Abstract
Eukaryotic gene expression is tightly regulated transcriptionally and post-transcriptionally by a host of noncoding (nc)RNAs. The best-studied class of short ncRNAs, microRNAs, mainly repress gene expression post-transcriptionally. Long noncoding (lnc)RNAs, which comprise RNAs differing widely in length and function, can regulate gene transcription as well as post-transcriptional mRNA fate. Collectively, ncRNAs affect a broad range of age-related physiologic deteriorations and pathologies, including reduced cardiovascular vigor and age-associated cardiovascular disease. This review presents an update of our understanding of regulatory ncRNAs contributing to cardiovascular health and disease as a function of advancing age. We will discuss (1) regulatory ncRNAs that control aging-associated cardiovascular homeostasis and disease, (2) the concepts, approaches, and methodologies needed to study regulatory ncRNAs in cardiovascular aging and (3) the challenges and opportunities that age-associated regulatory ncRNAs present in cardiovascular physiology and pathology. This article is part of a Special Issue entitled “CV Aging”.
Keywords: MicroRNA, Long noncoding RNA, Aging, Cardiovascular disease
1. Introduction
1.1. Gene regulation by ncRNAs
The adaptation of mammalian cells to external and internal signals requires precise transcriptional and post-transcriptional modulation of gene expression patterns. Transcription is controlled mainly through changes in chromatin composition and the recruitment of transcription factors, while post-transcriptional control is elicited through pre-mRNA splicing as well as mRNA transport, stability, storage, and translation. Noncoding (nc)RNAs, the main focus of this review, influence all of these gene regulatory levels [1]. Nuclear ncRNAs control transcription by associating with chromatin and transcriptional activators and repressors, and they control early post-transcriptional steps in gene regulation, such as pre-mRNA splicing [2]. Cytoplasmic ncRNAs regulate mRNA turnover rate, mRNA storage and localization, and mRNA recruitment to ribosomes [3,4]. Since aberrant gene control underlies the age-related decline in physiologic function and age-associated diseases, it is critically important to understand how gene expression is altered in aging. Gene regulation by DNA- and RNA-binding proteins affecting aging pathology and physiology has been studied for decades. However, the deep and broad impact of regulatory ncRNAs in aging is only now coming into view.
ncRNAs comprise a large and heterogeneous family. It includes many RNAs that have been known for decades and mainly have housekeeping functions, such as ribosomal RNAs (rRNAs), small nuclear RNAs (snRNAs), small nucleolar RNAs (snoRNAs), and transfer RNAs (tRNAs). However, the advent of tiling arrays and RNA-sequencing over the past ~15 years has revealed that large stretches of chromosomes previously believed not to be transcribed, are actually transcribed and encode vast numbers of ncRNAs [5]. Arbitrarily divided into long (lncRNAs, >200 nt), and short (<200 nt) ncRNAs, these regulatory transcripts display distinct temporal and spatial expression patterns. As details of the impact of ncRNAs on molecular and cellular processes are becoming better understood, their roles in aging-associated physiologic processes and disease conditions are also starting to emerge [6].
1.1.1. microRNAs
Although short RNAs also include piwi-interacting and small interfering RNAs (piRNAs, siRNAs), microRNAs (miRNAs) have been most actively studied in different contexts, including aging [7,8]. MicroRNAs are single-stranded, ~22 nt-long ncRNAs that regulate gene expression mainly by forming partial hybrids with target mRNAs and thereby lowering their translation and/or stability [9]. A microRNA is transcribed as a long, primary miRNA (pri-miRNA) which is cleaved by the microprocessor complex (containing the ribonuclease Drosha) to generate a miRNA precursor (pre-miRNA) that is exported to the cytoplasm [4]. In the cytoplasm, the ribonuclease Dicer cleaves the pre-miRNA to yield a ~22-bp-long duplex RNA; one strand of the duplex, the mature miRNA, is loaded onto the RNA-induced silencing complex (RISC), which contains Argonaute (Ago) proteins [10,11]. Each Ago-microRNA complex targets a subset of mRNAs forming partial hybrids, often at the 3′-untranslated region (UTR) of the mRNAs and frequently relying on the miRNA ‘seed’ region (nucleotides 2–7). Several microRNAs often work in concert to lower the expression of a shared target mRNA. MicroRNAs have been implicated in virtually all areas of mammalian homeostatic gene regulation, and disruption of miRNA function has been causally linked to a variety of cardiovascular diseases (CVDs) that rise with advancing age [7,8,12,13].
1.1.2. LncRNAs
LncRNAs can be classified according to their genomic localization and biogenesis: lincRNAs are expressed from intergenic regions, antisense lncRNAs are expressed from the opposite strand of mRNAs and lncRNAs, pseudogene-encoded lncRNAs are transcribed from vestigial genes that lost their coding potential, long intronic ncRNAs are present in introns of annotated genes, promoter-associated lncRNAs are transcribed from the promoter regions of coding mRNAs, and circular RNAs are often generated by the splicing machinery [14].
This large class of ncRNAs can also be classified according to their molecular mechanism of action. (1) Epigenetic regulation: some nuclear lncRNAs regulate gene expression by serving as scaffolds, bridges, and tethers of factors that regulate the state of the chromatin. By recruiting chromatin-modification enzymes, lncRNAs can transiently or permanently activate/inactivate genes and chromosomal regions. The ability to recognize DNA sequences uniquely enables lncRNAs to elicit highly precise, sequence-specific actions on transcription. Examples of chromatin-remodeling lncRNAs are XIST and HOTAIR [15,16]. (2) Transcriptional regulation: lncRNAs can also assemble transcriptional activators and repressors to modulate the rates of RNA polymerase II initiation and elongation. Examples of transcription-modulatory lncRNAs include NEAT1, ANRIL (CDKN2B-AS), GAS5, and MALAT1 [16,17]. (3) Nuclear compartmentalization: some nuclear lncRNAs have been implicated in maintaining nuclear structures, including nuclear speckles, paraspeckles, and interchromatin granules; examples include TUG1, MALAT1, NEAT1, and FIRRE [17]. (4) Post-transcriptional gene regulation: lncRNAs that basepair with mRNAs can modulate the translation and/or the stability of target mRNAs (e.g., 1/2-sbsRNAs, LINCRNAP21 [18,19]) while lncRNAs that do not basepair can affect precursor mRNA splicing and translation by acting as cofactors or competitors of RNA-binding proteins [20,21]. (5) Competing endogenous RNAs (ceRNAs): although lncRNAs are generally low-abundance transcripts, some lncRNAs accumulate because they are highly stable (e.g., circular RNAs) and can function as decoys or sponges for microRNAs (e.g., LINCMD1 [22]) and possibly other regulatory factors [23]. (6) Post-translational gene regulation on protein turnover has also been reported for the lncRNA HOTAIR, which scaffolds pairs of E3 ubiquitin ligases and their substrates [24]. As reviewed comprehensively elsewhere [25,26], lncRNA-modulated gene expression patterns are relevant to a variety of cell functions with impact upon many age-associated conditions.
1.2. Aging and CVDs
Age is the most important risk factor for CVDs, the most prominent cause of death worldwide [http://www.who.int/cardiovascular_diseases/resources/atlas/en/.]. Advancing age increases exposure to cardiovascular risk factors, and elicits intrinsic cardiovascular changes that compromise both the cardiovascular reserve capacity and the threshold for primary diseases to manifest [27,28].
The aging heart is characterized by several detrimental changes such as left ventricular hypertrophy, diastolic dysfunction, valve degeneration, increased cardiac fibrosis, increased prevalence of atrial fibrillation, and decreased maximal exercise capacity [29,30]. Endothelial function, particularly endothelium-dependent dilation [31,32], also decreases with aging; this decline is due, at least in part, to the reduced bioavailability of nitric oxide (NO) following inflammation and oxidative stress in the elderly [33,34]. Dysfunctional endothelium correlates with higher systolic and pulse pressure, and, consequently, with increased risk of age-associated conditions such as atherosclerosis, hypertension, acute myocardial infarction (AMI), and stroke [35]. Additionally, the walls of the arteries and arterioles become thicker and less elastic, making the vessels stiffer and less resilient [35,36].
2. Noncoding RNA in cardiovascular aging and age-associated CVDs
Our understanding of ncRNA function in the molecular mechanisms underpinning the age-associated changes in the cardiovascular systems is much more advanced for miRNAs than for lncRNAs. Thus, a provisional but already delineated scenario is emerging, with specific miRNAs regulating a variety of cardiovascular functions in both homeostasis and disease. By contrast, for lncRNAs, in most circumstances, we have just started scratching the surface, and, at the moment, the existing data linking individual lncRNAs and age-associated CVDs are mainly correlative. While many more studies specifically aimed at dissecting ncRNA functions in cardiovascular system aging are needed, the ncRNAs already identified in age-related CVDs are summarized in Table 1.
Table 1.
ncRNAs and age-related cardiovascular diseases.
| ncRNA | Disease/condition | Modulation | Refs |
|---|---|---|---|
| miR-146 | Heart failure | Up | [54] |
| miR-17-92 cluster | Heart failure (mouse) | Down | [69] |
| miR-1 | Heart failure, hypertrophy (mouse) | Up | [79] |
| miR-216a | Heart failure | Up | [188] |
| miR-29 family | Aorta aneurysm | Up | [53] |
| miR-155 | Atherosclerotic lesions | Up | [59] |
| miR-21 | Atherosclerotic plaques | Up | [96] |
| miR-216a | Atherosclerotic plaques | Up | [103] |
| miR-217 | Atherosclerotic plaques | Up | [109] |
| miR-1 | Myocardial infarction (mouse) | Up | [132] |
| miR-34a | Myocardial infarction (mouse) | Up | [61] |
| miR-200c | Peripheral ischemia | Up | [90] |
| miR-210 | Peripheral ischemia | Up | [99] |
| miR-145 | Ischemia/reperfusion | Down | [97] |
| Mkk7 | Heart failure (mouse) | Down | [126] |
| Mhrt | Heart failure (mouse) | Down | [132] |
| H19 | Heart failure | Up | [189] |
| Alc1-AS | Hypertrophy (mouse) | Up | [190] |
| Chrf | Hypertrophy (mouse) | Down | [125] |
| Mirt1/Mirt2 | Myocardial infarction (mouse) | Up | [137] |
2.1. miRNAs
MiRNAs are involved in virtually all processes of physiology and disease, including aging [11,37,38]. Some 65 miRNAs have been identified that display significantly different expression levels between young adult heart and older adult heart [39]. Here, we will review the miRNAs related to the dysfunction of several well-recognized age-associated processes affecting the cardiovascular system, including senescence, nutrient sensing, oxidative stress response, inflammation, and silent mating type information regulation 2 homolog (SIRT1)-regulated events.
2.1.1. Senescence
Cells from normal animal tissue placed in culture divide for a finite number of times but eventually stop proliferating and enter a state of terminal growth arrest named senescence [40]. Replicative senescence is triggered by the progressive shortening of the telomeres that results from cell division and by exposure to a range of damaging conditions [41]. Senescent cells do not divide, but they can remain viable and metabolically active for a long time, displaying a large and flat morphology, cytoplasmic vacuoles, enhanced autophagy, and elevated activity of lysosomal β-galactosidase [11,42]. Many senescent cells also display a characteristic senescence-associated secretory phenotype (SASP) [43], whereby they secrete many cytokines and chemokines (e.g., GM-CSF, IL-6, IL-8, IL-1α), and matrix metalloproteases (e.g., MMP-1, MMP-3). Another hallmark of senescence is the activation of tumor suppression pathways, mainly those controlled by p53/p21 and pRB/p16 [41]. Finally, since senescent cells frequently experience oxidative and genotoxic damage, they generally express DNA-damage response proteins, such as γ-H2AX, NIBRIN (NBS), MDC1, and 53BP1 [41].
Cellular senescence has been studied most extensively in cultured cells, but there is broad recognition that senescence occurs in vivo and affects aging phenotypes profoundly [41]. Since senescent cells accumulate in tissues as the organism ages, their metabolic behavior and their signature gene expression profile have been linked to a number of age-related physiologic and pathologic changes including CVDs [44, 45]. For instance, accumulation of senescent endothelial cells (ECs) in atherosclerotic lesions is an important factor contributing to age-associated arterial dysfunction [46,47].
Several transcription factors, such as p53 and proteins in the AP-1, E2F, Id and Ets families, have been implicated in driving senescence. In addition, several RNA-binding proteins can regulate senescence via post-transcriptional gene regulation, including human antigen R (HuR), AU-binding factor 1 (AUF1), and tristetraprolin (TTP), through their association with target mRNAs that encode senescence factors [48,49]. However, a growing number of regulatory miRNAs also influence senescence-associated gene expression patterns by regulating key molecular pathways (Fig. 1):
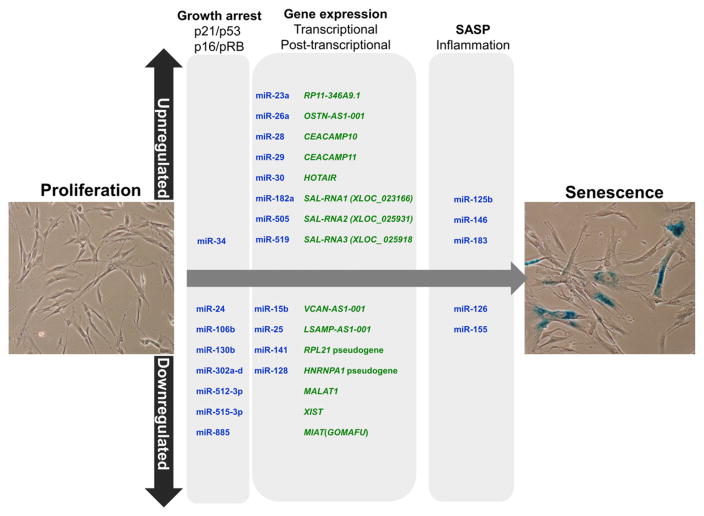
Fig. 1.
Senescence-associated ncRNAs. Several studies have identified ncRNAs upregulated and downregulated in senescence, but only few have been shown to modulate senescence experimentally. The figure shows microRNAs (blue) and lncRNAs (green) directly implicated in molecular pathways that govern cell senescence.
pRB/p16 pathway. Senescent cells have active (hypophosphorylated) pRB, resulting from risen levels of two potent inhibitors of cyclin-dependent kinases (cdks, which are pRB kinases): p21 (in part upregulated via p53) and p16. Some miRNAs that reduce p21 abundance (e.g., miR-106b, miR-130b) decline during senescence [50,51].
p53/p21 pathway. Senescent cells have elevated levels of p53, a protein subject to negative regulation by MCM5 (which is repressed by miR-885-5p) [52]. Elevated p53 transcriptionally upregulates miR-34a, which in turn reduces the levels of several proliferative proteins (E2F, c-Myc, cyclins and cdks) and anti-apoptotic proteins (Bcl-2 and SIRT1).
