Abstract
Purpose
To determine whether the addition of standardized uptake value (SUV) from PET scans to CT lung texture features could improve a radiomics‐based model of radiation pneumonitis (RP) diagnosis in patients undergoing radiotherapy.
Methods and materials
Anonymized data from 96 esophageal cancer patients (18 RP‐positive cases of Grade ≥ 2) were collected including pre‐therapy PET/CT scans, pre‐/post‐therapy diagnostic CT scans and RP status. Twenty texture features (first‐order, fractal, Laws' filter and gray‐level co‐occurrence matrix) were calculated from diagnostic CT scans and compared in anatomically matched regions of the lung. Classifier performance (texture, SUV, or combination) was assessed by calculating the area under the receiver operating characteristic curve (AUC). For each texture feature, logistic regression classifiers consisting of the average change in texture feature value and the pre‐therapy SUV standard deviation (SUVSD) were created and compared with the texture feature as a lone classifier using ANOVA with correction for multiple comparisons (P < 0.0025).
Results
While clinical parameters (mean lung dose, smoking history, tumor location) were not significantly different among patients with and without symptomatic RP, SUV and texture parameters were significantly associated with RP status. AUC for single‐texture feature classifiers alone ranged from 0.58 to 0.81 and 0.53 to 0.71 in high‐dose (≥ 30 Gy) and low‐dose (< 10 Gy) regions of the lungs, respectively. AUC for SUVSD alone was 0.69 (95% confidence interval: 0.54–0.83). Adding SUVSD into a logistic regression model significantly improved model fit for 18, 14 and 11 texture features and increased the mean AUC across features by 0.08, 0.06, and 0.04 in the low‐, medium‐, and high‐dose regions, respectively.
Conclusions
Addition of SUVSD to a single‐texture feature improves classifier performance on average, but the improvement is smaller in magnitude when SUVSD is added to an already effective classifier using texture alone. These findings demonstrate the potential for more accurate assessment of RP using information from multiple imaging modalities.
Keywords: CT, PET, radiation pneumonitis, radiomics, SUV, texture analysis
1. Introduction
Radiation pneumonitis (RP) is a symptomatic lung toxicity caused by an inflammatory response to radiation.1 This response allocates a cascade of cytokines to the radiation‐damaged tissue2 and can lead to the development of RP with varying severity. Patients with thoracic malignancies who undergo radiation therapy (RT) can thus develop a range of RP symptoms, including cough, dyspnea, fever, and even death.3 Therefore, development of a reliable method to predict future onset of RP is critical to assess patient‐specific risk associated with thoracic RT. Such an approach could help designate at‐risk patients and facilitate earlier intervention or earlier RP diagnosis and treatment by modifying the radiation treatment plan or initiating steroid administration to reduce the severity of eventual symptoms.
RP onset has been correlated with treatment variables such as dose and the volume of lung irradiated,4 as well as lung density or texture change as quantified by computed tomography (CT). Our laboratory previously5 analyzed the dose‐dependent change in 20 CT texture features as potential predictors of RP. Linear modeling showed a significant relationship between the change in texture feature values and development of grade ≥ 2 RP for 12 of these 20 features, even when controlling for dose. Earlier studies indicated that CT‐based texture features show promise as a means to distinguish between healthy and diseased lung tissue. Chabat et al.6 illustrated textural differences in CT images between three forms of obstructive lung disease and normal lung tissue using a Bayesian classifier. Mattonen et al.7 demonstrated the ability of texture features in CT images to predict cancer recurrence in non‐small cell lung cancer (NSCLC) patients undergoing stereotactic ablative radiotherapy.
Although CT texture provides quantitative assessment of structural changes in the lung induced by RT, the inflammatory roots of RP have prompted a search for additional biological predictors of RP, including cytokines and other immune response factors. Oh et al.8 and Craft et al.9 found alpha‐2‐macroglobulin (α2M), an acute‐phase protein involved in the inflammatory response, to be the best candidate for a predictive biomarker of early RP onset. Naqa et al.10 determined the post‐ to pre‐therapy ratios of α2M and interleukin‐6 (IL‐6) to be predictive of RP development in NSCLC patients. Castillo et al.11, 12 hypothesized that patients with naturally stronger immune responses would also be more susceptible to RP development and examined [18F]‐2‐fluoro‐2‐deoxyglucose (18F‐FDG) uptake levels in the lungs from pre‐therapy positron emission tomography (PET) scans of NSCLC patients11 and esophageal cancer patients.12 The functional data conveyed by the parameter SUV95, indicative of pre‐RT “background” lung inflammation, was found to be predictive of subsequent symptomatic RP.11, 12
The present study examined the association between development of symptomatic RP in esophageal cancer patients following RT and measures of the distribution of standardized 18F‐FDG uptake values in the lungs of those patients prior to RT. It also assessed the improvement of a new model for RP diagnosis, which combines dose‐dependent texture feature changes in CT as well as 18F‐FDG uptake in pre‐therapy PET scans, over a model that incorporates only CT texture feature changes.
2. Methods and materials
2.A. Patient population
A retrospective database of 106 esophageal cancer patients who received curative RT at The University of Texas M.D. Anderson Cancer Center was compiled as previously reported in a study that assessed the utility of CT texture features alone to study RP development.5 Each patient had a pre‐treatment CT scan, a treatment‐planning scan and at least one post‐treatment CT scan available. Only 96 of these patients also had pre‐treatment standardized uptake value (SUV) data and could be included in the present study.
