Abstract
The visual photoreceptor rhodopsin is a prototypical G-protein-coupled receptor (GPCR) that stabilizes its inverse agonist ligand, 11-cis-retinal (11CR), by a covalent, protonated Schiff base linkage. In the visual dark adaptation, the fundamental molecular event after photobleaching of rhodopsin is the recombination reaction between its apoprotein opsin and 11CR. Here we present a detailed analysis of the kinetics and thermodynamics of this reaction, also known as the “regeneration reaction”. We compared the regeneration of purified rhodopsin reconstituted into phospholipid/detergent bicelles with rhodopsin reconstituted into detergent micelles. We found that the lipid bilayer of bicelles stabilized the chromophore-free opsin over the long timescale required for the regeneration experiments, and also facilitated the ligand reuptake binding reaction. We utilized genetic code expansion and site-specific bioorthogonal labeling of rhodopsin with Alexa488 to enable, to our knowledge, a novel fluorescence resonance energy transfer-based measurement of the binding kinetics between opsin and 11CR. Based on these results, we report a complete energy diagram for the regeneration reaction of rhodopsin. We show that the dissociation reaction of rhodopsin to 11CR and opsin has a 25-pM equilibrium dissociation constant, which corresponds to only 0.3 kcal/mol stabilization compared to the noncovalent, tightly bound antagonist-GPCR complex of iodopindolol and β-adrenergic receptor. However, 11CR dissociates four orders-of-magnitude slower than iodopindolol, which corresponds to a 6-kcal/mol higher dissociation free energy barrier. We further used isothermal titration calorimetry to show that ligand binding in rhodopsin is enthalpy driven with –22 kcal/mol, which is 12 kcal/mol more stable than the antagonist-GPCR complex. Our data provide insights into the ligand-receptor binding reaction for rhodopsin in particular, and for GPCRs more broadly.
Introduction
The rod cell visual photoreceptor rhodopsin (Rho) found predominantly in rod outer segment (ROS) disk membranes consists of an apoprotein, opsin, and a chromophore, 11-cis-retinal (11CR) (1, 2, 3, 4). In the dark state, 11CR is covalently bound to opsin through a protonated Schiff base (PSB) linkage with Glu-113 acting as the counterion (5). Light illumination isomerizes the inverse agonist 11CR to an agonist all-trans-retinal (ATR), which activates the receptor to initiate the cGMP protein cascade. The deprotonated Schiff base linkage in the photoactivated rhodopsin (Meta-II) hydrolyzes, causing ATR to dissociate from the ligand binding pocket of Rho. To recycle the photoreceptor, the apoprotein opsin recombines with 11CR supplied from the retinal pigment epithelium, a process referred to as “regeneration” (6). Regeneration of Rho in the dark after photoactivation is the fundamental molecular reaction underlying visual dark adaptation. However, the energetics of the recombination reaction between opsin and 11CR remains to be understood. Because Rho is a prototypical member of the class-A G protein-coupled receptors (GPCRs), a mechanistic understanding of ligand binding in Rho should also provide insights into the structure-function relationship of this transmembrane receptor family.
Here we utilize fluorescence resonance energy transfer (FRET)-based assays to measure the kinetics of the recombination reaction between 11CR and opsin. We prepared expressed recombinant Rho with a genetically encoded p-azido-Phe (azF) (7) at the second intracellular loop that was labeled with Alexa488 using a robust bioorthogonal labeling reaction (8, 9, 10, 11). We utilize Alexa488-Rho and unlabeled Rho purified from bovine ROS to characterize the recombination of 11CR and opsin. We show that the1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine/3-[(3-cholamidopropyl) dimethylammonio]-1-propanesulfonate (POPC/CHAPS) bicelles serve as a suitable model system for characterizing chromophore binding in Rho. We then measure the binding kinetics at several temperatures. In an earlier report (12), we have obtained the temperature-dependent kinetics for its reverse reaction, i.e., the light-independent, spontaneous dissociation rate of 11CR from Rho. These kinetics data enable us to calculate the activation enthalpy and entropic terms from the Eyring plot. We also measure the overall enthalpy change of the reaction using isothermal titration calorimetry (ITC). Based on these kinetic and thermodynamic measurements, we are now able to derive an energy diagram for the reversible recombination reaction between 11CR and opsin (Fig. 1). We compared the energetics of ligand binding in Rho and adrenergic receptors. Interestingly, we found that the equilibrium dissociation constant (Kd) of 11CR for opsin is similar to that of certain high-affinity diffusible ligands for adrenergic receptors. Nonetheless, the ligand binding and dissociation for Rho is significantly slower, as manifested by the high activation barrier in the energy landscape. The energy diagram we derive here for the Rho regeneration reaction provides a useful guide for interpreting ligand binding modes for GPCRs in general.
Figure 1.
Deriving the binding energy landscape from kinetic and thermodynamic relations. To see this figure in color, go online.
Materials and Methods
Materials
n-Dodecyl-β-d-maltoside (DM) and 3-[(3-cholamidopropy l)dimethylammonio]-1-propanesulfonate (CHAPS) were obtained from Anatrace (Maumee, OH). 1-Palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) was obtained from Avanti Polar Lipids (Alabaster, AL). 11-cis-Retinal was a gift from Drs. P. Sorter and V. Toome, Hoffmann-La Roche (Basel, Switzerland).
Purifying the pigments
Rho wild-type (wt) samples were purified from bovine ROS. Opsin samples were prepared by photobleaching purified Rho. Alexa488-labeled Rho was prepared by labeling a genetically encoded azido-Phe with Alexa488 fluorophore in a quantitative bioorthogonal labeling reaction (8, 9, 10). The detailed methods for preparing these pigment samples are given in the Supporting Material.
Assessing the stability of opsin solubilized in DM micelles
The purified opsin (15 μL, 4.7 μM, solubilized in 0.1% (w/v) DM) was aliquoted into 1.5-mL Eppendorf tubes (Hamburg, Germany) and photobleached by irradiating with a 505-nm LED light source (Thorlabs, Newton, NJ) for 30 s. After photobleaching, the samples were kept in the dark at 28°C for varied durations before 135 μL POPC/CHAPS bicelle buffer A (1% (w/v) POPC, 1% (w/v) CHAPS, 125 mM KCl, 25 mM MES, 25 mM HEPES, 12.5 mM KOH, pH 6.0, 450 μL) supplemented with 11CR was added. The final concentration of opsin was 0.47 μM, and the molar ratio of 11CR to opsin was 1.5:1. The regeneration reaction was allowed to proceed overnight (>15 h) to reach completion. The dark and photobleached spectra of the regenerated samples were recorded on a Lambda 800 UV-Vis spectrophotometer (PerkinElmer, Waltham, MA). In the presence of 11CR, longer exposure to 505-nm LED light (1 min) was required to fully photobleach the photoreceptor. The extent of regeneration was evaluated based on the 500-nm absorbance of the difference spectra.
Assessing the stability of opsin solubilized in POPC/CHAPS bicelles
The purified opsin (15 μL, 4.7 μM, solubilized in 0.1% DM) was added to POPC/CHAPS bicelle buffer A (120 μL). After photobleaching and incubation, 11CR diluted in POPC/CHAPS bicelle buffer (15 μL was added to the opsin samples ([11CR]/[opsin] = 1.5:1). The regeneration reaction was allowed to reach completion and analyzed as described above.
Dynamic light scattering experiment to measure the hydrodynamic radius of POPC/CHAPS bicelles
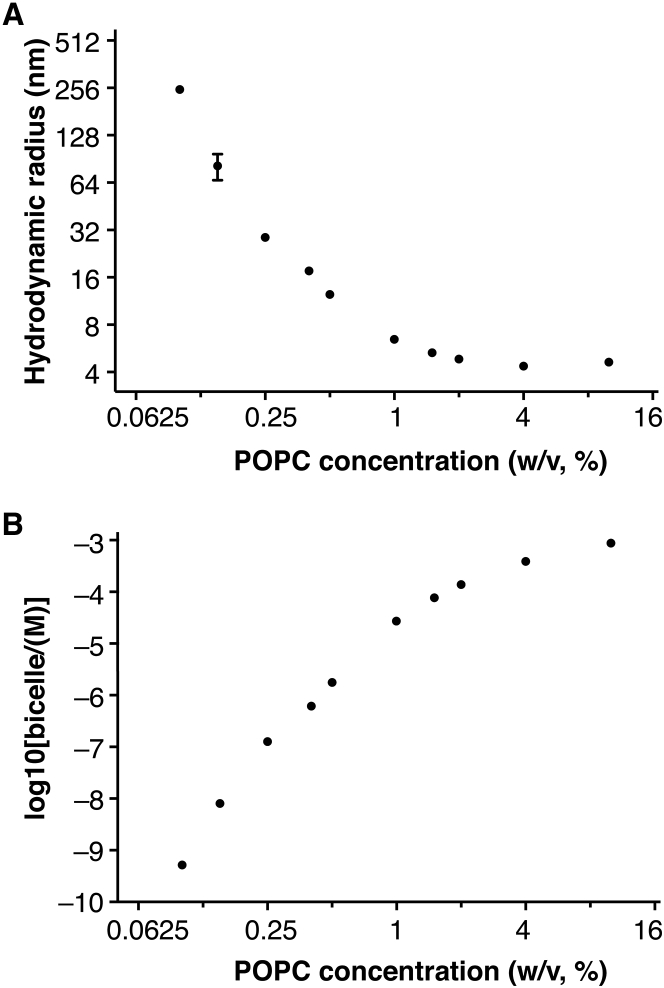
Various concentrations of 1:1 POPC/CHAPS (lipids w/v: 10, 4.0, 2.0, 1.5, 1.0, 0.50, 0.40, 0.25, and 0.10%) are prepared freshly. Before the dilution, the POPC/CHAPS stock solution and the buffer were filtered with an inorganic membrane filter (Anotop 10, 0.02 μm pore size, 10-mm diameter; Whatman, Maidstone, UK) to remove any contamination with scattering microparticles. Samples were added into a 384-well plate (60 μL each well, black, clear bottom; Greiner Bio-One, Kremsmünster, Austria), and then the plate was sealed on top with an Oracal soft PVC film (Orafol Americas, Avon, CT) to prevent evaporation. All the samples in this dynamic light scattering (DLS) experiment were measured in triplicate at 28°C, on a DynaPro Plate Reader II (Wyatt Technology, Goleta, CA). The hydrodynamic radii of bicelles were monitored for up to 14 h. The calculation of bicelle concentration is included in the Supporting Material. Each data point in Fig. 3 represents the average from 15 measurements, with the error bars indicated.
