Abstract
Spinocerebellar ataxia type 36 is a late-onset, slowly progressive cerebellar syndrome with motor neuron degeneration that is caused by expansions of a hexanucleotide repeat (GGCCTG) in the noncoding region of NOP56 gene, with a histopathological feature of RNA foci formation in postmortem tissues. Here, we report a cellular model using the spinocerebellar ataxia type 36 patient induced pluripotent stem cells (iPSCs). We generated iPSCs from spinocerebellar ataxia type 36 patients and differentiated them into neurons. The number of RNA-foci-positive cells was increased in patient iPSCs and iPSC-derived neurons. Treatment of the 2′-O, 4′-C-ethylene-bridged nucleic acid antisense oligonucleotides (ASOs) targeting NOP56 pre-mRNA reduced RNA-foci-positive cells to ∼50% in patient iPSCs and iPSC-derived neurons. NOP56 mRNA expression levels were lower in patient iPSCs and iPSC-derived neurons than in healthy control neurons. One of the ASOs reduced the number of RNA-foci-positive cells without altering NOP56 mRNA expression levels in patient iPSCs and iPSC-derived neurons. These data show that iPSCs from spinocerebellar ataxia type 36 patients can be useful for evaluating the effects of ASOs toward GGCCTG repeat expansion in spinocerebellar ataxia type 36.
Keywords: SCA36, iPSC, motor neuron, ASO, NOP56
Introduction
More than 20 repeat expansion diseases are reported in neurodegenerative disorders. Spinocerebellar ataxia type 36 (SCA36) is one of the repeat expansion diseases caused by GGCCTG hexanucleotide repeat expansion in intron 1 of NOP56.1 SCA36 was initially identified in Japan, and SCA36 causative mutation proved to be the same in a familial ataxia in Spain.2 SCA36 patients have also been found in France and China.3, 4, 5 The mean onset age of SCA36 is in the fifth decade, and the mean duration of the illness is over 10 years6. Cerebellar ataxia occurs in most cases, and motor neuron symptoms, including tongue/skeletal muscle atrophy or fasciculation, are also found in almost all cases with disease progression.2, 6 Thus, SCA36 has the characteristics of not only spinocerebellar ataxia but also motor neuron disease.
Although there is still no drug to cure repeat expansion diseases, an approach using nucleic acid medicine is expected to be promising. There are several kinds of antisense oligonucleotide (ASO) targeting pre-mRNA that have advantages of modification varieties and possible administration without carrier.7, 8, 9, 10 Several improvements have been added to ASOs. The 2′-O, 4′-C-ethylene-bridged nucleic acid (ENA) ASO, one of the modified ASOs, has a high-affinity advantage for the target and high-nuclease-resistant stability.11, 12, 13 As for the repeat expansion disease, the expected therapeutic mechanism of ASO is suppressing pre-mRNA, including repeat expansion, or affecting repeat RNA conformation. Modeling motor neuron disease using patient-induced pluripotent stem cells (iPSCs) for small-compound screening was reported.14, 15 ASO evaluation was also reported using iPSCs derived from patients with another hexanucleotide repeat expansion disease, C9orf72-related amyotrophic lateral sclerosis/frontotemporal lobar degeneration (ALS/FTLD). In C9orf72-related ALS/FTLD iPSC models, the ASO approach improved the pathology, including RNA gain of toxicity.16, 17 However, there exists no SCA36 model to test the ASO’s effectiveness.
Here, we generated iPSCs from SCA36 patients and differentiated them into motor neurons. We recapitulated RNA foci formation in both SCA36 patient iPSCs and iPSC-derived neurons (iPSNs). We tested the effects of five ENA ASOs and finally succeeded in identifying an ENA ASO that reduces RNA-foci-positive cells without altering NOP56 mRNA expression levels.
Results
Generation of iPSC Clones and Neural Differentiation
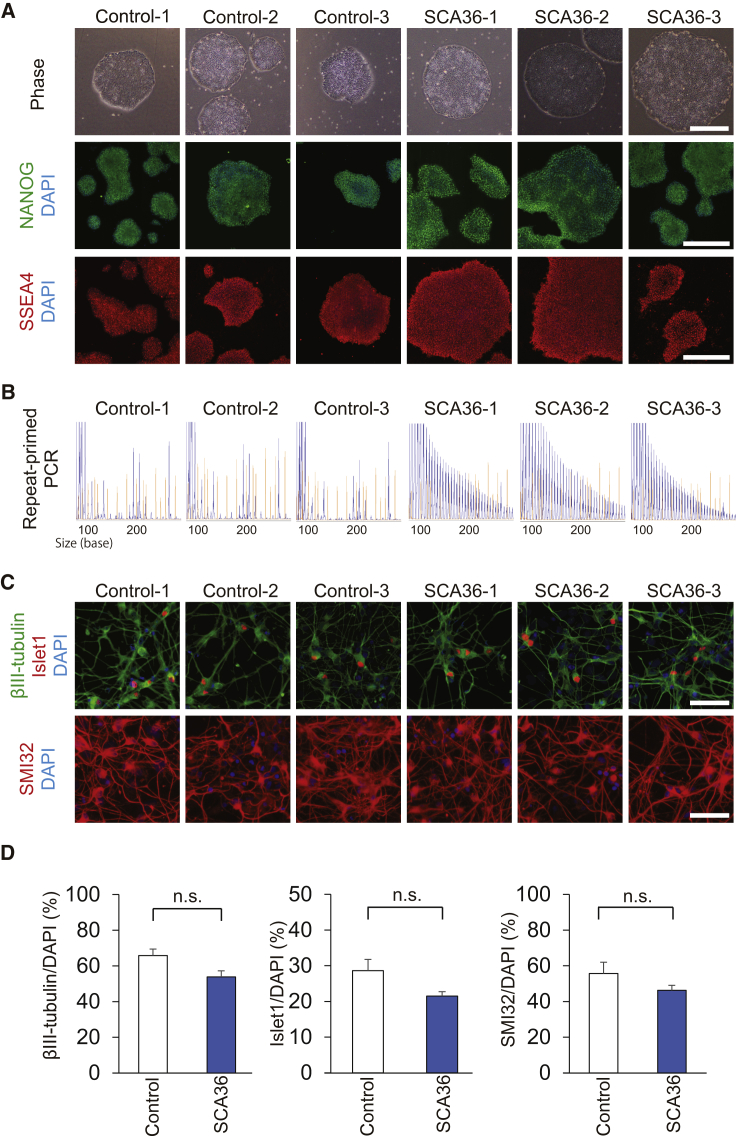
We generated iPSC clones from skin fibroblasts or peripheral blood mononuclear cells (PBMCs) of healthy controls and SCA36 patients (Table 1). We performed RNA sequencing (RNA-seq) analysis and confirmed that all iPSC clones were categorized in the same cluster, which was differentiated from human dermal fibroblast (HDF) or PBMC clusters (Figure S1). All six clones expressed the pluripotency markers SSEA4 and NANOG in immunostaining (Figure 1A). All iPSC clones were examined by repeat-primed PCR, and SCA36 patient iPSC clones were confirmed to retain the GGCCTG repeat expansion (Figure 1B). We performed Southern blot analysis of iPSCs and iPSNs (Figure S2) and found an expanded allele in SCA36 patient iPSCs and SCA36 patient iPSNs.
