Abstract
Abnormal telomerase activity is implicated in cancer initiation and development. The rs2736100 T > G polymorphism in the telomerase reverse transcriptase (TERT) gene, which encodes the telomerase catalytic subunit, has been associated with increased cancer risk. We conducted a meta-analysis to more precisely assess this association. After a comprehensive literature search of the PubMed and EMBASE databases up to November 1, 2016, 61 articles with 72 studies comprising 108,248 cases and 161,472 controls were included in our meta-analysis. Studies were conducted on various cancer types. The TERT rs2736100 polymorphism was associated with increased overall cancer risk in five genetic models [homozygous model (GG vs. TT): odds ratio (OR) = 1.39, 95% confidence interval (95% CI) = 1.26-1.54, P < 0.001; heterozygous model (TG vs. TT): OR = 1.16, 95% CI = 1.11-1.23, P < 0.001; dominant model (TG + GG vs. TT): OR = 1.23, 95% CI = 1.15-1.31, P < 0.001; recessive model (GG vs. TG + TT): OR = 1.25, 95% CI = 1.16-1.35, P < 0.001; and allele contrast model (G vs. T): OR = 1.17, 95% CI = 1.12-1.23, P < 0.001]. A stratified analysis based on cancer type associated the polymorphism with elevated risk of thyroid cancer, bladder cancer, lung cancer, glioma, myeloproliferative neoplasms, and acute myeloid leukemia. Our results confirm that the TERT rs2736100 polymorphism confers increased overall cancer risk.
Keywords: TERT, cancer, risk, meta-analysis, telomerase
INTRODUCTION
Cancer is a major public health problem worldwide, with an estimated 14.1 million new cancer cases and 8.2 million deaths in 2012 [1]. Carcinogenesis is a complex process, influenced by various genetic and environmental factors, such as smoking, poor diet, physical inactivity, reproductive changes and the growth and aging of the population [1, 2]. Telomeres, composed of the TTAGGG repeat sequence, are special chromatin structures located at each end of a chromosome. Telomeres maintain chromosomal integrity by protecting chromosome ends from DNA damage and end-to-end fusions [3]. Abnormally short telomeres may cause chromosomal instability, and consequentially contribute to cancer development. Telomerase (also known as terminal transferase), a reverse transcriptase enzyme, extends the 3′ end of chromosomal DNA by catalyzing the telomere synthesis reaction. Defects in telomerase activity have been observed in many human tumor cells, and telomere length was inversely associated with cancer incidence and mortality [4]. Telomerase reverse transcriptase (TERT), the telomerase catalytic subunit, maintains telomere stability [5]. In a previous genome-wide association study (GWAS), Shete, et al. discovered that certain TERT gene variants increase glioma susceptibility [6]. Since then, TERT variants have been associated with various cancers, including breast, lung, colorectal, ovarian, prostate, and gastric cancers [7, 8].
The TERT gene is located in 5p15.33. The rs2736100 T > G polymorphism in the second intron of the TERT gene has been associated with shortened telomere length in gastric cancer [9]. The association of this SNP with cancer susceptibility has been extensively explored, although the findings are as yet inconclusive. Several meta-analyses published in 2014 associated the TERT rs2736100 polymorphism with increased glioma and lung cancer susceptibility [10–14]. In 2012, Zou, et al. observed an association between this polymorphism and overall cancer risk [15], although their meta-analysis involved only 11 articles. However, between 2015 and 2016, more than 27 studies were published with large sample sizes [9, 16–37]. Thus, we performed an updated meta-analysis to more precisely assess the TERT rs2736100 polymorphism-cancer association, including 72 studies derived from 61 articles with 269,720 total subjects [6, 9, 16–74].
RESULTS
Study characteristics
We initially identified 432 records from the PubMed and EMBASE databases (Figure 1). After screening titles and abstracts, 268 articles were excluded and the full texts of the remaining 164 articles were further assessed. Articles were excluded for the following reasons: irrelevant association (87 articles), meta-analysis (7), and lacking sufficient raw data for further evaluation (12). Three additional articles were identified by manually screening the references of relevant articles. Finally, 72 studies extracted from 61 articles met our study inclusion criteria and were included in the current meta-analysis [6, 9, 16–74].
Figure 1. Flowchart of articles included in our meta-analysis.
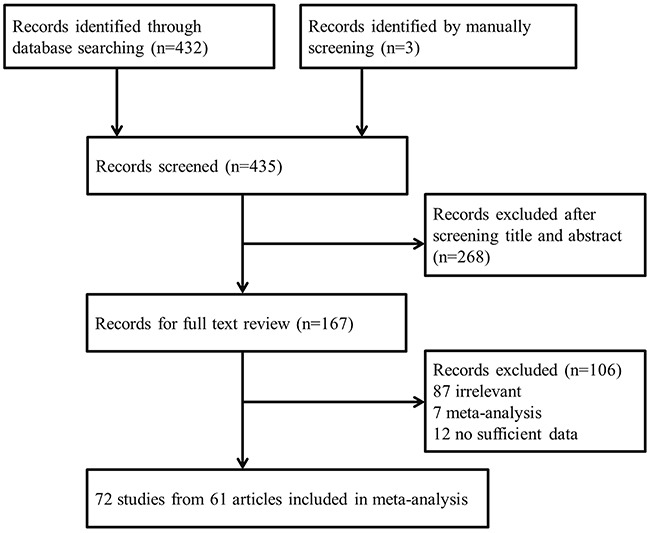
In most of the included studies, the TERT rs2736100 polymorphism genotypic distribution followed Hardy-Weinberg equilibrium (HWE) in controls, except for seven studies [6, 28, 43, 51, 63, 66, 72]. Since the genotype distributions of other polymorphisms were in compliance with HWE in these seven studies, we included these studies in the meta-analysis. In total, 72 studies with 108,248 cases and 161,472 controls were included in our pooled analysis. Studies were conducted on various cancer types, including lung (28 studies), glioma (5), colorectal (4), bladder (4), myeloproliferative neoplasms (MPN) (4), gastric (3), acute myeloid leukemia (AML) (2), breast (2), melanoma (2), and thyroid (2). The remaining 16 studies focused on different types of cancer, with one study for each type of cancer, and were grouped together as “other cancer” in our analyses. There were 37 studies conducted in Asians and 35 in Caucasians. Twenty-three studies included fewer than 500 controls, and 49 had 500 or more controls. Sixteen studies were categorized as low quality and 56 were high quality. The main characteristics of all the studies are summarized in Table 1.
Table 1. The main characteristics of all the studies included in the meta-analysis.
