Abstract
Terpenoids are a large and highly diverse group of natural products. All terpenoids are biosynthesized from isoprenyl diphosphate formed by the consecutive condensation of the five-carbon monomer isopentenyl diphosphate (IPP) to its isomer dimethylallyl diphosphate (DMAPP). Two distinct biosynthetic pathways produce the essential primary metabolites IPP and DMAPP: the 2-C-methylerythritol 4-phosphate pathway and the mevalonate pathway. The isoprenyl substrates can be cyclized by terpene cyclase into single-ring or multi-ring products, which can be further diversified by subsequent modification reactions, such as hydroxylation and glycosylation. This review article describes the biosynthetic pathways of terpenoids produced by Streptomyces and their related novel enzymes.
Introduction
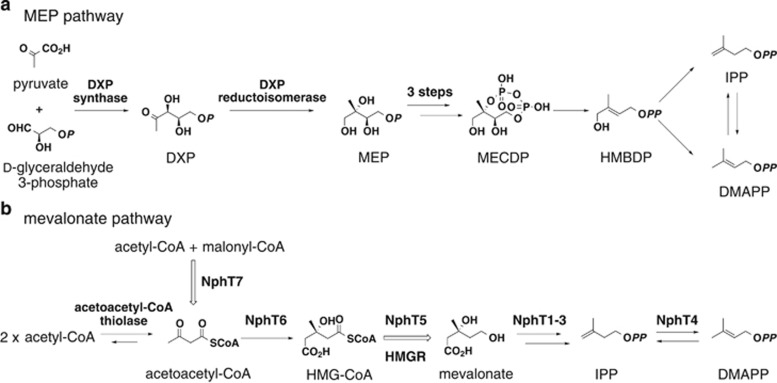
To date, over 50 000 terpenoids with diverse structures have been isolated from natural sources.1 Diverse terpenoid skeletons are generally biosynthesized from two simple five-carbon units: isopentenyl diphosphate (IPP) and dimethylallyl diphosphate (DMAPP).2, 3, 4, 5, 6, 7, 8 Multiple units are condensed and then undergo various modification reactions, including intricate cyclizations, hydroxylations and glycosylations, generating the structural diversity observed in terpenoids. Two completely distinct biosynthetic pathways produce the essential primary metabolites IPP and DMAPP: the 2-C-methylerythritol 4-phosphate (MEP) pathway and the mevalonate pathway (Figure 1).5, 6, 8 The distribution of these two pathways across the kingdoms of living organisms has largely been determined.
Figure 1.
MEP (a) and mevalonate pathways (b) for IPP and DMAPP. MECDP, 2-C-methyl-D-erythritol 2,4-cyclodiphosphate; HMBDP, 1-hydroxy-2-methyl-2-(E)-butenyl-4-diphosphate.
The mevalonate pathway was discovered in eukaryotes in the 1960s, and it was believed that bacteria also used this pathway.2, 3 However, 1-deoxy-D-xylulose 5-phosphate (DXP) synthase and DXP reductoisomerase, the first and second enzymes in the MEP pathway, were discovered in Escherichia coli in 1996 and 1998, respectively.9, 10, 11 Subsequently, it has been shown that most bacteria use the MEP pathway, and very few use the mevalonate pathway.6 This review gives an overview of the biosynthetic pathways of terpenoids produced by Streptomyces and their related novel enzymes.
Purification of the Mevalonate Pathway Rate-Limiting Enzyme
3-Hydroxy-3-methylglutaryl-CoA (HMG-CoA) reductase (HMGR) is the rate-limiting enzyme in the mevalonate pathway and is also a target molecule for statin drugs.2, 3, 7 For this reason, it has been cloned from many eukaryotes, including humans, and analyzed in detail. It has also been cloned from the archaebacteria Haloferax volcanii and Sulfolobus solfataricus, and its enzymatic properties have been characterized.12, 13 HMGR (EC 1.1.1.34) derived from these eukaryotes and archaebacteria reduces HMG-CoA using NADPH as a coenzyme, yielding mevalonate. However, an HMGR enzyme that similarly synthesizes mevalonate had not been identified in actinomycetes, a type of eubacteria. At the same time, the only HMGR that had been cloned from eubacteria was the HMGR (EC 1.1.1.88) from Pseudomonas mevalonii, which can grow using mevalonate as its sole carbon source.14 For this HMGR, equilibrium favors mevalonate conversion to HMG-CoA. This HMGR, which breaks down mevalonate, differs from HMGRs that synthesize mevalonate and is known to have a greater affinity for NAD(H) than NADP(H) as a coenzyme. P. mevalonii HMGR shares no more than approximately 25% amino-acid sequence identity with eukaryotic or archaebacterial HMGRs and lacks several conserved sequences, which might account for these enzyme differences.
Until 1995, it was believed that all actinomycetes possessed a mevalonate pathway similar to eukaryotes because terpenoid molecules such as terpentecin, furaquinocin, napyradiomycin and naphterpin are all produced by Streptomyces using the mevalonate pathway.15, 16, 17, 18, 19, 20 However, no research examining the enzymes involved in the Streptomyces mevalonate pathway had been reported at that time, although we were able to detect activity converting [3-14C]HMG-CoA to mevalonate in the presence of NADPH in a crude extract prepared from the cells of a naphterpin-producing Streptomyces sp. CL190.19 This means that CL190 encodes the same mevalonate-synthesizing HMGR as eukaryotes, prompting us to clone the mevalonate-synthesizing HMGR gene from CL190 by designing a variety of degenerate primers based on conserved sequences from relevant enzymes and performing PCR cloning. However, we were unable to obtain target DNA fragments, leading us to consider PCR cloning unsuitable, and we instead worked on purifying the enzyme. Subsequently, we successfully purified HMGR from a 10 liter culture of CL190 bacterial cells.19 Next, we determined the N-terminal amino-acid sequence and, using the colony hybridization method, obtained the complete nucleotide sequence of the HMGR gene. This was the first report of cloning the gene for mevalonate-synthesizing HMGR (EC 1.1.1.34) from eubacteria. By analyzing the sequence of HMGR from CL190 and the conserved sequences of mevalonate-synthesizing HMGRs, it became clear that HMGR enzymes from eukaryotes and Streptomyces differ slightly from one another, which likely led to PCR cloning failure.
