Abstract
Activation of the PTEN-PI3K-mTORC1 pathway consolidates metabolic programs that sustain cancer cell growth and proliferation1,2. Here we show that mTORC1 regulates polyamine dynamics, a metabolic route that is essential for oncogenicity. Through the use of integrative metabolomics in a mouse model3 and human biopsies4 of prostate cancer, we identified alterations in tumours impacting on the production of decarboxylated S-adenosylmethionine (dcSAM) and polyamine synthesis. Mechanistically, this metabolic rewiring stems from mTORC1-dependent regulation of S-adenosylmethionine decarboxylase 1 (AMD1) stability. This novel molecular regulation was validated in murine and human cancer specimens. AMD1 was upregulated in prostate cancer specimens with activated mTORC1. Conversely, samples from a clinical trial with the mTORC1 inhibitor everolimus5 exhibited a predominant decrease in AMD1 immunoreactivity that was associated to a decrease in proliferation, in line with the requirement of dcSAM production for oncogenicity. These findings provide fundamental information about the complex regulatory landscape controlled by mTORC1 to integrate and translate growth signals into an oncogenic metabolic program.
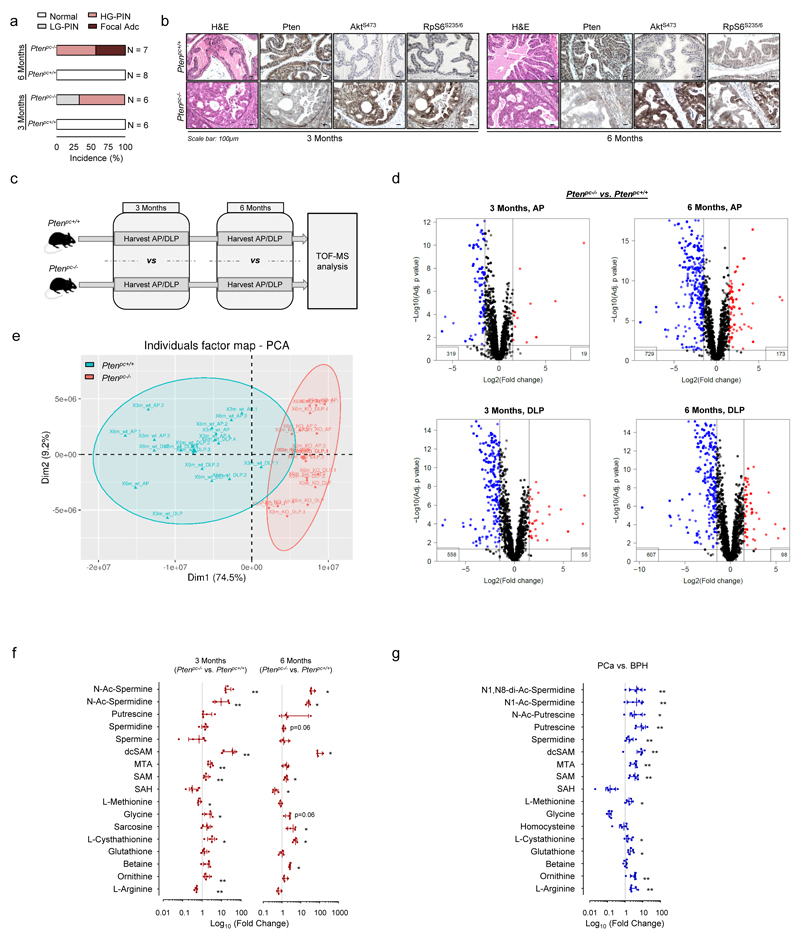
Alterations in the Phosphoinositide-3 kinase (PI3K) pathway have been reported in a high percentage of human cancers6,7. We sought to identify metabolic requirements of prostate cancer (PCa) taking advantage of a faithful genetically engineered mouse model (GEMM) of this disease driven by loss of Pten3, a negative regulator of the PI3K pathway that is frequently down-regulated in PCa6,8. First, we carried out high-throughput Time-Of-Flight mass spectrometry (TOF-MS) to examine metabolic alterations at two time points (3 and 6 months, onset of prostate intraepithelial neoplasia (PIN) and invasive prostate carcinoma, respectively) (Extended data Fig. 1a, b) in two different prostate lobes (Extended data Fig. 1c). From 7722 ions, we assigned metabolite identification (Human Metabolome Database (HMDB) score ≥ 40) to 632 (Supplementary Table 1). We did not observe significant influence of the prostate lobe or the time point of analysis, and after precluding significant alterations in candidate metabolic pathways, we focused on metabolites consistently altered in all conditions (Extended data Fig. 1d, e; Supplementary Table 2). We identified 72 unique metabolites (73 assigned ions) fulfilling the criteria (Fig. 1a; Supplementary Table 1). Pathway enrichment analysis in this set did not show significantly altered pathways including a considerable number of metabolites (Supplementary Table 3). Strikingly, we observed an increase in polyamine synthesis-related metabolites in Ptenpc-/- mice (Fig. 1b). These results were validated in mouse GEMM and human PCa tissues by quantitative Liquid Chromatography (LC)/MS (Extended data Fig. 1f, g; Supplementary Table 4).
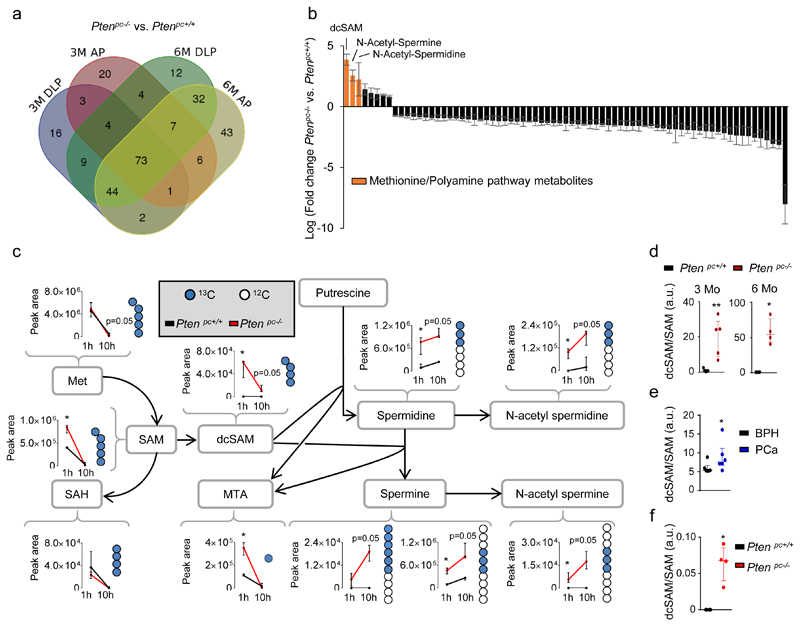
Figure 1. Integrative metabolomics in prostate cancer reveals a rewiring from methionine metabolism towards polyamine synthesis.
a-b, VENN diagram (anterior prostate - AP - and dorsolateral prostate - DLP, a), and Waterfall plot (b) from the analysis of altered metabolites in TOF-MS metabolomic analysis carried out in Ptenpc-/- and Ptenpc+/+ (6 months Ptenpc+/+ AP n=4 mice; rest of conditions n=5 mice) mouse prostate samples at the indicated age. Values in (b) represent the average of the Log (Fold change) with the s.e.m. of the two lobes and two time points (3 and 6 months of age) per metabolite. c, Incorporation of 13C from intravenously injected U-13C5-L-Methionine (100 mg/Kg) into the indicated metabolites at 3 months of age (AP). Peak area refers to natural abundance-corrected values (n=4 mice at 1 hour; n=3 mice at 10 hours). Data are represented as median with interquartile range. Blue dots: 13C; white dots: 12C; 1h: prostate samples extracted after 1-hour pulse with U-13C5-Methionine; 10h: prostate samples extracted after 10-hour pulse with U-13C5-Methionine. d, dcSAM/SAM ratios from Extended data Fig. 1f (n=4 as indicated by dots). e, dcSAM/SAM ratio from Extended data Fig. 1g (n=6 as indicated by dots). f, dcSAM/SAM ratio from Fig. 1c at 1 hour (n=4 as indicated by dots). a.u.: arbitrary units; 3M/6M: 3 Months / 6 Months; dcSAM: decarboxylated S-Adenosylmethionine; MTA: 5´ methylthioadenosine; SAM: S-Adenosylmethionine; SAH: S-Adenosylhomocysteine; Met: methionine; p, p-value; *, p<0.05; **, p<0.01. One tail (c-f) Mann-Whitney U test was used for data analysis.
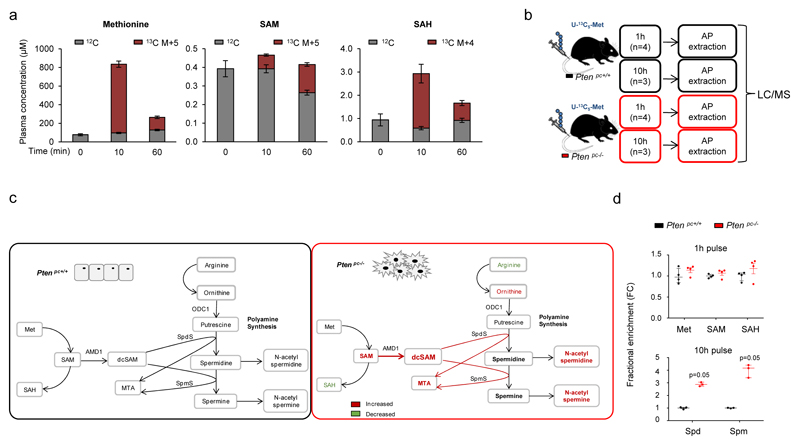
In order to determine how metabolic rewiring affects polyamine dynamics, we set up 13C-labelling metabolic analysis to trace the fate of methionine-derived carbons in vivo (Extended data Fig. 2a). Next, we injected U-13C5-methionine intravenously in Ptenpc+/+ and Ptenpc-/- mice (Extended data Fig. 2b). Prostate tissue analysis revealed an elevation in 13C-labelled decarboxylated S-adenosylmethionine (dcSAM), together with increased synthesis and fractional labelling of polyamines (Fig. 1c; Extended data Fig. 2c, d; Supplementary Table 5). Importantly, the increase of S-adenosylmethionine (SAM) decarboxylation (elevated dcSAM/SAM ratio) in both mouse and human pathological tissue strongly suggested that the enzyme which catalyses this reaction (S-adenosylmethionine decarboxylase 1, AMD1) is potentially responsible for the metabolic changes observed in PCa (Fig. 1d-f).
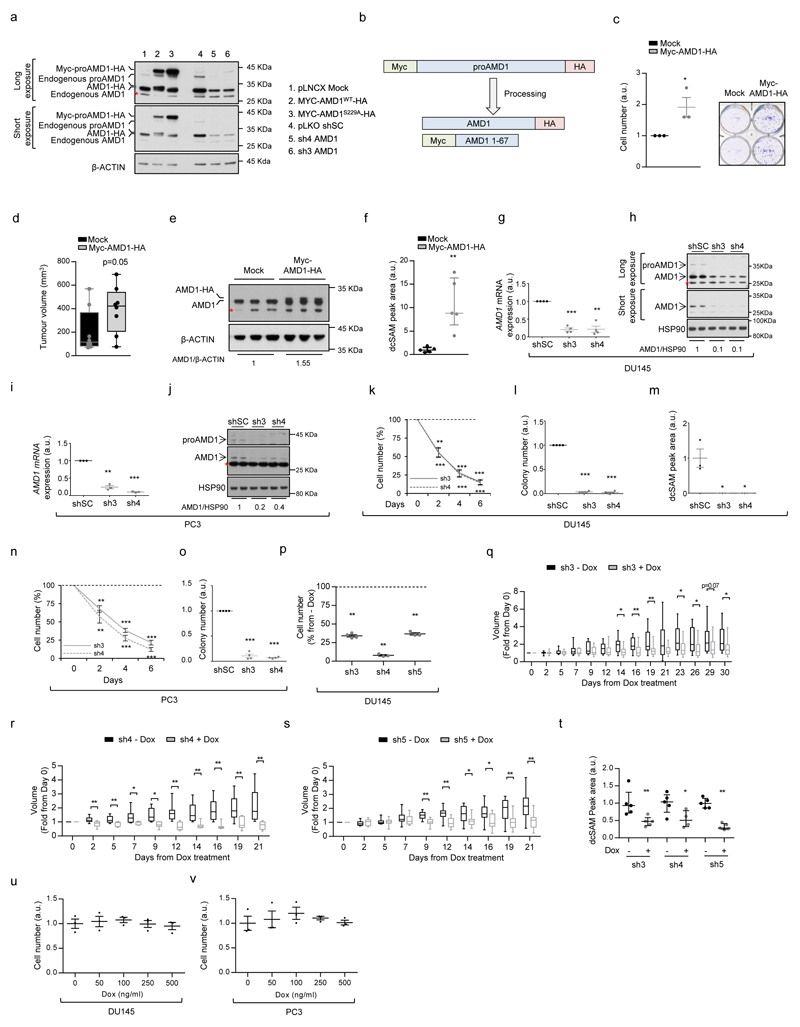
To address the contribution of dcSAM production to PCa cell oncogenicity, we ectopically expressed AMD1 in PCa cell lines. AMD1 is produced as a pro-enzyme, that is subject to self-cleavage and heterotetramerization, resulting in the active enzyme9. After validation of a polyclonal antibody for the detection of proAMD1 and AMD1 (Extended data Fig. 3a, b), we generated PCa cells in which the expression of AMD1 was up-regulated, which resulted in increased dcSAM abundance (Fig. 2a, b). Interestingly, this perturbation increased foci formation, anchorage-independent growth and in vivo tumour growth (Fig. 2c, d; Extended data Fig. 3c-f).
