Abstract
Background
Gastric microbiota along with Helicobacter pylori (HP) plays a key role in gastric disease. The aim of our study is to investigate the difference of human gastric microbiota between antrum and body according to disease (control vs. gastric cancer) and HP status.
Methods
Each antrum and body biopsy was collected from 12 subjects at Seoul National University Bundang Hospital. Gastric microbiota was analyzed by bar-coded 454 pyrosequencing of the 16S rRNA gene. Twelve subjects consisted of HP-negative control (n = 2), HP-negative cancer (n = 2), HP-positive control (n = 3), and HP-positive cancer (n = 5). The analysis was focused on non-HP urease-producing bacteria (UB) and non-HP nitrosating or nitroreducing bacteria (NB) between antrum and body.
Results
Gastric body samples showed higher diversity compared to gastric antrum mucosa samples but there was no significant difference. The mean of operational taxonomic units was higher in HP(−) cancer than HP(+) cancer (antrum, 273.5 vs. 228.2, P = 0.439; body, 585.5 vs. 183.2, P = 0.053). The number of non-HP UB and non-HP NB was higher in HP(−) cancer groups than the others. These differences were more pronounced in the body (P = 0.051 and P = 0.081, respectively). Analysis of overlap of non-HP UB and non-HP NB revealed the higher composition of Streptococcus pseudopneumoniae, S. parasanguinis, and S. oralis in HP(−) cancer groups than the others, only in the body (P = 0.030) but not in the antrum (P = 0.123).
Conclusions
Higher diversity and higher composition of S. pseudopneumoniae, S. parasanguinis, and S. oralis in HP(−) cancer group than the other groups in the body suggest that analysis of microbiota from body mucosa could be beneficial to identify a role of non-HP bacteria in the gastric carcinogenesis.
Keywords: Microbiota, Helicobacter pylori, Antrum mucosa, Body mucosa
INTRODUCTION
Human is constantly exposed to pathogenic microorganisms, such as bacteria, viruses, and fungi. The stomach was considered as a sterile organ due to acid production. However, Helicobacter pylori (HP) was found to be colonized in the gastric epithelium of more than half of the world’s human population.1 HP generates large quantities of urease, an enzyme capable of transiently buffering the acidic environment by the break-down of urea to generate ammonia and carbon dioxide.2 These two products could serve as substrates for other microbes and change the gastric microbiome.3 Additionally, HP urease is a major inducer of innate immune response in monocytes, macrophages, and neutrophils. Accumulation and activation of these cells is induced by the local production of chemokines, cytokine, and NO generation.4–6 A potent pro-inflammatory cytokines, such as interleukin (IL)-1β and TNF-α are produced during HP infection and IL-1β is also a powerful inhibitor of gastric acid secretion.7,8 HP infection is a risk factor for gastric cancer, which causes atrophic gastritis regulating inflammatory response or N-nitroso compounds (NOC) production.9,10 The product of NOC has been suggested to increase the risk of cancers.10 It has been known that many urease-producing bacteria (UB) and non-HP nitrosating or nitrate-reducing bacteria (NB) other than HP exist in stomach.
Recent advances in next-generation sequencing technology have revealed a complex gastric microbiome which may contribute to the development of gastric carcinogenesis. Our previous studies revealed that gastric microbiota were different according to HP infection status and presence or absence of gastric cancer in gastric mucosa by using a pyrosequencing method.11–13 We also conducted a research which suggested that gastric mucosa could be more effective than gastric fluid in the detection of meaningful gastric microbiome.11 On the other hand, gastric antrum and body are different in terms of acid secretion. Acid secretion depends on activation of the gastric H, K-ATPase, termed as the acid or proton pump.14 This enzyme was found uniquely in gastric parietal cells which are located at oxyntic gastric gland of the body. There is a close interaction or battle between this acid secretion and HP. On the contrary to the usual concept, HP is neutralophiles.15 That it, in the case of subjects with high acid-secretion, HP escapes from body and settles in the antrum leading to antrum-predominant gastritis.16 However, when HP succeeds in the colonization, it begins to dominant in the stomach resulting in decrease of microbiota diversity. However, when atrophy and intestinal metaplasia occur, then HP itself decreases to colonize in the stomach17 and eventually diversity of microbiota increases due to higher pH of gastric juice. From this background, we made a hypothesis that gastric microbiota could be different between in the antrum and in the body. Although we found a minor role of non-HP bacteria in the gastric carcinogenesis in the antrum,12 the microbiota analysis from body could be different. Thus the aim of our study is to investigate the difference of human gastric microbiota between antrum and body according to disease (control vs. gastric cancer) and HP status.
MATERIALS AND METHODS
1. Study subjects and gastric tissue specimen collection
Gastric biopsies were collected from 12 subjects who underwent standard endoscopy to screen for premalignant or malignant gastric mucosal lesions or received endoscopy due to dyspepsia. Gastric mucosal (antrum and body) biopsies and blood samples were obtained from each patient during endoscopy from October 2008 to March 2013 at Seoul National University Bundang Hospital. Ten biopsy specimens per subjects were obtained to perform HP tests and pyrosequencing as our previous study.11–13 Gastric biopsy specimens were assessed for the presence of HP and for the degree of inflammatory cell infiltration, atrophy, and intestinal metaplasia (H&E staining). Histological features of gastric mucosa were recorded as the updated Sydney scoring system (i.e., 0 = none, 1 = mild, 2 = moderate, 3 = marked).18 To avoid contamination, the endoscopes were washed and disinfected by immersing in a detergent solution containing 7% proteolytic enzymes and 2% glutaraldehyde and sterilized gastroscopy forceps were used while gaining another biopsy from the same patient. The biopsies were stored at −80°C. This study was approved by the ethics committee of Seoul National University Bundang Hospital (B-1112/141-007). Written informed consent was obtained from all of the participants.
2. Determination of H. pylori infection status
To determine the presence of current HP infection according to conventional tests: 1) rapid urease test (CLO test; Delta West, Bentley, Australia), 2) histologic examination (modified Giemsa staining), 3) culture for HP. Current HP infection was positive from any of the former three tests. In order to distinguish if the infection is an existing one, the following two methods were used: Serum HP immunoglobulin G (Genedia HP ELISA; Green Cross Medical Science Co., Eumseong, Korea), and a history of HP infection eradication treatment. If all the 5 tests were negative, we would have regarded the subject as HP-negative. Besides, by using a Latex-enhanced Turbidimetric Immunoassay (Shima Laboratories, Tokyo, Japan), serum concentrations of Pepsinogen I and II were evaluated, which are known to be associated with the severity of gastric atrophy.19
3. Bacterial genomic DNA extraction
The antrum and body mucosal samples from 12 subjects were subjected to pyrosequencing. Bacterial genomic DNA (gDNA) was extracted with the commercial kit (iNtRON Biotechnology, Seongnam, Korea).