The severity of RP for each patient at first presentation was determined retrospectively through consensus of three clinicians using the Common Toxicity Criteria for Adverse Events, version 4 (CTCAE v4), as described previously.5 Upon review of clinical notes including baseline respiratory function, treatment plan, and pre‐ and post‐RT imaging, each patient was assigned a binary value for RP status, which was evaluated up until six months after completion of RT or until esophagectomy: one indicated presence of symptomatic RP (Grade ≥ 2), and 0 indicated absence of symptomatic RP (Grade < 2).
2.B. PET images
A subset of the PET images used by Castillo et al.12 were acquired with calculated SUV values. For each patient, the raw PET images were converted to SUV maps on a pixel‐by‐pixel basis according to the following equation:
| (1) |
The voxels of the registered SUV map that were within the boundaries of the lung ROI were then used to generate a histogram of SUV values, from which the following statistics were calculated for each patient: the mean (SUVmean), maximum (SUVmax), standard deviation (SUVSD), and 50th, 60th, 70th, 80th, 90th and 95th percentile SUV values (SUV50–95). This resulted in a single value for each statistic to characterize the pre‐treatment tracer uptake in the lungs. The degree of overlap between high‐uptake (e.g., SUV95 or higher) and high‐dose regions of the lung was not evaluated, as it had been previously determined that this relationship did not contribute to the risk of RP in the parent database.12
2.C. CT images
Changes between pre‐treatment and post‐treatment diagnostic CT images, all of which were acquired with intravenous contrast, were analyzed as described previously.5 In summary, following application of in‐house automated lung segmentation and demons deformable registration between the pre‐ and post‐therapy diagnostic CT scans and treatment‐planning scans/dose maps of each patient, pairs of anatomically matched 32 × 32‐pixel ROIs were automatically placed in the lungs (mean: 703 ROI pairs per patient). The dose‐dependent change in each of 20 texture features distributed among first‐order, fractal, Laws' filter, and gray‐level co‐occurrence matrix (GLCM) classes, described elsewhere,13 was computed within each pair of ROIs. For each feature, a patient‐specific average change in feature value was calculated in three dose regions (0–10 Gy, 10–30 Gy, and > 30 Gy), according to:
| (2) |
where is the average change in that feature value over all ROIs located in dose region d of patient p, N p,d is the number of ROIs located in dose region d of patient p, and and are the computed feature values in ROI i of dose region d in the pre‐therapy and post‐therapy scan of patient p, respectively. While the prior study5 used a cohort of 106 patients, the present study used texture results from the subset of 96 patients who also had pre‐treatment SUV data.
2.D. Statistical analysis
2.D.1. Patient characteristic comparisons
Patient characteristics and treatment parameters were summarized using frequency tables. Associations with symptomatic RP were evaluated using the Chi‐squared test for categorical variables and the Mann–Whitney U‐test for continuous variables. Groups with an incidence of fewer than five patients were combined for Chi‐squared testing. A p‐value < 0.01 was used to assess significance.
2.D.2. SUV variable selection
SUV variables with the highest ability to distinguish between RP‐positive and RP‐negative patients were initially identified using Student's t‐tests (P < 0.05). Correlation among these SUV variable candidates was tested using Pearson's product moment correlation. Of the correlated variables, only the one with the lowest p‐value was chosen for inclusion in the logistic regression model.
2.D.3. ROC analysis of single variables
Receiver operating characteristic (ROC) analysis was used to evaluate the RP classification performance of mean lung dose (MLD) and volume of lung receiving more than 20 Gy (V20), which have both been previously used as dosimetric predictors of RP.4 The area under the ROC curve (AUC) was calculated for these variables. Additionally, ROC analysis was used to evaluate the performance of each CT texture feature and SUV variable individually. AUC values for CT texture features were computed using the average change in each feature from pre‐ to post‐therapy diagnostic CT scans in each dose region (low, medium, and high). AUC values were also calculated for all pre‐treatment SUV variables. Significance of AUC values was indicated by 95% confidence intervals (CIs) that did not overlap 0.5.
2.D.4. Regression modeling of multiple variables
Previous linear regression modeling on this database of cases indicated that texture feature change was significantly related to RP status, even when controlling for random patient effects and mean dose in each ROI.5 Thus, logistic regression models for RP as a function of two features were constructed according to:
| (3) |
where RP is the binary radiation pneumonitis status (grade ≥ 2 is positive), SUV is the SUV variable identified as described above, and ΔFV j is the mean dose‐dependent change in selected texture feature values between the pre‐ and post‐therapy CT scans. Models were created for each of 20 texture features (j) across low‐, medium‐, and high‐dose regions. Analysis of variance (ANOVA) was performed using a Chi‐squared test at an α = 0.05 level to determine whether addition of SUV to ΔFV significantly improved model fit when corrected for multiple comparisons using the Bonferroni approach (P < 0.0025). Only two features at a time were included in each regression model, as our previous modeling in this database indicated that over‐fitting occurs with more than two features.5, 14
2.D.5. ROC analysis of multiple variables
Patient data were divided into 50% training data and 50% test data by random sampling, maintaining the ratio of RP‐negative to RP‐positive cases [i.e., Fukunaga‐Hayes method].15 Following model training with the training data using Eq. (3), each model was used to assess RP diagnosis for each case in the test set, and an AUC value was calculated. This partitioning and calculation process was repeated 1000 times, and the average AUC value and confidence intervals over these iterations were obtained.
