Abstract
CXCR6 genetic variation was described for HIV-1-uninfected black (n=41) and Caucasian (n=40) South Africans. We also investigated the CXCR6 rs2234358 and rs2234355 single nucleotide polymorphisms in HIV-1 disease control in 124 HIV-1-infected drug-naïve black individuals [elite controllers (n=11), viraemic controllers (VCs, n=30), high viral load long-term nonprogressors (HVL LTNPs, n=11) and progressors (n=72)] compared to healthy controls (HCs; n=232). The rs2234358-T allele was underrepresented in VCs (40.0%) compared to HCs (59%, P=0.006), HVL LTNPs (72.7%, P=0.012) and progressors (59%, P=0.014). The rs2234358-TT genotype was underrepresented in VCs (7%) compared to progressors (32%; OR=6.57, P=0.006) and HCs (35%; OR=7.18, P=0.001, Pbonferroni=0.034). The rs2234355-GA genotype was overrepresented in VCs (80%) compared to HCs (50.4%; OR=0.25, P=0.003) and progressors (29.17%; OR=0.10, P=3.8x10−5, Pbonferroni=0.001). The combination of rs2234355-GA in the absence of rs2234358-TT was overrepresented in VCs (80%) compared to HCs (32.6%, OR=0.12, P=1x10−6, Pbonferroni =3.4x10−5) and to progressors (16.7%; OR=0.05, P<1x10−8, Pbonferroni <1x10−7).
Keywords: CXCR6, HIV-1 control, Single Nucleotide Polymorphisms, haplotypes
1. INTRODUCTION
The chemokine receptor, CXCR6 (formally known as STRL33, Bonzo or TYMSTR), is considered a secondary coreceptor used by all HIV-1 strains, HIV-2 and numerous SIV strains [1–6]. Although phylogenetically related to chemokine receptors, CXCR6 is structurally unique in that it lacks cysteine residues in the N terminus and the third extracellular loop that are conserved in all other receptors described to date [5].
Studies conducted in African green monkeys and sooty mangabeys, in which SIV infection is non-pathogenic despite robust viral replication, have shown efficient use of the CXCR6 co-receptor where CCR5 expression is low [7, 8]. These studies suggest that use of alternative entry pathways may enable infection of CCR5-negative target cell populations that can support high viral replication without causing loss of CD4+ T cell homeostasis [7, 8]. Interestingly, HIV-2-infected individuals remain AIDS free for prolonged periods without detectable levels of plasma viral RNA [9] and have been observed to have broad co-receptor usage and tropism, with CXCR6 having been identified as one of the major co-receptors used by HIV-2 [10].
A role for CXCR6 in long-term nonprogression (LTNP) to AIDS has been suggested following a genome wide association study (GWAS) set up to identify genetic variants which affect LTNP [11]. A single nucleotide polymorphism (SNP), rs2234358, located 42 base pairs (bp) downstream from the CXCR6 termination codon was significantly associated with long term nonprogression (LTNP), but not with elite controller (EC) status in HIV-1-infected Caucasian individuals. This association was replicated in three additional independent cohorts of European descent, and represented the first non-HLA-replicated association discovered using a GWAS approach [11]. Furthermore, this study highlights the importance of cohort selection and the valuable information that can be obtained by the study of extreme phenotypes [11].
A second CXCR6 SNP (rs2234355), CXCR6-E3K, which results in a non-conservative change within the N-terminus of CXCR6 (glutamic acid → lysine), is highly prevalent in African Americans (44%), and in HIV-1-infected highly active antiretroviral therapy (HAART)-naïve African Americans, has been associated with increased survival from Pneumocystis jiroveci pneumonia (PCP) [12]. A separate study involving a Caucasian cohort, demonstrated that patients with initial viral load suppression due to HAART showed a faster virological failure in the presence of the CXCR6-3K allele [13]. These two reports highlight interesting roles for the CXCR6-3K allele, however the population difference, the presence of HAART in the latter study and the different outcome measures (i.e. PCP survival and virological failure), make it difficult to come to any conclusions regarding the influence of the CXCR6-3K allele.
To our knowledge, there are no reported studies on the prevalence of the CXCR6 rs2234358 and rs2234355 SNPs in South African populations. In fact, few data exist regarding the SNP profile and haplotype structure of this gene in African populations, and no studies have explored the extent of variation of the CXCR6 gene within and between ethnically distinct populations. We therefore, firstly undertook a comprehensive comparison of CXCR6 gene variation in two South African populations (black and Caucasian). Secondly, we investigated the possible association between the two aforementioned CXCR6 SNPs and different phenotypes of HIV-1 control in HIV-1-infected black South African individuals.
2. MATERIALS AND METHODS
2.1 Study populations
This study was approved by the University of the Witwatersrand Committee for Research on Human Subjects, and informed written consent was obtained from all participants.
Characterization of the CXCR6 gene was carried out on 81 healthy, HIV-1-uninfected South African adult volunteers: 41 black and 40 Caucasian individuals. For association studies, an additional 193 healthy control black individuals were genotyped (rs2234355 and rs2234358 SNPs).
Black South African HIV-infected adults were recruited from sites in Soweto and Johannesburg, and were categorised into different groups according to their ability to control HIV-1 control in the absence of antiretroviral treatment (ART): elite controllers (ECs), HIV-1 viraemic controllers (VCs), high viral load LTNPs (HVL LTNPs), and HIV-1-infected progressors. ECs (n=11) had at least one viral load (VL) test yielding <50 RNA copies/ml and CD4+ T cell counts >500 cells/μl; VCs (n=30) were defined by low viral load set points (VL<2000 RNA copies/ml) with CD4+ T cell counts >500 cells/μl. HVL LTNPs (n=11) had VL>10 000 RNA copies/ml, CD4+ T cell counts >500 cells/μl without apparent CD4+ T cell decline for a period of ≥7 years. HIV-1-infected progressors (n=72) displayed a decline of CD4+ T cell counts from above 500 cells/μl to <350 cells/μl, were initiated on ART and had VL>10 000 RNA copies/ml at time of ART initiation. Information on the HIV-1-infected individuals, including the median length of time since HIV-1 diagnosis, is shown in Table 1.
Table 1.
Characteristics of HIV-1-infected black South African study participants classified according different clinical phenotypes of HIV-1 disease control
| Clinical Phenotype Group | n | Age (years) | Gender (%females) | CD4+ T cell counts(cells/μl)1 (Median and IQR) | Viral load(HIV RNA copies/ml)1 (Median and IQR) | logVL (Median and IQR) | Time since HIV-1 diagnosis (years) (Median and IQR) |
|---|---|---|---|---|---|---|---|
| Elite controllers (ECs) | 11 | 44 (19–54) | 81.8 | 853 (718 – 1022) | <40 (<40 (n=2), <20 (n=9)) | - | 11 9–12) |
| Viraemic controllers (VCs) | 30 | 36 (19–46) | 90.0 | 651 (555–832)2 | 495 (327 - 965)3 | 2.69 (2.51–2.98) | 3 (0–10.35) |
| High viral load long term nonprogressors (HVL LTNPs) | 11 | 41 (31–51) | 81.8 | 663 (635–749) | 54 375 (13 415–77 820) | 4.74 (4.12–4.89) | 8 (8–11) |
| Progressors | 72 | 38 (24–66) | 83.3 | 177 (146–210) | 38 444 (19–853–103 042) | 4.58 (4.30–5.01) | 6 (1–7) |
CD4+ T cell counts and viral loads of controllers used were from time of enrolment whereas for the progressors the last CD4+ T cell count and viral load prior to ART initiation was used.
Three individuals had CD4+ T cell counts <500 cells/μl at time of enrolment; however were included based on length of time of infection since diagnosis to time of enrolment, in the absence of ART (≥9 years).
One individual had a VL>2 000 (VL=6070), but was classified as a VC also due to length of infection since diagnosis with viral control in the absence of ART (9 years).
Stratification of HIV-1 infected individuals into the different groups was largely based upon data (i.e. CD4+ T cell counts and VLs) obtained over several time points and for the most part individuals fitted criteria, however four individuals did not strictly fit the defining criteria for the VC group but were included based on reasons listed in Table 1.