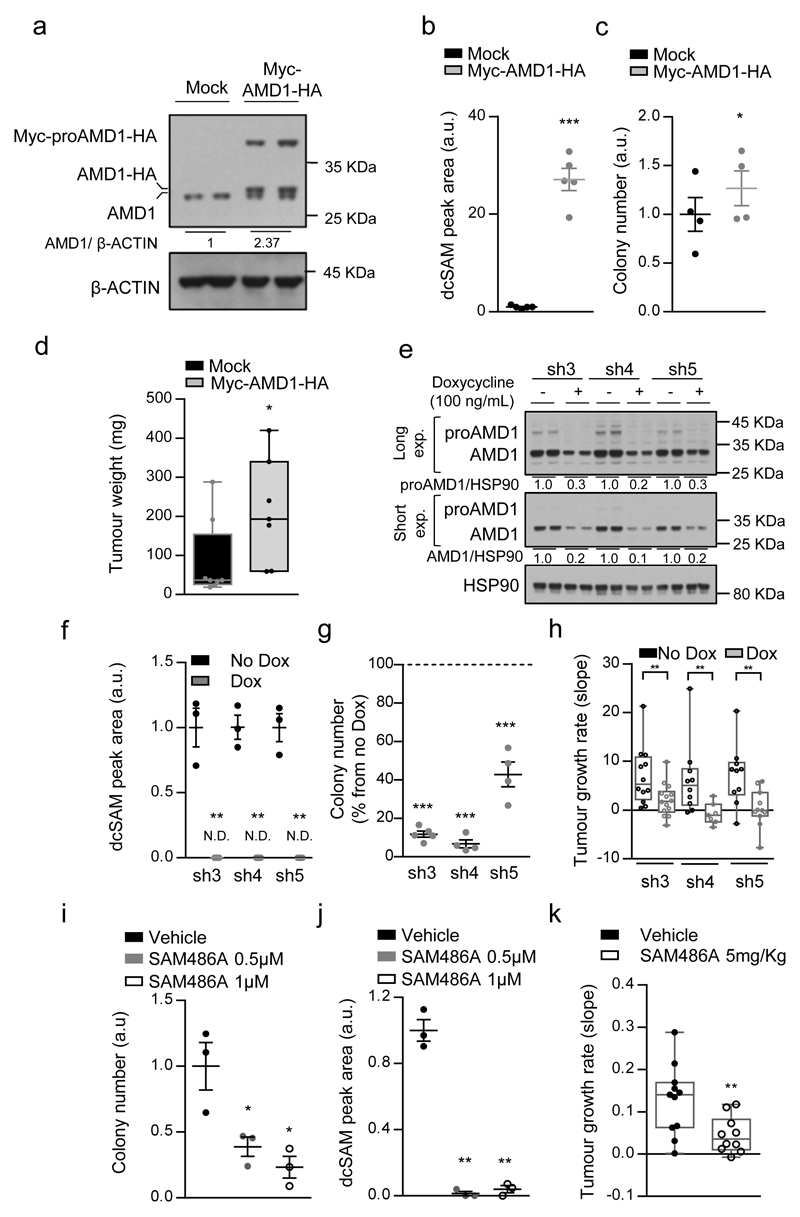
Figure 2. Genetic and pharmacological AMD1 modulation affects prostate cancer oncogenicity.
a-c, Impact of ectopic Myc-AMD1-HA expression (a, representative from 3 independent experiments) on dcSAM abundance (b, n=5 independent experiments) and anchorage-independent growth (c, n=4 independent experiments) in DU145 cells in vitro. Data are represented as mean with s.e.m. d, Impact of ectopic Myc-AMD1-HA expression on tumor weight in DU145 xenografts grown for 43 days (Mock n=8 tumours; Myc-AMD1-HA n=7 tumours). Data in is represented as box and whisker plot. e-g, Effect of doxycycline-inducible (100 ng/mL) AMD1 silencing (sh3-5) on AMD1 protein expression (e, representative from 3 independent experiments), dcSAM abundance (f, n=3 independent experiments) and anchorage-independent growth (g, n=4 independent experiments) in DU145 cells. Dash line indicates relative cell number of non-induced cells. Data are represented as mean with s.e.m. h, Impact of inducible AMD1 silencing on tumor growth rate of established DU145 xenografts (Tumour number: sh3 No Dox, n=12; sh3 Dox, n=14; sh4 No Dox, n=10; sh4 Dox, n=7; sh5 No Dox, n=10; sh5 Dox, n=11). Growth rate was inferred from the linear regression calculated for the progressive change in tumor volume of each individual tumor during the period depicted in Extended data Fig. 3q-s. Data is represented as box and whisker plot. i-j, Effect of pharmacological AMD1 inhibition with SAM486A on anchorage-independent growth (i, n=3 independent experiments) and dcSAM abundance (j, n=3 independent experiments) in DU145 cells. Dash line indicates relative cell number of vehicle-treated cells. Data is represented as mean with s.e.m. k, Impact of SAM486A treatment for 14 days (5mg/Kg/day, 5 days per week) on tumor growth rate of established DU145 xenografts (Vehicle, n=11 tumours; SAM4856A, n=10 tumours). Growth rate was inferred from the linear regression calculated for the progressive change in tumor volume of each individual tumor during the period depicted in Extended data Fig. 4i. Data is represented as box and whisker plot. a.u.: arbitrary units; dcSAM: decarboxylated S-adenosylmethionine; - Dox: non-induced condition; + Dox: doxycycline-induced (100 ng/mL) condition; Mock: Empty vector; Myc-AMD1-HA: Myc and HA-tagged AMD1 ectopic expression, sh: short hairpin RNA. p, p-value; *, p<0.05; **, p<0.01, ***, p<0.001. Statistic test: one tail Student T-test was used for cell line data analysis (b, c, f, g, i, j) and one tail Mann-Whitney U test for xenografts (d, h, k).
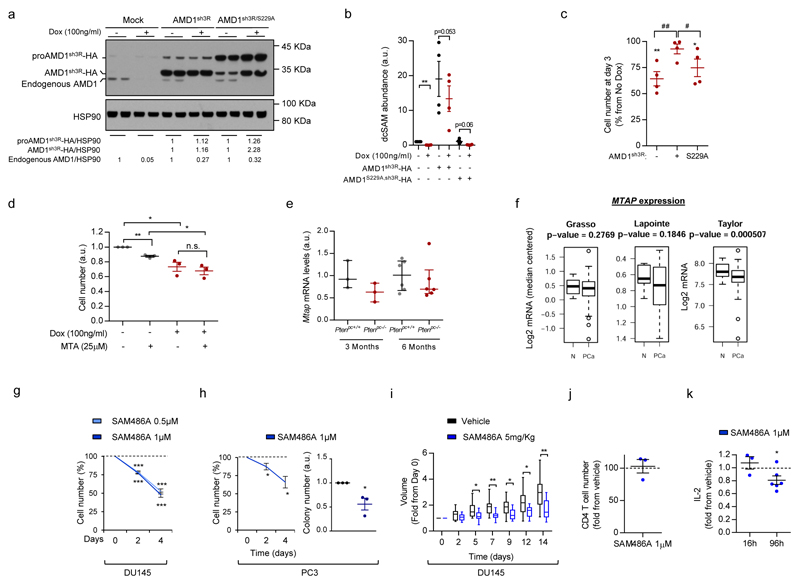
If AMD1 activity is essential for PCa cell function, targeting this enzyme would represent an attractive therapeutic strategy. To test this notion, we generated and validated three AMD1-targeting doxycycline-inducible and two constitutive shRNAs (Fig. 2e; Extended data Fig.3g-j), that resulted in a profound reduction in dcSAM levels, the inhibition of two-dimensional and anchorage-independent growth, and tumour growth in vivo (Fig. 2f-h; Extended data Fig. 3k-t). We excluded doxycycline-dependent (Extended data Fig. 3u, v) and off-target effects of the shRNA (by means of ectopic expression of shRNA-resistant wild-type – WT - or non-processable - S229A10 - AMD1 mutants) (Extended data Fig. 4a-c). Of note, we did not observe a contribution of MTAP11–14 nor 5´-Methylthioadenosine (MTA, a product of dcSAM metabolism to produce polyamines) to the effect of AMD1 inhibition (Extended data Fig. 4d-f).
A pharmacological inhibitor of AMD1, SAM486A, has been designed and evaluated in pre-clinical and clinical settings15–18. Pharmacological AMD1 inhibition recapitulated the biological consequences of genetic silencing, in the absence of overt toxicity in vivo (Fig. 2i-k; Extended data Fig. 4g-k; Supplementary Table 6). Our results collectively demonstrate that AMD1 activity is required for PCa oncogenicity.
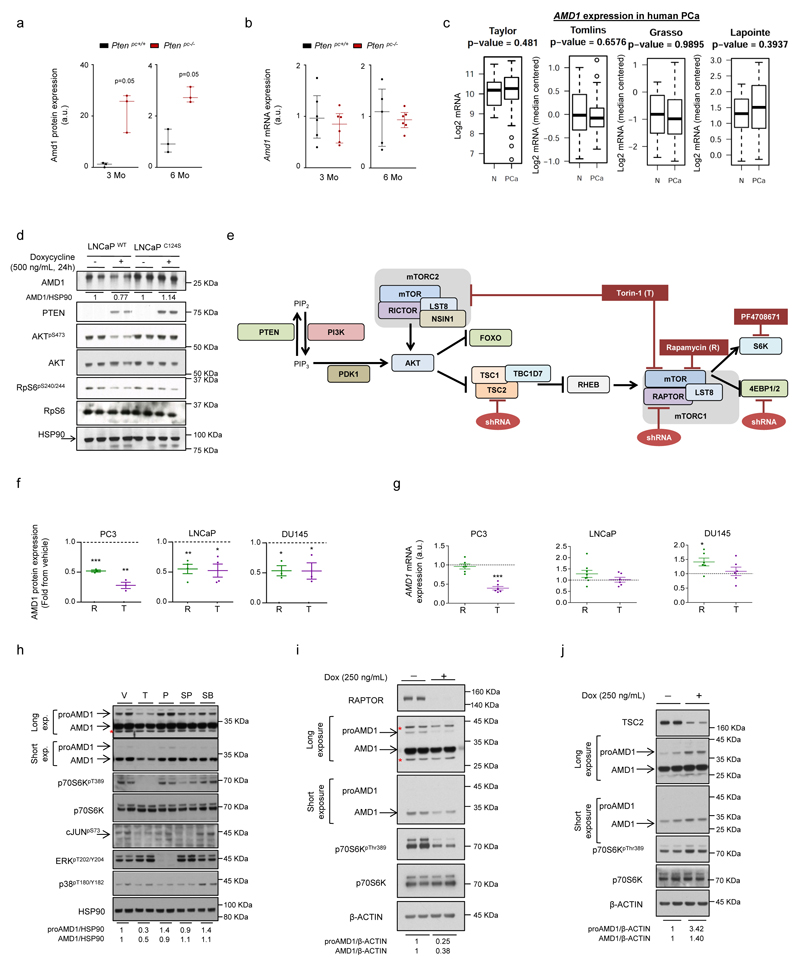
We next sought to elucidate the mechanism underlying the production of dcSAM. Interestingly, AMD1 protein levels were increased in prostate tissue from Ptenpc-/- mice in the absence of transcriptional modulation, consistent with mRNA analysis in human PCa datasets (Fig. 3a; Extended data Fig. 5a-c). To ascertain whether this phenotype was a direct consequence of the loss of PTEN, we analysed PTEN-deficient PCa cells (LNCaP). Re-expression of YFP-PTENWT, but not catalytically inactive YFP-PTENC124S, in these cells resulted in the reduction in AMD1 protein levels (Extended data Fig. 5d)19. Further dissection of the PI3K-mTORC1 pathway revealed that only mTORC1 blockers among various signalling inhibitors decreased proAMD1 and AMD1 protein abundance (without consistent effects on mRNA expression; Fig. 3b; Extended data Fig. 5e-h). The regulation of this enzyme by mTORC1 was validated upon genetic modulation of positive and negative regulators of the complex, RAPTOR and TSC2, respectively (Extended data Fig. 5e, i, j). Importantly, mTORC1 inhibitor-elicited AMD1 down-regulation was accompanied by a decrease in dcSAM production and polyamine synthesis (Fig. 3c, d). Of interest, spermidine (Spd) supplementation in rapamycin-treated PC3 cells (PTEN-deficient) elicited a significant (albeit small) increase in cell number (Extended data Fig. 6a).