Table 1.
List of iPSC Clones
| Experimental Clone | Clone Name at Establishment | Somatic Cell | Gender | Donor Age | Reprogramming Vector |
|---|---|---|---|---|---|
| Control | |||||
| Control-1 | HPS1042a | PBMC | male | 61 | episomal |
| Control-2 | HPS1044a | PBMC | female | 65 | episomal |
| Control-3 | hc6 NOR-B38 | PBMC | male | 47 | episomal |
| SCA36 | |||||
| SCA36-1 | SCD4 E2 | skin fibroblast | male | 64 | episomal |
| SCA36-2 | SCD10 9E | PBMC | male | 60 | episomal |
| SCA36-3 | SCD20 2E | PBMC | female | 65 | episomal |
RIKEN BioResource Center
Figure 1.
Generation of iPSC Clones and Neural Differentiation
(A) iPSCs from both healthy controls and SCA36 patients showing embryonic stem cell-like morphology (phase) and expressing pluripotent stem cell markers such as NANOG (green) and SSEA4 (red). Scale bars, 500 μm. (B) Sequence traces of repeat-primed PCR products are shown. A typical saw-tooth pattern by repeat expansion is observed in SCA36 patient iPSCs, but not in healthy control iPSCs. (C) Differentiation of neural cells containing motor neurons from iPSC clones. Generated neurons exhibited neuron marker βІІІ-tubulin and motor neuron markers Islet1 and SMI32. Scale bars, 50 μm. (D) There were no significant differences in the differentiation propensity of neurons or motor neurons between healthy control and SCA36 patient iPSC clones. Values are expressed as mean ± SEM from n = 3 clones. n.s., not significant.
All iPSC clones were differentiated into neural cells containing motor neurons by serum-free floating culture of embryoid-body-like aggregates by quick re-aggregation (SFEBq) method14, 18 (Figure 1C). We found no obvious difference in neuronal differentiation propensity between control and SCA36 iPSC clones (Figure 1D).
SCA36 iPSCs and iPSC-Derived Neurons Exhibit RNA Foci
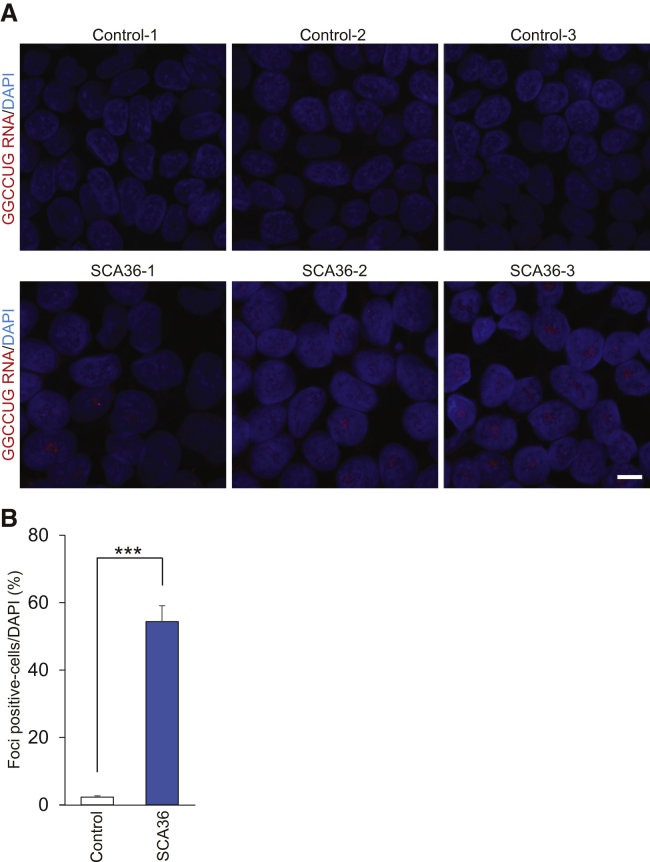
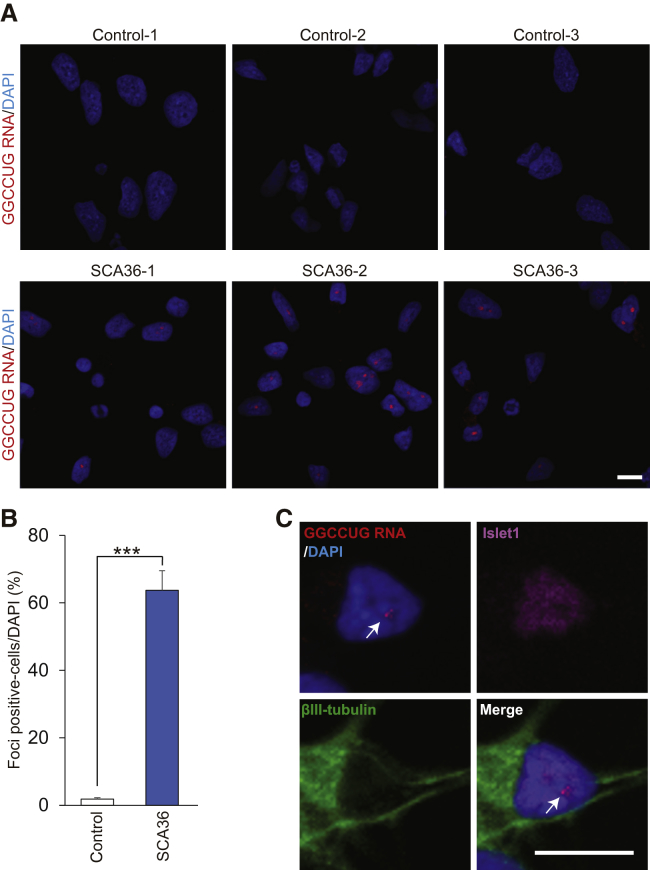
We previously detected intranuclear GGCCUG repeat RNA foci, which are sense RNA foci, in SCA36 patient brain, spinal cord, and lymphoblastoid cells by fluorescence in situ hybridization (FISH).1, 19 We used the same fluorescence-labeled locked nucleic acid oligonucleotide probes targeting the expanded repeat. The number of intranuclear sense RNA-foci-positive cells was significantly increased in SCA36 patient iPSC clones compared to healthy control iPSC clones (Figures 2A and 2B). The number of intranuclear RNA foci-positive cells was also significantly increased in the SCA36 patient iPSNs (Figures 3A and 3B). Co-immunostaining of Islet1 and βІІІ-tubulin with FISH showed that RNA foci existed in SCA36 patient iPSNs (Figure 3C). We also confirmed the specificity of the FISH probes by treating cells with RNase A, the results of which showed that these intranuclear inclusions are comprised of “GGCCUG” RNA and not DNA (Figure S3). In contrast to the sense RNA foci, antisense RNA-foci-positive cells were not detected in either SCA36 patient iPSCs or iPSNs (Figure S4), a result is corresponding to the previous report regarding SCA36 autopsied tissue.19
Figure 2.