2.2 PCR and sequencing of CXCR6
Genomic DNA was extracted from EDTA-anticoagulated whole blood using QIAamp DNA Mini Kit (QIAGEN, Dusseldorf, Germany) and stored at -20°C until use. A ~7.0 kb continuous region encompassing the CXCR6 open reading frame (ORF), and the 5 and 3 untranslated regions (UTRs) were polymerase chain reaction (PCR) amplified in 4 overlapping sections using Expand High Fidelity PCR System (Roche, Mannheim, Germany) and PCR conditions as recommended by the manufacturer (annealing temperature: 60°C). PCR and sequencing primers were designed using PRIMER DESIGNER for Windows (v. 2.0) (Supplementary Table 1) using the published sequences for CXCR6 (GenBank accession: EF064741.1) as a reference sequence.
All sequencing reactions were carried out using BigDye Terminator version 3.1 chemistry (Applied Biosystems, Foster City, CA, USA). Amplified fragments were sequenced using the 3100 Genetic Analyzer (Applied Biosystems).
2.3 Sequence analysis
Sequence data were assembled and analyzed for the presence of SNPs and indels using SEQUENCHER software version 4.5 (Gene Codes Corporation, Ann Arbor, MI, USA). Assembled sequences were aligned with each other and the published GenBank sequence (accession: EF064741.1) using SEQUENCHER, to identify polymorphisms. The GenBank NCBI SNP database (dbSNP) was searched for all reported SNPs in the CXCR6 gene to determine whether polymorphisms detected in this study had been previously reported. The first nucleotide of the CXCR6 translational start site was designated as +1 and the nucleotide immediately upstream from that as -1.
2.4 Hardy-Weinberg equilibrium
All polymorphic loci detected within the characterized CXCR6 gene region were tested for deviation from Hardy-Weinberg equilibrium using the conventional Monte Carlo exact test of Guo and Thompson (1992) implemented through the computer program TFPGA (Tools for Population Genetic Analyses version 1.3) [14, 15]. The black and Caucasian HIV-1-uninfected population groups were tested independently.
2.5 Inference of putative haplotypes
Analysis of the sequence data generated for CXCR6 revealed certain obvious patterns wherein the presence of a polymorphism at one position was consistently associated with polymorphisms at one or more other positions. These associations were identified as putative intragenic haplotypes. Haplotype estimation and pairwise linkage disequilibrium (LD) analysis were executed by Haploview software version 4.2 [16], and D and r2 pairwise values were calculated.
The frequencies of putative haplotypes were calculated by counting the number of haplotypic alleles and dividing by the total number of alleles. Counting of the haplotypes was irrespective of the presence of additional SNPs not forming part of the haplotypes in question.
2.6 CT shift assays for SNP detection
We developed real-time CT shift PCR assays to detect two single nucleotide polymorphisms (SNPs), rs2234355 and rs2234358, located in the CXCR6 ORF and 3 UTR, respectively. Briefly, the assays involve use of allele-specific PCR with two allele-specific primers [designed with their 3 -end bases complementary to one of the two SNP variants present and with a lock nucleic acid (LNA) modified 3 -end base] and a common pair primer. Two PCR reactions were thus conducted for each sample, each with the common primer and one of the allele-specific primers which bind specifically to either the “wild type” or “mutant” allele. CT shift PCR assays have been successfully used for SNP genotyping in previous studies [17]. PCR conditions and ΔCT calculations were conducted as per Picton et al. (2012). Supplementary Table 2 provides primer details and calculated ΔCT values.
2.7 Statistical tests
Fisher exact tests were performed using the Simple Interactive Statistical Analysis software [18] to calculate statistical significances and exact 95% confidence intervals (CI) of odds ratios (OR) of genotype frequency differences. Two-sided tests were used and statistical significance for analyses was set at P<0.05. We applied Bonferroni corrections to significant associations (indicated by Pbonf ) by multiplying the P values by number of hypotheses tested.
3. RESULTS
3.1 Genetic characterization of the CXCR6 gene in healthy HIV-1-uninfected black and Caucasian South Africans
3.1.1 Single nucleotide polymorphisms (SNPs)
Assembled sequences from 81 HIV-1 uninfected individuals were analyzed for DNA polymorphisms, SNPs and indels within the CXCR6 gene including the 5 flanking region, the ORF and the 3 UTR UTR region. Across the entire 7.0 kb region sequenced, 38 SNPs were identified. To the best of our knowledge, with comparison to the GenBank Gene and dbSNP database, three polymorphisms are newly identified and have been designated as newly identified (NI) in Table 2. However, it should be noted that the three NI SNPs were detected at low frequencies (1.2% population frequency for all three).
Table 2.
Frequencies of identified CXCR6 single nucleotide polymorphisms (SNPs) within the Black (n=41) and Caucasian (n=40) study populations as determined through full length sequencing of CXCR6
| Location on gene | SNP Position | Base change (wt/mut) | Accession numbera | n (Minor allele frequency)b | |
|---|---|---|---|---|---|
| Blacksc | Caucasiansc | ||||
| 5' flanking region (1 689 bp) | −4498 | T/A | rs55920693 | 0 | 7 (0.088) |
| −4495 | G/A | rs3774640 | 12 (0.146) | 16 (0.200) | |
| −4377 | C/G | NI-1 | 1 (0.012) | 0 | |
| −4320 | G/A | rs3774639 | 5 (0.061) | 34 (0.425) | |
| −4087 | C/T | rs3774638 | 12 (0.146) | 16 (0.200) | |
| −4077 | G/A | rs115192433 | 4 (0.049) | 0 | |
| −3996 | G/A | rs56354570 | 0 | 1 (0.013) | |
| −3877 | C/T | rs7627147 | 26 (0.317) | 1 (0.013) | |
| −3674 | G/C | rs2234350 | 6 (0.073) | 20 (0.250) | |
| −3651 | G/A | rs2234351 | 12 (0.146) | 16 (0.200) | |
| −3466 | A/G | rs2234352 | 7 (0.085) | 0 | |
| −3173 | T/C | rs5375415933 | 0 | 1 (0.013) | |
|
| |||||
| Intron (2 921bp) | −2851 | T/C | rs55642383 | 1 (0.012) | 0 |
| −2627 | C/G | rs6785091 | 40 (0.488) | 8 (0.100) | |
| −2376 | T/C | rs6802841 | 4 (0.049) | 0 | |
| −1922 | T/C | rs56196539 | 5 (0.061) | 0 | |
| −1755 | A/C | rs3774635 | 21 (0.256) | 34 (0.425) | |
| −1695 | G/A | rs56184849 | 1 (0.012) | 0 | |
| −1629 | A/G | NI-2 | 1 (0.012) | 0 | |
| −1532 | G/C | rs3774634 | 22 (0.268) | 34 (0.425) | |
| −1351 | C/A | rs936939 | 12 (0.146) | 16 (0.200) | |
| −1091 | C/T | rs146431762 | 3 (0.037) | 0 | |
| −975 | G/A | rs6766548 | 3 (0.037) | 1 (0.013) | |
| −678 | G/A | rs74427539 | 2 (0.024) | 0 | |
| −108 | G/C | rs2234353 | 1 (0.012) | 0 | |
|
| |||||
| ORF (1 029 bp) | +7 | G/A | rs2234355 | 42 (0.512) | 1 (0.013) |
|
| |||||
| 3' UTR (2 369 bp) | +1071 | G/T | rs2234358 | 45 (0.549) | 41 (0.500) |
| +1150 | T/C | rs2234359 | 15 (0.183) | 0 | |
| +1546 | T/C | rs17078463 | 3 (0.037) | 1 (0.013) | |
| +1603 | G/A | rs60514862 | 1 (0.012) | 0 | |
| +1605 | C/G | rs55698153 | 3 (0.037) | 0 | |
| +1622 | A/G | NI-3 | 0 | 1 (0.013) | |
| +1900 | C/T | rs56332428 | 4 (0.049) | 7 (0.088) | |
| +1923 | C/T | rs9825221 | 8 (0.098) | 0 | |
| +1948 | G/T | rs71325095 | 2 (0.024) | 7 (0.088) | |
| +2016 | T/C | rs114052321 | 10 (0.122) | 0 | |
| +2144 | A/G | rs17078464 | 25 (0.305) | 1 (0.013) | |
| +2244 | A/G | rs2054866 | 21 (0.256) | 34 (0.425) | |
Accession numbers of SNPs detected in this study which been previously reported in the SNP database (dbSNP) are listed here, NI indicates putative newly identified polymorphisms not found in dbSNP.
