Significance
When two populations interact, when does it pay to evolve rapidly, and can it ever be an advantage to evolve slowly? We address these questions using evolutionary game theory. In antagonistic interactions (e.g., host–parasite), we find that faster evolution by any means is beneficial—the “Red Queen” effect. In certain mutualisms, slower evolution is favored in the long run. This “Red King” effect is driven by differences in how efficiently natural selection acts in the two populations, rather than by differences in their generation times or mutation rates. Our results clarify the role of evolutionary rate in symbiont evolution.
Keywords: rate of evolution, symbiosis, mutualism, antagonism, Müllerian mimicry
Abstract
In antagonistic symbioses, such as host–parasite interactions, one population’s success is the other’s loss. In mutualistic symbioses, such as division of labor, both parties can gain, but they might have different preferences over the possible mutualistic arrangements. The rates of evolution of the two populations in a symbiosis are important determinants of which population will be more successful: Faster evolution is thought to be favored in antagonistic symbioses (the “Red Queen effect”), but disfavored in certain mutualistic symbioses (the “Red King effect”). However, it remains unclear which biological parameters drive these effects. Here, we analyze the effects of the various determinants of evolutionary rate: generation time, mutation rate, population size, and the intensity of natural selection. Our main results hold for the case where mutation is infrequent. Slower evolution causes a long-term advantage in an important class of mutualistic interactions. Surprisingly, less intense selection is the strongest driver of this Red King effect, whereas relative mutation rates and generation times have little effect. In antagonistic interactions, faster evolution by any means is beneficial. Our results provide insight into the demographic evolution of symbionts.
Antagonistic symbioses can be conceptualized well in the following simple, constant-sum game (1, 2):
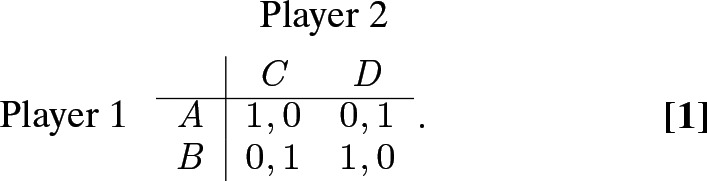 |
Here, player 1’s available strategies are and , player 2’s are and , the first payoff in each cell is player 1’s, and the second one is player 2’s. Interacting populations of “player 1s” and “player 2s” constitute an antagonistic symbiosis, which we expect to evolve according to arms-race dynamics (3). To see why, suppose we start with population 1 all playing strategy and population 2 all playing strategy , so that population 1 is doing well at the expense of population 2. Now, a mutant in population 2 who plays strategy does better (payoff ) than other members of population 2 (payoff ) and can take over, at population 1’s expense (their payoff decreases from to ). Population 1 is then expected to switch to strategy , after which population 2 switches to strategy , and so on. As the Red Queen said to Alice, “it takes all the running you can do, to keep in the same place” (ref. 4, p. 42).
Arms-race dynamics characterize many interactions both in the natural world and in human behavior. In a host–parasite interaction, for example, the host is selected to develop immunity to the parasite, which in turn selects for new “resistance” mutations in the parasite, which selects for the host to develop immunity to the new mutant parasite, and so on. In Batesian mimicry, a palatable species (of butterfly, for example) evolves to mimic the warning display of an unpalatable species, so that it is mistaken by predators for the unpalatable species. This selects for the unpalatable species to evolve a new display, to not be mistaken for the palatable species, which is then under selection to mimic the new display, and so on (5).
Antagonistic interactions play out at an intragenomic level as well. Examples, among many others (6, 7), are the X-linked male-meiotic driving gene Dox and its autosomal suppressor Nmy in Drosophila simulans (8, 9), centromeric repeat sequences seeking to drive in female meiosis and the centromere-histone genes that keep them in check (10, 11), and the mammalian recombination-specifying gene Prdm9 and the binding sequences of its protein [which are under positive selection to escape PRDM9 binding (12–15)]. Consistent with arms-race dynamics, the conflicting elements in these examples all show genetic evidence of rapid evolution (10, 11, 16–23).
Mutualistic symbioses with a degree of conflict (24, 25) can also be conceptualized in a simple game (26),
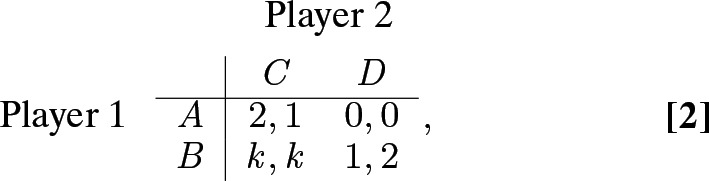 |
with . From either of the two mutualistic coordination states and , neither population is under selection to deviate to one of the noncoordination states and . But population 1 prefers coordination state , whereas population 2 prefers coordination state . Of most interest is the case , where game 2 is a true mutualism (both populations prefer the two coordination states to the two noncoordination states).
This game also describes many interactions in both nature and humans. Mutualisms often involve a division of labor (27), such as the production of different nutrients in a microalgal–microbial partnership (28), the separate production of a “poison” and its “antidote” by two linked genetic elements in a gamete-killing meiotic drive complex (6, 29), or the economic production of different goods by trading partners under increasing returns to scale (30). In these cases, it is beneficial to both interactants that all tasks be carried out, but there can be conflict over who should do which (31). For example, it is better to be the antidote producer than the poison producer in the meiotic drive complex, because the gains of their partnership are shared equally, but its disruption (by recombination) is costlier to the poison producer.
Another example, Müllerian mimicry, involves two poisonous species (of butterfly, say) evolving to share a common pattern, one pattern being easier for their predators to learn than two (32). But if the species’ original patterns evolved to suit differences in their respective habitats, behaviors, or genomic backgrounds, then there might be conflict over which pattern to converge on, with each species enjoying an evolutionary advantage if convergence is on its original pattern.
In both antagonistic and mutualistic symbioses, the relative rates of evolution of the participating populations are thought to be important determinants of their relative evolutionary success. A faster rate of evolution could be achieved through various means (3, 24–26). First, it could derive from a shorter generation time, allowing more generations over which to adapt (2). Second, it could derive from a higher rate of mutation, more rapidly generating new variants that allow a population to escape an unfavorable state of interaction (24, 33, 34). Finally, it could derive from more effective action of natural selection. This could be because the stakes are higher for one population than the other—“the rabbit is running for his life, while the fox is only running for his dinner” (ref. 3, p. 493)—or because selection acts more effectively in large populations, owing (roughly speaking) to a reduced effect of random drift (35) (a precise formulation of this statement is given in SI Appendix, section S5.2).
Common wisdom holds that a faster rate of evolution is advantageous in antagonistic interactions (36), the better to “keep ahead in the race” (ref. 3, p. 492). But it has not, to our knowledge, been clearly demonstrated which individual biological parameters drive this Red Queen effect in theory.
In an important paper, Bergstrom and Lachmann (26) (hereafter, B&L) demonstrated that a slower rate of evolution might be advantageous in some mutualistic interactions, an effect they called the Red King effect. They studied two populations interacting according to the mutualism game 2, with deterministic evolutionary dynamics operating in each of the two populations, both of infinite size and without the possibility of mutations.
In their model, is the proportion of population 1 playing strategy (so that the proportion play ), and is the proportion of population 2 playing ( play ). Then the expected payoff to a member of population 1 who plays when interacting with a random member of population 2 is, from the payoff matrix 2, . On the other hand, the payoff to a member playing is , and the average payoff in population 1 is . The analogous quantities in population 2, , , and , are calculated similarly. The strategy frequencies in the two populations evolve according to replicator dynamics:
| [3] |
Here are parameters that calibrate the relative rates of evolution of the two populations by determining how responsive their respective evolutionary dynamics are to fitness differences among strategies.
These dynamics are deterministic—evolutionary trajectories are fully determined once their starting points are known—and evolution necessarily leads to one of the two coordination equilibria, (; population 1’s preference) or (; population 2’s preference), where evolution then halts.
