Abstract
Divergence in phenotypic traits often contributes to premating isolation between lineages, but could also promote isolation at postmating stages. Phenotypic differences could directly result in mechanical isolation or hybrids with maladapted traits; alternatively, when alleles controlling these trait differences pleiotropically affect other components of development, differentiation could indirectly produce genetic incompatibilities in hybrids. Here, we determined the strength of 9 postmating and intrinsic postzygotic reproductive barriers among 10 species of Jaltomata (Solanaceae), including species with highly divergent floral traits. To evaluate the relative importance of floral trait diversification on the strength of these postmating barriers, we assessed their relationship to floral divergence, genetic distance, geographical context, and ecological differences, using conventional tests and a new linear mixed modeling approach. Despite close evolutionary relationships, all species pairs showed moderate to strong isolation. Nonetheless, floral trait divergence was not a consistent predictor of the strength of isolation; instead this was best explained by genetic distance, although we found evidence for mechanical isolation in one species, and an overall positive relationship between floral trait divergence and fruit set isolation across species pairs. Overall, our data indicate that intrinsic postzygotic isolation is more strongly associated with genome-wide genetic differentiation, rather than floral divergence.
Keywords: floral evolution, pollen-pistil, reinforcement, reproductive isolation, speciation, sympatry
Introduction
The evolution of reproductive barriers that reduce or prevent gene flow among diverging lineages is a key component of speciation (Coyne and Orr 2004). Numerous traits and underlying mechanisms can contribute to reproductive barriers (Coyne and Orr 2004; Baack et al. 2015). However, studies often focus on phenotypic traits that differ markedly between closely related taxa, and that could contribute to premating reproductive barriers between these lineages (Maan and Seehausen 2011), including aspects of mating and courtship displays (Masta and Maddison 2002; Shaw et al. 2007; Selz et al. 2016), and floral traits relating to pollinator discrimination between plant species (Kay and Sargent 2009). These traits also often mediate reproductive success within species (Higashi et al. 1999; Coyne and Orr 2004; Ritchie 2007; Yukilevich et al. 2016), and can be subject to divergent natural or sexual selection that accelerates their differentiation between lineages. Divergence in these traits can play a key role in speciation via direct effects on reducing gene flow prior to mating (Coyne and Orr 2004; Maan and Seehausen 2011; Mendelson and Shaw 2012), however the potential for this trait divergence to promote the expression of isolating barriers at other, later, reproductive stages remains unclear (Coyne and Orr 2004; Maan and Seehausen 2011). To address these potential collateral effects of rapid mating trait divergence, we require studies that examine multiple reproductive barriers among closely related taxa that differ in such trait(s), and are therefore able to simultaneously assess the relative importance of this divergence versus other axes of differentiation that might drive isolation between lineages (e.g. Jewell et al. 2012; Martin and Mendelson 2016).
Divergent mating traits are one such class of traits that might contribute to isolation, both by directly preventing or reducing the frequency of heterospecific mating, and via their later effects on postmating and postzygotic isolation. Species differences in elaborated mating traits have frequently been associated with premating reproductive barriers (Coyne and Orr 2004; Lowry et al. 2008; Baack et al. 2015), including behavioral isolation in diverse animal groups such as birds (Seddon et al. 2013), fish (Mendelson 2003; Selz et al. 2016), and especially insects (Mullen and Shaw 2014; Merrill et al. 2015), as well as pollinator isolation in plants (specifically angiosperms). Divergent floral traits can produce strong pollinator isolation via differential attraction or efficiency, and available data support a key direct role for pollinators in reducing gene flow between lineages via premating effects (e.g. Schemske and Bradshaw 1999; Hoballah et al. 2007; Kessler et al. 2015; and reviewed in Lowry et al. 2008; Kay and Sargent 2009). Nonetheless, divergent floral traits could also have collateral effects on later postmating and postzygotic reproductive barriers via both direct and indirect mechanisms, although the strength and importance of these effects remains much less well understood.
Floral differences could have direct effects on postmating barriers, for instance by mediating mechanical isolation or extrinsic postzygotic isolation. In the first case, pollen from taxa with short styles is unable to grow to the ovary of taxa with longer styles due to a mechanical mismatch between style length of one species and the maximal pollen tube length of the other (e.g., Williams and Rouse 1988; Lee et al. 2008). In the second case, hybrids – once formed between species – can have unattractive or unmanageable floral trait combinations, thereby reducing the attraction of or handling ability by pollinators of either parental species (e.g. Schemske and Bradshaw 1999). These direct forms of postmating isolation have been shown to reduce gene flow between some species (e.g. Lee et al. 2008; Tong et al. 2016), although direct empirical evidence for extrinsic postzygotic isolation due to floral traits is comparatively rare.
In contrast to direct effects on postmating and extrinsic postzygotic barriers, floral divergence might also contribute indirectly to additional isolating barriers. For instance, if genes underlying floral trait transitions have pleiotropic roles in other aspects of reproductive or morphological development, divergence in these mechanisms could result in dysfunctional development – and therefore reduced viability or fertility – in hybrids (Smaczniak et al. 2012; Haak et al. 2014). The resulting Dobzhansky-Muller Incompatibilities (DMIs) in hybrids are postzygotic barriers that increase isolation by preventing hybrid individuals from acting as conduits for gene flow across species boundaries. Specific developmental loci—such as the MIKCC-type MADS-box genes and their targets—are known to have functional roles in both flower and reproductive development in angiosperms (Smaczniak et al. 2012; Dreni and Zhang 2016), and reduced viability and fertility of hybrids has been observed in numerous plant groups (e.g. Moyle and Nakazato 2008; Oneal et al. 2016; reviewed in Baack et al. 2015). However whether such barriers are commonly due to pleiotropic consequences of floral trait differences between parental taxa remains unknown. Indeed, there is as yet no consensus about the mechanism(s) ultimately driving the association between species richness and floral diversity (Armbruster 2014), and floral trait variation could contribute to the evolution of reproductive isolating barriers via such effects on postmating and postzygotic stages of reproductive isolation.
In addition to specific components of mating trait divergence directly or indirectly driving the evolution of postmating and postzygotic isolation, other factors such as geographical or ecological context, as well as general genomic differentiation over time, could be important determinants of the type and strength of such barriers. For instance, differential adaption to spatially varying environmental factors could pleiotropically contribute to postzygotic barriers via DMIs (Coyne and Orr 2004) so that the strength of isolation scales with environmental divergence. Alternatively, geographical sympatry with other closely related species can directly select for increased premating isolation to reduce the frequency of low fitness matings between sympatric heterospecifics (i.e. reinforcement; Coyne and Orr 1989; Mendelson 2003; Hopkins 2013). Postmating and postzygotic barriers may also accumulate as the consequence of more general genomic differentiation between lineages over time. The resulting roughly monotonic increase in the strength of some stages of RI with increasing genetic distance (evolutionary time) has been observed in both animal (Coyne and Orr 1997; Presgraves 2002) and plant (Moyle et al. 2004; Scopece et al. 2007, 2008; Pinheiro et al. 2015) groups (reviewed in Coyne and Orr 2004; Baack et al. 2015).
One general approach to tease apart the relative importance of these effects is to examine multiple axes of divergence among a common set of taxa, so that their joint and individual effects on the accumulation of reproductive isolation can be simultaneously evaluated. For instance, pairwise data on genetic and geographical distance between numerous closely related species pairs, in conjunction with divergence in specific trait values and ecological factors, can be used to assess which axes of divergence are most strongly associated with the observed strength of reproductive isolation at one or more stages among these species pairs. While these kinds of comparative approaches have been used to identify key factors contributing to the evolution of reproductive isolation (Jewell et al. 2012; Martin and Mendelson 2016, and see above), they also have some limitations. Present methods do not allow inclusion of multiple potential predictors within the same statistical model, preventing direct assessment of their relative contributions to the strength of isolation. Further, compatibility data are typically non-independent, as the same species are used in multiple comparisons, but current methods to account for this non-independence also have shortcomings. Mantel tests (Mantel 1967) can potentially have elevated Type I error rates (Legendre 2000; Harmon and Glor 2010), whereas Phylogenetically-Independent Contrasts (PICs, Felsenstein 1985), which account for phylogenetic structure among included species, can have reduced power. To better understand which factors best explain patterns of isolation, a more robust statistical approach would allow multiple predictors (including both continuous and categorical variables), as well as account for phylogenetic structure among included species. Here we implement a linear mixed modeling approach that includes phylogenetic structure as a random effect matrix (Castillo 2016) to overcome several of these limitations of current methods.
