Abstract
The mitochondrial network is a major site of ATP production through the coupled integration of the electron transport chain (ETC) with oxidative phosphorylation. In melanoma arising from the V600E mutation in the kinase v-RAF murine sarcoma viral oncogene homolog B (BRAFV600E), oncogenic signaling enhances glucose-dependent metabolism while reducing mitochondrial ATP production. Likewise, when BRAFV600E is pharmacologically inhibited by targeted therapies (e.g. PLX-4032/vemurafenib), glucose metabolism is reduced, and cells increase mitochondrial ATP production to sustain survival. Therefore, collateral inhibition of oncogenic signaling and mitochondrial respiration may help enhance the therapeutic benefit of targeted therapies. Honokiol (HKL) is a well tolerated small molecule that disrupts mitochondrial function; however, its underlying mechanisms and potential utility with targeted anticancer therapies remain unknown. Using wild-type BRAF and BRAFV600E melanoma model systems, we demonstrate here that HKL administration rapidly reduces mitochondrial respiration by broadly inhibiting ETC complexes I, II, and V, resulting in decreased ATP levels. The subsequent energetic crisis induced two cellular responses involving cyclin-dependent kinases (CDKs). First, loss of CDK1-mediated phosphorylation of the mitochondrial division GTPase dynamin-related protein 1 promoted mitochondrial fusion, thus coupling mitochondrial energetic status and morphology. Second, HKL decreased CDK2 activity, leading to G1 cell cycle arrest. Importantly, although pharmacological inhibition of oncogenic MAPK signaling increased ETC activity, co-treatment with HKL ablated this response and vastly enhanced the rate of apoptosis. Collectively, these findings integrate HKL action with mitochondrial respiration and shape and substantiate a pro-survival role of mitochondrial function in melanoma cells after oncogenic MAPK inhibition.
Keywords: apoptosis, mitochondria, mitochondrial respiratory chain complex, oncogene, respiration, mitochondrial dynamics, targeted therapy
Introduction
Oncogenic MAPK mutations (i.e. a valine to glutamate amino acid substitution at residue 600 in v-RAF murine sarcoma viral oncogene homolog B, BRAFV600E) occur within numerous tumor types, including malignant melanoma, colorectal, and ovarian cancers (1). The presence of the BRAFV600E mutation results in hyperactivation of the MAPK pathway, leading to continuous cell cycle progression and proliferation. Consequently, inhibition of oncogenic MAPK signaling is a rational therapeutic strategy that can capture the majority of MAPK-driven cancers. For example, several small molecule inhibitors that target either BRAFV600E (i.e. PLX-4032/vemurafenib) or downstream effector proteins, such as MEK (i.e. GSK-1120212/Trametinib), have been approved for use in melanoma (2–6). Nevertheless, despite promising initial responses, melanoma patients invariably relapse from MAPK, signaling inhibition within a year of treatment commencement (7, 8). Several studies have now demonstrated reactivation of MAPK signaling as one of the primary causes for the development of treatment resistance (9–11). Therefore, the development of alternative strategies to eradicate cancer cells before treatment resistance is urgently required.
Oncogenic reprogramming of cellular metabolism is a hallmark of many cancers whereby altered utilization of glucose and glutamine supports rapid proliferation. Indeed, a key feature of BRAFV600E melanoma cells is the metabolic switch from mitochondrial respiration to glycolysis, which is termed the Warburg effect (12). Several mechanisms have been discovered to describe the Warburg effect in BRAFV600E cancers, including increased glucose uptake and expression of glycolytic enzymes (12). However, recent work has demonstrated that changes in mitochondrial dynamics (i.e. mitochondrial fusion and fragmentation) play a critical role in regulating mitochondrial metabolism in both normal and cancer cells. For example, loss of function of the mitochondrial GTPase dynamin-related protein 1 (DRP1)2 causes fusion of the mitochondrial network and increases respiration (13, 14). Conversely, oncogenic MAPK signaling increases the expression and activity of DRP1, resulting in mitochondrial fragmentation and decreased respiration (13, 15). Inhibition of BRAFV600E in melanoma cells results in mitochondrial fusion and up-regulation of oxidative phosphorylation genes and increases mitochondrial biogenesis and respiration (13, 16). These studies indicate that mitochondrial dynamics, oncogenic MAPK signaling, and cancer metabolism are intricately linked to tumorigenesis (17, 18). Moreover, it is becoming evident that melanoma cells are dependent on mitochondria after inhibition of oncogenic MAPK signaling and, therefore, may be vulnerable to compounds that disrupt mitochondrial function.
Honokiol (HKL) is a small molecule compound that has anti-cancer properties (19, 20). Previous reports have demonstrated proapoptotic and anti-migratory effects of HKL in cancer cell lines and xenograft models of various malignancies, including breast (21), leukemia (22), and melanoma (23). Interestingly, recent work has shown that HKL can regulate mitochondrial function in normal and transformed cells (24, 25). However, the underlying molecular action of HKL on mitochondrial function and morphology has not yet been characterized. In the current study we demonstrate that HKL rapidly disrupts mitochondrial respiration by affecting complexes I, II, and V of the mitochondrial electron transport chain (ETC). The resulting energetic crisis causes distinct cellular phenotypes, including decreases in cyclin-dependent kinase 1 (CDK1)-mediated phosphorylation of DRP1, resulting in mitochondrial fusion, whereas decreases in CDK2 activity are associated with G1 cell cycle arrest. Furthermore, HKL can act as a single agent and in combination strategies with inhibition of oncogenic MAPK signaling to promote the mitochondrial pathway of apoptosis.