| Surname | Year | Country | Ethnicity | Cancer type | Cases | Controls | HWE | Score | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| All | TT | TG | GG | All | TT | TG | GG | |||||||
| Zhou | 2016 | China | Asian | ESCC | 588 | 165 | 275 | 148 | 600 | 215 | 287 | 108 | 0.472 | 11 |
| Zhang | 2016 | China | Asian | NC | 855 | 265 | 428 | 162 | 1036 | 365 | 516 | 155 | 0.211 | 13 |
| Yuan | 2016 | China | Asian | UTUC | 212 | 83 | 81 | 48 | 289 | 86 | 144 | 59 | 0.928 | 10 |
| 2016 | China | Asian | Lung cancer | 418 | 216 | 164 | 38 | 410 | 268 | 124 | 18 | 0.452 | 10 | |
| Wang | 2016 | China | Asian | Lung cancer | 500 | 131 | 257 | 112 | 500 | 178 | 242 | 80 | 0.881 | 11 |
| Trifa | 2016 | Romania | Caucasian | MPN | 529 | 76 | 255 | 198 | 433 | 124 | 213 | 96 | 0.802 | 13 |
| Krahling 1 | 2016 | Hungary | Caucasian | PMN | 584 | 77 | 282 | 225 | 400 | 111 | 188 | 101 | 0.235 | 8 |
| Krahling 2 | 2016 | Hungary | Caucasian | CML | 86 | 25 | 43 | 18 | 400 | 111 | 188 | 101 | 0.235 | 8 |
| Krahling 3 | 2016 | Hungary | Caucasian | AML | 308 | 71 | 153 | 84 | 400 | 111 | 188 | 101 | 0.235 | 7 |
| Gong | 2016 | China | Asian | Thyroid cancer | 452 | 142 | 214 | 96 | 452 | 156 | 222 | 74 | 0.738 | 11 |
| Ge | 2016 | China | Asian | Thyroid cancer | 2300 | 644 | 1093 | 563 | 2300 | 875 | 1056 | 369 | 0.093 | 12 |
| Dahlstrom 1 | 2016 | Sweden | Caucasian | MPN | 126 | 15 | 64 | 47 | 756 | 167 | 377 | 212 | 0.980 | 9 |
| Dahlstrom 2 | 2016 | China | Asian | MPN | 101 | 17 | 52 | 32 | 101 | 33 | 50 | 18 | 0.722 | 8 |
| Bayram | 2016 | Turkey | Caucasian | Gastric cancer | 104 | 16 | 44 | 44 | 209 | 61 | 82 | 66 | 0.002 | 9 |
| Li | 2016 | China | Asian | Lung cancer | 391 | 109 | 201 | 81 | 337 | 117 | 159 | 61 | 0.587 | 9 |
| Shiraishi | 2016 | Japan | Asian | Lung cancer | 6830 | 2057 | 3386 | 1387 | 15155 | 5723 | 7133 | 2299 | 0.323 | 13 |
| Wei | 2015 | China | Asian | Lung cancer | 702 | 190 | 353 | 159 | 2520 | 814 | 1269 | 437 | 0.130 | 12 |
| Shadrina 1 | 2015 | Russia | Caucasian | Prostate cancer | 360 | 102 | 183 | 75 | 358 | 105 | 165 | 88 | 0.150 | 11 |
| Shadrina 2 | 2015 | Russia | Caucasian | Breast cancer | 642 | 192 | 310 | 140 | 523 | 132 | 280 | 111 | 0.097 | 12 |
| Mosrati | 2015 | Sweden | Caucasian | AML | 226 | 48 | 113 | 65 | 788 | 201 | 406 | 181 | 0.382 | 10 |
| Liu | 2015 | China | Asian | Lung cancer | 288 | 72 | 139 | 77 | 317 | 92 | 173 | 52 | 0.052 | 9 |
| Du | 2015 | China | Asian | Gastric cancer | 1105 | 360 | 557 | 188 | 994 | 346 | 464 | 184 | 0.197 | 11 |
| de Martino | 2015 | Austria | Caucasian | RCC | 241 | 61 | 120 | 60 | 375 | 97 | 181 | 97 | 0.502 | 10 |
| Choi | 2015 | South Korea | Asian | Gastric cancer | 243 | 34 | 107 | 102 | 246 | 38 | 122 | 86 | 0.625 | 8 |
| Campa | 2015 | Germany | Caucasian | Pancreatic cancer | 1724 | 445 | 861 | 418 | 3512 | 817 | 1763 | 932 | 0.764 | 13 |
| Campa | 2015 | Germany | Caucasian | Multiple myeloma | 2052 | 535 | 958 | 559 | 2633 | 634 | 1285 | 714 | 0.237 | 13 |
| Adel Fahmideh | 2015 | Sweden | Caucasian | Brain tumor | 240 | 61 | 103 | 76 | 478 | 109 | 256 | 113 | 0.120 | 12 |
| Yin | 2014 | China | Asian | Lung cancer | 524 | 139 | 273 | 112 | 524 | 186 | 255 | 83 | 0.777 | 11 |
| Wang | 2014 | China | Asian | Lung cancer | 1552 | 455 | 764 | 333 | 1605 | 549 | 780 | 276 | 0.971 | 12 |
| Liorca-Cardenosa | 2014 | Spain | Caucasian | Melanoma | 629 | 146 | 297 | 186 | 371 | 94 | 177 | 100 | 0.380 | 9 |
| Zhao | 2013 | China | Asian | Lung cancer | 1759 | 596 | 1163a | 1163a | 1804 | 674 | 1130a | 1130a | / | 9 |
| Sheng | 2013 | China | Asian | ALL | 569 | 178 | 270 | 121 | 656 | 233 | 323 | 100 | 0.490 | 13 |
| Pellatt | 2013 | USA | Caucasian | Breast cancer | 3698 | 1450 | 1934 | 314 | 3534 | 1179 | 1674 | 681 | 0.047 | 11 |
| Pellatt 1 | 2013 | USA | Caucasian | Colon cancer | 1555 | 410 | 798 | 347 | 1956 | 493 | 956 | 507 | 0.321 | 12 |
| Pellatt 2 | 2013 | USA | Caucasian | Rectal cancer | 754 | 214 | 356 | 184 | 959 | 270 | 465 | 224 | 0.386 | 12 |
| Myneni | 2013 | China | Asian | Lung cancer | 352 | 122 | 141 | 89 | 447 | 157 | 212 | 78 | 0.659 | 8 |
| Ma | 2013 | China | Asian | Bladder Cancer | 177 | 55 | 87 | 35 | 961 | 340 | 455 | 166 | 0.516 | 10 |
| Lan | 2013 | China | Asian | Lung cancer | 193 | 43 | 109 | 41 | 197 | 70 | 103 | 24 | 0.137 | 9 |
| Wang | 2012 | China | Asian | Cervical Cancer | 1010 | 322 | 462 | 226 | 1006 | 352 | 480 | 174 | 0.637 | 11 |
| Rajaraman b | 2012 | USA | Caucasian | Glioma | 1854 | / | / | / | 4949 | / | / | / | / | 12 |
| Kinnersley | 2012 | UK | Caucasian | Colorectal cancer | 16039 | 4191 | 8105 | 3743 | 16430 | 4090 | 8082 | 4258 | 0.039 | 12 |
| Ito | 2012 | Japan | Asian | Lung cancer | 716 | 248 | 340 | 128 | 716 | 279 | 329 | 108 | 0.496 | 12 |
| Hofer | 2012 | Austria | Caucasian | Colorectal cancer | 137 | 38 | 68 | 31 | 1705 | 458 | 859 | 388 | 0.700 | 11 |
| Chen | 2012 | China | Asian | Lung cancer | 196 | 45 | 101 | 50 | 229 | 69 | 112 | 48 | 0.838 | 10 |
| Shiraishi | 2012 | Japan | Asian | Lung cancer | 4648 | 1386 | 2265 | 997 | 12364 | 4650 | 5856 | 1858 | 0.838 | 13 |
| Bae | 2012 | Korea | Asian | Lung cancer | 1094 | 402 | 501 | 191 | 1100 | 422 | 522 | 156 | 0.790 | 10 |
| Pande b | 2011 | USA | Caucasian | Lung cancer | 1681 | / | / | / | 1235 | / | / | / | / | 10 |
| Nan 1 | 2011 | USA | Caucasian | Melanoma | 210 | 55 | 91 | 64 | 831 | 215 | 399 | 217 | 0.252 | 11 |
| Nan 2 | 2011 | USA | Caucasian | SCC | 277 | 57 | 125 | 95 | 831 | 215 | 399 | 217 | 0.252 | 11 |
| Nan 3 | 2011 | USA | Caucasian | BCC | 274 | 68 | 116 | 90 | 831 | 215 | 399 | 217 | 0.252 | 11 |
| Kohno | 2011 | Japan | Asian | Lung cancer | 377 | 142 | 175 | 53 | 325 | 116 | 165 | 39 | 0.090 | 9 |
| Hu | 2011 | China | Asian | Lung cancer | 8559 | 2393 | 4294 | 1872 | 9378 | 3231 | 4533 | 1614 | 0.724 | 13 |
| Ding | 2011 | China | Asian | HC | 1269 | 428 | 633 | 208 | 1322 | 449 | 651 | 222 | 0.591 | 12 |
| Chen | 2011 | China | Asian | Glioma | 953 | 244 | 515 | 194 | 1036 | 334 | 542 | 160 | 0.014 | 10 |
| Jaworowsk 1 | 2011 | Poland | Caucasian | Lung cancer | 855 | 247 | 403 | 205 | 844 | 263 | 425 | 156 | 0.494 | 11 |
| Jaworowsk 2 | 2011 | Poland | Caucasian | Bladder Cancer | 431 | 134 | 216 | 81 | 439 | 134 | 226 | 79 | 0.335 | 10 |
| Jaworowsk 3 | 2011 | Poland | Caucasian | Laryngeal cancer | 413 | 124 | 211 | 78 | 406 | 130 | 199 | 77 | 0.956 | 10 |
| Gago-Dominguez 1 | 2011 | USA | Caucasian | Bladder Cancer | 471 | 86 | 239 | 146 | 547 | 127 | 262 | 158 | 0.361 | 11 |
| Gago-Dominguez 2 | 2011 | USA | Asian | Bladder Cancer | 499 | 141 | 260 | 98 | 525 | 174 | 274 | 77 | 0.064 | 10 |
| Wang | 2010 | UK | Caucasian | Lung cancer | 239 | 42 | 115 | 82 | 553 | 136 | 259 | 158 | 0.146 | 8 |
| Turnbull | 2010 | UK | Caucasian | TGCT | 1588 | 520 | 767 | 301 | 7683 | 1904 | 3718 | 2061 | 0.005 | 10 |
| Miki | 2010 | Japan | Asian | Lung cancer | 2086 | 622 | 1048 | 416 | 1103 | 4093 | 5246 | 1695 | 0.835 | 13 |
| Kohno | 2010 | Japan | Asian | Lung cancer | 1656 | 488 | 796 | 372 | 968 | 373 | 460 | 135 | 0.719 | 13 |
| Hsiung | 2010 | China | Asian | Lung cancer | 2308 | 599 | 1187 | 522 | 2321 | 852 | 1132 | 337 | 0.211 | 12 |
| Yoon | 2010 | Korea | Asian | Lung cancer | 1425 | 467 | 696 | 262 | 3011 | 1187 | 1405 | 419 | 0.921 | 11 |
| Truong 1 | 2010 | France | Caucasian | Lung cancer | 9126 | 1878 | 4526 | 2722 | 11812 | 2853 | 5817 | 3142 | 0.116 | 13 |
| Truong 2 | 2010 | France | Asian | Lung cancer | 1686 | 538 | 836 | 312 | 2101 | 775 | 1014 | 312 | 0.506 | 12 |
| Schoemaker | 2010 | UK | Caucasian | Glioma | 216 | 30 | 114 | 72 | 241 | 54 | 127 | 60 | 0.397 | 9 |
| Shete | 2009 | USA | Caucasian | Glioma | 4344 | 781 | 2213 | 1350 | 6457 | 1623 | 3122 | 1712 | 0.008 | 11 |
| Landi b | 2009 | USA | Caucasian | Lung cancer | 5739 | / | / | / | 5848 | / | / | / | / | 11 |
| Jin | 2009 | China | Asian | Lung cancer | 1212 | 353 | 627 | 232 | 1339 | 450 | 658 | 231 | 0.719 | 13 |
| Wrensch | 2009 | USA | Caucasian | Glioma | 691 | 95 | 354 | 242 | 3981 | 1021 | 1904 | 1056 | 0.006 | 12 |
Abbreviations: ESCC: esophageal squamous cell carcinoma; NC: nasopharyngeal carcinoma; UTUC: upper tract urothelial carcinomas; MPN: myeloproliferative neoplasms; CML: chronic myeloid leukemia; AML: acute myeloid leukemia; RCC: renal cell carcinoma; ALL: acute lymphoblastic leukemia; SCC: squamous cell carcinoma; BCC: basal cell carcinoma; HC: hepatocellular carcinoma; TGCT: testicular germ cell tumor; HWE: Hardy-Weinberg equilibrium
a: Number of cases and controls for TG and GG genotypers. b: The allele frequence in the three studies was provided to estimate the association under allele contrast model (G vs. T).