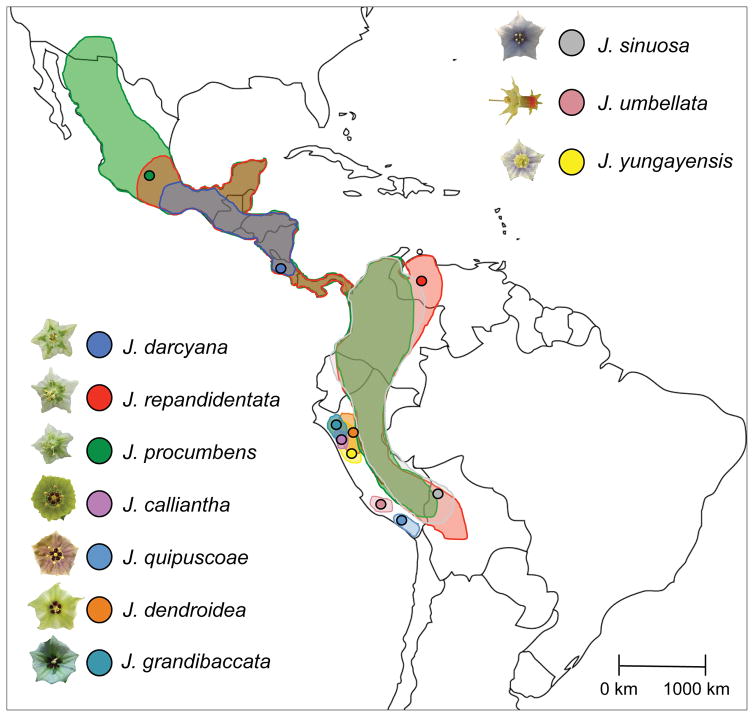
We examined the strength and accumulation of postmating reproductive isolation among 10 species in a plant genus with extensive floral diversity: Jaltomata (Solanaceae). Notably, this floral variation – including novel variation in petal and nectar color, overall size, and shape of floral organs (Figure 1), appears to have arisen within the last 5 MY (Sarkinen et al. 2013), likely in response to pollinator preferences (e.g. Fenster et al. 2004; and see Discussion). Moreover, these traits appear to be highly phylogenetically labile, indicating multiple independent transitions to similar floral forms (Miller et al. 2011). Jaltomata therefore provides an excellent opportunity to examine whether floral diversification influences the strength of postmating stages of reproductive isolation among multiple closely related species, while simultaneously considering genetic, geographical, and ecological relationships. In this study, we quantified reproductive isolation (RI) at up to 9 reproductive barriers (1 postmating and 8 intrinsic postzygotic) among these 10 species, and characterized floral trait variation and pairwise genetic, geographic, and ecological distance. Overall, our goals were: 1) to determine the relative contributions of floral trait divergence, geographical context/spatial distance, ecological divergence, and genetic distance to observed levels of RI between our species pairs, and, 2) to specifically assess evidence for the collateral (pleiotropic) effects of floral divergence on the strength of postmating and intrinsic postzygotic isolation barriers, in this florally diverse system.
Figure 1.
Jaltomata is native to Central and South America, with ranges for the 10 species included in this study shown, along with collection locations (filled circles) and representative flowers.
Materials and Methods
Study system
Jaltomata (Schlechtendal; Solanaceae) is the sister genus to the large and economically important Solanum (Olmstead et al. 2008; Sarkinen et al. 2013), and includes approximately 60–80 species distributed from the Southwestern United States to the Andes in South America, in addition to several species endemic to the Greater Antilles and the Galapagos Islands (Miller et al. 2011; T. Mione, pers. comm.). Although highly florally diverse, species exhibit one of three general floral morphologies: rotate (calyx and corolla are flat), campanulate (corolla is at least partially fused and shaped like a bell or cup), or tubular (corolla is mostly fused into a tube). Nectar color also ranges from essentially colorless to deep red, and may function as an honest signal to pollinators (Hansen et al. 2007). Current phylogenetic treatment of the genus (Miller et al. 2011; M. Wu, J.L. Kostyun, and L.C. Moyle, unpub.) indicates these variable floral traits are also highly evolutionarily labile, with evidence for multiple independent transitions among rotate, campanulate, and tubular floral forms. Therefore, species pairs can have similar floral forms either via shared ancestry, or via evolutionary convergence.
Species of Jaltomata also live in a variety of habitats, including tropical rainforests, rocky foothills, and lomas formations (patches of vegetation within desert supplied by fog). Although many Jaltomata species are sympatric, naturally occurring hybrids have yet to be observed (T. Mione, pers. comm.), suggesting the presence of reproductive barriers that reduce or prevent their formation. All examined Jaltomata species are self-compatible (Mione 1992; J.L. Kostyun and T. Mione, unpub.), unlike most other genera within the Solanaceae that exhibit genetically-determined self-incompatibility in some or all species (Goldberg et al. 2010).
Quantifying Floral Trait Variation
Wild-collected seeds from 10 Jaltomata species (Figure 1; Supp. Table 1) were germinated and plants cultivated under standard greenhouse conditions in our university research greenhouse. To characterize floral trait variation, we measured 15 floral traits on at least 3 individuals per species: 12 morphological, 2 color (petal and nectar), and 1 physiological (nectar volume). Our morphological traits were inflorescence size (number of flowers/buds), calyx diameter, sepal length, corolla diameter, corolla depth, corolla fusion, petal length, stamen length, anther length, ovary diameter, style length, and herkogamy (Supp. Figure 1), which were measured with a hand-held caliper. Nectar volume per flower was measured to the nearest 1μL using a pipette. Petal and nectar color were quantified from digital photographs (Kendal et al. 2013; Garcia et al. 2014): dissected petals and nectar drops were photographed on a standard background along with white and black color standards. Light conditions were standardized for all images using RAW Therapee (RAW Therapee Development Team 2012), and color space attributes were measured in ImageJ (Schneider et al. 2012). Because RGB (red-green-blue) color attributes are device-dependent, color values were also converted into device-independent L*a*b color attributes, using the ImageJ Color Space Converter plugin (Schwartzwald 2012) (Supp. Table 2).
We used two approaches to characterize floral trait differences for each species pair. First, species pairs were categorized based on whether they had the same general floral form (i.e. rotate, campanulate, or tubular) or not. Pairs with different general floral forms were categorized as “Divergent” (n=29 pairs), and pairs with the same general floral form were categorized as “Similar” (n=15 pairs). Within this second class, “Similar” species pairs were further classified based on current inferred phylogenetic relationships as “Shared” (share morphology from a common ancestor, n=5 pairs) vs. “Convergent” (similar morphology, but from different origins, n=10 pairs) (Supp. Table 3). Because different alleles may be responsible for phenotypically convergent floral traits, crosses involving these two different classes of ‘Similar” species pairs might be expected to have different outcomes for the expression of postmating (especially postzygotic) species barriers.
Second, we generated a composite quantitative estimate of pairwise multi-trait floral divergence (‘floral distance’) between species. We first used Principle Component Analysis (PCA; R statistical environment, R Core Team 2005) to summarize differences in floral trait axes explaining the relevant variation among species, as well as to standardize variance across traits (especially those with different units of measurement e.g. corolla diameter vs. nectar volume vs. color attributes). Then we calculated the pairwise Euclidean distance between species means in PC space for all species pairs. For comparison, we also calculated a separate ‘morphological floral distance’ and ‘color floral distance’ (using PCA on just the relevant traits for these subsets of floral divergence; Supp. Table 3), because different alleles controlling floral morphology, may be more likely to contribute to intrinsic incompatibilities in hybrids, compared to floral color differences (Haak et al. 2014; D. Castillo 2016).
Genetic Distance and Phylogenetic Relationships
Pair-wise genetic distance among species was estimated using a set of 60 orthologous transcripts (expressed loci) assembled from RNA-seq data (M. Wu, J.L. Kostyun, and L.C. Moyle, unpub.). Two RNA libraries (vegetative and reproductive tissues) were sequenced from each species, except for J. procumbens and J. grandibaccata for which only vegetative RNA was obtained. The vegetative libraries contained pooled (equi-molar) RNA from roots, leaf buds, immature leaves, and mature leaves; the reproductive libraries contained pooled RNA from 3 floral bud stages, pistillate flowers, hermaphroditic (mature) flowers, pollinated flowers, and early fruit. Tissues were collected from the same developmental stage under equivalent conditions (i.e. same temperature, lighting, and watering regimes) across species.
To identify orthologous transcripts for estimating genetic distance, reproductive and vegetative transcripts were first pooled for each species, and then aligned to the domestic tomato genome (Tomato Genome Consortium 2012) via all-by-all blastn and MCL clustering following van Dongen and Abreu-Goodger (2012) and Yang and Smith (2014). The 60 loci chosen to characterize genetic distance here were distributed across all 12 chromosomes (5 per chromosome, based on assumed synteny with the tomato genome; Supp. Table 4) and met the following criteria: all 10 Jaltomata species expressed transcripts at that locus, and reads from all species were in the central 50% of transcript length distribution. A concatenated sequence alignment of all 60 transcripts was used to calculate Ks (number of synonymous substitutions per synonymous site) in PAML v4.8a (Yang 2007), with codeml parameters runmode = −2, seqtype = 1, and codonFreq = 2 (Supp. Table 5). This concatenated alignment was also used to reconstruct phylogenetic relationships among species, using maximum likelihood (GTRCAT model) in RAxML (Stamatakis 2014) with 100 bootstraps. Short-read sequence data are available in NCBI Short Read Archive (SRX2676115-SRX2676141; BioProject PRJNA380644).
Geographical and Ecological Distances
Collection locations (latitude and longitude) of the 10 Jaltomata accessions were mapped in Google Earth (Figure 1; Supp. Table 1), and pairwise geographical distances (km) among these locations were calculated using the online program Movable Type Scripts (Veness 2002–2015) (Supp. Table 6). Because geographic distance contains information about both physical separation and potential ecological differences between species, a separate ‘ecological distance’ was also calculated between lineages. Nineteen bioclimatic variables for species collection locations were obtained from the WorldClim database (Hijmans et al. 2005) and, similar to ‘floral distance’, ‘ecological distance’ was calculated by using PCA to summarize variation in these climatic variables among each collection location, followed by taking the pairwise Euclidean distance between species PC values (Supp. Table 6). For completeness, we also calculated a ‘species-average ecological distance’ using a parallel approach but based instead on averages of WorldClim variables extracted for all known collection locations within each of our focal species (T. Mione, pers. comm.; J.L. Kostyun, unpub.; Supp. Table 7). Our results did not differ between these alternative metrics and, as individual population locations are more directly characteristic of the specific environmental conditions of the genotypes we examined here, we report analyses using ‘ecological distance’ in the main text and ‘species-average ecological distance’ in supplementary material (Supp. Tables 10–11, 16). In addition, species pairs were classified as allopatric or sympatric; allopatric species pairs were those with no known range overlap, while sympatric were those where the accession collection location occurred within the other species known range (Supp. Table 6) (T. Mione and S. Leiva G., pers. comm; J.L. Kostyun, unpub.).
