Abstract
Nucleosomes play important structural and regulatory roles by tightly wrapping the DNA that constitutes the metazoan genome. The Polycomb group (PcG) proteins modulate nucleosomes to maintain repression of key developmental genes, including Hox genes whose temporal and spatial expression is tightly regulated to guide patterning of the anterior-posterior body axis. CBX2, a component of the mammalian Polycomb Repressive Complex 1 (PRC1), contains a ‘compaction region’ that has the biochemically-defined activity of bridging adjacent nucleosomes. Here we demonstrate that a functional compaction region is necessary for proper body patterning, as mutating this region leads to homeotic transformations similar to those observed with PcG loss-of-function mutations. We propose that CBX2-driven nucleosome compaction is a key mechanism by which PcG proteins maintain gene silencing during mouse development.
Cell fate specification of many cell types requires the repression of specific genes and the maintenance of this gene silencing over time. Polycomb Repressive Complexes 1 and 2 (PRC1 and PRC2) are key developmental regulators that act on the chromatin of target genes to stably repress them. PRC1 and PRC2 each have four core subunits, whose activities have been individually characterized (1). PRC2 methylates histone H3 at lysine 27 (H3K27me3), which is required for the silencing of Hox genes. Hox genes are master regulators of body patterning and hence mis-expression leads to defects in patterning (2–4). How H3K27me3 leads to silencing remains unclear. Canonical PRC1 has a chromodomain-containing subunit that binds to H3K27me3 (1). Thus, it is thought that PRC1 is the effector of silencing. Individual subunits of PRC1, including Ring1B and Phc2, have been studied to determine which of their activities are required for silencing and proper body patterning. Whereas the histone H2A ubiquitylating function of Ring1B appears dispensable (5, 6), the self-polymerizing activity of Phc2 is required for patterning (7). Here, we examined the requirement for a third activity: the ability of canonical PRC1 to compact adjacent nucleosomes into globular structures (8–10).
The compaction function of PRC1 has been mapped to an intrinsically disordered region with high positive charge within the complex (10). In Drosophila, the Posterior Sex Comb (Psc) subunit carries this compaction region which, when truncated, leads to Hox gene mis-expression (9, 11). The extent of deletion of this region in Psc mutant alleles correlates with the severity of the mutant phenotypes, suggesting that compaction is critical for PRC1 function during Drosophila development. In mammals, compaction is driven by the