Abstract
Telomerase is an RNA–protein complex that extends the 3′ ends of linear chromosomes, using a unique telomerase reverse transcriptase (TERT) and template in the telomerase RNA (TR), thereby helping to maintain genome integrity. TR assembles with TERT and species-specific proteins, and telomerase function in vivo requires interaction with telomere-associated proteins. Over the past two decades, structures of domains of TR and TERT as well as other telomerase- and telomere-interacting proteins have provided insights into telomerase function. A recently reported 9-Å cryo–electron microscopy map of the Tetrahymena telomerase holoenzyme has provided a framework for understanding how TR, TERT, and other proteins from ciliate as well as vertebrate telomerase fit and function together as well as unexpected insight into telomerase interaction at telomeres. Here we review progress in understanding the structural basis of human and Tetrahymena telomerase activity, assembly, and interactions.
Keywords: telomerase RNA, electron microscopy, telomerase reverse transcriptase, CST, replication protein A, H/ACA RNP
INTRODUCTION AND OVERVIEW
Telomeres and telomerase are the nucleoprotein complexes that together protect the ends of linear chromosomes and provide a solution to the end replication problem (10, 11). First detected ~30 years ago in the ciliated protozoan Tetrahymena thermophila as a terminal transferase activity (52, 53), telomerase was subsequently found to extend the 3′ ends of linear chromosomes by reverse transcription of telomeric repeats (TTAGGG in vertebrates and TTGGGG in Tetrahymena) through the use of an integral RNA template (10) (Figure 1a). Since its discovery, telomerase has emerged as a major player in tumorigenesis, stem cell renewal, and cellular aging. Shortened telomeres can lead to cellular senescence, and a majority of cancers require upregulated telomerase activity to maintain immortal cell replication (4, 5, 9, 44, 100, 146). Additionally, telomerase insufficiency or dysregulation due to mutations in telomerase proteins, telomerase RNA, and telomere-associated proteins are linked to a wide variety of inherited diseases (4, 42, 68, 112, 137, 139, 146, 160). The catalytic core of telomerase comprises an integral telomerase RNA [TR; also called hTR or TERC (human), TER (ciliate), and TLC1 (yeast)], identified in 1989 (54), and the telomerase reverse transcriptase (TERT), identified in 1997 (92). Since then, a host of additional proteins that aid in assembly, recruitment, regulation, activity, and nucleic acid handling have been identified (38, 112, 140, 150).
Figure 1.
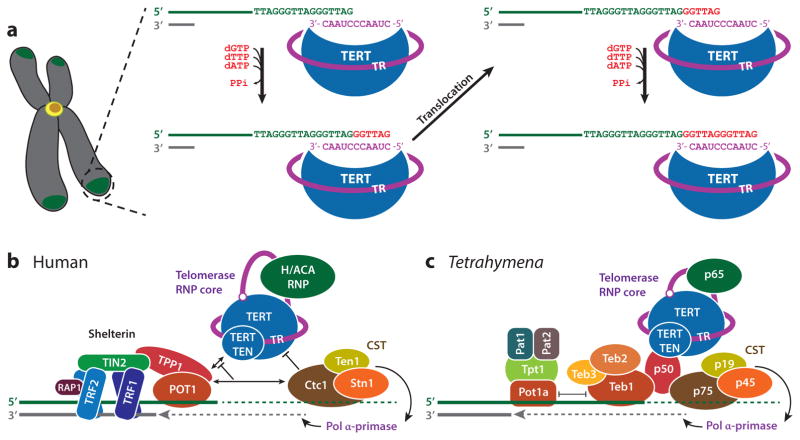
Schematics of telomere extension by telomerase and of human and Tetrahymena telomerase and telomerase-associated proteins at the telomeres. (a) Telomerase uses its integral RNA template for the reverse transcription of telomeric repeats onto the 3′ ends of chromosomes. The 3′ end of the DNA aligns with the 3′ end of the template in register to allow synthesis of the telomere repeat. After synthesis of a repeat (GGTTAG in humans), the telomerase template must shift register (translocate) by one repeat before nucleotide addition can reinitiate. (b) The human telomerase RNP core (TERT, TR, H/ACA scaRNP proteins) is recruited to telomeres by the shelterin complex. CST interaction with TPP1–POT1 inhibits telomerase activity. (c) Tetrahymena telomerase RNP core (TERT, TR, p65) is constitutively assembled with the p50, TEB, and CST accessory proteins. Abbreviations: RNP, ribonucleoprotein; scaRNP, small Cajal body ribonucleoprotein; TERT, telomerase reverse transcriptase; TR, telomerase RNA.
The protein components beyond TERT that constitute the telomerase holoenzyme have historically been difficult to define. Many associate with the telomere and catalytic core transiently as a function of cell cycle and lack obvious homology among different organisms (89, 91, 140). In addition, TRs are divergent between species in sequence and size, ranging from 147 to 205 nucleotides (nt) in ciliates, 312 to 559 nt in vertebrates, and 928 to >2,425 nt in yeasts (121, 122), and interact with apparently species-specific proteins. However, biochemical, genetic, and recent structural studies of TR, TERT, and other telomere- and telomerase-associated proteins have enhanced our understanding of their functions and revealed that these components often share commonalities of structure, function, and evolutionary themes (91, 112, 122).
These commonalities were highlighted in the recent 9-Å cryo–electron microscopy (EM) structure of Tetrahymena telomerase; nearly 30 years after the discovery, there is now a near complete model of the Tetrahymena telomerase holoenzyme (69). An unexpected finding was that components of the constitutively assembled Tetrahymena telomerase holoenzyme share homologies with human, yeast, and plant protein complexes that interact with telomerase at telomeres. This study has allowed three decades of data on telomerase function and interaction at telomeres from diverse organisms to be placed in a structural context (41, 69). Here we review structural advances in the field for ciliate (primarily Tetrahymena) and vertebrate (primarily human) telomeres with respect to TERT and TR domains, other proteins that bind to TR, and the proteins involved in G-strand single-stranded telomere repeat DNA (sstDNA) handling.
Overview of Human and Tetrahymena Telomerase and Interaction at Telomeres
The minimal components for telomerase catalytic activity in vitro are TERT and TR. Early studies established the basic mechanism for telomerase activity whereby TERT processively synthesizes multiple repeats of the G-strand telomeric sequence using the template, contained within the larger TR and comprising ~1.5–1.8 repeats of a sequence complementary to the telomeric repeat (53, 122). The 3′ end of the template aligns and base pairs with the 3′ end of the G-strand overhang, leaving template bases complementary to a single telomere repeat unpaired, and nucleotides are processively added until the 5′ end of the template is reached. Upon completion of one telomere repeat, the template has to realign with the newly formed end of the telomere, a process known as translocation, so that subsequent telomere repeats can be added (Figure 1a). The complete steps in the enzymatic cycle have been the subject of intense investigations that are beyond the scope of this review (14, 119, 175), but many details especially for the translocation step remain to be defined. At some point, the synthesis of the G-strand stops, and DNA-polymerase α-primase is recruited to synthesize the complementary C-strand (Figure 1b,c).
Telomerase contains multiple proteins in addition to TERT that bind directly or indirectly to the single TR. Here we define the telomerase ribonucleoprotein (RNP) core as TR, TERT, and additional proteins that bind directly to the TR and stay associated at least temporarily after assembly (Figures 1 and 2). Depending on the organism, the telomerase ribonucleoprotein (RNP) core can be constitutively associated or transiently associated via cell cycle regulation with other accessory proteins. In humans, the telomerase core RNP includes, in addition to TERT, the H/ACA small Cajal body RNP (scaRNP) complex proteins: dyskerin, NOP10, NHP2, and likely GAR1 [although it contacts dyskerin and not TR (84, 130)], as well as TCAB1 (166) [also called WDR79 (161)]—the gene product of WRAP53 (97), which directs the telomerase RNP to Cajal bodies (Figure 2a) (39, 140). Human telomeric DNA is protected by the six-component shelterin protein complex composed of the following: RAP1; the double-stranded telomeric DNA-binding TRF1 and TRF2; the sstDNA-binding POT1; and TIN2 and TPP1, which serve as mediators for shelterin assembly and telomerase recruitment (Figure 1b) (34, 60, 150, 169, 180, 189). Telomerase is recruited to telomeres primarily by a direct TPP1 interaction with TERT (111, 145, 189), and telomerase activity requires a switch in TPP1–POT1 function from telomere end protection to telomerase processivity factor (144, 169). Association of human telomerase to shelterin is mutually exclusive with and inhibited by a TPP1–POT1 interaction with the Ctc1–Stn1–Ten (CST) complex (24). In humans, the sstDNA-binding CST inhibits telomerase activity and recruits the DNA polymerase α-primase complex (Pol α-primase) for C-strand synthesis (Figure 1b) (22, 24). CST is related to the trimeric replication protein A (RPA) complex, the major single-stranded DNA–binding protein that plays numerous roles in DNA replication and repair (123, 153).
Figure 2.
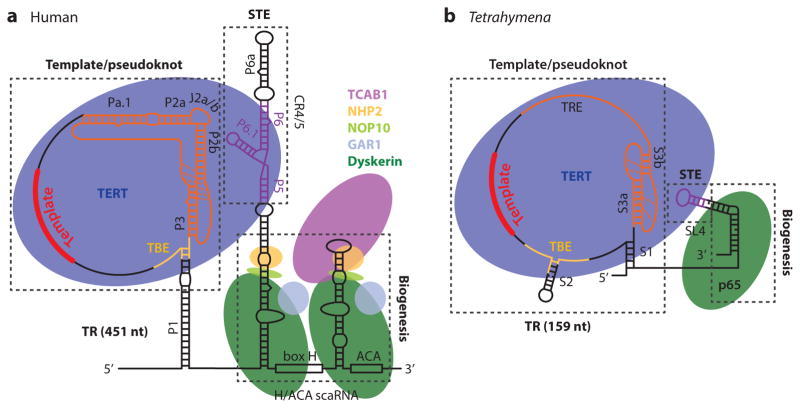
Schematics of human and Tetrahymena telomerase RNP cores illustrating TR secondary structure and known protein components. TR contains three major structural and functional domains: template/pseudoknot (t/PK), stem terminus element (STE), and biogenesis. (a) Human telomerase core RNP includes TERT, TR, and the H/ACA proteins. Human TERT (blue) interacts with the t/PK and the STE [conserved regions 4 and 5 (CR4/5)] (violet). In the t/PK, the template, pseudoknot, and template boundary element (TBE) are highlighted in red, orange, and yellow, respectively. Helical regions are labeled as P (20). The biogenesis domain comprises an H/ACA scaRNA motif that interacts with the H/ACA proteins dyskerin (dark green), NOP10 (light green), NHP2 (gold), GAR1 (pale blue), and TCAB1 (light purple). (b) Tetrahymena telomerase RNP core includes TR, TERT (blue), and p65 (dark green). Domains of the t/PK and STE are colored as in panel a, with the addition of the single-stranded template recognition element (TRE) in orange. The biogenesis domain at the 3′ end of TR is bound by p65; its N-terminal La module binds the TR polyU tail for biogenesis, and the C-terminal xRRM binds stem 4 around the GA bulge for assembly. Abbreviations: nt, nucleotide; RNP, ribonucleoprotein; scaRNA, small Cajal body ribonucleoprotein; TR, telomerase RNA.
In contrast to human telomerase, Tetrahymena telomerase is constitutively assembled (104, 165). TERT, TR, and the La-related protein p65 comprise the Tetrahymena telomerase RNP core (Figures 1c and 2b), as revealed by the cryo-EM structure (69). p50 links TERT to the two recently structurally characterized RPA-like sstDNA-binding complexes TEB (which comprises Teb1, Teb2, and Teb3) and CST (61, 69, 70). TEB is important for proper recruitment of telomerase to telomeres (69, 165), whereas Tetrahymena CST, comprising p75, p45, and p19 (69), is linked to C-strand synthesis (Figure 1c) (168). The G-overhang is capped by a four-protein complex—Pat1, Pat2, Tpt1, and Pot1a—and Pot1a is proposed to be mutually exclusive with TEB/telomerase binding to the G-overhang (Figure 1c) (124).
Although there are many seminal structural studies of telomere- and telomerase-associated proteins from yeast, specifically the Pot1 and CST proteins (48, 81, 108, 155), there are few structures from yeast TRs (17) and yeast telomerase RNP cores. For this reason and also space limitations, this review focuses on human and ciliate Tetrahymena telomerase. We refer the interested reader to excellent reviews on yeast and plant telomerase (28, 77, 89, 94, 98, 120, 172, 173). Advances in single molecule and fluorescence resonance energy transfer studies of telomerase are covered in this volume by Parks & Stone.
