Abstract
Residual DNA methylation in the gastric mucosa after Helicobacter pylori (H. pylori) eradication may have a role in gastric carcinogenesis. We examined the association between morphologic features and promoter methylation status of non‐neoplastic gastric mucosa especially after H. pylori eradication. A total of 140 gastric specimens from 99 participants who had at least 6 months of post‐eradication period were examined. The magnifying narrow‐band imaging (NBI) endoscopic feature of gastric mucosa was divided into two types: restored‐small, round pits, accompanied with honeycomb‐like subepithelial capillary networks; atrophic‐well‐demarcated oval or tubulovillous pits with clearly visible coiled or wavy vessels. Methylation status of five candidate genes (MYOD1, SLC16A12, IGF2, RORA, and PRDM5) were examined by bisulfite pyrosequencing. The atrophic type, informative endoscopic features of intestinal metaplasia, demonstrated higher methylation levels in all five genes compared to the restored type (all P < 0.0001). In the restored type, methylation levels were significantly lower among the samples with longer post‐eradication period (for all genes, P < 0.0001), which was not observed in atrophic type (for all genes, P > 0.1). Multivariate analysis demonstrated that atrophic type or presence of intestinal held an independent factor for hyper methylation (odds ratio: 24.69, 95% confidence interval: 6.95–87.76, P < 0.0001). The atrophic type by the magnifying NBI and presence of intestinal metaplasia are the morphologic characteristics of residual DNA methylation of after H. pylori eradication, regardless of the post‐eradication period and it might be considered as the epigenetic irreversible point with H. pylori eradication.
Keywords: DNA methylation, gastric mucosa, H. pylori eradication, narrow‐band imaging endoscopy
Introduction
Promoter CpG island methylation and subsequent transcriptional repression are important mechanisms in many types of cancers, while the aberrant methylation is also observed in non‐neoplastic tissues with aging and chronic inflammation 1, 2. Helicobacter pylori (H. pylori) infection plays an important role in gastric cancer development 3, 4, 5. H. pylori‐infected gastric mucosa is characterized as chronic inflammation and atrophy 5, leading to epigenetic changes characterized by the promoter methylation of multiple genes 6, 7. This phenomenon can be explained by the concept of an “epigenetic‐field‐defect” which is linked to gastric cancer predisposition.
Several studies have reported that H. pylori eradication prevents gastric cancer 8, 9, which might be attributed to the improvement of chronic inflammation or atrophy 10. The data also demonstrated that the therapy could reverse the hypermethylation in certain genes 11, 12. Even after successful H. pylori eradication, on the other hand, gastric cancers are sometimes identified 13, 14. The residual methylation in gastric mucosa after eradication might be relevant to this process 12, 15.
It is well known that the pathologic state of gastritis, such as severe gastric mucosal atrophy or intestinal metaplasia, is closely associated with the risk of gastric cancer among H. pylori‐infected patients 5. Recent studies have also suggested that the severe gastric atrophy after successful eradication is associated with increased risk of gastric cancer especially in gastric fundic area 16. We have reported that the morphologic appearances of gastric mucosa seen by the magnifying narrow‐band imaging (NBI) endoscopy are clearly distributed into atrophic or non‐atrophic mucosa, and correlating with gastric cancer occurrence after H. pylori eradication (Tahara et al., submitting). Various genotoxic and epigenetic changes in gastric mucosa which reflect past exposure of H. pylori infection might associate with the risk of gastric cancer after eradication. Morphologic identification of “field defect” has important implications for preventing gastric cancer. We examined the promoter methylation levels of candidate genes in non‐neoplastic gastric mucosa among patients with various post‐eradicated periods. We investigated the association among methylation status, magnifying NBI and histologic features.
