Abstract
Purpose
The molecular features that account for the distinct histology and aggressive biological behavior of Gleason pattern 4 (Gp4) versus Gp3 prostate cancer (PCa), and whether Gp3 tumors progress directly to Gp4, remain to be established.
Experimental Design
Whole exome sequencing and transcriptome profiling of laser-capture microdissected adjacent Gp3 and cribiform Gp4 were used to determine the relationship between these entities.
Results
Sequencing confirmed that adjacent Gp3 and Gp4 were clonal based on multiple shared genomic alterations. However, large numbers of unique mutations in the Gp3 and Gp4 tumors showed that the Gp4 were not derived directly from the Gp3. Remarkably, the Gp3 tumors retain their indolent appearing morphology despite acquisition of multiple genomic alterations including tumor suppressor losses. Although there were no consistent genomic alterations that distinguished Gp3 from Gp4, pairwise transcriptome analyses identified increased c-Myc and decreased p53 activity in Gp4 versus adjacent clonal Gp3 foci.
Conclusions
These findings establish that at least a subset of Gp3 and aggressive Gp4 tumors have a common origin, and support a branched evolution model wherein the Gp3 and Gp4 tumors emerge early from a common precursor and subsequently undergo substantial divergence. Genomic alterations detectable in the Gp3 may distinguish these tumors from truly indolent Gp3. Screening for a panel of these genomic alterations in men who have prostate biopsies showing only Gp3 (Gleason score 6, Gs6) may allow for more precise selection of men who can be safely managed by active surveillance versus those who may benefit from further intervention.
Keywords: prostate cancer, Gleason score, tumor progression, whole exome sequencing, tumor suppressor genes
Introduction
Prostate cancer (PCa) in many men will remain clinically insignificant, and identification of those who may benefit from radical prostatectomy (RP) or radiation therapy is currently a major challenge. Pathological evaluation of PCa uses the Gleason grading system, which is based on histological features with Gleason pattern 3, 4, and 5 (Gp3, Gp4, and Gp5) roughly corresponding to low, intermediate and high grades. Many cases contain more than one pattern, and this heterogeneity is captured using the Gleason score, which is sum of the two major patterns in the sample. Importantly, tumors containing only Gp3 glands (and thereby having a Gleason score 3+3=6, Gs6) have limited metastatic potential, as only a very small fraction of Gs6 patients who undergo RP, and who are confirmed to have no Gp4 or Gp5 upon pathological examination of the RP specimen, will relapse with metastatic PCa (1-3).
These observations have led to increased acceptance of active surveillance (AS) for men with Gs6 on biopsy, wherein treatment is deferred and men undergo interval repeat biopsies to detect potentially more aggressive disease. Higher grade tumors are detected upon subsequent biopsies in about 20-40% of these patients, who may then go on to RP or radiation therapy. Significantly, this tumor upgrading in many cases reflects incomplete sampling on the original biopsies rather than progression to Gp4 or Gp5, as higher grades are detected at comparable frequencies when patients undergo RP shortly after the initial biopsies (4-8). Unfortunately, concerns that more aggressive tumors may have been missed on biopsies, or that Gp3 tumors may progress to metastatic Gp4 or Gp5 tumors during surveillance, lead many patients who are AS candidates to opt for immediate surgery or radiation, from which they do not benefit.
A particularly controversial question with clinical implications has been whether Gp3 tumors represent a molecularly distinct entity that may be clinically insignificant, versus being related to Gp4 or Gp5 tumors with potential to progress to these higher grades over time. Gp4 glands in many Gs7 tumors are found adjacent to Gp3, but these may be independent tumor clones as many RP specimens contain two or more tumor foci that are genetically distinct (9-12). However, recent data have challenged the view that all Gp3 tumors are clinically insignificant, as at least a subset are clonally-related to higher grade tumors (13-17). Using anti-ERG immunohistochemistry (IHC) to assess for the TMPRSS2:ERG gene fusion, which occurs very early in approximately half of PCa and results in ERG expression (18-20), we found marked concordance for ERG expression in adjacent Gp3 and Gp4 foci. Moreover, in four cases we identified the TMPRSS2:ERG fusion breakpoints and showed they were identical in the adjacent Gp3 and Gp4 components (13).
In this study we used whole exome sequencing (WES) and transcriptome profiling to identify mechanisms mediat ing development and progression of these clonal Gp3 and Gp4 tumor components. Significantly, the data do not show a straightforward linear progression from Gp3 to Gp4, but instead indicate that both emerge early from a common precursor and then undergo extensive independent evolution, including acquisition of tumor suppressor losses in Gp3 foci. Molecular analyses of Gp3 in Gs6 biopsies may identify a subset that is likely to be associated with a higher grade tumor, or in which a higher grade tumor is likely to emerge.
Materials and Methods
Tissue Collection
Collection of tissues from RP specimens was in compliance with IRB-approved protocols. Prospectively-collected tissues were fixed in PAXgene, which preserves RNA integrity as well as tissue architecture after embedding in paraffin, and these samples were used for RNA isolation (21). Informed consent was given by all patients, and all samples were de-identified per institutional policies. See Supplementary Table 1 for clinical characteristics. Three of these men have to date relapsed with metastatic disease, but we have not included this in the analysis as the sample size is too small and follow up time is too short to draw conclusions with respect to prognostic biomarkers.
Histology
Samples were stained with H&E, ERG, and PIN-4, and laser capture microdissected (LCM) as previously described (13). No differences were observed between H&E, ERG, and PIN-4 staining for tissues fixed in formalin versus PAXgene. For c-Myc and MAD automated immunohistochemical staining, 5 μm FFPE sections were baked for 1 hour at 60°C. After cooling to room temperature, antigen retrieval was performed using PT Link (Dako). Staining was with EnVision FLEX High pH kit (Dako, cat# K8000021-2) following the manufacturer's recommendations. Sections were incubated with 300 μL primary antibody (anti-c-Myc, Abcam cat# ab32072, 1:200 dilution; anti-MAD, Sigma-Aldrich cat# HPA001599, 1:1000 dilution) for 60 minutes at room temperature.
RNA and DNA Purification
RNA and DNA were extracted simultaneously using the AllPrep DNA/RNA FFPE Kit (Qiagen) following the manufacturer's protocol with modifications. The film to which LCM cells adhered was removed using a disposable blade and immersed into buffer PKD with proteinase K. Following centrifugation and separation of the DNA fraction (the pellet including the polymer film), digestion with buffer ATL and proteinase K was performed at 56°C for 16 hours. All elution steps were 10-minute on-column incubations with water or buffer. DNA and RNA quantifications were with PicoGreen and RiboGreen (Life Technologies). RNA quality was assessed with the RNA ScreenTape (Agilent Technologies) and RNA with RIN values ≥5 were retained.
Gene Expression Analysis
100 ng of RNA from PAXgene-fixed tissues was prepared for microarray analysis using the Ovation Pico WTA System (NuGEN). 3 μg of cDNA was converted into sense-strand cDNA using WT-Ovation Exon (NuGEN) and 5 μg of sense-strand cDNA was fragmented and labeled using Encore Biotin (NuGEN). Labeled libraries were hybridized to Affymetrix Human Exon 1.0 GeneChips. Each microarray .CEL file was self-normalized using the SCAN.UPC application (22) for R/Bioconductor. SCAN “estimates” were used for subsequent analyses. Version 18 of the Affymetrix Human Exon Array 1.0 annotation was downloaded from BrainArray (23). Unsupervised hierarchical clustering was performed with the Multi-Experiment Viewer using the Pearson Correlation metric (24). Correlation with signatures of existing gene sets was performed with Gene Set Enrichment Analysis.
