Abstract
A microwell droplet approach provided high-quality samples in ≥90% yield of the “crystalline sponge”, which was exhibited previously as a revolutionary organic structure determination method. The new protocol, from crystal growth to guest soaking, was conducted in 1–7 days (depending on the guest) and was robust toward user errors, marking improvements over existing protocols. Unit cell determination was used as a practical crystal screening metric. These advances improve the practicality of the crystalline sponge technique for characterizing unknown organic molecules.
Graphical Abstract
The “crystalline sponge” method for X-ray crystallographic analysis represents a potential paradigm shift in the structural characterization of organic molecules. By soaking of target molecules as guests into the pores of a crystalline sponge host, in principle crystallographic analysis can be conducted without requiring crystallization of the analyte itself. Fujita and co-workers showcased the various capabilities of the crystalline sponge method in a seminal 2013 publication1 and published a detailed protocol in 2014.2 Subsequently, Fujita’s group and others have used the crystalline sponge method for small-molecule structure determination,3–6 fractional metabolite analysis,7 absolute configuration assignment,8–10 and reaction intermediate characterization.11
Despite the clear potential of this new method, its use as a routine characterization tool by nonexperts is hampered by several practical considerations, some of which are highlighted here. First and foremost, as evidenced by inspection of all of the published uses of the crystalline sponge method referenced above, a significant amount of trial-and-error is typically necessary for successful host–guest complexation and structure determination even when the method is employed by experts. The original Fujita protocol2 using the versatile sponge [(ZnI2)3(L)2]n·(solvent)x (1·solvent, L = 2,4,6-tris(4-pyridyl)-1,3,5-triazene) calls for 7 days of crystal growth, 7 days of solvent exchange, and 2 days of guest soaking; the resulting 16 day protocol is incompatible with convenient trial-and-error iterations. Furthermore, the original crystallization method produces multiple crystal morphologies, only ≤5% of which are suitable for guest soaking. Because distinguishing between suitable and unsuitable crystals is difficult for nonexperts, once again trial-and-error is arduous and successful structure determination can become improbable. Clardy and co-workers published a 5 day protocol by modification of the crystallization solvent to chloroform12 and also increased the fraction of suitable crystals grown by use of ZnCl2 or ZnBr2 in place of ZnI2.13 Further improvements to the practical usability of the crystalline sponge method will aid in the adoption of this revolutionary technique by a broader community of researchers.14
Here we report improvements to the crystal growth and guest soaking procedures that allow for a single pass of the protocol to be complete within 1–7 days using the most versatile sponge, 1, allowing for more rapid throughput in experimentation. The new crystallization method reproducibly generates the crystal morphology suitable for guest soaking in ≥90% yield, largely removing a main cause of trial-and-error and source of irreproducibility encountered with the original protocol. The potential for using unit cell determination to aid in rapid crystal screening for some guests also is discussed.
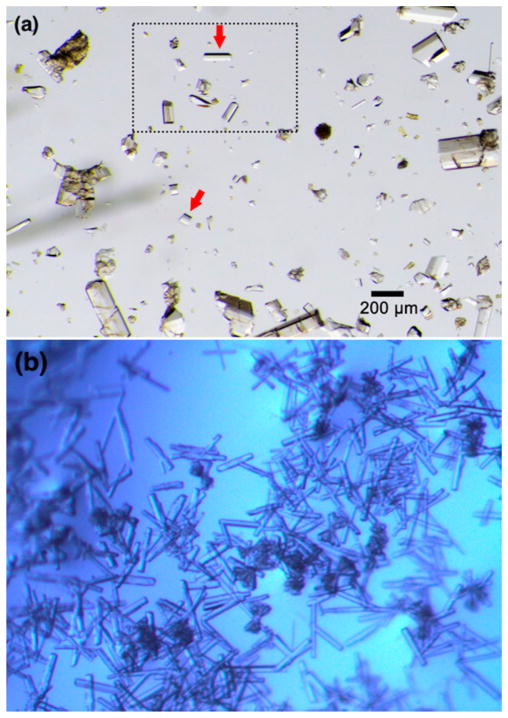
The original Fujita protocol calls for large-scale batch crystallization of 1·PhNO2 from layered PhNO2/MeOH mixtures, which takes 7 days and produces ≤5% suitable crystalline sponge crystals.2 These crystals must be identified and manually separated from each batch (after cyclohexane exchange, vide infra) for further experimentation. Although the rod shape of the crystals can be used as a guide for visual crystal selection (Figure 1a), the authors recommended the use of face indexing (which is difficult for nonexperts) as a more reliable selection criterion. During our own attempts to use the Fujita protocol, we found crystal selection and separation to be quite difficult and irreproducible. However, we did notice that suitable crystals reliably gave the reported C2/c unit cell during unit cell determination, while unsuitable crystals often gave other cell parameters. We thus used unit cell determination (which is easier for nonexperts) as a guide for crystal selection in many subsequent experiments.