Results
HKL induced mitochondrial dysfunction in melanoma cells by disrupting respiration
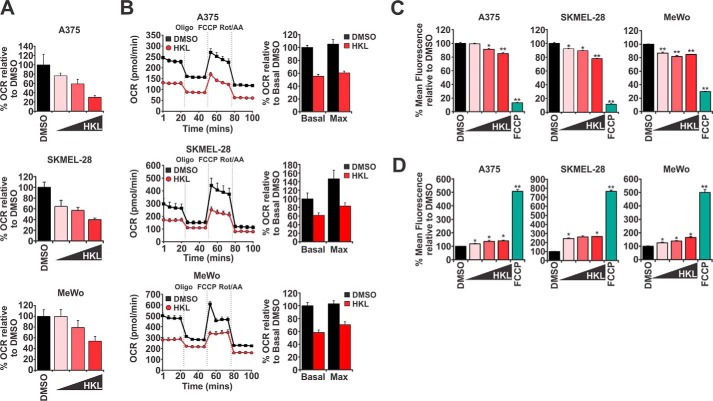
Currently there is little mechanistic information regarding the action of HKL in mitochondria. However, two independent studies have indicated that HKL is able to rapidly enter mitochondria after administration to cells (24, 25). We, therefore, began this study by first establishing if HKL has an immediate effect on mitochondrial respiration in melanoma cells. We used a Seahorse Bioanalyzer to measure the basal oxygen consumption rates (OCR), a surrogate readout of mitochondrial respiration in A375 (BRAFV600E), SKMEL-28 (BRAFV600E), and MeWo (wild-type BRAF) human malignant melanoma cell lines. Increasing doses of HKL resulted in a dose-dependent decrease in OCR in all cell lines tested (Fig. 1A). At the highest concentration of HKL (40 μm), OCR levels were reduced by 70%, 60%, and 46% in A375, SKMEL-28, and MeWo cells, respectively. Impressively, an HKL-mediated decrease in mitochondrial respiration occurred within 5 min of drug administration to cells, indicating rapid cellular uptake and targeting of mitochondrial function. To further elucidate the time-dependent effects HKL has on mitochondrial function, we assessed changes to OCR in cells treated with 40 μm HKL for 8 h. We found that HKL-treated cells exhibited significantly reduced basal OCR (between 38 and 45%) compared with control treated cells (Fig. 1B). Sequential administration of ETC complex inhibitors during these experiments allows for real-time assessment of different parameters associated with mitochondrial respiration. For example, oligomycin (complex V inhibitor) determines the amount of ATP production linked to respiration, and FCCP (carbonyl cyanide-4-(trifluoromethoxy)phenylhydrazone, mitochondrial uncoupler) enables measurement of maximal respiratory capacity, whereas a combination of rotenone (complex I inhibitor) and antimycin A (complex III inhibitor) determines the reserved respiratory capacity. We observed that HKL impaired all of these parameters compared with controls, indicating that HKL may inhibit multiple components of the ETC to repress overall cellular respiration (Fig. 1B).
Figure 1.
HKL promoted mitochondrial dysfunction by inhibiting cellular respiration. A, OCR were measured in A375, SKMEL-28, and MeWo melanoma cells using a Seahorse Bioanalyzer. DMSO or increasing doses (10, 20, or 40 μm) of HKL were directly administered to cells and incubated for ∼5 min followed by recording of OCR measurements. B, melanoma cells were treated with DMSO or HKL (40 μm) for 8 h before measuring OCR. Four baseline measurements were taken before sequential administration of oligomycin (1 μm), FCCP (1 μm), and a combination of rotenone (Rot) and antimycin A (AA) (0.5 μm). Bar graphs represent the effect HKL has on basal and maximal OCR. C and D, melanoma cells were treated with DMSO and increasing doses (10, 20, or 40 μm) of HKL for 8 h or FCCP (50 μm) for 2 h. Media was changed, and cells were loaded with TMRE (100 nm) or MitoSox for 30 min and analyzed by flow cytometry. Significance denoted by p < 0.05 (*) and p < 0.01 (**) by Student's t test. All data are representative of at least three independent experiments and are reported as the mean ± S.E.
To further interrogate HKL-mediated mitochondrial dysfunction, we used tetramethylrhodamine ethyl ester (TMRE; ThermoFisher Scientific) and MitoSox dyes to evaluate mitochondrial membrane potential and mitochondrial reactive oxygen species (ROS) levels after treatment with HKL. Flow cytometry analysis revealed HKL dose-dependently decreased in mitochondrial membrane potential with the highest concentration of HKL (40 μm)-reducing membrane potential between 15 and 22% after 8 h (Fig. 1C). Conversely, HKL treatment resulted in a dose-dependent increase in ROS production (140–160% increase at 40 μm HKL) in all cell lines tested (Fig. 1D). As expected, treatment with the mitochondrial uncoupler FCCP resulted in the maximal loss of membrane potential (70–89% decrease) and increase in ROS production (400–500%) in all cell lines (Fig. 1D). Importantly, the doses and timing of HKL treatment used in these experiments did not induce apoptosis (i.e. annexin V positivity) or cell cycle arrest (supplemental Fig. 1, A–B), indicating that HKL-mediated decreases in mitochondrial function are direct and are not due to secondary effects on the cell cycle or induction of cell death pathways.