Senescent cell-specific gene expression programs. Numerous proteins modulate senescence-associated gene transcription, such as PCGF4, HMGA2, RARγ and B-Myb (MYBL2), while other proteins affect senescence-associated gene expression post-transcriptionally, including the splicing factor ASF/SF2 and the mRNA stability and translation regulator HuR. Each of these proteins is also subject to control by senescence-associated miRNAs [11], and some of these miRNAs cross-talk with the p53/p21 and the p16/RB pathways (e.g., HMGA2 represses p21 transcription and Bmi-1 represses p16 transcription). Importantly, senescence factors can also directly modulate the expression of senescence-regulatory miRNAs; for example, pRB transcriptionally increases miR-29/miR-30 levels and this regulation can impact upon the deposition of extracellular matrix (ECM). In aged aortas, increased levels of miR-29 family members were causally associated to the downregulation of ECM components, linking miR-29 to a decrease in ECM proteins and the age-associated formation of aorta aneurysms [53].
SASP. Secretion of some SASP factors (e.g., IL-6, IL-8) is under control of IRAK1, a protein whose levels are reduced by the senescence-associated miR-146. Factors secreted through the SASP cause inflammation, contributing to a general increase of inflammation also termed “inflammageing”. Accordingly, miR-146 has been shown to be increased in senescent ECs [54] as well as in endothelial progenitor cells (EPCs) from heart failure (HF) patients [55].
The fact that miR-126 controls endothelial inflammation, at least in part by modulating the expression of VCAM-1 cell adhesion proteins [56], helps to explain why low levels of circulating miR-126 were found associated with in CVDs and diabetes [57], two conditions characterized by EC activation. These proinflammatory effects may also be linked to miR-126-mediated downregulation of NF-κBIB (IκBβ), a potent inhibitor of NF-κB signaling [58]. High levels of miR-155 were found in proinflammatory macrophages and atherosclerotic lesions, although the effects of miR-155 appear to differ between early and advanced atherosclerosis [59]. Interestingly, miR-155 lowers angiotensin II type 1 receptor (AT1R) activity leading to endothelial dysfunction, structural remodeling, and vascular inflammation [60].
Certain senescence-associated miRNAs function at the convergence of different pathways. For instance, miR-34a levels are increased in aged hearts, but since cardiomyocytes are mostly post-mitotic, miR-34a likely does not influence proliferation in these cells. Instead, cardiomyocyte miR-34a lowers Ppp1r10 (PNUTS) [61], a protein that interacts with the telomere regulator TRF2 to control DNA repair and reduce telomere attrition. PNUTS protects cardiomyocytes from oxidative damage and death and both increased PNUTS and reduced miR-34a prevent the deterioration of cardiac contractile function after AMI [61].
2.1.2. Fibrosis
Aging hearts are characterized by increased expression of ECM proteins [62], fibronectin [63], thrombospondin-2 (TSP-2) [64] and connective tissue growth factor (CTGF) [65]. These elevated levels are due, at least in part, to the age-dependent accumulation of senescent cells in these tissues, which promote inflammation and cardiac fibrosis [66–68]. Notably, miR-18a and miR-19a/b, from the miR-17–92 cluster, are causally linked to age-related fibrotic remodeling of the heart upon HF [69]. TSP-1 and CTGF, which contribute to the fibrotic process, are targets of miR-18a and miR-19a and their levels are inversely correlated.
Increased levels of miR-29 family members were causally associated to the downregulation of ECM components in aged aortas, linking miR-29 to the decrease in ECM proteins and the age-associated formation of aorta aneurysms [53]. Fibrotic myocardium also shows high levels of miR-21; by lowering the abundance of target protein SPRY2, miR-21 activates signaling through ERK/MAPK, leading to increased fibrosis and decreased apoptosis [70]. The role of miR-21 in age-associated fibrosis has not been elucidated, but it is worth noting that miR-21 levels increase in aged hearts [39]. Another microRNA linked to fibrosis, miR-30, targets the profibrotic growth factor CTGF. The downregulation of miR-30 in left ventricle hypertrophy is associated to increased production of ECM components [71].
2.1.3. Nutrient sensing pathways
Major metabolic pathways include those governed by insulin, insulin-like growth factor 1 (IGF1), and target of rapamycin (TOR). Excessive food intake leading to secretion of insulin, which facilitates the uptake of glucose to be stored as fat, the accumulation of visceral fat, and insulin resistance, are major risk factors for CVDs [72,73], including atherosclerosis [74–76]. The tendency of centenarians to maintain high insulin sensitivity [77] suggests that systemic insulin/IGF1 pathway activation itself confers protection from CVD in the absence of obesity.
The influence of miR-1 in IGF1 signaling in cardiomyocytes is well documented. miR-1 directly represses IGF1 mRNA in cardiac and skeletal muscle [78], and miR-1 levels are inversely correlated with IGF1 protein levels in models of cardiac hypertrophy and failure [79]. In this regard, Shan et al. [80] reported that the levels of IGF1 repressors miR-1 and miR-206 increased in the ischemic zone in rat AMI. Overexpression of miR-1 or silencing of IGF1 in H9C2 rat cardiomyoblasts upon serum withdrawal or hypoxia elevated caspase-3 activity and mitochondrial potential, in agreement with the anti-apoptotic function of IGF1 [81,82].
Moderate caloric restriction (CR) without malnutrition is a dietary regimen recognized to delay aging and extend lifespan [83]. In aged ECs, miR-144 abundance increases while that of its predicted target NF-E2-related factor 2 (Nrf2) decreases. This influence is potentially important, since activation of Nrf2 upon CR has potent anti-oxidative, pro-angiogenic, and anti-inflammatory effects in mouse ECs [84].
2.1.4. Oxidative stress
Mitochondrial dysfunction is another hallmark of aging-associated diseases. As cells and organisms age, the efficacy of the respiratory chain declines, causing increased electron leakage and reduced ATP generation [85]. Additionally, the production of reactive oxygen species (ROS) by dysfunctional mitochondria can trigger cellular events culminating in a wide range of CVDs [86,87]. Different miRNAs modulated by oxidative stress are involved in endothelial and vascular dysfunction and are associated to CVDs caused by excessive ROS production [88].
miR-200, miR-141
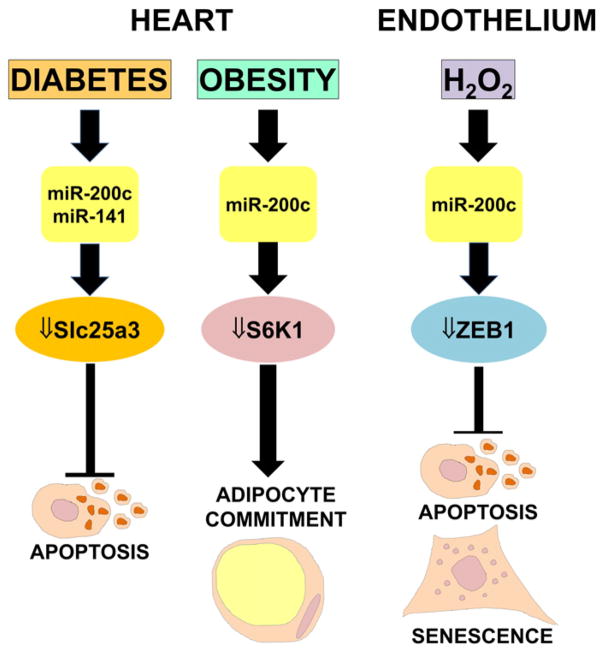
miRNA profiling experiments have revealed that the miR-200 family members miR-200c and miR-141 are upregulated in hydrogen peroxide (H2O2)-treated ECs [89] (Fig. 2). Moreover, the miR-200 family is also induced following acute ischemia, which generates oxidative stress [90]. Accordingly, in p66ShcA −/− mice, which display lower levels of oxidative stress upon ischemia, miR-200c and miR-200b are less upregulated [89]. The pro-survival protein ZEB1 was identified as a direct target of miR-200c, leading to the discovery that ZEB1 knockdown recapitulated miR-200c-triggered responses and that miR-200-mediated inhibition of ZEB1 was a key effector of ROS-induced apoptosis and senescence. In keeping with the increased oxidative stress levels observed in diabetes mellitus, miR-200c and miR-141 are among the most upregulated miRNAs in diabetic mouse heart [91]. The levels of miR-141-target SLC25A3 (solute carrier family 25 member 3), which provides inorganic phosphate to the mitochondrial matrix and is essential for ATP production, declines in type 1 diabetes, affecting adversely ATP production and cell viability. Finally, miR-200c, which is not normally expressed in heart, is readily detectable in the heart of Zucker obese rats, where it participates in adaptive mechanisms compensating excessive activation of the nutrient sensor kinase S6K1 [92].
Fig. 2.
miR-200 regulates endothelial dysfunction and cardiovascular complications linked to diabetes and obesity. ROS production or pathologies associated to elevated ROS production play a causal role in endothelial and cardiovascular diseases. (Left) MiR-200c and miR-141 are among the most highly upregulated miRNAs in diabetic mouse heart; accordingly, the target of miR-200 SLC25A3, a protein essential for ATP production, declines in type 1 diabetes, reducing ATP production and cell viability. (Center) In Zucker obese rats, a genetic model for obesity, hypertension and cardiac dysfunction, elevated miR-200c activates a compensatory mechanism to down-regulate excessive activation of the nutrient sensor kinase S6K1, involved in adipocyte lineage commitment. (Right) MiR-200c levels rise following oxidative stress; the ensuing inhibition of its target ZEB1 induces cell growth arrest, apoptosis and cellular senescence.
miR-21
Treatment of rat vascular smooth muscle cells (VSMCs) with H2O2 elevated miR-21, which in turn protected VSMCs from H2O2-dependent apoptosis and death. A major target of miR-21-mediated repression is PDCD4 (Programmed cell death 4), a pro-apoptotic protein that inhibits the activity of the transcription factor AP-1 [93], providing a plausible mechanism whereby elevated miR-21 protects ECs from oxidative injury. Among the factors triggering the development of atherosclerotic lesions are disturbances in flow dynamics [94]. Shear stress increases miR-21 levels, which contributes to protecting ECs by increasing endothelial nitric oxide synthase (eNOS) and nitric oxide (NO) production [95]. However, miR-21 function is complex and context-dependent; in this regard, miR-21 has detrimental effects in atherosclerotic plaques, where high levels of miR-21 decreases the functions of the mitochondrial defense proteins SPRY2 and superoxide dismutase-2 (SOD-2). These effects, in turn, lead to ERK/MAPK activation, resulting in increased ROS levels and EPC migratory defects [96].
miR-145
After AMI, during ischemia–reperfusion injury, ROS typically causes oxidative stress in cardiomyocytes. Recently, Li et al. found reduced miR-145 levels in both ischemia/reperfused heart and H2O2-treated cardiomyocytes [97]. Given that miR-145 protected against oxidative stress-induced cardiomyocyte apoptosis by inhibiting expression of its target BNIP3 and by preventing ROS generation, miR-145 was proposed to function as a cardioprotective molecule capable of counteracting ROS toxicity.
miR-210
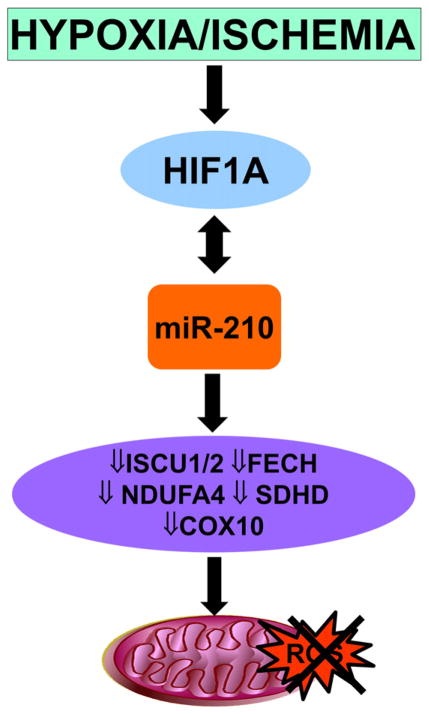
Another potent inducer of mitochondrial dysfunction and oxidative stress is hypoxia [98]. miR-210 is robustly induced by hypoxia, contributing to the oxidative phosphorylation decline observed in low oxygen by repressing ISCU1, ISCU2, COX10 and FECH mRNAs. These transcripts, in turn, encode proteins which directly or indirectly affect the mitochondrial respiratory chain [98]. Accordingly, miR-210 inhibition triggered mitochondrial dysfunction, oxidative stress and increased tissue damage upon ischemia [99] (Fig. 3).
Fig. 3.
miR-210 regulates mitochondrial activity. miR-210 represses target ISCU1, ISCU2, COX10 and FECH mRNAs, encoding proteins that directly or indirectly affect the mitochondrial respiratory chain and reduce ROS production.
2.1.5. Autophagy
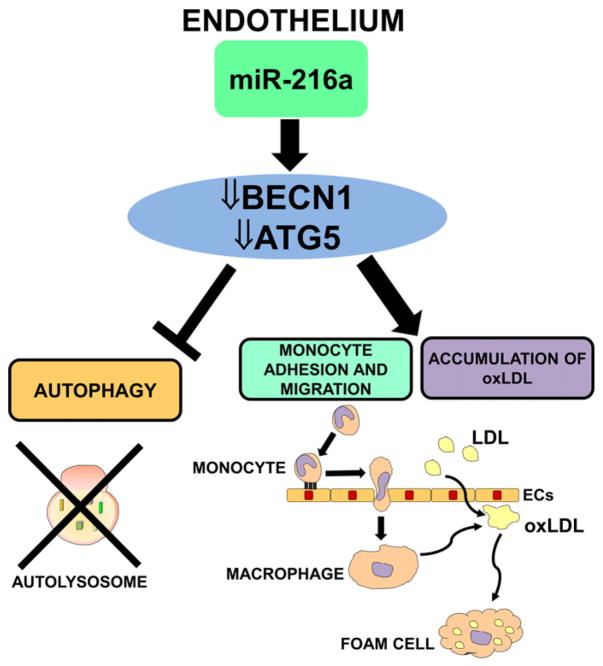
Autophagy is a recycling mechanism whereby cells degrade unnecessary cytoplasmic material and organelles in lysosomes [100]. A decline of the autophagy factors has been observed in aging tissues and in age-related disorders [101,102]. Autophagy seems to play a protective role during atherosclerosis, because it stabilizes plaques through the processing of oxidatively modified proteins, whereas acquired defects in plaques autophagy exacerbate atherosclerosis [100]. Menghini et al. [103] showed that expression of BECN1, ATG5 and LC3B-II/MAP1LC3B, key autophagy-related proteins, decreased during EC senescence, suggesting that the autophagic process is impaired with aging (Fig. 4). They also showed that miR-216a was induced during endothelial aging and that it was able to reduce the expression of BECN1 and ATG5. As a consequence, miR-216 also regulated autophagy induced by oxidized low-density lipoprotein (ox-LDL) treatment, stimulated ox-LDL accumulation, and promoted monocyte adhesion in ECs. The role of miR-216a seems to be particularly important not only in the development of atherosclerosis, but also in HF, since miR-216a expression in human failing hearts was inversely correlated with ejection fraction and with expression levels of BECN1 and ATG5 mRNAs [103].