Each Gp3 and Gp4 pair was compared collectively and individually to select gene sets enriched on a per-patient basis, and recurrently enriched gene sets (enriched in >50% of pairs) with q-values (false discovery rates) less than 0.25 were selected. Genes differentially regulated by ±1.5-fold between Gp3 and Gp4 were submitted to Ingenuity (Qiagen) for core pathway analysis and upstream regulator analysis. Recurrently upregulated pathways or upstream regulators (strong activation or inactivation in >25% of pairs) were identified. Microarray data has been deposited in the Gene Expression Omnibus under accession ID GSE52560.
Exome Sequencing and DNA Sequence Analysis
100 ng of genomic DNA from LCM tissue was prepared into libraries and sequenced as previously described (13,25). Alignment, deduplication, local realignment, and quality score recalibration were performed with BWA, PICARD and GATK. Mutation calling and annotation was performed using muTect, SomaticIndelDetector, and Oncotator (26,27). For any mutation called by MuTect or SomaticIndelDetector, mutations were confirmed by VarScan (28) requiring no greater than 90% strand bias, with mutations present in at least 5 reads and a minimum variant allele frequency of 20%. It was also required that the benign sample (obtained by LCM of an area on the block that was histologically benign) had at least 15× coverage with no more than 1 mutant read at the same position. It should be noted that low level detection of a mutation in the benign control could reflect contamination with tumor or a field effect. While none of the high allele frequency mutations detected in tumor foci were found at low frequency in the control samples (not shown), field effects cannot be ruled out (particularly if related to tumor suppressor losses). Only mutations covered or adjacent to hybrid capture probe target regions are reported. To be considered exclusive to a focus, it was required that the same position be covered at least 15× with no more than 1 mutant read at the same position in the other tumor focus.
To identify SCNAs, pre-processed and duplicate-removed BAM files were processed by the Python ngCGH package, comparing pairs of tumor BAM files to their matched benign control to generate pseudo-CGH probe estimates. Each probe consisted of a variable-length genomic window per 1,000 unique reads. Copy number change events were called from the median-centered log2 ratios of these values using the FASST2 circular binary segmentation algorithm and a hidden Markov model (HMM) with systemic quadratic correction in Nexus Copy Number (BioDiscovery, El Segundo, CA). To mark a region as gain or loss for a segment of the genome, 50 contiguous probes were required to be gained or lost at an HMM significance threshold of P < 10-6. Probes were required to be within 200 kilobases of each other for a contiguous gain or loss event. Thresholds were log2 ratios of −0.25 for shallow deletions, −0.5 for deep deletions and +0.25 for gains.
Cancer cell fraction (CCF) computation as described previously (29) required ploidy, purity, and variant allele fraction values. Ploidy was determined by copy number call from each segment of the genome as described above. A conservative estimate of 0.75 was presumed for the purity of all tumor foci based on review of the LCM slides. Only mutations with raw variant allele fractions of approximately 20% (corresponding to CCF ≥~35%) are reported. In cases where a mutation meets this criteria in one focus, the requirement for it to be considered to be shared was that the other focus/foci must harbor that mutation within 50% of the CCF for the first focus.
Mutations were individually inspected using the Integrative Genome Viewer (30). Sequence data has been deposited in the Sequence Read Archive under accession ID SRP010648. A matrix of GEO and SRA accession IDs is in Supplementary Table S8.
Phylogenetic Trees
Somatic mutation calling was performed on each tumor focus, considering the number of mutations marked as shared or unshared as defined by the above criteria. Each tip is drawn proportionally to the number of unshared somatic mutations between each focus, with branch and trunk lengths representing the number of shared somatic mutations.
Computer-aided Image Analysis
Serially stained slides were scanned using Pannoramic SCAN (3DHISTECH). Prior to scanning, and blinded to immunoreactivity to c-Myc and MAD staining, areas of Gp3 and G4 were marked on H&E slides by a board-certified pathologist. Using Pannoramic Viewer (3DHISTECH), corresponding regions marked on the H&E slide were selected on the c-Myc and MAD stained slides, cropped and exported as a TIFF file of either Gp3 or Gp4 regions. A composite training file containing randomly selected regions of Gp3 and Gp4 from c-Myc- or MAD-stained slides were imported into Definiens Developer XD 64 for automated image analysis (details in Supplementary Methods).
Statistics
Analyses for single comparisons between groups, or multiple comparisons within groups, were performed using either Student's t-test or ANOVA, respectively. P values were computed using GraphPad Prism 6.0 for MacOS X.
Results
Whole exome sequencing identifies shared mutations and SCNAs in adjacent Gp3 and Gp4 tumor foci
For this study we selected 18 RP specimens with Gs7 or higher that contained foci of adjacent Gp3 and Gp4 (with all Gp4 foci having cribriform morphology to clearly establish they were distinct from the Gp3), and used LCM to purify these foci in conjunction with an area of nonmalignant prostate (Fig. 1 and Supplementary Fig. S1). In 12 of the microdissected cases that yielded adequate DNA, we then performed whole exome sequencing. In each case we identified multiple mutations that were present at high allelic frequency in both the Gp3 and Gp4 foci, confirming their clonal relationship (Supplementary Table S2). This conclusion was further verified by targeted sequencing for a subset of these mutations (Supplementary Table S3). Some of the shared mutations were in genes known to function as oncogenes or tumor suppressors, and presumably represent truncal alterations that occurred in a common precursor (Fig. 2A). Amongst these were an activating H-Ras mutation (Q61R), which is uncommon in PCa and has not been described previously in Gp3 tumors. Other potentially-significant mutated genes included those encoding the Wnt antagonist SFRP2, the steroid metabolizing protein AKR1B15, the transcription factors E2F3 and HOXA5, and nonsense mutations in PDE8B and DDX21.
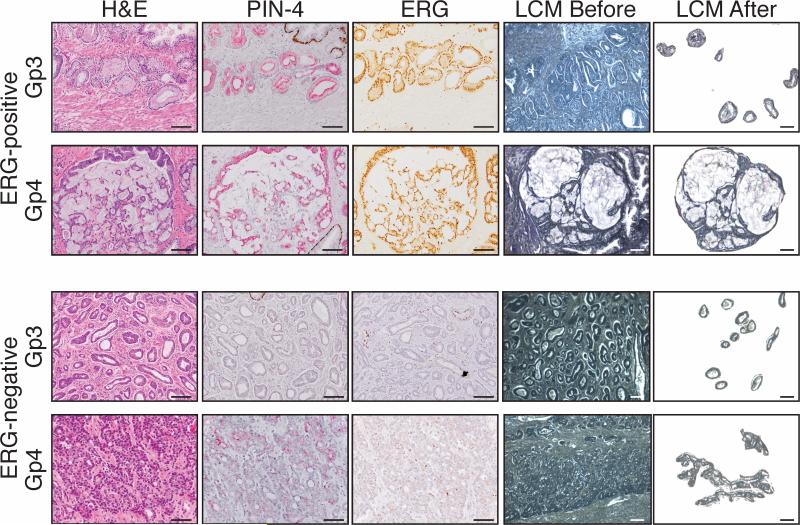
Figure 1. Representative microscopic images of laser-captured tissue.
