Abstract
Many RNA-binding proteins have modular structures, being composed of multiple repeats of just a few basic domains that are arranged in a variety of ways to satisfy their diverse functional requirements. Recent studies have investigated how different modules cooperate in regulating the RNA binding specificity and the biological activity of these proteins. They have also investigated how multiple modules cooperate with enzymatic domains to regulate the catalytic activity of enzymes acting upon RNA. These studies have shown how multiple modules define, for many RNA-binding proteins, the fundamental structural unit that is responsible for their biological function.
Introduction
RNA is rarely at a loss for companions; as soon as RNA is transcribed, ribonucleoproteins (RNPs) form co-transcriptionally on the nascent transcript and participate in processing, nuclear export, transport and localization1. The dynamic association of these proteins with RNA defines the lifetime, cellular localization, processing and the rate at which a specific mRNA is translated.
The diversity of functions of RNA-binding proteins would suggest a correspondingly large diversity in the structures that are responsible for RNA recognition. However, most RNA-binding proteins are built from relatively few RNA-binding modules (Table 1). The large structural diversity of substrates is accommodated instead by the presence of multiple copies of these RNA-binding domains presented in a variety of structural arrangements to expand the functional repertoire of these proteins (Figure 1)2. Modules of the same or different structural type combine to create versatile macromolecular binding surfaces to define the specificity of these proteins and combine with enzymatic domains to define the enzymes’ target and regulate catalytic activity (Figure 2). In order to understand the function of RNA-binding proteins, it is therefore important to know how these domains function together as RNA recognition units.
Table 1.
Common RNA binding domains and general features of their RNA binding sites and RNA recognition properties.
| Topology | RNA Recognition Surface | Protein-RNA interactions | Representative Structures (PDB ID) | |
|---|---|---|---|---|
| RRM | αβ | Surface of β–sheet | Interacts with 4 nucleotides of ssRNA through stacking, electrostatics and hydrogen bonding | U1A N-terminal RRM18(1URN) |
| KH (Type I and Type II) | αβ | Hydrophobic cleft formed by variable loop between β2 and β3 and GXXG loop; Type II: Same as type I, except variable loop is between α2 and β2 | Recognizes 4 nucleotides of ssRNA through hydrophobic interactions between non-aromatic residues and the bases; sugar-phosphate backbone contacts from GXXG loop, and hydrogen bonding to bases | Nova-1 KH3 (Type I)41 (1EC6) NusA (Type II)37(2ASB) |
| dsRBD | αβ | α-helix 1, N-terminal portion of α-helix 2, and loop between β1–β2 | Shape specific recognition of dsRNA’s minor-major-minor groove pattern through contacts to sugar-phosphate backbone; specific contacts from N-terminal α-helix to RNA in some proteins | dsRBD3 from Staufen48(1EKZ) |
| ZnF-C2H2 | αβ | Primarily residues in α-helices | Protein side chain contacts to bulged bases in loops and through electrostatic interactions between side chains and RNA backbone | Fingers 4–6 of TFIIIA56(1UN6) |
| ZnF-CCCH | Little regular secondary structure | Aromatic side chains form hydrophobic binding pockets for bases that make direct H-bonds to protein backbone | Stacking interactions between aromatic residues and bases create kink in RNA that allows for direct recognition of Watson-Crick edges of the bases by the protein backbone | Fingers 1 and 2 of Tis11d57 (1RGO) |
| S1 | β | Core formed by two β-strands with contributions from surrounding loops | Stacking interactions between base and aromatic residues and hydrogen bonding to the bases | Ribonuclease II118(2IX1), Exosome99(2NN6) |
| PAZ | αβ | Hydrophobic pocket formed by OB-like β-barrel and small αβ motif | Recognizes single-stranded 3′ overhangs of siRNA through stacking interactions and hydrogen bonds | PAZ73(1SI3), Argonaute76 (1U04), Dicer72(2FFL) |
| PIWI | αβ | Highly conserved pocket including a metal ion that is bound to the exposed C-terminal carboxylate | Recognizes the defining 5′ phosphate group in siRNA guide strand with highly conserved binding pocket that includes a metal ion | AfPIWI75(1YTU), Argonaute (1U04) |
| TRAP | β | Edges of β-sheets between each of the 11 subunits that form the entire protein structure | Recognizes GAG triplet through stacking interactions and hydrogen bonding to bases; limited contacts to the backbone | TRAP119 (1C9S) |
| Pumilio | α | Two repeats combine to form binding pocket for individual bases; helix α2 provides specificity-determining residues | Binding pockets for bases provided by stacking interactions; specificity dictated by hydrogen-bonds to Watson-Crick face of base by two amino acids in helix α2 | Pumilio84(1M8Y) |
| SAM | α | Hydrophobic cavity between three helices surrounded by an electropositive region | Shape-dependent recognition of RNA stem-loop, mainly through interactions with sugar-phosphate backbone and a single base in loop. | Vts1p120(2ESE) |
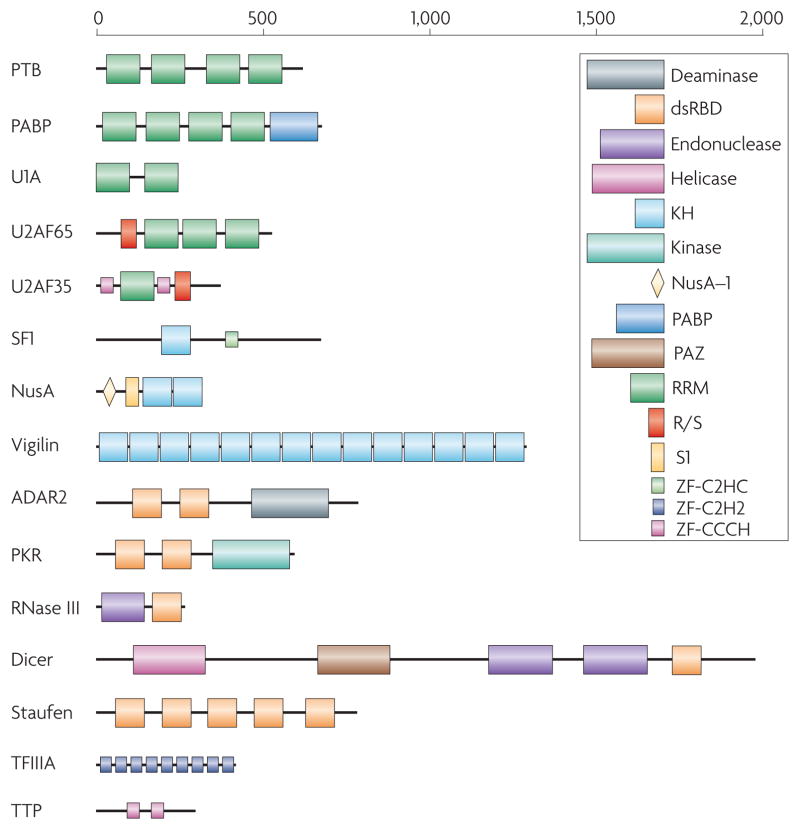
Figure 1. Many RNA-binding proteins have a modular structure.
Representative examples from some of the most common RNA-binding protein families, as illustrated here, demonstrate the variability in the number of copies (as many as 14 in vigilin) and arrangements that exist in RNA-binding proteins. This variability has direct functional implications. For example, Dicer and RNase III both contain an endonuclease catalytic domain followed by an RNA-binding dsRBD; thus, both proteins recognize double-stranded RNA, but Dicer has evolved to interact specifically with RNA species produced through the RNA interference pathway through additional domains that recognize the unique structural features of these RNAs. Different domains are schematically represented in colored boxes, including the RRM (RNA Recognition Motif; green), by far the most common RNA-binding protein module; the KH (K-Homology) domain, blue, capable of binding both single stranded RNA and DNA; the dsRBD (double-stranded RNA-binding domain), a sequence-independent dsRNA binding module (red); RNA-binding zinc finger domains (light blue or pink). Enzymatic domains and less common functional modules are indicated in a variety of colors.
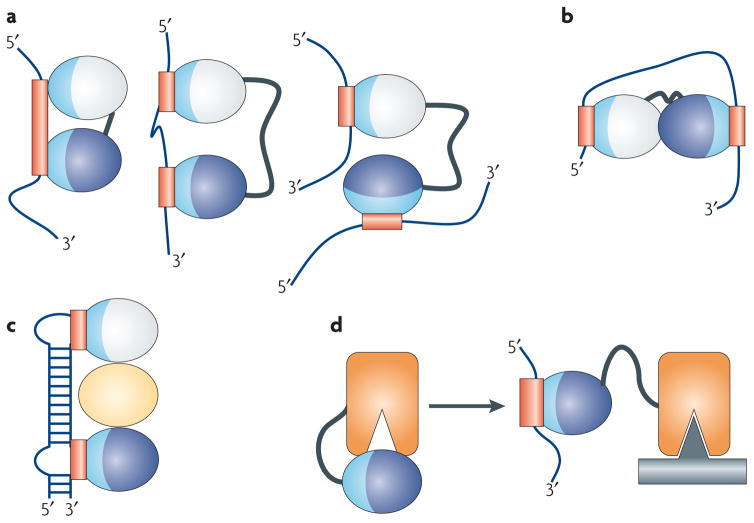
Figure 2. RNA-binding modules are combined to perform multiple functional roles.
RNA-binding domains function in a variety of ways. (a) They recognize RNA sequences with a specificity and affinity that would not be possible through a single domain or if multiple domains did not cooperate. Multiple domains combine to recognize a longer RNA sequence (left), sequences separated by many nucleotides (centre), or RNAs belonging to different molecules altogether (right). (b) RNA-binding domains can organize mRNAs topologically by interacting simultaneously with multiple RNA sequences or (c) they can function as spacers to properly position other modules for recognition. (d) They can combine with enzymatic domains to define the substrate specificity for catalysis or regulate enzymatic activity. In this figure, the RNA-binding modules are represented as ellipses with their RNA-binding surfaces colored in light blue, with the corresponding binding sites within the RNA colored in red; individual domains are colored differently.