Fig. 4.
miR-216a regulates autophagy. miR-216a activity on its targets BECN1 and ATG5 inhibits autophagy and stimulates ox-LDL accumulation in EC as well as monocyte adhesion and migration.
2.1.6. SIRT1 pathway
The “Silent mating type information regulation 2 homolog” (sirtuin 1 or SIRT1) is an NAD+-dependent class III histone deacetylase (HDAC) that can extend the lifespan of organisms and its levels are elevated with CR [104,105]. Via deacetylation, SIRT1 activates a myriad of stress-responsive transcription factors, co-regulators and enzymes, thus playing a direct role in metabolic control [106]. In ECs, SIRT1 upregulation and/or activation is associated with beneficial effects on ECs, while excessive ROS and aging decreases SIRT1 expression leading to endothelial dysfunction [107].
2.1.6.1. miR-217
This miRNA shows increased plasma levels with advancing age and decreased levels by CR in mouse [108]. Endothelial senescence is affected by this age-associated increase in miR-217, which lowers SIRT1 levels. In turn, decreasing SIRT1 levels promotes the acetylation state of Forkhead box protein O1 (FoxO1) and eNOS, reducing their activity [109] (Fig. 5). Accordingly, miR-217 correlates negatively with SIRT1 expression levels in human atherosclerotic plaques and with FOXO1 acetylation status [109].
Fig. 5.
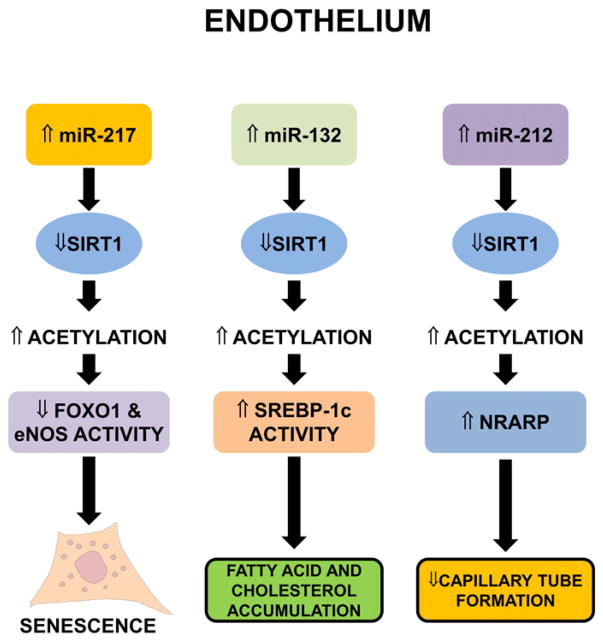
SIRT1 regulation by miRNAs in ECs. miR-217 downregulates SIRT1 expression. This leads to lower deacetylation of its targets FOXO1 and eNOS, rendering them inactive and triggering senescence (left). By targeting SIRT1 expression, miR-132 decreases SREBP-1c acetylation and increases its activity, promoting the accumulation of fatty acids and cholesterol (center). In TGF-β-treated ECs, miR-212 reduces SIRT1 abundance, inhibits endothelial migration and capillary tube formation, and stimulates Notch signaling via NRARP (right).
2.1.6.2. miR-132
In ECs overexpressing miR-132, SIRT1 levels were reduced [110] (Fig. 5). By lowering SIRT1 abundance, miR-132 promotes the accumulation of fatty acids and cholesterol in HUVEC cells. In fact, SIRT1 was shown to deacetylate and hence inhibit the sterol regulatory element-binding protein (SREBP)-1c, decreasing its association with lipogenic target genes [111]. In view of these data, SIRT1 may be involved in the fine-tuning of vascular endothelial proinflammatory processes triggered by fatty acid accumulation in vessels.
MiR-132 belongs to the pro-hypertrophic miR-212/132 cluster. In TGF-β-treated ECs, miR-212 expression levels increase, while the expression of its target SIRT1 decreases [112]. Inhibiting miR-212, but not miR-132, restores endothelial migration and capillary tube formation upon TGF-β challenge, while overexpression of SIRT1 rescues.tube formation capacity inmiR-212 overexpressing ECs [112]. Morever, SIRT1 deacetylates Notch, thereby inhibiting signaling through Notch, which negatively regulates angiogenic functions [113]. The inhibition of miR-212 partly normalizes TGF-β-induced overexpression of NRARP (Notch-Regulated Ankyrin Repeat Protein), indicating that TGF-β suppresses SIRT1 and activates Notch signaling in ECs, at least in part, via miR-212 (Fig. 5).
2.1.6.3. miR-34a
This pleiotropic microRNA also regulates cellular senescence and angiogenesis by reducing SIRT1 levels. miR-34a levels increase and SIRT1 levels decrease in aging hearts and senescent ECs [114]. The repression of SIRT1 by miR-34a also decreases the resistance of Bone Marrow Cells (BMCs) to oxidative stress and lowers the ability of EPCs to rescue heart function after AMI [115]. In EPCs, decreased miR-34a due to statin treatment rescues SIRT1 levels, possibly contributing to the beneficial effects of statins on endothelial function in coronary artery disease (CAD) [116].
2.1.6.4. miR-195
In cardiomyocytes, the free fatty acid palmitate increases miR-195 levels, which in turn lowers SIRT1 production and promotes apoptosis, causing lipotoxic cardiomyopathy [117] (Fig. 6).
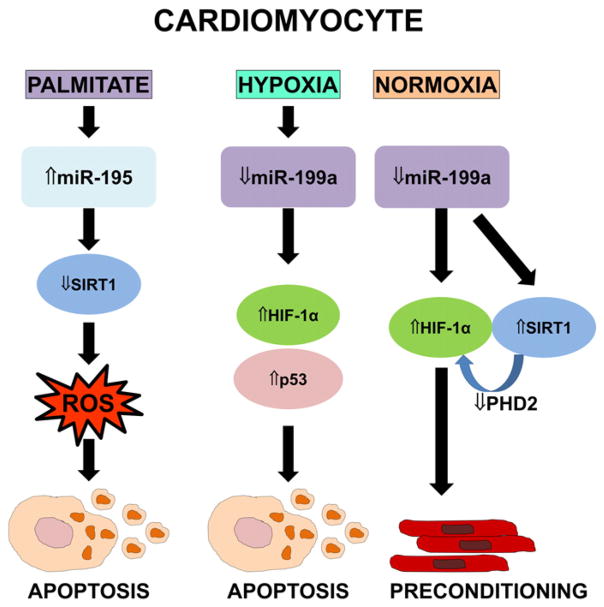
Fig. 6.
SIRT1 regulation by miRNAs in cardiomyocytes. In cardiomyocytes, the free fatty acid palmitate increases miR-195 levels, which in turn inhibits SIRT1 and promotes ROS-triggered apoptosis (left). In hypoxic cardiomyocytes, downregulation of miR-199a increases HIF-1α levels and p53-mediated cardiomyocyte apoptosis (center). In normoxic cardiomyocytes, low miR-199a raises SIRT1 and HIF-1α levels, simulating preconditioning (right).
2.1.6.5. miR-199
As shown in cardiomyocytes, the hypoxia-regulated miR-199a is another repressor of SIRT1 [118] (Fig. 6). Hypoxia acutely lowers miR-199a abundance, causing rapid upregulation of its target hypoxia inducible factor-1α (HIF-1α) and triggering p53-dependent apoptosis. Similarly, inhibition of miR-199a during normoxia induces HIF-1α and SIRT1, which in turn downregulates PHD2, required for HIF-1α stabilization, and thus recapitulates hypoxia preconditioning.
2.2. lncRNAs
As previously noted, very little is known about lncRNAs and aging. However, given the variety of functions and biological mechanisms regulated by lncRNAs and considering the rapid progress in our knowledge of lncRNAs, it is likely that lncRNAs will prove to affect aging broadly. Here, we provide some emerging examples of lncRNAs implicated in regulating key features of aging and their dysregulation in a variety of age-related CVDs.
2.2.1. Senescence-associated long noncoding (SAL)RNAs
RNA-seqencing analysis of proliferating and senescent fibroblasts revealed numerous senescence-associated lncRNAs (SAL-RNAs) that were differentially expressed in senescent cells; some lncRNAs had been previously annotated, including antisense transcripts and pseudogene-encoded transcripts, while many others were novel lncRNAs [119] (Fig. 1). However, just as we have vastly incomplete knowledge of lncRNA function in general, the impact of lncRNAs on senescence is also almost entirely unknown. Some lncRNAs that are beginning to be implicated in senescence are:
2.2.1.1. Naturally occurring antisense lncRNAs (NA-SAL-RNAs)
Some NASAL-RNAs showing higher expression levels in senescent cells were those targeting the mRNAs that encode metallopeptidase ADAMTS19 and the secreted protein osteocrin. On the other hand, NA-SAL-RNAs less abundant in senescent cells targeted the mRNAs encoding versican (an extracellular matrix proteoglycan) and the limbic system-associated membrane protein (LSAMP).
2.2.1.2. LncRNAs encoded by pseudogenes (PE-SAL-RNAs)
PE-SAL-RNAs more highly expressed in senescent cells included the carcinoembryonic antigen-related cell adhesion molecules CEACAMP10 and CEACAMP11, while PE-SAL-RNAs lower in senescent cells include transcripts expressed from the ribosomal protein L21 pseudogene and from the heterogeneous nuclear ribonucleoprotein A1 (HNRNPA1) pseudogene [120].
2.2.1.3. Other previously annotated lncRNAs
Among the previously annotated senescence-upregulated lncRNAs [119], HOTAIR functions as a scaffold factor for the ubiquitination and subsequent degradation of Ataxin-1 and Snurportin-1 mediated by E3 ubiquitin ligases [24]. By contrast, the well-known lncRNAs MALAT1, XIST, and MIAT/GOMAFU were less abundant in senescent cells. Silencing MALAT1 or MIAT enhanced cellular senescence, providing direct evidence that these lncRNAs were implicated in senescence [119]. Additionally, DNA-damage-activated p53 was necessary for the cell cycle arrest induced by MALAT1 [121]. The inhibition of cellular senescence by lncRNA 7SL was attributed, at least in part, to the repression of p53 translation by 7SL [120].
2.2.1.4. Novel differentially abundant SAL-RNAs
Among the novel differentially abundant SAL-RNAs identified in this screen, lncRNAs XLOC_023166, XLOC_025931 and XLOC_025918 modulated the onset of senescence and protected the viability of senescent cells [120].
2.2.2. LncRNAs and telomere shortening
Telomere shortening is a hallmark of aging and stress-induced senescence [122]. Telomere length critically affects cell senescence and organism life span. Accordingly, prevention of age-associated telomere shortening by telomerase activation increases both health and longevity [123]. A key component of the complex machinery regulating telomere length and erosion is the lncRNA family TERRA. Transcribed from the telomeres, TERRA lncRNAs associate with RNA-binding proteins such as hnRNPA1 and regulate telomerase activity. Only when the correct stoichiometry between TERRA and hnRNPA1 is achieved, telomerase is free to elongate telomeres and avoid erosion [124]. However, the impact of this mechanism upon age and age-associated CVDs remains unknown.
2.2.3. LncRNAs and age-associated CVDs
Even though the attention of the scientific community has only started to focus on lncRNAs in the past few years, several lncRNAs have been associated or causally linked to age-associated CVDs. Many further studies will be needed to discriminate lncRNAs that drive aging CVDs from those implicated only indirectly, but some information is already beginning to accumulate.
Sustained cardiac hypertrophy is often followed by maladaptive cardiac remodeling, which increases risk for HF. Recently, increased levels of CHRF were found in cardiomyocytes (CMs) treated with Angiotensin II (Ang II) to induce hypertrophy [125]. CHRF sponged and hence reduced the function of miR-489, lowering its activity in hypertrophic CMs; conversely, ectopic miR-489 expression in CMs and in transgenic mice reduced the hypertrophic influence of Ang II treatment.
In Pdk1-null mice, a different model of HF, Liu et al. identified several dysregulated lncRNAs [126]. Network and pathway analyses of these lncRNAs highlighted the involvement of the MAPK signaling pathway, which is causally involved in myocardial hypertrophy and HF [127]. Moreover, they identified MKK7, a sense overlapping lncRNA in the proximity of MAP2K7 (implicated in both HF and hypertrophy [126, 128]), as being downregulated in CMs in Pdk1-null mice.
Concerning naturally occurring antisense lncRNAs, cardiac troponin I (cTNI) is essential for normal sarcomere function in adult CMs and its expression appears to be regulated by cTNI sense-antisense duplexes [129]. Interestingly, cTNI expression levels correlate with ischemia and risk of HF, but the role of the antisense transcript in disease has not yet been evaluated.
Another interesting antisense lncRNA, NPPA-AS, modulates the alternative splicing of the NPPA (natriuretic peptide precursor A) pre-mRNA [130], which is usually expressed only in fetal atrial and ventricular myocardium. NPPA is re-expressed in patients exhibiting hypertrophy and HF [131], and is considered to be a marker for heart disease.
Very recently, an elegant study in mice identified a cluster of nuclear, cardiac-specific lncRNAs expressed from the Myh7 locus termed myosin heavy-chain-associated (Myheart or Mhrt) RNAs [132]. While abundant in adult mouse myocardium, Mhrt RNAs were downregulated in transaortic constriction (TAC)-induced HF, and this reduction coincided with a characteristic Myh6-to-Myh7 isoform switch. Mhrt transcription was inhibited by the Brg1–Hdac–Parp chromatin repressor complex 3, was activated by stress, and was essential for cardiomyopathy development. Accordingly, restoring Mhrt prevented heart hypertrophy and failure, indicating that Mhrt has a protective role in CVDs [132].
Reperfusion therapy is frequently used as treatment for AMI [133, 134]. However, after reperfusion therapy, patients often suffer from myocardial ischemia/reperfusion injury and oxidative stress [135]. The levels of several lncRNAs are modulated at early stages of reperfusion following ischemia [136]. Zangrando et al. [137] showed that AMI in mice was associated with modulation of 30 lncRNAs; among these, myocardial infarction-associated transcript 1 (MIRT1) and 2 (MIRT2) lncRNAs showed robust upregulation. MIRT1 and MIRT2 expression levels were negatively correlated with infarct size and positively correlated with ejection fraction (EF). In addition, MIRT1 and MIRT2 correlated with the expression of multiple genes known to be involved in processes affecting left ventricular remodeling, such as inflammation, extracellular matrix turnover, fibrosis and apoptosis.
Finally, the chromosomal locus 9p21, which contains one of the strongest genetic susceptibility locus for CVDs and type 2 diabetes, spans 50 kb of DNA that do not contain protein-coding genes but do contain the lncRNA ANRIL (CDKN2A/INK4 locus) [138–141]. Accordingly, ANRIL expression is tightly regulated by the identified SNPs [142] and ANRIL expression levels positively correlate with atherosclerosis severity [143]. However, as a word of caution, multiple ANRIL transcripts have been identified (17 annotated in Ensembl so far, http://www.ensembl.org/Homo_sapiens/Gene/Summary?db=core;g=ENSG00000240498;r=9:21994778-22121097) and the results may differ among isoforms.