All statistical analysis was performed using Revolution R v. 6.0.
3. Results
Patient characteristics are summarized in Table 1. Of the 96 patients, 19% developed RP grade ≥ 2. Patients with tumor histology other than adenocarcinoma were more likely to develop RP. Incidence of RP was not related to smoking history, RT modality, MLD, V20, or the time interval between CT scans and RT in our database (P > 0.01).
Table 1.
Patient, treatment, and scan characteristics
| Parameter total (N) | Parameter total (%) | Symptomatica (N) | Symptomatica (%) | p‐valueb | |
|---|---|---|---|---|---|
| No of patients | 96 | 100% | 18 | 19% | N/A |
| Gender | 0.74 | ||||
| Male | 83 | 86% | 16 | 19% | |
| Female | 13 | 14% | 2 | 15% | |
| Median age (range) | 62 yrs (29–81) yrs | N/A | 65.5 yrs (48.8–81) yrs | N/A | 0.27 |
| Histology | 0.005c | ||||
| Adenocarcinoma | 80 | 83% | 11 | 14% | |
| Squamous cell carcinoma | 13 | 14% | 5 | 38% | |
| Neuroendocrine | 2 | 2% | 1 | 50% | |
| Sarcoma | 1 | 1% | 1 | 100% | |
| Sum of Squamous, Neuroendocrine, Sarcoma | 16 | 17% | 7 | 44% | |
| Smoking history | 0.55 | ||||
| Current | 13 | 14% | 1 | 8% | |
| Former | 64 | 67% | 13 | 20% | |
| Never | 19 | 20% | 4 | 21% | |
| Tumor location | 0.43d | ||||
| Distal | 56 | 58% | 9 | 16% | |
| GEJ | 27 | 28% | 4 | 15% | |
| Middle | 11 | 11% | 4 | 36% | |
| Proximal | 2 | 2% | 1 | 50% | |
| Sum of GEJ, middle, proximal | 40 | 42% | 9 | 23% | |
| Treatment modality | 0.77 | ||||
| IMRT | 55 | 57% | 9 | 16% | |
| 3D‐CRT | 17 | 18% | 4 | 24% | |
| Proton | 24 | 25% | 5 | 21% | |
| Treatment dose parameters | |||||
| Median prescribed dose (range) | 50.4 Gy (36–59.4) Gy | N/A | 50.4 Gy (45–50.4) Gy | N/A | 0.30 |
| Median number of fractions (range) | 28 (12–30) | N/A | 28 (25–28) | N/A | 0.37 |
| Median MLD (range) | 10.0 Gy (1.6–18.3) Gy | N/A | 11.3 Gy (1.6–15.7) Gy | N/A | 0.10 |
| Median lung V20% (range) | 17.6% (3.5–34.8) % | N/A | 20.9% (3.6–32.3) % | N/A | 0.16 |
| Median interval between diagnostic CT scan and treatment | |||||
| Pre‐treatment scan to RT start (range) | 25 days (0–45) days | N/A | 13 days (7–41) days | N/A | 0.03 |
| Post‐treatment scan from RT end (range) | 38 days (21–75) days | N/A | 40 days (21–75) days | N/A | 0.07 |
| Pre‐ and post‐therapy CT scan parameters (N = 192) | |||||
| Kilovoltage = 120 kVp | 189 | ||||
| Kilovoltage = 140 kVp | 3 | ||||
| Average slice thickness (range) | 2.5 mm (2.0–4.0) mm | ||||
| Average in‐plane pixel resolution (range) | 0.79 mm (0.66–0.98) mm | ||||
| Incidence of RP | |||||
| Grade 0 | 36 | 38% | |||
| Grade 1 | 42 | 44% | |||
| Grade 2 | 10 | 10% | |||
| Grade 3 | 5 | 5% | |||
| Grade 4 | 2 | 2% | |||
| Grade 5 | 1 | 1% | |||
Symptomatic RP grade ≥ 2.
Significance assessed at P < 0.01 using Chi‐squared or Mann–Whitney tests.
Adenocarcinoma versus sum of Squamous, Neuroendocrine, Sarcoma.
Distal versus sum of GEJ, Middle, Proximal.
Figure 1 compares the pre‐ and post‐therapy CT scans and the pre‐therapy SUV map of a patient who did not develop symptomatic RP with those of a patient who developed grade 5 (fatal) RP.
Figure 1.
