Summary
The second most commonly mutated gene in primary microcephaly (MCPH) patients is wd40-repeat protein 62 (wdr62), but the relative contribution of WDR62 function to the growth of major brain lineages is unknown. Here, we use Drosophila models to dissect lineage-specific WDR62 function(s). Interestingly, although neural stem cell (neuroblast)-specific depletion of WDR62 significantly decreased neuroblast number, brain size was unchanged. In contrast, glial lineage-specific WDR62 depletion significantly decreased brain volume. Moreover, loss of function in glia not only decreased the glial population but also non-autonomously caused neuroblast loss. We further demonstrated that WDR62 controls brain growth through lineage-specific interactions with master mitotic signaling kinase, AURKA. Depletion of AURKA in neuroblasts drives brain overgrowth, which was suppressed by WDR62 co-depletion. In contrast, glial-specific depletion of AURKA significantly decreased brain volume, which was further decreased by WDR62 co-depletion. Thus, dissecting relative contributions of MCPH factors to individual neural lineages will be critical for understanding complex diseases such as microcephaly.
Keywords: neuroblast, glia, WD-repeat protein, WDR62, AURKA, mitosis, Drosophila, brain, microcephaly
Graphical Abstract
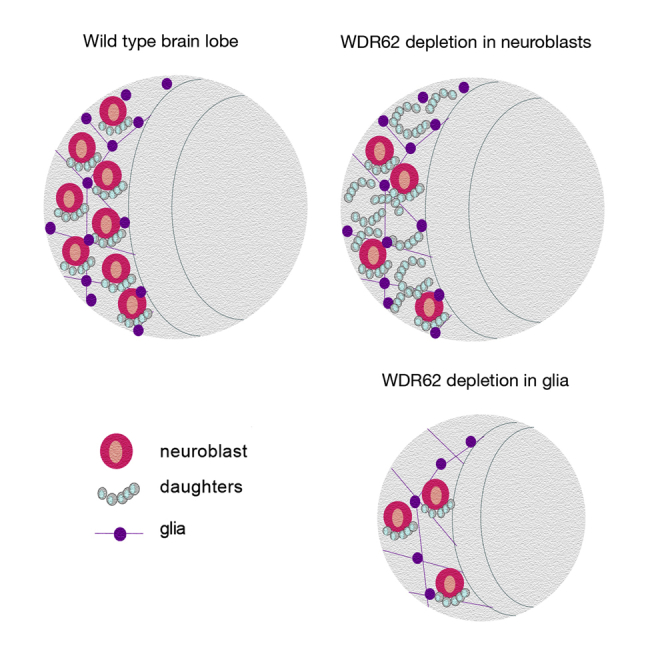
Highlights
-
•
Glial-specific depletion of microcephaly protein WDR62 impairs brain growth
-
•
wdr62 depletion in glia causes neural stem cell (NSC) loss
-
•
WDR62 is required cell extrinsically in the NSC niche
-
•
Lineage-specific interactions between WDR62 and AURKA control brain growth
Quinn and colleagues use Drosophila models to demonstrate that depletion of microcephaly protein WDR62 specifically in the glial lineage impairs brain growth. Moreover, wdr62 depletion in glia not only impairs brain growth by decreasing the glial population but also leads to neural stem cell (NSC) loss. Thus, MCPH proteins are essential not only intrinsically for glial cell proliferation but are also required cell extrinsically for establishing the NSC niche.
Introduction
Genome-wide exome sequencing of microcephaly (MCPH) patients identified wd40-repeat protein 62 (wdr62) as the second most commonly mutated gene (Bilguvar et al., 2010, Nicholas et al., 2010, Yu et al., 2010). WDR62 is a ubiquitously expressed cytoplasmic protein in interphase and localizes to the spindle pole in mitosis (Nicholas et al., 2010, Yu et al., 2010). A feature of many WDR62 MCPH-associated alleles is an inability to localize to the mitotic spindle pole (Lim et al., 2015, Nicholas et al., 2010, Yu et al., 2010), and wdr62 depletion is also associated with defects in spindle and centrosomal integrity, mitotic delay, and reduced brain growth in rodents (Chen et al., 2014, Xu et al., 2014). The neural stem cell (NSC) population gives rise to all neuronal cells in the adult brain. NSC behavior is governed by both cell intrinsic factors and extrinsic factors from the supporting stem cell niche, including the glial lineage, which acts non-autonomously to control stem cell renewal and differentiation of daughters (Cunningham et al., 2013, Florio and Huttner, 2014, Pollen et al., 2015).
The connection between NSCs and their niche, and the importance of spindle integrity to asymmetric division, has been best defined for Drosophila NSCs/neuroblasts (NB) (Homem and Knoblich, 2012, Smith et al., 2007, Wirtz-Peitz et al., 2008). WDR62 scaffolds kinases that are important mitotic regulators including c-Jun N-terminal kinase (JNK) members of the mitogen-activated protein kinase superfamily and Aurora kinase A (AURKA) (Chen et al., 2014, Lim et al., 2015, Xu et al., 2014). In flies, AURKA regulates NB proliferation and is required for the localization of mitotic NB polarity complex protein Bazooka (mammalian Par3) to the apical Par complex (comprising the Par anchor, Inscuteable [Insc] adaptor protein, and Gαi/Pins/Mud complex). This establishes the apical-basal NB axis essential for self-renewal and differentiation. As a consequence, mutant aurka causes NB overproliferation and tissue overgrowth (Atwood and Prehoda, 2009, Smith et al., 2007, Wirtz-Peitz et al., 2008). The WDR62 ortholog in Drosophila (dWDR62) is required for brain growth (Nair et al., 2016), but whether signaling between WDR62 and AURKA modulates brain development has not been reported.
