Abstract
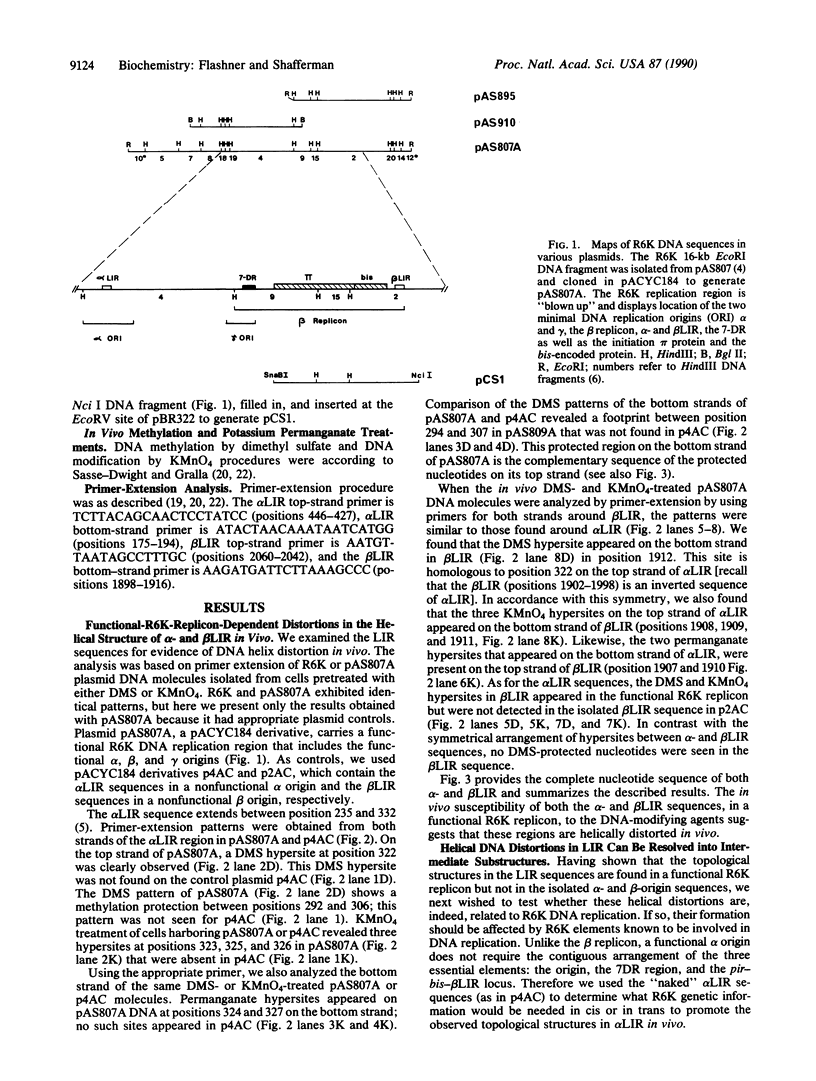
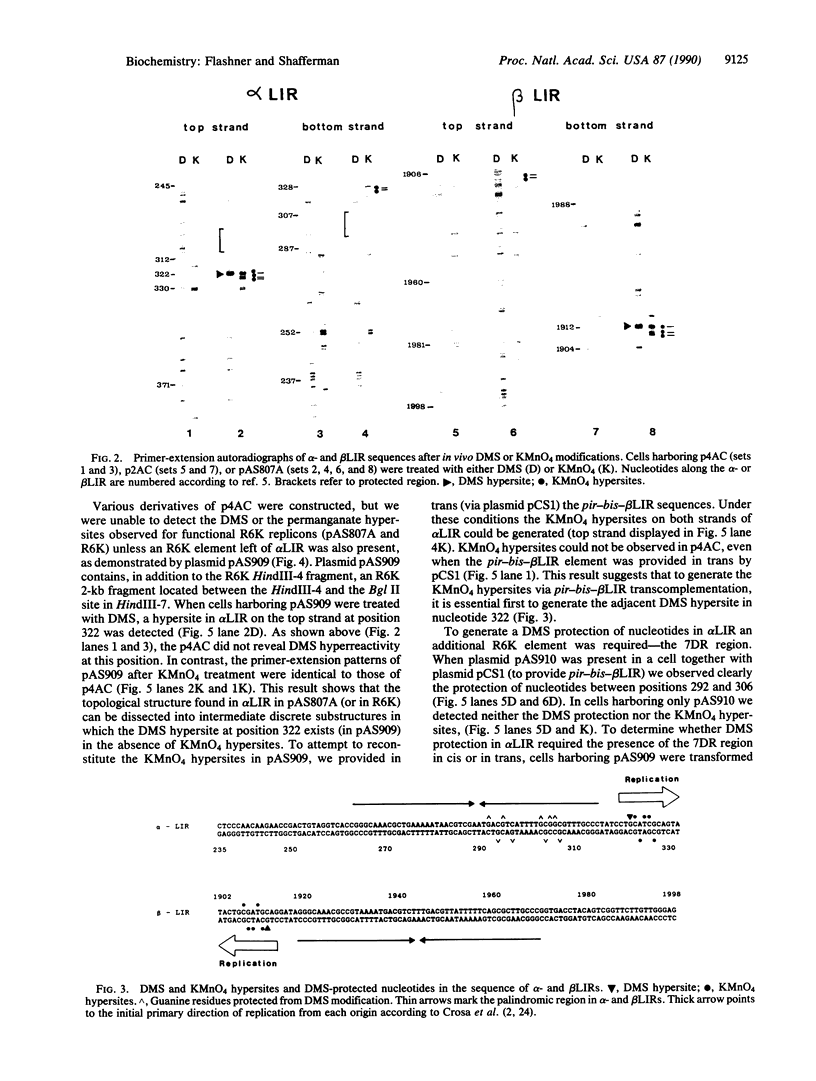
Plasmid R6K contains inverted repeats of an approximately 100-base-pair sequence separated by 5.5 kilobases. These long inverted repeats (LIRs) occur within the alpha and beta origins of replication and are essential for origin function. In this study, primer-extension analysis of DNA modified in vivo by dimethyl sulfate or KMnO4 revealed that both alpha and beta LIRs acquire similar structural distortions of the DNA helix in a functional R6K replicon. These distortions were not seen in plasmids containing isolated LIR sequences. In the functional replicon, the dimethyl sulfate and KMnO4 hyperreactive sites appear on complementary strands and are located to one side of an internal palindromic sequence within the LIRs. This asymmetry coincides with the primary direction of DNA replication from alpha and beta origins in vivo. We also observed two intermediate structures when certain R6K cis- or trans-acting elements are missing. Sequences near the alpha origin are required for generation of the dimethyl sulfate hyperreactive sites, whereas sequences near the beta origin are responsible for the appearance of KMnO4 hyperreactive sites. We suggest that these structures represent a hierarchy that leads to a "locked" preinitiation complex, which functions to synchronize and determine the direction of replication from the alpha and beta replication origins in vivo.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alfano C., McMacken R. Ordered assembly of nucleoprotein structures at the bacteriophage lambda replication origin during the initiation of DNA replication. J Biol Chem. 1989 Jun 25;264(18):10699–10708. [PubMed] [Google Scholar]
- Borowiec J. A., Gralla J. D. High-resolution analysis of lac transcription complexes inside cells. Biochemistry. 1986 Sep 9;25(18):5051–5057. doi: 10.1021/bi00366a012. [DOI] [PubMed] [Google Scholar]
- Borowiec J. A., Hurwitz J. Localized melting and structural changes in the SV40 origin of replication induced by T-antigen. EMBO J. 1988 Oct;7(10):3149–3158. doi: 10.1002/j.1460-2075.1988.tb03182.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bramhill D., Kornberg A. Duplex opening by dnaA protein at novel sequences in initiation of replication at the origin of the E. coli chromosome. Cell. 1988 Mar 11;52(5):743–755. doi: 10.1016/0092-8674(88)90412-6. [DOI] [PubMed] [Google Scholar]
