody blockade is expected to be highly individual, depending on a match between the particular MHC alleles expressed and the specificity of the antibody. Nevertheless, these experiments provided no evidence to support unconventional restriction of these responses.
Figure 4.
Testing IE-1 peptide–specific CD8 T cells from Towne/Toledo HCMV-vaccinated subjects for MHC II and HLA-E restriction. (A–C) PBMCs were preincubated with 100 µg/ml each of G46-6 and Tu39 MHC II–blocking antibodies or 200 µg/ml of isotype control antibody for 1 h before the indicated peptides were added. (A) Inhibition of CD4 responses by MHC II–blocking antibodies in naturally infected HCMV-seropositive controls stimulated with overlapping peptide pools covering the indicated antigens. Plots are gated on live CD3+CD4+CD8− lymphocytes. (B and C) The peptides used were the 15mers or core epitopes shown in Table S1. Plots are gated on live CD3+CD4−CD8+ lymphocytes. (D and E) HLA-E single transfectants were pulsed with the peptides shown in Table S1, washed extensively, and then used to stimulate PBMCs from the indicated subjects. In the same experiment, RK9-pulsed HLA-A3 single transfectants were used as a positive control (subject 28). (F) MHC I expression on HLA-E and HLA-A3 single transfectants used in this experiment. Data show FACS plots from every subject analyzed and depict results of one experiment (A–C) or are representative of two independent experiments (D–F).
CD8 T cell responses to IE-1 peptides result from Towne/Toledo HCMV vaccination and were not preexisting
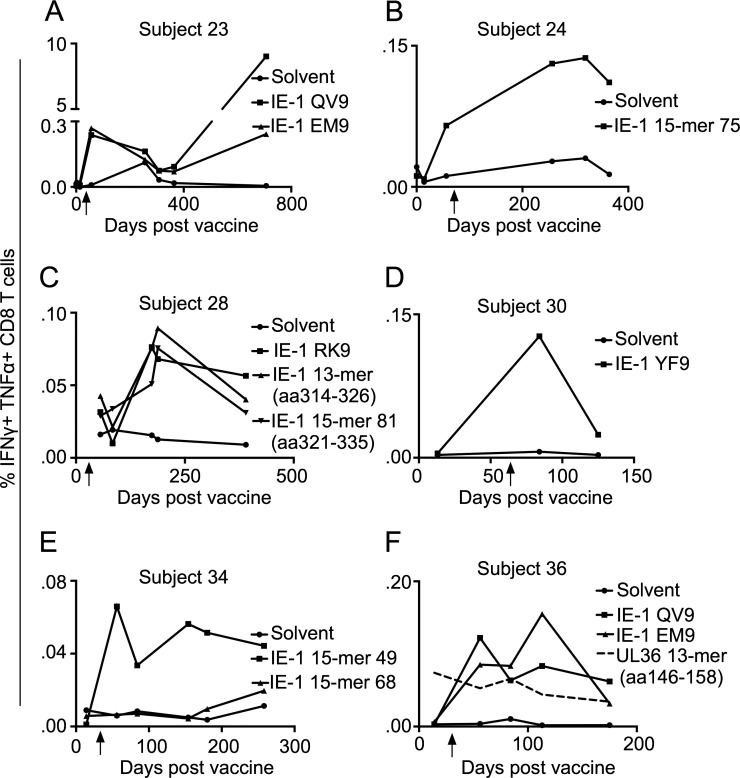
The CD8 T cell responses to the Towne/Toledo chimeras were very small, consistent with the extremely low vaccine dose. We considered the possibility that these small responses could represent preexisting cross-reactive responses, rather than de novo responses to Towne/Toledo HCMV vaccination. We assessed kinetics of the defined IE-1 peptide responses, and in all cases, these responses did not exist before HCMV seroconversion (Fig. 5). This contrasts with a preexisting response to a UL36 peptide in subject 36, which did not change after Towne/Toledo vaccination, and presumably represents a cross-reactive response (Fig. 5 F). Further, consistent with a specific vaccine-induced T cell response, subject 24 did not respond to the commercial Ad169 IE-1 peptide pool but did respond to a pool generated from the Toledo-derived IE-1 sequence present in the chimera 4 vaccine administered to this subject (Fig. 2 A). As previously reported (Adler et al., 2016), subject 23 was apparently exposed to wild-type CMV between 1 and 2 yr after vaccination. However, responses to both epitopes were clearly present after the vaccination and were then boosted by the infection. We conclude that these conventional IE-1–specific CD8 T cell responses were generated de novo in response to vaccination with the Towne/Toledo chimera vaccines.