Microscopic images of laser-captured tissue from a representative ERG fusion-positive, formalin-fixed case (patient 11-02, top) or ERG fusion-negative, PAXgene-fixed case (patient 12-03, bottom) of Gp3 and Gp4 PCa. For both cases, a representative image of tissue before and after laser-capture microdissection is shown. PIN-4 is a cocktail of four antibodies to distinguish invasive PCa from intraductal carcinoma and high grade prostatic intraepithelial neoplasia (PIN). Brown chromagen: p63, cytokeratin 5 and cytokeratin 14; red chromogen: alpha-methylacyl-CoA racemase (AMACR). Endothelial cells that express wild-type ERG were an internal control for ERG immunostaining. Bar: 50 μm.
Figure 2. Rapid emergence of Gleason pattern 4 following tumor development.
(A) Tumor phylogenies are shown for each focus individually sequenced. Phylogenetic tree branch lengths are proportional to the number of distinct mutations. Adjacent to each tree, shared and unshared copy number alteration to genes known to be altered in prostate cancers (34,35) are shown. SD: shallow deletion; DD: deep deletion. All nonsilent point mutations shared by all foci are shown for each case. Unshared point mutations are shown for genes frequently mutated in cancers (34,35,51) at sites overlapping with COSMIC or for nonsense mutations with clear functional significance. The computed breakpoints for the shallow deletions of 1BRCA1 in case 11-02 and 2TP53 in case 12-03 are distinct between foci, indicating that these SCNAs arose independently. (B) Whole genome copy number analysis is depicted. The entire genome was divided into 100-kilobase segments, and read depth within each segment was calculated and normalized to the coverage of the reference sample. Blue: gain; red loss.
Segmented read depth analysis to identify SCNAs further revealed shared events between adjacent Gp3 and Gp4 foci (Fig. 2B). These included shallow deletions of PTEN (which we previously reported in clonally-related Gp3 and Gp4 tumors (13)), BRCA1, BRCA2, CDKN1B, CDKN2A, CDKN2B, FANCA, MSH3, NCOR1, NKX3-1, PHLPP2, PIK3R1, SMAD4, TP53, and ZNRF3 (Fig. 2A,B and Supplementary Fig. S2A). Many of the shared structural events included larger, arm-level deletions encompassing a subset of these tumor suppressors (such as NKX3-1 in chromosome 8p) as well as shared deletions in regions previously found to be deleted in PCa (31), such as chromosome 6q.
Shared focal deletions of chromosome 21 were also detected in a subset of cases with TMPRSS2:ERG fusion (based on detection of the fusion breakpoint and/or ERG IHC), consistent with about half of these fusions being due to interstitial deletions (Supplementary Fig. S2B). In cases where we were able to extract adequate RNA for gene expression analysis (see below), there was also concordance between TMPRSS2:ERG fusion and increased ERG mRNA (Supplementary Fig. S2B).
High frequency of unique mutations and SCNAs in adjacent Gp3 versus Gp4 foci reflects early divergence and tumor microheterogeneity
For each pair of adjacent Gp3 and Gp4 foci, there was an average of two shared somatic nonsilent protein-coding alterations. When accounting for the additional coverage of intronic regions and noncoding RNAs, the number of shared somatic mutations ranged between 2 and 9 (see Supplementary Table S2). However, each case had many additional mutations/indels that were present at high cancer cell fraction in only the Gp3 or Gp4 foci, with comparable numbers in the Gp3 and Gp4 (Supplementary Table S4). Targeted re-sequencing for a subset of these mutations further confirmed that most were unique to the Gp3 or Gp4 foci (Supplementary Table S5).
To determine whether the relatively large number of unique mutations may be related to the Gp3 versus Gp4 morphology, versus reflecting tumor microheterogeneity, we LCM purified and sequenced DNA from a second Gp4 focus on an adjacent tissue block in two cases (cases 11-02 and 12-03). As expected, these second Gp4 foci contained the presumed truncal mutations and SCNAs shared between the other Gp4 and adjacent Gp3 foci (Fig. 2), as well as a discrete set of mutations that were in the adjacent Gp4 but not Gp3 tumors (Supplementary Table S6). Significantly, we also found that large numbers of high-confidence coding mutations were unique to each Gp4 focus (mean ~500, see Supplementary Table S4), indicating that these mutations did not generally mediate progression to Gp4. Instead these mutations presumably reflect tumor microheterogeneity evident due to microdissection of small tumor foci versus analysis of bulk tumor.
Significantly, the comparable large numbers of unique mutations in both the Gp3 and corresponding Gp4 tumors indicates that both Gp3 and Gp4 foci underwent substantial divergence from their common precursor, and argue against a straightforward linear progression from Gp3 to Gp4. Moreover, assuming that most of these are passenger mutations that accumulate at a relatively constant rate, this would suggest that these Gp3 and Gp4 tumors emerged from common precursors at approximately the same time and that subclones continued to emerge within each focus following divergence. Integrating all genomic events in each case to construct phylogenetic trees, the patterns that emerge indicate that these tumors progressed via branched clonal evolution from common progenitors that contained a relatively small number of genomic alterations (Fig. 2A).
Sections from one RP specimen with a large right-sided tumor (case 11-10) showed extracellular mucin secretion in tumor from the right base, but not from the right apex (Supplementary Fig. S3A and S3B). Therefore, we isolated and sequenced paired Gp3 and Gp4 foci from the right apex and right base. While the Gp3 and Gp4 foci at both locations were diffusely positive for ERG by immunohistochemistry, sequencing of the ERG locus revealed a novel translocation with CPEB3 in the Gp3 and Gp4 glands from the base (Supplementary Fig. S3C), while the apex had the typical TMPRSS2:ERG translocation. CPEB3 is normally expressed by prostate cells and this particular in-frame fusion would retain amino acids 314-486 of ERG (Supplementary Fig. S3D) and was therefore recognized by the C-terminal anti-ERG antibody. Because this fusion retained the Ets DNA-binding domain, it may have retained ERG function (32,33). This case further supports a branched evolution model for adjacent Gp3 and Gp4 tumors, with tumors in the apex and base being independent or possibly emerging from a very early common precursor prior to the gene fusion events.
Genomic events driving progression to Gp4
We next asked whether there were any genomic alterations in the Gp4 tumors, and absent in their adjacent clonal Gp3 tumors, that may account for their distinct morphology and biological behavior. We focused on mutations, indels, and SCNAs in genes previously identified as likely drivers of PCa (34,35) and/or reported in COSMIC. These potential drivers of Gp4 biology in single Gp4 focus included mutations to SMARCA4 (SWI/SNF Related, Matrix Associated 4), SETD2 (KMT3A, lysine methyltransferase), PTPRJ (protein tyrosine phosphatase, receptor type, J), MLLT4 (myeloid/lymphoid or mixed-lineage leukemia 4), UBR5 (E3 ubiquitin-protein ligase), and PDGFRB (platelet derived growth factor receptor beta). Consistent with our previous report (13), we observed progressive loss of PTEN in one case (11-03), with shallow deletion of PTEN in the Gp3 and deep deletion of PTEN in the Gp4. Amongst other tumor suppressor genes, shallow deletions in Gp4 tumors that were not found in their clonal Gp3 tumors included BRCA2, CDKN1B, CDKN2A, CDKN2B, FANCA, MLH3, NCOR1, NCOR2, NKX3-1, PHLPP1, PIK3R1, RB1, SMAD4, and TP53 (see Fig. 2 and Supplementary Fig. S2). Finally, copy number gains of the linked PIK3CA and PIK3CB genes were observed in the Gp4 from one case, and gain of CDK4 was observed in another case.