Materials and Methods
Study population
Study participants were prospectively enrolled from patients attending the endoscopy Center of Fujita Health University from January 2013 to March 2016. A total of 99 participants were invited and all agreed to participate. These 99 participants had a history of successful H. pylori eradication therapy for the various reasons; gastric or duodenal ulcer, chronic gastritis or after endoscopic resection (ER) of early gastric cancer. For H. pylori eradication, triple therapy using one of following three regimens were used for all the participants. (1) 10 mg of rabeprazole sodium b.d., 200 mg of clarithromycin b.d., and 750 mg of amoxicillin b.d., (2) 30 mg of lansoprazole b.d., 200 mg of clarithromycin b.d., and 750 mg of amoxicillin b.d. or (3) 30 mg of lansoprazole b.d., 250 mg of metronidazole b.d., and 750 mg of amoxicillin b.d. For all the participants, success of H. pylori eradication was confirmed based on the C‐urea breath test at least 12 weeks after the triple therapy. Median period after eradication therapy was 41 months (ranging from 6 to 330 months). In all, 34 participants visited to our hospital for the treatment of early gastric cancer diagnosed after H. pylori eradication and seven of these cases were metachronous gastric cancers, diagnosed after ER and subsequent H. pylori eradication. A total of 23 healthy participants without history of H. pylori infection were also enrolled from patients who underwent upper gastroscopy for yearly checkup examination or for the complaints of abdominal discomfort (median age: 53 years, female/male, 11/12). Fujita Health University of Medicine approved the protocol, and written informed consent was obtained from all participants.
Endoscopic procedure and sample collection
All participants underwent esophagogastroduodenoscopy (EGD) using a magnifying video endoscope (Olympus GIF‐H260Z and a CV260SL/CV290SL endoscopic system (Olympus Medical Systems). For the evaluation of endoscopic feature of gastric mucosa, the non‐pathologic mucosa of the gastric body was carefully evaluated with complete magnification coupled with a NBI light source.
The endoscopic feature of gastric mucosal morphology after H. pylori eradication has recently been reported (Tahara et al., submitting). The magnifying NBI patterns of gastric body can be divided into the following two types: restored and atrophic types (Fig. 1). The restored type is characterized as small, round pits, accompanied with honeycomb‐like subepithelial capillary networks, which resemble to healthy gastric body without history of H. pylori infection (Tahara et al., submitting). The atrophic type is characterized as well‐demarcated oval or tubulovillous pits with clearly visible coiled or wavy vessels, which is tightly linked to the presence of histologic intestinal metaplasia (Tahara et al., submitting). The classification of mucosal patterns among each case was based on the most predominant magnifying NBI pattern. The most predominant magnifying NBI pattern was taken as endoscopic pictures, and we obtained target biopsy specimens from that site. In case there were both restored and atrophic types in the same patient, endoscopic pictures and target biopsy specimens were obtained from both sites. At least, two biopsy specimens were obtained from targeted sites. We use one biopsy specimen to investigate the presence of intestinal metaplasia by the histologic analysis. The other biopsy was immediately frozen and stored at −80°C for the DNA methylation analysis. If there were obvious lesions, such as polyps, erosions, or cancers were seen, NBI scanning and target biopsy were performed getting enough distance from these pathologic lesions to avoid their influences. Endoscopic observations, classification of NBI patterns, and targeted biopsies were all performed by one expert endoscopist (T. T.). The interobserver concordance using representative endoscopic pictures from all participants demonstrated good κ coefficient values greater than 0.80 for both restored and atrophic patterns across an additional three expert endoscopists (M. O. N. H., and T. S., Tahara et al., submitting). Among 99 patients who have history of H. pylori eradication, one patient was taken five specimens, 30 patients were taken two specimens and 62 patients were taken one specimen for the DNA methylation analysis, respectively. One patient, who had taken annual follow‐up examination third times, was taken three specimens totally; two patients, who had taken annual follow‐up examination twice, was taken two specimens totally. One specimen was taken from each examination for the remaining patients. Thus, 140 biopsy specimens were obtained in total from 99 participants after H. pylori eradication. For the 23 healthy participants without history of H. pylori infection, one specimen was taken from each examination from greater curvature of uninvolved gastric corpus. Together, 163 biopsy specimens were stored for the molecular study.
Figure 1.