Frequency was calculated for both populations using total number of alleles, i.e., n=82 for Black individuals and n=80 for Caucasian individuals.
Grey shading highlights poylmorphisms where frequencies differ significantly (P<0.05) between the two study populations.
The distribution and frequency of identified CXCR6 SNPs differed between the two control South African populations as shown in Table 2. Significantly different SNP frequencies (P<0.05) have been highlighted. Twenty two and 34 SNPs were detected in Caucasian and black individuals, respectively. Several SNPs were found to occur exclusively in either population groups. Among these, three SNPs occurred at relatively high allelic frequencies in black South Africans and not in Caucasians: rs2234359 (18.3%), rs9825221 (9.8%) and rs114052321 (12.2%). Interestingly, these SNPs are all located within the CXCR6 3 UTR region.
Only one SNP, rs2234355, was detected within the ORF. This polymorphism was largely restricted to black individuals (with the rs2234355-A allele occurring at a high frequency of 51.2% in black individuals whose CXCR6 gene was fully sequenced), while in Caucasians it was detected at a frequency of 1.2%. The rs2234358 SNP, located within the CXCR6 3 UTR, was found to occur at similar frequencies in the two study populations through CXCR6 gene sequencing (54.9% and 50.0% in black and Caucasian individuals, respectively, Table 2).
Since the distribution of both possible alleles at position +1071 of CXCR6 (rs2234358) was approximately equal for both population groups (Table 2), it was not immediately apparent which allele to consider as the ‘wild type’ or ancestral allele. Due to the low mutation rate since the human-chimpanzee divergence, the human allele almost always corresponds to the allele present in chimpanzees [19]. Thus, we selected the ‘G’ allele as this corresponds to the Pan troglodytes sequence at this position. The ‘G’ allele is also considered the ancestral allele in the 1000 genomes project, however it is interesting to note that in all African-origin populations on 1000 genomes project (seven in total) the ‘T’ allele is the predominant allele, whereas for all the remaining populations (American, East and South Asian as well as European), the ‘G’ allele is the major allele [20]. The South African black population thus corresponds to other African-origin populations with having higher ‘T’ allele representation.
No significant deviations from Hardy-Weinberg equilibrium were noted for any of the SNP loci detected in this study in both the study populations.
3.1.2 Indels
Approximately 2 300 bp upstream from the CXCR6 ORF-containing exon, in the CXCR6 intron, there is a region consisting of numerous consecutive repeats of the dinucleotide “TG”, immediately followed by numerous consecutive repeats of the dinucleotide “TA”. Several indels have been documented to occur within this region, both “TG” (rs533226003, rs10562426, rs575276232 and rs57390542) and “TA” (rs546239448 and rs377271591) deletions have been reported. “TG” and “TA” deletions within this region appeared to be highly prevalent in both our South African study populations. These indels were found to occur in various different combinations in different individuals, thus, making it impossible to fully resolve this region. In order to fully resolve this region it would be necessary to clone the region of interest from both alleles for all individuals harbouring indels within this region.
Alignment of reference human CXCR6 sequences to corresponding Pan troglodytes sequences, demonstrated a high degree of homology between the two sequences with the exception of the region just upstream from this indel rich region. Interestingly, sequences of approximately 320 bp immediately upstream from this region are present in the Pan troglodytes but absent in the Homo sapiens 5 UTR regulatory region.
3.1.3 CXCR6 haplotypes
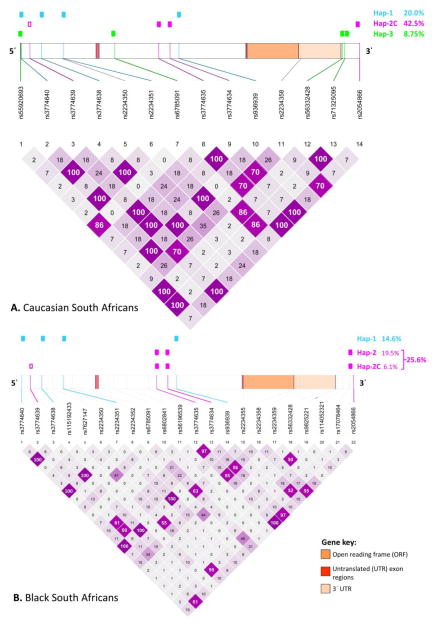
Three putative haplotypes spanning the CXCR6 gene were detected. These have been assigned numerical names, i.e., CXCR6 Hap-1, Hap-2 and Hap-3. The frequency and distribution of these haplotypes was found to differ between black and Caucasian individuals (Figure 1).
Figure 1.
Pairwise linkage disequilibrium (LD) among single nucleotide polymorphisms (SNPs) across the CXCR6 gene in South African Caucasian (A) and black (B) individuals. Only SNPs occurring at frequencies >5% in the two populations are represented. The significance of association (r2) between SNP pair-wise LD values are given in each diamond. A value of 100 represents maximum r2 values and LD. Relative positions of SNPs on the CXCR6 gene as well as identified haplotypes and their prevalence in the two populations are shown.
CXCR6 Hap-1 comprised four SNPs, rs3774640, rs3774638, rs2234351 and rs936939 (r2=1 for all pairwise comparisons), all of which are located upstream from the CXCR6 ORF. Hap-1, was found at similar frequencies in the two study populations (14.6% in black and 20.0% in Caucasian individuals, P=0.410). However, haplotype frequency of the other detected haplotypes differed significantly between the two populations. Hap-3, a four SNP haplotype (rs55920693, rs6785091, rs56332428 and rs71325095) spanning both the CXCR6 5 and 3 flanking regions, was present in Caucasian individuals at a frequency of 8.75% but was not detected in any of the black individuals sequenced (P=0.006). In Caucasian individuals, a single individual was heterozygous for one of the Hap-3 component SNPs (rs6785091) but was homozygous ‘wild type’ for the other 3 Hap-3 SNPs. Thus, the LD between rs6785091 and the other three Hap-3 component SNPs was lower than that between the latter 3 SNPs (r2=0.86 vs. r2=1.0). One of the Hap-3 component SNPs, rs55920693, was absent in the black individuals CXCR6 genotyped in this study, and is a very low frequency SNP in African populations (1% in total African population in 1000 Genomes project) [20]. The remaining Hap-3 component SNPs, rs6785091, rs56332428 and rs71325095, were detected at variable frequencies (48.8%, 4.9% and 2.4%, respectively) and were not in LD in black individuals.
For CXCR6 Hap-2, we have described two haplotypes that appear to be related, namely a three (Hap-2) and a four (Hap-2C) SNP haplotype, with the representation of these differing significantly between the two populations. The apparent root haplotype, Hap-2, consisting of two SNPs located within the CXCR6 intron (rs3774635 and rs3774634) and the third SNP (rs2054866) in the 3 flanking region, was prevalent in black individuals (19.5%) and absent in Caucasians (P<0.0001). The four SNP haplotype, Hap-2C, which comprised the rs3774639 SNP in LD with the Hap-2 SNPs, was found to be highly prevalent in Caucasian individuals (42.5%), and although detected in the black population (6.1%), the two populations differed significantly with respect to representation of this haplotype (P<1x10−7).
CXCR6 gene sequencing failed to detect complete LD between the 3 UTR rs2234358 SNP and other SNPs within CXCR6. However, partial linkage between rs2234358 and Hap-2/2C was observed. In Caucasian individuals, calculated LD between rs2234358 and the four Hap-2C component SNPs were equivalent (r2=0.70). In black individuals, lower LD between rs2234358 and the three Hap-2 component SNPs was observed (r2=0.55 for rs3774635 and rs2054866, r2=0.56 for rs3774634).
