Abstract
Sulfomenaquinone (SMK) is a recently identified metabolite that is unique to the Mycobacterium tuberculosis (M. tuberculosis) complex and is shown to modulate its virulence. Here, we report the identification of the SMK biosynthetic operon that, in addition to a previously identified sulfotransferase stf 3, includes a putative cytochrome P450 gene (cyp128) and a gene of unknown function, rv2269c. We demonstrate that cyp128 and stf 3 are sufficient for the biosynthesis of SMK from menaquinone and rv2269c exhibits promoter activity in M. tuberculosis. Loss of Stf3 expression, but not that of Cyp128, is correlated with elevated levels of menaquinone-9, an essential component in the electron-transport chain in M. tuberculosis. Finally, we showed in a mouse model of infection that the loss of cyp128 exhibits a hypervirulent phenotype similar to that in previous studies of the stf 3 mutant. These findings provide a platform for defining the molecular basis of SMK’s role in M. tuberculosis pathogenesis.
Keywords: Mycobacterium tuberculosis, menaquinone, cell wall, cytochrome P450, lipids, mass spectrometry
Graphical abstract
Mycobacterium tuberculosis has a unique cell envelope composed of multiple distinct lipid layers outside of the plasma membrane. The complex lipids in the M. tuberculosis cell wall serve multiple roles in bacteria. It is an effective protective barrier against oxidative and osmotic stress.1 Many of the lipids have been shown to modulate the host immune response.2,3 Furthermore, bacterial lipid synthesis alleviates the toxicity from the degradation of cholesterol and odd-chain fatty acids.4,5 With the aid of high-resolution mass spectrometry, we are discovering the range and complexity of lipids produced by M. tuberculosis. It has been very challenging to identify the function and regulation of the lipids. Accordingly, defining a lipid’s biosynthetic pathways is a crucial first step toward elucidating its role in M. tuberculosis pathogenesis.
Previous work from our laboratory identified and structurally characterized a novel sulfated lipid metabolite in M. tuberculosis, which we initially named S881 on the basis of its mass.6,7 Using mass spectrometry, we characterized the structure of S881 as an unprecedented terminally sulfated menaquinone.8 We renamed S881 as sulfomenaquinone (SMK), which is more descriptive of its structure. To date, SMK is the only reported example of a sulfated menaquinone derivative in any organism. SMK was discovered in a screen of M. tuberculosis sulfotransferase mutants. We found that the disruption of the sulfotransferase stf 3 (Δstf 3) resulted in a loss of this metabolite, indicating that stf 3 was required for SMK biosynthesis. Intriguingly, the Δstf 3 mutant strain exhibited hypervirulence in a mouse infection model, one of the first examples of this highly unusual phenotype. The Δstf 3-infected mice had an increased time to death compared to those infected with the wild-type (WT) M. tuberculosis strain. Although the Δstf 3 mutant showed no growth phenotype in vitro, mice infected with the Δstf 3 mutant had higher CFU in the lungs compared to WT.6,9
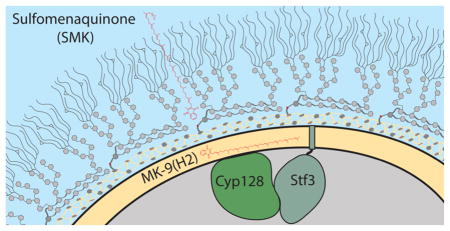
Elucidating the machinery underlying SMK biosynthesis is an important first step toward understanding its role in M. tuberculosis pathogenesis. To this end, we analyzed the M. tuberculosis genomic region surrounding stf 3. We observed a predicted operon that contains three genes: stf 3 (rv2267c), cyp128 (rv2268c, a putative cytochrome P450), and rv2269c (a conserved hypothetical protein)10 (Figure 1A). We propose that the biosynthetic pathway for SMK involves the hydroxylation of menaquinone, a reaction type commonly mediated by P450 enzymes, followed by Stf3-mediated sulfation (Figure 1B).
Figure 1.
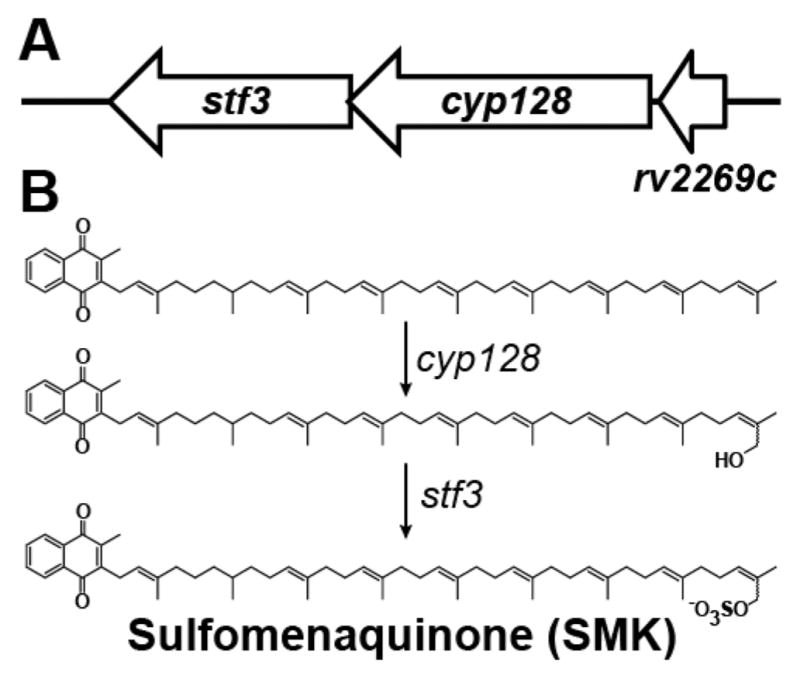
Proposed SMK biosynthesis in M. tuberculosis. (A) The stf 3 operon contains three genes: stf 3 (rv2297c), the previously identified sulfotransferase required for SMK production; cyp128 (rv2298), a putative cytochrome P450; and rv2269c, a conserved hypothetical protein of unknown function. The annotated start site of stf 3 is 6 bp before the annotated stop of cyp128. (B) Proposed biosynthesis of SMK. Menaquinone is hydroxylated by cyp128 followed by stf 3 sulfation.