4. 16S rRNA sequencing
PCR amplification was done by using primers targeting the V1 to V3 regions of bacterial 16S rRNA gene with bacterial gDNA. For bacterial amplification, barcoded primers of 9F (5′-CCTATCCC-CTGTGTGCCTTGGCAGTC-TCAG-AC-AGAGTTTGATCMTGGCTCAG-3′; underlined sequence indicates the target region primer) and 541R (5′-CCATCTCATCCCTGCGTGTCTCCGAC-TCAG-X-AC-ATTACCGCGGCTGCTGG-3′; ‘X’ presents the unique barcode for each subject) (http://oklbb.ezbiocloud.net/content/1001) as previous study were shown. The sequencing was performed at Chunlab Inc. (Seoul, Korea) with GS Junior Sequencing system, the modified laboratory benchtop form of 454 sequencing systems (Roche, Branford, CT, USA) as stated in the manufacturer’s directions.
5. Pyrosequencing data analysis
The primary analysis was conducted as described above. Reads taken from different samples were classified by unique barcodes of each PCR product. After identifying the target region in barcoded primers (9F or 541R), all of the linked sequences including adapter, barcode and linker were eliminated. Low quality sequences, such as reads containing two or more indefinite nucleotides, reads with a low quality score (average score < 25), or reads shorter than 300 bp, were eliminated. Potential chimeric sequences were confirmed by the Bellerophon formula, which compares the BLASTN search conclusions between the forward half and reverse half sequences.16 After removing the chimeric sequences, the taxonomic sorting of each read was assigned against the EzTaxon-e database (http://eztaxon-e.ezbiocloud.net),17 which has the 16S rRNA gene sequence of type strains that have valid published names and representative species level phylotypes of either cultured or uncultured entries in the GenBank database with complete hierarchical taxonomic classification from the phylum to the species. Phylogenetic trees were not created as we assigned reads into operational taxonomic units (OTUs) according to BLAST results. The raw 16S rRNA gene sequence originated from our study was deposited in NCBI’s SRA (GSE61493).
6. Evaluation of species richness and diversity
The species richness of samples was determined using the CLcommunity program (Chunlab Inc.). Random subsampling was conducted to equalize the read size of samples to compare the different read size within samples. To compare the OTUs between samples, shared OTUs were obtained with the XOR analysis of the CLcommunity program (Chunlab Inc.).
7. Statistical analysis
Comparisons between continuous parameters were performed by Kruskal-Wallis test and Mann-Whitney test. Statistical analyses were done by Prism 5 (GraphicPad Software Inc., La Jolla, CA, USA) and PASW 18.0 (IBM, Somers, NY, USA). Results with a P-values < 0.05 were considered statistically significant.
RESULTS
1. Baseline characteristics
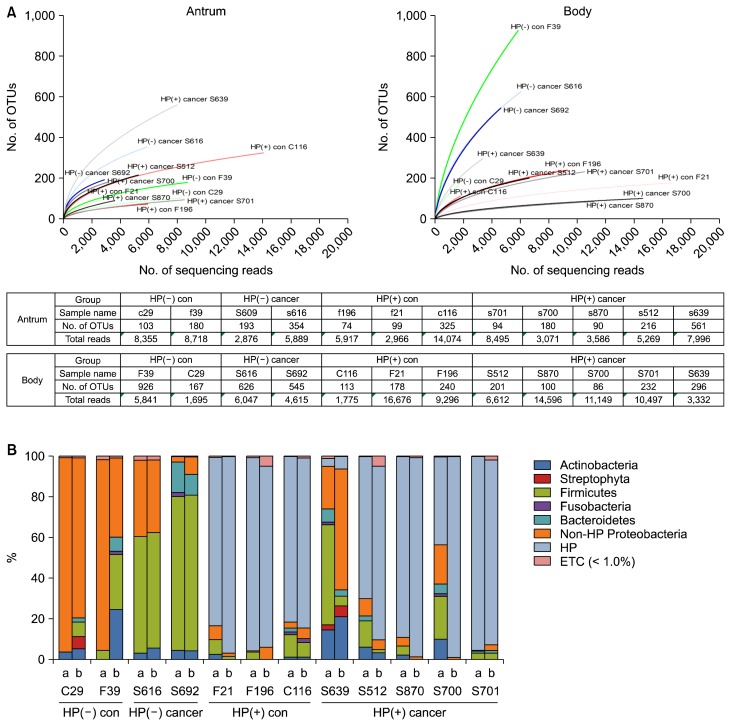
A total of 12 subjects were enrolled in this study, two HP(−) controls, two HP(−) cancer, three HP(+) controls and five HP(+) cancer patients. Baseline characteristics of clinical results of gastric antrum and body mucosa samples are shown in Table 1. The mean age of subjects was higher in the HP(−) groups than in the HP(+) groups (60.8 years vs. 52 years; P = 0.174). However, there was no significant difference between the two groups. Pepsinogen I/II ratio reflecting gastric atrophy was no significant difference between the two groups. (3.4 vs. 2.3, P = 0.173; Table 1). The grades of neutrophils and monocytes infiltration were lower in the HP(−) groups compared to those in the HP(+) groups (antrum, P = 0.006, P = 0.037; body, P = 0.001, P = 0.041 respectively). Additionally, the grades of neutrophils and monocytes infiltration were significantly different between HP (+) cancer group and the others (antrum, P = 0.046, P = 0.184; body, P = 0.013, P = 0.162, respectively).
Table 1.
