Abstract
Previous research has shown that adverse social conditions may promote a conserved transcriptional response to adversity (CTRA) involving up-regulation of proinflammatory gene expression and down-regulation of Type 1 interferon anti-viral genes in circulating blood cells. However, the impact of social conditions on lymphoid tissue gene regulation remains largely unexplored. This project assessed how social instability in adult male rhesus macaques (N = 10, 5 in unstable, and 5 in stable social conditions) might regulate gene expression within secondary lymphoid tissue (lymph nodes; LN). Unstable social conditions down-regulated axillary LN expression of genes involved in Type I interferon anti-viral responses. Transcript origin analyses implicated monocytes and B cells as cellular mediators of these effects, and promoter-based bioinformatics analyses indicated reduced activity of AP-1, NF-κB, IRF, and CREB transcription factors within the axillary LN microenvironment. Although the current study is limited in sample size, these results suggest that social influences on immune cell gene regulation extend beyond the circulating leukocyte pool to alter generalized transcriptome profiles in secondary lymphoid tissue, and they do so in a regulatory program that resembles the pattern of antiviral inhibition previously observed in circulating leukocytes.
Keywords: social genomics, social stress, inflammation, health, rhesus macaque
1. Introduction
Social stress has been established as a risk factor for a variety of diseases (Capitanio et al., 1998; Caspi et al., 2006; Cohen et al., 1997; Eng et al., 2002; Kroenke et al., 2006; Reynolds & Kaplan, 1990; Soler-Vila et al., 2003). The biological basis of this risk has previously been linked in part to a pattern of gene expression known as the conserved transcriptional response to adversity (CTRA), which is characterized by up-regulation of pro-inflammatory gene transcription and down-regulation of Type I interferon-mediated antiviral response genes (Slavich & Cole, 2013). These transcriptional shifts are hypothesized to transiently improve immune response to physical injury by enhancing acute phase responses to limit bacterial infection and accelerate wound healing (Irwin & Cole, 2011). However, chronic activation of the CTRA in response to extended social stressors may contribute to negative health outcomes by promoting inflammation-related chronic diseases and impairing immune responses to viral infections (Cole, 2013, 2014; Cole et al., 2007).
Our understanding of CTRA development and resolution is limited by the fact that most previous studies have mapped these transcriptome dynamics only in the circulating leukocyte pool. However, circulating leukocytes represent a small minority of the body’s total pool of immune cells (typically < 2%) and are selectively mobilized out of primary and secondary lymphoid tissues (which generally contain > 98% of the total leukocyte pool) (Westermann & Pabst, 1990). As such, it remains to be determined whether the core immunoregulatory dynamics observed in the circulating leukocyte CTRA are also present in the lymphoid tissues that contain the majority of immune cells and which may not be well represented by the small minority of selectively circulating leukocytes (Murphy et al., 2012).
Previous studies have implicated myeloid cell population dynamics as a key mediator in CTRA transcriptome skewing (Cole et al., 2015; Cole et al., 2012; Cole et al., 2011; Powell et al., 2013). Myeloid cells show complex trafficking patterns and developmental processes such that the majority of circulating monocytes and dendritic cells are relatively immature and quiescent cells, whereas myeloid cells that become activated through danger signals selectively exit from the blood and traffic into tissues, after which they may further transit to secondary lymphoid organs through the lymphatics. Thus it remains poorly understood whether the myeloid cell-related CTRA transcriptome dynamics observed in the circulating leukocyte pool are also observed within the secondary lymphoid organs that contain a more selected mix of myeloid cell types and that mediate myeloid-lymphoid interactions (e.g,, antigen presentation, T- and B-cell differentiation, etc.) that do not occur in circulating blood.
To address our lack of insight into secondary lymphoid tissue responses to social stress, we conducted genome-wide transcriptional profiling of axillary lymph node (LN) tissues obtained from adult male rhesus macaques randomized to either stable social conditions (control) or unstable social conditions involving chronic low-grade social threat. This social instability paradigm has been used extensively to examine the broader physiological effects of chronic social adversity (Capitanio et al., 2008; Capitanio & Cole, 2015; Capitanio et al., 1999; Capitanio et al., 1998; Cole et al., 2009; Sloan et al., 2008; Sloan et al., 2007; Sloan et al., 2006), and 3 studies have previously examined focal effects on specific gene transcripts in axillary LN tissues (Capitanio & Cole, 2015; Sloan et al., 2008; Sloan et al., 2007). However, previous studies of chronic social threat have not collected genome-wide transcriptional data from LNs and this distinction is important because the comprehensive and unbiased nature of such transcriptome profiles allows for bioinformatic analyses of transcription factor mediators (Cole et al., 2007; Cole et al., 2005) and cellular mechanisms (Cole et al., 2015; Cole et al., 2011) that cannot be derived from more focal assessments of a small number of target genes. LNs are primary sites of interaction between peripherally activated myeloid cells and resident lymphoid cells that subsequently undergo cellular activation and differentiation (Murphy et al., 2012). As such, our transcriptome-wide bioinformatics analyses focused on this tissue system in order to gauge the “downstream” effects on LN immunoregulatory dynamics that ensue from the CTRA dynamics previously observed in circulating leukocytes. We focused in particular on assessing, 1) whether axillary LNs show the same cardinal profiles of gene regulation observed in peripheral leukocyte CTRA (i.e., up-regulated inflammatory signaling and down-regulated antiviral / Type I interferon signaling), 2) whether axillary LNs showed evidence of transcription factor activity alterations previously observed in circulating leukocytes (e.g., activation of pro-inflammatory factors such as NF-κB and AP-1, inhibition of interferon response factors (IRFs), and activation of neurally regulated factors such as CREB), and 3) whether the observed tissue-wide transcriptome dynamics might take place predominately within a specific cellular compartment (e.g., in myeloid lineage monocytes/macrophages or dendritic cells that enter the LN to stimulate lymphoid lineage cells, or in the lymphoid lineage T and B cells that respond to myeloid stimulation, etc.). Results show that some components of the peripheral CTRA gene regulatory dynamic are mirrored closely in the axillary LN tissue transcriptome, whereas others are much less evident and may reflect shifting cellular differentiation states as peripheral circulating/patrolling myeloid cells mature into LN-resident cells involved in antigen presentation and other aspects of lymphoid tissue biology.