Cloning of the Mevalonate Pathway Gene Cluster
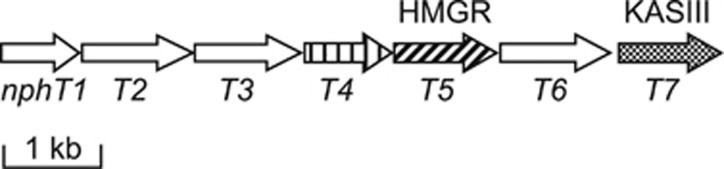
After we obtained the HMGR nucleotide sequence from the CL190 strain, we also cloned its flanking genes. Surprisingly, we found that seven genes, including five genes for enzymes involved in the mevalonate pathway, form a gene cluster oriented in the same direction (Figure 2).20 The five genes encoding HMG-CoA synthase (NphT6), HMG-CoA reductase (HMGR, NphT5), mevalonate kinase (NphT1), phosphomevalonate kinase (NphT3) and diphosphomevalonate decarboxylase (NphT2) form a gene cluster. This is the first mevalonate pathway gene cluster cloned from prokaryote. Interestingly, a gene for an enzyme (NphT4) with unknown function is included in this gene cluster, and the functional assignment of this gene is described below. The furthest downstream gene in this cluster is the gene for β-ketoacyl-[acyl carrier protein (ACP)] synthase (KAS III, later NphT7). KAS III typically catalyzes the initial reaction in fatty acid biosynthesis, condensing malonyl-ACP and acetyl-CoA to synthesize acetoacetyl-ACP. The function of NphT7, a KAS III homolog, is also described below.
Figure 2.
Gene cluster for the mevalonate pathway in Streptomyces sp. CL190. HMGR, 3-hydroxy-3-methylglutaryl-CoA reductase; KAS III, β-ketoacyl-[acyl carrier protein] synthase.
When 6.7 kb DNA fragments encoding six of these seven genes (nphT1–6, with nphT7 excluded) were inserted downstream of the lac promoter (lacP) in the E. coli plasmid pUC118, the transformants were able to use the mevalonate pathway.20 In other words, although E. coli originally uses the MEP pathway to synthesize IPP and DMAPP, this transformant used the introduced mevalonate pathway in addition to the endogenous MEP pathway.
As the details are described in separate review articles, the E. coli transformant harboring the introduced mevalonate pathway genes made a substantial contribution clarifying the MEP pathway, which at that time was elusive.5, 6, 8 In brief, three genes involved in the MEP pathway leading to MECDP from MEP were identified by selecting E. coli mutants that grew when the mevalonate pathway was induced but died when it was not; each function of the complementary genes was thus elucidated (Figure 1).21, 22, 23
Discovery of Type 2 IPP Isomerase
As mentioned above, a gene for an enzyme with unknown function (NphT4) was found in the mevalonate pathway gene cluster cloned from CL190 (Figure 2). Although homologs were identified in many organisms through homology searches using the Basic Local Alignment Search Tool, none of these data directly suggested a possible function of NphT4. Nonetheless, given that the nphT4 gene is located in the CL190 mevalonate pathway gene cluster, it seems highly likely that NphT4 is involved in the mevalonate pathway. As neither the gene for acetoacetyl-CoA thiolase nor IPP isomerase, which catalyze the first and final reactions of the mevalonate pathway, respectively, was identified in this gene cluster, NphT4 might catalyze one of these reactions. Interestingly, a homology search revealed that the Gram-positive bacteria Staphylococcus aureus and Enterococcus faecalis have NphT4 homologs. Although these Gram-positive bacteria use the mevalonate pathway, an IPP isomerase necessary for growth had not been identified in these bacteria. However, acetoacetyl-CoA thiolase had already been identified in these bacteria.7 Thus we hypothesized that NphT4 functions as an IPP isomerase and therefore attempted to measure the activity of recombinant NphT4.
The established method for the measurement of IPP isomerase activity is to react IPP isomerase with [1-14C]IPP in the presence of Mg2+ ions, treat the reaction mixture with acid and extract a derivative of the reaction product, [1-14C]DMAPP, with toluene.24 The extracted [1-14C]DMAPP derivative is detected with a scintillation counter. Using this method, IPP isomerase activity from E. coli was readily detected. However, the enzymatic activity of the recombinant NphT4 was unclear. Greater activity was detected than in the negative control, to which no protein was added, but the activity was extremely weak. When we tried to concentrate the NphT4 protein, we noticed that it was yellow and realized that NphT4 is a flavin enzyme.25 We therefore added flavin adenine dinucleotide and flavin mononucleotide to the reaction mixture and tried to measure the reaction again, but the activity remained low. We tested various coenzymes and finally succeeded in clearly detecting IPP isomerase activity when NADPH or NADH was added to the reaction system.25 This meant that NphT4 is a novel IPP isomerase that does not show amino-acid sequence homology with known IPP isomerases and requires both flavin mononucleotide and NAD(P)H. On this basis, we proposed designating the known IPP isomerase and the NphT4 homolog as ‘type 1 IPP isomerase’ and ‘type 2 IPP isomerase,’ respectively.25
Discovery of Acetoacetyl-CoA Synthase
As mentioned, the gene for KAS III homolog (NphT7) was the furthest downstream in the mevalonate pathway gene cluster cloned from CL190 (Figure 2). This mevalonate pathway gene cluster was subsequently identified in bacteria that produce terpentecin, furaquinocin and napyradiomycin, and each structure of the gene clusters was nearly identical.15, 16, 17 Moreover, the KAS III homologs appeared the furthest downstream in all of these gene clusters. We therefore hypothesized that the KAS III homolog (NphT7) is also involved in the mevalonate pathway.