Figure 5.
CD8 T cell responses to IE-1 epitopes were generated de novo upon vaccination with Towne/Toledo HCMV and were not preexisting. (A–F) PBMCs from the indicated time points were stimulated with the indicated peptides and assessed as in Fig. 2. Unless otherwise noted, the peptides used were the 15mers or core epitopes shown in Table S1. Arrows indicate the date of seroconversion. (A) Subject 23 acquired a natural CMV infection between days 365 and 707, as assessed by epithelial cell–neutralizing titers that rose from undetectable (<1:10) at day 365 to 1:6,600 at day 700. (C) Subject 28 was tested for responses to a peptide truncated from 15mer peptide 79 (aa 314–326: CYVLEETSVMLAK) because removing the N′ cysteine and C′ arginine gave more robust responses. No PBMCs were available from this subject before seroconversion, but responses to all three IE-1 epitopes continued to rise until ∼200 d after vaccination, which strongly suggests that these were elicited de novo by Towne/Toledo vaccination. This contrasts with a UL36 peptide response in subject 36 that was preexisting (dashed line; F), which did not change upon vaccination. Data show responses from every subject analyzed and depict results from one experiment.
Overall, our results show that CD8 T cell responses to vaccination with Towne/Toledo fibroblast-adapted HCMV vaccines are very similar in breadth and MHC restriction to those elicited by natural HCMV infection. We positively identified the classical MHC I restriction element for the majority of responses, and none of the remaining responses appeared to be MHC II or HLA-E restricted. Irrespective of whether attenuated fibroblast-adapted HCMV vaccines may elicit rare MHC II– or HLA-E–restricted CD8 T cells, it is clear that the majority of CD8 T cell responses are conventional, and several map to previously identified epitopes from natural CMV infection. This contrasts starkly with the rhesus vaccine, which elicited only MHC II– and MHC-E–restricted T cell responses.
Why do attenuated fibroblast-adapted CMV vaccines elicit such different CD8 T cell responses in macaques versus humans? It is possible that the rhesus immune system has a greater propensity to mount these unusually restricted CD8 T cell responses than does the human. Clearly, this is not the rule with all infections in macaques, as previously described CD8 T cell responses in macaques have been conventional. However, the rhesus MHC is much more polygenic than either the human or the mouse complex because of major duplication events in the MHC locus in old world monkeys since the most recent common ancestor with humans (Gibbs et al., 2007). Whereas individual humans and mice express up to six classical MHC I isoforms, an individual macaque can express as many as 22 classical MHC I isoforms, with Mamu-B being the most polygenic (Daza-Vamenta et al., 2004; Otting et al., 2005). Likewise, an individual macaque haplotype can include many versions of Mamu-DR. The number of MHC isoforms expressed by individual macaques nears the theoretical maximum above which negative selection is predicted to either excessively delete the TCR repertoire or fail to negatively select autoreactive TCRs (Woelfing et al., 2009). Thus, it is possible that the unusual conditions for negative selection in the rhesus thymus could result in enhanced selection of CD8 T cells with ability to recognize peptide presented on MHC II or with nonclassical MHC I. Nonetheless, rare instances of MHC II–reactive CD8 T cells have been reported in humans, including in HIV-infected individuals (Ranasinghe et al., 2016), and HLA-E–restricted responses are common in response to Mycobacterium tuberculosis (Heinzel et al., 2002).