Significantly, while these gains and losses likely contributed to development of the respective Gp4 tumors, shallow deletions were also observed for most of these genes in one or more Gp3 tumors (see Fig. 2). This indicates that these tumor suppressor losses alone are not sufficient for progression to Gp4. Therefore, we next assessed gene expression differences in clonal Gp4 versus Gp3 tumor foci.
Differences in gene expression that distinguish adjacent clonal Gp3 and Gp4 tumors
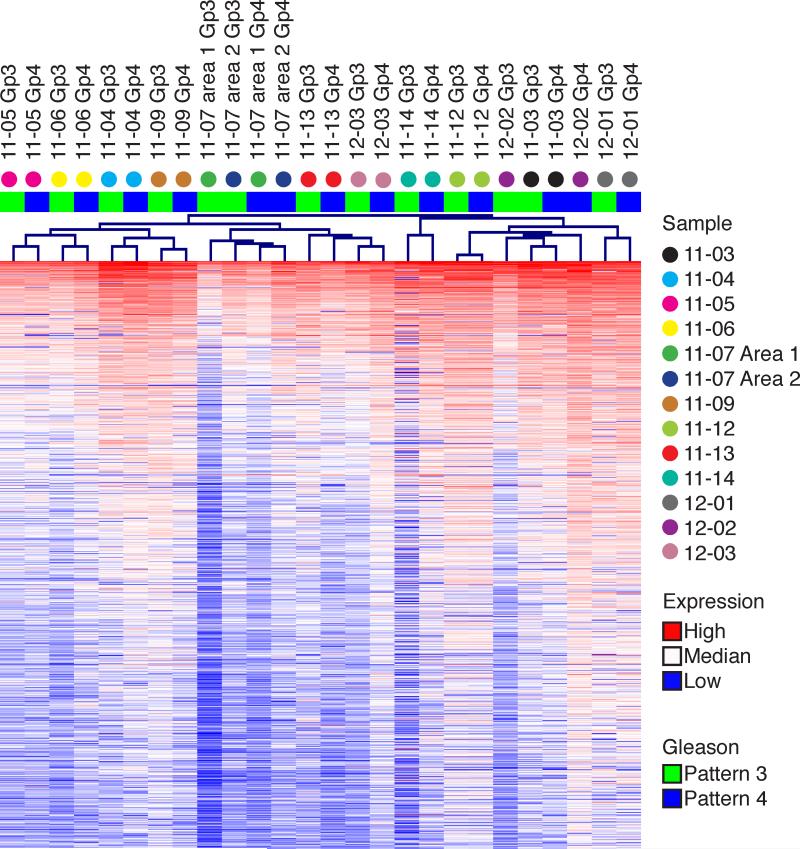
Adequate RNA was obtained from LCM in 13 Gp3/Gp4 pairs for analysis on Affymetrix Human Exon 1.0 microarrays. Unsupervised hierarchical clustering based on the 1000 genes with most variance across the entire dataset grouped the samples based on patients rather than on Gp3 versus Gp4 histology in 11 of 13 cases (Fig. 3). Moreover, within each pair, gene expression values were strongly and positively correlated (Supplementary Fig. S4). This correlation extended to a gene set used to assess aggressive potential based on PCa biopsies (OncotypeDX) (36). Expression of the OncotypeDX gene set by each Gp3 and Gp4 focus tracked together, with ten of the thirteen cases having Genomic Prostate Scores in their Gp3 and Gp4 foci within 20% of each other (Supplementary Fig. S5).
Figure 3. Overlap in gene expression between Gp3 and Gp4 tumors.
Unsupervised hierarchical clustering on the 1000 genes demonstrating the most variance across the entire dataset clustered the data by patient in 11 out of 13 cases. Color on heatmap represents intensity of expression value, color above tree indicates Gleason pattern, and colored dot identifies each column by patient sample.
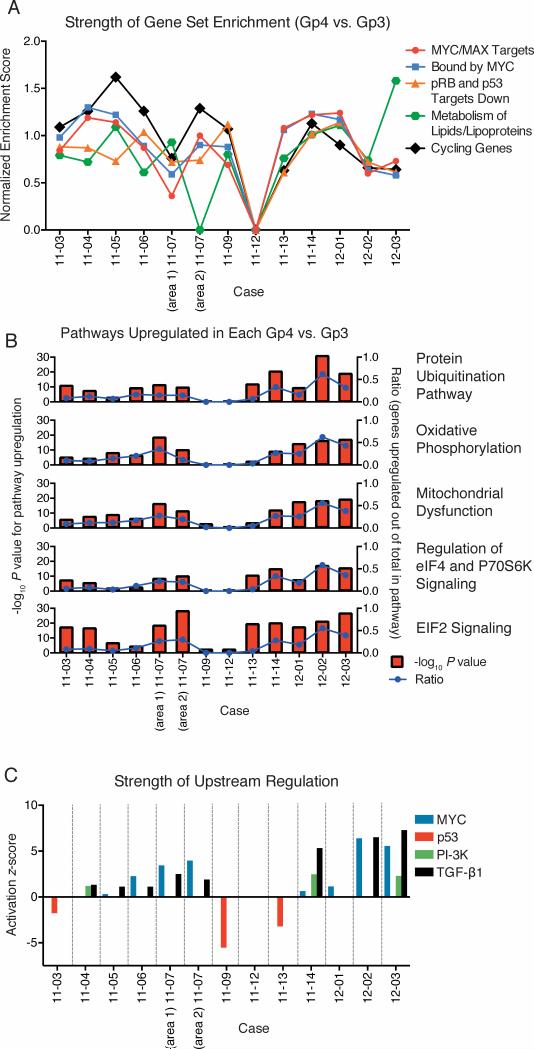
We next used transcriptome analysis to identify genes and biological processes that may be altered in Gp4 versus Gp3. Consistent with the marked gene expression overlap between Gp3 and Gp4, only 21 genes showed statistically significant differences across Gp3 versus Gp4 (Supplementary Table S7). Two of these genes (HSD17B4, and RAB18), as well as MAOA (P=0.0820), were previously found to be upregulated in Gp4 (37). However, neither Ingenuity Pathway Analysis (IPA) nor Gene Set Enrichment Analysis (GSEA) identified any pathways significantly associated with these 21 genes. Consequently, we next performed pairwise analyses within each sample using GSEA. Due to the noise intr insic to a paired analysis of single samples, many pathways showed enrichment in only 1 or 2 cases. We therefore focused on gene sets with a Normalized Enrichment Score >1 in multiple cases. This analysis showed that the majority of Gp4, relative to their adjacent Gp3, were enriched for genes involved in cell cycle and lipid metabolism (Fig. 4A). The Gp4 tumors also were enriched for two gene sets indicating increased expression of c-Myc targets and gene sets suggesting decreased activity of the tumor suppressors pRb and p53 (Fig. 4A).
Figure 4. Common upregulated pathways in Gp4.
(A) For the five most common gene sets identified from pairwise (Gp4 vs. Gp3) gene set enrichment analysis of microarray data, the strength of enrichment is indicated by the normalized enrichment score, which reflects correlation between ranked order of genes and the size of the gene set. Scores are shown for gene sets with a false discovery (q) rate less than 0.25, and scores greater than 1.0 suggest moderate enrichment. The most frequently enriched gene sets are targets of c-Myc and Max identified by ChIP in a Burkitt's lymphoma cell line (52), genes with promoters bound by Myc (53), genes downregulated in both Rb1 and Tp53 knockout mice (54), genes involving lipids and lipoproteins metabolism, and genes showing cell-cycle stage-specific expression (52). (B) Gene upregulation ratios and activation P value for recurrently-upregulated pathways in Gp4 (vs. Gp3) in each case. For the five most common pathways identified from pairwise Ingenuity Pathway Analysis, the statistical significance of pathway upregulation (negative log P value) ratio is given on the left y axis (red bars), and the ratio of upregulated genes (out of total within each pathway) is given on the right y axis (blue lines). (C) Activation (or negative: inactivation) z-scores for four recurrently-identified upstream regulators of gene expression in each case.