Baseline characteristics of 12 subjects
| Group | Subject No. | Sex/age (yr) | Site | Intestinal metaplasia | Neutrophil infiltration | Monocyte infiltration | Atrophy | CLO | H&E | HP IgG | PG I/II ratio | Eradication history | Helicobacter pylori (%) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HP(−) control | C29 | F/40 | Antrum | Mild | No | Mild | INA | - | - | 0.286 | 5.0 | No | 0.192 |
| Body | No | No | Mild | 0 | - | - | 0.000 | ||||||
| F39 | F/67 | Antrum | No | No | Mild | 0 | - | - | 0.095 | 4.8 | No | 0.023 | |
| Body | No | No | Mild | 0 | - | - | 0.839 | ||||||
| HP(−) cancer | S692 | F/75 | Antrum | No | No | Moderate | INA | - | - | 0.028 | 0.4 | No | 0.035 |
| Body | Mild | Mild | Moderate | INA | - | - | 0.043 | ||||||
| S616 | M/61 | Antrum | Moderate | No | Mild | INA | - | - | 0.029 | 3.3 | No | 0.187 | |
| Body | Moderate | No | Mild | 0 | - | - | 0.149 | ||||||
| HP(+) control | F21 | M/55 | Antrum | Mild | Moderate | Moderate | 1 | + | Mild | 3.338 | 2.0 | No | 82.873 |
| Body | No | Moderate | Moderate | 0 | + | Moderate | 96.288 | ||||||
| F196 | F/56 | Antrum | Mild | Moderate | Moderate | INA | + | Moderate | N/A | 2.3 | No | 94.997 | |
| Body | No | Moderate | Moderate | 0 | + | Moderate | 89.436 | ||||||
| C116 | M/41 | Antrum | No | Moderate | Marked | INA | + | Marked | 2.341 | 2.0 | No | 81.540 | |
| Body | No | Moderate | Marked | 2 | + | Moderate | 83.606 | ||||||
| HP(+) cancer | S512 | F/36 | Antrum | Mild | Moderate | Marked | INA | + | Moderate | 2.301 | 3.0 | No | 69.842 |
| Body | No | Moderate | Moderate | 0 | + | Moderate | 85.466 | ||||||
| S700 | F/54 | Antrum | Moderate | Moderate | Marked | 2 | + | Marked | N/A | 2.9 | No | 43.308 | |
| Body | No | Moderate | Marked | INA | + | Marked | 98.780 | ||||||
| S701 | M/57 | Antrum | No | Moderate | Moderate | INA | + | Moderate | N/A | 1.8 | No | 95.515 | |
| Body | No | Moderate | Moderate | INA | + | Marked | 91.245 | ||||||
| S870 | M/53 | Antrum | No | Moderate | Moderate | 0 | + | Mild | N/A | 1.9 | No | 88.957 | |
| Body | Mild | Moderate | Moderate | 1 | + | Marked | 97.986 | ||||||
| S639 | F/64 | Antrum | Moderate | No | Mild | INA | - | - | 0.059 | 2.8 | No | 3.902 | |
| Body | No | Moderate | Mild | INA | - | Mild | 6.182 |
CLO, Campylobacter-like organism; HP IgG, H. pylori immunoglobulin G; PG, pepsinogen; F, female; M, male; INA, inadequate to assess atrophy; N/A, not assessed.
2. Gastric antrum versus body mucosa
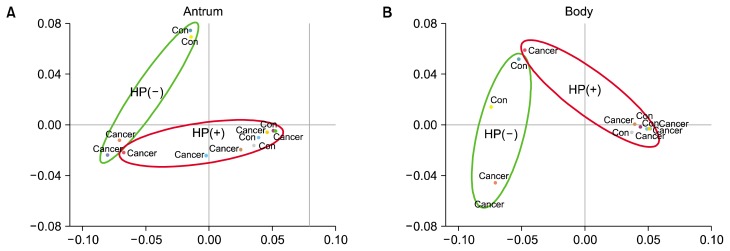
The means of reads and OTUs were lower in gastric antral mucosa samples than gastric body mucosa samples (Fig. 1A). Gastric body mucosa samples showed higher diversity compared to antrum mucosa samples (Fig. 1B). The unweighted UniFrac analysis indicated that there was very little separation between control and cancer groups under the HP infection status in both of gastric antral and body mucosa (Fig. 2). However, HP(−) groups showed a separation between control and cancer group in both of antrum and body. The bacterial communities at the phylum level among four groups showed that the proportion of Proteobacteria of the HP(+) groups was more greater than that of the HP(−) cancer group (Table 2). In HP(−) control group, Pseudomonas hibiscicola (44.87% vs. 0%) and Ralstonia pickettii (45.90% vs.0.03%) were more abundant in antrum than in body mucosa (Table 2). In HP(−) control group, Bradyrhizobium sp. (Bradyrhizobium jicamae, B. pachyrhizi, B. denitrificans, and B. g1 uc) were more abundant in body (8.66%, 5.33%, 1.14%, and 0.72%, respectively) than in antrum mucosa (all 0%, Table 2). Actinobacteria of HP(+) cancer group was more greater than that of the HP(−) cancer group. However, Propionibacterium acnes was more abundant in body than antrum mucosa (8.49% vs. 1.43%, Table 2). The proportion of Firmicutes in the HP(−) groups was more greater than the HP(+) groups. These differences were more pronounced in the antral mucosa. The bacterial communities at the species level among four groups in gastric antral and body mucosa showed that the proportion of Streptococcus sp. in the HP(−) cancer group was more greater than the others (Table 2). The proportion of Streptococcus mitis group,20–22 such as S. pseudopneumoniae, S. mitis, S. infantis, S. oralis, and S. tigurinus, in the HP(−) cancer group was more higher than in that of the HP(+) cancer group (antrum, 33.9 vs. 8.4, P = 0.076; body, 34.1 vs. 0.33, P = 0.009; Table 2). These results suggest a role of S. mitis in the gastric carcinogenesis despite the absence of HP.
Figure 1.
Bacterial diversity in gastric antrum and body mucosa samples. (A) The graph shows refraction curves indicating the number of assigned bacterial genera in relation to the number of 16S rRNA sequences, grouped by individual. (B) Taxonomic assignment of the 24 samples at the level of bacterial phylum. OTU, stands for operational taxonomic units; HP, Helicobacter pylori; con, control; a, antrum; b, body; ETC, et cetera.
Figure 2.
(A) Unweighted UniFrac-based principal coordinates analysis of gastric antrum and (B) body microbiome. There was very little separation between control and cancer groups under the same HP infection status in both of gastric antrum and body mucosa. HP, Helicobacter pylori; con, control.
Table 2.