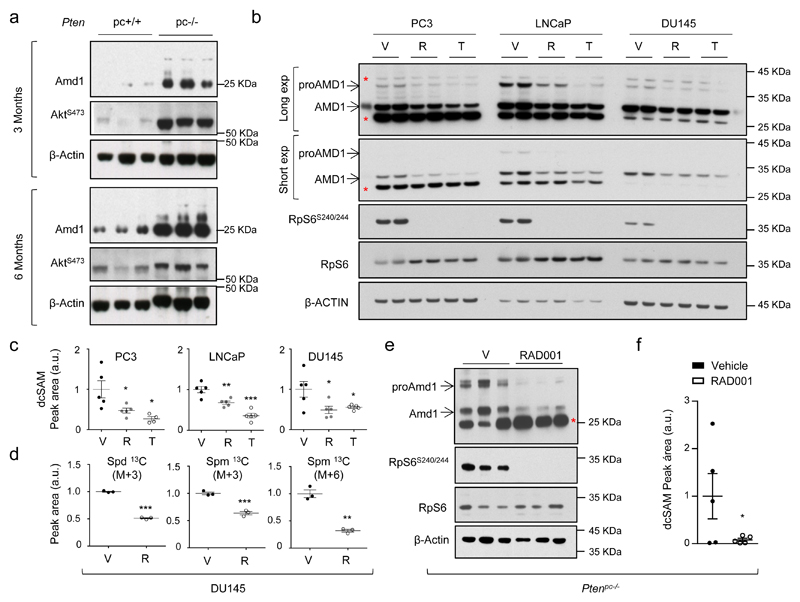
Figure 3. mTORC1 regulates AMD1 expression, dcSAM production and polyamine dynamics.
a, Amd1 protein abundance in Ptenpc-/- and Ptenpc+/+ of the indicated age (n=3 mice). AKTS473 is shown as control of PI3K pathway over-activation. b, Representative western blot (out of 3) depicting the changes in expression of the indicated proteins upon 24 hour treatment of PC3, DU145 and LNCaP cells with vehicle (V, DMSO), rapamycin (R, 20 nM) and Torin-1 (T, 250 nM for PC3 and DU145, 125 nM for LNCaP). c, dcSAM abundance in PC3 (n=4-5 independent experiments as indicated by dots), LNCaP (n=5 independent experiments) and DU145 (n=5 independent experiments), upon 24 hour treatment with vehicle (V, DMSO), rapamycin (R, 20 nM) and Torin-1 (T, 250 nM for PC3 and DU145, 125 nM for LNCaP). Data are represented as mean with s.e.m. d, Incorporation of 13C from U-13C5-L-Methionine (2h pulse) into the indicated metabolites after 30 hour treatment with vehicle (V, DMSO) or rapamycin (R, 20 nM) in DU145 cells (n=3 independent experiments). Data are represented as mean with s.e.m. e-f, Effect of 4-week RAD001 treatment on mTORC1 activity (RpS6S240/4) and Amd1 protein expression (e, n=3 mice), and dcSAM abundance (f, n=5 mice) in prostate tissue extracts from Ptenpc-/- mice. Data in (f) are represented as mean with s.e.m. a.u.: arbitrary units; 3Mo: 3 month-old mouse prostate analysis; 6Mo: 6 month-old prostate analysis; V: Vehicle; dcSAM: decarboxylated S-adenosylmethionine; Spd 13C (M+3): Spermidine labeled in three carbons; Spm 13C (M+3): Spermine labeled in three carbons; Spm 13C (M+6): Spermine labeled in six carbons; p, p-value; *, p<0.05; **, p<0.01; ***, p<0.001. Red asterisk in western blot indicates non-specific band; arrow: specific band. Statistic test: one tail Student T-test (c, d, f) was used.
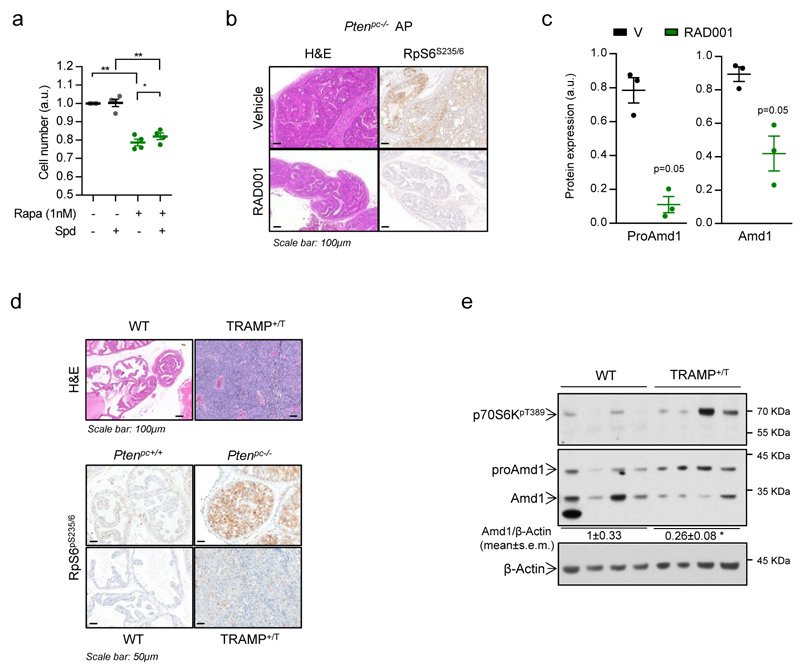
In order to ascertain the requirement of mTORC1 activation for dcSAM accumulation in vivo, we treated Ptenpc-/- mice with the rapamycin-derivative RAD001 and found that Amd1 and dcSAM abundance was reduced in line with the inhibition of mTORC1 in prostate tissue (Fig. 3e, f; Extended data Fig. 6b, c). Of note, a second PCa GEMM based on the expression of the TRAMP transgene20, which presented low mTORC1 activation, did not exhibit an increase in Amd1 nor dcSAM abundance, in support of our postulated mTORC1-AMD1 regulation (Extended data Fig. 6d, e; Supplementary Table 7).
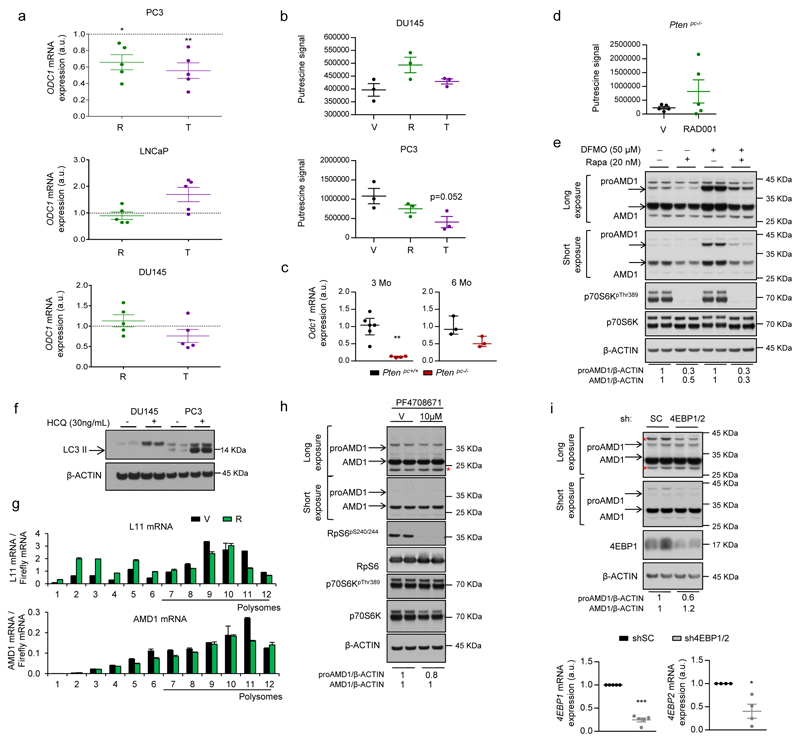
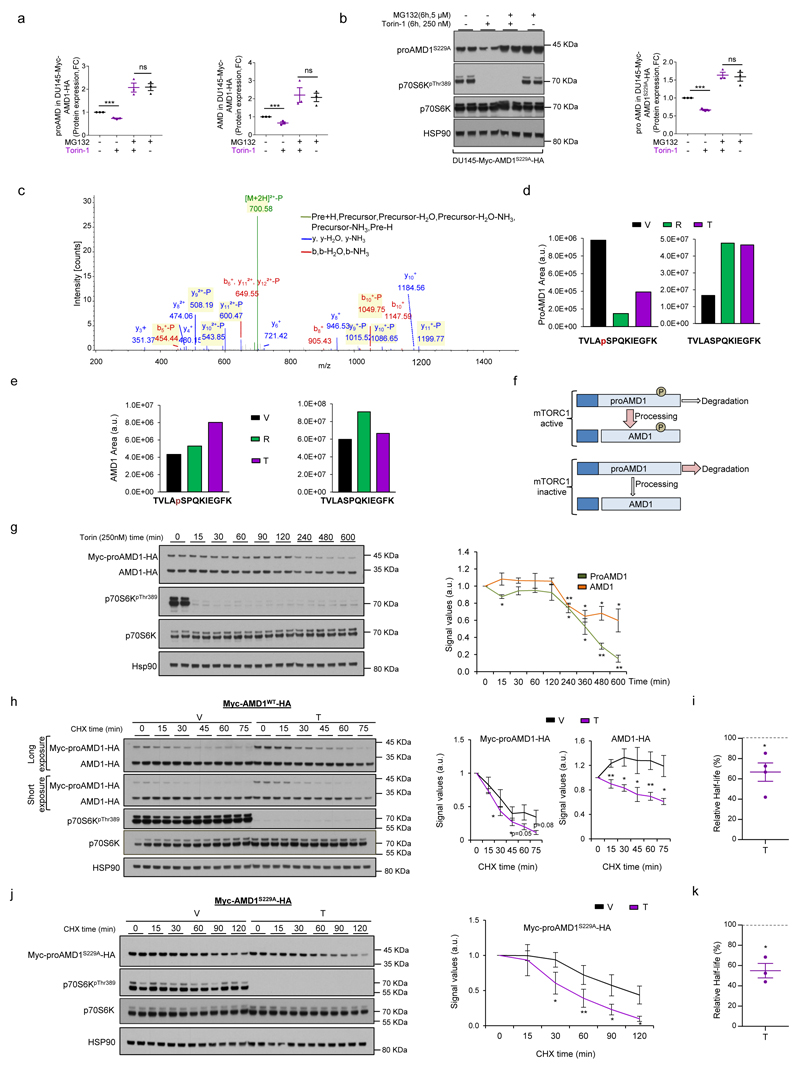
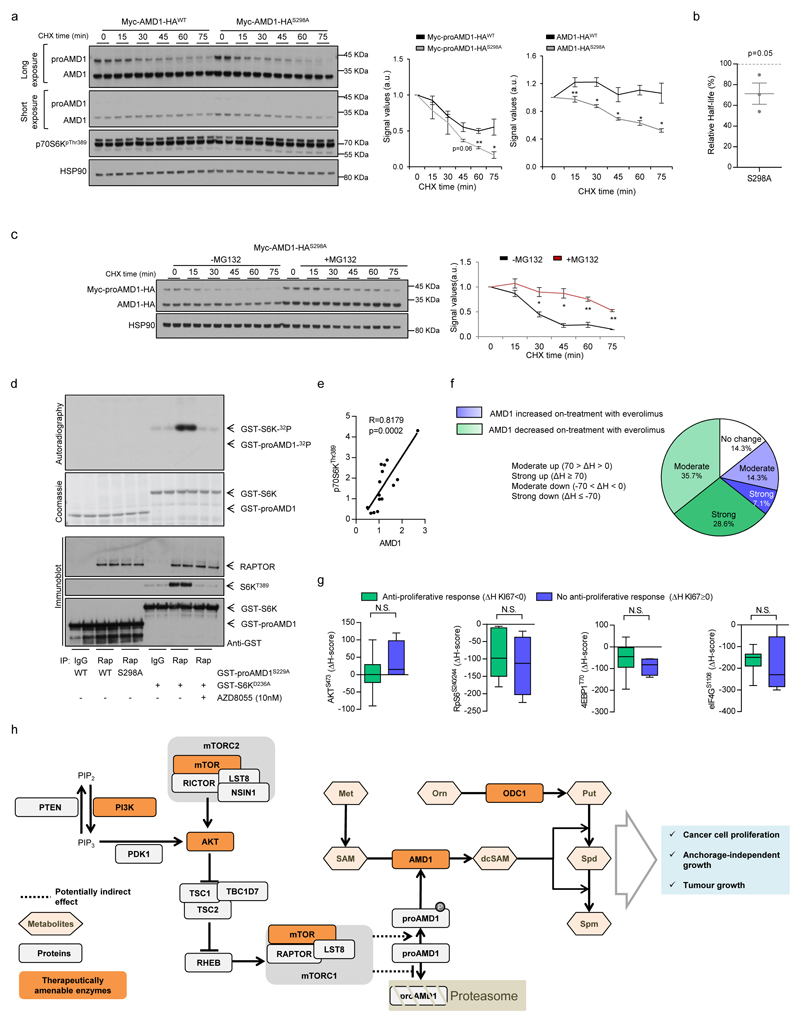
Mechanistically, we excluded the contribution of ornithine decarboxylase 121 (ODC1; Extended data Fig. 7a-e) and canonical mTORC1 downstream effectors and pathways1,22,23, including translation initiation, p70S6K, 4EBP and macro-autophagy (since DU145 cells lack functional ATG524 but retain the mTORC1-dependent regulation of AMD1) (Extended data Fig. 7f-i). Interestingly, we found that mTORC1 inhibition-dependent decrease in AMD1 (WT and S229A) protein levels was rescued by the proteasome inhibitor MG132 (Fig. 4a; Extended data Fig. 8a, b). To elucidate the molecular link between mTORC1 activity and proAMD1 stability, we carried out a phosphoproteomic analysis on ectopic proAMD1/AMD1 and identified a single phosphorylated residue (S298) on the pro-enzyme and enzyme (TVLApSPQKIEGFK) (Extended data Fig.8c) that was compatible with a consensus mTORC1 site25. Importantly, 6-hour treatment with rapamycin or Torin-1 reduced the phosphorylation of S298 in the pro-enzyme (and the ratio phospho-proAMD1/total proAMD1) but not the enzyme, leading us to hypothesize that S298 phosphorylation is controlled by mTORC1 and promotes proAMD1 stability (Fig. 4b and Extended data Fig. 8d-f). We evaluated the stability of proAMD1 after a 2 hour treatment with Torin-1 (prior to the detection of any effect on pro-enzyme abundance) and found reduced half-life upon mTORC1 inhibition (Extended data Fig. 8g-k). To establish the contribution of S298 phosphorylation in the regulation of proAMD1 stability, we inactivated this phosphorylation site (S298A). As predicted, non-phosphorylated proAMD1S298A exhibited decreased half-life, and this parameter was augmented upon inhibition of the proteasome (Extended data Fig. 9a-c). These results support the notion that mTORC1 activity promotes proAMD1 stability, at least in part, through the regulation of its phosphorylation in S298, hence allowing enzyme processing and activity. In vitro mTORC1 kinase assay with GST-proAMD1S229A did not show significant activity towards proAMD1 phosphorylation in these conditions, suggesting either that mTORC1 does not directly phosphorylate S298, or that additional cellular conditions (e.g. biochemical conditions, adaptor or intermediary proteins, sub-cellular compartments) are required for mTORC1 to phosphorylate proAMD1 (Extended data Fig. 9d). It is worth noting that our data does not rule out additional mechanisms downstream mTORC1 regulating proteasome-mediated protein degradation23,26. In order to extend this mechanistic link to human PCa, we extracted protein from well-diagnosed benign prostate hyperplasia (BPH) and PCa specimens4 (Supplementary Table 4). The results revealed that AMD1 was selectively more abundant in PCa specimens exhibiting high mTORC1 activity, and that the phosphorylation of p70S6K significantly correlated with the levels of AMD1 (correlation coefficient R =0.81; Fig. 4c; Extended data Fig. 9e).