Mitochondrial dysfunction caused by HKL occurred through loss of complex I, II, and V activity
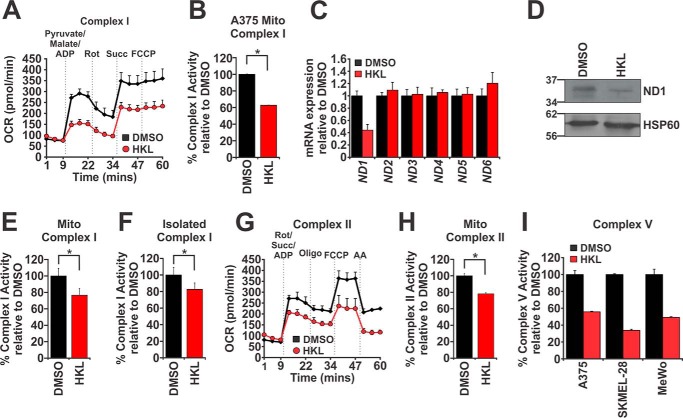
Given the potent inhibitory effect HKL has on mitochondrial respiration, we hypothesized that HKL acts to inhibit 1 or more ETC complexes. To investigate this, we utilized the Seahorse Bioanalyzer and used OCR as a surrogate readout of individual ETC complex activity after the addition of ETC metabolites and complex inhibitors (26). A375 cells were treated with either DMSO or 40 μm HKL for 8 h followed by permeabilization of the plasma membrane with recombinant perfringolysin O to ensure sufficient delivery of metabolites to mitochondria. Complex I activity was assessed by the addition of pyruvate, malate, and ADP. Pyruvate activates dehydrogenase subunits within complex I, whereas malate prevents the accumulation of acetyl CoA, which at high concentrations can inhibit pyruvate dehydrogenase activity (26). The inclusion of ADP maintains the mitochondria in a state of energy-stimulated oxygen consumption. In control-treated cells, the addition of complex I substrates increases OCR, whereas the subsequent addition of rotenone inhibits complex I activity and consequently reduces OCR (Fig. 2A). This reduction was reversed after the administration of the complex II metabolite succinate. Finally, to determine the maximal respiratory capacity of these cells, we administered FCCP and found no additional increase in OCR, indicating that the concentration of succinate (10 mm) used in these experiments was sufficient to cause maximal respiration (Fig. 2A). Likewise, we observed that HKL-treated A375 cells followed the same pattern of OCR responses after the addition of each compound (Fig. 2A). Importantly, however, the magnitude of responses was significantly lower compared with control-treated cells, indicating that complex I activity was impaired and that the addition of succinate was not sufficient to rescue respiration (Fig. 2A). These findings were confirmed in SKMEL-28 cells (supplemental Fig. 2A).
Figure 2.
HKL-mediated mitochondrial dysfunction occurred through inhibition of electron transport chain complexes I, II, and V. A, A375 cells were treated with DMSO or HKL (40 μm) for 8 h. Plasma membranes were permeabilized with recombinant perfringolysin O (1 nm) for 30 min at 37 °C. Basal OCR were measured in a Seahorse bioanalyzer before sequential administration of a combination of pyruvate (10 mm), malate (0.5 mm), and ADP (4 mm), succinate (Succ, 10 mm), and FCCP (1 μm). Rot, rotenone. B, complex I enzymatic activity was measured from A375 cells treated with DMSO or HKL (40 μm) for 24 h. Mitochondrial fractions were isolated and assessed for complex I activity. C, quantitative real-time PCR analysis of nicotinamide dehydrogenase (ND) subunits 1–6 from mRNA extracted from A375 cells treated with DMSO or HKL (40 μm) for 24 h. Gene expression was normalized to ACTB. D, Western blot analysis of ND1 protein levels from isolated mitochondrial fractions from A375 cells treated with either DMSO or HKL (40 μm) for 24 h. Hsp60 was used as a loading control. E, complex I enzymatic activity was measured from mouse liver mitochondrial extracts treated with DMSO or HKL (40 μm) for 30 min at room temperature. F, mouse liver mitochondria were lysed in complex I extraction buffer, and complex I proteins were immunoprecipitated on ELISA plates in the presence of DMSO or HKL (40 μm). G, same as in A except after basal OCR measurements cells were sequentially treated with a combination of rotenone (1 μm), succinate (10 mm), and ADP (4 mm), oligomycin (1 μm), FCCP (1 μm), and antimycin A (0.5 μm). H, same as in E except complex II enzymatic activity was analyzed. I, ATP-linked respiration was determined using a Seahorse bioanalyzer. Melanoma cells treated with DMSO or HKL (40 μm) for 8 h were treated with oligomycin (1 μm), and OCR measurements were recorded. Significance was denoted by p < 0.05 (*) by Student's t test. All data are representative of three independent experiments and are reported as the mean ± S.E.