B&L (26) showed that, when in game 2 and population 1 evolves slower than population 2 (), the set of starting points from which evolution proceeds to population 1’s favored equilibrium (the equilibrium’s “basin of attraction”) is larger than the basin of attraction of the equilibrium . In this sense, slower evolution is beneficial. When , the opposite result holds: If population 1 evolves slower, the basin of attraction of is smaller than that of .
Here, we construct a finite-population model of symbiosis evolution, incorporating all of the biological determinants of evolutionary rate. This model allows us to extend B&L’s (26) results for mutualisms in several ways and to apply a similar analysis to antagonistic symbioses, leading to a richer picture of the evolutionary dynamics of symbioses.
First, our model allows for an explicit characterization of which evolutionary rate parameters influence the relative success of the interacting populations. This is unknown for mutualisms [B&L’s (26) general rate parameters and have no clear biological interpretation], has not been fully disentangled for antagonistic symbioses, and in both cases is crucial for standard empirical measures of evolutionary rate [such as the substitution rate at neutral genetic loci, which depends on mutation rate and generation time, but is insensitive to changes in population size (37)].
Second, our model allows us to uncover a key influence of evolutionary timescale on symbiosis evolution. In the mutualism game 2, we show that the short-run behavior of our stochastic evolutionary dynamics is very similar to that of the replicator dynamics studied by B&L (26): From a given starting state, the dynamics rapidly converge to, or near to, one of the two equilibria of the game. Which equilibrium is most likely to be approached depends critically on the starting point, just as in the dynamics of B&L (26). On a longer timescale, however, we show the evolutionary dynamics of this game to be of a very different nature. The long-run dynamics involve transitions between equilibria, driven by sporadic mutation, and eventually become independent of where the dynamics first started. As we show, in mutualisms, many of the conclusions concerning the short-run dynamics are either annulled or reversed in the long run.
Our main results are summarized in Table 1.
Table 1.
The biological parameters that drive the Red Queen and King effects in antagonistic and mutualistic symbioses
| Antagonistic symbiosis | Mutualism, small | Mutualism, large | ||||
| Short run | Long run | Short run | Long run | Short run | Long run | |
| /generation time | ♕Q | ♕Q | ♕Q | No effect | ♔K | No effect |
| Mutation rate | No effect | ♕Q | No effect | No effect | No effect | No effect |
| Selection strength | ♕Q | ♕Q | ♕Q | ♔K | ♔K | ♕Q |
| Population size | ♕Q | ♕Q | ♕Q | ♔K | ♔K | ♕Q |
For each of the parameters that determine evolutionary rate, we ask whether the population with the larger parameter value, which therefore evolves faster, is more successful (♕Q, a Red Queen effect) or less successful (♔K, a Red King effect) in the interaction, holding the other rate parameters constant and equal between the two populations. The short-run results are numerically computed for particular parameter values (Figs. 1–3). The long-run results are for the weak-mutation limit and are exact (see text for details). For selection strength and population size in the mutualisms, (i) we set one population’s selection strength to w (or both populations’, when studying the effect of population size) and assume that the larger parameter value is larger by a small amount, (ii) we define “k small” as k < 1/(1 + w) and “k large“ as k > 1/(1 + w), and (iii) we assume the populations to be sufficiently large
A Finite-Population Model of Symbiosis Evolution
Populations 1 and 2 are of sizes and and interact according to a two-player, two-strategy game such as games 1 and 2. The “populations” here can be broadly construed: They could be all of the individuals of two species in a symbiosis, for example, or all of the alleles at two distinct loci among the genomes of a single species.
The evolutionary process occurs in discrete time steps. Each time step, individuals in each population receive their average payoffs from interacting with a random member of the opposite population. An individual’s payoff in population is translated to a nonnegative fitness value , so that the “selection strengths” calibrate the effectiveness of natural selection in the two populations.
In each time step, a “birth–death event” occurs in one of the populations. Individuals in the two populations have relative generation times and : In a given time step, the birth–death event occurs in population with probability proportional to , independently across time steps.
A birth–death event in a population involves choosing an individual to reproduce, with probability proportional to fitness, and an individual to die, with each one equally likely. These can be the same individual. A single offspring of the reproducing individual replaces the individual that was chosen to die. This within-population process, the Moran process (38, 39), has been used as a model both of biological evolution (40, 41) and of imitation learning (42–44). In the Moran process, one “generation” of a population typically corresponds to the number of birth–death events that is about the same as the population’s size (41). In our two-population framework, we label the number of time steps equal to the sum of the populations’ sizes as a common generation: Each individual experiences on average one birth–death event per generation.
Finally, to account for the possibility of mutation, we assume that, in a birth–death event in population , the offspring inherits its parent’s strategy with probability or mutates to the alternative strategy with probability . Thus, and represent the relative mutation rates of the two populations (they are unitless), whereas the parameter calibrates the overall frequency of mutations in the two populations (in units “per replication”).
In sum, given a payoff matrix such as [1] and [2] and rate parameters , , , , , , , , and and provided , the above defines an ergodic Markov chain over a state space comprising all possible strategy compositions of the two populations. If is the number of strategists in population 1 at some point in time (so that individuals play ), and is the number of strategists in population 2 ( play ), then the population state is . The behavior of the evolutionary process is characterized by the probability of moving from state to in one time step, for all such pairs of states—these probabilities are provided in SI Appendix, section S1.
Two regimes are of interest in these dynamics, corresponding roughly to their short-run and long-run behavior.
Given some initial population state , the short-run dynamics (i) are not affected much by mutations, instead being governed mostly by selection strengths, population sizes, and generation times; (ii) depend critically on the starting point ; (iii) have trajectories that are similar to those of the replicator dynamics, especially when the populations are large; and (iv) in coordination games, like game 2, converge rapidly to or near one of the pure population states (in which each population is monomorphic) associated with the equilibria of the game. For illustration, Figs. 2 A–D, Upper and 3 A–D, Upper and especially SI Appendix, Fig. S1 display the similarity of the short-run behavior of our dynamics to that of the replicator dynamics, in the context of the mutualism game 2 [compare figure 2 of B&L (26)].
Fig. 2.
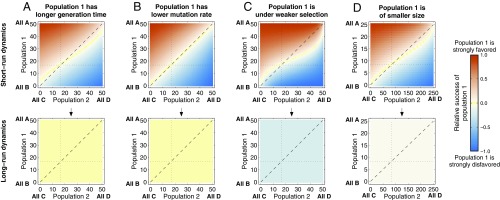
Evolutionary dynamics of mutualisms when is small, and population 1 evolves slower than population 2 owing to (A) a longer generation time, (B) a lower mutation rate, (C) weaker selection, and (D) a smaller population size. Population parameters are the same as in Fig. 1, and . A short-run Red Queen effect is observed for generation time, selection strength (especially), and population size. Relative differences in mutation rate again have no discernible short-run effect. In the long run, differences in mutation rate and generation time again have no effect, whereas a Red King effect is found for selection strength and population size.
Fig. 3.
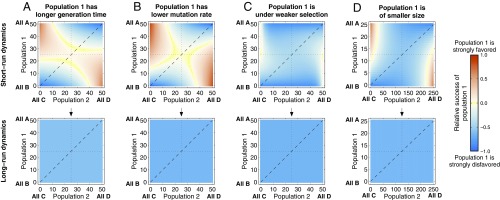
Evolutionary dynamics of mutualisms when is large and population 1 evolves slower than population 2 owing to (A) a longer generation time, (B) a lower mutation rate, (C) weaker selection, and (D) a smaller population size. Population parameters are the same as in Fig. 2, but now . A short-run Red King effect is observed for generation time, selection strength, and population size, with that for selection strength most pronounced. These short-run Red King effects are analogous to those found by B&L (26). Relative differences in mutation rate have no discernible short-run effect. In the long run, differences in mutation rate and generation time have no effect on the relative success of the populations, whereas a Red Queen effect is found for selection strength and population size.
The long-run dynamics are of a very different nature. Because the evolutionary process, as we have defined it, is ergodic, the probability that the system is in some state in the future eventually becomes independent of where the system started (45). This effect is illustrated for the antagonistic symbiosis game in Fig. 1 A–D, Lower and for the mutualism game in Figs. 2 A–D, Lower and 3 A–D, Lower. The state of the system comes to depend not on the early dynamics that emanate from the starting point, but instead on infrequent transitions between equilibria, driven by mutations.