We similarly conducted pairwise analyses using the canonical pathway and upstream regulator functions of IPA for genes with >1.5 fold change between Gp3 and Gp4 pairs. The former identified five pathways upregulated in a majority of the Gp4 versus their adjacent clonal Gp3 (Fig. 4B). The IPA upstream regulator analysis then implicated c-Myc activation as driving these pathways, and identified increased PI-3K and TGF-β1 signaling and inactivation of p53 as drivers of the Gp4 phenotype in subsets of cases (Fig. 4C). Significantly, examination of the c-Myc gene set showed substantial variability in the genes that were increased in the Gp4 versus the corresponding Gp3, including in expression of MYC itself (Supplementary Fig. S6A). This likely reflects variable c-Myc pathway activity in each Gp3 focus and explains why increased c-Myc pathway activity was not clearly observed when groups of Gp3 and Gp4 were compared. There was similarly variability in the p53 gene set (Supplementary Fig. S6B).
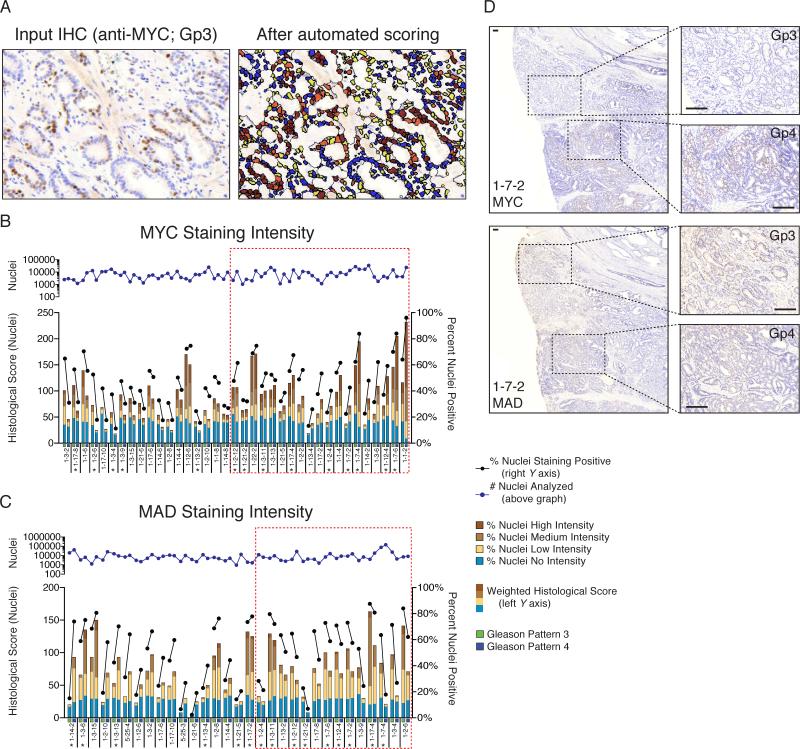
We did not find recurrent MYC gene copy number gain in Gp4, and MYC mRNA was not consistently increased in the Gp4 relative to paired Gp3 foci. However, as c-Myc protein is regulated at multiple levels (38), we next investigated c-Myc by immunohistochemistry in an independent cohort of ~35 Gs7 tumors. Since c-Myc levels can be very heterogeneous, we used automated image analysis to determine the fraction of positive nuclei in adjacent Gp3 and Gp4 foci, and to quantify intensity of nuclear staining (histological score) (Fig. 5A). As expected, c-Myc was higher in the Gp3 and Gp4 foci relative to normal glands (not shown). Based on the fraction of positive nuclei, c-Myc was greater in the Gp4 versus adjacent Gp3 foci in approximately 50% cases (19 of 37 evaluable samples) (Fig. 5B, D). There were no consistent differences in the distribution of staining intensities between adjacent Gp3 and Gp4 foci, and differences in the histological scores were primarily driven by the fraction of positive nuclei. We similarly examined the levels of MAD protein, a c-Myc negative regulator. MAD protein levels, based on percent positive nuclei, were lower in Gp4 versus adjacent Gp3 foci in 14 of 31 evaluable cases (Fig. 5C, 5D). As with c-Myc, there were no consistent differences in the distribution of staining intensities. Nine of the 14 Gp4 foci with lower MAD also had higher c-Myc, while c-Myc was lower in the remaining 5 cases. These findings suggest that increased c-Myc and/or decreased MAD may mediate increased c-Myc pathway activity in slightly over half of cases. Interestingly, the lncRNA PCGEM1, which has been reported to coactivate c-Myc (39), was increased in Gp4 relative to the Gp3 in 12 of the 13 cases we analyzed, providing another possible mechanism for c-Myc pathway activation (not shown). Taken together these results indicate that increased c-Myc pathway activity, mediated by diverse mechanisms, contributes to the morphology and presumably more aggressive behavior of Gp4 versus Gp3.
Figure 5. Differential c-Myc status in Gp4 vs. Gp3.
(A) Representative immunohistochemical staining used as input (left) for automated image analysis with Definiens, and the processed file used for quantification of staining intensity (right). (B) Quantified intensity of c-Myc protein staining of the Gp3 and Gp4 components in an independent cohort of Gs7 tumors. Red boxes indicate the cases showing increase in c-Myc staining from Gp3 to Gp4. Asterisks indicate those cases where changes in levels of MAD decrease from Gp3 to Gp4. (C) Quantified intensity of MAD protein staining of the Gp3 and Gp4 components in an independent cohort of Gs7 tumors. Red boxes indicate the cases showing decrease in MAD staining from Gp3 to Gp4. Asterisks indicate those cases where changes in levels of c-Myc increase from Gp3 to Gp4. For (B) and (C), each case is indicated below the x-axis with green boxes indicating Gp3 and blue boxes indicating Gp4. The histologic score for cells analyzed by Definiens is plotted on the left y axis in multi-colored vertical bars, and this score was used to define an increase or decrease within each Gp3/Gp4 pair. Each color in the vertical bar corresponds to the percentage of cells in each Gp3 or Gp4 with variable staining intensity as indicated, out of 100% nuclei within each column. The percentage of positive nuclei is plotted on the right y axis as a black line. The number of nuclei considered for the histological score is given above each chart, plotted as a blue line. (D) Representative stains from case 1-7-2 showing from Gp3 to Gp4 an increase in c-Myc staining (top), and a decrease in MAD staining (bottom). Bar: 200 μm.
Discussion
The molecular features that account for the distinct histology and biological behavior of Gp4 versus Gp3 tumors, whether Gp3 tumors progress to Gp4, and whether there are subsets of Gp3 tumors that reflect a truly indolent disease process, remain to be established. In this study, we used WES and transcriptome profiling of LCM purified adjacent Gp3 and Gp4 tumors to address the molecular relationship between these entities. In all cases the adjacent Gp3 and Gp4 tumors shared a series of mutations and SCNAs, confirming that they were derived from a common clone. However, both the Gp3 and Gp4 tumors also had substantially larger numbers of mutations that were not shared, which does not support a straightforward model of linear progression from Gp3 to Gp4. Moreover, several Gp3 tumors had likely oncogenic genomic alterations, including tumor suppressor losses, which were not found in the adjacent Gp4 tumors. Finally, the Gp3 and Gp4 tumors had comparable numbers of unique high allele frequency mutations. These findings indicate that Gp3 tumors do not directly progress to Gp4, and instead support a branched evolution model with the Gp3 and Gp4 tumors emerging early from a common precursor and subsequently undergoing substantial divergence.