In this report, we demonstrate that cyp128 is essential to SMK biosynthesis in M. tuberculosis and in Mycobacterium smegmatis strains engineered to produce SMK de novo. In M. tuberculosis, the disruption of either stf 3 or cyp128 results in the loss of SMK production, as determined by mass spectrometry and radiolabeling analysis. We found that the steady-state level of menaquinone is higher in the Δstf 3 mutant than in WT, but no such difference was observed in the Δcyp128 mutant. We hypothesized that changes in SMK and menaquinone levels would affect bacterial respiration and overall growth fitness. SMK mutants were similarly susceptible to electron-transport chain inhibitors as WT. Although SMK production had no effect on bacterial growth or respiration in vitro, levels of SMK appear to be regulated both transcriptionally and by active transport to the bacterial surface. In the mouse model of infection, we compared the Δcyp128 mutant to the Δstf 3 mutant and their respective complemented strains. We found that the Δcyp128 mutant has increased growth in the lungs of mice compared to WT, further suggesting that SMK plays a role in host immune modulation.
RESULTS AND DISCUSSION
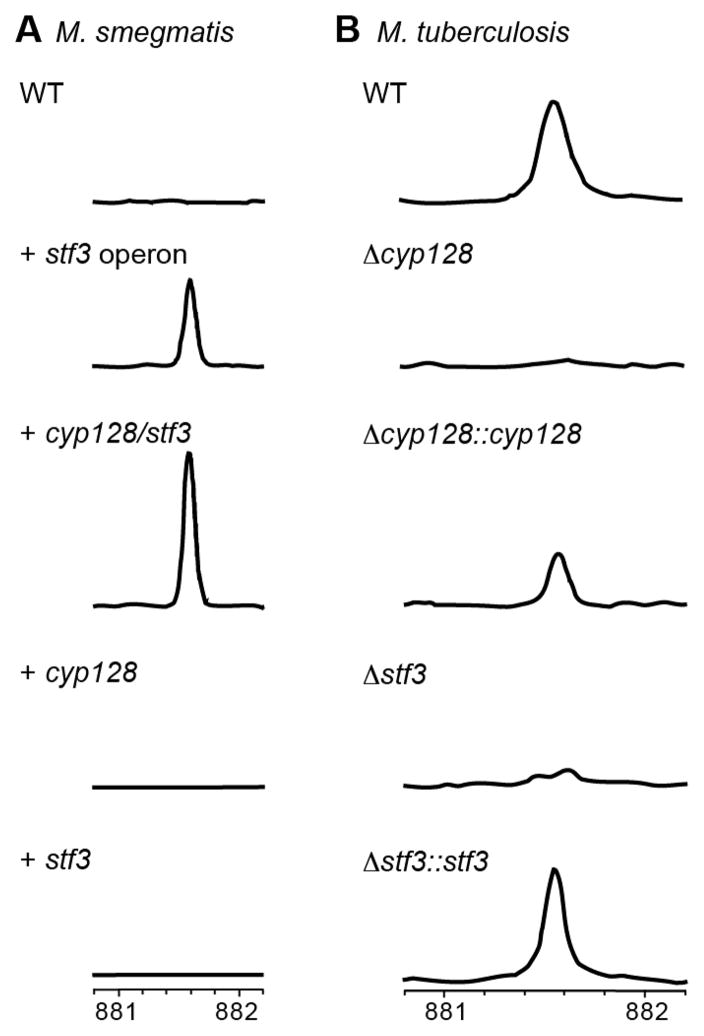
In the proposed operon that includes stf 3, which is required for the production of SMK,6 there are two additional genes: rv2269c, previously annotated as a protein of unknown function, and cyp128, a putative cytochrome P450 (Figure 1A). To establish whether cyp128 and rv2269c are also necessary for SMK biosynthesis, we engineered M. smegmatis, a fast-growing relative of M. tuberculosis, to express the genes in various combinations. Like most mycobacteria, M. smegmatis utilizes menaquinone as a respiratory cofactor11,12 but does not encode homologues for any of the genes in the stf 3 operon. We introduced rv2269c, cyp128, or stf 3 individually; cyp128 and stf 3 only; or the entire stf 3 operon (rv2269c, cyp128, and stf 3) into M. smegmatis on an episomal plasmid with a constitutive promoter (Table S2). To assess SMK production, total lipid extracts from all strains were analyzed by high-resolution mass spectroscopy. As expected, WT M. smegmatis and the strains expressing each gene individually did not produce SMK (Figure 2A and Figure S1). By contrast, the M. smegmatis strains expressing the entire stf 3 operon or cyp128 and stf 3 together produced SMK (Figure 2A and Figure S1). We confirmed the identity of the corresponding SMK ion by fragmentation analysis as previously reported.8 Thus, we conclude that SMK biosynthesis from menaquinone requires only two genes, cyp128 and stf 3 in M. smegmatis. Furthermore, because the addition of rv2269c to M. smegmatis did not influence SMK production, we hypothesize that rv2269c is indispensable to SMK biosynthesis in M. smegmatis. All attempts to biochemically characterize Cyp128 and Stf3 proved unsuccessful because we were unable to express full-length soluble protein in either E. coli or M. smegmatis.