Fig. 1.
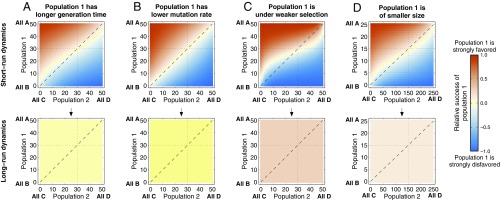
Evolutionary dynamics of antagonistic symbioses, when population 1 evolves slower than population 2 owing to (A) a longer generation time, (B) a lower mutation rate, (C) weaker selection, and (D) a smaller population size. Each panel shows the numerically computed dynamics, assuming that both populations coincide in their evolutionary rate parameters except for the parameter explicitly varied. A–D, Upper show, for each initial population state, population 1’s relative success after generations. Except when the populations differ only in their mutation rates, a short-run Red Queen effect operates in all cases: As the blue area covers more than 50% of the square, population 1’s slower evolution leads to its typically being disfavored after 50 generations for a larger fraction of initial population states. The short-run Red Queen effect is strongest for selection strength and population size. A–D, Lower show population 1’s relative success after generations, by which time the starting configuration no longer influences the dynamics: The panels have a uniform color. A strong long-run Red Queen effect is observed in all cases: Population 1’s slower evolution causes a larger fraction of long-run time to be spent in states unfavorable to it. Baseline parameters are , , , , and . (A) ; (B) ; (C) ; (D) , . A generation is elementary time steps of the Moran process.
The object of interest in these long-run dynamics is their stationary distribution, the proportion of time spent in each population state in the long run. Equivalently, the stationary distribution tells us, were we to observe many independent instances of equivalent symbioses evolving, what proportion of these symbioses we should expect to find in each possible state at some fixed point in time in the long run (45).
Whereas we numerically study the stationary distribution of our evolutionary process for large mutation rates and selection strengths, we also invoke recent methodological advances in evolutionary game theory (46, 47) to study it analytically in certain limits. Chief among these is the “weak-mutation limit,” , which approximates the case where mutations are very infrequent: (44, 46). This is a common assumption in the population genetics and evolutionary game theory literatures (44, 48, 49) and is realistic when populations are small or when the individual mutation rate is very small. For example, in genetical evolution, the mutations we are considering might be single-nucleotide substitutions, which occur at rates of order or smaller per generation for most organisms (50); if multiple nucleotide substitutions are required to change strategies, then the relevant mutation rates are even lower. Therefore, whereas the case of infrequent mutations is certainly not general, it is a relevant and interesting case to consider.
In the weak-mutation limit, the dynamics converge to an embedded dynamical process over just the four pure states (42, 46); the stationary distribution collapses to a probability distribution over these pure states, . In the embedded dynamics, transitions between pure states occur with probabilities determined by the relative frequency of appearance of mutants in the two populations and the probabilities of fixation of these mutants (46) (details in SI Appendix, section S4).
In this paper, we examine the influence of individual rate parameters—mutation rate, generation time, selection strength, and population size—on the outcomes of antagonistic and mutualistic symbioses. For the majority of our analysis, we take the following “all-else equal” approach: For each rate parameter, we hold equal and constant for the two populations all but that parameter and then ask whether the population with the larger value of that parameter is, by some relevant criterion, evolutionarily more successful.
This all-else equal approach is motivated by two considerations: First, it allows for simple mathematical characterization of the long-run dynamics in the weak-mutation limit. Second, it allows us to ask in a clear way, “What is the contribution of a given parameter to the success of the populations?” For illustration, suppose we see that a large population of parasites does well against a small host population. We might wonder whether it is the parasite’s large population size or its rapid generation time that is the main reason for its success. To get at an answer, we would ask what the effect of the parasite’s large population size would be if we eliminated any benefit from a faster generation time—i.e., by setting the parasite’s generation time equal to its host’s. We would then do the same for generation time, by setting the parasite and the host population sizes equal.
This approach ignores possible interactions between rate parameters—for example, if the parasite’s larger population size is beneficial only if the parasite has a faster generation time than its host’s. For antagonistic symbioses, in the weak-mutation limit, we will be able to relax the all-else equal simplification to prove more general results about the long-run effect of the various rate parameters. For mutualisms, our long-run weak-mutation analytical results do require the all-else equal assumption. We give some numerical suggestion that they hold qualitatively when we relax this assumption.
Antagonistic Symbioses
In this section, we study the evolutionary dynamics of populations interacting according to the antagonistic symbiosis game 1. An appropriate measure of the relative success of population 1 in a given population state is the proportion of and matchings (favorable to population 1) minus the proportion of and matchings (unfavorable to population 1). If this quantity is positive, then population 1 has a larger average payoff than population 2 in that state.
Strong Mutation, Strong Selection.
We begin by numerically studying the short- and long-run dynamics, when mutations are not very rare and selection is not very weak.
We first set the two populations’ sizes, selection strengths, and mutation rates to be equal (, , ) and vary their relative generation times and . For the parameter values that we consider, we find a weak Red Queen effect in the short run (Fig. 1A): When population 1 has a longer generation time, the set of initial states from which, after 50 generations, population 1 is on average more successful is greater than the set of initial states for which the reverse is true. We find a more pronounced Red Queen effect in the long run, when the dynamics have become independent of where they started (Fig. 1A).
We then set the populations’ generation times equal (), as well as their sizes () and mutation rates (), and allow their selection strengths and to vary. We now find very strong Red Queen effects in both the short run and the long run (Fig. 1C). We find similar Red Queen effects, especially in the long run, when populations differ in their mutation rates (Fig. 1B) or in their sizes (Fig. 1D).
We have spoken loosely of the “short run” and “long run.” Whereas 50 generations are certainly short run, the long run should be more precisely defined as the time by which the distribution over population states is close to the stationary distribution. In the language of Markov chains, this is known as the “mixing time” of the dynamical process (51).
For small population sizes, the time evolution of the probability distribution over states can feasibly be computed given any starting point. SI Appendix, Fig. S6 shows that, in the antagonistic dynamics, for the parameter values used in Fig. 1 (with small populations, of size 50), we may speak of the long run as being after 100–1,000 generations. For larger population sizes, it is not computationally feasible to compute the time evolution of the probability distribution over states, and we must resort to approximate analytical arguments.
We give such arguments in SI Appendix, section S3. Here, we summarize their conclusions. We assume that mutations are infrequent. When selection acts weakly in at least one of the populations (), the mixing time is approximately proportional to . When selection acts strongly in both populations (), then the mixing time is proportional to . In this latter case, the mixing time decreases with increasing population size, owing to a higher substitution rate of beneficial mutations (which drive the arms-race dynamics) in larger populations (SI Appendix, section S5.2). That the mixing time does not increase rapidly with increasing population size, and in fact decreases in the case of strong selection, indicates that the stationary distribution will be relevant on realistic timescales.
Weak Mutation.
To gain a greater understanding of the above results, we study the analytically tractable case of rare mutations, , using the methodology developed in ref. 46.
In this “weak-mutation limit,” the long-run stationary distribution collapses to a distribution over just the four pure states, . The relative success of population 1, as defined above, then simplifies to , which, by symmetry of the underlying states [, ], is proportional to .
First, we examine the influence of generation time and mutation rate on the success of population 1. We fix and and write . Define , the relative arrival rate of mutations in population 1. The stationary distribution of the evolutionary process, , is then
| [4] |
where ensures that sums to one (calculations in SI Appendix, section S5.4). The relative success of population 1 is
| [5] |
which is increasing in . Because both a higher mutation rate and a shorter generation time for population 1 increase , they are both associated with greater evolutionary success in Red Queen interactions. In fact, this result can be shown to hold far more generally: It does not require that the population sizes and selection strengths be set equal for the two populations and holds for all standard evolutionary dynamics, including those exhibiting frequency dependence (39, 40, 47, 52, 53) (proof in SI Appendix, section S5.3).
A larger population size and a stronger selection strength of population 1 can also be shown to increase its relative success, , although the results cannot be written as neatly as in Eq. 4 (SI Appendix, sections S5.1 and S5.2). Again, these results hold more generally: For many evolutionary processes, including the Moran and Wright–Fisher processes, they hold even if the generation times, mutation rates, and selection strengths are not set equal for the two populations.