Comparison of species frequency of gastric antrum and body mucosal samples
| Phylum | Species | Antrum (%) | Body (%) | ||||||
|---|---|---|---|---|---|---|---|---|---|
|
|
|
||||||||
| HP(−) control (n = 2) | HP(−) cancer (n = 2) | HP(+)control (n = 3) | HP(+)cancer (n = 5) | HP(−) control (n = 2) | HP(−) cancer (n = 2) | HP(+)control (n = 3) | HP(+)cancer (n = 5) | ||
| Firmicutes | Streptococcus pseudopneumoniae | 0.14 | 18.68a | 3.37a | 6.04a | 3.80a | 23.11a | 0.98 | 0.20 |
| S. mitis | 0.02 | 9.01a | 0.51 | 0.82 | 0.45 | 3.09a | 0.23 | 0.00 | |
| S. salivarius | 0.01 | 4.00a | 0.11 | 0.40 | 0.27 | 6.37a | 0.03 | 0.02 | |
| S. infantis | 0.32 | 3.39a | 0.24 | 1.10a | 3.07a | 5.38a | 0.08 | 0.12 | |
| Veillonella atypica | 0.03 | 2.97a | 0.23 | 0.64 | 0.32 | 1.22a | 0.03 | 0.01 | |
| V. dispar | 0.03 | 2.37a | 0.18 | 0.77 | 0.53 | 1.25a | 0.05 | 0.01 | |
| Granulicatella adiacens | 0.04 | 3.33a | 0.08 | 0.71 | 0.51 | 1.98a | 0.03 | 0.02 | |
| Gemella haemolysans | 0.00 | 2.09a | 0.19 | 0.20 | 0.03 | 1.16a | 0.05 | 0.01 | |
| S. australis | 0.05 | 2.19a | 0.14 | 0.68 | 0.59 | 2.01a | 0.10 | 0.01 | |
| S. parasanguinis | 0.14 | 1.51a | 0.27 | 0.60 | 0.86 | 3.36a | 0.14 | 0.04 | |
| S._uc | 0.10 | 1.14a | 0.20 | 0.65 | 0.76 | 2.96a | 0.06 | 0.06 | |
| GQ130066_s | 0.01 | 0.90 | 0.21 | 0.54 | 0.00 | 0.00 | 0.00 | 0.00 | |
| S. oralis | 0.02 | 1.72a | 0.14 | 0.30 | 0.50 | 2.07a | 0.08 | 0.01 | |
| Lactobacillus salivarius | 0.00 | 1.93a | 0.00 | 0.05 | 0.01 | 0.67 | 0.00 | 0.00 | |
| S. tigurinus | 0.01 | 1.06a | 0.03 | 0.17 | 0.25 | 0.43 | 0.00 | 0.00 | |
| Solobacterium moorei | 0.02 | 0.17 | 0.03 | 0.54 | 0.01 | 0.13 | 0.00 | 0.02 | |
| S. lactarius | 0.02 | 0.88 | 0.05 | 0.08 | 0.14 | 0.65 | 0.02 | 0.00 | |
| Megasphaera micronuciformis | 0.00 | 0.70 | 0.02 | 0.15 | 0.03 | 0.04 | 0.00 | 0.00 | |
| Streptococcaceae_uc_s | 0.02 | 0.41 | 0.10 | 0.11 | 0.57 | 1.90a | 0.12 | 0.05 | |
| Lactobacillales_uc_s | 0.01 | 0.19 | 0.02 | 0.08 | 0.82 | 2.09a | 0.07 | 0.05 | |
| Proteobacteria | HP | 0.11 | 0.11 | 86.47a | 60.30a | 0.42 | 0.10 | 89.78a | 75.93a |
| Haemophilus parainfluenzae | 0.15 | 3.84a | 0.46 | 0.96 | 2.15a | 2.41a | 0.13 | 0.02 | |
| Escherichia coli group | 0.03 | 0.00 | 0.00 | 1.46a | 0.00 | 0.01 | 0.00 | 0.05 | |
| H. paraphrohaemolyticus | 0.00 | 1.60a | 0.08 | 0.60 | 0.03 | 1.10a | 0.02 | 0.01 | |
| Methylobacterium adhaesivum | 0.06 | 2.22a | 0.12 | 0.56 | 0.06 | 0.00 | 0.00 | 0.05 | |
| Bradyrhizobium jicamae | 0.00 | 0.07 | 0.01 | 0.22 | 8.66a | 0.68 | 0.24 | 1.24a | |
| Neisseria perflava | 0.00 | 4.24a | 1.14a | 0.17 | 0.04 | 5.12a | 0.04 | 0.02 | |
| Pseudomonas beteli | 1.05a | 0.05 | 0.01 | 0.20 | 0.00 | 0.00 | 0.00 | 0.00 | |
| P. hibiscicola | 44.87a | 0.06 | 0.24 | 0.17 | 0.00 | 0.02 | 0.00 | 0.01 | |
| Helicobacteraceae_uc_s | 0.01 | 0.00 | 0.08 | 0.07 | 0.06 | 0.03 | 1.28a | 0.49 | |
| H._uc | 0.00 | 1.00a | 0.07 | 0.11 | 0.09 | 0.86 | 0.04 | 0.00 | |
| Aggregatibacter segnis | 0.02 | 1.07a | 0.00 | 0.05 | 0.00 | 0.44 | 0.00 | 0.01 | |
| H. haemolyticus | 0.00 | 0.61 | 0.10 | 0.04 | 0.04 | 0.53 | 0.04 | 0.00 | |
| Ralstonia pickettii | 45.90a | 0.05 | 0.02 | 0.04 | 0.03 | 0.00 | 0.00 | 0.00 | |
| Pasteurellaceae_uc_s | 0.01 | 0.19 | 0.02 | 0.03 | 0.36 | 0.69 | 0.02 | 0.01 | |
| Pasteurellales_uc_s | 0.00 | 0.04 | 0.01 | 0.02 | 0.12 | 0.63 | 0.00 | 0.01 | |