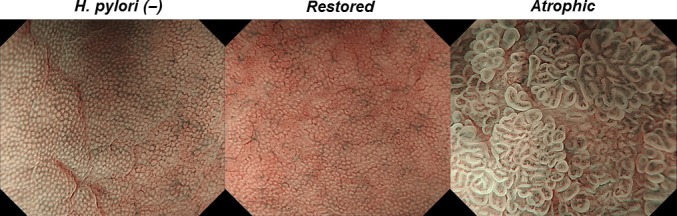
Magnifying NBI patterns of gastric body. Left, healthy gastric body without history of Helicobacter pylori (H. pylori) infection; Uniform small, round pits, accompanied with regular honeycomb‐like subepithelial capillary networks are seen. Center, restored type; small, round pits, accompanied with honeycomb‐like subepithelial capillary networks are also shown. Right, atrophic type; well‐demarcated oval or tubulovillous pits with clearly visible coiled or wavy vessels are seen.
DNA methylation analysis by bisulfite pyrosequencing
DNA was extracted using the standard protein precipitation method. Bisulfite pyrosequencing was used to quantify the promoter methylation of five genes (MYOD1, SLC16A12, IGF2, RORA, and PRDM5). Their selection was based on the frequency of methylation in gastric cancer (RORA and PRDM5) 17 or H. pylori‐infected gastric mucosa (MYOD1, SLC16A12, and IGF2) 18. Methylation of all these genes were also well correlated with magnifying NBI features of H. pylori‐infected patients (Tahara et al., submitting).
Bisulfite‐treated genomic DNA was used to evaluate the methylation status by bisulfite pyrosequencing. Bisulfite treatment of DNA was carried out using an EZ DNA Methylation Kit (Zymo Research) according to the manufacturer's protocol. Pyrosequencing was carried out using a PSQ24 system with Pyro‐Gold reagent Kit (QIAGEN, Tokyo, Japan), and the results were analyzed using PyroMark Q24 software (QIAGEN). The primers used for pyrosequencing are listed in Table S1. To evaluate the quality of absolute methylation quantification of each assay, we used methylation positive and negative control DNAs. CpG Methylase (Ipswich, MA) treated genomic DNA was used as the methylation positive control and the genomic DNA amplified using GenomePlex® Complete Whole Genome Amplification Kit (Tokyo, Japan) was used as the methylation negative control.
Data analysis
Continuous variables between un‐matched two groups were assessed using the student's t‐test. Continuous variables between matched two groups were assessed using the Wilcoxon Signed Rank test. The correlation of continuous variables between two groups was assessed using the Spearman correlation analysis. An unsupervised hierarchical clustering analysis was used to identify distinct subgroups based on the methylation status of the selected five genes. Univariate and multivariate analyses were also performed to assess the factors related to DNA methylation. P value <0.05 was considered statistically significant.
Results
DNA methylation status of gastric mucosa, in relation to the magnifying NBI features
The clinic‐pathologic characteristics of 99 participants are shown in Table 1.
Table 1.
Clinic‐pathologic characteristics of 99 participants
| Variables | |
|---|---|
| Age: median (range) | 68 (44–86) |
| Gender: male % (n) | 68.7% (68) |
| Duration after H. pylori eradication: median months (range) | 41 (6–360) |
| Reason for H. pylori eradication: | |
| Gastric or duodenal ulcer % (n) | 23.2% (23) |
| After ER for early gastric cancer | 27.3% (27) |
| Others | 49.5% (49) |
| Gastric cancer occurrence | |
| Gastric cancer diagnosed before H. pylori eradication | 27.3% (27) |
| Gastric cancer diagnosed after H. pylori eradication | 34.3% (34)a |
| Cancer free | 45.4% (45) |
ER, endoscopic resection.
For seven cases, metachronous gastric cancer was detected after subsequent H. pylori eradication after ER.
We analyzed the methylation levels of five genes (MYOD1, SLC16A12, IGF2, RORA, and PRDM5) in 140 gastric mucosa biopsies from 99 participants after H. pylori eradication. The five genes we analyzed are frequently methylated in gastric cancer (RORA and PRDM5) 10 or H. pylori‐infected gastric mucosa (MYOD1, SLC16A12, and IGF2) 11. Initially, we investigated the association between methylation status of those five genes and gastric mucosal morphologic features seen by the magnifying NBI endoscopy (Fig. 1).