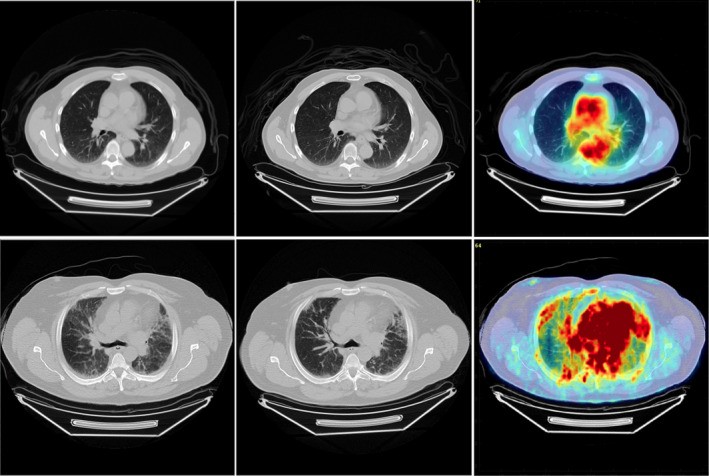
(From left to right) Spatially registered pre‐therapy CT scans, post‐therapy CT scans and pre‐therapy SUV maps for two different patients. The patient in the top row did not develop symptomatic RP. The patient in the bottom row developed grade 5 (fatal) RP. SUV color maps are presented on the same scale. [Color figure can be viewed at wileyonlinelibrary.com]
SUV parameter values for the 96 patients are summarized in Fig. 2. SUVSD differed the most between the RP‐negative and RP‐positive groups (P = 0.015), while SUVmax differed second most (P = 0.027). Because SUVSD was significantly correlated with SUVmax (r = 0.806), SUVSD alone was selected for inclusion in the model. The AUC values obtained from RP status classification based on each SUV parameter alone are depicted in Fig. 3, demonstrating that SUVmax and SUVSD are the only parameters with AUC values significantly different from 0.5, equaling 0.71 (95% CI: 0.56–0.85) and 0.69 (95% CI: 0.54–0.83), respectively.
Figure 2.
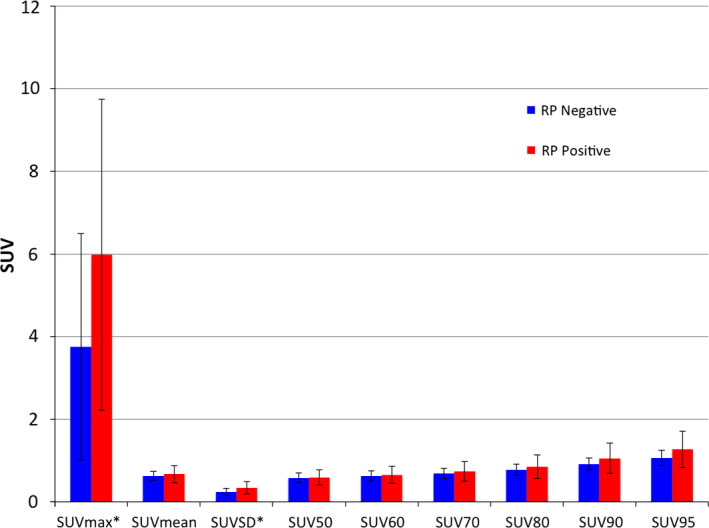
Mean SUV parameter values across patients, separated by RP status. Error bars indicate one standard deviation. Significant differences between groups using Student's t‐test (p < 0.05) are indicated with an asterisk (*). [Color figure can be viewed at wileyonlinelibrary.com]
Figure 3.
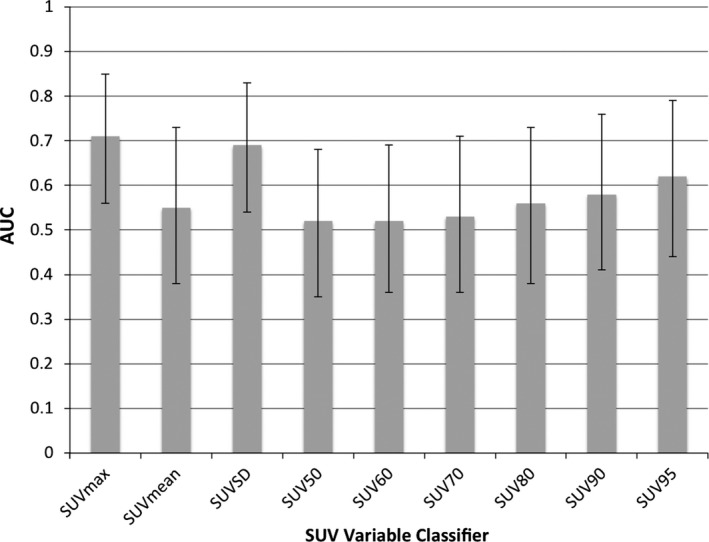
AUC values for SUV variables as lone classifiers. (Error bars indicate 95% CI of AUC). SUVmax and SUVSD are the only variables whose AUC values are significantly different from 0.5.
Our previous work5 identified 12 CT texture features distributed among four feature classes that were associated with RP status, even when controlling for mean dose in each ROI using linear regression modeling (indicated with ‘a’ in Table 2). In the present study, ROC analysis for each CT texture feature in each dose region resulted in feature‐averaged AUC values > 0.5 as listed in Table 2. For 17 of these features, AUC values differed significantly from 0.5 in at least one dose region (indicated with ‘b’ in Table 2). However, with the exception of the low‐dose regions, these values were generally higher (by 0.02 and 0.03 for medium‐ and high‐dose regions on average, respectively) than those obtained previously,5 likely due to the reduced database size. ROC curves created for MLD and V20 resulted in AUCs of 0.625 (95% CI: 0.469–0.782) and 0.615 (95% CI: 0.469–0.761), respectively, indicating no significant differences from 0.5. This demonstrates that, unlike SUVSD or texture features, MLD and V20 did not correlate with RP in our database. To combine two discriminators, logistic regression models comprising one CT texture feature and SUVSD were used, and AUC values of the classifiers were computed. With the addition of SUVSD, AUC values improved by 0.08, 0.06, and 0.04 on average in the low‐, medium‐, and high‐dose regions, respectively, over classification based on the single‐texture feature alone. ANOVA comparisons of these logistic regression models using Chi‐squared tests corrected for multiple testing (P < 0.0025) showed SUVSD significantly improved model fit when added to 19 of the 20 CT texture features in at least one dose region (indicated by ‘c’ in Table 2). Adding SUVSD significantly improved model fit for 18 texture features calculated in low‐dose regions, where the single‐texture feature average AUC was lower in value, compared with 11 texture features calculated in high‐dose regions.