A nonmutually exclusive possibility is that, despite similar viral tropism, genetic differences between RhCMV68-1 and the Town/Toledo chimera vaccines contribute to the disparate CD8 T cell responses elicited in macaques versus humans. Unconventional CD8 T cell responses were only elicited by RhCMV vectors that lack the pentameric complex (Hansen et al., 2013a), and parsimony argues that the altered viral tropism resulting from loss of the pentameric complex causes unconventional CD8 T cell responses in macaques. However, another possibility raised by Hansen et al. (2013a) is that one or more of the proteins encoded in UL128-131 has an additional function that affects generation of CD8 T cell responses. RhCMV68-1 lacks the rhesus orthologues of UL128 and UL130 (Malouli et al., 2012), whereas the Towne/Toledo chimeras used in our study have a defect only in UL128 (Suárez et al., 2017). Thus, despite similar cellular tropism (Fig. 1), genetic differences between RhCMV68-1 and the Towne/Toledo HCMV vaccines could be responsible for the disparate CD8 T cell responses elicited in macaques versus humans. These differences could involve the pentameric complex genes or other genes that differ between these viruses (GenBank accession nos. KX101021–KX101024). RhCMV and Towne/Toledo chimeras all encode intact immune evasion genes known to impact the MHC I pathway (US2, US3, US6, and US11). Furthermore, the UL40 homologue that impacts trafficking of MHC-E in CMV-infected cells is intact in RhCMV and in chimera 2 but disrupted in chimera 4 (GenBank accession nos. KX101021–KX101024; Suárez et al., 2017). However, with a genome of >200 kB containing possibly 200 open reading frames, there are many other genes whose functions may be important in shaping the CD8 T cell response in macaques and/or humans. Until we understand the mechanisms by which the RhCMV68-1 vector elicits these unusual CD8 T cell responses in macaques, it is difficult to speculate which genes might be important.
Because of CMVs’ strict species specificity and marked coevolution with their hosts’ immune systems, translating information gained from CMV vaccine studies in nonhuman primates to a human vaccine is a daunting task. Overall, our results show that vaccination with fibroblast-adapted Towne/Toledo HCMV did not elicit the unusual MHC II– and MHC-E–restricted responses predicted by RhCMV vaccine studies in macaques. If these unconventional responses are needed for protection against SIV/HIV, it is important to know how to elicit them in humans. If CMV cellular tropism is the primary determinant of unconventional responses in monkeys, eliciting them in humans could be difficult or impossible. However, if specific viral genes are the primary determinant, the genetic sequences of the Towne/Toledo chimeras may provide useful information in designing HCMV vaccines.
Materials and methods
Human subjects and study design
36 HCMV-seronegative male subjects received one of four live attenuated Towne/Toledo chimeric HCMV vaccines in a phase I dose escalation trial at Virginia Commonwealth University. Generation of vaccine strains, study design, study subject characteristics, and clinical assessment have been previously described (Heineman et al., 2006; Adler et al., 2016). This study was performed in accordance with the principles of the Helsinki Declaration. The Institutional Review Board of Virginia Commonwealth University approved the study, and informed written consent was obtained from all subjects. Naturally infected HCMV-seropositive healthy control subjects were recruited at Oregon Health & Science University. The Institutional Review Board of Oregon Health & Science University approved the study, and informed written consent was obtained from all subjects.
Viruses and infectivity assay
Generation of the vaccine strains has been previously described (Heineman et al., 2006). The HCMV clinical isolate TR-BAC–derived virus and the TRΔ4 virus lacking the UL-b′ region have been described previously (Ryckman et al., 2006). All virus stocks were made by infecting monolayers of normal human dermal fibroblasts (NHDFs) at an MOI of 0.1 PFU/cell. At 7–10 d after infection, infected cell supernatants were harvested, clarified by centrifugation at 5,000 g for 5 min, and stored at −80°C. The titers of virus stocks were determined by plaque assay on NHDFs. Propagation of NHDFs, human retinal pigmented epithelial cells (ARPE-19), and HUVECs have been described previously (Vanarsdall et al., 2008). THP-1 (monocytic) cells were from ATCC and were grown as recommended (TIB-202). To assess viral infectivity, monolayers plated in 24-well dishes were incubated with virus stocks at MOIs of 3 for NHDFs, 10 for ARPE-19 cells and HUVECs, and 100 for THP-1 cells. Cells were incubated for 24 h at 37°C, fixed with 4% formaldehyde, permeabilized, and assayed by immunofluorescent microscopy to detect the presence of the HCMV IE-1 antigen as described previously (Vanarsdall et al., 2012). Virus entry was quantified by imaging at least three random fields and comparing the number of IE-1–positive cells to the total number of DAPI-stained cells.