In addition to the NB lineage, Drosophila studies suggest that the glial lineage governs overall brain volume (Pereanu et al., 2005) through regulation of cell-cycle re-entry and neuroepithelial expansion of NBs (Chell and Brand, 2010, Morante et al., 2013). However, potential contribution(s) of individual brain lineage(s) (NB or glia) to the defective brain growth associated with global depletion of wdr62 or aurka is currently unclear. Here, we confirm that WDR62 is required for spindle orientation in NBs (Nair et al., 2016), however, wdr62 depletion specifically in NBs does not significantly retard brain growth. Rather, control of brain growth predominantly depends upon glial lineage function, as depletion of either aurka or wdr62 specifically in the glial lineage significantly reduces brain volume. Moreover, although wdr62 depletion suppressed brain overgrowth associated with aurka depletion in NBs, wdr62 knockdown specifically in the glial lineage enhanced the small brain phenotype associated with aurka depletion. Collectively, our data suggest that WDR62 function is negatively regulated by AURKA in NBs but positively regulated by AURKA in glia, and thus demonstrates that lineage-specific signaling functions of AURKA-WDR62 in Drosophila orchestrate larval brain growth and development.
Results
WDR62 Is Required for Spindle Orientation and Mitotic Progression in NBs
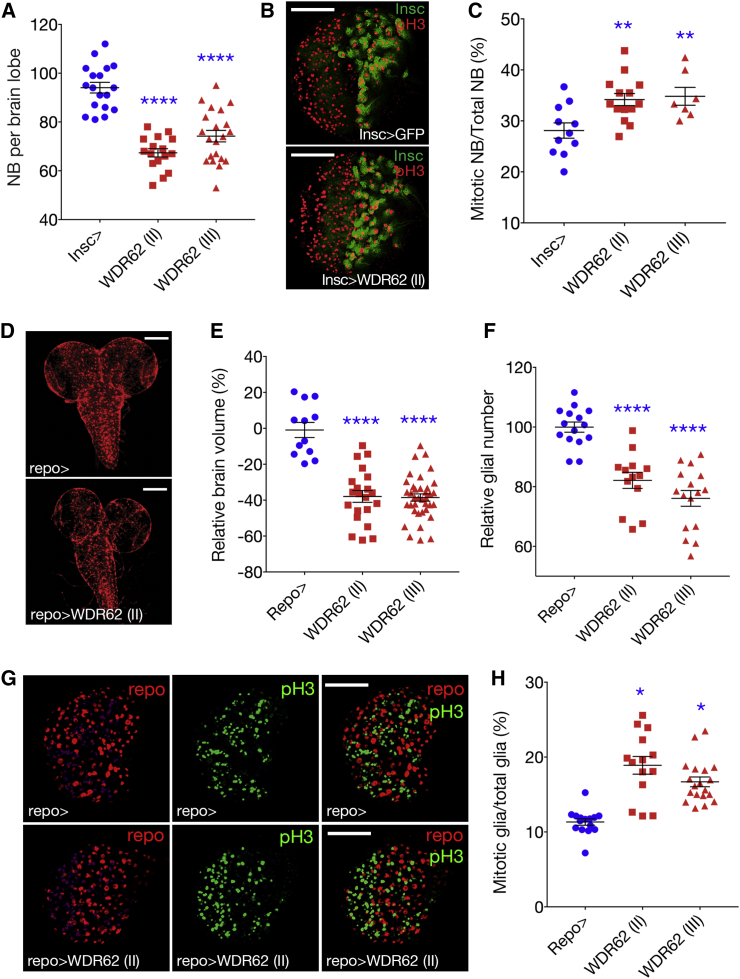
To dissect the contribution of WDR62 to the NB and glial lineage during brain development, we used two alternate UAS-wdr62 RNAi lines (to non-overlapping regions of cg7337, VDRC #110764 [on chromosome 2], NIG 7337-R1 [on chromosome 3] referred to here as WDR62 [II] and [III]). qPCR revealed a significant decrease in mRNA abundance following ubiquitous wdr62 knockdown (Figure S1A). In the developing Drosophila brain, NBs give rise to intermediate progenitors and ganglion mother cells that differentiate into neurons (Betschinger et al., 2006, Homem and Knoblich, 2012, Knoblich, 2008, Lancaster and Knoblich, 2012). Depletion of wdr62 in type I and II NB and intermediate progenitors (INP), using Insc-GAL4 (Betschinger et al., 2006), decreased NB number (Figure 1A) but did not significantly alter brain volume (Figure S1B). wdr62 knockdown was not associated with altered timing of major developmental stages (Figure S1C). Thus, analyses conducted on brains of equivalent developmental stages revealed that a significant decrease in NBs does not necessarily manifest in a global reduction in brain growth.
Figure 1.
Glial-Driven Knockdown of WDR62 Decreases Brain Volume
(A) NBs per third instar larval brain lobe (96 hr after larval hatching [ALH]) for control and WDR62 RNAi (II) or (III) knockdown (ANOVA, p < 0.0001).
(B) Mitotic cells in brain lobes marked with anti-phosphohistone H3 (pH3). Insc-GAL4 in NBs and INPs marked with GFP.
(C) Quantification of mitotic NBs as a percentage of total NB (ANOVA, p = 0.0043) (no. of brains: Insc-GAL4 = 11, siWDR62 (II) = 14, siWDR62(III) = 7).
(D) Control and WDR62 knockdown Repo-GAL4 in glia marked with GFP.
(E) Brain lobe volume (ANOVA, p = 0.0022).
(F) Total glial volume (ANOVA, p < 0.0001).
(G) anti-pH3 and Repo.
(H) Mitotic glia as a percentage of total glia (ANOVA, p < 0.0001). At least 12 brains were quantified across three independent experiments.
In control third instar larval brains, 28.1% of total NBs are mitotic and wdr62 depletion significantly increased the mitotic index (Figures 1B and 1C), which suggests a mitotic delay. Decreased NBs are unlikely a consequence of increased cell death, as cleaved caspase-3-positive NBs were unchanged (Figure S1D). The bipolar mitotic spindle must be properly orientated, bisecting the apical and basal crescents of the polarity factors inherited by daughter cells, to ensure timely mitotic progression and asymmetric division of NBs (Knoblich, 2008). Worniu-GAL4 driven depletion of wdr62 caused spindle misorientation (Figure S1E), consistent with previous reports of spindle alignment defects in wdr62 loss-of-function mutants (Nair et al., 2016). However, further studies are required to determine if this contributes to the increased mitotic index in wdr62 loss-of-function NBs as spindle misorientation alone is not sufficient to cause cell-cycle delay (Homem and Knoblich, 2012).