2. Material and Methods
2.1 Subjects and living arrangements
Subjects were 10 adult male rhesus monkeys (mean age = 6.0 years) born in any of 17 outdoor 0.2-hectare enclosures at the California National Primate Research Center (CNPRC). Each enclosure contained up to 150 animals of all ages and both sexes, approximately reflecting the composition of a troop of wild rhesus monkeys. The current subjects (n=10) were randomly selected from a larger study (n=24, 12-unstable) involving the administration of methamphetamine and all animals were eventually inoculated with SIV (Capitanio & Cole, 2015). Subjects for the current study were controls for the methamphetamine manipulation, receiving daily saline doses beginning 8–13 (mean = 10.1) days after the first socialization period. All measures for the current study were collected prior to SIV inoculation. Animals were relocated from outdoor social groups to individual housing in 0.74 m2 (floor area) cages where they lived throughout the experiment. Animals were provided water ad libitum and were fed laboratory monkey chow (Lab Diet #5038, Purina Mills International, St. Louis, MO) twice daily in the morning between 0715 and 0745 and in the afternoon between 1345 and 1415. Fruits and vegetable supplements were provided twice weekly and trained animal care technicians checked on the animals’ health daily.
2.2 Procedure
All procedures were conducted according to the Guidelines for Use and Care of Laboratory Animals of the National Research Council supported by the National Institutes of Health and according to CNPRC SOPs. Experimental protocols were approved by the University of California, Davis IACUC. Animals were randomly assigned to unstable or stable social conditions and socialization occurred during a 22–27 day window. At the end of the socialization period, axillary LNs were collected and stored for later analysis.
2.2.1 Social Conditions
Socialization occurred up to 4 days per week for 100 min per day during the 22–27 days prior to LN biopsy (Section 2.3.1). All animals experienced 13 socialization sessions during the socialization period, with the exception of one animal that experienced 12 sessions. The number and duration of socializations was selected based on previous studies showing that this duration alters lymphoid tissue biology (Sloan et al., 2007) and natural killer cell activity (Capitanio & Cole, 2015). The socialization cage was constructed of pipe and chain link and measured 1.8 x 3.1 x 2.2 m. Each animal was randomly assigned to either the stable social condition (i.e., socializing with the same two other animals each day; i.e., a stable group of three; n=5), or the unstable social condition (i.e., socializing with a different randomly generated set of one to three partner animals on any given day, with the number and identity of partners changing on a daily basis; average of daily group size over time remained three; n=5). There was 24 subjects in the larger study (Capitanio & Cole, 2015). Subjects in the current study were randomly selected from any of the control animals (n=12; which received saline, not methamphetamine) from the 4 stable groups formed during the study. Animals in each social group (whether stable or unstable) were unfamiliar with each other on the first day of socialization, and were unrelated based upon examination of the CNPRC microsatellite-based parentage data. All subjects would become increasingly familiar with each other over time; but whereas the same animals in the stable groups met daily, the growing familiarity among any two animals in the unstable groups was affected by the number and identities of the other animals present on any given day. The unpredictability of the composition of the unstable groups required daily re-establishment of social relationships, which was stressful, as evidenced by the behavioral indicators described below (see also references listed in the Introduction from prior studies that utilized this social manipulation).
Trained observers collected behavioral data using the Observer 3.0 (Noldus Information Technology, Wageningen, Netherlands). For all animals, with the exception of one, over the socialization period, ad lib data were collected for a total of 260 min (20 min per group per observation day), while focal observations were collected for a total of 65 min (5 min per animal per observation day). The single animal that had one fewer daily social session experienced a total of 240 minutes of ad lib data and a total of 60 minutes of focal observations. Behavior categories used were those commonly used for this species (Capitanio, 1985), which included threat displays, lipsmacks, and social contact. Bites and injury were minimal; on the rare occasions that threats and chases escalated to the point of potential injury, however, data collection was paused and the fight was interrupted using a variety of means, including shouting, banging on the cage, and if needed, spraying a hose in the direction of the fighting animals. Detailed logbook records indicate that, throughout the duration of the study, fights were interrupted on 13 occasions: 6 times in the stable groups and 7 times in the unstable groups. There were no injuries that required veterinary intervention. To accommodate the single animal that had one fewer day of socialization, behavioral outcome measures were normalized based on number of observation days by dividing durations or frequencies by the number of observation days for a given animal. Nonparametric tests were used to compare differences between groups for behavioral data.