In this review, we begin illustrating general themes as to how modularity facilitates function. We then briefly summarize the principles of RNA recognition by individual RNA-binding domains as a necessary prologue to the subsequent discussion of how specific combinations of modules cooperate functionally and structurally. The reader is referred to several excellent reviews that discuss the molecular mechanisms used by individual domains to recognize specific RNAs in greater detail3–6. The focus of this review is on how RNA-binding modules are combined and arranged to facilitate a myriad of different interactions and regulatory events.
Modularity facilitates function
Many cellular processes, for example intracellular signaling and the extracellular matrix7–9, rely on proteins that are constructed through multiple repeats of a few basic modular units. The advantages to constructing a protein with a modular architecture arise from the resulting versatility. By existing in multiple copies (Figure 1), these modules endow a protein with the ability to bind RNA with increased specificity and affinity than would be possible with individual domains, which often bind short RNA stretches with relatively weak affinity. Thus, by constructing an interaction surface through multiple modules, high affinity and specificity for a particular target can be obtained by combining multiple weak interactions. These weak interactions make it easier to regulate formation of these complexes by disassembling them when needed. Furthermore, these multiple binding sites have the ability to evolve independently. The modular architecture is also ideally suited to construct proteins that match in their RNA specificity the relatively poorly conserved sequence features observed in splicing and 3′-end processing sites of eukaryotic mRNAs10–12.
The first effect of providing a protein with multiple domains is therefore that the protein itself can recognize a much longer stretch of nucleic acids than would be possible with a single domain (Figure 2A, left). This modularity also allows proteins to recognize sequences that are separated either by an intervening stretch of nucleotides (Figure 2A, centre) or that belong to different RNAs (Figure 2A, right).
The specificity of individual domains within a protein is obviously functionally important, but so is the way in which domains are arranged relative to each other. This is reflected in evolution: higher levels of conservation are often found between domains that occupy the same position in orthologous proteins, as opposed to domains within the same protein but in different position. For example, in both the splicing factor U2AF65 and in the poly(A) binding protein (PABP), RRM1 in yeast is more similar to RRM1 of the human protein compared to RRM3 or RRM4 of the yeast protein.
Much of the ability of these proteins to recognize RNA specifically is dependent upon the linker between the two domains. Long linkers are generally disordered and allow two domains to recognize a diverse set of targets, as seen in the centre and right panels of Figure 2A, while short linkers predispose the domains to bind to a contiguous stretch of nucleic acids (Figure 2A, left side). When this occurs, the linker domain generally becomes ordered, forming a short α-helix in response to RNA binding that positions the two domains relative to one another and sometimes contacts RNA directly13–16. In these situations, inter-domain sequences are as well conserved as the domains themselves17, or better, because the precise positioning of domains facilitates their function.
The modular architecture allows a protein to topologically arrange the generally flexible RNA for a particular function (Figure 2B). Conversely, the proteins themselves can be topologically organized to interact with a particular RNA structure (Figure 2C), for example by utilizing additional domains (yellow oval, Figure 2C) to organize the RNA-binding domains.
Finally, the combination of enzymatic and RNA binding domains provide ways to regulate catalytic activity. In Figure 2D, we outline a situation where the active site of an enzyme is occluded by the presence of an RNA binding domain. In the presence of the substrate RNA, the RNA binding domain binds its target, thereby releasing the enzyme from its inactive state.
RNA recognition by RNA-binding modules
RRM
The RNA recognition motif (RRM, also known as the RNA binding domain RBD or ribonucleoprotein motif RNP), is by far the most common and best characterized of the RNA-binding modules. In this review, we will refer to it as RRM, and use the term RNA binding domain for any domain that binds to RNA. The RRM is composed of 80–90 amino acids that form a four-stranded anti-parallel β-sheet with two helices packed against it, giving the domain the split αβ (βαββαβ) topology18 (Figure 3A). More than 9,000 RRMs have been identified that function in most, if not all, post-transcriptional gene expression processes; in humans, ~0.5–1% of genes contains an RRM, often in multiple copies within the same polypeptide19.
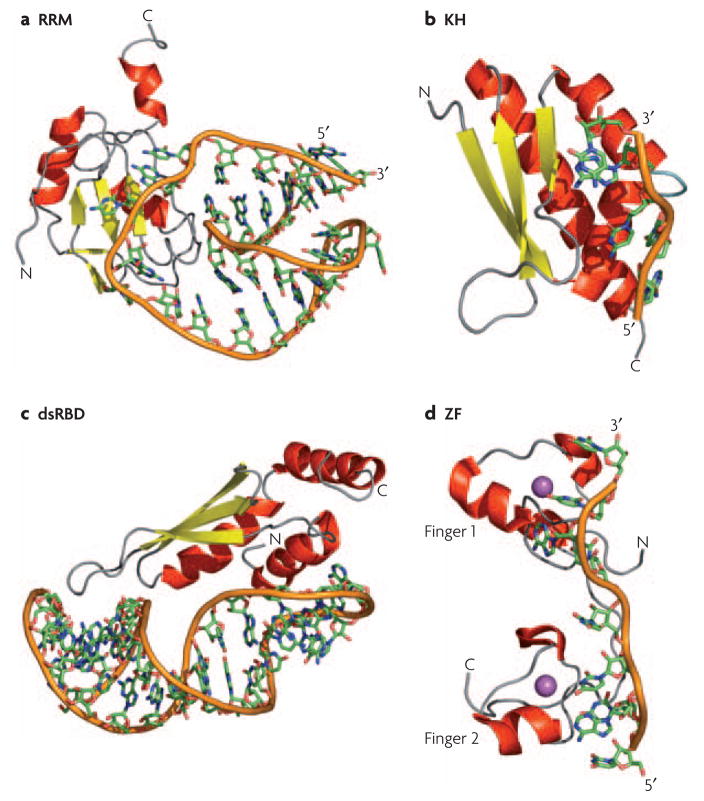
Figure 3. How RNA-binding modules recognize RNA.
a) Structure of the N-terminal RNA-recognition motif (RRM) of human U1A bound to RNA18; in this structure, and in many other RRM-RNA complexes, single stranded bases are specifically recognized through the protein β-sheet and two loops connecting the secondary structure elements. b) The hnRNP K homology 3 (KH3) domain of Nova-2 bound to 5′-AUCAC-3′41; these domains bind to both single-stranded DNA and RNA through a conserved GXXG sequence located in an exposed loop (light blue). c) The Rnt1 double-stranded RNA-binding domain (dsRBD) bound to a RNA helix capped by an AGNN tetraloop50; a conserved protein loop (left-most part of the structure) interacts with 2′-OH groups in the RNA minor groove while highly conserved Lys and Arg residues at the end of the longer helix recognize the position of phosphate atoms characteristic of an A-form helix. d) The two zinc fingers of Tis11d bound to an AU-rich RNA element57; the identity of the single stranded RNA is recognized by the protein backbone through hydrogen bonds with the Watson-Crick face of each base. In all panels, the RNA backbone is represented with an orange ribbon, α-helices are in red and β-sheets in yellow; the Zn atom in the Tis11d structure is in magenta.
In the about 20 structures of RRM–RNA complexes, RNA recognition usually occurs on the surface of the β-sheet13–16, 18, 20–28. Binding is mediated in most cases by three conserved residues, an Arg/Lys that forms a salt bridge to the phosphodiester backbone, and two aromatic residues that make stacking interactions with the nucleobases. These amino acids reside in the two highly conserved motives, termed ribonucleoprotein motif 1 and 2 (RNP1 and RNP2), that define the motif at the sequence level and are located in the two central β-strands18. This conserved platform allows for recognition of two nucleotides in the center of the β-sheet, and two additional nucleotides on either side6. However, a single RRM can recognize anywhere from 4 to 8 nucleotides by using exposed loops and additional secondary structure elements that are not present in the canonical structure3, 6. This general mechanism of recognition is found in many RRMs, but not all22, 28; some of these domains even interact with proteins and not RNA29–35. Thus, some individual RRMs can bind to RNA with great specificity, but in many cases multiple domains are needed to define specificity because the number of nucleotides recognized by an individual RRM is generally too small to define a unique binding sequence3.
KH
The hnRNP K homology domain (KH domain) is a domain that binds to both ssDNA and ssRNA36–42 and is ubiquitously found in eukaryotes, eubacteria and archaea43. The domain is composed of ~70 amino acids with a signature sequence of (I/L/V)-I-G-X-X-G-X-X-(I/L/V) near the center of the domain that is functionally very important. Mutations within this region of the Fmr1 protein cause Fragile X mental retardation syndrome44. All KH domains form a three-stranded β-sheet packed against three α–helices, but can be separated in two subfamilies on the basis of their topology45 (type I: βααββα topology; type II: αββααβ topology). Four nucleotides are recognized for both classes in a cleft formed by the GXXG loop, the flanking helices, the β-strand that follows helix 2 (type I) or 3 (type II), and the variable loop between β2 and β3 in (type I) or between α2 and β2 in (type II; Figure 3B). Quite unlike the RRM, this binding platform is free of aromatic amino acids; recognition is achieved instead by hydrogen bonding, electrostatic interactions and shape complementarity.
dsRBD
The double-stranded RNA-binding domain (dsRBD) is another small αβ domain of 70–90 amino acids that is widely found in both bacteria and eukaryotes. However, it interacts with double-stranded (ds)RNA without making specific contacts with the nucleobases. The protein binds across two successive minor grooves and the intervening major groove on one face of the dsRNA helix (Figure 3C)46. Unlike the RRM or KH domains, the majority of intermolecular contacts are sequence independent and involve 2′-OH groups and the phosphate backbone46. The presence of multiple dsRBDs can impart specificity for certain structures because of their ability to recognize certain arrangements of RNA helices49, 51, 52. In addition, the specificity of at least some dsRBDs is mediated in part by an N-terminal helix that binds to irregular helical elements within A-form RNA such as stem-loops, base mismatches and bulges (Figure 3C)47–50.
Zinc Fingers
Zinc fingers are classical DNA-binding proteins that can also bind to RNA53, 54, as eloquently demonstrated by several recent structures55–57. They are typically classified based on the residues used to coordinate zinc: Cys2His2 (C2H2), CCCH, or CCHC and are generally present in multiple repeats within a protein. Thus, TFIIIA (where the motif was first identified) contains nine C2H2 zinc fingers: fingers 1–3, 5 and 7–9 interact with DNA, while fingers 4–6 interact with 5S RNA58, 59. C2H2 zinc fingers interact with DNA primarily by forming direct hydrogen bonds to Watson–Crick base pairs in the major groove, using residues within their recognition α-helix60, while TFIIIA binds RNA by making specific contacts to two RNA loops through the recognition helices of fingers 4 and 6. Thus, zinc fingers can use some of the same residues to recognize both nucleic acids, but the different DNA and RNA structures dictate a distinct structural arrangement of the zinc fingers on the nucleic acid template.