How can brain size remain unchanged despite the number of NBs decreasing? One possible explanation could be compensatory proliferation of progeny cells. To determine progenitors in G1, S, or G2M, fluorescence ubiquitination cell-cycle indicator (FUCCI) cell-cycle profiles for deadpan positive G1, S, or G2M cells were subtracted from the total Insc-G4 population, which includes NBs and progenitors (Figure S1F). Interestingly, increased S-phase cells were observed in the Wdr62 knockdown, which suggests that compensatory proliferation of NB daughters might maintain brain size.
Knockdown of WDR62 in the Glial Lineage Decreases Brain Volume
Depletion of wdr62 decreased NB number but did not change brain size (Figures 1 and S1F). Pan-glial depletion of wdr62, using Repo-GAL4 (Sepp et al., 2001), resulted in pupal lethality but did not disrupt larval developmental timing (Figure S1C). Wdr62 depletion significantly decreased third instar larval brain lobe volume (Figures 1D and 1E), decreased glia (Figure 1F), and increased the mitotic index for remaining glia (Figures 1G and 1H). Thus, wdr62 loss of function likely results in a mitotic delay in both NBs and glia but is only associated with a significant reduction in brain volume when depleted from the glial lineage.
WDR62 Is Essential for Brain Overgrowth Driven by the Mitotic Kinase AURKA
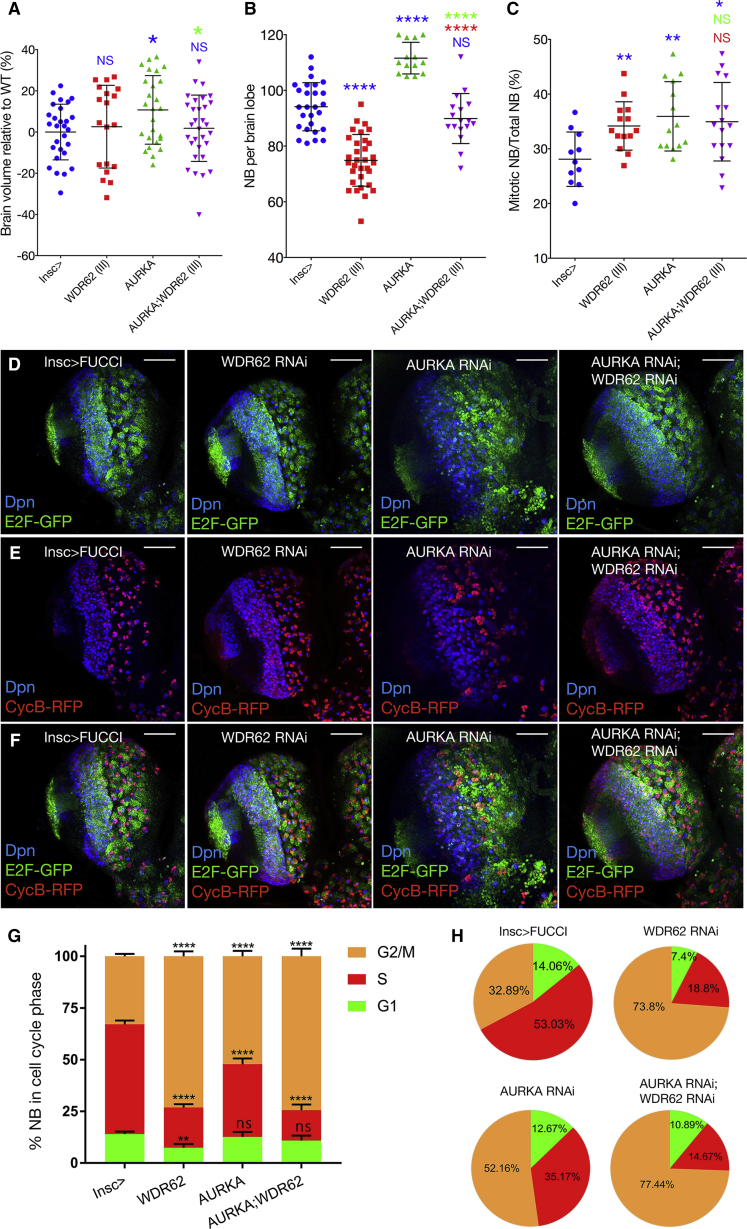
Previous ex vivo studies suggest AURKA binds WDR62 and regulates its trafficking to the mitotic spindle pole and, thus, spindle integrity (Lim et al., 2015, Lim et al., 2016). To explore potential connections between the AURKA signaling nexus and WDR62 in vivo, we took advantage of the WDR62 brain phenotypes. As qPCR following global aurka depletion revealed a relative decrease of 65% for RNAi II and 26% for III, respectively (Figure S1A), RNAi II was used for all subsequent studies. Insc-driven depletion of aurka significantly increased brain volume (Figure 2A), consistent with brain overgrowth reported for aurka mutants (Lee et al., 2006, Wang et al., 2006). Interestingly, brain overgrowth associated with aurka depletion in NBs depends on endogenous levels of wdr62 as co-knockdown brought brain volume back to within the control range (Figure 2A). In line with the increased brain volume, aurka knockdown significantly increased NB, and co-depletion of wdr62 restored NB numbers (Figure 2B).
Figure 2.
WDR62 Is Required for NB Expansion and Brain Overgrowth Associated with Depletion of AURKA
(A–C) Brain lobe volume (A) (ANOVA, p = 0.0883), neuroblast number per brain lobe (B) (ANOVA, p < 0.0001), mitotic neuroblast as a percentage of total NB (C) (ANOVA, p < 0.01) for genotypes marked.