To expand upon these findings we assessed the effect of HKL on complex I activity using an enzymatic ELISA-based assay. A375 cells were treated with either DMSO or 40 μm HKL for 24 h. We purified the mitochondrial fractions as previously described (27), immunoprecipitated complex I, and measured the rate of oxidation of NADH to NAD+ (i.e. complex I activity). There was a 37.2% reduction in the rate of complex I activity in HKL-treated cells compared with control, suggesting that indeed HKL targets complex I to reduce respiration (Fig. 2B). Given that this assay specifically measures NADH dehydrogenase activity, we hypothesized that HKL-mediated complex I impairment could be due to decreased expression of NADH dehydrogenase (ND) subunits. Quantitative real-time PCR analysis revealed that ND1 gene expression was reduced by 2.3-fold in HKL-treated A375 cells compared with controls, whereas the expression of all other ND subunits (ND2–6) remained unchanged (Fig. 2C). This decrease in ND1 mRNA expression correlated with a similar decrease in ND1 protein levels (Fig. 2D).
The above findings indicate that HKL mediates disruption of complex I activity through loss of expression of ND1. However, our earlier results indicate that either immediate administration (Fig. 1A) or short-term treatment (i.e. 8 h; Figs. 1B and 2A) of HKL reduces respiration, suggesting that HKL may directly affect complex I activity. To answer this possibility we isolated heavy membrane fractions from mouse livers and treated them with either DMSO or 40 μm HKL for 30 min at room temperature followed by measurement of complex I activity. The rate of complex I enzymatic activity was attenuated by 23% in HKL-treated mitochondria compared with controls (Fig. 2E). The ETC complexes are located within the inner mitochondrial membrane and are organized in a solid state system where individual complexes can assemble to form supercomplexes, which increases the efficiency of electron transfer and respiration (28, 29). We, therefore, asked if the context of complex I within the inner mitochondrial membrane, proximity to other ETC complexes, and other mitochondrial co-factors is essential for HKL-mediated inhibition of complex I activity. Mouse liver mitochondria were lysed, and complex I was separated from other mitochondrial components by immunoprecipitation either in the presence or absence of HKL. After 3 h of incubation, HKL was able to inhibit isolated complex I activity by 17.5% compared with control, indicating that HKL directly targets complex I activity (Fig. 2F).
Although complex I is a possible target for HKL, the decrease in complex I activity appears to be relatively minor when compared with overall loss in mitochondrial respiration as assessed by our earlier Seahorse analysis (Fig. 1, A and B), which indicates other ETC complexes may be involved. Mitochondrial respiration can be maintained via the donation of electrons from FADH2 to complex II. Indeed, our initial cell permeabilization experiments demonstrated ineffective succinate-mediated recovery of OCR in HKL-treated cells (Fig. 2A), indicating that HKL may act downstream of complex I. To assess if complex II is also affected by HKL, we permeabilized DMSO or HKL pretreated A375 and SKMEL-28 cells and administered a combination of rotenone, succinate, and ADP to specifically assess complex II activity independently of complex I. The addition of succinate increased OCR by 3.5-fold in control-treated cells compared with a 2.3-fold increase in HKL-treated cells (Fig. 2G, supplemental Fig. 2B), suggesting that HKL impairs complex II activity. The addition of oligomycin, FCCP, and antimycin A further demonstrates that all other respiration parameters are impaired in HKL-treated cells, which is most likely a consequence of HKL acting on multiple components of the ETC (Fig. 2G). To confirm complex II is directly inhibited by HKL, we conducted complex II ELISA-based enzymatic activity assays using mouse liver mitochondria treated with either DMSO or HKL for 30 min at room temperature. These experiments revealed that HKL reduced the rate of complex II enzymatic activity by 21.9% compared with control (Fig. 2H). Finally, we examined the effect inhibition of complex I and II has on ATP production, which is a measure of complex V activity. To answer this, we treated melanoma cells with or without HKL for 8 h and measured OCR after the addition of oligomycin and found that HKL decreased ATP production between 44–66% in all cell lines tested (Fig. 2I). Collectively, these data show that HKL specifically affects the activity of ETC complexes I, II, and V to reduce mitochondrial respiration.
HKL-mediated energetic crisis resulted in mitochondrial fusion via loss of CDK1-mediated DRP1 phosphorylation at serine 616
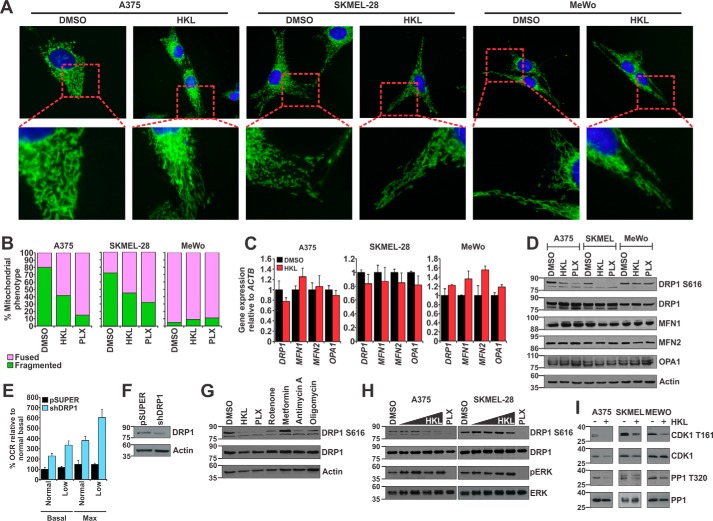
Thus far we have shown that HKL acts as an inhibitor of mitochondrial respiration, but an effect of HKL on mitochondrial function still remains unknown. Mitochondria are dynamic organelles capable of altering their shape, size, and subcellular localization to meet specific energetic and biological requirements (17). As such, a structural-functional relationship exists between specific mitochondrial shapes and their energetic states. For example, activation of the mitochondrial division protein DRP1 is essential for mitochondrial fragmentation during mitosis to ensure equal distribution of mitochondria to daughter cells (30). Fusion of the outer mitochondrial membrane is mediated by mitofusin 1 and 2 (MFN1 and MFN2), whereas inner mitochondrial membrane fusion is regulated by OPA1. Given that our findings show HKL induces an energetic crisis via ETC inhibition, we hypothesized