Mutualistic Symbioses
In this section, we study the evolutionary dynamics of populations interacting according to the mutualism game 2. We take the measure of relative success of population 1 in a given population state to be the proportion of matchings in that state minus the proportion of matchings. If this quantity is positive, then population 1 has a larger average payoff than population 2 in that state.
Strong Mutation, Strong Selection.
As we did for the antagonistic game in the previous section, we begin by numerically studying the short- and long-run dynamics, when mutations are not rare and selection is not weak. We consider small and large values of , specifically and .
When , and we set the populations’ sizes, selection strengths, and mutation rates equal (, , ) and vary their relative generation times and , we find a weak Red Queen effect in the short run for the parameter values we consider: When population 1 has a longer generation time, the set of initial states from which, after 50 generations, the proportion of matchings is expected to be less than that of matchings is larger than the set of initial states for which the reverse is true (Fig. 2A). This is similar to B&L’s (26) general replicator-dynamics result for .
In the long run, however, the average proportion of matchings relative to matchings becomes independent of where the dynamics started; this effect is clear after 50,000 generations (Fig. 2 A–D, Lower). In this case, we find that differences in generation time between the two populations have almost no effect on their relative success (Fig. 2A).
We also find that differences in the populations’ relative mutation rates and , when their other rate parameters are equal, confer almost no advantage to either population, in both the short run and the long run (Fig. 2B).
If, instead, we allow the populations’ selection strengths and to differ, setting their other rate parameters equal, we observe stronger effects. Now, there is a pronounced Red Queen effect in the short run, which reverses to a strong Red King effect in the long run (Fig. 2C). A similar result is found when the populations have equal rate parameters except for their population sizes: When population 1 is smaller (so that it evolves slower—see SI Appendix, section S5.2 for a precise statement of this), it suffers a small disadvantage in the short run (a Red Queen effect), but an advantage in the long run (a Red King effect) (Fig. 2D).
When , these results reverse (Fig. 3). We now find a Red King effect in the short run, which is largest when rate differences between the populations derive from differences in selection strength (Fig. 3C). This is similar to B&L’s (26) general replicator-dynamics result for , and indeed, Fig. 3 A, B, and D, Upper and SI Appendix, Fig. S1 look very similar to B&L’s figure 2 (26). Again, generation time and mutation rate differences have little effect on the long-run dynamics (Fig. 3 A and B). In contrast, a strong Red Queen effect is found in the long run when the populations differ in their selection strengths (Fig. 3C), with population size having a weak Red Queen effect in the long run (Fig. 3D).
To summarize, when mutation rates are not very small, we find, for , a Red Queen effect in the short run and a Red King effect in the long run; for , we find a Red King effect in the short run and a Red Queen effect in the long run. The long-run effects are driven by selection strengths and population sizes; generation times and mutation rates have little effect on the long-run dynamics.
Again, the appropriate definition of the long run is how long it takes for the dynamics to get close to their stationary distribution. This can be computed exactly for small population sizes. In the mutualism game 2, for the parameter values we have considered in Figs. 2 and 3, SI Appendix, Fig. S6 suggests that the long run could be considered any time after about 1,000–10,000 generations.
For larger population sizes, such computations are not feasible, and we must again resort to approximate analytical arguments. These are detailed in SI Appendix, section S3. We summarize their conclusions here. Mutations are assumed to be infrequent. When selection acts weakly in both populations (), the mixing time is approximately proportional to . When selection acts strongly in at least one population , then the mixing time is approximately proportional to .
The exponential term in this last expression is a result of requiring substitutions against selection for the process to mix. It means that the mixing time will be prohibitively long when populations are large and selection acts strongly in them [mixing times increasing exponentially with population size have also been observed in single-population coordination games (54, 55), where transitions between equilibria require evolution against selection too]. In these cases, our stationary distributions will not be empirically relevant; evolution over realistic timescales will involve movement to an equilibrium and then stasis there, as in B&L’s (26) analysis. When populations are not large and selection is not very strong, then the stationary distribution will still be reached on a realistic timescale. This will also be true when the effective sizes of the populations are not large (56) or when the populations are subdivided into small subpopulations (57, 58), properties that hold for many mutualistic symbionts (59–61).
Weak Mutation.
Again, analytical results can be obtained for the long-run dynamics in the weak-mutation limit (), using the methodology developed in ref. 46. In this limit, the stationary distribution collapses to a distribution over just the pure states, , and population 1’s relative success, as defined above, simplifies to the quantity .
We first examine the influence of generation time and mutation rate. Setting and , but not specifying , , , , or , the stationary distribution of the evolutionary process is
| [6] |
where ensures that sums to one (calculations in SI Appendix, section S6.1). As expected, most time in the long run is spent in the two coordination states and , because and . Less intuitively, for small values of (), more time is spent in the worse noncoordination state than in the better noncoordination state .
Similar to the numerical results we obtained for the case where mutations are not very rare, neither the generation times nor the relative mutation rates of the two populations have any influence on the stationary distribution in the weak-mutation limit: If populations 1 and 2 are of the same size, and if natural selection acts equally efficiently in them, then they will be equally successful in the long run, no matter their relative generation times or mutation rates. This weak-mutation result in fact holds for all standard evolutionary processes, including those exhibiting within-population frequency dependence, such as the dynamics that govern Müllerian mimicry (SI Appendix, section S6.1).
It turns out (mathematical details in SI Appendix, section S6.1) that transitions against selection play an important role in this weak-mutation result. For example, whereas a higher relative mutation rate (or shorter generation time) in population 1 renders the transition from to more likely than that from to , they also make the reverse transition, from to , more likely than that from to . Because of the symmetry of the fitness changes in these two sets of transitions, these mutation effects (and, similarly, generation effects) cancel out exactly. In words, with a higher mutation rate, population 1 is typically the one that evolves to a coordination equilibrium (in the direction of selection), but it is also typically the one that evolves back out of it (against the direction of selection).
Numerical calculations suggest that this weak-mutation result is broadly robust to inequalities in population sizes and selection strengths (SI Appendix, Fig. S10 C and D).
The directional effects of selection strength and population size limit are more subtle. For selection strength, we equate population 1 and 2’s sizes at and fix population 2’s selection strength at . Then, if is sufficiently large, and population 1 has a slightly higher selection strength than population 2, population 1 is less successful when and more successful when (proofs in SI Appendix, section S6.3). Numerical calculations suggest that the “sufficiently large” population size is not very large: Results for agree with those above (SI Appendix, Fig. S10A).
The results are similar if we equate the selection strengths in the two populations at and set population 1’s size slightly higher than that of population 2: Population 1 is less successful when and more successful when (proofs in SI Appendix, section S6.4). Again, numerical calculations suggest that these results are robust to inequality in mutation rates and generation times and larger differences in the two populations’ sizes when both are sufficiently large (SI Appendix, Fig. S10 B, E, and F).
Intuition for these results can be gained by noting that any switch from one coordination state to the other involves two transitions: one transition out of the original equilibrium (against the direction of selection) and a subsequent transition into the new equilibrium state (in the direction of selection). Whether weaker selection (or a smaller population size) favors population 1 depends on which nonequilibrium state is more often passed during these two transitions. This is driven predominantly by the relative probabilities of the transitions against selection, because these probabilities are much more sensitive to population size, selection strength, and the payoffs involved.
Of the transitions against selection, transitions to always involve the fixation of mutants of payoff in populations of payoff , whereas transitions to always involve the fixation of mutants of payoff in populations of payoff .
Therefore, when is large, the passed nonequilibrium state is usually [indeed, note that has a higher weight than in the stationary distribution 6 precisely when ]. Focusing therefore on transitions through , if population 1 experiences weaker selection (or is of smaller size), the transition from to occurs more easily than that from to . So we expect more time to be spent in population 1’s disfavored equilibrium state in the long run—a Red Queen effect.
Contrariwise, if is small, the passed nonequilibrium state is usually . So, if selection is less effective in population 1, more time is spent in its favored equilibrium state —a Red King effect—because transitions from this state to , involving fixation of a selected-against mutant in population 2, are relatively very rare.