Table 2.
AUC values and 95% CIs for ΔFV with and without SUVSD in three dose ranges to assess feature performance in determining RP status
| Low dose (0–10 Gy) | Medium dose (10–30 Gy) | High dose (> 30 Gy) | ||||
|---|---|---|---|---|---|---|
| Feature alone | Feature + SUVSD | Feature alone | Feature + SUVSD | Feature alone | Feature + SUVSD | |
| First‐order features | ||||||
| 70% quantilea | 0.71b [0.58, 0.85] | 0.79c [0.67, 0.90] | 0.78b [0.66, 0.91] | 0.82 [0.72, 0.93] | 0.80b [0.68, 0.92] | 0.83 [0.72, 0.94] |
| Mediana | 0.70b [0.56, 0.84] | 0.78c [0.66, 0.90] | 0.75b [0.61, 0.89] | 0.80c [0.69, 0.92] | 0.78b [0.65, 0.91] | 0.83 [0.72, 0.93] |
| Meana | 0.68b [0.54, 0.83] | 0.77c [0.63, 0.89] | 0.76b [0.62, 0.89] | 0.81c [0.70, 0.92] | 0.79b [0.67, 0.90] | 0.83c [0.72, 0.94] |
| Binned entropya | 0.66b [0.51, 0.80] | 0.74c [0.61, 0.85] | 0.74b [0.61, 0.87] | 0.78 [0.66, 0.90] | 0.77b [0.64, 0.90] | 0.80 [0.68, 0.91] |
| 30% quantilea | 0.68b [0.53, 0.83] | 0.76c [0.64, 0.89] | 0.72b [0.57, 0.87] | 0.78c [0.66, 0.90] | 0.76b [0.62, 0.90] | 0.82c [0.71, 0.94] |
| Unbinned entropy | 0.64b [0.50, 0.78] | 0.70c [0.56, 0.83] | 0.68b [0.54, 0.82] | 0.73c [0.61, 0.85] | 0.73b [0.60, 0.85] | 0.77c [0.67, 0.88] |
| 5% quantile | 0.63 [0.48, 0.79] | 0.71c [0.58, 0.83] | 0.67b [0.52, 0.82] | 0.73c [0.60, 0.85] | 0.69b [0.55, 0.84] | 0.76c [0.63, 0.89] |
| Minimum | 0.61 [0.45, 0.77] | 0.68c [0.53, 0.80] | 0.60 [0.44, 0.76] | 0.68c [0.55, 0.81] | 0.70b [0.55, 0.84] | 0.70c [0.56, 0.83] |
| Fractal features | ||||||
| Brownian dimensiona | 0.67b [0.51, 0.83] | 0.71 [0.57, 0.85] | 0.74b [0.61, 0.87] | 0.79 [0.68, 0.89] | 0.81b [0.70, 0.92] | 0.79c [0.67, 0.91] |
| Box‐counting dimension | 0.53 [0.36, 0.69] | 0.65c [0.51, 0.78] | 0.55 [0.39, 0.71] | 0.66c [0.52, 0.79] | 0.64 [0.48, 0.80] | 0.72c [0.58, 0.84] |
| Fine box‐counting dimension | 0.55 [0.39, 0.71] | 0.65c [0.51, 0.77] | 0.54 [0.37, 0.70] | 0.65c [0.51, 0.77] | 0.58 [0.41, 0.75] | 0.67c [0.53, 0.80] |
| Laws' filter features | ||||||
| E5L5 entropya | 0.62 [0.48, 0.76] | 0.72c [0.56, 0.84] | 0.74b [0.62, 0.86] | 0.80c [0.70, 0.90] | 0.80b [0.70, 0.90] | 0.83c [0.73, 0.92] |
| R5L5 entropya | 0.63 [0.49, 0.77] | 0.73 [0.60, 0.86] | 0.74b [0.61, 0.86] | 0.78 [0.68, 0.89] | 0.80b [0.68, 0.91] | 0.81 [0.70, 0.91] |
| S5L5 entropya | 0.62 [0.49, 0.76] | 0.71c [0.56, 0.83] | 0.73b [0.61, 0.86] | 0.79c [0.68, 0.89] | 0.79b [0.69, 0.90] | 0.82 [0.72, 0.91] |
| W5L5 entropya | 0.63b [0.50, 0.76] | 0.73c [0.61, 0.83] | 0.74b [0.61, 0.86] | 0.79c [0.70, 0.89] | 0.79b [0.68, 0.90] | 0.81 [0.71, 0.91] |
| GLCM features | ||||||
| Sum averagea | 0.68b [0.54, 0.83] | 0.77c [0.65, 0.88] | 0.75b [0.62, 0.88] | 0.81 [0.70, 0.92] | 0.79b [0.67, 0.90] | 0.83 [0.73, 0.93] |
| Sum of squares variancea | 0.69b [0.55, 0.83] | 0.77c [0.65, 0.88] | 0.76b [0.63, 0.89] | 0.81 [0.70, 0.92] | 0.79b [0.68, 0.90] | 0.83 [0.73, 0.94] |
| Sum entropy | 0.66b [0.52, 0.80] | 0.71c [0.58, 0.83] | 0.70b [0.56, 0.83] | 0.75c [0.62, 0.87] | 0.75b [0.63, 0.86] | 0.78 [0.68, 0.89] |
| Difference entropy | 0.60 [0.45, 0.75] | 0.67c [0.53, 0.80] | 0.64 [0.49, 0.79] | 0.70c [0.57, 0.82] | 0.68b [0.54, 0.82] | 0.73c [0.61, 0.85] |
| Entropy | 0.57 [0.42, 0.72] | 0.67c [0.48, 0.81] | 0.59 [0.44, 0.75] | 0.68c [0.54, 0.80] | 0.61 [0.46, 0.77] | 0.68c [0.53, 0.80] |
| Average AUC | 0.64 | 0.72 | 0.70 | 0.76 | 0.74 | 0.78 |
Significant relationship between change in feature value (ΔFV) and grade ≥ 2 RP (P < 0.0025).