TELOMERASE RNA DOMAIN STRUCTURES
The TR secondary structure and associated proteins that comprise the human and Tetrahymena telomerase RNP core are illustrated in Figure 2. TR variability in size, sequence, and domains can be explained by its identification as a rapidly evolving long noncoding RNA, with regions of structural homology attributed to its roles in TERT interaction and function (113, 122). Two nearly universally present structural elements that are required together for TERT binding and catalysis in vitro have been identified: a template/pseudoknot (t/PK) (also called core or pseudoknot) domain and a stem terminus element (STE) [also called activating domain and, in vertebrates, conserved regions 4 and 5 (CR4/5)] (122, 158, 187). In addition, TRs from diverse organisms contain a 3′ species-specific RNA stability/biogenesis element (Figure 2). Differences between organisms in the requirements for 3′-end processing, stabilization, and nuclear compartment localization account for much of the diversity in TR and TR binding proteins (Figure 2) (39, 122, 167).
Template/Pseudoknot Domain
The t/PK includes the template, a 5′, template-adjacent template boundary element (TBE) (21, 78, 162), and a pseudoknot connected to the 3′ end of the template by a single-stranded region, usually enclosed in a circle by a stem (Figure 2). Ciliates have the smallest and simplest H-type pseudo-knots, with ~30 nt, whereas vertebrates and yeasts have pseudoknots with additional subdomain(s) in their longer stem 1 (P2 in humans). In vertebrate telomerase, the full-length pseudoknot includes a minimal pseudoknot (P2b–P3), essentially equivalent to the complete ciliate pseudoknot, containing most of the conserved bases (Figures 2 and 3a); an adjacent helical region that includes a 5–6-nt bulge conserved in location but not sequence (P2a–J2a/b–P2b); and, at least in mammals, a helical extension (P2a.1) (Figures 2a and 3b) (121). The structure of the human telomerase minimal pseudoknot (Figure 3a) (74, 157) established that the pseudoknot is stabilized by tertiary interactions between the loops and stems and, in particular, a run of U · A · U base triples that has become known as the triple helix (127, 147). A direct correlation between pseudoknot stability and telomerase activity was demonstrated by thermodynamic analysis, through the use of nucleotide substitutions and compensatory mutations (157). Biochemical, mutational, and structural studies of yeast and medaka fish pseudoknots have revealed a conserved structure and confirmed the importance of the base triples for pseudoknot stability and telomerase function (Figure 3a) (17, 127, 147, 163, 171). A recent structure of the Tetrahymena pseudoknot revealed a more limited set of base triples, explaining in part its lower stability (Figure 3a) (18, 69).
Figure 3.
Structures of TR domains. (a) Solution structures of minimal (P2b–P3) pseudoknots from human (Homo sapiens) (PDB ID 2K95), yeast (Kluyveromyces lactis) (PDB ID 2M8K), and Tetrahymena (Tetrahymena thermophila) (PDB ID 2N6Q). Secondary structure schematic of the human P2b–P3 pseudoknot is shown on far left (18, 74). (b) Sequence and secondary structure of the human TR t/PK domain. Subdomains for which NMR structures have been determined are boxed with dashed lines: P2a–J2a.1–P2a.1 (purple–black–orange), P2a–J2a/b–P2b (PDB ID 2L3E) (orange–green–red), P2b–P3 pseudoknot (red–blue–gold). P1b (tan) is the closing helix of the TBE (187). (c) Model structure of the human t/PK. For the model, the subdomain structures were computationally combined to make the P2–P3 pseudoknot (188). The single-stranded region (gray) containing the template (black) is shown with a DNA primer (cyan) bound. (d) Sequence and secondary structures of medaka (Oryzias latipes) STE (CR4/5), NMR structure (PDB ID 2MHI), and crystal structure of the complex with medaka TERT TRBD (PDB ID 4O26). Nucleotides proposed to interact with TRBD are colored green. (e) Sequence and secondary structures of Tetrahymena STE (stem-loop 4), NMR structure (PDB ID 2FEY), and structure of the complex with Tetrahymena p65 and TERT TRBD [modeled from crystal structure of p65 xRRM-stem 4 (PDB ID 4ERD), NMR structure of loop 4 (PDB ID 2M21) (149)], and TRBD from the cryo–electron microscopy model (69). The GA bulge nucleotides involved in p65 binding are colored cyan. (f) Sequence and secondary structure of human TR biogenesis domain (H/ACA) and secondary structure and solution NMR structure of the 3′ apical stem-loop (CR7) containing the CAB and BIO boxes. Open boxes within the CR7 indicate the location of the CAB box (magenta) and BIO box (green). Nucleotides important for TCAB1 and putative NHP2 binding are highlighted in magenta and green, respectively, in the secondary structure schematic and are colored by residue type on the surface of the NMR solution structure of the CR7 hairpin (PDB ID 2QH2): A (orange), U (green), G (blue), C (red). Human TR domains are named P for paired regions and J for junction regions (20, 159). Abbreviations: CR4/5; conserved regions 4 and 5; NMR, nuclear magnetic resonance; STE, stem terminus element; TBE, template boundary element; TERT, telomerase reverse transcriptase; t/PK, template/pseudoknot; TR, telomerase RNA; TRBD, telomerase RNA binding domain.
Structures and dynamics of the additional subdomains of the pseudoknots of human and of medaka, the latter of which contains the smallest known vertebrate TR (312 nt) (178), have also been studied by nuclear magnetic resonance (NMR) (73, 171, 188). The structure of the human P2a–J2a/b–P2b subdomain [P2ab] showed that the 5-nt J2a/b bulge induces a large (89°) and somewhat flexible bend between P2a and P2b (Figure 3b,c) that defines the overall topology of the full-length pseudoknot (188). In mammals, P2a.1 is connected to P2a by an internal loop that forms a series of mismatched base pairs, so P2a and P2a.1 [P2a1a] essentially form a single helix as shown for human TR. In medaka, this region was predicted to be single stranded but appears to form a small hairpin stacked on P2a, extending the similarity between mammalian and nonmammalian vertebrate TR full-length pseudoknots (171). The subdomain structures of the full-length pseudoknot were computationally combined, and the single-stranded template and adjacent nucleotides were included to make models of the t/PK for human TR (Figure 3c) and medaka TR (171, 188). These models approximate the structure of the t/PK when bound to TERT; t/PK may form alternate structures when not bound to TERT (30, 47, 59, 114).
The t/PK also contains the TBE, a short helix flanked by single-stranded DNA. In human TR, this helix also closes the t/PK into a circle, whereas in Tetrahymena, it is a hairpin located a few nucleotides 5′ of the template (Figure 2) (21, 67, 78, 133, 187). The TBE binds with high affinity to the TERT TRBD (telomerase RNA binding domain) (117), providing an anchor point that prevents TR nucleotides beyond the 5′ end of the template from being pulled into the active site (discussed below) (67, 69).
Stem Terminus Element
In vertebrate TR, the STE is the CR4/5, which is a helical region extending from the 5′ hairpin of the H/ACA scaRNA domain (Figures 2 and 3d). This region of TR forms a three-way junction (P5, P6, and P6.1 emanating from an internal loop); the P6.1 hairpin has a 3-nt loop and is highly conserved in vertebrates (135). The binding site was mapped to TRBD by cross-linking (Figure 3d) (12). The structure of medaka STE (CR4/5) has been solved both free from (73) and bound to TERT TRBD (Figure 3d) (63). Binding to the TRBD induces a large conformational change in the position of P6.1 relative to P6 and a rearrangement of base interactions in the three-way junction. P6.1 is almost parallel to P6 in the free RNA, but in complex with TRBD, it is approximately perpendicular to and on the opposite side of P6. TRBD helices between the QFP and T motifs insert deep into the cleft between the two stems, forming an extensive interface (Figure 3d). Although the nucleotides of the P6.1 hairpin loop are not visible in the medaka TRBD–CR4/5 crystal structure, the P6.1 apical loop is proposed to insert at the interface between the TRBD and the C-terminal extension (CTE) (12, 63, 69). Two potential pseudouridine (ψ) modification sites were identified in human P6.1 by N-cyclohexyl-N′-β-(4-methylmorpholinium) ethylcarbodiimide p-tosylate probing at the highly conserved U307 and the loop closing U306, and structural studies indicated that these ψ affected the loop conformation (72). More recent genome-wide mapping by ψ-seq has confirmed the U307 modification site (143), and it will be interesting to see if it is important in the P6.1–CTE interaction.
In ciliates, the STE is simply a stem-loop, equivalent to P6.1. The structures of Tetrahymena stem-loop 4 (25), loop 4 (134), and stem 4 bound to the C-terminal xRRM of p65 have been solved (Figure 3e) (149). Binding of p65 xRRM at a conserved GA bulge in stem-loop 4 induces a large conformational change that is important for hierarchal assembly of p65–TR with TERT (Figure 3e) (116, 149, 152). Comparing the medaka CR4/5–TRBD structure with the pseudoatomic cryo-EM model of stem-loop 4–TRBD explains why the interaction of loop 4 with the TRBD is much weaker than CR4/5 with the TRBD, because the single stem would not have the additional contacts afforded by P6 in vertebrates, and explains why p65 is required to help position loop 4 interact with TERT.
Telomerase RNA Biogenesis Element and Protein Interactions
The biogenesis element in vertebrate telomerase is an H/ACA scaRNA domain and is located at the 3′ end of the mature TR (Figure 2). Like other eukaryotic H/ACA RNAs, it contains two hairpins with large internal loops, a 5′ and 3′ hairpin followed, respectively, by an H box and ACA box (Figure 3f). These hairpins each bind a set of H/ACA RNP proteins (dyskerin, NOP10, NHP2, GAR1) (Figure 2a) (37). However, unlike classical H/ACA scaRNAS, the internal loops do not serve as guide sequences for site-specific pseudouridylation of snRNAs; rather, the TR H/ACA RNP is important for TR stability, localization to Cajal bodies, and possibly assembly (31, 39, 167). The 3′ hairpin apical loop contains the CAB box sequence (132) that binds TCAB1 (161, 166), which targets telomerase to Cajal bodies (159, 166). Structures of single hairpin archaeal H/ACA proteins, complexes, and H/ACA RNPs with bound substrates (56, 87, 99) and yeast H/ACA dyskerin/Cbf5-NOP10-L7Ae-GAR1 complexes (35, 85, 86) as well as NHP2 alone (76) have been reported and provide insight into how these proteins might assemble with TR (reviewed in 57, 75, 183). Archaeal H/ACA RNPs have the protein L7Ae, which specifically binds a kink-turn, in place of NHP2.
The 5′ hairpin of the vertebrate TR H/ACA domain is extended by the STE, the CR4/5 element that contains the three-way junction discussed above. The 3′ hairpin apical loop contains both the CAB box on the 5′ side and a region referred to as the BIO box on the 3′ side (Figure 3f) (38). Structural and functional studies of wild-type and mutant hairpins established the sequence and structural elements important for targeting to Cajal bodies and RNP biogenesis, respectively (38, 159). The upper stem and conserved bulge U in the BIO box correspond to the predicted binding site for NHP2 (Figure 3f) (76, 159). It is interesting to note that an equivalent site with a bulge U does not appear to be present in the 5′ hairpin. The BIO box has been shown to contribute to the enhanced assembly of the dyskerin–NOP10–NHP2–NAF1 complex with the H/ACA domain that is required for biological accumulation of TR (38). Association of the dyskerin complex has also been shown to protect the 3′ end from exonucleolytic digestion (39, 148). Thus, the 3′ H/ACA RNP contributes to biogenesis, stabilization, and localization. The H/ACA RNP may also contribute to positioning of the STE, but this remains to be determined.
In Tetrahymena TR, stem 4 and its 3′ UUUU single-stranded end, in complex with p65, function as the biogenesis element (Figure 2b). In contrast to the vertebrate TR transcribed by RNAP II, ciliate TR is transcribed by RNAP III (39, 54). The polyU tail at the 3′ end of RNAP III transcripts are cotranscriptionally bound and protected by the La module (composed of La and RRM domains) of La protein. The N-terminal La module of p65, a LARP7 (La-related protein group 7), replaces that of La on the 3′ end of stem-loop 4, while its C-terminal xRRM domain binds and bends stem 4 (Figure 3e) (149). Thus, p65 plays a dual role in TR stability and RNP assembly.