(D–F) Insc-GAL4-driven FUCCI for control or WDR62 (III) and/or AURKA (II) RNAi. (D) E2F-GFP marks G1 cells counterstained with deadpan for NBs, (E) CycB-RFP marks S phase, (F) overlap of CycB and E2F (yellow) marks G2/M.
(G) Quantification of FUCCI in neuroblasts (ANOVA, p < 0.0059).
(H) Cell-cycle distribution of NBs. At least 11 brains were quantified across three independent experiments.
As observed for wdr62, aurka knockdown also significantly increased mitotic NBs, however, co-depletion of wdr62 did not further significantly alter mitotic NBs compared with depletion of aurka alone (Figure 2C). To further dissect cell-cycle defects and monitor the timing of gap phases, we used FUCCI to identify G1, G2/M, and S-phase NBs and INP daughters (Zielke et al., 2014). Insc-driven FUCCI for individual depletion of wdr62 or aurka revealed increased NBs in G2/M (Figures 2D–2H, control 33%, wdr62 74% or aurka 52%). Co-depletion resulted in a further significant increase in G2/M NBs compared with individual depletion of aurka, but not wdr62. These data together with observations that co-depletion did not significantly increase mitotic NBs, compared with individual depletion of wdr62 or aurka (Figure 2C), suggests that wdr62 depletion extends G2, whether alone or in the aurka background.
S-phase NBs (normally 53%) were significantly reduced for both wdr62 (19%) or aurka knockdown alone (35%), and the effect was much more pronounced for wdr62 RNAi (i.e., compared with aurka alone), which brought S phases in the aurka RNAi background to within the range of wdr62 knockdown alone (15%; Figures 2G and 2H). The reduced proportion of S-phase cells further suggested that wdr62 depletion reduces NB expansion and prevents brain overgrowth associated with aurka knockdown by increasing the G2 delay in NBs. Conversely, the observation that NB loss associated with wdr62 depletion is reversed by aurka co-knockdown is consistent with AURKA behaving as a negative regulator of cell-cycle progression in NBs and WDR62 being required for maintenance of NB proliferation.
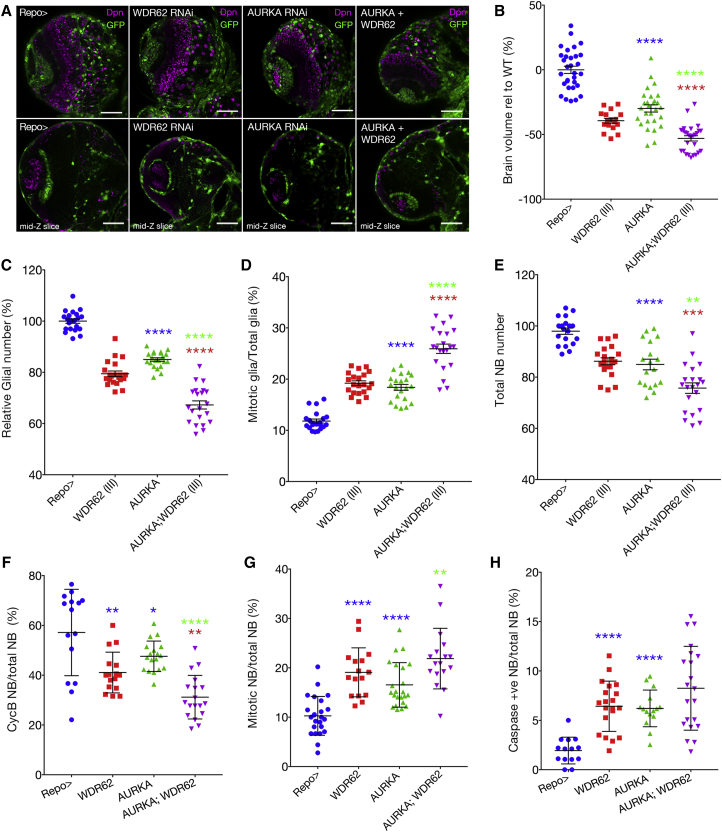
Glial Depletion of AURKA Reduces Brain Size and Enhances the WDR62 Phenotype
As the decreased NB number associated with WDR62 loss of function can be restored by co-depletion of aurka, we sought to test if AURKA might modify the glial-dependent decreases in brain size following wdr62 depletion. In contrast to the increased brain size associated with aurka depletion in NBs, aurka knockdown in glia significantly decreased larval brain volume (Figures 3A and 3B), as observed for WDR62 (also Figure 2E). Moreover, co-depletion of wdr62 and aurka further decreased brain lobe volume and glial number compared with depletion of either wdr62 or aurka alone (Figures 3A–3C). Glial cell apoptosis was significantly increased following individual knockdown and further increased following co-depletion compared with aurka (p = 0.003) but not wdr62 knockdown in isolation (Figure S2).
Figure 3.
Glial-Specific Depletion of AURKA Decreases NBs, Glia, and Brain Volume Alone and Enhances the WDR62 Phenotype
(A) Repo-driven GFP control, WDR62 (III) and/or AURKA (II) RNAi with Dpn (mid-section of the brain lobe in the lower panel).
(B) Brain lobe volume.
(C–H) Relative glial number (C), neuroblast number (D), mitotic glia (E), CycB-positive neuroblast (F), mitotic neuroblast (G), caspase-positive neuroblast (H). ANOVA, p < 0.0001 for all datasets in this figure. At least 15 brains were quantified across three independent experiments unless stated otherwise.