Acetoacetyl-CoA is a mevalonate pathway precursor. Only acetoacetyl-CoA thiolase, a typical enzyme of the thiolase superfamily, was believed to synthesize de novo acetoacetyl-CoA from two molecules of acetyl-CoA. This enzymatic reaction is an equilibrium reaction that favors decomposition of acetoacetyl-CoA and is therefore not useful to synthesize acetoacetyl-CoA. An enzyme that favors production of acetoacetyl-CoA would be desirable to produce useful compounds derived from acetoacetyl-CoA. We showed that the NphT7 recombinant enzyme synthesizes acetoacetyl-CoA from acetyl-CoA and malonyl-CoA and also found that this new enzyme does not exhibit acetoacetyl-CoA thiolysis activity, thus we named the enzyme acetoacetyl-CoA synthase.26
We cloned the nphT7 gene into a Streptomyces lividans TK23 host together with the HMG-CoA synthase gene (nphT5) and the HMG-CoA reductase gene (nphT6) for mevalonate production. Mevalonate synthesis was approximately 250% greater than when nphT7 was not included.26 This was likely because introduction of nphT7, the gene for acetoacetyl-CoA synthase, increased the amount of acetoacetyl-CoA synthesis within the TK23 host, leading to increased mevalonate production. This experimental result could potentially inform the increased production of useful acetoacetyl-CoA-derived compounds, such as 1-butanol (biofuel) and 3-hydroxybutyric acid (raw material for biodegradable plastics), and useful terpenoids, such as carotenoids (pigment, antioxidant), taxol (anticancer agent), artemisinin (antimalarial drug) and coenzyme Q10 (antioxidant). Actual examples of the nphT7 gene being used to produce useful compounds include the heterologous production of 1-butanol in Synechococcus elongatus PCC 7942 and the heterologous production of hydroxyalkanoic acid in Synechocystis sp. PCC 6803.27, 28
Discovery of Aromatic Substrate Prenyltransferase NphB
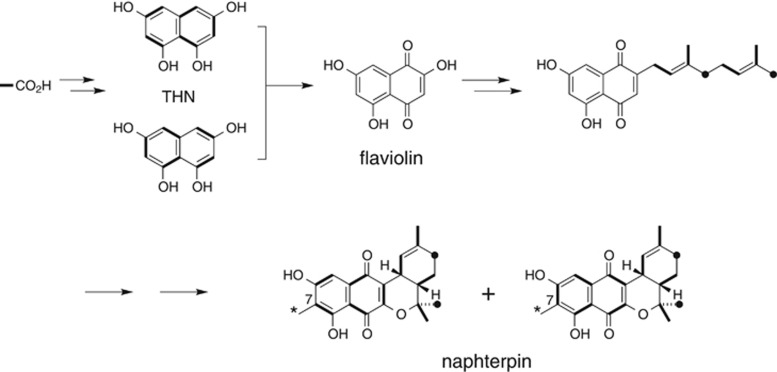
Naphterpin was isolated from the metabolites of the actinomycete Streptomyces sp. CL190 during lipid peroxidation inhibitor screening (Figure 3).29 Tracer experiments using precursors labeled with stable isotopes have shown that naphterpin is a terpenoid–polyketide hybrid compound with both terpenoid and polyketide moieties.18 The polyketide moiety is hypothesized to be biosynthesized via tetrahydroxynaphthalene (THN), a compound with bilateral symmetry. It has also been shown that the terpenoid moiety derives from geranyl diphosphate (GPP), a 10-carbon compound that is biosynthesized in the mevalonate pathway, and that the methyl group at C7 is derived from methionine. From these experimental results, the putative biosynthetic pathway, shown in Figure 3, has been proposed.18
Figure 3.
Putative biosynthetic pathway for naphterpin. Asterisks represent the methyl group derived from methionine. THN, tetrahydroxynaphthalene.
Identification of the naphterpin biosynthetic gene cluster
When the nucleotide sequences upstream and downstream of the mevalonate pathway gene cluster of the naphterpin-producing CL190 were characterized and a homology search was performed, a possible gene cluster encoding 14 genes (nphA to nphN) involved in naphterpin biosynthesis was discovered.30 Among these, NphB shows homology (albeit low, at approximately 20%) with hydroxyphenyl pyruvate:dimethylallyl transferases (CloQ and NovQ), which are involved in the biosynthesis of the aminocoumarin antibiotics clorobiocin and novobiocin, respectively.31 Both enzymes catalyze dimethylallyl group transfer to hydroxyphenyl pyruvate, the intermediate in the biosynthesis of these antibiotics.
Given that NphB shows homology with hydroxyphenyl pyruvate:dimethylallyl transferase and that an NphB-disruption mutant does not produce naphterpin, we considered NphB to be the aromatic substrate prenyltransferase that catalyzes the condensation reaction between the terpenoid and polyketide moieties. Next, we examined the NphB-catalyzed reaction in vitro.
Characterization of NphB prenyltransferase
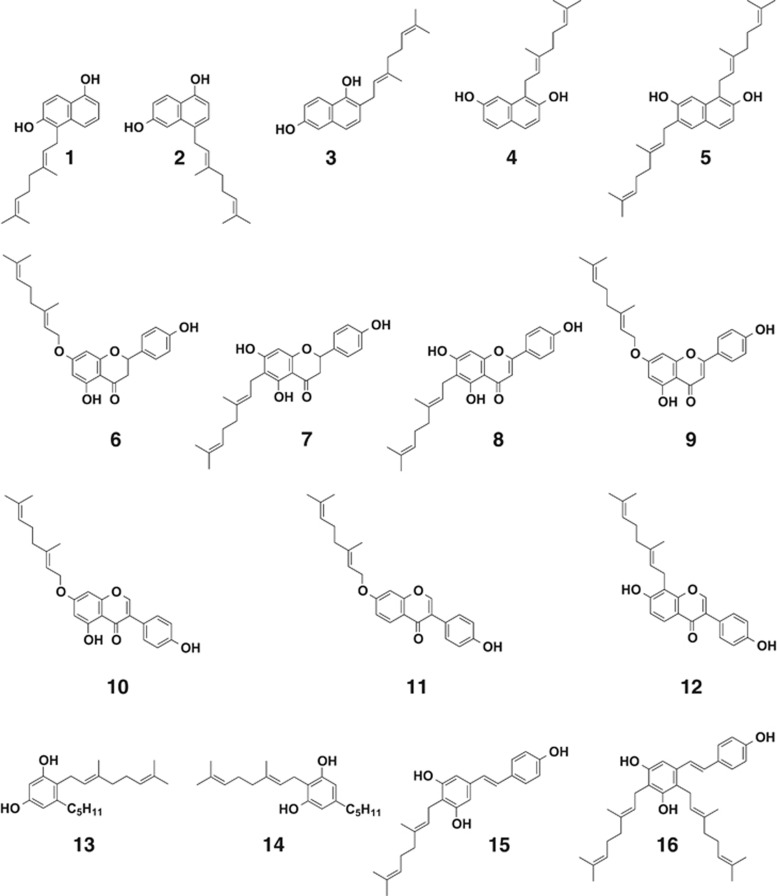
Recombinant NphB was purified from the E. coli transformant overexpressing the nphB gene. NphB enzyme activity was investigated with 1,3-dihydroxynaphthalene (DHN), 1,6-DHN and 2,7-DHN, all of which structurally resemble THN but are more stable, as prenyl group acceptors and DMAPP, GPP and farnesyl diphosphate (FPP) as prenyl group donors. When 1,6-DHN and 2,7-DHN were added as substrates, the reaction progresses only in the presence of magnesium ions (Figure 4).30 GPP (10 carbons) was the optimum prenyl group donor, whereas very little activity was observed with FPP (15 carbons). With regard to metal ions, magnesium was essential for the reaction. These findings suggest that, as expected, NphB is a geranyl transferase and displays promiscuous substrate specificity against geranyl group acceptors.30
Figure 4.
Prenylated compounds synthesized by NphB prenyltransferase. 1, 5-geranyl 1,6-DHN; 2, 4-geranyl 1,6-DHN; 3, 2-geranyl 1,6-DHN; 4, 1-geranyl 2,7-DHN; 5, 1,6-digeranyl 2,7-DHN; 6, 7-O-geranyl naringenin; 7, 6-geranyl naringenin; 8, 6-geranyl apigenin; 9, 7-O-geranyl apigenin; 10, 7-O-geranyl genistein; 11, 7-O-geranyl daidzein; 12, 8-geranyl daidzein; 13, 2-geranyl olivetol; 14, 4-geranyl olivetol; 15, 4-geranyl resveratrol; 16, 2,4-digeranyl resveratrol.