2.3 Measures
2.3.1 Lymph Node Biopsies and Transcriptome Profiling
One to three days following the last social session (mean=1.5 days, mode=1 day, group difference was non-significant [p=0.306]), animals were immobilized with ketamine hydrochloride (10 mg/kg) and medetomidine (0.03 mg/kg) intramuscularly, and a subcutaneous injection of bupivacaine was administered proximal and medial to the biopsy site. One to three axillary LNs were surgically removed and frozen in liquid nitrogen. LN biopsies were performed on three animals per day, always between 0800 and 1000 hrs. Following the procedures of previous genome-wide transcriptome analyses in rhesus macaques (Tung et al., 2012), RNA was extracted from tissues (Qiagen RNeasy), tested for suitable mass and integrity, and subject to genome-wide transcriptional profiling utilizing Illumina HumanRef-8 v3 arrays in the UCLA Neuroscience Genomics Core Laboratory following the manufacturer’s standard protocol. Rhesus macaques and humans have largely similar genome sequences, particularly in coding regions (~96.5%). Our study utilized comparisons of gene expression levels within a species, thus mismatches with respect to the array probes are not expected to generate biased estimates of gene expression levels (Gilad et al., 2005). Previous work has demonstrated that within species comparisons utilizing an array designed for a closely related species (particularly human microarrays with macaque samples) does not produce bias in analyses, or a reduction in power (Oshlack et al., 2007). Resulting gene expression values were quantile-normalized and log2-transformed. Data are posted as Gene Expression Omnibus GSE83629. Bioinformatics analyses examined all gene transcripts showing ≥ 1.25-fold differential expression between axillary LN from unstable vs. stable social conditions to assess: 1) transcription factor activity, as indicated by TELiS database analyses quantifying relative prevalence of pro-inflammatory (AP-1, NF-κB), interferon-related (IRF), neurally-related (CREB and glucocorticoid receptor, GR) response elements in promoters of differentially expressed genes (Cole et al., 2005); and 2) predominate cellular origins of differentially expressed genes, using Transcript Origin Analysis (TOA; Cole et al., 2011). We used an effect size-based cut-off for identifying differentially expressed genes because gene lists derived from effect-size thresholds have been shown in previous studies to be more replicable and reliable than gene lists derived from statistical testing at the level of individual genes (Shi et al., 2010; Shi et al., 2008; Witten & Tibshirani, 2007). To avoid Type 1 errors, gene lists are controlled for in the high-level statistical analysis that tests whether specific gene features (e.g., promoter transcription factor binding sites) differ in prevalence in up- vs. down-regulated gene sets. For more information on statistical background, see Cole (2010) & Fredrickson et al. (2013) for supporting information analyses. Nonparametric bootstrapping of linear model residuals was used to derive standard errors and p-values for these analyses, and residuals were vector-sampled (i.e., across genes within individuals) in order to account for any potential correlations across genes (Efron & Tibshirani, 1993).
2.3.1.1 Transcription Control Pathway Analysis
A two-sample variant of TELiS (www.telis.ucla.edu) was used to compare the prevalence of transcription factor binding motifs (TFBM) from the TRANSFAC database (V$AP1_Q6, V$NFKB_C, V$IRF2_01, V$CREB_01, V$GR_Q6) in promoters of genes overexpressed in cells from unstable vs. stable social conditions. Analyses averaged results derived from nine parametric variations of promoter length (−300 bp relative to RefSeq transcription start site, −600, and −1000 bp to +200) and target TFBM match stringency (MatSim = 0.80, 0.90, 0.95) (Cole et al., 2005). Differential TFBM prevalence was assessed by two tailed p values derived from comparing mean (log2-transformed) prevalence ratios with bootstrap-derived standard errors as described above.
2.3.1.2 Transcript Origin Analysis
To identify the predominate cellular sources of tissue-level transcriptional differences between axillary LNs from unstable vs. stable social groups, we conduced TOA on all of the genes showing ≥1.25-fold difference between groups as previously described (Cole et al., 2011). Reference data on basal expression of all human genes in distinct leukocyte subsets were derived from the publically available Human Gene Atlas (GEO GSE1133). Briefly, this analysis involves deriving a cell type diagnosticity score for each gene identifying the extent to which it is predominately expressed by each major leukocyte cell type in the reference study (monocyte, plasmacytoid dendritic cell, CD4+ T cell, CD8+ T cell, B cell, natural killer cell). Given a set of differentially expressed genes (e.g., in axillary LNs from unstable vs. stable groups), the mean diagnosticity score for each candidate cell type of origin can be compared with the null diagnosticity value of 0 using bootstrap standard errors derived as described above. To the extent that average diagnosticity scores are significantly greater than the null hypothesis value, the observed experimental differences in the aggregate transcriptome can be attributed at least in part to that candidate cell type (see Cole et al., 2011 for more details).