Functional studies showed that ANRIL expression stimulates cell proliferation, promotes adhesion, and decreases apoptosis, providing a potential disease mechanism, at least for atherosclerosis [144]. From a molecular perspective, ANRIL like several other lncRNAs, recruits Polycomb repressive complexes 1 and 2 and Polycomb-associated activating proteins RYBP and YY1, influencing gene expression both in cis and in trans [141].
3. Circulating ncRNAs
As illustrated in the previous sections, significant changes of tissue ncRNA ‘signatures’ occur in various diseases, including CVDs. However, routine biopsies for miRNA and/or lncRNA profiling are not clinically feasible. Given that some miRNAs are released from the cells of origin and can be measured in bodily fluids [145,146], extracellular miRNAs are remarkably stable in the circulation [147–150], and disease-specific miRNA signatures can be identified in fluids [151,152], investigators are turning to less invasive approaches such as miRNA biomarkers in circulation (e.g., in plasma or serum) [153–159]. Since distal tissues can take up circulating miRNAs, they may represent an important form of cell-to-cell communication [61,160,161]. Furthermore, very recent evidence show that lncRNAs are present in plasma/serum as well, and may potentially be used as biomarkers [146,162]. Plasma or serum ncRNA identified in age-related CVD are summarized in Table 2.
Table 2.
Circulating ncRNAs and ncRNAs in age-related cardiovascular diseases.
| Disease/condition | Modulation | Refs | |
|---|---|---|---|
| miR-1 | Myocardial infarction | Up | [191–194] |
| miR-133 | Myocardial infarction | Up | [159,192] |
| miR-208a | Myocardial infarction | Up | [193] |
| miR-208b | Myocardial infarction | Up | [173,193] |
| miR-499 | Myocardial infarction | Up | [166,193,194] |
| miR-328 | Myocardial infarction | Up | [195] |
| miR-27b | Myocardial infarction | Down | [196] |
| miR-126 | Myocardial infarction | Down | [197] |
| miR-126, miR-92a, miR-17, miR-155, and miR-145 | Coronary artery disease | Down | [198] |
| miR-147 | Coronary artery disease | Down | [171] |
| miR-135 | Coronary artery disease | Up | [171] |
| miR-337-5p, miR-433, and miR-485-3p | Coronary artery disease | Up | [170] |
| miR-134, miR-198, and miR-370 | Unstable angina vs stable angina | Up | [171] |
| miR-106b, miR-25, miR-92a, miR-21, miR-590-5p, miR-126*, and miR-451 | Unstable angina vs stable angina | Up | [162] |
| miR-423-3p | Heart failure | Up | [172] |
| miR-499, -122 | Heart failure | Up | [173] |
| miR-126 | Heart failure | Down | [57] |
| miR-107, miR-139, miR-142-5p, miR-125b, and miR-497 | Heart failure | Down | [174] |
| miR-142-3p, miR-29b | Heart failure | Up | [174] |
| miR-503 | t2dm + critical limb ischemia | Up | [175] |
| ANRIL | Myocardial infarction | Down | [179] |
| aHIF | Myocardial infarction | Up | [179] |
| ANRIL, KCNQ1OT1 | Myocardial infarction | Down | [179] |
| LIPCAR | Myocardial infarction | Down | [181] |
3.1. Circulating microRNAs
3.1.1. MI
Altered levels of circulating miRNAs were detected in patients with AMI, some elevated (miR-1, miR-133, miR-208a/b, miR-499, miR-328), and some reduced (let-7b, miR-126) [153,163,164]. Many upregulated plasma miRNAs were highly expressed in myocytes and correlated with plasma cardiac troponin T (cTnT), suggesting that they were released from injured cardiomyocytes.
In an interesting commentary, Cui and Zao highlighted the importance of selecting the proper control group in this kind of studies and in general in all investigations aimed at identifying diagnostic markers [165]. Using healthy people as control group might artifactually increase the specificity of the diagnostic tests. Indeed, in clinical practice, patients often suffer from various other diseases, some of which might impact upon the biomarker under investigation.
Only a few studies have explored the specific. circulating microRNAs in geriatric patients. In acute non-ST segment elevation myocardial infarction (NSTEMI) of the elderly, circulating miR-499-5p displayed a diagnostic accuracy superior to that of cTnT in patients with modest elevation at presentation [166]. Furthermore, circulating miR-499-5p levels were associated with 12-month cardiovascular mortality after NSTEMI in elderly subjects [167].
3.1.2. CAD
Plasma levels of EC-enriched miRNAs (miR-126, miR-17, and miR-92a), inflammation-associated miR-155, and smooth muscle-enriched miR-145 were reported to be significantly reduced in stable CAD patients compared to healthy controls [153,154,158,168,169]. In addition, miR-135a and miR-147 levels were found to be increased and decreased, respectively, in peripheral blood mononuclear cells (PBMCs) from CAD patients. Recently, miR-337-5p, miR-433, and miR-485-3p expression levels were shown to be higher in CAD patients [170]. Interestingly, increased levels of miR-134, miR-198 and miR-370 [171], as well as miR-106b, miR-25, miR-92a, miR-21, miR-590-5p, miR-126* and miR-451 [162] were able to discriminate unstable from stable angina pectoris, suggesting that this miRNA signature could be used to identify patients at risk for acute coronary syndromes.
3.1.3. HF
A plethora of miRNAs have been found to be dysregulated in HF [153,154,158,168,169]: a) miR-423-5p, which correlates significantly with brain natriuretic peptide (BNP) levels and left ventricular ejection fraction [172]; b) elevated concentrations of miR-499 and miR-122 in acute HF [173]; and c) decreased levels of miR-126 in chronic HF, which correlated inversely with age and disease severity [57]. In addition, profiling PBMC miRNAs in both ischemic (ICM) and nonischemic dilated cardiomyopathy (NIDCM) patients showed that miR-107, miR-139, and miR-142-5p levels were low in both HF classes, miR-142-3p and miR-29b levels increased only in NIDCM patients, and miR-125b and miR-497 levels decreased only in ICM patients [174].
3.1.4. Impaired peripheral angiogenesis
miR-503 contributed to the impaired peripheral angiogenic signaling in patients with type 2 diabetes mellitus (T2DM) and miR-503 plasma levels were elevated in diabetic patients with critical limb ischemia [175].
Finally, studies of the effects of CR on serum miRNAs in young and aged control mice have identified sets of miRNAs displaying increased levels with age and antagonization of this increase by CR [108]. Interestingly, the proteins predicted to be repressed by this set of age-modulated miRNAs are implicated in age-relevant biological processes, including metabolic changes [176–178]. It is tempting to speculate that serum miRNAs may participate in age-induced changes in physiology and pathology, and that their modulation may underlie, at least in part, the anti-aging effects of CR.
3.2. Circulating lncRNAs
Data on circulating lncRNAs are still emerging, and some of them are related to CVDs; however, so far, only one lncRNA was linked to aging. Vausort et al. [179] identified 5 dysregulated lncRNAs in peripheral blood cells of MI patients: aHIF, ANRIL, KCNQ1OT1, MIAT and MALAT1. The levels of aHIF, KCNQ1OT1 and MALAT1 were higher in AMI patients than in healthy volunteers, while the levels of ANRIL were lower in AMI patients. Patients with ST-elevation myocardial infarction (STEMI) had lower levels of ANRIL, KCNQ1OT1, MIAT and MALAT1 compared to patients with non-STEMI patients. Lower levels of ANRIL were associated with advancing age, diabetes, and hypertension, while ANRIL and KCNQ1OT1 improved the prediction of left ventricular dysfunction [179].
Using microarrays, Li et al. [180] found several lncRNAs modulated in a mouse model of HF; 32 among these were simultaneously expressed in the heart, whole blood, and plasma, indicating their potential usefulness as HF biomarkers.
In global transcriptomic analyses on plasma RNA from patients with and without left ventricular remodeling after AMI, the mitochondrial lncRNA uc022bqs.1 (renamed LIPCAR) was found downregulated early after AMI but upregulated at later times following AMI [181]. Measurement of LIPCAR levels was successfully used to stratify patients with respect to their risk of developing cardiac remodeling, and to predict cardiac remodeling and cardiovascular death after HF.
4. Challenges and opportunities
Although the clinical value of ncRNAs is only beginning to surface, the available data already highlight opportunities for ncRNA-based therapies in age-related CVDs. MicroRNAs represent particularly attractive therapeutic targets, since they can be easily synthesized in both sense and antisense (inhibitory) orientation, modified for stability, and attached to medical devices and biomaterials. Strategies can be envisioned using ncRNAs as the therapeutic agents to be delivered ectopically. Reciprocally, the aim can also be to repress or neutralize pathologic endogenous ncRNAs. Preclinical studies adopting these approaches are summarized in Table 3.
Table 3.
Effects of the administration of miRNAs and anti-miRNAs in animal models of CVDs.
| miR | Intervention | Effects | Refs |
|---|---|---|---|
| miR-15 | Inhibition | Infarct size reduction, cardiac remodeling, cardiac function improvement | [199] |
| miR ~ 17–92 | KO | Reduced proliferation | [200] |
| miR-24 | Inhibition | Infarct size reduction, cardiac and vascular function improvement | [201] |
| miR-29b | Inhibition | Reduced fibrosis | [202] |
| miR-320 | Inhibition | Infarct size reduction | [203] |
| miR-590/199a | Overexpression | Infarct size reduction, cardiac function improvement | [204] |
| miR-208a | KO, transgenic Inhibition | Hypertrophy reduction after trans-aorta binding | [205] |
| Cardiac remodeling reduction in Dahl rats | [206] | ||
| [207] | |||
| miR-132/212 family | Inhibition, KO | Protection from pressure overload-induced heart failure | [208] |
| miR-133 | Inhibition | Induction of cardiac hypertrophy | [209] |
| miR-21 | Inhibition | Fibrosis reduction after pressure overload | [70] |
| miR-101a/b | Overexpression | Fibrosis reduction after infarction | [210] |
| miR-24 | Inhibition | Prevention of decompensated hypertrophy | [211] |
| miR-22 | KO | Prevention of hypertrophy and remodeling | [212] |
| miR-199b | Inhibition | Prevention of hypertrophy and fibrosis in HF mouse model | [213] |
| miR-378 | Overexpression | Hypertrophy reduction after thoracic aortic constriction | [214] |
While no human experimentation targeting miRNAs has been started yet for CVDs, a phase I clinical trial for cancer therapy is ongoing using a mimic of miR-34a named MRX34 (ClinicalTrials.gov Identifier: NCT01829971). MRX34 is a liposome-formulated mimic of miR-34a, a microRNA showing decreased levels in many tumor types [182]. Moreover, the applicability of miRNA inhibition as a therapeutic tool is confirmed by the successful completion of a phase IIa clinical trial based on anti-miR-122 (NCT01200420) [183]. This study showed that locked nucleic acid (LNA)-anti-miRNA developed for hepatitis C therapy is effective, safe, and well tolerated.
LNA modifications might also be useful for inhibiting lncRNAs with GapmeRs, antisense oligonucleotides that contain a central stretch (gap) of DNA monomers flanked by blocks of LNA-modified nucleotides. The LNA-nucleotides increase the affnity for a target RNA and stability of the oligonucleotide, while the DNA forms a duplex DNA-RNA that is cleaved by RNase H. This tool is particularly useful to target nuclear transcripts that are poor RNAi substrates [184].
Many hurdles must be overcome to achieve clinically feasible strategies for ncRNA-based therapy, including the optimization of delivery systems and the chemistry of the therapeutic molecule. These obstacles are particularly challenging for lncRNAs, since they are longer, bulkier, and more labile. However, some of the lessons learned with mRNA-based gene therapy might prove useful for lncRNA-based interventions [185–187].
Delivering miRNAs as therapeutic agents offers a powerful tool for fine-tuning target proteins or pathways. A major potential drawback of this approach is the fact that one miRNA can theoretically affect many biological processes, not just a single gene product. However, this feature of microRNAs could be exploited for therapeutic gain in aging dysfunction and disease, if we wish to intervene in the broader molecular pathways regulated by the specific miRNA. To take full advantage of this trait, we must first understand in detail the actions of the miRNA in a relevant pathological context, before progressing to clinical application.
Acknowledgments
This study was supported by Ministero della Salute (SG and FM), Fondazione Cariplo grant #2013-0887 (SG and FM) and Telethon-Italy GGP14092 (FM). KM Kim assisted with illustrations. MG was supported by the NIA-IRP, NIH (AG000393-07).
Abbreviations
- 1/2-sbsRNAs
half-STAU1-binding site RNAs
- 53BP1
tumor suppressor p53-binding protein 1
- ANRIL
antisense noncoding RNA in the INK4 locus
- AP-1
activator protein 1
- ASF1/SF2
alternative splicing factor 1, pre-mRNA-splicing factor
- ATG5
autophagy-related 5
- BCL-2
B-cell lymphoma 2
- BECN
BECLIN 1, autophagy-related
- B-MYB
v-myb avian myeloblastosis viral oncogene homolog-like 2
- BNIP3
BCL2/adenovirus E1B 19 kDa protein-interacting protein 3
- Brg1 (Smarca4)
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 4
- CDKN2A/INK4
cyclin-dependent kinase inhibitor 2A
- CEACAMP
carcinoembryonic antigen-related cell adhesion molecule pseudogene
- CHFR
cardiac hypertrophy related factor
- COX10
cytochrome c oxidase assembly homolog 10
- ERK
extracellular signal-regulated kinase
- FECH
Ferrochelatase
- FIRRE
functional intergenic repeating RNA element
- GAS5
growth arrest-specific 5
- GMC-SF
granulocyte macrophage colony-stimulating factor
- HDAC
histone deacetylase
- HMGA2
high-mobility group AT-hook 2
- HOTAIR
HOX transcript antisense RNA
- Id
DNA-binding protein, inhibitor
- IGF1
Insulin-linke growth factor 1
- IL1α
interleukin 1α
- IL-6
interleukin 6
- IL-8
interleukin 8
- IRAK1
interleukin-1 receptor-associated kinase 1
- ISCU
iron–sulfur cluster assembly enzyme 1
- KCNQ1OT1
potassium voltage-gated channel, KQT-like subfamily, member 1 opposite strand 1
- LC3B-II/MAP1LC3B
microtubule-associated protein 1 light chain 3 beta-II
- LincRNA
long intervening noncoding RNA
- LINCMD1
long noncoding RNA, muscle differentiation
- MALAT1
metastasis associated lung adenocarcinoma transcript 1
- MAPK
mitogen-activated protein kinase
- MCM5
minichromosome maintenance complex component 5
- MDC1
mediator of DNA-damage checkpoint protein 1
- MI
myocardial infarction
- MIAT/GOMAFU
myocardial infarction-associated transcript
- MKK7
MAP kinase kinase 7
- MMP-1
matrix metalloproteinase-1
- MMP-3
matrix metalloproteinase-3
- MYH6
myosin, heavy chain 6, cardiac muscle, alpha
- MYH7
myosin, heavy chain 7, cardiac muscle, beta
- NAD+
nicotinamide adenine dinucleotide
- NEAT1
nuclear enriched abundant transcript 1
- NFκBIB
nuclear factor of kappa light polypeptide gene enhancer in B cells inhibitor, beta
- NF-κB
nuclear factor of kappa light polypeptide gene enhancer in B cells
- NPPA
natriuretic peptide A
- NPPA-AS
natriuretic peptide A antisense RNA
- NRARP
notch-regulated ankyrin repeat protein
- Nrf2
NF-E2 related factor 2
- PARP
poly (ADP-ribose) polymerase
- PCGF4
polycomb group RING finger protein 4
- Pdk1
pyruvate dehydrogenase kinase, isozyme 1
- Ppp1r10
protein phosphatase 1, regulatory subunit 10
- RAR-γ
retinoic acid receptor gamma
- RB
retinoblastoma protein
- S6K1
ribosomal protein S6 kinase 1
- ShcA
Src homology 2 domain-containing
- SIRT1
silent mating type information regulation 2 homolog
- SPRY2
sprouty homolog 2
- SREBP-1c
sterol regulatory element-binding protein
- TERRA
telomeric repeat-containing RNA
- TRF2
telomeric repeat binding factor 2
- TUG1
taurine upregulated gene 1
- XIST
X-inactive specific transcript
- ZEB1
zinc finger E-box binding homeobox 1
- γ-H2AX
gamma-H2A histone family, member X
Footnotes
Disclosures
None.