Quantifying Reproductive Isolation
We quantified up to 9 metrics of isolation (1 postmating prezygotic, and 8 intrinsic postzygotic (Table 1)), among all species in a full reciprocal diallel design, with at least 3 biological replicates per crossing direction per species combination for each relevant reproductive stage. For species pairs that produced viable F1 hybrids, hybrid fertility was scored on at least 3 adult F1 individuals.
Table 1.
Mean values of reproductive isolation at 9 examined barriers (1 postmating prezygotic [pollen-pistil], and 8 postzygotic) and 2 cumulative measures, along with their absolute and relative contributions to total isolation.
| Pollen-Pistil (n=42) | Fruit Set (n=44) | Fruit Size (n=33) | Seed Set (n=33) | Seed Viability (n=33) | F1 Pollen Viability (n=9) | F1 Fruit Set (n=9) | F1 Fruit Size (n=6) | F1 Seed Set (n=6) | Total Post-Zygotic (n=44) | Total (n=44) | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Mean RI | 0.070 | 0.682 | 0.429 | 0.449 | 0.879 | 0.322 | 0.388 | 0.205 | 0.408 | 0.971 | 0.971 |
| Mean Absolute Contribution to Total | 0.079 | 0.589± | 0.155 | 0.077 | 0.133 | 0.012 | −0.025 | −0.057 | 0.044 | 0.921 | 1.000 |
| Mean Relative Contribution to Total | 0.079 | 0.588± | 0.148 | 0.083 | 0.228 | 0.012 | −0.062 | −0.121 | 0.084 | 0.921 | 1.000 |
| Mean Asymmetry | 0.066* | 0.496*** | 0.463** | 0.562** | 0.153* | 0.057 | 0.292* | 0.506* | 0.168 | 0.064 | 0.064 |
Fruit Set has the highest absolute and relative contribution to total isolation (ANOVA, p< 0.00001; Tukey HSD p< 0.00001).
Sample sizes refer to number of species pairs. RI values between reciprocal crosses were often highly asymmetric:
p<0.05,
p<0.001;
p<0.0001 (one-sample t-test).
Controlled intra- and interspecific pollinations were performed among individuals, and successful fruit set (mature fruit produced over total pollinations), mature fruit size (average mass [g]), seed set per fruit, and viable seed set per fruit (based on the proportion that germinated in a subset of seeds), were scored. These data were used to calculate 4 metrics of intrinsic postzygotic compatibility (fruit set, fruit size, seed set, and seed viability). For species combinations that produced viable F1 hybrids, we calculated three additional metrics of F1 hybrid female fertility (successful fruit set, fruit size, and seed set per fruit, following back-cross pollinations with parental pollen) and one of F1 hybrid male fertility (proportional viable pollen grains). The latter was quantified using established protocols (Jewell et al. 2012); briefly, undehisced anthers from 3 flowers per individual were individually collected into aniline blue pollen stain, each sample was ground to release pollen, and pollen grains were counted and evaluated for viability (darkly stained cytoplasm) under an EVOS FL Digital Inverted Fluorescence Microscope (Fisher Scientific).
In species combinations that did not produce fruit in our initial inter-specific pollinations, we further assessed evidence for pollen-pistil compatibility by examining pollen tube growth within heterospecific styles using epi-fluorescence microscopy. Additional pollinations were performed on emasculated undehisced flowers, and styles were collected 24 hours following pollination (except J. umbellata styles, which were collected after 48 hours), as 24 hours is sufficient for intra-specific pollen to reach the ovary in all species (except J. umbellata, which requires ~48 hours). Pollinated styles were collected into 3:1 95% ethanol: glacial acetic acid, and stored at 4°C for at least 24 hours prior to analysis. Styles were then softened in 5M NaOH for 20 hours, stained with Aniline Blue Fluorochrome (BioSupplies) for 3.5 hours in the dark, and examined under the DAPI setting on an EVOS scope. Pollen-pistil compatibility was calculated as the ratio of pollen tube length to style length, following measurement in ImageJ (Schneider et al. 2012). For species pairs with evidence of fruit production from our initial crosses, pollen-pistil compatibility was scored as 1 (pollen tubes reach ovary).
Reproductive isolation (RI) was calculated for each species pair combination at each barrier, following Sobel and Chen (2014) (Supp. Table 8–9). This metric is similar to previous measures (e.g. RI = 1- inter-specific value/intra-specific value; Coyne and Orr 1989), but ranges from −1 to 1 (and therefore allows cases where interspecific success exceeds intraspecific success), in addition to better allowing comparison across reproductive barriers and study systems. All results reported below use these RI values, however for completeness we provide equivalent analyses based on RI values calculated following Coyne and Orr (1989), in supplementary information (Supp. Tables 10–11). Note that our qualitative inferences from these alternative approaches do not differ. Because reproductive barriers are often asymmetric (Tiffin et al. 2001; Turelli and Moyle 2007), RI values were calculated separately for each direction of the cross, and also averaged to produce a joint value for each species pair. In instances where RI could only be measured in one direction of the cross, this value was used as the joint value. Following Sobel and Chen (2014), the cumulative strength of each barrier (which takes into account the effect of earlier-acting barriers on how much later-acting barriers can further limit gene flow), total cumulative isolation of the 9 quantified barriers, absolute vs. relative (i.e. cumulative effect/total isolation) contributions of each barrier to estimated isolation, were calculated for each species pair (Supp. Table 12).
Statistical Analyses
Comparisons among reproductive barriers
We compared the relative strength of different barriers across species pairs using an ANOVA, followed by Tukey’s HSD. Following Sobel and Chen (2014, and above), we used the absolute sequential contribution of each barrier in order to take into account the effect of earlier-acting barriers on later-acting ones. To assess asymmetry in the strength of isolation between reciprocal crosses, the magnitude (i.e. absolute value) of their difference was calculated for each barrier for each species pair (Supp. Table 13). A one-sample t-test (null: differences between reciprocals are not different from 0) was used to determine whether individual barriers were significantly asymmetric across species pairs. Under some speciation models, the magnitude of isolation asymmetry between reciprocal species combinations can vary depending upon time since divergence (Turelli and Moyle 2007), so we also evaluated the relationship between magnitude of asymmetry and genetic distance, using linear regression (Supp. Table 14). Because RI values among developmentally related reproductive stages (e.g. fruit size and seed number) might be correlated, we assessed the magnitude of correlations among RI values at all examined stages (Supp. Table 15). Note that because fruit size and seed set per fruit were highly correlated, we calculated Total RI and Total Postzygotic RI and performed all analyses with fruit size both included and excluded (Supp. Tables 10, 11, 14, 16, 17); results of the alternative analyses were qualitatively indistinguishable so only analyses including fruit size are reported in the Results. All analyses were run within the R statistical environment (R Core Team 2005).
Relationship between RI and other metrics of divergence
In conventional comparative analyses, the relationship between RI values and other metrics of pairwise divergence are assessed using Mantel tests (e.g. Jewell et al. 2012) (matrix regression, Mantel 1967), which were designed to account for non-independence among pair-wise data. However, these tests have both statistical and inferential limitations: under some conditions they produce elevated Type I errors (Legendre 2000; Harmon and Glor 2010), and they can fail to appropriately account for autocorrelation within data (e.g. phylogenetic signal) (Guillot and Rousset 2013). In addition, they cannot appropriately accommodate categorical predictors (such as allopatric vs. sympatric range), including those with more than 2 levels (such as different floral divergence categories), and they do not support more than one potential predictor within a single model, preventing robust simultaneous assessment of the relative contributions of multiple factors to RI values. Therefore, to assess the relationship between RI values and potential factors associated with their accumulation (genetic, geographical, ecological, and floral distances), we used a linear mixed model approach which allows inclusion of categorical predictors and more than one predictor in a single model, while accounting for phylogenetic structure among species pairs by specifying this as a random effect matrix (Castillo 2016). Specifically, we generated a standardized matrix of ranked relatedness based on the estimated topology of species relationships (see Figure 2); although this topology was inferred from the same loci as our genetic distance metric (Supp. Table 4), our relatedness matrix solely specifies relative ranking of shared ancestry but does not include data on branch lengths (Supp. Table 5).