| B. pachyrhizi | 0.00 | 0.01 | 0.00 | 0.03 | 5.33a | 0.49 | 0.11 | 0.70 | |
| Klebsiella pneumoniae | 0.00 | 0.07 | 0.00 | 1.05a | 0.00 | 0.02 | 0.00 | 0.03 | |
| Neisseriaceae_uc_s | 0.00 | 0.22 | 0.00 | 0.02 | 0.05 | 0.60 | 0.00 | 0.00 | |
| Neisseriales_uc_s | 0.01 | 0.05 | 0.01 | 0.01 | 0.08 | 0.80 | 0.00 | 0.00 | |
| FJ269053_s | 0.02 | 0.08 | 0.00 | 0.01 | 14.23a | 1.38a | 0.46 | 2.94a | |
| B. denitrificans | 0.00 | 0.07 | 0.00 | 0.00 | 1.14a | 0.09 | 0.03 | 0.10 | |
| U87765_s | 0.02 | 0.02 | 0.00 | 0.00 | 2.97a | 0.27 | 0.08 | 0.51 | |
| Bradyrhizobiaceae_uc_s | 0.00 | 0.00 | 0.00 | 0.01 | 0.98 | 0.20 | 0.04 | 0.13 | |
| DQ532251_g_uc | 0.00 | 0.00 | 0.00 | 0.00 | 0.79 | 0.09 | 0.03 | 0.18 | |
| Pelomonas saccharophila | 0.00 | 0.00 | 0.00 | 0.00 | 3.26a | 0.47 | 0.22 | 0.95 | |
| Rhizobium hainanense | 0.00 | 0.00 | 0.00 | 0.00 | 1.10a | 0.11 | 0.04 | 0.24 | |
| M. longum | 0.00 | 0.00 | 0.00 | 0.00 | 1.91a | 0.09 | 0.06 | 0.40 | |
| M. radiotolerans | 0.01 | 0.00 | 0.00 | 0.00 | 3.08a | 0.18 | 0.11 | 0.62 | |
| B._g1_uc | 0.00 | 0.00 | 0.00 | 0.00 | 0.72 | 0.09 | 0.02 | 0.09 | |
| Bacteroidetes | Prevotella histicola | 0.01 | 3.08a | 0.11 | 0.43 | 0.11 | 1.85a | 0.08 | 0.00 |
| P. melaninogenica | 0.01 | 1.56a | 0.06 | 0.84 | 0.14 | 0.92 | 0.04 | 0.02 | |
| P._uc | 0.01 | 1.07a | 0.23 | 0.31 | 0.21 | 0.98 | 0.06 | 0.01 | |
| P. pallens | 0.02 | 0.65 | 0.08 | 0.33 | 0.03 | 0.31 | 0.00 | 0.02 | |
| P. salivae | 0.00 | 0.78 | 0.06 | 0.14 | 0.05 | 0.46 | 0.02 | 0.01 | |
| EF123551_g_uc | 0.00 | 0.00 | 0.00 | 0.00 | 1.45a | 0.11 | 0.07 | 0.51 | |
| Actinobacteria | Propionibacterium acnes | 1.43a | 0.42 | 0.82 | 4.42a | 8.49a | 1.39a | 0.27 | 4.34a |
| Actinomyces odontolyticus | 0.05 | 1.31a | 0.17 | 0.89 | 0.44 | 0.95 | 0.12 | 0.03 | |
| Rothia mucilaginosa | 0.09 | 0.26 | 0.07 | 0.12 | 0.90 | 0.09 | 0.05 | 0.01 | |
| Propionibacterium_uc | 0.02 | 0.01 | 0.01 | 0.04 | 0.53 | 0.07 | 0.01 | 0.07 | |
| Propionibacteriaceae_uc_s | 0.04 | 0.00 | 0.00 | 0.02 | 0.79 | 0.18 | 0.04 | 0.16 | |
| Viridiplantae | Prunus persica | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.03 | 0.83 |
| Nicotiana tabacum | 0.00 | 0.00 | 0.00 | 0.00 | 1.09a | 0.00 | 0.00 | 0.05 | |
| Ipomoea purpurea | 0.00 | 0.00 | 0.00 | 0.00 | 1.59a | 0.00 | 0.00 | 0.00 | |
| Fusobacteria | Fusobacterium nucleatum | 0.09 | 0.92 | 0.26 | 0.11 | 0.33 | 0.16 | 0.08 | 0.03 |
Values are presented as mean percent. HP, Helicobacter pylori.
This means cut off > 1.0.
3. H. pylori(−) cancer vs. H. pylori(+) cancer
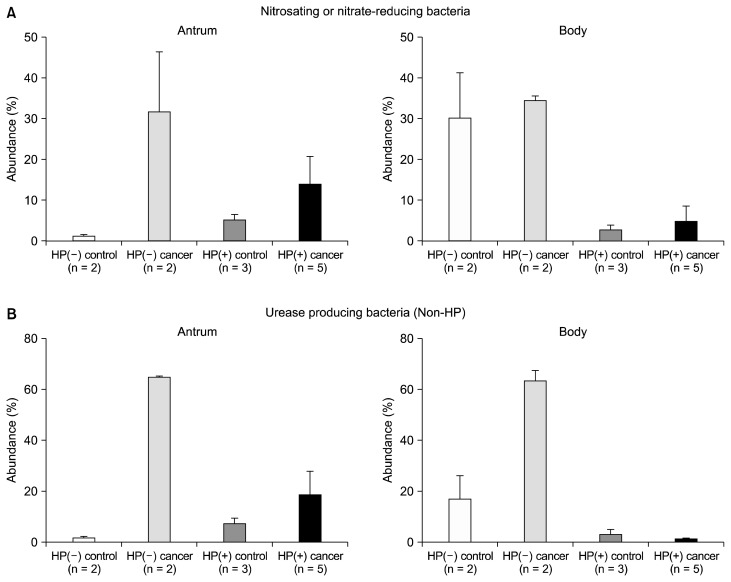
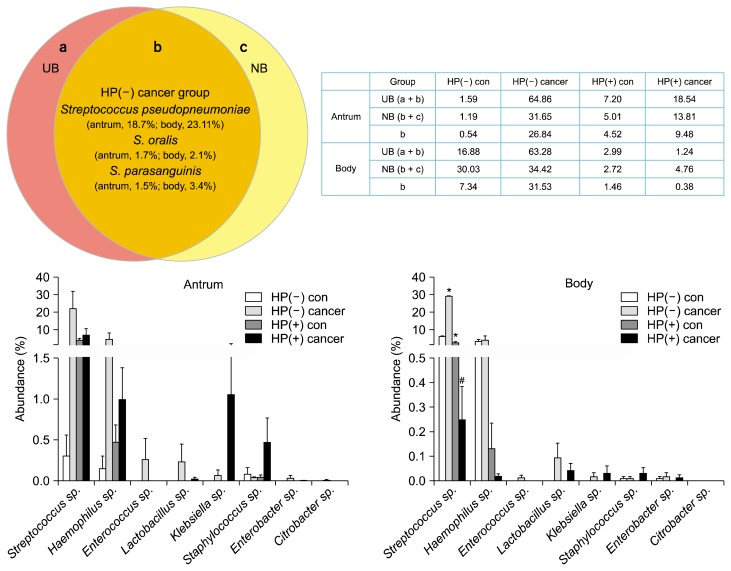
The composition of UB and NB12 was higher in HP(−) cancer and HP(+) groups than each control group (Supplementary 1 and 2). The proportion of non-HP-UB and non-HP-NB was higher in the HP(−) cancer group than in that of the HP(+) cancer group, especially body mucosa (antrum, P = 0.053, P = 0.121; body, P = 0.053, P = 0.051, respectively; Fig. 3). The overlap of non-HP-UB and non-HP-NB was presented at Figure 4 and Table 3. When we assessed the overlap of non-HP-UB and non-HP-NB, it revealed that Streptococcus sp. occupied high proportion in the HP(−) cancer group except S. pneumoniae. As these strains are urease and produce NOC in gastric mucosa (Fig. 4), S. pseudopneumoniae, S. parasanguinis, and S. oralis are pathogens (antrum, P = 0.123; body, P = 0.030, respectively; Kruskal-Wallis test).
