Abstract
Previous studies have shown that diets can alter the metabolism of drugs; however, it is difficult to compare the effects of multiple diets on drug metabolism among different experimental settings. Phase-I related genes play a major role in the biotransformation of pro-drugs and drugs.
In the current study, effects of nine diets on the mRNA expression of phase-I drug-metabolizing enzymes in livers of mice were simultaneously investigated. Compared to the AIN-93M purified diet (control), 73 of the 132 critical phase-I drug metabolizing genes were differentially regulated by at least one diet. Diet restriction produced the most number of changed genes (51), followed by the atherogenic diet (27), high-fat diet (25), standard rodent chow (21), western diet (20), high-fructose diet (5), EFA deficient diet (3), and low n-3 FA diet (1). The mRNAs of the Fmo family changed most, followed by Cyp2b and 4a subfamilies, as well as Por (From 1121 to 21-fold increase of theses mRNAs). There were 59 genes not altered by any of these diets.
The present results may improve the interpretation of studies with mice and aid in determining effective and safe doses for individuals with different nutritional diets.
Keywords: Disposition, mRNA level, microarray
Introduction
Cardiovascular and neuropsychiatric disorders are estimated to be the top two leading causes for increasing disability the next ten years, and nutritional inputs are recognized as important modifiers of these lifestyle diseases (Bhatia et al., 2011; Prottey et al., 1975). In the western countries, processed, low-nutritional, and high-caloric foods are a significant part of the daily diet. According to the U.S. Department of Health, eating healthier diets could decrease at least $71 billion per year in medical costs, lost productivity, and deaths.
Alteration of laboratory animal diets is a good method to investigate the influence of nutrients on various parameters and the phenotype of lifestyle disease caused by an imbalance of nutrition intake, especially to exclude complex genetic and husbandry variables. High-fat, atherogenic, western and high-fructose diets mimic the consumption of diets high in fat, cholesterol and carbohydrates in humans, and subsequently cause metabolic syndrome, diet-induced obesity, diabetes, and cardiovascular disease in animal models (Guo et al., 1999). However, insufficient fat will also damage biological functions of organisms, and the most commonly investigated are essential fatty acids (EFA) and docosahexaenoic acid (DHA, n-3 FA). Low EFA diets highlight the importance of certain nutrients that must be ingested because the body requires but cannot synthesize them, and almost all cells express receptors that mediate EFA-based signaling pathways. Deficiency of EFAs have been reported to be associated with cardiovascular, neuropsychiatric, immune-inflammatory disorders, cancer proliferation, and increasing the length of stay in hospitals (Lands, 2012). DHA is a “conditionally essential” omega-3 fatty acid (n-3 FA), and is made naturally in small amounts, but also must be obtained from food or supplements. DHA plays an important role in the development of the nervous system (Guesnet and Alessandri, 2011). In order to find a more healthy balance of energy intake and consumption, diet restriction has been used, and reported to increase insulin sensitivity and lifespan in mammals (Honkakoski and Negishi, 2000).
Nutrition may have a global impact on genes involved in metabolism (Osada, 2013), and lifestyle diseases due to imbalance of nutrition intake are often attempted to be treated by long-term polypharmacotherapy. In addition, drugs are used in people that are on various diets. Therefore, it is important to determine whether differences in diet affect the metabolism of drugs.
Phase-I drug-metabolizing enzymes in the liver are very essential for increasing the water solubility of drugs and other chemicals. These phase-I enzymes include carboxylesterases (Cess), paraoxonases (Pons), alcohol dehydrogenases (Adhs), aldehyde dehydrogenases (Aldhs), NAD(P)H:quinone oxidoreductases (Nqos), flavin-containing monooxygenases (Fmos), carbonyl reductases (Cbrs), epoxide hydrolases (Ephxs), aldehyde oxidases (Aoxs), xanthine dehydrogenases (Xdhs), Aldo-keto reductases (Akr), P450 reductase (Por), and cytochrome P450s (Cyps). Many phase-I enzymes are regulated by transcription factors, such as the aryl hydrocarbon receptor (AhR), constitutive androstane receptor (CAR, Nr1l3), pregnane X receptor (PXR, Nr1l2), peroxisome proliferator-activated receptor (PPAR, Nr1c1), farnesoid X receptor (FXR, NrIh4), hepatic nuclear factors (HNF), liver X receptor (LXR), and Nuclear factor (erythroid-derived 2)-like 2 (Nrf2) (Klaassen and Aleksunes, 2010; Klaassen and Lu, 2008).
Various amounts, types and lengths of dietary lipids and carbohydrates have been examined to determine their influence on phase-I drug metabolism enzymes and their transcriptional regulators (Osada, 2013). High-fat diet induced PPARα, β, γ1, and γ2, HNF4, LXRα and their target Cyp mRNAs (Anderson et al., 2009), and reduced Cyp3a11, Cyp2b10, Cyp2a4, PXR, and CAR mRNAs (Ghose et al., 2011). An atherogenic diet for 3 months decreased Pon1 mRNA and activity (Costa et al., 2005). A western diet induced LXR and PPARα expression in mice (Osada, 2013). A diet rich in polyunsaturated fatty acids (including DHA), activated CYP4a and 4f subfamilies (Comba et al., 2011). Short-chain fructo-oligosaccharides altered the expression of PPARα and FXR target genes in rat liver (Fukasawa et al., 2010). Caloric restriction reversed the changes in PPARα and LXR gene expression (Cao et al., 2001; Osada, 2013).
However, all the studies described above were done separately and with different animal models; thus it is impossible to compare these data directly. In the current study, a standard AIN-93M purified diet (Duffy et al., 2002) was used as a control to compare with 8 other classic laboratory diet formulations in C57BL/6 mice for 3 weeks, and genes involved in phase-I drug metabolizing enzymes as well as transcription factors regulating these enzymes were quantified by microarray and validated by real-time PCR (RT-PCR), in order to determine which diets are most likely to alter the expression of phase-I metabolism related genes, and thus might change the effectiveness and adverse effect of drugs.
Methods
Ethics Statement
The housing facility is an American Animal Associations Laboratory Animal Care-accredited facility at the University of Kansas Medical Center, and all procedures were approved in accordance with the Institutional Animal Care and Use Committee guidelines.