Similarly, complete LD between the ORF mutation, E3K (rs2234355), and other CXCR6 SNPs was not detected. However, partial linkage between E3K and the SNPs rs7627147 (r2=0.55), rs6785091 (r2=0.63) and rs17078464 (r2=0.52) was found.
3.1.4 Comparison of the CXCR6 rs2234358 and rs2234355 SNP frequencies to reference populations
We compared the frequencies of the two CXCR6 SNPs (rs2234358 and rs2234355) in our healthy black (larger group post CT shift genotyping of additional individuals) and Caucasian controls to two sub-Saharan African populations (Yoruba from Nigeria and Luhya from Kenya), and two populations of European ancestry (Utah, USA and Tuscany, Italy), respectively (Table 3). The Caucasian populations showed very similar allele frequencies with no significant differences between the populations compared. However, the Yoruba population from Nigeria differed significantly from the black South African population in both the rs2234355 and rs2234358 SNPs, with the Yoruba population having significantly more of the minor alleles and consequently more of the major alleles for both SNPs (Table 3). With respect to genotype comparisons, only heterozygosity for both SNPs did not differ significantly between the Yoruba population and black SA population, whereas both major and minor allele homozygosity differed significantly between the two populations with the Yoruba population having lower major allele homozygosity and higher minor allele homozygosity compared to the black South Africans for both SNPs (Table 3). The sub-Saharan Luhya population did not differ significantly from the black South African population for either of the two SNPs.
Table 3.
Comparison of CXCR6 single nucleotide polymorphism, rs2234355 and rs2234358, prevalence between populations investigated in this study and other populations of similar ancestry
| Populations with sub-Saharan African ancestry | Populations with European ancestry | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
|
|
||||||||||
| Black South Africans (n) | Nigeria- Yoruba (n)a | Black SAs vs.Yorouba P | Kenya- Luhya (n) a | Black SAs vs.Luhya P | Caucasian South Africans (n) | Caucasians (Utah) (n) a | Caucasian SAs vs. Utah P | Tuscans in Italy (n) a | Caucasian SAs vs. Tuscans P | |
| CXCR6 SNP rs2234358 (3 UTR) | ||||||||||
|
| ||||||||||
| Allele | ||||||||||
| G | 0.409 (190) | 0.245 (53) | <0.001 | 0.364 (72) | 0.298 | 0.513 (41) | 0.510 (101) | 1 | 0.565 (121) | 0.432 |
| T | 0.591 (274) | 0.755 (163) | 0.636 (126) | 0.488 (39) | 0.490 (97) | 0.435 (93) | ||||
| Genotype | ||||||||||
| GG | 0.164 (38) | 0.037 (4) | 0.001 | 0.141 (14) | 0.627 | 0.300 (12) | 0.263 (26) | 0.678 | 0.355 (38) | 0.564 |
| GT | 0.491 (114) | 0.417 (45) | 0.202 | 0.444 (44) | 0.471 | 0.425 (17) | 0.495 (49) | 0.574 | 0.421 (45) | 1 |
| TT | 0.345 (80) | 0.546 (59) | <0.001 | 0.414 (41) | 0.262 | 0.275 (11) | 0.242 (24) | 0.829 | 0.224 (24) | 0.664 |
|
| ||||||||||
| CXCR6 SNP rs2234355 (3EK) | ||||||||||
|
| ||||||||||
| Allele | ||||||||||
| G | 0.517 (242) | 0.398 (86) | 0.004 | 0.581 (115) | 0.149 | 0.988 (79) | 1.000 (198) | 0.288 | 0.991 (212) | 1 |
| A | 0.483 (226) | 0.602 (130) | 0.419 (83) | 0.013 (1) | 0 | 0.009 (2) | ||||
| Genotype | ||||||||||
| GG | 0.265 (62) | 0.148 (16) | 0.018 | 0.354 (35) | 0.114 | 0.975 (39) | 1.000 (99) | 0.288 | 0.981 (105) | 1 |
| GA | 0.504 (118) | 0.500 (54) | 1 | 0.455 (45) | 0.472 | 0.025 (1) | 0 | 0.288 | 0.019 (2) | 1 |
| AA | 0.231 (54) | 0.352 (38) | 0.025 | 0.192 (19) | 0.471 | 0 | 0 | 1 | 0 | 1 |
Data from the 1000 Genomes project [20]
SAs: South Africans
Grey shading indicates significant difference (P <0.05) between South African healthy population SNP frequency and reference population
3.2 The effect of CXCR6 rs2234358 and rs2234355 SNPs on HIV-1 control
Successful design of CT shift assays for genotyping the CXCR6 rs2234358 and rs2234355 SNPs allowed us to genotype additional black healthy controls as well drug-naïve HIV-1-infected controllers and progressors demonstrating different degrees of control. The roles of these two SNPs in HIV-1 control were thus determined by comparing the prevalence of these SNPs between respective groups.
3.2.1 CXCR6 rs2234358
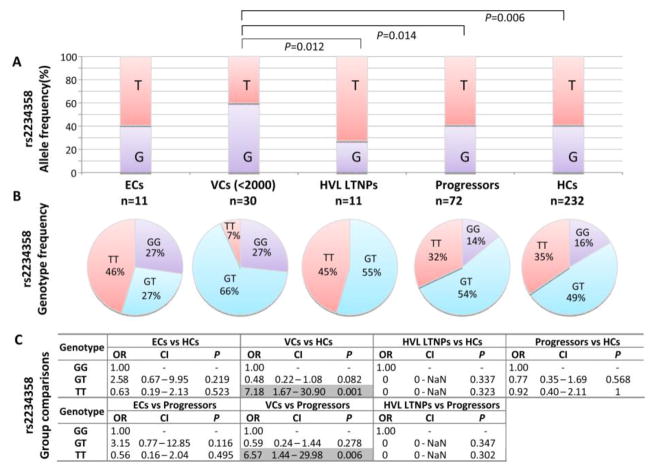
Allele frequency of the 3 UTR rs2234358-T allele was almost identical in HCs and HIV-1-infected individuals, when the HIV-1-infected individuals were grouped together (59.1% and 59.6%, respectively). The frequency of this allele did not significantly differ between HVL LTNP individuals (72.7%) and progressors (59.0%, P=0.348) (Figure 2A). Furthermore rs2234358-T allele frequency did not differ between EC individuals (59.1%) and both progressing and HC individuals. However, the rs2234358-T allele was less prevalent in VC individuals (40.0%) compared to HVL LTNPs (OR=4 [1.37–11.67], P=0.012, Pbonf= 0.3), progressors (OR=2.16 [1.17–3.99], P=0.014, Pbonf=0.50), as well as HCs (OR=2.16 [1.25–3.74], P=0.006, Pbonf =0.20) (Figure 2A).
Figure 2.
Distribution and comparison between CXCR6 rs22345358 allele (A) and genotype frequencies (B & C) within black South African study groups: HIV-1 infected elite controllers (ECs), viraemic controllers (VCs), high viral load long term nonprogressors (HVL LTNPs), progressors and healthy HIV-1-uninfected control individuals (HCs). Where significant (P<0.05), comparisons have been indicated or highlighted.
Comparison of rs2234358 genotype frequencies between the different groups revealed homozygosity for the rs2234358-T allele (TT) to be markedly underrepresented in the VC group (6.7%) in compared to all other study groups (Figure 2B). This was particularly evident when VCs were compared to HCs (34.5% TT, OR=7.19 [1.67–30.90], P=0.001, Pbonf=0.036) and progressors (31.9% TT, OR=6.57 [1.44–29.98], P=0.006, Pbonf=0.22) (Figure 2C). The distribution of rs2234358 genotypes, however, was similar between EC and HC individuals.
It is interesting that in the small group of HVL LTNP individuals (n=11), the rs2234358-T allele frequency was higher (72.7%) than all other study groups. Notably, within the HVL LTNP group no individuals were homozygous for the rs2234358-G allele (Figure 2B).
3.2.2 CXCR6 rs2234355 (E3K)
Through full length sequencing of healthy control individuals we detected only one SNP within the CXCR6 ORF, rs2234355 or E3K. Given that this mutation appeared to be present at significantly different frequencies in the two populations (Table 2) and that, as an ORF non-synonymous mutation it is likely to affect protein function, we designed a SNP detection assay and assessed the prevalence of this SNP in a larger cohort of HC as well as in the different groups of HIV-1-infected individuals.
