Summary
The inflammatory bowel diseases (IBD) are chronic gastrointestinal inflammatory disorders that affect millions worldwide. Genome-wide association studies have identified 200 IBD-associated loci, but few have been conclusively resolved to specific functional variants. Here we report fine-mapping of 94 IBD loci using high-density genotyping in 67,852 individuals. We pinpointed 18 associations to a single causal variant with >95% certainty, and an additional 27 associations to a single variant with >50% certainty. These 45 variants are significantly enriched for protein-coding changes (n=13), direct disruption of transcription factor binding sites (n=3) and tissue specific epigenetic marks (n=10), with the latter category showing enrichment in specific immune cells among associations stronger in CD and in gut mucosa among associations stronger in UC. The results of this study suggest that high-resolution fine-mapping in large samples can convert many GWAS discoveries into statistically convincing causal variants, providing a powerful substrate for experimental elucidation of disease mechanisms.
The inflammatory bowel diseases (IBD) are a group of chronic, debilitating disorders of the gastrointestinal tract with peak onset in adolescence and early adulthood. More than 1.4 million people are affected in the USA alone1, with an estimated direct healthcare cost of $6.3 billion/year. IBD affects millions worldwide, and is rising in prevalence, particularly in pediatric and non-European ancestry populations2. IBD has two subtypes, ulcerative colitis (UC) and Crohn’s disease (CD), which have distinct presentations and treatment courses. To date, 200 genomic loci have been associated with IBD3,4, but only a handful have been conclusively ascribed to a specific causal variant with direct insight into the underlying disease biology. This scenario is common to all genetically complex diseases, where the pace of identifying associated loci outstrips that of defining specific molecular mechanisms and extracting biological insight from each association.
The widespread correlation structure of the human genome (known as linkage disequilibrium, or LD) often results in similar evidence for association among many neighboring variants. However, unless LD is perfect (r2 = 1), it is possible, with a sufficiently large sample size, to statistically resolve causal variants from neighbors even at high levels of correlation (Extended Data Figure 1 and ref 5). Novel statistical approaches applied to very large datasets that address this problem6 require that the highly correlated variants are directly genotyped or imputed with certainty. Truly high-resolution mapping data, when combined with increasingly sophisticated and comprehensive public databases annotating the putative regulatory function of DNA variants, are likely to reveal novel insights into disease pathogenesis7–9 and the mechanisms of disease-associated variants.
Genetic architecture of associated loci
We genotyped 67,852 individuals of European ancestry, including 33,595 IBD (18,967 CD and 14,628 UC) and 34,257 healthy controls using the Illumina™ Immunochip (Extended Data Table 1). This genotyping array was designed to include all known variants from European individuals in the February 2010 release of the 1000 Genomes Project10,11 in 187 high-density regions known to be associated to one or more immune-mediated diseases12. Because fine-mapping uses subtle differences in strength of association between tightly correlated variants to infer which is most likely to be causal, it is particularly sensitive to data quality. We therefore performed stringent quality control to remove genotyping errors and batch effects (Methods). We imputed into this dataset from the 1000 Genomes reference panel13,14 to fill in variants missing from the Immunochip, or filtered out by our quality control (Extended Data Figure 2). We then evaluated the 97 high-density regions that had previous IBD associations3 and contained at least one variant that showed significant association (Methods) in this data set. The major histocompatibility complex was excluded from these analyses as fine-mapping has been reported elsewhere15.
We applied three complementary Bayesian fine-mapping methods that used different priors and model selection strategies to identify independent association signals within a region, and to assign a posterior probability of causality to each variant (Supplementary Methods and Extended Data Figure 2). For each independent signal detected by each method, we sorted all variants by the posterior probability of association, and added variants to the ‘credible set’ of associated variants until the sum of their posterior probability exceeded 95% – that is, the credible set contains the minimum list of DNA variants that are >95% likely to contain the causal variant (Figure 1). These sets ranged in size from one to > 400 variants. We merged these results and subsequently focused only on signals where an overlapping credible set of variants was identified by at least two of the three methods and all variants were either directly genotyped or imputed with INFO score > 0.4 (Methods and Figure 1).
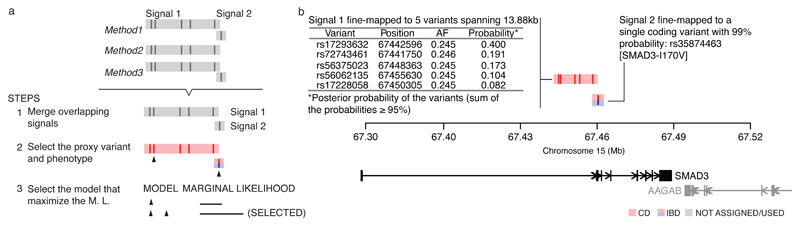
Figure 1. Fine-mapping procedure and output using the SMAD3 region as an example.
a, 1) We merge overlapping signals across methods; 2) select a lead variant (black triangle) and phenotype (color); and 3) choose the best model. Details for each step are available in Methods. b, Example fine-mapping output. This region has been mapped to two independent signals. For each signal, we report the phenotype it is associated with (colored), the variants in the credible set, and their posterior probabilities.
In three out of 97 regions, a consistent credible set could not be identified; when multiple independent effects exist in a region of very high LD, multiple distinct fine-mapping solutions may not be distinguishable (Supplementary Note). Sixty-eight of the remaining 94 regions contain a single association, while 26 harbor two or more independent signals, for a total of 139 independent associations defined across the 94 regions (Figure 2a). Only IL23R and NOD2 (both previously established to contain multiple associated protein-coding variants16) contain more than three independent signals. Consistent with previous reports3, the vast majority of signals are associated with both CD and UC, though many of these have a significantly stronger association with one subtype. For the purposes of enrichment analyses below, we compared 79 signals that are more strongly associated with CD to 23 signals that are more strongly associated with UC (the remaining 37 were equally associated with both subtypes, Supplementary Table 1).
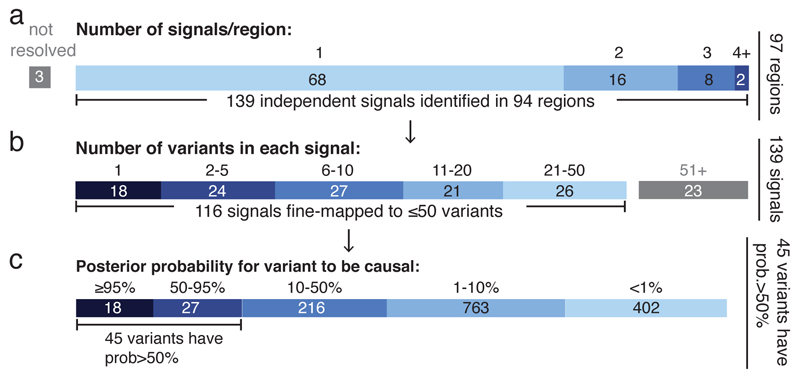
Figure 2. Summary of fine-mapped associations.
a, Independent signals. Sixty-eight loci containing one association and 26 loci containing multiple associations. b, Number of variants in credible sets. 18 associations were fine-mapped to a single variant, and 116 to ≤ 50 variants. c, Distribution of the posterior probability of the variants in credible sets having ≤ 50 variants.
Using a restricted maximum likelihood mixed model approach17, we evaluated the proportion of total variance in disease risk attributed to these 94 regions and how much of that is explained by the 139 specific associations. We estimated that 25% of CD risk was explained by the specific associations described here, out of a total of 28% explained by these loci (correspondingly for UC: 17% out of 22%). The single strongest signals in each region contribute 76% of this variance explained and the remaining associations contribute 24% (Extended Data Figure 3), highlighting the importance of secondary and tertiary associations in GWAS results15,18.
Associations mapped to a single variant
For 18 signals, the 95% credible set consisted of a single variant (‘single variant credible sets’), and for 24 others the credible set consisted of two to five variants (Figure 2b). The single variant credible sets included five previously reported coding variants: three in NOD2 (fs1007insC, R702W, G908R), a rare protective allele in IL23R (V362I) and a splice variant in CARD9 (c.IVS11+1G>C) 16,19. The remaining single variant credible sets were comprised of three missense variants (I170V in SMAD3, I923V in IFIH1 and N289S in NOD2), four intronic variants (in IL2RA, LRRK2, NOD2 and RTEL1/TNFRSF6B) and six intergenic variants (located 3.7kb downstream of GPR35; 3.9kb upstream of PRDM1; within a EP300 binding site 39.9 kb upstream of IKZF1; 500 bp before the transcription start site of JAK2; 9.4kb upstream of NKX2-3; and 3.5kb downstream from HNF4A) (Table 1). Of note, while physical proximity does not guarantee functional relevance, the credible set of variants for 30 associated loci now implicates a specific gene either because it resides within 50 kb of only that gene or has a coding variant with >50% probability – improved from only 3 so refined using an earlier HapMap-based definition. Using the same definitions, the total number of potential candidate genes was reduced from 669 to 233. Examples of IBD candidate genes clearly prioritized in our data are described in the Supplementary Box, and a customizable browser (http://finemapping.broadinstitute.org/) is available to review the detailed fine-mapping results.
Table 1. Variants having posterior probability >50%.