Animals
Male C57BL/6 mice (22±2 g, 8-weeks old, n=5) were obtained from Charles River Laboratories, Inc. (Wilmington, MA). Animals were housed in a temperature-, light-, and humidity controlled environment. Lab chow (#8604; teklad rodent diet; 14% calories from fat), EFA deficient diet (#84224), high-fat diet (#97070; 59.9% calories from fat), western diet (42% calories from fat, and 0.2% cholesterol, #88137), atherogenic rodent diet (#02028, 42.6% calories from fat; cholesterol 1.3%, 0.5% cholic acid), high-fructose diet (#89247 60% fructose diet), AIN-93M purified diet (#94048; 10.2% calories from fat), and low n-3 FA diet (#00235 + 7% sunflower oil) (Levant et al., 2006) were all purchased from Harlan Laboratories (Madison, WI), and diet restriction performed by providing 75% of the #8604 diet consumed by ad libitum feeding (Varady et al., 2007) were given to mice for 3 weeks, and all mice drank water ad lib. The ad libitum daily feed intake was approximately 4 g/mouse. Therefore, for diet restriction, each mouse was given approximately 2.7–3.0 g of food per day. All mice were euthanized in the morning (8:00–10:00 A.M.) and blood and tissue samples were collected. Mice were not fasted before liver sample isolation. Livers were collected and frozen in liquid nitrogen, and stored at 80°C before use. These mouse livers were previous used to examine the influence of diets on expression of genes involved in lipid metabolism, oxidative stress, and inflammation (Renaud et al., 2014).
Total RNA isolation
Total liver RNA was isolated using RNAzol Bee reagent (Tel-Test Inc., Friendswood, TX) per the manufacturer’s protocol, and concentrations were quantified with a NanoDrop Spectrophotometer (NanoDrop Technologies, Wilmington, DE) at 260 nm. Formaldehyde-agarose gel electrophoresis was used to evaluate the integrity of these total RNA samples, which were confirmed by visualization of the 18s and 28s rRNA bands.
Microarray and data analysis
Gene expression in livers of mice in each group consuming one of the nine diets was determined using Affymetrix Mouse 430.20 arrays at the KUMC Microarray Core Facility. The cRNAs of three mice of each diet were individually hybridized to an array. Raw data CEL files were imported into the ‘R’ program using the ‘‘Affy’’ package, normalized by the Robust Multichip Averaging (GCRMA) package, with the output data being log2 transformed. The probes with intensities higher than log2100 in at least one group were selected for further analysis. Gene annotations were obtained using GeneSpring (Agilent Technologies, Santa Clara, CA), and gene symbols were obtained from the mouse 4302 package (Wu et al., 2012).
Quantification of Cyp2b10, 2d22, 3a11, 4a14 and Fmo3 mRNA expression by RT-PCR assay
Diethyl pyrocarbanate (DEPC)-treated double-distilled water was used to dilute each RNA sample to 50 ng/μl for real-time PCR quantification. Reverse transcription of RNA to cDNA was performed using the Applied Biosystems High Capacity Reverse Transcriptase kit (Applied Biosystems, Foster City, CA). Primers for RT-PCR were synthesized by Integrated DNA Technologies (Coralville, IA). Reactions were seeded in 384-well optical reaction plates (Applied Biosystems) and fluorescence was quantified using Applied Biosystems 7900 Real Time PCR System. Data from the mice fed the standard AIN-93M purified diet was considered the control diet. The mRNAs for the phase-I genes were normalized to Gapdh, the most commonly used housekeeping gene, and the mRNA levels of Gapdh remained relatively constant in mice fed these 9 diets for 3 weeks.
Statistical analysis
Differential expression of microarray data was determined using the “limma” package in ‘R’ (compared to data from mice fed AIN purified diet, P<0.05). All values are expressed as mean±S.E.M. Hierarchical clustering of phase-I drug-metabolism enzyme mRNAs that exhibited changes in at least 1 diet was compared to the AIN-93M purified diet by using ‘R’ program using the ‘‘Pheatmap’’ package. Average values of three replicates per age are given by colored squares. High mRNA abundance is represented in red, whereas low mRNA abundance is in blue. Validation data of RT-PCR are expressed as mean±S.E.M. (n=5). Differences between mean values were tested for statistical significance (P<0.05) by one-way analysis of variance followed by Duncan’s post hoc test.
Results
Cytochrome P450 phase-I drug-metabolizing enzymes
Cyp1 family
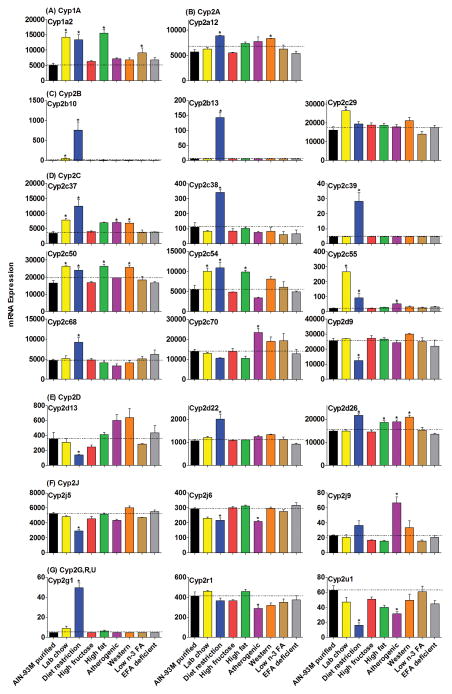
Cyp1a2 was the only enzyme significantly changed in the Cyp1 family by the various diets (Fig. 1A). The mRNA of Cyp1a2 increased 185% in mice fed lab chow, 170% with diet restriction, 213% with a high-fat diet, and 82% in mice fed a low n-3 FA diet.
Figure 1.
The mRNAs of enzymes in the Cyp1 and 2 families fed various diets. “AIN-93M purified” represents AIN-93M purified diet (#94048); “Lab chow” represents teklad rodent diet (#8604); Diet restriction performed by providing 75% of the diet (#8604) consumed by ad libitum feeding; “high fructose” represents 60% fructose diet (#89247); “high fat” represents high-fat diet (#97070); “atherogenic” represents atherogenic rodent diet (#02028); “western” represents adjusted calories diet (western diet, #88137); “low n-3 FA” represents DHA deficient diet (#00235 + 7% sunflower oil); “EFA deficient” represents EFA-deficient diet (#84224). The same abbreviations used in all the figures of the current study. Data are presented as means ± S.E.M of three mice, * represent P<0.05.
Cyp2 family
The mRNAs of the Cyp2 gene family were altered markedly by the various diets (Fig. 1B–1G). In the Cyp2a subfamily, Cyp2a12 mRNA was up-regulated 56% by diet restriction and 48% by the western diet (Fig. 1B). The mRNAs of the Cyp2b subfamily increased markedly in mice fed the restricted diet and the lab chow (Fig. 1C). More specifically, Cyp2b10 mRNA increased in mice fed lab chow (10-fold) and diet restriction (153-fold), and Cyp2b13 mRNA was higher in mice on the restricted diet (28-fold) than the purified diet.