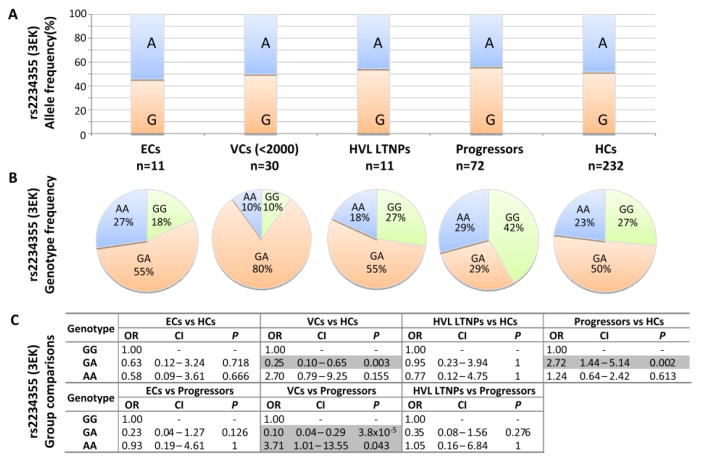
CXCR6-E3K was found to be present at high allelic frequencies within control black individuals (48.3%) and virtually absent in the Caucasian individuals genotyped. This is in agreement with data from 1000 Genomes project that reports a 49% rs2234355-A allele frequency in the total African population and a 1% rs2234355-A allele frequency in the European population [20]. No differences in allele frequency were observed between the groups of HIV-1-uninfected and HIV-1-infected individuals (Figure 3A). Furthermore, no significant allelic differences were noted between individuals demonstrating HIV-1 control and progressing individuals. When the data were stratified and analysed with respect to genotypic distribution of this SNP, marked differences were noted in comparisons between VCs and both HC and progressor groups (Figure 3B and 3C).
Figure 3.
Distribution and comparison between CXCR6 rs22345355 allele (A) and genotype frequencies (B & C) within black South African study groups: HIV-1 infected elite controllers (ECs), viraemic controllers (VCs), high viral load long term nonprogressors (HVL LTNPs), progressors and healthy HIV-1-uninfected control individuals (HCs). Where significant (P<0.05), comparisons have been indicated or highlighted.
Among all the groups of individuals in this study, the lowest proportion of CXCR6-E3K heterozygous (GA) individuals was observed in the progressor group (Figure 3B). The proportion of heterozygous EC individuals was comparable to that observed in the group of healthy controls (54.5% vs. 50.4%, OR=0.63, P=0.718). The proportion of heterozygous individuals was underrepresented in the group of progressors (29.2%) when compared to HCs (OR=2.72 [1.44–5.14], P=0.002, Pbonf=0.07) (Figure 3B and 3C).
Heterozygosity for the E3K SNP was overrepresented (80.0%) in VCs compared to progressing individuals (OR=0.10 [0.04–0.29], P=3.8x10−5, Pbonf=0.001) (Figure 3B and 3C). Similarly, VCs had significantly higher proportions of individuals heterozygous for the E3K SNP compared to HCs (OR=0.25 [0.10–0.65], P=0.003, Pbonf=0.10). Taken together, our results indicate that heterozygosity for the CXCR6 E3K mutation is likely to contribute towards viraemic control of HIV-1, i.e. VCs, who have demonstrated viraemic control through maintaining HIV-1 viral loads at <2000 HIV RNA copies/ml, demonstrate a significantly higher prevalence of the rs2234355-GA genotype compared to individuals who have progressed to disease and relative to the distribution of this genotype in healthy HIV-1-uninfected individuals.
Interestingly, the proportion of individuals homozygous for the E3K mutations (i.e. AA genotype) was underrepresented in VCs compared to progressors (10.0% vs. 29.2%, OR=3.71 [1.01–13.55], P=0.043, Pbonf=1.0).
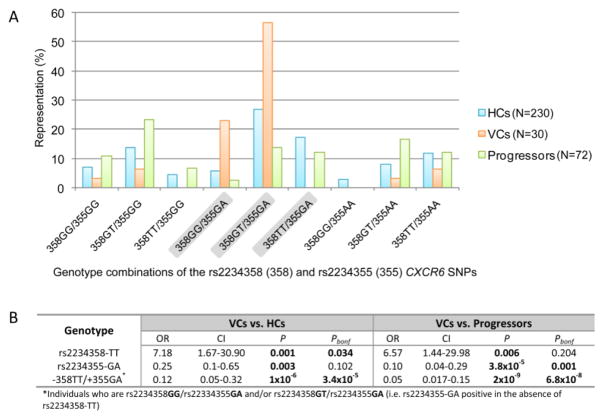
3.2.3 The effect of the combination of CXCR6 rs2234358 and rs2234355 SNPs
Since both the CXCR6 rs2234358 and rs2234355 SNPs were associated with control in the VC group, i.e. underrepresentation of rs2234358-TT and an overrepresentation of rs2234355-GA in VCs ( compared to both HCs and progressors, we investigated whether there was a stronger association with the presence of advantageous rs2234355 heterozygosity in the absence of the deleterious rs2234358-TT genotype (designated as -358TT/+355GA). Representation of all the genotypic combinations of the two SNPs in the HCs, VCs and progressors is shown in Figure 4A. Comparison of the VCs (80%) with both HCs (33%) and progressors (16.7%) revealed a highly significant overrepresentation of -358TT/+355GA in VCs compared to HCs (OR=0.12 [0.05–0.32], P=1x10−6, Pbonf =3.4x10−5) and compared to progressors (OR=0.05 [0.017–0.15], P=2x10−9, Pbonf =6.8x10−8) (Figure 4B). These associations were thus stronger than comparison of either genotype alone, even after Bonferroni correction, suggesting an additive effect of the presence of rs2234355-GA and absence of rs2234358-TT in natural control of HIV-1 in the presence of detectable viraemia (50>VL<2000 RNA copies/ml plasma). The progressors also had significant underrepresentation of -358TT/+355GA combination compared to HCs, however this association did not withstand Bonferroni correction (OR=0.41[0.21–0.80], P=0.011, Pbonf=0.374).
Figure 4.
Bar graph showing representation (percentage) of the combined rs2234358 (designated 358) and rs2234355 (designated 355) genotypes in the Healthy controls (HCs), viraemic controllers (VCs) and progressors (A). The three highlighted (grey shading) genotypes are the combined genotypes that are distinctly differentially represented in VCs compared to both HCs and progressors. (B) Comparison of the rs2234358-TT, rs2234355-GA as well as the combination of the absence of rs2234358-TT and the presence of rs2234355-GA (-358TT/+355GA) between VCs and HCs and progressors showing Bonferroni corrected P values for the comparisons (Pbonf). Significant P values (P<0.05) are shown in bold text.
4. DISCUSSION
The population frequency of two CXCR6 SNPs, rs2234358 and rs2234355, previously found to play a role in HIV-1 disease in independent studies [11–13] was determined for HIV-1-unifected black and Caucasian South Africans. While allele and genotype frequencies were similar between Caucasian South Africans and other populations with European ancestry, frequencies of both these SNPs in black South Africans differed markedly from that reported for the sub-Saharan Yoruba population from Nigeria. The East sub-Saharan African Luhya population from Kenya however was more similar to the black South African population and also differed markedly from the Yoruba population (data not shown). In a study testing the hypothesis that exposure to simian immunodeficiency virus (SIV) may have led to genetic selection for alleles conferring resistance to immunodeficiency viruses within populations living in SIV endemic regions, the CXCR6 E3K mutation was among the postulated HIV-1 protective alleles found to occur at significantly higher frequencies in Biaka Western Pygmies compared to Mbuti Eastern Pygmies [21]. The former population group historically resided in communities in the same geographical region as the central African chimpanzee (Pan troglodytes troglodytes), natural hosts for the SIV strains from which HIV-1 is believed to have originated [21]. Interestingly, we found the frequency of the E3K mutation to be higher in black South Africans (48.3%) than in both the Biaka (38%) and Mbuti (12%) as well as the Luhya (41.9%), however the Yoruba population had the highest prevalence of both the E3K (60%) and the rs2234358 3 UTR (76%) mutations. These data suggest that CXCR6 in Western sub-Saharan African populations has likely experienced considerable evolutionary pressure and also highlight the importance of assessing genetic variation of individual genes within different populations.