References
- 1.Cech TR, Steitz JA. The noncoding RNA revolution-trashing old rules to forge new ones. Cell. 2014;157:77–94. doi: 10.1016/j.cell.2014.03.008. [DOI] [PubMed] [Google Scholar]
- 2.Salmanidis M, Pillman K, Goodall G, Bracken C. Direct transcriptional regulation by nuclear microRNAs. Int J Biochem Cell Biol. 2014;54c:304–11. doi: 10.1016/j.biocel.2014.03.010. [DOI] [PubMed] [Google Scholar]
- 3.Yoon JH, Abdelmohsen K, Gorospe M. Posttranscriptional gene regulation by long noncoding RNA. J Mol Biol. 2013;425:3723–30. doi: 10.1016/j.jmb.2012.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet. 2010;11:597–610. doi: 10.1038/nrg2843. [DOI] [PubMed] [Google Scholar]
- 5.Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, et al. Landscape of transcription in human cells. Nature. 2012;489:101–8. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li L, Chang HY. Physiological roles of long noncoding RNAs: insight from knockout mice. Trends Cell Biol. 2014;24:594–602. doi: 10.1016/j.tcb.2014.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Small EM, Olson EN. Pervasive roles of microRNAs in cardiovascular biology. Nature. 2011;469:336–42. doi: 10.1038/nature09783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen LH, Chiou GY, Chen YW, Li HY, Chiou SH. MicroRNA and aging: a novel modulator in regulating the aging network. Ageing Res Rev. 2010;9(Suppl 1):S59–66. doi: 10.1016/j.arr.2010.08.002. [DOI] [PubMed] [Google Scholar]
- 9.Fabian MR, Sonenberg N, Filipowicz W. Regulation of mRNA translation and stability by microRNAs. Annu Rev Biochem. 2010;79:351–79. doi: 10.1146/annurev-biochem-060308-103103. [DOI] [PubMed] [Google Scholar]
- 10.Ha M, Kim VN. Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol. 2014;15:509–24. doi: 10.1038/nrm3838. [DOI] [PubMed] [Google Scholar]
- 11.Gorospe M, Abdelmohsen K. MicroRegulators come of age in senescence. Trends Genet. 2011;27:233–41. doi: 10.1016/j.tig.2011.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jansson MD, Lund AH. MicroRNA and cancer. Mol Oncol. 2012;6:590–610. doi: 10.1016/j.molonc.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fiore R, Siegel G, Schratt G. MicroRNA function in neuronal development, plasticity and disease. Biochim Biophys Acta. 2008;1779:471–8. doi: 10.1016/j.bbagrm.2007.12.006. [DOI] [PubMed] [Google Scholar]
- 14.Kung JT, Colognori D, Lee JT. Long noncoding RNAs: past, present, and future. Genetics. 2013;193:651–69. doi: 10.1534/genetics.112.146704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Spitale RC, Tsai MC, Chang HY. RNA templating the epigenome: long noncoding RNAs as molecular scaffolds. Epigenetics. 2011;6:539–43. doi: 10.4161/epi.6.5.15221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee JT. Epigenetic regulation by long noncoding RNAs. Science. 2012;338:1435–9. doi: 10.1126/science.1231776. [DOI] [PubMed] [Google Scholar]
- 17.Bergmann JH, Spector DL. Long non-coding RNAs: modulators of nuclear structure and function. Curr Opin Cell Biol. 2014;26:10–8. doi: 10.1016/j.ceb.2013.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gong C, Maquat LE. lncRNAs transactivate STAU1-mediated mRNA decay by duplexing with 3′ UTRs via Alu elements. Nature. 2011;470:284–8. doi: 10.1038/nature09701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yoon JH, Abdelmohsen K, Srikantan S, Yang X, Martindale JL, De S, et al. LincRNA-p21 suppresses target mRNA translation. Mol Cell. 2012;47:648–55. doi: 10.1016/j.molcel.2012.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mattick JS. Long noncoding RNAs in cell and developmental biology. Semin Cell Dev Biol. 2011;22:327. doi: 10.1016/j.semcdb.2011.05.002. [DOI] [PubMed] [Google Scholar]
- 21.Anko ML, Neugebauer KM. Long noncoding RNAs add another layer to pre-mRNA splicing regulation. Mol Cell. 2010;39:833–4. doi: 10.1016/j.molcel.2010.09.003. [DOI] [PubMed] [Google Scholar]
- 22.Cesana M, Cacchiarelli D, Legnini I, Santini T, Sthandier O, Chinappi M, et al. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell. 2011;147:358–69. doi: 10.1016/j.cell.2011.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tay Y, Rinn J, Pandolfi PP. The multilayered complexity of ceRNA crosstalk and competition. Nature. 2014;505:344–52. doi: 10.1038/nature12986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yoon JH, Abdelmohsen K, Kim J, Yang X, Martindale JL, Tominaga-Yamanaka K, et al. Scaffold function of long non-coding RNA HOTAIR in protein ubiquitination. Nat Commun. 2013;4:2939. doi: 10.1038/ncomms3939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wapinski O, Chang HY. Long noncoding RNAs and human disease. Trends Cell Biol. 2011;21:354–61. doi: 10.1016/j.tcb.2011.04.001. [DOI] [PubMed] [Google Scholar]
- 26.Harries LW. Long non-coding RNAs and human disease. Biochem Soc Trans. 2012;40:902–6. doi: 10.1042/BST20120020. [DOI] [PubMed] [Google Scholar]
- 27.Mozaffarian D, Afshin A, Benowitz NL, Bittner V, Daniels SR, Franch HA, et al. Population approaches to improve diet, physical activity, and smoking habits: a scientific statement from the American Heart Association. Circulation. 2012;126:1514–63. doi: 10.1161/CIR.0b013e318260a20b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lozano R, Naghavi M, Foreman K, Lim S, Shibuya K, Aboyans V, et al. Global and regional mortality from 235 causes of death for 20 age groups in 1990 and 2010: a systematic analysis for the Global Burden of Disease Study 2010. Lancet. 2012;380:2095–128. doi: 10.1016/S0140-6736(12)61728-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shioi T, Inuzuka Y. Aging as a substrate of heart failure. J Cardiol. 2012;60:423–8. doi: 10.1016/j.jjcc.2012.07.015. [DOI] [PubMed] [Google Scholar]
- 30.Dai DF, Rabinovitch PS, Ungvari Z. Mitochondria and cardiovascular aging. Circ Res. 2012;110:1109–24. doi: 10.1161/CIRCRESAHA.111.246140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Csiszar A, Ungvari Z, Edwards JG, Kaminski P, Wolin MS, Koller A, et al. Aging-induced phenotypic changes and oxidative stress impair coronary arteriolar function. Circ Res. 2002;90:1159–66. doi: 10.1161/01.res.0000020401.61826.ea. [DOI] [PubMed] [Google Scholar]
- 32.Lakatta EG. Arterial and cardiac aging: major shareholders in cardiovascular disease enterprises: part III: cellular and molecular clues to heart and arterial aging. Circulation. 2003;107:490–7. doi: 10.1161/01.cir.0000048894.99865.02. [DOI] [PubMed] [Google Scholar]
- 33.Gimbrone MA, Jr, Topper JN, Nagel T, Anderson KR, Garcia-Cardena G. Endothelial dysfunction, hemodynamic forces, and atherogenesis. Ann N Y Acad Sci. 2000;902:230–9. doi: 10.1111/j.1749-6632.2000.tb06318.x. discussion 9–40. [DOI] [PubMed] [Google Scholar]
- 34.Ungvari Z, Sonntag WE, Csiszar A. Mitochondria and aging in the vascular system. JMol Med. 2010;88:1021–7. doi: 10.1007/s00109-010-0667-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lakatta EG, Levy D. Arterial and cardiac aging: major shareholders in cardiovascular disease enterprises: part I: aging arteries: a “set up” for vascular disease. Circulation. 2003;107:139–46. doi: 10.1161/01.cir.0000048892.83521.58. [DOI] [PubMed] [Google Scholar]
- 36.Lee HY, Oh BH. Aging and arterial stiffness. Circ J. 2010;74:2257–62. doi: 10.1253/circj.cj-10-0910. [DOI] [PubMed] [Google Scholar]
- 37.Kato M, Slack FJ. Ageing and the small, non-coding RNA world. Ageing Res Rev. 2013;12:429–35. doi: 10.1016/j.arr.2012.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Harries LW. MicroRNAs as mediators of the ageing process. Genes. 2014;5:656–70. doi: 10.3390/genes5030656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang X, Azhar G, Wei JY. The expression of microRNA and microRNA clusters in the aging heart. PLoS ONE. 2012;7:e34688. doi: 10.1371/journal.pone.0034688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hayflick L. The limited in vitro lifetime of human diploid cell strains. Exp Cell Res. 1965;37:614–36. doi: 10.1016/0014-4827(65)90211-9. [DOI] [PubMed] [Google Scholar]
- 41.Kuilman T, Michaloglou C, Mooi WJ, Peeper DS. The essence of senescence. Genes Dev. 2010;24:2463–79. doi: 10.1101/gad.1971610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Campisi J. Senescent cells, tumor suppression, and organismal aging: good citizens, bad neighbors. Cell. 2005;120:513–22. doi: 10.1016/j.cell.2005.02.003. [DOI] [PubMed] [Google Scholar]
- 43.Rodier F, Campisi J. Four faces of cellular senescence. J Cell Biol. 2011;192:547–56. doi: 10.1083/jcb.201009094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tyner SD, Venkatachalam S, Choi J, Jones S, Ghebranious N, Igelmann H, et al. p53 mutant mice that display early ageing-associated phenotypes. Nature. 2002;415:45–53. doi: 10.1038/415045a. [DOI] [PubMed] [Google Scholar]
- 45.Baker DJ, Wijshake T, Tchkonia T, LeBrasseur NK, Childs BG, van de Sluis B, et al. Clearance of p16Ink4a-positive senescent cells delays ageing-associated disorders. Nature. 2011;479:232–6. doi: 10.1038/nature10600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fenton M, Barker S, Kurz DJ, Erusalimsky JD. Cellular senescence after single and repeated balloon catheter denudations of rabbit carotid arteries. Arterioscler Thromb Vasc Biol. 2001;21:220–6. doi: 10.1161/01.atv.21.2.220. [DOI] [PubMed] [Google Scholar]
- 47.Minamino T, Miyauchi H, Yoshida T, Ishida Y, Yoshida H, Komuro I. Endothelial cell senescence in human atherosclerosis: role of telomere in endothelial dysfunction. Circulation. 2002;105:1541–4. doi: 10.1161/01.cir.0000013836.85741.17. [DOI] [PubMed] [Google Scholar]
- 48.Abdelmohsen K, Kuwano Y, Kim HH, Gorospe M. Posttranscriptional gene regulation by RNA-binding proteins during oxidative stress: implications for cellular senescence. Biol Chem. 2008;389:243–55. doi: 10.1515/BC.2008.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–33. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Borgdorff V, Lleonart ME, Bishop CL, Fessart D, Bergin AH, Overhoff MG, et al. Multiple microRNAs rescue from Ras-induced senescence by inhibiting p21(Waf1/Cip1) Oncogene. 2010;29:2262–71. doi: 10.1038/onc.2009.497. [DOI] [PubMed] [Google Scholar]
- 51.Marasa BS, Srikantan S, Masuda K, Abdelmohsen K, Kuwano Y, Yang X, et al. Increased MKK4 abundance with replicative senescence is linked to the joint reduction of multiple microRNAs. Sci Signal. 2009;2:ra69. doi: 10.1126/scisignal.2000442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Afanasyeva EA, Mestdagh P, Kumps C, Vandesompele J, Ehemann V, Theissen J, et al. MicroRNA miR-885-5p targets CDK2 and MCM5, activates p53 and inhibits proliferation and survival. Cell Death Differ. 2011;18:974–84. doi: 10.1038/cdd.2010.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Boon RA, Seeger T, Heydt S, Fischer A, Hergenreider E, Horrevoets AJ, et al. MicroRNA-29 in aortic dilation: implications for aneurysm formation. Circ Res. 2011;109:1115–9. doi: 10.1161/CIRCRESAHA.111.255737. [DOI] [PubMed] [Google Scholar]
- 54.Olivieri F, Lazzarini R, Recchioni R, Marcheselli F, Rippo MR, Di Nuzzo S, et al. MiR-146a as marker of senescence-associated pro-inflammatory status in cells involved in vascular remodelling. Age (Dordr) 2013;35:1157–72. doi: 10.1007/s11357-012-9440-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Olivieri F, Spazzafumo L, Santini G, Lazzarini R, Albertini MC, Rippo MR, et al. Age-related differences in the expression of circulating microRNAs: miR-21 as a new circulating marker of inflammaging. Mech Ageing Dev. 2012;133:675–85. doi: 10.1016/j.mad.2012.09.004. [DOI] [PubMed] [Google Scholar]
- 56.Asgeirsdottir SA, van Solingen C, Kurniati NF, Zwiers PJ, Heeringa P, van Meurs M, et al. MicroRNA-126 contributes to renal microvascular heterogeneity of VCAM-1 protein expression in acute inflammation. Am J Physiol Renal Physiol. 2012;302:F1630–9. doi: 10.1152/ajprenal.00400.2011. [DOI] [PubMed] [Google Scholar]
- 57.Fukushima Y, Nakanishi M, Nonogi H, Goto Y, Iwai N. Assessment of plasma miRNAs in congestive heart failure. Circ J. 2011;75:336–40. doi: 10.1253/circj.cj-10-0457. [DOI] [PubMed] [Google Scholar]
- 58.Feng X, Wang H, Ye S, Guan J, Tan W, Cheng S, et al. Up-regulation of microRNA-126 may contribute to pathogenesis of ulcerative colitis via regulating NF-kappaB inhibitor IkappaBalpha. PLoS ONE. 2012;7:e52782. doi: 10.1371/journal.pone.0052782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wei Y, Nazari-Jahantigh M, Neth P, Weber C, Schober A. MicroRNA-126, -145, and -155: a therapeutic triad in atherosclerosis? Arterioscler Thromb Vasc Biol. 2013;33:449–54. doi: 10.1161/ATVBAHA.112.300279. [DOI] [PubMed] [Google Scholar]
- 60.Kim S, Iwao H. Molecular and cellular mechanisms of angiotensin II-mediated cardiovascular and renal diseases. Pharmacol Rev. 2000;52:11–34. [PubMed] [Google Scholar]