The expression of 9 genes in the Cyp2c subfamily changed significantly with at least one diet when compared to the purified diet (Fig. 1D). The lab chow up-regulated the mRNAs of Cyp2c29 (65%), 2c37 (100%), 2c50 (58%), 2c54 (83%) and 2c55 (1100%). Diet restriction increased mRNA levels for Cyp2c37 (300%), 2c38 (200%), 2c50 (44%), 2c54 (98%), 2c55 (300%), and 2c68 (96%) in livers of mice. The high-fat diet increased the mRNA of Cyp2c50 (58%) and 2c54 (80%) in livers of mice. The atherogenic diet induced the expression of Cyp2c37 (100%), 2c55 (100%), and 2c70 (67%) in livers of mice. The western diet increased the mRNA of Cyp2c37 (96%) and 2c50 (54%) in livers of mice.
The Cyp2d family was not altered markedly by the various diets (Fig. 1E). Diet restriction decreased the mRNAs of Cyp2d9 and 2d13 about 50%, but increased Cyp2d22 mRNA (89%) in livers of mice. A small increase in Cyp2d26 expression (less than 50%) was noted in mice fed the following diets: diet restriction, high-fat diet, atherogenic diet, and western diet.
The expression of other enzymes in the Cyp2 family is shown in Fig. 1F–1G. Cyp2j9 mRNA increased 100% in livers of mice fed the atherogenic diet, while Cyp2g1 mRNA increased 900% in livers of mice fed the restricted diet. The mRNAs of Cyp2j5, 2j6 and 2u1 were slightly decreased in mice fed the restricted diet, which was similar to changes of the mRNAs of Cyp2j6, 2r1 and 2u1 in mice fed the atherogenic diet.
Cyp3 family
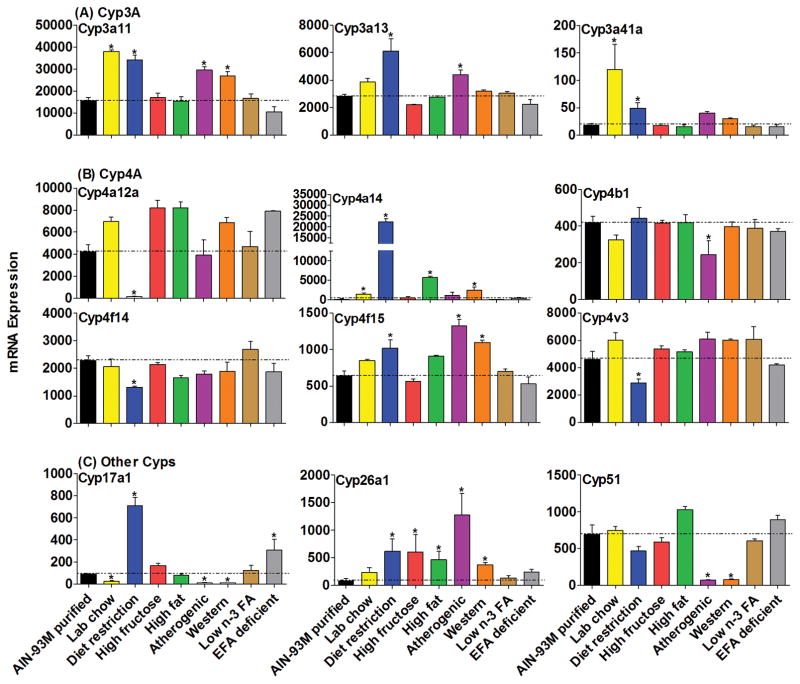
The most abundant Cyp3 enzymes, namely Cyp3a11 and 3a13, and Cyp3a41a mRNAs were increased by various diets (Fig. 2A). The Cyp3a11 mRNA increased about 100% in livers of mice fed lab chow or diet restriction, while it increased 80% with the atherogenic diet and western diet. An increase in the mRNA of Cyp3a13 was observed in livers of mice fed lab chow (100%) and the atherogenic diet (55%). Cyp3a41a mRNA was increased in livers of mice fed lab chow (500%) and also by diet restriction (200%).
Figure 2.
The mRNAs of enzymes in the Cyp3, 4 and other Cyp families in livers of mice fed 8 different diets compared with the AIN-93M purified diet. Data are presented as means ± S.E.M of three mice, * represent P<0.05.
Cyp4 family
Diet restriction had a marked influence on the effects of two members of the Cyp4 family (Fig. 2B), as Cyp4a12a mRNA expression was almost silenced whereas Cyp4a14 mRNA increased 173-fold. The mRNA of Cyp4a14 also increased in livers of mice fed the lab chow (10-fold), high-fat diet (44-fold), and western diet (18-fold). Cyp4b1 mRNA decreased 41% in mice fed the atherogenic diet, while Cyp4f14 and 4v3 mRNAs were decreased about 40% by the restricted diet. The mRNA of Cyp4f15 increased somewhat in mice fed the restricted diet (59%), atherogenic diet (100%) and western diet (71%).
Other Cyps
Three Cyp genes were altered markedly other than Cyp1, 2, 3 and 4 families (Fig. 2C), which are often considered the drug metabolizing Cyps. The mRNA of the steroid-synthetic enzyme Cyp17a1 decreased in livers of mice fed the lab chow (−72%), atherogenic diet (−89%) and western diet (−88%), but increased in mice on restricted diet (700%) and the EFA-deficient diet (200%). Interestingly, the mRNA of retinoic acid related enzyme Cyp26a1 was increased by five of the diets, including diet restriction (7-fold), high-fructose diet (7-fold), high-fat diet (5-fold), atherogenic diet (15-fold), and western diet (4-fold). The mRNA of Cyp51, which is involved in sterol biosynthesis, was almost abolished in livers of mice fed the atherogenic diet (−90% decrease) and the western diet (−89% decrease).
Non-Cyp phase-I drug-metabolizing enzymes
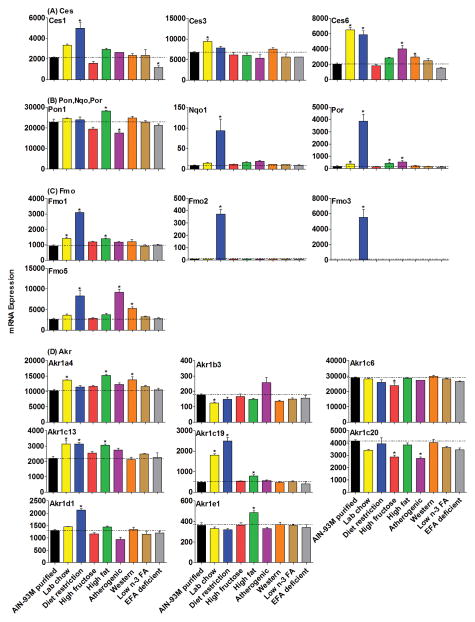
The mRNA expression of the Ces family of enzymes in the liver was increased (Fig. 3A) in mice fed lab chow (Ces3, 41%; Ces6, 200%), diet restriction (Ces1, 100%; Ces6, 200%), atherogenic diet (Ces6, 99%) and western diet (Ces6, 45%).