3. Results
3.1 Effects of Social Instability on Behavior
Animals were randomly assigned to 3–4 weeks of daily social interaction in either stable, or unstable social conditions. Unstable social conditions have been shown to induce neuroendocrine and behavioral indications of stress in previous studies (Capitanio et al., 1998) and behavioral effects of unstable social groups was confirmed in the current study. In the current study, animals in the unstable social condition showed higher numbers of threat displays (mean ± SE, 1.9 ± 0.6 per 20 min vs. 0.4 ± 0.2 for controls, p = .028) and lipsmacks (1.0 ± 0.3 vs. 0.2 ± 0.1 per 20 min for controls, p = .028), as well as less time in contact (mean ± SE, 5.7 sec ± 3.1 vs. 22.2 ± 7.8 for controls per 5 min, p = .047). In addition, to test whether animals in the unstable group habituated to socialization procedures, we analyzed whether frequency or duration of behaviors summarized during the first 6 days of observations differed from the last 7 days. There were no significant differences between all behaviors (p > .223) for unstable animals.
3.2 Regulation of the Axillary Lymph Node Transcriptome
To determine whether social instability altered genome-wide transcriptional profiles in axillary LNs, we identified all transcripts showing ≥ 1.25 fold differential expression in tissues collected from animals under unstable social condition vs. control stable social conditions. Results identified 112 up-regulated and 82 down-regulated genes (listed in Table S1). Prominently represented among the down-regulated genes were transcripts involved in Type I interferon innate anti-viral responses (IFITM2, IFI6, IFI27, IFI30, IFIT1L, MX1, ISG15, ISG20). No obvious functional pattern associated with inflammation was characterized in the set of genes up-regulated by social instability.
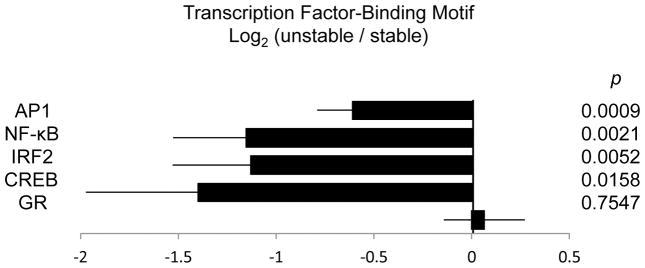
Promoter-based bioinformatics analysis of differentially expressed genes (Fig 1) indicated that down-regulated genes showed an enrichment of transcription factor-binding motifs for the pro-inflammatory factors, AP-1 (mean log2 ratio = −0.61, SE = 0.18) and NF-κB (mean log2 ratio = −1.16, SE = 0.37), the interferon responsive factor, IRF2 (mean log2 ratio = −1.13, SE = 0.40), and the neural-related factor, CREB (mean log2 ratio = −1.40, SE = 0.57). Results showed no indication of differential GR activity (mean log2 ratio = 0.06, SE = 0.21).
Figure 1.
Transcriptional activity of AP-1, NF-κB, IRF2, CREB, and GR as assessed by TELiS bioinformatics analysis of transcription factor binding motif prevalence in promoter DNA sequences of genes showing ≥ 1.25 fold differential expression in unstable vs. stable social conditions. Two-tailed p values were derived from comparing mean (log2-transformed) prevalence ratios for each transcription factor (i.e., relative frequency of promoter sites among up-regulated genes / frequency among down-regulated genes) using bootstrap-derived standard errors.
3.3 Cellular Origins of Differentially Expressed Genes
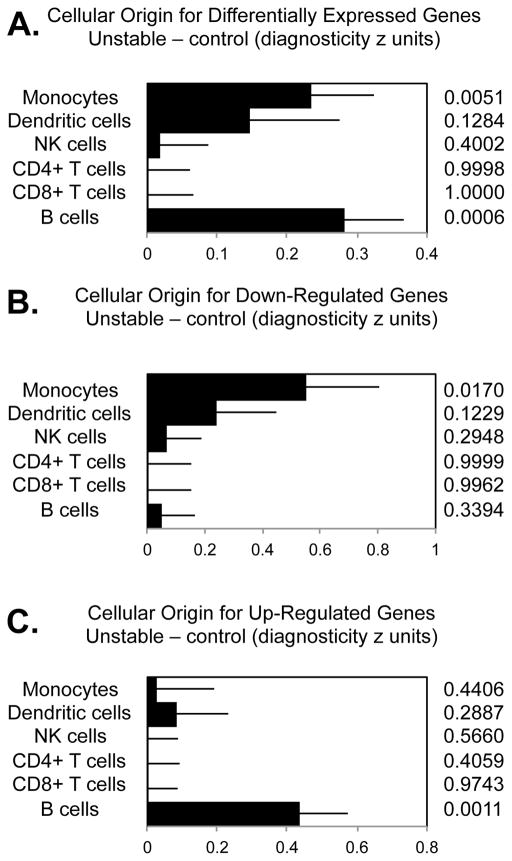
To assess whether the aggregate axillary LN transcriptome effects of unstable social conditions were mediated predominately by myeloid lineage cells, we conducted TOA on all genes showing ≥ 1.25-fold differential expression. Results identified monocytes and B cells as primary sources of differentially expressed genes (Fig 2A). Down-regulated transcripts derived predominately from monocytes (Fig 2B) whereas up-regulated transcripts were associated with B cells (Fig 2C).