Figure 1.
(a) Representative batch of 1·cyclohexane crystals obtained using the large-scale batch method of Fujita.2 Red arrows indicate suitable crystals. This figure was reproduced from the original publication (copyright 2014 Nature Publishing Group). (b) Representative batch of 1·PhNO2 crystals obtained from the microwell droplet method reported here. Most of the crystals are suitable for guest soaking.
We grew crystals of 1·PhNO2 by layering a MeOH droplet of ZnI2 over a PhNO2 droplet of L in multiwell microplates. With this method, crystallization occurs more rapidly than in a batch method because of the evaporation of a small solvent volume15,16 with a large surface area through a shorter reservoir distance.17 (A detailed protocol with photographs is included in the Supporting Information.) One crucial benefit of this method in this case is that suitable crystals reliably grew within 10 h, which is a major asset for rapid throughput of experimentation. Furthermore, crystals grown by this method were almost exclusively rod-shaped, to the near exclusion of other morphologies (Figure 1b). Randomly selected diffraction-quality crystals reproducibly exhibited the desired C2/c unit cell (Table 1, entry 1) and were reliably suitable for guest soaking. In one run, 13 out of 14 crystals sampled were deemed to be high enough in quality on the basis of unit cell determination and examination of the observed reflections on the reciprocal lattice. On the basis of this observation, we estimate that this method produces ≥90% suitable crystalline sponge crystals, again a dramatic improvement in comparison with batch crystallization.
Table 1.
Unit Cell Parameters
| entry | sample | [DIPA]c | a (Å) | b (Å) | c (Å) | β (deg) |
|---|---|---|---|---|---|---|
| 1a | 1·PhNO2 | – | 34.37 ± 0.77 | 14.99 ± 0.10 | 30.55 ± 0.41 | 100.93 ± 0.83 |
| 2a | 1·DIPA | neat | 36.39 ± 0.21 | 14.70 ± 0.03 | 30.53 ± 0.12 | 101.90 ± 0.32 |
| 3b | 1·PhNO2 | – | 34.219(2) | 15.002(1) | 30.331(2) | 100.567(5) |
| 4 | 1·DIPA | 48 M | 36.966(7) | 14.695(8) | 30.906(4) | 102.626(4) |
| 5 | 1·DIPA | 23 M | 37.085(3) | 14.666(1) | 30.858(2) | 102.546(5) |
| 6 | 1·DIPA | 12 M | 37.024(5) | 14.718(2) | 30.883(9) | 102.706(13) |
| 7 | 1·DIPA | 5.3 M | 37.018(11) | 14.668(2) | 30.809(5) | 102.471(6) |
| 8 | 1·DIPA | 1.3 M | 37.174(19) | 14.764(2) | 30.539(6) | 102.885(21) |
| 9 | 1·DIPA | 0.1 M | 37.266(9) | 14.801(1) | 30.471(6) | 103.229(14) |
| 10 | 1·DIPA | 0.05 M | 37.167(11) | 14.799(2) | 30.481(5) | 102.987(17) |
To further test the robustness of the crystallization method, we varied the ZnI2:L ratio incrementally between 1:1 and 1:10 and were delighted to find that suitable crystals formed regardless of the metal:ligand stoichiometry (e.g., Table 1, entry 3). In other words, the method is “foolproof” and robust toward users measuring the metal:ligand stoichiometry inaccurately or incorrectly. Overall, reliably obtaining suitable samples of the crystalline sponge is greatly enhanced by this microwell droplet approach to crystal growth of 1·PhNO2, both because of the significantly shorter crystal growth stage (<1 day as opposed to 3–7 days) and because of the reliably near-quantitative formation of the suitable crystal morphology (90% as opposed to 5%).