Figure 3.
The mRNAs of Ces, Pon, Nqo, Por, Fmo and Akr families in livers of mice fed 8 different diets compared with the AIN-93M purified diet. Data are presented as means ± S.E.M of three mice, * represent P<0.05.
The mRNA of Pon1 (Fig. 3B) was relatively resistant to alteration by the various diets, in that it slightly increased in mice fed a high-fat diet (24%) and slightly decreased by the atherogenic diet (−23%). More dramatically, Nqo1 mRNA was increased 10-fold by the restricted diet. The mRNA of Por increased markedly in livers of mice given the restricted diet (21-fold), and smaller increases were observed in mice fed lab chow (100%), high-fat (100%), and atherogenic rodent diets (200%) (Fig. 3B).
The Fmo family (Fig. 3C) was altered the most when placed on the restricted diet with a 1121-fold increase in Fmo3, a 47-fold increase in Fmo2, and 2-fold increases in Fmo1 and 5. The mRNA of Fmo1 was increased by lab chow (52%) and the high-fat diet (51%), while Fmo5 mRNA was increased by the atherogenic diet (300%) and western diet (100%).
Among the changes in the Akr family (Fig. 3D), Akr1c19 mRNA was altered the most with increases in mice fed the lab chow (300%), diet restriction (400%) and high-fat diets (60%), while Akr1c13 were increased about 40% with these three diets. The mRNA alterations of other genes included: increases in Akr1a4 in livers of mice fed the lab chow diet (34%), high-fat diet (49%), and western diet (35%); increase of Akr1d1 in mice on diet restriction (64%); increase of Akr1e1 (34%) in mice on the high-fat diet; decrease of Akr1b3 in mice on lab chow (30%); decrease of Akr1c6 in mice on the high-fructose diet (18%); decrease of Akr1c20 in mice on the high-fructose (30%) and atherogenic diet (30%).
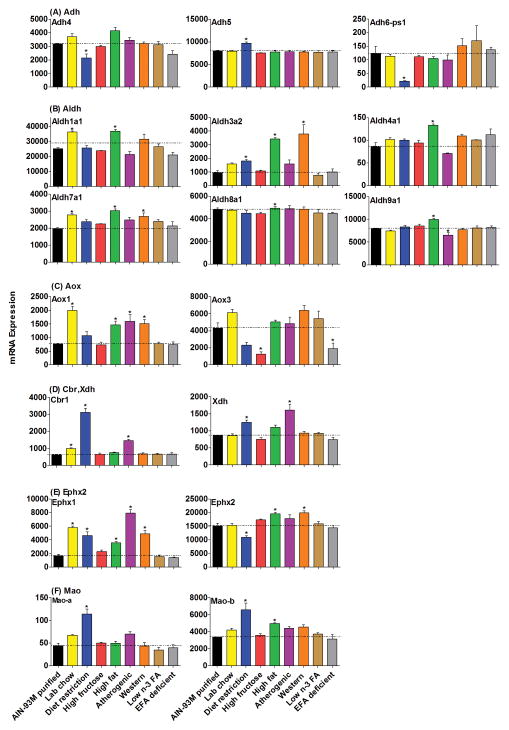
In general, the mRNAs of the Adh and Aldh families were not altered markedly by the various diets (Fig. 4A–4B). However, there was an increase of Aldh3a2 in mice on the restricted diet (90%), high-fat diet (300%), and western diet (300%). The other alterations include a decrease of Adh4 in livers of mice on the restricted diet (33%), a decrease of Aldh9a1 in livers of mice on the atherogenic diet (26%), and an increase of Adh5 in livers of mice on the restricted diet (21%). Aldh mRNAs had some minor changes also, with an increase of Aldh1a1 in the mice fed the lab chow (44%) and high-fat diet (46%), an increase of Aldh4a1 fed the high-fat diet (56%), an increase of Aldh7a1 on the lab chow (41%), high-fat (54%), and western diets (37%), an increase of Aldh8a1 on the high-fat diet (31%), and an increase of Aldh9a1 on the high-fat diet (26%).
Figure 4.
The mRNAs of Adh, Aldh, Aox, Cbr, Xdh, Ephx and Mao families in livers of mice fed 8 different diets compared with AIN-93M purified diet. Data are presented as means ± S.E.M of three mice, * represent P<0.05.
Aox, Cbr, Xdh, Ephx, and Mao gene families also had diverse expression patterns in the current study (Fig. 4C–4F). Aox1 mRNA was increased about 100% by the high-fat diet, atherogenic diet and western diet, as well as 200% by the lab chow. In contrast, another altered gene in this family, Aox3, was decreased 71% by the high-fructose diet and 56% by the EFA-deficient diet (Fig. 4C). Cbr1 mRNA was higher in mice fed a restricted diet (400%), atherogenic diet (100%), and lab chow diet (58%) (Fig. 4D). The mRNA level of Xdh was 45% higher in livers of mice fed the restricted diet and 86% higher in mice fed the atherogenic diet (Fig. 4D). Changes in Ephx1 mRNA were more obvious than Ephx2 with the various diets, as it was about 100% higher with the high-fat diet; 200% higher in lab chow, diet restriction and western diets; and 400% higher with the atherogenic diet. Ephx2 mRNA was 28% lower in livers of mice fed the restricted diet, 29% higher on the high-fat diet, and 33% higher on the western diet (Fig. 4E). Mao-a and b mRNA levels were increased about 100–200% in mice fed the restricted diet, and Mao-b was also increased 47% in livers of mice on the high-fat diet (Fig. 4F).