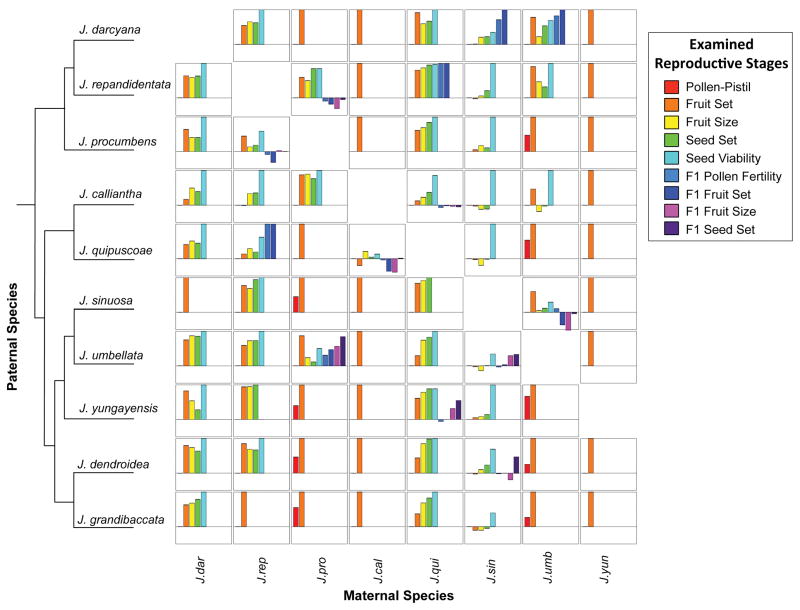
Figure 2.
Reproductive isolation for examined species combinations, across 9 postmating stages. Columns represent species acting as the mother while rows represent species acting as the father, arranged in phylogenetic order as shown in the cladogram. Cladogram based on the 60 transcripts used to estimate genetic distance (see Methods). Within each cell, different barriers are arranged along the x-axis, and RI values ranging from −0.5 to 1 along the y-axis.
These models were applied in two parallel analyses: First, we analyzed our data using separate RI values from each direction of reciprocal cross (where maternal species:paternal species interaction was also included as a random effect); and second, we analyzed species pair average RI values (averaged between both directions of the cross). Starting with the least complex model (i.e. RI ~ (1|random effect)), we determined the model of best fit for each barrier by step-wise adding potential predictors and comparing these nested models with likelihood ratio tests. All linear mixed model analyses were performed within the R package nlme (Pinheiro et al. 2016). For completeness, and to facilitate comparison with other prior comparative analyses, we also analyzed our data with conventional Mantel tests, with significance levels determined based on 10000 permutations, as implemented in the R package vegan (Oksanen et al. 2016); results for these are reported in supplementary material (Supp. Table 16).
Results
Floral traits are highly divergent among species, although not associated with genetic, geographic, or ecological measures of divergence
As expected, overall floral and morphological distances were associated with our qualitative floral morphology categories: “Divergent” species pairs had significantly higher trait–based distance than either “Shared” or “Convergent” pairs (F=36.06; F=38.72, both p < 0.00001; Tukey HSD, p < 0.00001), while “Convergent” and “Shared” pairs were not significantly different (Tukey HSD, p < 0.066). Nonetheless, floral trait differentiation among species was not associated with genetic, geographic or ecological distance, with one exception (a significant positive correlation between genetic distance and morphological floral distance (Mantel’s r = 0.251, p = 0.048; Supp. Table 16)). Genetic distance also did not significantly differ between qualitative floral morphology categories, for either “Similar” vs. “Divergent” (t = 0.233, p = 0.817), or “Shared” vs. “Convergent” vs. “Divergent” (F = 1.282, p = 0.288) comparisons, and neither did geographic distance (“Similar” vs. “Divergent” (t = 0.279, p = 0.782); “Shared” vs. “Convergent” vs. “Divergent” (F = 0.113, p = 0.893)).
Jaltomata species pairs exhibit a broad range of isolating barriers, that are often asymmetric
Reproductive isolation was quantified for a total of 74 unique species combinations (44 species pairs) for up to 9 postmating isolation metrics (Figure 2; Supp. Tables 8–9), in addition to two cumulative estimates (Table 1; Supp. Table 12). Levels of RI varied greatly among species pairs, ranging from species isolated by strong pollen-pistil barriers through to pairs that successfully produced backcross fruits and seeds. RI values among some developmentally associated stages (e.g. fruit size and seed set) were significantly correlated, while values among unrelated measures (e.g. seed viability and F1 hybrid pollen viability) were not (Supp. Table 15). F1 pollen viability and F1 fruit set were also significantly correlated, indicating that both pollen and seed fertility problems have accumulated among some of the relatively closely related species examined here.
As the majority of species combinations (44 of 74) produced at least some fruit from pollinations, most of the species isolating barriers we detected appear to be postzygotically acting. Of the 30 species combinations that did not produce fruit from interspecific pollinations, we observed pollen tubes successfully reached the ovary in 12 combinations, consistent with other later acting reproductive barriers, such as incompatible pollen-ovule signaling (postmating prezygotic RI) or very early seed failure (postzygotic RI), preventing successful fruit production. Because at least some of these crosses produced noticeably swollen ovaries that then failed to develop, the barrier(s) are more likely to be early acting postzygotic. The operation of pollen-pistil barriers in nine additional combinations, involving a single maternal species (J. yungayensis), could not be directly tested because this species stopped flowering during the course of the experiment. We directly observed that pollen tubes did not reach the ovary in the remaining 9 combinations, all of which involve just two maternal species (J. procumbens and J. umbellata) (Figure 2; Supp. Table 8; and see further below). Note that because pollen-pistil isolation was only directly evaluated in crosses that failed to produce fruits, partial or incomplete isolation at this stage—for example, that reduced the frequency with which fruits were formed—may be present in other untested species pairs, especially those with low levels of fruit production. Therefore our assessment of pollen-pistil isolation can be considered a minimum quantitative estimate of this species barrier.
Of the 44 species combinations (33 pairs) that produced fruit, 14 (9 pairs) produced at least some viable F1 hybrid seeds (Figure 2). All F1 hybrids were vegetatively robust and produced flowers, although in four cases these flowers were sterile (produced non-functional gametes). Together, the examined 9 postmating metrics contribute 38 – 100% reproductive isolation among species pairs (ranging from −55 – 100% among species combinations; Supp. Tables 9 and 12). Of these barriers, RI at the fruit set stage (i.e. the proportion of successful fruit following pollination) was the strongest across species pairs (F=48.09, p < 0.00001; Tukey HSD, p < 0.00001; Table 1; Figure 2; Supp. Figure 2).
In addition to variation in the strength of isolating barriers among species pairs, RI was also frequently asymmetric within individual species pairs (i.e. differed depending on the direction of the cross) (Figure 3; Supp. Table 13). When considering all species pairs, reproductive isolation values were significantly asymmetric for 7 of the 11 examined isolation metrics (9 measured traits and 2 cumulative isolation estimates; Table 1). Nonetheless, the magnitude of asymmetry in RI was not significantly associated with genetic distance (as a proxy of divergence time) for any of our reproductive barriers, including total RI (Supp. Table 14).
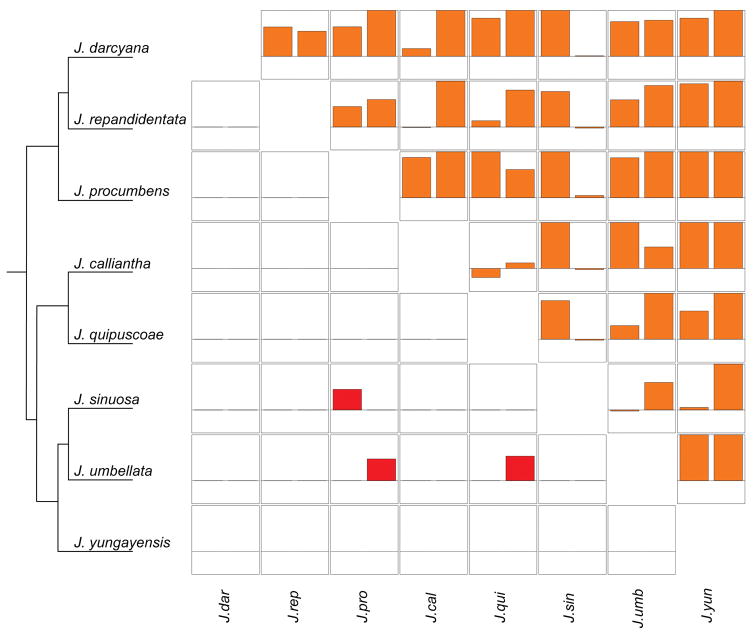
Figure 3.
Reproductive isolation in both reciprocal directions for two focal stages, Pollen-Pistil (bottom diagonal) and Fruit Set (top diagonal). Each cell represents a single inter-specific pair with RI values from Species A × Species B and Species B × Species A plotted adjacently. The y-axis represents RI values ranging from −0.5 to 1. Species are arranged phylogenetically following Figure 2, although J. dendroidea and J. grandibaccata are not included because they were primarily used as pollen donors (therefore asymmetry could not be calculated).
Mechanical isolation is not a widespread species barrier
Differences in style length specifically (rather than overall floral differences) could contribute directly to postmating mechanical isolation via incomplete pollen tube growth. However our data support this as a potential mechanism of postmating prezygotic isolation for only one species. Of the species pairs that were directly tested, we detected pollen-pistil isolation in two species when acting as the maternal parent, J. procumbens and J. umbellata (Figure 2; and see above), which are the shortest-styled and the longest-styled species, respectively, in our dataset (Supp. Table 2). It is therefore likely that mechanical isolation contributes to observed RI in the latter case (J. umbellata); indeed, of the interspecific pollen that did not reach the ovary of J. umbellata, pollen from species with shorter styles tended to result in higher levels of pollen-pistil RI, although this was not significant (F = 2.86, p = 0.094) (Supp. Figure 3). Although mechanical isolation likely contributes to pollen-pistil RI in this species, it is not a common explanatory mechanism for RI among our Jaltomata species pairs more broadly. Indeed, neither linear mixed models nor Mantel tests on separate reciprocal RI values (which account for the direction of style length difference), support significant associations between style length difference and level of pollen-pistil RI (Table 2; Supp. Table 16). Note that mechanical isolation also does not appear to be contributing to quantitative reductions in fruit set from crosses for which we did not directly assess pollen-pistil interactions; in particular, fruit set RI was not associated with style length difference between species (Mantel r = 0.084, p = 0.372; lmm p = 0.952), suggesting that pollen-pistil mechanical isolation is not driving patterns detected for this later stage of isolation.