Figure 2.
cyp128 and stf 3 are required for SMK production. (A) Mass spectra of TLE from M. smegmatis strains expressing SMK biosynthetic genes. Only strains containing both cyp128 and stf 3 produced SMK. The introduction of each of gene individually is not sufficient for SMK biosynthesis. (B) Mass spectra of TLE from M. tuberculosis WT and SMK deletion mutants. The deletion of cyp128 from M. tuberculosis H37Rv (Δcyp128) resulted in a loss of SMK production similar to that of the previously characterized stf 3 deletion mutant (Δstf 3). Complementation of Δcyp128 (Δcyp128::cyp128) restores SMK production. A representative scan from one experiment of FT-ICR mass spectra in region m/z 881– 882 is shown.
After determining that cyp128 and stf 3 are sufficient for SMK biosynthesis in M. smegmatis, we made a cyp128 gene deletion mutant in M. tuberculosis strain H37Rv (Δcyp128) (Figure S2). Total lipid extracts from the Δcyp128 mutant did not contain SMK as determined by high-resolution mass spectrometry (Figure 2B and Figure S4). For comparison, total lipid extracts of the previously characterized Δstf 3 mutant6 were made in parallel and similarly showed a loss of SMK. The complementation of Δcyp128 (Δcyp128::cyp128) with WT cyp128 restored SMK production. The loss of SMK in Δcyp128 was further confirmed by growing the SMK mutants (Δcyp128 and Δstf 3) on radioactive 35S-containing sulfate (Figure S3A). WT, Δcyp128, Δcyp128::cyp128, and Δstf 3 strains were incubated overnight in the presence of 35S-sulfate in PBS, and total lipid extracts were analyzed by thin-layer chromatography (TLC). The Δcyp128 and Δstf 3 mutants did not produce a spot corresponding to SMK, but complementation with each respective gene restored SMK production (Figure S3A). In addition to cyp128, there is another cytochrome (cyp124) adjacent to stf 3 in the M. tuberculosis genome that has been shown to have activity on structures similar to SMK in vitro.13 Our data confirms the role of cyp128 in SMK biosynthesis and suggests that Cyp128 is the only cytochrome P450 of the 20 encoded in the M. tuberculosis genome involved in this pathway.
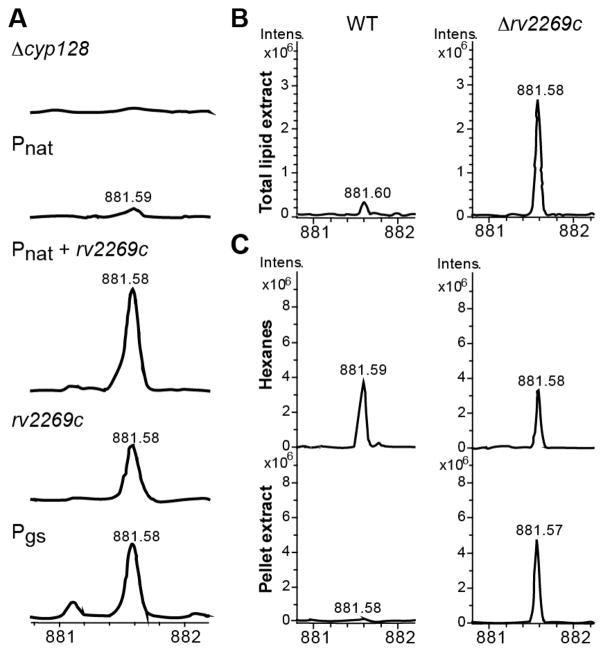
In addition to cyp128 and stf 3, there is a small 333 bp predicted ORF rv2269c of unknown function.10 Bioinformatic analysis of this ORF shows very little secondary structure and no homology to any known protein. We demonstrated that rv2269c is not required for SMK biosynthesis in M. smegmatis (Figure 2A). One study of transcriptional start site mapping indicates that there is a predicted transcriptional start site of 28 nucleotides before the annotated start site of cyp128.14 We hypothesized that rv2269c is not a functional coding gene but may play a regulatory role in SMK biosynthesis. To determine if it had promoter activity, we made a series of plasmids that encoded cyp128 along with potential promoter sequences: either one kilobase (kb) of DNA upstream of rv2269c (Pnat), 1 kb upstream of cyp128 (including the gene rv2269c, Pnat+rv2269c), or the 333 bp of the rv2269c DNA sequence alone (Prv2269c) (depicted in Figure S5). We transformed each of these plasmids into Δcyp128 and examined the ability of each of these constructs to restore SMK production by mass spectrometry. Complementation using the promoter Prv2269c successfully restored SMK production in the Δcyp128 mutant, whereas Pnat (lacking the rv2269c sequence) did not (Figure 3A and Figure S6). The expression of cyp128 by the promoter Pnat+rv2269c produced an ion with the same nominal mass as SMK, but this could not be confirmed by fragmentation because of the low ion intensity. As a control, we used a strong constitutive promoter for glutamine synthase to drive the expression of cyp128 and found WT levels of SMK. For all future experiments, we used Rv2269c as the promoter for the cyp128 complement strain (Δcyp128::cyp128).
Figure 3.
Rv2269c exhibits promoter activity. (A) Mass spectra of TLE from Δcyp128 complementation strains with cyp128 under the control of three different promoters. Only promoters containing rv2269c restored SMK production. The glutamine synthase promoter (Pgs) was used as a positive control. FT-ICR mass spectra scans in region m/z 881–882 are shown. (B) Comparison of mass spectra of TLE from WT M. tuberculosis and the Δrv2269c mutant. (C) Mass spectra of WT M. tuberculosis cell envelope fractionation are compared to the mass spectra from the Δrv2269c strain. Representative scans from one experiment of FT-ICR mass spectra scans in region m/z 881–882 are shown.
We created an rv2269c deletion strain (Δrv2269c) in M. tuberculosis to confirm that it was not essential to SMK biosynthesis. To make the deletion strain, we replaced from nucleotides 94 to 280 with a hygromycin cassette. The hygromycin cassette was inserted in the same orientation as cyp128 and stf 3, instead of in the opposite orientation as is usually done. The promoter for hygromycin drove the transcription of cyp128 and stf 3, leading to increased levels of mRNA (data not shown) and resulted in increased levels of SMK (Figure 3B). The increased production of SMK in the Δrv2269c mutant enabled us to study the result of increased SMK production in our experiments.