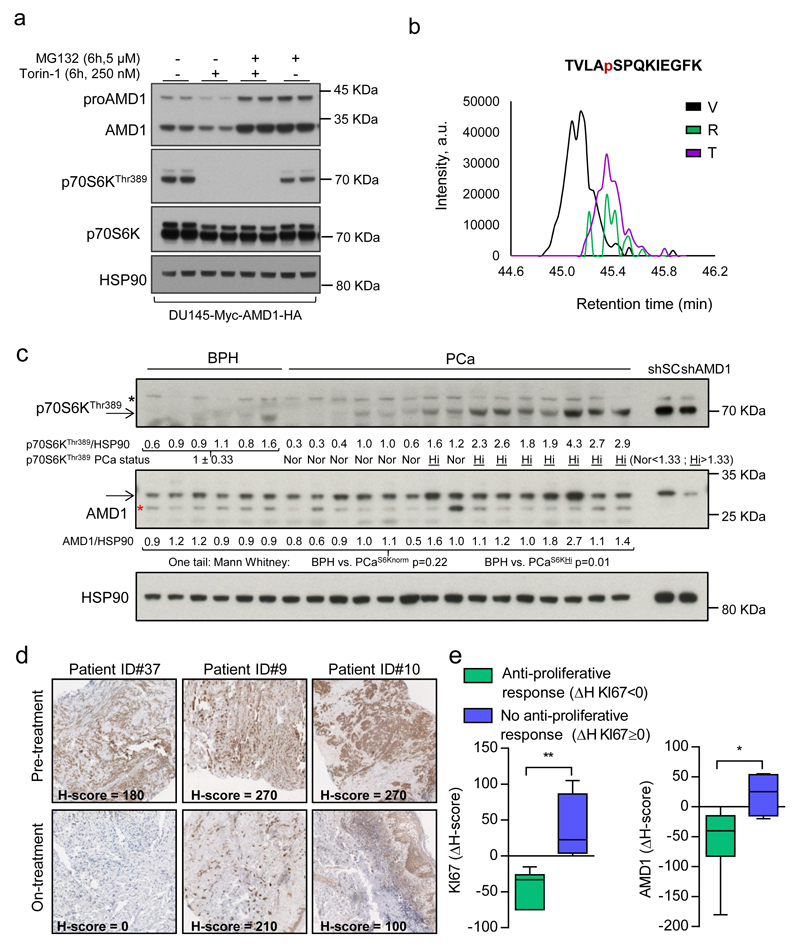
Figure 4. mTORC1 regulates AMD1 stability and this molecular regulation is recapitulated in vivo.
a, Representative western blot (out of 3) of DU145 cells expressing Myc-AMD1-HA treated with vehicle or Torin-1 (250 nM, 6h) in the presence or absence of MG132 (5 µM, 6h). b, Extracted ion chromatogram (XIC) of the TVLASPQKIEGFK phosphorylated proAMD1 peptide. c, Western blot analysis (individual tissue specimens are presented) of AMD1 and p70S6KThr389 in prostate tissue samples of BPH and PCa. Densitometry values of AMD1 and p70S6KThr389 are provided below the scans (corrected by HSP90 immunoreactivity). p70S6KThr389 PCa status was defined as normal (Nor; PCaS6K Nor) when the densitometry values of the PCa sample were lower than [mean + standard deviation] of the BPH specimens, and high (Hi, PCaS6K Hi) when the densitometry values of the PCa samples were greater than [mean + standard deviation] of the BPH specimens. The statistical analysis related to differential AMD1 immunoreactivity was performed analysing separately PCaS6K Nor (n=7 specimens) and PCaS6K Hi (n=8 specimens) vs. the BPH specimens (n=6 specimens). d, Representative AMD1 immunoreactivity images of three patient specimens before (pre-treatment) or after (on treatment) therapy with everolimus (n=14 specimen pairs). e, Box and whisker plot of the immunoreactivity of KI67 and AMD1 in cancer patients with (ΔH score for KI67 < 0) or without (ΔH score for KI67 ≥ 0) anti-proliferative tumour response upon everolimus treatment. a.u.: arbitrary units; ΔH: Differential H-Score (ΔH=[H score on-therapy] - [H score pre-therapy]); p, p-value; BPH, benign prostate hyperplasia; PCa, prostate cancer; *, p<0.05; **, p<0.01. Arrows indictate specific immunoreactive bands. Red asterisk in western blot indicates non-specific band. Statistic test: Mann Whitney test (c, e).
mTORC1 inhibitors are currently used to treat certain tumours (despite the unpredicted inefficacy in many others)27, and prior work by us contributed to define the pharmaco-dynamic properties of everolimus in individuals with advanced cancers of different origin5,28,29. Strikingly, we observed a predominant decrease in AMD1 immunoreactivity in 14 biopsies obtained from patients treated with this drug relative to a biopsy of the same lesion prior to treatment (Fig. 4d; Extended data Fig. 9f; Supplementary Table 8). When we stratified patients on the basis of the anti-proliferative response achieved after everolimus therapy (responders: differential KI67 H-score: ΔH < 0, n = 6 specimen pairs; non-responders: differential KI67 H-score: ΔH ≥ 0, n = 4 specimen pairs), we found that only AMD1 (among all targets analysed) presented significantly decreased immunoreactivity in responders (Fig. 4e; Extended data Fig. 9g).
Polyamine production is a hallmark of highly proliferating cells30, but their regulation by oncogenic signals remains largely unknown. Our results demonstrate that increased polyamine synthesis is associated to oncogenic signalling in PCa. The regulation of AMD1 production and dcSAM synthesis downstream mTORC1 described herein provides a mechanistic explanation for the control of this metabolic program (Extended data Fig. 9h). AMD1 is an unprecedented metabolic target of this protein complex and supports its role in cancer cell proliferation. Importantly, the control of dcSAM and polyamine synthesis is relevant beyond the cancer scenario, and suggests that physiological and developmental processes that require active cell proliferation might be tightly associated to the regulation of AMD1 and polyamine synthesis downstream of mTORC1.
Methods
Patient samples
All prostate specimens were obtained upon informed consent and with evaluation and approval from the corresponding ethics committee (CEIC code OHEUN11-12 and OHEUN14-14)4. A table providing clinic-pathological information is included as Supplementary Table 4. The details of the clinical trial with everolimus are described in Ref. 5 and in Supplementary Table 8.
Animals
All mouse experiments were carried out following the ethical guidelines established by the Biosafety and Animal Welfare Committee at CIC bioGUNE (under protocol P-CBG-CBBA-0715). The procedures employed were carried out following the recommendations from AAALAC. Xenograft experiments were performed as previously described (maximum total tumour volume per mouse 1.5 cm3)31, injecting 4x106 (AMD1 silencing) or 4x106 (AMD1 ectopic expression) cells with Matrigel (BD Biosciences) per condition in two flanks per mouse. GEMM experiments were carried out in a mixed background as reported32. The Ptenlox conditional knockout allele has been described elsewhere3. Prostate epithelium-specific deletion was effected by the Pb-Cre43. Mice were fasted for 6h prior to tissue harvest (9am-3pm) in order to prevent metabolic alterations due to immediate food intake. The TRAMP mice string was originally obtained from The Jackson Laboratory repository. Animals were maintained at the Animal Facility (awarded with the AAALAC accreditation) of the Spanish National Cancer Research Centre (CNIO) in accordance with the guidelines stated in the International Guiding Principles for Biomedical Research Involving Animals, developed by the Council for International Organizations of Medical Sciences (CIOMS). All animal experiments were approved by the Competent Authority of the Comunidad de Madrid. The generation and characterisation of TRAMP mice was have been previously described20. At CNIO TRAMP mice originally provided in FVB/NJ genetic background were backcrossed to a C57BL/6 background by successive mating of (T/+) male-mice to (+/+) C57BL/6 female-mice and then maintained in a C57BL/6 background.
To address the potential undesirable effects of systemic AMD1 inhibition, we administered intraperitoneally SAM486A (5mg/Kg/day, 5-days per week) for 17 days in immunocompetent C57BL/6 mice. We measured body and organ weight, blood biochemistry, haematocrit and white blood cell count (information provided in Supplementary Table 6). Terminal blood harvest was performed intracardially after CO2-inhalation based euthanasia. For non-terminal harvest, a facial vein blood sample was obtained by puncture with a sterile 4 mm lancet (MEDIpoint Inc., USA). For plasma preparation, blood was deposited in tubes with Dipotassium EDTA (Microtainer®, Becton-Dickinson, Franklin, NJ, USA) (for haematocrit and FACS analysis) or heparinized tubes (10 µL,1 U/µL) (for plasma metabolomics). For haematocrit analysis blood samples were analysed using an Abacus Junior Vet analyser (Diatron, Hungary) according to manufacturers´ guidelines. For blood biochemistry, a Selectra Junior Spinlab 100 analyser (Vital Scientific, Dieren, The Netherlands) was employed. A calibrated control was run before each use and was within established ranges prior to analysing samples. For white blood cell analysis, the spleen of SAM486A or saline-treated mice was grinded using a syringe plunger and passing the cells through a 70 µm cell strainer, and cellular composition was evaluated by flow cytometry, using the following antibodies: CD4, CD8, B220, Ly6C, F4/80, GR-1, CD25, CD11b, CD44, CD73, FR4, Nrp-1 (Miltenyi Biotec).
Purification and activation of murine splenic CD4+ T cells
In order to address the toxicity of SAM486A, we purified CD4 T cells from the spleen of C57BL/6 mice by negative selection using the CD4 purification kit following the manufacturer´s instructions (Miltenyi Biotec, Auburn, CA). 5 × 105 CD4 T cells were activated in TexMACS Medium (Miltenyi Biotec) with plate-bound anti-CD3 (5 µg/mL) and soluble anti-CD28 (1 µg/mL) in the presence of vehicle or SAM486A (1µM) for 16 and 96 h and assessed for IL-2 production by capture ELISA (R&D Systems, MAB702). To analyse the effect of SAM486A on T cell proliferation, purified CD4 T cells were recovered 4 days after activation and treatment and counted in a hemocytometer chamber.