A second family of RNA-binding zinc fingers contains CCCH motivess61. Remarkably, in the structure of Tis11d bound to an AU-rich RNA element (ARE), sequence-specific RNA recognition occurs primarily through hydrogen bonding to the protein backbone (Figure 3D)57. Thus, the shape of the protein is the primary determinant of specificity by providing a rigid hydrogen-bonding template. This mode of recognition is reminiscent a third type of zinc fingers with a CCHC-zinc binding motif that is found in the nucleocapsid domain of the retroviral Gag proteins and in the HIV-1 nucleocapsid protein62–63.
S1 domain
S1 domains were first identified in ribosomal protein S1 (hence the name), but have since been found in other RNA-binding proteins, including several exonucleases64. The domain is composed of approximately 70 amino acids arranged in a 5-stranded antiparallel β-barrel capped by a short 310 helix65. The fold is similar to the oligonucleotide/oligosaccharide binding (OB) fold superfamily, which also contains the related RNA-binding Cold Shock Domain66. The S1 domain uses the common OB-fold binding surface to recognize nucleic acids through two β-strands surrounded by several loops67. Thus, RNA binding by the S1 domain is somewhat reminiscent of RNA recognition by the RRM, where a two-stranded β-sheet core contributes several conserved aromatic residues for stacking interactions with the nucleic acid bases, that are augmented by interactions provided by the surrounding loops and secondary structure elements65, 68.
PAZ and PIWI domains
RNA processing during RNAi and microRNA biogenesis generate species with unique structural and chemical features that must be recognized specifically but in a sequence-independent manner. These functional requirements are fulfilled by a specialized set of domains encountered in proteins involved in processing microRNA (miRNA) and small interfering RNA (siRNA) precursors.
The 110-amino-acid PAZ domain contains a β–barrel domain that resembles an OB or S1 fold juxtaposed to a small αβ domain that forms a clamp-like structure where RNA binds (Table 1)69–71. It selectively binds to the 2-nucleotide overhangs and probably serves as an anchor to position the miRNA for proper cleavage by Dicer72, 73. PAZ domains in Argonaute proteins facilitate cleavage of the target strand by the RISC complex responsible for degradation of the RNA targeted for silencing. The additional PIWI domain in Argonaute adopts instead an RNase H fold and anchors the unique 5′ end of the guide strand to position the target strand for degradation (Table 1)74–78.
Expanding conventional RNA-binding surfaces
The type of RNA that can be recognized by RNA-binding domains is increased not only by providing multiple domains within a protein (as discussed in the next section), but also by expanding a canonical RNA-binding surface through additional secondary structures or loops6, 50. In the reverse situation, a canonical recognition surface can be occluded by secondary structure elements, leading to the regulation of the RNA-binding activity. Thus, many proteins that are involved in spliceosome assembly have RNA-binding modules that differ from their canonical structure. For example, the SF1 protein that binds to the branch-point sequence has an additional QUA2 domain that defines an enlarged KH domain by making extensive hydrophobic interactions with the KH domain itself. By increasing the recognition surface, SF1 is able to bind to the seven single-stranded nucleotides that define the branch-point sequence42.
The structures of the first two quasi-RRMs from heterogeneous ribonucleoprotein (hnRNP) F demonstrate instead how an RRM can use a different surface for RNA recognition when the β-sheet surface is occluded79. This member of the hnRNP family is involved in the recognition of G-rich sequences (G-tracts) that are often found at recognition elements responsible for 5′ splice site recognition80–82. In the structure of the hnRNP F protein bound to the G-tract in Bcl-x pre-mRNA, each domain resembles a canonical RRM despite the absence of the RNP1 and RNP2 motifs normally used to bind RNA. Furthermore, the β-sheet surface is occluded by the presence of a C-terminal α–helix packed against it. Thus, the first two qRRMs of hnRNP F recognize RNA through a novel surface composed of a small β-hairpin between α2 and β4 and the β1–α1 and β2–β3 loops79. Perhaps the requirement for binding through a different surface in this complex stems from the necessity to recognize G-quadruplex RNA while at the same time preventing nonspecific binding to single stranded RNAs normally recognized by RRM proteins.
An additional α–helix C-terminal to the canonical domain is common in RRMs. The La protein C-terminal domain, Cleavage Stimulation Factor 64 (CstF-64) and U1A, all have a helix at the C terminus of the domain (Figure 3A)12, 20, 83. Many other domains form such an helix when bound to RNA, for example Hrp1, HuD and Polyadenylate Binding Protein14, 16, 25. The C-terminal RRM of La does not interact with RNA at all and, in the U1A and CstF-64 structures, the helix moves away from the β–sheet to allow RNA recognition using the canonical site (Figure 3A), suggesting that these helices perform primarily a regulatory role.
Multiple domains specify RNA recognition
Tandem domains
Isolated RNA-binding domains generally have limited ability to interact with RNA in a sequence-specific manner because their recognition sequences are too short6. Thus, multiple domains (typically two) are tethered together on a single polypeptide to create a much larger binding interface that recognizes a longer sequence. Perhaps the most extreme example of this concept comes from the Pumilio (Puf) family of proteins. Each domain recognizes a single nucleotide on its own, but by combining multiple repeats, the protein can bind with high affinity and specificity to as many as eight nucleotides (Table 1, Figure 4A)84. In fact, the three amino acids that recognize a particular nucleotide provide a reasonably predictive recognition code that can be exploited to engineer proteins that recognize different RNA sequences from those specified by the wild-type proteins84,85.
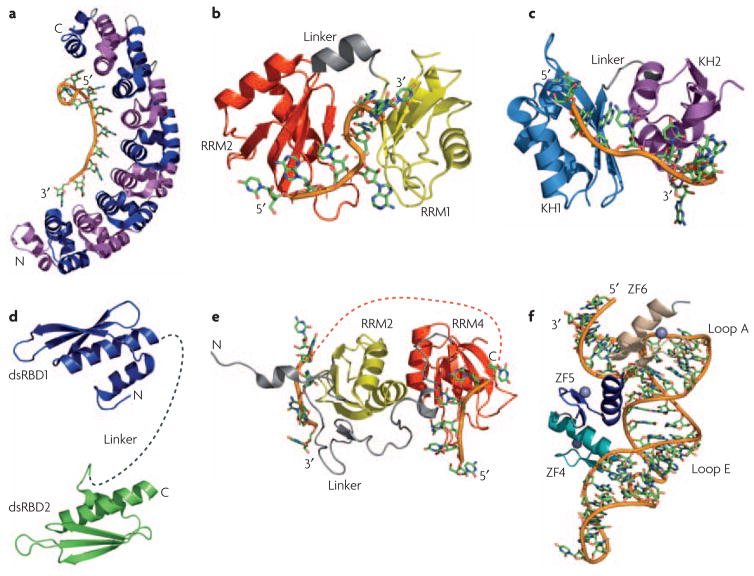
Figure 4. RNA-binding modules function together or alone to recognize a specific RNA.
a) The structure of human Pumilio provides an example of how multiple repeats (eight in this case) that individually recognize a few nucleotides (one in this case) combine to specifically recognize a much longer RNA sequence. Repeats are alternatively colored in magenta and blue; the RNA is colored similarly in all other structures with the backbone shown as an orange tube. b) In the structures of the two RNA recognition motifs (RRMs) from Hrp116 and c) the two K-homology (KH) domains of NusA37, a short linker (in gray) allows the two domains to position themselves with respect to one another upon binding RNA; for Hrp1, the first RRM is in yellow and the second in red; for NusA, the first KH domain is cyan and the second is purple. d) Flexibility within the linker between two double-stranded RNA-binding domains (dsRBDs) allow recognition of separated binding sites. The two dsRBDs of ADAR2 are connected by a flexible linker (dashed gray line) that may allow the protein to interact with a variety of targets of different structure49. e) The RNA-recognition motifs RRM3 (yellow) and RRM4 (red) of PTB form interdomain interactions involving the face of the protein opposite to the β-sheet involved in RNA recognition. This interaction positions the two domains in such away that interacting RNA sequences are looped away from each other, as indicated by the orange dotted line connecting the two RNAs28. f) The structure of the TFIIIA-RNA complex illustrates how zinc finger ZF5 (blue) functions as a spacer that properly positions zinc fingers 4 (teal) and 6 (tan) for recognition of loops E and A, respectively, within 5S rRNA55.
Inter-domain arrangement
Multiple domains associate with each other in a variety of ways to generate extended RNA recognition interfaces. The recent structure of Hrp1 (Figure 4B) exemplifies the structural principles involved in RNA recognition by two RRMs in tandem. In the free protein, both domains function as independent, rigid structures separated by a short flexible linker. Upon binding, both protein and RNA undergo significant changes in structure, with the linker forming a short helix and several inter-domain contacts creating a compact surface for recognition of adjacent stretches in the RNA16 (Figure 4B). The same is observed in Sxl, PABP, nucleolin and HuD proteins13–15, 25.
In contrast, when the zinc finger protein Tis11d binds to AU-rich RNA, there are few inter-domain interactions. However, a pre-organized linker between the two zinc fingers orients the two domains for recognition of an eight-nucleotide RNA by the protein main chain with little side chain involvement57 (Figure 3D). In a third example, in the structure of NusA bound to RNA, the two KH domains make extensive inter-domain contacts with each other, burying 1270Å2 86. This association of the KH domains creates an extended RNA-binding surface that allows the two domains to recognize an 11-nucleotide RNA37 (Figure 4C). Thus, each of the KH domains of NusA specifically recognizes four nucleotides, as is canonical for KH domains; their separation by a three-nucleotide linker that also makes interactions with the protein generates the complete recognition sequence37. This binding interface is further extended by an S1 domain N-terminal to the first KH domain that makes extensive inter-domain contacts and, in doing so, may provide an additional surface for RNA recognition.