RNA Foci Formation in SCA36 Patient iPSCs
Fluorescence in situ hybridization (FISH) analysis was performed for each iPSC clone using a cy3-conjugated C(CAGGCC)2CAG locked nucleic acid oligonucleotide probe. (A) Sense RNA foci were detected in SCA36 patient iPSCs, but not in healthy control iPSCs. Most of the RNA foci were located in nucleus. Scale bar, 10 μm. (B) Graph shows the percentage of RNA-foci-positive cells. ***p < 0.001. Values are expressed as mean ± SEM from n = 3 clones.
Figure 3.
RNA Foci Formation in SCA36 Patient iPSNs
(A) Sense RNA foci were detected in nucleus of SCA36 patient iPSNs by FISH analysis. Scale bar, 10 μm. (B) The RNA-foci-positive cell ratio. ***p < 0.001. Values are expressed as mean ± SEM from n = 3 clones. (C) Representative image showing RNA foci (arrows) and Islet1-positive motor neurons. Scale bar, 10 μm.
ASOs Reduce SCA36 RNA Foci
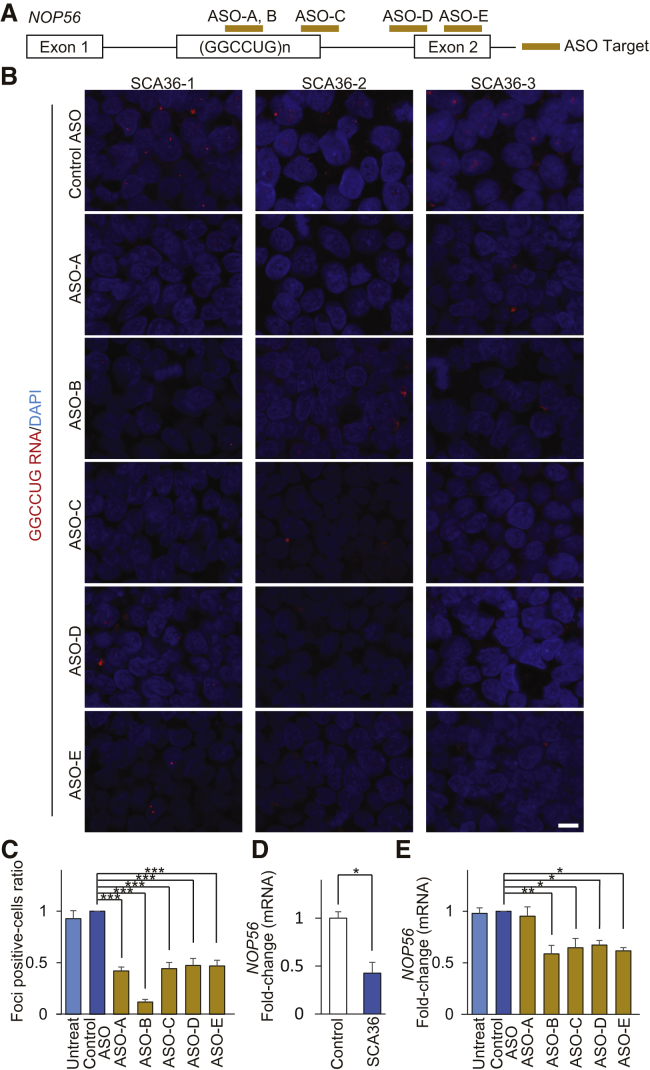
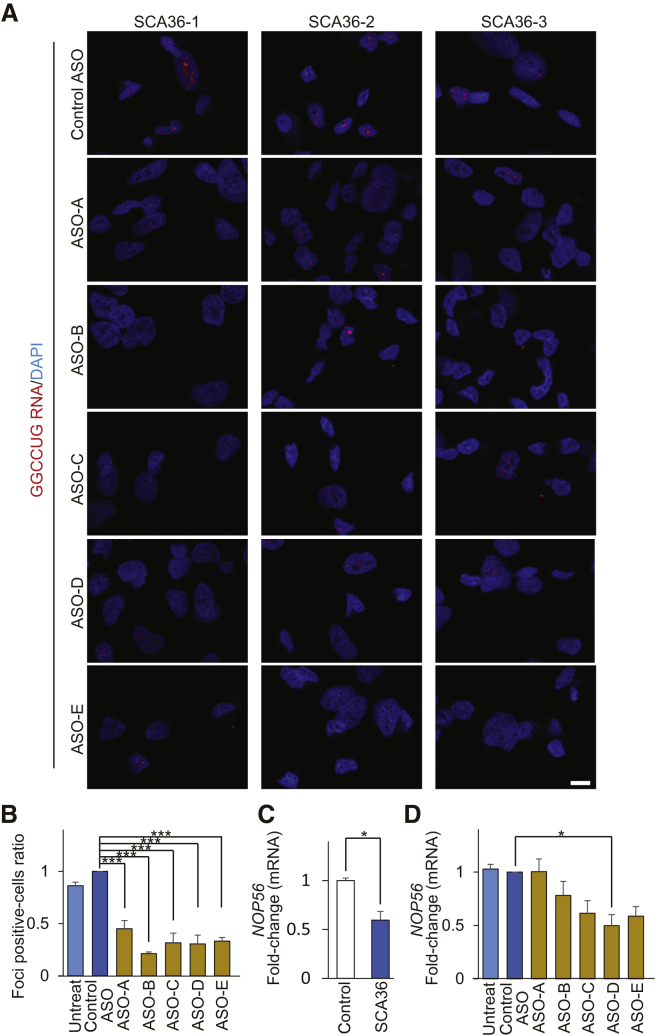
Regarding ASO development, although a large number of sequence candidates could be designed for the target, the effect validation would be difficult in silico. Thus, we tested the efficacy of five ENA ASOs (Figure 4A; Table 2) using SCA36 patient iPSCs and iPSNs to identify effective ASOs. As in previous reports,16 we designed ASO-A for blocking the GGCCUG RNA structure without RNase H-activation12 and ASO-B, C, D, and E for activating RNase H with the ENA/DNA/ENA gapmer structure.10 We validated the relative reduction effect of these ASOs compared to control ASO. All five ENA ASOs significantly reduced the number of cells containing GGCCUG RNA foci to ∼50% in SCA36 patient iPSCs (Figures 4B, 4C, and S5A). Furthermore, NOP56 mRNA expression levels were significantly lower in SCA36 patient iPSCs than in healthy controls (Figure 4D). Then, with our tested ENA ASOs, NOP56 mRNA expression levels in SCA36 patient iPSCs were maintained by ASO-A but were significantly decreased by the other four ASOs (Figures 4E and S5B).
Figure 4.