In contrast to the minor influence of mutation rates and generation times in the long run, the effects of population size and selection strength are very large. For example, when and , a small increase in from to increases (another measure of population 1’s relative success, useful for extreme values) by 35% when and decreases it by 30% when (SI Appendix, Fig. S10A). Changes of similar magnitude are seen when holding and increasing from 100 to 105 (SI Appendix, Fig. S10B). These effects, caused by small changes in population size and selection strength, are larger than those caused by varying relative mutation rates and generation times across 10 orders of magnitude (SI Appendix, Fig. S10 C and D). Larger changes in population size and selection strength have enormous effects on the long-run relative success of the two populations (SI Appendix, Fig. S10 E and F).
Weak Selection.
Returning to the effect of mutation rate and generation time, we have shown analytically that these have no effect on the long-run stationary distribution in the weak-mutation limit. To analytically study the long-run effect of mutation rates when they need not be negligibly small, we make use of another recent advance in evolutionary game theory (47). We impose that the population sizes , selection strengths , and generation times of the two populations be equal. Their mutation rates, and , need not be equal, and we do not impose that . Instead, we assume ; that is, we assume that selection operates very weakly in the two populations. Long-run Red King and Queen effects consistent with those we have found above for selection strength and population size would have that slower evolution (via a smaller mutation rate) is favored when and disfavored when [the threshold for that was previously relevant, , tends to as ].
Unlike in the weak-mutation case, the stationary distribution of this process places nonnegligible weight on every possible population state, including those where one or both populations are polymorphic. Again, the relative success of population 1 depends on the long-run proportion of and matchings, which, following the notation in ref. 47, we denote by and , respectively. Using equation 29 in ref. 47, we calculate the long-run advantage to population 1 in the weak-selection limit,
| [7] |
where (calculations in SI Appendix, section S7). So, when , population 1 does better when it evolves slower (); when , population 1 does better when it evolves faster (). We have thus recovered long-run Red King and Queen effects of mutation consistent with those for selection strength and population size. When mutation rates are small (), the two populations do approximately equally well, consistent with our weak-mutation results above.
Discussion
We have placed the evolution of symbioses, both antagonistic and mutualistic, into a simple finite-population model and studied the effect of parameters that influence the rate of evolution of the participating populations on those populations’ relative success.
Among these “rate parameters,” mutation rate and generation time are perhaps the most responsive (34, 62–68). Selection strengths, although also a clear determinant of evolutionary rate, seem more of a fixed property of an interaction, but could change over time if one population reduces its dependence on the interaction (3) or actively increases the dependence of its interactant (69). Population size is the most complicated of the rate parameters, because changes in the relative success of interacting populations (possibly driven by differences in their population sizes) are expected to manifest themselves in subsequent changes in the populations’ sizes (70).
It is important to realize that, although we have studied differences in the relative success of interacting populations caused by different evolutionary rate parameters, our results do not say anything directly about how the rate parameters themselves should evolve. For example, it is conceivable that a slower generation time leads to greater success for a population (i.e., that a Red King effect holds), but that a faster generation time is selected for within that population. In this example, slow evolution could be interpreted as a public good.
This has important implications for how results such as ours and those of B&L (26) should be interpreted in studies of molecular evolution (e.g., ref. 71). Although generation times and mutation rates determine the rate of nucleotide substitution at neutral sites (37), were we to find that a slower generation time or a lower mutation rate leads to greater success for a population in a mutualism, this would not necessarily imply that species involved in mutualisms should exhibit lower neutral substitution rates.
Examining the evolution of the rate parameters themselves would require a more complicated model than the one we have studied. In a population genetic setting, one could posit in each population two distinct genetic loci: a “strategy locus,” the alleles at which determine which game strategies their bearers play, and a “rate locus,” the alleles at which determine the value of a rate parameter such as mutation rate. The evolutionary dynamics would then inform how both the success of the populations and their evolutionary rates evolve. One setting where we might expect the evolutionary rate within a population to evolve to increase that population’s relative success is if the interacting populations are subdivided into many isolated symbioses (on “islands”), between which migration occurs [this model is similar to the “haystack model” considered by B&L (26)]. A rate parameter value that improves a subpopulation’s success on an island causes that subpopulation to send out more migrants—who bear the rate parameter value—to colonize other islands. Thus, in symbioses with a high degree of population structure, we might expect the results we have found in this paper to be informative of how evolutionary rate will evolve within populations.
In antagonistic symbioses, we have shown that faster evolution through any of the rate parameters leads to greater evolutionary success in our model, in both the short run and the long run—a clear Red Queen effect. Mutation rate and generation time play similar roles in determining the long-run outcome of antagonistic interactions when mutations are infrequent. As far as our results have implications for evolution within populations, this suggests that one population might compensate for a longer generation time by evolving an elevated mutation rate. Consistent with this, bacterial strains subjected to antagonistic interactions with bacteriophages often exhibit much higher mutation rates than control strains (72). A similar explanation has been suggested for the finding that, in mammals, generation time is positively correlated with the rate of intrachromosomal recombination (73), which, like mutation, is a generator of variation (1). With particular respect to selection strength, the “life–dinner principle” (3) holds in our model: In an antagonistic interaction, the population for whom the interaction matters more (the rabbit, not the fox) is evolutionarily more successful.
In mutualistic symbioses, we have uncovered an important influence of evolutionary timescale on the relative success of interacting populations, with results in the short run and long run often being in opposition. In the short run, the stochastic evolutionary dynamics that we have studied are similar to the deterministic dynamics studied by B&L (26), and our short-run results replicate theirs: We find a short-run Red Queen effect when is small in game 2 and a short-run Red King effect when is large. Our analysis extends B&L’s (26) by allowing us to determine which of the biological rate parameters drive these effects: to wit, generation times, population sizes, and (especially) selection strengths.
In the long run, we find a Red King effect when is small and a Red Queen effect when is large, contrary to the short-run results. Among the rate parameters that could drive these long-run results, mutation rate and generation time in fact have little to no effect, unless mutation is frequent. This is a surprising result, given that mutation rate and generation time are perhaps the most prominent determinants of evolutionary rate. This result depends on the possibility of evolution against selection, always present in stochastic evolutionary dynamics. It could not be discovered using deterministic evolutionary dynamics, because in such dynamics, evolution always proceeds in the direction of selection. Deterministic dynamics admit little notion of the strength of selection: Selection is either “positive” or “negative.” In reality, selection is a continuum—as the Red Queen said to Alice, “I could show you hills, in comparison with which you’d call that a valley” (ref. 4, p. 37)—and deleterious mutations do sometimes fix because of random drift (37, 74–77).
The long-run Red King and Queen effects in mutualisms, for small and large , respectively, instead operate predominantly in our model through the efficiency of selection (which increases with selection strength and population size), not the generation of variation (mutation and generation rates).
Which results, short-run or long-run, are relevant to a specific case depends on which evolutionary timescale is appropriate. Again, this question is treated quantitatively in SI Appendix, section S3. In mutualisms, the long-run dynamics are characterized by transitions between the equilibria—involving evolution first against, and then with, selection—so the applicability of our long-run results depends on the timescale of these transitions. When both populations are very large and under strong selection, the time it takes for a transition between equilibria to occur can be so long as to be empirically irrelevant; here, the short-run dynamics are of more interest. When the populations have small effective population sizes (owing to small census size or population structure, for example) or selection acts weakly in them, equilibrium transitions can be frequent, and the long-run dynamics apply.
In our analysis, we have made several modeling decisions and simplifying assumptions, described below.
We have assumed that interactions are pairwise, for simplicity. The consideration of multiplayer games (2, 78–83) is a desirable extension and has been shown to influence the outcome of mutualism games in infinite populations (80) and antagonism and Snowdrift games in finite populations (2).