Antibodies, peptides, tetramers, and cytokines
The following antibodies were used for flow cytometry analysis: CD3 (SK7; BD), CD4 (SK3; eBioscience), CD8 (RPA-T8; BD), CD69 (FN50; BioLegend), TNF (MAb11; eBioscience), IFN-γ (4S.B3; eBioscience), and MHC I (W6/32; Bio X Cell). The following functional grade antibodies were used to stimulate T cells in culture: CD28 (L293; BD), CD49d (L25; BD), and CD3 (SK7; eBioscience). The pan–anti–MHC II–blocking antibody Tu39 and isotype control IgG2a κ were from BioLegend. The anti–HLA-DR–blocking antibody G46-6 was from BD. The 15mer-overlapping peptide pool covering Ad169 IE-1 and custom-made 15mer-overlapping peptide pools covering the Towne and Toledo IE-1 sequences were from JPT Peptide Technologies. Individual peptides used for truncation studies and for pulsing HLA transfectants were synthesized by JPT or Genemed Synthesis Inc. HLA-B08 tetramers folded with the EM9 and QV9 peptides were from the National Institutes of Health tetramer core facility. Recombinant human IL-2, IL-4, and GM-CSF were from Immunex.
PBMC isolation
PBMCs were isolated via leukapheresis or from peripheral blood via Ficoll-Hypaque density gradient centrifugation. PBMCs were frozen in FBS/10%DMSO and stored and shipped in liquid nitrogen. PBMCs for all analyses except the kinetic data displayed in Fig. 5 were obtained at the following time points after vaccination: subject 23, days 56 (Figs. 2 A and 3) and 707 (Fig. 2 B only); subject 24, days 256 and 319; subject 28, day 188; subject 30, day 125; subject 34, day 154; and subject 36, day 113.
Media
Cells were cultured in RPMI 1640 medium (Thermo Fisher Scientific) supplemented with 2 mM l-glutamine (Thermo Fisher Scientific), 100 U/ml penicillin (Invitrogen), and 100 µg/ml streptomycin (Invitrogen). RPMI medium was supplemented with 10% FBS (RPMI-10) for intracellular cytokine staining (ICS) and 10% or 2% human AB serum (RPMI-hu10 and RPMI-hu2, respectively) for T cell line generation.
ICS and flow cytometric analysis
Frozen PBMCs were rapidly thawed at 37°C, washed twice with warm RPMI-10 medium, plated at 0.5–1 × 105 cells per well in a 96-well plate, and rested overnight at 37°C and 5% CO2 before peptide stimulation. Freshly isolated PBMCs were not rested before stimulation. PBMCs were stimulated for 8 h with 2 µg/ml anti-CD28/CD49d and 5 µg/ml of overlapping peptide pools, 20 µg/ml of single peptides, or peptide solvent to assess background. 3 µg/ml brefeldin A (eBioscience) was added at the time of stimulation or, when CD69 was to be assessed, after 2 h. PBMCs were stained with Live/Dead-Fixable Aqua (Thermo Fisher Scientific), followed by surface marker staining. Cells were fixed and permeabilized with a Cytofix/Cytoperm kit (BD) and stained for intracellular cytokines. Cells were run on an LSRII or LSRFortessa flow cytometer (BD) and analyzed in FlowJo (v. 9; Tree Star). CD8 T cells were gated as CD3+CD4−CD8+ live lymphocytes. Positive CD8 T cell responses were defined as the IFNγ+TNF+ proportion that was >0.03% and more than three standard deviations above background.
In some experiments, PBMCs were stimulated with peptide-pulsed single HLA-transfected K562 cells. K562 cells were cultured in RPMI-10 medium overnight at 27°C to increase MHC expression and then pulsed with 10 µg/ml peptide for 1.5 h at 37 or 27°C and washed three times with PBS and once with RPMI-10 medium. Then, PBMCs were stimulated with anti-CD28/CD49d and peptide-pulsed K562 at a 1:10 ratio and assessed for cytokine production by ICS, as described in the previous paragraph.