To date, however, we have been unable to identify the physiological substrate of NphB. Considering that NphC synthesizes THN and that NphD converts THN to flaviolin, we expected flaviolin to be an NphB substrate. Contrary to expectation, flaviolin was not a good substrate for NphB. In future, it might be possible to identify the physiological substrate for NphB by performing functional analysis of other genes in the naphterpin biosynthetic gene cluster.
Chemoenzymatic synthesis of prenylated flavonoids and polyketides
Prenylated flavonoids are produced by plants and have a range of activity, including antioxidant, antibacterial and antitumor activities as well as controlling multi-drug resistance.32 Only small amounts of prenylated flavonoids can be isolated naturally, however, and many aspects of their bioactivity and mechanisms of action have not been clarified. Methods to synthesize prenylated flavonoids artificially using enzymatic reactions might lead to the discovery of more useful biological activity and enable research into the mode of action. We therefore attempted prenylation of flavonoids using NphB as a biocatalyst. As shown in Figure 4, NphB attached a geranyl group to the 6-position carbon atom or the oxygen atom to the 7-position hydroxyl group of the flavonoids naringenin, apigenin, genistein and daidzein. NphB is the first example of an enzyme that attaches a geranyl group to an oxygen atom.33
In addition to flavonoids, we investigated the prenylation of plant-derived polyketides with aromatic rings. The results established that NphB attached a prenyl group to olivetol, an intermediate in cannabinoid biosynthesis, and resveratrol, an antioxidant found in red wine (Figure 4).33 Geranylated olivetol synthesized by NphB is a precursor of tetrahydrocannabinol, the psychoactive ingredient of cannabis, but geranylated olivetol does not itself have any action. There have been cases of geranylated resveratrol isolated from African teak destined for timber, but no physiological activity has been reported for this compound.
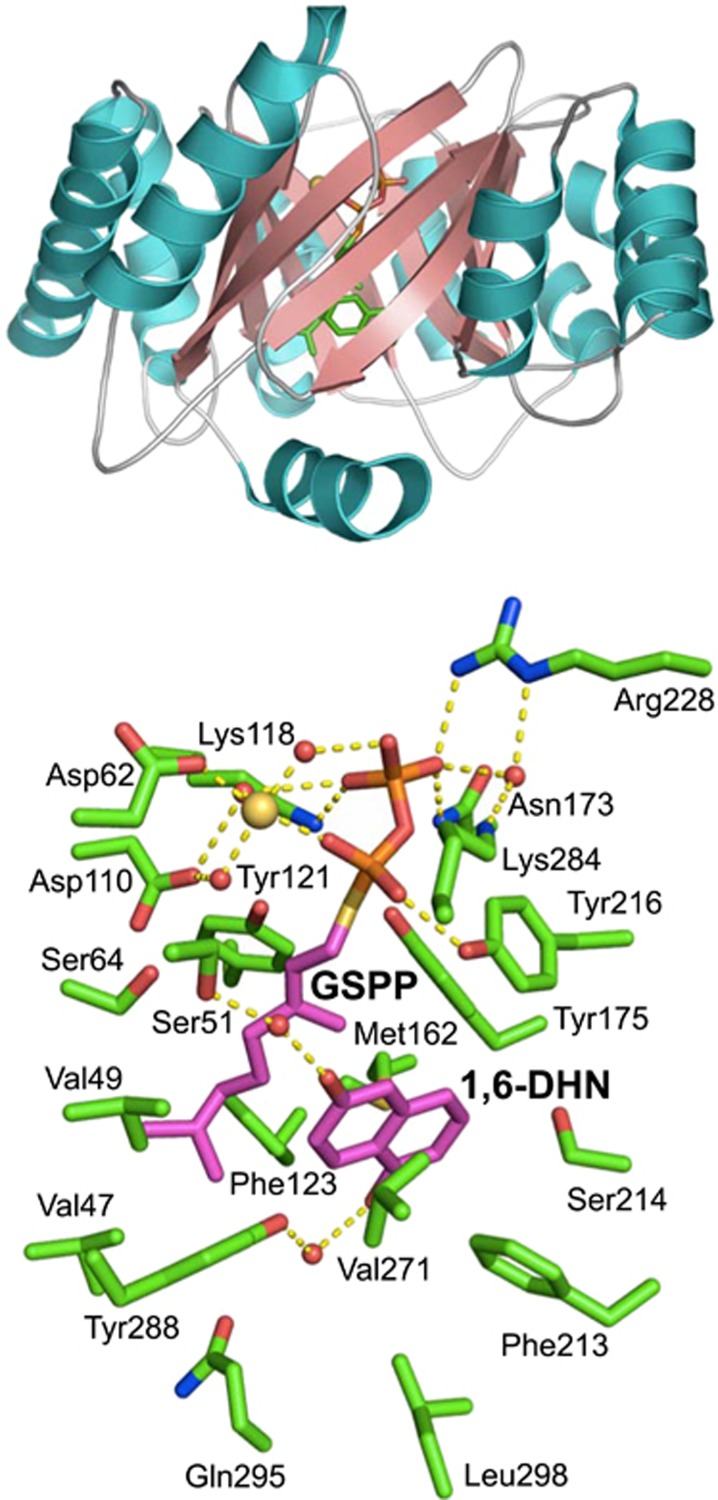
Crystal structure of NphB prenyltransferase
To analyze the 3-D structure of NphB, which dictates the mechanism of its reaction and its substrate specificity, crystals of the enzyme were prepared, and the crystal structure was determined at 1.55 Å resolution.30 The overall structure is shown in Figure 5. At first sight, the enzyme appears to have a common ‘TIM barrel’ protein fold structure. It has two significant differences, however, and appears to be a novel protein fold termed the ‘PT barrel.’ First, typical TIM barrels are formed by linkages of β-strands and α-helices, but this enzyme contains an (ααββ)4-(αββα) structural motif. Second, the barrel portion of TIM barrels contains parallel β-strands, whereas the barrel of the present enzyme is formed from anti-parallel β-strands. Owing to the αββα architecture of this protein fold, this group of enzymes was later designated as ABBA prenyltransferases.34
Figure 5.
3-D structures of NphB prenyltransferase. ABBA-fold structure (upper). Active site pocket with the 1,6-DHN and GSPP substrates (lower).