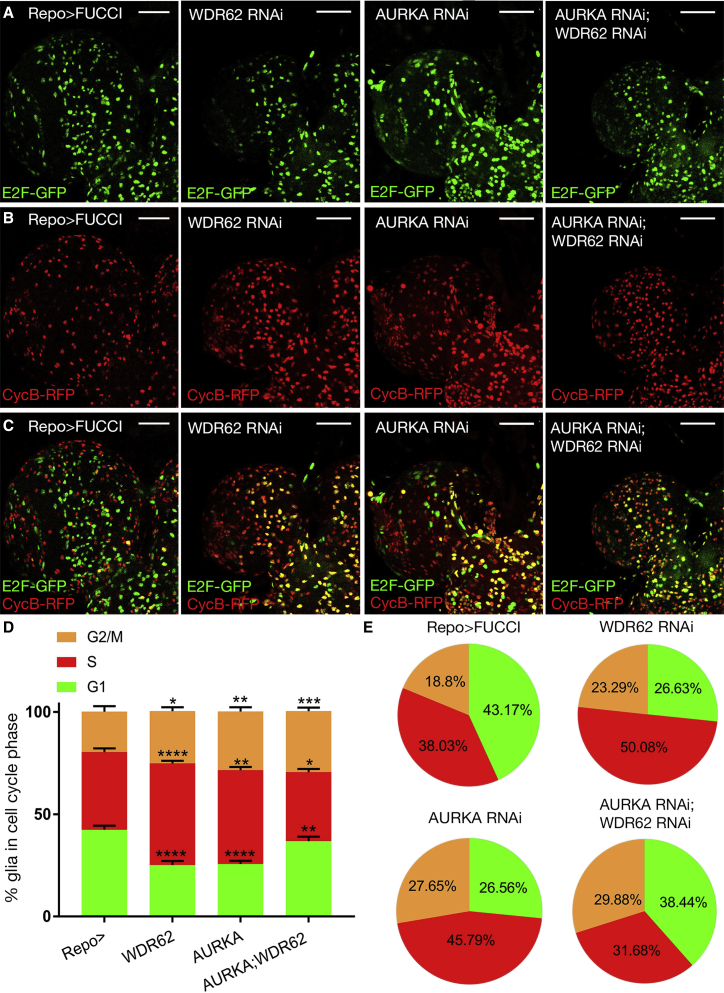
Glial depletion of aurka increased mitosis, and co-depletion with wdr62 further increased the mitotic index (Figure 3D) and the G2/M delay (Figures 4A–4E). Although G1 was truncated following wdr62 or aurka depletion alone, the proportion of S-phase glia increased. Interestingly, co-depletion increased the proportion of G1 cells and reduced S phase compared with individual knockdown, suggesting potential roles for the WDR62 and AURKA nexus in interphase (Figures 4D and 4E). In contrast to NBs, where AURKA antagonizes WDR62 function in NB proliferation, our data suggest that AURKA likely cooperates with WDR62 to regulate glial cell number.
Figure 4.
WDR62 and AURKA Kinase Modulate Cell-Cycle Progression in Glia Cells. Repo-driven FUCCI control, WDR62 (III), and/or AURKA (II) RNAi
(A–C) E2F-GFP (A), CycB-RFP (B), and overlap (C).
(D) FUCCI as a percentage of glial cells. G1 ANOVA, p < 0.0001; G2-M ANOVA, p < 0.0001; S phase ANOVA, p < 0.0091.
(E) Cell-cycle distribution of NBs. At least 20 brains were quantified across three independent experiments.
Quantified data represented in all graphs are shown with means and SEM. Statistical tests between two specific datasets are performed with non-paired Student’s t test with 95% confidence level. Multiple comparison across a given dataset was performed with one-way ANOVA, with Tukey's post-test. Scale bars in all figures represent 100 μM (ns, p > 0.05, ∗p ≤ 0.05, ∗∗p ≤ 0.01, ∗∗∗p ≤ 0.001, ∗∗∗∗p ≤ 0.0001).
In support of the observation that the glial lineage provides a supportive environment for NB renewal and differentiation, wdr62 depletion in glia significantly decreased NBs (Figures 3A and 3E). Similarly, aurka depletion in glia decreased NB number, and co-depletion further significantly reduced NBs compared with individual depletion of wdr62 or aurka. Thus, the associated decrease in brain size may also occur (at least in part) through indirect effects on the NB lineage. To better understand the changes to brain size, we conducted cell-cycle and apoptotic analysis of NBs following manipulation of wdr62 and/or aurka in glia (Figures 3F–3H, Figure S3). Interestingly, depletion of either wdr62 or aurka in glia significantly increased the NB mitotic index compared with control (p > 0.0001). Although the mitotic index was further significantly decreased following wdr62 and aurka co-depletion in glia compared with aurka knockdown alone (p = 0.003), it was not significantly different to wdr62 knockdown in isolation (Figures 3G and S3). In the same context, a reduction in CycB-positive NBs was observed after knocking down wdr62 (p = 0.0023) or aurka (p = 0.0363), which was further significantly decreased in the double knockdown compared with wdr62 (p = 0.0019) or aurka alone (p > 0.0001, Figures 3F and S3).
With regard to apoptosis, following individual knockdown of wdr62 or aurka in glia, we observed a significant increase in apoptotic cell death (marked by cleaved caspase) in NBs compared with control (p > 0.0001), although co-knockdown did not further significantly increase NB apoptosis (Figures 3H and S3). Together these data suggest that the overall decrease in NBs and reduced brain growth, following either wdr62 or aurka knockdown in glia, is due to shortened/accelerated progression through G2 phase and a delay in mitosis, which is accompanied by increased apoptosis that prevents NB expansion.
Using Wrapper-Gal4 for manipulation of wdr62 and/or aurka in the cortical glial subpopulation, we demonstrated that although individual depletion of wdr62 or aurka did not alter cell death, co-depletion significantly increased apoptosis compared with wdr62 (p = 0.004) or aurka alone (p = 0.05) (Figure S4). Moreover, individual wdr62 or aurka knockdown in the cortical glial subtype did not significantly change proliferation within these glia, NB count, or brain volume, but co-knockdown of wdr62 and aurka significantly decreased both cortical glia (p = 0.0177) and NB number (p = 0.003) compared with control. This was associated with increased caspase-positive cortical glia (p = 0.0074) without a change in brain volume or mitotic cortical glia (Figure S4). Although the phenotypes are less severe, these data demonstrate that interaction between WDR62 and AURKA is required autonomously for maintaining the cortical glial subpopulation, and within these cells to support neighboring NB stem cell populations.