Mutations within the CXCR6 chemokine receptor ORF could affect CXCR6 function by causing conformational changes in the protein, affecting cell surface density of the coreceptor, altering binding affinity to its ligand, CXCL16, altering affinity of binding to HIV-1 and/or affecting CXCR6 signalling and chemotactic ability. In a study that assessed the role of negatively charged amino acid residues within the CXCR6 protein on its interaction with its chemokine ligand, CXCL16, substitution of the negatively charged glutamic acid at position three of the CXCR6 protein with its neutral counterpart, glutamine (E3Q), resulted in markedly reduced CXCR6 surface expression [22]. The authors hence postulated that the naturally occurring CXCR6-E3K mutation, where glutamic acid is replaced with the positively charged lysine, may have a similar effect on cell surface expression of the chemokine receptor.
Previous reports suggest that there exists a complex relationship in the association between the CXCR6 E3K (rs2234355) mutation and HIV-1 disease progression: the CXCR6 3KK genotype was demonstrated to associate with longer survival among HIV-1-infected African Americans infected with PCP, a prevalent opportunistic infection, in comparison to individuals with the CXCR6 3EK and 3EE genotypes [12]. The presence of the CXCR6 3K gene in Caucasian HIV-1-infected individuals with initial viral load suppression due to ART associated with faster virological failure compared with individuals who lacked this allele [13]. These are however different populations with different phenotypic outcomes. Our results demonstrated a very strong association with rs2234355 heterozygosity (CXCR6 3EK) and HIV-1 control. If the E3K mutation does indeed lead to reduced surface expression as predicted [22], then the results suggest that balance in CXCR6 expression may be a key factor in HIV-1 control, highlighting the importance of a better understanding of CXCR6 function.
It is not always clear how SNPs within the regulatory region of a gene, such as rs2234358 located 42 bp downstream from the CXCR6 stop codon, may affect gene expression. SNPs within regulatory genes can influence gene expression in a number of ways such as through altered RNA splicing, transcription factor binding, DNA methylation and microRNA (miRNA) recruitment [23]. Determining the role of a particular SNP in disease pathology is further complicated when it is in high linkage disequilibrium with other genetic polymorphisms, making it unclear which particular SNP or SNP interaction is conferring a particular disease phenotype. Thus, in this study, in addition to calculating the population frequency of the above mentioned CXCR6 SNPs, we characterised genetic polymorphisms and intragenic haplotypes found within the CXCR6 gene in two divergent study populations through sequencing of the entire gene in healthy control individuals.
In line with expected greater genetic diversity and lower levels of linkage disequilibrium within African populations in comparison to European-originating populations [24, 25], variation within the CXCR6 gene was found to be greater in black individuals in comparison to Caucasians, with a larger number of SNPs identified in black individuals. In addition, CXCR6 gene haplotype structure and distribution was found to differ between the two populations. The two CXCR6 SNPs of particular interest in the context of HIV-1 disease progression were not found to be in complete linkage disequilibrium with other identified SNPs. However, partial linkage between rs2234358 and CXCR6 Hap-2 and Hap-2C was found. The relationship between rs2234358 and Hap-2 was strongest in Caucasians. Our findings, however, differ from what has been previously reported for Caucasians. Limou et al. (2010) report high linkage disequilibrium with rs2234358 and 2-SNP haplotypes composed of the rs2234350 SNP and either one of the following SNPs: rs3774640, rs3774638 or rs2234351. The latter three SNPs form part of the CXCR6 Hap-1 haplotype identified in this study and were found to always occur together irrespective of population (r2=1). The rs2234350 SNP, however, exhibited low linkage with rs2234358 in our Caucasian individuals (r2=0.35) and showed no LD in black individuals (r2=0.08).
The CXCR6 3 UTR SNP, rs2234358, may exert an effect on disease progression either by directly altering CXCR6 expression or through interaction/association with Hap-2 SNPs within the CXCR6 promoter region (5 regulatory region). Since the discovery of miRNAs in 1993 [26], these regulatory elements have been demonstrated to play a key role in gene regulation mainly through specific binding to complementary regions within the 3 UTR of genes. Thus, to test for a direct effect on gene regulation, we carried out in silico analysis using PITA software [27] to predict miRNA binding to the rs2234358-G and rs2234358-T alleles. Results indicate that the two alleles differ both in miRNA profiles and binding affinity of common miRNAs thus indicating that mutations at this point might affect CXCR6 expression (data not shown). This however, is only predictive and would need to be experimentally validated. If the influence of rs2234358 is through its’ association with other SNPs upstream, then individuals harbouring this mutation could be divided into two groups, i.e., individuals with the rs2234358 and Hap-2 and individuals with rs2234358 in the absence of Hap-2. Studies could thus be structured to test whether individuals harbouring the rs2234358 mutation exhibit two different disease phenotypes by genotyping individuals within study cohorts for both rs2234358 and Hap-2. Different LD patterns between populations could also explain why different population groups often display variable disease phenotypes despite similar allele frequencies of a given SNP. Our results indicate that the rs2234358-G allele associates with HIV-1 virus control in black South Africans, an observation which is in agreement with that reported for European Caucasians [11]. Furthermore, Limou et al. (2010) found the association with HIV-1 disease progression to be strongest with rs2234358 compared to other SNPs, in partial LD with rs2234358, which were also detected in our study. Taken together, it seems more probable that the rs2234358 SNP, rather than the Hap-2 SNPs, might affect CXCR6 expression. Further experiments are necessary to explore this hypothesis. What was interesting from the full gene sequencing data generated for CXCR6 is that out of the twelve SNPs detected in the 3 UTR (both black and Caucasian populations), four of these SNPs (rs2234359, rs9825221, rs114052321 and rs17078464) were differentially represented in two populations with relatively high prevalence in the black South Africans and zero to low prevalence in the Caucasians. Preliminary in silico analysis using both PITA [27] and TargetScan [28] software indicated that these SNPs may be located in miRNA binding sites and consequently involved in CXCR6 expression (data not shown).
When analysed independently, we found both rs2234358 and E3K associate with HIV-1 viraemic but not elite or long term control with high viraemia (HVL LTNP). Furthermore these two SNPs exert an additive effect in the VCs in that the proportion of individuals who have the protective rs2234355- GA genotype and lack the deleterious rs2234355-TT genotype is highly enriched in VCs compared to both HCs and progressors (Pbonf<0.0001 for both comparisons). Although these two CXCR6 SNPs exhibit an association with HIV-1 disease control, they do not associate with elite controller status in black South Africans. Our results reflect similar findings for the rs2234358 SNP, where this SNP associates with HIV-1 long-term non-progression but not elite control in HIV-1-infected Caucasian individuals [11]. Criteria for LTNP status in the Limou et al. study (2010) included VLs>100 RNA copies/ml of plasma, and although our VCs were defined by 50>VLs<2000 RNA copies/ml plasma, only 2/30 of our VCs had VLs<100 RNA copies/ml plasma and 1/30 had a VL<400 RNA copies/ml plasma (LOD of assay) at the time of inclusion. Thus the LTNPs in the Limou et al. (2010) study and the VCs in this study are comparable groups. When considering other genetic loci associated with HIV-1 disease control, other studies have demonstrated a similar distinction between LTNP and EC individuals. For example, enrichment of the killer immunoglobulin-like receptor, KIR3DS1, has been demonstrated in LTNP individuals [29], however, this protective allele was reported to occur at low frequencies in a study of elite controllers [30]. Similarly, the -35C variant located 35 kb upstream from the HLA-C gene has been demonstrated to be overrepresented in a cohort of LTNPs, but not in elite controllers [31]. Taken together, these studies emphasise that while ECs, VCs and HVL LTNPs all demonstrate HIV-1 disease control, the defining characteristics of each group differ, demonstrating that some of the underlying determinants of resistance to HIV-1 disease progression are likely to be different.