Table 2.
Linear Mixed Model results for examining relationships between RI values and predictors across species pairs, for each barrier.
| Pollen-Pistil RI (n=65; 42) | Fruit Set RI (n=74; 44) | Fruit Size RI (n=44; 33) | Seed Set RI (n=44; 33) | Seed Viability RI (n=44; 33) | F1 Pollen Viability RI (n=13; 9) | F1 Fruit Set RI (n=13; 9) | F1 Fruit Size RI (n=9; 6) | F1 Seed Set RI (n=9; 6) | Total Postzygotic RI (n=74; 44) | Total RI (n=74; 44) | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Separate Reciprocal Data | |||||||||||
| Fixed Effects | |||||||||||
| Intercept | 0.059 (0.038) | 0.475 (0.118) | 0.090 (0.132) | 0.120 (0.134) | 0.895 (0.066) | −0.047 (0.137) | −0.372 (0.118) | −0.347 (0.196) | 0.365 (0.162) | 0.664 (0.127) | 0.664 (0.127) |
| Genetic Distance | 7.445 (4.676) | 13.905** (3.828) | 11.928* (4.824) | 37.580* (6.240) | 54.432*** (6.204) | 57.790 (24.785) | 12.868* (5.214) | 12.868* (5.214) | |||
| Geograph. Distance | −0.213* (0.047) | −0.246* (0.072) | −0.267 (0.175) | ||||||||
| Eco. Distance | |||||||||||
| Morph. Distance | |||||||||||
| Style Distance | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | |
| Random Effects | |||||||||||
| Phylogenetic Structure | 0.000 (0.000) | 0.003 (0.057) | 0.000 (0.000) | 0.000 (0.000) | 0.000 (0.019) | 0.000 (0.000) | 0.000 (0.000) | 0.000 (0.000) | 0.012 (0.034) | 0.028 (0.167) | 0.028 (0.167) |
| Mother:Father | 0.009 (0.096) | 0.139 (0.371) | 0.043 (0.208) | 0.116 (0.341) | 0.026 (0.161) | 0.047 (0.217) | 0.000 (0.000) | 0.000 (0.000) | 0.000 (0.000) | 0.007 (0.084) | 0.007 (0.084) |
| Pair Average Data | |||||||||||
| Fixed Effects | |||||||||||
| Intercept | 0.079 (0.026) | 0.315 (0.137) | 0.022 (0.110) | 0.102 (0.128) | 0.591 (0.150) | −0.206 (0.137) | −0.345 (0.157) | ND | ND | 0.618 (0.129) | 0.618 (0.129) |
| Genetic Distance | 5.932 (3.739) | 14.958** (3.929) | 12.520* (4.589) | 41.772* (7.276) | 51.099* (8.201) | 15.325* (5.400) | 15.325* (5.400) | ||||
| Geograph. Distance | −0.199* (0.078) | −0.224* (0.083) | |||||||||
| Eco. Distance | |||||||||||
| Morph. Distance | 0.045 (0.024) | ||||||||||
| Style Distance | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | |
| Random Effects | |||||||||||
| Phylogenetic Structure | 0.000 (0.000) | 0.000 (0.000) | 0.000 (0.000) | 0.000 (0.000) | 0.032 (0.178) | 0.004 (0.066) | 0.011 (0.105) | ND | ND | 0.034 (0.185) | 0.034 (0.185) |
Samples sizes indicated for each barrier: number of unique species pair combinations (i.e. separate reciprocals), followed by number of species pairs (i.e. pair averages). Shown coefficients for each RI barrier were included in the model of best fit (based on likelihood ratio tests), standard error in parentheses. Bold indicates significant contribution of predictor:
p<0.05,
p<0.001;
p<0.0001;
bold without asterisks indicates marginally significant p<0.07. (ND) not determined, as the sample size was too small to run these analyses.
Accumulation of intrinsic postzygotic reproductive isolation is best explained by overall genetic distance, rather than floral differentiation or geographical and ecological factors
We found little evidence for a general association between quantitative divergence in floral traits and the strength of intrinsic postzygotic isolation. Floral morphological distance was marginally significant as a predictor in the linear mixed model of best fit for fruit set RI (p < 0.067; Table 2) in the pair averaged dataset. A similar result was obtained with a partial Mantel test controlling for genetic distance, of this relationship (Supp. Table 16). However, neither quantitative measures of floral distance nor qualitative categories were significant predictors for any other barriers, regardless of statistical approach.
In contrast, analyses support a strong association between genetic distance and most of our metrics of postzygotic RI, based on both linear mixed models and Mantel tests (Table 2; Supp. Table 16). Indeed, genetic distance is the strongest predictor of overall patterns of intrinsic postzygotic isolation, regardless of specific statistical approach, or whether analyses were based on separate reciprocal RI values or on species pair average RI values. Consistent with this, genetic distance was also significantly associated with total RI based on both approaches, and with both reciprocal and averaged RI datasets. In comparison, genetic distance was clearly not associated with pollen-pistil RI, based on either of the statistical approaches or datasets (Table 2; Supp. Table 16).
Compared with genetic distance, geographical or ecological factors explained little of the observed variation in isolation metrics, with the exception that both genetic and geographic distances were significantly associated with F1 pollen viability and F1 fruit set, based on linear mixed model analyses (Table 2). No significant associations between RI values and either geographic or ecological distance were detected with partial Mantel tests, once genetic distance was controlled for (Supp. Table 16). Moreover, the categorical predictor of species ranges (allopatric vs. sympatric) was not significant for any of the examined RI barriers in linear mixed models. Consistent with this, the strength of isolation did not differ between allopatric and sympatric species pairs, for any of the examined barriers, including total isolation (Tukey HSD, p = 0.99).
Discussion
Differences in showy phenotypic traits are often thought to contribute disproportionately to reproductive isolation between species via their effects on premating barriers. These traits could also contribute to the evolution of postmating barriers, however this potential mechanistic relationship remains comparatively less studied (Haak et al. 2014; Baack et al. 2015). Here we assessed the presence and strength of multiple postmating reproductive barriers among 10 florally diverse Jaltomata species (44 pairs). Our analysis revealed that moderate to strong, often asymmetric postmating prezygotic and/or intrinsic postzygotic reproductive barriers isolate all examined species pairs. Moreover, the predominant predictor of intrinsic postzygotic isolation accumulation was not floral differentiation, or geographical or ecological context, but instead overall genome-wide divergence. Together, these results suggest the rapid accumulation of multiple postmating and intrinsic postzygotic barriers over a relatively brief evolutionary period in this group but, interestingly, little evidence for a direct or indirect role of floral trait divergence in the erection of the specific isolation barriers examined here.
Strong, diverse, and asymmetric postmating and postzygotic reproductive isolation isolate these closely related species
All examined species pairs showed evidence of postmating or intrinsic postzygotic isolation, with substantial diversity in the type and magnitude of RI separating individual species pairs, suggesting that multiple mechanisms contribute to reproductive isolation in this system. Moreover, most species pairs (37 pairs) appear to be completely isolated when considering the cumulative effects of all the examined barriers. For the remaining 7 species pairs, total estimated isolation ranged from ~40 to 99.9%. This is, of course, a minimum estimate of total isolation as these species pairs likely have additional prezygotic or extrinsic postzygotic reproductive barriers that were not examined here. Pre-mating or extrinsic postzygotic isolation via differential pollinator behavior has not yet been directly experimentally evaluated in this group. However, both opportunistic field observations of different pollinators visiting species with divergent floral traits (e.g. hymenopterans vs. hummingbirds; T. Mione, pers. comm.; J.L. Kostyun, unpub.), and floral trait suites that are consistent with distinct pollinator syndromes (i.e. Fenster et al. 2004), suggest the presence of such barriers among some species pairs, as well as the potential for pollinator-mediated extrinsic postzygotic isolation. Regardless, the moderate to very strong postmating and intrinsic postzygotic reproductive barriers identified here appear to have evolved relatively recently (i.e. <5 MY; Sarkinen et al. 2013), and could act as effective mechanisms to reduce gene flow between extant lineages, over and above the direct effects of floral trait divergence on pollinator mediated isolation.
We also found that isolation was often asymmetric depending on the direction of the cross (Figures 2–3; Supp. Table 13). Such asymmetry in RI appears to be common across taxa (including both animals and plants), and is likely caused by nuclear-cytoplasmic interactions in hybrids themselves, or by unidirectional DMIs which can arise during imbalanced interactions of maternal and paternal genomes at earlier reproductive or developmental stages (i.e. pollen-pistil interactions or seed development in angiosperms) (Turelli and Moyle 2007; Lafon-Placette and Köhler 2016). While additional experiments would be needed to dissect the specific nature and relative contributions of bidirectional and unidirectional DMIs in these cases, we did find that RI at these stages was significantly asymmetric across Jaltomata species pairs (Table 1), consistent with some contribution of unidirectional DMIs to these observed patterns of asymmetry. The fact that these barriers were often strong even among closely related species is also consistent with the rapid observed accumulation of improper parental genome dosage in some hybrids, especially during seed development (Lafon-Placette and Köhler 2016).