We classified the 140 biopsy specimens into restored type (95 specimens) and atrophic type (45 specimens). We found that methylation levels of all five genes in the atrophic type were significantly higher than those of restored type (all P < 0.0001: Fig. 2). We next compared the methylation levels of restored and atrophic types in 30 patients who had both restored and atrophic types in the individual stomach. Since one patient was taken five biopsies at endoscopic examination, 34 matched samples from these patients were included for this analysis. The significant higher methylation levels in the atrophic type were also confirmed in all five genes (all P < 0.0001: Fig. 3).
Figure 2.
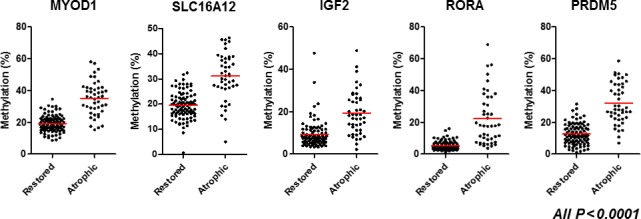
Methylation status of five gene promoters (MYOD1, SLC16A12, IGF2, RORA, and PRDM5) among restored and atrophic types. Horizontal bars represent mean methylation percentage. Statistical analysis was performed using Student's t‐test.
Figure 3.
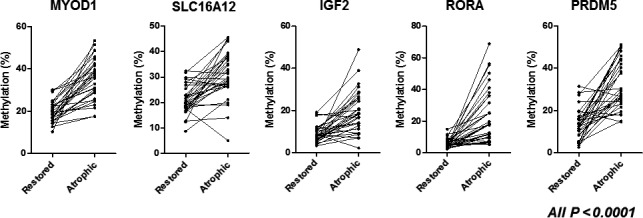
Methylation status of five gene promoters (MYOD1, SLC16A12, IGF2, RORA, and PRDM5) among restored and atrophic types among matched samples in patients who had both restored and atrophic types in the individual stomach. Statistical analysis was performed using Wilcoxon Signed Rank test.
Association between DNA methylation and duration after H. pylori eradiation
DNA methylation levels in the gastric mucosa improved after H. pylori eradiation by the time‐dependent manner 12. We next investigated the association between methylation levels and post‐eradication period. In the restored type, methylation levels were significantly decreased in patients with longer post‐eradication period (all P < 0.0001, Fig. 4). We also compared the methylation status of five genes in the restored type and the 23 healthy gastric mucosae without history of H. pylori infection. For three genes (IGF2, RORA, and PRDM5), DNA methylation levels of restored type after 10 years of post‐eradication period were similar to those of the healthy gastric mucosa without H. pylori infection (all P > 0.1), while it remained higher for two genes (MYOD1, SLC16A12, both P < 0.0001, Fig. S1). Concerning the atrophic type, we did not observe any correlation between methylation status of all five genes and duration after H. pylori eradiation. Moreover, the correlation analysis of methylation status between restored and atrophic types among patients who had both restored and atrophic types in the individual stomach only showed significant correlation only in one gene (MYOD1, P = 0.03, Fig. S2). It is suggested that gastric mucosa has renewal capacity regarding the methylation levels in the restored type, but it would be lost when it turned into the atrophic type.
Figure 4.
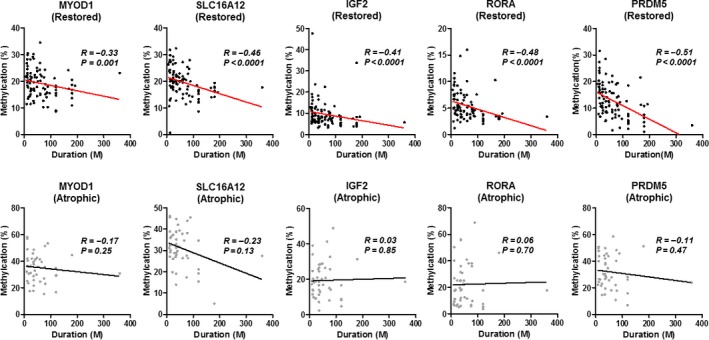
Association between methylation status and duration after H. pylori eradication among five gene promoters (MYOD1, SLC16A12, IGF2, RORA, and PRDM5). Upper, restored type; Lower, atrophic type; Statistical analysis was performed using Spearman correlation analysis.