| Variant | Chr | Position | Ns | Phe | AF | Prob | INFO | Func | Annotation |
|---|---|---|---|---|---|---|---|---|---|
| Signals mapped to a single variant | |||||||||
| rs7307562 | 12 | 40724960 | 2 | CD | 0.398 | 0.999 | 1 | LRRK2(intronic) | |
| rs2066844 | 16 | 50745926 | 10 | CD | 0.063 | 0.999 | 0.8 | C | NOD2(R702W) |
| rs2066845 | 16 | 50756540 | 10 | CD | 0.022 | 0.999 | 1 | C | NOD2(G908R) |
| rs6017342 | 20 | 43065028 | 2 | UC | 0.544 | 0.999 | 1 | E |
HNF4A (downstream), Gut_H3K27ac |
| rs61839660 | 10 | 6094697 | 2 | CD | 0.094 | 0.999 | 1 | E |
IL2RA(intronic), Immune_H3K4me1 |
| rs5743293 | 16 | 50763781 | 10 | CD | 0.964 | 0.999 | 1 | C | NOD2(fs1007insC) |
| rs6062496 | 20 | 62329099 | 1 | IBD | 0.587 | 0.996 | 1 | T |
RTEL1-TNFRSF6B(ncRNA_intronic), EBF1 TFBS |
| rs141992399 | 9 | 139259592 | 3 | IBD | 0.005 | 0.995 | 1 | C | CARD9(1434+1G>C) |
| rs35667974 | 2 | 163124637 | 1 | UC | 0.021 | 0.994 | 1 | C | IFIH1(I923V) |
| rs74465132 | 7 | 50304782 | 3 | IBD | 0.034 | 0.994 | 1 | T,E |
IKZF1(upstream), EP300 TFBS, Immune_H3K4me1 |
| rs4676408 | 2 | 241574401 | 1 | UC | 0.508 | 0.994 | 0.99 | GPR35(downstream) | |
| rs5743271 | 16 | 50744688 | 10 | CD | 0.007 | 0.993 | 1 | C | NOD2(N289S) |
| rs10748781 | 10 | 101283330 | 2 | IBD | 0.55 | 0.990 | 1 | E |
NKX2-3(upstream), Gut_H3K27ac |
| rs35874463 | 15 | 67457698 | 2 | IBD | 0.054 | 0.989 | 1 | C,E |
SMAD3(I170V), Gut_H3K27ac |
| rs72796367 | 16 | 50762771 | 10 | CD | 0.023 | 0.983 | 1 | NOD2(intronic) | |
| rs1887428 | 9 | 4984530 | 1 | IBD | 0.603 | 0.974 | 0.97 | JAK2(upstream) | |
| rs41313262 | 1 | 67705900 | 5 | CD | 0.014 | 0.973 | 1 | C | IL23R(V362I) |
| rs28701841 | 6 | 106530330 | 2 | CD | 0.116 | 0.971 | 1 | PRDM1(upstream) | |
| Signals mapped to 2-50 variants and the lead variant has posterior probability > 50% | |||||||||
| rs76418789 | 1 | 67648596 | 5 | CD | 0.006 | 0.937 | 0.59 | C | IL23R(G149R) |
| rs7711427 | 5 | 40414886 | 3 | CD | 0.633 | 0.919 | 1 | ||
| rs1736137 | 21 | 16806695 | 2 | CD | 0.407 | 0.879 | 1 | ||
| rs104895444 | 16 | 50746199 | 10 | CD | 0.003 | 0.865 | 1 | C | NOD2(V793M) |
| rs56167332 | 5 | 158827769 | 2 | IBD | 0.353 | 0.845 | 1 | IL12B | |
| rs104895467 | 16 | 50750810 | 10 | CD | 0.002 | 0.833 | 1 | C | NOD2(N852S) |
| rs630923 | 11 | 118754353 | 2 | CD | 0.153 | 0.820 | 0.98 | ||
| rs3812565 | 9 | 139272502 | 3 | IBD | 0.402 | 0.815 | 1 | Q | eQTL of INPP5E in CD4 and CD8; CARD9 in CD14 |
| rs4655215 | 1 | 20137714 | 3 | UC | 0.763 | 0.784 | 1 | E | Gut_H3K27ac |
| rs145530718 | 19 | 10568883 | 3 | CD | 0.023 | 0.762 | 0.97 | ||
| rs6426833 | 1 | 20171860 | 3 | UC | 0.555 | 0.752 | 1 | ||
| chr20: 43258079 |
20 | 43258079 | 2 | CD | 0.041 | 0.736 | 0.88 | ||
| rs17229679 | 2 | 199560757 | 2 | UC | 0.028 | 0.716 | 1 | ||
| rs4728142 | 7 | 128573967 | 1 | UC | 0.448 | 0.664 | 1 | E | Immune_H3K4me1 |
| rs2143178 | 22 | 39660829 | 2 | IBD | 0.157 | 0.662 | 1 | T,E | NFKB TFBS, Gut_H3K27ac |
| rs34536443 | 19 | 10463118 | 3 | CD | 0.038 | 0.649 | 1 | C | TYK2(P1104A) |
| rs138425259 | 16 | 50663477 | 10 | UC | 0.009 | 0.648 | 0.92 | ||
| rs146029108 | 9 | 139329966 | 3 | CD | 0.036 | 0.643 | 0.92 | ||
| rs12722504 | 10 | 6089777 | 2 | CD | 0.26 | 0.615 | 1 | ||
| rs60542850 | 19 | 10488360 | 3 | IBD | 0.17 | 0.591 | 0.89 | ||
| rs2188962 | 5 | 131770805 | 1 | CD | 0.44 | 0.590 | 1 | E,Q | Gut_H3K27ac, eQTL of SLC22A5 in CD14, CD15 and IL |
| rs2019262 | 1 | 67679990 | 5 | IBD | 0.4 | 0.586 | 1 | ||
| rs3024493 | 1 | 206943968 | 2 | IBD | 0.171 | 0.537 | 1 | E | Immune_H3K4me1 |
| rs7915475 | 10 | 64381668 | 3 | CD | 0.304 | 0.528 | 1 | ||
| rs77981966 | 2 | 43777964 | 1 | CD | 0.077 | 0.521 | 1 | ||
| rs9889296 | 17 | 32570547 | 1 | CD | 0.264 | 0.512 | 1 | ||
| rs2476601 | 1 | 114377568 | 1 | CD | 0.908 | 0.508 | 1 | C | PTPN22(W620R) |
Ns: number of independent signals in the locus. Phe: phenotype. AF: allele frequency. Prob: posterior probability for being a causal variant. INFO: imputation. Func: functional annotations -- coding (C), disrupting transcription factor binding sites (T), overlapping epigenetic peaks (E) and colocalization with eQTL (Q).
Associated protein coding variants
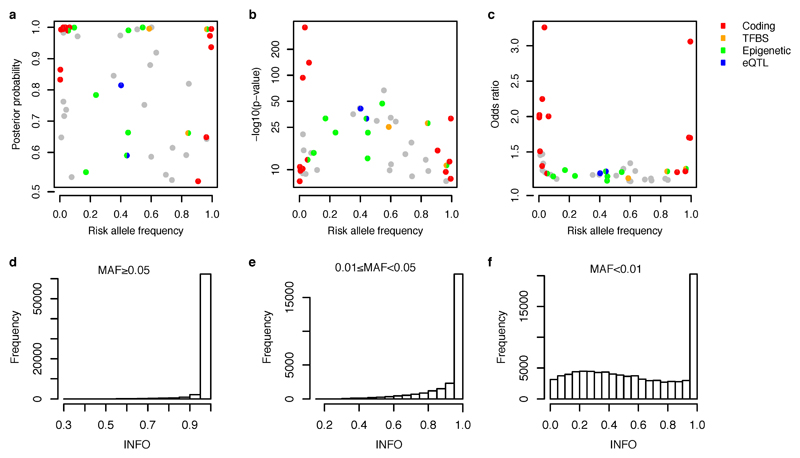
We first annotated the possible functional consequences of the IBD variants by their effect on the amino acid sequences of proteins. Thirteen out of 45 variants (Figure 2c) that have >50% posterior probability are non-synonymous (Table 1), an 18-fold enrichment (enrichment P=2x10-13, Fisher’s exact test) relative to randomly drawn variants in our regions (Figure 3a). By contrast, only one variant with >50% probability is synonymous (enrichment P=0.42). All common coding variants previously reported to affect IBD risk are included in a 95% credible set including: IL23R (R381Q, V362I and G149R); CARD9 (c.IVS11+1G>C and S12N); NOD2 (S431L, R702W, V793M, N852S and G908R, fs1007insC); ATG16L1 (T300A); PTPN22 (R620W); and FUT2 (W154X). While this enrichment of coding variation (Figure 3a) provides assurance about the accuracy of our approach, it does not suggest that 30% of all associations are caused by coding variants; rather, it is almost certainly the case that associated coding variants have stronger effect sizes, making them easier to fine-map.
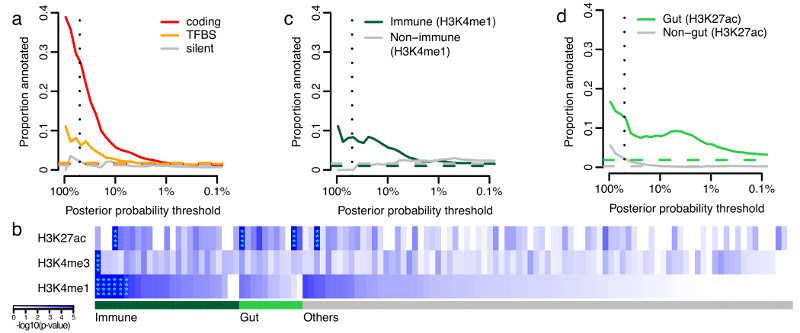
Figure 3. Functional annotation of causal variants.
a, Proportion of credible variants that are protein coding, disrupt/create transcription factor binding motif sites (TFBS) or are synonymous, sorted by posterior probability. b, Epigenetic peaks overlapping credible variants in cell and tissue types from the Roadmap Epigenomics Consortium39. Significant enrichment has been marked with asterisks. Proportion of credible variants that overlap (c) core immune peaks for H4K4me1or (d) core gut peaks for H3K27ac (Methods). In panels a, c and d, the vertical dotted lines mark 50% posterior probability and the horizontal dashed lines show the background proportions of each functional category.
Associated non-coding variants
We next examined conserved nucleotides in high confidence binding site motifs of 84 transcription factor (TF) families20 (Methods). There was a significant positive correlation between TF motif disruption and IBD association posterior probability (P=0.006, logistic regression) (Figure 3a), including three variants with >50% probability (two >95%). In the RTEL1/TNFRSF6B region, rs6062496 is predicted to disrupt a TF binding site (TFBS) for EBF1, a TF involved in the maintenance of B cell identity and prevention of alternative fates in committed cells21. A low frequency (3.6%) protective allele at rs74465132 creates a binding site for EP300 less than 40kbp upstream of IKZF1. The third notable example of TFBS disruption, although not in a single variant credible set, is detailed in the Supplementary Box for the association at SMAD3.
Recent studies have shown that trait associated variants are enriched for epigenetic marks highlighting cell type specific regulatory regions9,22,23. We compared our credible sets with ChIPseq peaks corresponding to chromatin immunoprecipitation with H3K4me1, H3K4me3 and H3K27ac (shown previously22,23 to highlight enhancers, promoters and active regulatory elements, respectively) in 120 adult and fetal tissues, assayed by the Roadmap Epigenomics Mapping Consortium24 (Figure 3b). Using a threshold of P=1.3x10-4 (0.05 corrected for 360 tests), we observed significant enrichment of H3K4me1 in 6 immune cell types and for H3K27ac in 2 gastrointestinal (gut) samples (sigmoid colon and rectal mucosa) (Figure 3b and Supplementary Table 2). The subset of signals that are more strongly associated with CD overlap more with immune cell chromatin peaks, whereas UC signals overlap more with gut chromatin peaks (Supplementary Table 2).
These three chromatin marks are correlated both within tissues (we observe additional signal in other marks in the tissues described above) and across related tissues. We therefore defined a set of “core immune peaks” for H3K4me1 and “core gut peaks” for H3K27ac as the set of overlapping peaks in all enriched immune cell and gut tissue types, respectively. These two sets of peaks are independently significant and capture the observed enrichment compared to “control peaks” made up of the same number of ChIPseq peaks across our 94 regions in non-immune and non-gut tissues (Figure 3c,d). These two tracks summarize our epigenetic-GWAS overlap signal, and the combined excess over the baseline suggests that a substantial number of regions, particularly those not mapped to coding variants, may ultimately be explained by functional variation in recognizable enhancer/promoter elements.