ASOs Reduce the Number of RNA-Foci-Positive Cells in SCA36 iPSCs
(A) Schema of NOP56 pre-mRNA structure and the ASO target (gold bars). (B) Representative images of RNA FISH analysis in SCA36 iPSCs treated with five kinds of ASOs. Scale bar, 10 μm. (C) The graph shows the number of RNA-foci-positive cells. The mean number of RNA-foci-positive cells in the control ASO treatment group was standardized as 1.0. Compared to the control ASO, all five ENA ASOs significantly reduced the percentage of RNA-foci-positive cells in SCA36 patient iPSCs. ***p < 0.001. Data are presented as mean ± SEM from n = 3 clones. (D) The graph shows the NOP56 mRNA expression levels in healthy control and SCA36 patient iPSCs. The mean NOP56 mRNA expression level of control iPSCs was standardized to be 1.0. NOP56 mRNA expression levels were significantly lower in SCA36 patient iPSCs than in control iPSCs. *p < 0.05. Data are presented as mean ± SEM from n = 3 clones. (E) The graph shows the NOP56 mRNA expression levels in SCA36 patient iPSCs after ASO treatment. The mean NOP56 mRNA expression level of the control ASO treatment group was standardized as 1.0. Compared to the control ASO, NOP56 mRNA expression levels were significantly decreased by ASO-B, ASO-C, ASO-D, and ASO-E but maintained by ASO-A. *p < 0.05, **p < 0.01. Values are expressed as mean ± SEM from n = 3 clones.
Table 2.
ASO Information
| ASO Sequence (5′ to 3′) | Modification | Function | |
|---|---|---|---|
| ASO-A | CCCAGGCCCAGGCCCAGGCC | ENA and 2′-O-methyl RNA | block |
| ASO-B | CCCAGGCCCAGGCCCAGGCC | gapmer (ENA-DNA-ENA) | RNaseH |
| ASO-C | CGCAGGCGCAGGCCCAGGCC | gapmer (ENA-DNA-ENA) | RNaseH |
| ASO-D | GTGCAACAGCACCTGGAAGG | gapmer (ENA-DNA-ENA) | RNaseH |
| ASO-E | CGCGTGCTCAAACAGCACGT | gapmer (ENA-DNA-ENA) | RNaseH |
| Control ASO | CGCATGCGAAGCCTCTGGGCC | gapmer (ENA-DNA-ENA) | five-pair mismatch of ASO-C |
Besides the iPSCs, all five ENA ASOs also significantly reduced the number of cells containing GGCCUG RNA foci to approximately 50% in the SCA36 patient iPSNs (Figures 5A, 5B, and S5C). In addition, NOP56 mRNA expression levels were significantly lower in SCA36 patient iPSNs than in healthy controls (Figure 5C). NOP56 mRNA expression levels in SCA36 patient iPSNs were also maintained by ASO-A but decreased with ASO-D and showed a tendency to decrease with other three ASOs when compared to the control ASO (Figures 5D and S5D). Figure S6 shows NOP56 mRNA expression levels in healthy control iPSCs and iPSNs with the five ENA ASOs.
Figure 5.
ASOs Reduce the Number of RNA-Foci-Positive Cells in SCA36 iPSNs
(A) Representative images of RNA FISH in SCA36 patient iPSNs treated with five ASOs. Scale bar, 10 μm. (B) The graph shows the number of RNA-foci-positive cells. Compared to the control ASO, all five ENA ASOs significantly reduced the number of RNA-foci-positive cells in SCA36. ***p < 0.001. Data are presented as mean ± SEM from n = 3 clones. (C) The graph shows NOP56 mRNA expression levels in iPSNs of healthy controls and SCA36 patients. NOP56 mRNA expression levels were significantly lower in SCA36 patient iPSNs than in healthy control iPSNs. *p < 0.05. Data presented as mean ± SEM from n = 3 clones. (D) The graph shows NOP56 mRNA expression levels in the SCA36 patient iPSNs after ASO treatment. Compared to control ASO, NOP56 mRNA expression levels were decreased by ASO-D. *p < 0.05. Values are expressed as mean ± SEM from n = 3 clones.
Discussion
In this study, we established iPSC clones from three SCA36 patients and showed that their iPSCs and iPSNs both contain intranuclear GGCCUG RNA foci. We designed five ENA ASOs targeting the NOP56 pre-mRNA sequence and tested the efficacy of reducing RNA-foci-positive cells using SCA36 patient iPSCs and iPSNs. Finally, we found that one of the ENA ASOs reduced RNA-foci-positive cells in both SCA36 patient iPSCs and iPSNs without altering NOP56 mRNA expression levels.
RNA foci are characteristic histopathological hallmarks of the pathogenesis of some repeat expansion diseases.21 The expanded repeat type, repeat expression level, and repertoire and abundance of expressed proteins are considered to influence foci size, shape, and colocalization with other proteins. The repeat expansion transcript detected as RNA foci by FISH is known to form microscopic RNA structures and interact with several proteins, inducing cellular abnormality, including apoptosis activation or aberrant alternative splicing.22, 23, 24, 25, 26 Thus, reducing the number of RNA foci is considered to be a target for repeat expansion disease treatment, although there is still a debate as to whether RNA foci are a cause of the pathology or epiphenomena.16, 21, 27, 28
As this therapy has developed, several models have been presented to verify the effects of ASOs.16, 17, 29, 30 Regarding hexanucleotide repeat expansion disease, the C9orf72-related ALS/FTLD, ASOs validated in patient iPSC models16, 17 were also effective in Drosophila31 and mouse27 models. These ASOs were designed to activate RNase-H-mediated C9orf72 RNA degradation or alter repeat RNA conformation by blocking the GGGGCC repeat RNA. As shown in the previous studies of C9ORF72 repeat expansion ALS/FTLD,16, 17 we also designed four kinds of different targeting ENA ASOs with gapmer structure that activate RNase H to the GGCCUG repeat expansion containing pre-mRNA in SCA36 and one ENA ASO that has high affinity to pre-mRNA and blocks the GGCCUG RNA structure without RNase H activation. All five ENA ASOs significantly decreased the number of RNA-foci-positive cells in SCA36 patient iPSCs and iPSNs. Only ASO-A preserved NOP56 mRNA expression levels in both SCA36 patient iPSCs and iPSNs. We speculate that this result is dependent on the respective functions of each ASO. NOP56 functions as a component of small nucleolar RNA proteins32 and is highly conserved from yeast to humans.33 In a Drosophila model, NOP56 has been reported to be required for the growth and proliferation of neuroepithelial stem cells in the optic lobe.34 In addition, NOP56 mRNA expression levels were decreased in spinal motor neurons of ALS model mice in the early phase of the disease.35 Our data also showed that NOP56 mRNA expression was significantly lower in SCA36 patient iPSCs and iPSNs than in healthy controls. Thus, the reduction of NOP56 mRNA expression levels may contribute to the pathogenesis of neurodegenerative disease, and we suggest that ASO-A might be useful for the treatment of SCA36, because it reduced the number of GGCCUG RNA-foci-positive cells without altering NOP56 mRNA expression levels.
Although the ASOs in the current study succeeded in reducing the number of RNA-foci-positive cells in vitro by ∼50%, it is unclear whether this level of effectiveness is sufficient for the clinical treatment of SCA36 patients, and validation of ASO effectiveness in vivo will be needed for clinical applications. Furthermore, several pathologies, such as non-ATG-initiated peptides or loss of function of NOP56, may be involved in SCA36 as well as some repeat expansion diseases.36, 37 Further validation of the effectiveness of ASOs for these pathologies may be required.