Using the SMK mutants, we wanted to study its role in M. tuberculosis biology. We observed that Δcyp128, Δstf 3, and Δrv2269c showed no in vitro growth phenotype (Figure S3B and ref 6). Previous analyses of SMK suggested that the sulfated variant is primarily situated in the outer cell envelope,6 unlike menaquinone, which is located in the plasma membrane.15 We confirmed this by the fractionation of the M. tuberculosis cell wall using hexanes, which selectively removes surface lipids from the covalently attached cell wall lipids.16,17 The majority of SMK was found in the hexanes fraction with minimal amounts detectable in the remaining cell pellet (Figure 3C). Despite having increased levels of SMK overall, the Δrv2269c mutant had an accumulation of SMK in the cell pellet, but cell surface levels remained similar to those found in WT (Figure 3C). This observation suggests that the exporting of SMK to the outer envelope is via active transport and is not mediated simply by passive diffusion from the cell interior.
To determine if SMK plays a role in the cell wall integrity, we subjected cells to chemical oxidative stress (H2O2, HClO3, and NO), detergent stress (SDS), and cell wall biosynthesis inhibitors (isoniazid, ethambutol, and ethionamide) and measured the effects of these treatments on viability. Under all conditions, the SMK mutants showed no difference in survival from WT (Table S1), indicating that SMK does not play a role in the cell wall integrity.
Although SMK is not necessary for cell wall integrity, we hypothesized that this metabolite may play a role in the regulation of M. tuberculosis respiration. Menaquinone is essential to M. tuberculosis viability because of its role in the electron transport chain. Levels of menaquinone have been implicated in the activation of genes necessary for entry into and exit from an anaerobic culture.18–20 Mycobacteria primarily utilize menaquinone-9(II-H2) (MK-9(II-H2), Figure 1B), which has the second isoprene unit saturated. Analogue MK-9 (Figure S7), which has all isoprene units unsaturated, is found at lower levels than MK-9(II-H2).11 Enzyme MenJ that is responsible for converting MK-9 to MK-9(II-H2) was recently identified in M. tuberculosis.20 When MenJ is deleted in M. smegmatis, the bacteria increase the synthesis of MK-9 and utilize 3 times less oxygen than WT. Furthermore, when M. tuberculosis growth was inhibited using a menaquinone biosynthesis inhibitor under aerobic conditions, an analogue of MK-9 rescued the growth better than an analogue of MK-9(II-H2).18 Both studies support an important role for menaquinone, specifically MK-9, in regulating respiration.
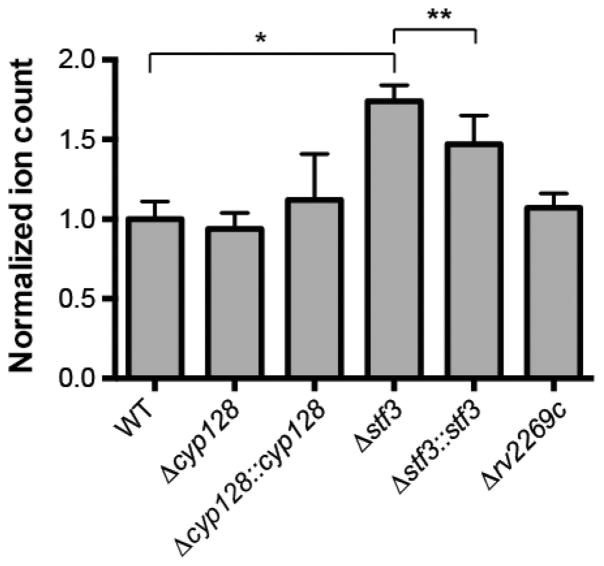
Because menaquinone is the precursor to SMK, SMK biosynthesis could alter menaquinone levels, leading to changes in the respiration of M. tuberculosis. We measured levels of MK-9 in the SMK mutants by LC-MS. We found that the Δstf 3 mutant had significantly higher levels of MK-9 than WT, whereas the Δstf 3::stf 3 strain partially complemented the phenotype (Figure 4). By contrast, both the Δcyp128 mutant and Δcyp128::cyp128 complement strains had WT levels of MK-9. We also measured levels of menaquinone in the Δrv2269c mutant that had significantly increased levels of SMK. Notably, the increased production of SMK did not significantly change menaquinone levels as compared to those in WT (Figure 4). The significant increase of MK-9 in the Δstf 3 mutant suggests a regulatory link between menaquinone levels and SMK sulfotransferase stf 3.
Figure 4.
Levels of MK-9 as measured by the total ion count from targeted LC-MS/MS lipidomics from WT and SMK mutant strains from M. tuberculosis. Error bars indicate the standard deviation, n = 5. * P < 0.01 and ** P <0.05.
The altered levels of MK-9 in the Δstf 3 mutant suggested a role for SMK in the regulation of the electron transport chain in M. tuberculosis. We tested the susceptibility of the SMK mutants to known inhibitors of the electron transport chain (scheme of the inhibitors and their targets, Figure S8). Each compound was serially diluted in media and inoculated with WT, the SMK mutants, or the complemented strains. Surprisingly, SMK mutants showed no distinguishing phenotype with respect to growth, ATP levels, or NADH/NAD+ ratio when treated with each compound. This suggests that SMK production does not alter M. tuberculosis growth or respiration rates under aerobic conditions.