Meta-analysis results
Heterogeneity among studies was observed for all five genetic models. Consequently, the random effect model was applied to calculate odds ratios (ORs). Risk estimates indicated that the TERT rs2736100 polymorphism was associated with overall cancer risk via all five genetic models [homozygous model (GG vs. TT): OR=1.39, 95% confidence interval (CI)=1.26–1.54, P<0.001; heterozygous model (TG vs. TT): OR=1.16, 95% CI=1.11–1.23, P<0.001; dominant model (TG + GG vs. TT): OR=1.23, 95% CI=1.15–1.31, P<0.001; recessive model (GG vs. TG + TT): OR=1.25, 95% CI=1.16–1.35, P<0.001; and allele contrast model (G vs. T): OR=1.17, 95% CI=1.12–1.23, P<0.001 (Figure 2, Table 2)]. The stratified analysis by cancer type associated the TERT rs2736100 polymorphism with lung cancer risk (homozygous model: OR=1.60, 95% CI=1.49–1.71, P<0.001; heterozygous model: OR=1.25, 95% CI=1.20–1.31, P=0.008; dominant model: OR=1.33, 95% CI 1.26–1.39, P<0.001; recessive model: OR=1.40, 95% CI=1.32–1.48, P<0.001; and allele contrast model: OR=1.24, 95% CI=1.17–1.31, P<0.001). This polymorphism was also associated with increased risk for thyroid cancer, bladder cancer, glioma, MPN and AML. Inversely, the TERT rs2736100 polymorphism was associated with decreased colorectal cancer risk (homozygous model: OR=0.86, 95% CI=0.82–0.91, P=0.512; dominant model: OR=0.94, 95% CI=0.90–0.98, P=0.970; recessive model: OR=0.88, 95% CI=0.82–0.96, P=0.279; and allele contrast model: OR=0.93, 95% CI=0.90–0.96, P=0.548). Stratified analysis was also performed by patient ethnicity, sample size of controls, and quality score. Elevated cancer risk was found among Asians in all five genetic models and among Caucasians under all five genetic models except for the recessive model. Our results also associated the TERT rs2736100 polymorphism with elevated overall cancer risk in all subgroups divided by sample size of controls and quality score in all the five genetic models.
Figure 2. Forest plot of the association between the TERT rs2736100 polymorphism and overall cancer susceptibility in the allele contrast model.
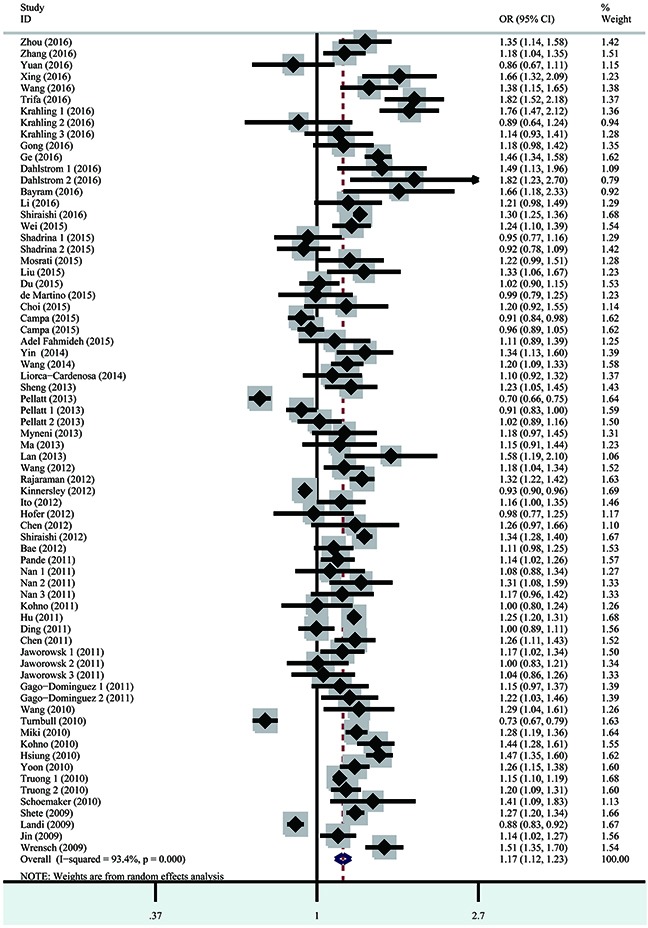
Table 2. Meta-analysis of TERT rs2736100 T>G polymorphism on cancer risk.