Overlap with expression QTLs
Variants that change enhancer or promoter activity might change gene expression, and baseline expression of many genes has been found to be regulated by genetic variation25–27. Indeed, it has been suggested that these so-called expression quantitative trait loci (eQTLs) underlie a large proportion of GWAS associations25,28. We therefore searched for variants that are both in an IBD-associated credible set with 50 or fewer variants, and the most significantly associated eQTL variant for a gene in a study29 of peripheral blood mononuclear cells (PBMC) from 2,752 twins. Sixty-eight of the 76 regions with signals fine-mapped to ≤ 50 variants harbor at least one significant eQTL (affecting a gene within 1 Mb with P < 10-5). Despite this abundance of eQTLs in fine-mapped regions, only 3 credible sets include the most significantly associated eQTL variants, compared with 3.7 expected by chance (Methods). Data from a more recent study30 using PBMCs from 8,086 individuals did not yield a substantively different outcome, demonstrating a modest but non-significant enrichment (8 observed overlaps, 4.2 expected by chance, P=0.06). Using a more lenient definition of overlap which requires the lead eQTL variant to be in LD (R2 > 0.4) with an IBD credible set variant increased the number of potential overlaps but again these numbers were not greater than chance expectation.
As PBMCs are a heterogeneous collection of immune cell populations, cell type-specific signals or signals corresponding to genes expressed most prominently in non-immune tissues may be missed. We therefore tested the enrichment of eQTLs that overlap credible sets in five primary immune cell populations (CD4+, CD8+, CD19+, CD14+ and CD15+), platelets, and three distinct intestinal locations (rectum, colon and ileum) isolated from 350 healthy individuals (Methods). We observed a significant enrichment of credible SNP/eQTL overlaps in CD4+ cells and ileum (Extended Data Table 2): three and two credible sets overlapped eQTLs, respectively, compared to 0.4 and 0.3 expected by chance (P=0.005 and 0.020). An enrichment was also observed for the naïve CD14+ cells from another study31: eight overlaps observed compared to 2.7 expected by chance (P=0.001). We did not observe enrichment of overlaps in stimulated (with interferon or lipopolysaccharide) CD14+ cells from the same source (Extended Data Table 2).
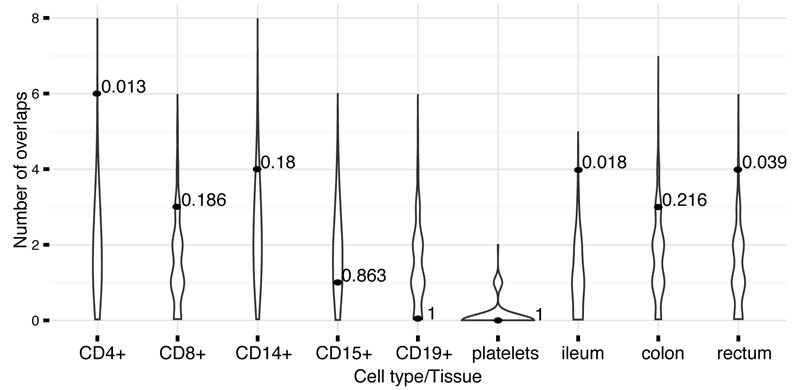
We investigated eQTL overlaps more deeply by applying two colocalization approaches (one frequentist, one Bayesian, Methods) to the our cell-separated dataset where primary genotype and expression data were available. We confirmed greater than expected overlap with eQTLs in CD4+ and ileum described above (Figure 4 and Extended Data Table 2). These CD4+ colocalized eQTLs also had stronger overlap with CD4+ ChIPseq peaks than our other credible sets, further supporting a regulatory causal mechanism. The number of colocalizations in other purified cell types and tissues was largely indistinguishable from what we expect under the null using either method, except for moderate enrichment in rectum (4 observed and 1.4 expected, P=0.039, Frequentist approach) and colon (3 observed and 0.8 expected, P=0.04, Bayesian approach). Only two of these colocalizations correspond to an IBD variant with causal probability > 50% (Table 1 and Extended Data Figure 4a).
Figure 4. Number of credible sets that colocalize eQTLs.
Distributions of the number of colocalizations by chance (violins) and observed number of colocalizations with p-values (dots). Both the background and the observed numbers were calculated using the “Frequentist colocalization using conditional P values” approach (Methods).
Discussion
We have performed fine-mapping of 94 previously reported genetic risk loci for IBD. Rigorous quality control followed by an integration of three novel fine-mapping methods generated lists of genetic variants accounting for 139 independent associations across these loci. Our methods are concordant with an existing fine-mapping method6 (67 of 68 credible sets in single signal regions overlap, including exact matches for all single variant credible sets), and provide extensions to support the phenotype assignment (CD, UC or IBD) and the conditional estimation of multiple credible sets in loci with multiple independent signals. The use of multiple methods allowed us to focus our downstream analyses on loci where the choice of fine-mapping method did not substantially alter conclusions about the biology of IBD. Our results improve on previous fine-mapping efforts using a preset LD threshold32 (e.g. r2> 0.6) (Extended Data Figure 5) by formally modeling the posterior probability of association of every variant. Much of this resolution derives from the very large sample size we employed, because the number of variants in a credible set decreases with increasing significance (P=0.0069).
The high-density of genotyping also aids in improved resolution. For instance, the primary association at IL2RA has now been mapped to a single variant associated with CD, rs61839660. This variant was not present in the Hapmap 3 reference panel and was therefore not reported in earlier studies3,33 (nearby tagging variants, rs12722489 and rs12722515, were reported instead). Imputation using the 1000 genomes reference panel and the largest assembled GWAS dataset3 did not separate rs61839660 from its neighbors (unpublished results), due to the loss of information in imputation using the limited reference. Only direct genotyping, available in the immunochip high-density regions, permitted the conclusive identification of the causal variant.
Accurate fine-mapping should, in many instances, ultimately point to the same variant across diseases in shared loci. Among our single-variant credible sets, we fine-mapped a UC association to a rare missense variant (I923V) in IFIH1, which is also associated with type 1 diabetes (T1D)37 with an opposite direction of effect (Supplementary Box). The intronic variant noted above (rs61839660, AF=9%) in IL2RA was also similarly associated with T1D, again with a discordant directional effect38 (Supplementary Box). Simultaneous high-resolution fine-mapping in multiple diseases should therefore better clarify both shared and distinct biology.
Resolution of fine-mapping can be further improved by leveraging LD from other ethnicities34. However, the sample size from other ethnicities we have collected is small compared with European samples (9,846 across East-Asian, South-Asian and Middle-Eastern). Limited access to matched imputation reference panels from all cohorts and the fact that the smaller non-European sets are not from populations (e.g., African-derived) with narrower LD also suggest that gains in fine-mapping accuracy would be limited at this time. Ultimately this effort will be aided by more substantial investment in genotyping non-European population samples and by developing and applying more robust trans-ethnic fine-mapping algorithms.
A new release of the 1000 genomes (phase 3)35 and the UK10K36 project have introduced new variants that were not present in the reference panel in our study. Our major findings remain the same using this new reference panel: the 18 single-variant credible sets are not in high LD (r2 > 0.95) with any new variants in either new dataset, and the 1,426 variants in IBD associations mapped to ≤50 variants are in high LD with only 47 new variants (3.3% of the total size of these credible sets, Supplementary Table 1). Given that this release represents a near complete catalogue of variants with minor allele frequency (MAF) > 1% in European populations, we believe our current fine mapping results are likely to be robust, especially for common variant associations. High-resolution fine-mapping demonstrates that causal variants are significantly enriched for variants that alter protein coding variants or disrupt transcription factor binding motifs. Enrichment was also observed in H3K4me1 marks in immune related cell types and H3K27ac marks in sigmoid colon and rectal mucosal tissues, with CD loci demonstrating a stronger immune signature and UC loci more enriched for gut tissues (P values are 0.014, 0.0005 and 0.0013 respectively for H3K4me1, H3K27ac and H3K4me3; chi-square test). By contrast, overall enrichment of eQTLs is quite modest compared with prior reports and not seen strongly in excess of chance in our well-refined credible sets (≤ 50 variants). This result underscores the importance of high-resolution mapping and the careful incorporation of the high background rate of eQTLs. It is worth noting that evaluating the overlap between two distinct mapping results is fundamentally different than comparing genetic mapping results to fixed genomic features, and depends on both mappings being well resolved. While these data challenge the paradigm that easily surveyed baseline eQTLs explain a large proportion of non-coding GWAS signals, the modest excesses observed in smaller but cell-specific data sets suggest that much larger tissue or cell-specific studies (and under the correct stimuli or developmental time points) will resolve the contribution of eQTLs to GWAS hits.
Resolving multiple independent associations may often help target the causal gene more precisely. For example, the SMAD3 locus hosts a non-synonymous variant and a variant disrupting the conserved transcription factor binding site (also overlapping the H3K27ac marker in gut tissues), unambiguously articulating a role in disease and providing an allelic series for further experimental inquiry. Similarly, the TYK2 locus has been mapped to a non-synonymous variant and a variant disrupting a conserved transcription factor binding site (http://finemapping.broadinstitute.org/).
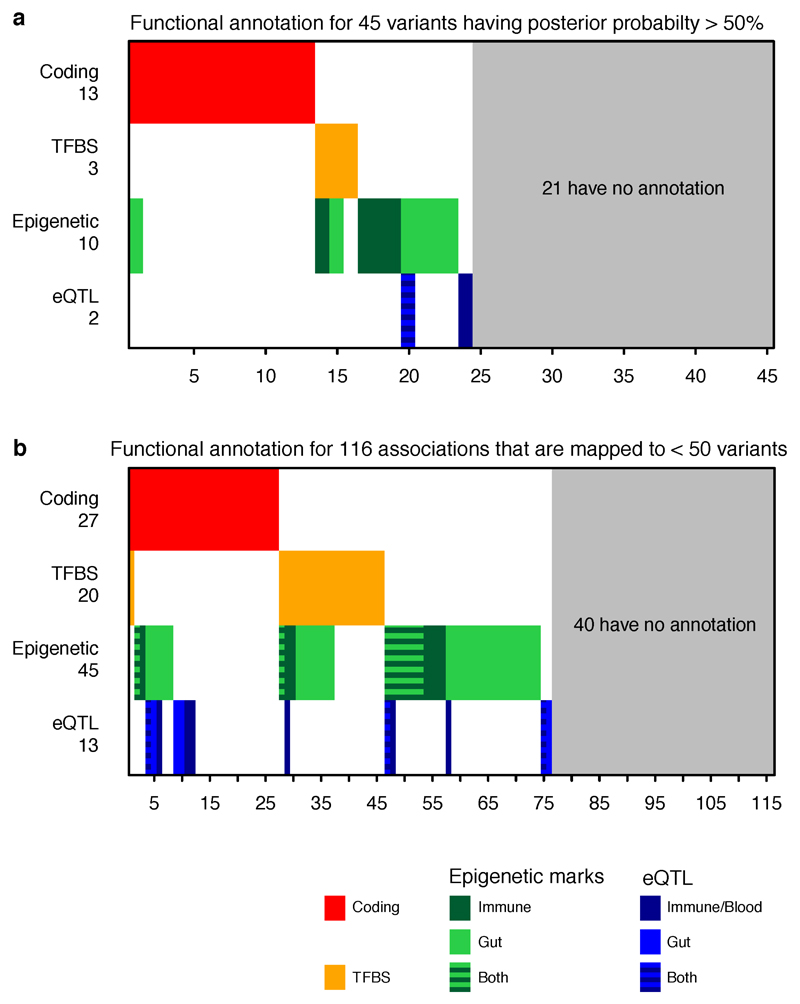
One-hundred and sixteen associations have been fine-mapped to ≤ 50 variants. Among them, 27 associations contain coding variants, 20 contain variants disrupting transcription factor binding motifs, and 45 are within histone H3K4me1 or H3K27ac marked DNA regions. The best-resolved associations - 45 variants having >50% posterior probabilities for being causal (Table 1) – are similarly significantly enriched for variants with known or presumed function from genome annotation. Of these, 13 variants cause non-synonymous change in amino acids, three disrupt a conserved TF binding motif, ten are within histone H3K4me1 or H3K27ac marked DNA regions in disease-relevant tissues, and two colocalize with a significant cis-eQTL (Extended Data Figure 4a). Risk alleles of these variants can be found throughout the allele frequency spectrum, with protein coding variants having somewhat larger effects and more extreme risk allele frequencies (Extended Data Figure 6a-c).