Expression of xenobiotic-sensing receptors
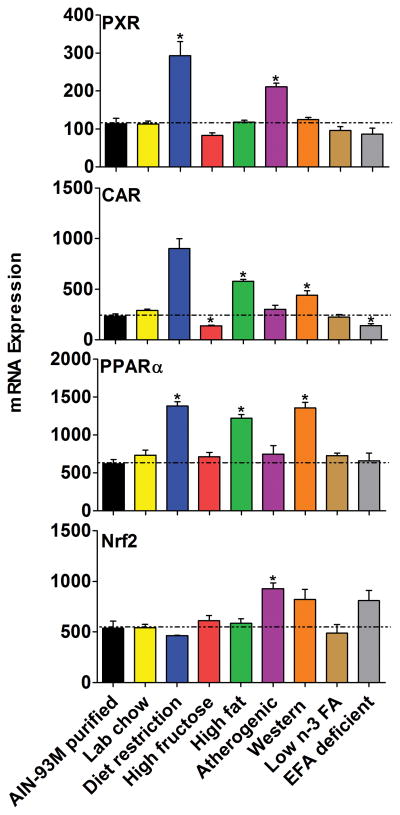
The expression of transcription factors known to regulate phase-I drug metabolism genes were altered by 6 of the 8 diets, when compared to the AIN-93M purified diet (Fig. 5). Diet restriction up-regulated CAR (300%), PXR (200%) and PPARα (100%) mRNA in livers of mice. In livers of mice fed the high-fat and atherogenic diet, CAR and PPARα increased about 100%. The atherogenic diet also increased PXR (86%) and Nrf2 (73%) mRNA expression.
Figure 5.
The mRNAs of transcription factors important in phase-I metabolism in livers of mice fed 8 different diets compared with the AIN-93M purified diet. Data are presented as means ± S.E.M of three mice, * represent P<0.05.
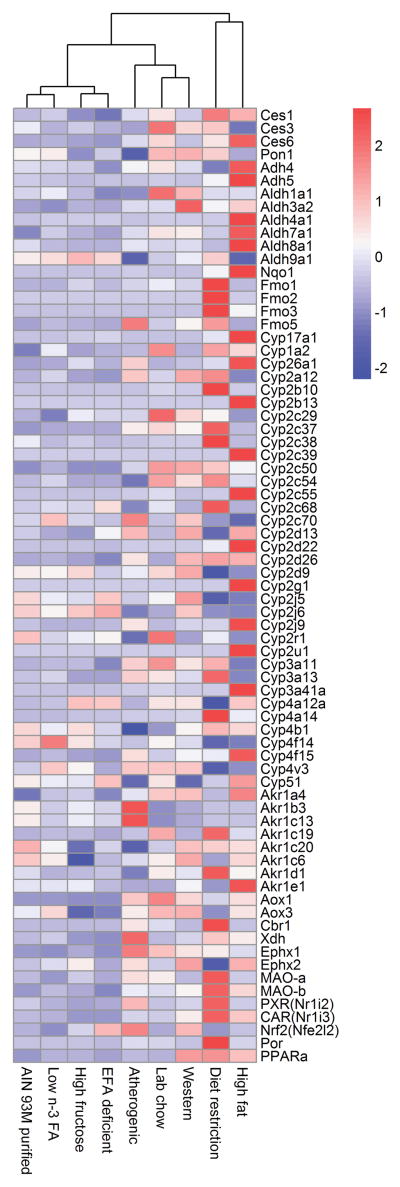
Hierarchical cluster analysis of mRNA profiles for phase-I drug metabolizing enzymes
In the current study, 132 phase-I related genes were examined, and the expression of 73 genes were changed by at least one diet (Fig. 6). Diet restriction altered the expression of 51 genes, followed by the atherogenic diet (27 genes), high-fat diet (25 genes), lab chow (21 genes), western diet (20 genes), high-fructose diet (5 genes), EFA deficient diet (3 genes), and low n-3 FA diet (1 gene). The other 59 genes that were not altered by any of these diets include Cyp1a1, 1b1, 2b19, 2c44, 2c65, 2d10, 2d34, 2e1, 2f2, 2j11, 2j13, 2s1, 2w1, 3a16, 3a44, 4a29, 4a31, 4f13, 4f16, 4f39, 4f41, 4x1, 11a1, 11b1, 11b2, 19a1, 20a1, 21a1, 24a1, 26b1, 27b1, and 46a1; Adh1, 6a, 6b, 7, fe1; Aldh 1a2, 1a3, 1a7, 1b1, 1l1, 1l2, 2, 3a1, 3b1, 3b2, 5a1, 6a1, 16a1, and 18a1; Ces 5, 7, and 8; Nqo2, Fmo4, Pon2, Pon3, and AhR.
Figure 6.
Hierarchical cluster analysis of mRNA profiles for phase-I drug metabolizing enzymes. Compared with the AIN-93M purified diet 73 phase-I related genes changed in at least one diet treatment. Diet restriction altered the mRNA of 51 genes, followed by the atherogenic rodent diet that altered the mRNA expression of 27 phase-I drug metabolizing genes. Lab chow, high-fructose diet, high-fat diet, western diet, low n-3 FA diet and EFA deficient diet, altered the mRNA expression of 21, 6, 25, 20, 1, and 3 genes, respectively. Average values of three replicates per diet are given by colored squares. High mRNA abundance is represented in red, whereas low mRNA abundance is in blue.
The mRNA alterations of Cyp2b10, 2d22, 3a11, 4a14 and Fmo3 quantified by microarray were validated with real-time PCR, because the changes of these genes are the most obvious in the present study. In general, the trend of the changes for these 6 genes was similar by these 2 methods (Supplemental Fig.1).
Discussion
The present study characterizes the mRNA profiles of phase-I-related genes in the livers of mice fed 9 various diets. Compared with the AIN-93M purified diet, changes in phase-I related gene expression produced by 8 classic laboratory diets were examined simultaneously, which yielded more information on these genes within diverse nutrition conditions than in previous separate reports (Anderson et al., 2009; Dawson et al., 2013; Duffy et al., 2002; Ghose et al., 2011; Guesnet and Alessandri, 2011; Kanoski and Davidson, 2011; Vergnes et al., 2003).
Many phase-I related genes changed markedly in livers of mice on the restricted diet (51 genes). Diet restriction is a regimen based on lower dietary intake relative to the subject's previous intake before intentionally restricting calories, or, to an average intake of a person of similar body type (Anderson et al., 2009; Fontana, 2004; Witte, 2009). The present study indicates the mRNAs of 42 genes were increased by calorie restriction. If the data from the mice fed the calorie restricted diet would have been compared to the data from the mice fed the lab chow, the statistics would have been similar except for a few mRNAs that are also increased by the lab chow (Cyp1a2, 2c29, 2c50, 2c54, 2c55, 3a11, 3a41a, Ces6, Ephx1).