The substrate 1,6-DHN and the substrate analog geranyl thiodiphosphate (GSPP) were introduced to the enzyme crystal using the soaking method, and the structure of the substrate–enzyme complex was determined (Figure 5). First, from the structure of the complex, it was observed that lysine at amino-acid residue 118 in the NphB sequence (Lys 118), Asn 173 and Arg 228 tether the β-phosphate of GSPP. The α-phosphate of GSPP forms a hydrogen bond with Tyr 216 and Lys 284 and coordinates to a magnesium ion. The carboxyl group of Asp 62 bonds with α-phosphate through coordinate bonds with a magnesium ion. The hydrophobic geranyl bases are surrounded by the side chains of Val 49, Phe 123, Met 162, Tyr 175 and Tyr 216. At the same time, the two aromatic rings of 1,6-DHN are suspended between the side chains of Met 162 and Phe 213. The short α-helix at the C-terminus of NphB forms the ‘cap’ of the barrel, and the side chains of Gln 295 and Leu 298 on this cap form one side of the wall of the aromatic substrate-binding pocket. In addition, Ser 51 and Tyr 288 bond to the hydroxyl groups of 1,6-DHN via water molecules. Thus most of the amino-acid residues involved in substrate binding were identified from the structural analysis.
Based on the 3-D structure of NphB, we hypothesize that the reaction mechanism involves the three tyrosine residues Tyr 121, Tyr 175 and Tyr 216 that surround the C1 atom adjacent to the sulfur atom of GSPP (Figure 5). From this spatial arrangement, it appears that the π-electrons of the aromatic rings of the tyrosine bases interact with the geranyl carbocation produced by GPP heterolysis (cation–π interaction), thus stabilizing the geranyl carbocation. Presumably, an electrophilic substitution reaction occurs, in which the electron pair of the prenyl receptor aromatic ring attacks the geranyl carbocation, forming geranyl compounds via carbocation intermediates. This reaction mechanism adequately explains the promiscuous substrate specificity of NphB.
Other prenyltransferases
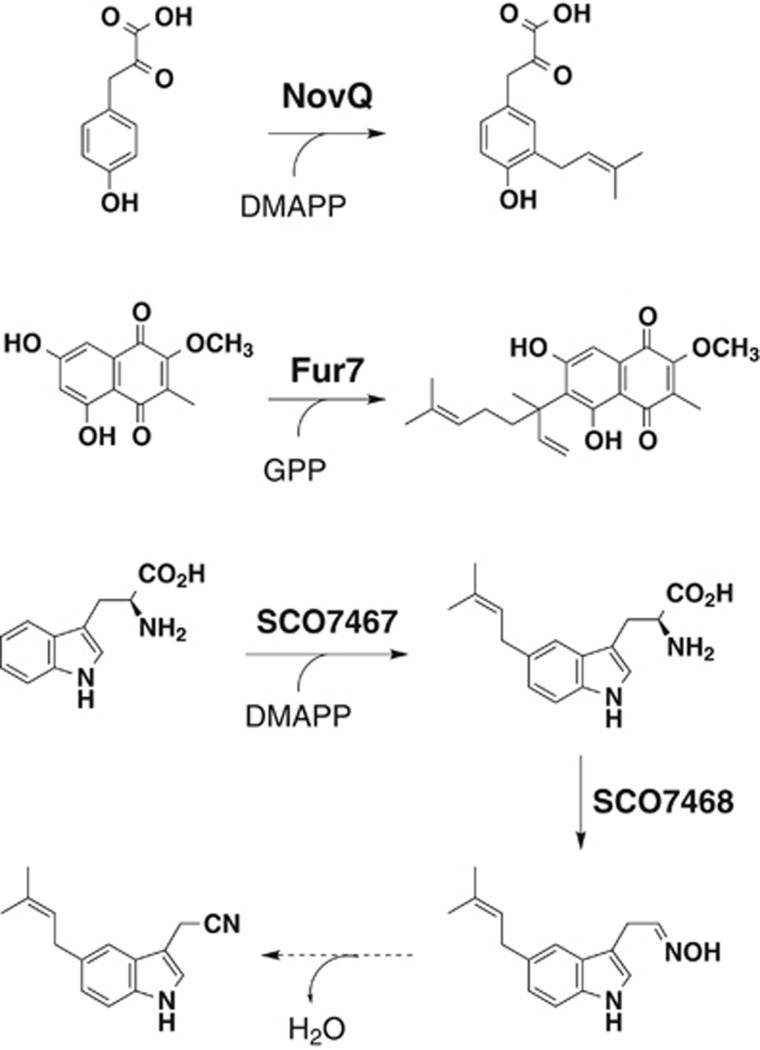
We characterized two NphB homologs: NovQ prenyltransferase from a novobiocin-producing Streptomyces niveus 16259 and Fur7 prenyltransferase from a furaquinocin-producing Streptomyces sp. KO-3988 (Figure 6).35, 36 NovQ catalyzes the transfer of a dimethylallyl group to 4-hydroxyphenylpyruvate (4-HPP) to yield 3-dimethylallyl-4-HPP, an intermediate of novobiocin.35 In addition to the prenylation of 4-HPP, NovQ catalyzes carbon–carbon-based and carbon–oxygen-based prenylations of a diverse collection of phenylpropanoids, such as p-coumaric acid and caffeic acid, flavonoids and DHNs. Fur7 catalyzes the transfer of a geranyl group to 2-methoxy-3-methyl-flaviolin to yield 6-prenyl-2-methoxy-3-methyl-flaviolin, an intermediate of furaquinocin.36 In addition, characterization of SCO7467 prenyltransferase, which catalyzes the prenylation of L-tryptophan to form 5-dimethylallyl-L-tryptophan, revealed novel L-tryptophan metabolism leading to 5-dimethylallylindole-3-acetonitrile in Streptomyces coelicolor A3(2) (Figure 6).37
Figure 6.
Other prenyltransferases from Streptomyces. NovQ catalyzes dimethylallyl transfer to 4-HPP to give 3-dimethylallyl-4-HPP. Fur7 catalyzes the transfer of a geranyl group to 2-methoxy-3-methyl-flaviolin to yield 6-prenyl-2-methoxy-3-methyl-flaviolin. SCO7467 catalyzes dimethylallyl transfer to tryptophan to give 5-dimethylallyl tryptophan, which is further transformed to 5-dimethylallylindole-3-acetaldoxime by a flavin-dependent monooxygenase SCO7468. Finally, dehydration of the acetaldoxime presumably occurs to yield 5-dimethylallylindole-3-acetonitrile.
Reaction Mechanisms of Terpene Cyclases
Lavanducyanin biosynthesis
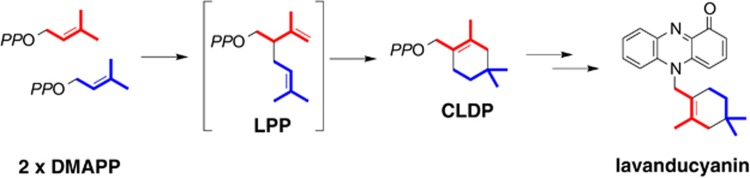
In addition to naphterpin, Streptomyces sp. CL190 produces lavanducyanin (Figure 7).38 Lavanducyanin has a unique structure in which a characteristic cyclolavandulyl group (a cyclic terpenoid) is linked to a phenazine skeleton. The cyclolavandulyl skeleton was discovered in nature in the 1960s, but there were no reports of the biosynthetic mechanism. The cyclolavandulyl skeleton was assumed to be biosynthesized by a cyclase following the formation of branched-chain lavandulyl diphosphate (LPP), which is derived from bimolecular DMAPP with a homolog of the condensation enzyme isoprenyl diphosphate synthase (IDS) (Figure 7).39 However, no such cyclase has been identified.