subsequent changes in mitochondrial shape as a means of energetic compensation. A previous study demonstrated that oncogenic BRAFV600E signaling promotes a fragmented mitochondrial network through ERK-mediated phosphorylation of DRP1 at serine 616 (DRP1 Ser-616), a marker of active DRP1 (13). The specific BRAFV600E inhibitor PLX-4032 (PLX) effectively silences this pathway and results in mitochondrial fusion (13). We assessed mitochondrial morphology within A375, SKMEL-28, and MeWo cells using live cell fluorescent microscopy after treatment with DMSO and HKL, whereas PLX was used as a control drug that induces changes in mitochondrial morphology. Control-treated A375 and SKMEL-28 cells contain a fragmented mitochondrial phenotype, whereas the mitochondrial network in MeWo cells was more fused and interconnected (Fig. 3A). Treatment with HKL resulted in mitochondrial fusion in A375 and SKMEL-28 cells but did not affect MeWo cells (Fig. 3, A and B). As expected, PLX induced mitochondrial fusion in the BRAFV600E-positive A375 and SKMEL-28 cells. As MeWo cells are the wild type for BRAF, there was no effect of PLX on mitochondrial shape in these cells (Fig. 3, A and B, supplemental Fig. 3A).
Figure 3.
HKL caused mitochondrial fusion by down-regulating CDK1-mediated DRP1 phosphorylation at serine 616. A, melanoma cells were treated with DMSO or HKL (40 μm) for 24 h. Cells were loaded with MitoTracker Green and Hoechst 33342 (nuclei) and imaged. B, quantification of mitochondrial phenotypes in A375, SKMEL-28, and MeWo cells. In each treatment group 200 cells were counted and scored as containing either fused or fragmented mitochondria. C, quantitative real-time PCR analysis of mitochondrial dynamics genes from melanoma cells treated with DMSO or HKL (40 μm) for 24 h. Gene expression was normalized to ACTB. D, Western blot analysis of mitochondrial dynamics proteins from melanoma cells treated with DMSO, HKL (40 μm), or PLX-4032 (1 μm) for 24 h. β-Actin was used as a loading control. E, SKMEL-28 shDRP1 (or pSUPER) cells were analyzed for basal and maximal OCR in the presence of normal (35 mm) or low (5 mm) glucose. F, Western blot analysis of DRP1 protein levels in control pSUPER or shDRP1 SKMEL-28 cells. β-Actin was used as a loading control. G, A375 cells were treated with DMSO, HKL (40 μm), or 1 μm concentrations of either PLX-4032, rotenone, metformin, antimycin A, or oligomycin for 24 h. Protein lysates were analyzed by Western blot for phosphorylated DRP1 (DRP1 S616) and total DRP1. β-Actin was used as a loading control. H, A375 and SKMEL-28 cells were treated with DMSO, increasing doses (10, 20, 30, or 40 μm) HKL, or PLX-4032 (1 μm) for 24 h. Protein lysates were analyzed by Western blot for pDRP1 S616, total DRP1, phosphorylated ERK (pERK), and total ERK. I, melanoma cells were treated with DMSO or HKL (40 μm) for 24 h. Protein lysates were analyzed by Western blot for phosphorylated CDK1 (CDK1 T161), total CDK1, phosphorylated PP1 (PP1 T320), and total PP1. All data are representative of at least three independent experiments and are reported as the mean ± S.E.
To investigate the mechanism through which HKL treatment induces fusion of the mitochondrial network, we screened for changes within the mitochondrial dynamics machinery at both the mRNA transcript and protein levels. HKL treatment did not significantly alter the expression of the mitochondrial machinery nor were there any changes in total DRP1, MFN1, MFN2, and OPA1 protein levels (Fig. 3, C and D). However, DRP1 Ser-616 levels were significantly lower in HKL-treated cells (Fig. 3D). As expected, inhibition of BRAFV600E with PLX resulted in a decrease in DRP1 Ser-616 levels (Fig. 3D). We further integrated these results by determining how HKL affects the colocalization of DRP1 Ser-616 with fused or fragmented mitochondria. Using immunofluorescence, SKMEL-28 cells were stained for DRP1 Ser-616 and HSP60, a mitochondrial matrix marker. Under control conditions, DRP1 Ser-616 protein colocalized with fragmented mitochondria (supplemental Fig. 3B). Cells treated with HKL displayed reduced DRP1 Ser-616 associated with mitochondria, whereas PLX completely eliminated the colocalization between DRP1 Ser-616 and fused mitochondria (supplemental Fig. 3B). Collectively, these data indicate that HKL-mediated mitochondrial fusion occurs through inhibition of DRP1 and decreased mitochondrial fragmentation in BRAFV600E melanoma cells. A previous study demonstrated that oncogenic MAPK signaling promotes both increased glucose uptake and DRP1 activity (13). Given this relationship between oncogenic signaling, cellular metabolism, and mitochondrial shape, we were interested in assessing the effect glucose depletion and DRP1 knockdown has on mitochondrial respiration. SKMEL-28 cells were infected with pSUPER or shDRP1 and cultured for 72 h. Depletion of glucose results in a moderate increase in both basal and maximal OCR (Fig. 3E, black bars), an effect that was exacerbated after knockdown of DRP1 (Fig. 3, E and F), suggesting that DRP1 plays an important role in regulating mitochondrial respiration. Given that HKL inhibits both mitochondrial respiration and DRP1 phosphorylation, we wanted to assess if this was a common phenotype exhibited by other ETC inhibitors. Indeed, Western blot analysis demonstrated that inhibition of complex I (rotenone), complex III (antimycin A), or complex V (oligomycin) all reduce DRP1 Ser-616 levels, albeit to a lesser degree than HKL or PLX (Fig. 3G). We did not observe any decrease in DRP1 Ser-616 levels with metformin, a complex I inhibitor, which may be due to the fact that it has a lower potency at inhibiting complex I compared with rotenone (31).