Figure 2.
Transcript origin analysis identifying major leukocyte subset origins of (A) all differentially expressed 194 gene transcripts; (B) 82 down-regulated transcripts; (C) 112 up-regulated transcripts that showed ≥ 1.25 fold differential expression in unstable vs. stable social conditions. P values were derived by comparing the mean diagnosticity score for each candidate cell type of origin with the null diagnosticity value of 0, with standard errors estimated by bootstrap resampling.
4. Discussion
The present study shows that social stress can modulate genome-wide transcriptional profiles in secondary lymphoid tissue (LN) in ways that partially resemble the CTRA transcriptome shift previously observed in circulating leukocytes. Specifically, the aggregate axillary LN transcriptome in rhesus macaques exposed to 3–4 weeks of unstable social conditions resembled the peripheral blood CTRA profile in showing reduced expression of Type I interferon-related genes, whereas it diverged from the peripheral blood CTRA profile in showing no evidence of up-regulated pro-inflammatory gene expression. Consistent with these observations, promoter-based bioinformatic analysis of differentially expressed genes showed indications of reduced activity of interferon response factors (IRF), as well as reduced activity of key pro-inflammatory transcription factors, AP-1 and NF-κB. Results also suggested reduced activity of CREB, and no indication of any differential activity of the GR. However, axillary LN gene expression paralleled previous studies in circulating leukocytes in identifying monocytes as primary cellular source of transcripts down-regulated by unstable social conditions and B cells as primary source of up-regulated transcripts. These results suggest that social influences on immune cell gene regulation are sufficiently pronounced to alter transcriptome profiles in secondary lymphoid tissue, but these effects only partially recapitulate the overall CTRA transcriptome shift previously observed in circulating leukocytes. We hypothesize that the pro-inflammatory component of the CTRA may fail to emerge in axillary LNs due to the highly selective trafficking pattern of activated myeloid cells. Specifically, monocyte/macrophage and dendritic cell trafficking to the LN is generally dependent on myeloid cell activation processes (i.e., myeloid cells do not typically show constitutive patrolling through LNs under basal conditions) (Teijeira et al., 2013). As such, the LN may selectively contain only those myeloid cells that have reached some minimal degree of pro-inflammatory activation (as opposed to circulating or peripheral tissue myeloid populations that maintain a mix of quiescent and activated myeloid lineage cells). Indeed, the down-regulation observed across both pro-inflammatory and Type I interferon-related transcription factor pathways may potentially indicate a global reduction in activation and/or recruitment of myeloid lineage cells into the LN tissue environment in the context of social stress. While this hypothesis remains to be confirmed in future studies in which LN cells are available for direct enumeration (e.g., by flow cytometry), this hypothesis would provide a parsimonious explanation for a wide range of previously observed effects of social stress on primary immune responses (e.g., due to reduced antigen presentation) and impaired responses to viral infections in particular (Chen et al., 2009; O’Donovan et al., 2011; Slavich & Cole, 2013).
4.1 Effects of Social Instability on Behavior
In the current study, we found evidence of behavioral stress in response to social instability; however, we did not obtain concurrent physiological measures of stress during the axillary LN biopsies. Although we acknowledge this limitation, our previous work using this model involved, as this study did, examining physiological measures during basal conditions, at 2- to 4-week intervals (depending on the study), on days when the animals did not experience their social conditions. Our initial study (Capitanio et al., 1998) demonstrated that HPA regulation does change as a result of social instability, but as measured under basal conditions, requires several weeks to be evident; the present study was too short to see this effect. A more recent study (Capitanio & Cole, 2015), in which we obtained measures of urinary metabolites of epinephrine and norepinephrine, demonstrated that by the second week of unstable social conditions, there were significant elevations in metabolites of both epinephrine and norepinephrine. Thus, in the present study we found evidence of behavioral effects of social instability, occurring during a period of time when a previous study demonstrated physiological indicators of stress; we acknowledge that a limitation of the current study is the lack of concurrent physiological measures of stress obtained during the axillary LN biopsies.
4.2 Social Regulation of Lymph Node Transcriptome
Altered activity of inflammation- and interferon-related transcription factors in monocytes and B cells provides a potential mechanism within secondary lymphoid tissue for the association of social stress with negative health outcomes (Capitanio et al., 1998; Caspi et al., 2006; Cohen et al., 1997; Eng et al., 2002; Kroenke et al., 2006; Reynolds & Kaplan, 1990; Soler-Vila et al., 2003). Stress-induced alterations in gene expression can significantly affect the primary physiological function of the LN as a mediator of the immune response to infection. Myeloid cells (i.e. monocytes) have been associated both with inflammation (Gordon & Taylor, 2005; Robbins & Swirski, 2010) and several forms of psychosocial stress including low social economic status (Powell et al., 2013), perceived social isolation (Cole et al., 2015), and depressive symptoms in patients with coronary artery disease (Serfozo et al., 2016). In addition, secondary lymphoid tissue is an important initiator of the immune response, contains a high density of functional B cells (von Andrian & Mempel, 2003), and is an area of rapid turnover of B cells (Thomas et al., 2016). Given the functional importance of both monocytes and B cells within lymphoid tissue, the stress-induced alterations in gene regulation within these cell populations and inflammation- and interferon-related transcription factors give us an idea of the mechanism of how these cells populations are responding within the axillary LN.