CBX2 subunit (10); CBX2 is one of five homologs of Polycomb (Pc), each of which forms different versions of PRC1 (1). To test the role of the CBX2 compaction region in gene silencing and body pattern formation, we introduced point mutations that disrupt the in vitro ability of CBX2 to compact nucleosomes, as characterized in (10), into mouse embryonic stem cells (mESCs) and mice, and assessed the resultant phenotypes.
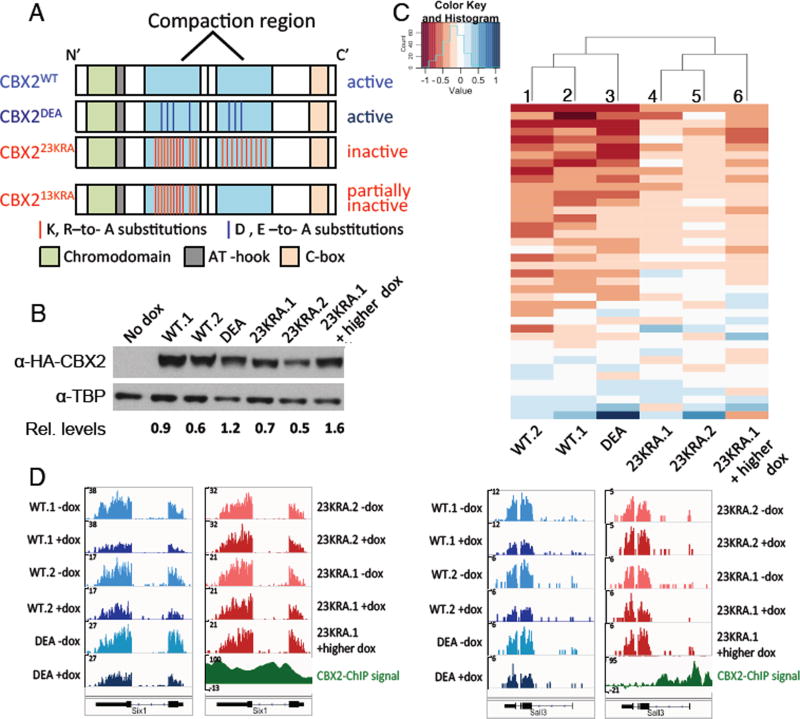
We first verified CBX2 is a component of PRC1 in wild-type mESCs (fig. S1). To examine the role of compaction, we expressed, using doxycycline induction in Cbx2−/− mESCs, wild-type CBX2 (CBX2WT) or one of two previously characterized CBX2 variants, CBX223KRA and CBX2DEA. CBX223KRA has 23 lysine (K) and arginine (R) residues mutated to alanine (A) in its compaction region, which should render it compaction inactive, whereas CBX2DEA is a compaction proficient control where aspartic acid (D) and glutamic acid (E) residues are mutated to alanine in the same region (Fig. 1A and fig. S2A) (10). If compaction regions are important for gene silencing in mESCs, we expect CBX2DEA- and CBX2WT –expressing mESC lines to be similar to each other and distinct from CBX223KRA-expressing lines. We obtained two independent CBX2WT and CBX223KRA lines, respectively, and one CBX2DEA line that express CBX2 protein to similar levels (Fig. 1B). The CBX223KRA variant did not affect the protein levels of other PRC1 subunits (fig. S2B) and co-immunoprecipitated with the core PRC1 component Ring1B (fig. S2C), indicating appropriate complex formation. This is consistent with previous findings that mutations in the compaction region do not affect CBX2-Ring1B interaction (10, 12). In addition, CBX7, which is the dominant Pc homolog in mESCs, also co-immunoprecipitated with Ring1B in the presence of CBX223KRA, suggesting that CBX223KRA does not interfere with the formation of an alternate form of canonical PRC1 (fig. S2C).
Fig. 1.
Gene repression by CBX2 in mESCs correlates with in vitro compaction ability. (A) Schematics of wild-type and variant CBX2 expressed in mESCs (top three) and CBX213KRA, used in mutant mice (see Fig. 3). (B) Western blot of CBX2 in mESC lines following doxycycline-induction with protein levels relative to TBP loading control. (C) Heatmap and hierarchical clustering of gene expression changes at candidate CBX2 target genes (see Methods for identification of candidates). Heatmap values are log2(fold change[+dox/−dox]). Red represents repression by CBX2. (D) RNA-seq tracks of CBX2-bound genes before and after induction of CBX2 expression using doxycycline (−dox and +dox, respectively). y-axis indicates normalized read counts.