The original Fujita protocol prescribed a solvent exchange step prior to guest soaking, involving incubation of 1·PhNO2 crystals for 7 days at 50 °C in cyclohexane to produce 1· cyclohexane.2 These 1·cyclohexane crystals have typically been used as versatile hosts for the various guests referenced above, with the cyclohexane solvent acting as a labile leaving group. In our hands, many of the crystals would become damaged during this solvent exchange stage, whose behavior was sensitive to many factors including temperature fluctuations, vessel size, vessel shape, and rate of temperature ramping. These observations motivated us to revisit guest soaking directly with 1 ·PhNO2. As a test molecule, we chose to examine 2,6-diisopropylaniline (DIPA), which was one of the guests used in Fujita’s initial report.1 Once again, we used microliter quantities of neat DIPA for soaking into 1·PhNO2 in multiwell microplates, allowing for rapid evaporation of volatiles. To our delight, we found that DIPA reliably replaced PhNO2 under these conditions to produce 1·DIPA, which was confirmed by structure solution and refinement (Figure 2). The soaking stage was successful in as little as 6 h, making the overall protocol (from crystal growth to guest soaking) take just 16 h.
Figure 2.
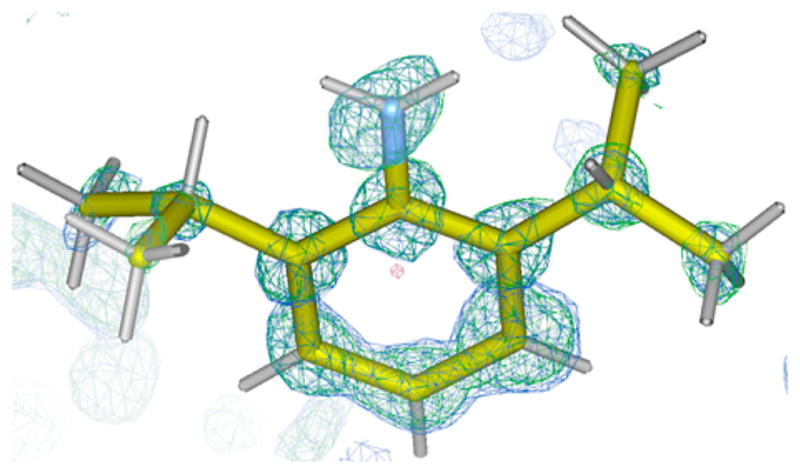
Electron density maps, 1.0σ for the F0 map (blue) and 4.5σ for the difference electron density map (green), of 2,6-diisopropylani-line determined by the crystalline sponge method using the protocol reported here.
Interestingly, although 1·PhNO2 and 1·DIPA both maintain the well-known C2/c morphology, their unit cell parameters are slightly different. Table 1 shows the unit cell parameters for 1· PhNO2 (entry 1) and 1·DIPA (entry 2) averaged over 10 crystals each. Two of the unit cell parameters, a and β, give statistically significant differences (with 2σ confidence) as a function of the guest. Therefore, in the case of DIPA, simple unit cell determination can be used as a reliable method for crystal screening. If this were found to be more general, one could imagine using unit cell parameters measured with an in-house instrument to determine whether a guest has successfully soaked into the crystalline sponge, thereby streamlining data collection at a synchrotron and avoiding needlessly collecting data on 1·PhNO2. Unfortunately, analysis of the unit cell parameters for other 1·guest data sets available in the literature (see Table S3) shows that this screening method would work for some guests but is by no means general. In the case of DIPA, the orientation of the guest within the pore apparently causes the unit cell expansion. The bulky isopropyl groups of the DIPA guest apparently push against the pyridine groups of 1 along the a axis in comparison with their position in 1·PhNO2 (Figure 3).