This analysis, however, leaves 21 non-coding variants (Extended Data Figure 4b), all of which have >50% probabilities to be causal (five have >95%), that are not located within known motifs, annotated elements, nor in any experimentally determined ChIPseq peaks or eQTL credible sets yet discovered. While we have identified a statistically compelling set of genuine associations (often intronic or within 10 kb of strong candidate genes), we can make little inference about function. For example, the intronic single-variant credible set of LRRK2 has no annotation, eQTL or ChIPseq peak of note. This underscores the incompleteness of our knowledge regarding the function of non-coding DNA and its role in disease, and calls for comprehensive studies on transcriptome and epigenome in a wide range of cell lines and stimulation conditions. That the majority of the best-refined non-coding associations have no available annotation is perhaps sobering with respect to how well we may currently be able to interpret non-coding variation in medical sequencing efforts. It does suggest, however, that detailed fine-mapping of GWAS signals down to single variants, combined with emerging high-throughput genome-editing methodology, may be among the most effective ways to advance to a greater understanding of the biology of the non-coding genome.
Methods
Genotyping and QC
We genotyped 35,197 unaffected and 35,346 affected individuals (20,155 CD and 15,191 UC) using the Immunochip array. Genotypes were called using optiCall40 for 192,402 autosomal variants before QC. We removed variants with missing data rate >2% across the whole dataset, or >10% in any one batch, and variants that failed (FDR < 10-5 in either the whole dataset or at least two batches) tests for: a) Hardy-Weinberg equilibrium in controls; b) differential missingness between cases and controls; c) different allele frequency across different batches in controls, CD or UC. We also removed non-coding variants that were present in the 1000 Genomes pilot stage but were not in the subsequent Phase I integrated variant set (March 2012 release) and had not been in releases 2 or 3 of HapMap as these mostly represent false positives from the 1000 Genomes pilot, which often genotype poorly. Where a variant failed in exactly one batch we set all genotypes to missing for that batch (to be reimputed later) and included the site if it passed in the remainder of the batches. We removed individuals that had >2% missing data, had significantly higher or lower (defined as FDR<0.01) inbreeding coefficient (F), or were duplicated or related (PI_HAT ≥ 0.4, calculated from the LD pruned dataset described below), by sequentially removing the individual with the largest number of related samples until no related samples remain. We projected all remaining samples onto principal component axes generated from HapMap 3, and classified their ancestry using a Gaussian mixture model fitted to the European (CEU+TSI), African (YRI+LWK+ASW+MKK), East Asian (CHB+JPT) and South Asian (GIH) HapMap samples. We removed all samples that were classified as non-European, or that lay more than 8 standard deviations from the European cluster. After QC, there were 67,852 European-derived samples with valid diagnosis (healthy control, CD or UC), and 161,681 genotyped variants available for downstream analyses.
Linkage-disequilibrium pruning and principal components analysis
From the clean dataset we removed variants in long range LD41 or with MAF < 0.05, and then pruned 3 times using the ‘--indep’ option in PLINK (with window size of 50, step size of 5 and VIF threshold of 1.25). Principal component axes were generated within controls using this LD pruned dataset (18,123 variants). The axes were then projected to cases to generate the principal components for all samples. The analysis was performed using our in-house C code (https://github.com/hailianghuang/efficientPCA) and LAPACK package42 for efficiency.
Controlling for population structure, batch effects and other confounders
We used 2,853 “background SNPs” present on the Immunochip but not known to be associated with immune disorders to calculate the genomic inflation factor λGC. After including the first five principal components calculated above as covariates, λGC = 1.29, 1.25 and 1.31 for CD, UC and IBD (adding additional principal components did not further reduce λGC, Extended Data Table 3a). Because our genotype data were processed in 15 batches with variable ratios of cases to controls, we conducted two analyses to ensure possible batch effects were adequately controlled. First, we split the samples into a “balanced” cohort with studies that have both cases and controls and an “imbalanced” cohort with studies that have exclusively cases or controls (Extended Data Table 1). As λGC under polygenic inheritance scales with the sample size43, we randomly down-sampled the full dataset to match the sample size of the balanced and the imbalanced cohorts respectively. We tested for association in these subsets of our data (and included batch ID as a covariate in the balanced cohort), and found the λGC from the balanced and imbalanced cohorts to be within the 95% confidence interval of size matched values from our full data, suggesting that batch effects are not systematically inflating our association statistics (Extended Data Table 3b). We also performed a heterogeneity test for the odds ratio (OR) of lead variants in each credible set using the balanced and imbalanced cohorts, and observed no significant heterogeneity after Bonferroni correction (Supplementary Table 3).
We next sought to disentangle the contributions of polygenic inheritance and uncorrected population structure in our observed λGC. LD score regression44 is able to differentiate these two effects, but requires genome-wide data, so is not possible in our Immunochip dataset. Instead, we compared λGC and λ1000 values calculated using the same set of background SNPs from the largest IBD meta-analysis with genome-wide data45. For both CD and UC the λ1000 values in our Immunochip study (1.012 and 1.012) were equal or less than those in the genome-wide study (1.016 and 1.012). Furthermore, LD score regression on the genome-wide data shows that the majority of inflation is caused by polygenic risk (LD score intercept = 1.09 for both CD and UC, compared to λGC = 1.23 and 1.29). Together, these results show that our residual inflation is consistent with polygenic signal and modest residual confounding. We tested what effect correcting for the LD score intercept of 1.09 would have on posterior probabilities and credible sets and found no major differences compared to uncorrected values. The full comparison of λ values is shown in Extended Data Table 3c.
Imputation
Imputation was performed separately in each Immunochip autosomal high-density region (185 total) from the 1000 Genomes Phase I integrated haplotype reference panel. To prevent the edge effect, we extended each side of the high density regions by 50kbp. Two imputations were performed sequentially (Extended Data Figure 2) using software and parameters as described below. The first imputation was performed immediately after the quality control, from which the major results were manually inspected (Manual cluster plot inspection, Methods). The second imputation was performed after removing variants that failed the manual cluster plot inspection. We used SHAPEIT46,47 (versions: first imputation: v2.r644, second imputation: v2.r769) to pre-phase the genotypes, followed by IMPUTE213,14 (versions: first: 2.2.2, second: 2.3.0) to perform the imputation. The reference panels were downloaded from the IMPUTE2 website (first: Mar 2012 release, second: Dec 2013 release). After the second imputation, there were 388,432 variants with good imputation quality (INFO > 0.4). These include 99.9% of variants with MAF ≥0.05, 99.3% of variants with 0.05>MAF ≥0.01, and 63.0% of variants with MAF < 0.01 (Extended Data Figure 6d-f), with similar success rates for both coding and non-coding variants, making it unlikely that missing variants substantially affect our fine-mapping conclusions.
Manual cluster plot inspection
Variants that had posterior probability greater than 50% or in credible sets mapped to ≤ 10 variants were manually inspected using Evoker v2.248. Each variant was inspected by three independent reviewers (ten reviewers participated) and scored as pass, fail or maybe. Reviewers were blinded to the posterior probability of these variants. We removed variants that received one or more fails, or received less than 2 passes. 220 out of 276 inspected variants passed this inspection, and 53 of 56 failed variants were restored by imputation. There is no difference in MAF between the failed and the passed variants (P=0.66). A further cluster plot inspection flagged two additional failed variants after removing the failed variants from the first inspection and redoing the imputation and analysis. Dramatic clustering errors accounted for 27/58 flagged variants, which were eliminated from final credible sets. The remaining 31 had only minor issues, and the imputed data for these remained in our final credible sets, with marginally smaller posteriors (mean of the difference: 9.8%, P=0.06, paired t test).
Establishing a P value threshold
We used a multiple testing corrected P value threshold for associations of 1.35 x 10-6, which was established by permutation. We generated 200 permuted datasets by randomly shuffling phenotypes across samples and carried out association analyses for each permutation across all variants in high-density regions that overlap IBD-associated loci3. We stored (i) all the point-wise P values (αS), as well as (ii) the “best” P values (αB) of each of the 200 permuted datasets. We then computed the empirical, experiment-wide P value (αM)(corrected for multiple testing) for each of the tests as its rank/200 with respect to the 200 αB. We then estimated the number of independent tests performed in the studied regions, n, as the slope of the regression of log(1-αM) on log(1-αS), knowing that αM = 1 − (1−αS)n, yielding a value of 37,056. The P value threshold was determined as 0.05/n ≈ 1.35 x 10-6.
Detecting and fine-mapping association signals
We used three fine-mapping methods (Supplementary Methods) to detect independent signals and create credible sets across 97 Immunochip autosomal high-density regions that contained at least one variant with p < 1.35 x 10-6. Our process for merging the results of the three methods is described below and illustrated in Figure 1a.
We merged signals from different methods if their credible sets overlapped. To ensure a conservative credible set, this new merged credible set included all variants from all merged signals (the union of constituent credible sets). We assigned each variant in the merged credible sets a posterior probability equal to the average of the probabilities from the methods that reported this signal. To filter out technical artifacts we required genotyped variants in small credible sets to pass manual cluster plot inspection (see above) and all imputed variants to have INFO > 0.4. For signals reported by only one or two methods that contain only imputed variants (i.e. no directly genotyped variants), we additionally required at least one variant with INFO > 0.8 and MAF > 0.01.
We next assigned each signal to a provisional combination of lead variant and phenotype (CD, UC or IBD) that maximized the marginal likelihood of equation 8 in Supplementary Methods.
At loci with >1 signals, we built a multivariate model with all signals reported by all three methods, and tested all possible combinations of adding signals reported by one or two methods, as long as they still had p < 1.35 x 10-6 when jointly fitted in the multi-signal model. We selected the combination with the highest joint marginal likelihood (equation 8 in Supplementary Methods).