Figure 3.
The comparison of gastric microbiome in gastric antrum and body mucosa. (A) The proportion of non-HP nitrosating or nitrate-reducing bacteria between the gastric antrum and body mucosa. (B) The proportion of non-HP urease-producing bacteria between the gastric antrum and body mucosa. HP, Helicobacter pylori.
Figure 4.
The comparison of urease and N-nitroso compounds-producing bacteria in gastric antrum and body mucosa. The strains of the overlap of non-HP-UB and non-HP-NB between the gastric antrum and body mucosa. Kruskal-Wallis test was used to determine statistical significance existed between the four groups. HP, Helicobacter pylori; NB, nitrosating or nitrate-reducing bacteria; UB, urease-producing bacteria; con, control; *P < 0.05 compared to HP(−) control; #P < 0.05 compared to HP(−) cancer.
Table 3.
Comparison of urease and N-nitroso compounds-producing bacteria between antrum and body
| Species | Antrum (%) | Body (%) | ||||||
|---|---|---|---|---|---|---|---|---|
|
|
|
|||||||
| HP(−) control (n = 2) | HP(−) cancer (n = 2) | HP(+)control (n = 3) | HP(+)cancer (n = 5) | HP(−) control (n = 2) | HP(−) cancer (n = 2) | HP(+)control (n = 3) | HP(+)cancer (n = 5) | |
| Streptococcus pseudopneumoniae | 0.139 | 18.68a | 3.375 | 6.038 | 3.797 | 23.11a | 0.983 | 0.196 |
| Haemophilus parainfluenzae | 0.155 | 3.842 | 0.457 | 0.959 | 2.154 | 2.413 | 0.130 | 0.018 |
| S. oralis | 0.023 | 1.724a | 0.145 | 0.297 | 0.500 | 2.073a | 0.078 | 0.009 |
| S. parasanguinis | 0.138 | 1.506a | 0.275 | 0.596 | 0.855 | 3.361a | 0.135 | 0.042 |
| H. influenzae | 0.000 | 0.433 | 0.016 | 0.034 | 0.000 | 0.408 | 0.000 | 0.000 |
| Enterococcus hirae | 0.000 | 0.261 | 0.000 | 0.000 | 0.000 | 0.011 | 0.000 | 0.000 |
| Lactobacillus fermentum | 0.000 | 0.209 | 0.000 | 0.000 | 0.000 | 0.084 | 0.000 | 0.030 |
| HP | 0.107 | 0.111 | 86.470 | 60.305 | 0.419 | 0.096 | 89.777 | 75.932 |
| Klebsiella pneumoniae | 0.000 | 0.068 | 0.000 | 1.054 | 0.000 | 0.017 | 0.000 | 0.030 |
| Staphylococcus epidermidis | 0.084 | 0.043 | 0.047 | 0.468 | 0.009 | 0.008 | 0.000 | 0.030 |
| Enterobacter aerogenes | 0.000 | 0.035 | 0.000 | 0.004 | 0.000 | 0.000 | 0.000 | 0.012 |
| L. gasseri | 0.000 | 0.026 | 0.000 | 0.025 | 0.000 | 0.008 | 0.000 | 0.011 |
| Citrobacter rodentium | 0.000 | 0.008 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 |
| S. pneumoniae | 0.006 | 0.008 | 0.201 | 0.008 | 0.017 | 0.019 | 0.133 | 0.000 |
| E. mori | 0.000 | 0.000 | 0.000 | 0.000 | 0.009 | 0.017 | 0.000 | 0.000 |
Values are expressed as mean percent. HP, Helicobacter pylori.
This means cut off > 1.0.
DISCUSSION
In recent years, high throughput techniques for studying microbiome have been developed which provide more comprehensive data on microbiome. The goal of these techniques is to identify key microbial players between health and disease outcome. It has a clear potential to benefit clinical part. Bacterial infection has been linked to cancer through two mechanisms; 1) chronic inflammation and 2) production of carcinogenic metabolites such as HP infection.23
Gastric acidity is a barrier to bacterial overgrowth.24,25 The bacterial colonization in stomach increases under the condition such as acid-reducing drug, atrophic gastritis, and gastric surgery.10,25 In addition, decreased gastric acid secretion is responsible for an increased risk of infection.20 The antrum and body of stomach are distinct niches for microbial colonization owing to differential ability to secrete gastric acid.26 Thus, comparison of gastric microbiota between gastric antrum and body will be useful. Li et al.27 investigated the gastric microbiota of five non-HP and non-NSAID (non-steroidal anti-inflammatory drug) antral gastritis individuals and five normal individuals by pyrosequencing, and they identified potential pathogens (S. pneumonia, S. mitis and S. salivarius) were high in antral gastritis stomach. However, there was little difference in gastric microbiota between antrum and body in normal control group except Prevotella.27 Similarly, our results also showed that HP(−) cancer group showed high proportion of Streptococcus (phylum Firmicutes) in both gastric antrum (41.3%) and body (49.5%). However, the proportion of Streptococcus sp. was more pronounced in the body.