Immunization with ovalbumin
To address the effect of SAM486A on immune cell proliferation in vivo, potential we administered intraperitoneally SAM486A (5 mg/Kg/day, 5-days per week) for 17 days in immunocompetent C57BL/6 mice, and then immunized them subcutaneously 2 weeks later with 50 µg of ovalbumin (OVA) in Complete Freund´s adjuvant and kept on treatment. After 2 weeks, the mice were analysed for ovalbumin-specific serum IgG and IgM levels by ELISA33.
Reagents
Cell lines were purchased from Leibniz-Institut DSMZ - Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH (DSMZ) and tested negative for mycoplasma. An authentication certificate was provided by DSMZ for cell lines. Rapamycin (prepared in DMSO, final concentration 20 nM), Torin-1 (prepared in DMSO, final concentration 125-250 nM), Dimethylfluorornithine (DFMO, prepared in water, final concentration 50 µM), PF-4708671 (PF47, prepared in DMSO, final concentration 10 µM) Hydroxychloroquine (HCQ, prepared in water, final concentration 30 ng/mL), MG132 (5 µM, prepared in DMSO), PD0325901 (100 nM, prepared in DMSO), SB203580 (5 µM, prepared in DMSO), SP600125 (10 µM, prepared in DMSO), Spermidine (Spd, 0.5-1 µM, prepared in water), 5'-Deoxy-5'-(methylthio)adenosine (MTA) and cycloheximide (CHX, prepared in ethanol, final concentration 5 µg/mL) were purchased from LC laboratories (Rapamycin), Sigma (CHX, PF47, HCQ, Spd, MTA), Calbiochem (SB203580) and Tocris (Torin-1, DFMO, SP600125). RAD001 was purchased from Selleckchem and was administered 6-days per week by oral gavage (prepared in 1.5% NMP–98.5% PEG) at 10mg/Kg. SAM486A was kindly provided by Novartis and prepared in water (in vitro) or saline solution (in vivo, 5 mg/Kg/day intraperitoneally Monday-Friday). U-13C5-L-Methionine was purchased from Cambridge Isotope laboratories and administered intravenously at a final concentration of 100 mg/Kg in vivo and at 30 µg/mL in vitro (with dialyzed FBS). Doxycycline was purchased from Sigma and used at 500 ng/mL for over-expression of YFP-PTEN, 100 ng/mL for silencing of AMD1 and 250 ng/mL for silencing of RAPTOR and TSC2. shRNAs against AMD1 were purchased from Sigma (TRCN0000078462: sh3; TRCN0000078460: sh4, TRCN0000078461: sh5) and control shRNA sequence is included (CCGGCAACAAGATGAAGAGCACCAACTCGAGTTGGTGCTCTTCATCTTGTTG)34. shRNAs against 4EBP1 and 2 were kindly provided by Dr. Nahum Sonenberg35. Sub-cloning of shRNA AMD1 into pLKO-Tet-On vector was done introducing AgeI and EcoRI in the 5´end of top and bottom shRNA oligos respectively (TET-pLKO puro was a gift from Dr. Dmitri Wiederschain36, Addgene plasmid # 21915). Myc-AMD1-HA expressing vector was generated starting from the ORF obtained from PlasmID Harvard (https://plasmid.med.harvard.edu; HsCD00321977) and cloned into a modified retroviral pLNCX vector that harbours BglII-SalI sites (cloned with BamHI-SalI). RNAi-resistant versions of AMD1 were generated using overlap extension PCR and cloned into a lentiviral backbone derived from vector pLenti-Cas9-blast (Cas9 removed; lentiCas9-Blast was a gift from Feng Zhang, Addgene #52962; Ref. 37) using HiFi Assembly Kit (NEB). The resulting vectors express AMD1-HA-2A-blast (WT or S229A) with AMD1-HA portion being excisable using BshT1-BamH1. The target of AMD1 shRNA#3 (5’-gtctccaagagacgtttcatt-3’) was changed to an RNAi-resistant version (5’-gtGAGcaaACGTAGAttTatCtt-3’). Cloning details are available upon request. All clones were sequence-validated. Site-directed mutagenesis for generation of AMD1S229A and AMD1S298A was performed using Agilent QuikChange II Site-Directed Mutagenesis Kit. YFP-PTEN-expressing lentiviral constructs were described in Ref.19.
Cellular and molecular assays
Cell number quantification was done with crystal violet29. Western blot was performed as previously described38, run in Nupage gradient precast gels (Life Technologies) in MOPS or MES buffer (depending on the proteins analysed; please note that the migration pattern of molecular weight markers varies in these two buffers). Anti-AMD1 was from Proteintech (11052-1-AP). Anti-RpS6S240/244, anti-RpS6, anti-S6KT389, anti-S6K, anti-LC3B, anti-HSP90, anti-PTEN, anti-AKTS473, anti-AKT, anti-4EBP1 and anti-RAPTOR antibodies were from Cell Signalling Technologies. Anti-β-actin antibody was from Sigma and anti-TSC2 from Thermo Scientific (MA5-15004). Densitometry-based quantification was carried out using ImageJ software. For half-life assays, DU145 cells stably expressing the indicated constructs were challenged with CHX (5 µg/mL) and protein was extracted at the indicated time points (cells were treated with vehicle – DMSO, MG132 – 5 µM, or Torin-1 – 250 nM – 120 minutes prior to CHX challenges when indicated). Anchorage independent growth assays were performed as previously described39, seeding 3000 (PC3) or 5000 (DU145) cells/well. RNA was extracted using NucleoSpin® RNA isolation kit from Macherey-Nagel (ref: 740955.240C). 1 μg of total RNA was used for cDNA synthesis using qScript cDNA Supermix from Quanta (ref. 95048). Quantitative Real Time PCR (q-RTPCR) was performed as previously described38. Applied biosystems TaqMan probes: Amd1/AMD1 (Mm04207265, Hs00750876s1), β-ACTIN/β-Actin (Hs99999903_m1/Mm00607939_s1) and GAPDH/Gapdh (Hs02758991_g1/Mm99999915_g1). Universal Probe Library (UPL, Roche) probes: AMD1 (Probe 72, primer F: cagacctcctatgatgacctga; primer R: tcaggtcacgaattccactct), Odc1 (Probe 80, primer F: gctaagtcgaccttgtgagga; primer R: agctgctcatggttctcgat), ODC1 (Probe 34, primer F: aaaacatgggcgcttacact; primer R: tggaattgctgcatgagttg) and Mtap (Probe 12, primer F: ccatggcaaccgactatgat; primer R: aaaccccatccactgacact). Foci assays were performed seeding 500 cells per well (6-well plate) and staining and counting them by crystal violet after 10 days. Lentiviral and retroviral transductions were performed as previously described34,38.
Kinase assay
Human AMD1 variants (C-terminal HA tag, non-processing mutant S229A; S298 (WT) or S298A; details available upon request) were prepared by overlapping PCR and cloned as BamH1-Not1 into pGEX-6P-1 (GE Healthcare). Sequence-confirmed clones were induced with 1mM IPTG for 16hrs at 20°C in C41 (DE3) pLysS (Lucigen). GST fusion proteins were purified first by glutathione affinity chromatography (eluted in 40mM reduced glutathione; 25mM Hepes pH 8; 50mM KCl; 0,1% BME buffer) and then were separated by a gel filtration chromatography. Proteins were concentrated by ultrafiltration (Vivaspin 5K MWCO cutoff; Sartorius) and used for kinase assays.
Endogenous mTORC1 complex was immunoprecipitated from HEK293 cells using anti-Raptor antibody (S682B, 4th bleed, https://mrcppureagents.dundee.ac.uk/) coupled to Protein G Sepharose beads (Amersham). The cells were stimulated with IGF (50 ng/ml) for 20 min prior to lysis in mTORC1 lysis buffer (40 mM HEPES pH 7.4, 120 mM NaCl, 1 mM EDTA, 0.3% (w/v), CHAPS, 10 mM Na-pyrophosphate, 10 mM Na-glycerophosphate, 1 mM Na-orthovanadate, protease inhibitor cocktail (Roche)). The immunoprecipitate was washed twice with mTORC1 lysis buffer, containing 0.5 M NaCl, twice with mTORC1 lysis buffer and twice with mTORC1 kinase assay buffer (25 mM HEPES pH 7.4, 50 mM KCl). The substrates were added to immunoprecipitate in kinase assay buffer (15 μl) before adding the 10 μl of the ATP mixture (10 mM MnCl2, 100 μM ATP, 1 μCi 32P γ-ATP in kinase buffer). The reaction was carried out in thermomixer at 30ºC, for 30min and was terminated by adding the 4x sample buffer (NuPAGE LDS sample buffer, Life Technologies). The reaction mixture was loaded on gel. Dried gel was exposed to X-ray films (Amersham). 1 μL of reaction mixture was loaded on gel for immunoblot analysis. GST-S6KD236A (DU32609, https://mrcppureagents.dundee.ac.uk/) was purified from HEK293 cells pretreated with 0.1 μM AZD-8055. The protein was purified using GST-Sepharose beads (Amersham) according to manufacturer’s instructions. Anti-Raptor (S682B, 4th bleed https://mrcppureagents.dundee.ac.uk/), anti-phospho-S6K1 Thr389 (#9205, Cell Signaling Technologies), anti-GST (S902A, 3rd bleed, https://mrcppureagents.dundee.ac.uk/).
Immunohistochemical analysis
Histochemical analysis by Haematoxylin-Eosin, anti-RpS6S235/6, anti-AktS473 (Cell Signalling Technologies) and Pten (51-2400) immunostaining was performed as previously described40,41. Immunohistochemical analysis of AMD1 (Proteintech, dilution 1/100) was performed using DAKO envision Flex High pH (DAKO). The scoring system was based on the quantification of the % of cells negative, low (1+), medium (2+) or high (3+) immunoreactivity. Subsequently, H-score was calculated as follows: H = [percentage of cells 1+] + [2 x (percentage of cells 2+)] + [3 x (percentage of cells 3+)]. Differential H score was calculated as follows: ΔH = H[on treatment] – H[pre-treatment].
Metabolomic analysis
For in vitro metabolomic analysis, growing cells were washed with ammonium carbonate pH 7.4 and snap-frozen in liquid nitrogen. Metabolites were extracted from cells or tissues with cold 80/20 (v/v) methanol/water. Samples were then dried and stored at -80°C until MS analysis. High-throughput Time-Of-Flight analysis was conducted using flow injection analysis as previously described42. In short, samples were re-suspended and injected on an Agilent 1100 coupled with an Agilent 6520 QToF mass spectrometer with an electrospray ionization source. Mobile phase consisted of 60/40 methanol/water with 0.1% formic acid and was used to deliver 2 µL of each sample to the MS, flowing at 150 µL/min. Data was collected in positive mode with 4 GHz HiRes resolving power with internal lock masses. Data processing was conducted with Matlab R2010b. Relative cell number or protein amount was used for normalization.
Quantitative LC/MS was conducted as previously described43. A Thermo Accela 1250 pump delivered a gradient of 0.025% heptafluorobutyric acid, 0.1% formic acid in water and acetonitrile at 400µL/min. Stationary phase was an Atlantis T3, 3 µm, 2.1x150 mm column. A QExactive Mass Spectrometer was used at 70.000 resolving power to acquire data in full-scan mode. Data analysis was conducted in MAVEN44 and Spotfire. Peak areas derived from stable isotope labelling experiments have been corrected for naturally occurring isotope abundance.