Figure 3.
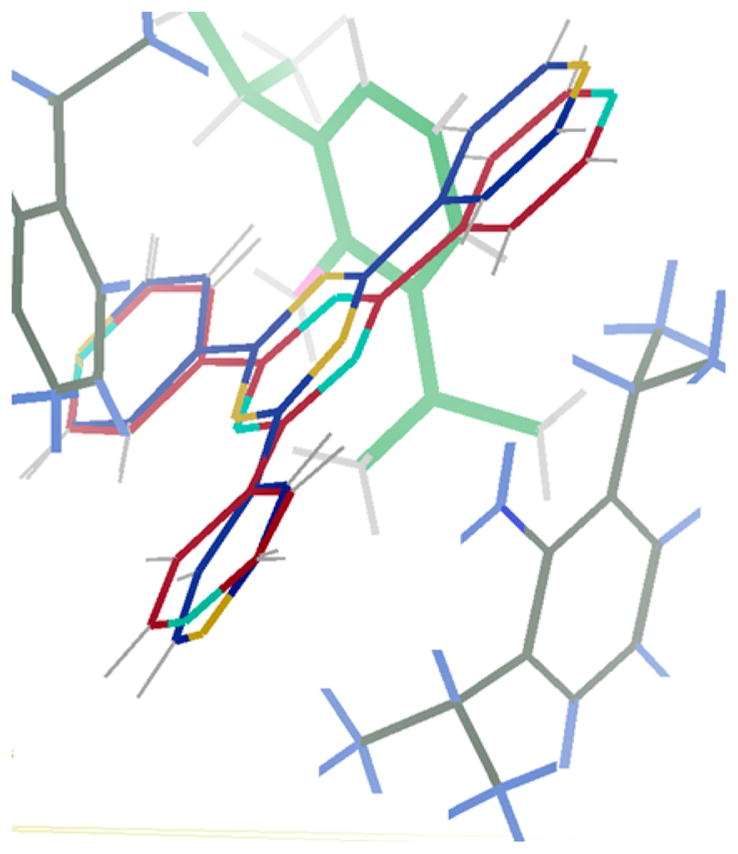
Overlaid structures of 1·PhNO2 and 1·DIPA, both viewed down the crystallographic b axis. The 2,4,6-tris(4-pyridyl)-1,3,5-triazene carbon atoms are shown in red for 1·PhNO2 and in blue for 1·DIPA. The DIPA guest is shown with gray carbon atoms. ZnI2 and PhNO2 moieties have been omitted for clarity. The crystallographic a axis is oriented vertically and the c axis horizontally.
Next, we examined introducing DIPA as a solute rather than in neat form. The 1·PhNO2 crystals were exposed to solutions of DIPA in nBu2O with concentrations ranging from 0.05 to 48 M. In all cases, DIPA had soaked into the pores as judged by unit cell determination and comparison to the known parameters for 1·DIPA; selected data are shown in Table 1 (entries 4–10). Selected data sets were further analyzed to confirm that DIPA had indeed soaked into the pores. Clearly, at least for DIPA, pre-exchange of PhNO2 for cyclohexane is not necessary with the new protocol, even when DIPA is introduced as a dilute solution. Overall, crystal growth and guest soaking can thus be accomplished in <1 day, marking a dramatic improvement over previous protocols.