The zinc-finger domains of TFIIIA provide another example of how linkers between RNA-recognition domains play a crucial role in substrate recognition. Quite remarkably, the linker in this case is a zinc-finger module! In the TFIIIA-5S RNA complex, fingers 4 and 6 interact extensively with the RNA, while finger 5 acts as a spacer that makes sequence-independent contacts involving the side-chains of its α-helix and the RNA backbone. Effectively, it serves as a bridge between loops E and A within 5S RNA, that are directly recognized by fingers 4 and 6, respectively56 (Figure 4F).
While the previous examples illustrate the importance of an ordered linker, the presence of a long flexible linker can be favored (Figure 2A) because it allows RNA-binding proteins to recognize sites that have a variable number of nucleotides between them, that are quite separated from each other on the same RNA or on different RNA molecules altogether. In these cases, ordering of the linker upon binding RNA is not likely to occur. A good illustration of this situation is provided by the two dsRBDs of the RNA-editing enzyme ADAR2, where the two domains do not interact and are separated by a flexible linker in the free or bound protein49 (Figure 4D). Since ADAR2 is required to edit multiple RNAs, interdomain flexibility allows each dsRBD to bind to its preferred site within RNAs of varying length and structure.
Yet another example of the potential advantages of connecting domains with flexible linkers can be found in complexes where conformational flexibility is required for function. In the FBP–FUSE complex, a 30-residue linker separates the KH3 and KH4 domains of FBP, so that they can move independently of each other even when the protein is bound to DNA39. This property is likely to be functionally important because FBP binds to and modulates the helicase activity of the general transcription factor TFIIH. Since this protein might function as a torque-generating machine, it is important for FBP to bind to the dynamic TFIIH molecule while maintaining its interaction with DNA.
This theme is observed even in proteins containing RRM domains, a departure from the common and canonical arrangement described above for Hrp1 and other proteins13–16, 25. The structure of polypyrimidine tract binding (PTB) protein shows that RRMs 3 and 4 are connected by a long linker and interact with each other in a way that forces their respective RNA-binding surfaces to face in opposite direction28. This orientation is essentially the opposite of what is observed in many di-domain proteins, yet may be functionally critical in splicing regulation by causing the exon or branch-point sequence to loop out, preventing binding of spliceosomal components and repressing splicing (Figure 4E).
The linker length is important
The considerations of the previous paragraph indicate that one of the major determinants for the affinity and specificity of RNA-binding proteins containing multiple domains resides within the amino acids linking the domains. The length and rigidity of the linker can have dramatic effects on RNA affinity87 and may influence whether a protein binds a single RNA or multiple RNAs (Figure 2A, right). Using the assumption that the free energy of binding individual domains is additive, we would expect the affinity of a protein with multiple RNA binding domains to be the product of the affinity of the individual domains. However, because the linker remains flexible in hnRNP A1, the affinity of the two-domain protein is 1000-fold less than the product of the affinities of the individual domains88. When the first RNA binding domain is bound, the second RNA binding domain sweeps a volume proportional to the length of the linker. Within this sphere, the effective concentration of the second domain is different than in the free solution, leading to altered affinity. A simple model was developed to calculate how the length of the linker affects affinity; using this model, long linkers (more than 50–60 residues) are predicted to have a negligible impact on affinity, because the two domains act independently of each other. As the linker gets shorter, the affinity for RNA increases between 10- and 1000-fold, when compared to the affinity of individual RRMs added together87.
This simple model assumes that the linker does not contact the RNA, but in many cases the linker becomes ordered upon binding RNA. In the example of nucleolin, the model would have predicted a 100-fold increase in affinity compared to that of the two individual nucleolin RRMs, but an increase of between 1000- and 100,000-fold was observed depending on the RNA sequence tested89. Part of the increase in affinity was attributable to the ordering of the linker into an α-helix to effectively shorten its length by half. When the prediction was repeated with this correction, predicted and measured affinities agreed to within 10 fold for some RNAs. However, because of direct interactions between the linker and target RNAs, even this calculation could not account for the 1000-fold difference between predicted and observed affinities for other RNAs 89.
Protein-protein interactions and RNA recognition
Homo- and hetero-dimerization of RNA-binding proteins
In addition to expanding the ways in which RNA can be recognized, multiple modules also allow RNA-binding proteins to interact simultaneously with other proteins and with RNA. The simplest example of this is dimerization. Two proteins involved in the viral response to RNA silencing provide exquisite examples of how dimerization allows specific interactions to be established that would not be possible in the isolated proteins.
The p19 protein is required for tombusvirus virulence in plants, and can also provide this activity when expressed in both Drosophila and human cells90, 91. It functions by specifically binding to siRNAs and preventing its loading into the RISC complex92. Two structures of p19 proteins bound to 21-nucleotides siRNA demonstrate that the protein adopts an αβ topology and binds RNA as a homodimer. The RNA binding surface is formed by a continuous 8-stranded β-sheet formed by the two monomers. Each monomer measures the length of the siRNA by providing a Trp that forms stacking interactions with the bases at the 5′ and 3′ end of the siRNA; the position of the Trp is defined by the structure of the homodimer. Thus, dimerization of p19 allows this protein to measure the length of the siRNA with great precision by positioning the two critical Trp side chains92, 93.
Another potent viral suppressor of RNAi is the Flock House Virus B2 protein. Its structure is composed of three α-helices that dimerize to create a four-helix bundle that recognizes RNA along one face of an A-form helix94, 95. Structural and biochemical evidence demonstrated that this protein suppresses silencing in two ways: by binding to siRNAs and preventing loading into RISC, and by coating longer dsRNA precursors and protecting them from cleavage by Dicer. For both p19 and Flock House Virus B2, the conserved features of the siRNAs (their size and double helical character)92–95 are recognized because dimerization generates extended binding sites out of small protein domains and because it establishes the relative position of amino acids involved in RNA recognition.
These two examples illustrate the role of dimerization in RNA recognition, but there are other examples of RNA binding domains that function by dimerization or by forming protein-protein interactions. In the structure of the N-terminal RRM of U1A bound an RNA regulatory element within its own 3′-untranslated region (UTR), two separate RRMs interact through their C-terminal helices to form a homodimer after binding to the RNA. This cooperative binding event can only occur in the presence of RNA because the C-terminal helix is associated with the β-sheet surface of the RRM in the free protein. Interestingly, this dimerization also creates an interface that inhibits polyadenylation by direct interaction with poly(A) polymerase24. In the Nova-1 KH3 domain, changes in the rigidity of the protein are observed upon dimerization, and this stiffening of the entire protein may aid in nucleic acid recognition by reducing the entropic cost of binding to RNA. Furthermore, dimerization presents two recognition sites for RNA binding and thus can provide a cooperative interaction that strengthens the affinity of the protein for the RNA96.
The formation heterodimers through interactions between an RNA binding domain and another protein can increase the specificity of RNA interaction. For example, the binding of the spliceosomal U2B″ RRM to a stem-loop within U2 snRNA requires an interaction with the U2A′ protein23. In a different example, the CBP80 subunit of the cap-binding complex must interact with the RRM of CBP20 if this RRM is to bind with high affinity to the 7-methylguanosine cap of mRNA22, 97. The recent structures of the archaeal and eukaryotic exosomes have revealed extensive protein-protein interactions between proteins containing both KH and S1 domains with the core of the protein complex98, 99. These interactions may position the S1 domains of specific exosome subunits to recognize the RNAs targeted for exosomal degradation.
Protein-protein interactions define RNA specificity
RNA-binding domains from different proteins can cooperate to recognize an RNA through a combination of weak protein-RNA and protein-protein interactions. The recent dissection of a complex derived from the spliceosome demonstrates this principle and illustrates how even relatively small sequence and structural alterations in RNA-binding domains can modulate their RNA recognition properties indirectly by altering protein-protein interactions (Figure 5).
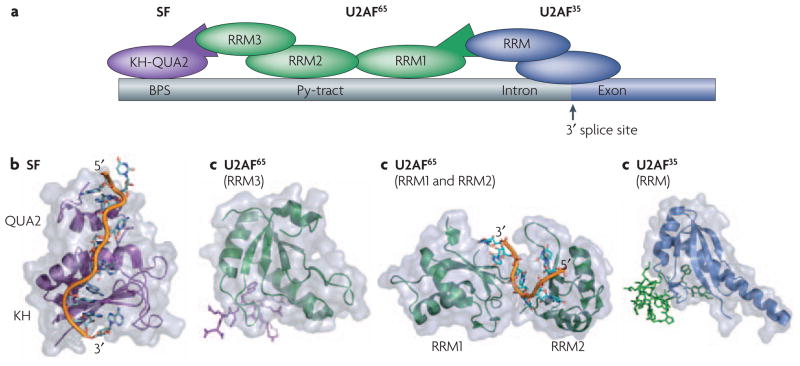
Figure 5. Protein-protein interactions and protein-RNA interactions define the site of spliceosomal assembly.
Proper definition of the splicing site requires a number of cooperative binding events that are mediated by both protein-protein and protein-RNA contacts between various RNA binding modules. a) Schematic of the interactions between various proteins and RNA at the splicing site. Some of the key domains involved in these interactions whose structures are shown below are labeled. Within the RNA the branch-point sequence (BPS), pyrimidine tract (Py-tract), and the 3′ splice site (3′ ss) are labeled with the intron colored gray and the exon colored dark blue. b) SF1 recognizes the BPS through its KH-QUA2 domains. The additional QUA2 domain creates an extended KH domain that can recognize the full BPS sequence RNA42. c) This interaction is strengthened by protein-protein interactions between it’s N-terminus and the non-canonical RRM3 of U2AF6535, d) which is bound to the pyrmidine tract through it’s first two canonical RRMs10. Finally, the U2AF65 interaction is also aided by protein-protein interactions between it’s N-terminus and the non-canonical RRM of U2AF35 bound at the 3′ splice site33. All of the protein and peptide structures are colored as shown in the schematic (a).