For plasma 13C-methionine analysis, blood samples from mice were extracted at indicated times, transferred at room temperature to heparinized collection tubes and centrifuged at 13,000 rpm 4ºC during 10 minutes. Plasma was transferred to fresh tubes and processed for UPLC-MS analysis. Briefly, to 40 µL aliquots of mouse plasma, 40 µL of water/0.15% FA was added. Subsequently, proteins were precipitated by addition of 120 µL of acetonitrile. In order to get optimum extraction, after addition of acetonitrile, the samples were sonicated for 10 minutes at 4ºC and agitated at 1,400 rpm for 30 min at 4ºC. Next, the samples were centrifuged at 14,000 rpm for 30 min at 4ºC. The supernatant was transferred to a fresh vial and measured with an UPLC system (Acquity, Waters, Manchester) coupled to a Time of Flight mass spectrometer (ToF MS, SYNAPT G2, Waters). A 2.1 x 100 mm, 1.7 µm BEH AMIDE column (Waters), thermostated at 40 °C. Solvent A (aqueous phase) consisted of 99.5% water, 0.5% formic acid and 20 mM ammonium formate while solvent B (organic phase) consisted of 29.5% water, 70% MeCN, 0.5% formic acid and 1 mM Ammonium formate. In order to obtain a good separation of the analytes the following gradient was used: from 5% A to 50% A in 2.4 minutes in curved gradient ( #8, as defined by Waters), from 50% A to 99.9% A in 0.2 minutes constant at 99.9% A for 1.2 minutes, back to 5% A in 0.2 minutes. The flow rate was 0.250 mL/min and the injection volume was 2 µL. All samples were injected randomly and analytes were measured in enhanced duty cycle (EDC) mode, optimized for the mass of the analyte in question. Methionine and 13C5-methionine was measured in scan function 1 (EDC at 152), SAH and 13C4-SAH was measured in scan function 2 (EDC at 387), SAM and 13C5-SAM was measured in scan function 3 (EDC at 402). Extracted ion traces were obtained for methionine (m/z = 150.0589), 13C5-methionine nine (m/z = 155.0756), SAH (m/z = 385.1294), 13C4-SAH (m/z =389.1428), SAM (m/z =399.145) and 13C5-SAM (m/z =404.1618) in a 20 mDa window and subsequently smoothed (2 points, 2 iterations) and integrated with QuanLynx software (Waters, Manchester). For quantitation, stock solutions of 10 mM in water for each of the analytes were prepared. Stock solutions were pooled and diluted in order to obtain a mixture including all analytes. The mixture was further diluted in water in order to obtain the concentrations as used in the calibration curve. The calibration range for all analytes included the following concentrations: 100, 50, 25, 10, 5, 2.5, 1, 0.5, 0.25, 0.1, 0.05 and 0.025 µM.
Targeted metabolomics
Levels of dcSAM in cell cultures and tissues were analysed by ultra-high performance liquid-chromatography coupled to mass spectrometry (UPLC-MS). Briefly, extraction and homogenization was done in methanol/acetic acid (80/20 %v/v) Speed-vacuum-dried metabolites were solubilized in 100 µl of a mixture of water/acetonitrile (40/60 %v/v) and injected onto the UPLC/MS system (Acquity and SYNAPT G2, Waters, Manchester). The extracted ion traces were obtained for dcSAM (RT = 3.0’, m/z 355.16). Corrected signals were normalized to relative cell number.
Polysome profiling
Distribution of mRNAs across sucrose gradients was performed as described earlier45, except for minor modifications.
Phosphoproteomic analysis of AMD1
DU145 cells stably expressing Myc-AMD1-HA were plated in 2-3 150 mm plates per condition to ensure a final density no higher than 50-60% and sufficient protein amount to immunoprecipitate ectopic AMD1 (using agarose HA-beads, Sigma, according to manufacturers´ instructions). Cells were treated for 6 hours with rapamycin (20 nM) and Torin-1 (250 nM) prior to immunoprecipitation. Protein eluates from the immunoprecipitated samples were separated by SDS-PAGE. The bands corresponding to AMD1 and the proenzyme were visualized using Sypro-Ruby (Invitrogen), excised and in-gel digested with trypsin. The resulting peptides were analyzed by LC-MS/MS using a LTQ Orbitrap Velos mass spectrometer (Thermo Scientific). Raw files were searched against a Uniprot Homo sapiens database (20,187 sequences) using Sequest-HT as the search engine through the Proteome Discoverer 1.4 (Thermo Scientific) software. Peptide identifications were filtered by Percolator at 1% FDR using the target-decoy strategy. Label-free quantification was performed with MaxQuant and extracted ion chromatograms for AMD1 phosphopeptides were manually validated in Xcalibur 2.2 (Thermo).
Bioinformatic analysis
All the datasets used for the data mining analysis46–49 were downloaded from GEO, and subjected to background correction, log2 transformation and quartile normalization. In the case of using a pre-processed dataset, this normalization was reviewed and corrected if required.
Statistical analysis
No statistics were applied to determine sample size. The experiments were not randomized. The investigators were not blinded to allocation during experiments and outcome assessment. Data analysed by parametric tests are represented by the mean ± s.e.m. of pooled experiments and for non-parametric tests median with interquartile range is depicted, unless otherwise stated. n values represent the number of independent experiments performed or the number of individual mice or patient specimens. For each independent in vitro experiment, at least three technical replicates were used and a minimum number of three experiments were performed to ensure adequate statistical power. ANOVA test was used for multi-component comparisons and Student's t-test for two component comparisons. In the in vitro experiments, normal distribution was confirmed or assumed (for n<5) Two-tailed statistical analysis was applied for experimental design without predicted result, and one-tail for validation or hypothesis-driven experiments. The confidence level used for all the statistical analyses was of 0.95 (alpha value=0.05).
Extended Data
Extended data Figure 1. Metabolomics characterization of murine and human prostate cancer.
a, Incidence of pathological alterations observed in Ptenpc+/+ and Ptenpc-/- mice. Number of mice as indicated. b, Representative immunohistochemical images of prostate tissue stained with Haematoxylin-Eosin (H&E), Pten, AktS473 and RpS6S235/6 from mice at three and six months of age (representative from 3 mice per condition). c, Experimental design of the Time-of-flight mass spectrometry (TOF-MS) metabolomics analysis. AP, anterior prostate; DLP, dorsolateral prostate. d-e, Volcano plot (d) and principal component analysis (PCA, e) from altered metabolites in TOF-MS metabolomic analysis carried out in Ptenpc-/- and Ptenpc+/+ mouse prostate samples at the indicated age (6 months Ptenpc+/+ AP n=4 mice; rest of conditions n=5 mice). Grey dots: not significantly altered; red dots: significantly increased in Ptenpc-/- prostate extracts; blue dots: significantly decreased in Ptenpc-/- prostate extracts. f, Liquid chromatography/mass spectrometry (LC/MS) analysis of methionine cycle and polyamine pathway metabolites from Ptenpc-/- vs. Ptenpc+/+ mouse prostate samples at the indicated age (AP, 3 months n=5 mice; 6 months n=4 mice). Data are represented as median with interquartile range. g, LC/MS analysis of methionine cycle and polyamine pathway metabolites from PCa vs. BPH human specimens (6 prostate specimens per condition). Data are represented as median with interquartile range. p, p-value; *, p<0.05; **, p<0.01; ***; PIN: Prostatic Intraepithelial Neoplasia; LG-PIN: Low Grade PIN; HG-PIN: High Grade PIN; Focal Adc: Focal adenocarcinoma; BPH, benign prostate hyperplasia; PCa: Prostate Cancer; dcSAM: decarboxylated S-adenosylmethionine; SAM: S-adenosylmethionine; SAH: S-adenosylhomocysteine; MTA: 5´ Methylthioadenosine. Statistical analysis: One tail Mann-Whitney U test (f, g) was used for data analysis.
Extended data Figure 2. Metabolic tracing of 13C-Methionine in Pten-prostate specific knockout mice.
a, Plasma liquid chromatography/mass spectrometry (LC/MS) analysis of the indicated metabolite concentration after intravenous injection of U-13C5-L-Methionine (100mg/Kg) in C57BL/6 mice at 3 months of age (Time 0 min n=7 mice; Time 10 min/60 min n=6 mice). The unlabelled (M+0, 12C) and major labelled (13C, M+4 or M+5) metabolite concentration is presented in the histogram. Error values depict s.e.m. b, Experimental design of the U-13C5-L-Methionine (100mg/Kg) in vivo. c, Summary schematic of the alterations observed in the metabolomic analysis in Ptenpc-/-. d, Fractional labeling of the indicated metabolites from Fig. 1e. Values are represented as median with interquartile range (1 hour –upper panel - n=4; 10 hour – lower panel - n=3). p, p-value; Spm: Spermine; dcSAM: decarboxylated S-Adenosylmethionine; Spd: Spermidine; Met: Methionine; SAM: S-Adenosylmethionine; SAH: S-Adenosylhomocysteine; MTA: 5´ Methylthioadenosine; ODC1: Ornithine Decarboxylase 1; SpdS: Spermidine Synthase; SpmS: Spermine Synthase; U-13C5-Met: L-Methionine labeled with13C in five carbons; 1h: prostate samples extracted after 1h pulse with U-13C5-L-Methionine; 10h: prostate samples extracted after 10 hour pulse with U-13C5-L-Methionine; FC: fold change; a.u.: arbitrary units.One tail Mann-Whitney U test (d) was used for data analysis.
Extended data Figure 3. Genetic modulation of AMD1 in prostate cancer cells.
a, Technical setup of anti-AMD1 antibody using the indicated constructs or shRNAs in DU145 cells. AMD1S229A mutant lacks processing ability and is expressed exclusively as a pro-enzyme (representative western blot out of 3 independent experiments). b, Schematic representation of tagged ectopic AMD1 processing. c, Impact of ectopic Myc-AMD1-HA expression on foci number in DU145 cells in vitro (n=3 independent experiments). d-f, Impact of ectopic Myc-AMD1-HA expression on tumour volume (d, n=8 tumours per condition), AMD1 protein levels (e, n=3 tumours per condition) and dcSAM abundance (f, n=5 tumours per condition) in DU145 xenografts grown for 43 days. Data in d is represented as box and whisker plot. Panel f shows a dot plot with the median and the interquartile range. g-o, Effect of constitutive silencing of AMD1 (g, i, mRNA levels in g-i and protein expression in h-j) on cell number (k, n), anchorage-independent growth (l, o) and dcSAM abundance (m) with two different hairpins in DU145 (g, h, k, l, m) and PC3 (i, j, n, o) cells (n=3-4 independent experiments as indicated by dots). Values are represented as mean with s.e.m. p, Effect of doxycycline-inducible (100 ng/mL) AMD1 silencing on cell number in DU145 cells (sh3 n=3; sh4-5 n=4 independent experiments as indicated by dots). q-s, Impact of AMD1 inducible silencing in tumor volume from DU145 xenografts (Tumours analysed: sh3 No Dox, n=12; sh3 Dox, n=14; sh4 No Dox, n=10; sh4 Dox, n=7; sh5 No Dox, n=10; sh5 Dox, n=11). Values are represented as box and whisker plot. t, Impact of AMD1 inducible silencing in dcSAM abundance in DU145 xenografts from panels q-s (n=5 tumours). Values are represented as median with interquartile range. u-v, Dose-dependent effect of doxycycline on cell number in DU145 (u) and PC3 (v) cells (cell number measured at day 6) (n=3 independent experiments as indicated by dots). shSC: scramble short hairpin; sh3 and sh4: two different short-hairpins targeting AMD1; dox: doxycycline; p, p-value; *, p<0.05; **, p<0.01; ***, p<0.001. Red asterisk in western blot indicates non-specific band. Dashed line indicates cell number in shC-transduced cells. Statistic test: one tail T-test (c, g, i, k-p, u-v), and one-tail Mann Whitney U test (d, f, q-t).
Extended data Figure 4. Genetic and pharmacological manipulation of AMD1 in prostate cancer cells.
a-c, DU145 cells carrying doxycycline-inducible shRNA against AMD1 (sh3) were transduced with empty (Mock), sh3-resistant wildtype (AMD1sh3R) or processing-deficient (AMD1S229A,sh3R) AMD1 constructs. AMD1 protein (a, representative experiment out of 4), dcSAM abundance (b) and cell number expression (c) in the aforementioned cells (n=4 independent experiments as indicated by dots). Asterisks indicate significant differences compared to the corresponding DU145 cells in the absence of doxycycline, and hash indicates significant differences in the indicated comparison. Values are represented as mean with s.e.m. d, Effect of 5´ Methylthioadenosine (MTA, 25 µM) on AMD1 silencing (sh3)-elicited antiproliferative activity. MTA was administered at day 0 and cells were analyzed at day 3 (n=3 independent experiments as indicated by dots). e, Mtap gene expression levels in Ptenpc+/+and Ptenpc-/- mice at the indicated time points (see Extended data Fig. 1a) (3 months n=3 mice; 6 months n=6 mice). Values are represented as median with interquartile range. f, MTAP gene expression analysis in publicly available datasets (see methods; N: Normal, PCa: Prostate cancer; Number of patients analised: Taylor et al., Normal n=29, PCa n=150; Grasso et al., Normal n=12, PCa n=76; Lapointe et al., Normal n=9, PCa n=17). g-h, Effect of pharmacological AMD1 inhibition with SAM486A on cell number (g and left h panels ; DU145 n=5; PC3 n=4 independent experiments as indicated by dots), and anchorage-independent growth (righ h panel, n=3 independent experiments as indicated by dots) in PC3 or DU145 cells as indicated. Data is represented as mean with s.e.m. i, Effect of pharmacological AMD1 inhibition with SAM486A in established DU145 xenograft tumor volume (Vehicle n=11 tumours, SAM486A n=10 tumours). Values are represented as box and whisker plot. j-k, Effect of pharmacological AMD1 inhibition with SAM486A in activated T CD4 cell number (96h, j, n=3 independent experiments as indicated by dots) or IL-2 production (k, n=3-6 independent experiments as indicated by dots). a.u.: arbitrary units, Dox: doxycycline,p, p-value; */#, p<0.05; **/##, p<0.01; ***, p<0.001. Dashed line indicates cell number (g, h) or IL-2 abundance (k) in vehicle-treated cells. Statistic test:Student T-test (b, c, d, f,g, h, j, k) and one tail Mann Whitney test (e, i).