In deriving our main analytical results, we assumed that mutations are infrequent. This is a realistic assumption in many cases, but not always. SI Appendix, Fig. S5 shows that mutation rates in the two populations do not have to be exceedingly small for our weak-mutation analytical results to be an excellent quantitative match (46, 84, 85), and in SI Appendix, Figs. S2–S5 suggest that, even for very large mutation rates, the long-run relative success of the populations qualitatively matches our weak-mutation results in most cases. The only notable exceptions, all for the mutualism game 2, are (i) for large , when the populations have different selection strengths, a strong long-run Red Queen effect for small mutation rates reverses to a weak Red King effect when mutation rates are very large (SI Appendix, Figs. S2C and S5C); (ii) when the populations have different mutation rates and both their mutation rates are very large, then, contrary to our weak-mutation result where differences in mutation rates have no effect, we find a long-run Red King effect for small (SI Appendix, Figs. S3B and S5B) and a Red Queen effect for large (SI Appendix, Figs. S2B and S5C) (exception ii is consistent with our weak-selection result, Eq. 7); and (iii) for small , generation time has a Red Queen effect for intermediate and large mutation rates (SI Appendix, Figs. S3A and S5B). A very recent methodological advance in stochastic evolutionary dynamics (86) suggests that tractable analysis of the stationary distribution outside the weak-mutation regime may soon be possible.
In finite-population evolutionary game theory, game payoffs must be translated to positive fitnesses (40), with this translation calibrated by a strength of selection . We have used the commonly used linear translation , but others are also possible. The exponential translation has the advantage that it guarantees fitnesses always to be positive, no matter the range of game payoffs, and also sometimes allows for neater characterization of long-run dynamics (87–90). Results are expected to be similar for the two translations, and they are identical in the weak-selection limit . SI Appendix, Fig. S5 suggests that our results are essentially unchanged when we use either translation.
The dynamics in our model, based on the Moran process, are highly stylized, especially in their assumption of constant population sizes. It would be interesting to study evolution in games 1 and 2, using more complicated intergenerational population dynamics (91–93).
Finally, the games themselves are particular simplifications of more complex interactions. One key simplification in games 1 and 2 is that strategies are discrete: , , , . This will be relevant for many antagonistic symbioses; for example, it is probably a reasonable summary that a pathogen is either resistant or not to a host’s defenses. For mutualisms, we have motivated the discrete game 2 with examples that exhibit such discreteness, such as division of labor and Müllerian mimicry. However, many mutualisms are likely to involve continuous strategies: for example, how much energy to expend on a cooperative task, as in the degree to which an ant colony protects from herbivores the plant that houses and feeds it (94) and in turn the energy the plant dedicates to housing and feeding the ants. For such cases, an alternative, continuous-strategy model specification is appropriate.
An attractive option is a simple modification of a Nash bargaining game (95). Players 1 and 2 choose activity levels and . If , then both receive zero payoff. If , then player 1 receives payoff , and player 2 receives . calibrates the degree to which players’ payoffs depend on their own actions. As long as , both players’ payoffs increase if either player increases its activity, so that the game is mutualistic. But when is small, each player would prefer to have a lower activity than the other, whereas when is large, each would prefer to have the larger activity. In contrast to the discrete-strategy mutualism game, where there were two discrete equilibria, here there is a continuous path of equilibria (the line ). In SI Appendix, section S8, we study the coevolutionary dynamics of populations of player 1s and 2s. The short-run dynamics in this game involve evolution to the equilibrium line, whereas the long-run dynamics involve drift-like movement along and around it.
In both the short and long runs, the faster-evolving population is at an advantage when is large (a Red Queen effect; SI Appendix, Fig. S11), but at a disadvantage when is small (a Red King effect; SI Appendix, Fig. S12). The short-run effects are driven by all rate parameters. But similar to what we found for the discrete mutualism game, the long-run effects are driven predominantly by selection strength and population size—differences in mutation rate and generation time have little effect. This is because selection strength and population size contribute differentially here to a case of drift-induced selection: Stochastic jumps off the equilibrium path return with an average directional bias. Drift-induced selection is a newly recognized phenomenon that has recently gained attention in the stochastic dynamics literature (96–98). It can be studied analytically using diffusion approximations and separations of timescales (99). Such a study in the context of continuous-strategy mutualisms would be an important extension of our preliminary analysis.
In this vein, studying the finite-population dynamics of other games in which slow evolution has been hypothesized to be beneficial (100) would also be desirable.
Supplementary Material
Acknowledgments
C.V. is grateful to Philipp Altrock for introducing him to the Red King literature. We thank Carl Bergstrom, Kirill Borusyak, Kooskia Burns-Edelman, Richard Childers, Michael Doebeli, David Haig, Jonathan Libgober, Alex McAvoy, Sam Sinai, Luis Zaman, and participants at the Harvard Economics Department’s Games and Markets lunch for helpful comments.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. S.A.F. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1702020114/-/DCSupplemental.
References
- 1.Nee S. Antagonistic co-evolution and the evolution of genotypic randomization. J Theor Biol. 1989;140:499–518. doi: 10.1016/s0022-5193(89)80111-0. [DOI] [PubMed] [Google Scholar]
- 2.Damore J, Gore J. A slowly evolving host moves first in symbiotic interactions. Evolution. 2011;65:2391–2398. doi: 10.1111/j.1558-5646.2011.01299.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dawkins R, Krebs J. Arms races between and within species. Proc Biol Sci. 1979;205:489–511. doi: 10.1098/rspb.1979.0081. [DOI] [PubMed] [Google Scholar]
- 4.Carroll L. Through the Looking-Glass. Macmillan; London: 1871. [Google Scholar]
- 5.Turner J. The evolutionary dynamics of Batesian and Muellerian mimicry: Similarities and differences. Ecol Entomol. 1987;12:81–95. [Google Scholar]
- 6.Burt A, Trivers R. Genes in Conflict. Harvard Univ Press; Cambridge, MA: 2006. [Google Scholar]
- 7.McLaughlin RN, Malik HS. Genetic conflicts: The usual suspects and beyond. J Exp Biol. 2017;220:6–17. doi: 10.1242/jeb.148148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tao Y, Masly J, Araripe L, Ke Y, Hartl D. A sex-ratio meiotic drive system in Drosophila simulans. I: An autosomal suppressor. PLoS Biol. 2007;5:e292. doi: 10.1371/journal.pbio.0050292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tao Y, et al. A sex-ratio meiotic drive system in Drosophila simulans. II: An X-linked distorter. PLoS Biol. 2007;5:e293. doi: 10.1371/journal.pbio.0050293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Henikoff S, Ahmad K, Malik H. The centromere paradox: Stable inheritance with rapidly evolving DNA. Science. 2001;293:1098–1102. doi: 10.1126/science.1062939. [DOI] [PubMed] [Google Scholar]
- 11.Malik H, Henikoff S. Conflict begets complexity: The evolution of centromeres. Curr Opin Genet Dev. 2002;12:711–718. doi: 10.1016/s0959-437x(02)00351-9. [DOI] [PubMed] [Google Scholar]
- 12.Boulton A, Myers R, Redfield R. The hotspot conversion paradox and the evolution of meiotic recombination. Proc Natl Acad Sci USA. 1997;94:8058–8063. doi: 10.1073/pnas.94.15.8058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baudat F, et al. PRDM9 is a major determinant of meiotic recombination hotspots in humans and mice. Science. 2010;327:836–840. doi: 10.1126/science.1183439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Parvanov E, Petkov P, Paigen K. Prdm9 controls activation of mammalian recombination hotspots. Science. 2010;327:835–835. doi: 10.1126/science.1181495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Úbeda F, Wilkins J. The Red Queen theory of recombination hotspots. J Evol Biol. 2011;24:541–553. doi: 10.1111/j.1420-9101.2010.02187.x. [DOI] [PubMed] [Google Scholar]