Sulfated metabolites from other organisms, both bacterial and eukaryotic, often play roles in cell–cell communication or host–pathogen interactions.21,22 Sulfolipids from M. tuberculosis and the M. avium complex have known immunomodulatory effects.23,24 It is possible that SMK serves such a function for M. tuberculosis. Notably, stf 3, cyp128, and rv2269c have no obvious counterparts in mycobacteria outside the M. tuberculosis complex. Thus, the function of their product, SMK, may have a role specific for M. tuberculosis pathogenesis.
The earlier observation that Δstf 3 is hypervirulent in mice supports the SMK-mediated modulation of M. tuberculosis virulence. The different levels of menaquinone in the Δcyp128 and Δstf 3 mutants suggest that Δcyp128 may have a different phenotype in the mouse model of infection. We infected mice with the Δcyp128 mutant in parallel with the Δstf 3 mutant and their respective complement strains. BALB/c mice were infected via aerosol infection (200–300 CFU per lung) of each strain. Lungs were harvested after 3 weeks of infection, the first time point that a difference in growth between WT and the Δstf 3 mutant was previously identified.6 Mice infected with the Δcyp 128 mutant had significantly higher CFU in the lungs compared to WT, although not as many as the Δstf 3 mutant (Figure 5). Both complemented strains showed WT levels of growth in the lungs.
Figure 5.
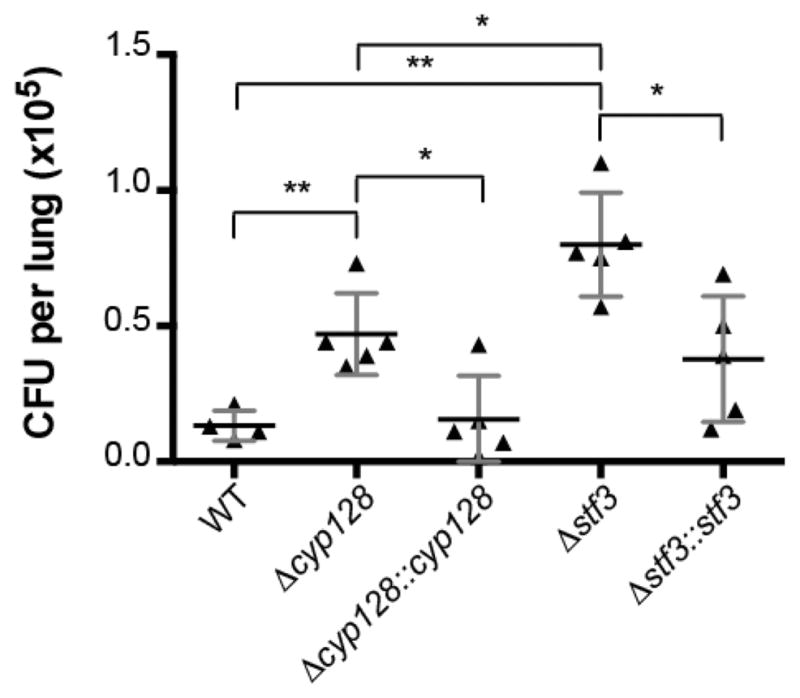
CFU from the lungs of mice. BALB/c mice were infected via aerosol infection with low-dose infection. Error bars indication the standard deviation, n = 5. Data are shown from one of two experiments. * P < 0.005 and ** P < 0.05.
Further studies are needed to understand why the SMK mutants have increased growth in mice. As we demonstrate here, SMK seems to play a negligible role in the growth or cell wall integrity of M. tuberculosis in vitro. Despite the difference in menaquinone levels between the Δcyp128 mutant and the Δstf 3 mutant, it is unlikely that the menaquinone levels can explain a change in respiration or growth rate to account for the increased number of bacteria in the lungs. Although this may contribute to the increase in growth of the Δstf 3 mutant, it cannot account for the increase in growth of both mutants. An alternative hypothesis is that SMK alters the ability of the mouse to control bacterial growth during the acute phase of infection. As shown by Cambier et al., M. tuberculosis produces lipids to mask more immunostimulatory molecules on the surface.25 SMK may be masking or altering the ability of macrophages to mount an effective response to M. tuberculosis infection. An alternative hypothesis is that SMK activates a specific immune response that is necessary for the control of M. tuberculosis growth. Further studies on the specific immune modulations of SMK are necessary to tease out the role of SMK. The elucidation of SMK biosynthesis and its role in M. tuberculosis pathogenesis adds to the repertoire of M. tuberculosis lipids known to have immunomodulatory effects in vivo.
METHODS
Bacterial Strains and Growth Media
The mycobacterial strains used in these studies were M. tuberculosis H37Rv and M. smegmatis mc2155. M. tuberculosis was grown in 7H9 (liquid medium) or 7H11 (solid medium) with 10% oleate/albumin/dextrose/catalase supplement, 0.5% glycerol, and 0.05% Tween 80 unless stated otherwise. M. smegmatis was grown in 7H9 (liquid) or 7H10 (solid) medium with 10% albumin/dextrose/catalase, 0.5% glycerol, and 0.05% Tween 80 unless stated otherwise. Media and supplements were from BD Biosciences. Antibiotics were used for selection at 20 μg/mL kanamycin or 50 μg/mL hygromycin for mycobacteria. Cloning and plasmid propagation were performed in E. coli TOP10 and XL-1 blue strains. M. tuberculosis genes were amplified from H37Rv genomic DNA. All E. coli cultures were grown in LB medium with 50μg/mL kanamycin or 100 μg/mL hygromycin for selection.
Construction of M. tuberculosis Mutant Strains
The Δcyp128 mutant strain was created by homologous recombination as previously described.26 Briefly, specialized transduction phage phKMS109 (rv2269c) or phKMS110 (cyp128) was incubated with concentrated M. tuberculosis H37Rv for 4 h at 37 °C. Cells were then plated on 7H11 plates containing hygromycin. Colonies were picked and screened for gene disruption by PCR (Figure S2), which confirmed the replacement of 1279 bp of cyp128 (amino acids 32–457) with a hygromycin resistance cassette. The Δcyp128::cyp128 complementation strain was created by cloning the cyp128 gene plus 333 bp upstream of the gene from M. tuberculosis strain H37Rv into mycobacterial expression vector pMV306, a derivative of the pMV361 vector27 with a multiple cloning site in place of the expression cassette. The resulting plasmid was electroporated into Δcyp128 cells, and transformants were selected on 7H11 kanamycin-containing plates.