- 61.Boon RA, Iekushi K, Lechner S, Seeger T, Fischer A, Heydt S, et al. MicroRNA-34a regulates cardiac ageing and function. Nature. 2013;495:107–10. doi: 10.1038/nature11919. [DOI] [PubMed] [Google Scholar]
- 62.Bradshaw AD, Baicu CF, Rentz TJ, Van Laer AO, Bonnema DD, Zile MR. Age-dependent alterations in fibrillar collagen content and myocardial diastolic function: role of SPARC in post-synthetic procollagen processing. Am J Physiol Heart Circ Physiol. 2010;298:H614–22. doi: 10.1152/ajpheart.00474.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Burgess ML, McCrea JC, Hedrick HL. Age-associated changes in cardiac matrix and integrins. Mech Ageing Dev. 2001;122:1739–56. doi: 10.1016/s0047-6374(01)00296-2. [DOI] [PubMed] [Google Scholar]
- 64.Swinnen M, Vanhoutte D, Van Almen GC, Hamdani N, Schellings MW, D’Hooge J, et al. Absence of thrombospondin-2 causes age-related dilated cardiomyopathy. Circulation. 2009;120:1585–97. doi: 10.1161/CIRCULATIONAHA.109.863266. [DOI] [PubMed] [Google Scholar]
- 65.Wang M, Zhang J, Walker SJ, Dworakowski R, Lakatta EG, Shah AM. Involvement of NADPH oxidase in age-associated cardiac remodeling. J Mol Cell Cardiol. 2010;48:765–72. doi: 10.1016/j.yjmcc.2010.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.St John Sutton M, Pfeffer MA, Moye L, Plappert T, Rouleau JL, Lamas G, et al. Cardiovascular death and left ventricular remodeling two years after myocardial infarction: baseline predictors and impact of long-term use of captopril: information from the Survival and Ventricular Enlargement (SAVE) trial. Circulation. 1997;96:3294–9. doi: 10.1161/01.cir.96.10.3294. [DOI] [PubMed] [Google Scholar]
- 67.Gould KE, Taffet GE, Michael LH, Christie RM, Konkol DL, Pocius JS, et al. Heart failure and greater infarct expansion in middle-aged mice: a relevant model for postinfarction failure. Am J Physiol Heart Circ Physiol. 2002;282:H615–21. doi: 10.1152/ajpheart.00206.2001. [DOI] [PubMed] [Google Scholar]
- 68.Minamino T, Komuro I. Vascular cell senescence: contribution to atherosclerosis. Circ Res. 2007;100:15–26. doi: 10.1161/01.RES.0000256837.40544.4a. [DOI] [PubMed] [Google Scholar]
- 69.van Almen GC, Verhesen W, van Leeuwen RE, van de Vrie M, Eurlings C, Schellings MW, et al. MicroRNA-18 and microRNA-19 regulate CTGF and TSP-1 expression in age-related heart failure. Aging Cell. 2011;10:769–79. [Google Scholar]
- 70.Thum T, Gross C, Fiedler J, Fischer T, Kissler S, Bussen M, et al. MicroRNA-21 contributes to myocardial disease by stimulating MAP kinase signalling in fibroblasts. Nature. 2008;456:980–4. doi: 10.1038/nature07511. [DOI] [PubMed] [Google Scholar]
- 71.Duisters RF, Tijsen AJ, Schroen B, Leenders JJ, Lentink V, van der Made I, et al. miR-133 and miR-30 regulate connective tissue growth factor: implications for a role of microRNAs in myocardial matrix remodeling. Circ Res. 2009;104:170–8. 6–8. doi: 10.1161/CIRCRESAHA.108.182535. [DOI] [PubMed] [Google Scholar]
- 72.Oldham S. Obesity and nutrient sensing TOR pathway in flies and vertebrates: Functional conservation of genetic mechanisms. Trends Endocrinol Metab. 2011;22:45–52. doi: 10.1016/j.tem.2010.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Curtis B, Lage MJ. Glycemic control among patients with type 2 diabetes who initiate basal insulin: a retrospective cohort study. J Med Econ. 2014;17:21–31. doi: 10.3111/13696998.2013.862538. [DOI] [PubMed] [Google Scholar]
- 74.Bornfeldt KE. 2013 Russell Ross memorial lecture in vascular biology: cellular and molecular mechanisms of diabetes mellitus-accelerated atherosclerosis. Arterioscler Thromb Vasc Biol. 2014;34:705–14. doi: 10.1161/ATVBAHA.113.301928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yu Q, Gao F, Ma XL. Insulin says NO to cardiovascular disease. Cardiovasc Res. 2011;89:516–24. doi: 10.1093/cvr/cvq349. [DOI] [PubMed] [Google Scholar]
- 76.Cersosimo E, DeFronzo RA. Insulin resistance and endothelial dysfunction: the road map to cardiovascular diseases. Diabetes Metab Res Rev. 2006;22:423–36. doi: 10.1002/dmrr.634. [DOI] [PubMed] [Google Scholar]
- 77.Curtis R, Geesaman BJ, DiStefano PS. Ageing and metabolism: drug discovery opportunities. Nat Rev Drug Discov. 2005;4:569–80. doi: 10.1038/nrd1777. [DOI] [PubMed] [Google Scholar]
- 78.Yu XY, Song YH, Geng YJ, Lin QX, Shan ZX, Lin SG, et al. Glucose induces apoptosis of cardiomyocytes via microRNA-1 and IGF-1. Biochem Biophys Res Commun. 2008;376:548–52. doi: 10.1016/j.bbrc.2008.09.025. [DOI] [PubMed] [Google Scholar]
- 79.Elia L, Contu R, Quintavalle M, Varrone F, Chimenti C, Russo MA, et al. Reciprocal regulation of microRNA-1 and insulin-like growth factor-1 signal transduction cascade in cardiac and skeletal muscle in physiological and pathological conditions. Circulation. 2009;120:2377–85. doi: 10.1161/CIRCULATIONAHA.109.879429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Shan ZX, Lin QX, Fu YH, Deng CY, Zhou ZL, Zhu JN, et al. Upregulated expression of miR-1/miR-206 in a rat model of myocardial infarction. Biochem Biophys Res Commun. 2009;381:597–601. doi: 10.1016/j.bbrc.2009.02.097. [DOI] [PubMed] [Google Scholar]
- 81.Li Y, Higashi Y, Itabe H, Song YH, Du J, Delafontaine P. Insulin-like growth factor-1 receptor activation inhibits oxidized LDL-induced cytochrome C release and apoptosis via the phosphatidylinositol 3 kinase/Akt signaling pathway. Arterioscler Thromb Vasc Biol. 2003;23:2178–84. doi: 10.1161/01.ATV.0000099788.31333.DB. [DOI] [PubMed] [Google Scholar]
- 82.Delafontaine P, Brink M. The growth hormone and insulin-like growth factor 1 axis in heart failure. Ann Endocrinol. 2000;61:22–6. [PubMed] [Google Scholar]
- 83.Ungvari Z, Parrado-Fernandez C, Csiszar A, de Cabo R. Mechanisms underlying caloric restriction and lifespan regulation: implications for vascular aging. Circ Res. 2008;102:519–28. doi: 10.1161/CIRCRESAHA.107.168369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Valcarcel-Ares MN, Gautam T, Warrington JP, Bailey-Downs L, Sosnowska D, de Cabo R, et al. Disruption of Nrf2 signaling impairs angiogenic capacity of endothelial cells: implications for microvascular aging. J Gerontol A: Biol Med Sci. 2012;67:821–9. doi: 10.1093/gerona/glr229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Green DR, Galluzzi L, Kroemer G. Mitochondria and the autophagy-inflammation-cell death axis in organismal aging. Science. 2011;333:1109–12. doi: 10.1126/science.1201940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Napoli C, de Nigris F, Palinski W. Multiple role of reactive oxygen species in the arterial wall. J Cell Biochem. 2001;82:674–82. doi: 10.1002/jcb.1198. [DOI] [PubMed] [Google Scholar]
- 87.Tsutsui H, Kinugawa S, Matsushima S. Oxidative stress and heart failure. Am J Physiol Heart Circ Physiol. 2011;301:H2181–90. doi: 10.1152/ajpheart.00554.2011. [DOI] [PubMed] [Google Scholar]
- 88.Magenta A, Greco S, Gaetano C, Martelli F. Oxidative stress and microRNAs in vascular diseases. Int J Mol Sci. 2013;14:17319–46. doi: 10.3390/ijms140917319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Magenta A, Cencioni C, Fasanaro P, Zaccagnini G, Greco S, Sarra-Ferraris G, et al. miR-200c is upregulated by oxidative stress and induces endothelial cell apoptosis and senescence via ZEB1 inhibition. Cell Death Differ. 2011;18:1628–39. doi: 10.1038/cdd.2011.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zaccagnini G, Martelli F, Fasanaro P, Magenta A, Gaetano C, Di Carlo A, et al. p66ShcA modulates tissue response to hindlimb ischemia. Circulation. 2004;109:2917–23. doi: 10.1161/01.CIR.0000129309.58874.0F. [DOI] [PubMed] [Google Scholar]
- 91.Baseler WA, Thapa D, Jagannathan R, Dabkowski ER, Croston TL, Hollander JM. miR-141 as a regulator of the mitochondrial phosphate carrier (Slc25a3) in the type 1 diabetic heart. Am J Physiol Cell Physiol. 2012;303:C1244–51. doi: 10.1152/ajpcell.00137.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Pulakat L, Aroor AR, Gul R, Sowers JR. Cardiac insulin resistance and microRNA modulators. Exp Diabetes Res. 2012;2012:654904. doi: 10.1155/2012/654904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lin Y, Liu X, Cheng Y, Yang J, Huo Y, Zhang C. Involvement of MicroRNAs in hydrogen peroxide-mediated gene regulation and cellular injury response in vascular smooth muscle cells. J Biol Chem. 2009;284:7903–13. doi: 10.1074/jbc.M806920200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Hulsmans M, Holvoet P. The vicious circle between oxidative stress and inflammation in atherosclerosis. J Cell Mol Med. 2010;14:70–8. doi: 10.1111/j.1582-4934.2009.00978.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Weber M, Baker MB, Moore JP, Searles CD. MiR-21 is induced in endothelial cells by shear stress and modulates apoptosis and eNOS activity. Biochem Biophys Res Commun. 2010;393:643–8. doi: 10.1016/j.bbrc.2010.02.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Fleissner F, Jazbutyte V, Fiedler J, Gupta SK, Yin X, Xu Q, et al. Short communication: asymmetric dimethylarginine impairs angiogenic progenitor cell function in patients with coronary artery disease through a microRNA-21-dependent mechanism. Circ Res. 2010;107:138–43. doi: 10.1161/CIRCRESAHA.110.216770. [DOI] [PubMed] [Google Scholar]
- 97.Li R, Yan G, Li Q, Sun H, Hu Y, Sun J, et al. MicroRNA-145 protects cardiomyocytes against hydrogen peroxide (H(2)O(2))-induced apoptosis through targeting the mitochondria apoptotic pathway. PLoS ONE. 2012;7:e44907. doi: 10.1371/journal.pone.0044907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Greco S, Gaetano C, Martelli F. HypoxamiR regulation and function in ischemic cardiovascular diseases. Antioxid Redox Signal. 2014;21:1202–19. doi: 10.1089/ars.2013.5403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Zaccagnini G, Maimone B, Di Stefano V, Fasanaro P, Greco S, Perfetti A, et al. Hypoxia-induced miR-210 modulates tissue response to acute peripheral ischemia. Antioxid Redox Signal. 2014;21:1177–88. doi: 10.1089/ars.2013.5206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Martinet W, De Meyer GR. Autophagy in atherosclerosis: a cell survival and death phenomenon with therapeutic potential. Circ Res. 2009;104:304–17. doi: 10.1161/CIRCRESAHA.108.188318. [DOI] [PubMed] [Google Scholar]
- 101.Cuervo AM. Autophagy and aging: keeping that old broom working. Trends Genet. 2008;24:604–12. doi: 10.1016/j.tig.2008.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kang HT, Lee KB, Kim SY, Choi HR, Park SC. Autophagy impairment induces premature senescence in primary human fibroblasts. PLoS ONE. 2011;6:e23367. doi: 10.1371/journal.pone.0023367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Menghini R, Casagrande V, Marino A, Marchetti V, Cardellini M, Stoehr R, et al. MiR-216a: a link between endothelial dysfunction and autophagy. Cell Death Dis. 2014;5:e1029. doi: 10.1038/cddis.2013.556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Rogina B, Helfand SL. Sir2 mediates longevity in the fly through a pathway related to calorie restriction. Proc Natl Acad Sci U S A. 2004;101:15998–6003. doi: 10.1073/pnas.0404184101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Bordone L, Guarente L. Calorie restriction, SIRT1 and metabolism: understanding longevity. Nat Rev Mol Cell Biol. 2005;6:298–305. doi: 10.1038/nrm1616. [DOI] [PubMed] [Google Scholar]
- 106.Schwer B, Verdin E. Conserved metabolic regulatory functions of sirtuins. Cell Metab. 2008;7:104–12. doi: 10.1016/j.cmet.2007.11.006. [DOI] [PubMed] [Google Scholar]
- 107.Potente M, Dimmeler S. Emerging roles of SIRT1 in vascular endothelial homeostasis. Cell Cycle. 2008;7:2117–22. doi: 10.4161/cc.7.14.6267. [DOI] [PubMed] [Google Scholar]
- 108.Dhahbi JM, Spindler SR, Atamna H, Yamakawa A, Guerrero N, Boffelli D, et al. Deep sequencing identifies circulating mouse miRNAs that are functionally implicated in manifestations of aging and responsive to calorie restriction. Aging. 2013;5:130–41. doi: 10.18632/aging.100540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Menghini R, Casagrande V, Cardellini M, Martelli E, Terrinoni A, Amati F, et al. MicroRNA 217 modulates endothelial cell senescence via silent information regulator 1. Circulation. 2009;120:1524–32. doi: 10.1161/CIRCULATIONAHA.109.864629. [DOI] [PubMed] [Google Scholar]
- 110.Zhang L, Huang D, Wang Q, Shen D, Wang Y, Chen B, et al. MiR-132 inhibits expression of SIRT1 and induces pro-inflammatory processes of vascular endothelial inflammation through blockade of the SREBP-1c metabolic pathway. Cardiovasc Drugs Ther. 2014;28:303–11. doi: 10.1007/s10557-014-6533-x. [DOI] [PubMed] [Google Scholar]
- 111.Ponugoti B, Kim DH, Xiao Z, Smith Z, Miao J, Zang M, et al. SIRT1 deacetylates and inhibits SREBP-1C activity in regulation of hepatic lipid metabolism. J Biol Chem. 2010;285:33959–70. doi: 10.1074/jbc.M110.122978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Kumarswamy R, Volkmann I, Beermann J, Napp LC, Jabs O, Bhayadia R, et al. Vascular importance of the miR-212/132 cluster. Eur Heart J. 2014;35:3224–31. doi: 10.1093/eurheartj/ehu344. [DOI] [PubMed] [Google Scholar]