Discussion
In the mammalian brain, radial glia behave as NSCs (Florio and Huttner, 2014) that are supported by outer radial glia through cell-cell contact and secretion of growth factors required for maintenance of a stem cell niche (Pollen et al., 2015). Another class of glial cells, the microglia population, regulates neural precursor cell numbers to govern final neuronal numbers residing in the cortex (Cunningham et al., 2013). However, whether MCPH genes such as wdr62 are important for glial cell fate is unclear. Here, we dissected the lineage-specific contribution of WDR62 to brain development and revealed that loss of WDR62 function specifically in the glial, but not the NB lineage, profoundly altered brain growth. Moreover, wdr62 depletion in glia likely impairs brain growth autonomously (i.e., through depletion of glia), and also results in NSC loss, suggesting that a focus on WDR62 function in glia will be integral to elucidating how wdr62 loss of function contributes to MCPH. That depletion of wdr62 in the NB lineage was not associated with reduced brain volume, despite reducing NB number, also provides a likely explanation for the recent observation that NB defects associated with global wdr62 depletion fail to account for reduced brain size (Nair et al., 2016).
Hypomorphic wdr62 mutant mice have reduced brain size, with associated mitotic defects and an overall decrease in neural progenitor cells (Chen et al., 2014). In Drosophila, spindle orientation defects following wdr62 loss of function likely underlie the G2 delay and increased mitotic figures in NBs (this work and Nair et al., 2016). This phenotype is also reminiscent of the cleavage plane misorientation observed in NSCs in wdr62-depleted rat brains (Xu et al., 2014). In Drosophila NBs, WDR62 regulates the interphase localization of Centrosomin (CNN, mammalian CDK5RAP2) to the apical centrosome, and thus centrosomal maturation and positioning (Nair et al., 2016). Interestingly, CNN is also an AURKA target that governs spindle orientation independently from cortical polarity establishment during mitosis (Bowman et al., 2006, Lee et al., 2006). Similar to the phenotype observed for wdr62 depletion in NBs, cnn loss of function is associated with spindle orientation defects and reduced NB number (Bowman et al., 2006). Thus, it is tempting to speculate that WDR62 and CNN function in the same AURKA-dependent signaling complex during mitosis.
Ex vivo studies have demonstrated that AURKA phosphorylation of WDR62 promotes spindle pole localization during mitosis (Lim et al., 2015, Lim et al., 2016). Mouse models suggest AURKA and WDR62 interact in vivo to control brain growth (Chen et al., 2014). Compound heterozygous wdr62+/−;aurka+/− mice have a much smaller body size than single heterozygotes but, although the mitotic index of the cerebral cortex was significantly increased and NSCs were reduced, consistent with a mitotic delay radial glia, potential changes to brain volume were not measured (Chen et al., 2014). Here, we demonstrate that the brain overgrowth associated with aurka depletion specifically in NBs was suppressed by co-depletion of wdr62, bringing brain volume to within the control range. In contrast, the small brain phenotypes, due to glial-specific depletion of either aurka or wdr62, were further reduced by co-knockdown. Thus, in the context of normal brain development, AURKA likely acts to promote WDR62-dependent glial proliferation, but antagonizes WDR62 function in the NB lineage.
Our findings indicate that WDR62 likely functions in AURKA-mediated regulation of spindle orientation but not in the establishment of cortical polarity. One reason for the differential output of AURKA regulation of WDR62 (between NB and glia) could stem from the symmetrical nature of glial division, where there is no evidence for cortical polarization. In contrast to the in vivo mammalian studies and previous Drosophila studies, which employed global depletion strategies for wdr62, our studies have enabled dissection of the relative contribution of wdr62 loss of- function from each of the major brain lineages. In particular, the observation that depletion in either the NB or glial lineage is associated with reduced cell number, but an overall reduction in brain size was only observed when wdr62 was reduced in glia, places great interest in examining the relative contribution of glial-specific depletion of wdr62 in mice to brain size. Moreover, future studies of the pathogenic wdr62 mutations, and identified AURKA phosphorylation sites on WDR62, in the glial lineage are likely to inform on the contribution of this lineage to impaired brain growth in microcephaly.
Experimental Procedures
Fly Strains and Crosses
Fly strains were obtained from the following stock collections: Bloomington: Tub-GAL80ts, Tub-GAL4, Insc-GAL4, Wor-GAL4-UAS-Mira-GFP, Repo-GAL4, UAS-aurka-RNAi (III) (31704), FUCCI (UAS-E2F-GFP, UAS-CycB-RFP); VDRC: UAS-wdr62 RNAi II (110764), UAS-aurka-RNAi (II) (108446); NIG: UAS-wdr62 RNAi II (7337-R1). For developmental timing, embryonic lays were conducted at 25°C for 2 hr, prior to a shift to 29°C.
Immunofluorescence
Developmentally staged late third instar larval heads were fixed in 4% paraformaldehyde/PBS (40 min), washed in PBT (PBS containing 0.1% Tween-20), blocked with 5 mg/mL BSA/PBT, and incubated overnight with appropriate primary antibodies (4°C). After staining with fluorophore-conjugated secondary antibody, samples were counterstained with DAPI and mounted in 80% glycerol for confocal microscopy. 5-Ethynyl-2′-deoxyuridine (EdU) labeling was conducted with the Click-iT EdU Cell Proliferation Assay kit (Life Technologies, cat. no. C10340). Antibodies included mouse anti-α-tubulin (1:200, Sigma DM1A, cat. no. T9026), rabbit anti-cleaved Dcp-1 (1:100, Cell Signaling, cat. no. 9578), rat anti-deadpan (1:100, Abcam, cat. no. ab195173), rabbit anti-pH3 (1:300, Merck, cat. no. 06-570), mouse anti-repo (1:500, DSHB 8D12, cat. no. ab-528448), rabbit anti-pH3 (1:300, Abcam, cat. no. ab47297), rabbit anti-mcherry (1:300, Abcam, cat. no. ab167453), chicken anti-GFP (1:1,000, Invitrogen, cat. no. A10262), and rabbit anti-cycB (DSHB, 1/20).