During initial steps in spliceosome assembly, the splicing factor 1 (SF1) and U2 auxiliary factor (U2AF) proteins cooperatively bind to sequences at the 3′ splice site and upstream of it (Figure 5A). Recognition of RNA cis-acting elements by the two U2AF subunits, U2AF65 and U2AF35, commits the pre-mRNA to the splicing reaction. Specifically, U2AF65 recognizes the polypyrimidine tract within the pre-mRNA primarily through its two central canonical RRMs(Figure 5A, D); this interaction is strengthened by the interaction between a third non-canonical RRM in this protein and SF1 protein (Figure 5A, C), which is instead bound at the branch-point sequence through a KH domain (Figure 5A, B). Additional cooperativity in the assembly of this complex is provided by protein–protein interactions between a non-canonical RRM in U2AF35 (Figure 5A, E), bound at the 3′ splice site, and the N terminus of U2AF65.
Protein-protein interaction surfaces
As described in the previous paragraph, RRM domains can form protein-protein as well as protein-RNA interactions. The protein-protein interactions occur via non-canonical RRM domains within both U2AF65 and U2AF35 that have a much longer α1 helix compared to other RRMs; this helix is the primary mediator of the protein–protein interactions observed in this complex33, 35 (Figure 5C, E). Closer inspection of these U2AF structures reveals a few common themes that may indicate whether an RRM binds to protein or to RNA: poor conservation of the RNP motifs, an Arg-X-Phe motif in the last loop of the RRM, and conserved acidic residues in the α1 helix100. These features define a novel functional class, the U2AF-homology motifs (UHMs), that are capable of forming protein–protein interactions.
The UHM class does not exhaust all possible ways in which two RRMs can interact. The interactions of other RRMs with proteins (for example the Y14-Magoh structure from the exon-junction complex and the Upf2–Upf3 RNA surveillance complexes29, 31, 32, 34, 101, 102) occur on the surface of the β-sheet through residues that are involved in RNA binding in other RRMs. Until more structures of such protein-protein complexes become available, the sequence and structural features in such RRMs that allow them to bind to other proteins rather than RNA will remain unclear.
RNA-binding domains other than the RRM have the ability to participate in protein-protein interactions. As previously described, a number of the KH domains can dimerize, and dsRBD domains form protein-protein interactions that regulate the assembly of complexes involved in RNA localization and the catalytic activity of enzymes acting upon double-stranded RNA. One dsRBD example is illustrated by Staufen, a protein involved in RNA localization in early development and in neurons. Staufen proteins contain up to 5 dsRBDs; some domains are capable of binding dsRNA48, while other domains bind other proteins during embryogenesis103. Remarkably, surface-exposed amino acids involved in RNA recognition are conserved among Staufen dsRBDs that bind to dsRNA, but not in protein-binding dsRBDs. For these domains it is the surface opposite to dsRNA in the canonical dsRBD-dsRNA structure that is conserved instead48. Thus, the ability of these proteins to bind to other proteins can be as important functionally as its RNA-binding activity.
Catalytic domains acting upon RNA
Positioning catalytic domains onto their substrate
Modularity allows RNA-binding domains to target a substrate, and to promote or repress the enzymatic activity of catalytic domains within the same polypeptide (Figure 2D). The way in which RNA-binding and enzymatic modules are positioned within a protein can define how a particular protein recognizes RNA. However, the enzymatic activity can also be enhanced or repressed through mutually exclusive or cooperative interactions between RNA-binding domains, catalytic domains and RNA.
An elegant example of how domain positioning facilitates enzymatic function comes from the RNAi pathway. In the first step of the cascade leading to gene silencing, Drosha and Pasha process primary miRNAs to stem-loops of ~70 nucleotides; Dicer subsequently binds to these miRNA precursors by recognizing two 3′-terminal nucleotides overhangs generated by Drosha104. A minimal Dicer structure from Giardia (lacking the N-terminal helicase and the C-terminal dsRBD, Figure 1) demonstrates that Dicer likely functions as a molecular ruler that positions the catalytic RNase III domains ~25 nucleotides from where the 3′ overhanging nucleotides are recognized by its PAZ domain72, the approximate length of siRNAs.
Another particularly beautiful example of this principle is found in the recent structure of a complete archaeal Box H/ACA small nucleolar RNP (snoRNP)105. These particles are responsible for the catalytic conversion of uracil to pseudouridine in ribosomal and other RNAs106. In this structure, the site of pseudouridylation is juxtaposed to the catalytic center of the protein enzyme Cbf5/dyskerin by two protein clamps at either end of the RNA. The 3′-terminal ‘clamp’ (the ACA sequence motif that defines this class of non-coding RNAs) is recognized by the PUA domain of Cbf5, while the second clamp (the apical loop of the non coding RNA) is recognized by a complex of Cbf5 with two other protein components of the particle.
Activating and repressing enzymes acting on dsRNA
The dsRNA-dependent protein kinase PKR (Figure 6A) and the RNA-editing enzyme ADAR2 (Figure 6B) provide examples of how RNA-binding domains can modulate enzymatic activity by interacting with both the substrate RNA and with the catalytic domain (Figure 2D). PKR is an interferon-induced kinase that plays a key role in controlling viral infections and maintaining cellular homeostasis by becoming activated in response to double-stranded viral RNAs. In the active form, it phosphorylates the α subunit of eukaryotic initiation factor 2 (eIF-2), thereby inhibiting translation and suppressing viral spread107. ADARs act on dsRNA to catalyze the conversion of adenosine to inosine, which is then recognized as guanosine, affecting both the primary sequence and the structure of the edited RNA108.
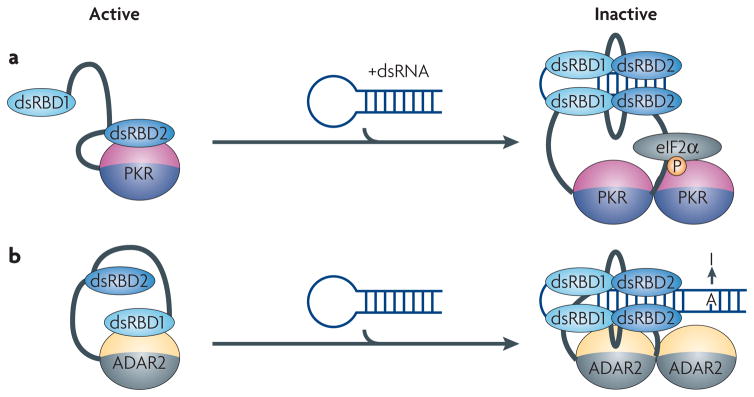
Figure 6. Modular architecture allows for regulation of the catalytic activity of enzymatic domains.
In both PKR and ADAR proteins, inter-domain interactions between the RNA-binding module and the catalytic domain maintain the proteins in an inactive state. a) The kinase domain of PKR is inhibited by an interaction with the double-stranded RNA-binding domain (dsRBD2). Binding to dsRNA releases the kinase from its inactive state allowing it to inhibit translation by phosphorylating eIF2α. b) The activity of ADAR2 is controlled by a mechanism similar to PKR, but in this case dsRBD1 is involved in the inactivation of the catalytic domain. When double-stranded RNA (dsRNA) binds to both dsRBDs, the protein dimerizes and the catalytic domain becomes exposed to convert adenosine to inosine.
Both proteins have two N-terminal dsRBDs that bind to dsRNA; in each case, the dsRBDs function both as an RNA-recognition unit and as an auto-inhibitor of the catalytic domain109, 110. In PKR, the second dsRBD masks the kinase domain by binding to it directly, thereby maintaining its inactive state (Figure 6A)109, 111, 112. In ADAR2, the proposed inhibitory element is the first dsRBD110 (Figure 6B). In both proteins, however, RNA binding causes enzyme activation by relieving the auto-inhibition caused by the interactions between the RNA-binding and catalytic domains (Figure 6A, B). Since both ADAR and PKR require RNA of sufficient length for activation, the two dsRBDs may be necessary for fully de-repressing the catalytic activity110. In PKR, the presence of a sufficiently long dsRNA (for example, viral RNAs such as HIV TAR) allows both dsRBDs to cooperatively bind to RNA113, 114, relieving the structural block and allowing the kinase domain to be activated through autophosphorylation and dimerization115–117. The initial event in this cascade is likely to be binding of the first dsRBD to dsRNA, because this domain has much higher affinity for RNA compared to the second domain114. Only in the presence of a sufficiently long dsRNA can the second dsRBD bind as well, thereby releasing the kinase from its inactive state.
Conclusions
Many RNA-binding proteins are composed of relatively few modules of conserved structure but often limited sequence specificity. By combining these motifs in a variety of structural arrangements, evolution has generated proteins that are capable of recognizing RNA with the affinity and selectivity required to find cognate RNAs in the cellular medium, while at the same time retaining the versatility required to regulate, assemble and disassemble RNA-processing complexes. Structural biology has provided the molecular details concerning how individual domains recognize RNA, but many of these proteins require multiple copies of one of several common domains to function (Figure 1). It is therefore important to understand how multiple modules bind RNA, and how the modular nature of these proteins specifies their biological function. We have described here some of the structural principles of how multiple domains recognize an RNA(Figure 2), but there are still relatively few structures of proteins containing multiple RNA binding domains. Recent studies have also led to the observation that RNA binding modules can regulate the biological activity of enzymes acting upon RNAs in ways that go beyond the identification of the target RNA, but full understanding of these regulatory mechanisms will require detailed structural characterization that is not yet available. We expect that future structural analysis will expand upon the diverse ways in which combinations of RNA binding domains augment protein function.
Acknowledgments
Work in our laboratories is supported by grants from NIH-NIGMS (GV and CM). We apologize to the many colleagues whose work could not be properly referenced due to lack of space.
Glossary Terms
- Ribonucleoprotein (RNP)
Complexes that contain both proteins and RNA. The ribonucleoprotein motif refers to the two conserved sequence elements found within the RNA Recognition Motif (within its two central β-strands) that participate in RNA recognition and identify the RRM domain at the sequence level.
- Zinc finger
A class of DNA- and RNA-binding proteins characterized by a Cys- and His-rich domain that chelate a Zinc ion. Different classes of zinc-finger proteins contain different combination of metal binding amino acids; thus, C2H2 zinc finger contain two Cys and two His residues, while CCCH and CCHC zinc-binding motifs contain three Cys and a single His in a different topological arrangement.
- AU-rich element (ARE)
Sequences rich in A and U nucleotides found in the 3′-untranslated regions of mRNAs that promote stability or degradation of their associated RNAs, thus providing a mechanism for the control of gene expression.