Southern blot analysis revealed that SCA36-1 iPSNs had different GGCCTG repeat numbers while the repeat numbers in SCA36-2 or SCA36-3 iPSNs were the same as those observed in iPSCs, which suggests that GGCCTG repeat instability during neuronal differentiation of iPSCs occurs in SCA36-1, but not in SCA36-2 and SCA36-3. In C9orf72-related ALS/FTLD, GGGGCC repeat instability during neuronal differentiation of iPSCs was reported in some clones.17, 20 GGCCTG repeat instability between generations has also been reported,2 and it is considered that the GGCCTG repeat in SCA36 may have similar instability as the GGGGCC repeat in C9orf72-related ALS/FTLD, although the pathomechanisms are as yet unclear.
Overall, we first modeled RNA foci formation in SCA36 using patient iPSCs and finally discovered the ASOs that reduced RNA foci. These data then provided basic information for moving toward clinical applications using ASOs for the treatment of SCA36 and other repeat expansion diseases.
Materials and Methods
Human Subjects
This study was approved by the Ethics Committees of Kyoto University and Tokushima University, and it obtained exempted approval from the institutional review board based on our guidelines. Informed consent was obtained from all participants.
iPSC Generation
Human healthy control38 and SCA36 iPSCs were generated from fibroblasts or PBMCs using episomal vectors for OCT3/4, Sox2, Klf4, L-Myc, Lin28, and dominant-negative p53 or OCT3/4, Sox2, Klf4, L-Myc, Lin28, and shRNA for p53, respectively, as previously reported.39 They were cultured by the feeder-free and xeno-free culture systems with StemFit (Ajinomoto) with penicillin/streptomycin.40
Genotyping
The expansions of the hexanucleotide repeat GGCCTG in intron 1 of NOP56 were examined by a repeat-primed PCR method. The repeat-primed PCR experiment was performed as previously described.1 We used three primers: fluorescent-dye-conjugated forward primer corresponding to the upstream of the repeat region, the first reverse primer consisting of four units of the GGCCTG repeat, and the second reverse primer as an anchor. PCR was performed with TaKaRa LA Taq with GC buffer (Takara Bio). The sequence of the forward primer was 5′-TTTCGGCCTGCGTTCGGG-3′, that of the first reverse primer was 5′-TACGCATCCCAGTTTGAGACGCAGGCCCAGGCCCAGGCCCAGGCC-3′, and that of the second reverse primer was 5′-TACGCATCCCAGTTTGAGACG-3′.
Southern Blot
The size of expanded GGCCTG repeats in our model was estimated by Southern blot analysis. Southern blotting was performed as previously described, with minor modifications.1 Briefly, genome DNA (10 μg) was digested overnight with 100 U of AvrІІ (New England Biolabs). Digestion products were separated by electrophoresis on a 0.7% agarose gel, depurinated with 0.25 M HCl, denaturized with 1 M NaOH, and transferred with 20× saline sodium citrate buffer (SSC) onto a positively charged nylon membrane (Roche Applied Science). DNA was immobilized on the membrane by UV cross-linking. A 452-bp probe, covering exon 4 of NOP56, was synthesized from genomic DNA by PCR using the PCR DIG Probe Synthesis Kit (Roche Applied Science) and primers. Forward primer was 5′-TTTAAGAGCTTCCAAGGCTGA-3′ and reverse primer was 5′-AGTGCCCACAAGGAAACGTTA-3′. The probe was labeled with digoxigenin. The labeled probe was denatured at 95°C for 5 min and added to the DIG Easy Hyb (Roche Applied Science). Hybridization with labeled probe was carried out overnight at 46.8°C. After the recommended treatment with DIG Wash and Block Buffer Set (Roche Applied Science), the digoxigenin-labeled probe was detected with anti-digoxigenin antibody and CDP-Star reagent using DIG Luminescent Detection Kit (Roche Applied Science).
RNA-Seq Analysis
Total RNA was extracted using the RNeasy Mini Kit (QIAGEN), and the sequencing libraries were prepared from 100 ng RNA using the TruSeq Stranded mRNA LT Sample Prep Kit (Illumina) following the manufacturer’s instructions. The resulting libraries were sequenced (75 bp single end) on a NextSeq 500 system (Illumina). The trimming adaptor sequences and low-quality bases were performed by cutadapt-1.8.1.41 Then, the sequenced reads were mapped to the human reference genome (hg38) by tophat-2.1.0 with the aligner Bowtie2-2.2.5.42, 43 The number of reads mapped to each gene was calculated by HTSeq-0.6.1 software44 and then normalized with the DESeq2 R/Bioconductor package (v. 1.12.3).45 Hierarchical clustering analysis and principal-component analysis (PCA) were performed by R software version 3.3.1.
Neuronal Differentiation from iPSCs
Neural cells containing motor neurons were differentiated from human iPSCs as previously described.14, 18 Briefly, iPSCs were dissociated to single cells and quickly reaggregated in low-cell-adhesion U-shaped 96-well plates (Lipidule-Coated Plate A-U96, NOF Corporation). Aggregations were cultured in Dulbecco’s modified Eagle’s medium/Ham’s F12 (Thermo Fisher Scientific) containing 5% KSR (Invitrogen), minimum essential medium-nonessential amino acids (Invitrogen), L-glutamine (Sigma-Aldrich), 2-mercaptoethanol (Wako Pure Chemicals Industries), 2 μM dorsomorphin (Sigma-Aldrich), 10 μM SB431542 (Cayman), 3 μM CHIR99021 (Cayman), and 12.5 ng/mL fibroblast growth factor (Wako Pure Chemicals Industries) in a neural inductive stage for 11 days. 100 nM retinoic acid (Sigma-Aldrich) and 500 nM Smoothened ligand (Enzo Life Sciences) were added on day 4. After patterning with neurobasal medium (Thermo Fisher Scientific) supplemented with B27 Supplement (Thermo Fisher Scientific), 100 nM retinoic acid, 500 nM Smoothened ligand, and 10 μM DAPT (Selleck), the aggregates were separated by Accumax (Innovative Cell Technologies), dissociated into single cells, and adhered to Matrigel-coated (BD Biosciences) dishes on day 16. Adhesive cells were cultured in neurobasal medium with 10 ng/mL brain-derived neurotrophic factor (R&D Systems), 10 ng/mL glial cell line-derived neurotrophic factor (R&D Systems), and 10 ng/mL neurotrophin-3 (R&D Systems) for 8 days.