Limited evidence for a strong role of floral trait divergence in postmating prezygotic isolation
Even though exaggerated floral trait (i.e. style length) divergence could directly elevate postmating prezygotic isolation, we detected direct evidence for pollen-pistil isolation in only two species; in one instance, pollen-pistil isolation appears to follow simple mechanical isolation, while in the other, evidence suggests that pollen-pistil incompatibility is due to other molecular or physiological mechanisms. Note that this potentially underestimates the frequency of quantitative or partial pollen-pistil barriers, especially in species pairs with low fruit set, as we only directly assessed pollen-pistil isolation in species pairs that did not produce fruit. However our data do indicate that complete pollen-pistil RI does not appear to be a frequent direct pleiotropic effect of rapid floral divergence among the species pairs examined here.
For one of our species with frequent pollen-pistil isolation, patterns are consistent with simple mechanical isolation: J. umbellata has the longest style of the examined species and, in interspecific crosses where pollen does not reach the J. umbellata ovary, we found a roughly linear relationship between parental style length difference and how far pollen travels down the J. umbellata style after 48 hours (Supp. Figure 3). These observations indicate mismatched style length/pollen tube growth differences between species (i.e. mechanical isolation) likely explain the patterns of postmating isolation observed for this species, suggesting a direct role for floral divergence (i.e. lengthening of the style) in the expression of postmating isolation in this case.
In contrast, several lines of evidence suggest that the other clear instance of pollen-pistil RI involves molecular mechanisms of pollen-pistil incompatibility, rather than mechanical isolation. In our dataset, J. procumbens has the shortest style, yet we observed that it rejected pollen from most other species when used as the pistil parent. In contrast, in reciprocal crosses its pollen tubes reach the ovary of other species, indicating that the pollen-pistil barrier involves pistil-side rejection specifically by J. procumbens, rather than generalized bilateral ‘incongruity’ between heterospecific pollen and pistil signals (de Nettancourt 2001). Unilateral heterospecific pollen rejection is a common postmating reproductive barrier in several plant groups, including other Solanaceous systems (Lee et al. 2008; Bedinger at al. 2011); however, most of these instances appear to involve proteins that also function in self-incompatibility rejection mechanisms (Murfett et al. 1996; McClure et al. 2011; Tovar-Mendez et al. 2014), such that pistils of SI species reject pollen from self-compatible (SC) species but the reciprocal cross is successful. Because all examined Jaltomata species including J. procumbens are self-compatible (J.L. Kostyun and T. Mione, unpub.), unilateral pollen-pistil barriers are unlikely to be related to factors involved in self-incompatibility, although they might involve S-RNase independent mechanisms which have also been described in the Solanaceae (Bedinger et al. 2011). Regardless, it is possible that the lack of functional SI within Jaltomata might explain the relatively modest overall contribution of strong pollen-pistil barriers to reproductive isolation in this group, despite being widespread in other related genera.
Intrinsic postzygotic isolation is best explained by genome-wide genetic distance rather than floral divergence, geographic context, or ecological differences
Another goal of this study was to examine evidence for indirect effects of recent rapid floral divergence on the expression of intrinsic postzygotic isolation. Interestingly, we found limited evidence for a strong association between the strength of intrinsic postzygotic isolation and the magnitude of floral trait divergence between species, although our analysis of floral variation confirmed that these traits are highly labile among species, including highly divergent floral traits between very closely related species. Nonetheless, we found that levels of intrinsic postzygotic isolation among examined Jaltomata species pairs are by far best explained by overall genome-wide genetic distance. This relationship was significant for most of the examined postzygotic barriers, and by both statistical approaches (Table 2; Supp. Table 16), suggesting a monotonic or roughly ‘clock-like’ accumulation of isolation over evolutionary divergence. (Alternatively, the strength of postmating and postzygotic barriers could itself determine the extent of genome-wide divergence although, in the absence of evidence for substantial recent or ongoing gene flow between species, this explanation for the observed relationship is less likely.) A positive relationship between RI and genetic distance has been found in numerous other studies across diverse taxa (Coyne and Orr 1989, 1997; Presgraves 2002; Mendelson 2003; Moyle et al. 2004), indicating DMIs might frequently accumulate in concert with genome-wide evolutionary divergence.
Nonetheless, we detected one suggestive instance where intrinsic postzygotic isolation might be related to floral trait differences — an association between floral morphological distance and fruit set RI — which was significant based on partial Mantel tests controlling for genetic distance, and marginally so in linear mixed model analyses (p < 0.067) for averaged species pair values. This relationship is fairly weak, so interpreting its significance for species isolation requires caution. However, if this relationship were to hold with additional data from Jaltomata, it suggests that genetic changes underlying differences in floral morphological traits among species (for which multivariate floral distance is a proxy here) might exacerbate the failure of interspecific crosses to successfully produce fruit (the strongest barrier observed across species pairs; Table 1). We observed aborted fruits in many of these crosses, so that isolation at this stage is likely predominantly due to early seed failure. For hybrid inviability via seed failure to be mechanistically associated with floral trait differences between parents, divergent alleles controlling these floral differences would also need to function during seed development, and genetic differences in these loci would need to pleiotropically interfere with hybrid seed development. (The alternative—that these alleles are tightly linked to loci which produce genetic incompatibilities, e.g. Wright et al. 2013—is unlikely to lead to a general association between floral divergence across multiple species pairs.). There are a priori reasons to expect that these different reproductive stages are developmentally associated and therefore could influence each other pleiotropically. For example, the MIKCC-type MADS-box genes (which encode transcription factors) and their targets function in both flower and seed development (Smaczniak et al. 2012; Dreni and Zhang 2016). In addition, because many MADS-box proteins are obligate hetero-dimers (especially in core eudicots, such as Solanaceae; Bartlett et al. 2016), hybrids inheriting protein partners from different parents could experience functional disruption if those partners no longer dimerize. Further, these genes have undergone extensive duplication (Kim et al. 2004; Smacziak et al. 2012), and several studies have found functional divergence among paralogs (Sharma and Kramer 2013), including within other Solanaceous systems (de Martino et al. 2006; Geuten and Irish 2010).
Finally, while we cannot exclude the possible influence of other finer-scale ecological or geographical factors than those examined here, we observed little independent role for geographical or ecological (abiotic climatic) factors in the evolution of the isolating barriers examined here or, indeed, of floral trait divergence. Although genetic distance was strongly correlated with both geographic and ecological distances, partial Mantel tests and linear mixed models consistently supported genetic distance as the best explanatory predictor (Table 2; Supp. Table 16). Similarly, we also found no evidence that stronger postmating pollen-pistil barriers were associated with geographical sympatry. This is inconsistent with a strong role for direct selection to increase the strength of this barrier in response to detrimental heterospecific reproductive interactions (i.e. reinforcement). It remains possible that reinforcement could be operating on other prezygotic (especially premating) barriers not examined here (e.g. several sympatric species pairs differ in at least one floral trait that could contribute to pollinator discrimination); however, more generally, there was a trend for sympatric species pairs to have less diverged floral traits compared to allopatric pairs (lower floral distance for both morphological and color traits, including marginally significantly for morphological floral distance t = 2.212, p = 0.051). Therefore, in contrast to several studies in animals (Coyne and Orr 2004; Mendelson 2003), as well as some in angiosperms (Hopkins 2013), we did not find patterns of RI strongly consistent with reinforcement on pollen-pistil (or floral trait) isolation. This in turn suggests that reproductive interactions with heterospecifics (i.e. avoidance of heterospecific gene flow) is not the factor predominantly driving floral diversification in this group.
Conclusions
Although florally diverse species are typically thought to be maintained primarily by differential pollinator attraction (Kay and Sargent 2009; Moyle et al. 2014), we have demonstrated that Jaltomata species have numerous diverse (and often strong) postmating barriers. In two instances, these isolating barriers might be related to floral trait differences: pollen-pistil isolation via mechanical isolation in one species, and a positive (although weak) relationship between divergence in morphological floral traits and level of fruit set RI. However, our data generally suggest that intrinsic postzygotic isolation accumulates with overall genome-wide divergence, rather than being strongly shaped by geographic context, ecological differentiation, or rapid recent evolution of floral trait differences in this florally diverse plant group. In the latter case, this finding suggests that divergence in these mating traits has few strong pleiotropic effects on later reproductive compatibility between lineages, despite potential physiological, developmental, and genetic connections between these isolation stages.
Supplementary Material
Acknowledgments
We thank Dean Castillo for statistical advice, Meng Wu for assistance with genetic distance estimates, Jeremy Davis for assistance with WorldClim data extraction, members of the Moyle and Hahn labs for helpful feedback, undergraduate research assistants for help with data collection, IU Greenhouse staff for plant care, and Thomas Mione for kindly sharing seed material and unpublished data. This work was supported by the IU Biology Department, National Science Foundation Award (NSF DEB 1136707), National Science Foundation Graduate Research Fellowship Program (NSF DEB 1342962) and Doctoral Dissertation Improvement Grant (NSF DEB 1601078), and the National Institutes of Health Genetics, Cellular, and Molecular Sciences training grant (No. 5T32GM007757). Any opinions, findings, and conclusions or recommendations expressed in this material are those of the author(s) and do not necessarily reflect the views of the National Science Foundation or the National Institutes of Health. The authors declare no conflicts of interest.