TELOMERASE REVERSE TRANSCRIPTASE DOMAIN STRUCTURES
The Telomerase Reverse Transcriptase Ring
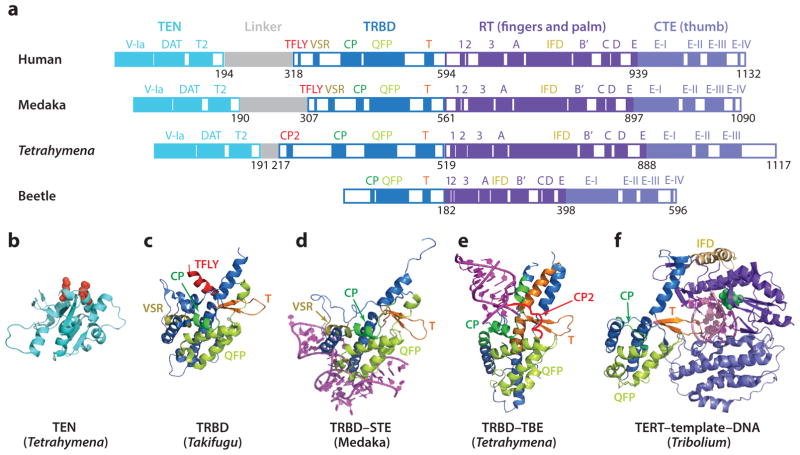
TERT contains four evolutionarily conserved functional domains: TERT-essential N-terminal (TEN) domain, TRBD, reverse transcriptase (RT) domain, and CTE (Figure 4a) (6, 71, 102, 121, 122, 177). Crystal structures of Tribolium castaneum TERT without and with an RNA DNA hybrid hairpin that mimics the template–DNA complex revealed that TRBD, RT, and CTE form a ring (Figure 4f) (49, 106). Because Tribolium TERT lacks the TEN domain and the TR has not yet been found, questions have been raised about whether it might be another type of RT (83); regardless, its ring shape is clearly consistent with the EM maps for both human and Tetrahymena telomerase (69, 70, 105, 138). The RT includes the enzyme active site where the TR template is located, and the CTE promotes telomerase processivity; these two conserved domains correspond to the palm and fingers and the thumb domains, respectively, found in other polymerases (6, 177). The RT domain has two telomerase-specific regions—motif 3, which facilitates the realignment between DNA and RNA (179), and the insertions of fingers domain (IFD), which mediates telomerase processivity and facilitates telomerase recruitment to telomeres (26, 27, 49, 96). The Tribolium IFD is much smaller than that in human and Tetrahymena, and there is no good homology model in those organisms. The TRBD has specific binding sites for the TBE and the STE (1, 12, 58, 67, 79). Tribolium TRBD is also much smaller than that of Tetrahymena and vertebrates (Figure 4a).
Figure 4.
Domains and structures of TERT. (a) Domains and conserved motifs of human, medaka, Tetrahymena, and beetle TERTs, aligned at the RT domains. Crystal structures of (b) Tetrahymena TEN (PDB ID 2B2A) (c) Takifugu TRBD (PDB ID 4LMO), (d) medaka TRBD–STE complex (PDB ID 4O26), (e) Tetrahymena TRBD–TBE complex (PDB ID 5C9H), and (f) flour beetle TERT–template–DNA complex (PDB ID 3KYL). In panel b, residues that are proposed to be involved in interaction of the TEN domain with p50 (and TPP1 in human) are red spacefill. RNA and DNA are magenta and pink, respectively. The active site in panel e is green spacefill. Abbreviations: CTE, C-terminal extension; RT, reverse transcriptase; STE, stem terminus element; TEN, TERT-essential N-terminal; TERT, telomerase reverse transcriptase; TRBD, telomerase RNA binding domain.
Telomerase Reverse Transcriptase Domain Structures and Telomerase RNA Interactions
Two Tetrahymena and two vertebrate TRBD structures have been determined (Figure 4c–e) (58, 63, 67, 136). The recently reported crystal structure of the complete Tetrahymena TRBD with the TBE revealed the interface between TRBD CP2 (1) and the TBE (SL2 and its adjacent single strands) (Figure 4e) (67). In vertebrates, the TFLY motif (equivalent to CP2 in ciliates) (Figure 4c) within the N-terminus of the Takifugu rubripes TRBD has been shown to interact with P1 and adjacent single strands (vertebrate TBE) (58). The crystal structure of medaka TRBD with CR4/5 defined the TR interaction with this second TRBD high-affinity binding site (Figure 4d), consistent with cross-linking results (12). The TEN domain, which is connected by a linker of variable length to the TRBD, is proposed to interact with DNA–template duplex and is essential for processivity (Figure 4b) (66, 175, 184). The Tetrahymena TEN domain has a unique fold, with phylogenetically conserved residues that are important for activity and implicated in sstDNA binding (66). The location of the TEN domain relative to the TERT ring and TR was revealed in the cryo-EM model structure of Tetrahymena holoenzyme (Figure 5) (69).
Figure 5.
Cryo–electron microscopy (EM) structure of Tetrahymena telomerase and model of human TERT–t/PK. (a) Front view of the 9.4-Å cryo-EM map with the RNP core (blue), TEB (gold), CST (tan), and p50 (red). Arrows indicate location of protein C-terminus (or N-terminus for p50) and TR SL2 determined by Fab labeling and MS2 coat protein labeling, respectively, in negative stain EM (70). (b) Same color designation and labeling scheme as panel a but with pseudoatomic models of the RNP core, TEB trimer of OB folds, and CST trimer of OB folds. Additional domains of Teb1, Teb2, and p45, not visible in the cryo-EM map, are shown as crystal structures [Teb1A (PDB ID 3U4V), Teb1B (PDB ID 3U4Z), p45C (PDB ID 5DFN)], homology models (Teb2C based on PDB ID 1DPU), or ovals (70). (c) Schematic of Tetrahymena telomerase, 180° rotation from panel b, illustrating DNA bound to template and exit path. (d) Schematic illustrating movement of the template through the active site for one round of telomere repeat synthesis, and template boundary definition by the TBE. (e) Pseudoatomic model of Tetrahymena telomerase catalytic core, showing the path of TR on TERT. Inset shows schematic of the t/PK. TR elements in panels e and f are colored as in Figure 2. (f) Model of model of human telomerase catalytic core, based on the model of the human t/PK (171, 188), medaka CR4/5 with TRBD (PDB ID 4O26) (63), and the positions of homologous domains of Tetrahymena TR in the cryo-EM map. Inset shows schematic of the t/PK with TR elements colored as in the model. Abbreviations: CR4/5, conserved regions 4 and 5; CTE, C-terminal extension; OB, oligonucleotide/oligosaccharide binding; RNP, ribonucleoprotein; RT, reverse transcriptase; sstDNA, single-stranded telomere repeat DNA; STE, stem terminus element; TBE, template boundary element; TEN, TERT-essential N-terminal domain; TERT, telomerase reverse transcriptase; t/PK, template/pseudoknot; TR, telomerase RNA.
ELECTRON MICROSCOPY STRUCTURES OF TELOMERASE
Negative Stain Electron Microscopy Structures
Negative stain EM structures at ~25-Å resolution of human (138) and Tetrahymena (70) telomerase were first reported in 2013. A dimeric human telomerase was affinity purified using a DNA primer from cells that had been transiently transfected with TR and TERT (32), and mass spectrometry indicated it contained TERT, TR, dyskerin, and NOP10 (138). The three-dimensional reconstruction of human telomerase showed a dumbbell shape with TERT fit into the density at each of the knobs; the connecting rod was attributed to a duplex region of TR.
There has been conflicting data on whether human telomerase is a monomer (3, 37, 39, 176) or obligate dimer (29, 138, 142, 174). A recent study established that in vitro human TERT–TR reconstitution yields active monomers or dimers depending on the purification method, and the dimerization can be suppressed both in vitro and in vivo by removing or modifying the linker between TRBD and TEN (176). Tetrahymena and yeast telomerase have been clearly shown to be monomers (7, 16).
In the negative stain EM study of Tetrahymena telomerase, samples with affinity tags on the subunits were used to identify the location of six of the seven then-known proteins and of TR domains (Figure 5a) (70). A surprise of the negative stain EM map was the identification of p50 as a central hub that links the catalytic core, Teb1, and p75–p45–p19, and is essential for the processivity enhancement conferred by Teb1.
Cryo–Electron Microscopy Structure of Tetrahymena Telomerase
In the cryo-EM map at 9-Å resolution (69) protein α-helices, β-barrels, β-hairpins (but not β-sheets), and RNA helices can be readily identified when fit with known structures, but structures cannot be built de novo. Thus, the identification of subunit locations in the negative stain EM structure (70) and the NMR, crystal, and homology model structures of telomerase subunit domains (13, 49, 66, 69, 106, 136, 149, 169, 186) were crucial in constructing a pseudoatomic model of most of the holoenzyme (Figure 5a) (41, 69). The pseudoatomic model of the telomerase RNP core was built starting from fitting TR, TERT, and p65 domain structures discussed above into the EM map (Figure 5b) (49, 66, 106, 136, 149). Of the TEB proteins, only Teb1 had been previously discovered (104); Teb2 and Teb3 were identified by mass spectrometry after discovering that the crystal structure of the three interacting oligosaccharide/oligonucleotide binding (OB) folds from the paralogous RPA complex (13) fit into a region of the EM map that could not be accounted for by the known proteins. Similarly, crystal structures of p19 and p45C that showed structural homology to Ten1 (OB fold) and Stn1C tandem winged-helix (WH) domains suggested that the p75–p45–p19 complex is a CST, and the RPA complex also fit into a second site corresponding to the known locations of p75 and p19 (69). Despite its identification as a central hub, no homology or threading model of p50 could be identified that fit into the cryo-EM map, although it too appears to be a β-barrel, consistent with an OB fold. Of note, several protein domains are not visible in the cryo-EM map, specifically Teb1N, Teb1A, Teb1B, Teb2C, p50C, p65N, and p45C. These domains are apparently connected by flexible linkers and therefore are averaged out in the class averages because of flexible positioning. This explains why the location of p45 was not identified by Fab labeling in the negative stain EM (illustrated in Figure 5b). In addition, the entire CST complex hinges as a unit around p50, as could be seen in the class averages (69, 70). The biological significance, if any, of this motion remains to be determined. The 9-Å resolution cryo-EM structure of the Tetrahymena telomerase holoenzyme provides a structural framework for understanding telomerase catalytic core assembly and TR interactions and interaction of telomerase with telomere-associated proteins. In the sections below, we compare and contrast Tetrahymena and human telomerase catalytic core and accessory protein complexes.
THE CATALYTIC CORE OF HUMAN AND TETRAHYMENA TELOMERASE
Pseudoatomic Model of Tetrahymena Telomerase Ribonucleoprotein Core
The cryo-EM structure of Tetrahymena telomerase provided the first view of the path of TR on TERT for the apo-enzyme and indicated specific locations of the TR elements important for function (Figure 5b–e), thus providing a physical basis for interpreting and/or corroborating previous biochemical and mutational data on function. The TRBD-RT-CTE domains form the expected TERT ring structure, and the TEN domain is stacked over the CTE on the template side of the ring. The t/PK encircles the TERT ring approximately perpendicular to the plane of the ring, and loop 4 of the STE inserts between the TRBD and CTE. Of particular significance is the location of the conserved pseudoknot element on the opposite side of the TERT ring from the template, near the CTE, as there was little biochemical data on its location (Figure 5b,e) (128). The location of the pseudoknot, far from the active site, suggests a role in assembly of TR on TERT, essentially locking the t/PK circle into place (18, 69), rather than a direct role in catalysis as previously suggested (127). The TRBD has high-affinity binding sites for the TBE and STE and threads through the t/PK circle to contact them (Figure 5d,e). The location of the TBE relative to the template and TERT active site suggest that template boundary definition is determined by physical anchoring of the TBE to the TRBD, with the TR between the end of the template and the TBE maximally extended when the last DNA nucleotide is added to the template (Figure 5d) (67, 69). This model is at least partially consistent with the TR accordion model for template positioning (8). However, how the 3′ end of the template is repositioned into the active site remains unknown. The single-stranded region on the 3′ side of the template wraps around the CTE (thumb domain), threading between the CTE and TEN domain but has no apparent anchor (Figure 5b,e). The TEN domain is positioned such that the sstDNA–template duplex could butt up against it at the end of a telomere repeat synthesis and play a role in strand separation (Figure 5c,d) (2, 36). The STE of Tetrahymena, distal stem-loop 4, inserts between the TRBD and the CTE (Figure 5b–e) (69, 70). A remarkable feature of the telomerase structure is the large hole at the so-called bottom, where stem 1–stem-loop 4 forms a U shape and only loop 4 interacts with TERT (Figure 5b). Bending of stem 4 to ~105° by the C-terminal xRRM domain of p65 (Figure 3e), apparently positions loop 4 to interact with TERT. In the absence of p65 xRRM, loop 4 would point away from TERT (Figures 3e and 5).
Modeling the Vertebrate Telomerase Catalytic Core
Recently, model structures of vertebrate TERT–t/PK complexes have been proposed (Figure 5f) (171) on the basis of the positions of TR (e.g., TBE, pseudoknot) and TERT domains in the cryo-EM model structure of Tetrahymena telomerase (69, 70) and the NMR model structures of medaka and human TR t/PK (171, 188). The vertebrate pseudoknot has an extended helical region where Tetrahymena has a single-stranded region (Figure 2). The bend at the human 5-nt bulge (J2ab) (Figure 3c) allows the full-length pseudoknot to wrap around the TERT, with the end (P2a.1 helix) abutting the TEN domain (Figure 5f). NMR dynamics studies of human P2a–J2a/b–P2b showed that the 5-nt bulge allows a limited range of motion of the two helices relative to each other, which may allow further bending at the J2a/b bulge in the complex and/or may allow the RNA to flex during the telomere repeat cycle as the template moves through the active site (187, 188). The region of mismatched base pairs between P2a.1 and P2a (Figure 2b,c) may provide a second hinge site for TR bending. For this review, we also modeled the human STE (CR4/5) (Figure 2), which is positioned on the TRBD based on the medaka TRBD–CR4/5 structure (Figure 5f). P6.1 is predicted to insert to the TRBD–CTE interface (12, 63), similar to Tetrahymena loop 4 (69, 70). However, to avoid steric clash with the CTE, the position of the P6.1 loop would have to be changed. The vertebrate STE may be positioned for interaction with TERT by a bend induced in the RNA by binding of the H/ACA proteins (Figure 2a) and/or by the conformational change in the CR4/5 three-way junction upon interaction with the TRBD (Figure 3b).