| Variables | Homozygous | Heterozygous | Recessive | Dominant | Allele | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GG vs. TT | TG vs. TT | GG vs. (TG + TT) | (TG +GG) vs. TT | G vs. T | |||||||||||
| OR (95% CI) | Phet | I2 (%) | OR (95% CI) | Phet | I2 (%) | OR (95% CI) | Phet | I2 (%) | OR (95% CI) | Phet | I2 (%) | OR (95% CI) | Phet | I2 (%) | |
| All | 1.39 (1.26-1.54) | <0.001 | 93.3 | 1.16 (1.11-1.23) | <0.001 | 80.0 | 1.25 (1.16-1.35) | <0.001 | 91.1 | 1.23 (1.15-1.31) | <0.001 | 88.9 | 1.17 (1.12-1.23) | <0.001 | 93.4 |
| Cancer type | |||||||||||||||
| Lung | 1.60 (1.49-1.71) | <0.001 | 65.7 | 1.25 (1.20-1.31) | 0.008 | 45.5 | 1.40 (1.32-1.48) | <0.001 | 61.2 | 1.33 (1.26-1.39) | <0.001 | 58.6 | 1.24 (1.17-1.31) | <0.001 | 89.4 |
| MPN | 3.17 (2.51-4.00) | 0.854 | 0.0 | 2.03 (1.64-2.51) | 0.972 | 0.0 | 1.89 (1.59-2.24) | 0.616 | 0.0 | 2.40 (1.97-2.94) | 0.957 | 0.0 | 1.74 (1.56-1.95) | 0.679 | 0.0 |
| AML | 1.40 (1.04-1.88) | 0.631 | 0.0 | 1.22 (0.94-1.59) | 0.744 | 0.0 | 1.23 (0.97-1.56) | 0.411 | 0.0 | 1.28 (1.00-1.64) | 0.970 | 0.0 | 1.18 (1.02-1.37) | 0.658 | 0.0 |
| Thyroid | 1.79 (1.25-2.56) | 0.076 | 68.3 | 1.26 (0.96-1.65) | 0.085 | 66.2 | 1.62 (1.37-1.92) | 0.266 | 19.3 | 1.38 (1.02-1.88) | 0.041 | 76.0 | 1.33 (1.08-1.64) | 0.040 | 76.4 |
| Gastric | 1.39 (0.82-2.33) | 0.028 | 72.1 | 1.22 (0.90-1.66) | 0.204 | 37.2 | 1.19 (0.83-1.70) | 0.044 | 68.1 | 1.31 (0.90-1.90) | 0.085 | 59.4 | 1.22 (0.94-1.58) | 0.023 | 73.5 |
| Breast | 0.56 (0.25-1.28) | <0.001 | 95.0 | 0.88 (0.73-1.07) | 0.158 | 49.8 | 0.63 (0.24-1.64) | <0.001 | 97.3 | 0.78 (0.71-0.85) | 0.892 | 0.0 | 0.80 (0.61-1.04) | 0.003 | 88.8 |
| Melanoma | 1.18 (0.90-1.54) | 0.890 | 0.0 | 1.00 (0.78-1.27) | 0.444 | 0.0 | 1.18 (0.95-1.47) | 0.700 | 0.0 | 1.06 (0.85-1.33) | 0.570 | 0.0 | 1.09 (0.95-1.26) | 0.922 | 0.0 |
| Colorectal | 0.86 (0.82-0.91) | 0.512 | 0.0 | 0.98 (0.93-1.03) | 0.989 | 0.0 | 0.88 (0.82-0.96) | 0.279 | 21.9 | 0.94 (0.90-0.98) | 0.970 | 0.0 | 0.93 (0.90-0.96) | 0.548 | 0.0 |
| Bladder | 1.31 (1.08-1.59) | 0.481 | 0.0 | 1.15 (0.98-1.34) | 0.498 | 0.0 | 1.18 (1.00-1.39) | 0.598 | 0.0 | 1.19 (1.02-1.38) | 0.436 | 0.0 | 1.13 (1.03-1.25) | 0.507 | 0.0 |
| Glioma | 1.89 (1.52-2.35) | 0.028 | 67.0 | 1.55 (1.30-1.84) | 0.055 | 60.0 | 1.35 (1.21-1.49) | 0.241 | 28.5 | 1.65 (1.37-1.99) | 0.020 | 69.4 | 1.33 (1.25-1.42) | 0.089 | 50.4 |
| Others | 1.09 (0.89-1.32) | <0.001 | 86.7 | 0.97 (0.88-1.07) | 0.002 | 58.4 | 1.11 (0.95-1.29) | <0.001 | 84.3 | 1.01 (0.89-1.13) | <0.001 | 78.2 | 1.04 (0.94-1.15) | <0.001 | 87.0 |
| Ethnicity | |||||||||||||||
| Asian | 1.56 (1.46-1.67) | <0.001 | 65.0 | 1.22 (1.17-1.28) | 0.001 | 49.5 | 1.39 (1.32-1.46) | <0.001 | 50.4 | 1.30 (1.28-1.36) | <0.001 | 62.1 | 1.25 (1.20-1.29) | <0.001 | 67.7 |
| Caucasian | 1.22 (1.04-1.44) | <0.001 | 94.4 | 1.12 (1.02-1.22) | <0.001 | 83.6 | 1.11 (0.99-1.25) | <0.001 | 92.5 | 1.16 (1.04-1.29) | <0.001 | 90.7 | 1.11 (1.03-1.19) | <0.001 | 94.2 |
| Sample Size | |||||||||||||||
| ≥ 500 | 1.34 (1.19-1.51) | <0.001 | 95.1 | 1.16 (1.09-1.23) | <0.001 | 83.7 | 1.22 (1.11-1.33) | <0.001 | 93.6 | 1.21 (1.13-1.30) | <0.001 | 91.5 | 1.15 (1.09-1.22) | <0.001 | 95.1 |
| <500 | 1.52 (1.26-1.82) | <0.001 | 72.5 | 1.19 (1.04-1.37) | <0.001 | 66.2 | 1.34 (1.19-1.51) | 0.001 | 55.1 | 1.29 (1.11-1.49) | <0.001 | 73.3 | 1.23 (1.12-1.35) | <0.001 | 74.1 |
| Score | |||||||||||||||
| High | 1.33 (1.18-1.48) | <0.001 | 94.5 | 1.15 (1.09-1.21) | <0.001 | 82.7 | 1.22 (1.12-1.33) | <0.001 | 92.8 | 1.20 (1.12-1.28) | <0.001 | 90.8 | 1.15 (1.09-1.21) | <0.001 | 94.5 |
| Low | 1.72 (1.40-2.10) | 0.001 | 60.5 | 1.30 (1.10-1.54) | 0.003 | 57.0 | 1.41 (1.26-1.59) | 0.154 | 27.4 | 1.40 (1.20-1.63) | <0.001 | 65.7 | 1.30 (1.18-1.43) | <0.001 | 60.5 |
Abbreviations: MPN, Myeloproliferative neoplasms; AML, Acute myeloid leukemia
Heterogeneity and sensitivity analyses
Heterogeneity was detected amongst studies with respect to the association between the TERT rs2736100 polymorphism and overall cancer risk (homozygous model: P<0.001; heterozygous model: P<0.001; dominant model: P<0.001; recessive model: P<0.001; and allele contrast model: P<0.001). Therefore, we used the random effects model to generate pooled ORs and 95% CIs. Sensitivity analyses indicated that no single study could change the between-study heterogeneity and the results of meta-analysis.
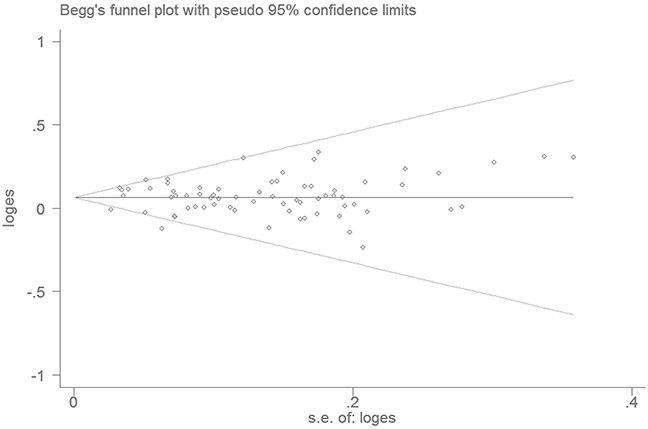
Publication bias
The Begg's funnel plot and Egger's linear regression analysis did not reveal any evidence of publication bias in the meta-analysis (homozygous model: P=0.183; heterozygous model: P=0.805; dominant model: P=0.406; recessive model: P=0.085; and allele model: P=0.122; Figure 3).
Figure 3. Funnel plot analysis to evaluate publication bias.
False positive report probability (FPRP) analyses
We calculated FPRP values for associations between the TERT rs2736100 T>G polymorphism and overall cancer risk using the five genetic models. FPRP values were all <0.20, suggesting that these associations were noteworthy (Table 3).
Table 3. False-positive report probability values for associations between the TERT rs2736100 T>G polymorphism and overall cancer risk.
| Genetic models | OR (95% CI) | P | Power | Prior Probability | |||||
|---|---|---|---|---|---|---|---|---|---|
| 0.25 | 0.1 | 0.01 | 0.001 | 0.0001 | 0.00001 | ||||
| Homozygous (GG vs. TT) | 1.39 (1.26-1.54) | <0.001 | 0.555 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 |
| Heterozygous (TG vs. TT) | 1.16 (1.11-1.23) | <0.001 | 0.872 | 0.000 | 0.000 | 0.000 | 0.001 | 0.008 | 0.073 |
| Recessive (GG vs. TG + TT) | 1.25 (1.16-1.35) | <0.001 | 0.841 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.002 |
| Dominant (TG +GG vs. TT) | 1.23 (1.15-1.31) | <0.001 | 0.957 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 |
| Allele (G vs. T) | 1.17 (1.12-1.23) | <0.001 | 0.839 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 |
DISCUSSION
Telomeres are special structures at the ends of eukaryotic chromosomes, and are responsible for protecting chromosomes from degradation, end-to-end fusion, and rearrangement [10]. Telomerase maintains proper telomere length by adding repetitive telomeric sequences to the 3′ ends of telomeres. Abnormal telomerase activity is implicated in the initiation and development of cancer and other age-associated diseases [75]. The TERT subunit of telomerase consists of three highly conserved domains: the RNA-binding domain (TRBD), the reverse transcriptase domain, and a carboxy-putative extension (CTE) proposed to constitute the putative thumb domain [75]. TERT is overexpressed in many human cancers [76]. The TERT rs2736100 polymorphism, localized in the second intron of the TERT gene, has been wildly studied with respect to cancer risk [7, 8]. However, the functional significance of the TERT rs2736100 polymorphism was not clear. Preliminary studies in gastric cancer suggested that this SNP is associated with decreased telomere length [9].