- RISC complex
A protein complex responsible for degradation of RNA species targeted by small interfering RNAs. Argonaute protein is the catalytic component of RISC.
- Exon junction complex
This is a multi-subunit protein complex that is deposited on the mRNA during the splicing reaction near the splice site. It remains bound to the RNA during subsequent gene expression events, and serves as a platform to recruit nuclear and cytoplasmic factors that influence mRNA localization, transport, stability and translation.
- Orthology
Orthologous proteins are direct evolutionary counterparts that retain the same function in different organisms and that have arisen due to speciation events but not through the process of gene duplication (paralogy).
References
- 1.Dreyfuss G, Kim VN, Kataoka N. Messenger-RNA-binding proteins and the messages they carry. Nat Rev Mol Cell Biol. 2002;3:195–205. doi: 10.1038/nrm760. [DOI] [PubMed] [Google Scholar]
- 2.Burd CG, Dreyfuss G. Conserved structures and diversity of functions of RNA-binding proteins. Science. 1994;265:615–21. doi: 10.1126/science.8036511. [DOI] [PubMed] [Google Scholar]
- 3.Auweter SD, Oberstrass FC, Allain FH. Sequence-specific binding of single-stranded RNA: is there a code for recognition? Nucleic Acids Res. 2006;34:4943–59. doi: 10.1093/nar/gkl620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chang KY, Ramos A. The double-stranded RNA-binding motif, a versatile macromolecular docking platform. Febs J. 2005;272:2109–17. doi: 10.1111/j.1742-4658.2005.04652.x. [DOI] [PubMed] [Google Scholar]
- 5.Hall TM. Multiple modes of RNA recognition by zinc finger proteins. Curr Opin Struct Biol. 2005;15:367–73. doi: 10.1016/j.sbi.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 6.Maris C, Dominguez C, Allain FH. The RNA recognition motif, a plastic RNA-binding platform to regulate post-transcriptional gene expression. Febs J. 2005;272:2118–31. doi: 10.1111/j.1742-4658.2005.04653.x. [DOI] [PubMed] [Google Scholar]
- 7.Pawson T, Nash P. Assembly of cell regulatory systems through protein interaction domains. Science. 2003;300:445–52. doi: 10.1126/science.1083653. [DOI] [PubMed] [Google Scholar]
- 8.Doolittle RF. The multiplicity of domains in proteins. Annu Rev Biochem. 1995;64:287–314. doi: 10.1146/annurev.bi.64.070195.001443. [DOI] [PubMed] [Google Scholar]
- 9.Bork P, Downing AK, Kieffer B, Campbell ID. Structure and distribution of modules in extracellular proteins. Q Rev Biophys. 1996;29:119–67. doi: 10.1017/s0033583500005783. [DOI] [PubMed] [Google Scholar]
- 10.Sickmier EA, et al. Structural basis for polypyrimidine tract recognition by the essential pre-mRNA splicing factor U2AF65. Mol Cell. 2006;23:49–59. doi: 10.1016/j.molcel.2006.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deka P, Rajan PK, Perez-Canadillas JM, Varani G. Protein and RNA dynamics play key roles in determining the specific recognition of GU-rich polyadenylation regulatory elements by human Cstf-64 protein. J Mol Biol. 2005;347:719–33. doi: 10.1016/j.jmb.2005.01.046. [DOI] [PubMed] [Google Scholar]
- 12.Perez Canadillas JM, Varani G. Recognition of GU-rich polyadenylation regulatory elements by human CstF-64 protein. Embo J. 2003;22:2821–30. doi: 10.1093/emboj/cdg259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Allain FH, Bouvet P, Dieckmann T, Feigon J. Molecular basis of sequence-specific recognition of pre-ribosomal RNA by nucleolin. Embo J. 2000;19:6870–81. doi: 10.1093/emboj/19.24.6870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deo RC, Bonanno JB, Sonenberg N, Burley SK. Recognition of polyadenylate RNA by the poly(A)-binding protein. Cell. 1999;98:835–45. doi: 10.1016/s0092-8674(00)81517-2. [DOI] [PubMed] [Google Scholar]
- 15.Handa N, et al. Structural basis for recognition of the tra mRNA precursor by the Sex-lethal protein. Nature. 1999;398:579–85. doi: 10.1038/19242. [DOI] [PubMed] [Google Scholar]
- 16.Perez-Canadillas JM. Grabbing the message: structural basis of mRNA 3′UTR recognition by Hrp1. Embo J. 2006;25:3167–78. doi: 10.1038/sj.emboj.7601190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Birney E, Kumar S, Krainer AR. Analysis of the RNA-recognition motif and RS and RGG domains: conservation in metazoan pre-mRNA splicing factors. Nucleic Acids Res. 1993;21:5803–16. doi: 10.1093/nar/21.25.5803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oubridge C, Ito N, Evans PR, Teo CH, Nagai K. Crystal structure at 1.92 A resolution of the RNA-binding domain of the U1A spliceosomal protein complexed with an RNA hairpin. Nature. 1994;372:432–8. doi: 10.1038/372432a0. [DOI] [PubMed] [Google Scholar]
- 19.Finn RD, et al. Pfam: clans, web tools and services. Nucleic Acids Res. 2006;34:D247–51. doi: 10.1093/nar/gkj149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Allain FH, et al. Specificity of ribonucleoprotein interaction determined by RNA folding during complex formulation. Nature. 1996;380:646–50. doi: 10.1038/380646a0. [DOI] [PubMed] [Google Scholar]
- 21.Ding J, et al. Crystal structure of the two-RRM domain of hnRNP A1 (UP1) complexed with single-stranded telomeric DNA. Genes Dev. 1999;13:1102–15. doi: 10.1101/gad.13.9.1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mazza C, Segref A, Mattaj IW, Cusack S. Large-scale induced fit recognition of an m(7)GpppG cap analogue by the human nuclear cap-binding complex. Embo J. 2002;21:5548–57. doi: 10.1093/emboj/cdf538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Price SR, Evans PR, Nagai K. Crystal structure of the spliceosomal U2B″-U2A′ protein complex bound to a fragment of U2 small nuclear RNA. Nature. 1998;394:645–50. doi: 10.1038/29234. [DOI] [PubMed] [Google Scholar]
- 24.Varani L, et al. The NMR structure of the 38 kDa U1A protein - PIE RNA complex reveals the basis of cooperativity in regulation of polyadenylation by human U1A protein. Nat Struct Biol. 2000;7:329–35. doi: 10.1038/74101. [DOI] [PubMed] [Google Scholar]
- 25.Wang X, Tanaka Hall TM. Structural basis for recognition of AU-rich element RNA by the HuD protein. Nat Struct Biol. 2001;8:141–5. doi: 10.1038/84131. [DOI] [PubMed] [Google Scholar]
- 26.Auweter SD, et al. Molecular basis of RNA recognition by the human alternative splicing factor Fox-1. Embo J. 2006;25:163–73. doi: 10.1038/sj.emboj.7600918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hargous Y, et al. Molecular basis of RNA recognition and TAP binding by the SR proteins SRp20 and 9G8. Embo J. 2006;25:5126–37. doi: 10.1038/sj.emboj.7601385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oberstrass FC, et al. Structure of PTB bound to RNA: specific binding and implications for splicing regulation. Science. 2005;309:2054–7. doi: 10.1126/science.1114066. [DOI] [PubMed] [Google Scholar]
- 29.Bono F, Ebert J, Lorentzen E, Conti E. The crystal structure of the exon junction complex reveals how it maintains a stable grip on mRNA. Cell. 2006;126:713–25. doi: 10.1016/j.cell.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 30.Bono F, et al. Molecular insights into the interaction of PYM with the Mago-Y14 core of the exon junction complex. EMBO Rep. 2004;5:304–10. doi: 10.1038/sj.embor.7400091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fribourg S, Gatfield D, Izaurralde E, Conti E. A novel mode of RBD-protein recognition in the Y14-Mago complex. Nat Struct Biol. 2003;10:433–9. doi: 10.1038/nsb926. [DOI] [PubMed] [Google Scholar]
- 32.Kadlec J, Izaurralde E, Cusack S. The structural basis for the interaction between nonsense-mediated mRNA decay factors UPF2 and UPF3. Nat Struct Mol Biol. 2004;11:330–7. doi: 10.1038/nsmb741. [DOI] [PubMed] [Google Scholar]
- 33.Kielkopf CL, Rodionova NA, Green MR, Burley SK. A novel peptide recognition mode revealed by the X-ray structure of a core U2AF35/U2AF65 heterodimer. Cell. 2001;106:595–605. doi: 10.1016/s0092-8674(01)00480-9. [DOI] [PubMed] [Google Scholar]
- 34.Lau CK, Diem MD, Dreyfuss G, Van Duyne GD. Structure of the Y14-Magoh core of the exon junction complex. Curr Biol. 2003;13:933–41. doi: 10.1016/s0960-9822(03)00328-2. [DOI] [PubMed] [Google Scholar]
- 35.Selenko P, et al. Structural basis for the molecular recognition between human splicing factors U2AF65 and SF1/mBBP. Mol Cell. 2003;11:965–76. doi: 10.1016/s1097-2765(03)00115-1. [DOI] [PubMed] [Google Scholar]
- 36.Backe PH, Messias AC, Ravelli RB, Sattler M, Cusack S. X-ray crystallographic and NMR studies of the third KH domain of hnRNP K in complex with single-stranded nucleic acids. Structure. 2005;13:1055–67. doi: 10.1016/j.str.2005.04.008. [DOI] [PubMed] [Google Scholar]
- 37.Beuth B, Pennell S, Arnvig KB, Martin SR, Taylor IA. Structure of a Mycobacterium tuberculosis NusA-RNA complex. Embo J. 2005;24:3576–87. doi: 10.1038/sj.emboj.7600829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Braddock DT, Baber JL, Levens D, Clore GM. Molecular basis of sequence-specific single-stranded DNA recognition by KH domains: solution structure of a complex between hnRNP K KH3 and single-stranded DNA. Embo J. 2002;21:3476–85. doi: 10.1093/emboj/cdf352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Braddock DT, Louis JM, Baber JL, Levens D, Clore GM. Structure and dynamics of KH domains from FBP bound to single-stranded DNA. Nature. 2002;415:1051–6. doi: 10.1038/4151051a. [DOI] [PubMed] [Google Scholar]