PROTEINS INVOLVED IN sstDNA HANDLING AND TELOMERASE RECRUITMENT TO TELOMERES
Telomeric DNA ends in a 3′ single-stranded G-strand overhang that requires protection from exonuclease activity as well as DNA repair enzymes (118). Telomere end–protection proteins also play a dual role in the regulation of telomerase recruitment and activation. sstDNA-binding proteins can sequester the telomeric 3′ end, rendering the DNA inaccessible to telomerase and other DNA acting enzymes (101). However, some of these same proteins are required to recruit telomerase to telomeres and stimulate activity and processivity (112).
TPP1 and POT1
In humans, TPP1 is the major facilitator of telomerase activation and recruitment (112, 129, 180). It binds directly with the TERT TEN domain (111, 141, 144, 185, 189), TIN2, and the sstDNA-binding POT1, simultaneously bridging telomerase to shelterin components and the 3′ single-stranded G-overhang (Figure 1b) (34, 93, 181, 185). TPP1 can also interact with the sstDNA-binding complex CST, which occludes telomerase recruitment and access to telomeres (24). Although the structural basis for how all of these factors cooperate to regulate telomerase activity is unknown, structures of individual domains and apparently homologous complexes have provided important insights.
Among the first studied telomere end–binding proteins were the Sterkiella nova (formerly Oxytricha nova) telomere end–binding proteins, TEBPα and TEBPβ. Seminal structures of the TEBPα–TEBPβ–sstDNA complex revealed that TEBPα consists of three OB folds, with the two N-terminal OB folds important for sstDNA binding, whereas the N-terminus of TEBPβ consists of a single OB fold with a C-terminal extension important for complex formation (Figure 6a,b,f) (62). Structures of POT1AB OB folds bound to sstDNA and of the free TPP1 N-terminal OB fold revealed structural homologies to TEBPα and TEBPβ, respectively (Figure 6a–c) (82, 169). The TPP1 N-terminal OB fold structurally aligns with the N-terminal domain of TEBPβ (Figure 6a) whereas the two N-terminal POT1 OB folds bound to sstDNA are similar to the corresponding TEBPα N-terminal OB folds (Figure 6b,c,f). Although not apparent in the crystal structure of TPP1, a C-terminal region of the OB fold in both TPP1 and TEBPβ is important for interaction with the C-terminal OB fold of POT1 (Figure 6a,f) (62, 169, 180). These striking similarities between the domain organizations suggest that POT1–TPP1 may structurally and functionally interact with each other in a similar manner to TEBPα–TEBPβ. However, consistent with data that TEBPα–TEBPβ inhibits in vitro telomerase activity (43), the TEBPα–TEBPβ–sstDNA complex structure is in a conformation that occludes the 3′ sstDNA end (62). This is in contrast to observations that TPP1–POT1 activates telomerase and increases processivity in vitro (169). It is possible that when bound to TERT, a distinct conformation of TPP1–POT1 exists that allows access to the 3′ sstDNA end.
Figure 6.
Domains, interactions, and structures of telomere G-strand binding and telomerase recruitment proteins. (a) Superposition of Sterkiella (Sterkiella nova) TEBPβN-terminal OB (PDB ID 1OTC) with human TPP1 N-terminal OB (PDB ID 2I46). (b) Side-by-side comparison of the two N-terminal OB folds of Sterkiella TEBPα and human POT1, both bound to sstDNA. (c) Structural alignment of the most N-terminal sstDNA-binding OB fold from Sterkiella TEBPα (PDB ID 1OTC), human POT1 (PDB ID 1XJV), and Tetrahymena Teb1 (PDB ID 3U4V). (d) Schematic representation of interaction network between TERT, p50, and TEB proteins in Tetrahymena telomerase, based on the cryo-EM structure (69). (e) Region of 9-Å cryo-EM map with the p50 density and interactions with the TEN domain (PDB ID 2B2A), TEB [modeled from RPA heterotrimer (PDB ID 1L1O) with Teb1C (PDB ID 3U50) substituted for RPA70C], and the IFD. (f) Schematic of domains and interactions of Sterkiella TEBPα–TEBPβ, human POT1–TPP1, and Tetrahymena Teb1–p50. Abbreviations: EM, electron microscopy; IFD, insertions of fingers domain; OB, oligonucleotide/oligosaccharide binding; RPA, replication protein A; sstDNA, single-stranded telomere repeat DNA; TEN, TERT-essential N-terminal; TERT, telomerase reverse transcriptase.
Recruitment of telomerase to telomeres is primarily facilitated by the TPP1 OB fold, which binds to TERT at the TEN domain (144, 185, 189). A region of Glu- and Leu-rich residues have been identified as the so-called TEL patch on TPP1 and have been shown to interact directly with the TEN domain, and corresponding residues on the TEN domain have been shown to participate in this interaction (Figure 4b) (111, 141, 185). Separation of function experiments support the notion that in addition to facilitating interaction with TERT, these TEL patch residues can exert a biochemical effect on telomerase that aids translocation independently of preventing primer dissociation (33, 111). A mechanistic explanation for these observations will likely require high-resolution structural understanding of translocation as well as of the TPP1–TEN interaction.
Tetrahymena Telomerase Recruitment and Telomere End Protection
In Tetrahymena, recruitment of telomerase to telomeres requires p50 and the sstDNA-binding protein Teb1 (165), which is part of the RPA-related TEB complex (69). Crystal structures of Teb1A and Teb1B showed that these domains and their mode of sstDNA binding are homologous to those of POT1A and POT1B (Figure 6c) (186). Teb1C is structurally homologous to RPA70C, and Teb2 and Teb3 are RPA32 and RPA14 homologs, respectively. Intriguingly, a recent study establishes that Teb2 and Teb3 are actually shared subunits with Tetrahymena RPA (164). The domain topology of TEB is the same as that of RPA (Figure 7c). Of the three TEB proteins, only Teb1 has been shown to bind directly to sstDNA (186), whereas Teb2 and Teb3 enhance telomerase activity likely by stabilizing association of Teb1 to telomerase (69, 164). Similar to POT1–TPP1, Teb1 can stimulate processivity of the catalytic core but only in the presence of p50 (61, 70). The recent cryo-EM structure of Tetrahymena telomerase provided the structural basis for this observation. p50 interacts directly with the TERT TEN domain and Teb1C, mediating the association of TEB with TERT (Figure 6d,e) (69). Significantly, p50 interacts at the region on the TEN domain that corresponds with the human TEN–TPP1 interface (141). On the basis of this, p50–TEB is proposed to be functionally equivalent to TPP1–POT1, enhancing processivity and bridging telomerase to telomeres (69). Interestingly and consistent with recent studies implicating a TPP1–IFD interaction (26, 27), in the cryo-EM model the predicted IFD region of Tetrahymena TERT is next to p50 (69). Although there is no high-resolution structure of p50, the EM density shows that the region identified as p50N is a β-barrel similar in shape to the TPP1N OB fold (Figure 6e) (69). If p50 is indeed a TPP1 homolog, then the cryo-EM structure provides a first glimpse of a TERT–TPP1–POT1 interaction (Figure 6e).
Figure 7.
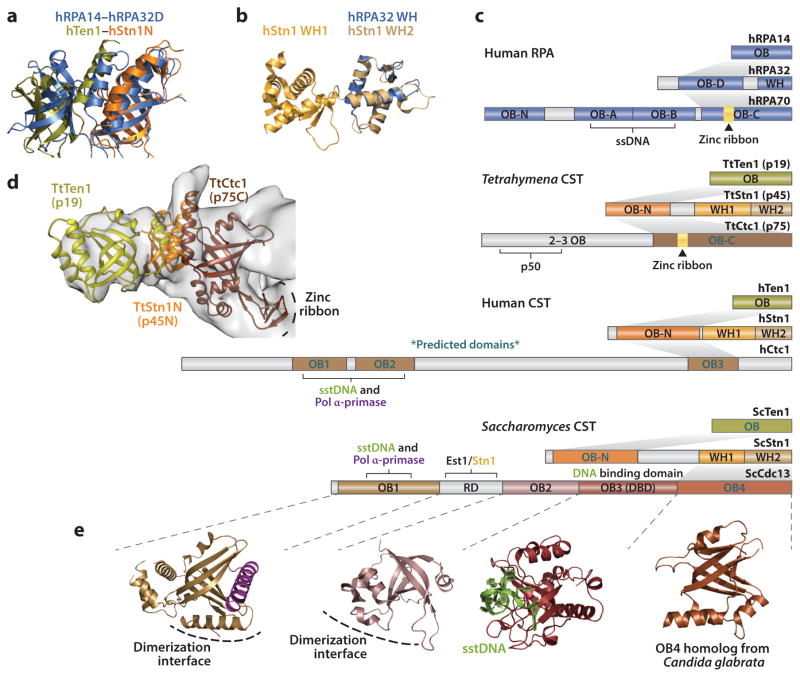
Domains, interactions, and structures of RPA and CST proteins. (a) Superpositions of structures of human RPA14–RPA32D (PDB ID 1QUQ) and Ten1–Stn1N complexes. (b) Structure of human Stn1 C-terminal tandem winged-helix domain (PDB ID 4JQF). Human RPA32 C-terminal winged-helix (PDB ID 1DPU) superpositions with Stn1 WH2. (c) Comparison of interactions and domain topologies of the human RPA, Tetrahymena CST, human CST, and Saccharomyces CST trimeric complexes. (d) Region of Tetrahymena telomerase cryo-EM density comprising the CST trimeric core with structures of Ten1–Stn1N (PDB ID 5DOI) and homology model of p75C [based on RPA70C (PDB ID 1L1O)] fit in (69). (e) Structures of Saccharomyces Cdc13 OB1 complexed to pol α-peptide (PDB ID 3OIQ), OB2 (PDB ID 4HCE), and OB3–sstDNA complex (PDB ID 1S40) and Candida Cdc13 OB4 (PDB ID 3RMH). Abbreviations: EM, electron microscopy; OB, oligonucleotide/oligosaccharide binding; RPA, replication protein A; ssDNA, single-stranded DNA; sstDNA, single-stranded telomere repeat DNA; WH, winged helix.
The discovery that the POT1 homolog Teb1 is part of a heterotrimeric RPA–related complex TEB, (Figures 5b,c and 6d–f) suggests a closer relationship between POT1 and RPA than previously appreciated. The POT1AB OB folds are structurally similar to the RPA70AB OB folds and bind DNA similarly. However, POT1C is not expected to be similar to RPA70C, nor is there evidence for the presence of POT1 binding RPA14/RPA32 homologs. Perhaps the smaller subunits of RPA were lost during evolution of POT1. The recurring presence of RPA as an evolutionary theme in telomerase (83) is also seen with the RPA-like CST complex (22), discussed in the next section.
It should be noted that POT1–TPP1 homologs (Pot1a–Tpt1) have previously been identified in Tetrahymena (Figure 1c) (64, 90). The Pot1a–Tpt1 proteins are the single-stranded telomere capping proteins for these ciliates (90), although no evidence suggests that they are involved in the recruitment or stimulation of telomerase. Because the 3′ overhang in Tetrahymena is short (~2 to 3 telomeric repeats) (65), it is likely that the association of telomerase to telomeres via p50–Teb1 is mutually exclusive with Tpt1–Pot1 bound to the 3′ sstDNA overhang (124). In humans, TPP1–POT1 switches from being inhibitory to activating upon telomerase recruitment (169, 180). In Tetrahymena, displacement of Tpt1–Pot1 by p50–TEB could be the first step in telomere extension.
CST PROTEINS INVOLVED IN C-STRAND SYNTHESIS AND TELOMERASE REGULATION
CST was originally identified in S. cerevisiae as the sstDNA-binding protein complex, comprising Cdc13, Stn1, and Ten1 (50, 51, 115). In Saccharomyces, it is the major capping mechanism for 3′ single-stranded telomere ends, and Cdc13 fulfills the roles of human POT1 in binding sstDNA and aiding recruitment of telomerase to telomeres (28, 88). Initially thought to be unique to yeast, because of the low sequence homology of these proteins, CST was subsequently identified in vertebrates and plants as Ctc1–Stn1–Ten1 (110, 125, 156). In plants, yeasts, and mammals, CST negatively regulates telomerase activity (24, 51, 80, 115), perhaps serving as a stop point for the subsequent recruitment of Pol α-primase by CST for C-strand synthesis (19, 24, 126). More recently, mammalian CST has been proposed to have extra telomeric roles, being implicated in genome-wide replication restart as well as recovery from DNA damage and replication stress (151, 170).