One limitation of this protocol is that other guests we have examined, such as cyclohexanone, menthyl acetate, and pentamethylbenzene, do not soak into the pores of 1·PhNO2 to an appreciable extent under the conditions used for DIPA, which as an electron-rich aromatic compound has a strong affinity for the pores of 1. It is likely that a solvent exchange stage would need to be introduced, in analogy to the Fujita protocol,2 in order to have more generality for guests with weaker affinity for the pores of 1. This is not surprising, as it is well-known that crystalline sponge methodology typically needs to be re-evaluated and reoptimized for each individual guest molecule that one encounters.14
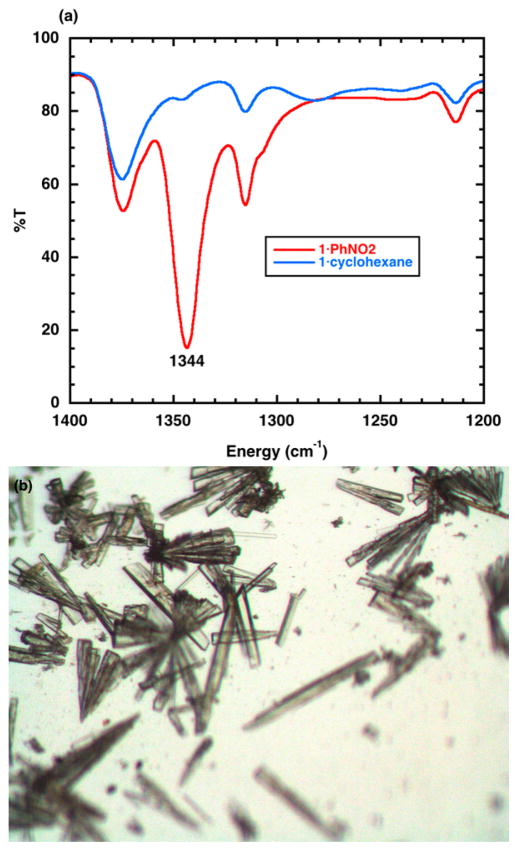
We used our microwell droplet approach to examine conversion of 1·PhNO2 to 1·cyclohexane and 1·CHCl3 by soaking 1·PhNO2 crystals in droplets of the appropriate solvent at room temperature. We monitored the displacement of the nitrobenzene solvent by IR spectroscopy (Figure 4a), as was demonstrated previously by Fujita.2 We found that soaking for 6 days was required to observe complete disappearance of PhNO2 in both cases (see the Supporting Information for a detailed protocol with photos). In the case of 1·CHCl3, a significant fraction of the crystals were damaged during the solvent exchange process, although the remaining rod-shaped crystals are easily identifiable by a nonexpert (see the Supporting Information). In the case of 1·cyclohexane, relatively fewer crystals were damaged during the solvent exchange process (Figure 4b), which represents an improvement over the original 50 °C incubation (vide supra). The versatility of 1·cyclohexane and 1·CHCl3 for use with a range of guest molecules has been established in the existing crystalline sponge literature of Fujita and Clardy, respectively. Overall, our protocol allows these two crystalline sponges to be accessible in at most 7 days, with improvements to the fraction of sponge-quality crystals and to the reproducibility of the entire process.
Figure 4.
(a) FT-IR spectra of 1·PhNO2 and 1·cyclohexane. The 1344 cm−1 feature is a signature for the PhNO2 guest. (b) Representative batch of 1·cyclohexane crystals obtained from the microwell droplet method reported here.
Supplementary Material
Acknowledgments
This work was financed by Grant U41 AT008706 from the NIH (NCCIH and ODS). Additional funds to N.P.M. were provided by the UIC Department of Chemistry and by an Alfred P. Sloan Research Fellowship. This work is based upon research conducted at the Northeastern Collaborative Access Team beamlines, which are funded by the National Institute of General Medical Sciences of the National Institutes of Health (P41 GM103403). The Pilatus 6M detector on beamline 24-ID-C is funded by an NIH-ORIP HEI grant (S10 RR029205). This research used resources of the Advanced Photon Source, a U.S. Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract DE-AC02-06CH11357. Use of the LS-CAT Sector 21 was supported by the Michigan Economic Development Corporation and the Michigan Technology Tri-Corridor (Grant 085P1000817). We also acknowledge use of the UIC Research Resources Center (Macromolecular Structure Facility).