- 113.Guarani V, Deflorian G, Franco CA, Kruger M, Phng LK, Bentley K, et al. Acetylation-dependent regulation of endothelial Notch signalling by the SIRT1 deacetylase. Nature. 2011;473:234–8. doi: 10.1038/nature09917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Ito T, Yagi S, Yamakuchi M. MicroRNA-34a regulation of endothelial senescence. Biochem Biophys Res Commun. 2010;398:735–40. doi: 10.1016/j.bbrc.2010.07.012. [DOI] [PubMed] [Google Scholar]
- 115.Xu Q, Seeger FH, Castillo J, Iekushi K, Boon RA, Farcas R, et al. Micro-RNA-34a contributes to the impaired function of bone marrow-derived mononuclear cells from patients with cardiovascular disease. J Am Coll Cardiol. 2012;59:2107–17. doi: 10.1016/j.jacc.2012.02.033. [DOI] [PubMed] [Google Scholar]
- 116.Tabuchi T, Satoh M, Itoh T, Nakamura M. MicroRNA-34a regulates the longevity-associated protein SIRT1 in coronary artery disease: effect of statins on SIRT1 and microRNA-34a expression. Clin Sci (Lond) 2012;123:161–71. doi: 10.1042/CS20110563. [DOI] [PubMed] [Google Scholar]
- 117.Zhu H, Yang Y, Wang Y, Li J, Schiller PW, Peng T. MicroRNA-195 promotes palmitate-induced apoptosis in cardiomyocytes by down-regulating Sirt1. Cardiovasc Res. 2011;92:75–84. doi: 10.1093/cvr/cvr145. [DOI] [PubMed] [Google Scholar]
- 118.Rane S, He M, Sayed D, Vashistha H, Malhotra A, Sadoshima J, et al. Downregulation of miR-199a derepresses hypoxia-inducible factor-1alpha and Sirtuin 1 and recapitulates hypoxia preconditioning in cardiac myocytes. Circ Res. 2009;104:879–86. doi: 10.1161/CIRCRESAHA.108.193102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Abdelmohsen K, Panda A, Kang MJ, Xu J, Selimyan R, Yoon JH, et al. Senescence-associated lncRNAs: senescence-associated long noncoding RNAs. Aging Cell. 2013;12:890–900. doi: 10.1111/acel.12115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Abdelmohsen K, Panda AC, Kang MJ, Guo R, Kim J, Grammatikakis I, et al. 7SL RNA represses p53 translation by competing with HuR. Nucleic Acids Res. 2014;42:10099–111. doi: 10.1093/nar/gku686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Tripathi V, Shen Z, Chakraborty A, Giri S, Freier SM, Wu X, et al. Long noncoding RNA MALAT1 controls cell cycle progression by regulating the expression of oncogenic transcription factor B-MYB. PLoS Genet. 2013;9:e1003368. doi: 10.1371/journal.pgen.1003368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Hewitt G, Jurk D, Marques FD, Correia-Melo C, Hardy T, Gackowska A, et al. Telomeres are favoured targets of a persistent DNA damage response in ageing and stress-induced senescence. Nat Commun. 2012;3:708. doi: 10.1038/ncomms1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Bernardes de Jesus B, Vera E, Schneeberger K, Tejera AM, Ayuso E, Bosch F, et al. Telomerase gene therapy in adult and old mice delays aging and increases longevity without increasing cancer. EMBO Mol Med. 2012;4:691–704. doi: 10.1002/emmm.201200245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Redon S, Zemp I, Lingner J. A three-state model for the regulation of telomerase by TERRA and hnRNPA1. Nucleic Acids Res. 2013;41:9117–28. doi: 10.1093/nar/gkt695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Wang K, Liu F, Zhou LY, Long B, Yuan SM, Wang Y, et al. The long noncoding RNA CHRF regulates cardiac hypertrophy by targeting miR-489. Circ Res. 2014;114:1377–88. doi: 10.1161/CIRCRESAHA.114.302476. [DOI] [PubMed] [Google Scholar]
- 126.Liu H, Song G, Zhou L, Hu X, Liu M, Nie J, et al. Compared analysis of LncRNA expression profiling in pdk1 gene knockout mice at two time points. Cell Physiol Biochem. 2013;32:1497–508. doi: 10.1159/000356586. [DOI] [PubMed] [Google Scholar]
- 127.Ravingerova T, Barancik M, Strniskova M. Mitogen-activated protein kinases: a new therapeutic target in cardiac pathology. Mol Cell Biochem. 2003;247:127–38. doi: 10.1023/a:1024119224033. [DOI] [PubMed] [Google Scholar]
- 128.Wang Y, Su B, Sah VP, Brown JH, Han J, Chien KR. Cardiac hypertrophy induced by mitogen-activated protein kinase kinase 7, a specific activator for c-Jun NH2-terminal kinase in ventricular muscle cells. J Biol Chem. 1998;273:5423–6. doi: 10.1074/jbc.273.10.5423. [DOI] [PubMed] [Google Scholar]
- 129.Podlowski S, Bramlage P, Baumann G, Morano I, Luther HP. Cardiac troponin I sense-antisense RNA duplexes in the myocardium. J Cell Biochem. 2002;85:198–207. [PubMed] [Google Scholar]
- 130.Annilo T, Kepp K, Laan M. Natural antisense transcript of natriuretic peptide precursor A (NPPA): structural organization and modulation of NPPA expression. BMC Mol Biol. 2009;10:81. doi: 10.1186/1471-2199-10-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Horsthuis T, Houweling AC, Habets PE, de Lange FJ, el Azzouzi H, Clout DE, et al. Distinct regulation of developmental and heart disease-induced atrial natriuretic factor expression by two separate distal sequences. Circ Res. 2008;102:849–59. doi: 10.1161/CIRCRESAHA.107.170571. [DOI] [PubMed] [Google Scholar]
- 132.Han P, Li W, Lin CH, Yang J, Shang C, Nuernberg ST, et al. A long noncoding RNA protects the heart from pathological hypertrophy. Nature. 2014;514:102–6. doi: 10.1038/nature13596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Eagle KA, Nallamothu BK, Mehta RH, Granger CB, Steg PG, Van de Werf F, et al. Trends in acute reperfusion therapy for ST-segment elevation myocardial infarction from 1999 to 2006: we are getting better but we have got a long way to go. Eur Heart J. 2008;29:609–17. doi: 10.1093/eurheartj/ehn069. [DOI] [PubMed] [Google Scholar]
- 134.Windecker S, Bax JJ, Myat A, Stone GW, Marber MS. Future treatment strategies in ST-segment elevation myocardial infarction. Lancet. 2013;382:644–57. doi: 10.1016/S0140-6736(13)61452-X. [DOI] [PubMed] [Google Scholar]
- 135.White HD, Chew DP. Acute myocardial infarction. Lancet. 2008;372:570–84. doi: 10.1016/S0140-6736(08)61237-4. [DOI] [PubMed] [Google Scholar]
- 136.Liu Y, Li G, Lu H, Li W, Li X, Liu H, et al. Expression profiling and ontology analysis of long noncoding RNAs in post-ischemic heart and their implied roles in ischemia/ reperfusion injury. Gene. 2014;543:15–21. doi: 10.1016/j.gene.2014.04.016. [DOI] [PubMed] [Google Scholar]
- 137.Zangrando J, Zhang L, Vausort M, Maskali F, Marie PY, Wagner DR, et al. Identification of candidate long non-coding RNAs in response to myocardial infarction. BMC Genomics. 2014;15:460. doi: 10.1186/1471-2164-15-460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Broadbent HM, Peden JF, Lorkowski S, Goel A, Ongen H, Green F, et al. Susceptibility to coronary artery disease and diabetes is encoded by distinct, tightly linked SNPs in the ANRIL locus on chromosome 9p. Hum Mol Genet. 2008;17:806–14. doi: 10.1093/hmg/ddm352. [DOI] [PubMed] [Google Scholar]
- 139.Cugino D, Gianfagna F, Santimone I, de Gaetano G, Donati MB, Iacoviello L, et al. Type 2 diabetes and polymorphisms on chromosome 9p21: a meta-analysis. Nutr Metab Cardiovasc Dis. 2012;22:619–25. doi: 10.1016/j.numecd.2010.11.010. [DOI] [PubMed] [Google Scholar]
- 140.Jeck WR, Siebold AP, Sharpless NE. Review: a meta-analysis of GWAS and age-associated diseases. Aging Cell. 2012;11:727–31. doi: 10.1111/j.1474-9726.2012.00871.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Holdt LM, Teupser D. From genotype to phenotype in human atherosclerosis—recent findings. Curr Opin Lipidol. 2013;24:410–8. doi: 10.1097/MOL.0b013e3283654e7c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Holdt LM, Teupser D. Recent studies of the human chromosome 9p21 locus, which is associated with atherosclerosis in human populations. Arterioscler Thromb Vasc Biol. 2012;32:196–206. doi: 10.1161/ATVBAHA.111.232678. [DOI] [PubMed] [Google Scholar]
- 143.Holdt LM, Beutner F, Scholz M, Gielen S, Gabel G, Bergert H, et al. ANRIL expression is associated with atherosclerosis risk at chromosome 9p21. Arterioscler Thromb Vasc Biol. 2010;30:620–7. doi: 10.1161/ATVBAHA.109.196832. [DOI] [PubMed] [Google Scholar]
- 144.Holdt LM, Hoffmann S, Sass K, Langenberger D, Scholz M, Krohn K, et al. Alu elements in ANRIL non-coding RNA at chromosome 9p21 modulate atherogenic cell functions through trans-regulation of gene networks. PLoS Genet. 2013;9:e1003588. doi: 10.1371/journal.pgen.1003588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Cortez MA, Calin GA. MicroRNA identification in plasma and serum: a new tool to diagnose and monitor diseases. Expert Opin Biol Ther. 2009;9:703–11. doi: 10.1517/14712590902932889. [DOI] [PubMed] [Google Scholar]
- 146.Huang X, Yuan T, Tschannen M, Sun Z, Jacob H, Du M, et al. Characterization of human plasma-derived exosomal RNAs by deep sequencing. BMC Genomics. 2013;14:319. doi: 10.1186/1471-2164-14-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Chen X, Ba Y, Ma L, Cai X, Yin Y, Wang K, et al. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008;18:997–1006. doi: 10.1038/cr.2008.282. [DOI] [PubMed] [Google Scholar]
- 148.Hunter MP, Ismail N, Zhang X, Aguda BD, Lee EJ, Yu L, et al. Detection of microRNA expression in human peripheral blood microvesicles. PLoS ONE. 2008;3:e3694. doi: 10.1371/journal.pone.0003694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A. 2008;105:10513–8. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Gilad S, Meiri E, Yogev Y, Benjamin S, Lebanony D, Yerushalmi N, et al. Serum microRNAs are promising novel biomarkers. PLoS ONE. 2008;3:e3148. doi: 10.1371/journal.pone.0003148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Weber JA, Baxter DH, Zhang S, Huang DY, Huang KH, Lee MJ, et al. The microRNA spectrum in 12 body fluids. Clin Chem. 2010;56:1733–41. doi: 10.1373/clinchem.2010.147405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.van Empel VP, De Windt LJ, da Costa Martins PA. Circulating miRNAs: reflecting or affecting cardiovascular disease? Curr Hypertens Rep. 2012;14:498–509. doi: 10.1007/s11906-012-0310-7. [DOI] [PubMed] [Google Scholar]
- 153.Gupta SK, Bang C, Thum T. Circulating microRNAs as biomarkers and potential paracrine mediators of cardiovascular disease. Circ Cardiovasc Genet. 2010;3:484–8. doi: 10.1161/CIRCGENETICS.110.958363. [DOI] [PubMed] [Google Scholar]
- 154.Xu J, Zhao J, Evan G, Xiao C, Cheng Y, Xiao J. Circulating microRNAs: novel bio-markers for cardiovascular diseases. J Mol Med. 2012;90:865–75. doi: 10.1007/s00109-011-0840-5. [DOI] [PubMed] [Google Scholar]
- 155.da Silva Gomes AM, Silbiger VN. miRNAs as biomarkers of atrial fibrillation. Biomarkers. 2014:1–6. doi: 10.3109/1354750X.2014.954001. [DOI] [PubMed] [Google Scholar]
- 156.Zampetaki A, Willeit P, Drozdov I, Kiechl S, Mayr M. Profiling of circulating microRNAs: from single biomarkers to re-wired networks. Cardiovasc Res. 2012;93:555–62. doi: 10.1093/cvr/cvr266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Fasanaro P, D’Alessandra Y, Magenta A, Pompilio G, Capogrossi MC. MicroRNAs: promising biomarkers and therapeutic targets of acute myocardial ischemia. Curr Vasc Pharmacol. 2013 doi: 10.2174/15701611113119990011. [DOI] [PubMed] [Google Scholar]
- 158.Di Stefano V, Zaccagnini G, Capogrossi MC, Martelli F. microRNAs as peripheral blood biomarkers of cardiovascular disease. Vasc Pharmacol. 2011;55:111–8. doi: 10.1016/j.vph.2011.08.001. [DOI] [PubMed] [Google Scholar]
- 159.D’Alessandra Y, Devanna P, Limana F, Straino S, Di Carlo A, Brambilla PG, et al. Circulating microRNAs are new and sensitive biomarkers of myocardial infarction. Eur Heart J. 2010;31:2765–73. doi: 10.1093/eurheartj/ehq167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Chen X, Liang H, Zhang J, Zen K, Zhang CY. Secreted microRNAss: a new form of intercellular communication. Trends Cell Biol. 2012;22:125–32. doi: 10.1016/j.tcb.2011.12.001. [DOI] [PubMed] [Google Scholar]
- 161.Loyer X, Vion AC, Tedgui A, Boulanger CM. Microvesicles as cell–cell messengers in cardiovascular diseases. Circ Res. 2014;114:345–53. doi: 10.1161/CIRCRESAHA.113.300858. [DOI] [PubMed] [Google Scholar]
- 162.Ren J, Zhang J, Xu N, Han G, Geng Q, Song J, et al. Signature of circulating microRNAs as potential biomarkers in vulnerable coronary artery disease. PLoS ONE. 2013;8:e80738. doi: 10.1371/journal.pone.0080738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Wang GK, Zhu JQ, Zhang JT, Li Q, Li Y, He J, et al. Circulating microRNA: a novel potential biomarker for early diagnosis of acute myocardial infarction in humans. Eur Heart J. 2010;31:659–66. doi: 10.1093/eurheartj/ehq013. [DOI] [PubMed] [Google Scholar]