- Campbell J. L. Eukaryotic DNA replication. Annu Rev Biochem. 1986;55:733–771. doi: 10.1146/annurev.bi.55.070186.003505. [DOI] [PubMed] [Google Scholar]
- Crosa J. H., Luttropp L. K., Falkow S. Mode of replication of the conjugative R-plasmid RSF1040 in Escherichia coli. J Bacteriol. 1976 Apr;126(1):454–466. doi: 10.1128/jb.126.1.454-466.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crosa J. H., Luttropp L. K., Falkow S. Molecular cloning of replication and incompatibility regions from the R-plasmid R6K. J Mol Biol. 1978 Sep 25;124(3):443–468. doi: 10.1016/0022-2836(78)90181-x. [DOI] [PubMed] [Google Scholar]
- Crosa J. H. Three origins of replication are active in vivo in the R plasmid RSF1040. J Biol Chem. 1980 Dec 10;255(23):11075–11077. [PubMed] [Google Scholar]
- Dodson M., McMacken R., Echols H. Specialized nucleoprotein structures at the origin of replication of bacteriophage lambda. Protein association and disassociation reactions responsible for localized initiation of replication. J Biol Chem. 1989 Jun 25;264(18):10719–10725. [PubMed] [Google Scholar]
- Filutowicz M., Appelt K. The integration host factor of Escherichia coli binds to multiple sites at plasmid R6K gamma origin and is essential for replication. Nucleic Acids Res. 1988 May 11;16(9):3829–3843. doi: 10.1093/nar/16.9.3829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filutowicz M., Uhlenhopp E., Helinski D. R. Binding of purified wild-type and mutant pi initiation proteins to a replication origin region of plasmid R6K. J Mol Biol. 1986 Jan 20;187(2):225–239. doi: 10.1016/0022-2836(86)90230-5. [DOI] [PubMed] [Google Scholar]
- Flashner Y., Gralla J. D. Dual mechanism of repression at a distance in the lac operon. Proc Natl Acad Sci U S A. 1988 Dec;85(23):8968–8972. doi: 10.1073/pnas.85.23.8968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Germino J., Bastia D. Interaction of the plasmid R6K-encoded replication initiator protein with its binding sites on DNA. Cell. 1983 Aug;34(1):125–134. doi: 10.1016/0092-8674(83)90142-3. [DOI] [PubMed] [Google Scholar]