CXCR6 is largely expressed on effector memory CD4+ T cells, rather than longer-lived central memory CD4+ T cell subsets [32]. Many scenarios of well-controlled infection or disease support a skewing of SIV/HIV infection towards effector memory CD4+ T cells that may be more dispensable, resulting in a more protected CD4+ T cell memory pool. These include non-pathogenic infection of nonhuman primates such as sooty mangabeys and African green monkeys [33], human LTNP/ECs [34, 35], post-treatment controllers [36], and unique groups - adult viraemic nonprogressors [37] and paediatric nonprogressors [38] - who have high viral loads, low immune activation and conserved CD4+ T cell counts. Furthermore, the expression of CXCR6 is critical for antigen-specific memory liver NK cells [39] , and CXCX6 correlates with CD56 expression on T cells which associates with cytotoxic potential [32]. These attributes, together with the immunogenetics findings from this study which provide additional support for a pivotal role of the CXCR6 coreceptor in HIV-1 disease, irrespective of host ethnic background, highlight the importance of a better understanding of the CXCR6-CXCL16 immune axis to inform the development of preventative and therapeutic vaccines as well as novel therapeutics for HIV cure.
Supplementary Material
Highlights.
Sequencing and variant characterization of complete CXCR6 gene in two populations
Black and Caucasian South Africans differ markedly in SNP and haplotype profiles
Two CXCR6 SNPs (rs2234358; rs2234355) studied with respect to HIV-1 disease control Both CXCR6 SNPs play an additive role in viraemic control in black South Africans
CXCR6, an HIV-1, HIV-2 and SIV co-receptor, is a key molecule natural control
Acknowledgments
This work is based on the research supported by grants awards from South African Research Chairs Initiative of the Department of Science and Technology and National Research Foundation of South Africa, the South African Medical Research Council (Self-Initiated Research Grant), the Strategic Health Innovation Partnerships (SHIP) Unit of the South African Medical Research Council (a grantee of the Bill & Melinda Gates Foundation), the Poliomyelitis Research Foundation (PRF) and the Claude Leon Foundation (ACP: Postdoctoral Fellowship). The Longitudinal Study of HIV-Associated Lung Infections in Soweto (progressor group) was funded by the National Institutes of Health, USA (R01HL090312 and P30AI094189: R. E. Chaisson).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Alkhatib G, Liao F, Berger EA, Farber JM, Peden KW. A new SIV co-receptor, STRL33. Nature. 1997;388:238. doi: 10.1038/40789. [DOI] [PubMed] [Google Scholar]
- 2.Deng HK, Unutmaz D, KewalRamani VN, Littman DR. Expression cloning of new receptors used by simian and human immunodeficiency viruses. Nature. 1997;388:296–300. doi: 10.1038/40894. [DOI] [PubMed] [Google Scholar]
- 3.Edinger AL, Hoffman TL, Sharron M, Lee B, O'Dowd B, Doms RW. Use of GPR1, GPR15, and STRL33 as coreceptors by diverse human immunodeficiency virus type 1 and simian immunodeficiency virus envelope proteins. Virology. 1998;249:367–378. doi: 10.1006/viro.1998.9306. [DOI] [PubMed] [Google Scholar]
- 4.Isaacman-Beck J, Hermann EA, Yi Y, Ratcliffe SJ, Mulenga J, Allen S, Hunter E, Derdeyn CA, Collman RG. Heterosexual transmission of human immunodeficiency virus type 1 subtype C: Macrophage tropism, alternative coreceptor use, and the molecular anatomy of CCR5 utilization. Journal of virology. 2009;83:8208–8220. doi: 10.1128/JVI.00296-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liao F, Alkhatib G, Peden KW, Sharma G, Berger EA, Farber JM. STRL33, A novel chemokine receptor-like protein, functions as a fusion cofactor for both macrophage-tropic and T cell line-tropic HIV-1. The Journal of experimental medicine. 1997;185:2015–2023. doi: 10.1084/jem.185.11.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tscherning-Casper C, Vodros D, Menu E, Aperia K, Fredriksson R, Dolcini G, Chaouat G, Barre-Sinoussi F, Albert J, Fenyo EM. Coreceptor usage of HIV-1 isolates representing different genetic subtypes obtained from pregnant Cameroonian women. European Network for In Utero Transmission of HIV-1. Journal of acquired immune deficiency syndromes. 2000;24:1–9. doi: 10.1097/00126334-200005010-00001. [DOI] [PubMed] [Google Scholar]
- 7.Riddick NE, Hermann EA, Loftin LM, Elliott ST, Wey WC, Cervasi B, Taaffe J, Engram JC, Li B, Else JG, Li Y, Hahn BH, Derdeyn CA, Sodora DL, Apetrei C, Paiardini M, Silvestri G, Collman RG. A novel CCR5 mutation common in sooty mangabeys reveals SIVsmm infection of CCR5-null natural hosts and efficient alternative coreceptor use in vivo. PLoS pathogens. 2010;6:e1001064. doi: 10.1371/journal.ppat.1001064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wetzel KS, Yi Y, Elliott ST, Romero D, Jacquelin B, Hahn BH, Muller-Trutwin M, Apetrei C, Pandrea I, Collman RG. CXCR6-mediated SIVagmSab Entry into Sabaeus African Green Monkey Lymphocytes Implicates Widespread Use of Non-CCR5 Pathways in Natural Host Infections. Journal of virology. 2016 doi: 10.1128/JVI.01626-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marlink R, Kanki P, Thior I, Travers K, Eisen G, Siby T, Traore I, Hsieh CC, Dia MC, Gueye EH, et al. Reduced rate of disease development after HIV-2 infection as compared to HIV-1. Science. 1994;265:1587–1590. doi: 10.1126/science.7915856. [DOI] [PubMed] [Google Scholar]
- 10.Blaak H, Boers PH, Gruters RA, Schuitemaker H, van der Ende ME, Osterhaus AD. CCR5, GPR15, and CXCR6 are major coreceptors of human immunodeficiency virus type 2 variants isolated from individuals with and without plasma viremia. Journal of virology. 2005;79:1686–1700. doi: 10.1128/JVI.79.3.1686-1700.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Limou S, Coulonges C, Herbeck JT, van Manen D, An P, Le Clerc S, Delaneau O, Diop G, Taing L, Montes M, van't Wout AB, Gottlieb GS, Therwath A, Rouzioux C, Delfraissy JF, Lelievre JD, Levy Y, Hercberg S, Dina C, Phair J, Donfield S, Goedert JJ, Buchbinder S, Estaquier J, Schachter F, Gut I, Froguel P, Mullins JI, Schuitemaker H, Winkler C, Zagury JF. Multiple-cohort genetic association study reveals CXCR6 as a new chemokine receptor involved in long-term nonprogression to AIDS. The Journal of infectious diseases. 2010;202:908–915. doi: 10.1086/655782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Duggal P, An P, Beaty TH, Strathdee SA, Farzadegan H, Markham RB, Johnson L, O'Brien SJ, Vlahov D, Winkler CA. Genetic influence of CXCR6 chemokine receptor alleles on PCP-mediated AIDS progression among African Americans. Genes and immunity. 2003;4:245–250. doi: 10.1038/sj.gene.6363950. [DOI] [PubMed] [Google Scholar]
- 13.Passam AM, Sourvinos G, Krambovitis E, Miyakis S, Stavrianeas N, Zagoreos I, Spandidos DA. Polymorphisms of Cx(3)CR1 and CXCR6 receptors in relation to HAART therapy of HIV type 1 patients. AIDS research and human retroviruses. 2007;23:1026–1032. doi: 10.1089/aid.2006.0248. [DOI] [PubMed] [Google Scholar]
- 14.Guo SW, Thompson EA. Performing the exact test of Hardy-Weinberg proportion for multiple alleles. Biometrics. 1992;48:361–372. [PubMed] [Google Scholar]
- 15.Miller MP. Tools for population genetic analyses version 1.3. 1997. [Google Scholar]
- 16.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 17.Picton AC, Shalekoff S, Paximadis M, Tiemessen CT. Marked differences in CCR5 expression and activation levels in two South African populations. Immunology. 2012;136:397–407. doi: 10.1111/j.1365-2567.2012.03592.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Uitenbrook DG. SISA-Binomial. 1997 http://quantitativeskills.com/sisa/distributions/binomial.