The Fmos catalyze chemical reactions via the bound cofactor flavin, which oxidize heteroatoms, particularly nucleophilic atoms such as the nitrogen of amines (Phillips and Shephard, 2008). The current study showed that the Fmo family was markedly induced with diet restriction (Fig. 3C), especially Fmo3 and Fmo2, which is consistent with about a 1000-fold higher level of enzyme activity in guinea pigs on a restricted diet (Brodfuehrer and Zannoni, 1987). Moreover, activation of CAR (increase of Cyp2b10), lipid metabolism (Cyp4a14 and PPARα), oxidative stress (Nqo1) (Fig. 1, 2, 5, and 3) and Nrf2 might have occurred in mice with 3-week diet restriction, and the data are consistent with previous studies (Cao et al., 2001; Osada, 2013). Cyp17a1 and 26a1 (Fig. 1) were induced 7-fold in the mice on diet restriction, indicating the influence on the synthesis of retinoic acid and steroids. About 4- to 9-fold increases in the mRNAs of Cyp2g1, Akr1c19, and Cbr1 (Fig. 1, 3, 4) were observed in diet-restricted mice. Cyp2g1 metabolizes chemicals including coumarin (Gu et al., 1998); Akr1c19 reduces isatin, which is a pharmacologically active molecule produced by intestinal bacteria (Ishikura et al., 2005); Cbr1 reduces doxorubicin in humans and detoxifies reactive aldehydes (Doorn et al., 2004; Kassner et al., 2008). Therefore, induction of the mRNAs of these genes by diet restriction may alter the corresponding biotransformation. Por is the only electron donor for all microsomal P450s, and the 21-fold induction of the mRNA expression by diet restriction might increase the function of Cyps (Fig. 3B). In contrast, Cyp4a12a (Fig. 2B) was almost abolished with diet restriction, which further indicates the influence of restricted diet on the metabolism of fat. Mao-a (oxidizes de-amination of amines), Ces1, and Ces6 (biotransformation of ester- or amide-type prodrugs) had 1- to 2-fold increases of their mRNA in livers of mice fed the restricted diet (Fig. 3 and 4), and thus might alter the metabolism of circulating serotonin, norepinephrine, as well as cocaine, temocapril, meperidine and delapril. The high-fat, atherogenic and western diet ranked as the second most influential diets in regulating phase-I metabolism genes (atherogenic diet: 27 genes, high-fat diet: 25 genes, western diet: 20 genes). The similarity among these 3 diets is the high fat content. Specifically, the high-fat diet supplies high triglycerides, the atherogenic diet provides high cholesterol, bile acids and fat, and the western diet includes high fat and carbohydrates. Therefore, it is not surprising that the major changes observed were related to lipid metabolism, such as induction of Cyp4a14 and PPARα mRNA (Fig. 2 and 5), as well as the marked decrease in expression of Cyp17a1 and 51 (Cyp17a1 catalyses the oxidative removal of the 14α-methyl group of lanosterol or eburicol, and Cyp51 required for sterol biosynthesis) (Mullins et al., 2011), which is consistent with results in previous studies (Anderson et al., 2009; Osada, 2013). However, other genes such as Cyp26a1, which metabolizes retinoic acid, also had 4- to 14-fold increases in mice fed these three diets, but especially the atherogenic diet, which suggest a strong interaction in the synthesis for retinoic acid, cholesterol and bile acids (Fig. 1C). The decrease of Cyp3a11, 2b10, 2a4, PXR and CAR in mice fed a high-fat diet compared to a low-fat diet was not reproduced in the present study, which might be due to difference in control diet (Ghose et al., 2011). In the current study, Ephx1, which plays an important role in both activation and detoxification of exogenous chemicals, such as polycyclic aromatic hydrocarbons (Nguyen et al., 2013), was obviously up-regulated in livers of mice fed the atherogenic diet (Fig. 4E). Fmo5 is expressed lowly in livers of mice but highly in livers of humans, and was increased 3-fold in livers of mice fed the atherogenic diet (Fig. 3C). In addition, the decrease of Pon1 mRNA in livers of mice fed the atherogenic diet (Fig. 3B) was consistent with a previous study (Costa et al., 2005). Aldh3a2 catalyzes the oxidation of a variety of saturated and unsaturated aliphatic aldehydes, and converts hexadecenal to hexadecenoic acid (Koppaka et al., 2012). The mRNA of Aldh3a2 was 3-times higher in mice fed a high-fat or western diet (Fig. 4B), which indicates extensive oxidation of long-chain aliphatic aldehydes to fatty acids. Cyp1a2, 2c37, 2c55, 2j9, 4f5, Axo1, Ces6, PPARα, and Por were also increased slightly by these 3 diets, and the significance of these findings needs further research.
Lab chow revealed differences from the AIN-93M Purified Diet. The lab chow resulted in about a 10-fold induction of Cyp2b10, Cyp2c55 and 4a14 (Konno et al., 2010) (Fig. 1 and 2), which are CAR, PXR and PPARα target genes (Fig. 5); thus the increase might be due to phytochemicals that activates these nuclear receptors. In addition, Cyp3a41a, 17a1 (Fig. 2), and Akr1c19 (Fig. 3D) were altered by the lab chow diet.
The high-fructose diet altered the expression of fewer genes related to retinoic acid, steroid hormones, and lipid metabolism. Cyp26a1 mRNAs was 7-times higher in mice fed a high-fructose diet (Fig. 2), indicating a marked acceleration of retinoic acid synthesis may occur. Aox oxidizes organic molecules containing aldehyde functionality into the corresponding carboxylic acid, and hydroxylates aza- and oxo-heterocycles (Coelho et al., 2012). Aox3 is the only member of the Aox family that was significantly changed in livers of mice fed the high-fructose diet (Fig. 4C), and the 71% decrease in expression suggests that inactivation of drugs as well as other xenobiotics metabolized by Aox3 is likely because it is the prevalent form of Aox in livers of mice. The constitutively active transcriptional regulator CAR (Fig. 5) was decreased 42% in livers of mice fed the high-fructose diet, and therefore, drug metabolism, glucose metabolism and bilirubin clearance may be changed (Wada et al., 2009). The influence of other down-regulated genes (Fig. 3D), including Akr1c6 (−18%), and 1c20 (−31%) may change the metabolism of steroid hormones and lipids in livers of mice fed a high-fructose diet. The expression of phase-I drug metabolizing genes was not markedly altered in mice fed the low n-3 FA diet and the EFA-deficient diet. Cyp1a2 was slightly increased (Fig. 1) in mice fed the two diets, but the biological significance of these changes is not obvious. The mRNA of CAR (Fig. 5) and Aox3 (Fig. 4C) were decreased, and Cyp17a1 (Fig. 2) was increased by the EFA-deficient diet, therefore, the metabolism of Aox3 substrates and steroidogenesis (Cyp17a1 involved) might be changed (Pryde et al., 2010).
Caution is needed when interpreting these mRNA results to corresponding enzyme activity and function. However, for many of the enzymes there are no specific substrates or specific antibodies to determine the similarities of the changes in mRNA and enzyme activities. Because of the large number of pathways examined in the present study, it will require a massive effort to quantify enzyme activity, once specific substrates are known for each pathway.