Evaluation of clinic‐pathologic factors in relation to the methylation status in the gastric mucosa after H. pylori eradication
We investigated association between the clinic‐pathologic factors and the methylation status in the gastric mucosa after H. pylori eradication. We performed unsupervised hierarchical clustering analysis based on the methylation status of the selected five genes (Fig. 5). This analysis demonstrated that the atrophic types were mainly clustered as hypermethylated samples. The samples with intestinal metaplasia were also clustered as hypermethylated samples and tightly linked to the atrophic type, which was in line with recent our study (Tahara et al., submitting). We also found that the samples obtained from lesser curvature were clustered as hypermethylated samples. On the other hand, there was no clear association between cancer occurrence, duration after H. pylori eradication and the methylation status. We performed univariate and multivariate analyses to assess the factors related to DNA methylation statistically. We calculated mean Z‐score methylation of five genes to define hypermethylated samples. The mean Z‐score methylation of all five PCGIs in the gastric mucosa presented an approximately Gaussian distribution, with over represented of methylation‐high cases, we set cutoff value of 0.006 (mean Z‐score methylation) for the definition of methylation‐high cases. Univariate analysis revealed that location (lesser curvature, odds ratio: 13.05, 95% confidence interval: 4.16–40.92, P < 0.0001), longer duration after H. pylori eradication (odds ratio: 0.99, 95% confidence interval: 0.98–1.00, P = 0.016), and presence of atrophic type or intestinal metaplasia (odds ratio: 64.92, 95% confidence interval: 10.00–26.36, P < 0.0001) were significantly associated with methylation‐high (Table 2). Multivariate analysis of these factors revealed that longer duration after H. pylori eradication significantly reduced the risk of DNA methylation (odds ratio: 0.99, 95% confidence interval: 0.98–1.00, P = 0.019), and the presence of atrophic type or intestinal metaplasia held a strong risk for DNA methylation (odds ratio: 24.69, 95% confidence interval: 6.95–87.76, P < 0.0001) as an independent factor.
Figure 5.
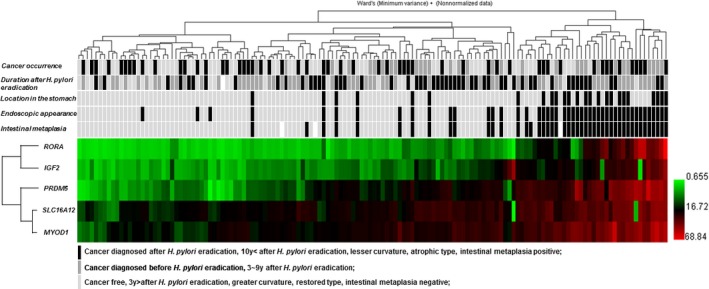
Unsupervised hierarchical clustering analysis of five gene promoters (MYOD1, SLC16A12, IGF2, RORA, and PRDM5) among 140 samples. Black, cancer diagnosed after H. pylori eradication, 10 years < after H. pylori eradication, lesser curvature, atrophic type, intestinal metaplasia positive; gray, cancer diagnosed before H. pylori eradication, 3–9 years after H. pylori eradication; light gray, cancer free, 3 years > after H. pylori eradication, restored type, intestinal metaplasia negative; white, not determined; Note that seven metachronous gastric cancer cases diagnosed after endoscopic resection and subsequent H. pylori eradication were treated as cancer diagnosed after H. pylori eradication.
Table 2.
Univariate analysis assessing the factors related to methylation‐high
| Variables | Odds ratio (95% confidence interval) | P value |
|---|---|---|
| Age | 1.04 (0.99–1.08) | 0.12 |
| Gender (male) | 1.69 (0.76–3.80) | 0.2 |
| Location (lesser curvature) | 13.05 (4.16–40.92) | <0.0001 |
| Duration | 0.99 (0.98–1.00) | 0.016 |
| Cancer occurrence | 1.97 (0.96–4.06) | 0.066 |
| Atrophic type or IM positive | 64.92 (10.00–25.35) | <0.0001 |
IM, intestinal metaplasia.