- 19.Cargill M, Altshuler D, Ireland J, Sklar P, Ardlie K, Patil N, Shaw N, Lane CR, Lim EP, Kalyanaraman N, Nemesh J, Ziaugra L, Friedland L, Rolfe A, Warrington J, Lipshutz R, Daley GQ, Lander ES. Characterization of single-nucleotide polymorphisms in coding regions of human genes. Nat Genet. 1999;22:231–238. doi: 10.1038/10290. [DOI] [PubMed] [Google Scholar]
- 20.C. The Genomes Project. A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhao K, Ishida Y, Oleksyk TK, Winkler CA, Roca AL. Evidence for selection at HIV host susceptibility genes in a West Central African human population. BMC evolutionary biology. 2012;12:237. doi: 10.1186/1471-2148-12-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Petit SJ, Chayen NE, Pease JE. Site-directed mutagenesis of the chemokine receptor CXCR6 suggests a novel paradigm for interactions with the ligand CXCL16. European journal of immunology. 2008;38:2337–2350. doi: 10.1002/eji.200838269. [DOI] [PubMed] [Google Scholar]
- 23.Huang Q. Genetic Study of Complex Diseases in the Post-GWAS Era. Journal of genetics and genomics = Yi chuan xue bao. 2015;42:87–98. doi: 10.1016/j.jgg.2015.02.001. [DOI] [PubMed] [Google Scholar]
- 24.Tishkoff SA, Reed FA, Friedlaender FR, Ehret C, Ranciaro A, Froment A, Hirbo JB, Awomoyi AA, Bodo JM, Doumbo O, Ibrahim M, Juma AT, Kotze MJ, Lema G, Moore JH, Mortensen H, Nyambo TB, Omar SA, Powell K, Pretorius GS, Smith MW, Thera MA, Wambebe C, Weber JL, Williams SM. The genetic structure and history of Africans and African Americans. Science. 2009;324:1035–1044. doi: 10.1126/science.1172257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tishkoff SA, Verrelli BC. Patterns of human genetic diversity: implications for human evolutionary history and disease. Annu Rev Genomics Hum Genet. 2003;4:293–340. doi: 10.1146/annurev.genom.4.070802.110226. [DOI] [PubMed] [Google Scholar]
- 26.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 27.Kertesz M, Iovino N, Unnerstall U, Gaul U, Segal E. The role of site accessibility in microRNA target recognition. Nat Genet. 2007;39:1278–1284. doi: 10.1038/ng2135. [DOI] [PubMed] [Google Scholar]
- 28.Agarwal V, Bell GW, Nam JW, Bartel DP. Predicting effective microRNA target sites in mammalian mRNAs. eLife. 2015;4 doi: 10.7554/eLife.05005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jiang Y, Chen O, Cui C, Zhao B, Han X, Zhang Z, Liu J, Xu J, Hu Q, Liao C, Shang H. KIR3DS1/L1 and HLA-Bw4-80I are associated with HIV disease progression among HIV typical progressors and long-term nonprogressors. BMC infectious diseases. 2013;13:405. doi: 10.1186/1471-2334-13-405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.O'Connell KA, Han Y, Williams TM, Siliciano RF, Blankson JN. Role of natural killer cells in a cohort of elite suppressors: low frequency of the protective KIR3DS1 allele and limited inhibition of human immunodeficiency virus type 1 replication in vitro. Journal of virology. 2009;83:5028–5034. doi: 10.1128/JVI.02551-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ballana E, Ruiz-de Andres A, Mothe B, Ramirez de Arellano E, Aguilar F, Badia R, Grau E, Clotet B, del Val M, Brander C, Este JA. Differential prevalence of the HLA-C -- 35 CC genotype among viremic long term non-progressor and elite controller HIV+ individuals. Immunobiology. 2012;217:889–894. doi: 10.1016/j.imbio.2011.12.012. [DOI] [PubMed] [Google Scholar]
- 32.Kim CH, Kunkel EJ, Boisvert J, Johnston B, Campbell JJ, Genovese MC, Greenberg HB, Butcher EC. Bonzo/CXCR6 expression defines type 1-polarized T-cell subsets with extralymphoid tissue homing potential. The Journal of clinical investigation. 2001;107:595–601. doi: 10.1172/JCI11902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Paiardini M, Cervasi B, Reyes-Aviles E, Micci L, Ortiz AM, Chahroudi A, Vinton C, Gordon SN, Bosinger SE, Francella N, Hallberg PL, Cramer E, Schlub T, Chan ML, Riddick NE, Collman RG, Apetrei C, Pandrea I, Else J, Munch J, Kirchhoff F, Davenport MP, Brenchley JM, Silvestri G. Low levels of SIV infection in sooty mangabey central memory CD(4)(+) T cells are associated with limited CCR5 expression. Nature medicine. 2011;17:830–836. doi: 10.1038/nm.2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Descours B, Avettand-Fenoel V, Blanc C, Samri A, Melard A, Supervie V, Theodorou I, Carcelain G, Rouzioux C, Autran B, Group AACS. Immune responses driven by protective human leukocyte antigen alleles from long-term nonprogressors are associated with low HIV reservoir in central memory CD4 T cells. Clinical infectious diseases : an official publication of the Infectious Diseases Society of America. 2012;54:1495–1503. doi: 10.1093/cid/cis188. [DOI] [PubMed] [Google Scholar]
- 35.Potter SJ, Lacabaratz C, Lambotte O, Perez-Patrigeon S, Vingert B, Sinet M, Colle JH, Urrutia A, Scott-Algara D, Boufassa F, Delfraissy JF, Theze J, Venet A, Chakrabarti LA. Preserved central memory and activated effector memory CD4+ T-cell subsets in human immunodeficiency virus controllers: an ANRS EP36 study. Journal of virology. 2007;81:13904–13915. doi: 10.1128/JVI.01401-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Saez-Cirion A, Bacchus C, Hocqueloux L, Avettand-Fenoel V, Girault I, Lecuroux C, Potard V, Versmisse P, Melard A, Prazuck T, Descours B, Guergnon J, Viard JP, Boufassa F, Lambotte O, Goujard C, Meyer L, Costagliola D, Venet A, Pancino G, Autran B, Rouzioux C, Group AVS. Post-treatment HIV-1 controllers with a long-term virological remission after the interruption of early initiated antiretroviral therapy ANRS VISCONTI Study. PLoS pathogens. 2013;9:e1003211. doi: 10.1371/journal.ppat.1003211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Klatt NR, Bosinger SE, Peck M, Richert-Spuhler LE, Heigele A, Gile JP, Patel N, Taaffe J, Julg B, Camerini D, Torti C, Martin JN, Deeks SG, Sinclair E, Hecht FM, Lederman MM, Paiardini M, Kirchhoff F, Brenchley JM, Hunt PW, Silvestri G. Limited HIV infection of central memory and stem cell memory CD4+ T cells is associated with lack of progression in viremic individuals. PLoS pathogens. 2014;10:e1004345. doi: 10.1371/journal.ppat.1004345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Muenchhoff M, Adland E, Karimanzira O, Crowther C, Pace M, Csala A, Leitman E, Moonsamy A, McGregor C, Hurst J, Groll A, Mori M, Sinmyee S, Thobakgale C, Tudor-Williams G, Prendergast AJ, Kloverpris H, Roider J, Leslie A, Shingadia D, Brits T, Daniels S, Frater J, Willberg CB, Walker BD, Ndung'u T, Jooste P, Moore PL, Morris L, Goulder P. Nonprogressing HIV-infected children share fundamental immunological features of nonpathogenic SIV infection. Science translational medicine. 2016;8:358ra125. doi: 10.1126/scitranslmed.aag1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Paust S, Gill HS, Wang BZ, Flynn MP, Moseman EA, Senman B, Szczepanik M, Telenti A, Askenase PW, Compans RW, von Andrian UH. Critical role for the chemokine receptor CXCR6 in NK cell-mediated antigen-specific memory of haptens and viruses. Nature immunology. 2010;11:1127–1135. doi: 10.1038/ni.1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.