A breakthrough in understanding the structural biology of CST was the prediction that these proteins have a domain organization akin to the trimeric RPA complex (45); this was later confirmed when the first structures of Stn1 and Ten1 from yeast were solved (48, 155). Most recently, the Tetrahymena telomerase proteins p75, p45, and p19 were identified as a ciliate CST complex (69, 168). Although the association of CST is transient in most other organisms, in Tetrahymena, it is a stable part of the holoenzyme (Figure 1c) (69) and thus provides an unprecedented opportunity for structural studies of CST–telomerase interactions (22, 125).
Stn1–Ten1 Structure and Similarity to RPA
The RPA complex contains a trimeric core of three OB folds, comprising RPA14, RPA32D, and RPA70C (Figure 7c). Along with a three-helix bundle, formed at the C-terminus of each participating OB fold, RPA32D bridges the interaction between RPA14 and RPA70C. RPA32 contains an additional WH domain at its C-terminus, whereas RPA70, the largest subunit, possesses three additional N-terminal OB folds (123, 153). The most well-conserved components of the CST complex, Stn1 and Ten1, also bear the most similarity to their RPA counterparts, RPA32 and RPA14, respectively. Ten1 comprises a single OB fold that binds to the Stn1N OB fold (Figure 7a,c,d). Despite a lack of primary sequence similarity, structures of the human (15), Tetrahymena (168), Candida, and Schizosaccharomyces Ten1–Stn1N (155) complex all superpose with high similarity to each other as well as to the structure of the human RPA14–RPA32D complex (Figure 7a) (13).
Like RPA32, Stn1 contains a C-terminal domain connected to the N-terminal OB fold via a flexible linker. However, whereas RPA32 contains a single WH, Stn1 possesses a tandem WH domain (Figure 7b,c) (15, 48, 69, 155, 168). The relative orientation of the two WH domains with respect to each other is similar between Tetrahymena and Saccharomyces but different from human (15, 48, 69, 155, 168). However, the short linker and large buried area between the tandem domains in all three organisms suggest that indeed the two WH domains in Stn1 function as a structured unit (15, 48, 69, 155, 168). Interestingly, RPA32 WH superposes with both domains but much more closely with the human Stn1 WH2 than with the WH1 (Figure 7b). The same is true for Tetrahymena but not for Saccharomyces, where its Stn1 WH2 possesses a much more divergent wing motif, comprising significantly longer β-strands (48, 69, 155, 168). Although much work has shown the involvement of RPA32 WH in the recruitment of proteins for DNA repair (153), recent work has implicated the Stn1 WH2 in the recruitment and activation of Pol α-primase for C-strand synthesis (46, 95).
Ctc1/Cdc13 Structure and Interactions
Numerous studies and the comparison to RPA suggest that Stn1 bridges the trimeric interaction between the CST proteins (22). However, a high-resolution structure of the trimeric CST or of the Ctc1/Cdc13–Stn1 detailing this interaction remains to be determined. The 9-Å cryo-EM structure of Tetrahymena telomerase provided the first direct structural evidence of and insight into a trimeric CST interaction (Figure 7d) (69). In the model, p45N sits in between and interacts directly with p75C and p19, similar to the RPA trimeric core (13). Although p75C is based on a homology model from RPA70C, the cryo-EM map shows clear density for the three-helix bundle as well as a zinc-ribbon motif in p75C (Figure 7d) (69). Primary sequence analysis of p75 suggests that the zinc-ribbon motif does indeed exist in the putative p75C (Figure 7c), strongly suggesting that the C-terminal domain of p75 is homologous to RPA70C. In contrast, the C-terminal OB fold of Cdc13 is not predicted to contain a zinc ribbon, and its overall structure is different from RPA70C (Figure 7e) (182). These differences highlight the apparent divergence of the large subunit of CST.
Structures of Saccharomyces Cdc13OB2 and Candida Cdc13OB4 do not superpose with their RPA counterparts. The main β-barrel of Sacharromyces Cdc13OB1 superposes with RPA70N and has been shown to interact with a small peptide of Pol α-primase as well as display sstDNA binding (Figure 7e) (107, 154). Cdc13OB1 and OB2 are expected to facilitate Cdc13 dimerization, which is important for the proper functioning of Cdc13 as well as formation of the trimeric CST complex (103). Unlike RPA70, which displays cooperative ssDNA binding between its three C-terminal OB folds, the sstDNA-binding activity of Cdc13 is performed primarily by OB1 and OB3 (Figure 7e) (107–109). Tetrahymena p75 and human Ctc1 have also been shown to bind sstDNA (23, 24, 55, 168), but the structural basis for this or high-resolution structures for these subunits or their subdomains have yet to been determined.
Overall, structural and functional investigations of CST proteins have revealed both similarities and differences between different organisms. There are now structures of Stnt1 and Ten1 from many organisms, but for the largest and most divergent subunit, only yeast Cdc13 has been structurally characterized. A structural basis for the large body of functional genetic and biochemical data that has been gathered for Ctc1 (human, plants, Tetrahymena) (23, 24, 55, 131) will be vital for understanding the role of CST in telomerase regulation and telomere maintenance.
FUTURE PROSPECTS
More than a quarter century after the discovery of telomerase, we now have a snapshot of how the components of telomerase fit and function together at telomeres. Problems of solubility, expression, and heterogeneity inherent to telomerase complexes remain a challenge for structural biologists. With recent advances in structural biology methods and biochemical characterization of telomerase, in the next few years, we can expect the first structures of human and yeast telomerase, higher-resolution structures of the telomerase RNP core with sstDNA, and details of telomerase interaction at telomeres.
Acknowledgments
Telomerase research in the Feigon laboratory is supported by grants from NIH (RO1 GM48123) and NSF (MCB 1517629) to J.F. Additional support for UCLA-DOE X-ray and NMR facilities is provided by DOE grant DE-FC02-023463421. H.C. is partially supported by the Ruth L. Kirschstein National Research Service Award GM007185.
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- 1.Akiyama BM, Gomez A, Stone MD. A conserved motif in Tetrahymena thermophila telomerase reverse transcriptase is proximal to the RNA template and is essential for boundary definition. J Biol Chem. 2013;288:22141–49. doi: 10.1074/jbc.M113.452425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Akiyama BM, Parks JW, Stone MD. The telomerase essential N-terminal domain promotes DNA synthesis by stabilizing short RNA–DNA hybrids. Nucleic Acids Res. 2015;43:5537–49. doi: 10.1093/nar/gkv406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alves D, Li H, Codrington R, Orte A, Ren X, et al. Single-molecule analysis of human telomerase monomer. Nat Chem Biol. 2008;4:287–89. doi: 10.1038/nchembio.82. [DOI] [PubMed] [Google Scholar]
- 4.Armanios M, Blackburn EH. The telomere syndromes. Nat Rev Genet. 2012;13:693–704. doi: 10.1038/nrg3246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Artandi SE, DePinho RA. Telomeres and telomerase in cancer. Carcinogenesis. 2010;31:9–18. doi: 10.1093/carcin/bgp268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Autexier C, Lue NF. The structure and function of telomerase reverse transcriptase. Annu Rev Biochem. 2006;75:493–517. doi: 10.1146/annurev.biochem.75.103004.142412. [DOI] [PubMed] [Google Scholar]
- 7.Bajon E, Laterreur N, Wellinger RJ. A single templating RNA in yeast telomerase. Cell Rep. 2015;12:441–48. doi: 10.1016/j.celrep.2015.06.045. [DOI] [PubMed] [Google Scholar]
- 8.Berman AJ, Akiyama BM, Stone MD, Cech TR. The RNA accordion model for template positioning by telomerase RNA during telomeric DNA synthesis. Nat Struct Mol Biol. 2011;18:1371–75. doi: 10.1038/nsmb.2174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bernardes de Jesus B, Blasco MA. Telomerase at the intersection of cancer and aging. Trends Genet. 2013;29:513–20. doi: 10.1016/j.tig.2013.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Blackburn EH, Collins K. Telomerase: an RNP enzyme synthesizes DNA. Cold Spring Harb Perspect Biol. 2011;3:a003558. doi: 10.1101/cshperspect.a003558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Blackburn EH, Greider CW, Szostak JW. Telomeres and telomerase: the path from maize, Tetrahymena and yeast to human cancer and aging. Nat Med. 2006;12:1133–38. doi: 10.1038/nm1006-1133. [DOI] [PubMed] [Google Scholar]
- 12.Bley CJ, Qi X, Rand DP, Borges CR, Nelson RW, Chen JJ-L. RNA–protein binding interface in the telomerase ribonucleoprotein. PNAS. 2011;108:20333–38. doi: 10.1073/pnas.1100270108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bochkareva E, Korolev S, Lees-Miller SP, Bochkarev A. Structure of the RPA trimerization core and its role in the multistep DNA-binding mechanism of RPA. EMBO J. 2002;21:1855–63. doi: 10.1093/emboj/21.7.1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brown AF, Podlevsky JD, Qi X, Chen Y, Xie M, Chen JJ-L. A self-regulating template in human telomerase. PNAS. 111:11311–16. doi: 10.1073/pnas.1402531111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bryan C, Rice C, Harkisheimer M, Schultz DC, Skordalakes E. Structure of the human telomeric Stn1-Ten1 capping complex. PLOS ONE. 2013;8:e66756. doi: 10.1371/journal.pone.0066756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bryan TM, Goodrich KJ, Cech TR. Tetrahymena telomerase is active as a monomer. Mol Biol Cell. 2003;14:4794–804. doi: 10.1091/mbc.E03-07-0474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cash DD, Cohen-Zontag O, Kim N-K, Shefer K, Brown Y, et al. Pyrimidine motif triple helix in the Kluyveromyces lactis telomerase RNA pseudoknot is essential for function in vivo. PNAS. 2013;110:10970–75. doi: 10.1073/pnas.1309590110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cash DD, Feigon J. Structure and folding of the Tetrahymena telomerase RNA pseudoknot. Nucleic Acids Res. 45:482–95. doi: 10.1093/nar/gkw1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Casteel DE, Zhuang S, Zeng Y, Perrino FW, Boss GR, et al. A DNA polymerase α · primase cofactor with homology to replication protein A-32 regulates DNA replication in mammalian cells. J Biol Chem. 2009;284:5807–18. doi: 10.1074/jbc.M807593200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen J-L, Blasco MA, Greider CW. Secondary structure of vertebrate telomerase RNA. Cell. 2000;100:503–14. doi: 10.1016/s0092-8674(00)80687-x. [DOI] [PubMed] [Google Scholar]
- 21.Chen J-L, Greider CW. Template boundary definition in mammalian telomerase. Genes Dev. 2003;17:2747–52. doi: 10.1101/gad.1140303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen L-Y, Lingner J. CST for the grand finale of telomere replication. Nucleus. 2013;4:277–82. doi: 10.4161/nucl.25701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen L-Y, Majerská J, Lingner J. Molecular basis of telomere syndrome caused by CTC1 mutations. Genes Dev. 2013;27:2099–108. doi: 10.1101/gad.222893.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen L-Y, Redon S, Lingner J. The human CST complex is a terminator of telomerase activity. Nature. 2012;488:540–44. doi: 10.1038/nature11269. [DOI] [PubMed] [Google Scholar]
- 25.Chen Y, Fender J, Legassie JD, Jarstfer MB, Bryan TM, Varani G. Structure of stem-loop IV of Tetrahymena telomerase RNA. EMBO J. 2006;25:3156–66. doi: 10.1038/sj.emboj.7601195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chu TW, D’Souza Y, Autexier C. The insertion in fingers domain in human telomerase can mediate enzyme processivity and telomerase recruitment to telomeres in a TPP1-dependent manner. Mol Cell Biol. 2016;36:210–22. doi: 10.1128/MCB.00746-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chu TW, MacNeil DE, Autexier C. Multiple mechanisms contribute to the cell growth defects imparted by human telomerase insertion in fingers domain mutations associated with premature aging diseases. J Biol Chem. 2016;291:8374–86. doi: 10.1074/jbc.M116.714782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Churikov D, Corda Y, Luciano P, Géli V. Cdc13 at a crossroads of telomerase action. Front Oncol. 2013;3:39. doi: 10.3389/fonc.2013.00039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cohen SB, Graham ME, Lovrecz GO, Bache N, Robinson PJ, Reddel RR. Protein composition of catalytically active human telomerase from immortal cells. Science. 2007;315:1850–53. doi: 10.1126/science.1138596. [DOI] [PubMed] [Google Scholar]