Phenotype assignment of signals
The provisional phenotype assignment carried out during the signal merging described above is merely a point estimate, and does not capture the uncertainty associated with the phenotypic assignment. We therefore recomputed the assignment of each signal as CD-specific, UC-specific or shared using the Bayesian multinomial model from fine-mapping method 2, Empirical covariance prior with Laplace approximation49, as it is designed to assess evidence of sharing in the presence of potentially correlated effect sizes. For the lead variant for each credible set, we calculated the marginal likelihoods as in equation 13 from Supplementary Methods, restricting either βUC = 0 (for the CD-only model) or βCD = 0 (for the UC-only model), as well as using the unconstrained prior (for the associated-to-both model). We then calculated the log Bayes factor in favor of sharing, i.e. the log of the ratio of marginal likelihoods between the associated-to-both model and the best of the single-phenotype associated models. These sharing log Bayes factors are given in Supplementary Table 1 (column ‘sharingBF’), and are a probabilistic assessment of phenotype assignment: for instance, the log Bayes factor of 97.4 for the primary signal at IL23R suggests a very high certainty that this signal is shared across both CD and UC, whereas the log Bayes factor of 0.4 for the primary signal at FUT2 is more ambiguous. In addition to providing the log Bayes factor itself, we also applied a log Bayes factor cut-off of 10 to select variants with strong evidence of being shared across phenotypes.
Final filters
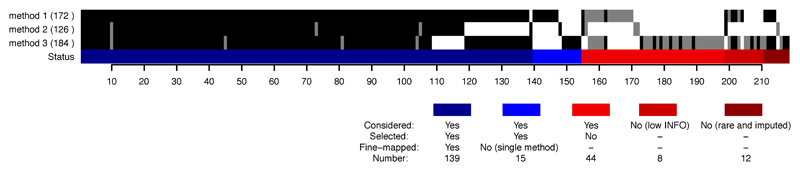
These procedures generated some signals where all three methods largely agreed, and some where they differed. While the signals where the methods disagree are of interest for methods development, here we chose to focus on the most concordant signals, as they are most straightforward to interpret biologically. We therefore discarded all signals found by only one method (which completely removed one locus), and two loci where the ratio of marginal likelihoods (equation 8 in Supplementary Methods) for the best model and the second-best model was < 10 (Supplementary Notes). After these filters (Extended Data Figure 7) we considered 139 signals from 94 regions (containing a total of 181,232 variants) to be confidently fine-mapped, and took them forward for subsequent analysis.
Estimating the variance explained by the fine-mapping
We used a mixed model framework to estimate the total risk variance attributable to the IBD risk loci, and to the signals identified in the fine-mapping. We used the GCTA software package50 to compute a gametic relationship matrix (G-matrix) using genotype dosage information for the genotyped variants in the high-density regions (which we will call GHD). We then fit a variety of variance component models by restricted maximum likelihood analysis using an underlying liability threshold model implemented with the DMU package51. The first model is a standard heritability mixed-model that includes fixed effects for five principal components (to correct for stratification) and a random effect summarizing the contribution of all variants in the fine-mapping regions, such that the liabilities across all individuals are distributed according to
where λ1 is thus the variance explained by all variants in fine-mapping regions, which we estimate. We then fitted a model that included an additional random effect for the contribution of the lead variants that have been specifically identified (with G-matrix Gsignals), such that the liability is distributed as
The variance explained by the signals under consideration is then given by the reduction in the variance explained by all variants in the fine-mapping regions between the two models (λ1−λ′1). We used this approach to estimate what fraction of this variance was accounted for by (i) the single strongest signals in each region (as would be typically done prior to fine-mapping), or (ii) all signals identified in fine-mapping. We used Cox and Snell’s method52 to estimate the variance explained across individual signals (Extended Data Figure 3b) for computational efficiency.
Overlap between transcription factor binding motifs and causal variants
For each motif in the ENCODE TF ChIP-seq data (http://compbio.mit.edu/encode-motifs/, accessed Nov 2014)20, we calculated the overall information content (IC) as the sum of IC for each position53, and only considered motifs with overall IC ≥ 14 bits (equivalent to 7 perfectly conserved positions). For every variant in a high-density region we determined whether it creates or disrupts a motif at a high-information site (IC ≥ 1.8).
Overlap between epigenetic signatures and causal variants
For each combination of 120 tissues and three histone marks (H3K4me1, H3K4me3 and H3K27ac) from the Roadmap Epigenome Project we calculated an overlap score, equal to the sum of fine-mapping posterior probabilities for all variants in peaks of that histone mark in that tissue. We generated a null distribution of this score for each tissue/mark by shifting chromatin marks randomly (between 0bp and 44.53Mbp, the length of all high-density regions) and circularly (peaks at the end of the region shifted to the beginning of the region) over the high-density regions while keeping the same inter-peak distances. To summarize these correlated results across many cell and tissue types we defined a set of “core” H3K4me1 immune and H3K27ac gut peaks as sets of overlapping peaks in cells that showed the strongest enrichment. Intersects were made using bedtools v2.24.0 default settings54. We selected 6 immune cell types for H3K4me1 and 3 gut cell types for H3K27ac (Supplementary Table 2). We also chose controls (Supplementary Table 2) from non-immune and non-gut cell types with similar density of peaks in the fine-mapped regions as compared to immune/gut cell types to confirm the tissue-specificity of the overlap. We used the phenotype assignments (described above) in dissecting the enrichment for the CD and UC signals. Sixty-five CD and 21 UC signals that were mapped to ≤ 50 variants were used in this analysis.
Published eQTL summary statistics
We used eQTL summary statistics from three published studies:
Peripheral blood eQTLs from the GODOT study29 of 2,752 twins, reporting loci with MAF>0.5%. Imputation was performed using the 1000 genomes reference panel11.
Peripheral blood eQTLs from the Westra et al. study30 of 8,086 individuals, including variants with MAF>5%. Imputation was performed using the HapMap 2 CEU population reference panel55.
CD14+ monocyte eQTLs from Table S2 in Fairfax et al.31, comprised of 432 European individuals, measured in a naïve state and after stimulation with interferon-γ or lipopolysaccharide (for 2 or 24 hours), reporting loci with MAF>4% and FDR<0.05. Imputation was performed using the 1000 genomes reference panel10.
Processing and quality control of new eQTL ULg dataset
A detailed description of the ULg dataset is in preparation (Momozawa et al., in preparation). Briefly, we collected venous blood and intestinal biopsies at three locations (ileum, transverse colon and rectum) from 350 healthy individuals of European descent, average age 54 (range 17-87), 56% female. SNPs were genotyped on Illumina Human OmniExpress v1.0 arrays interrogating 730,525 variants, and SNPs and individuals were subject to standard QC procedures using call rate, Hardy-Weinberg equilibrium, MAF ≥ 0.05, and consistency between declared and genotype-based sex as criteria. We further imputed genotypes at ~7 million variants on the entire cohort using the Impute2 software package13 and the 1,000 Genomes Project as reference population (Phase 3 integrated variant set, released 12 Oct 2014) 11,14. From the blood, we purified CD4+, CD8+, CD19+, CD14+ and CD15+ cells by positive selection, and platelets (CD45-negative) by negative selection. RNA from all leucocyte samples and intestinal biopsies was hybridized on Illumina Human HT-12 arrays v4. After standard QC, raw fluorescent intensities were variance stabilized56 and quantile normalized57 using the lumi R package58, and were corrected for sex, age, smoking status, number of probes with expression level significantly above background as fixed effects and array number (sentrix id) as random effect. For each probe with measureable expression (detection P value < 0.05 in >25% of samples) we tested for cis-eQTLs at all variants within a 500 kbp window. The nominal P value of the best SNP within a cis-window was Sidak-corrected for the window-specific number of independent tests. The number of independent test in each window was estimated exactly in the same manner as for the number of independent test for fine-mapping methods (Establishing a P value threshold, Methods). We estimated false discovery rates (q-values) from the resulting P values across all probes using the qvalue R package59. 480 cis-eQTL with FDR ≤ 0.10 for which the lead SNPs (i.e. the SNP yielding the best P value for the cis-eQTL) mapped within the 97 high-density regions (94 fine-mapped plus 3 unresolved) were retained for further analyses.
Naïve colocalization using lead SNPs
We calculated the number of IBD credible sets that contain a lead eQTL variant in a particular tissue (“observed”). This number is then compared to the background number of overlaps (“expected”):
where Ni is the total number of variants in region i in 1000 genomes with an allele frequency greater than a certain threshold (equal to the threshold used for the original eQTL study), Ci is the number of these variants that lie in IBD credible sets, and S is a set of regions that have at least one significant eQTL. We simulated 1,000 trials per region with binomial probability equal to the regional background overlap rate: Empirical P values were estimated by comparing the observed number of overlaps with the simulated number of the overlaps. More specifically, P value is defined as the proportion of trails that have equal or more overlaps in the simulations than the observed.
Frequentist colocalization using conditional P values
We next used conditional association to test for evidence of colocalization, as described in Nica et al.25. This method compares the P value of association for the lead SNP of an eQTL before and after conditioning on the SNP with the highest posterior in the credible set, and measures the drop in –log(P). An empirical P value for this drop is then calculated by comparing it to the drop for all variants in the high-density region. Because this method requires full genotypes we could only apply it to the ULg dataset (MAF > 5%). An empirical P value ≤ 0.05 was considered as evidence that the corresponding credible set is colocalized with the corresponding cis-eQTL. To evaluate whether our fine-mapping associations colocalized with cis-eQTL more often than expected by chance we counted the number of credible sets affecting at least one cis-eQTL with P≤0.05, and compared how often this number was matched or exceeded by 1,000 sets of variants that were randomly selected yet distributed amongst the loci in accordance with the real credible sets. The number of variants per set is same as the number of credible sets in this eQTL analysis (MAF matched, size≤50), shown in Extended Data Table 2.
Bayesian colocalization using Bayes factors
Finally, we used the Bayesian colocalization methodology described by Giambartolomei et al.60, modified to use the credible sets and posteriors generated by our fine-mapping methods (similarly only applicable to the ULg full genotype data). The method takes as input a pair of IBD and eQTL signals, with corresponding credible sets SIBD and SeQTL, and posteriors for each variant (with ). Credible sets and posteriors were generated for eQTL signals using the Bayesian quantitative association mode in SNPTest (with default parameters), with credible sets in regions with multiple independent signals generated conditional on all other signals. Our method calculates a Bayes factor (BF) summarizing the evidence in favor of a colocalized model (i.e. a single underlying causal variant between the IBD and eQTL signals) compared to a non-colocalized model (where different causal variants are driving the two signals), given by the ratio of marginal likelihoods
The marginal likelihood for the colocalized model (i.e. hypothesis H4 in Giambartolomei et al.) is given by
and the marginal likelihood for the model where the signals are not colocalized (i.e., hypothesis H3) is given by:
In both cases, N is the total number of variants in the region. We only count towards N variants that have r2 > 0.2 with either the lead eQTL variant or the lead IBD variant.
To measure enrichment in colocalization BFs compared to the null, we carried out a permutation analysis. In this analysis, we randomly reassigned eQTL signals to new fine-mapping regions to generate a set of simulated null datasets. This is carried out using the following scheme on variants and credible sets with the same MAF cut-off as the eQTL dataset (ULg, MAF > 5%):
Estimate the standarized effect size βg for each eQTL signal g, equal to standard deviation increase in gene expression for each dose of the minor allele.