HP causes atrophic gastritis modulating inflammatory responses and making NOC.9,10 NOC can be formed from nitrite and secondary amines by nitrosating bacteria of stomach, such as Clostridium, Veillonella, Haemophilus, Staphylococcus, Streptococcus, and Neisseria.12,28 NOC formation has been suggested to increase the risk of gastric cancer.12 Urease is a major inducer of innate immune response.4–6 Urease-producing non-HP microbes including Actinomyces, Clostridium, Corynebacterium, member of the Enterobacteriaceae (Citrobacter, Enterobacter, Klebsiella, Morganella, Providencia, and Proteus), Enterococcus, Gardnerella, Haemophilus, Lactococcus, Mycobacterium, Streptococcus, Staphylococcus, Ureaplasma, and Yersinia were detected in the oral cavity, gastrointestinal tract, urethrogenital tract and skin.29–31 Gastric pH modifications induced by UB may modify bacterial substrate availability and local immune responses by relationship between their members. NB concentrations were significantly higher when pH was > 4.32 Nitrosating capacity was higher in a range from pH 3 to pH 6.33 UB-NB interaction was able to produce a pro-carcinogenic inflammatory response like as HP. Surprisingly, the proportion of UB and NB was significantly higher in HP(−) cancer group, especially in the body mucosa. When we assessed the overlap of non-HP-UB and non-HP-NB, it revealed that the composition of S. pseudopneumoniae, S. parasanguinis, and S. oralis was higher in HP(−) cancer groups than the others. S. mitis group, such as S. mitis, S. pseudopneumoniae, S. oralis, S. infantis, and S. tigurinus strains were associated with serious invasive infections, pneumonia, and endocarditis.34–36 In addition, S. mitis was significantly more prevalent within oesophageal carcinoma tissues.37 Importantly, S. mitis could induce the expression of CXC chemokine genes (IL-8 and GROa), which recruitment and activation of neutrophils and monocytes could be stimulated during cancer progression.37 Additionally, S. parasanguinis strain was associated with cystic fibrosis.38 Taken together, these species could be a significant human pathogen. Actually, we missed this point in the previous report using 63 samples in the antrum 12, thus, further analysis is planned in the future. Anyway, our analysis using 12 samples in the body added another clue for the role of bacteria other than HP to gastric carcinogenesis. Actually, it has been suggested by a number of researches using pyrosequencing.39,40 However, this study has a limitation due to small sample size, and further research using more samples are needed.
Supplementary Information
ACKNOWLEDGMENTS
This work was supported by the National Research Foundation of Korea (NRF) grant for the Global Core Research Center (GCRC) funded by the Korea government (MSIP) (No. 2011-0030001).
Footnotes
CONFLICT OF INTEREST
No potential conflicts of interest were disclosed.
REFERENCES
- 1.Parsonnet J. Helicobacter pylori: the size of the problem. Gut 1998;43 Suppl 1:S6–9. 10.1136/gut.43.2008.S6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fahey JW, Stephenson KK, Wade KL, Talalay P. Urease from Helicobacter pylori is inactivated by sulforaphane and other isothiocyanates. Biochem Biophys Res Commun 2013;435:1–7. 10.1016/j.bbrc.2013.03.126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yu H, Zeng J, Liang X, Wang W, Zhou Y, Sun Y, et al. Helicobacter pylori promotes epithelial-mesenchymal transition in gastric cancer by downregulating programmed cell death protein 4 (PDCD4). PLoS One 2014;9:e105306. 10.1371/journal.pone.0105306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gobert AP, Mersey BD, Cheng Y, Blumberg DR, Newton JC, Wilson KT. Cutting edge: urease release by Helicobacter pylori stimulates macrophage inducible nitric oxide synthase. J Immunol 2002; 168:6002–6. 10.4049/jimmunol.168.12.6002 [DOI] [PubMed] [Google Scholar]
- 5.Suerbaum S, Michetti P. Helicobacter pylori infection. N Engl J Med 2002;347:1175–86. 10.1056/NEJMra020542 [DOI] [PubMed] [Google Scholar]
- 6.MacMicking J, Xie QW, Nathan C. Nitric oxide and macrophage function. Annu Rev Immunol 1997;15:323–50. 10.1146/annurev.immunol.15.1.323 [DOI] [PubMed] [Google Scholar]
- 7.Noach LA, Bosma NB, Jansen J, Hoek FJ, van Deventer SJ, Tytgat GN. Mucosal tumor necrosis factor-alpha, interleukin-1 beta, and interleukin-8 production in patients with Helicobacter pylori infection. Scand J Gastroenterol 1994;29:425–9. 10.3109/00365529409096833 [DOI] [PubMed] [Google Scholar]
- 8.El-Omar EM, Rabkin CS, Gammon MD, Vaughan TL, Risch HA, Schoenberg JB, et al. Increased risk of noncardia gastric cancer associated with proinflammatory cytokine gene polymorphisms. Gastroenterology 2003;124:1193–201. 10.1016/S0016-5085(03)00157-4 [DOI] [PubMed] [Google Scholar]
- 9.Sanduleanu S, Jonkers D, De Bruïne A, Hameeteman W, Stockbrügger RW. Double gastric infection with Helicobacter pylori and non-Helicobacter pylori bacteria during acid-suppressive therapy: increase of pro-inflammatory cytokines and development of atrophic gastritis. Aliment Pharmacol Ther 2001;15: 1163–75. 10.1046/j.1365-2036.2001.01029.x [DOI] [PubMed] [Google Scholar]
- 10.Mowat C, Williams C, Gillen D, Hossack M, Gilmour D, Carswell A, et al. Omeprazole, Helicobacter pylori status, and alterations in the intragastric milieu facilitating bacterial N-nitrosation. Gastroenterology 2000;119:339–47. 10.1053/gast.2000.9367 [DOI] [PubMed] [Google Scholar]
- 11.Sung J, Kim N, Kim J, Jo HJ, Park JH, Nam RH, et al. Comparison of gastric microbiota between gastric juice and mucosa by next generation sequencing method. J Cancer Prev 2016;21:60–5. 10.15430/JCP.2016.21.1.60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jo HJ, Kim J, Kim N, Park JH, Nam RH, Seok YJ, et al. Analysis of gastric microbiota by pyrosequencing: minor role of bacteria other than Helicobacter pylori in the gastric carcinogenesis. Helicobacter 2016;21:364–74. 10.1111/hel.12293 [DOI] [PubMed] [Google Scholar]
- 13.Kim J, Kim N, Jo HJ, Park JH, Nam RH, Seok YJ, et al. An appropriate cutoff value for determining the colonization of Helicobacter pylori by the pyrosequencing method: comparison with conventional methods. Helicobacter 2015;20:370–80. 10.1111/hel.12214 [DOI] [PubMed] [Google Scholar]