Extraction and Mass Spectrometry Analysis of Lipids
Extraction and mass spectrometry analysis of lipids was performed as previously described.8 Briefly, for total lipid extracts (TLEs), mid-log-phase cultures of M. tuberculosis strains or M. smegmatis strains were washed and diluted in Tween-free media to an OD600 of 0.2 and grown for two doublings. Cells were harvested via centrifugation, resuspended in 4 mL of 1:1 chloroform/methanol, and extracted overnight at RT. For surface lipid extractions, the pellet was resuspended in 2 mL of hexanes per 50 mL of culture and spun for 2 min at 3500 rpm. The upper organic phase was removed and added to an equal volume of 1:1 chloroform/methanol. The remaining cell pellet and aqueous phase were extracted with 4 mL of 1:1 chloroform/methanol per pellet overnight at RT. Cell debris was removed by centrifugation, and the supernatant was stored at −20 °C until analysis. All extractions were repeated in at least three independent experiments.
High-resolution FT-ICR mass spectra were obtained on an Apex II FT-ICR mass spectrometer equipped with a 7 T actively shielded superconducting magnet (Bruker Daltonics, Billerica, MA) as described previously.8 Briefly, samples were introduced into the ion source via direct injection at a rate of 2 μL/min. Ions were generated with an Apollo pneumatically assisted electrospray ionization source (Bruker Daltonics) operating in negative ion mode and were accumulated in an rf-only external hexapole for 0.5–1 s before being transferred to the ICR cell for mass analysis. Mass spectra consisted of 512 000 data points and were an average of 50–100 scans. The spectra were acquired using XMASS version 7.0.8 (Bruker Daltonics). For accurate mass measurements, spectra were internally calibrated with known compounds.
Metabolic Labeling of M. tuberculosis and M. smegmatis with 35S-Sulfate and Lipid Analysis by ThinLayer Chromatography (TLC)
M. tuberculosis or M. smegmatis strains were grown in 7H9 to late log phase. Two generations prior to labeling, strains were diluted to an OD600 of 0.3. For 35S-sulfate labeling, cells were resuspended at OD600 ≈ 1 in 10 mL of PBS with 1% sodium acetate and 100 μCi of 35S-sulfate (PerkinElmer). After overnight incubation, cell pellets were extracted with 1:1 chloroform/methanol as described above. Solvent was removed by evaporation, and the extracts were resuspended with 1:1 chloroform/methanol at 1/10 or 1/20 of the original volume. An equal volume of each fraction was spotted on silica plates (HPTLC silica gel 60, EMD Chemicals) and developed in 60:12:1 chloroform/methanol/water. Plates were analyzed by phosphorimaging (GE Biosciences Typhoon).
Quantification of Menaquinone Levels by LC-MS Analysis
Total lipid extracts from each strain were obtained as previously described. Using an authentic standard of menaquinone-9 (MK-9, Figure S3), we determined the unique fragmentation pattern used to identify MK-9 in complex samples for quantification by targeted metabolomics. We quantified ion counts for three independent transitions (m/z 785 to 109, 785 to 187, and 802 to 81; collision energy 12 V) from five replicates for each sample. All mass transitions showed the same trend in MK-9 levels for each M. tuberculosis strain. As an external standard, coenzyme Q9 (CoQ9, 1 ng) was added to total lipid extracts immediately after bacterial lysis. Each sample was filtered and transferred to a glass vial and stored at −80 °C until analysis. MS analysis was performed as previously described.28 Briefly, an aliquot of the extract (10 μL) was analyzed using an Agilent G6430 QQQ instrument. Metabolite separation by liquid chromatography (LC) for lipophilic metabolites was achieved using a Gemini reverse-phase C5 column from Phenomonex. Mobile phase A consisted of 95:5 water/methanol, and mobile phase B consisted of 60:35:5 isopropanol/methanol/water. Formic acid (0.1%) was included to assist in ion formation in positive ionization mode. The flow rate for each run started at 0.1 mL/min with 0% B. At 5 min, the solvent was immediately changed to 60% B with a flow rate of 0.4 mL/min and increased linearly to 100% B over 15 min. This was followed by an isocratic gradient of 100% B for 8 min at 0.5 mL/min before equilibrating for 3 min at 0% B at 0.5 mL/min. Metabolites were detected using single reaction and were quantified by integrating the area under the curve and normalizing to the amount of external standard recovered and the OD600 at the time of harvest. All values presented are averages of five replicates per strain with the standard deviation.
Minimum Inhibitor Concentration (MIC) Determination Using Chemical Inhibitors
M. tuberculosis was grown to mid log phase in 50 mL of 7H9 media in roller bottles. Clumps were removed by centrifugation, and the culture suspension was diluted to an OD600 of 0.2. Two-fold serial dilutions of each chemical inhibitor were set up in 96 well plates, in concentrations starting with 4-fold over the literature MICs and in a total volume of 90 μL. Each well was inoculated with 10 μL of the designated strain in triplicate. Sterile PBS was added to the remaining wells. Plates were placed in a sealed Tupperware container containing damp towels to prevent evaporation. Plates were incubated for 7 days at 37 °C. Plates were fixed with 100 μ L of 10% formalin in buffered PBS, and the cell density was quantified using a VersaMax UV/vis plate reader (Molecular Devices). MIC was determined by the concentration of treatment that inhibited 90% of growth compared to untreated conditions. Experiments were done in triplicate.