Figure 7.
Biosynthesis of lavanducyanin. CLDP is biosynthesized via the condensation of two molecules of DMAPP and the subsequent cyclization by the actions of CLDS. The cyclolavandulyl moiety is linked to a phenazine skeleton by unidentified prenyltransferase.
The genes for the biosynthesis of secondary metabolites in actinomycetes almost certainly form clusters, suggesting that if the LPP synthase genes could be identified, the cyclase gene would be found nearby. We therefore searched for the IDS homolog by examining a draft genome sequence of the CL190 strain from which we discovered at least 10 IDS homologs.40 Many of the gene products exhibited high homology to known IDSs that synthesize acyclic terpenoids such as GPP, FPP and geranylgeranyl diphosphate (GGPP) and were therefore unlikely to be the LPP synthase. IDS3, however, exhibited low homology to IDSs with known function and relatively high homology to an enzyme called Mcl22 of unknown function. Mcl22 is an IDS coded by a gene in the cluster that synthesizes merochlorin, a meroterpenoid with a branched chain terpenoid skeleton.41 We therefore considered whether IDS3 was the LPP synthase. We prepared recombinant IDS3 protein and incubated it with DMAPP in the presence of magnesium ion. Surprisingly, the IDS3 enzyme reaction progressed as far as a cyclization reaction, and the product was cyclolavandulyl diphosphate.40 In other words, IDS3 catalyzed not only the condensation of bimolecular DMAPP but also its cyclization (Figure 7). IDS3 is thus cyclolavandulyl diphosphate synthase (CLDS), a new, multifunctional terpene synthase with only an IDS condensation enzyme domain.40 How CLDS catalyzes both the condensation of bimolecular DMAPP and subsequent cyclization to form cyclolavandulyl diphosphate is the next issue we will address.
Cyclooctatin biosynthesis
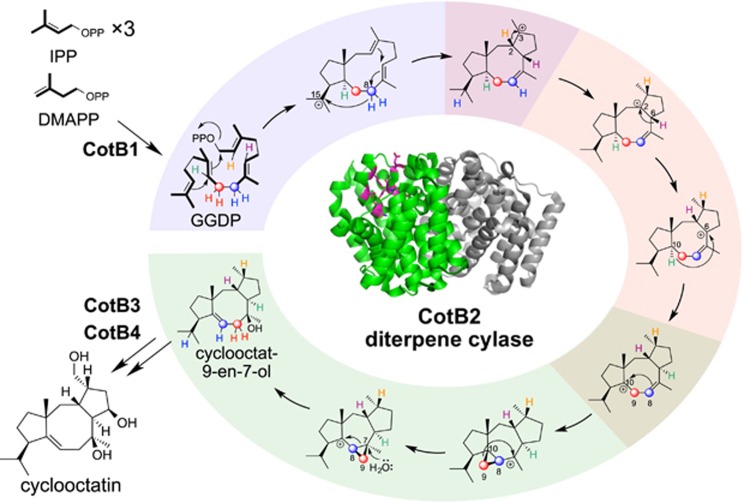
Cyclooctatin is a diterpene produced by Streptomyces melanosporofaciens MI614-43F2 and contains the first 5-8-5-membered ring structure to be isolated from prokaryotes (Figure 8).42 We reported that cyclooctatin is biosynthesized by four enzymes, CotB1–CotB4.43 We showed that CotB1 synthesizes GGPP, and CotB2 catalyzes the cyclization of GGPP to form cyclooctat-9-en-7-ol, which has a 5-8-5-membered ring. Next, the cytochrome P450 hydroxylases CotB3 and CotB4 attaches hydroxyl groups to C5 and C18, respectively, to yield cyclooctatin. Of these enzymes, CotB2 is unique in that it forms cyclooctat-9-en-7-ol, which has six chiral centers on a 5-8-5 membered ring skeleton, from GGPP, an acyclic substrate with no chiral centers, in a single catalytic action. Thus, we were interested in the mechanism by which CotB2 constructs the ring structure while precisely controlling the stereochemistry.
Figure 8.
Biosynthesis of diterpene cyclooctatin. Cyclooctatin is biosynthesized from GGPP by the successive actions of CotB1 to CotB4.
We conducted tracer experiments using [U-13C]glucose, a stable isotope-labeled glucose in which all six carbon atoms are labeled with carbon-13. The unexpected finding was that C8 and C9 of GGPP were rearranged during the CotB2-catalyzed reaction.44
For in vitro analysis, we chemically synthesized GGPP with hydrogen atoms on C2, C6, C8, C9 or C10 replaced with deuterium. We then incubated each regiospecifically deuterated GGPP with CotB2 and used NMR to analyze which position the deuterium was transferred to in the reaction product. The results were as follows: (1) C2 deuterium in GGPP transferred to C3 in cyclooctat-9-en-7-ol; (2) C6 deuterium transferred to C2; (3) the two C8 deuterium atoms transferred to C9 and C15; (4) the two C9 deuterium atoms both transferred to C8; and (5) C10 deuterium transferred to C6. Finally, by integrating these experimental results to consider the CotB2 reaction mechanism, we deduced the mechanism shown in Figure 6. We showed that CotB2 acts on the GGPP substrate to initiate the reaction by dissociating the diphosphate group at the terminus. Then there is an electrophilic ring closure reaction and proton transfer from C8 to C15, from C2 to C3, from C6 to C2 and from C10 to C6. The carbon–carbon bond between C8 and C9 is then rearranged by the formation and opening of a cyclopropane ring to assemble the 5-8-5 membered ring skeleton with precise stereochemical control (Figure 8).44, 45
We intend to determine the crystal structure of CotB2 and to use this structure to identify the amino-acid residues important to catalysis. By substituting some of these amino acids, we would be able to manufacture artificial enzymes with the ability to synthesize terpenes with new skeletons. In addition, detailed analysis of the reaction mechanism of other terpene cyclases will lead to the development of new biocatalysts and the discovery of diverse compounds exhibiting biological activity.
Conclusions and Perspectives
Actinomycetes differ greatly from model organisms such as E. coli, Bacillus subtilis and Saccharomyces cerevisiae in that they are capable of producing a range of structurally diverse compounds. Actinomycetes therefore represent a storehouse of diverse biosynthetic enzymes and machinery. It has been shown that there are 20–30 biosynthetic gene clusters for secondary metabolites present in each actinomycete genome.46, 47, 48 However, these gene clusters do not appear to be expressed under normal culture conditions. Perhaps as yet unknown stimuli present only in complex ecosystems are needed to stimulate these biosynthetic gene clusters to produce secondary metabolites. Discovering these stimuli might reveal novel biosynthetic machinery in microorganisms living in diverse ecosystems, thus establishing a methodology to obtain novel biologically active compounds.