- 16.Kingan SB, Garrigan D, Hartl DL. Recurrent selection on the Winters sex-ratio genes in Drosophila simulans. Genetics. 2010;184:253–265. doi: 10.1534/genetics.109.109587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Myers S, et al. Drive against hotspot motifs in primates implicates the PRDM9 gene in meiotic recombination. Science. 2010;327:876–879. doi: 10.1126/science.1182363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ponting C. What are the genomic drivers of the rapid evolution of PRDM9? Trends Genet. 2011;27:165–171. doi: 10.1016/j.tig.2011.02.001. [DOI] [PubMed] [Google Scholar]
- 19.Jeffreys AJ, Cotton VE, Neumann R, Lam KW. Recombination regulator PRDM9 influences the instability of its own coding sequence in humans. Proc Natl Acad Sci USA. 2013;110:600–605. doi: 10.1073/pnas.1220813110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schwartz J, Roach D, Thomas J, Shendure J. Primate evolution of the recombination regulator PRDM9. Nat Commun. 2014;5:4370. doi: 10.1038/ncomms5370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lesecque Y, Glémin S, Lartillot N, Mouchiroud D, Duret L. The Red Queen model of recombination hotspots evolution in the light of archaic and modern human genomes. PLoS Genet. 2014;10:e1004790. doi: 10.1371/journal.pgen.1004790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Davies B, et al. Re-engineering the zinc fingers of PRDM9 reverses hybrid sterility in mice. Nature. 2016;530:171–176. doi: 10.1038/nature16931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baker Z, et al. Repeated losses of PRDM9-directed recombination despite the conservation of PRDM9 across vertebrates. eLife. 2017;6:e24133. doi: 10.7554/eLife.24133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Doebeli M, Knowlton N. The evolution of interspecific mutualisms. Proc Natl Acad Sci USA. 1998;95:8676–8680. doi: 10.1073/pnas.95.15.8676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Herre E, Knowlton N, Mueller U, Rehner S. The evolution of mutualisms: Exploring the paths between conflict and cooperation. Trends Ecol Evol. 1999;14:49–53. doi: 10.1016/s0169-5347(98)01529-8. [DOI] [PubMed] [Google Scholar]
- 26.Bergstrom C, Lachmann M. The Red King effect: When the slowest runner wins the coevolutionary race. Proc Natl Acad Sci USA. 2003;100:593–598. doi: 10.1073/pnas.0134966100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Leigh E. The evolution of mutualism. J Evol Biol. 2010;23:2507–2528. doi: 10.1111/j.1420-9101.2010.02114.x. [DOI] [PubMed] [Google Scholar]
- 28.Hom E, Aiyar P, Schaeme D, Mittag M, Sasso S. A chemical perspective on microalgal–microbial interactions. Trends Plant Sci. 2015;20:689–693. doi: 10.1016/j.tplants.2015.09.004. [DOI] [PubMed] [Google Scholar]
- 29.Haig D, Grafen A. Genetic scrambling as a defence against meiotic drive. J Theor Biol. 1991;153:531–558. doi: 10.1016/s0022-5193(05)80155-9. [DOI] [PubMed] [Google Scholar]
- 30.Krugman P. Increasing returns, monopolistic competition, and international trade. J Int Econ. 1979;9:469–479. [Google Scholar]
- 31.Wahl L. Evolving the division of labour: Generalists, specialists and task allocation. J Theor Biol. 2002;219:371–388. doi: 10.1006/jtbi.2002.3133. [DOI] [PubMed] [Google Scholar]
- 32.Sherratt T. The evolution of Müllerian mimicry. Naturwissenschaften. 2008;95:681–695. doi: 10.1007/s00114-008-0403-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Taddei F, et al. Role of mutator alleles in adaptive evolution. Nature. 1997;387:700–702. doi: 10.1038/42696. [DOI] [PubMed] [Google Scholar]
- 34.Sniegowski P, Gerrish P, Johnson T, Shaver A. The evolution of mutation rates: Separating causes from consequences. BioEssays. 2000;22:1057–1066. doi: 10.1002/1521-1878(200012)22:12<1057::AID-BIES3>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 35.Lanfear R, Kokko H, Eyre-Walker A. Population size and the rate of evolution. Trends Ecol Evol. 2014;29:33–41. doi: 10.1016/j.tree.2013.09.009. [DOI] [PubMed] [Google Scholar]
- 36.Van Valen L. A new evolutionary law. Evol Theor. 1973;1:1–30. [Google Scholar]
- 37.Kimura M. The Neutral Theory of Molecular Evolution. Cambridge Univ Press; Cambridge, UK: 1983. [Google Scholar]
- 38.Moran P. Random processes in genetics. Math Proc Camb Philos Soc. 1958;54:60–71. [Google Scholar]
- 39.Taylor C, Fudenberg D, Sasaki A, Nowak M. Evolutionary game dynamics in finite populations. Bull Math Biol. 2004;66:1621–1644. doi: 10.1016/j.bulm.2004.03.004. [DOI] [PubMed] [Google Scholar]
- 40.Nowak M. Evolutionary Dynamics. Harvard Univ Press; Cambridge, MA: 2006. [Google Scholar]
- 41.Hartl DL, Clark AG. Principles of Population Genetics. 4th Ed Sinauer; Sunderland, MA: 2007. [Google Scholar]
- 42.Fudenberg D, Imhof LA. Imitation processes with small mutations. J Econ Theor. 2006;131:251–262. [Google Scholar]
- 43.Hauert C, Traulsen A, De Silva H, Nowak M, Sigmund K. Public goods with punishment and abstaining in finite and infinite populations. Biol Theor. 2008;3:114–122. doi: 10.1162/biot.2008.3.2.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fudenberg D, Nowak MA, Taylor C, Imhof LA. Evolutionary game dynamics in finite populations with strong selection and weak mutation. Theor Popul Biol. 2006;70:352–363. doi: 10.1016/j.tpb.2006.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Karlin S, Taylor H. A First Course in Stochastic Processes. Academic; New York: 1975. [Google Scholar]
- 46.Veller C, Hayward L. Finite-population evolution with rare mutations in asymmetric games. J Econ Theor. 2016;162:93–113. [Google Scholar]
- 47.Ohtsuki H. Stochastic evolutionary dynamics of bimatrix games. J Theor Biol. 2010;264:136–142. doi: 10.1016/j.jtbi.2010.01.016. [DOI] [PubMed] [Google Scholar]
- 48.McCandlish D, Stoltzfus A. Modeling evolution using the probability of fixation: History and implications. Q Rev Biol. 2014;89:225–252. doi: 10.1086/677571. [DOI] [PubMed] [Google Scholar]
- 49.Waxman D, Gavrilets S. 20 questions on adaptive dynamics. J Evol Biol. 2005;18:1139–1154. doi: 10.1111/j.1420-9101.2005.00948.x. [DOI] [PubMed] [Google Scholar]
- 50.Lynch M. Evolution of the mutation rate. Trends Genet. 2010;26:345–352. doi: 10.1016/j.tig.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Levin D, Peres Y, Wilmer E. Markov Chains and Mixing Times. Am Math Soc; Providence, RI: 2009. [Google Scholar]
- 52.Nowak M, Sasaki A, Taylor C, Fudenberg D. Emergence of cooperation and evolutionary stability in finite populations. Nature. 2004;428:646–650. doi: 10.1038/nature02414. [DOI] [PubMed] [Google Scholar]
- 53.Sekiguchi T, Ohtsuki H. Fixation probabilities of strategies for bimatrix games in finite populations. Dyn Games Appl. 2015;7:93–111. [Google Scholar]
- 54.Black A, Traulsen A, Galla T. Mixing times in evolutionary game dynamics. Phys Rev Lett. 2012;109:028101. doi: 10.1103/PhysRevLett.109.028101. [DOI] [PubMed] [Google Scholar]
- 55.Kandori M, Mailath G, Rob R. Learning, mutation, and long run equilibria in games. Econometrica. 1993;61:29–56. [Google Scholar]
- 56.Charlesworth B. Fundamental concepts in genetics: Effective population size and patterns of molecular evolution and variation. Nat Rev Genet. 2009;10:195–205. doi: 10.1038/nrg2526. [DOI] [PubMed] [Google Scholar]
- 57.Whitlock MC. Fixation probability and time in subdivided populations. Genetics. 2003;164:767–779. doi: 10.1093/genetics/164.2.767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ellison G. Learning, local interaction, and coordination. Econometrica. 1993;61:1047–1071. [Google Scholar]
- 59.Moran NA, Wernegreen JJ. Lifestyle evolution in symbiotic bacteria: Insights from genomics. Trends Ecol Evol. 2000;15:321–326. doi: 10.1016/s0169-5347(00)01902-9. [DOI] [PubMed] [Google Scholar]
- 60.Cook J, Rasplus J. Mutualists with attitude: Coevolving fig wasps and figs. Trends Ecol Evol. 2003;18:241–248. [Google Scholar]
- 61.Caldera E, Currie C. The population structure of antibiotic-producing bacterial symbionts of Apterostigma dentigerum ants: Impacts of coevolution and multipartite symbiosis. Am Nat. 2012;180:604–617. doi: 10.1086/667886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sniegowski P, Gerrish P, Lenski R. Evolution of high mutation rates in experimental populations of E. coli. Nature. 1997;387:703–705. doi: 10.1038/42701. [DOI] [PubMed] [Google Scholar]
- 63.Williams G. Pleiotropy, natural selection, and the evolution of senescence. Evolution. 1957;11:398–411. [Google Scholar]
- 64.Hamilton W. The moulding of senescence by natural selection. J Theor Biol. 1966;12:12–45. doi: 10.1016/0022-5193(66)90184-6. [DOI] [PubMed] [Google Scholar]
- 65.Bull J. Virulence. Evolution. 1994;48:1423–1437. doi: 10.1111/j.1558-5646.1994.tb02185.x. [DOI] [PubMed] [Google Scholar]
- 66.Nowak M, May R. Superinfection and the evolution of parasite virulence. Proc Biol Sci. 1994;255:81–89. doi: 10.1098/rspb.1994.0012. [DOI] [PubMed] [Google Scholar]
- 67.Stearns S, Ackermann M, Doebeli M, Kaiser M. Experimental evolution of aging, growth, and reproduction in fruitflies. Proc Natl Acad Sci USA. 2000;97:3309–3313. doi: 10.1073/pnas.060289597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Vérin M, Menu F, Rajon E. 2016. The biased evolution of generation time. arXiv:1502.05508.