- 30.Cole DI, Legassie JD, Bonifacio LN, Sekaran VG, Ding F, et al. New models of Tetrahymena telomerase RNA from experimentally derived constraints and modeling. J Am Chem Soc. 2012;134:20070–80. doi: 10.1021/ja305636u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Collins K. The biogenesis and regulation of telomerase holoenzymes. Nat Rev Mol Cell Biol. 2006;7:484–94. doi: 10.1038/nrm1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cristofari G, Lingner J. Telomere length homeostasis requires that telomerase levels are limiting. EMBO J. 2006;25:565–74. doi: 10.1038/sj.emboj.7600952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dalby AB, Hofr C, Cech TR. Contributions of the TEL-patch amino acid cluster on TPP1 to telomeric DNA synthesis by human telomerase. J Mol Biol. 2015;427:1291–303. doi: 10.1016/j.jmb.2015.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.de Lange T. Shelterin: the protein complex that shapes and safeguards human telomeres. Genes Dev. 2005;19:2100–10. doi: 10.1101/gad.1346005. [DOI] [PubMed] [Google Scholar]
- 35.Duan J, Li L, Lu J, Wang W, Ye K. Structural mechanism of substrate RNA recruitment in H/ACA RNA-guided pseudouridine synthase. Mol Cell. 2009;34:427–39. doi: 10.1016/j.molcel.2009.05.005. [DOI] [PubMed] [Google Scholar]
- 36.Eckert B, Collins K. Roles of telomerase reverse transcriptase N-terminal domain in assembly and activity of Tetrahymena telomerase holoenzyme. J Biol Chem. 2012;287:12805–14. doi: 10.1074/jbc.M112.339853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Egan ED, Collins K. Specificity and stoichiometry of subunit interactions in the human telomerase holoenzyme assembled in vivo. Mol Cell Biol. 2010;30:2775–86. doi: 10.1128/MCB.00151-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Egan ED, Collins K. An enhanced H/ACA RNP assembly mechanism for human telomerase RNA. Mol Cell Biol. 2012;32:2428–39. doi: 10.1128/MCB.00286-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Egan ED, Collins K. Biogenesis of telomerase ribonucleoproteins. RNA. 2012;18:1747–59. doi: 10.1261/rna.034629.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Errington TM, Fu D, Wong JM, Collins K. Disease-associated human telomerase RNA variants show loss of function for telomere synthesis without dominant-negative interference. Mol Cell Biol. 2008;28:6510–20. doi: 10.1128/MCB.00777-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Feigon J, Chan H, Jiang J. Integrative structural biology of Tetrahymena telomerase—insights into catalytic mechanism and interaction at telomeres. FEBS J. 2016;283:2044–50. doi: 10.1111/febs.13691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fogarty PF, Yamaguchi H, Wiestner A, Baerlocher GM, Sloand E, et al. Late presentation of dyskeratosis congenita as apparently acquired aplastic anaemia due to mutations in telomerase RNA. Lancet. 2003;362:1628–30. doi: 10.1016/S0140-6736(03)14797-6. [DOI] [PubMed] [Google Scholar]
- 43.Froelich-Ammon SJ, Dickinson BA, Bevilacqua JM, Schultz SC, Cech TR. Modulation of telom-erase activity by telomere DNA-binding proteins in Oxytricha. Genes Dev. 1998;12:1504–14. doi: 10.1101/gad.12.10.1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fujii H, Shao L, Colmegna I, Goronzy JJ, Weyand CM. Telomerase insufficiency in rheumatoid arthritis. PNAS. 2009;106:4360–65. doi: 10.1073/pnas.0811332106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gao H, Cervantes RB, Mandell EK, Otero JH, Lundblad V. RPA-like proteins mediate yeast telomere function. Nat Struct Mol Biol. 2007;14:208–14. doi: 10.1038/nsmb1205. [DOI] [PubMed] [Google Scholar]
- 46.Gasparyan HJ, Xu L, Petreaca RC, Rex AE, Small VY, et al. Yeast telomere capping protein Stn1 overrides DNA replication control through the S phase checkpoint. PNAS. 2009;106:2206–11. doi: 10.1073/pnas.0812605106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gavory G, Symmons MF, Krishnan Ghosh Y, Klenerman D, Balasubramanian S. Structural analysis of the catalytic core of human telomerase RNA by FRET and molecular modeling. Biochemistry. 2006;45:13304–11. doi: 10.1021/bi061150a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gelinas AD, Paschini M, Reyes FE, Heroux A, Batey RT, et al. Telomere capping proteins are structurally related to RPA with an additional telomere-specific domain. PNAS. 2009;106:19298–303. doi: 10.1073/pnas.0909203106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gillis AJ, Schuller AP, Skordalakes E. Structure of the Tribolium castaneum telomerase catalytic subunit TERT. Nature. 2008;455:633–37. doi: 10.1038/nature07283. [DOI] [PubMed] [Google Scholar]
- 50.Grandin N, Damon C, Charbonneau M. Ten1 functions in telomere end protection and length regulation in association with Stn1 and Cdc13. EMBO J. 2001;20:1173–83. doi: 10.1093/emboj/20.5.1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Grandin N, Reed SI, Charbonneau M. Stn1, a new Saccharomyces cerevisiae protein, is implicated in telomere size regulation in association with Cdc13. Genes Dev. 1997;11:512–27. doi: 10.1101/gad.11.4.512. [DOI] [PubMed] [Google Scholar]
- 52.Greider CW, Blackburn EH. Identification of a specific telomere terminal transferase activity in Tetrahymena extracts. Cell. 1985;43:405–13. doi: 10.1016/0092-8674(85)90170-9. [DOI] [PubMed] [Google Scholar]
- 53.Greider CW, Blackburn EH. The telomere terminal transferase of Tetrahymena is a ribonucleo-protein enzyme with two kinds of primer specificity. Cell. 1987;51:887–98. doi: 10.1016/0092-8674(87)90576-9. [DOI] [PubMed] [Google Scholar]
- 54.Greider CW, Blackburn EH. A telomeric sequence in the RNA of Tetrahymena telomerase required for telomere repeat synthesis. Nature. 1989;337:331–37. doi: 10.1038/337331a0. [DOI] [PubMed] [Google Scholar]
- 55.Gu P, Chang S. Functional characterization of human CTC1 mutations reveals novel mechanisms responsible for the pathogenesis of the telomere disease Coats plus. Aging Cell. 2013;12:1100–9. doi: 10.1111/acel.12139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hamma T, Ferré-D’Amaré AR. Structure of protein L7Ae bound to a K-turn derived from an archaeal box H/ACA sRNA at 1.8 Å resolution. Structure. 2004;12:893–903. doi: 10.1016/j.str.2004.03.015. [DOI] [PubMed] [Google Scholar]
- 57.Hamma T, Ferré-D’Amaré AR. The box H/ACA ribonucleoprotein complex: interplay of RNA and protein structures in post-transcriptional RNA modification. J Biol Chem. 2010;285:805–9. doi: 10.1074/jbc.R109.076893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Harkisheimer M, Mason M, Shuvaeva E, Skordalakes E. A motif in the vertebrate telomerase N-terminal linker of TERT contributes to RNA binding and telomerase activity and processivity. Structure. 2013;21:1870–78. doi: 10.1016/j.str.2013.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hengesbach M, Kim N-K, Feigon J, Stone MD. Single-molecule FRET reveals the folding dynamics of the human telomerase RNA pseudoknot domain. Angew Chem Int Ed. 2012;51:5876–79. doi: 10.1002/anie.201200526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hockemeyer D, Collins K. Control of telomerase action at human telomeres. Nat Struct Mol Biol. 2015;22:848–52. doi: 10.1038/nsmb.3083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hong K, Upton H, Miracco EJ, Jiang J, Zhou ZH, et al. Tetrahymena telomerase holoenzyme assembly, activation, and inhibition by domains of the p50 central hub. Mol Cell Biol. 2013;33:3962–71. doi: 10.1128/MCB.00792-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Horvath MP, Schweiker VL, Bevilacqua JM, Ruggles JA, Schultz SC. Crystal structure of the Oxytricha nova telomere end binding protein complexed with single strand DNA. Cell. 1998;95:963–74. doi: 10.1016/s0092-8674(00)81720-1. [DOI] [PubMed] [Google Scholar]
- 63.Huang J, Brown AF, Wu J, Xue J, Bley CJ, et al. Structural basis for protein-RNA recognition in telomerase. Nat Struct Mol Biol. 2014;21:507–12. doi: 10.1038/nsmb.2819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jacob NK, Lescasse R, Linger BR, Price CM. Tetrahymena POT1a regulates telomere length and prevents activation of a cell cycle checkpoint. Mol Cell Biol. 2007;27:1592–601. doi: 10.1128/MCB.01975-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jacob NK, Skopp R, Price CM. G-overhang dynamics at Tetrahymena telomeres. EMBO J. 2001;20:4299–308. doi: 10.1093/emboj/20.15.4299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jacobs SA, Podell ER, Cech TR. Crystal structure of the essential N-terminal domain of telomerase reverse transcriptase. Nat Struct Mol Biol. 2006;13:218–25. doi: 10.1038/nsmb1054. [DOI] [PubMed] [Google Scholar]
- 67.Jansson LI, Akiyama BM, Ooms A, Lu C, Rubin SM, Stone MD. Structural basis of template-boundary definition in Tetrahymena telomerase. Nat Struct Mol Biol. 2015;22:883–88. doi: 10.1038/nsmb.3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jaskelioff M, Muller FL, Paik J-H, Thomas E, Jiang S, et al. Telomerase reactivation reverses tissue degeneration in aged telomerase-deficient mice. Nature. 2011;469:102–6. doi: 10.1038/nature09603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jiang J, Chan H, Cash DD, Miracco EJ, Ogorzalek Loo RR, et al. Structure of Tetrahymena telomerase reveals previously unknown subunits, functions, and interactions. Science. 2015;350:aab4070. doi: 10.1126/science.aab4070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jiang J, Miracco EJ, Hong K, Eckert B, Chan H, et al. The architecture of Tetrahymena telomerase holoenzyme. Nature. 2013;496:187–92. doi: 10.1038/nature12062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kelleher C, Teixeira MT, Forstemann K, Lingner J. Telomerase: biochemical considerations for enzyme and substrate. Trends Biochem Sci. 2002;27:572–79. doi: 10.1016/s0968-0004(02)02206-5. [DOI] [PubMed] [Google Scholar]
- 72.Kim N-K, Theimer CA, Mitchell JR, Collins K, Feigon J. Effect of pseudouridylation on the structure and activity of the catalytically essential P6.1 hairpin in human telomerase RNA. Nucleic Acids Res. 2010;38:6746–56. doi: 10.1093/nar/gkq525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kim N-K, Zhang Q, Feigon J. Structure and sequence elements of the CR4/5 domain of medaka telomerase RNA important for telomerase function. Nucleic Acids Res. 2014;42:3395–408. doi: 10.1093/nar/gkt1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kim N-K, Zhang Q, Zhou J, Theimer CA, Peterson RD, Feigon J. Solution structure and dynamics of the wild-type pseudoknot of human telomerase RNA. J Mol Biol. 2008;384:1249–61. doi: 10.1016/j.jmb.2008.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kiss T, Fayet-Lebaron E, Jády BE. Box H/ACA small ribonucleoproteins. Mol Cell. 2010;37:597–606. doi: 10.1016/j.molcel.2010.01.032. [DOI] [PubMed] [Google Scholar]
- 76.Koo B-K, Park C-J, Fernandez CF, Chim N, Ding Y, et al. Structure of H/ACA RNP protein Nhp2p reveals Cis/Trans isomerization of a conserved proline at the RNA and Nop10 binding interface. J Mol Biol. 2011;411:927–42. doi: 10.1016/j.jmb.2011.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kupiec M. Biology of telomeres: lessons from budding yeast. FEMS Microbiol Rev. 2014;38:144–71. doi: 10.1111/1574-6976.12054. [DOI] [PubMed] [Google Scholar]
- 78.Lai CK, Miller MC, Collins K. Template boundary definition in Tetrahymena telomerase. Genes Dev. 2002;16:415–20. doi: 10.1101/gad.962602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lai CK, Mitchell JR, Collins K. RNA binding domain of telomerase reverse transcriptase. Mol Cell Biol. 2001;21:990–1000. doi: 10.1128/MCB.21.4.990-1000.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Leehy KA, Lee JR, Song X, Renfrew KB, Shippen DE. MERISTEM DISORGANIZATION1 encodes TEN1, an essential telomere protein that modulates telomerase processivity in Arabidopsis. Plant Cell. 2013;25:1343–54. doi: 10.1105/tpc.112.107425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lei M, Podell ER, Baumann P, Cech TR. DNA self-recognition in the structure of Pot1 bound to telomeric single-stranded DNA. Nature. 2003;426:198–203. doi: 10.1038/nature02092. [DOI] [PubMed] [Google Scholar]
- 82.Lei M, Podell ER, Cech TR. Structure of human POT1 bound to telomeric single-stranded DNA provides a model for chromosome end-protection. Nat Struct Mol Biol. 2004;11:1223–29. doi: 10.1038/nsmb867. [DOI] [PubMed] [Google Scholar]