Figure 3.
Dynamic light scattering (DLS) experiment to measure the hydrodynamic radius of POPC/CHAPS bicelles. (A) The hydrodynamic radii of bicelles change with the lipid concentration. (B) Bicelle concentration as a function of the lipid concentration. For 1% POPC/CHAPS, the hydrodynamic radius of bicelles is 6.46 ± 0.05 nm and the concentration is 27 μM.
The measurement of retinal uptake kinetics based on quenching of tryptophan or Alexa488 fluorescence
Fluorescence measurement was done on a SPEX Fluorolog tau-311 spectrofluorometer (Horiba Instruments, Irvine, CA) in photon-counting mode. An aliquot of purified Rho (30 μL) was added to the POPC/CHAPS bicelle buffer A (1% (w/v) POPC, 1% (w/v) CHAPS, 125 mM KCl, 25 mM MES, 25 mM HEPES, 12.5 mM KOH, pH 6.0, 450 μL) under constant stirring. In the tryptophan (Trp)-based experiment, the excitation wavelength was 295 nm with a 0.6-nm bandpass, and the emission was measured at 330 nm with a 15-nm bandpass. The concentration of Rho was typically 0.25–0.30 μM. In the Alexa488-based experiment, the sample was excited at 488 nm with a 0.2-nm bandpass, and the emission was measured at 520 nm with a 15-nm bandpass. In addition to small excitation bandpass to reduce the intensity of the exciting beam, photobleaching was further minimized by integrating the fluorescence signal for 2 s in 30-s intervals, keeping the excitation beam closed using an automatic shutter. Because Alexa488 has a greater quantum yield than Trp, the concentrations of Alexa488-Rho in these experiments were lower, normally in the range of 5–50 nM. After recording the fluorescence signal of dark-state samples, 20 μL of 11CR working dilution (ethanolic stock of 11CR diluted in POPC/CHAPS bicelle buffer A) was added to the cuvette to give a final concentration in the range of 1.5–2.0 μM. For each measurement, the concentration of freshly diluted 11CR was determined by UV-Vis spectroscopy (ε378 nm = 25,600 M−1 s−1). The decrease of Trp or Alexa488 fluorescence was fitted with a pseudo first-order exponential decay model to derive the apparent regeneration rate (kobs). The second-order rate constant (k2) for the recombination reaction between opsin and retinal was calculated as k2 = kobs/[retinal].
Determination of the binding enthalpy of 11CR and opsin using ITC
For each measurement, opsin (9 ± 1 μM) and 11CR (250 μM) bicelle solutions were prepared fresh in the POPC/CHAPS buffer B (1% (w/v) POPC, and 1% (w/v) CHAPS, 137.5 mM NaCl, 0.25 mM EDTA, 25 mM MES, 25 mM HEPES hemisodium salt, pH 6.0). To avoid the dilution heat of ethanol in water, the ethanolic solution of 11CR was evaporated with dry argon under red light and redissolved in POPC/CHAPS buffer. The solution was centrifuged (14,000 × g, 10 min) to remove any insoluble fraction. The baseline curve was generated by sampling the intermediate time points between each injection. In these plots, the signals were corrected by subtracting the baseline. Due to the very slow binding kinetics compared with typical ITC studies of a ligand binding to a protein, the experiments are approximately 10 times longer and the corresponding differential power signals are severalfold smaller.
Results
POPC/CHAPS bicelles improve the thermal stability of photobleached Rho
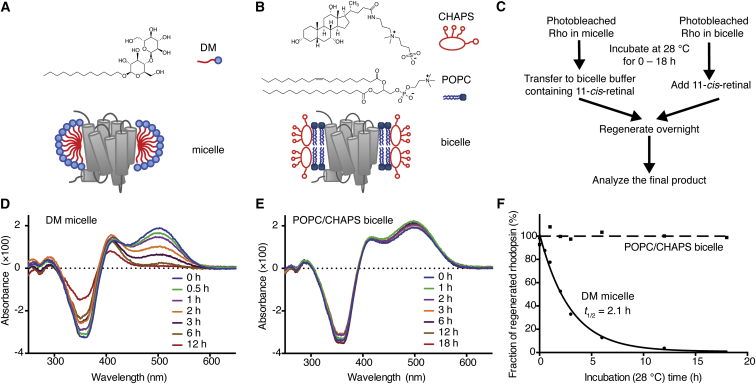
Bicelles are model membranes that consist of long-chain phospholipids, which form the planar lipid bilayer segment, and detergents or short-chain phospholipids, which stabilize the edge of the bilayer. In an earlier report (12), we showed that dark-state Rho was stable in 1% (w/v) POPC/CHAPS bicelles over the timescale of weeks. Here, we evaluated the stability of opsin in POPC/CHAPS bicelles (Fig. 2). Purified Rho was solubilized in POPC/CHAPS bicelles, photobleached, and subsequently incubated at 28°C for various lengths of time. Then the samples were supplemented with fresh 11CR and kept in the dark until regeneration was complete. As a comparison with the bicelle system, we also assessed opsin stability in DM detergent micelles. The photobleached samples were first incubated for various lengths of time, then transferred into POPC/CHAPS bicelles to quench the denaturation process. After incubation of excess 11CR, the extent of 11CR binding was assessed based on the recovery of 500-nm absorbance (Fig. 2, D and E). The denatured, misfolded fraction would lose the ability to bind with 11CR to recover the 500-nm peak. We found that the micelle-solubilized opsin lost the ability to bind with 11CR with a half-life of 2.1 h, whereas 98% of the bicelle-solubilized receptor retained the potential for regeneration after 18 h of incubation (Fig. 2 F). We further showed that in bicelles the binding between opsin and 11CR yielded high-purity Rho, and we determined the accurate extinction coefficients of opsin and Rho (Fig. S1; Table S1).
Figure 2.
Photobleached Rho is more stable in POPC/CHAPS bicelles than it is in DM micelles. (A) The structures of DM and a cartoon of Rho reconstituted in DM micelles. (B) The structures of CHAPS and POPC, and a cartoon of Rho reconstituted in POPC/CHAPS bicelle. (C) The procedures for assaying the stability of bleached Rho in DM micelles or POPC/CHAPS bicelles. (D) The dark–light end-point difference spectral time course of Rho in DM showed a gradual loss of the 500 nm absorbance, indicating the increase of denatured opsin that failed to bind with 11CR. (E) The spectral time course shows that bleached Rho in POPC/CHAPS bicelles after incubation could be regenerated close to completion. (F) The time course of the thermal denaturation of photobleached Rho in DM micelles and POPC/CHAPS bicelles.
The size and concentration of bicelles changes as a function of lipid concentration
In the bicelle/water system, the more hydrophilic CHAPS can exist both as monomeric free detergents at a concentration close to its critical micelle concentration and as associated structures in bicelles. The more hydrophobic POPC molecules essentially completely associate as bicelles. When POPC/CHAPS are diluted together, more CHAPS molecules will dissociate from the bicelles, and hence the ratio of CHAPS to POPC in the bicelle structure would decrease. The number of CHAPS molecules in the rim scales with the radius of the bicelle, whereas the number of POPC molecules in the planar center scales with the square of the radius. Therefore, dilution of POPC/CHAPS bicelle would result in an enlarged planar center. When the ratio of CHAPS to POPC is sufficiently low, the bicelle structure would become unstable, begin to aggregate, and form vesicles.