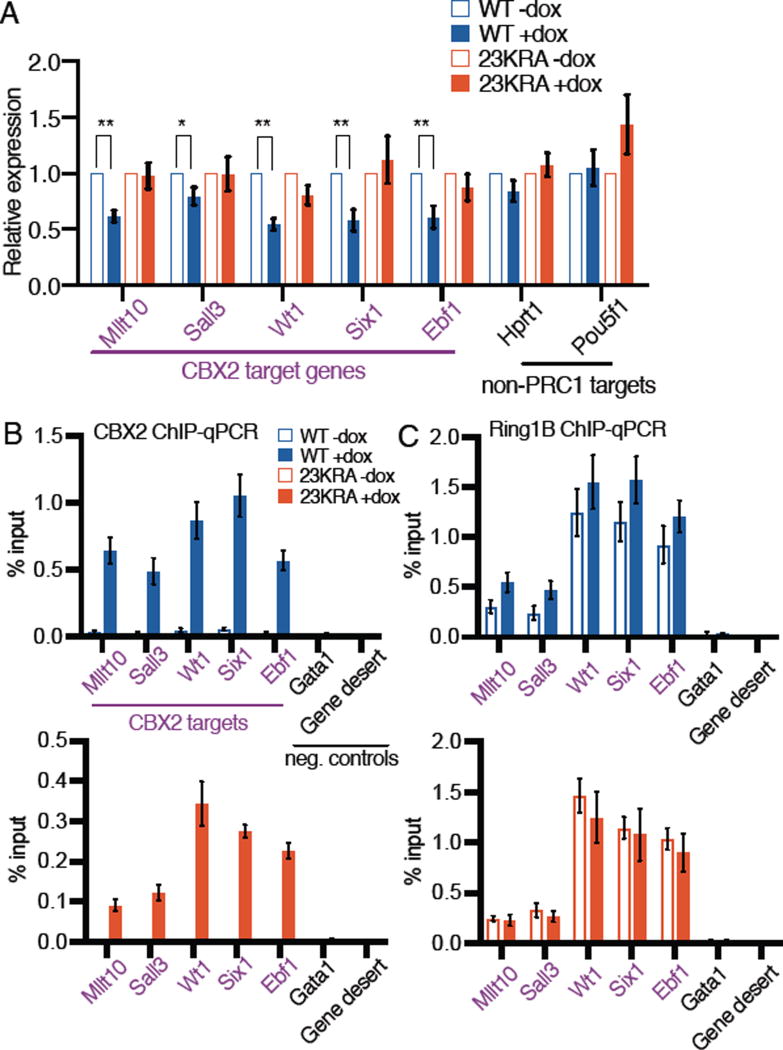
To determine whether mutations that disrupt the in vitro compaction activity of CBX2 affect gene expression, we measured RNA levels genome-wide in mESC lines before and after induction of wild-type and variant CBX2. We used the mESC lines described above that express equivalent levels of the various CBX2 proteins, and a condition in which CBX223KRA expression was induced to a higher level (Fig. 1B) to determine whether more CBX223KRA would lead to CBX2WT-like activity. Many CBX2-bound genes and known PRC1 targets were repressed following introduction of CBX2WT (Fig. 1C, columns 1 and 2; fig. S3A). Notably, this repression was weaker in the presence of CBX223KRA (columns 4 and 5), even when CBX223KRA was expressed at a higher level than CBX2WT (column 6; fig. S3A). In contrast, expression of CBX2DEA resulted in repression similar to that observed with CBX2WT (Fig. 1C, column 3; fig. S3A). Hierarchical clustering analysis of the gene expression changes revealed that the CBX2WT and CBX2DEA mESC lines clustered together, whereas the CBX223KRA lines formed a separate cluster (Fig. 1C). At the single gene level, CBX2-bound genes displayed decreased mRNA levels following CBX2WT and CBX2DEA expression (Fig. 1D, blue tracks), but remained unchanged following CBX223KRA expression (red tracks). In contrast, non-PRC1 targets were unaffected by the expression of any of the CBX2 variants (fig. S3B). Gene expression changes were verified by reverse transcription-qPCR analyses (Fig. 2A). We conclude that CBX223KRA has less repressive activity than CBX2WT, suggesting a correlation between repression and compaction activity.
Fig. 2.
PRC1 component binding is unperturbed at genes affected by CBX223KRA. (A) Gene expression changes by RT-qPCR analyses in mESC lines. Mean±SD, n=4 biological replicates. **P<0.01, *P<0.05, t-test. (B–C) ChIP-qPCR analyses for CBX2 (B) and Ring1B (C) binding. Mean±SD, n≥3 biological replicates. For (C), there are no statistically significant differences between –dox and +dox pairs, or between WT+dox and 23KRA+dox samples.