95% CI of AUC shown in brackets does not overlap 0.5.
Significant improvement of logistic regression model fit with addition of SUVSD using ANOVA (P < 0.0025).
4. Discussion
Although the means and medians of SUV values for pixels within the lungs do not appear to vary much between RP‐positive and RP‐negative patients, the RP‐positive group tends to exhibit a greater frequency of high SUV values (associated with higher FDG uptake) in the lung (Fig. 1). This tendency leads to selection of SUVSD as a viable candidate for model improvement, while this variable's correlation with SUVmax justifies the exclusion of SUVmax from model building. Since the logistic regression models for RP assessment improve significantly in at least one dose region when SUVSD is individually paired with 19 of the 20 CT texture features, it is highly likely that SUVSD provides information independent from that provided by texture analysis. This finding was expected, as SUV measures baseline lung inflammation prior to RT, while CT texture feature changes are indicative of radiation‐induced reactionary biologic processes. Although texture change demonstrated in post‐therapy CT scans may be influenced both by radiation dose and by the development of RP (which itself is affected by radiation dose), our prior work has shown that CT texture change is related to RP development even when controlling for dose.5 A more thorough approach to determine independence of SUV variables from CT texture changes would involve linear modeling of texture feature change as a function of several variables, including SUV parameters, to determine whether these radiation‐induced changes are affected by pre‐RT lung inflammation. Our previous work demonstrated that use of three features in this limited database does not significantly improve the model fit, likely due to over‐fitting.5, 14 This result may change if tested on a larger sample size with more positive cases.
The AUC values obtained from this study should be validated using an independent patient cohort because this data set was also used to select the SUV variable included in the analysis. While the texture classifiers were selected in an independent database,13 repeating ROC analysis on an independent data set is still recommended due to the low prevalence of positive cases among these patients. This is particularly important given that a relatively slight but representative reduction in this data set (two RP‐positive and eight RP‐negative cases out of the initial 106 cases) resulted in higher AUC values across features compared with the results obtained by Cunliffe et al.5 Nevertheless, the AUC value for ΔFV was significantly higher than 0.5 for 11, 15, and 17 texture features for low‐, medium‐, and high‐dose regions, respectively. Although there was an insufficient number of RP events to parse the data set into training and validation sets for selecting the SUV parameter, such a partitioning was done when calculating confidence intervals for the AUC values in Table 2. The bivariate model built using the 50%/50% data partition resulted in the smallest 95% CIs for AUC values when compared with a 75%/25% partition or leave‐one‐out cross validation. Because it resulted in similar or slightly lower AUC values, the 50%/50% data partition also ensured a more conservative estimate of the AUC, which was prudent given the small number of positive cases in our database.
This study analyzed a subset of data reported on by Castillo et al.,12 who demonstrated that SUV95 had AUC values of 0.676 for classifying RP, which is comparable to the AUC calculated in this study using SUDSD (0.69). In the present study, SUVSD improved the AUC in a regression model primarily when the CT texture feature under consideration was a poor classifier by itself. That is, the utility of SUVSD was limited in the presence of other good classifiers of RP. This finding was demonstrated by the less dramatic increase in feature‐averaged AUC of 0.04 for the high‐dose regions, where texture feature changes were more likely to occur and provide diagnostic information. Significant improvements in model fitting with the addition of SUVSD also occurred less frequently in high‐dose versus low‐dose regions. On the other hand, SUVSD improved AUC more frequently and by a wider margin for low‐ and medium‐dose measurements, where texture feature value change was less pronounced and thus a poorer classifier. SUVSD had this effect despite the fact that, on its own, its AUC value (0.69) was less than or equal to the average AUC value across many texture features, which attests to the independence of the SUV data from the CT texture data. Furthermore, SUV data were gathered from pre‐treatment scans alone, thus potentially providing prognostic information to clinicians prior to the design of the radiotherapy plan.