The present meta-analysis, comprising 108,248 cases and 161,472 controls, found that the TERT rs2736100 polymorphism increased overall cancer risk by 16–39%, suggesting that this SNP may contribute to carcinogenesis. A previous meta-analysis conducted by Zou, et al. in 2012 [15] also concluded that this polymorphism was associated with increased cancer risk. However, this analysis included only 11 case-control articles with 23,032 cases and 38,274 controls, which studied only lung cancer, glioma, and bladder cancer. Our stratified analysis by cancer type showed that the TERT rs2736100 polymorphism correlated with increased risk of lung cancer and glioma. Such associations were also observed in lung cancer- and glioma-specific meta-analyses published in 2014 [10–15, 77]. Between 2015 and 2016, at least 27 studies (6 studies on lung cancer) were published investigating the association between the TERT rs2736100 polymorphism and overall cancer susceptibility. To the best of our knowledge, ours is the largest meta-analysis of this association, with the strongest statistical power. Apart from lung cancer, glioma, and bladder cancer, our meta-analysis also investigated the association between the TERT rs2736100 polymorphism and risk of colorectal cancer (4 studies), MPN (4), gastric cancer (3), AML (2), breast cancer (2), melanoma (2), and thyroid cancer (2) as well as “other cancers” (16). We observed that this polymorphism was associated with decreased colorectal cancer risk. Since only four colorectal cancer studies were included in our meta-analysis, such an association might be a false positive, and validation will require further study.
The current meta-analysis had several limitations. First, there were substantial heterogeneities in the pooled study investigating the association between the TERT rs2736100 polymorphism and overall cancer risk. We reduced the degree of heterogeneity through stratified analyses by cancer type, patient ethnicity, sample size, and study quality score. Some cross-study heterogeneity might be attributed to differences among ethnic groups [78]. However, other sources of heterogeneity were not identified, such as control sources and genotyping methods. Second, the studies in this meta-analysis focused on Asian and Caucasian populations only, so we may not have had sufficient statistical power to evaluate associations based on ethnicity. Third, our results were based on unadjusted ORs due to the unavailability of confounding factor information for cases and controls (e.g., age, sex, smoking status, drinking status, and environmental exposure). Finally, lacking the original data from eligible studies limited our ability to explore gene-environment interactions.
In conclusion, our meta-analysis indicated that the TERT rs2736100 polymorphism was associated with increased overall cancer risk, especially lung cancer risk. Larger studies involving patients of different ethnicities are needed to confirm our findings.
MATERIALS AND METHODS
Identification of eligible studies
A comprehensive literature search of the PubMed and EMBASE databases was performed up to November 1, 2016. To find all eligible case-control studies that assessed the association between the TERT rs2736100 polymorphism and cancer risk, we used the following keywords: “TERT or telomerase reverse transcriptase”, “polymorphism or variant”, and “cancer or tumor or neoplasm or carcinoma”. We also evaluated additional studies by manually screening the references of both primary articles and reviews.
Inclusion and exclusion criteria
Eligible studies included in our analysis met the following criteria: (i) the TERT rs2736100 polymorphism-cancer risk association was assessed; (ii) case-control studies or cohort studies; (iii) sufficient data to calculate an OR with 95% CI; (iv) studies in English. Exclusion criteria were as follows: (i) case only studies; (ii) overlapping publications; (iii) abstract, case report, editorial comment, and review. Studies that deviated from HWE in controls were excluded, unless further evidence showed that another polymorphism was in HWE.
Data extraction
Two investigators independently extracted available data from each eligible study. The following information was collected: first author's surname, year of publication, country of origin, patient ethnicity, cancer type, numbers of cases and controls, genotype counts of cases and controls, results of the HWE test, and quality scores (low quality studies with score ≤9, high quality studies with score >9) [79]. Any disagreements were solved by discussion until a consensus was reached between the two investigators. If no consensus was reached, another investigator joined the discussion, and a final decision was made by a majority.
FPRP analysis
FPRP values were applied to assess the statistical power of our significant findings [80, 81]. An FPRP value of 0.20 was set as the criterion for noteworthiness. A prior probability of 0.1 was assigned to detect an OR of 0.67/1.50 (protective/risk effects) for an association with genotypes under investigation.
Statistical analysis
HWE in control subjects was assessed by chi-squared test. The strength of association between the TERT rs2736100 polymorphism and cancer risk was estimated by calculating crude ORs and their 95% CIs using all five genetic models: homozygous (GG vs. TT), heterozygous (TG vs. TT), dominant (GG vs. TG + TT), and recessive (TG + GG vs. TT), as well as the allele contrast model (G vs. T). Q-test was used to quantify heterogeneity among all eligible studies, and P>0.10 suggested a lack of heterogeneity among studies. Generally, the fixed effects model (Mantel–Haenszel method) or the random effects model (DerSimonian–Laird method) was employed in the absence (P≥0.10) or presence (P<0.10) of heterogeneity, respectively [82–84]. Heterogeneity was also estimated using the I2 test [85]. Subgroup analyses were conducted by patient ethnicity, cancer type, and study sample size. The Begg's funnel plot and the Egger's linear regression test were used to evaluate publication bias [86]. All statistical analyses were performed using STATA version 12.0 software (STATA Corporation, College Station, TX). All statistical analyses were two-sided. P<0.05 was considered statistically significant.
Footnotes
CONFLICTS OF INTEREST
The authors declare no conflicts of interest.
REFERENCES
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Song M, Lee KM, Kang D. Breast cancer prevention based on gene-environment interaction. Mol Carcinog. 2011;50:280–90. doi: 10.1002/mc.20639. [DOI] [PubMed] [Google Scholar]
- 3.Blasco MA. Telomeres and human disease: ageing, cancer and beyond. Nat Rev Genet. 2005;6:611–22. doi: 10.1038/nrg1656. [DOI] [PubMed] [Google Scholar]
- 4.Willeit P, Willeit J, Mayr A, Weger S, Oberhollenzer F, Brandstatter A, Kronenberg F, Kiechl S. Telomere length and risk of incident cancer and cancer mortality. JAMA. 2010;304:69–75. doi: 10.1001/jama.2010.897. [DOI] [PubMed] [Google Scholar]
- 5.Blackburn EH. Switching and signaling at the telomere. Cell. 2001;106:661–73. doi: 10.1016/s0092-8674(01)00492-5. [DOI] [PubMed] [Google Scholar]