Image Acquisition and Data Quantification
Images were acquired on a Zeiss Imager Z confocal microscope with ZEN interface. Analysis of non-deconvoluted images was conducted with Imaris (Bitplane). For brain volume analysis, brain area for each confocal plane was manually demarcated, and volume calculated using compiled area and plane depth. NBs were identified via antibody staining against the NB marker Deadpan, or in the Insc-GAL4 background by co-expression of UAS-GFP. Similarly, glial cells were identified via staining with anti-repo, or in the Repo-GAL4 background by co-expression of UAS-GFP. The number of glia or NBs in a confocal stack prepared by an overlapping z series from a third instar brain lobe were quantified automatically using the “spot” function in Imaris quantification software, after applying radius filters according to average cell size. To ensure accuracy of automated data outputs, datasets were selected at random for manual quantification. For co-localization studies (e.g., quantification of mitotic glia) the ImarisColoc function was employed. For Repo FUCCI experiments, glia in G2/M phase were determined by quantifying co-localized E2F-GFP glia and CycB-RFP glia; glia in G1 or S phase were determined by subtracting GFP or RFP glial numbers from the G2/M count, respectively. For Insc FUCCI experiments, co-localized GFP and RFP NBs were quantified to determine NBs in G2/M. This number was subtracted from NBs expressing only GFP or RFP to determine G1 or S phase, respectively. In all figures, measurements are represented as a percentage of wild-type measurements.
qRT-PCR
Late third instar larval heads were dissected and RNA isolated (SV Total RNA Isolation System, Promega), cDNA synthesis performed (Bioline, Tetro cDNA Synthesis Kit) for qPCR (Bioline, SensiFAST SYBR Hi-ROX) performed as described previously (Guo et al., 2016). Briefly reactions were run using Applied Biosystems ViiA7 system in 384-well plates, in technical and biological triplicate. Amplicon specificity was verified by melt curve analysis, average Ct values were calculated for each sample, and fold change was normalized to Cyp1 and B-tubulin housekeeper genes via the 2-ΔΔCT method (Guo et al., 2016). The primers used for the analysis of mRNA levels are as follows:
cg7337 primer 1: fwd 5′-TGGGGCCTTATCCATACCCA-3′
rev 5′-GTCATCTGTTGACTGGGCGA-3′
cg7337 primer 2 fwd 5′-AAGGATCAGGGCTCGTCTCT-3′
rev 5′-TGATCTTGATGCACCGCACT-3′
aur primer 1 fwd 5′-AGTATGCGCCACAAGGAA-3′
rev 5′-CCTGAATATAGGTGGCCGACTGG-3′
aur primer 2 fwd 5′-AGCCCAACAGCGAGAATATGG-3′
rev 5′-GGAAGCTATGGAATTGGAGCCT-3′
Statistical Analysis
For an experiment dataset, the data were collected from at least three independent experiments. Statistical significance was calculated with GraphPad Prism 6 using unpaired Student's t test with 95% confidence interval to compare two specific datasets. Multiple comparison across a given dataset was performed with one-way ANOVA, with Tukey's post-test. Scale bars in all figures represent 100 μM. In all graphs, the error bars represent SEM. According to the GraphPad classification of significance points: ∗p < 05, ∗∗p < 0.01, ∗∗∗p < 0.001, and ∗∗∗∗p < 0.0001.
Author Contributions
N.R.L., B.S., O.Z., and N.M. conceived and conducted experiments and assisted with drafting the manuscript. N.R.L. and B.S. contributed equally as first authors. S.S.M., D.C.H.N., and L.M.Q. conceived experiments and drafted the manuscript. D.C.H.N. and L.M.Q. contributed equally as senior authors.
Published: June 15, 2017
Footnotes
Supplemental Information includes four figures and can be found with this article online at http://dx.doi.org/10.1016/j.stemcr.2017.05.015.
Supplemental Information
References
- Atwood S.X., Prehoda K.E. aPKC phosphorylates Miranda to polarize fate determinants during neuroblast asymmetric cell division. Curr. Biol. 2009;19:723–729. doi: 10.1016/j.cub.2009.03.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betschinger J., Mechtler K., Knoblich J.A. Asymmetric segregation of the tumor suppressor brat regulates self-renewal in Drosophila neural stem cells. Cell. 2006;124:1241–1253. doi: 10.1016/j.cell.2006.01.038. [DOI] [PubMed] [Google Scholar]
- Bilguvar K., Oztürk A.K., Louvi A., Kwan K.Y., Choi M., Tatli B., Yalnizoğlu D., Tüysüz B., Cağlayan A.O., Gökben S. Whole-exome sequencing identifies recessive WDR62 mutations in severe brain malformations. Nature. 2010;467:207–210. doi: 10.1038/nature09327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman S.K., Neumüller R.A., Novatchkova M., Du Q., Knoblich J.A. The Drosophila NuMA homolog Mud regulates spindle orientation in asymmetric cell division. Dev. Cell. 2006;10:731–742. doi: 10.1016/j.devcel.2006.05.005. [DOI] [PubMed] [Google Scholar]
- Chell J.M., Brand A.H. Nutrition-responsive glia control exit of neural stem cells from quiescence. Cell. 2010;143:1161–1173. doi: 10.1016/j.cell.2010.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J.-F., Zhang Y., Wilde J., Hansen K.C., Lai F., Niswander L. Microcephaly disease gene Wdr62 regulates mitotic progression of embryonic neural stem cells and brain size. Nat. Commun. 2014;5:3885. doi: 10.1038/ncomms4885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham C.L., Martínez-Cerdeño V., Noctor S.C. Microglia regulate the number of neural precursor cells in the developing cerebral cortex. J. Neurosci. 2013;33:4216–4233. doi: 10.1523/JNEUROSCI.3441-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florio M., Huttner W.B. Neural progenitors, neurogenesis and the evolution of the neocortex. Development. 2014;141:2182–2194. doi: 10.1242/dev.090571. [DOI] [PubMed] [Google Scholar]