- 40.Du Z, et al. Crystal structure of the first KH domain of human poly(C)-binding protein-2 in complex with a C-rich strand of human telomeric DNA at 1.7 A. J Biol Chem. 2005;280:38823–30. doi: 10.1074/jbc.M508183200. [DOI] [PubMed] [Google Scholar]
- 41.Lewis HA, et al. Sequence-specific RNA binding by a Nova KH domain: implications for paraneoplastic disease and the fragile X syndrome. Cell. 2000;100:323–32. doi: 10.1016/s0092-8674(00)80668-6. [DOI] [PubMed] [Google Scholar]
- 42.Liu Z, et al. Structural basis for recognition of the intron branch site RNA by splicing factor 1. Science. 2001;294:1098–102. doi: 10.1126/science.1064719. [DOI] [PubMed] [Google Scholar]
- 43.Siomi H, Matunis MJ, Michael WM, Dreyfuss G. The pre-mRNA binding K protein contains a novel evolutionarily conserved motif. Nucleic Acids Res. 1993;21:1193–8. doi: 10.1093/nar/21.5.1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.De Boulle K, et al. A point mutation in the FMR-1 gene associated with fragile X mental retardation. Nat Genet. 1993;3:31–5. doi: 10.1038/ng0193-31. [DOI] [PubMed] [Google Scholar]
- 45.Grishin NV. KH domain: one motif, two folds. Nucleic Acids Res. 2001;29:638–43. doi: 10.1093/nar/29.3.638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ryter JM, Schultz SC. Molecular basis of double-stranded RNA-protein interactions: structure of a dsRNA-binding domain complexed with dsRNA. Embo J. 1998;17:7505–13. doi: 10.1093/emboj/17.24.7505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Leulliot N, et al. A new alpha-helical extension promotes RNA binding by the dsRBD of Rnt1p RNAse III. Embo J. 2004;23:2468–77. doi: 10.1038/sj.emboj.7600260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ramos A, et al. RNA recognition by a Staufen double-stranded RNA-binding domain. Embo J. 2000;19:997–1009. doi: 10.1093/emboj/19.5.997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stefl R, Xu M, Skrisovska L, Emeson RB, Allain FH. Structure and specific RNA binding of ADAR2 double-stranded RNA binding motifs. Structure. 2006;14:345–55. doi: 10.1016/j.str.2005.11.013. [DOI] [PubMed] [Google Scholar]
- 50.Wu H, Henras A, Chanfreau G, Feigon J. Structural basis for recognition of the AGNN tetraloop RNA fold by the double-stranded RNA-binding domain of Rnt1p RNase III. Proc Natl Acad Sci U S A. 2004;101:8307–12. doi: 10.1073/pnas.0402627101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stephens OM, Haudenschild BL, Beal PA. The binding selectivity of ADAR2’s dsRBMs contributes to RNA-editing selectivity. Chem Biol. 2004;11:1239–50. doi: 10.1016/j.chembiol.2004.06.009. [DOI] [PubMed] [Google Scholar]
- 52.Xu M, Wells KS, Emeson RB. Substrate-dependent contribution of double-stranded RNA-binding motifs to ADAR2 function. Mol Biol Cell. 2006;17:3211–20. doi: 10.1091/mbc.E06-02-0162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Carballo E, Lai WS, Blackshear PJ. Feedback inhibition of macrophage tumor necrosis factor-alpha production by tristetraprolin. Science. 1998;281:1001–5. doi: 10.1126/science.281.5379.1001. [DOI] [PubMed] [Google Scholar]
- 54.Picard B, Wegnez M. Isolation of a 7S particle from Xenopus laevis oocytes: a 5S RNA-protein complex. Proc Natl Acad Sci U S A. 1979;76:241–5. doi: 10.1073/pnas.76.1.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lee BM, et al. Induced fit and “lock and key” recognition of 5S RNA by zinc fingers of transcription factor IIIA. J Mol Biol. 2006;357:275–91. doi: 10.1016/j.jmb.2005.12.010. [DOI] [PubMed] [Google Scholar]
- 56.Lu D, Searles MA, Klug A. Crystal structure of a zinc-finger-RNA complex reveals two modes of molecular recognition. Nature. 2003;426:96–100. doi: 10.1038/nature02088. [DOI] [PubMed] [Google Scholar]
- 57.Hudson BP, Martinez-Yamout MA, Dyson HJ, Wright PE. Recognition of the mRNA AU-rich element by the zinc finger domain of TIS11d. Nat Struct Mol Biol. 2004;11:257–64. doi: 10.1038/nsmb738. [DOI] [PubMed] [Google Scholar]
- 58.Clemens KR, et al. Molecular basis for specific recognition of both RNA and DNA by a zinc finger protein. Science. 1993;260:530–3. doi: 10.1126/science.8475383. [DOI] [PubMed] [Google Scholar]
- 59.Searles MA, Lu D, Klug A. The role of the central zinc fingers of transcription factor IIIA in binding to 5 S RNA. J Mol Biol. 2000;301:47–60. doi: 10.1006/jmbi.2000.3946. [DOI] [PubMed] [Google Scholar]
- 60.Wolfe SA, Nekludova L, Pabo CO. DNA recognition by Cys2His2 zinc finger proteins. Annu Rev Biophys Biomol Struct. 2000;29:183–212. doi: 10.1146/annurev.biophys.29.1.183. [DOI] [PubMed] [Google Scholar]
- 61.Lai WS, Carballo E, Thorn JM, Kennington EA, Blackshear PJ. Interactions of CCCH zinc finger proteins with mRNA. Binding of tristetraprolin-related zinc finger proteins to Au-rich elements and destabilization of mRNA. J Biol Chem. 2000;275:17827–37. doi: 10.1074/jbc.M001696200. [DOI] [PubMed] [Google Scholar]
- 62.D’Souza V, Summers MF. Structural basis for packaging the dimeric genome of Moloney murine leukaemia virus. Nature. 2004;431:586–90. doi: 10.1038/nature02944. [DOI] [PubMed] [Google Scholar]
- 63.De Guzman RN, et al. Structure of the HIV-1 nucleocapsid protein bound to the SL3 psi-RNA recognition element. Science. 1998;279:384–8. doi: 10.1126/science.279.5349.384. [DOI] [PubMed] [Google Scholar]
- 64.Subramanian AR. Structure and functions of ribosomal protein S1. Prog Nucleic Acid Res Mol Biol. 1983;28:101–42. doi: 10.1016/s0079-6603(08)60085-9. [DOI] [PubMed] [Google Scholar]
- 65.Bycroft M, Hubbard TJ, Proctor M, Freund SM, Murzin AG. The solution structure of the S1 RNA binding domain: a member of an ancient nucleic acid-binding fold. Cell. 1997;88:235–42. doi: 10.1016/s0092-8674(00)81844-9. [DOI] [PubMed] [Google Scholar]
- 66.Murzin AG. OB(oligonucleotide/oligosaccharide binding)-fold: common structural and functional solution for non-homologous sequences. Embo J. 1993;12:861–7. doi: 10.1002/j.1460-2075.1993.tb05726.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Arcus V. OB-fold domains: a snapshot of the evolution of sequence, structure and function. Curr Opin Struct Biol. 2002;12:794–801. doi: 10.1016/s0959-440x(02)00392-5. [DOI] [PubMed] [Google Scholar]
- 68.Schubert M, et al. Structural characterization of the RNase E S1 domain and identification of its oligonucleotide-binding and dimerization interfaces. J Mol Biol. 2004;341:37–54. doi: 10.1016/j.jmb.2004.05.061. [DOI] [PubMed] [Google Scholar]
- 69.Lingel A, Simon B, Izaurralde E, Sattler M. Structure and nucleic-acid binding of the Drosophila Argonaute 2 PAZ domain. Nature. 2003;426:465–9. doi: 10.1038/nature02123. [DOI] [PubMed] [Google Scholar]
- 70.Lingel A, Simon B, Izaurralde E, Sattler M. Nucleic acid 3′-end recognition by the Argonaute2 PAZ domain. Nat Struct Mol Biol. 2004;11:576–7. doi: 10.1038/nsmb777. [DOI] [PubMed] [Google Scholar]
- 71.Yan KS, et al. Structure and conserved RNA binding of the PAZ domain. Nature. 2003;426:468–74. doi: 10.1038/nature02129. [DOI] [PubMed] [Google Scholar]
- 72.Macrae IJ, et al. Structural basis for double-stranded RNA processing by Dicer. Science. 2006;311:195–8. doi: 10.1126/science.1121638. [DOI] [PubMed] [Google Scholar]
- 73.Ma JB, Ye K, Patel DJ. Structural basis for overhang-specific small interfering RNA recognition by the PAZ domain. Nature. 2004;429:318–22. doi: 10.1038/nature02519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yuan YR, et al. Crystal structure of A. aeolicus argonaute, a site-specific DNA-guided endoribonuclease, provides insights into RISC-mediated mRNA cleavage. Mol Cell. 2005;19:405–19. doi: 10.1016/j.molcel.2005.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ma JB, et al. Structural basis for 5′-end-specific recognition of guide RNA by the A. fulgidus Piwi protein. Nature. 2005;434:666–70. doi: 10.1038/nature03514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Song JJ, Smith SK, Hannon GJ, Joshua-Tor L. Crystal structure of Argonaute and its implications for RISC slicer activity. Science. 2004;305:1434–7. doi: 10.1126/science.1102514. [DOI] [PubMed] [Google Scholar]