Randomly reassign each eQTL signal to a new fine-mapping region, and then select a new causal variant with a MAF within 1 percentage point of the lead variant from the real signal. If multiple such variants exist, select one at random. If no such variants exist, pick the variant with the closest MAF.
Generate new simulated gene expression signals for each individual from Normal where xj is the individual’s minor allele dosage at the new causal variant and f is the minor allele frequency.
Carry out fine-mapping and calculate colocalization BFs for each pair of (real) IBD signal and (simulated) eQTL signal.
Repeat stages 2-4 1000 times for each tissue type
We can use these permuted BFs to calculate P values for each IBD credible set, given by the proportion of time the permuted BFs were as large or greater than the one observed in the real dataset. To generate a high-quality set of colocalized eQTL and IBD signals, we take all IBD signals that have the colocalization BF > 2, P < 0.01 and r2 (with the eQTL variant) >0.8.
Extended Data
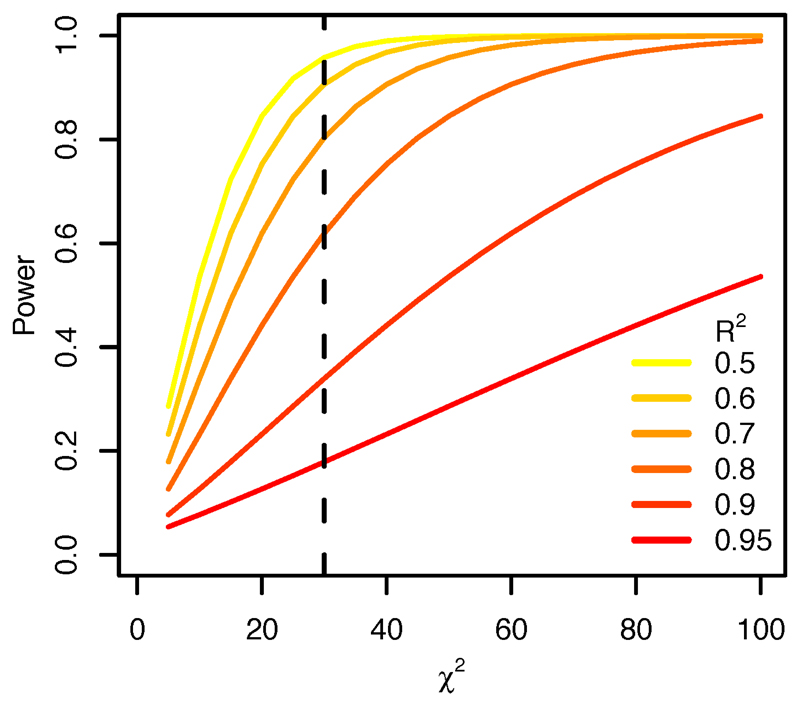
Extended Data Figure 1. Power of the fine-mapping analysis.
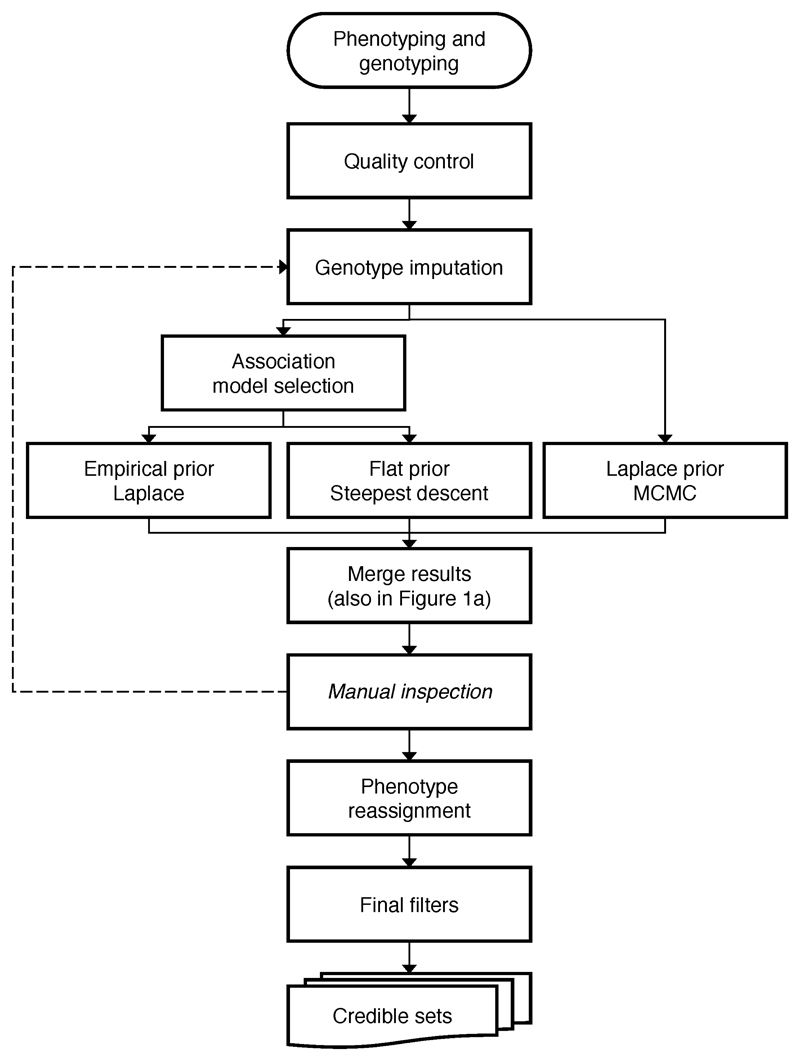
Extended Data Figure 2. Procedures in the fine-mapping analysis.
Details for each stage are described in Methods. The dashed line means the imputation was performed only once after the manual inspection (not iteratively).
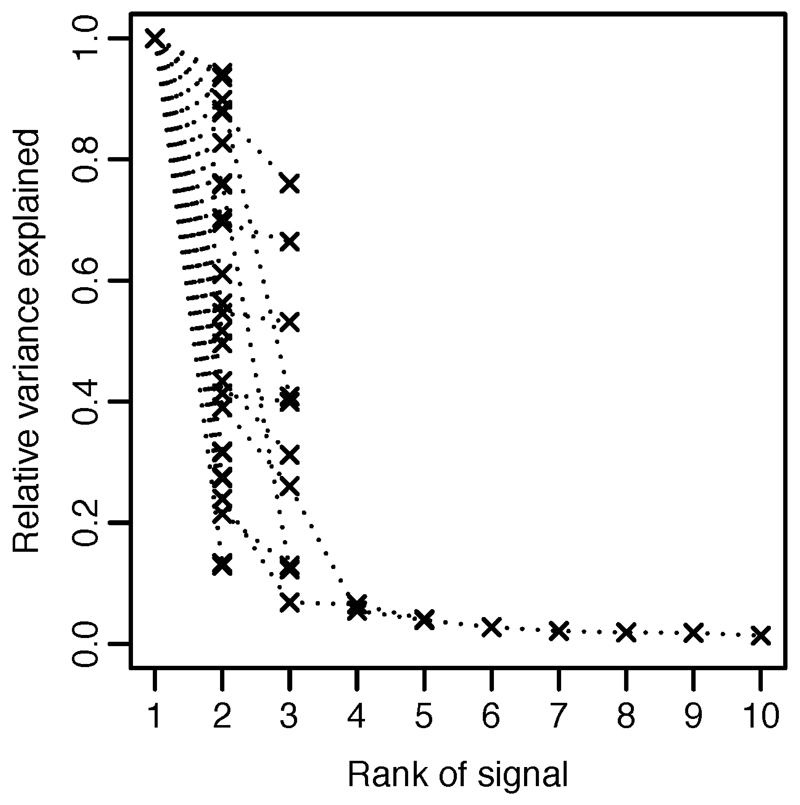
Extended Data Figure 3. Variance explained.
Variance explained by secondary, tertiary, … variants as a fraction of the primary signal at each locus.
Extended Data Figure 4. Functional annotations.
a, Functional annotation for 45 variants having posterior probability > 50%. b, Functional annotation for 116 association signals that are fine-mapped to ≤ 50 variants. Annotations are defined in Methods. We additionally grouped eQTLs into “Immune/Blood” (CD4+, CD8+, CD19+, CD14+ CD15+, platelets) and “Gut” (ileum, transverse colon and rectum). The eQTLs were generated from the ULg dataset using the “frequentist colocalization using conditional P values” approach (Methods).
Extended Data Figure 5. Size of credible sets.
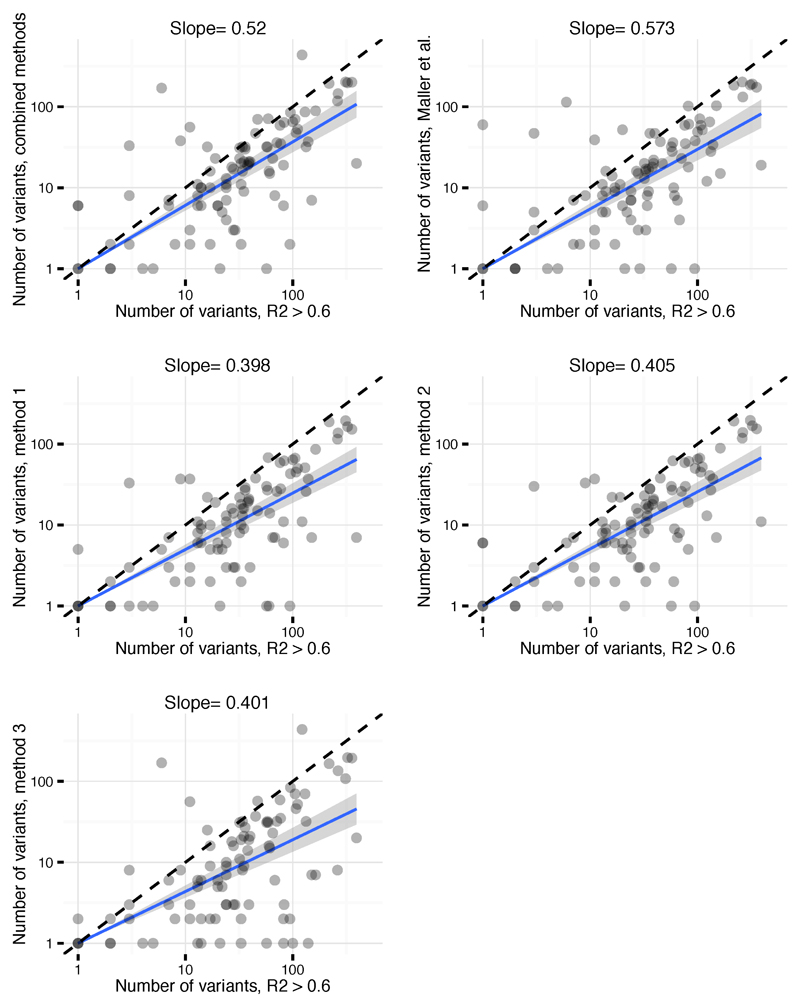
Comparison of credible set sizes for primary signals using each of our fine-mapping methods (methods 1, 2 and 3), the combined approach (as adopted in final results) and the approach described in Maller et al.6 (y axis) and the R2 > 0.6 cut-off (x axis). Fine-mapping maps most signals to smaller numbers of variants.