HKL is a potent inhibitor of various oncogenic signaling pathways, including PI3K/mTORC and MAPK signaling in glioblastoma cells (32, 33). Given that HKL and PLX both act to decrease DRP1 Ser-616 levels, we asked if the mechanism of HKL-mediated inhibition of DRP1 was through blocking MAPK signaling. Using ERK phosphorylation (pERK) as a readout of active MAPK signaling, we demonstrated that treatment with PLX for 24 h significantly decreased pERK levels in A375 and SKMEL-28 cells, respectively (Fig. 3H). However, we did not observe any decrease in pERK or total ERK with increasing HKL concentrations, suggesting HKL does not inhibit oncogenic MAPK signaling in these melanoma cells. Nonetheless, HKL did reduce DRP1 Ser-616 levels, indicating that HKL may act on DRP1 through alternative mechanisms (Fig. 3H). It has been previously documented that during mitosis, DRP1 is phosphorylated at Ser-616 by CDK1 (34, 35). We, therefore, assessed if HKL affected CDK1 activity via phosphorylation of threonine 161 (CDK1 T161), which is representative of active CDK1. Western blot analysis showed that HKL reduced CDK1 Thr-161 levels in all melanoma cell lines (Fig. 3I). Total CDK1 was slightly lower in HKL-treated A375 cells but remained unaffected in SKMEL-28 and MeWo cells (Fig. 3I). Finally, to further integrate the inhibitory effect of HKL on CDK1 activity, we examined the phosphorylation status of protein phosphatase 1 (PP1), a known CDK1 substrate (36). Western blot analysis revealed that HKL-treated melanoma cells have decreased phosphorylation at threonine residue 320 (PP1 T320; Fig. 3I), indicating that HKL targets CDK1 and affects its downstream substrates. Collectively, these data suggest that HKL induction of mitochondrial fusion occurs through loss of CDK1-mediated activation of DRP1.
HKL promoted CDK2-mediated cell cycle arrest and induced the mitochondrial pathway of apoptosis
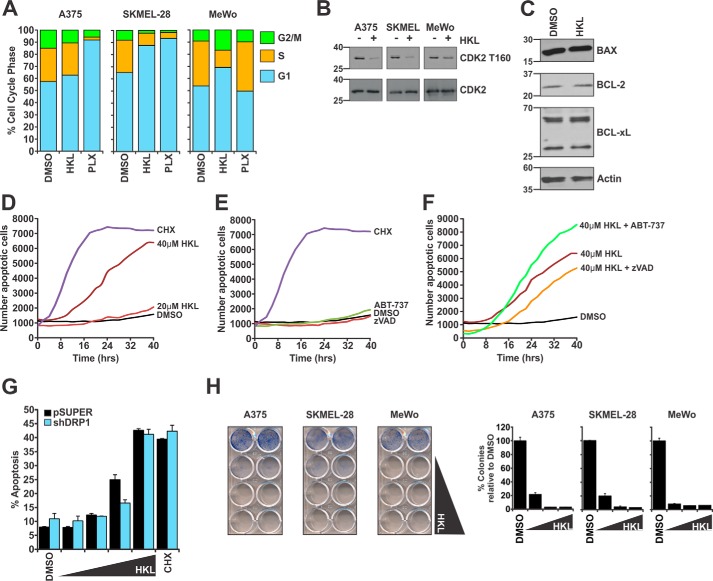
The finding that CDK1 activity is targeted by HKL prompted us to hypothesize that HKL treatment would result in G2-M arrest of the cell cycle, as this phase of the cell cycle is regulated by CDK1. Surprisingly, cell cycle analysis demonstrated that HKL-treated cells arrested in the G1 phase of the cell cycle (Fig. 4A, Table 1). As a control, PLX treatment induced G1 cell cycle arrest in A375 and SKMEL-28 but not MeWo cells as expected. These data suggest that HKL might target a regulator of G1-S transition. To elucidate the underlying mechanism for the G1-arrest phenotype, we examined the phosphorylation status of CDK2, a known regulator of the G1/S transition. HKL treatment decreased phosphorylation of CDK2 at threonine residue 160 (CDK2 T160), which is indicative of active CDK2, whereas total CDK2 levels were unaffected (Fig. 4B). Collectively, these findings suggest that loss of CDK2 activity mediates G1 cell cycle arrest, whereas the decrease in CDK1 prevents the phosphorylation of DRP1.