Footnotes
Data archive:
Raw data are available in Dryad: http://dx.doi.org/10.5061/dryad.3vr00
Short-read Illumina data are available in NCBI Short Read Archive: SRX2676115-SRX2676141 (BioProject PRJNA380644)
Author Contributions
J.L.K and L.C.M. designed the study and experiments, J.L.K. conducted the experiments and analyzed the data, and J.L.K. and L.C.M. wrote the paper.
Literature Cited
- Armbruster WS. Floral specialization and angiosperm diversity: phenotypic divergence, fitness trade-offs and realized pollination accuracy. Aob Plants. 2014:6. doi: 10.1093/aobpla/plu003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baack E, Melo MC, Rieseberg LH, Ortiz-Barrientos D. The origins of reproductive isolation in plants. New Phytologist. 2015;207:968–984. doi: 10.1111/nph.13424. [DOI] [PubMed] [Google Scholar]
- Bartlett M, Thompson B, Brabazon H, Del Gizzi R, Zhang T, Whipple C. Evolutionary Dynamics of Floral Homeotic Transcription Factor Protein-Protein Interactions. Molecular Biology and Evolution. 2016;33:1486–1501. doi: 10.1093/molbev/msw031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillo DM. Factors contributing to the accumulation of reproductive isolation: a mixed model approach. bioRxiv. 2016 doi: 10.1002/ece3.3093. doi: http://dx.doi.org/10.1101/072264. [DOI] [PMC free article] [PubMed]
- Coyne JA, Orr HA. Patterns of speciation in Drosophila. Evolution. 1989;43:362–381. doi: 10.1111/j.1558-5646.1989.tb04233.x. [DOI] [PubMed] [Google Scholar]
- Coyne JA, Orr HA. “Patterns of speciation in Drosophila” revisited. Evolution. 1997;51:295–303. doi: 10.1111/j.1558-5646.1997.tb02412.x. [DOI] [PubMed] [Google Scholar]
- Coyne JA, Orr HA. Speciation. Sinauer Publishers; Sunderland, MA: 2004. [Google Scholar]
- de Martino G, Pan I, Emmanuel E, Levy A, Irish VF. Functional analyses of two tomato APETALA3 genes demonstrate diversification in their roles in regulating floral development. Plant Cell. 2006;18:1833–1845. doi: 10.1105/tpc.106.042978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Nettancourt D. Incompatibility and incongruity in wild and cultivated plants. Berlin: Springer; 2001. [Google Scholar]
- Dreni L, Zhang DB. Flower development: the evolutionary history and functions of the AGL6 subfamily MADS-box genes. Journal of Experimental Botany. 2016;67:1625–1638. doi: 10.1093/jxb/erw046. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. Phylogenies and the comparative method. American Naturalist. 1985;125:1–15. doi: 10.1086/703055. [DOI] [PubMed] [Google Scholar]
- Fenster CB, Armbruster WS, Wilson P, Dudash MR, Thomson JD. Pollination syndromes and floral specialization. Annual Review of Ecology Evolution and Systematics. 2004;35:375–403. [Google Scholar]
- Garcia JE, Greentree AD, Shrestha M, Dorin A, Dyer AG. Flower Colours through the Lens: Quantitative Measurement with Visible and Ultraviolet Digital Photography. Plos One. 2014:9. doi: 10.1371/journal.pone.0096646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geuten K, Irish V. Hidden Variability of Floral Homeotic B Genes in Solanaceae Provides a Molecular Basis for the Evolution of Novel Functions. Plant Cell. 2010;22:2562–2578. doi: 10.1105/tpc.110.076026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg EE, Kohn JR, Lande R, Robertson KA, Smith SA, Igic B. Species Selection Maintains Self-Incompatibility. Science. 2010;330:493–495. doi: 10.1126/science.1194513. [DOI] [PubMed] [Google Scholar]
- Guillot G, Rousset F. Dismantling the Mantel tests. Methods in Ecology and Evolution. 2013;4:336–344. [Google Scholar]
- Haak DC, Kostyun JL, Moyle LC. Merging Ecology and Genomics to Dissect Diversity in Wild Tomatoes and Their Relatives. In: Landry CR, AubinHorth N, editors. Ecological Genomics: Ecology and the Evolution of Genes and Genomes. 2014. pp. 273–298. [DOI] [PubMed] [Google Scholar]
- Hansen DM, Olesen JM, Mione T, Johnson SD, Muller CB. Coloured nectar: distribution, ecology, and evolution of an enigmatic floral trait. Biological Reviews. 2007;82:83–111. doi: 10.1111/j.1469-185X.2006.00005.x. [DOI] [PubMed] [Google Scholar]
- Harmon LJ, Glor RE. Poor statistical performance of the Mantel test in phylogenetic comparative analyses. Evolution. 2010;64:2173–2178. doi: 10.1111/j.1558-5646.2010.00973.x. [DOI] [PubMed] [Google Scholar]
- Higashi M, Takimoto G, Yamamura N. Sympatric speciation by sexual selection. Nature. 1999;402:523–526. doi: 10.1038/990087. [DOI] [PubMed] [Google Scholar]
- Hijmans RJ, Cameron SE, Parra JL, Jones PG, Jarvis A. Very high resolution interpolated climate surfaces for global land areas. International Journal of Climatology. 2005;25:1965–1978. [Google Scholar]
- Hoballah ME, Gubitz T, Stuurman J, Broger L, Barone M, Mandel T, Dell’Olivo A, Arnold M, Kuhlemeier C. Single gene-mediated shift in pollinator attraction in Petunia. Plant Cell. 2007;19:779–790. doi: 10.1105/tpc.106.048694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopkins R. Reinforcement in plants. New Phytologist. 2013;197:1095–1103. doi: 10.1111/nph.12119. [DOI] [PubMed] [Google Scholar]
- Jewell C, Papineau AD, Freyre R, Moyle LC. Patterns of reproductive isolation in Nolana (Chilean bellflower) Evolution. 2012;66:2628–2636. doi: 10.1111/j.1558-5646.2012.01607.x. [DOI] [PubMed] [Google Scholar]
- Kay KM, Sargent RD. The Role of Animal Pollination in Plant Speciation: Integrating Ecology, Geography, and Genetics. Annual Review of Ecology Evolution and Systematics 2009:637–656. [Google Scholar]
- Kendal D, Hauser CE, Garrard GE, Jellinek S, Giljohann KM, Moore JL. Quantifying Plant Colour and Colour Difference as Perceived by Humans Using Digital Images. Plos One. 2013;8 doi: 10.1371/journal.pone.0072296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler D, Kallenbach M, Diezel C, Rothe E, Murdock M, Baldwin IT. How scent and nectar influence floral antagonists and mutualists. Elife. 2015:4. doi: 10.7554/eLife.07641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim ST, Yoo MJ, Albert VA, Farris JS, Soltis PS, Soltis DE. Phylogeny and diversification of B-function MADS-box genes in angiosperms: Evolutionary and functional implications of a 260-million-year-old duplication. American Journal of Botany. 2004;91:2102–2118. doi: 10.3732/ajb.91.12.2102. [DOI] [PubMed] [Google Scholar]
- Lafon-Placette C, Kohler C. Epigenetic mechanisms of postzygotic reproductive isolation in plants. Current Opinion in Plant Biology. 2015;23:39–44. doi: 10.1016/j.pbi.2014.10.006. [DOI] [PubMed] [Google Scholar]
- Lee CB, Page LE, McClure BA, Holtsford TP. Post-pollination hybridization barriers in Nicotiana section Alatae. Sexual Plant Reproduction. 2008;21:183–195. [Google Scholar]
- Legendre P. Comparison of permutation methods for the partial correlation and partial Mantel tests. Journal of Statistical Computation and Simulation. 2000;67:37–73. [Google Scholar]
- Lowry DB, Modliszewski JL, Wright KM, Wu CA, Willis JH. The strength and genetic basis of reproductive isolating barriers in flowering plants. Philosophical Transactions of the Royal Society B-Biological Sciences. 2008;363:3009–3021. doi: 10.1098/rstb.2008.0064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maan ME, Seehausen O. Ecology, sexual selection and speciation. Ecology Letters. 2011;14:591–602. doi: 10.1111/j.1461-0248.2011.01606.x. [DOI] [PubMed] [Google Scholar]
- Mantel N. Detection of disease clustering and a generalized regression approach. Cancer Research. 1967;27:209–&. [PubMed] [Google Scholar]
- Martin MD, Mendelson TC. The accumulation of reproductive isolation in early stages of divergence supports a role for sexual selection. Journal of Evolutionary Biology. 2016;29:676–689. doi: 10.1111/jeb.12819. [DOI] [PubMed] [Google Scholar]
- Masta SE, Maddison WP. Sexual selection driving diversification in jumping spiders. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:4442–4447. doi: 10.1073/pnas.072493099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClure B, Cruz-Garcia F, Romero C. Compatibility and incompatibility in S-RNase-based systems. Annals of Botany. 2011;108:647–658. doi: 10.1093/aob/mcr179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendelson TC. Sexual isolation evolves faster than hybrid inviability in a diverse and sexually dimorphic genus of fish (Percidae : Etheostoma) Evolution. 2003;57:317–327. doi: 10.1111/j.0014-3820.2003.tb00266.x. [DOI] [PubMed] [Google Scholar]
- Mendelson TC, Shaw KL. The (mis)concept of species recognition. Trends in Ecology & Evolution. 2012;27:421–427. doi: 10.1016/j.tree.2012.04.001. [DOI] [PubMed] [Google Scholar]
- Merrill RM, Dasmahapatra KK, Davey JW, Dell’Aglio DD, Hanly JJ, Huber B, Jiggins CD, Joron M, Kozak KM, Llaurens V, Martin SH, Montgomery SH, Morris J, Nadeau NJ, Pinharanda AL, Rosser N, Thompson MJ, Vanjari S, Wallbank RWR, Yu Q. The diversification of Heliconius butterflies: what have we learned in 150 years? Journal of Evolutionary Biology. 2015;28:1417–1438. doi: 10.1111/jeb.12672. [DOI] [PubMed] [Google Scholar]
- Miller RJ, Mione T, Phan HL, Olmstead RG. Color by Numbers: Nuclear Gene Phylogeny of Jaltomata (Solanaceae), Sister Genus to Solanum, Supports Three Clades Differing in Fruit Color. Systematic Botany. 2011;36:153–162. [Google Scholar]
- Mione T. 1992. The systematics and evolution of Jaltomata (Solanaceae) Available from ProQuest Dissertations & Theses Global. (303964972) [Google Scholar]
- Moyle LC, Nakazato T. Comparative genetics of hybrid incompatibility: Sterility in two Solanum species crosses. Genetics. 2008;179:1437–1453. doi: 10.1534/genetics.107.083618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyle LC, Olson MS, Tiffin P. Patterns of reproductive isolation in three angiosperm genera. Evolution. 2004;58:1195–1208. doi: 10.1111/j.0014-3820.2004.tb01700.x. [DOI] [PubMed] [Google Scholar]
- Moyle LC, Jewell CP, Kostyun JL. Fertile approaches to dissecting mechanisms of premating and postmating prezygotic reproductive isolation. Curr Opin Plant Biol. 2014;18:16–23. doi: 10.1016/j.pbi.2013.12.005. [DOI] [PubMed] [Google Scholar]
- Mullen SP, Shaw KL. Insect Speciation Rules: Unifying Concepts in Speciation Research. In: Berenbaum MR, editor. Annual Review of Entomology. 2014. pp. 339–U1007.pp. 59 [DOI] [PubMed] [Google Scholar]
- Murfett J, Strabala TJ, Zurek DM, Mou BQ, Beecher B, McClure BA. S-RNase and interspecific pollen rejection in the genus Nicotiana: Multiple pollen-rejection pathways contribute to unilateral incompatibility between self-incompatible and self-compatible species. Plant Cell. 1996;8:943–958. doi: 10.1105/tpc.8.6.943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oksanen J, Blanchet FG, Kindt R, Legendre P, Minchin PR, O’Hara RB, Simpson GL, Solymos P, Henry M, Stevens H, Wagner H. vegan: Community Ecology Package. R package version 2.3–3. 2016 http://CRAN.R-project.org/package=vegan.