Extended data Figure 5. PTEN-PI3K-mTORC1-dependent regulation of AMD1 in prostate cancer.
a-b, Amd1 protein quantification from Fig. 3a (a, n=3 mice) and mRNA expression (b, n=5-6 mice as indicated by dots) in Ptenpc-/- and Ptenpc+/+ mice of the indicated age. Data are represented as median with interquartile range. c, AMD1 gene expression analysis in publicly available datasets (see methods; N: Normal, PCa: Prostate cancer; Number of patients analysed: Taylor et al., Normal n=29, PCa n=150; Grasso et al., Normal n=12, PCa n=76; Lapointe et al., Normal n=9, PCa n=17). d, Representative western blot showing the expression of the indicated proteins upon doxycycline-inducible expression (24h) of YFP-PTENWT or catallitically inactive YFP-PTENC124S in PTEN-deficient LNCaP prostate cancer cells (n=3 independent experiments). e, Schematic representation of the PI3K signaling pathway and the pharmacological/genetic tools employed in this study. f, ProAMD1 and AMD1 protein quantification from Fig. 3b (sample number as indicated by dots). g, AMD1 gene expression upon treatment (24h) with vehicle (V, DMSO), Rapamycin (R, 20nM) and Torin-1 (T, 250nM for PC3 and DU145, 125nM for LNCaP) (LNCaP n=8 for T; R and n=6 for PC3 and DU145 n=6 independent experiments as indicated by dots). Values are represented as mean with s.e.m. h, Representative western blot analysis of AMD1 levels upon 24-hour treatment of DU145 cells with vehicle (V), Torin-1 (mTORC1/2 inhibitor, T; 250 nM), PD032901 (ERK-MAPK inhibitor, PD; 100 nM), SP600125 (JNK-SAPK inhibitor, SP; 10 µM) and SB203580 (p38-MAPK inhibitor, SB; 5 µM) (n=3 independent experiments). i, Impact of inducible RAPTOR silencing in DU145 cells on proAMD1 protein levels (doxycycline-induced, 250 ng/mL) (Representative experiment out of n=6). j, Impact of inducible TSC2 silencing in DU145 cells on proAMD1 protein levels (doxycycline-induced, 250 ng/mL) (Representative experiment out of n=6). a.u.: arbitrary units, Dox: doxycycline, N: Normal, PCa: Prostate cancer; p, p-value; *, p<0.05; **, p<0.01; ***, p<0.001. Red asterisk in western blot indicates non-specific band. Arrows indicate specific immunoreactive bands. Statistic test: Student T-test (c, f, g) and Mann Whitney test (a, b).
Extended data Figure 6. mTORC1-dependent AMD1 regulation in vivo.
a, Effect of spermidine (Spd, 0.75 µM) on PC3 cell number upon rapamycin treatment (1 nM, outcome measured 72h after treatment). Drugs were administered at day 0 and only Spd was additionally administered at day 1 (n=4 independent experiments as indicated by dots). b,Effect of RAD001 treatment on prostate pathological features and mTORC1 activity (Haematoxylin-Eosin - H&E - and RpS6S235/6 staining by IHC) (n=3 mice). c, ProAmd1 and Amd1 protein abundance quantification from Fig. 3e (right panel; number of mice as indicated by dots). d, Representative immunohistochemical images of prostate tissue from wildtype or TRAMP mice (+/T, 28-32 weeks old) stained with H&E (top panels) and RpS6S235/6 (lower panels, Ptenpc+/+ and Ptenpc-/- prostate tissues are presented as comparison of the RpS6 phosphorylation levels) (n=3 mice). e, Evaluation of AMD1 expression by western blot in prostate tissues from wildtype or TRAMP mice (+/T, 28-32 weeks old) (n=4 mice). a.u.: arbitrary units, p, p-value; *, p<0.05; **, p<0.01. Arrows indicate specific immunoreactive bands. Statistic test: Student T-test (a) and Mann Whitney test (c, e).
Extended data Figure 7. Contribution of mTORC1 effector pathways and targets on the regulation of AMD1.
a, ODC1 gene expression upon treatment (24h) of Vehicle (V, DMSO), Rapamycin (R, 20 nM) and Torin-1 (T, 250 nM for PC3 and DU145, 125 nM for LNCaP) in PC3, LNCaP and DU145 cells (n=5 independent experiments as indicated by dots).Values are represented as mean with s.e.m. b, Putrescine abundance upon treatment (24h) of Vehicle (V, DMSO), Rapamycin (R, 20 nM) and Torin-1 (T, 250 nM) in DU145 and PC3 cells (n=3 independent experiments as indicated by dots). Values are represented as mean with s.e.m. c, Odc1 gene expression in 3-month and 6-month old Ptenpc+/+ and Ptenpc-/- mice (n=3-6 as indicated by dots). Values are represented as median with interquartile range. d, Putrescine abundance in 12 week-old Ptenpc-/- mice upon treatment with vehicle (V) or RAD001 (10 mg/kg, 6-days per week) for four weeks (n=5 mice). Values are represented as mean with s.e.m. e, Representative western blot (n=3 independent experiments) depicting the changes in expression of the indicated proteins upon 24 hour treatment of DU145 cells with Rapamycin (R, 20 nM) and/or DFMO (an inhibitor of ODC1, 50 µM) with the corresponding vehicles. f, Representative westenblot showing LC3 lipidation in Hydroxycloroquine (HCQ)-treated (6h) DU145 and PC3 cells, as a readout of macro-autophagy (n=3 independent experiments). Arrow indicates LC3-II. g, For the analysis of translation initiation, polysome profiling analysis of AMD1 and L11 as positive control in DU145 cells treated with vehicle or Rapamycin (20 nM, 8h) is shown. Error bars depicts standard deviation from technical replicates. h, Effect of pharmacological p70S6K inhibition with PF4708671 (10 µM) in DU145 cells on AMD1 protein expression (Representative experiment out of 5). i, Effect of 4EBP1/2 silencing on proAMD1 and AMD1 protein expression (upper panel) (Representative experiment out of 3). Lower panels show 4EBP1 (n=5 independent experiments) and 4EBP2 (n=4 independent experiments) gene expression in shRNA-transduced DU145 cells. a.u.: arbitrary units; DFMO, Difluoromethylornithine; Rapa, Rapamycin; HCQ, Hydroxychloroquine; p, p-value; *, p<0.05; **, p<0.01; ***, p<0.001. Red asterisk in western blot indicates non-specific band. Arrows indicate specific immunoreactive bands. Statistic test: Student T-test (a, b, i) and Mann Whitney U test (c, d).
Extended data Figure 8. Regulation of proAMD1 stability by mTORC1.
a, ProAmd1 (left) and Amd1 (right) protein abundance quantification from Fig. 4a (n=3 independent experiments as indicated by dots). b, Representative western blot of DU145 cells expressing Ser-229-Ala (S229A) mutant Myc-AMD1-HA treated with vehicle or Torin-1 (250 nM, 6h) in the presence or absence of MG132 (5 µM, 6h) (n=3 independent experiments). Quantification is provided in right panel. c, Representative MS/MS spectrum of the TVLASPQKIEGFK peptide in proAMD1 and AMD1, in which phosphorylation was unambiguously assigned to S298 residue. d, Calculated areas under the curves from extracted ion chromatogram (XIC) of the TVLASPQKIEGFK peptide for phosphorylated (highlighted in red in the sequence, left panel) and total (right panel) proAMD1. e, Calculated areas under the curves from extracted ion chromatogram (XIC) of the TVLASPQKIEGFK peptide for phosphorylated (highlighted in red in the sequence, middle panel) and total (right panel) AMD1. f, Schematic representation of the working hypothesis of proAMD1 regulation by mTORC1-dependent phosphorylation. g, Effect of Torin (250 nM) on proAMD1 and AMD1 protein at different time points in Myc-AMD1-HA-expressing DU145 cells (representative western blot out of 3 independent experiments, right panel presents the densitometric quantification). Error bars depict s.e.m. h-i, Representative western blot depicting the stability of ectopic proAMD1 and AMD1 in DU145 cells challenged with vehicle (V) or Torin-1 (T, 250 nM, 2h) upon CHX treatment (densitometry of proAMD1 and AMD1 levels is represented in right panels, h) and half-life reduction in proAMD1 (i) (n=4 independent experiments). j-k, Representative western blot depicting the stability of S229A mutant AMD1 construct in DU145 cells challenged with vehicle (V) or Torin-1 (T, 250 nM, 2h) upon CHX treatment (densitometry of proAMD1 is represented in right panels, j) and half-life reduction in proAMD1 (k) (n=3 independent experiments). Data are represented as mean with s.e.m. a.u.: arbitrary units; CHX, Cycloheximide; n.s.: not significant p, p-value; *, p<0.05; **, p<0.01; ***, p<0.001. Statistic test: Student T-test.
Extended data Figure 9. Contribution of proAMD1 phosphorylation downstream mTORC1 to the stability of the enzyme.
a-b, Representative western blot depicting the stability of WT and S298A phospho-mutant AMD1 constructs in DU145 cells upon cycloheximide treatment (densitometry of proAMD1 and AMD1 is represented in right panels, a) and half-life reduction in proAMD1 (b) (n=3 independent experiments). Data are represented as mean with s.e.m. c, Representative western blot depicting the stability of S298A mutant AMD1 construct in DU145 cells challenged with vehicle (V) or MG132 (5 µM) upon CHX treatment (densitometry of proAMD1 is represented in right panel, n=3 independent experiments). Data are represented as mean with s.e.m. d, mTORC1 kinase activity (by means of RAPTOR immunoprecipitation from HEK293 cells) on GST-proAMD1S229A or GST-proAMD1S229A/S298A, using bacteria-purified proteins. GST-p70S6K is presented as positive control. AZD8055 is employed as control of mTORC1 inhibition. e, Correlation analysis between p70S6KThr389 and AMD1 densitometry values in PCa specimens from Fig. 4c (n=15 patient specimens). f, Quantification of AMD1 immunoreactivity in response to everolimus in tumor biopsies, based on the ΔH score (n=14 specimen pairs). g, Box and whisker plot of the immunoreactivity of mTOR downstream effectors (AKTS473, RpS6S240/244, 4EBP1/2T70, eIF4GS1108) in cancer patients with (ΔH score for KI67 < 0; n=6 specimen pairs) or without (ΔH score for KI67 ≥ 0; n=4 specimen pairs) anti-proliferative tumour response upon everolimus treatment. h, Schematic representation of the main findings of this study. Met, methionine; SAM, S-Adenosylmethionine; dcSAM, decarboxylated S-Adenosylmethionine; Orn, ornithine; Put, putrescine; Spd, spermidine; Spm, spermine; AMD1, S-Adenosylmethionine decarboxylase 1; ODC1, ornithine decarboxylase 1; PIP2, Phosphatidyl Inositol biphospate; PIP3, Phosphatidyl Inositol triphospate; a.u.: arbitrary units; ΔH: Differential H-Score (ΔH = H[on treatment] – H[pre-treatment]); p, p-value; CHX, Cycloheximide; *, p<0.05; **, p<0.01; n.s.: not significant. Arrows indicate specific immunoreactive/autoradiographic bands. Statistic test: one tail Student T-test (a, b, c), two-tail Mann Whitney test (g) and Spearman correlation analysis (e).
Supplementary Material
Acknowledgements
Apologies to those whose related publications were not cited due to space limitations. Thanks to Dr. Nahum Sonenberg for providing 4EBP1 and 4EBP2-targeting shRNAs, Dr. Dario Alessi for critical discussions and technical advice, and Novartis for providing SAM486A. Funding: Ramón y Cajal award (To AC, AE, JM, DO), Juan de la Cierva (To EC, AC-M), BFU grant (To RB: BFU2014-52282-P and BFU2011-25986), SAF grant (To AC: SAF2016-79381-R,FEDER/EU; MLM-C:SAF2014-54658-R; JMF-P: SAF2015-66312; JMM: SAF 2014-52097R; AE: SAF2015-67538-R; JA: SAF2015-65327R; GT: SAF2011-24967) from MINECO; European Union (To AC: ERC-StG-336343, PoC754627; AC-M: CIG 660191; JA: 602272; AE: ERC-2014-STG-638891). Basque Government of Health (To VT: 2016111109; JMF-P: 2015111149), education (To AC: PI2012/03 and IKERTALDE IT1106-16; RB: PI2012/42; MLM-C: 2013) and PhD grants (To A.A-A and L.V-J). AECC (To VT: 2016 JP Bizkaia; NM-M: 2011 JP Bizkaia; MLM-C). ISCIII (To AC: PI10/01484, PI13/00031; JM: Proteored PRB2 and grant PT13/0001; RF: PI15/209; VS: PI13/01714, CP14/00228). Ramón Areces foundation (To JMF-P). Basque Department of Industry, Tourism and Trade – Etortek (To AC). FERO foundation (To AC, VS). Fundación Vasca de Innovación e Investigación Sanitarias, BIOEF (To VT: BIO15/CA/052). BBVA foundation (To AC; PN team). NIH (To CCC and MC: P01CA087497; JMM: R01AT001576). Fundación CRIS contra el Cáncer (To DO team). 2014 Stewart Rahr - Young Investigator Award from Prostate Cancer Foundation (To DO). FPU predoctoral fellowship (To YC: 15/05126). Catalan Agency AGAUR (To VS: 2014 SGR 1331). Medical Research Council (To RBago - Dario Alessi lab: grant number MC_UU_12016/2). The activity of CIBERONC was co-funded with FEDER funds.