Immunocytochemistry
For immunostaining of iPSCs and iPSNs, cells were fixed in 4% paraformaldehyde (pH 7.4) for 30 min at room temperature and rinsed with PBS. The cells were permeabilized in PBS containing 0.2% Triton X-100 for 10 min at room temperature, followed by rinsing with PBS. After blocking with 5% bovine serum albumin for 60 min at room temperature, cells were incubated with primary antibodies overnight at 4°C. The following primary antibodies were used: Nanog (1:500; REPROCELL), SSEA-4 (1:1,000; EMD Millipore), βІІІ-tubulin (1:2,000; Cell Signaling Technology), Islet1 (1:200; Developmental Studies Hybridoma Bank), and SMI32 (1:2,500; Covance). After three rinses with PBS, cells were incubated with appropriate Alexa-Fluor-conjugated secondary antibodies for 1 hr at room temperature, and DAPI (Life Technologies) was used to label nuclei. Cell images were acquired with the IN CELL Analyzer 6000 (GE Healthcare). The number of cells was automatically quantified with IN CELL Analyzer 6000 and IN CELL Developer toolbox software 1.92 (GE Healthcare).
qRT-PCR
Total RNA was isolated using the miRNeasy Mini Kit (QIAGEN), and RNA were reverse transcribed to cDNA using the ReverTra Ace α kit (TOYOBO). qPCR was performed with SYBR Green Master Mix (Applied Biosystems) using the ΔΔCT method. The sequence of the NOP56 forward primer was 5′-AAATTCCACAGCATCGTTCGT-3′ and that of the reverse primer was 5′-TGTATTGCGGCACCAATCTTG-3′. Ct values for each sample were normalized with GAPDH. The sequence of the GAPDH forward primer was 5′-TCCACTGGCGTCTTCACC-3′ and that of the reverse primer was 5′-GGCAGAGATGATGACCCTTTT-3′.
Fluorescence In Situ Hybridization
All solutions were made with diethylpyrocarbonate (DEPC)-treated water. In order to detect RNA foci in cells, FISH was performed using two kinds of 5′ fluorescein (Cy3) labeled locked nucleic acid oligonucleotide probes by Gene Design. One was C(CAGGCC)2CAG for detecting sense RNA foci and the other was CUG(GGCCUG)2G for detecting antisense RNA foci. The cells were fixed in 4% paraformaldehyde for 10 min, permeabilized in 70% ethanol on ice, equilibrated in 50% formamide/2× SSC for 30 min at 66°C, and hybridized for 3 hr at 52°C with denatured locked nucleic acid probe (40 nM), which were previously heated at 80°C for 10 min, in hybridization buffer consisting of 50% formamide, 2× SSC, and 10% dextran sulfate. The cells were washed twice with 50% formamide/2× SSC for 20 min at 52°C and three times with 1× SSC at room temperature. Finally, incubation with DAPI was performed, and the slides were mounted with ProLong Gold antifade reagent. For RNase treatment, after fixation, cells were incubated with 50 μg/mL DNase-free RNase A (Roche Applied Science) for 1.5 hr at 37°C.
In order to combine FISH with immunofluorescence staining, after washing with 1× SSC, the sections were blocked with blocking buffer (5% bovine serum albumin and 0.2% Triton X-100) for 20 min at room temperature. They were then incubated overnight at 4°C with primary antibodies in the blocking buffer. The following primary antibodies were used: rabbit βІІІ-tubulin (1:2,000; Cell Signaling Technology) and mouse Islet1 (1:200; Developmental Studies Hybridoma Bank). After three rinses with PBS, cells were incubated with the appropriate Alexa-Fluor-conjugated secondary antibodies for 1 hr at room temperature. After three rinses with PBS, the sections were counterstained with DAPI and mounted with ProLong Gold antifade reagent. Cell images were acquired with LSM710 microscope (Carl Zeiss) and IN CELL Analyzer 6000. In the analysis, the Cy3 signal with a diameter over 0.60 μm was captured as RNA foci, and the number of RNA-foci-positive cells was automatically quantified with IN CELL Developer toolbox software 1.92.
ASO Treatment
We designed five ENA ASOs, and the target sequences are listed in Table 2. The function of ASO-A was blocking, while that of the other four ASOs was activating RNase H by gapmer structure. ASO-A and ASO-B targeted the same genome sequence, the GGCCUG repeat. ASO-C targeted the GGCCUG repeat and the intron 1 overlapping genome sequence. ASO-D targeted intron 1 and the exon 2 overlapping genome sequence. ASO-E targeted exon 2. Control ASO was designed as a five-pair mismatch of ASO-C. All ENA ASO was purchased from KNC Laboratories. ASO was added at a final concentration of 400 nM, which was introduced to the iPSCs with Lipofectamine LTX Reagent (Thermo Fisher Scientific) and to iPSNs by electroporation using Amaxa 4D-Nucleofector (Lonza). Before the assay, iPSCs were co-cultured with the ASO for 48 hr, and iPSNs were subsequently cultured for 48 hr after ASO treatment.
Statistical Analysis
Values are expressed as mean ± SEM. Significant differences among multiple groups were determined by one-way analysis of variance (ANOVA) followed by Tukey’s post hoc test or Student’s t test. Differences were considered significant when p < 0.05, and calculations were performed with the statistical software package SPSS version 21.0 for Windows (IBM SPSS Statistics).
Author Contributions
H.I. conceived the project. K.M., K.I., and H.I. designed the experiment. K.M., K.T., K.I., and N.M. performed the experiments. K.M., K.I., T. Yamamoto, and H.I. analyzed the data. Y.I., R.K., Y.O., T. Yamashita, and K.A. provided patient samples and information. K.M., K.I., and H.I. wrote the manuscript.
Conflicts of Interest
The authors declare that they have no competing interests.
Acknowledgments
We would like to express our sincere gratitude to all our coworkers and collaborators; Takako Enami, Misato Funayama, Ran Shibukawa, Kasumi Nakao, and Mitsuyo Kawada for their technical support; and Noriko Endo and Ruri Taniguchi for their administrative support. This research was funded in part by a grant for the Program for Intractable Diseases Research, which utilizes disease-specific iPSCs from Japan Agency for Medical Research and Development (AMED), a grant from AMED for the Core Center for iPS Cell Research of Research Center Network for Realization of Regenerative Medicine, the Research Project for Practical Applications of Regenerative Medicine from AMED, and The Daiichi Sankyo Foundation of Life Science (all to H.I.).
Footnotes
Supplemental Information includes six figures and can be found with this article online at http://dx.doi.org/10.1016/j.omtn.2017.06.017.