- 14.Shin JM, Kim N. Pharmacokinetics and pharmacodynamics of the proton pump inhibitors. J Neurogastroenterol Motil 2013;19:25–35. 10.5056/jnm.2013.19.1.25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weeks DL, Eskandari S, Scott DR, Sachs G. A H+-gated urea channel: the link between Helicobacter pylori urease and gastric colonization. Science 2000;287:482–5. 10.1126/science.287.5452.482 [DOI] [PubMed] [Google Scholar]
- 16.Kusters JG, van Vliet AH, Kuipers EJ. Pathogenesis of Helicobacter pylori infection. Clin Microbiol Rev 2006;19:449–90. 10.1128/CMR.00054-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kang HY, Kim N, Park YS, Hwang JH, Kim JW, Jeong SH, et al. Progression of atrophic gastritis and intestinal metaplasia drives Helicobacter pylori out of the gastric mucosa. Dig Dis Sci 2006;51:2310–5. 10.1007/s10620-006-9276-0 [DOI] [PubMed] [Google Scholar]
- 18.Dixon MF, Genta RM, Yardley JH, Correa P. Classification and grading of gastritis. The updated Sydney System. International Workshop on the Histopathology of Gastritis, Houston 1994 Am J Surg Pathol 1996;20:1161–81. [DOI] [PubMed] [Google Scholar]
- 19.Yoon H, Kim N, Lee HS, Shin CM, Park YS, Lee DH, et al. Helicobacter pylori-negative gastric cancer in South Korea: incidence and clinicopathologic characteristics. Helicobacter 2011; 16:382–8. 10.1111/j.1523-5378.2011.00859.x [DOI] [PubMed] [Google Scholar]
- 20.Kawamura Y, Whiley RA, Shu SE, Ezaki T, Hardie JM. Genetic approaches to the identification of the mitis group within the genus Streptococcus. Microbiology 1999;145:2605–13. 10.1099/00221287-145-9-2605 [DOI] [PubMed] [Google Scholar]
- 21.Kawamura Y, Hou XG, Todome Y, Sultana F, Hirose K, Shu SE, et al. Streptococcus peroris sp. nov. and Streptococcus infantis sp. nov., new members of the Streptococcus mitis group, isolated from human clinical specimens. Int J Syst Bacteriol 1998; 48:921–7. 10.1099/00207713-48-3-921 [DOI] [PubMed] [Google Scholar]
- 22.Dhotre SV, Mehetre GT, Dharne MS, Suryawanshi NM, Nagoba BS. Isolation of Streptococcus tigurinus: a novel member of Streptococcus mitis group from a case of periodontitis. FEMS Microbiol Lett 2014;357:131–5. [DOI] [PubMed] [Google Scholar]
- 23.Parsonnet J. Bacterial infection as a cause of cancer. Environ Health Perspect 1995;103 Suppl 8:263–8. 10.1289/ehp.95103s8263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Giannella RA, Broitman SA, Zamcheck N. Gastric acid barrier to ingested microorganisms in man: studies in vivo and in vitro. Gut 1972;13:251–6. 10.1136/gut.13.4.251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martinsen TC, Bergh K, Waldum HL. Gastric juice: a barrier against infectious diseases. Basic Clin Pharmacol Toxicol 2005; 96:94–102. 10.1111/j.1742-7843.2005.pto960202.x [DOI] [PubMed] [Google Scholar]
- 26.Sachs G, Weeks DL, Melchers K, Scott DR. The gastric biology of Helicobacter pylori. Annu Rev Physiol 2003;65:349–69. 10.1146/annurev.physiol.65.092101.142156 [DOI] [PubMed] [Google Scholar]
- 27.Li XX, Wong GL, To KF, Wong VW, Lai LH, Chow DK, et al. Bacterial microbiota profiling in gastritis without Helicobacter pylori infection or non-steroidal anti-inflammatory drug use. PLoS One 2009;4:e7985. 10.1371/journal.pone.0007985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ayanaba A, Alexander M. Microbial formation of nitrosamines in vitro. Appl Microbiol 1973;25:862–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Osaki T, Mabe K, Hanawa T, Kamiya S. Urease-positive bacteria in the stomach induce a false-positive reaction in a urea breath test for diagnosis of Helicobacter pylori infection. J Med Microbiol 2008;57:814–9. 10.1099/jmm.0.47768-0 [DOI] [PubMed] [Google Scholar]
- 30.Brandi G, Biavati B, Calabrese C, Granata M, Nannetti A, Mattarelli P, et al. Urease-positive bacteria other than Helicobacter pylori in human gastric juice and mucosa. Am J Gastroenterol 2006;101:1756–61. 10.1111/j.1572-0241.2006.00698.x [DOI] [PubMed] [Google Scholar]
- 31.Mobley HL, Hausinger RP. Microbial ureases: significance, regulation, and molecular characterization. Microbiol Rev 1989;53: 85–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Verdu E, Viani F, Armstrong D, Fraser R, Siegrist HH, Pignatelli B, et al. Effect of omeprazole on intragastric bacterial counts, nitrates, nitrites, and N-nitroso compounds. Gut 1994;35:455–60. 10.1136/gut.35.4.455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stockbrügger R, Armbrecht U. Bacterial overgrowth in the upper gastrointestinal-tract and possible consequences: report of a workshop in Brussels, Belgium, 9–10 February, 1990 Microb Ecol Health Dis 1991;4: i–vii. [Google Scholar]
- 34.Shelburne SA, Sahasrabhojane P, Saldana M, Yao H, Su X, Horstmann N, et al. Streptococcus mitis strains causing severe clinical disease in cancer patients. Emerg Infect Dis 2014;20: 762–71. 10.3201/eid2005.130953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Musher DM, Roig IL, Cazares G, Stager CE, Logan N, Safar H. Can an etiologic agent be identified in adults who are hospitalized for community-acquired pneumonia: results of a one-year study. J Infect 2013;67:11–8. 10.1016/j.jinf.2013.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zbinden A, Mueller NJ, Tarr PE, Eich G, Schulthess B, Bahlmann AS, et al. Streptococcus tigurinus, a novel member of the Streptococcus mitis group, causes invasive infections. J Clin Microbiol 2012;50:2969–73. 10.1128/JCM.00849-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Narikiyo M, Tanabe C, Yamada Y, Igaki H, Tachimori Y, Kato H, et al. Frequent and preferential infection of Treponema denticola, Streptococcus mitis, and Streptococcus anginosus in esophageal cancers. Cancer Sci 2004;95:569–74. 10.1111/j.1349-7006.2004.tb02488.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maeda Y, Elborn JS, Parkins MD, Reihill J, Goldsmith CE, Coulter WA, et al. Population structure and characterization of viridans group streptococci (VGS) including Streptococcus pneumoniae isolated from adult patients with cystic fibrosis (CF). J Cyst Fibros 2011;10:133–9. 10.1016/j.jcf.2010.11.003 [DOI] [PubMed] [Google Scholar]
- 39.Bik EM, Eckburg PB, Gill SR, Nelson KE, Purdom EA, Francois F, et al. Molecular analysis of the bacterial microbiota in the human stomach. Proc Natl Acad Sci U S A 2006;103:732–7. 10.1073/pnas.0506655103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Delgado S, Cabrera-Rubio R, Mira A, Suarez A, Mayo B. Microbiological survey of the human gastric ecosystem using culturing and pyrosequencing methods Microb Ecol 2013;65:763–72. 10.1007/s00248-013-0192-5 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.