Several recent studies examining multiple tumor foci in prostate similarly found divergence from a common clone and extensive microheterogeneity (40,41), and one study also found substantial divergence between clonal Gp3 and Gp4 foci (17). This latter study of four subjects further found that alterations in likely driver genes were only in the Gp4 foci. In contrast, we found likely driver events such as tumor suppressor losses in many of the Gp3 tumors, which appeared to be truncal in some cases and unique to the Gp3 in other cases. Remarkably, the Gp3 tumors retain their indolent-appearing morphological features despite acquisition of these multiple genomic alterations.
Sequencing studies to characterize the genomic landscape of primary PCa have generally concluded that there is a relatively low mutation rate of ~10-30 non-synonymous, protein-coding mutations per tumor sample, with a small set of genes (such as PTEN, SPOP, FOXA1 and MED12) that are recurrently altered (35,42,43). The number of high allele frequency shared mutations we found in our Gp3 and Gp4 tumors are consistent with this mutation rate, and we presume that the much higher number of unique mutations we find in LCM purified Gp3 and Gp4 tumor foci reflects tumor microheterogeneity that is not observed when larger tissue samples containing multiple tumor foci are examined. This conclusion was supported by our analysis of a second Gp4 focus in two cases, which showed extensive divergence between the clonal Gp4 foci. This phenomenon of tumor microheterogeneity has been described in other cancers (44), and higher apparent mutation rates have been reported in PCa when purer samples are analyzed (including PCa explant xenografts and metastases) (45,46).
Notably, by directly comparing adjacent Gp3 and Gp4 tumors, we could identify a number of genomic alterations that may have driven progression (from an earlier Gp3 or common precursor) to Gp4, and account for the aggressive cribriform histology in these tumors. Most notable were losses in the Gp4 foci, but not the adjacent Gp3 focus, of tumor suppressor genes including PTEN, BRCA1, BRCA2, NKX3-1, RB1, and TP53. However, it is clear that these losses by themselves are not adequate to mediate development of Gp4, as we also found losses of these genes in one or more Gp3 tumors. As an alternative approach to identify mechanisms that may be driving development of Gp4, we globally assessed gene expression in adjacent Gp3 versus Gp4 foci. Significantly, gene expression differences between adjacent Gp3 and Gp4 foci were modest, but pairwise analyses of each Gp4 against its clonal Gp3 indicated increased PI-3K activation, decreased p53 and pRb activity, and increased c-Myc activity, with the latter being the most consistent finding. These processes have been implicated in PCa development, but they have not previously been correlated with Gleason pattern in individual Gs7 tumors. Immunohistochemistry suggested increased c-Myc and decreased MAD as one possible mechanism for increased MYC pathway activation in about half of cases, but other mechanisms including increases in the lncRNA PCGEM1 are also likely regulating MYC pathway activity (39). We have not yet further explored mechanisms that may be altering PI-3K, p53, or pRb activities in Gp3 versus Gp4, and certainly cannot exclude further mechanisms including alternative splicing or epigenetic alterations.
In summary, this study establishes that a subset of Gp3 tumors develop and extensively diverge from a common precursor lesion (which may be high grade PIN, an earlier Gp3, or another entity) that can also give rise to Gp4, and hence that these Gp4 tumors are not immediate descendants of their associated Gp3. This study further indicates that once tumors emerge as Gp3, their morphology may be relatively fixed despite an accumulation of genomic alterations. Nonetheless, direct progression of Gp3 to Gp4 or Gp5 may certainly occur in some cases (such as Gs6 with tertiary Gp4), and may indeed have occurred at some early point in the evolution of the tumors examined here. Finally, although it does not appear that any single recurrent genomic alteration drives the distinct morphology and biological behavior of these cribriform Gp4 versus Gp3 tumors, the paired transcriptome analyses indicated that increased c-Myc activity may be a common driver.
An important unanswered question is whether there is a molecularly distinct subset of Gp3 tumors that emerge de novo or from precursor lesions that have little or no potential to give rise to Gp4. The observation that some RP specimens contain relatively high volume Gp3 with no higher grades (Gs6) suggests that these may be molecularly distinct, but this could be stochastic, and many of these cases on careful examination are found to have very small foci of Gp4 (47,48). Further molecular analyses of these potentially indolent Gp3 tumors will be needed to address whether there exists a distinct subset. In any case, it is noteworthy that many of the Gp3 tumors examined in this study had one or more oncogenic genomic alterations in addition to TMPRSS2-ERG fusion. Supported by recent findings that PTEN loss and chromosome 8 alterations in Gp3 tumor biopsies are associated with detection of higher grade tumor in RP specimens (49,50), we hypothesize that detection of certain oncogenic alterations (including but not restricted to PTEN loss) in biopsies that show only Gp3 may be indicative of a higher grade tumor that was missed on biopsy or increased risk that a higher grade tumor will emerge. Therefore, from a clinical perspective, consideration should be given to molecular analyses of Gs6 biopsies for tumor suppressor losses or for gene expression alterations that may reflect these underlying genomic alterations.
Supplementary Material
Statement of Translational Relevance.
There is increasing acceptance of active surveillance (AS) for men with Gleason pattern 3 (Gp3) prostate cancer (PCa) on biopsy, but concerns that more aggressive tumors may have been missed, or that these tumors may progress to higher grades, lead many patients to opt for therapy. This study provides a molecular characterization of Gp3 that are associated with Gp4, confirms that they are clonally related, and shows that these Gp3 have a spectrum of genomic alterations including tumor suppressor losses. These results establish that at least a subset of Gp3 emerge from a common precursor that also gives rise to Gp4, and indicate that genomic alterations in these Gp3 may distinguish them from truly indolent Gp3. These findings suggest that assessing for a spectrum of genomic alterations in Gp3 tumors from biopsies may improve our ability to identify AS candidates who are at increased risk of harboring higher grade tumors.
Acknowledgments
The authors wish to express their gratitude to the patients and the families of patients who contributed to this study. The authors acknowledge excellent technical assistance from Cesar Vazquez, Bonnie Wong, Dr. Victoria Petkova, and Olga Voznesensky.
Financial Support: This work was supported by the National Institutes of Health (T32 CA081156 to AGS, P01 CA163227 to SPB, DF/HCC-Prostate Cancer SPORE P50 CA090381 to SPB and AGS, R01 HG007538 to WL), the Department of Defense Prostate Cancer Research Program (Postdoctoral Training Award W81XWH-13-1-0267 to AGS, Idea Development Award W81XWH-15-1-0710 to AGS, Exploration-Hypothesis Development Award W81XWH-15-1-0136 to AGS, Idea Development Awards W81XWH-11-1-0295, W81XWH-08-1-0414, and W81XWH-07-1-0443 to SPB), CPRIT (RP110471 and RP150292 to WL), the Prostate Cancer Foundation (Young Investigator Awards to AGS, HTK and HY, and a Challenge Award to SPB), a High Impact Mazzone Award from the Dana-Farber/Harvard Cancer Center (SPB), a Translational Grant from the V Foundation for Cancer Research (SBP), and the Intramural Research Program of the NIH, National Cancer Institute.
Footnotes
Conflicts of Interest: The authors have no conflicts to report.