Figure 4.
HKL induced G1 cell cycle arrest and the mitochondrial pathway of apoptosis. A, melanoma cells were treated with DMSO, HKL (40 μm), or PLX-4032 (1 μm) for 24 h and analyzed for cell cycle distribution by flow cytometry. B, Melanoma cells treated the same as in A were analyzed by Western blot for phosphorylated CDK2 (CDK2 T160) and total CDK2 protein levels. C, A375 cells were treated with DMSO or HKL (40 μm) for 24 h. Protein lysates were analyzed by Western blot for components of the apoptotic machinery. β-Actin was used as a loading control. D–F, kinetic apoptosis assays were performed using an IncuCyte Zoom. A375 cells were treated with DMSO, HKL (20 or 40 μm), CHX (25 μg/ml), or ABT-737 (0.5 μm). Where indicated, cells were treated with zVAD-fmk (50 μm) for 2 h before treatment with HKL. Data represent the number of annexin V-positive events captured every 2 h over a time course of 40 h. G, pSUPER or shDRP1 SKMEL-28 cells were treated with DMSO or increasing doses (20, 30, 40, or 50 μm) HKL for 24 h. Apoptosis was measured by annexin V-staining by flow cytometry. CHX (25 μg/ml) was used as a positive control for apoptosis. H, colony formation assays were performed with melanoma cells plated at a density of 8000 cells/well and treated every 2 days with DMSO or increasing doses (40, 50, and 60 μm) HKL. Once control cells reached confluency, treatments were removed, and cells were stained with methylene blue. Graphs represent the percentage of colonies relative to DMSO. All data represent at least three independent experiments and are reported as the mean ± S.E.
Table 1.
Effect of HKL and PLX-4032 on cell cycle profiles
Data represent the mean percentage of cells in each cell cycle phase ± S.E.
| Cell line and treatment | %G1 ± S.E. | %S ± S.E. | %G2/M ± S.E. |
|---|---|---|---|
| A375 | |||
| DMSO | 57 ± 0.27 | 28 ± 1.1 | 15 ± 0.48 |
| 40 μm HKL | 63 ± 1.8 | 27 ± 2.2 | 11 ± 0.68 |
| 1 μm PLX | 92 ± 0.13 | 2 ± 2.9 | 6 ± 0.42 |
| SKMEL-28 | |||
| DMSO | 65 ± 0.53 | 27 ± 0.64 | 8 ± 0.88 |
| 40 μm HKL | 88 ± 2.2 | 10 ± 3 | 2.2 ± 0.89 |
| 1 μm PLX | 93 ± 0.78 | 5 ± 0.09 | 2 ± 0.70 |
| MeWo | |||
| DMSO | 54 ± 1.3 | 37 ± 1.1 | 9.1 ± 0.39 |
| 40 μm HKL | 69 ± 0.28 | 14 ± 3.7 | 17 ± 3.9 |
| 1 μm PLX | 50 ± 0.41 | 41 ± 0.35 | 10 ± 0.17 |
Previously published literature on the effects of HKL in cancer cells confirms a cell cycle arrest phenotype but also indicates that HKL is a potent inducer of cell death (19, 20). Although the mechanism of HKL-induced cell death remains unclear, several studies have indicated that HKL can down-regulate the anti-apoptotic proteins BCL-2 and BCL-xL (37, 38). However, Western blot analysis of A375 cells treated with DMSO or HKL found no change in BCL-xL protein levels or changes in protein levels other pro- and anti-apoptotic BCL-2 proteins (Fig. 4C). To characterize the cell death response, we performed kinetic apoptosis assays as measured by annexin V staining, which demonstrate that 40 μm HKL is capable of inducing cell death over the course of 40 h (Fig. 4D). To determine if this cell death phenotype is apoptosis, we pretreated cells with zVAD-fmk, a pan-caspase inhibitor, or co-treated with HKL and ABT-737, a small-molecule inhibitor to several anti-apoptotic BCL-2 proteins (i.e. BCL-2, BCL-xL, and BCL-W). These compounds are well tolerated by cells as single agents (Fig. 4E). Pretreatment with zVAD-fmk reduced HKL-mediated cell death, whereas combination of HKL with ABT-737 increased annexin V staining, indicating that the mechanism of cell death is the mitochondrial pathway of apoptosis (Fig. 4F). Given the effects of HKL on mitochondrial shape, we asked if the presence of DRP1 was important for HKL-mediated apoptosis to proceed. Knockdown of DRP1 in SKMEL-28 did not significantly alter the induction of HKL-mediated apoptosis (Fig. 4G), indicating that DRP1 does not play any significant role in mediating HKL-induced apoptosis. Finally, colony formation assays indicate that increasing doses of HKL causes a marked reduction in long-term clonogenic survival of melanoma cell lines (Fig. 4H). Collectively, these data indicate that HKL is an inhibitor of the cell cycle by reducing the activity of CDK2 while also inducing the mitochondrial pathway of apoptosis.