To evaluate the stability of POPC/CHAPS bicelles, we used DLS to monitor the hydrodynamic radius of POPC/CHAPS (w/w) bicelles for different lipid concentrations (Fig. 3 A) over a time course of 15 h. We found that at the tested concentrations, the hydrodynamic radii of the POPC/CHAPS bicelles were stable during the measured period. The critical micelle concentration for CHAPS is between 6 and 10 mM, corresponding to a w/v proportion from 0.3 to 0.6%. As a result, a dramatic increase in the bicelle size was observed when POPC/CHAPS was diluted below 1% (w/v), suggesting aggregation of lipids as liposomes. We calculated bicelle concentration as a function of the lipid concentration (Fig. 3 B). For 1% POPC/CHAPS, the concentration of bicelles was 27 μM. Based on the above results, we conclude that 1% POPC/CHAPS results in well-dispersed bicelles that maintain the stability of ligand-free opsin. We used this condition throughout the study to characterize the binding between opsin and 11CR.
FRET-based assays to measure the kinetics of 11CR binding to opsin
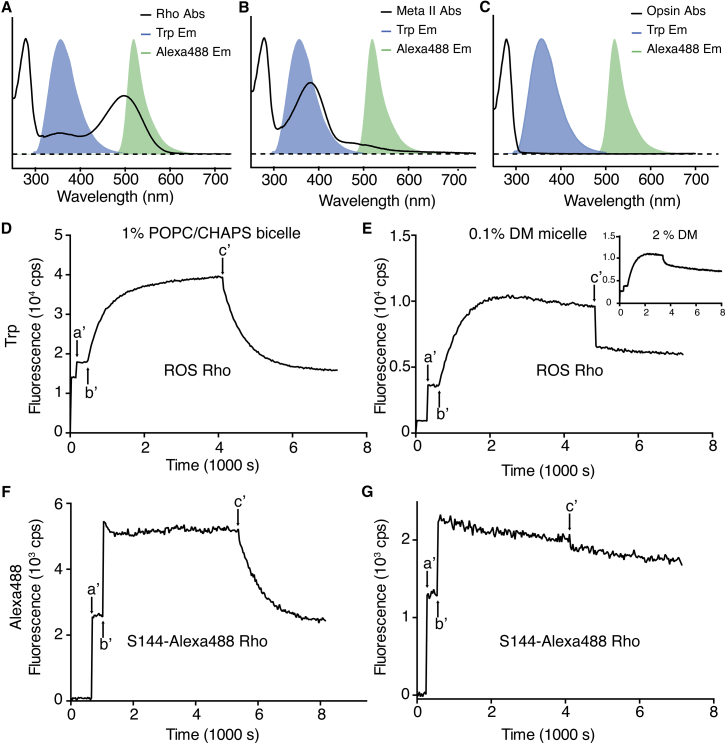
In the dark-state Rho, the apoprotein and 11CR are linked together through a positively charged PSB linkage, which gives rise to its characteristic 500-nm peak. In the photoactivated Meta-II, the retinal absorption peak shifts from 500 to 380 nm. As ATR is released from the binding pocket, the ligand-free opsin shows no retinal peak. Taking advantage of the photochemistry of Rho, we employed two FRET-based assays to measure regeneration kinetics, both involving a fluorescent reporter as the energy donor and retinal as the acceptor (Fig. 4, A–C).
Figure 4.
FRET-based measurement of Rho regeneration in different reconstitution systems. The absorption spectra (solid line) of Rho (A), Meta-II state Rho (B), and opsin (C) are overlaid with the fluorescence emission peaks of Trp (shade, maximum at 330 nm) and Alexa488 (shade, maximum at 520 nm). The intensities of the absorption peaks and fluorescence emission peaks are normalized. Fluorescence time course traces are presented in (D)–(G). In all cases, pigment (either wt Rho or S144-Alexa488 Rho) was added into the assay buffer at arrow a′. The sample was then photobleached at arrow b′ to form the Meta-II state. After Meta-II decay was close to completion, the ligand 11CR was added to initiate pigment regeneration at arrow c′. (D) Purified ROS Rho reconstituted in bicelles (1% POPC/CHAPS). (E) ROS Rho in DM micelles (0.1% DM; inset: ROS Rho in 2% DM). (F) Purified S144azF mutant Rho labeled with an Alexa488 fluorophore (S144-Alexa488 Rho) in 1% POPC/CHAPS bicelles. The FRET efficiency between Alexa488 and dark-state Rho was calculated to be 0.55±0.04 (53). (G) S144-Alexa488 Rho in 0.1% DM micelles. To see this figure in color, go online.
The first assay based on the intrinsic tryptophan (Trp) fluorescence of Rho has been described in Tian et al. (12) and Farrens and Khorana (13). In brief, Trp fluorescence can be quenched by 11CR in the dark-state Rho (Fig. 4 A), and by ATR even more efficiently in Meta-II Rho (13) (Fig. 4 B). Only when retinal dissociates from opsin is there no energy transfer between retinal and Trp (Fig. 4 C). The Trp-based assay indicates whether retinal is present in the ligand binding pocket.
The second assay utilizes an extrinsic fluorescent reporter Alexa488, which was attached to a Rho mutant at a genetically encoded azF residue in the second intracellular loop (S144-Alexa488 Rho) using a bioorthogonal azide-alkyne [3 + 2] cycloaddition reaction (10). The energy transfer between Alexa488 and retinal is critically dependent on the overlap of the 500-nm absorption peak of dark-state Rho and the Alexa488 emission spectrum (Fig. 4 A). Upon photoactivation of Rho, the deprotonation of PSB results in the instant loss of spectral overlap, and consequently a decrease in energy transfer efficiency (Fig. 4 B). Thus the Alexa488-based assay distinguishes between a mature pigment with a PSB bond and a noncovalent complex of opsin and 11CR.
We reconstituted immunopurified wt Rho or S144-Alexa488 Rho into POPC/CHAPS bicelles (Fig. 4, D and F) or DM micelles (Fig. 4, E and G). We then photobleached the samples and allowed the resulting Meta-II photoproducts to decay to completion. For each of the samples reconstituted into bicelles (wt Rho or S144-Alexa488 Rho), after photobleaching and decay, the addition of exogenous 11CR initiated a robust monoexponential decrease in the Trp or Alexa488 fluorescence signals (Fig. 4, D and F). Previously, we have shown that this monoexponential decay of fluorescence signal resulted from pigment regeneration, and that Alexa488-labeled Rho regenerated in POPC/CHAPS bicelles can be repeatedly photoactivated (10).
By contrast, the photobleached pigments could not be effectively regenerated in DM micelles at room temperature (Fig. 4, E and G), and we were not able to observe similar monoexponential fluorescence quenching for wt Rho or S144-Alexa488 Rho. For wt Rho in 0.1% DM (Fig. 4 E), a pronounced drop of Trp fluorescence signal occurred immediately after the addition of 11CR. We hypothesized that this fast-phase quenching reflected the nonspecific energy transfer between Trp and retinal that partitioned into the hydrophobic compartment of DM micelles (14), rather than the formation of mature pigment. We then reasoned that increasing the concentration of DM might reduce such nonspecific quenching. Indeed, in 2% DM the fast-phase quenching of Trp fluorescence by retinal was only 1/6 as much as in 0.1% DM (Fig. 4 E: inset, corrected for dilution and inner filter effects).
We further tested S144-Alexa488 Rho in 0.1% DM (Fig. 4 G). The monotonic decrease both before and after the addition of 11CR was different from what was observed in POPC/CHAPS bicelle buffer (Fig. 4 F). Because this assay unambiguously distinguishes between nonspecific quenching and real pigment regeneration, the result suggested opsin denaturation in DM micelles rather than real regeneration. There was no initial fast quenching phase in the Alexa488-based assay, supporting our conclusion that the quenching of Trp fluorescence in DM results from retinal that partitioned into micelles.
We also found that the DM-solubilized photobleached Rho, after transfer into POPC/CHAPS bicelle, could be partially regenerated at 28°C (Fig. S2). While at room temperature, opsin in 0.1% DM micelles cannot be regenerated by 11CR; when the temperature was lowered to 4°C, opsin can recombine with 11CR to form a mature pigment (Fig. S3). Taken together, these results show that the lipid bilayer environment of bicelles is important for the recombination of opsin and 11CR at room temperature.