- Gralla J. D. Rapid "footprinting" on supercoiled DNA. Proc Natl Acad Sci U S A. 1985 May;82(10):3078–3081. doi: 10.1073/pnas.82.10.3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inuzuka M., Helinski D. R. Requirement of a plasmid-encoded protein for replication in vitro of plasmid R6K. Proc Natl Acad Sci U S A. 1978 Nov;75(11):5381–5385. doi: 10.1073/pnas.75.11.5381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inuzuka N., Inuzuka M., Helinski D. R. Activity in vitro of three replication origins of the antibiotic resistance plasmid RSF1040. J Biol Chem. 1980 Dec 10;255(23):11071–11074. [PubMed] [Google Scholar]
- Kolter R., Helinski D. R. Activity of the replication terminus of plasmid R6K in hybrid replicons in Escherichia coli. J Mol Biol. 1978 Sep 25;124(3):425–441. doi: 10.1016/0022-2836(78)90180-8. [DOI] [PubMed] [Google Scholar]
- Kolter R., Helinski D. R. Construction of plasmid R6K derivatives in vitro: characterization of the R6K replication region. Plasmid. 1978 Sep;1(4):571–580. doi: 10.1016/0147-619x(78)90014-8. [DOI] [PubMed] [Google Scholar]
- Kolter R., Helinski D. R. Plasmid R6K DNA replication. II. Direct nucleotide sequence repeats are required for an active gamma-origin. J Mol Biol. 1982 Oct 15;161(1):45–56. doi: 10.1016/0022-2836(82)90277-7. [DOI] [PubMed] [Google Scholar]
- Kolter R., Inuzuka M., Helinski D. R. Trans-complementation-dependent replication of a low molecular weight origin fragment from plasmid R6K. Cell. 1978 Dec;15(4):1199–1208. doi: 10.1016/0092-8674(78)90046-6. [DOI] [PubMed] [Google Scholar]
- Lovett M. A., Sparks R. B., Helinski D. R. Bidirectional replication of plasmid R6K DNA in Escherichia coli; correspondence between origin of replication and position of single-strand break in relaxed complex. Proc Natl Acad Sci U S A. 1975 Aug;72(8):2905–2909. doi: 10.1073/pnas.72.8.2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee S., Erickson H., Bastia D. Enhancer-origin interaction in plasmid R6K involves a DNA loop mediated by initiator protein. Cell. 1988 Feb 12;52(3):375–383. doi: 10.1016/s0092-8674(88)80030-8. [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay P., Filutowicz M., Helinski D. R. Replication from one of the three origins of the plasmid R6K requires coupled expression of two plasmid-encoded proteins. J Biol Chem. 1986 Jul 15;261(20):9534–9539. [PubMed] [Google Scholar]