- 77.Parker JS, Roe SM, Barford D. Crystal structure of a PIWI protein suggests mechanisms for siRNA recognition and slicer activity. Embo J. 2004;23:4727–37. doi: 10.1038/sj.emboj.7600488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Parker JS, Roe SM, Barford D. Structural insights into mRNA recognition from a PIWI domain-siRNA guide complex. Nature. 2005;434:663–6. doi: 10.1038/nature03462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Dominguez C, Allain FH. NMR structure of the three quasi RNA recognition motifs (qRRMs) of human hnRNP F and interaction studies with Bcl-x G-tract RNA: a novel mode of RNA recognition. Nucleic Acids Res. 2006;34:3634–45. doi: 10.1093/nar/gkl488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Swanson MS, Dreyfuss G. Classification and purification of proteins of heterogeneous nuclear ribonucleoprotein particles by RNA-binding specificities. Mol Cell Biol. 1988;8:2237–41. doi: 10.1128/mcb.8.5.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.McCullough AJ, Berget SM. G triplets located throughout a class of small vertebrate introns enforce intron borders and regulate splice site selection. Mol Cell Biol. 1997;17:4562–71. doi: 10.1128/mcb.17.8.4562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Garneau D, Revil T, Fisette JF, Chabot B. Heterogeneous nuclear ribonucleoprotein F/H proteins modulate the alternative splicing of the apoptotic mediator Bcl-x. J Biol Chem. 2005;280:22641–50. doi: 10.1074/jbc.M501070200. [DOI] [PubMed] [Google Scholar]
- 83.Jacks A, et al. Structure of the C-terminal domain of human La protein reveals a novel RNA recognition motif coupled to a helical nuclear retention element. Structure. 2003;11:833–43. doi: 10.1016/s0969-2126(03)00121-7. [DOI] [PubMed] [Google Scholar]
- 84.Wang X, McLachlan J, Zamore PD, Hall TM. Modular recognition of RNA by a human pumilio-homology domain. Cell. 2002;110:501–12. doi: 10.1016/s0092-8674(02)00873-5. [DOI] [PubMed] [Google Scholar]
- 85.Cheong CG, Hall TM. Engineering RNA sequence specificity of Pumilio repeats. Proc Natl Acad Sci U S A. 2006;103:13635–9. doi: 10.1073/pnas.0606294103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Worbs M, Bourenkov GP, Bartunik HD, Huber R, Wahl MC. An extended RNA binding surface through arrayed S1 and KH domains in transcription factor NusA. Mol Cell. 2001;7:1177–89. doi: 10.1016/s1097-2765(01)00262-3. [DOI] [PubMed] [Google Scholar]
- 87.Shamoo Y, Abdul-Manan N, Williams KR. Multiple RNA binding domains (RBDs) just don’t add up. Nucleic Acids Res. 1995;23:725–8. doi: 10.1093/nar/23.5.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Shamoo Y, et al. Both RNA-binding domains in heterogenous nuclear ribonucleoprotein A1 contribute toward single-stranded-RNA binding. Biochemistry. 1994;33:8272–81. doi: 10.1021/bi00193a014. [DOI] [PubMed] [Google Scholar]
- 89.Finger LD, Johansson C, Rinaldi B, Bouvet P, Feigon J. Contributions of the RNA-binding and linker domains and RNA structure to the specificity and affinity of the nucleolin RBD12/NRE interaction. Biochemistry. 2004;43:6937–47. doi: 10.1021/bi049904d. [DOI] [PubMed] [Google Scholar]
- 90.Lakatos L, Szittya G, Silhavy D, Burgyan J. Molecular mechanism of RNA silencing suppression mediated by p19 protein of tombusviruses. Embo J. 2004;23:876–84. doi: 10.1038/sj.emboj.7600096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Dunoyer P, Lecellier CH, Parizotto EA, Himber C, Voinnet O. Probing the microRNA and small interfering RNA pathways with virus-encoded suppressors of RNA silencing. Plant Cell. 2004;16:1235–50. doi: 10.1105/tpc.020719. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 92.Vargason JM, Szittya G, Burgyan J, Tanaka Hall TM. Size selective recognition of siRNA by an RNA silencing suppressor. Cell. 2003;115:799–811. doi: 10.1016/s0092-8674(03)00984-x. [DOI] [PubMed] [Google Scholar]
- 93.Ye K, Malinina L, Patel DJ. Recognition of small interfering RNA by a viral suppressor of RNA silencing. Nature. 2003;426:874–8. doi: 10.1038/nature02213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lingel A, Simon B, Izaurralde E, Sattler M. The structure of the flock house virus B2 protein, a viral suppressor of RNA interference, shows a novel mode of double-stranded RNA recognition. EMBO Rep. 2005;6:1149–55. doi: 10.1038/sj.embor.7400583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Chao JA, et al. Dual modes of RNA-silencing suppression by Flock House virus protein B2. Nat Struct Mol Biol. 2005;12:952–7. doi: 10.1038/nsmb1005. [DOI] [PubMed] [Google Scholar]
- 96.Ramos A, et al. Role of dimerization in KH/RNA complexes: the example of Nova KH3. Biochemistry. 2002;41:4193–201. doi: 10.1021/bi011994o. [DOI] [PubMed] [Google Scholar]
- 97.Calero G, et al. Structural basis of m7GpppG binding to the nuclear cap-binding protein complex. Nat Struct Biol. 2002;9:912–7. doi: 10.1038/nsb874. [DOI] [PubMed] [Google Scholar]
- 98.Buttner K, Wenig K, Hopfner KP. Structural framework for the mechanism of archaeal exosomes in RNA processing. Mol Cell. 2005;20:461–71. doi: 10.1016/j.molcel.2005.10.018. [DOI] [PubMed] [Google Scholar]
- 99.Liu Q, Greimann JC, Lima CD. Reconstitution, activities, and structure of the eukaryotic RNA exosome. Cell. 2006;127:1223–37. doi: 10.1016/j.cell.2006.10.037. [DOI] [PubMed] [Google Scholar]
- 100.Kielkopf CL, Lucke S, Green MR. U2AF homology motifs: protein recognition in the RRM world. Genes Dev. 2004;18:1513–26. doi: 10.1101/gad.1206204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Andersen CB, et al. Structure of the exon junction core complex with a trapped DEAD-box ATPase bound to RNA. Science. 2006;313:1968–72. doi: 10.1126/science.1131981. [DOI] [PubMed] [Google Scholar]
- 102.Stroupe ME, Tange TO, Thomas DR, Moore MJ, Grigorieff N. The three-dimensional arcitecture of the EJC core. J Mol Biol. 2006;360:743–9. doi: 10.1016/j.jmb.2006.05.049. [DOI] [PubMed] [Google Scholar]
- 103.Irion U, Adams J, Chang CW, St Johnston D. Miranda couples oskar mRNA/Staufen complexes to the bicoid mRNA localization pathway. Dev Biol. 2006;297:522–33. doi: 10.1016/j.ydbio.2006.05.029. [DOI] [PubMed] [Google Scholar]
- 104.Collins RE, Cheng X. Structural domains in RNAi. FEBS Lett. 2005;579:5841–9. doi: 10.1016/j.febslet.2005.07.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Li L, Ye K. Crystal structure of an H/ACA box ribonucleoprotein particle. Nature. 2006;443:302–7. doi: 10.1038/nature05151. [DOI] [PubMed] [Google Scholar]
- 106.Reichow SL, Hamma T, Ferre-D’Amare AR, Varani G. The structure and function of small nucleolar ribonucleoproteins. Nucleic Acids Res. 2007 doi: 10.1093/nar/gkl1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Williams BR. PKR; a sentinel kinase for cellular stress. Oncogene. 1999;18:6112–20. doi: 10.1038/sj.onc.1203127. [DOI] [PubMed] [Google Scholar]
- 108.Bass BL. RNA editing by adenosine deaminases that act on RNA. Annu Rev Biochem. 2002;71:817–46. doi: 10.1146/annurev.biochem.71.110601.135501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Nanduri S, Rahman F, Williams BR, Qin J. A dynamically tuned double-stranded RNA binding mechanism for the activation of antiviral kinase PKR. Embo J. 2000;19:5567–74. doi: 10.1093/emboj/19.20.5567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Macbeth MR, Lingam AT, Bass BL. Evidence for auto-inhibition by the N terminus of hADAR2 and activation by dsRNA binding. Rna. 2004;10:1563–71. doi: 10.1261/rna.7920904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Gelev V, et al. Mapping of the Auto-inhibitory Interactions of Protein Kinase R by Nuclear Magnetic Resonance. J Mol Biol. 2006 doi: 10.1016/j.jmb.2006.08.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Li S, et al. Molecular basis for PKR activation by PACT or dsRNA. Proc Natl Acad Sci U S A. 2006;103:10005–10. doi: 10.1073/pnas.0602317103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Bevilacqua PC, Cech TR. Minor-groove recognition of double-stranded RNA by the double-stranded RNA-binding domain from the RNA-activated protein kinase PKR. Biochemistry. 1996;35:9983–94. doi: 10.1021/bi9607259. [DOI] [PubMed] [Google Scholar]
- 114.Kim I, Liu CW, Puglisi JD. Specific recognition of HIV TAR RNA by the dsRNA binding domains (dsRBD1-dsRBD2) of PKR. J Mol Biol. 2006;358:430–42. doi: 10.1016/j.jmb.2006.01.099. [DOI] [PubMed] [Google Scholar]
- 115.Carpick BW, et al. Characterization of the solution complex between the interferon-induced, double-stranded RNA-activated protein kinase and HIV-I trans-activating region RNA. J Biol Chem. 1997;272:9510–6. doi: 10.1074/jbc.272.14.9510. [DOI] [PubMed] [Google Scholar]
- 116.Romano PR, et al. Autophosphorylation in the activation loop is required for full kinase activity in vivo of human and yeast eukaryotic initiation factor 2alpha kinases PKR and GCN2. Mol Cell Biol. 1998;18:2282–97. doi: 10.1128/mcb.18.4.2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Zhang F, et al. Binding of double-stranded RNA to protein kinase PKR is required for dimerization and promotes critical autophosphorylation events in the activation loop. J Biol Chem. 2001;276:24946–58. doi: 10.1074/jbc.M102108200. [DOI] [PubMed] [Google Scholar]
- 118.Frazao C, et al. Unravelling the dynamics of RNA degradation by ribonuclease II and its RNA-bound complex. Nature. 2006;443:110–4. doi: 10.1038/nature05080. [DOI] [PubMed] [Google Scholar]
- 119.Antson AA, et al. Structure of the trp RNA-binding attenuation protein, TRAP, bound to RNA. Nature. 1999;401:235–42. doi: 10.1038/45730. [DOI] [PubMed] [Google Scholar]
- 120.Oberstrass FC, et al. Shape-specific recognition in the structure of the Vts1p SAM domain with RNA. Nat Struct Mol Biol. 2006;13:160–7. doi: 10.1038/nsmb1038. [DOI] [PubMed] [Google Scholar]