Mouse Infections
Female BALB/c mice were purchased from the Jackson Laboratory. Six week old mice were infected via the aerosol route. Bacteria were aerosolized by using the Inhalation Exposure System (Glas-col, Terre Haute, IN) to deliver 100 to 200 bacilli per mouse lung. To evaluate the initial inoculum, lungs from five mice were harvested 24 h after infection to determine the number of bacteria seeded. At 21 days after infection, bacterial numbers were enumerated by plating serial dilutions of lung homogenates from five mice for each group on 7H10-OADC. Colonies were counted after 21 days. The data represent one of two experiments.
Ethics Statement
All procedures involving the use of mice were approved by the University of California, Berkeley IACUC, the Animal Care and Use Committee (protocol number R353-1113B). All protocols conform to federal regulations, the National Research Council’s Guide for the Care and Use of Laboratory Animals, and the Public Health Service’s (PHS’s) Policy on Humane Care and Use of Laboratory Animals.
Supplementary Material
Acknowledgments
We thank Drs. Sarah Gilmore, Jessica Seeliger, and Sarah Stanley for technical advice and their critical reading of the manuscript. This work was supported by National Institutes of Health grant R01 AI051622 and Howard Hughes Medial Institute to C.R.B., NCI R01 CA172667 to D.K.N., and R01 GM47356 to J.A.L.
ABBREVIATIONS
- SMK
sulfomenaquinone
- MIC
minimum inhibitory concentration
- H2O2
hydrogen peroxide
- HClO3
chloric acid
- NO
nitric oxide
- SDS
sodium dodecyl sulfate
- TLE
total lipid extract
- TLC
thin layer chromatography
Footnotes
The authors declare no competing financial interest.
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acsinfecdis.6b00106.
Validation of cyp128 deletion, TLC of radiolabeled sulfated compound from WT and mutants strains, structure of menaquinone-9, schematic of M. tuberculosis electron transport chain with inhibitors tested, MIC values for inhibitors of cell wall biosynthesis, and plasmids and primers used in the study (PDF)
References
- 1.Mestre O, Hurtado-Ortiz R, Dos Vultos T, Namouchi A, Cimino M, Pimentel M, Neyrolles O, Gicquel B. High Throughput Phenotypic Selection of Mycobacterium tuberculosis Mutants with Impaired Resistance to Reactive Oxygen Species Identifies Genes Important for Intracellular Growth. PLoS One. 2013;8:e53486. doi: 10.1371/journal.pone.0053486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stanley SA, Cox JS. Host-pathogen interactions during Mycobacterium tuberculosis infections. Curr Top Microbiol Immunol. 2013;374:211–241. doi: 10.1007/82_2013_332. [DOI] [PubMed] [Google Scholar]
- 3.Jain M, Petzold CJ, Schelle MW, Leavell MD, Mougous JD, Bertozzi CR, Leary JA, Cox JS. Lipidomics reveals control of Mycobacterium tuberculosis virulence lipids via metabolic coupling. Proc Natl Acad Sci U S A. 2007;104:5133–5138. doi: 10.1073/pnas.0610634104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Griffin JE, Pandey AK, Gilmore SA, Mizrahi V, McKinney JD, Bertozzi CR, Sassetti CM. Cholesterol catabolism by Mycobacterium tuberculosis requires transcriptional and metabolic adaptations. Chem Biol. 2012;19:218–227. doi: 10.1016/j.chembiol.2011.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee W, VanderVen BC, Fahey RJ, Russell DG. Intracellular Mycobacterium tuberculosis Exploits Host-derived Fatty Acids to Limit Metabolic Stress. J Biol Chem. 2013;288:6788–6800. doi: 10.1074/jbc.M112.445056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mougous JD, Senaratne RH, Petzold CJ, Jain M, Lee DH, Schelle MW, Leavell MD, Cox JS, Leary JA, Riley LW, Bertozzi CR. A sulfated metabolite produced by stf3 negatively regulates the virulence of Mycobacterium tuberculosis. Proc Natl Acad Sci U S A. 2006;103:4258–4263. doi: 10.1073/pnas.0510861103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mougous JD, Leavell MD, Senaratne RH, Leigh CD, Williams SJ, Riley LW, Leary JA, Bertozzi CR. Discovery of sulfated metabolites in mycobacteria with a genetic and mass spectrometric approach. Proc Natl Acad Sci U S A. 2002;99:17037–17042. doi: 10.1073/pnas.252514899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Holsclaw CM, Sogi KM, Gilmore SA, Schelle MW, Leavell MD, Bertozzi CR, Leary JA. Structural characterization of a novel sulfated menaquinone produced by stf3 from Mycobacterium tuberculosis. ACS Chem Biol. 2008;3:619–624. doi: 10.1021/cb800145r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.ten Bokum AMC, Movahedzadeh F, Frita R, Bancroft GJ, Stoker NG. The case for hypervirulence through gene deletion in Mycobacterium tuberculosis. Trends Microbiol. 2008;16:436–441. doi: 10.1016/j.tim.2008.06.003. [DOI] [PubMed] [Google Scholar]
- 10.Cole ST, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, Gordon SV, Eiglmeier K, Gas S, Barry CE, Tekaia F, Badcock K, Basham D, Brown D, Chillingworth T, Connor R, Davies R, Devlin K, Feltwell T, Gentles S, Hamlin N, Holroyd S, Hornsby T, Jagels K, Krogh A, McLean J, Moule S, Murphy L, Oliver K, Osborne J, Quail MA, Rajandream MA, Rogers J, Rutter S, Seeger K, Skelton J, Squares R, Squares S, Sulston JE, Taylor K, Whitehead S, Barrell BG. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 11.Collins MD, Pirouz T, Goodfellow M, Minnikin DE. Distribution of menaquinones in actinomycetes and corynebacteria. J Gen Microbiol. 1977;100:221–230. doi: 10.1099/00221287-100-2-221. [DOI] [PubMed] [Google Scholar]