Although our transcript origin analysis revealed significant findings for B cells, we did not find any specific indication of altered T cell regulation. This is unsurprising because LNs contain a heterogeneous population of T cells that are quiescent, and it may be difficult for any whole genome analysis of the aggregate tissue to detect alterations occurring within the subpopulations of T cells. To test specific hypotheses relating to T cell subpopulations, future research may need to physically isolate specific T cell subsets (e.g., using flow cytometry), to provide a more sensitive test of social influences on T lymphocyte responses to social stress. Indeed, the alterations observed here in monocyte and dendritic cell-related gene expression suggests that subpopulations of T cells may play a role in axillary LNs in response to social stress, and may simply have been missed in this study due to this study’s focus on whole tissue-level influences. Furthermore, it is possible we may see different effects in different secondary lymphoid organs. Future research will be required to clarify whether similar effects are observed in other secondary lymphoid organs (e.g., the spleen). As an example of that potential, we also note that repeated social defeat in mice has shown profound increases in the myeloid cell population in the spleen (Powell et al., 2013; Wohleb et al., 2014).
Although our findings are consistent with epidemiological data linking social stress to disease risk (Capitanio et al., 1998; Caspi et al., 2006; Cohen et al., 1997; Eng et al., 2002; Kroenke et al., 2006; Reynolds & Kaplan, 1990; Soler-Vila et al., 2003), these results should be reviewed with caution because of the small sample size. In addition, the present sample size was too small to support any exploratory/discovery analyses and thus we confined ourselves to a small number of a priori hypotheses at the level of sets of genes (i.e., pro-inflammatory gene sets, antiviral gene sets, transcription factor gene set analyses, and cell type gene set analyses). However, future studies involving much larger numbers of tissue samples, as well as pre and post measurements, may allow for discovery of many new biological effects that were overlooked in the targeted hypothesis testing approach utilized here. In addition, these results do not necessarily reflect changes in gene expression within specific subsets of cells (i.e. on a per-cell basis), and could potentially stem from changes in the relative prevalence of the (per-cell fixed) transcriptome within the overall LN. Future research will be required to confirm the specific neural-immune interactions mediating these monocyte and B cell dynamics and to verify the present indications at the level of protein and cellular function read-outs, as well as to examine their implications for health outcomes. However, the overall pattern of the present genomic results is consistent with previous findings showing that social stress can undermine host defense against viral infections (Capitanio et al., 1998; Cohen et al., 1997; Cole et al., 2015; Slavich & Cole, 2013).
Supplementary Material
Table S1. List of differential expression values for up- and down-regulated genes comparing unstable vs. stable social conditions.
Acknowledgments
The authors are grateful to Erna Tarara for project coordination and behavioral data collection and to the support of Primate Services at the California National Primate Research Center for animal handling, care, and coordination. The authors also thank the reviewers for helpful comments on an earlier version of the manuscript. This project was funded by P51 OD011107, R01 DA024441, K07CA188237-01A1, R01 CA160890, AI52737, R01 AG033590, R01 AG043404, and the Norman Cousins Center for Psychoneuroimmunology.
References
- Capitanio JP. Early experience and social processes in rhesus macaques (Macaca mulatta): II. Complex social interaction. J Comp Psychol. 1985;99(2):133–144. [PubMed] [Google Scholar]
- Capitanio JP, Abel K, Mendoza SP, Blozis SA, McChesney MB, Cole SW, et al. Personality and serotonin transporter genotype interact with social context to affect immunity and viral set-point in simian immunodeficiency virus disease. Brain Behav Immun. 2008;22(5):676–689. doi: 10.1016/j.bbi.2007.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capitanio JP, Cole SW. Social instability and immunity in rhesus monkeys: the role of the sympathetic nervous system. Philos Trans R Soc Lond B Biol Sci. 2015;370(1669) doi: 10.1098/rstb.2014.0104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capitanio JP, Mendoza SP, Baroncelli S. The relationship of personality dimensions in adult male rhesus macaques to progression of simian immunodeficiency virus disease. Brain Behav Immun. 1999;13(2):138–154. doi: 10.1006/brbi.1998.0540. [DOI] [PubMed] [Google Scholar]