Dosimetric parameters such as V20 and MLD, which have been shown to correlate with RP in large analyses utilizing pooled, multi‐institutional data for both standard fractionated16, 17 and hypofractionated photon treatments18 delivered with 3DCRT, IMRT or proton treatments,19 did not reach significance in the current data set. While other dosimetric parameters such as V5 could have been studied, it is unlikely that results would differ because dosimetric parameters tend to be highly correlated.17 Furthermore, analysis of the parent data set from which the data set in the present study was obtained indicated no correlation of V5, V10, V20, V30, or MLD with RP development, a finding that is not expected to change in this smaller subset.12
Contrary to other studies,11, 12 the present work demonstrated that CT texture change correlates with RP development. Other groups compared pre‐treatment CT Hounsfield unit (HU) statistics calculated over the entire lung volume to SUV and determined that CT values were not associated with RP. The present study calculated texture in many (> 700) small regions of the lung and tracked the planned dose to these regions, instead of characterizing CT texture throughout the lung using a single HU statistic. Results demonstrated that texture changes in high‐dose regions are more strongly associated with RP status. Furthermore, changes in texture before and after treatment were quantified, thus controlling for patient‐dependent effects that may be indicative of underlying co‐morbidities or differences in CT acquisition parameters.13 Finally, half of the features reported here were higher order features that are mathematically derived (i.e., agnostic features that do not correlate with changes that are readily identified by eye), thus harnessing the full power of radiomics.20
Future work would incorporate a larger and independent patient cohort with a greater number of positive cases, particularly since PET‐based features were identified and tested in the same database. Further AUC analysis could include linear modeling of AUC as a function of the number of variables used to verify the effect of SUVSD as a predictor. Other SUV variables could also be evaluated as predictors, although Castillo et al. showed that SUV variables were highly correlated, thus conjecturing that predictive models would not improve with addition of more than a single SUV parameter.12 The most relevant features could then be combined into a single classifier, enabling evaluation of false‐positive/false‐negative rates once a single cut‐off value along the ROC curve is identified. The time interval effects on sensitivity and specificity could potentially be evaluated in a larger data set which prospectively tracks RP development. Such effects could not be studied in the current data set due to the uncertainty in identifying the exact time of RP development that resulted from retrospective data collection.5
The texture of SUV maps themselves could also be analyzed as a next step in texture analysis for prediction of RP. Several studies have already examined relationships between FDG‐PET texture features and cancer outcomes. El Naqa et al. have found texture‐based features from SUV images to be predictive of tumor response in head and neck and cervical cancers21 and more correlated with local control in NSCLC patients than CT texture features.22 Yip et al. found the temporal change in PET texture features before and after chemo‐radiation to be more predictive of patient response than SUVmean or SUVmax.23 Such PET‐based radiomics could easily be applied toward prediction of RP. However, texture features from SUV images in the thorax suffer from errors attributable to motion,22 and there is an evident need to standardize the texture analysis methodology applied to SUV images.24
This pilot study demonstrated that quantitative image analysis (i.e., radiomics) has the potential to assess development of symptomatic RP, particularly when a patient's baseline CT scan is used as a control. Similar to other studies of this clinical endpoint,5, 12, 17 our work is limited by the uncertainty in RP diagnosis that is associated with retrospective identification through the medical record. It remains to be determined, ideally in a prospective clinical trial, whether these quantitative techniques could impact clinical care. Given the emergence of trials testing immunotherapy drugs such as PD‐1 inhibitors in conjunction with RT, for which there is an increased incidence of RP induction,25 automated techniques that could identify patients requiring closer clinical management could be used as a secondary endpoint in such trials.
5. Conclusion
This study found SUVSD to be significantly correlated with development of symptomatic (Grade ≥ 2) RP. When distinguishing patients with symptomatic RP from those without, inclusion of SUVSD in a logistic regression model significantly improved model fit and increased the AUC over the use of dose‐dependent CT texture feature changes as lone classifiers; this improvement was most pronounced in lower dose regions. To our knowledge, this is the first study to examine lung CT texture features together with whole‐lung FDG PET information as potential predictors of treatment outcomes following radiotherapy.
Conflicts of interest
Dr. Armato III reports grants from NIH/NIBIB, during the conduct of the study; personal fees from Aduro Biotech, Inc., outside the submitted work; and royalties and licensing fees related to computer‐aided diagnosis technology through The University of Chicago. Dr. Al‐Hallaq reports royalties for computer‐aided diagnosis software for breast cancer detection, licensed from The University of Chicago.
Acknowledgments
Supported, in part, by the U.S. National Institute of Biomedical Imaging and Bioengineering of the National Institutes of Health under grant number T32 EB002103. Dr Castillo was supported in part by a NIH research scientist development award K01CA181292. We thank Sang Mee Lee, Ph.D., Biostatistics Lab, Department of Public Health Sciences at The University of Chicago for providing statistical guidance.