Extended Data Figure 6. Distributions of the allele frequency and the imputation quality.
Panels a-c: distribution of the risk allele frequency for 45 variants having > 50% posterior probability plotted against (a) posterior probability, (b) significance of the association as –log10(P), and (c) odds ratio of the association. Variants are color coded according to their functions. Odds ratio for IBD associations was the larger of odds ratios for CD and UC. Panels d-f: distribution of imputation quality (INFO measure from the IMPUTE2 program) for variants having MAF ≥5% (d), between 5% and 1% (e) and <1% (f).
Extended Data Figure 7. Merging and adjudicating signals across methods.
The number of signals for each method is shown in the brackets, and for each method a black bar indicates a signal with p < 1.35 x 10–6, and a grey bar a signal that does not reach that threshold. The colored bar shows the final status of each signal after merging and model selection (Methods). “Low info” corresponds to INFO < 0.8 (the threshold used for signals reported by 1 or 2 methods) and “rare and imputed” to MAF < 0.01 and no genotyped variants in the credible set, regardless of INFO (Methods).
Extended Data Table 1. Study samples.
Genotyped samples in each batch for healthy controls (Control), Crohn’s disease (CD) and ulcerative colitis (UC). Batches were grouped into cohorts for further analysis (Controlling for population structure, batch effects and other confounders, Methods).
| Batch | Control | CD | UC | Cohort |
|---|---|---|---|---|
| IMSGC | 5740 | 0 | 0 | imbalanced |
| NIDDK | 1786 | 3653 | 3020 | balanced |
| D. Ellinghaus | 4559 | 2696 | 1006 | balanced |
| E. Theatre | 713 | 1109 | 559 | balanced |
| H. Huang | 3 | 551 | 316 | imbalanced |
| J. Barrett | 4397 | 2715 | 2835 | balanced |
| K. Fransen | 1598 | 1234 | 430 | balanced |
| L. Jostins | 1354 | 1252 | 1063 | balanced |
| P. Gregersen | 1611 | 0 | 0 | imbalanced |
| R. Duerr | 1696 | 321 | 1611 | balanced |
| S. Rich | 4259 | 0 | 0 | imbalanced |
| S. Sommeren | 107 | 77 | 201 | balanced |
| S. Vermeire | 922 | 1539 | 838 | balanced |
| T. Balschun | 5511 | 1882 | 1683 | balanced |
| T. Haritunians | 1 | 1938 | 1066 | imbalanced |
Extended Data Table 2. Colocalization with eQTL.
The number of IBD credible sets that colocalize with eQTLs using the naïve, frequentist and Bayesian approaches. Significant observations are boldfaced. ‘Number of credible sets’ reports the number of credible sets that have MAF above the cut-off.
| Tissue/cell line | Method | Overlaps observed | Overlaps Expected | P value | Dataset | MAF cut-off | Number of credible sets |
|---|---|---|---|---|---|---|---|
| whole blood | Naïve | 3 | 3.7 | 0.746 | GODOT | 0.005 | 113 |
| whole blood | 8 | 4.2 | 0.060 | Westra | 0.05 | 95 | |
| CD14 naïve | 8 | 2.7 | 0.001 | Fairfax | 0.04 | 98 | |
| CD14 IFN stimulated | 4 | 3.2 | 0.398 | Fairfax | 0.04 | 98 | |
| CD14 LPS 2h stimulated | 1 | 2.1 | 0.869 | Fairfax | 0.04 | 98 | |
| CD14 LPS 24h stimulated | 5 | 2.5 | 0.106 | Fairfax | 0.04 | 98 | |
| CD4 | 3 | 0.4 | 0.005 | ULg | 0.05 | 95 | |
| CD8 | 1 | 0.3 | 0.306 | ULg | 0.05 | 95 | |
| CD14 | 0 | 0.2 | 1.000 | ULg | 0.05 | 95 | |
| CD15 | 1 | 0.2 | 0.199 | ULg | 0.05 | 95 | |
| CD19 | 0 | 0.1 | 1.000 | ULg | 0.05 | 95 | |
| platelets | 0 | 0.0 | 1.000 | ULg | 0.05 | 95 | |
| ileum | 2 | 0.3 | 0.020 | ULg | 0.05 | 95 | |
| colon | 1 | 0.2 | 0.202 | ULg | 0.05 | 95 | |
| rectum | 1 | 0.2 | 0.189 | ULg | 0.05 | 95 | |
| CD4 | Frequentist | 6 | 1.9 | 0.013 | ULg | 0.05 | 95 |
| CD8 | 3 | 1.5 | 0.186 | ULg | 0.05 | 95 | |
| CD14 | 4 | 2.3 | 0.180 | ULg | 0.05 | 95 | |
| CD15 | 1 | 1.8 | 0.863 | ULg | 0.05 | 95 | |
| CD19 | 0 | 1.4 | 1.000 | ULg | 0.05 | 95 | |
| platelets | 0 | 0.1 | 1.000 | ULg | 0.05 | 95 | |
| ileum | 4 | 1.1 | 0.018 | ULg | 0.05 | 95 | |
| colon | 3 | 1.7 | 0.216 | ULg | 0.05 | 95 | |
| rectum | 4 | 1.4 | 0.039 | ULg | 0.05 | 95 | |
| CD4 | Bayesian | 4 | 1.0 | 0.010 | ULg | 0.05 | 95 |
| CD8 | 1 | 0.8 | 0.566 | ULg | 0.05 | 95 | |
| CD14 | 1 | 0.9 | 0.595 | ULg | 0.05 | 95 | |
| CD15 | 0 | 0.7 | 1.000 | ULg | 0.05 | 95 | |
| CD19 | 0 | 0.6 | 1.000 | ULg | 0.05 | 95 | |
| platelets | 0 | 0.1 | 1.000 | ULg | 0.05 | 95 | |
| ileum | 2 | 0.4 | 0.069 | ULg | 0.05 | 95 | |
| colon | 3 | 0.8 | 0.040 | ULg | 0.05 | 95 | |
| rectum | 2 | 0.6 | 0.124 | ULg | 0.05 | 95 | |
Extended Data Table 3. Genomic inflation.
Genomic inflation factors and LD score regression intercept for Crohn’s disease (CD), ulcerative colitis (UC) and both (IBD). a, Genomic inflation factors using the first four, five and six principal components. The factors were calculated using 2,853 background variants from the Immunochip. b, Genomic inflation factors for subsets of the data (using five principal components for the same 2,853 background variants). Balanced, imbalanced and down-sampled cohorts are defined in Methods. Numbers in brackets indicate the 95% confidence interval for the inflation factors (only estimated for the down-sampled cohorts). c, LD score regression intercept and genomic inflation factors (λGC and λ1000) from the largest IBD meta-analyses with genome-wide data (CD:GWAS and UC:GWAS).
| a | ||||
|---|---|---|---|---|
| CD | UC | IBD | ||
| PC 1-4 | 1.41 | 1.31 | 1.38 | |
| PC 1-5 | 1.29 | 1.25 | 1.31 | |
| PC 1-6 | 1.28 | 1.25 | 1.32 | |
| b | ||||
|---|---|---|---|---|
| CD | UC | IBD | ||
| Balanced cohort | 1.24 | 1.21 | 1.20 | |
| Down-sampled (balanced) | 1.24 (1.14-1.36) | 1.21 (1.11-1.31) | 1.23 (1.14-1.36) | |
| Imbalanced cohort | 1.02 | 1.08 | 1.00 | |
| Down-sampled (imbalanced) | 1.08 (0.97-1.23) | 1.04 (0.96-1.16) | 1.07 (0.95-1.21) | |
| All samples | 1.29 | 1.25 | 1.31 | |
| c | ||||||
|---|---|---|---|---|---|---|
| genome-wide | “background SNPS” | |||||
| LDscore intercept | λGC | λ1000 | λGC | λ1000 | ||
| CD: lmmunochip (this study) | - | - | - | 1.29 | 1.012 | |
| CD: GWAS | 1.09 | 1.23 | 1.014 | 1.28 | 1.016 | |
| UC: lmmunochip (this study) | - | - | - | 1.25 | 1.012 | |
| UC: GWAS | 1.09 | 1.29 | 1.016 | 1.21 | 1.012 | |
Supplementary Material
Acknowledgements
We thank Mariya Khan and Bang Wong for their assistance in designing illustrations, and Katie de Lange for helpful comments on the Supplementary Methods. Support from the following grants: M.J.D. and R.J.X.: P30DK43351, U01DK062432, R01DK64869, Helmsley grant 2015PG-IBD001 and CCFA. C.A.A. and J.C.B: Wellcome Trust grant 098051. M.G.:WELBIO (CAUSIBD), BELSPO (BeMGI), Fédération Wallonie-Bruxelles (ARC IBD@Ulg), and Région Wallonne (CIBLES, FEDER). H.H.: ASHG/Charles J. Epstein Trainee Award. J.L.: Wellcome Trust 098759/Z/12/Z. D.M.: Olle Engkvist Foundation and Swedish Research Council (grants 2010-2976 and 2013-3862). R.K.W.: VIDI grant (016.136.308) from the Netherlands Organization for Scientific Research. J.D.R.: Canada Research Chair, NIDDK grants DK064869 and DK062432, CIHR #GPG-102170 from the Canadian Institutes of Health, GPH-129341 from Genome Canada and Génome Québec, and Crohn’s Colitis Canada. J.H.C.: DK062429, DK062422, DK092235, DK106593, and the Sanford J. Grossman Charitable Trust. R.H.D.: Inflammatory Bowel Disease Genetic Research Chair at the University of Pittsburgh, U01DK062420 and R01CA141743. E.D.: Marie-Curie Fellowship. A-S.G: FNRS and Fonds Léon Fredericq fellowships. J.H.: Örebro University Hospital Research Foundation and the Swedish Research Council (grant no. 521 2011 2764). C.G.M. and M.P.: NIHR Biomedical Research Centre awards to Guy's & St Thomas' NHS Trust / King's College London and to Addenbrooke’s Hospital / University of Cambridge School of Clinical Medicine. D.E.: German Federal Ministry of Education and Research (SysInflame grant 01ZX1306A), DFG Excellence Cluster No. 306 “Inflammation at Interfaces”. A.F: professorship of Foundation for Experimental Medicine (Zuerich, Switzerland). D.P.B.M.: DK062413, AI067068, U54DE023789-01, 305479 from the European Union, and The Leona M. and Harry B. Helmsley Charitable Trust. Additional acknowledgements for the original data are in the Supplementary Information.