- 6.Shete S, Hosking FJ, Robertson LB, Dobbins SE, Sanson M, Malmer B, Simon M, Marie Y, Boisselier B, Delattre JY, Hoang-Xuan K, El Hallani S, Idbaih A, et al. Genome-wide association study identifies five susceptibility loci for glioma. Nat Genet. 2009;41:899–904. doi: 10.1038/ng.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang C, Doherty JA, Burgess S, Hung RJ, Lindstrom S, Kraft P, Gong J, Amos CI, Sellers TA, Monteiro AN, Chenevix-Trench G, Bickeboller H, Risch A, et al. Genetic determinants of telomere length and risk of common cancers: a Mendelian randomization study. Hum Mol Genet. 2015;24:5356–66. doi: 10.1093/hmg/ddv252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu T, Yuan X, Xu D. Cancer-Specific Telomerase Reverse Transcriptase (TERT) Promoter Mutations: Biological and Clinical Implications. Genes (Basel) 2016:7. doi: 10.3390/genes7070038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Choi BJ, Yoon JH, Kim O, Choi WS, Nam SW, Lee JY, Park WS. Influence of the hTERT rs2736100 polymorphism on telomere length in gastric cancer. World J Gastroenterol. 2015;21:9328–36. doi: 10.3748/wjg.v21.i31.9328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhou P, Wei L, Xia X, Shao N, Qian X, Yang Y. Association between telomerase reverse transcriptase rs2736100 polymorphism and risk of glioma. J Surg Res. 2014;191:156–60. doi: 10.1016/j.jss.2014.03.043. [DOI] [PubMed] [Google Scholar]
- 11.Yuan Y, Lu C, Xue L, Ge D. Association between TERT rs2736100 polymorphism and lung cancer susceptibility: evidence from 22 case-control studies. Tumour Biol. 2014;35:4435–42. doi: 10.1007/s13277-013-1583-4. [DOI] [PubMed] [Google Scholar]
- 12.Yang J, Jiao S. Increased lung cancer risk associated with the TERT rs2736100 polymorphism: an updated meta-analysis. Tumour Biol. 2014;35:5763–9. doi: 10.1007/s13277-014-1765-8. [DOI] [PubMed] [Google Scholar]
- 13.Wu H, Zhu R. Quantitative assessment of common genetic variants on chromosome 5p15 and lung cancer risk. Tumour Biol. 2014;35:6055–63. doi: 10.1007/s13277-014-1802-7. [DOI] [PubMed] [Google Scholar]
- 14.Nie W, Zang Y, Chen J, Xiu Q. TERT rs2736100 polymorphism contributes to lung cancer risk: a meta-analysis including 49,869 cases and 73,464 controls. Tumour Biol. 2014;35:5569–74. doi: 10.1007/s13277-014-1734-2. [DOI] [PubMed] [Google Scholar]
- 15.Zou P, Gu A, Ji G, Zhao L, Zhao P, Lu A. The TERT rs2736100 polymorphism and cancer risk: a meta-analysis based on 25 case-control studies. BMC Cancer. 2012;12:7. doi: 10.1186/1471-2407-12-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou L, Fu G, Wei J, Shi J, Pan W, Ren Y, Xiong X, Xia J, Shen Y, Li H, Yang M. The identification of two regulatory ESCC susceptibility genetic variants in the TERT-CLPTM1L loci. Oncotarget. 2016;7:5495–506. doi: 10.18632/oncotarget.6747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang Y, Zhang X, Zhang H, Zhai Y, Wang Z, Li P, Yu L, Xia X, Zhang Y, Zeng Y, He F, Zhou G. Common variations in TERT-CLPTM1L locus are reproducibly associated with the risk of nasopharyngeal carcinoma in Chinese populations. Oncotarget. 2016;7:759–70. doi: 10.18632/oncotarget.6397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yuan X, Meng Y, Li P, Ge N, Kong F, Yang L, Bjorkholm M, Zhao S, Xu D. The association between the TERT rs2736100 AC genotype and reduced risk of upper tract urothelial carcinomas in a Han Chinese population. Oncotarget. 2016;7:31972–31979. doi: 10.18632/oncotarget.7777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xing YL, Liu F, Li JF, Lin JC, Zhu GD, Li M, Zhang CR, Niu YY. Case-Control Study on Impact of the Telomerase Reverse Transcriptase Gene Polymorphism and Additional Single Nucleotide Polymorphism (SNP)- SNP Interaction on Non-Small Cell Lung Cancers Risk in Chinese Han Population. J Clin Lab Anal. 2016 doi: 10.1002/jcla.21982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang X, Ma K, Chi L, Cui J, Jin L, Hu JF, Li W. Combining Telomerase Reverse Transcriptase Genetic Variant rs2736100 with Epidemiologic Factors in the Prediction of Lung Cancer Susceptibility. J Cancer. 2016;7:846–53. doi: 10.7150/jca.13437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Trifa AP, Banescu C, Tevet M, Bojan A, Dima D, Urian L, Torok-Vistai T, Popov VM, Zdrenghea M, Petrov L, Vasilache A, Murat M, Georgescu D, et al. TERT rs2736100 A>C SNP and JAK2 46/1 haplotype significantly contribute to the occurrence of JAK2 V617F and CALR mutated myeloproliferative neoplasms - a multicentric study on 529 patients. Br J Haematol. 2016;174:218–26. doi: 10.1111/bjh.14041. [DOI] [PubMed] [Google Scholar]
- 22.Shiraishi K, Okada Y, Takahashi A, Kamatani Y, Momozawa Y, Ashikawa K, Kunitoh H, Matsumoto S, Takano A, Shimizu K, Goto A, Tsuta K, Watanabe S, et al. Association of variations in HLA class II and other loci with susceptibility to EGFR-mutated lung adenocarcinoma. Nat Commun. 2016;7:12451. doi: 10.1038/ncomms12451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li X, Xu X, Fang J, Wang L, Mu Y, Zhang P, Yao Z, Ma Z, Liu Z. Rs2853677 modulates Snail1 binding to the TERT enhancer and affects lung adenocarcinoma susceptibility. Oncotarget. 2016;7:37825–37838. doi: 10.18632/oncotarget.9339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Krahling T, Balassa K, Kiss KP, Bors A, Batai A, Halm G, Egyed M, Fekete S, Remenyi P, Masszi T, Tordai A, Andrikovics H. Co-occurrence of Myeloproliferative Neoplasms and Solid Tumors Is Attributed to a Synergism Between Cytoreductive Therapy and the Common TERT Polymorphism rs2736100. Cancer Epidemiol Biomarkers Prev. 2016;25:98–104. doi: 10.1158/1055-9965.EPI-15-0805. [DOI] [PubMed] [Google Scholar]
- 25.Gong L, Xu Y, Hu YQ, Ding QJ, Yi CH, Huang W, Zhou M. hTERT gene polymorphism correlates with the risk and the prognosis of thyroid cancer. J Back Musculoskelet Rehabil. 2016 doi: 10.3233/BMR-160631. [DOI] [PubMed] [Google Scholar]
- 26.Ge M, Shi M, An C, Yang W, Nie X, Zhang J, Lv Z, Li J, Zhou L, Du Z, Yang M. Functional evaluation of TERT-CLPTM1L genetic variants associated with susceptibility of papillary thyroid carcinoma. Sci Rep. 2016;6:26037. doi: 10.1038/srep26037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dahlstrom J, Liu T, Yuan X, Saft L, Ghaderi M, Wei YB, Lavebratt C, Li P, Zheng C, Bjorkholm M, Xu D. TERT rs2736100 genotypes are associated with differential risk of myeloproliferative neoplasms in Swedish and Chinese male patient populations. Ann Hematol. 2016 doi: 10.1007/s00277-016-2787-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bayram S, Ulger Y, Sumbul AT, Kaya BY, Genc A, Rencuzogullari E, Dadas E. Polymorphisms in human telomerase reverse transcriptase (hTERT) gene and susceptibility to gastric cancer in a Turkish population: Hospital-based case-control study. Gene. 2016;585:84–92. doi: 10.1016/j.gene.2016.03.030. [DOI] [PubMed] [Google Scholar]
- 29.Wei R, Cao L, Pu H, Wang H, Zheng Y, Niu X, Weng X, Zhang H, Favus MJ, Zhang L, Jia W, Zeng Y, Amos CI, et al. TERT Polymorphism rs2736100-C Is Associated with EGFR Mutation-Positive Non-Small Cell Lung Cancer. Clin Cancer Res. 2015;21:5173–80. doi: 10.1158/1078-0432.CCR-15-0009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shadrina AS, Boyarskikh UA, Oskina NA, Sinkina TV, Lazarev AF, Petrova VD, Filipenko ML. TERT polymorphisms rs2853669 and rs7726159 influence on prostate cancer risk in Russian population. Tumour Biol. 2015;36:841–7. doi: 10.1007/s13277-014-2688-0. [DOI] [PubMed] [Google Scholar]
- 31.Mosrati MA, Willander K, Falk IJ, Hermanson M, Hoglund M, Stockelberg D, Wei Y, Lotfi K, Soderkvist P. Association between TERT promoter polymorphisms and acute myeloid leukemia risk and prognosis. Oncotarget. 2015;6:25109–20. doi: 10.18632/oncotarget.4668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu SG, Ma L, Cen QH, Huang JS, Zhang JX, Zhang JJ. Association of genetic polymorphisms in TERT-CLPTM1L with lung cancer in a Chinese population. Genet Mol Res. 2015;14:4469–76. doi: 10.4238/2015.May.4.4. [DOI] [PubMed] [Google Scholar]
- 33.Du J, Zhu X, Xie C, Dai N, Gu Y, Zhu M, Wang C, Gao Y, Pan F, Ren C, Ji Y, Dai J, Ma H, et al. Telomere length, genetic variants and gastric cancer risk in a Chinese population. Carcinogenesis. 2015;36:963–70. doi: 10.1093/carcin/bgv075. [DOI] [PubMed] [Google Scholar]