Here I have overviewed the biosynthesis of terpenoids in Streptomyces. In our laboratory, we are conducting rigorous research on the biosynthesis of terpenoids and other biologically active compounds with a variety of structures. I believe such work will help demonstrate the beauty and elegance of nature.
Acknowledgments
Research was conducted in the Institute of Molecular and Cellular Biosciences (formerly Institute of Applied Microbiology) and the Laboratory of Cell Biotechnology at the Biotechnology Research Center, The University of Tokyo, Tokyo, Japan. Regarding winning the Sumiki-Umezawa Memorial Award 2016 from Japan Antibiotics Research Association, I appreciate all my students, colleagues and mentors, many of whom I cannot list here. I especially express sincere thanks to late Professor Haruo Seto (winner of the 1987 Sumiki-Umezawa Memorial Award), who was my supervisor in the Graduate School of Agricultural and Life Sciences, The University of Tokyo, Tokyo, Japan. This work was supported by JSPS KAKENHI (Grant nos. 26292058 and 16H06453).
Footnotes
Tomohisa Kuzuyama was awarded Japan Antibiotics Research Association’s Sumiki-Umezawa Memorial Award in 2016. This review article is based on his award-winning research.
The authors declare no conflict of interest.
References
- Sacchettini, J. C. & Poulter, C. D. Creating isoprenoid diversity. Science 277, 1788–1789 (1997). [DOI] [PubMed] [Google Scholar]
- Qureshi, N. & Porter, J. W. Biosynthesis of Isoprenoid Compounds Vol. 1, 47–94 Wiley, New York, USA, (1981). [Google Scholar]
- Bochar, D. A., Friesen, J. A., Stauffacher, C. V. & Rodwell, V. W. Comprehensive Natural Product Chemistry Vol. 2, 15–44 Pergamon, Oxford, UK, (1999). [Google Scholar]
- Rohmer, M. Comprehensive Natural Product Chemistry Vol. 2, 45–68 Pergamon, Oxford, UK, (1999). [Google Scholar]
- Kuzuyama, T. Mevalonate and nonmevalonate pathways for the biosynthesis of isoprene units. Biosci. Biotechnol. Biochem. 66, 1619–1627 (2002). [DOI] [PubMed] [Google Scholar]
- Kuzuyama, T. & Seto, H. Diversity of the biosynthesis of the isoprene units. Nat. Prod. Rep. 20, 171–183 (2003). [DOI] [PubMed] [Google Scholar]
- Kuzuyama, T., Hemmi, H. & Takahashi, S. Comprehensive Natural Products Chemistry II Vol. 1, 493–516 Elsevier, The Netherland, (2010). [Google Scholar]
- Kuzuyama, T. & Seto, H. Two distinct pathways for essential metabolic precursors for isoprenoid biosynthesis. Proc. Jpn Acad. Ser. B Phys. Biol. Sci. 88, 41–52 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohmer, M., Seemann, M., Horbach, S., Bringer-Meyer, S. & Sahm, H. Glyceraldehyde 3-phosphate and pyruvate as precursors of isoprenic units in an alternative non-mevalonate pathway for terpenoid biosynthesis. J. Am. Chem. Soc. 118, 2564–2566 (1996). [Google Scholar]
- Kuzuyama, T., Takahashi, S., Watanabe, H. & Seto, H. Direct formation of 2-C-methyl-D-erythritol 4-phosphate from 1-deoxy-D-xylulose 5-phosphate by 1-deoxy-D-xylulose 5-phosphate reductoisomerase, a new enzyme in the non-mevalonate pathway to isopentenyl diphosphate. Tetrahedron Lett. 39, 4509–4512 (1998). [Google Scholar]
- Takahashi, S., Kuzuyama, T., Watanabe, H. & Seto, H. A 1-deoxy-D-xylulose 5-phosphate reductoisomerase catalyzing the formation of 2-C-methyl-D-erythritol 4-phosphate in an alternative nonmevalonate pathway for terpenoid biosynthesis. Proc. Natl Acad. Sci. USA 95, 9879–9884 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischoff, K. M. & Rodwell, V. W. 3-Hydroxy-3-methylglutaryl-coenzyme A reductase from Haloferax volcanii: purification, characterization, and expression in Escherichia coli. J. Bacteriol. 178, 19–23 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bochar, D. A. et al. 3-hydroxy-3-methylglutaryl coenzyme A reductase of Sulfolobus solfataricus: DNA sequence, phylogeny, expression in Escherichia coli of the hmgA gene, and purification and kinetic characterization of the gene product. J. Bacteriol. 179, 3632–3638 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill, J. F. Jr, Beach, M. J. & Rodwell, V. W. Mevalonate utilization in Pseudomonas sp. M. Purification and characterization of an inducible 3-hydroxy-3-methylglutaryl coenzyme A reductase. J. Biol. Chem. 260, 9393–9398 (1985). [PubMed] [Google Scholar]
- Dairi, T. et al. Eubacterial diterpene cyclase genes essential for production of the isoprenoid antibiotic terpentecin. J. Bacteriol. 183, 6085–6094 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawasaki, T. et al. Biosynthesis of a natural polyketide-isoprenoid hybrid compound, furaquinocin A: identification and heterologous expression of the gene cluster. J. Bacteriol. 188, 1236–1244 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter, J. M. et al. Molecular basis for chloronium-mediated meroterpene cyclization: cloning, sequencing, and heterologous expression of the napyradiomycin biosynthetic gene cluster. J. Biol. Chem. 282, 16362–16368 (2007). [DOI] [PubMed] [Google Scholar]
- Shin-ya, K., Furihata, K., Hayakawa, Y. & Seto, H. Biosynthetic studies of naphterpin, a terpenoid metabolite of Streptomyces. Tetrahedron Lett. 31, 6025–6026 (1990). [Google Scholar]
- Takahashi, S., Kuzuyama, T. & Seto, H. Purification, characterization, and cloning of a eubacterial 3-hydroxy-3-methylglutaryl coenzyme A reductase, a key enzyme involved in biosynthesis of terpenoids. J. Bacteriol. 181, 1256–1263 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagi, M., Kuzuyama, T., Takahashi, S. & Seto, H. A gene cluster for the mevalonate pathway from Streptomyces sp. strain CL190. J. Bacteriol. 182, 4153–4157 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzuyama, T., Takagi, M., Kaneda, K., Dairi, T. & Seto, H. Formation of 4-(cytidine 5′-diphospho)- 2-C-methyl-D-erythritol from 2-C-methyl-D-erythritol 4-phosphate by 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase, a new enzyme in the nonmevalonate pathway. Tetrahedron Lett. 41, 703–706 (2000). [Google Scholar]