Methylation‐high, mean Z‐score methylation > −0.006.
Table 3.
Multivariate analysis assessing the factors related to methylation‐high
| Variables | Odds ratio (95% confidence interval) | P value |
|---|---|---|
| Location | 1.30 (0.26–6.52) | 0.75 |
| Duration | 0.99 (0.98–1.00) | 0.019 |
| Atrophic type or IM positive | 24.69 (6.95–87.76) | <0.0001 |
IM, intestinal metaplasia.
Methylation‐high, mean Z‐score methylation > −0.006.
Discussion
We showed that the methylation status of gastric mucosa after H. pylori eradication is closely associated with its endoscopic and histologic features. The atrophic type, which is informative magnifying NBI appearance of intestinal metaplasia, demonstrated significantly higher methylation levels in all five genes, compared to the restored type.
The five genes analyzed in this study are frequently methylated in gastric cancer (RORA and PRDM5) 17 or H. pylori‐infected gastric mucosa (MYOD1, SLC16A12, and IGF2) 18. Methylation of all these genes are also well correlated with magnifying NBI and histologic features of H. pylori‐infected gastric mucosa (Tahara et al., submitting). DNA methylation after H. pylori eradication is thought to be associated with gastric cancer predisposition 15, but the morphologic features of this residual methylation have not been clearly described. Our current result provided the evidence that morphologic features related to the intestinal metaplasia are associated with residual methylation in gastric mucosa after H. pylori eradication. We classified the morphologic features of gastric body (fundic area) using magnifying NBI endoscopy, assuming that the inter‐individual difference in atrophic status after H. pylori eradication would be more enhanced in the gastric body rather than in the pyloric region (antrum). We have shown that the incidence as well as the portion of atrophic area were not associated with post‐eradication period, while the spread of atrophic area was an indicator for gastric cancer occurrence (Tahara et al., submitting). Other studies have also suggested that the severe gastric atrophy especially in the fundic area might be a risk of gastric cancer 16. The atrophic type with hyper‐DNA methylation in the gastric fundic area might reflect the past exposure of H. pylori infection. Several studies showed that H. pylori eradication could reverse DNA methylation by time‐dependent manner 12, 15. In fact, among restored type, methylation levels were significantly lower in patients with longer post‐eradication period in this study. The methylation levels of IGF2, RORA, and PRDM5 genes among restored type with more than 10 years of post‐eradication period demonstrated almost same as healthy gastric mucosa without history of H. pylori infection. On the other hand, this issue was not applied to the atrophic type; there was no association between the duration and methylation levels. The atrophic type also presented significantly higher methylation levels comparing to the restored type in all genes among matched samples from patients who had both restored and atrophic types in the individual stomach, but they had no correlation with each other except MYOD1 gene. This suggests that the atrophic type represents the epigenetic irreversible point with H. pylori eradication.
To evaluate the association between DNA methylation status and several clinic‐pathologic factors, we performed unsupervised hierarchical clustering analysis and multivariate analysis. Atrophic type was very tightly linked with intestinal metaplasia and clustered as hypermethylated samples. The multivariate analysis showed that atrophic type or intestinal metaplasia was the strong independent risk for DNA methylation after H. pylori eradication. Although DNA hypermethylation might be a risk of gastric cancer regardless of H. pylori infection status 6, 19, 20, 21, in our study, the most important factor of DNA hypermethylation was magnifying NBI and histologic features. This indicates that the methylation change is closely linked to the magnifying NBI and histologic features within the focal points. It might also be informative for estimating risk of gastric cancer to assess the spread of NBI patterns in the stomach (Tahara et al., submitting). Morphologic features of aberrant methylation have described as reflecting its distinct clinic‐pathologic features in colorectal cancer [22]. Our current result also demonstrated the reliability of the magnifying NBI features of gastric mucosa after H. pylori eradication to estimate the “field defect” from a molecular view point. Gastric cancer incidence after H. pylori eradication is increasing recently, but it is difficult to find it because of its ambiguity. 14. Our result might provide more appropriate clinical implementation for gastric cancer surveillance after eradication, reflecting individual cancer risk. We believe that our findings provide salient information for many endoscopists to further improve gastric cancer risk stratification among patients after H. pylori eradication.