The kinetics of the recombination reaction between opsin and 11CR
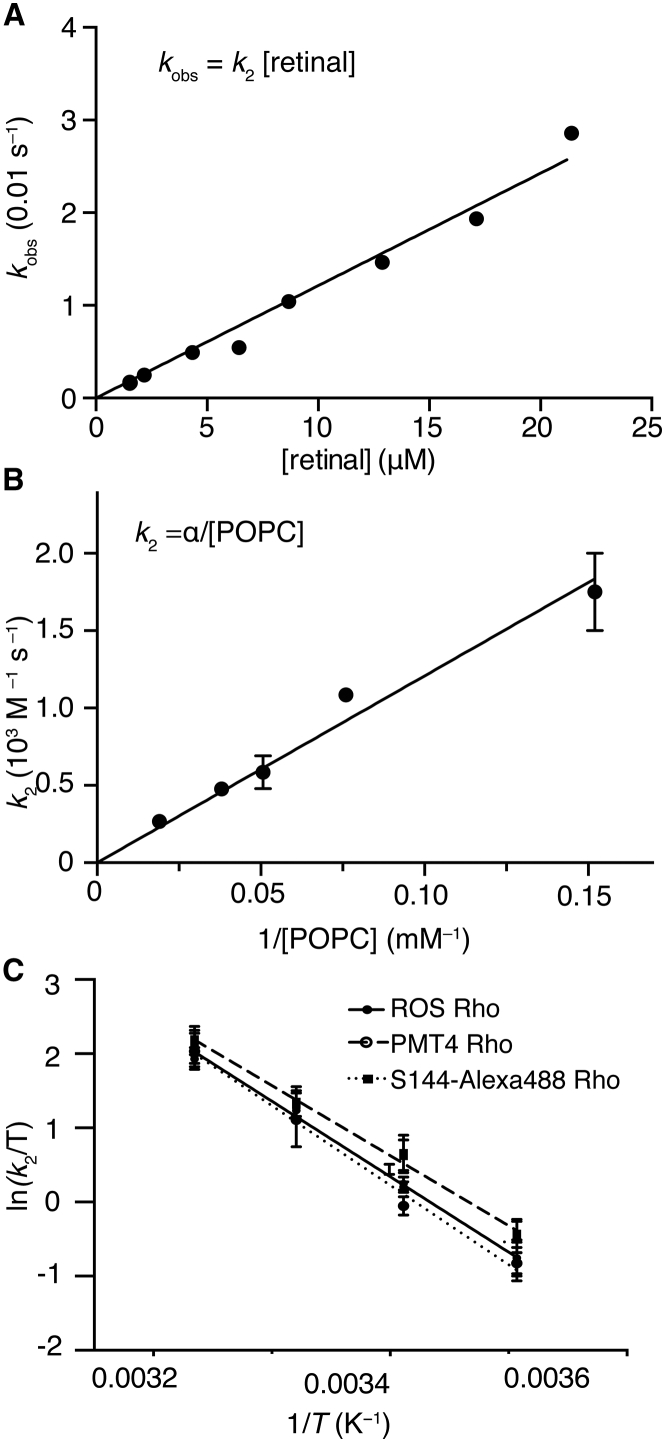
To test whether the recombination reaction of Rho and 11CR in bicelles is bimolecular, we conducted the Trp-based FRET assay for purified wt Rho at various retinal concentrations. The pseudo first-order reaction rate (kobs) was linearly correlated with the nominal retinal concentration, demonstrating concordance with a second-order rate law within the tested range of retinal concentrations (Fig. 5 A).
Figure 5.
The kinetics of the recombination reaction between 11CR and opsin. (A) The kobs of Rho regeneration was measured at various concentrations of 11CR (molar ratio of retinal to the receptor: 8.0 to 64). The slope of the plot corresponds to the k2 ((1.21 ± 0.04) × 103 M−1 s−1 at 28°C). (B) The measured k2 of Rho regeneration is inversely correlated with the concentrations of lipids (α = (1.21±0.05)×104 s−1). (C) Eyring plot for the second-order rate constants of three purified Rho samples: 1) Rho from bovine ROS (ROS Rho, Trp-based assay), 2) Rho heterologously expressed in HEK293F cells (PMT4 Rho, Trp-based assay), and 3) S144azF mutant Rho labeled with an Alexa488 fluorophore (S144-Alexa488 Rho, Alexa488-based assay).
In the regeneration experiment using 1% POPC/CHAPS, the ratio of bicelle, retinal, and Rho is ∼140:8:1, and hence not every bicelle necessarily contains one molecule of retinal and one molecule of the receptor at the same time. A prerequisite of regeneration is that a molecule of retinal diffuses into a receptor-containing bicelle. Thus, we asked whether the measured kinetics reflects retinal entry into opsin binding pocket or rather a retinal diffusion among bicelles. We reasoned that when the ratio of retinal to bicelles decreases, it should take longer for retinal to find an opsin-containing bicelle. Thus the measured second-order rate constant (k2) should increase as the concentration of retinal decreases. On the contrary, Fig. 5 A showed that k2 did not change with the concentration of retinal.
The DLS experiment (compare to Fig. 3) enabled us to estimate the concentration of bicelles as a function of lipids. If retinal diffusion were rate limiting, then when the concentration of retinal was lower than that of bicelles, and it exceeded the bicelle concentration, the chromophore binding rate should exhibit two phases. The concentration of retinal was 1.5–2 μM. Therefore, k2 should be dramatically faster in 0.5% POPC/CHAPS (2 μM) than it is in 1% POPC/CHAPS (27 μM bicelles). Instead, we found an inverse correlation between the k2 and the concentration of lipids (Fig. 5 B). A simple model can explain this inverse correlation: retinal molecules are practically solubilized in bicelles due to its high partition coefficient into lipids (15). Because retinal diffusion among bicelles is fast, the timescale for reaching equilibrium is negligible compared to that of ligand binding. Thus the effect of increasing lipids could be treated as a decrease in the effective concentration of retinal, even if the nominal concentration of retinal remains unchanged. Taken together, retinal diffusion among bicelles is not the rate-limiting step, and the measured kinetics in the bicelle system reflects the actual ligand-binding process.
The activation energy (Ea,on) for 11CR binding in ROS Rho was 20.8 ± 0.9 kcal mol−1 (Table S2). To our knowledge, the activation energy for 11CR diffusing in lipids has not been determined. When compared to an earlier study that reported an Ea of 8.8 ± 0.3 kcal mol−1 for a hydrophobic drug in POPC (16), 11CR binding with opsin requires an activation energy higher by 12 kcal mol−1. This distinctly higher energy barrier suggests that the chromophore binding process measured here is rate limited by protein dynamics instead of 11CR diffusion.
Taken together, retinal diffusion among bicelles is not the rate-limiting step, and the measured kinetics in the bicelle system reflects the actual ligand-binding process.
The reaction enthalpy of the recombination reaction between opsin and 11CR
We conducted an ITC experiment to measure the overall change in enthalpy (ΔHo) between the two steady states. Opsin was directly purified into POPC/CHAPS bicelles. Three independent ITC measurements (Fig. 6, A–C) gave a stoichiometry of 1.15 ± 0.14 for retinal binding to opsin, slightly greater than the expected 1.0. We obtained an average ΔH° of –21.6 ± 1.3 kcal mol−1 (Fig. 6 D). Due to the limitations of the sensitivity of the ITC instrument (17), we could not determine Kd from the shape of the titration curve.
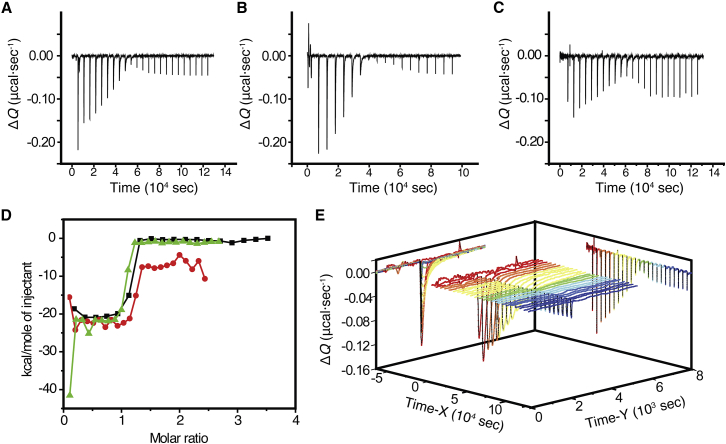
Figure 6.
Reaction enthalpy for the binding of 11CR to opsin was determined from ITC experiments. (A–C) Raw ITC data plots from three independent experiments. The signals were corrected by subtracting the baseline. (D) The integrated reaction heat per mole of injectant versus the stoichiometry for three independent experiments (A, dots; B, squares, and C, triangles). The calculated ΔH° were −24.2 ± 1.5 kcal mol−1, −19.4 ± 0.5 kcal mol−1, and −21.2 ± 1.6 kcal mol−1, giving an average ΔH° of –21.6 ± 1.3 kcal mol−1. (E) Given here is a 3D projection of a representative ITC experiment (C). Time X represents the entire time course of the experiment; time Y represents the time course of each injection.
In the ITC experiment, the ligand binding kinetics for opsin was comparable to the binding rates measured in the fluorescence quenching experiments. The reaction time for each injection increased as the concentration of ligand-free opsin decreased, and correspondingly the maximal differential power for each injection decreased with slower reaction kinetics. The differential power peaks after the titration point showed faster decay time with a negligible integrated area (Fig. 6 E), which were likely due to the mixing heat.