Footnotes
Author Contributions Overall project supervision and management: M.J.D. J.C.B, M.G. Fine-mapping algorithms: H.H., M.F., L.J. TFBS analyses: H.H., K.F. Epigenetic analyses: M.U.M., G.T. eQTL dataset generation: E.L., E.T., J.D., E.D., M.E., R.M., M.M., Y.M., V.D., A.G. eQTL analyses: M.F., J.D., L.J., A.C. Variance component analysis: T.M., M.F. Contribution to overall statistical analyses: G.B. Primary drafting of the manuscript: M.J.D., J.C.B, M.G., H.H., L.J. Major contribution to drafting of the manuscript: M.F., M.U.M., J.H.C., D.P.B.M., J.D.R., C.G.M., R.H.D., R.K.W. The remaining authors contributed to the study conception, design, genotyping QC and/or writing of the manuscript. All authors saw, had the opportunity to comment on, and approved the final draft.
Author Information The study protocols were approved by the institutional review board (IRB) at each center involved with recruitment. Informed consent and permission to share the data were obtained from all subjects, in compliance with the guidelines specified by the recruiting center's IRB.
The authors declare no competing financial interests.
Code availability
Computer code used in this study is provided in the ‘Software availability’ sections in Supplementary Methods.
Data availability
The data that support the findings of this study are available from the international IBD Genetics Consortium but restrictions apply to the availability of these data, which were used under license for the current study, and so are not publicly available. Data are however available from the authors upon reasonable request and with permission of the international IBD Genetics Consortium.
References
- 1.Kappelman MD, et al. Direct health care costs of Crohn's disease and ulcerative colitis in US children and adults. Gastroenterology. 2008;135:1907–1913. doi: 10.1053/j.gastro.2008.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Molodecky NA, et al. Increasing incidence and prevalence of the inflammatory bowel diseases with time, based on systematic review. Gastroenterology. 2012;142:46–54.e42. doi: 10.1053/j.gastro.2011.10.001. quiz e30. [DOI] [PubMed] [Google Scholar]
- 3.Jostins L, et al. Host–microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 2012;491:119–124. doi: 10.1038/nature11582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu JZ, et al. Association analyses identify 38 susceptibility loci for inflammatory bowel disease and highlight shared genetic risk across populations. Nat Genet. 2015 doi: 10.1038/ng.3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van de Bunt M, et al. Evaluating the Performance of Fine-Mapping Strategies at Common Variant GWAS Loci. PLoS Genet. 2015;11:e1005535. doi: 10.1371/journal.pgen.1005535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maller JB, et al. Bayesian refinement of association signals for 14 loci in 3 common diseases. Nat Genet. 2012;44:1294–1301. doi: 10.1038/ng.2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang J, et al. FTO genotype is associated with phenotypic variability of body mass index. Nature. 2012;490:267–272. doi: 10.1038/nature11401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.International Multiple Sclerosis Genetics Consortium (IMSGC) et al. Analysis of immune-related loci identifies 48 new susceptibility variants for multiple sclerosis. Nat Genet. 2013;45:1353–1360. doi: 10.1038/ng.2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Onengut-Gumuscu S, et al. Fine mapping of type 1 diabetes susceptibility loci and evidence for colocalization of causal variants with lymphoid gene enhancers. Nat Genet. 2015;47:381–386. doi: 10.1038/ng.3245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.1000 Genomes Project Consortium et al. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.1000 Genomes Project Consortium. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jostins L. Using Next-Generation Genomic Datasets In Disease Association. The University of Cambridge; 2012. [Google Scholar]
- 13.Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5:e1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Howie B, Marchini J, Stephens M. Genotype imputation with thousands of genomes. G3 (Bethesda) 2011;1:457–470. doi: 10.1534/g3.111.001198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goyette P, et al. High-density mapping of the MHC identifies a shared role for HLA-DRB1*01:03 in inflammatory bowel diseases and heterozygous advantage in ulcerative colitis. Nat Genet. 2015 doi: 10.1038/ng.3176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rivas MA, et al. Deep resequencing of GWAS loci identifies independent rare variants associated with inflammatory bowel disease. Nat Genet. 2011;43:1066–1073. doi: 10.1038/ng.952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang J, et al. Genome partitioning of genetic variation for complex traits using common SNPs. Nat Genet. 2011;43:519–525. doi: 10.1038/ng.823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang H, Chanda P, Alonso A, Bader JS, Arking DE. Gene-based tests of association. PLoS Genet. 2011;7:e1002177. doi: 10.1371/journal.pgen.1002177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Momozawa Y, et al. Resequencing of positional candidates identifies low frequency IL23R coding variants protecting against inflammatory bowel disease. Nat Genet. 2011;43:43–47. doi: 10.1038/ng.733. [DOI] [PubMed] [Google Scholar]
- 20.Kheradpour P, Kellis M. Systematic discovery and characterization of regulatory motifs in ENCODE TF binding experiments. Nucleic Acids Res. 2014;42:2976–2987. doi: 10.1093/nar/gkt1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nechanitzky R, et al. Transcription factor EBF1 is essential for the maintenance of B cell identity and prevention of alternative fates in committed cells. Nat Immunol. 2013;14:867–875. doi: 10.1038/ni.2641. [DOI] [PubMed] [Google Scholar]
- 22.Trynka G, et al. Chromatin marks identify critical cell types for fine mapping complex trait variants. Nat Genet. 2013;45:124–130. doi: 10.1038/ng.2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Farh KK-H, et al. Genetic and epigenetic fine mapping of causal autoimmune disease variants. Nature. 2015;518:337–343. doi: 10.1038/nature13835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bernstein BE, et al. The NIH Roadmap Epigenomics Mapping Consortium. 2010;28:1045–1048. doi: 10.1038/nbt1010-1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nica AC, et al. Candidate causal regulatory effects by integration of expression QTLs with complex trait genetic associations. PLoS Genet. 2010;6:e1000895. doi: 10.1371/journal.pgen.1000895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lappalainen T, et al. Transcriptome and genome sequencing uncovers functional variation in humans. Nature. 2013;501:506–511. doi: 10.1038/nature12531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wallace C, et al. Statistical colocalization of monocyte gene expression and genetic risk variants for type 1 diabetes. Human Molecular Genetics. 2012;21:2815–2824. doi: 10.1093/hmg/dds098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dubois PCA, et al. Multiple common variants for celiac disease influencing immune gene expression. Nat Genet. 2010;42:295–302. doi: 10.1038/ng.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wright FA, et al. Heritability and genomics of gene expression in peripheral blood. Nat Genet. 2014;46:430–437. doi: 10.1038/ng.2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Westra H-J, et al. Systematic identification of trans eQTLs as putative drivers of known disease associations. Nat Genet. 2013;45:1238–1243. doi: 10.1038/ng.2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fairfax BP, et al. Innate immune activity conditions the effect of regulatory variants upon monocyte gene expression. 2014;343 doi: 10.1126/science.1246949. 1246949–+ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ripke S, et al. Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511:421–427. doi: 10.1038/nature13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Franke A, et al. Genome-wide meta-analysis increases to 71 the number of confirmed Crohn's disease susceptibility loci. Nat Genet. 2010;42:1118–1125. doi: 10.1038/ng.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Spain SL, Barrett JC. Strategies for fine-mapping complex traits. Human Molecular Genetics. 2015;24:R111–9. doi: 10.1093/hmg/ddv260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.1000 Genomes Project Consortium et al. A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.UK10K Consortium et al. The UK10K project identifies rare variants in health and disease. Nature. 2015 doi: 10.1038/nature14962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nejentsev S, Walker N, Riches D, Egholm M, Todd JA. Rare variants of IFIH1, a gene implicated in antiviral responses, protect against type 1 diabetes. 2009;324:387–389. doi: 10.1126/science.1167728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huang J, Ellinghaus D, Franke A, Howie B, Li Y. 1000 Genomes-based imputation identifies novel and refined associations for the Wellcome Trust Case Control Consortium phase 1 Data. Eur J Hum Genet. 2012;20:801–805. doi: 10.1038/ejhg.2012.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Roadmap Epigenomics Consortium et al. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317–330. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shah TS, et al. optiCall: a robust genotype-calling algorithm for rare, low-frequency and common variants. 2012;28:1598–1603. doi: 10.1093/bioinformatics/bts180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Price AL, et al. Long-Range LD Can Confound Genome Scans in Admixed Populations. Am J Hum Genet. 2008;83:132–135. doi: 10.1016/j.ajhg.2008.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Anderson E, et al. LAPACK Users' Guide. Society for Industrial and Applied Mathematics; 1999. [Google Scholar]
- 43.Yang J, et al. Genomic inflation factors under polygenic inheritance. European Journal of Human Genetics. 2011;19:807–812. doi: 10.1038/ejhg.2011.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bulik-Sullivan BK, et al. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat Genet. 2015 doi: 10.1038/ng.3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.de Lange KM, et al. Genome-wide association study implicates immune activation of multiple integrin genes in inflammatory bowel disease. Nat Genet. 2017 doi: 10.1038/ng.3760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Delaneau O, Marchini J, Zagury J-F. A linear complexity phasing method for thousands of genomes. Nature Methods. 2011;9:179–181. doi: 10.1038/nmeth.1785. [DOI] [PubMed] [Google Scholar]
- 47.Delaneau O, Zagury J-F, Marchini J. Improved whole-chromosome phasing for disease and population genetic studies. Nature Methods. 2012;10:5–6. doi: 10.1038/nmeth.2307. [DOI] [PubMed] [Google Scholar]
- 48.Morris JA, Randall JC, Maller JB, Barrett JC. Evoker: a visualization tool for genotype intensity data. 2010;26:1786–1787. doi: 10.1093/bioinformatics/btq280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jostins L, McVean G. Trinculo: Bayesian and frequentist multinomial logistic regression for genome-wide association studies of multi-category phenotypes. Bioinformatics. 2016 doi: 10.1093/bioinformatics/btw075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yang J, Lee SH, Goddard ME, Visscher PM. GCTA: A Tool for Genome-wide Complex Trait Analysis. The American Journal of Human Genetics. 2011;88:76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Madsen P, Su G, Labouriau R, Christensen F. DMU-a package for analyzing multivariate mixed models. Proceedings of the Ninth World Congress on Genetics Applied to Livestock Production. 2010 [Google Scholar]
- 52.Cox DR, Snell EJ. Analysis of Binary Data, Second Edition. CRC Press; 1989. [Google Scholar]
- 53.D'haeseleer P. What are DNA sequence motifs? 2006;24:423–425. doi: 10.1038/nbt0406-423. [DOI] [PubMed] [Google Scholar]
- 54.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gibbs RA, et al. The International HapMap Project. Nature. 2003;426:789–796. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- 56.Lin SM, Du P, Huber W, Kibbe WA. Model-based variance-stabilizing transformation for Illumina microarray data. Nucleic Acids Res. 2008;36:e11–e11. doi: 10.1093/nar/gkm1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 58.Du P, Kibbe WA, Lin SM. lumi: a pipeline for processing Illumina microarray. 2008;24:1547–1548. doi: 10.1093/bioinformatics/btn224. [DOI] [PubMed] [Google Scholar]
- 59.Dabney A, Storey JD, Warnes G. Q-value estimation for false discovery rate control. R package version. 2006;1 [Google Scholar]
- 60.Giambartolomei C, et al. Bayesian test for colocalisation between pairs of genetic association studies using summary statistics. PLoS Genet. 2014;10:e1004383. doi: 10.1371/journal.pgen.1004383. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.