- 69.Wyatt G, Kiers E, Gardner A, West S. Restricting mutualistic partners to enforce trade reliance. Nat Comm. 2016;7:10322. doi: 10.1038/ncomms10322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Schenk H, Traulsen A, Gokhale C. 2016. Chaotic provinces in the kingdom of the red queen. arXiv:1607.01564.
- 71.Rubin B, Moreau C. Comparative genomics reveals convergent rates of evolution in ant–plant mutualisms. Nat Commun. 2016;7:12679. doi: 10.1038/ncomms12679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pal C, Maciá M, Oliver A, Schachar I, Buckling A. Coevolution with viruses drives the evolution of bacterial mutation rates. Nature. 2007;450:1079–1081. doi: 10.1038/nature06350. [DOI] [PubMed] [Google Scholar]
- 73.Burt A, Bell G. Mammalian chiasma frequencies as a test of two theories of recombination. Nature. 1987;326:803–805. doi: 10.1038/326803a0. [DOI] [PubMed] [Google Scholar]
- 74.Ohta T. Slightly deleterious mutant substitutions in evolution. Nature. 1973;246:96–98. doi: 10.1038/246096a0. [DOI] [PubMed] [Google Scholar]
- 75.Ohta T. The nearly neutral theory of molecular evolution. Annu Rev Ecol Systemat. 1992;23:263–286. [Google Scholar]
- 76.Lynch M. The Origins of Genome Architecture. Sinauer; Sunderland, MA: 2007. [Google Scholar]
- 77.McCandlish D, Epstein C, Plotkin J. The inevitability of unconditionally deleterious substitutions during adaptation. Evolution. 2014;68:1351–1364. doi: 10.1111/evo.12350. [DOI] [PubMed] [Google Scholar]
- 78.Gokhale C, Traulsen A. Evolutionary games in the multiverse. Proc Natl Acad Sci USA. 2010;107:5500–5504. doi: 10.1073/pnas.0912214107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gokhale C, Traulsen A. Strategy abundance in evolutionary many-player games with multiple strategies. J Theor Biol. 2011;283:180–191. doi: 10.1016/j.jtbi.2011.05.031. [DOI] [PubMed] [Google Scholar]
- 80.Gokhale C, Traulsen A. Mutualism and evolutionary multiplayer games: Revisiting the red king. Proc Biol Sci. 2012;279:4611–4616. doi: 10.1098/rspb.2012.1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wu B, Traulsen A, Gokhale C. Dynamic properties of evolutionary multi-player games in finite populations. Games. 2013;4:182–199. [Google Scholar]
- 82.Gokhale C, Traulsen A. Evolutionary multiplayer games. Dyn Games Appl. 2014;4:468–488. [Google Scholar]
- 83.McAvoy A, Hauert C. Structure coefficients and strategy selection in multiplayer games. J Math Biol. 2016;72:203–238. doi: 10.1007/s00285-015-0882-3. [DOI] [PubMed] [Google Scholar]
- 84.Wu B, Gokhale C, Wang L, Traulsen A. How small are small mutation rates? J Math Biol. 2012;64:803–827. doi: 10.1007/s00285-011-0430-8. [DOI] [PubMed] [Google Scholar]
- 85.Rand D, Tarnita C, Ohtsuki H, Nowak M. Evolution of fairness in the one-shot anonymous Ultimatum Game. Proc Natl Acad Sci USA. 2013;110:2581–2586. doi: 10.1073/pnas.1214167110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Vasconcelos V, Santos F, Santos F, Pacheco J. Stochastic dynamics through hierarchically embedded Markov chains. Phys Rev Lett. 2017;118:058301. doi: 10.1103/PhysRevLett.118.058301. [DOI] [PubMed] [Google Scholar]
- 87.Blume L. The statistical mechanics of strategic interaction. Games Econ Behav. 1993;5:387–424. [Google Scholar]
- 88.Traulsen A, Nowak M, Pacheco J. Stochastic dynamics of invasion and fixation. Phys Rev E. 2006;74:011909. doi: 10.1103/PhysRevE.74.011909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Traulsen A, Shoresh N, Nowak M. Analytical results for individual and group selection of any intensity. Bull Math Biol. 2008;70:1410–1424. doi: 10.1007/s11538-008-9305-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Cooney D, Allen B, Veller C. Assortment and the evolution of cooperation in a Moran process with exponential fitness. J Theor Biol. 2016;409:38–46. doi: 10.1016/j.jtbi.2016.08.026. [DOI] [PubMed] [Google Scholar]
- 91.Gokhale C, Papkou A, Traulsen A, Schulenburg H. Lotka–Volterra dynamics kills the red queen: Population size fluctuations and associated stochasticity dramatically change host-parasite coevolution. BMC Evol Biol. 2013;13:254. doi: 10.1186/1471-2148-13-254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Song Y, Gokhale C, Papkou A, Schulenburg H, Traulsen A. Host-parasite coevolution in populations of constant and variable size. BMC Evol Biol. 2015;15:212. doi: 10.1186/s12862-015-0462-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Papkou A, Gokhale C, Traulsen A, Schulenburg H. Host–parasite coevolution: Why changing population size matters. Zoology. 2016;119:330–338. doi: 10.1016/j.zool.2016.02.001. [DOI] [PubMed] [Google Scholar]
- 94.Frederickson M, et al. The direct and ecological costs of an ant-plant symbiosis. Am Nat. 2012;179:768–778. doi: 10.1086/665654. [DOI] [PubMed] [Google Scholar]
- 95.Nash J., Jr The bargaining problem. Econometrica. 1950;18:155–162. [Google Scholar]
- 96.Constable G, McKane A, Rogers T. Stochastic dynamics on slow manifolds. J Phys Math Theor. 2013;46:295002. [Google Scholar]
- 97.Constable G, Rogers T, McKane A, Tarnita C. Demographic noise can reverse the direction of deterministic selection. Proc Natl Acad Sci USA. 2016;113:E4745–E4754. doi: 10.1073/pnas.1603693113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Veller C, Muralidhar P, Constable G, Nowak M. 2017. Drift-induced selection between male and female heterogamety. bioRxiv:141929. [DOI] [PMC free article] [PubMed]
- 99.Parsons T, Rogers T. 2015. Dimension reduction via timescale separation in stochastic dynamical systems. arXiv:1510.07031.
- 100.Hilbe C, Nowak M, Sigmund K. Evolution of extortion in Iterated Prisoner’s Dilemma games. Proc Natl Acad Sci USA. 2013;110:6913–6918. doi: 10.1073/pnas.1214834110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.