- Roberts J. M. Simian virus 40 (SV40) large tumor antigen causes stepwise changes in SV40 origin structure during initiation of DNA replication. Proc Natl Acad Sci U S A. 1989 Jun;86(11):3939–3943. doi: 10.1073/pnas.86.11.3939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasse-Dwight S., Gralla J. D. KMnO4 as a probe for lac promoter DNA melting and mechanism in vivo. J Biol Chem. 1989 May 15;264(14):8074–8081. [PubMed] [Google Scholar]
- Schnos M., Zahn K., Inman R. B., Blattner F. R. Initiation protein induced helix destabilization at the lambda origin: a prepriming step in DNA replication. Cell. 1988 Feb 12;52(3):385–395. doi: 10.1016/s0092-8674(88)80031-x. [DOI] [PubMed] [Google Scholar]
- Shafferman A., Flashner Y., Hertman I. Genetic instability of an artificial palindrom DNA sequence. J Biomol Struct Dyn. 1983 Dec;1(3):729–742. doi: 10.1080/07391102.1983.10507478. [DOI] [PubMed] [Google Scholar]
- Shafferman A., Flashner Y., Hertman I., Lion M. Identification and characterization of the functional alpha origin of DNA replication of the R6K plasmid and its relatedness to the R6K beta and gamma origins. Mol Gen Genet. 1987 Jun;208(1-2):263–270. doi: 10.1007/BF00330452. [DOI] [PubMed] [Google Scholar]
- Shafferman A., Helinski D. R. Structural properties of the beta origin of replication of plasmid R6K. J Biol Chem. 1983 Apr 10;258(7):4083–4090. [PubMed] [Google Scholar]
- Shafferman A., Kolter R., Stalker D., Helinski D. R. Plasmid R6K DNA replication. III. Regulatory properties of the pi initiation protein. J Mol Biol. 1982 Oct 15;161(1):57–76. doi: 10.1016/0022-2836(82)90278-9. [DOI] [PubMed] [Google Scholar]
- Stalker D. M., Kolter R., Helinski D. R. Nucleotide sequence of the region of an origin of replication of the antibiotic resistance plasmid R6K. Proc Natl Acad Sci U S A. 1979 Mar;76(3):1150–1154. doi: 10.1073/pnas.76.3.1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stalker D. M., Kolter R., Helinski D. R. Plasmid R6K DNA replication. I. Complete nucleotide sequence of an autonomously replicating segment. J Mol Biol. 1982 Oct 15;161(1):33–43. doi: 10.1016/0022-2836(82)90276-5. [DOI] [PubMed] [Google Scholar]