For MHC II–blocking experiments, 100 µg/ml each of Tu39 and G46-6 anti–HLA-DR or 200 µg/ml of isotype control (IgG2a κ) were preincubated with PBMCs for 1 h before addition of peptide. For tetramer staining, PBMCs were used immediately upon thawing. Cells were costained with allophycocyanin (APC) and FITC-labeled HLA-B08:01 tetramers concurrently with all other surface markers. CD8 T cells were gated as CD3+CD4−CD8+ live lymphocytes and were considered tetramer binding if positive for either APC or FITC.
HLA transfectants
Stably transduced HLA-E–expressing K562 cells were provided by Jonah Sacha (Oregon Health & Science University Vaccine and Gene Therapy Institute, Beaverton, OR; Hansen et al., 2016). HLA-A1:01, A2:01, A11:01, B8:01, and B44:03 were transduced into K562 cells. HLA-A3:01, A24:02, A26:01, A33:03, B14:02, B44:02, Cw01:02, and Cw03:03 were transfected into K562 cells and grown in selection media containing 250 µg/ml G418 (Sigma-Aldrich). MHC expression was assessed regularly by flow cytometry with W6/32 pan–anti–MHC I antibody.
Generation of T cell line
An IE-1–specific CD8 T cell line was generated from subject 34. PBMCs were magnetically enriched for CD8 T cells by EasySep negative selection (STEMCELL Technologies). T cells were cultured with 5 ng/ml IL-2 in RPMI-h10 medium and stimulated at a 10:1 ratio with autologous monocyte-derived DCs pulsed overnight with 2.5 µg/ml of the HCMV IE-1–overlapping peptide pool. On day 8, T cells were labeled with CFSE (Invitrogen) and restimulated at a 1:10 ratio with 2.5 µg/ml autologous peptide-pulsed monocyte-derived macrophages plus 5 ng/ml IL-2. 7 d later, CFSE-low cells were FACS sorted and rapidly expanded after overnight rest in 0.5 ng/ml IL-2. T cells were expanded with 30 ng/ml soluble anti-CD3 and a 10–20-fold excess of irradiated feeder cells. Feeder cells consisted of a 5:1 ratio of autologous PBMCs (30 Gy) to allogeneic lymphoblastoid cell lines (60 Gy). On day 5, cells were washed and given fresh RPMI-h10 medium with 2 ng/ml IL-2. IL-2 was replenished every 2–3 d thereafter. The T cell line was assessed functionally beginning on 11 d after CFSE-low sort, and some cells were frozen for subsequent analysis. Upon thawing, the T cell line was rapidly expanded before functional analysis.
Monocyte-derived DCs and macrophages for generation of T cell line
The protocol for differentiating autologous monocyte-derived DCs was adapted from Romani et al. (1994). In brief, PBMCs were incubated in RPMI-h2 with 60 µg/ml DNase for 1 h. Media was aspirated, and attached monocytes were washed two times with PBS. Monocytes were differentiated to DCs in RPMI-hu10 supplemented with 10 ng/ml GM-CSF and 10 ng/ml IL-4 for 8 d. Autologous macrophages were differentiated from magnetically isolated CD14+ PBMCs (autoMacs Pro Separator; Miltenyi Biotec) by plating at 106/ml on tissue culture–treated plates and culturing 5 d in RPMI-h10 medium.
HLA typing
HLA typing was performed on DNA isolated from PBMCs by ProImmune.
Online supplemental material
Table S1 shows a summary of vaccine chimeras and doses administered, epitopes and HLA restriction of T cell responses, and HLA typing for each subject.
Supplementary Material
Acknowledgments
We thank Jonah Sacha for HLA-E transfectants, Jo Cox for numerous insightful discussions and critical review of the manuscript, Erin Meermeier for technical help generating T cell lines, and Mark Slifka and Chris Snyder for critical review of the manuscript.
This work was supported by the National Institutes of Health (grants R21AI116107-01 and R01AI047206 to A.B. Hill) and M.J. Murdock Charitable Trust (grant 2013269 to S.E. Murray). The clinical work was partially funded by IAVI with the generous support of USAID and other donors. A full list of IAVI donors is available at www.iavi.org. The contents of this manuscript are the responsibility of the authors and do not necessarily reflect the views of the USAID or the US Government.
The authors declare no competing financial interests.