Conclusion
compared with the AIN-93M purified diet, the mRNA of 73 phase-I related genes were altered by at least one diet in 8 various diets. Diet restriction had the most dramatic effects in that it altered the expression of 51 genes, followed by the atherogenic diet (27 genes), high-fat diet (25 genes), lab chow (21 genes), western diet (20 genes), high-fructose diet (5 genes), EFA deficient diet (3 genes), and low n-3 FA diet (1 gene). Diet restriction altered the mRNA of the Fmo gene family the most (Fmo3: 1121-fold increase, Fmo2: 47-fold increase) followed by Cyp2b (Cyp2b10: 153-fold increase, Cyp2b13: 28-fold increase) and 4a (Cyp4a12a: 97% decrease, Cyp4a14: 173-fold increase) subfamilies, as well as Por (21-fold increase). Changes in the mRNA of other phase I enzymes were also produced by changing the diets. Thus the diet is another parameter that should be considered when determining the dose of drugs to produce the desirable but not adverse effects.
Supplementary Material
Acknowledgments
The authors would like to thank all the graduate students, postdoctoral fellows, and Xiaohong Lei in Dr. Klaassen's lab for technical support of the experiments.
Abbreviations
- Adhs
alcohol dehydrogenases
- AhR
aryl hydrocarbon receptor
- Aldhs
aldehyde dehydrogenases
- Aoxs
aldehyde oxidases
- CAR
constitutive androstane receptor
- Cbrs
carbonyl reductases
- Cess
carboxylesterases
- Cyp
Cytochromes P450
- DHA
docosahexaenoic acid
- Ephxs
epoxide hydrolases
- EFA
essential fatty acids
- Fmos
flavin-containing monooxygenases
- FXR
farnesoid X receptor
- HNF
hepatic nuclear factors
- LXR
liver X receptor
- Por
P450 reductase
- Pons
paraoxonases
- PPAR
peroxisome proliferator-activated receptor
- PXR
pregnane X receptor
- Nrf2
Nuclear factor (erythroid-derived 2)-like 2
- Nqos, NAD(P)H
quinone oxidoreductases
- Xdhs
xanthine dehydrogenases
Footnotes
Declaration of interest
The authors report no declarations of interest. This work was supported by the National Institutes of Health [Grants ES009649, ES019487]; and China Postdoctoral Science Foundation [Grant 2013M542147].
References
- Anderson RM, Shanmuganayagam D, Weindruch R. Caloric restriction and aging: studies in mice and monkeys. Toxicol Pathol. 2009;37:47–51. doi: 10.1177/0192623308329476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatia HS, Agrawal R, Sharma S, Huo YX, Ying Z, Gomez-Pinilla F. Omega-3 fatty acid deficiency during brain maturation reduces neuronal and behavioral plasticity in adulthood. PLoS One. 2011;6:e28451. doi: 10.1371/journal.pone.0028451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodfuehrer JI, Zannoni VG. Modulation of the flavin-containing monooxygenase in guinea pigs by ascorbic acid and food restriction. J Nutr. 1987;117:286–90. doi: 10.1093/jn/117.2.286. [DOI] [PubMed] [Google Scholar]
- Cao SX, Dhahbi JM, Mote PL, Spindler SR. Genomic profiling of short- and long-term caloric restriction effects in the liver of aging mice. Proc Natl Acad Sci U S A. 2001;98:10630–5. doi: 10.1073/pnas.191313598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coelho C, Mahro M, Trincao J, Carvalho AT, Ramos MJ, Terao M, Garattini E, Leimkuhler S, Romao MJ. The first mammalian aldehyde oxidase crystal structure: insights into substrate specificity. J Biol Chem. 2012;287:40690–702. doi: 10.1074/jbc.M112.390419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comba A, Lin YH, Eynard AR, Valentich MA, Fernandez-Zapico ME, Pasqualini ME. Basic aspects of tumor cell fatty acid-regulated signaling and transcription factors. Cancer Metastasis Rev. 2011;30:325–42. doi: 10.1007/s10555-011-9308-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa LG, Vitalone A, Cole TB, Furlong CE. Modulation of paraoxonase (PON1) activity. Biochem Pharmacol. 2005;69:541–50. doi: 10.1016/j.bcp.2004.08.027. [DOI] [PubMed] [Google Scholar]
- Dawson DW, Hertzer K, Moro A, Donald G, Chang HH, Go VL, Pandol SJ, Lugea A, Gukovskaya AS, Li G, et al. High-fat, high-calorie diet promotes early pancreatic neoplasia in the conditional KrasG12D mouse model. Cancer Prev Res (Phila) 2013;6:1064–73. doi: 10.1158/1940-6207.CAPR-13-0065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doorn JA, Maser E, Blum A, Claffey DJ, Petersen DR. Human carbonyl reductase catalyzes reduction of 4-oxonon-2-enal. Biochemistry. 2004;43:13106–14. doi: 10.1021/bi049136q. [DOI] [PubMed] [Google Scholar]
- Duffy PH, Lewis SM, Mayhugh MA, McCracken A, Thorn BT, Reeves PG, Blakely SA, Casciano DA, Feuers RJ. Effect of the AIN-93M purified diet and dietary restriction on survival in Sprague-Dawley rats: implications for chronic studies. J Nutr. 2002;132:101–7. doi: 10.1093/jn/132.1.101. [DOI] [PubMed] [Google Scholar]
- Fontana L. Long-term calorie restriction is highly effective in reducing the risk for atherosclerosis in humans. Proceedings of the National Academy of Sciences. 2004;101:6659–63. doi: 10.1073/pnas.0308291101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukasawa T, Kamei A, Watanabe Y, Koga J, Abe K. Short-chain fructooligosaccharide regulates hepatic peroxisome proliferator-activated receptor alpha and farnesoid X receptor target gene expression in rats. J Agric Food Chem. 2010;58:7007–12. doi: 10.1021/jf1006616. [DOI] [PubMed] [Google Scholar]
- Ghose R, Omoluabi O, Gandhi A, Shah P, Strohacker K, Carpenter KC, McFarlin B, Guo T. Role of high-fat diet in regulation of gene expression of drug metabolizing enzymes and transporters. Life Sci. 2011;89:57–64. doi: 10.1016/j.lfs.2011.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu J, Zhang QY, Genter MB, Lipinskas TW, Negishi M, Nebert DW, Ding X. Purification and characterization of heterologously expressed mouse CYP2A5 and CYP2G1: role in metabolic activation of acetaminophen and 2,6-dichlorobenzonitrile in mouse olfactory mucosal microsomes. J Pharmacol Exp Ther. 1998;285:1287–95. [PubMed] [Google Scholar]