Footnotes
Author Contributions
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
The authors declare no competing financial interest.
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.or-glett.6b03119.
Experimental protocols and unit cell data (PDF)
Crystallographic data for 1·PhNO2 (CIF)
Crystallographic data for 1·DIPA (CIF)
References
- 1.Inokuma Y, Yoshioka S, Ariyoshi J, Arai T, Hitora Y, Takada K, Matsunaga S, Rissanen K, Fujita M. Nature. 2013;495:461. doi: 10.1038/nature11990. [DOI] [PubMed] [Google Scholar]
- 2.Inokuma Y, Yoshioka S, Ariyoshi J, Arai T, Fujita M. Nat Protoc. 2014;9:246. doi: 10.1038/nprot.2014.007. [DOI] [PubMed] [Google Scholar]
- 3.Vinogradova EV, Müller P, Buchwald SL. Angew Chem, Int Ed. 2014;53:3125. doi: 10.1002/anie.201310897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.O’Brien AG, Maruyama A, Inokuma Y, Fujita M, Baran PS, Blackmond DG. Angew Chem, Int Ed. 2014;53:11868. doi: 10.1002/anie.201407948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cuenca AB, Zigon N, Duplan V, Hoshino M, Fujita M, Fernández E. Chem - Eur J. 2016;22:4723. doi: 10.1002/chem.201600392. [DOI] [PubMed] [Google Scholar]
- 6.Sanna E, Escudero-Adán EC, Bauzá A, Ballester P, Frontera A, Rotger C, Costa A. Chem Sci. 2015;6:5466. doi: 10.1039/c5sc01838b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Inokuma Y, Ukegawa T, Hoshino M, Fujita M. Chem Sci. 2016;7:3910. doi: 10.1039/c6sc00594b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yoshioka S, Inokuma Y, Hoshino M, Sato T, Fujita M. Chem Sci. 2015;6:3765. doi: 10.1039/c5sc01681a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zigon N, Hoshino M, Yoshioka S, Inokuma Y, Fujita M. Angew Chem, Int Ed. 2015;54:9033. doi: 10.1002/anie.201502302. [DOI] [PubMed] [Google Scholar]
- 10.Urban S, Brkljača R, Hoshino M, Lee S, Fujita M. Angew Chem, Int Ed. 2016;55:2678. doi: 10.1002/anie.201509761. [DOI] [PubMed] [Google Scholar]
- 11.Duplan V, Hoshino M, Li W, Honda T, Fujita M. Angew Chem, Int Ed. 2016;55:4919. doi: 10.1002/anie.201509801. [DOI] [PubMed] [Google Scholar]
- 12.Ramadhar TR, Zheng SL, Chen YS, Clardy J. Acta Crystallogr, Sect A: Found Adv. 2015;71:46. doi: 10.1107/S2053273314019573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ramadhar TR, Zheng SL, Chen YS, Clardy J. Chem Commun. 2015;51:11252. doi: 10.1039/c5cc03840e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hoshino M, Khutia A, Xing H, Inokuma Y, Fujita M. IUCrJ. 2016;3:139. doi: 10.1107/S2052252515024379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Santarsiero BD, Yegian DT, Lee CC, Spraggon G, Gu J, Scheibe D, Uber DC, Cornell EW, Nordmeyer RA, Kolbe WF, Jin J, Jones AL, Jaklevic JM, Schultz PG, Stevens RC. J Appl Crystallogr. 2002;35:278. [Google Scholar]
- 16.Diller DJ, Hol WGJ. Acta Crystallogr, Sect D: Biol Crystallogr. 1999;55:656. doi: 10.1107/s0907444998014589. [DOI] [PubMed] [Google Scholar]
- 17.Luft JR, Albright DT, Baird JK, DeTitta GT. Acta Crystallogr, Sect D: Biol Crystallogr. 1996;52:1098. doi: 10.1107/S0907444996006932. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.