Conflict of Interest
The authors declare that they have no conflicts of interest.
Supporting information
Table S1. Primer sequences used for pyrosequencing.
Figure S1. Methylation status of five gene promoters (MYOD1, SLC16A12, IGF2, RORA and PRDM5) among restored type and healthy gastric mucosa without history of H. pylori infection. Statistical analysis was performed using Student's t‐test.
Figure S2. Correlation of methylation status of five gene promoters (MYOD1, SLC16A12, IGF2, RORA and PRDM5) among restored and atrophic types in matched samples in patients who had both restored and atrophic types in the individual stomach. Statistical analysis was performed using Spearman correlation analysis.
Cancer Medicine 2017; 6(7):1730–1737
References
- 1. Issa, J. P. 2014. Aging and epigenetic drift: a vicious cycle. J Clin Invest. 124:24–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kang, G. H. , Lee H. J., Hwang K. S., Lee S., Kim J. H., and Kim J. S.. 2003. Aberrant CpG island hypermethylation of chronic gastritis, in relation to aging, gender, intestinal metaplasia, and chronic inflammation. Am J Pathol. 163:1551–1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Parsonnet, J. , Friedman G. D., Vandersteen D. P., Chang Y., Vogelman J. H., Orentreich N., et al. 1991. Helicobacter pylori infection and the risk of gastric carcinoma. N Endl J Med. 325:1127–1131. [DOI] [PubMed] [Google Scholar]
- 4. Huang, J. Q. , Sridhar S., Chen Y., and Hunt R. H.. 1998. Meta‐analysis of the relationship between Helicobacter pylori seropositivity and gastric cancer. Gastroenterology. 114:1169–1179. [DOI] [PubMed] [Google Scholar]
- 5. Uemura, N. , Okamoto S., Yamamoto S., Matsumura N., Yamaguchi S., Yamakido M., et al. 2001. Helicobacter pylori infection and the development of gastric cancer. N Engl J Med. 345:784–789. [DOI] [PubMed] [Google Scholar]
- 6. Maekita, T. , Nakazawa K., Mihara M., Nakajima T., Yanaoka K., Iguchi M., et al. 2006. High levels of aberrant DNA methylation in Helicobacter pylori‐infected gastric mucosae and its possible association with gastric cancer risk. Clin Cancer Res. 12:989–995. [DOI] [PubMed] [Google Scholar]
- 7. Zou, X. P. , Zhang B., Zhang X. Q., Chen M., Cao J., and Liu W. J.. 2009. Promoter hypermethylation of multiple genes in early gastric adenocarcinoma and precancerous lesions. Hum Pathol. 40:1534–1542. [DOI] [PubMed] [Google Scholar]
- 8. Uemura, N. , Mukai T., Okamoto S., Yamaguchi S., Mashiba H., Taniyama K., et al. 1997. Effect of Helicobacter pylori eradication on subsequent development of cancer after endoscopic resection of early gastric cancer. Cancer Epidemiol Biomarkers Prev. 6:639–642. [PubMed] [Google Scholar]
- 9. Fukase, K. , Kato M., Kikuchi S., Inoue K., Uemura N., Okamoto S., et al. 2008. Effect of eradication of Helicobacter pylori on incidence of metachronous gastric carcinoma after endoscopic resection of early gastric cancer: an open‐label, randomized controlled trial. Lancet. 372:392–397. [DOI] [PubMed] [Google Scholar]
- 10. Asaka, M. , Mabe K., Matsushima R., and Tsuda M.. 2015. Helicobactor pylori eradication to eliminate gastric cancer: the Japanese strategy. Gastroenterol Clin North Am. 44:639–648. [DOI] [PubMed] [Google Scholar]
- 11. Chan, A. O. , Peng J. Z., Lam S. K., Lai K. C., Yuen M. F., Cheung H. K., et al. 2006. Eadication of Helicobacter pylori infection reverses E‐cadherin promoter hypermethylation. Gut. 55:463–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Shin, C. M. , Kim N., Lee H. S., Park J. H., Ahn S., Kang G. H., et al. 2013. Changes in aberrant DNA methylation after Helicobacter pylori eradication: a long‐term follow‐up study. Int J Cancer. 133:2034–2042. [DOI] [PubMed] [Google Scholar]