- Capitanio JP, Mendoza SP, Lerche NW, Mason WA. Social stress results in altered glucocorticoid regulation and shorter survival in simian acquired immune deficiency syndrome. Proc Natl Acad Sci U S A. 1998;95(8):4714–4719. doi: 10.1073/pnas.95.8.4714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspi A, Harrington H, Moffitt TE, Milne BJ, Poulton R. Socially isolated children 20 years later: risk of cardiovascular disease. Arch Pediatr Adolesc Med. 2006;160(8):805–811. doi: 10.1001/archpedi.160.8.805. [DOI] [PubMed] [Google Scholar]
- Chen E, Miller GE, Walker HA, Arevalo JM, Sung CY, Cole SW. Genome-wide transcriptional profiling linked to social class in asthma. Thorax. 2009;64(1):38–43. doi: 10.1136/thx.2007.095091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen S, Doyle WJ, Skoner DP, Rabin BS, Gwaltney JM., Jr Social ties and susceptibility to the common cold. JAMA. 1997;277(24):1940–1944. [PubMed] [Google Scholar]
- Cole SW. Elevating the perspective on human stress genomics. Psychoneuroendocrinology. 2010;35(7):955–962. doi: 10.1016/j.psyneuen.2010.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole SW. Social regulation of human gene expression: mechanisms and implications for public health. Am J Public Health. 2013;103(Suppl 1):S84–92. doi: 10.2105/AJPH.2012.301183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole SW. Human social genomics. PLoS Genet. 2014;10(8):e1004601. doi: 10.1371/journal.pgen.1004601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole SW, Capitanio JP, Chun K, Arevalo JM, Ma J, Cacioppo JT. Myeloid differentiation architecture of leukocyte transcriptome dynamics in perceived social isolation. Proc Natl Acad Sci U S A. 2015;112(49):15142–15147. doi: 10.1073/pnas.1514249112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole SW, Conti G, Arevalo JM, Ruggiero AM, Heckman JJ, Suomi SJ. Transcriptional modulation of the developing immune system by early life social adversity. Proc Natl Acad Sci U S A. 2012;109(50):20578–20583. doi: 10.1073/pnas.1218253109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole SW, Hawkley LC, Arevalo JM, Cacioppo JT. Transcript origin analysis identifies antigen-presenting cells as primary targets of socially regulated gene expression in leukocytes. Proc Natl Acad Sci U S A. 2011;108(7):3080–3085. doi: 10.1073/pnas.1014218108. 1014218108 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole SW, Hawkley LC, Arevalo JM, Sung CY, Rose RM, Cacioppo JT. Social regulation of gene expression in human leukocytes. Genome Biol. 2007;8(9):R189. doi: 10.1186/gb-2007-8-9-r189. gb-2007-8-9-r189 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole SW, Mendoza SP, Capitanio JP. Social stress desensitizes lymphocytes to regulation by endogenous glucocorticoids: insights from in vivo cell trafficking dynamics in rhesus macaques. Psychosom Med. 2009;71(6):591–597. doi: 10.1097/PSY.0b013e3181aa95a9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole SW, Yan W, Galic Z, Arevalo J, Zack JA. Expression-based monitoring of transcription factor activity: the TELiS database. Bioinformatics. 2005;21(6):803–810. doi: 10.1093/bioinformatics/bti038. [DOI] [PubMed] [Google Scholar]
- Efron B, Tibshirani RJ. An introduction to the bootstrap. New York: Chapman & Hall; 1993. [Google Scholar]
- Eng PM, Rimm EB, Fitzmaurice G, Kawachi I. Social ties and change in social ties in relation to subsequent total and cause-specific mortality and coronary heart disease incidence in men. Am J Epidemiol. 2002;155(8):700–709. doi: 10.1093/aje/155.8.700. [DOI] [PubMed] [Google Scholar]
- Fredrickson BL, Grewen KM, Coffey KA, Algoe SB, Firestine AM, Arevalo JM, et al. A functional genomic perspective on human well-being. Proc Natl Acad Sci U S A. 2013;110(33):13684–13689. doi: 10.1073/pnas.1305419110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilad Y, Rifkin SA, Bertone P, Gerstein M, White KP. Multi-species microarrays reveal the effect of sequence divergence on gene expression profiles. Genome Res. 2005;15(5):674–680. doi: 10.1101/gr.3335705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon S, Taylor PR. Monocyte and macrophage heterogeneity. Nat Rev Immunol. 2005;5(12):953–964. doi: 10.1038/nri1733. [DOI] [PubMed] [Google Scholar]
- Irwin MR, Cole SW. Reciprocal regulation of the neural and innate immune systems. Nat Rev Immunol. 2011;11(9):625–632. doi: 10.1038/nri3042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroenke CH, Kubzansky LD, Schernhammer ES, Holmes MD, Kawachi I. Social networks, social support, and survival after breast cancer diagnosis. J Clin Oncol. 2006;24(7):1105–1111. doi: 10.1200/JCO.2005.04.2846. [DOI] [PubMed] [Google Scholar]
- Murphy Kenneth, Travers Paul, Walport Mark, Janeway Charles. Janeway’s immunobiology. New York: Garland Science; 2012. [Google Scholar]
- O’Donovan A, Sun B, Cole S, Rempel H, Lenoci M, Pulliam L, et al. Transcriptional control of monocyte gene expression in post-traumatic stress disorder. Dis Markers. 2011;30(2–3):123–132. doi: 10.3233/DMA-2011-0768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshlack Alicia, Chabot Adrien E, Smyth Gordon K, Gilad Yoav. Using DNA microarrays to study gene expression in closely related species. Bioinformatics. 2007;23(10):1235–1242. doi: 10.1093/bioinformatics/btm111. [DOI] [PubMed] [Google Scholar]