- 34.de Martino M, Taus C, Lucca I, Hofbauer SL, Haitel A, Shariat SF, Klatte T. Association of human telomerase reverse transcriptase gene polymorphisms, serum levels, and telomere length with renal cell carcinoma risk and pathology. Mol Carcinog. 2015 doi: 10.1002/mc.22388. [DOI] [PubMed] [Google Scholar]
- 35.Campa D, Rizzato C, Stolzenberg-Solomon R, Pacetti P, Vodicka P, Cleary SP, Capurso G, Bueno-de-Mesquita HB, Werner J, Gazouli M, Butterbach K, Ivanauskas A, Giese N, et al. TERT gene harbors multiple variants associated with pancreatic cancer susceptibility. Int J Cancer. 2015;137:2175–83. doi: 10.1002/ijc.29590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Campa D, Martino A, Varkonyi J, Lesueur F, Jamroziak K, Landi S, Jurczyszyn A, Marques H, Andersen V, Jurado M, Brenner H, Petrini M, Vogel U, et al. Risk of multiple myeloma is associated with polymorphisms within telomerase genes and telomere length. Int J Cancer. 2015;136:E351–8. doi: 10.1002/ijc.29101. [DOI] [PubMed] [Google Scholar]
- 37.Adel Fahmideh M, Lavebratt C, Schuz J, Roosli M, Tynes T, Grotzer MA, Johansen C, Kuehni CE, Lannering B, Prochazka M, Schmidt LS, Feychting M. CCDC26, CDKN2BAS, RTEL1 and TERT Polymorphisms in pediatric brain tumor susceptibility. Carcinogenesis. 2015;36:876–82. doi: 10.1093/carcin/bgv074. [DOI] [PubMed] [Google Scholar]
- 38.Yin Z, Cui Z, Ren Y, Zhang H, Yan Y, Zhao Y, Ma R, Wang Q, He Q, Zhou B. Genetic polymorphisms of TERT and CLPTM1L, cooking oil fume exposure, and risk of lung cancer: a case-control study in a Chinese non-smoking female population. Med Oncol. 2014;31:114. doi: 10.1007/s12032-014-0114-5. [DOI] [PubMed] [Google Scholar]
- 39.Wang F, Fu P, Pang Y, Liu C, Shao Z, Zhu J, Li J, Wang T, Zhang X, Liu J. TERT rs2736100T/G polymorphism upregulates interleukin 6 expression in non-small cell lung cancer especially in adenocarcinoma. Tumour Biol. 2014;35:4667–72. doi: 10.1007/s13277-014-1611-z. [DOI] [PubMed] [Google Scholar]
- 40.Llorca-Cardenosa MJ, Pena-Chilet M, Mayor M, Gomez-Fernandez C, Casado B, Martin-Gonzalez M, Carretero G, Lluch A, Martinez-Cadenas C, Ibarrola-Villava M, Ribas G. Long telomere length and a TERT-CLPTM1 locus polymorphism association with melanoma risk. Eur J Cancer. 2014;50:3168–77. doi: 10.1016/j.ejca.2014.09.017. [DOI] [PubMed] [Google Scholar]
- 41.Zhao Z, Li C, Yang L, Zhang X, Zhao X, Song X, Li X, Wang J, Qian J, Yang Y, Jin L, Chen H, Lu D. Significant association of 5p15.33 (TERT-CLPTM1L genes) with lung cancer in Chinese Han population. Exp Lung Res. 2013;39:91–8. doi: 10.3109/01902148.2012.762436. [DOI] [PubMed] [Google Scholar]
- 42.Sheng X, Tong N, Tao G, Luo D, Wang M, Fang Y, Li J, Xu M, Zhang Z, Wu D. TERT polymorphisms modify the risk of acute lymphoblastic leukemia in Chinese children. Carcinogenesis. 2013;34:228–35. doi: 10.1093/carcin/bgs325. [DOI] [PubMed] [Google Scholar]
- 43.Pellatt AJ, Wolff RK, Torres-Mejia G, John EM, Herrick JS, Lundgreen A, Baumgartner KB, Giuliano AR, Hines LM, Fejerman L, Cawthon R, Slattery ML. Telomere length, telomere-related genes, and breast cancer risk: the breast cancer health disparities study. Genes Chromosomes Cancer. 2013;52:595–609. doi: 10.1002/gcc.22056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pellatt AJ, Wolff RK, Herrick J, Lundgreen A, Slattery ML. TERT's role in colorectal carcinogenesis. Mol Carcinog. 2013;52:507–13. doi: 10.1002/mc.21885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Myneni AA, Chang SC, Niu R, Liu L, Ochs-Balcom HM, Li Y, Zhang C, Zhao B, Shi J, Han X, Li J, Su J, Cai L, et al. Genetic polymorphisms of TERT and CLPTM1L and risk of lung cancer—a case-control study in a Chinese population. Lung Cancer. 2013;80:131–7. doi: 10.1016/j.lungcan.2013.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ma Z, Hu Q, Chen Z, Tao S, Macnamara L, Kim ST, Tian L, Xu K, Ding Q, Zheng SL, Sun J, Xia G, Xu J. Systematic evaluation of bladder cancer risk-associated single-nucleotide polymorphisms in a Chinese population. Mol Carcinog. 2013;52:916–21. doi: 10.1002/mc.21932. [DOI] [PubMed] [Google Scholar]
- 47.Lan Q, Cawthon R, Gao Y, Hu W, Hosgood HD, 3rd, Barone-Adesi F, Ji BT, Bassig B, Chow WH, Shu X, Cai Q, Xiang Y, Berndt S, et al. Longer telomere length in peripheral white blood cells is associated with risk of lung cancer and the rs2736100 (CLPTM1L-TERT) polymorphism in a prospective cohort study among women in China. PLoS One. 2013;8:e59230. doi: 10.1371/journal.pone.0059230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang S, Wu J, Hu L, Ding C, Kan Y, Shen Y, Chen X, Shen H, Guo X, Hu Z. Common genetic variants in TERT contribute to risk of cervical cancer in a Chinese population. Mol Carcinog. 2012;51(Suppl 1):E118–22. doi: 10.1002/mc.21872. [DOI] [PubMed] [Google Scholar]
- 49.Shiraishi K, Kunitoh H, Daigo Y, Takahashi A, Goto K, Sakamoto H, Ohnami S, Shimada Y, Ashikawa K, Saito A, Watanabe S, Tsuta K, Kamatani N, et al. A genome-wide association study identifies two new susceptibility loci for lung adenocarcinoma in the Japanese population. Nat Genet. 2012;44:900–3. doi: 10.1038/ng.2353. [DOI] [PubMed] [Google Scholar]
- 50.Rajaraman P, Melin BS, Wang Z, McKean-Cowdin R, Michaud DS, Wang SS, Bondy M, Houlston R, Jenkins RB, Wrensch M, Yeager M, Ahlbom A, Albanes D, et al. Genome-wide association study of glioma and meta-analysis. Hum Genet. 2012;131:1877–88. doi: 10.1007/s00439-012-1212-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kinnersley B, Migliorini G, Broderick P, Whiffin N, Dobbins SE, Casey G, Hopper J, Sieber O, Lipton L, Kerr DJ, Dunlop MG, Tomlinson IP, Houlston RS, et al. The TERT variant rs2736100 is associated with colorectal cancer risk. Br J Cancer. 2012;107:1001–8. doi: 10.1038/bjc.2012.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ito H, McKay JD, Hosono S, Hida T, Yatabe Y, Mitsudomi T, Brennan P, Tanaka H, Matsuo K. Association between a genome-wide association study-identified locus and the risk of lung cancer in Japanese population. J Thorac Oncol. 2012;7:790–8. doi: 10.1097/JTO.0b013e3182475028. [DOI] [PubMed] [Google Scholar]
- 53.Hofer P, Baierl A, Bernhart K, Leeb G, Mach K, Micksche M, Gsur A. Association of genetic variants of human telomerase with colorectal polyps and colorectal cancer risk. Mol Carcinog. 2012;51(Suppl 1):E176–82. doi: 10.1002/mc.21911. [DOI] [PubMed] [Google Scholar]
- 54.Chen XF, Cai S, Chen QG, Ni ZH, Tang JH, Xu DW, Wang XB. Multiple variants of TERT and CLPTM1L constitute risk factors for lung adenocarcinoma. Genet Mol Res. 2012;11:370–8. doi: 10.4238/2012.February.16.2. [DOI] [PubMed] [Google Scholar]
- 55.Bae EY, Lee SY, Kang BK, Lee EJ, Choi YY, Kang HG, Choi JE, Jeon HS, Lee WK, Kam S, Shin KM, Jin G, Yoo SS, et al. Replication of results of genome-wide association studies on lung cancer susceptibility loci in a Korean population. Respirology. 2012;17:699–706. doi: 10.1111/j.1440-1843.2012.02165.x. [DOI] [PubMed] [Google Scholar]
- 56.Pande M, Spitz MR, Wu X, Gorlov IP, Chen WV, Amos CI. Novel genetic variants in the chromosome 5p15.33 region associate with lung cancer risk. Carcinogenesis. 2011;32:1493–9. doi: 10.1093/carcin/bgr136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nan H, Qureshi AA, Prescott J, De Vivo I, Han J. Genetic variants in telomere-maintaining genes and skin cancer risk. Hum Genet. 2011;129:247–53. doi: 10.1007/s00439-010-0921-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kohno T, Kunitoh H, Mimaki S, Shiraishi K, Kuchiba A, Yamamoto S, Yokota J. Contribution of the TP53, OGG1, CHRNA3, and HLA-DQA1 genes to the risk for lung squamous cell carcinoma. J Thorac Oncol. 2011;6:813–7. doi: 10.1097/JTO.0b013e3181ee80ef. [DOI] [PubMed] [Google Scholar]