We sought to determine whether the mutations in CBX223KRA might have impacted the targeting of PRC1 to chromatin, thereby indirectly affecting gene repression. We determined CBX223KRA binding sites using a standard ChIP-seq protocol, which involves crosslinking between DNA and lysine residues in the target protein. As CBX223KRA and CBX2WT differ significantly in the number of lysine residues, the respective ChIP signal intensities are not comparable (fig. S4A and B), but the locations for binding can be compared. We found that the binding sites of CBX223KRA and CBX2WT highly overlap (85%) (fig. S4C). We verified by ChIP-qPCR that CBX223KRA is present at genes that it failed to repress (Fig. 2B). We also assessed the occupancy of other PRC1 components and found that Ring1B, a core component of all PRC1 complexes, and CBX7 are both bound at equivalent levels in CBX223KRA and CBX2WT mESCs (Fig. 2C and fig. S5A). Therefore, CBX7, which does not compact nucleosomes in vitro, is insufficient for full repression by PRC1 without wild-type CBX2. The PRC1- and PRC2-deposited histone modifications H2AK119Ub and H3K27me3 were also present at similar levels in CBX223KRA and CBX2WT mESCs (fig. S5B and C). These data suggest that mutations that are expected to inhibit CBX2-mediated nucleosome compaction result in disrupted gene repression without affecting the assembly of PRC1 on chromatin.
Since mutations in the compaction region of CBX2 disrupted gene repression in mESCs, we hypothesized that they would also impact PRC1 function during embryogenesis. We therefore generated mouse lines that carried either the Cbx223KRA or Cbx213KRA mutations (fig. S6 to 8). The CBX213KRA mutant contains a subset of the mutations in CBX223KRA (Fig. 1A and fig. S6A) and has in vitro compaction activity lower than that of CBX2WT but higher than that of CBX223KRA (fig. S6B). Cbx223KRA/23KRA and Cbx213KRA/13KRA animals were viable, and the respective mutant alleles were expressed normally in vivo (fig. S6C). Thus, we generated two distinct mouse lines containing alleles of compaction-deficient Cbx2 with distinct compaction abilities.
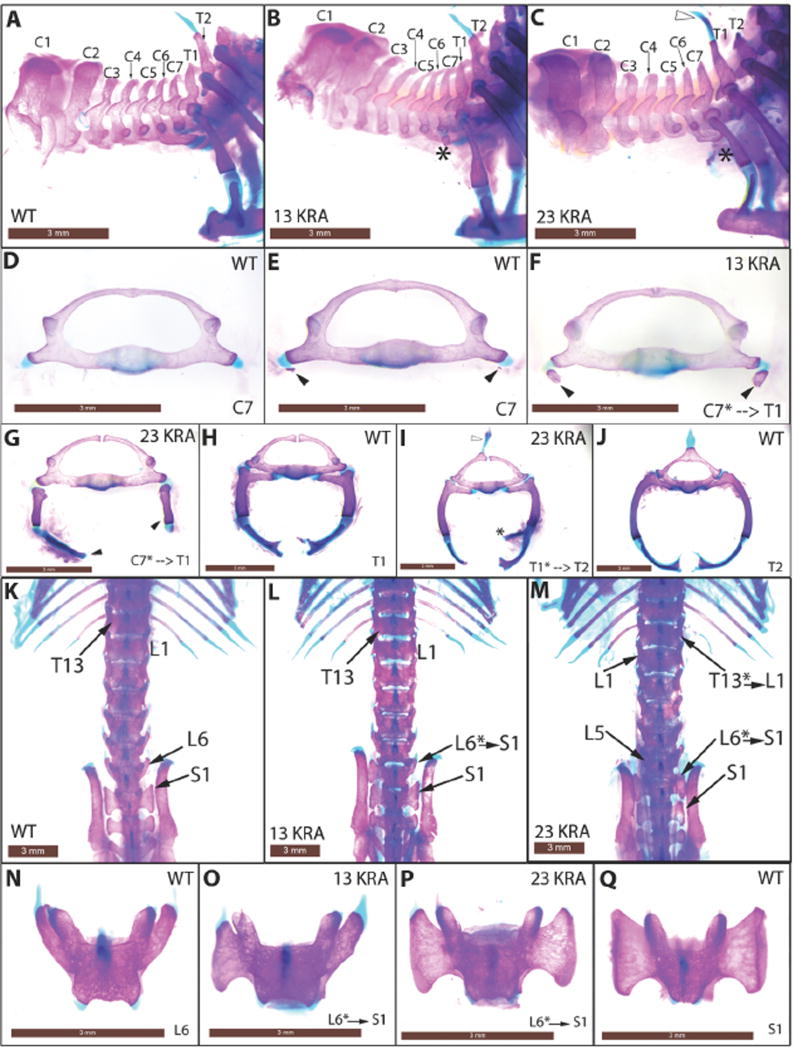
If the compaction activity of CBX2 is important for PRC1 function, we would expect the Cbx223KRA and Cbx213KRA homozygotes to exhibit anterior-to-posterior transformations spanning the axial skeleton, the classic PcG phenotype found in Cbx2−/− mice (13, 14). In fact, Cbx223KRA/23KRA and Cbx213KRA/13KRA animals exhibited posteriorization across the vertebral column, similar to that observed in Cbx2−/−, with C7-to-T1, T1-to-T2, T7-to-T8, T13-to-L1 and L6-to-S1 transformations (Fig. 3 and fig. S9). This indicated that both Cbx223KRA/23KRA and Cbx213KRA/13KRA mimic Cbx2−/− in axial patterning. The transformations and their penetrance resemble the null phenotypes of two other PRC1 subunits, Phc2 and Bmi1 (Table 1) (7, 15).