REFERENCES
- 1.Draisma G, Boer R, Otto SJ, van der Cruijsen IW, Damhuis RA, Schroder FH, et al. Lead times and overdetection due to prostate-specific antigen screening: estimates from the European Randomized Study of Screening for Prostate Cancer. Journal of the National Cancer Institute. 2003;95(12):868–78. doi: 10.1093/jnci/95.12.868. [DOI] [PubMed] [Google Scholar]
- 2.Albertsen PC, Hanley JA, Fine J. 20-year outcomes following conservative management of clinically localized prostate cancer. JAMA. 2005;293(17):2095–101. doi: 10.1001/jama.293.17.2095. [DOI] [PubMed] [Google Scholar]
- 3.Eggener SE, Scardino PT, Walsh PC, Han M, Partin AW, Trock BJ, et al. Predicting 15-year prostate cancer specific mortality after radical prostatectomy. J Urol. 2011;185(3):869–75. doi: 10.1016/j.juro.2010.10.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Epstein JI, Walsh PC, Carter HB. Dedifferentiation of prostate cancer grade with time in men followed expectantly for stage T1c disease. J Urol. 2001;166(5):1688–91. [PubMed] [Google Scholar]
- 5.Sheridan TB, Carter HB, Wang W, Landis PB, Epstein JI. Change in prostate cancer grade over time in men followed expectantly for stage T1c disease. J Urol. 2008;179(3):901–4. doi: 10.1016/j.juro.2007.10.062. discussion 04-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Klotz L, Zhang L, Lam A, Nam R, Mamedov A, Loblaw A. Clinical results of long-term follow-up of a large, active surveillance cohort with localized prostate cancer. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2010;28(1):126–31. doi: 10.1200/JCO.2009.24.2180. [DOI] [PubMed] [Google Scholar]
- 7.Tosoian JJ, Trock BJ, Landis P, Feng Z, Epstein JI, Partin AW, et al. Active surveillance program for prostate cancer: an update of the Johns Hopkins experience. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2011;29(16):2185–90. doi: 10.1200/JCO.2010.32.8112. [DOI] [PubMed] [Google Scholar]
- 8.Porten SP, Whitson JM, Cowan JE, Cooperberg MR, Shinohara K, Perez N, et al. Changes in prostate cancer grade on serial biopsy in men undergoing active surveillance. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2011;29(20):2795–800. doi: 10.1200/JCO.2010.33.0134. [DOI] [PubMed] [Google Scholar]
- 9.Kobayashi M, Ishida H, Shindo T, Niwa S, Kino M, Kawamura K, et al. Molecular analysis of multifocal prostate cancer by comparative genomic hybridization. The Prostate. 2008;68(16):1715–24. doi: 10.1002/pros.20832. [DOI] [PubMed] [Google Scholar]
- 10.Cheng L, Bostwick DG, Li G, Wang Q, Hu N, Vortmeyer AO, et al. Allelic imbalance in the clonal evolution of prostate carcinoma. Cancer. 1999;85(9):2017–22. doi: 10.1002/(sici)1097-0142(19990501)85:9<2017::aid-cncr20>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- 11.Cheng L, Song SY, Pretlow TG, Abdul-Karim FW, Kung HJ, Dawson DV, et al. Evidence of independent origin of multiple tumors from patients with prostate cancer. Journal of the National Cancer Institute. 1998;90(3):233–7. doi: 10.1093/jnci/90.3.233. [DOI] [PubMed] [Google Scholar]
- 12.Macintosh CA, Stower M, Reid N, Maitland NJ. Precise microdissection of human prostate cancers reveals genotypic heterogeneity. Cancer Res. 1998;58(1):23–8. [PubMed] [Google Scholar]
- 13.Sowalsky AG, Ye H, Bubley GJ, Balk SP. Clonal Progression of Prostate Cancers from Gleason Grade 3 to Grade 4. Cancer Research. 2013;73(3):1050–55. doi: 10.1158/0008-5472.CAN-12-2799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kovtun IV, Cheville JC, Murphy SJ, Johnson SH, Zarei S, Kosari F, et al. Lineage relationship of Gleason patterns in Gleason score 7 prostate cancer. Cancer Res. 2013;73(11):3275–84. doi: 10.1158/0008-5472.CAN-12-2803. [DOI] [PubMed] [Google Scholar]
- 15.Haffner MC, Mosbruger T, Esopi DM, Fedor H, Heaphy CM, Walker DA, et al. Tracking the clonal origin of lethal prostate cancer. The Journal of clinical investigation. 2013;123(11):4918–22. doi: 10.1172/JCI70354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lotan TL, Carvalho FL, Peskoe SB, Hicks JL, Good J, Fedor HL, et al. PTEN loss is associated with upgrading of prostate cancer from biopsy to radical prostatectomy. Mod Pathol. 2014 doi: 10.1038/modpathol.2014.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.VanderWeele DJ, Brown CD, Taxy JB, Gillard M, Hatcher DM, Tom WR, et al. Low-grade prostate cancer diverges early from high grade and metastatic disease. Cancer Sci. 2014;105(8):1079–85. doi: 10.1111/cas.12460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Esgueva R, Perner S, C JL, Scheble V, Stephan C, Lein M, et al. Prevalence of TMPRSS2-ERG and SLC45A3-ERG gene fusions in a large prostatectomy cohort. Mod Pathol. 2010;23(4):539–46. doi: 10.1038/modpathol.2009.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barry M, Perner S, Demichelis F, Rubin MA. TMPRSS2-ERG fusion heterogeneity in multifocal prostate cancer: clinical and biologic implications. Urology. 2007;70(4):630–3. doi: 10.1016/j.urology.2007.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tomlins SA, Rhodes DR, Perner S, Dhanasekaran SM, Mehra R, Sun XW, et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310(5748):644–8. doi: 10.1126/science.1117679. [DOI] [PubMed] [Google Scholar]
- 21.Mathieson W, Marcon N, Antunes L, Ashford DA, Betsou F, Frasquilho SG, et al. A Critical Evaluation of the PAXgene Tissue Fixation System: Morphology, Immunohistochemistry, Molecular Biology, and Proteomics. Am J Clin Pathol. 2016;146(1):25–40. doi: 10.1093/ajcp/aqw023. [DOI] [PubMed] [Google Scholar]
- 22.Piccolo SR, Sun Y, Campbell JD, Lenburg ME, Bild AH, Johnson WE. A single-sample microarray normalization method to facilitate personalized-medicine workflows. Genomics. 2012;100(6):337–44. doi: 10.1016/j.ygeno.2012.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dai M, Wang P, Boyd AD, Kostov G, Athey B, Jones EG, et al. Evolving gene/transcript definitions significantly alter the interpretation of GeneChip data. Nucleic Acids Res. 2005;33(20):e175. doi: 10.1093/nar/gni179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N, et al. TM4: a free, open-source system for microarray data management and analysis. Biotechniques. 2003;34(2):374–8. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
- 25.Chen EJ, Sowalsky AG, Gao S, Cai C, Voznesensky O, Schaefer R, et al. Abiraterone Treatment in Castration-Resistant Prostate Cancer Selects for Progesterone Responsive Mutant Androgen Receptors. Clinical Cancer Research. 2015;21(6):1273–80. doi: 10.1158/1078-0432.CCR-14-1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cibulskis K, Lawrence MS, Carter SL, Sivachenko A, Jaffe D, Sougnez C, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol. 2013;31(3):213–9. doi: 10.1038/nbt.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ramos AH, Lichtenstein L, Gupta M, Lawrence MS, Pugh TJ, Saksena G, et al. Oncotator: cancer variant annotation tool. Human mutation. 2015;36(4):E2423–9. doi: 10.1002/humu.22771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Koboldt DC, Zhang Q, Larson DE, Shen D, McLellan MD, Lin L, et al. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome research. 2012;22(3):568–76. doi: 10.1101/gr.129684.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Landau DA, Tausch E, Taylor-Weiner AN, Stewart C, Reiter JG, Bahlo J, et al. Mutations driving CLL and their evolution in progression and relapse. Nature. 2015;526(7574):525–30. doi: 10.1038/nature15395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thorvaldsdottir H, Robinson JT, Mesirov JP. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Briefings in bioinformatics. 2012 doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Williams JL, Greer PA, Squire JA. Recurrent copy number alterations in prostate cancer: an in silico meta-analysis of publicly available genomic data. Cancer Genet. 2014;207(10-12):474–88. doi: 10.1016/j.cancergen.2014.09.003. [DOI] [PubMed] [Google Scholar]