The energy diagram of ligand binding in Rho
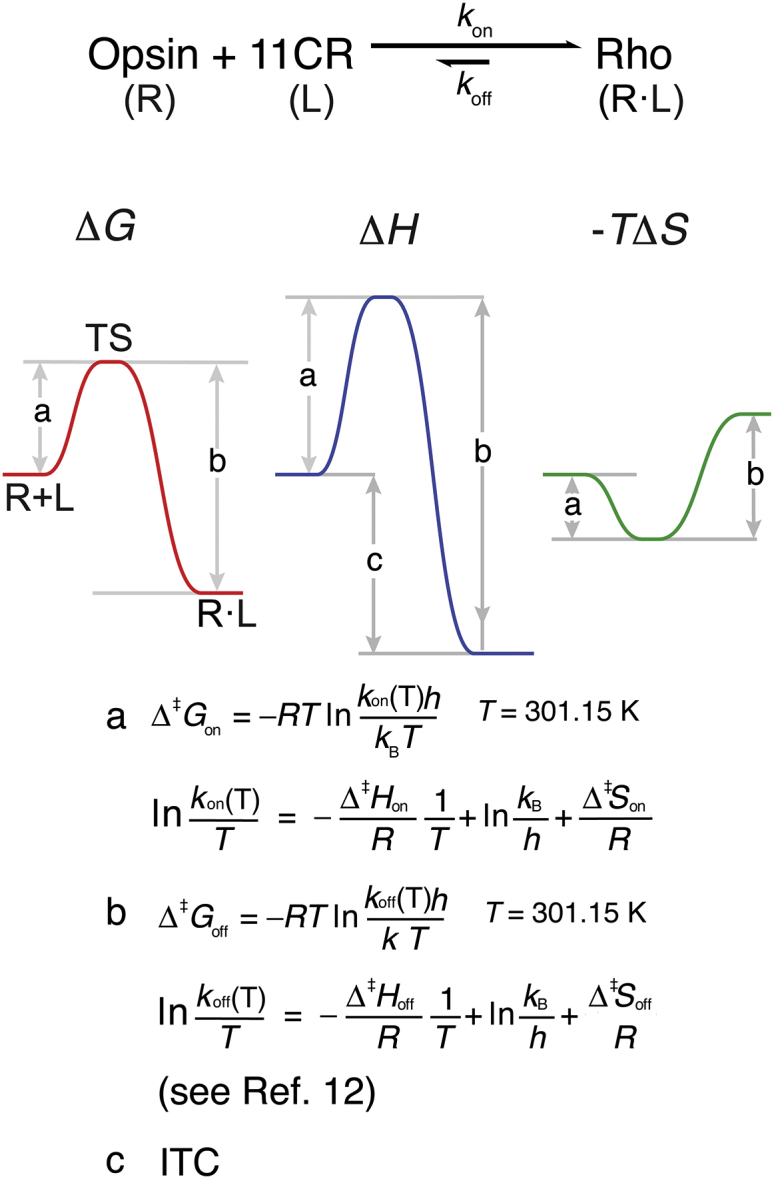
To describe an energy diagram for the ligand-receptor binding reaction with a single transition state, at least two of the following three parameters need to be determined: 1) the activation energy of the forward reaction Δ‡Xon (X = G, H, or S); 2) the activation energy of the reverse reaction Δ‡Xoff; and/or 3) the energy difference between the products and the reactants of a particular state ΔX°. We derived the energy diagram based on the forward and reverse kinetic studies and the calorimetry experiment (Fig. 7 A. See the Supporting Material for the calculation). We assumed a modified biochemical standard state: 1% POPC/CHAPS bicelles in aqueous solution, at 28°C, pH 6.0, under ambient atmospheric pressure, and with the concentrations of reactants and products all at 1 M.
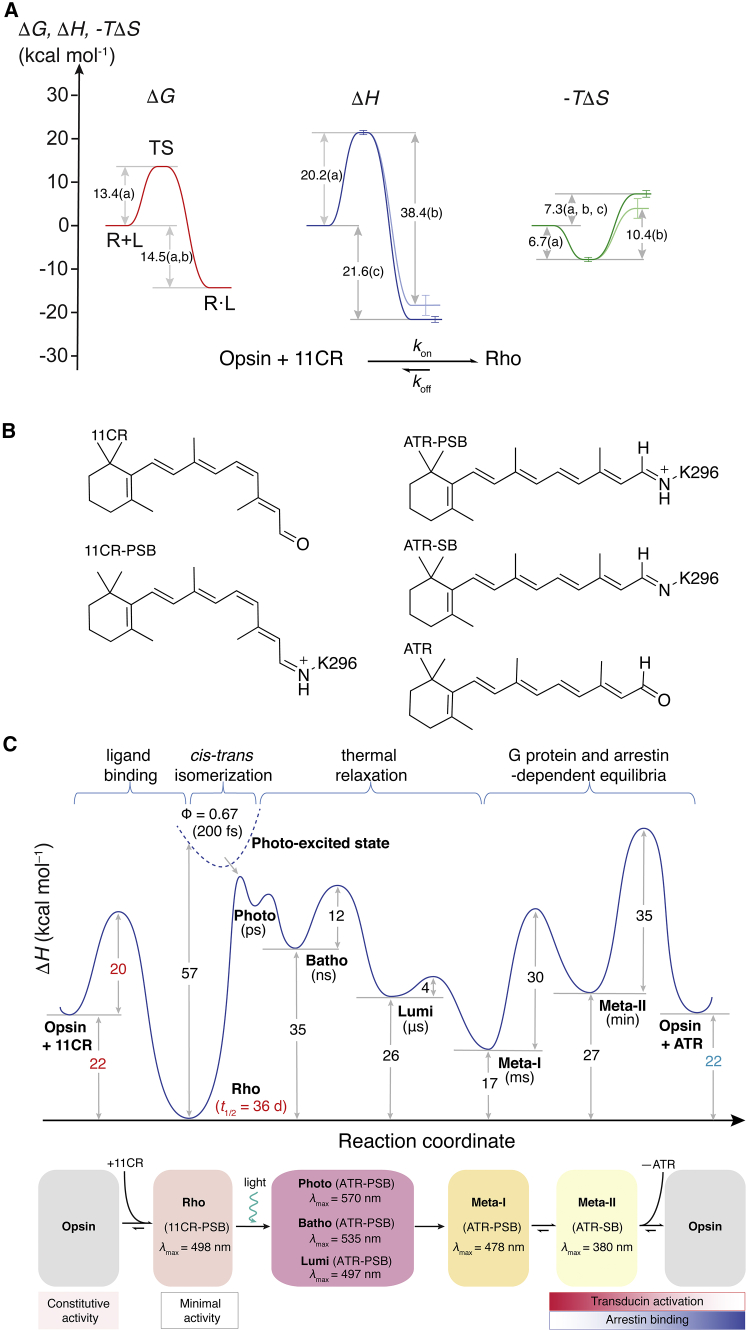
Figure 7.
(A) Energy diagram for the binding reaction between opsin and 11CR. The letter next to each value indicates the experimental approach used to derive the value: (a) the kinetics of retinal entry (Fig. 5); (b) the chromophore exchange experiment (12); (c) the ITC experiment (Fig. 6). Note that here we highlighted the energy landscape derived from the calorimetric data in solid color. The retinal release energetics was shown in grey color. (B) The structures of the retinal-derived species in the photocycle of Rho. (Note that the retinylidene chromophores in Rho are 6-s-cis-isomers.) (C) The complete enthalpy profile of Rho regeneration and photoactivation. The lifetimes of the intermediate species are shown in the parentheses. The values determined or corrected by the present study are shown in red and turquoise, respectively.
We present the complete enthalpy profile for the conversion of Rho in the photocycle (Fig. 7, B and C) based on our kinetic and calorimetric data and literature values (18). Because the cis-trans isomerization of retinal only has a small ΔH° of 0.15 kcal mol−1 (19), the changes of enthalpy in the photocycle essentially reflect the conversion between photon energy and chemical energy stored in the ligand-receptor complex. The enthalpy of formation (ΔH°) was –21.6 ± 1.3 kcal mol−1, and the entropic contribution (−TΔS°) was 7.3 ± 1.3 kcal mol−1. Thus, the recombination reaction between 11CR and opsin is enthalpy driven.
Discussion
Detergent micelles impede ligand binding in Rho
To analyze the binding of opsin and retinal, we need a reconstitution method to maintain the stability of opsin. Dodecylmaltoside (DM) micelles have been routinely employed in the purification of rhodopsin from recombinant sources (20). However, a previous report (21) and our results (Fig. 2 F) showed that detergent micelles are ineffective for stabilizing ligand-free opsin. By comparison, a bicelle system better mimics the native membrane environment and effectively stabilizes free opsin at its functional state (12, 22, 23). Compared with liposomes (24), bicelles are easier to prepare, show much less light scattering, and are more compatible with UV-Vis and fluorescence spectroscopy.
Based on the denaturation kinetics (compare to Fig. 3 F) at room temperature, there was >50% of opsin that remains functional within 2 h after photobleaching and could still regenerate after being transferred into bicelles (Fig. S2). This observation implies that limitations of the detergent system were not simply a lack of thermal stability of opsin at room temperature. Another relevant piece of evidence is that Rho with an engineered N2C/D282C disulfide bond, even when solubilized in DM, can be readily regenerated with 11CR (25, 26). This additional disulfide bond connects the N-terminus and the third extracellular loop but does not make contact with the chromophore binding pocket. It is improbable that the N2C/D282C mutation can dramatically alter the interaction between detergent and receptor. Therefore, the most plausible explanation is that DM might inhibit the binding of opsin and 11CR by modulating the conformation and energetics of opsin and that the disulfide bond alters the receptor conformational dynamics in a manner that offsets the effect of DM. In line with this notion, molecular dynamic simulation on an adenosine receptor suggested that DM and POPC gave slightly different receptor conformations (27). Two more experimental observations also support a crucial role of conformational energetics. First, the structure of opsin crystallized from a detergent with similar properties to DM (28) shows helix movements relative to the rhodopsin ground state that are characteristic of the light-activated state of rhodopsin (29, 30). Second, opsin in 1% DM could be regenerated at 4°C (compare to Fig. S3). Although we cannot rule out the possibility that detergent may penetrate into the ligand binding site of the receptor, which has been suggested by structure studies (31) and molecular dynamics simulations (27), and thus may compete with 11CR binding, the relevance of such a scenario awaits direct experimental evidence. We propose that POPC/CHAPS bicelles stabilize a conformation of opsin that is similar to the rhodopsin ground state, which facilitates binding of 11CR. Our findings in the context of previously published literature should prompt a rethinking of the validity of measurements of GPCR activity in detergent systems.