Supplemental Information
References
- 1.Kobayashi H., Abe K., Matsuura T., Ikeda Y., Hitomi T., Akechi Y., Habu T., Liu W., Okuda H., Koizumi A. Expansion of intronic GGCCTG hexanucleotide repeat in NOP56 causes SCA36, a type of spinocerebellar ataxia accompanied by motor neuron involvement. Am. J. Hum. Genet. 2011;89:121–130. doi: 10.1016/j.ajhg.2011.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.García-Murias M., Quintáns B., Arias M., Seixas A.I., Cacheiro P., Tarrío R., Pardo J., Millán M.J., Arias-Rivas S., Blanco-Arias P. ‘Costa da Morte’ ataxia is spinocerebellar ataxia 36: clinical and genetic characterization. Brain. 2012;135:1423–1435. doi: 10.1093/brain/aws069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Obayashi M., Stevanin G., Synofzik M., Monin M.L., Duyckaerts C., Sato N., Streichenberger N., Vighetto A., Desestret V., Tesson C. Spinocerebellar ataxia type 36 exists in diverse populations and can be caused by a short hexanucleotide GGCCTG repeat expansion. J. Neurol. Neurosurg. Psychiatry. 2015;86:986–995. doi: 10.1136/jnnp-2014-309153. [DOI] [PubMed] [Google Scholar]
- 4.Zeng S., Zeng J., He M., Zeng X., Zhou Y., Liu Z., Xia K., Pan Q., Jiang H., Shen L. Genetic and clinical analysis of spinocerebellar ataxia type 36 in Mainland China. Clin. Genet. 2016;90:141–148. doi: 10.1111/cge.12706. [DOI] [PubMed] [Google Scholar]
- 5.Lee Y.C., Tsai P.C., Guo Y.C., Hsiao C.T., Liu G.T., Liao Y.C., Soong B.W. Spinocerebellar ataxia type 36 in the Han Chinese. Neurol. Genet. 2016;2:e68. doi: 10.1212/NXG.0000000000000068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ikeda Y., Ohta Y., Kobayashi H., Okamoto M., Takamatsu K., Ota T., Manabe Y., Okamoto K., Koizumi A., Abe K. Clinical features of SCA36: a novel spinocerebellar ataxia with motor neuron involvement (Asidan) Neurology. 2012;79:333–341. doi: 10.1212/WNL.0b013e318260436f. [DOI] [PubMed] [Google Scholar]
- 7.Zellweger T., Miyake H., Cooper S., Chi K., Conklin B.S., Monia B.P., Gleave M.E. Antitumor activity of antisense clusterin oligonucleotides is improved in vitro and in vivo by incorporation of 2′-O-(2-methoxy)ethyl chemistry. J. Pharmacol. Exp. Ther. 2001;298:934–940. [PubMed] [Google Scholar]
- 8.Moschos S.A., Frick M., Taylor B., Turnpenny P., Graves H., Spink K.G., Brady K., Lamb D., Collins D., Rockel T.D. Uptake, efficacy, and systemic distribution of naked, inhaled short interfering RNA (siRNA) and locked nucleic acid (LNA) antisense. Mol. Ther. 2011;19:2163–2168. doi: 10.1038/mt.2011.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Surono A., Van Khanh T., Takeshima Y., Wada H., Yagi M., Takagi M., Koizumi M., Matsuo M. Chimeric RNA/ethylene-bridged nucleic acids promote dystrophin expression in myocytes of duchenne muscular dystrophy by inducing skipping of the nonsense mutation-encoding exon. Hum. Gene Ther. 2004;15:749–757. doi: 10.1089/1043034041648444. [DOI] [PubMed] [Google Scholar]
- 10.Chan J.H.P., Lim S., Wong W.S.F. Antisense oligonucleotides: from design to therapeutic application. Clin. Exp. Pharmacol. Physiol. 2006;33:533–540. doi: 10.1111/j.1440-1681.2006.04403.x. [DOI] [PubMed] [Google Scholar]
- 11.Morita K., Hasegawa C., Kaneko M., Tsutsumi S., Sone J., Ishikawa T., Imanishi T., Koizumi M. 2′-O,4′-C-ethylene-bridged nucleic acids (ENA) with nuclease-resistance and high affinity for RNA. Nucleic Acids Res. Suppl. 2001;2001:241–242. doi: 10.1093/nass/1.1.241. [DOI] [PubMed] [Google Scholar]
- 12.Morita K., Hasegawa C., Kaneko M., Tsutsumi S., Sone J., Ishikawa T., Imanishi T., Koizumi M. 2′-O,4′-C-ethylene-bridged nucleic acids (ENA): highly nuclease-resistant and thermodynamically stable oligonucleotides for antisense drug. Bioorg. Med. Chem. Lett. 2002;12:73–76. doi: 10.1016/s0960-894x(01)00683-7. [DOI] [PubMed] [Google Scholar]
- 13.Morita K., Takagi M., Hasegawa C., Kaneko M., Tsutsumi S., Sone J., Ishikawa T., Imanishi T., Koizumi M. Synthesis and properties of 2′-O,4′-C-ethylene-bridged nucleic acids (ENA) as effective antisense oligonucleotides. Bioorg. Med. Chem. 2003;11:2211–2226. doi: 10.1016/s0968-0896(03)00115-9. [DOI] [PubMed] [Google Scholar]
- 14.Egawa N., Kitaoka S., Tsukita K., Naitoh M., Takahashi K., Yamamoto T., Adachi F., Kondo T., Okita K., Asaka I. Drug screening for ALS using patient-specific induced pluripotent stem cells. Sci. Transl. Med. 2012;4:145ra104. doi: 10.1126/scitranslmed.3004052. [DOI] [PubMed] [Google Scholar]
- 15.Burkhardt M.F., Martinez F.J., Wright S., Ramos C., Volfson D., Mason M., Garnes J., Dang V., Lievers J., Shoukat-Mumtaz U. A cellular model for sporadic ALS using patient-derived induced pluripotent stem cells. Mol. Cell. Neurosci. 2013;56:355–364. doi: 10.1016/j.mcn.2013.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Donnelly C.J., Zhang P.W., Pham J.T., Haeusler A.R., Mistry N.A., Vidensky S., Daley E.L., Poth E.M., Hoover B., Fines D.M. RNA toxicity from the ALS/FTD C9ORF72 expansion is mitigated by antisense intervention. Neuron. 2013;80:415–428. doi: 10.1016/j.neuron.2013.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sareen D., O’Rourke J.G., Meera P., Muhammad A.K., Grant S., Simpkinson M., Bell S., Carmona S., Ornelas L., Sahabian A. Targeting RNA foci in iPSC-derived motor neurons from ALS patients with a C9ORF72 repeat expansion. Sci. Transl. Med. 2013;5:208ra149. doi: 10.1126/scitranslmed.3007529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maury Y., Côme J., Piskorowski R.A., Salah-Mohellibi N., Chevaleyre V., Peschanski M., Martinat C., Nedelec S. Combinatorial analysis of developmental cues efficiently converts human pluripotent stem cells into multiple neuronal subtypes. Nat. Biotechnol. 2015;33:89–96. doi: 10.1038/nbt.3049. [DOI] [PubMed] [Google Scholar]
- 19.Liu W., Ikeda Y., Hishikawa N., Yamashita T., Deguchi K., Abe K. Characteristic RNA foci of the abnormal hexanucleotide GGCCUG repeat expansion in spinocerebellar ataxia type 36 (Asidan) Eur. J. Neurol. 2014;21:1377–1386. doi: 10.1111/ene.12491. [DOI] [PubMed] [Google Scholar]