- 12.GALE PH, ARISON BH, TRENNER NR, PAGE AC, FOLKERS K. Characterization of vitamin K9(H) from Mycobacterium phlei. Biochemistry. 1963;2:200–203. doi: 10.1021/bi00901a038. [DOI] [PubMed] [Google Scholar]
- 13.Johnston JB, Kells PM, Podust LM, Ortiz de Montellano PR. Biochemical and structural characterization of CYP124: a methyl-branched lipid omega-hydroxylase from Mycobacterium tuberculosis. Proc Natl Acad Sci U S A. 2009;106:20687–20692. doi: 10.1073/pnas.0907398106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shell SS, Wang J, Lapierre P, Mir M, Chase MR, Pyle MM, Gawande R, Ahmad R, Sarracino DA, Ioerger TR, Fortune SM, Derbyshire KM, Wade JT, Gray TA, Viollier PH. Leaderless Transcripts and Small Proteins Are Common Features of the Mycobacterial Translational Landscape. PLoS Genet. 2015;11:e1005641. doi: 10.1371/journal.pgen.1005641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee SH, Sutherland TO, Deveś R, Brodie AF. Restoration of active transport of solutes and oxidative phosphorylation by naphthoquinones in irradiated membrane vesicles from Mycobacterium phlei. Proc Natl Acad Sci U S A. 1980;77:102–106. doi: 10.1073/pnas.77.1.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goren MB. Sulfolipid I of Mycobacterium tuberculosis, strain H37Rv. I Purification and properties. Biochim Biophys Acta, Lipids Lipid Metab. 1970;210:116–126. doi: 10.1016/0005-2760(70)90067-6. [DOI] [PubMed] [Google Scholar]
- 17.Goren MB, D’Arcy Hart P, Young MR, Armstrong JA. Prevention of phagosome-lysosome fusion in cultured macrophages by sulfatides of Mycobacterium tuberculosis. Proc Natl Acad Sci U S A. 1976;73:2510–2514. doi: 10.1073/pnas.73.7.2510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dhiman RK, Mahapatra S, Slayden RA, Boyne ME, Lenaerts A, Hinshaw JC, Angala SK, Chatterjee D, Biswas K, Narayanasamy P, Kurosu M, Crick DC. Menaquinone synthesis is critical for maintaining mycobacterial viability during exponential growth and recovery from non-replicating persistence. Mol Microbiol. 2009;72:85–97. doi: 10.1111/j.1365-2958.2009.06625.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Honaker RW, Dhiman RK, Narayanasamy P, Crick DC, Voskuil MI. DosS responds to a reduced electron transport system to induce the Mycobacterium tuberculosis DosR regulon. J Bacteriol. 2010;192:6447–6455. doi: 10.1128/JB.00978-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Upadhyay A, Fontes FL, Gonzalez-Juarrero M, McNeil MR, Crans DC, Jackson M, Crick DC. Partial Saturation of Menaquinone in Mycobacterium tuberculosis: Function and Essentiality of a Novel Reductase, MenJ. ACS Cent Sci. 2015;1:292–302. doi: 10.1021/acscentsci.5b00212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lerouge P, Roche P, Faucher C, Maillet F, Truchet G, Promé JC, Dénarié J. Symbiotic host-specificity of Rhizobium meliloti is determined by a sulphated and acylated glucosamine oligosaccharide signal. Nature. 1990;344:781–784. doi: 10.1038/344781a0. [DOI] [PubMed] [Google Scholar]
- 22.Chapman E, Best MD, Hanson SR, Wong CH. Sulfotransferases: structure, mechanism, biological activity, inhibition, and synthetic utility. Angew Chem, Int Ed. 2004;43:3526–3548. doi: 10.1002/anie.200300631. [DOI] [PubMed] [Google Scholar]
- 23.Gilmore SA, Schelle MW, Holsclaw CM, Leigh CD, Jain M, Cox JS, Leary JA, Bertozzi CR. Sulfolipid-1 biosynthesis restricts Mycobacterium tuberculosis growth in human macrophages. ACS Chem Biol. 2012;7:863–870. doi: 10.1021/cb200311s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Khoo KH, Jarboe E, Barker A, Torrelles J, Kuo CW, Chatterjee D. Altered expression profile of the surface glycopeptidolipids in drug-resistant clinical isolates of Mycobacterium avium complex. J Biol Chem. 1999;274:9778–9785. doi: 10.1074/jbc.274.14.9778. [DOI] [PubMed] [Google Scholar]
- 25.Cambier CJ, Takaki KK, Larson RP, Hernandez RE, Tobin DM, Urdahl KB, Cosma CL, Ramakrishnan L. Mycobacteria manipulate macrophage recruitment through coordinated use of membrane lipids. Nature. 2013;505:218–222. doi: 10.1038/nature12799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Glickman MS, Cox JS, Jacobs WR. A novel mycolic acid cyclopropane synthetase is required for cording, persistence, and virulence of Mycobacterium tuberculosis. Mol Cell. 2000;5:717–727. doi: 10.1016/s1097-2765(00)80250-6. [DOI] [PubMed] [Google Scholar]
- 27.Stover CK, de la Cruz VF, Fuerst TR, Burlein JE, Benson LA, Bennett LT, Bansal GP, Young JF, Lee MH, Hatfull GF. New use of BCG for recombinant vaccines. Nature. 1991;351:456–460. doi: 10.1038/351456a0. [DOI] [PubMed] [Google Scholar]
- 28.Nomura DK, Morrison BE, Blankman JL, Long JZ, Kinsey SG, Marcondes MCG, Ward AM, Hahn YK, Lichtman AH, Conti B, Cravatt BF. Endocannabinoid hydrolysis generates brain prostaglandins that promote neuro-inflammation. Science. 2011;334:809–813. doi: 10.1126/science.1209200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.