Several studies on Rho regeneration in DM micelles have reported a large decrease in Trp fluorescence immediately upon the addition of 11CR, followed by a slower phase of fluorescence decrease (24, 32, 33). Our experiments comparing high (2%) and low (0.1%) concentration of DM (Fig. 4 E) suggested that this initial fast quenching resulted from retinal partitioning into DM micelles. The implication of this fast-phase quenching was discussed in greater detail in the Supporting Material.
Rho regeneration in bicelles exhibited single-phase kinetics
Electrophysiological studies suggested that the pigment regeneration process involves a transient noncovalent complex between retinal and opsin, which is followed by the formation of the mature pigment with the PSB bond (34). The formation of the noncovalent complex can be reversible, i.e., there is a theoretical probability of retinal dissociation from the ligand binding pocket. In our earlier article, we have estimated the rate of spontaneous break of the PSB linkage to be as slow as 10−7 s−1 (12). Thus when the retinal binding reaction is concerned, the second step can be treated as virtually irreversible.
The tryptophan fluorescence assay indicated whether the retinal resides in the binding pocket or somewhere else in close proximity to opsin, whereas the Alexa488 fluorescence assay reflected overall kinetics from retinal entry to PSB bond formation. If there is significant reversible dissociation of retinal in the first step, the retinal binding rates measured by the Alexa488-based assay should be slower than the rates measured by the Trp-based assay. However, we found that these two assays gave statistically indistinguishable results (Fig. 5 C). Besides direct observations of a noncovalent complex, the presence of a noncovalent intermediate product should yield a nonlinear second-order kinetic plot (35). Nonetheless, we have not found any deviation from linearity of the overall reaction rate with the concentration of retinal. Thus the formation of the noncovalent intermediate and the formation of PSB bond are tightly coupled.
The ITC experiment corroborates the kinetic measurements
The principles of microscopic reversibility and detailed balance indicate that the forward and the reverse reactions traverse through the same energy barrier. The fluorescence quenching studies provided a value for the energy barrier for the recombination reaction (Δ‡Gon). Previously we obtained the activation energy for the reverse reaction (Δ‡Goff) by measuring the kinetics of 11CR dissociation from Rho (12). However, because more than one molecular pathway might contribute to retinal dissociation, it is important to verify whether these two measured barriers actually correspond to the same transition state.
Due to the technical challenge of measuring the reaction enthalpy (ΔH°) from the temperature dependence of the equilibrium constant (Kd) and the associated change in free energy (ΔG°) following van ’t Hoff’s relation, we chose to directly determine the enthalpy of formation by ITC. Ordinarily, opsin rapidly denatures in detergent micelles. Here the ITC experiment for ligand binding in Rho was made possible by utilizing POPC/CHAPS bicelles to stabilize the apoprotein over the timescale of the experiment (15∼30 h). The principle of microscopic reversibility predicts that the difference between the activation enthalpy of the forward and reverse reactions should have the same value as the enthalpy of formation (Δ‡Hon − Δ‡Hoff = ΔH°). Indeed, the kinetics experiments (Δ‡Hon − Δ‡Hoff = −18.2 ± 4.5 kcal mol−1) and ITC experiment (ΔH° = −21.6 ± 1.3 kcal mol−1) yielded consistent results; within the stated errors these two values are statistically indistinguishable. Therefore, the calorimetric measurement substantiated the accuracy of the kinetics data. More importantly, the ITC result supports our analysis that the direct breaking of the PSB linkage, rather than thermal isomerization of 11CR to ATR, is the primary pathway for the spontaneous dissociation of 11CR from Rho (12).
The ITC result was used to derive a complete enthalpy profile of Rho regeneration and photoactivation (Fig. 7 C). In our diagram, the change of enthalpy of (ATR + opsin) relative to Rho (+22 kcal mol−1 at pH 6.0) differs from an earlier study (+12 kcal mol−1 at pH 5.4) (18). We contend that the ITC experiment in this study gives a more accurate value because it directly measures the binding enthalpy and does not rely on complicated thermodynamic cycles involving hydroxylamine-dependent reaction steps.
The dissociation constant of Rho in disc membrane
Our kinetics measurement was done in 1% (w/v) of POPC (13 mM), whereas the concentration of lipids in the ROS disc membrane is ∼150 mM (36). The inverse correlation between regeneration kinetics and lipid concentration (Fig. 5 B) predicts that the k2 in ROS would be ∼8.7% of that in 1% POPC/CHAPS (60 M−1 s−1 at 25°C or 235 M−1 s−1 at 37°C). This value is close to the regeneration rate measured in ROS membrane (3.0 × 102 to 4.0 × 102 M−1 s−1 at 35°C) (37). To estimate the dissociation constant between opsin and 11CR in the dark-adapted retina, we assumed that Rho regeneration in ROS membrane has a k2 of 300 M−1 s−1 at 37°C. Based on the measured dissociation rate (koff of 2.2 × 10−7 s−1), the dissociation constant (Kd = koff /k2) for the 11CR-opsin covalent complex (i.e., Rho) is 0.73 nM at 37°C in the ROS membrane. Note that the dissociation constants in 1% POPC/CHAPS at 25 and 37°C were 25 and 82 pM, respectively (compare to Supporting Material). In a previous study of 11CR binding to opsin using surface plasmon resonance (SPR), a dissociation constant of 130 nM has been inferred (38). However, we contend that estimate because the binding times in the surface plasmon resonance study were orders-of-magnitude too short to ensure equilibration.
Previous psychophysical studies on dark adaptation showed that the exponential phase of Rho regeneration in human retina has a pseudo-first order kobs of 1.5 × 10−3 s−1, or 0.09 min−1 (6). Then the concentration of intracellular free 11CR is calculated to be 5 μM ([11CR] = kobs/k2), only a tiny fraction (0.1%) of the pigment content in rods (4.6 mM) (39). This value is consistent with the observation that Rho regeneration at high photobleaching level is rate limited by the diffusion of 11CR through retinal pigment epithelium cells (6). The estimated concentration of 11CR in the ROS is 6800-times larger than the dissociation constant. Following Le Chatelier’s principle, the concentration of 11CR is sufficient to drive the opsinRho equilibrium far to the side of Rho, and the concentration of opsin is only ∼0.015% of Rho (12).
Comparison of ligand binding in Rho and other GPCRs
Among GPCRs, visual pigments, including Rho, are unique in that they form covalent bonds with their inverse agonist (11CR) and agonist (ATR) ligands in the receptor active site. Protease-activated receptors are somewhat similar in that they encompass a tethered-ligand as a part of their N-terminal tails that is released upon the action of a serine protease. However, most other GPCRs, if not all, are activated by endogenous diffusible agonist ligands, or inhibited by exogenously added ligand drugs. One might assume that the covalent linkage between opsin and 11CR would dramatically alter the energetics of ligand binding in Rho as compared with ligand binding in other GPCRs.
Relevant literature values are available for three high-affinity ligands in complex with β-adrenergic receptor: β-adrenergic receptor (βAR)-carazolol (40), βAR-iodopindolol (41), and βAR-BI-167107 (42) (Table 1). We compared the values for the energetics of ligand binding to opsin with the published values for βAR. Surprisingly, the Kd values and the overall change of free energy (ΔG°) for the ligand-binding reaction for the two systems are on the same order of magnitude. Despite similar ΔG°, the change of enthalpy (ΔH°) is much higher for Rho, possibly due to the formation of the SB bond, which provides an example of enthalpy-entropy compensation in ligand-receptor binding (43). This comparison shows that the underlying energetics that defines high-affinity ligand-receptor interactions might be conserved among visual pigments and GPCRs utilizing diffusible ligands.
Table 1.
The Kinetic and Thermodynamic Parameters for βAR and Rho with Their Ligands at 25°C
| Receptor | β2AR | β2AR | βAR | Rho |
|---|---|---|---|---|
| Ligand | BI-167107a | Carazololb | Iodopindololc | 11CRd |
| k2 (M−1 s−1) | 7.6 × 104 | >6.3 × 104 | 8 × 106 | 6.9 × 102 |
| koff (s−1) | 6.4 × 10−6 | 6.3 × 10−6 | 4.2 × 10−4 | 1.7 × 10−8 |
| Kd (pM) | 84 | <100 | 53 | 25e |
| ΔG° (kcal mol−1) | −13.7 | <−13.6 | −14.1 ± 0.1 | −14.4 ± 0.04 |
| ΔH° (kcal mol−1) | — | — | −9.3 ± 0.1 | −21.6 ± 1.3 |
| −TΔS° (kcal mol−1) | — | — | −4.7 ± 0.1 | 7.2 ± 1.3 |
| Δ‡Gon (kcal mol−1) | 10.8 | >10.9 | 8.0 | 13.5 |
| Δ‡Goff (kcal mol−1) | 24.5 | 24.5 | 22.0 | 27.9 |
Kd and koff taken from Rasmussen et al. (42); other values are calculated based on the transition state theory.