- Kuzuyama, T. et al. Studies on the nonmevalonate pathway: conversion of 4-(cytidine 5′-diphospho)-2-C-methyl-D-erythritol to its 2-phospho derivative by 4-(cytidine 5′-diphospho)-2-C-methyl- D-erythritol kinase. Tetrahedron Lett. 41, 2925–2928 (2000). [Google Scholar]
- Takagi, M. et al. Studies on the nonmevalonate pathway: formation of 2-C-methyl-D-erythritol 2,4-cyclodiphosphate from 2-phospho-4-(cytidine 5′-diphospho)-2-C-methyl-D-erythritol. Tetrahedron Lett. 41, 3395–3398 (2000). [Google Scholar]
- Satterwhite, D. M. Isopentenyldiphosphate delta-isomerase. Methods Enzymol. 110, 92–99 (1985). [DOI] [PubMed] [Google Scholar]
- Kaneda, K., Kuzuyama, T., Takagi, M., Hayakawa, Y. & Seto, H. An unusual isopentenyl diphosphate isomerase found in the mevalonate pathway gene cluster from Streptomyces sp. strain CL190. Proc. Natl Acad. Sci. USA 98, 932–937 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura, E., Tomita, T., Sawa, R., Nishiyama, M. & Kuzuyama, T. Unprecedented acetoacetyl- coenzyme A synthesizing enzyme of the thiolase superfamily involved in the mevalonate pathway. Proc. Natl Acad. Sci. USA 107, 11265–11270 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan, E. I. & Liao, J. C. ATP drives direct photosynthetic production of 1-butanol in cyanobacteria. Proc. Natl Acad. Sci. USA 109, 6018–6023 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau, N. S., Foong, C. P., Kurihara, Y., Sudesh, K. & Matsui, M. RNA-Seq analysis provides insights for understanding photoautotrophic polyhydroxy-alkanoate production in recombinant Synechocystis sp. PLoS ONE 9, e86368 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin-ya, K. et al. Isolation and structural elucidation of an antioxidative agent, naphterpin. J. Antibiot. 43, 444–447 (1990). [DOI] [PubMed] [Google Scholar]
- Kuzuyama, T., Noel, J. P. & Richard, S. B. Structural basis for the promiscuous biosynthetic prenylation of aromatic natural products. Nature 435, 983–987 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pojer, F. et al. CloQ, a prenyltransferase involved in clorobiocin biosynthesis. Proc. Natl Acad. Sci. USA 100, 2316–2321 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Pietro, A. et al. Modulation by flavonoids of cell multidrug resistance mediated by P-glycoprotein and related ABC transporters. Cell. Mol. Life Sci. 59, 307–322 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumano, T., Richard, S. B., Noel, J. P., Nishiyama, M. & Kuzuyama, T. Chemoenzymatic syntheses of prenylated aromatic small molecules using Streptomyces prenyltransferases with relaxed substrate specificities. Bioorg. Med. Chem. 16, 8117–8126 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tello, M., Kuzuyama, T., Heide, L., Noel, J. P. & Richard, S. B. The ABBA family of aromatic prenyltransferases: broadening natural product diversity. Cell. Mol. Life Sci. 65, 1459–1463 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozaki, T., Mishima, S., Nishiyama, M. & Kuzuyama, T. NovQ is a prenyltransferase capable of catalyzing the addition of a dimethylallyl group to both phenylpropanoids and flavonoids. J. Antibiot. 62, 385–392 (2009). [DOI] [PubMed] [Google Scholar]
- Kumano, T., Tomita, T., Nishiyama, M. & Kuzuyama, T. Functional characterization of the promiscuous prenyltransferase responsible for furaquinocin biosynthesis: identification of a physiological polyketide substrate and its prenylated reaction products. J. Biol. Chem. 285, 39663–39671 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozaki, T., Nishiyama, M. & Kuzuyama, T. Novel tryptophan metabolism by a potential gene cluster that is widely distributed among actinomycetes. J. Biol. Chem. 288, 9946–9956 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai, S., Furihata, K., Hayakawa, Y., Noguchi, T. & Seto, H. Lavanducyanin, a new antitumor substance produced by Streptomyces sp. J. Antibiot. 42, 1196–1198 (1989). [DOI] [PubMed] [Google Scholar]
- Demissie, Z. A., Erland, L. A., Rheault, M. R. & Mahmoud, S. S. The biosynthetic origin of irregular monoterpenes in Lavandula: isolation and biochemical characterization of a novel cis-prenyl diphosphate synthase gene, lavandulyl diphosphate synthase. J. Biol. Chem. 288, 6333–6341 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozaki, T., Zhao, P., Shinada, T., Nishiyama, M. & Kuzuyama, T. Cyclolavandulyl skeleton biosynthesis via both condensation and cyclization catalyzed by an unprecedented member of the cis-isoprenyl diphosphate synthase superfamily. J. Am. Chem. Soc. 136, 4837–4840 (2014). [DOI] [PubMed] [Google Scholar]
- Kaysser, L. et al. Merochlorins A–D, cyclic meroterpenoid antibiotics biosynthesized in divergent pathways with vanadium-dependent chloroperoxidases. J. Am. Chem. Soc. 134, 11988–11991 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoyama, T., Naganawa, H., Muraoka, Y., Aoyagi, T. & Takeuchi, T. The structure of cyclooctatin, a new inhibitor of lysophospholipase. J. Antibiot. 45, 1703–1704 (1992). [DOI] [PubMed] [Google Scholar]
- Kim, S. Y. et al. Cloning and heterologous expression of the cyclooctatin biosynthetic gene cluster afford a novel diterpene cyclase and two cytochrome P450 hydroxylases. Chem. Biol. 16, 736–743 (2009). [DOI] [PubMed] [Google Scholar]
- Meguro, A. et al. An unusual terpene cyclization mechanism involving a carbon−carbon bond rearrangement. Angew. Chem. Int. Ed. 54, 4353–4356 (2015). [DOI] [PubMed] [Google Scholar]
- Sato, H. et al. ‘Cation-stitching cascade’: exquisite control of terpene cyclization in cyclooctatin biosynthesis. Sci. Rep. 5, 18471 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley, S. D. et al. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 417, 141–147 (2002). [DOI] [PubMed] [Google Scholar]
- Ikeda, H. et al. Complete genome sequence and comparative analysis of the industrial microorganism Streptomyces avermitilis. Nat. Biotechnol. 21, 526–531 (2003). [DOI] [PubMed] [Google Scholar]
- Ohnishi, Y. et al. Genome sequence of the streptomycin-producing microorganism Streptomyces griseus IFO 13350. J. Bacteriol. 190, 4050–4060 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]