- Powell ND, Sloan EK, Bailey MT, Arevalo JM, Miller GE, Chen E, et al. Social stress up-regulates inflammatory gene expression in the leukocyte transcriptome via beta-adrenergic induction of myelopoiesis. Proc Natl Acad Sci U S A. 2013;110(41):16574–16579. doi: 10.1073/pnas.1310655110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds P, Kaplan GA. Social connections and risk for cancer: prospective evidence from the Alameda County Study. Behav Med. 1990;16(3):101–110. doi: 10.1080/08964289.1990.9934597. [DOI] [PubMed] [Google Scholar]
- Robbins CS, Swirski FK. The multiple roles of monocyte subsets in steady state and inflammation. Cell Mol Life Sci. 2010;67(16):2685–2693. doi: 10.1007/s00018-010-0375-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serfozo G, Horvath T, Foldesi I, Rafael B, von Kanel R, Keresztes M. The Monocyte-to-Lymphocyte Ratio Correlates with Psycho-Neuro-Inflammatory Factors in Patients with Stable Coronary Artery Disease. Neuroimmunomodulation. 2016 doi: 10.1159/000443835. [DOI] [PubMed] [Google Scholar]
- Shi L, Campbell G, Jones WD, Campagne F, Wen Z, Walker SJ, et al. The MicroArray Quality Control (MAQC)-II study of common practices for the development and validation of microarray-based predictive models. Nat Biotechnol. 2010;28(8):827–838. doi: 10.1038/nbt.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi L, Jones WD, Jensen RV, Harris SC, Perkins RG, Goodsaid FM, et al. The balance of reproducibility, sensitivity, and specificity of lists of differentially expressed genes in microarray studies. BMC Bioinformatics. 2008;9(Suppl 9):S10. doi: 10.1186/1471-2105-9-S9-S10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slavich GM, Cole SW. The Emerging Field of Human Social Genomics. Clin Psychol Sci. 2013;1(3):331–348. doi: 10.1177/2167702613478594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloan EK, Capitanio JP, Tarara RP, Cole SW. Social temperament and lymph node innervation. Brain Behav Immun. 2008;22(5):717–726. doi: 10.1016/j.bbi.2007.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloan EK, Capitanio JP, Tarara RP, Mendoza SP, Mason WA, Cole SW. Social stress enhances sympathetic innervation of primate lymph nodes: mechanisms and implications for viral pathogenesis. J Neurosci. 2007;27(33):8857–8865. doi: 10.1523/JNEUROSCI.1247-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloan EK, Tarara RP, Capitanio JP, Cole SW. Enhanced replication of simian immunodeficiency virus adjacent to catecholaminergic varicosities in primate lymph nodes. J Virol. 2006;80(9):4326–4335. doi: 10.1128/JVI.80.9.4326-4335.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soler-Vila H, Kasl SV, Jones BA. Prognostic significance of psychosocial factors in African-American and white breast cancer patients: a population-based study. Cancer. 2003;98(6):1299–1308. doi: 10.1002/cncr.11670. [DOI] [PubMed] [Google Scholar]
- Teijeira A, Rouzaut A, Melero I. Initial afferent lymphatic vessels controlling outbound leukocyte traffic from skin to lymph nodes. Front Immunol. 2013;4:433. doi: 10.3389/fimmu.2013.00433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas SN, Rohner NA, Edwards EE. Implications of Lymphatic Transport to Lymph Nodes in Immunity and Immunotherapy. Annu Rev Biomed Eng. 2016 doi: 10.1146/annurev-bioeng-101515-014413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tung Jenny, Barreiro Luis B, Johnson Zachary P, Hansen Kasper D, Michopoulos Vasiliki, Toufexis Donna, et al. Social environment is associated with gene regulatory variation in the rhesus macaque immune system. Proceedings of the National Academy of Sciences. 2012;109(17):6490–6495. doi: 10.1073/pnas.1202734109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Andrian UH, Mempel TR. Homing and cellular traffic in lymph nodes. Nat Rev Immunol. 2003;3(11):867–878. doi: 10.1038/nri1222. [DOI] [PubMed] [Google Scholar]
- Westermann Jürgen, Pabst Reinhard. Lymphocyte subsets in the blood: a diagnostic window on the lymphoid system? Immunology Today. 1990;11:406–410. doi: 10.1016/0167-5699(90)90160-b. http://dx.doi.org/10.1016/0167-5699(90)90160-B. [DOI] [PubMed] [Google Scholar]
- Witten Daniela M, Tibshirani Robert. A comparison of fold-change and the t-statistic for microarray data analysis. Stanford, CA: 2007. [Google Scholar]
- Wohleb Eric S, McKim Daniel B, Shea Daniel T, Powell Nicole D, Tarr Andrew J, Sheridan John F, et al. Re-establishment of Anxiety in Stress-Sensitized Mice Is Caused by Monocyte Trafficking from the Spleen to the Brain. Biological psychiatry. 2014;75(12):970–981. doi: 10.1016/j.biopsych.2013.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. List of differential expression values for up- and down-regulated genes comparing unstable vs. stable social conditions.