Kd and koff taken from Rosenbaum et al. (40); other values are calculated based on the transition state theory.
k2, koffKd, ΔG°, and ΔH° taken from Contreras et al. (52); other values are calculated based on the transition state theory.
Calculated for 25°C from the Eyring plot (Fig. 4C), so the values are different from the values indicated in Fig. 7A.
Calculated as Kd = koff/k2.
The role of the PSB linkage in Rho is to lock retinal inside the ligand-binding pocket near to the center of the receptor. The buried binding pocket, in turn, protects the PSB from hydrolysis. Thus, Rho is evolved to increase the lifetime of the inactive ligand-receptor complex. The energy barrier for ligand dissociation (Δ‡Goff) in Rho is notably higher than in βAR that has no endogenous inverse agonist. The energy barrier for ligand binding (Δ‡Gon) is correspondingly higher in Rho, explaining the slow ligand-binding kinetics for Rho compared with other GPCRs.
Ligand binding in Rho was proposed to proceed through an intramembranous pathway (44, 45, 46), whereas in the case of the βAR, it occurs through the aqueous exposed extracellular surface (47). The higher Δ‡Gon for Rho suggests that retinal entry into the ligand-binding pocket of opsin involves greater conformational change, as inferred from the structures. The viscosity of lipid bilayer also contributes to the free energy barrier of ligand binding to GPCRs in an additive fashion (48). Therefore, the difference in Δ‡Gon between opsin and βAR might be correlated to the distinct ligand binding modes utilized by the two receptors.
GPCRs constitute the largest category of small-molecule drug targets (49). Pharmacologists are accustomed to using the equilibrium dissociation constant (Kd) as a guiding principle for drug design. Recently, there is increasing appreciation in the GPCR pharmacology field for the relevance of receptor residence time (= 1/koff) of ligand (50), and hence a growing need to develop new assays for quantifying ligand binding and dissociation kinetics. In this study, we harnessed the unique photochemistry of Rho to develop energy transfer-based kinetic assays. Although most GPCRs do not utilize chromophores as ligands, the rapid development of fluorescent ligands for GPCRs (51), combined with our site-specific labeling strategy, may facilitate the study of ligand binding kinetics for a variety of receptors.
Conclusions
We present here a comprehensive analysis of the kinetics and thermodynamics of the recombination reaction between opsin and 11CR to form the mature visual pigment, Rho. We found that the lipid bilayer environment is important for ligand binding in Rho. We also show that despite the covalent linkage between 11CR and opsin in Rho, the equilibrium dissociation constant of 11CR for opsin is similar to affinities of diffusible ligands for GPCRs. The higher energy barrier for ligand binding in Rho underlies the slower ligand binding and dissociation kinetics for Rho compared with other GPCRs. We suggest that the energy diagram derived in this study provides a useful guide for interpreting ligand binding modes for other GPCRs.
Author Contributions
H.T., T.P.S., and T.H designed the study, analyzed the results, and wrote the manuscript. H.T. and T.H. conducted the experiments.
Acknowledgments
We thank Prof. Klaus Peter Hofmann for discussions that inspired a part of this work. We thank Prof. King-Wai Yau for discussions of this work.
We acknowledge the generous support from the Crowley Family Fund and the Danica Foundation. This work has also been generously supported by an International Research Alliance with Prof. Thue W. Schwartz at The Novo Nordisk Foundation Center for Basic Metabolic Research (http://www.metabol.ku.dk) through an unconditional grant from the Novo Nordisk Foundation to the University of Copenhagen. We also acknowledge support from National Institutes of Health (NIH) grant R01 EY012049 (T.P.S. and T.H.), as well as the Tri-Institutional Training Program in Chemical Biology in supporting H.T.
Editor: Andreas Engel.
Footnotes
He Tian’s present address is Department of Chemistry and Chemical Biology, Harvard University, Cambridge, Massachusetts.
Supporting Materials and Methods, Supporting Data, three figures, and two tables are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(17)30575-1.
Contributor Information
Thomas P. Sakmar, Email: sakmar@rockefeller.edu.
Thomas Huber, Email: hubert@rockefeller.edu.
Supporting Material
References
- 1.Wald G. Molecular basis of visual excitation. Science. 1968;162:230–239. doi: 10.1126/science.162.3850.230. [DOI] [PubMed] [Google Scholar]
- 2.Schertler G.F., Villa C., Henderson R. Projection structure of rhodopsin. Nature. 1993;362:770–772. doi: 10.1038/362770a0. [DOI] [PubMed] [Google Scholar]
- 3.Palczewski K., Kumasaka T., Miyano M. Crystal structure of rhodopsin: a G protein-coupled receptor. Science. 2000;289:739–745. doi: 10.1126/science.289.5480.739. [DOI] [PubMed] [Google Scholar]
- 4.Menon S.T., Han M., Sakmar T.P. Rhodopsin: structural basis of molecular physiology. Physiol. Rev. 2001;81:1659–1688. doi: 10.1152/physrev.2001.81.4.1659. [DOI] [PubMed] [Google Scholar]
- 5.Sakmar T.P., Franke R.R., Khorana H.G. Glutamic acid-113 serves as the retinylidene Schiff base counterion in bovine rhodopsin. Proc. Natl. Acad. Sci. USA. 1989;86:8309–8313. doi: 10.1073/pnas.86.21.8309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lamb T.D., Pugh E.N., Jr. Dark adaptation and the retinoid cycle of vision. Prog. Retin. Eye Res. 2004;23:307–380. doi: 10.1016/j.preteyeres.2004.03.001. [DOI] [PubMed] [Google Scholar]
- 7.Ye S., Huber T., Sakmar T.P. FTIR analysis of GPCR activation using azido probes. Nat. Chem. Biol. 2009;5:397–399. doi: 10.1038/nchembio.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tian H., Sakmar T.P., Huber T. Site-specific labeling of genetically encoded azido groups for multicolor, single-molecule fluorescence imaging of GPCRs. Methods Cell Biol. 2013;117:267–303. doi: 10.1016/B978-0-12-408143-7.00015-3. [DOI] [PubMed] [Google Scholar]
- 9.Huber T., Naganathan S., Sakmar T.P. Unnatural amino acid mutagenesis of GPCRs using amber codon suppression and bioorthogonal labeling. Methods Enzymol. 2013;520:281–305. doi: 10.1016/B978-0-12-391861-1.00013-7. [DOI] [PubMed] [Google Scholar]
- 10.Tian H., Naganathan S., Huber T. Bioorthogonal fluorescent labeling of functional G-protein-coupled receptors. ChemBioChem. 2014;15:1820–1829. doi: 10.1002/cbic.201402193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tian H., Fürstenberg A., Huber T. Labeling and single-molecule methods to monitor G protein-coupled receptor dynamics. Chem. Rev. 2017;117:186–245. doi: 10.1021/acs.chemrev.6b00084. [DOI] [PubMed] [Google Scholar]
- 12.Tian H., Sakmar T.P., Huber T. Measurement of slow spontaneous release of 11-cis-retinal from rhodopsin. Biophys. J. 2017;112:153–161. doi: 10.1016/j.bpj.2016.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Farrens D.L., Khorana H.G. Structure and function in rhodopsin. Measurement of the rate of metarhodopsin II decay by fluorescence spectroscopy. J. Biol. Chem. 1995;270:5073–5076. doi: 10.1074/jbc.270.10.5073. [DOI] [PubMed] [Google Scholar]
- 14.McCaslin D.R., Tanford C. Effects of detergent micelles on the recombination reaction of opsin and 11-cis-retinal. Biochemistry. 1981;20:5207–5212. doi: 10.1021/bi00521a017. [DOI] [PubMed] [Google Scholar]
- 15.Frederiksen R., Boyer N.P., Cornwall M.C. Low aqueous solubility of 11-cis-retinal limits the rate of pigment formation and dark adaptation in salamander rods. J. Gen. Physiol. 2012;139:493–505. doi: 10.1085/jgp.201110685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gaede H.C., Gawrisch K. Lateral diffusion rates of lipid, water, and a hydrophobic drug in a multilamellar liposome. Biophys. J. 2003;85:1734–1740. doi: 10.1016/S0006-3495(03)74603-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pierce M.M., Raman C.S., Nall B.T. Isothermal titration calorimetry of protein-protein interactions. Methods. 1999;19:213–221. doi: 10.1006/meth.1999.0852. [DOI] [PubMed] [Google Scholar]