- Olmstead RG, Bohs L, Migid HA, Santiago-Valentin E, Garcia VF, Collier SM. A molecular phylogeny of the Solanaceae. Taxon. 2008;57:1159–1181. [Google Scholar]
- Oneal E, Willis JH, Franks RG. Disruption of endosperm development is a major cause of hybrid seed inviability between Mimulus guttatus and Mimulus nudatus. New Phytologist. 2016;210:1107–1120. doi: 10.1111/nph.13842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinheiro F, Cafasso D, Cozzolino S, Scopece G. Transitions between self-compatibility and self-incompatibility and the evolution of reproductive isolation in the large and diverse tropical genus Dendrobium (Orchidaceae) Annals of Botany. 2015;116:457–467. doi: 10.1093/aob/mcv057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinheiro J, Bates D, DebRoy S, Sarkar D RC Team. nlme: Linear and Nonlinear Mixed Effects Models. R package version 3.1-128. 2016 http://CRAN.R-project.org/package=nlme.
- Presgraves DC. Patterns of postzygotic isolation in Lepidoptera. Evolution. 2002;56:1168–1183. doi: 10.1111/j.0014-3820.2002.tb01430.x. [DOI] [PubMed] [Google Scholar]
- R Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing; Vienna, Austria: 2015. https://www.R-project.org/ [Google Scholar]
- Raw Therapee Development Team. Raw Therapee: cross-platform raw image processing program. 2012 http://rawtherapee.com/
- Ritchie MG. Sexual selection and speciation. Annual Review of Ecology Evolution and Systematics 2007:79–102. [Google Scholar]
- Sarkinen T, Bohs L, Olmstead RG, Knapp S. A phylogenetic framework for evolutionary study of the nightshades (Solanaceae): a dated 1000-tip tree. Bmc Evolutionary Biology. 2013:13. doi: 10.1186/1471-2148-13-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schemske DW, Bradshaw HD. Pollinator preference and the evolution of floral traits in monkeyflowers (Mimulus) Proceedings of the National Academy of Sciences of the United States of America. 1999;96:11910–11915. doi: 10.1073/pnas.96.21.11910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nature Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartzwald D. Color Space Converter Plugin for ImageJ. 2012 https://imagej.nih.gov/ij/plugins/color-space-converter.html.
- Scopece G, Musacchio A, Widmer A, Cozzolino S. Patterns of reproductive isolation in Mediterranean deceptive orchids. Evolution. 2007;61:2623–2642. doi: 10.1111/j.1558-5646.2007.00231.x. [DOI] [PubMed] [Google Scholar]
- Scopece G, Widmer A, Cozzolino S. Evolution of postzygotic reproductive isolation in a guild of deceptive orchids. American Naturalist. 2008;171:315–326. doi: 10.1086/527501. [DOI] [PubMed] [Google Scholar]
- Seddon N, Botero CA, Tobias JA, Dunn PO, MacGregor HEA, Rubenstein DR, Uy JAC, Weir JT, Whittingham LA, Safran RJ. Sexual selection accelerates signal evolution during speciation in birds. Proceedings of the Royal Society B-Biological Sciences. 2013;280:9. doi: 10.1098/rspb.2013.1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selz OM, Thommen R, Pierotti MER, Anaya-Rojas JM, Seehausen O. Differences in male coloration are predicted by divergent sexual selection between populations of a cichlid fish. Proceedings of the Royal Society B-Biological Sciences. 2016:283. doi: 10.1098/rspb.2016.0172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma B, Kramer E. Sub- and neo-functionalization of APETALA3 paralogs have contributed to the evolution of novel floral organ identity in Aquilegia (columbine, Ranunculaceae) New Phytologist. 2013;197:949–957. doi: 10.1111/nph.12078. [DOI] [PubMed] [Google Scholar]
- Shaw KL, Parsons YM, Lesnick SC. QTL analysis of a rapidly evolving speciation phenotype in the Hawaiian cricket Laupala. Molecular Ecology. 2007;16:2879–2892. doi: 10.1111/j.1365-294X.2007.03321.x. [DOI] [PubMed] [Google Scholar]
- Smaczniak C, Immink RGH, Angenent GC, Kaufmann K. Developmental and evolutionary diversity of plant MADS-domain factors: insights from recent studies. Development. 2012;139:3081–3098. doi: 10.1242/dev.074674. [DOI] [PubMed] [Google Scholar]
- Sobel JM, Chen GF. Unification of methods for estimating the strength of reproductive isolation. Evolution. 2014;68:1511–1522. doi: 10.1111/evo.12362. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiffin P, Olson MS, Moyle LC. Asymmetrical crossing barriers in angiosperms. Proceedings of the Royal Society B-Biological Sciences. 2001;268:861–867. doi: 10.1098/rspb.2000.1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomato Genome Consortium. The tomato genome sequence provides insights into fleshy fruit evolution. Nature. 2012;485:635–641. doi: 10.1038/nature11119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tovar-Mendez A, Kumar A, Kondo K, Ashford A, Baek YS, Welch L, Bedinger PA, McClure BA. Restoring pistil-side self-incompatibility factors recapitulates an interspecific reproductive barrier between tomato species. Plant Journal. 2014;77:727–736. doi: 10.1111/tpj.12424. [DOI] [PubMed] [Google Scholar]
- Turelli M, Moyle LC. Asymmetric postmating isolation: Darwin’s corollary to Haldane’s rule. Genetics. 2007;176:1059–1088. doi: 10.1534/genetics.106.065979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dongen S, Abreu-Goodger C. Using MCL to extract clusters from networks. Bacterial Molecular Networks: Methods and Protocols. 2012:281–295. doi: 10.1007/978-1-61779-361-5_15. [DOI] [PubMed] [Google Scholar]
- Williams EG, Rouse JL. Disparate style lengths contribute to isolation of species in Rhododendron. Australian Journal of Botany. 1988;36:183–191. [Google Scholar]
- Wright KM, Lloyd D, Lowry DB, Macnair MR, Willis JH. Indirect Evolution of Hybrid Lethality Due to Linkage with Selected Locus in Mimulus guttatus. Plos Biology. 2013:11. doi: 10.1371/journal.pbio.1001497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Smith SA. Orthology inference in nonmodel organisms using transcriptomes and low-coverage genomes: improving accuracy and matrix occupancy for phylogenomics. Mol Biol Evol. 2014;31:3081–3092. doi: 10.1093/molbev/msu245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang ZH. PAML 4: Phylogenetic analysis by maximum likelihood. Molecular Biology and Evolution. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- Yukilevich R, Aoki F. Is cascade reinforcement likely when sympatric and allopatric populations exchange migrants? Current Zoology. 2016;62:155–167. doi: 10.1093/cz/zow007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.