Fig. 3.
Anterior-to-posterior transformations in Cbx223KRA and Cbx213KRA homozygous mutant mice. (A–C) Lateral views of the cervical-thoracic boundary of the axial skeleton. Asterisks mark C7 rib anlage (B) or ectopic rib (C) that is indicative of C7-to-T1 transformation. Open arrowhead marks ectopic spinous process on T1 that characterizes T1-to-T2 transformation (C, also in I). (D–J) Anterior views of disarticulated C7 to T2 vertebrae. Black arrowheads mark rib anlages (E, F) or ectopic ribs (G) on C7. Asterisk marks a spurious second attachment site to the sternum (I) on T1. (K–M) Dorsal views of the thoracic-lumbar and lumbar-sacral transitions. Asterisks indicate positions of homeotic transformations. (N–Q) Dorsal views of disarticulated L6 and S1vertebrae. L6-to-S1 transformation is characterized by the sacral ala (large triangular surface) on L6 (O, unilateral left; P, bilateral). Scale bar = 3mm.
Table 1.
Frequencies of homeotic transformations
| Cbx2+/+ | Cbx223KRA/23KRA | Cbx2+/+ | Cbx213KRA/13KRA | |
|---|---|---|---|---|
| (n=9) | (n=9) | (n=9) | (n=11) | |
| C7-to-T1 (rib anlage only) | 9 (100%) | 4 (44%) | 4 (44%) | 9 (82%) |
|
| ||||
| C7-to-T1 (ectopic rib) | 0 | 5 (56%) | 0 | 0 |
|
| ||||
| T1-to-T2 | 0 | 5 (56%) | 0 | 3 (27%) |
|
| ||||
| T7-to-T8 unilateral | 0 | 2 ( 2 2 %) | 0 | 4 (36%) |
| bilateral | 0 | 4 (44%) | 0 | 0 |
|
| ||||
| T13-to-L1 | 0 | 2 (22%) | 0 | 0 |
|
| ||||
| L6-to-S1 unilateral | 1 (11%) | 1 (11%) | 0 | 3 (27%) |
| bilateral | 0 | 7 (78%) | 0 | 2 (18%) |
Given that CBX223KRA is less active in vitro than CBX213KRA, we compared the severity of the phenotypes between the two mutants. C7-to-T1 transformations were more dramatic in Cbx223KRA/23KRA than in Cbx213KRA/13KRA: Cbx223KRA/23KRA exhibited long ectopic C7 cervical ribs which articulated to the sternum or T1 rib (Fig. 3C and G), whereas Cbx213KRA/13KRA displayed increased incidence of C7 rib anlage when compared to Cbx2+/+ (Fig. 3A, B, E, F, and Table 1). The frequencies of the other transformations were consistently higher in Cbx223KRA/23KRA compared to Cbx213KRA/13KRA (Table 1). For L6-to-S1 (Fig. 3K–Q) and T7-to-T8 (fig. S9A–E), the occurrences of bilateral changes were higher in Cbx223KRA/23KRA (Table 1), and T13-to-L1 (Fig. 3M and fig. S9F–H) was only observed in Cbx223KRA/23KRA (Table 1). Thus, the skeletal abnormalities collectively indicated that the Cbx223KRA mutation resulted in more severe phenotypes than Cbx213KRA. Although we note that the two mutant alleles are in different genetic backgrounds, it is striking that the relative severity of the mutant phenotypes correlates with the in vitro activity of CBX223KRA and CBX213KRA.