- 18.Cooper A. Rhodopsin photoenergetics: lumirhodopsin and the complete energy profile. FEBS Lett. 1981;123:324–326. doi: 10.1016/0014-5793(81)80319-5. [DOI] [PubMed] [Google Scholar]
- 19.Hubbard R. The stereoisomerization of 11-cis-retinal. J. Biol. Chem. 1966;241:1814–1818. [PubMed] [Google Scholar]
- 20.Karnik S.S., Sakmar T.P., Khorana H.G. Cysteine residues 110 and 187 are essential for the formation of correct structure in bovine rhodopsin. Proc. Natl. Acad. Sci. USA. 1988;85:8459–8463. doi: 10.1073/pnas.85.22.8459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.De Grip W.J. Thermal stability of rhodopsin and opsin in some novel detergents. Methods Enzymol. 1982;81:256–265. doi: 10.1016/s0076-6879(82)81040-9. [DOI] [PubMed] [Google Scholar]
- 22.McKibbin C., Farmer N.A., Booth P.J. Opsin stability and folding: modulation by phospholipid bicelles. J. Mol. Biol. 2007;374:1319–1332. doi: 10.1016/j.jmb.2007.10.018. [DOI] [PubMed] [Google Scholar]
- 23.Reeves P.J., Hwa J., Khorana H.G. Structure and function in rhodopsin: kinetic studies of retinal binding to purified opsin mutants in defined phospholipid-detergent mixtures serve as probes of the retinal binding pocket. Proc. Natl. Acad. Sci. USA. 1999;96:1927–1931. doi: 10.1073/pnas.96.5.1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sánchez-Martín M.J., Ramon E., Garriga P. Improved conformational stability of the visual G protein-coupled receptor rhodopsin by specific interaction with docosahexaenoic acid phospholipid. ChemBioChem. 2013;14:639–644. doi: 10.1002/cbic.201200687. [DOI] [PubMed] [Google Scholar]
- 25.Xie G., Gross A.K., Oprian D.D. An opsin mutant with increased thermal stability. Biochemistry. 2003;42:1995–2001. doi: 10.1021/bi020611z. [DOI] [PubMed] [Google Scholar]
- 26.Piechnick R., Ritter E., Heck M. Effect of channel mutations on the uptake and release of the retinal ligand in opsin. Proc. Natl. Acad. Sci. USA. 2012;109:5247–5252. doi: 10.1073/pnas.1117268109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee S., Mao A., Vaidehi N. How do short chain nonionic detergents destabilize G-protein-coupled receptors? J. Am. Chem. Soc. 2016;138:15425–15433. doi: 10.1021/jacs.6b08742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Park J.H., Scheerer P., Ernst O.P. Crystal structure of the ligand-free G-protein-coupled receptor opsin. Nature. 2008;454:183–187. doi: 10.1038/nature07063. [DOI] [PubMed] [Google Scholar]
- 29.Sheikh S.P., Zvyaga T.A., Bourne H.R. Rhodopsin activation blocked by metal-ion-binding sites linking transmembrane helices C and F. Nature. 1996;383:347–350. doi: 10.1038/383347a0. [DOI] [PubMed] [Google Scholar]
- 30.Farrens D.L., Altenbach C., Khorana H.G. Requirement of rigid-body motion of transmembrane helices for light activation of rhodopsin. Science. 1996;274:768–770. doi: 10.1126/science.274.5288.768. [DOI] [PubMed] [Google Scholar]
- 31.Szczepek M., Beyrière F., Scheerer P. Crystal structure of a common GPCR-binding interface for G protein and arrestin. Nat. Commun. 2014;5:4801. doi: 10.1038/ncomms5801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Heck M., Schädel S.A., Hofmann K.P. Secondary binding sites of retinoids in opsin: characterization and role in regeneration. Vision Res. 2003;43:3003–3010. doi: 10.1016/j.visres.2003.08.011. [DOI] [PubMed] [Google Scholar]
- 33.Schadel S.A., Heck M., Hofmann K.P. Ligand channeling within a G-protein-coupled receptor. The entry and exit of retinals in native opsin. J. Biol. Chem. 2003;278:24896–24903. doi: 10.1074/jbc.M302115200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kefalov V.J., Crouch R.K., Cornwall M.C. Role of noncovalent binding of 11-cis-retinal to opsin in dark adaptation of rod and cone photoreceptors. Neuron. 2001;29:749–755. doi: 10.1016/s0896-6273(01)00249-5. [DOI] [PubMed] [Google Scholar]
- 35.Henselman R.A., Cusanovich M.A. Characterization of the recombination reaction of rhodopsin. Biochemistry. 1976;15:5321–5325. doi: 10.1021/bi00669a019. [DOI] [PubMed] [Google Scholar]
- 36.Liebman P.A., Jagger W.S., Bargoot F.G. Membrane structure changes in rod outer segments associated with rhodopsin bleaching. Nature. 1974;251:31–36. doi: 10.1038/251031a0. [DOI] [PubMed] [Google Scholar]
- 37.Kawaguchi T., Hamanaka T., Kito Y. Kinetic study of transfer of 11-cis-retinal between rod outer segment membranes using regeneration of rhodopsin. Biophys. Chem. 1986;24:5–12. doi: 10.1016/0301-4622(86)85053-0. [DOI] [PubMed] [Google Scholar]
- 38.Bieri C., Ernst O.P., Vogel H. Micropatterned immobilization of a G protein-coupled receptor and direct detection of G protein activation. Nat. Biotechnol. 1999;17:1105–1108. doi: 10.1038/15090. [DOI] [PubMed] [Google Scholar]
- 39.Nickell S., Park P.S., Palczewski K. Three-dimensional architecture of murine rod outer segments determined by cryoelectron tomography. J. Cell Biol. 2007;177:917–925. doi: 10.1083/jcb.200612010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rosenbaum D.M., Cherezov V., Kobilka B.K. GPCR engineering yields high-resolution structural insights into β2-adrenergic receptor function. Science. 2007;318:1266–1273. doi: 10.1126/science.1150609. [DOI] [PubMed] [Google Scholar]
- 41.Contreras M.L., Wolfe B.B., Molinoff P.B. Thermodynamic properties of agonist interactions with the β-adrenergic receptor-coupled adenylate cyclase system. II. Agonist binding to soluble β-adrenergic receptors. J. Pharmacol. Exp. Ther. 1986;237:165–172. [PubMed] [Google Scholar]
- 42.Rasmussen S.G., Choi H.J., Kobilka B.K. Structure of a nanobody-stabilized active state of the β2 adrenoceptor. Nature. 2011;469:175–180. doi: 10.1038/nature09648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Borea P.A., Dalpiaz A., Gilli G. Can thermodynamic measurements of receptor binding yield information on drug affinity and efficacy? Biochem. Pharmacol. 2000;60:1549–1556. doi: 10.1016/s0006-2952(00)00368-3. [DOI] [PubMed] [Google Scholar]
- 44.Huber T., Gunnison K.M., Sakmar T.P. Identification of the primary entry site in visual rhodopsins: an intramembranous pathway from mutagenesis and MD simulations. Biophys. J. 2005;88:507a–508a. [Google Scholar]
- 45.Wang T., Duan Y. Chromophore channeling in the G-protein coupled receptor rhodopsin. J. Am. Chem. Soc. 2007;129:6970–6971. doi: 10.1021/ja0691977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hildebrand P.W., Scheerer P., Heck M. A ligand channel through the G protein coupled receptor opsin. PLoS One. 2009;4:e4382. doi: 10.1371/journal.pone.0004382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Venkatakrishnan A.J., Deupi X., Babu M.M. Molecular signatures of G-protein-coupled receptors. Nature. 2013;494:185–194. doi: 10.1038/nature11896. [DOI] [PubMed] [Google Scholar]
- 48.Lee A.G. Lipids and their effects on membrane proteins: evidence against a role for fluidity. Prog. Lipid Res. 1991;30:323–348. doi: 10.1016/0163-7827(91)90002-m. [DOI] [PubMed] [Google Scholar]
- 49.Santos R., Ursu O., Overington J.P. A comprehensive map of molecular drug targets. Nat. Rev. Drug Discov. 2017;16:19–34. doi: 10.1038/nrd.2016.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Guo D., Heitman L.H., Ijzerman A.P. The role of target binding kinetics in drug discovery. ChemMedChem. 2015;10:1793–1796. doi: 10.1002/cmdc.201500310. [DOI] [PubMed] [Google Scholar]
- 51.Stoddart L.A., Kilpatrick L.E., Hill S.J. Probing the pharmacology of G protein-coupled receptors with fluorescent ligands. Neuropharmacology. 2015;98:48–57. doi: 10.1016/j.neuropharm.2015.04.033. [DOI] [PubMed] [Google Scholar]
- 52.Contreras M.L., Wolfe B.B., Molinoff P.B. Thermodynamic properties of agonist interactions with the β-adrenergic receptor-coupled adenylate cyclase system. I. High- and low-affinity states of agonist binding to membrane-bound β-adrenergic receptors. J. Pharmacol. Exp. Ther. 1986;237:154–164. [PubMed] [Google Scholar]
- 53.Tian H., Sakmar T.P., Huber T. Micelle-enhanced bioorthogonal labeling of genetically encoded azido groups on the lipid-embedded surface of a GPCR. ChemBioChem. 2015;16:1314–1322. doi: 10.1002/cbic.201500030. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.