- 83.Lewis KA, Wuttke DS. Telomerase and telomere-associated proteins: structural insights into mechanism and evolution. Structure. 2012;20:28–39. doi: 10.1016/j.str.2011.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Li L, Ye K. Crystal structure of an H/ACA box ribonucleoprotein particle. Nature. 2006;443:302–7. doi: 10.1038/nature05151. [DOI] [PubMed] [Google Scholar]
- 85.Li S, Duan J, Li D, Ma S, Ye K. Structure of the Shq1–Cbf5–Nop10–Gar1 complex and implications for H/ACA RNP biogenesis and dyskeratosis congenita. EMBO J. 2011;30:5010–20. doi: 10.1038/emboj.2011.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Li S, Duan J, Li D, Yang B, Dong M, Ye K. Reconstitution and structural analysis of the yeast box H/ACA RNA-guided pseudouridine synthase. Genes Dev. 2011;25:2409–21. doi: 10.1101/gad.175299.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Liang B, Xue S, Terns RM, Terns MP, Li H. Substrate RNA positioning in the archaeal H/ACA ribonucleoprotein complex. Nat Struct Mol Biol. 2007;14:1189–95. doi: 10.1038/nsmb1336. [DOI] [PubMed] [Google Scholar]
- 88.Lin J-J, Zakian VA. The Saccharomyces CDC13 protein is a single-strand TG1–3 telomeric DNA-binding protein in vitro that affects telomere behavior in vivo. PNAS. 1996;93:13760–65. doi: 10.1073/pnas.93.24.13760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lin KW, Zakian VA. 21st century genetics: mass spectrometry of yeast telomerase. Cold Spring Harb Symp Quant Biol. 2015;80:111–16. doi: 10.1101/sqb.2015.80.027656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Linger BR, Morin GB, Price CM. The Pot1a-associated proteins Tpt1 and Pat1 coordinate telomere protection and length regulation in Tetrahymena. Mol Biol Cell. 2011;22:4161–70. doi: 10.1091/mbc.E11-06-0551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Linger BR, Price CM. Conservation of telomere protein complexes: shuffling through evolution. Crit Rev Biochem Mol Biol. 2009;44:434–46. doi: 10.3109/10409230903307329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lingner J, Hughes TR, Shevchenko A, Mann M, Lundblad V, Cech TR. Reverse transcriptase motifs in the catalytic subunit of telomerase. Science. 1997;276:561–67. doi: 10.1126/science.276.5312.561. [DOI] [PubMed] [Google Scholar]
- 93.Liu D, Safari A, O’Connor MS, Chan DW, Laegeler A, et al. PTOP interacts with POT1 and regulates its localization to telomeres. Nat Cell Biol. 2004;6:673–80. doi: 10.1038/ncb1142. [DOI] [PubMed] [Google Scholar]
- 94.Lloyd NR, Dickey TH, Hom RA, Wuttke DS. Tying up the ends: plasticity in the recognition of single-stranded DNA at telomeres. Biochemistry. 2016;55:5326–40. doi: 10.1021/acs.biochem.6b00496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lue NF, Chan J, Wright WE, Hurwitz J. The CDC13-STN1-TEN1 complex stimulates Pol α activity by promoting RNA priming and primase-to-polymerase switch. Nat Commun. 2014;5:5762. doi: 10.1038/ncomms6762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lue NF, Lin Y-C, Mian IS. A conserved telomerase motif within the catalytic domain of telomerase reverse transcriptase is specifically required for repeat addition processivity. Mol Cell Biol. 2003;23:8440–49. doi: 10.1128/MCB.23.23.8440-8449.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Mahmoudi S, Henriksson S, Weibrecht I, Smith S, Söderberg O, et al. WRAP53 is essential for Cajal body formation and for targeting the survival of motor neuron complex to Cajal bodies. PLOS Biol. 2010;8:e1000521. doi: 10.1371/journal.pbio.1000521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Malyavko AN, Parfenova YY, Zvereva MI, Dontsova OA. Telomere length regulation in budding yeasts. FEBS Lett. 2014;588:2530–36. doi: 10.1016/j.febslet.2014.05.049. [DOI] [PubMed] [Google Scholar]
- 99.Manival X, Charron C, Fourmann J-B, Godard F, Charpentier B, Branlant C. Crystal structure determination and site-directed mutagenesis of the Pyrococcus abyssi aCBF5–aNOP10 complex reveal crucial roles of the C-terminal domains of both proteins in H/ACA sRNP activity. Nucleic Acids Res. 2006;34:826–39. doi: 10.1093/nar/gkj482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Marrone A, Walne A, Dokal I. Dyskeratosis congenita: telomerase, telomeres and anticipation. Curr Opin Genet Dev. 2005;15:249–57. doi: 10.1016/j.gde.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 101.Martínez P, Blasco MA. Replicating through telomeres: a means to an end. Trends Biochem Sci. 2015;40:504–15. doi: 10.1016/j.tibs.2015.06.003. [DOI] [PubMed] [Google Scholar]
- 102.Mason M, Schuller A, Skordalakes E. Telomerase structure function. Curr Opin Struct Biol. 2011;21:92–100. doi: 10.1016/j.sbi.2010.11.005. [DOI] [PubMed] [Google Scholar]
- 103.Mason M, Wanat JJ, Harper S, Schultz DC, Speicher DW, et al. Cdc13 OB2 dimerization required for productive Stn1 binding and efficient telomere maintenance. Structure. 2013;21:109–20. doi: 10.1016/j.str.2012.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Min B, Collins K. An RPA-related sequence-specific DNA-binding subunit of telomerase holoenzyme is required for elongation processivity and telomere maintenance. Mol Cell. 2009;36:609–19. doi: 10.1016/j.molcel.2009.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Miracco EJ, Jiang J, Cash DD, Feigon J. Progress in structural studies of telomerase. Curr Opin Struct Biol. 2014;24:115–24. doi: 10.1016/j.sbi.2014.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Mitchell MT, Gillis A, Futahashi M, Fujiwara H, Skordalakes E. Structural basis for telomerase catalytic subunit TERT binding to RNA template and telomeric DNA. Nat Struct Mol Biol. 2010;17:513–18. doi: 10.1038/nsmb.1777. [DOI] [PubMed] [Google Scholar]
- 107.Mitchell MT, Smith JS, Mason M, Harper S, Speicher DW, et al. Cdc13 N-terminal dimerization, DNA binding, and telomere length regulation. Mol Cell Biol. 2010;30:5325–34. doi: 10.1128/MCB.00515-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Mitton-Fry RM, Anderson EM, Hughes TR, Lundblad V, Wuttke DS. Conserved structure for single-stranded telomeric DNA recognition. Science. 2002;296:145–47. doi: 10.1126/science.1068799. [DOI] [PubMed] [Google Scholar]
- 109.Mitton-Fry RM, Anderson EM, Theobald DL, Glustrom LW, Wuttke DS. Structural basis for telomeric single-stranded DNA recognition by yeast Cdc13. J Mol Biol. 2004;338:241–55. doi: 10.1016/j.jmb.2004.01.063. [DOI] [PubMed] [Google Scholar]
- 110.Miyake Y, Nakamura M, Nabetani A, Shimamura S, Tamura M, et al. RPA-like mammalian Ctc1-Stn1-Ten1 complex binds to single-stranded DNA and protects telomeres independently of the Pot1 pathway. Mol Cell. 2009;36:193–206. doi: 10.1016/j.molcel.2009.08.009. [DOI] [PubMed] [Google Scholar]
- 111.Nandakumar J, Bell CF, Weidenfeld I, Zaug AJ, Leinwand LA, Cech TR. The TEL patch of telomere protein TPP1 mediates telomerase recruitment and processivity. Nature. 2012;492:285–89. doi: 10.1038/nature11648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Nandakumar J, Cech TR. Finding the end: recruitment of telomerase to telomeres. Nat Rev Mol Cell Biol. 2013;14:69–82. doi: 10.1038/nrm3505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Nelson AD, Shippen DE. Evolution of TERT-interacting lncRNAs: expanding the regulatory landscape of telomerase. Front Genet. 2015;6:277. doi: 10.3389/fgene.2015.00277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Niederer RO, Zappulla DC. Refined secondary-structure models of the core of yeast and human telomerase RNAs directed by SHAPE. RNA. 2015;21:254–61. doi: 10.1261/rna.048959.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Nugent CI, Hughes TR, Lue NF, Lundblad V. Cdc13p: a single-strand telomeric DNA-binding protein with a dual role in yeast telomere maintenance. Science. 1996;274:249–52. doi: 10.1126/science.274.5285.249. [DOI] [PubMed] [Google Scholar]
- 116.O’Connor CM, Collins K. A novel RNA binding domain in Tetrahymena telomerase p65 initiates hierarchical assembly of telomerase holoenzyme. Mol Cell Biol. 2006;26:2029–36. doi: 10.1128/MCB.26.6.2029-2036.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.O’Connor CM, Lai CK, Collins K. Two purified domains of telomerase reverse transcriptase reconstitute sequence-specific interactions with RNA. J Biol Chem. 2005;280:17533–39. doi: 10.1074/jbc.M501211200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.O’Sullivan RJ, Karlseder J. Telomeres: protecting chromosomes against genome instability. Nat Rev Mol Cell Biol. 2010;11:171–81. doi: 10.1038/nrm2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Parks JW, Stone MD. Coordinated DNA dynamics during the human telomerase catalytic cycle. Nat Commun. 5:4146. doi: 10.1038/ncomms5146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Pfeiffer V, Lingner J. Replication of telomeres and the regulation of telomerase. Cold Spring Harb Perspect Biol. 2013;5:a010405. doi: 10.1101/cshperspect.a010405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Podlevsky JD, Bley CJ, Omana RV, Qi X, Chen JJ-L. The telomerase database. Nucleic Acids Res. 2008;36:D339–43. doi: 10.1093/nar/gkm700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Podlevsky JD, Chen JJ-L. Evolutionary perspectives of telomerase RNA structure and function. RNA Biol. 2016;13:720–32. doi: 10.1080/15476286.2016.1205768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Prakash A, Borgstahl GE. The structure and function of replication protein A in DNA replication. Subcell Biochem. 2012;62:171–96. doi: 10.1007/978-94-007-4572-8_10. [DOI] [PubMed] [Google Scholar]
- 124.Premkumar VL, Cranert S, Linger BR, Morin GB, Minium S, Price C. The 3′ overhangs at Tetrahymena thermophila telomeres are packaged by four proteins, Pot1a, Tpt1, Pat1, and Pat2. Eukaryot Cell. 2014;13:240–45. doi: 10.1128/EC.00275-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Price CM, Boltz KA, Chaiken MF, Stewart JA, Beilstein MA, Shippen DE. Evolution of CST function in telomere maintenance. Cell Cycle. 2010;9:3157–65. doi: 10.4161/cc.9.16.12547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Qi H, Zakian VA. The Saccharomyces telomere-binding protein Cdc13p interacts with both the catalytic subunit of DNA polymerase αand the telomerase-associated Est1 protein. Genes Dev. 2000;14:1777–88. [PMC free article] [PubMed] [Google Scholar]
- 127.Qiao F, Cech TR. Triple-helix structure in telomerase RNA contributes to catalysis. Nat Struct Mol Biol. 2008;15:634–40. doi: 10.1038/nsmb.1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Qiao F, Goodrich KJ, Cech TR. Engineering cis-telomerase RNAs that add telomeric repeats to themselves. PNAS. 2010;107:4914–18. doi: 10.1073/pnas.0909366107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Rajavel M, Mullins MR, Taylor DJ. Multiple facets of TPP1 in telomere maintenance. Biochim Biophys Acta. 2014;1844:1550–59. doi: 10.1016/j.bbapap.2014.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Rashid R, Liang B, Baker DL, Youssef OA, He Y, et al. Crystal structure of a Cbf5-Nop10-Gar1 complex and implications in RNA-guided pseudouridylation and dyskeratosis congenita. Mol Cell. 2006;21:249–60. doi: 10.1016/j.molcel.2005.11.017. [DOI] [PubMed] [Google Scholar]
- 131.Rice C, Skordalakes E. Structure and function of the telomeric CST complex. Comput Struct Biotechnol J. 2016;14:161–67. doi: 10.1016/j.csbj.2016.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Richard P, Darzacq X, Bertrand E, Jády BE, Verheggen C, Kiss T. A common sequence motif determines the Cajal body-specific localization of box H/ACA scaRNAs. EMBO J. 2003;22:4283–93. doi: 10.1093/emboj/cdg394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Richards RJ, Theimer CA, Finger LD, Feigon J. Structure of the Tetrahymena thermophila telomerase RNA helix II template boundary element. Nucleic Acids Res. 2006;34:816–25. doi: 10.1093/nar/gkj481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Richards RJ, Wu H, Trantirek L, O’Connor CM, Collins K, Feigon J. Structural study of elements of Tetrahymena telomerase RNA stem-loop IV domain important for function. RNA. 2006;12:1475–85. doi: 10.1261/rna.112306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Robart AR, Collins K. Investigation of human telomerase holoenzyme assembly, activity, and processivity using disease-linked subunit variants. J Biol Chem. 2010;285:4375–86. doi: 10.1074/jbc.M109.088575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Rouda S, Skordalakes E. Structure of the RNA-binding domain of telomerase: implications for RNA recognition and binding. Structure. 2007;15:1403–12. doi: 10.1016/j.str.2007.09.007. [DOI] [PubMed] [Google Scholar]