Author contributions: S.E. Murray designed and performed experiments, analyzed data, and wrote the manuscript. P.A. Nesterenko performed experiments, analyzed data, and prepared figures. A.L. Vanarsdall performed experiments and prepared figures. M.W. Munks provided critical insight. S.M. Smart, E.M. Veziroglu, and L.C. Sagario performed experiments. R. Lee processed vaccine trial samples. F.H.J. Claas and I.I.N. Doxiadis generated HLA transfectants. M.A. McVoy and S.P. Adler designed the vaccine trial and provided critical insight. A.B. Hill conceived of the study, designed experiments, and wrote the manuscript.
Footnotes
Abbreviations used:
- APC
- allophycocyanin
- HCMV
- human CMV
- ICS
- intracellular cytokine staining
- MOI
- multiplicity of infection
- NHDF
- normal human dermal fibroblasts
- RhCMV
- rhesus CMV
- SIV
- simian immunodeficiency virus
References
- Adler S.P., Manganello A.-M., Lee R., McVoy M.A., Nixon D.E., Plotkin S., Mocarski E., Cox J.H., Fast P.E., Nesterenko P.A., et al. 2016. A phase 1 study of 4 live, recombinant human cytomegalovirus Towne/Toledo chimera vaccines in cytomegalovirus-seronegative men. J. Infect. Dis. 214:1341–1348. 10.1093/infdis/jiw365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui X., Meza B.P., Adler S.P., and McVoy M.A.. 2008. Cytomegalovirus vaccines fail to induce epithelial entry neutralizing antibodies comparable to natural infection. Vaccine. 26:5760–5766. 10.1016/j.vaccine.2008.07.092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daza-Vamenta R., Glusman G., Rowen L., Guthrie B., and Geraghty D.E.. 2004. Genetic divergence of the rhesus macaque major histocompatibility complex. Genome Res. 14:1501–1515. 10.1101/gr.2134504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elkington R., Walker S., Crough T., Menzies M., Tellam J., Bharadwaj M., and Khanna R.. 2003. Ex vivo profiling of CD8+-T-cell responses to human cytomegalovirus reveals broad and multispecific reactivities in healthy virus carriers. J. Virol. 77:5226–5240. 10.1128/JVI.77.9.5226-5240.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs R.A., Rogers J., Katze M.G., Bumgarner R., Weinstock G.M., Mardis E.R., Remington K.A., Strausberg R.L., Venter J.C., Wilson R.K., et al. Rhesus Macaque Genome Sequencing and Analysis Consortium . 2007. Evolutionary and biomedical insights from the rhesus macaque genome. Science. 316:222–234. 10.1126/science.1139247 [DOI] [PubMed] [Google Scholar]
- Hansen S.G., Ford J.C., Lewis M.S., Ventura A.B., Hughes C.M., Coyne-Johnson L., Whizin N., Oswald K., Shoemaker R., Swanson T., et al. 2011. Profound early control of highly pathogenic SIV by an effector memory T-cell vaccine. Nature. 473:523–527. 10.1038/nature10003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen S.G., Piatak M. Jr., Ventura A.B., Hughes C.M., Gilbride R.M., Ford J.C., Oswald K., Shoemaker R., Li Y., Lewis M.S., et al. 2013b Immune clearance of highly pathogenic SIV infection. Nature. 502:100–104. 10.1038/nature12519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen S.G., Sacha J.B., Hughes C.M., Ford J.C., Burwitz B.J., Scholz I., Gilbride R.M., Lewis M.S., Gilliam A.N., Ventura A.B., et al. 2013a Cytomegalovirus vectors violate CD8+ T cell epitope recognition paradigms. Science. 340:1237874 10.1126/science.1237874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen S.G., Wu H.L., Burwitz B.J., Hughes C.M., Hammond K.B., Ventura A.B., Reed J.S., Gilbride R.M., Ainslie E., Morrow D.W., et al. 2016. Broadly targeted CD8+ T cell responses restricted by major histocompatibility complex E. Science. 351:714–720. 10.1126/science.aac9475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heineman T.C., Schleiss M., Bernstein D.I., Spaete R.R., Yan L., Duke G., Prichard M., Wang Z., Yan Q., Sharp M.A., et al. 2006. A phase 1 study of 4 live, recombinant human cytomegalovirus Towne/Toledo chimeric vaccines. J. Infect. Dis. 193:1350–1360. 10.1086/503365 [DOI] [PubMed] [Google Scholar]