- 164.Sayed AS, Xia K, Yang TL, Peng J. Circulating microRNAs: a potential role in diagnosis and prognosis of acute myocardial infarction. Dis Markers. 2013;35:561–6. doi: 10.1155/2013/217948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Cui J, Zhao N. Consideration in selection of the control group in diagnostic tests. Int J Biol Sci. 2012;8:1418–9. doi: 10.7150/ijbs.5459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Olivieri F, Antonicelli R, Lorenzi M, D’Alessandra Y, Lazzarini R, Santini G, et al. Diagnostic potential of circulating miR-499-5p in elderly patients with acute non ST-elevation myocardial infarction. Int J Cardiol. 2013;167:531–6. doi: 10.1016/j.ijcard.2012.01.075. [DOI] [PubMed] [Google Scholar]
- 167.Olivieri F, Antonicelli R, Spazzafumo L, Santini G, Rippo MR, Galeazzi R, et al. Admission levels of circulating miR-499-5p and risk of death in elderly patients after acute non-ST elevation myocardial infarction. Int J Cardiol. 2014;172:e276–8. doi: 10.1016/j.ijcard.2013.12.203. [DOI] [PubMed] [Google Scholar]
- 168.Thum T, Mayr M. Review focus on the role of microRNA in cardiovascular biology and disease. Cardiovasc Res. 2012;93:543–4. doi: 10.1093/cvr/cvs085. [DOI] [PubMed] [Google Scholar]
- 169.Fichtlscherer S, Zeiher AM, Dimmeler S. Circulating microRNAs: biomarkers or mediators of cardiovascular diseases? Arterioscler Thromb Vasc Biol. 2011;31:2383–90. doi: 10.1161/ATVBAHA.111.226696. [DOI] [PubMed] [Google Scholar]
- 170.D’Alessandra Y, Carena MC, Spazzafumo L, Martinelli F, Bassetti B, Devanna P, et al. Diagnostic potential of plasmatic MicroRNA signatures in stable and unstable angina. PLoS ONE. 2013;8:e80345. doi: 10.1371/journal.pone.0080345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Hoekstra M, van der Lans CA, Halvorsen B, Gullestad L, Kuiper J, Aukrust P, et al. The peripheral blood mononuclear cell microRNA signature of coronary artery disease. Biochem Biophys Res Commun. 2010;394:792–7. doi: 10.1016/j.bbrc.2010.03.075. [DOI] [PubMed] [Google Scholar]
- 172.Tijsen AJ, Creemers EE, Moerland PD, de Windt LJ, van der Wal AC, Kok WE, et al. MiR423-5p as a circulating biomarker for heart failure. Circ Res. 2010;106:1035–9. doi: 10.1161/CIRCRESAHA.110.218297. [DOI] [PubMed] [Google Scholar]
- 173.Corsten MF, Dennert R, Jochems S, Kuznetsova T, Devaux Y, Hofstra L, et al. Circulating microRNA-208b and microRNA-499 reflect myocardial damage in cardiovascular disease. Circ Cardiovasc Genet. 2010;3:499–506. doi: 10.1161/CIRCGENETICS.110.957415. [DOI] [PubMed] [Google Scholar]
- 174.Voellenkle C, van Rooij J, Cappuzzello C, Greco S, Arcelli D, Di Vito L, et al. MicroRNA signatures in peripheral blood mononuclear cells of chronic heart failure patients. Physiol Genomics. 2010;42:420–6. doi: 10.1152/physiolgenomics.00211.2009. [DOI] [PubMed] [Google Scholar]
- 175.Caporali A, Meloni M, Vollenkle C, Bonci D, Sala-Newby GB, Addis R, et al. Deregulation of microRNA-503 contributes to diabetes mellitus-induced impairment of endothelial function and reparative angiogenesis after limb ischemia. Circulation. 2011;123:282–91. doi: 10.1161/CIRCULATIONAHA.110.952325. [DOI] [PubMed] [Google Scholar]
- 176.Dhahbi JM, Kim HJ, Mote PL, Beaver RJ, Spindler SR. Temporal linkage between the phenotypic and genomic responses to caloric restriction. Proc Natl Acad Sci U S A. 2004;101:5524–9. doi: 10.1073/pnas.0305300101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Pollack M, Phaneuf S, Dirks A, Leeuwenburgh C. The role of apoptosis in the normal aging brain, skeletal muscle, and heart. Ann N Y Acad Sci. 2002;959:93–107. doi: 10.1111/j.1749-6632.2002.tb02086.x. [DOI] [PubMed] [Google Scholar]
- 178.Higami Y, Shimokawa I. Apoptosis in the aging process. Cell Tissue Res. 2000;301:125–32. doi: 10.1007/s004419900156. [DOI] [PubMed] [Google Scholar]
- 179.Vausort M, Wagner DR, Devaux Y. Long noncoding RNAs in patients with acute myocardial infarction. Circ Res. 2014;115:668–77. doi: 10.1161/CIRCRESAHA.115.303836. [DOI] [PubMed] [Google Scholar]
- 180.Li D, Chen G, Yang J, Fan X, Gong Y, Xu G, et al. Transcriptome analysis reveals distinct patterns of long noncoding RNAs in heart and plasma of mice with heart failure. PLoS ONE. 2013;8:e77938. doi: 10.1371/journal.pone.0077938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Kumarswamy R, Bauters C, Volkmann I, Maury F, Fetisch J, Holzmann A, et al. Circulating long noncoding RNA, LIPCAR, predicts survival in patients with heart failure. Circ Res. 2014;114:1569–75. doi: 10.1161/CIRCRESAHA.114.303915. [DOI] [PubMed] [Google Scholar]
- 182.He X, He L, Hannon GJ. The guardian’s little helper: microRNAs in the p53 tumor suppressor network. Cancer Res. 2007;67:11099–101. doi: 10.1158/0008-5472.CAN-07-2672. [DOI] [PubMed] [Google Scholar]
- 183.Janssen HL, Reesink HW, Lawitz EJ, Zeuzem S, Rodriguez-Torres M, Patel K, et al. Treatment of HCV infection by targeting microRNA. N Engl J Med. 2013;368:1685–94. doi: 10.1056/NEJMoa1209026. [DOI] [PubMed] [Google Scholar]
- 184.Wheeler TM, Leger AJ, Pandey SK, MacLeod AR, Nakamori M, Cheng SH, et al. Targeting nuclear RNA for in vivo correction of myotonic dystrophy. Nature. 2012;488:111–5. doi: 10.1038/nature11362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Takahashi H, Carninci P. Widespread genome transcription: new possibilities for RNA therapies. Biochem Biophys Res Commun. 2014;452:294–301. doi: 10.1016/j.bbrc.2014.08.139. [DOI] [PubMed] [Google Scholar]
- 186.Kaufmann KB, Buning H, Galy A, Schambach A, Grez M. Gene therapy on the move. EMBO Mol Med. 2013;5:1642–61. doi: 10.1002/emmm.201202287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Verma IM, Weitzman MD. Gene therapy: twenty-first century medicine. Annu Rev Biochem. 2005;74:711–38. doi: 10.1146/annurev.biochem.74.050304.091637. [DOI] [PubMed] [Google Scholar]
- 188.Greco S, Fasanaro P, Castelvecchio S, D’Alessandra Y, Arcelli D, Di Donato M, et al. MicroRNA dysregulation in diabetic ischemic heart failure patients. Diabetes. 2012;61:1633–41. doi: 10.2337/db11-0952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Lee JH, Gao C, Peng G, Greer C, Ren S, Wang Y, et al. Analysis of transcriptome complexity through RNA sequencing in normal and failing murine hearts. Circ Res. 2011;109:1332–41. doi: 10.1161/CIRCRESAHA.111.249433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Ritter O, Haase H, Schulte HD, Lange PE, Morano I. Remodeling of the hypertrophied human myocardium by cardiac bHLH transcription factors. J Cell Biochem. 1999;74:551–61. doi: 10.1002/(sici)1097-4644(19990915)74:4<551::aid-jcb5>3.3.co;2-0. [DOI] [PubMed] [Google Scholar]
- 191.Ai J, Zhang R, Li Y, Pu J, Lu Y, Jiao J, et al. Circulating microRNA-1 as a potential novel biomarker for acute myocardial infarction. Biochem Biophys Res Commun. 2010;391:73–7. doi: 10.1016/j.bbrc.2009.11.005. [DOI] [PubMed] [Google Scholar]
- 192.Cheng Y, Tan N, Yang J, Liu X, Cao X, He P, et al. A translational study of circulating cell-free microRNA-1 in acute myocardial infarction. Clin Sci (Lond) 2010;119:87–95. doi: 10.1042/CS20090645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Gidlof O, Andersson P, van der Pals J, Gotberg M, Erlinge D. Cardiospecific microRNA plasma levels correlate with troponin and cardiac function in patients with ST elevation myocardial infarction, are selectively dependent on renal elimination, and can be detected in urine samples. Cardiology. 2011;118:217–26. doi: 10.1159/000328869. [DOI] [PubMed] [Google Scholar]
- 194.Li YQ, Zhang MF, Wen HY, Hu CL, Liu R, Wei HY, et al. Comparing the diagnostic values of circulating microRNAs and cardiac troponin T in patients with acute myocardial infarction. Clinics (Sao Paulo) 2013;68:75–80. doi: 10.6061/clinics/2013(01)OA12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Wang R, Li N, Zhang Y, Ran Y, Pu J. Circulating microRNAs are promising novel biomarkers of acute myocardial infarction. Intern Med J. 2011;50:1789–95. doi: 10.2169/internalmedicine.50.5129. [DOI] [PubMed] [Google Scholar]
- 196.Long G, Wang F, Duan Q, Yang S, Chen F, Gong W, et al. Circulating miR-30a, miR-195 and let-7b associated with acute myocardial infarction. PLoS ONE. 2012;7:e50926. doi: 10.1371/journal.pone.0050926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Long G, Wang F, Duan Q, Chen F, Yang S, Gong W, et al. Human circulating microRNA-1 and microRNA-126 as potential novel indicators for acute myocardial infarction. Int J Biol Sci. 2012;8:811–8. doi: 10.7150/ijbs.4439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Fichtlscherer S, De Rosa S, Fox H, Schwietz T, Fischer A, Liebetrau C, et al. Circulating microRNAs in patients with coronary artery disease. Circ Res. 2010;107:677–84. doi: 10.1161/CIRCRESAHA.109.215566. [DOI] [PubMed] [Google Scholar]
- 199.Hullinger TG, Montgomery RL, Seto AG, Dickinson BA, Semus HM, Lynch JM, et al. Inhibition of miR-15 protects against cardiac ischemic injury. Circ Res. 2012;110:71–81. doi: 10.1161/CIRCRESAHA.111.244442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Chen J, Huang ZP, Seok HY, Ding J, Kataoka M, Zhang Z, et al. mir-17-92 cluster is required for and sufficient to induce cardiomyocyte proliferation in postnatal and adult hearts. Circ Res. 2013;112:1557–66. doi: 10.1161/CIRCRESAHA.112.300658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Fiedler J, Jazbutyte V, Kirchmaier BC, Gupta SK, Lorenzen J, Hartmann D, et al. MicroRNA-24 regulates vascularity after myocardial infarction. Circulation. 2011;124:720–30. doi: 10.1161/CIRCULATIONAHA.111.039008. [DOI] [PubMed] [Google Scholar]
- 202.van Rooij E, Sutherland LB, Thatcher JE, DiMaio JM, Naseem RH, Marshall WS, et al. Dysregulation of microRNAs after myocardial infarction reveals a role of miR-29 in cardiac fibrosis. Proc Natl Acad Sci U S A. 2008;105:13027–32. doi: 10.1073/pnas.0805038105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Ren XP, Wu J, Wang X, Sartor MA, Qian J, Jones K, et al. MicroRNA-320 is involved in the regulation of cardiac ischemia/reperfusion injury by targeting heat-shock protein 20. Circulation. 2009;119:2357–66. doi: 10.1161/CIRCULATIONAHA.108.814145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Eulalio A, Mano M, Dal Ferro M, Zentilin L, Sinagra G, Zacchigna S, et al. Functional screening identifies miRNAs inducing cardiac regeneration. Nature. 2012;492:376–81. doi: 10.1038/nature11739. [DOI] [PubMed] [Google Scholar]
- 205.van Rooij E, Sutherland LB, Qi X, Richardson JA, Hill J, Olson EN. Control of stress-dependent cardiac growth and gene expression by a microRNA. Science. 2007;316:575–9. doi: 10.1126/science.1139089. [DOI] [PubMed] [Google Scholar]
- 206.Callis TE, Pandya K, Seok HY, Tang RH, Tatsuguchi M, Huang ZP, et al. MicroRNA-208a is a regulator of cardiac hypertrophy and conduction in mice. J Clin Invest. 2009;119:2772–86. doi: 10.1172/JCI36154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Montgomery RL, Hullinger TG, Semus HM, Dickinson BA, Seto AG, Lynch JM, et al. Therapeutic inhibition of miR-208a improves cardiac function and survival during heart failure. Circulation. 2011;124:1537–47. doi: 10.1161/CIRCULATIONAHA.111.030932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Ucar A, Gupta SK, Fiedler J, Erikci E, Kardasinski M, Batkai S, et al. The miRNA-212/132 family regulates both cardiac hypertrophy and cardiomyocyte autophagy. Nat Commun. 2012;3:1078. doi: 10.1038/ncomms2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Care A, Catalucci D, Felicetti F, Bonci D, Addario A, Gallo P, et al. MicroRNA-133 controls cardiac hypertrophy. Nat Med. 2007;13:613–8. doi: 10.1038/nm1582. [DOI] [PubMed] [Google Scholar]
- 210.Pan Z, Sun X, Shan H, Wang N, Wang J, Ren J, et al. MicroRNA-101 inhibited postinfarct cardiac fibrosis and improved left ventricular compliance via the FBJ osteosarcoma oncogene/transforming growth factor-beta1 pathway. Circulation. 2012;126:840–50. doi: 10.1161/CIRCULATIONAHA.112.094524. [DOI] [PubMed] [Google Scholar]
- 211.Li RC, Tao J, Guo YB, Wu HD, Liu RF, Bai Y, et al. In vivo suppression of microRNA-24 prevents the transition toward decompensated hypertrophy in aortic-constricted mice. Circ Res. 2013;112:601–5. doi: 10.1161/CIRCRESAHA.112.300806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Huang ZP, Chen J, Seok HY, Zhang Z, Kataoka M, Hu X, et al. MicroRNA-22 regulates cardiac hypertrophy and remodeling in response to stress. Circ Res. 2013;112:1234–43. doi: 10.1161/CIRCRESAHA.112.300682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.da Costa Martins PA, Salic K, Gladka MM, Armand AS, Leptidis S, el Azzouzi H, et al. MicroRNA-199b targets the nuclear kinase Dyrk1a in an auto-amplification loop promoting calcineurin/NFAT signalling. Nat Cell Biol. 2010;12:1220–7. doi: 10.1038/ncb2126. [DOI] [PubMed] [Google Scholar]
- 214.Ganesan J, Ramanujam D, Sassi Y, Ahles A, Jentzsch C, Werfel S, et al. MiR-378 controls cardiac hypertrophy by combined repression of mitogen-activated protein kinase pathway factors. Circulation. 2013;127:2097–106. doi: 10.1161/CIRCULATIONAHA.112.000882. [DOI] [PubMed] [Google Scholar]