- Guo L., Zaysteva O., Nie Z., Mitchell N.C., Amanda Lee J.E., Ware T., Parsons L., Luwor R., Poortinga G., Hannan R.D. Defining the essential function of FBP/KSRP proteins: drosophila Psi interacts with the mediator complex to modulate MYC transcription and tissue growth. Nucleic Acids Res. 2016;44:7646–7658. doi: 10.1093/nar/gkw461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homem C.C.F., Knoblich J.A. Drosophila neuroblasts: a model for stem cell biology. Development. 2012;139:4297–4310. doi: 10.1242/dev.080515. [DOI] [PubMed] [Google Scholar]
- Knoblich J.A. Mechanisms of asymmetric stem cell division. Cell. 2008;132:583–597. doi: 10.1016/j.cell.2008.02.007. [DOI] [PubMed] [Google Scholar]
- Lancaster M.A., Knoblich J.A. Spindle orientation in mammalian cerebral cortical development. Curr. Opin. Neurobiol. 2012;22:737–746. doi: 10.1016/j.conb.2012.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C.Y., Andersen R.O., Cabernard C., Manning L., Tran K.D., Lanskey M.J., Bashirullah A., Doe C.Q. Drosophila Aurora-A kinase inhibits neuroblast self-renewal by regulating aPKC/Numb cortical polarity and spindle orientation. Genes Dev. 2006;20:3464–3474. doi: 10.1101/gad.1489406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim N.R., Yeap Y.Y.C., Zhao T.T., Yip Y.Y., Wong S.C., Xu D., Ang C.S., Williamson N.A., Xu Z., Bogoyevitch M.A., Ng D.C.H. Opposing roles for JNK and Aurora A in regulating the association of WDR62 with spindle microtubules. J. Cell Sci. 2015;128:527–540. doi: 10.1242/jcs.157537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim N.R., Yeap Y.Y.C., Ang C.S., Williamson N.A., Bogoyevitch M.A., Quinn L.M., Ng D.C.H. Aurora A phosphorylation of WD40-repeat protein 62 in mitotic spindle regulation. Cell Cycle. 2016;15:413–424. doi: 10.1080/15384101.2015.1127472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morante J., Vallejo D.M., Desplan C., Dominguez M. Conserved miR-8/miR-200 defines a glial niche that controls neuroepithelial expansion and neuroblast transition. Dev. Cell. 2013;27:174–187. doi: 10.1016/j.devcel.2013.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair A.R., Singh P., Garcia D.S., Rodriguez-Crespo D., Egger B., Cabernard C. The microcephaly-associated protein Wdr62/CG7337 is required to maintain centrosome asymmetry in Drosophila neuroblasts. Cell Rep. 2016;14:1100–1113. doi: 10.1016/j.celrep.2015.12.097. [DOI] [PubMed] [Google Scholar]
- Nicholas A.K., Khurshid M., Désir J., Carvalho O.P., Cox J.J., Thornton G., Kausar R., Ansar M., Ahmad W., Verloes A. WDR62 is associated with the spindle pole and is mutated in human microcephaly. Nat. Genet. 2010;42:1010–1014. doi: 10.1038/ng.682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereanu W., Shy D., Hartenstein V. Morphogenesis and proliferation of the larval brain glia in Drosophila. Dev. Biol. 2005;283:191–203. doi: 10.1016/j.ydbio.2005.04.024. [DOI] [PubMed] [Google Scholar]
- Pollen A.A., Nowakowski T.J., Chen J., Retallack H., Sandoval-Espinosa C., Nicholas C.R., Shuga J., Liu S.J., Oldham M.C., Diaz A. Molecular identity of human outer radial glia during cortical development. Cell. 2015;163:55–67. doi: 10.1016/j.cell.2015.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sepp K.J., Schulte J., Auld V.J. Peripheral glia direct axon guidance across the CNS/PNS transition zone. Dev. Biol. 2001;238:47–63. doi: 10.1006/dbio.2001.0411. [DOI] [PubMed] [Google Scholar]
- Smith C.A., Lau K.M., Rahmani Z., Dho S.E., Brothers G., She Y.M., Berry D.M., Bonneil E., Thibault P., Schweisguth F. aPKC-mediated phosphorylation regulates asymmetric membrane localization of the cell fate determinant Numb. EMBO J. 2007;26:468–480. doi: 10.1038/sj.emboj.7601495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Somers G.W., Bashirullah A., Heberlein U., Yu F., Chia W. Aurora-A acts as a tumor suppressor and regulates self-renewal of Drosophila neuroblasts. Genes Dev. 2006;20:3453–3463. doi: 10.1101/gad.1487506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirtz-Peitz F., Nishimura T., Knoblich J.A. Linking cell cycle to asymmetric division: aurora-A phosphorylates the Par complex to regulate Numb localization. Cell. 2008;135:161–173. doi: 10.1016/j.cell.2008.07.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D., Zhang F., Wang Y., Sun Y., Xu Z. Microcephaly-associated protein WDR62 regulates neurogenesis through JNK1 in the developing neocortex. Cell Rep. 2014;6:104–116. doi: 10.1016/j.celrep.2013.12.016. [DOI] [PubMed] [Google Scholar]
- Yu T.W., Mochida G.H., Tischfield D.J., Sgaier S.K., Flores-Sarnat L., Sergi C.M., Topçu M., McDonald M.T., Barry B.J., Felie J.M. Mutations in WDR62, encoding a centrosome-associated protein, cause microcephaly with simplified gyri and abnormal cortical architecture. Nat. Genet. 2010;42:1015–1020. doi: 10.1038/ng.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zielke N., Korzelius J., van Straaten M., Bender K., Schuhknecht G.F.P., Dutta D., Xiang J., Edgar B.A. Fly-FUCCI: a versatile tool for studying cell proliferation in complex tissues. Cell Rep. 2014;7:588–598. doi: 10.1016/j.celrep.2014.03.020. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.