- Guesnet P, Alessandri JM. Docosahexaenoic acid (DHA) and the developing central nervous system (CNS) - Implications for dietary recommendations. Biochimie. 2011;93:7–12. doi: 10.1016/j.biochi.2010.05.005. [DOI] [PubMed] [Google Scholar]
- Guo W, Healey JH, Meyers PA, Ladanyi M, Huvos AG, Bertino JR, Gorlick R. Mechanisms of methotrexate resistance in osteosarcoma. Clin Cancer Res. 1999;5:621–7. [PubMed] [Google Scholar]
- Honkakoski P, Negishi M. Regulation of cytochrome P450 (CYP) genes by nuclear receptors. Biochem J. 2000;347:321–37. doi: 10.1042/0264-6021:3470321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikura S, Horie K, Sanai M, Matsumoto K, Hara A. Enzymatic properties of a member (AKR1C19) of the aldo-keto reductase family. Biol Pharm Bull. 2005;28:1075–8. doi: 10.1248/bpb.28.1075. [DOI] [PubMed] [Google Scholar]
- Kanoski SE, Davidson TL. Western diet consumption and cognitive impairment: links to hippocampal dysfunction and obesity. Physiol Behav. 2011;103:59–68. doi: 10.1016/j.physbeh.2010.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassner N, Huse K, Martin HJ, Godtel-Armbrust U, Metzger A, Meineke I, Brockmoller J, Klein K, Zanger UM, Maser E, et al. Carbonyl reductase 1 is a predominant doxorubicin reductase in the human liver. Drug Metab Dispos. 2008;36:2113–20. doi: 10.1124/dmd.108.022251. [DOI] [PubMed] [Google Scholar]
- Klaassen CD, Aleksunes LM. Xenobiotic, bile acid, and cholesterol transporters: function and regulation. Pharmacol Rev. 2010;62:1–96. doi: 10.1124/pr.109.002014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klaassen CD, Lu H. Xenobiotic transporters: ascribing function from gene knockout and mutation studies. Toxicol Sci. 2008;101:186–96. doi: 10.1093/toxsci/kfm214. [DOI] [PubMed] [Google Scholar]
- Konno Y, Kamino H, Moore R, Lih F, Tomer KB, Zeldin DC, Goldstein JA, Negishi M. The nuclear receptors constitutive active/androstane receptor and pregnane x receptor activate the Cyp2c55 gene in mouse liver. Drug Metab Dispos. 2010;38:1177–82. doi: 10.1124/dmd.110.032334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koppaka V, Thompson DC, Chen Y, Ellermann M, Nicolaou KC, Juvonen RO, Petersen D, Deitrich RA, Hurley TD, Vasiliou V. Aldehyde dehydrogenase inhibitors: a comprehensive review of the pharmacology, mechanism of action, substrate specificity, and clinical application. Pharmacol Rev. 2012;64:520–39. doi: 10.1124/pr.111.005538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lands B. Consequences of essential fatty acids. Nutrients. 2012;4:1338–57. doi: 10.3390/nu4091338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levant B, Ozias MK, Carlson SE. Diet (n-3) polyunsaturated fatty acid content and parity interact to alter maternal rat brain phospholipid fatty acid composition. J Nutr. 2006;136:2236–42. doi: 10.1093/jn/136.8.2236. [DOI] [PubMed] [Google Scholar]
- Mullins JG, Parker JE, Cools HJ, Togawa RC, Lucas JA, Fraaije BA, Kelly DE, Kelly SL. Molecular modelling of the emergence of azole resistance in Mycosphaerella graminicola. PLoS One. 2011;6:e20973. doi: 10.1371/journal.pone.0020973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen HL, Yang X, Omiecinski CJ. Expression of a novel mRNA transcript for human microsomal epoxide hydrolase (EPHX1) is regulated by short open reading frames within its 5'-untranslated region. RNA. 2013;19:752–66. doi: 10.1261/rna.037036.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osada J. The use of transcriptomics to unveil the role of nutrients in mammalian liver. ISRN Nutrition. 2013;2013:1–19. doi: 10.5402/2013/403792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips IR, Shephard EA. Flavin-containing monooxygenases: mutations, disease and drug response. Trends Pharmacol Sci. 2008;29:294–301. doi: 10.1016/j.tips.2008.03.004. [DOI] [PubMed] [Google Scholar]
- Prottey C, Hartop PJ, Press M. Correction of the cutaneous manifestations of essential fatty acid deficiency in man by application of sunflower-seed oil to the skin. J Invest Dermatol. 1975;64:228–34. doi: 10.1111/1523-1747.ep12510667. [DOI] [PubMed] [Google Scholar]
- Pryde DC, Dalvie D, Hu Q, Jones P, Obach RS, Tran TD. Aldehyde oxidase: an enzyme of emerging importance in drug discovery. J Med Chem. 2010;53:8441–60. doi: 10.1021/jm100888d. [DOI] [PubMed] [Google Scholar]
- Renaud HJ, Cui JY, Lu H, Klaassen CD. Effect of diet on expression of genes involved in lipid metabolism, oxidative stress, and inflammation in mouse liver-insights into mechanisms of hepatic steatosis. PLoS One. 2014;9:e88584. doi: 10.1371/journal.pone.0088584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varady KA, Roohk DJ, Loe YC, McEvoy-Hein BK, Hellerstein MK. Effects of modified alternate-day fasting regimens on adipocyte size, triglyceride metabolism, and plasma adiponectin levels in mice. J Lipid Res. 2007;48:2212–9. doi: 10.1194/jlr.M700223-JLR200. [DOI] [PubMed] [Google Scholar]
- Vergnes L, Phan J, Strauss M, Tafuri S, Reue K. Cholesterol and cholate components of an atherogenic diet induce distinct stages of hepatic inflammatory gene expression. J Biol Chem. 2003;278:42774–84. doi: 10.1074/jbc.M306022200. [DOI] [PubMed] [Google Scholar]
- Wada T, Gao J, Xie W. PXR and CAR in energy metabolism. Trends Endocrinol Metab. 2009;20:273–9. doi: 10.1016/j.tem.2009.03.003. [DOI] [PubMed] [Google Scholar]
- Witte AVFM, Gellner R, Knecht S, Floel A. From the Cover: Caloric restriction improves memory in elderly humans. Proceedings of the National Academy of Sciences. 2009;106:1255–60. doi: 10.1073/pnas.0808587106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu KC, Cui JY, Klaassen CD. Effect of graded Nrf2 activation on phase-I and -II drug metabolizing enzymes and transporters in mouse liver. PLoS One. 2012;7:e39006. doi: 10.1371/journal.pone.0039006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.