- 32.Carrère S, Verger A, Flourens A, Stehelin D, Duterque-Coquillaud M. Erg proteins, transcription factors of the Ets family, form homo, heterodimers and ternary complexes via two distinct domains. Oncogene. 1998;16(25):3261–8. doi: 10.1038/sj.onc.1201868. [DOI] [PubMed] [Google Scholar]
- 33.Salehi-Ashtiani K, Lupták A, Litovchick A, Szostak JW. A genomewide search for ribozymes reveals an HDV-like sequence in the human CPEB3 gene. Science. 2006;313(5794):1788–92. doi: 10.1126/science.1129308. [DOI] [PubMed] [Google Scholar]
- 34.Robinson D, Van Allen Eliezer M, Wu Y-M, Schultz N, Lonigro Robert J, Mosquera J-M, et al. Integrative Clinical Genomics of Advanced Prostate Cancer. Cell. 2015;161(5):1215–28. doi: 10.1016/j.cell.2015.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Network CGAR. The Molecular Taxonomy of Primary Prostate Cancer. Cell. 2015;163(4):1011–25. doi: 10.1016/j.cell.2015.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Knezevic D, Goddard AD, Natraj N, Cherbavaz DB, Clark-Langone KM, Snable J, et al. Analytical validation of the Oncotype DX prostate cancer assay - a clinical RT-PCR assay optimized for prostate needle biopsies. BMC Genomics. 2013;14:690. doi: 10.1186/1471-2164-14-690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.True L, Coleman I, Hawley S, Huang CY, Gifford D, Coleman R, et al. A molecular correlate to the Gleason grading system for prostate adenocarcinoma. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(29):10991–6. doi: 10.1073/pnas.0603678103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Koh CM, Bieberich CJ, Dang CV, Nelson WG, Yegnasubramanian S, De Marzo AM. MYC and Prostate Cancer. Genes Cancer. 2010;1(6):617–28. doi: 10.1177/1947601910379132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hung CL, Wang LY, Yu YL, Chen HW, Srivastava S, Petrovics G, et al. A long noncoding RNA connects c-Myc to tumor metabolism. Proceedings of the National Academy of Sciences of the United States of America. 2014;111(52):18697–702. doi: 10.1073/pnas.1415669112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cooper CS, Eeles R, Wedge DC, Van Loo P, Gundem G, Alexandrov LB, et al. Analysis of the genetic phylogeny of multifocal prostate cancer identifies multiple independent clonal expansions in neoplastic and morphologically normal prostate tissue. Nature genetics. 2015;47(4):367–72. doi: 10.1038/ng.3221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Boutros PC, Fraser M, Harding NJ, de Borja R, Trudel D, Lalonde E, et al. Spatial genomic heterogeneity within localized, multifocal prostate cancer. Nature genetics. 2015;47(7):736–45. doi: 10.1038/ng.3315. [DOI] [PubMed] [Google Scholar]
- 42.Barbieri CE, Baca SC, Lawrence MS, Demichelis F, Blattner M, Theurillat JP, et al. Exome sequencing identifies recurrent SPOP, FOXA1 and MED12 mutations in prostate cancer. Nature genetics. 2012;44(6):685–9. doi: 10.1038/ng.2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Berger MF, Lawrence MS, Demichelis F, Drier Y, Cibulskis K, Sivachenko AY, et al. The genomic complexity of primary human prostate cancer. Nature. 2011;470(7333):214–20. doi: 10.1038/nature09744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Barber LJ, Davies MN, Gerlinger M. Dissecting cancer evolution at the macro-heterogeneity and micro-heterogeneity scale. Curr Opin Genet Dev. 2014;30C:1–6. doi: 10.1016/j.gde.2014.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kumar A, White TA, MacKenzie AP, Clegg N, Lee C, Dumpit RF, et al. Exome sequencing identifies a spectrum of mutation frequencies in advanced and lethal prostate cancers. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(41):17087–92. doi: 10.1073/pnas.1108745108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Grasso CS, Wu YM, Robinson DR, Cao X, Dhanasekaran SM, Khan AP, et al. The mutational landscape of lethal castration-resistant prostate cancer. Nature. 2012;487(7406):239–43. doi: 10.1038/nature11125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Adam M, Hannah A, Budaus L, Steuber T, Salomon G, Michl U, et al. A tertiary Gleason pattern in the prostatectomy specimen and its association with adverse outcome after radical prostatectomy. J Urol. 2014;192(1):97–101. doi: 10.1016/j.juro.2014.01.029. [DOI] [PubMed] [Google Scholar]
- 48.Epstein JI, Feng Z, Trock BJ, Pierorazio PM. Upgrading and downgrading of prostate cancer from biopsy to radical prostatectomy: incidence and predictive factors using the modified Gleason grading system and factoring in tertiary grades. European urology. 2012;61(5):1019–24. doi: 10.1016/j.eururo.2012.01.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Trock BJ, Fedor H, Gurel B, Jenkins RB, Knudsen BS, Fine SW, et al. PTEN loss and chromosome 8 alterations in Gleason grade 3 prostate cancer cores predicts the presence of un-sampled grade 4 tumor: implications for active surveillance. Mod Pathol. 2016;29(7):764–71. doi: 10.1038/modpathol.2016.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lotan TL, Carvalho FL, Peskoe SB, Hicks JL, Good J, Fedor HL, et al. PTEN loss is associated with upgrading of prostate cancer from biopsy to radical prostatectomy. Mod Pathol. 2015;28(1):128–37. doi: 10.1038/modpathol.2014.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nik-Zainal S, Davies H, Staaf J, Ramakrishna M, Glodzik D, Zou X, et al. Landscape of somatic mutations in 560 breast cancer whole-genome sequences. Nature. 2016;534(7605):47–54. doi: 10.1038/nature17676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ben-Porath I, Thomson MW, Carey VJ, Ge R, Bell GW, Regev A, et al. An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nature genetics. 2008;40(5):499–507. doi: 10.1038/ng.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zeller KI, Jegga AG, Aronow BJ, O'Donnell KA, Dang CV. An integrated database of genes responsive to the Myc oncogenic transcription factor: identification of direct genomic targets. Genome Biol. 2003;4(10):R69. doi: 10.1186/gb-2003-4-10-r69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Martínez-Cruz AB, Santos M, Lara MF, Segrelles C, Ruiz S, Moral M, et al. Spontaneous squamous cell carcinoma induced by the somatic inactivation of retinoblastoma and Trp53 tumor suppressors. Cancer Res. 2008;68(3):683–92. doi: 10.1158/0008-5472.CAN-07-3049. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.