Cbx2−/− mice display other phenotypes in addition to the classic PcG homeotic transformations; these include post-natal lethality, reduced body weight, and male-to-female sex reversal (13, 14). Notably, we did not observe any of these phenotypes in Cbx223KRA/23KRA and Cbx213KR/13KRA animals (fig. S10), suggesting that the mutations in the compaction region of CBX2 specifically disrupted a function that is required for proper axial patterning, instead of creating a generically dysfunctional protein. We examined chromatin binding of CBX223KRA and CBX2WT in mouse embryos by native ChIP; readout from native ChIP (no crosslinking) reflects binding affinity and localized chromatin occupancy. We observed occupancy of CBX223KRA and CBX2WT on PRC1-regulated genes (fig. S11A); thus, CBX2 localization was not impacted by the mutations. The signal for CBX223KRA was lower compared to CBX2WT, consistent with its predicted lower affinity for binding when compaction is disrupted (see fig. S11A). Ring1B binding to chromatin was unaffected in Cbx223KRA/23KRA embryos (fig. S11B), suggesting that binding of all PRC1 complexes on chromatin is unperturbed by the CBX2 mutations. Together these observations indicate that the effects of the mutations in the compaction region of CBX2 are exerted in a specific manner, consistent with the hypothesis that nucleosome compaction is necessary for proper patterning of the body axis during development.
Here we show that the compaction region of CBX2 plays a key role in establishing proper axial patterning, thereby expanding the functional regions of this protein. The N-terminal chromodomain of CBX2 binds to H3K27me3, the histone modification that marks developmental genes for silencing. Thus, CBX2 has the dual function of targeting PRC1 to H3K27me3-marked genes and effecting PRC1-dependent gene repression via its compaction region. We note that the other Pc homologs CBX4, CBX6, CBX7 and CBX8 have weak or no compaction activity (10), and their null mutants do not exhibit posterior transformation phenotypes, unlike Cbx2−/− or Cbx223KRA and Cbx213KRA mutants (16). We infer from this that the regulation of Hox genes for patterning during development is particularly dependent on compaction that is mediated by CBX2; testing this proposed mechanism requires an assay for local compaction in the relevant cell types in mice, which is not currently available. PRC1 also mediates subnuclear clustering through its Polyhomeotic subunit, whose self-polymerizing function is also needed for axial patterning (7, 17). We therefore propose a model in which PRC1 organizes nucleosomes by coordinating local chromatin compaction and subnuclear clustering, thus creating structures that are refractory to transcription and thereby instilling stable silencing.
Supplementary Material
Acknowledgments
We thank W. Press, S. Miller for guidance on mouse work; R. Mostoslavsky, J. Lee, and R.E.K. lab for discussions; G. Zhou, L. Wu for advice on generating mutant mice; J. Cochrane, S. Miller, I. Tchasovnikarova for critical reading of the manuscript. Funding: Agency of Science, Research and Technology, Singapore (M.S.L), NIH (GM043901, to R.E.K; HD032443, to C.J.T; P30DK040561, to R.I.S).