References
- 1. Abratt RP, Morgan GW. Lung toxicity following chest irradiation in patients with lung cancer. Lung Cancer Amst Neth. 2002;35:103–109. [DOI] [PubMed] [Google Scholar]
- 2. Rubin P, Johnston CJ, Williams JP, et al. A perpetual cascade of cytokines postirradiation leads to pulmonary fibrosis. Int J Radiat Oncol. 1995;33:99–109. [DOI] [PubMed] [Google Scholar]
- 3. Inoue A, Kunitoh H, Sekine I, et al. Radiation pneumonitis in lung cancer patients: a retrospective study of risk factors and the long‐term prognosis. Int J Radiat Oncol. 2001;49:649–655. [DOI] [PubMed] [Google Scholar]
- 4. Rodrigues G, Lock M, D'Souza D, et al. Prediction of radiation pneumonitis by dose ‐ volume histogram parameters in lung cancer–a systematic review. Radiother Oncol J Eur Soc Ther Radiol Oncol. 2004;71:127–138. [DOI] [PubMed] [Google Scholar]
- 5. Cunliffe A, Armato SG III, Castillo R, et al. Lung texture in serial thoracic computed tomography scans: correlation of radiomics‐based features with radiation therapy dose and radiation pneumonitis development. Int J Radiat Oncol. 2015;91:1048–1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chabat F, Yang G‐Z, Hansell DM. Obstructive lung diseases: texture classification for differentiation at CT. Radiology. 2003;228:871–877. [DOI] [PubMed] [Google Scholar]
- 7. Mattonen SA, Palma DA, Haasbeek CJA, et al. Early prediction of tumor recurrence based on CT texture changes after stereotactic ablative radiotherapy (SABR) for lung cancer. Med Phys. 2014;41:033502. [DOI] [PubMed] [Google Scholar]
- 8. Oh JH, Craft JM, Townsend R, et al. A bioinformatics approach for biomarker identification in radiation‐induced lung inflammation from limited proteomics data. J Proteome Res. 2011;10:1406–1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Craft JM, Oh J, Ju M, et al. Quantitative mass spectroscopy and the identification of Alpha2macroglobulin as a potential biomarker for radiation pneumonitis. Int J Radiat Oncol. 2010;78:S498–S499. [Google Scholar]
- 10. Naqa IE, Bradley J, Oh J, et al. Investigating alpha‐2‐macroglobulin and its dosimetric interactions for predicting radiation pneumonitis. Int J Radiat Oncol Biol Phys. 2011;81:S756–S757. [Google Scholar]
- 11. Castillo R, Pham N, Ansari S, et al. Pre‐radiotherapy FDG PET predicts radiation pneumonitis in lung cancer. Radiat Oncol. 2014;9:74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Castillo R, Pham N, Castillo E, et al. Pre‐radiation therapy fluorine 18 fluorodeoxyglucose PET helps identify patients with esophageal cancer at high risk for radiation pneumonitis. Radiology. 2015;275:822–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cunliffe AR, Al‐Hallaq HA, Labby ZE, et al. Lung texture in serial thoracic CT scans: assessment of change introduced by image registration. Med Phys. 2012;39:4679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hua J, Xiong Z, Lowey J, et al. Optimal number of features as a function of sample size for various classification rules. Bioinforma Oxf Engl. 2005;21:1509–1515. [DOI] [PubMed] [Google Scholar]
- 15. Sahiner B, Chan H‐P, Hadjiiski L. Classifier performance prediction for computer‐aided diagnosis using a limited dataset. Med Phys. 2008;35:1559–1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Marks LB, Bentzen SM, Deasy JO, et al. Radiation dose‐volume effects in the lung. Int J Radiat Oncol Biol Phys. 2010;76:S70–S76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Palma DA, Senan S, Tsujino K, et al. Predicting radiation pneumonitis after chemoradiation therapy for lung cancer: an international individual patient data meta‐analysis. Int J Radiat Oncol Biol Phys. 2013;85:444–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhao J, Yorke ED, Li L, et al. Simple factors associated with radiation‐induced lung toxicity after stereotactic body radiation therapy of the thorax: a pooled analysis of 88 studies. Int J Radiat Oncol Biol Phys. 2016;95:1357–1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Remick JS, Schonewolf C, Gabriel P, et al. First clinical report of proton beam therapy for postoperative radiotherapy for non‐small‐cell lung cancer. Clin Lung Cancer. 2017; doi: 10.1016/j.cllc.2016.12.009. [DOI] [PubMed] [Google Scholar]
- 20. Gillies RJ, Kinahan PE, Hricak H. Radiomics: images are more than pictures they are data. Radiology. 2016;278:563–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. El Naqa I, Grigsby P, Apte A, et al. Exploring feature‐based approaches in PET images for predicting cancer treatment outcomes. Pattern Recognit. 2009;42:1162–1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Vaidya M, Creach KM, Frye J, et al. Combined PET/CT image characteristics for radiotherapy tumor response in lung cancer. Radiother Oncol J Eur Soc Ther Radiol Oncol. 2012;102:239–245. [DOI] [PubMed] [Google Scholar]
- 23. Yip SSF, Coroller TP, Sanford NN, et al. Relationship between the temporal changes in positron‐emission‐tomography‐imaging‐based textural features and pathologic response and survival in esophageal cancer patients. Front Oncol. 2016;6:72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Leijenaar RTH, Nalbantov G, Carvalho S, et al. The effect of SUV discretization in quantitative FDG‐PET radiomics: the need for standardized methodology in tumor texture analysis. Sci Rep. 2015;5:11075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Nishino M, Giobbie‐Hurder A, Hatabu H, et al. Incidence of programmed cell death 1 inhibitor–related pneumonitis in patients with advanced cancer: a systematic review and meta‐analysis. JAMA Oncol. 2016;2:1607–1616. [DOI] [PubMed] [Google Scholar]