- 137.Sarek G, Marzec P, Margalef P, Boulton SJ. Molecular basis of telomere dysfunction in human genetic diseases. Nat Struct Mol Biol. 2015;22:867–74. doi: 10.1038/nsmb.3093. [DOI] [PubMed] [Google Scholar]
- 138.Sauerwald A, Sandin S, Cristofari G, Scheres SH, Lingner J, Rhodes D. Structure of active dimeric human telomerase. Nat Struct Mol Biol. 2013;20:454–60. doi: 10.1038/nsmb.2530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Savage SA, Bertuch AA. The genetics and clinical manifestations of telomere biology disorders. Genet Med. 2010;12:753–64. doi: 10.1097/GIM.0b013e3181f415b5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Schmidt JC, Cech TR. Human telomerase: biogenesis, trafficking, recruitment, and activation. Genes Dev. 2015;29:1095–105. doi: 10.1101/gad.263863.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Schmidt JC, Dalby AB, Cech TR. Identification of human TERT elements necessary for telomerase recruitment to telomeres. eLife. 2014;3:e03563. doi: 10.7554/eLife.03563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Schnapp G, Rodi HP, Rettig WJ, Schnapp A, Damm K. One-step affinity purification protocol for human telomerase. Nucleic Acids Res. 1998;26:3311–13. doi: 10.1093/nar/26.13.3311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Schwartz S, Bernstein DA, Mumbach MR, Jovanovic M, Herbst RH, et al. Transcriptome-wide mapping reveals widespread dynamic-regulated pseudouridylation of ncRNA and mRNA. Cell. 2014;159:148–62. doi: 10.1016/j.cell.2014.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Sexton AN, Regalado SG, Lai CS, Cost GJ, O’Neil CM, et al. Genetic and molecular identification of three human TPP1 functions in telomerase action: recruitment, activation, and homeostasis set point regulation. Genes Dev. 2014;28:1885–99. doi: 10.1101/gad.246819.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Sexton AN, Youmans DT, Collins K. Specificity requirements for human telomere protein interaction with telomerase holoenzyme. J Biol Chem. 2012;287:34455–64. doi: 10.1074/jbc.M112.394767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Shay JW. Role of telomeres and telomerase in aging and cancer. Cancer Discov. 2016;6:584–93. doi: 10.1158/2159-8290.CD-16-0062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Shefer K, Brown Y, Gorkovoy V, Nussbaum T, Ulyanov NB, Tzfati Y. A triple helix within a pseudoknot is a conserved and essential element of telomerase RNA. Mol Cell Biol. 2007;27:2130–43. doi: 10.1128/MCB.01826-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Shukla S, Schmidt JC, Goldfarb KC, Cech TR, Parker R. Inhibition of telomerase RNA decay rescues telomerase deficiency caused by dyskerin or PARN defects. Nat Struct Mol Biol. 2016;23:286–92. doi: 10.1038/nsmb.3184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Singh M, Wang Z, Koo B-K, Patel A, Cascio D, et al. Structural basis for telomerase RNA recognition and RNP assembly by the holoenzyme La family protein p65. Mol Cell. 2012;47:16–26. doi: 10.1016/j.molcel.2012.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Stewart JA, Chaiken MF, Wang F, Price CM. Maintaining the end: roles of telomere proteins in end-protection, telomere replication and length regulation. Mutat Res. 2012;730:12–19. doi: 10.1016/j.mrfmmm.2011.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Stewart JA, Wang F, Chaiken MF, Kasbek C, Chastain PD, II, et al. Human CST promotes telomere duplex replication and general replication restart after fork stalling. EMBO J. 2012;31:3537–49. doi: 10.1038/emboj.2012.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Stone MD, Mihalusova M, O’Connor CM, Prathapam R, Collins K, Zhuang X. Stepwise protein-mediated RNA folding directs assembly of telomerase ribonucleoprotein. Nature. 2007;446:458–61. doi: 10.1038/nature05600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Sugitani N, Chazin WJ. Characteristics and concepts of dynamic hub proteins in DNA processing machinery from studies of RPA. Prog Biophys Mol Biol. 2015;117:206–11. doi: 10.1016/j.pbiomolbio.2014.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Sun J, Yang Y, Wan K, Mao N, Yu T-Y, et al. Structural bases of dimerization of yeast telomere protein Cdc13 and its interaction with the catalytic subunit of DNA polymerase α. Cell Res. 2011;21:258–74. doi: 10.1038/cr.2010.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Sun J, Yu EY, Yang Y, Confer LA, Sun SH, et al. Stn1–Ten1 is an Rpa2–Rpa3-like complex at telomeres. Genes Dev. 2009;23:2900–14. doi: 10.1101/gad.1851909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Surovtseva YV, Churikov D, Boltz KA, Song X, Lamb JC, et al. Conserved telomere maintenance component 1 interacts with STN1 and maintains chromosome ends in higher eukaryotes. Mol Cell. 2009;36:207–18. doi: 10.1016/j.molcel.2009.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Theimer CA, Blois CA, Feigon J. Structure of the human telomerase RNA pseudoknot reveals conserved tertiary interactions essential for function. Mol Cell. 2005;17:671–82. doi: 10.1016/j.molcel.2005.01.017. [DOI] [PubMed] [Google Scholar]
- 158.Theimer CA, Feigon J. Structure and function of telomerase RNA. Curr Opin Struct Biol. 2006;16:307–18. doi: 10.1016/j.sbi.2006.05.005. [DOI] [PubMed] [Google Scholar]
- 159.Theimer CA, Jády BE, Chim N, Richard P, Breece KE, et al. Structural and functional characterization of human telomerase RNA processing and Cajal body localization signals. Mol Cell. 2007;27:869–81. doi: 10.1016/j.molcel.2007.07.017. [DOI] [PubMed] [Google Scholar]
- 160.Townsley DM, Dumitriu B, Young NS. Bone marrow failure and the telomeropathies. Blood. 2014;124:2775–83. doi: 10.1182/blood-2014-05-526285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Tycowski KT, Shu M-D, Kukoyi A, Steitz JA. A conserved WD40 protein binds the Cajal body localization signal of scaRNP particles. Mol Cell. 2009;34:47–57. doi: 10.1016/j.molcel.2009.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Tzfati Y, Fulton TB, Roy J, Blackburn EH. Template boundary in a yeast telomerase specified by RNA structure. Science. 2000;288:863–67. doi: 10.1126/science.288.5467.863. [DOI] [PubMed] [Google Scholar]
- 163.Tzfati Y, Knight Z, Roy J, Blackburn EH. A novel pseudoknot element is essential for the action of a yeast telomerase. Genes Dev. 2003;17:1779–88. doi: 10.1101/gad.1099403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Upton HE, Chan H, Feigon J, Collins K. Shared subunits of Tetrahymena telomerase holoenzyme and replication protein A have different functions in different cellular complexes. J Biol Chem. 2017;292:217–28. doi: 10.1074/jbc.M116.763664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Upton HE, Hong K, Collins K. Direct single-stranded DNA binding by Teb1 mediates the recruitment of Tetrahymena thermophila telomerase to telomeres. Mol Cell Biol. 2014;34:4200–12. doi: 10.1128/MCB.01030-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Venteicher AS, Abreu EB, Meng Z, McCann KE, Terns RM, et al. A human telomerase holoen-zyme protein required for Cajal body localization and telomere synthesis. Science. 2009;323:644–48. doi: 10.1126/science.1165357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Vogan JM, Zhang X, Youmans DT, Regalado SG, Johnson JZ, et al. Minimized human telomerase maintains telomeres and resolves endogenous roles of H/ACA proteins, TCAB1, and Cajal bodies. eLife. 2016;5:e18221. doi: 10.7554/eLife.18221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Wan B, Tang T, Upton H, Shuai J, Zhou Y, et al. The Tetrahymena telomerase p75–p45–p19 subcomplex is a unique CST complex. Nat Struct Mol Biol. 2015;22:1023–26. doi: 10.1038/nsmb.3126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Wang F, Podell ER, Zaug AJ, Yang Y, Baciu P, et al. The POT1-TPP1 telomere complex is a telomerase processivity factor. Nature. 2007;445:506–10. doi: 10.1038/nature05454. [DOI] [PubMed] [Google Scholar]
- 170.Wang F, Stewart J, Price CM. Human CST abundance determines recovery from diverse forms of DNA damage and replication stress. Cell Cycle. 2014;13:3488–98. doi: 10.4161/15384101.2014.964100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Wang Y, Yesselman JD, Zhang Q, Kang M, Feigon J. Structural conservation in the template/pseudoknot domain of vertebrate telomerase RNA from teleost fish to human. PNAS. 2016;113:E5125–34. doi: 10.1073/pnas.1607411113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Watson JM, Riha K. Comparative biology of telomeres: where plants stand. FEBS Lett. 2010;584:3752–59. doi: 10.1016/j.febslet.2010.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Wellinger RJ, Zakian VA. Everything you ever wanted to know about Saccharomyces cerevisiae telomeres: beginning to end. Genetics. 2012;191:1073–105. doi: 10.1534/genetics.111.137851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Wenz C, Enenkel B, Amacker M, Kelleher C, Damm K, Lingner J. Human telomerase contains two cooperating telomerase RNA molecules. EMBO J. 2001;20:3526–34. doi: 10.1093/emboj/20.13.3526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Wu RA, Collins K. Human telomerase specialization for repeat synthesis by unique handling of primer-template duplex. EMBO J. 2014;33:921–35. doi: 10.1002/embj.201387205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Wu RA, Dagdas YS, Yilmaz ST, Yildiz A, Collins K. Single-molecule imaging of telomerase reverse transcriptase in human telomerase holoenzyme and minimal RNP complexes. eLife. 2015;4:e08363. doi: 10.7554/eLife.08363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Wyatt HD, West SC, Beattie TL. InTERTpreting telomerase structure and function. Nucleic Acids Res. 2010;38:5609–22. doi: 10.1093/nar/gkq370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Xie M, Mosig A, Qi X, Li Y, Stadler PF, Chen JJ-L. Structure and function of the smallest vertebrate telomerase RNA from teleost fish. J Biol Chem. 2008;283:2049–59. doi: 10.1074/jbc.M708032200. [DOI] [PubMed] [Google Scholar]
- 179.Xie M, Podlevsky JD, Qi X, Bley CJ, Chen JJ-L. A novel motif in telomerase reverse transcriptase regulates telomere repeat addition rate and processivity. Nucleic Acids Res. 2010;38:1982–96. doi: 10.1093/nar/gkp1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Xin H, Liu D, Wan M, Safari A, Kim H, et al. TPP1 is a homologue of ciliate TEBP-β and interacts with POT1 to recruit telomerase. Nature. 2007;445:559–62. doi: 10.1038/nature05469. [DOI] [PubMed] [Google Scholar]
- 181.Ye JZ-S, Hockemeyer D, Krutchinsky AN, Loayza D, Hooper SM, et al. POT1-interacting protein PIP1: a telomere length regulator that recruits POT1 to the TIN2/TRF1 complex. Genes Dev. 2004;18:1649–54. doi: 10.1101/gad.1215404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Yu EY, Sun J, Lei M, Lue NF. Analyses of Candida Cdc13 orthologues revealed a novel OB fold dimer arrangement, dimerization-assisted DNA binding, and substantial structural differences between Cdc13 and RPA70. Mol Cell Biol. 2012;32:186–98. doi: 10.1128/MCB.05875-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Yu Y-T, Meier UT. RNA-guided isomerization of uridine to pseudouridine—pseudouridylation. RNA Biol. 2014;11:1483–94. doi: 10.4161/15476286.2014.972855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Zaug AJ, Podell ER, Cech TR. Mutation in TERT separates processivity from anchor-site function. Nat Struct Mol Biol. 2008;15:870–72. doi: 10.1038/nsmb.1462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Zaug AJ, Podell ER, Nandakumar J, Cech TR. Functional interaction between telomere protein TPP1 and telomerase. Genes Dev. 2010;24:613–22. doi: 10.1101/gad.1881810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Zeng Z, Min B, Huang J, Hong K, Yang Y, et al. Structural basis for Tetrahymena telomerase processivity factor Teb1 binding to single-stranded telomeric-repeat DNA. PNAS. 2011;108:20357–61. doi: 10.1073/pnas.1113624108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Zhang Q, Kim N-K, Feigon J. Architecture of human telomerase RNA. PNAS. 2011;108:20325–32. doi: 10.1073/pnas.1100279108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Zhang Q, Kim N-K, Peterson RD, Wang Z, Feigon J. Structurally conserved five nucleotide bulge determines the overall topology of the core domain of human telomerase RNA. PNAS. 2010;107:18761–68. doi: 10.1073/pnas.1013269107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Zhong FL, Batista LF, Freund A, Pech MF, Venteicher AS, Artandi SE. TPP1 OB-fold domain controls telomere maintenance by recruiting telomerase to chromosome ends. Cell. 2012;150:481–94. doi: 10.1016/j.cell.2012.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]