Footnotes
References
- 1.Di Croce L, Helin K. Nat Struct Mol Biol. 2013;20:1147–1155. doi: 10.1038/nsmb.2669. [DOI] [PubMed] [Google Scholar]
- 2.Cao R, et al. Science. 2002;298:1039–1043. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- 3.Müller J, et al. Cell. 2002;111:197–208. doi: 10.1016/s0092-8674(02)00976-5. [DOI] [PubMed] [Google Scholar]
- 4.Pengelly AR, Copur Ö, Jäckle H, Herzig A, Müller J. Science. 2013;339:698–699. doi: 10.1126/science.1231382. [DOI] [PubMed] [Google Scholar]
- 5.Pengelly AR, Kalb R, Finkl K, Müller J. Genes & Development. 2015;29:1487–1492. doi: 10.1101/gad.265439.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Illingworth RS, et al. Genes & Development. 2015;29:1897–1902. doi: 10.1101/gad.268151.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Isono K, et al. Developmental Cell. 2013;26:565–577. doi: 10.1016/j.devcel.2013.08.016. [DOI] [PubMed] [Google Scholar]
- 8.Shao Z, et al. Cell. 1999;98:37–46. doi: 10.1016/S0092-8674(00)80604-2. [DOI] [PubMed] [Google Scholar]
- 9.Francis NJ, Kingston RE, Woodcock CL. Science. 2004;306:1574–1577. doi: 10.1126/science.1100576. [DOI] [PubMed] [Google Scholar]
- 10.Grau DJ, et al. Genes & Development. 2011;25:2210–2221. doi: 10.1101/gad.17288211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.King IFG, et al. Mol. Cell. Biol. 2005;25:6578–6591. doi: 10.1128/MCB.25.15.6578-6591.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schoorlemmer J, et al. EMBO J. 1997;16:5930–5942. doi: 10.1093/emboj/16.19.5930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Coré N, et al. Development. 1997;124:721–729. doi: 10.1242/dev.124.3.721. [DOI] [PubMed] [Google Scholar]
- 14.Katoh-Fukui Y, et al. Nature. 1998;393:688–692. doi: 10.1038/31482. [DOI] [PubMed] [Google Scholar]
- 15.van der Lugt NM, et al. Genes & Development. 1994;8:757–769. doi: 10.1101/gad.8.7.757. [DOI] [PubMed] [Google Scholar]
- 16.Gil J, O'Loghlen A. Trends in Cell Biology. 2014;24:632–641. doi: 10.1016/j.tcb.2014.06.005. [DOI] [PubMed] [Google Scholar]
- 17.Wani AH, et al. Nat Comms. 2016;7:10291. doi: 10.1038/ncomms10291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kharchenko PV, Tolstorukov MY, Park PJ. Nature Biotechnology. 2008;26:1351–1359. doi: 10.1038/nbt.1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brind’Amour J, et al. Nat Comms. 2015;6:6033. doi: 10.1038/ncomms7033. [DOI] [PubMed] [Google Scholar]
- 20.Trapnell C, Pachter L, Salzberg SL. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Anders S, Pyl PT, Huber W. Bioinformatics. 2014;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Robinson MD, McCarthy DJ, Smyth GK. Bioinformatics. 2009;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kundu S, et al. Mol. Cell. 2017;65:p432–446.e5. [Google Scholar]
- 24.Sander JD, et al. Nucleic Acids Research. 2010;38:W462–8. doi: 10.1093/nar/gkq319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Steger DJ, Eberharter A, John S, Grant PA, Workman JL. Proc. Natl. Acad. Sci. U.S.A. 1998;95:12924–12929. doi: 10.1073/pnas.95.22.12924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Côté J, Utley RT, Workman JL. Methods in Molecular Genetics. Vol. 6. Elsevier; 1995. pp. 108–128. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.