Footnotes
Authors´ Contributions
In vitro studies: AZ-L and AA-A with support from NM-M, SF-R, and LA. In vivo studies: AZ-L, AA-A and NM-M with support for tail vein from NB. Genotyping: PS-M. Metabolomics analysis: MClas, PQ, SM, AB, GL and DP, KMarks and KMarjon at AGIOS and DC, SMVL and JMF-P at CIC bioGUNE. Everolimus trial samples and staining: VS, JJ, LP, PN, IT, JT, TM and JB. Human prostate specimens: AU-O, AL-I, IL-V and MU. Histochemical analysis in mice: SF-R, ARev and AAlim. Generation of molecular AMD1 tools: JDS and RB. Immune cell analysis and GST-AMD1 purification; JT-C, IF-D and Jang. Phosphoproteomics: PX-E, AE and JM. Kinase assay: RBago. TRAMP mice material: Statistical supervision and bioinformatics: AR-C. Polysome profiling: AG and GT. GEMM Pathology: MC-M and CC-C. Technical support and critical discussions: BDM, JMM, MLM-C, MA, FE, IA, AMA, VT, LV-J, PZ-G, MP, AC-M, RF and YZ. KMarjon directed the metabolomics strategy, analysed the results and contributed to critical discussions and manuscript preparation and revision. AC directed the project, supervised data analysis and wrote the manuscript.
The authors declare no competing financial interest
Data availability. The authors declare that data supporting the findings of this study are available within the paper [and its supplementary information files]. Source data for unprocessed scans and Figure 2d and Extended data figures 3d, q-s and 4i are provided with the paper.
References
- 1.Efeyan A, Comb WC, Sabatini DM. Nutrient-sensing mechanisms and pathways. Nature. 2015;517:302–310. doi: 10.1038/nature14190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zoncu R, Efeyan A, Sabatini DM. mTOR: from growth signal integration to cancer, diabetes and ageing. Nat Rev Mol Cell Biol. 2011;12:21–35. doi: 10.1038/nrm3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen Z, et al. Crucial role of p53-dependent cellular senescence in suppression of Pten-deficient tumorigenesis. Nature. 2005;436:725–730. doi: 10.1038/nature03918. doi:nature03918 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ugalde-Olano A, et al. Methodological aspects of the molecular and histological study of prostate cancer: focus on PTEN. Methods. 2015;77–78:25–30. doi: 10.1016/j.ymeth.2015.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tabernero J, et al. Dose- and schedule-dependent inhibition of the mammalian target of rapamycin pathway with everolimus: a phase I tumor pharmacodynamic study in patients with advanced solid tumors. J Clin Oncol. 2008;26:1603–1610. doi: 10.1200/JCO.2007.14.5482. [DOI] [PubMed] [Google Scholar]
- 6.Carracedo A, Pandolfi PP. The PTEN-PI3K pathway: of feedbacks and cross-talks. Oncogene. 2008;27:5527–5541. doi: 10.1038/onc.2008.247. doi:onc2008247 [pii] [DOI] [PubMed] [Google Scholar]
- 7.Engelman JA, Luo J, Cantley LC. The evolution of phosphatidylinositol 3-kinases as regulators of growth and metabolism. Nat Rev Genet. 2006;7:606–619. doi: 10.1038/nrg1879. [DOI] [PubMed] [Google Scholar]
- 8.Song MS, Salmena L, Pandolfi PP. The functions and regulation of the PTEN tumour suppressor. Nat Rev Mol Cell Biol. 2012;13:283–296. doi: 10.1038/nrm3330. [DOI] [PubMed] [Google Scholar]
- 9.Pegg AE. S-Adenosylmethionine decarboxylase. Essays Biochem. 2009;46:25–45. doi: 10.1042/bse0460003. [DOI] [PubMed] [Google Scholar]
- 10.Xiong H, Pegg AE. Mechanistic studies of the processing of human S-adenosylmethionine decarboxylase proenzyme. Isolation of an ester intermediate. J Biol Chem. 1999;274:35059–35066. doi: 10.1074/jbc.274.49.35059. [DOI] [PubMed] [Google Scholar]
- 11.Kryukov GV, et al. MTAP deletion confers enhanced dependency on the PRMT5 arginine methyltransferase in cancer cells. Science. 2016;351:1214–1218. doi: 10.1126/science.aad5214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marjon K, et al. MTAP Deletions in Cancer Create Vulnerability to Targeting of the MAT2A/PRMT5/RIOK1 Axis. Cell reports. 2016;15:574–587. doi: 10.1016/j.celrep.2016.03.043. [DOI] [PubMed] [Google Scholar]
- 13.Mavrakis KJ, et al. Disordered methionine metabolism in MTAP/CDKN2A-deleted cancers leads to dependence on PRMT5. Science. 2016;351:1208–1213. doi: 10.1126/science.aad5944. [DOI] [PubMed] [Google Scholar]
- 14.Shlomi T, Fan J, Tang B, Kruger WD, Rabinowitz JD. Quantitation of cellular metabolic fluxes of methionine. Analytical chemistry. 2014;86:1583–1591. doi: 10.1021/ac4032093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eskens FA, et al. Phase I and pharmacological study of weekly administration of the polyamine synthesis inhibitor SAM 486A (CGP 48 664) in patients with solid tumors. European Organization for Research and Treatment of Cancer Early Clinical Studies Group. Clin Cancer Res. 2000;6:1736–1743. [PubMed] [Google Scholar]
- 16.Paridaens R, et al. A phase I study of a new polyamine biosynthesis inhibitor, SAM486A, in cancer patients with solid tumours. Br J Cancer. 2000;83:594–601. doi: 10.1054/bjoc.2000.1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Regenass U, et al. CGP 48664, a new S-adenosylmethionine decarboxylase inhibitor with broad spectrum antiproliferative and antitumor activity. Cancer Res. 1994;54:3210–3217. [PubMed] [Google Scholar]
- 18.Siu LL, et al. A phase I and pharmacokinetic study of SAM486A, a novel polyamine biosynthesis inhibitor, administered on a daily-times-five every-three-week schedule in patients with Advanced solid malignancies. Clin Cancer Res. 2002;8:2157–2166. [PubMed] [Google Scholar]
- 19.Serra H, et al. PTEN mediates Notch-dependent stalk cell arrest in angiogenesis. Nat Commun. 2015;6:7935. doi: 10.1038/ncomms8935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Greenberg NM, et al. Prostate cancer in a transgenic mouse. Proc Natl Acad Sci U S A. 1995;92:3439–3443. doi: 10.1073/pnas.92.8.3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Origanti S, et al. Ornithine decarboxylase mRNA is stabilized in an mTORC1-dependent manner in Ras-transformed cells. Biochem J. 2012;442:199–207. doi: 10.1042/BJ20111464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bale S, Ealick SE. Structural biology of S-adenosylmethionine decarboxylase. Amino Acids. 2010;38:451–460. doi: 10.1007/s00726-009-0404-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang Y, et al. Coordinated regulation of protein synthesis and degradation by mTORC1. Nature. 2014;513:440–443. doi: 10.1038/nature13492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ouyang DY, et al. Autophagy is differentially induced in prostate cancer LNCaP, DU145 and PC-3 cells via distinct splicing profiles of ATG5. Autophagy. 2013;9:20–32. doi: 10.4161/auto.22397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hsu PP, et al. The mTOR-regulated phosphoproteome reveals a mechanism of mTORC1-mediated inhibition of growth factor signaling. Science. 2011;332:1317–1322. doi: 10.1126/science.1199498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhao J, Zhai B, Gygi SP, Goldberg AL. mTOR inhibition activates overall protein degradation by the ubiquitin proteasome system as well as by autophagy. Proc Natl Acad Sci U S A. 2015;112:15790–15797. doi: 10.1073/pnas.1521919112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang Z, et al. Clinical efficacy of mTOR inhibitors in solid tumors: a systematic review. Future Oncol. 2015;11:1687–1699. doi: 10.2217/fon.15.70. [DOI] [PubMed] [Google Scholar]
- 28.O'Reilly KE, et al. mTOR inhibition induces upstream receptor tyrosine kinase signaling and activates Akt. Cancer Res. 2006;66:1500–1508. doi: 10.1158/0008-5472.CAN-05-2925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Carracedo A, et al. Inhibition of mTORC1 leads to MAPK pathway activation through a PI3K-dependent feedback loop in human cancer. J Clin Invest. 2008;118:3065–3074. doi: 10.1172/JCI34739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gerner EW, Meyskens FL., Jr Polyamines and cancer: old molecules, new understanding. Nat Rev Cancer. 2004;4:781–792. doi: 10.1038/nrc1454. [DOI] [PubMed] [Google Scholar]
- 31.Song MS, et al. Nuclear PTEN regulates the APC-CDH1 tumor-suppressive complex in a phosphatase-independent manner. Cell. 2011;144:187–199. doi: 10.1016/j.cell.2010.12.020. doi:S0092-8674(10)01473-X [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen Z, et al. Differential p53-independent outcomes of p19(Arf) loss in oncogenesis. Sci Signal. 2009;2:ra44. doi: 10.1126/scisignal.2000053. doi:2/84/ra44 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Motameni AR, et al. Delivery of the immunosuppressive antigen Salp15 to antigen-presenting cells by Salmonella enterica serovar Typhimurium aroA mutants. Infect Immun. 2004;72:3638–3642. doi: 10.1128/IAI.72.6.3638-3642.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Torrano V, et al. The metabolic co-regulator PGC1alpha suppresses prostate cancer metastasis. Nat Cell Biol. 2016;18:645–656. doi: 10.1038/ncb3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dowling RJ, et al. mTORC1-mediated cell proliferation, but not cell growth, controlled by the 4E-BPs. Science. 2010;328:1172–1176. doi: 10.1126/science.1187532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wiederschain D, et al. Single-vector inducible lentiviral RNAi system for oncology target validation. Cell Cycle. 2009;8:498–504. doi: 10.4161/cc.8.3.7701. [DOI] [PubMed] [Google Scholar]
- 37.Sanjana NE, Shalem O, Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods. 2014;11:783–784. doi: 10.1038/nmeth.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Carracedo A, et al. A metabolic prosurvival role for PML in breast cancer. The Journal of clinical investigation. 2012;122:3088–3100. doi: 10.1172/JCI62129. doi:62129 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Salazar M, et al. Loss of Tribbles pseudokinase-3 promotes Akt-driven tumorigenesis via FOXO inactivation. Cell Death Differ. 2015;22:131–144. doi: 10.1038/cdd.2014.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Alimonti A, et al. A novel type of cellular senescence that can be enhanced in mouse models and human tumor xenografts to suppress prostate tumorigenesis. J Clin Invest. 2010;120:681–693. doi: 10.1172/JCI40535. doi:40535 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Revandkar A, et al. Inhibition of Notch pathway arrests PTEN-deficient advanced prostate cancer by triggering p27-driven cellular senescence. Nat Commun. 2016;7:13719. doi: 10.1038/ncomms13719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fuhrer T, Heer D, Begemann B, Zamboni N. High-throughput, accurate mass metabolome profiling of cellular extracts by flow injection-time-of-flight mass spectrometry. Analytical chemistry. 2011;83:7074–7080. doi: 10.1021/ac201267k. [DOI] [PubMed] [Google Scholar]
- 43.Jha AK, et al. Network integration of parallel metabolic and transcriptional data reveals metabolic modules that regulate macrophage polarization. Immunity. 2015;42:419–430. doi: 10.1016/j.immuni.2015.02.005. [DOI] [PubMed] [Google Scholar]
- 44.Melamud E, Vastag L, Rabinowitz JD. Metabolomic analysis and visualization engine for LC-MS data. Analytical chemistry. 2010;82:9818–9826. doi: 10.1021/ac1021166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fumagalli S, Ivanenkov VV, Teng T, Thomas G. Suprainduction of p53 by disruption of 40S and 60S ribosome biogenesis leads to the activation of a novel G2/M checkpoint. Genes Dev. 2012;26:1028–1040. doi: 10.1101/gad.189951.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Grasso CS, et al. The mutational landscape of lethal castration-resistant prostate cancer. Nature. 2012;487:239–243. doi: 10.1038/nature11125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lapointe J, et al. Gene expression profiling identifies clinically relevant subtypes of prostate cancer. Proc Natl Acad Sci U S A. 2004;101:811–816. doi: 10.1073/pnas.0304146101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Taylor BS, et al. Integrative genomic profiling of human prostate cancer. Cancer Cell. 2010;18:11–22. doi: 10.1016/j.ccr.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tomlins SA, et al. Integrative molecular concept modeling of prostate cancer progression. Nat Genet. 2007;39:41–51. doi: 10.1038/ng1935. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.