- 13. Kamada, T. , Hata J., Sugiu K., Kusunoki H., Ito M., Tanaka S., et al. 2005. Clinical features of gastric cancer discovered after successful eradication of Helicobacter pylori: results from a 9‐year prospective follow‐up study in Japan. Aliment Pharmacol Ther. 21:1121–1126. [DOI] [PubMed] [Google Scholar]
- 14. Yamamoto, K. , Kato M., Takahashi M., Haneda M., Shinada K., Nishida U., et al. 2011. Clinicopathological analysis of early‐stage gastric cancers detected after successful eradication of Helicobacter pylori . Helicobacter. 16:210–216. [DOI] [PubMed] [Google Scholar]
- 15. Nakajima, T. , Enomoto S., Yamashita S., Ando T., Nakanishi Y., Nakazawa K., et al. 2010. Persistence of a component of DNA methylation in gastric mucosae after Helicobacter pylori eradication. J Gastroenterol. 45:37–44. [DOI] [PubMed] [Google Scholar]
- 16. Hanaoka, N. , Uedo N., Shiotani A., Inoue T., Takeuchi Y., Higashino K., et al. 2010. Autofluorescence imaging for predicting development of metachronous gastric cancer after Helicobacter pylori eradication. J Gastroenterol Hepatol. 25:1844–1849. [DOI] [PubMed] [Google Scholar]
- 17. Watanabe, Y. , Kim H. S., Castoro R. J., Chung W., Estecio M. R., Kondo K., et al. 2009. Sensitive and specific detection of early gastric cancer with DNA methylation analysis of gastric washes. Gastroentwrology. 136:2149–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tahara, T. , Shibata T., Okubo M., Kawamura T., Horiguchi N., Ishizuka T., et al. 2016. Demonstration of potential link between helicobacter pylori related promoter CpG island methylation and telomere shortening in human gastric mucosa. Oncotarget. 7:43989–43996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Tahara, T. , Arisawa T., Shibata T., Wang F. Y., Nakamura M., Sakata M., et al. 2007. Risk prediction of gastric cancer by analysis of aberrant DNA methylation in non‐neoplastic gastric epithelium. Digestion. 75:54–61. [DOI] [PubMed] [Google Scholar]
- 20. Suzuki, R. , Yamamoto E., Nojima M., Maruyama R., Yamano H. O., Yoshikawa K., et al. 2014. Aberrant methylation of microRNA‐34b/c is a predictive marker of metachronous gastric cancer risk. J Gastroenterol. 49:1135–1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Asada, K. , Nakajima T., Shimazu T., Yamamichi N., Maekita T., Yokoi C., et al. 2015. Demonstration of the usefulness of epigenetic cancer risk prediction by a multicentre prospective cohort study. Gut. 64:388–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kimura, T. , Yamamoto E., Yamano H. O., Suzuki H., Kamimae S., Nojima M., et al. 2012. A novel pit pattern identifies the precursor of colorectal cancer derived from sessile serrated adenoma. Am J Gastroenterol. 107:460–469. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Primer sequences used for pyrosequencing.
Figure S1. Methylation status of five gene promoters (MYOD1, SLC16A12, IGF2, RORA and PRDM5) among restored type and healthy gastric mucosa without history of H. pylori infection. Statistical analysis was performed using Student's t‐test.
Figure S2. Correlation of methylation status of five gene promoters (MYOD1, SLC16A12, IGF2, RORA and PRDM5) among restored and atrophic types in matched samples in patients who had both restored and atrophic types in the individual stomach. Statistical analysis was performed using Spearman correlation analysis.