- Heinzel A.S., Grotzke J.E., Lines R.A., Lewinsohn D.A., McNabb A.L., Streblow D.N., Braud V.M., Grieser H.J., Belisle J.T., and Lewinsohn D.M.. 2002. HLA-E–dependent presentation of Mtb-derived antigen to human CD8+ T cells. J. Exp. Med. 196:1473–1481. 10.1084/jem.20020609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kern F. 1999. Identification of CD8-T-cell epitopes in the non-structural HCMV major immediate early protein. J. Clin. Virol. 12:149 10.1016/S1386-6532(99)90500-0 [DOI] [Google Scholar]
- Malouli D., Nakayasu E.S., Viswanathan K., Camp D.G. II, Chang W.L.W., Barry P.A., Smith R.D., and Früh K.. 2012. Reevaluation of the coding potential and proteomic analysis of the BAC-derived rhesus cytomegalovirus strain 68-1. J. Virol. 86:8959–8973. 10.1128/JVI.01132-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otting N., Heijmans C.M.C., Noort R.C., de Groot N.G., Doxiadis G.G.M., van Rood J.J., Watkins D.I., and Bontrop R.E.. 2005. Unparalleled complexity of the MHC class I region in rhesus macaques. Proc. Natl. Acad. Sci. USA. 102:1626–1631. 10.1073/pnas.0409084102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranasinghe S., Lamothe P.A., Soghoian D.Z., Kazer S.W., Cole M.B., Shalek A.K., Yosef N., Jones R.B., Donaghey F., Nwonu C., et al. 2016. Antiviral CD8+ T cells restricted by human leukocyte antigen class II exist during natural HIV infection and exhibit clonal expansion. Immunity. 45:917–930. 10.1016/j.immuni.2016.09.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romani N., Gruner S., Brang D., Kämpgen E., Lenz A., Trockenbacher B., Konwalinka G., Fritsch P.O., Steinman R.M., and Schuler G.. 1994. Proliferating dendritic cell progenitors in human blood. J. Exp. Med. 180:83–93. 10.1084/jem.180.1.83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryckman B.J., Jarvis M.A., Drummond D.D., Nelson J.A., and Johnson D.C.. 2006. Human cytomegalovirus entry into epithelial and endothelial cells depends on genes UL128 to UL150 and occurs by endocytosis and low-pH fusion. J. Virol. 80:710–722. 10.1128/JVI.80.2.710-722.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suárez N.M., Lau B., Kemble G.M., Lee R., Mocarski E.S., Wilkinson G.W., Adler S.P., McVoy M.A., and Davison A.J.. 2017. Genomic analysis of chimeric human cytomegalovirus vaccine candidates derived from strains Towne and Toledo. Virus Genes. 10.1007/s11262-017-1452-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanarsdall A.L., Ryckman B.J., Chase M.C., and Johnson D.C.. 2008. Human cytomegalovirus glycoproteins gB and gH/gL mediate epithelial cell-cell fusion when expressed either in cis or in trans. J. Virol. 82:11837–11850. 10.1128/JVI.01623-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanarsdall A.L., Wisner T.W., Lei H., Kazlauskas A., and Johnson D.C.. 2012. PDGF receptor-α does not promote HCMV entry into epithelial and endothelial cells but increased quantities stimulate entry by an abnormal pathway. PLoS Pathog. 8:e1002905 10.1371/journal.ppat.1002905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wills M.R., Okecha G., Weekes M.P., Gandhi M.K., Sissons P.J.G., and Carmichael A.J.. 2002. Identification of naive or antigen-experienced human CD8+ T cells by expression of costimulation and chemokine receptors: analysis of the human cytomegalovirus-specific CD8+ T cell response. J. Immunol. 168:5455–5464. 10.4049/jimmunol.168.11.5455 [DOI] [PubMed] [Google Scholar]
- Woelfing B., Traulsen A., Milinski M., and Boehm T.. 2009. Does intra-individual major histocompatibility complex diversity keep a golden mean? Philos. Trans. R. Soc. Lond. B Biol. Sci. 364:117–128. 10.1098/rstb.2008.0174 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.