- 59.Jaworowska E, Trubicka J, Lener MR, Masojc B, Zlowocka-Perlowska E, McKay JD, Renard H, Oszutowska D, Wokolorczyk D, Lubinski J, Grodzki T, Serwatowski P, Nej-Wolosiak K, et al. Smoking related cancers and loci at chromosomes 15q25, 5p15, 6p22.1 and 6p21.33 in the Polish population. PLoS One. 2011;6:e25057. doi: 10.1371/journal.pone.0025057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hu Z, Wu C, Shi Y, Guo H, Zhao X, Yin Z, Yang L, Dai J, Hu L, Tan W, Li Z, Deng Q, Wang J, et al. A genome-wide association study identifies two new lung cancer susceptibility loci at 13q12.12 and 22q12.2 in Han Chinese. Nat Genet. 2011;43:792–6. doi: 10.1038/ng.875. [DOI] [PubMed] [Google Scholar]
- 61.Gago-Dominguez M, Jiang X, Conti DV, Castelao JE, Stern MC, Cortessis VK, Pike MC, Xiang YB, Gao YT, Yuan JM, Van Den Berg DJ. Genetic variations on chromosomes 5p15 and 15q25 and bladder cancer risk: findings from the Los Angeles-Shanghai bladder case-control study. Carcinogenesis. 2011;32:197–202. doi: 10.1093/carcin/bgq233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ding CY, Hu LM, Hu ZB, Shen HB. The relationship between gene polymorphism of telomerase reverse transcriptase and susceptibility to hepatocellular carcinoma. [Article in Chinese] Zhonghua Yu Fang Yi Xue Za Zhi. 2011;45:593–6. [PubMed] [Google Scholar]
- 63.Chen H, Chen Y, Zhao Y, Fan W, Zhou K, Liu Y, Zhou L, Mao Y, Wei Q, Xu J, Lu D. Association of sequence variants on chromosomes 20, 11, and 5 (20q13.33, 11q23.3, and 5p15.33) with glioma susceptibility in a Chinese population. Am J Epidemiol. 2011;173:915–22. doi: 10.1093/aje/kwq457. [DOI] [PubMed] [Google Scholar]
- 64.Yoon KA, Park JH, Han J, Park S, Lee GK, Han JY, Zo JI, Kim J, Lee JE, Takahashi A, Kubo M, Nakamura Y, Lee JS. A genome-wide association study reveals susceptibility variants for non-small cell lung cancer in the Korean population. Hum Mol Genet. 2010;19:4948–54. doi: 10.1093/hmg/ddq421. [DOI] [PubMed] [Google Scholar]
- 65.Wang Y, Broderick P, Matakidou A, Eisen T, Houlston RS. Role of 5p15.33 (TERT-CLPTM1L), 6p21.33 and 15q25.1 (CHRNA5-CHRNA3) variation and lung cancer risk in never-smokers. Carcinogenesis. 2010;31:234–8. doi: 10.1093/carcin/bgp287. [DOI] [PubMed] [Google Scholar]
- 66.Turnbull C, Rapley EA, Seal S, Pernet D, Renwick A, Hughes D, Ricketts M, Linger R, Nsengimana J, Deloukas P, Huddart RA, Bishop DT, Easton DF, et al. Variants near DMRT1, TERT and ATF7IP are associated with testicular germ cell cancer. Nat Genet. 2010;42:604–7. doi: 10.1038/ng.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Truong T, Hung RJ, Amos CI, Wu X, Bickeboller H, Rosenberger A, Sauter W, Illig T, Wichmann HE, Risch A, Dienemann H, Kaaks R, Yang P, et al. Replication of lung cancer susceptibility loci at chromosomes 15q25, 5p15, and 6p21: a pooled analysis from the International Lung Cancer Consortium. J Natl Cancer Inst. 2010;102:959–71. doi: 10.1093/jnci/djq178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Schoemaker MJ, Robertson L, Wigertz A, Jones ME, Hosking FJ, Feychting M, Lonn S, McKinney PA, Hepworth SJ, Muir KR, Auvinen A, Salminen T, Kiuru A, et al. Interaction between 5 genetic variants and allergy in glioma risk. Am J Epidemiol. 2010;171:1165–73. doi: 10.1093/aje/kwq075. [DOI] [PubMed] [Google Scholar]
- 69.Miki D, Kubo M, Takahashi A, Yoon KA, Kim J, Lee GK, Zo JI, Lee JS, Hosono N, Morizono T, Tsunoda T, Kamatani N, Chayama K, et al. Variation in TP63 is associated with lung adenocarcinoma susceptibility in Japanese and Korean populations. Nat Genet. 2010;42:893–6. doi: 10.1038/ng.667. [DOI] [PubMed] [Google Scholar]
- 70.Kohno T, Kunitoh H, Shimada Y, Shiraishi K, Ishii Y, Goto K, Ohe Y, Nishiwaki Y, Kuchiba A, Yamamoto S, Hirose H, Oka A, Yanagitani N, et al. Individuals susceptible to lung adenocarcinoma defined by combined HLA-DQA1 and TERT genotypes. Carcinogenesis. 2010;31:834–41. doi: 10.1093/carcin/bgq003. [DOI] [PubMed] [Google Scholar]
- 71.Hsiung CA, Lan Q, Hong YC, Chen CJ, Hosgood HD, Chang IS, Chatterjee N, Brennan P, Wu C, Zheng W, Chang GC, Wu T, Park JY, et al. The 5p15.33 locus is associated with risk of lung adenocarcinoma in never-smoking females in Asia. PLoS Genet. 2010;6 doi: 10.1371/journal.pgen.1001051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wrensch M, Jenkins RB, Chang JS, Yeh RF, Xiao Y, Decker PA, Ballman KV, Berger M, Buckner JC, Chang S, Giannini C, Halder C, Kollmeyer TM, et al. Variants in the CDKN2B and RTEL1 regions are associated with high-grade glioma susceptibility. Nat Genet. 2009;41:905–8. doi: 10.1038/ng.408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Landi MT, Chatterjee N, Yu K, Goldin LR, Goldstein AM, Rotunno M, Mirabello L, Jacobs K, Wheeler W, Yeager M, Bergen AW, Li Q, Consonni D, et al. A genome-wide association study of lung cancer identifies a region of chromosome 5p15 associated with risk for adenocarcinoma. Am J Hum Genet. 2009;85:679–91. doi: 10.1016/j.ajhg.2009.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jin G, Xu L, Shu Y, Tian T, Liang J, Xu Y, Wang F, Chen J, Dai J, Hu Z, Shen H. Common genetic variants on 5p15.33 contribute to risk of lung adenocarcinoma in a Chinese population. Carcinogenesis. 2009;30:987–90. doi: 10.1093/carcin/bgp090. [DOI] [PubMed] [Google Scholar]
- 75.Gillis AJ, Schuller AP, Skordalakes E. Structure of the Tribolium castaneum telomerase catalytic subunit TERT. Nature. 2008;455:633–7. doi: 10.1038/nature07283. [DOI] [PubMed] [Google Scholar]
- 76.Stewart SA, Weinberg RA. Telomeres: cancer to human aging. Annu Rev Cell Dev Biol. 2006;22:531–57. doi: 10.1146/annurev.cellbio.22.010305.104518. [DOI] [PubMed] [Google Scholar]
- 77.Wang HM, Zhang XY, Jin B. TERT genetic polymorphism rs2736100 was associated with lung cancer: a meta-analysis based on 14,492 subjects. Genet Test Mol Biomarkers. 2013;17:937–41. doi: 10.1089/gtmb.2013.0322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Xue WQ, He YQ, Zhu JH, Ma JQ, He J, Jia WH. Association of BRCA2 N372H polymorphism with cancer susceptibility: a comprehensive review and meta-analysis. Sci Rep. 2014;4:6791. doi: 10.1038/srep06791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.He J, Liao XY, Zhu JH, Xue WQ, Shen GP, Huang SY, Chen W, Jia WH. Association of MTHFR C677T and A1298C polymorphisms with non-Hodgkin lymphoma susceptibility: evidence from a meta-analysis. Sci Rep. 2014;4:6159. doi: 10.1038/srep06159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wacholder S, Chanock S, Garcia-Closas M, El Ghormli L, Rothman N. Assessing the probability that a positive report is false: an approach for molecular epidemiology studies. J Natl Cancer Inst. 2004;96:434–42. doi: 10.1093/jnci/djh075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.He J, Wang MY, Qiu LX, Zhu ML, Shi TY, Zhou XY, Sun MH, Yang YJ, Wang JC, Jin L, Wang YN, Li J, Yu HP, et al. Genetic variations of mTORC1 genes and risk of gastric cancer in an Eastern Chinese population. Mol Carcinog. 2013;52(Suppl 1):E70–9. doi: 10.1002/mc.22013. [DOI] [PubMed] [Google Scholar]
- 82.Mantel N, Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst. 1959;22:719–48. [PubMed] [Google Scholar]
- 83.DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7:177–88. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- 84.Tang J, Li H, Luo J, Mei H, Peng L, Li X. The LSP1 rs3817198 T > C polymorphism contributes to increased breast cancer risk: a meta-analysis of twelve studies. Oncotarget. 2016;7:63960–63967. doi: 10.18632/oncotarget.11741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Higgins JP, Thompson SG. Quantifying heterogeneity in a meta-analysis. Stat Med. 2002;21:1539–58. doi: 10.1002/sim.1186. [DOI] [PubMed] [Google Scholar]
- 86.Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315:629–34. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]