- 20.Almeida S., Gascon E., Tran H., Chou H.J., Gendron T.F., Degroot S., Tapper A.R., Sellier C., Charlet-Berguerand N., Karydas A. Modeling key pathological features of frontotemporal dementia with C9ORF72 repeat expansion in iPSC-derived human neurons. Acta Neuropathol. 2013;126:385–399. doi: 10.1007/s00401-013-1149-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wojciechowska M., Krzyzosiak W.J. Cellular toxicity of expanded RNA repeats: focus on RNA foci. Hum. Mol. Genet. 2011;20:3811–3821. doi: 10.1093/hmg/ddr299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Conlon E.G., Lu L., Sharma A., Yamazaki T., Tang T., Shneider N.A., Manley J.L. The C9ORF72 GGGGCC expansion forms RNA G-quadruplex inclusions and sequesters hnRNP H to disrupt splicing in ALS brains. eLife. 2016;5:e17820. doi: 10.7554/eLife.17820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Haeusler A.R., Donnelly C.J., Periz G., Simko E.A., Shaw P.G., Kim M.S., Maragakis N.J., Troncoso J.C., Pandey A., Sattler R. C9orf72 nucleotide repeat structures initiate molecular cascades of disease. Nature. 2014;507:195–200. doi: 10.1038/nature13124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Paul S., Dansithong W., Jog S.P., Holt I., Mittal S., Brook J.D., Morris G.E., Comai L., Reddy S. Expanded CUG repeats Dysregulate RNA splicing by altering the stoichiometry of the muscleblind 1 complex. J. Biol. Chem. 2011;286:38427–38438. doi: 10.1074/jbc.M111.255224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.White M.C., Gao R., Xu W., Mandal S.M., Lim J.G., Hazra T.K., Wakamiya M., Edwards S.F., Raskin S., Teive H.A. Inactivation of hnRNP K by expanded intronic AUUCU repeat induces apoptosis via translocation of PKCdelta to mitochondria in spinocerebellar ataxia 10. PLoS Genet. 2010;6:e1000984. doi: 10.1371/journal.pgen.1000984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wheeler T.M., Lueck J.D., Swanson M.S., Dirksen R.T., Thornton C.A. Correction of ClC-1 splicing eliminates chloride channelopathy and myotonia in mouse models of myotonic dystrophy. J. Clin. Invest. 2007;117:3952–3957. doi: 10.1172/JCI33355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jiang J., Zhu Q., Gendron T.F., Saberi S., McAlonis-Downes M., Seelman A., Stauffer J.E., Jafar-Nejad P., Drenner K., Schulte D. Gain of toxicity from ALS/FTD-linked repeat expansions in C9ORF72 is alleviated by antisense oligonucleotides targeting GGGGCC-containing RNAs. Neuron. 2016;90:535–550. doi: 10.1016/j.neuron.2016.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lagier-Tourenne C., Baughn M., Rigo F., Sun S., Liu P., Li H.R., Jiang J., Watt A.T., Chun S., Katz M. Targeted degradation of sense and antisense C9orf72 RNA foci as therapy for ALS and frontotemporal degeneration. Proc. Natl. Acad. Sci. USA. 2013;110:E4530–E4539. doi: 10.1073/pnas.1318835110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nizzardo M., Simone C., Salani S., Ruepp M.D., Rizzo F., Ruggieri M., Zanetta C., Brajkovic S., Moulton H.M., Müehlemann O. Effect of combined systemic and local morpholino treatment on the spinal muscular atrophy Δ7 mouse model phenotype. Clin. Ther. 2014;36 doi: 10.1016/j.clinthera.2014.02.004. 340–56.e5. [DOI] [PubMed] [Google Scholar]
- 30.Wheeler T.M., Leger A.J., Pandey S.K., MacLeod A.R., Nakamori M., Cheng S.H., Wentworth B.M., Bennett C.F., Thornton C.A. Targeting nuclear RNA for in vivo correction of myotonic dystrophy. Nature. 2012;488:111–115. doi: 10.1038/nature11362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang K., Donnelly C.J., Haeusler A.R., Grima J.C., Machamer J.B., Steinwald P., Daley E.L., Miller S.J., Cunningham K.M., Vidensky S. The C9orf72 repeat expansion disrupts nucleocytoplasmic transport. Nature. 2015;525:56–61. doi: 10.1038/nature14973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tran E., Brown J., Maxwell E.S. Evolutionary origins of the RNA-guided nucleotide-modification complexes: from the primitive translation apparatus? Trends Biochem. Sci. 2004;29:343–350. doi: 10.1016/j.tibs.2004.05.001. [DOI] [PubMed] [Google Scholar]
- 33.Gautier T., Bergès T., Tollervey D., Hurt E. Nucleolar KKE/D repeat proteins Nop56p and Nop58p interact with Nop1p and are required for ribosome biogenesis. Mol. Cell. Biol. 1997;17:7088–7098. doi: 10.1128/mcb.17.12.7088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang H., Chen X., He T., Zhou Y., Luo H. Evidence for tissue-specific Jak/STAT target genes in Drosophila optic lobe development. Genetics. 2013;195:1291–1306. doi: 10.1534/genetics.113.155945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miyazaki K., Yamashita T., Morimoto N., Sato K., Mimoto T., Kurata T., Ikeda Y., Abe K. Early and selective reduction of NOP56 (Asidan) and RNA processing proteins in the motor neuron of ALS model mice. Neurol. Res. 2013;35:744–754. doi: 10.1179/1743132813Y.0000000196. [DOI] [PubMed] [Google Scholar]
- 36.Gendron T.F., Cosio D.M., Petrucelli L. c9RAN translation: a potential therapeutic target for the treatment of amyotrophic lateral sclerosis and frontotemporal dementia. Expert Opin. Ther. Targets. 2013;17:991–995. doi: 10.1517/14728222.2013.818659. [DOI] [PubMed] [Google Scholar]
- 37.Richter J.D., Bassell G.J., Klann E. Dysregulation and restoration of translational homeostasis in fragile X syndrome. Nat. Rev. Neurosci. 2015;16:595–605. doi: 10.1038/nrn4001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mishima T., Ishikawa T., Imamura K., Kondo T., Koshiba Y., Takahashi R., Takahashi J., Watanabe A., Fujii N., Tsuboi Y., Inoue H. Cytoplasmic aggregates of dynactin in iPSC-derived tyrosine hydroxylase-positive neurons from a patient with Perry syndrome. Parkinsonism Relat. Disord. 2016;30:67–72. doi: 10.1016/j.parkreldis.2016.06.007. [DOI] [PubMed] [Google Scholar]
- 39.Okita K., Yamakawa T., Matsumura Y., Sato Y., Amano N., Watanabe A., Goshima N., Yamanaka S. An efficient nonviral method to generate integration-free human-induced pluripotent stem cells from cord blood and peripheral blood cells. Stem Cells. 2013;31:458–466. doi: 10.1002/stem.1293. [DOI] [PubMed] [Google Scholar]
- 40.Nakagawa M., Taniguchi Y., Senda S., Takizawa N., Ichisaka T., Asano K., Morizane A., Doi D., Takahashi J., Nishizawa M. A novel efficient feeder-free culture system for the derivation of human induced pluripotent stem cells. Sci. Rep. 2014;4:3594. doi: 10.1038/srep03594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal. 2011;17:10–12. [Google Scholar]
- 42.Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R., Salzberg S.L. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Anders S., Pyl P.T., Huber W. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.