MAPK inhibition increases respiration and sensitizes melanoma cells to HKL-induced apoptosis
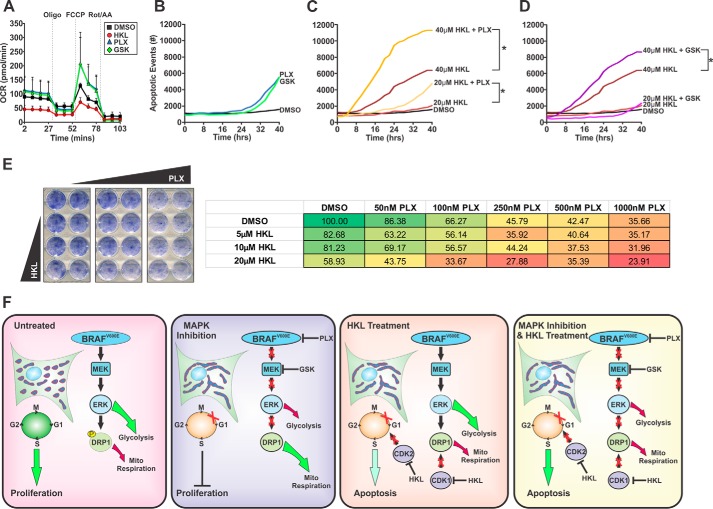
Oncogenic MAPK signaling promotes glycolysis and inhibits mitochondrial respiration (12, 13). Although MAPK pathway inhibitors are not potent cell death inducers as single agents, they effectively block MAPK signaling and induce cell cycle arrest (Fig. 3G and 4A). We hypothesize that an additional consequence of MAPK inhibition is increased respiration, which will sensitize cells to ETC inhibitors, such as HKL. Indeed, treatment with PLX or the MEK inhibitor GSK-1120212 (GSK) increased cellular respiration, whereas HKL decreased OCR levels consistent with our earlier data (Fig. 5A). We also increased the concentration of PLX (10 μm) and GSK (50 nm) and assessed apoptotic responses over time. Both drugs were able to induce a moderate apoptotic response after 30 h of treatment (Fig. 5B). Importantly, the combination of PLX with either 20 or 40 μm HKL significantly enhanced the rate of apoptosis, whereas a similar response was observed with the combination of GSK and HKL (Fig. 5, C–D). Finally, the combination of these drugs at lower doses significantly decreased clonogenic survival (Fig. 5E, supplemental Fig. 4). These data demonstrate that inhibition of MAPK signaling markedly increased mitochondrial respiration, which sensitizes melanoma cells to HKL-induced apoptosis.
Figure 5.
HKL-mediated mitochondrial dysfunction potentiates the apoptotic responses of MAPK inhibition. A, A375 cells were treated with DMSO, HKL (40 μm), PLX-4032 (1 μm), or GSK-1120212 (10 nm) for 24 h and then analyzed for changes in OCR using a Seahorse Bioanalyzer. Four basal OCR measurements were made before sequential treatment of oligomycin (1 μm), FCCP (1 μm), and a combination of rotenone and antimycin A (0.5 μm). B–D, kinetic apoptosis assays were performed in A375 cells treated with DMSO, PLX-4032 (10 μm), GSK-1120212 (10 nm), HKL (20 or 40 μm), or in combination. Annexin V-positive events were captured using an IncuCyte Zoom every 2 h over a period of 40 h. Significance was denoted by p < 0.05 (*) by Student's t test. E, colony formation assays were performed using A375 cells plated at a density of 8000 cells/well and treated with DMSO, increasing doses of HKL (5, 10, or 20 μm), PLX-4032 (50, 100, 250, 500, or 1000 nm), or in combination every 2 days. Cells were stained with methylene blue. Representative percentages of colonies are presented. All data are representative of three independent experiments and are reported as the mean ± S.E. F, model summarizing the effects of MAPK inhibitors and HKL on cellular metabolism, mitochondrial shape, cell cycle progression, and cell fate. Inhibition of MAPK signaling causes mitochondrial fusion, increased mitochondrial respiration, cell cycle arrest, and decreased proliferation. HKL inhibits mitochondrial respiration, increases mitochondrial fusion, cell cycle arrest, and apoptosis. Combined HKL and inhibition of MAPK signaling significantly enhances apoptotic responses.
Discussion
This study reveals several underlying mechanisms of how HKL disrupts mitochondrial function in melanoma cancer cells (Fig. 5F). In brief, we demonstrate that HKL is a potent inhibitor of mitochondrial respiration by decreasing the activities of ETC complexes I, II, and V (Fig. 1, A and B, and Fig. 2, A, B, E–H, and I), HKL-induced fusion of the mitochondrial network by inhibiting the CDK1-DRP1 axis (Fig. 3, A and I), and finally, HKL synergized with MAPK inhibitors to enhance apoptotic responses (Fig. 5, A–F). Collectively, these data represent novel HKL modalities that impinge on mitochondrial function and highlight the impact of small molecules that target mitochondrial ATP production as anti-cancer strategies.
Oncogenic signaling, such as BRAFV600E in melanoma, actively adjusts cellular metabolism toward glycolysis to support the high energetic and anabolic demands of rapidly proliferating cancer cells (12). Furthermore, glucose dependence within these cancers is the foundation by which clinical imaging techniques (i.e. positron emission tomography) map metabolic responses before and after treatment. Indeed, clinical results demonstrate a complete loss of glucose uptake by BRAFV600E positive melanomas after treatment with PLX-4032; however, there is considerable variation in responses with reduction in tumor burden, suggesting that alternative pathways are active to maintain ATP production and cancer cell survival (39, 40). In the current study we demonstrate that PLX-4032 and GSK-1120212 increase mitochondrial respiration in BRAFV600Emelanoma cells (Fig. 5A). Importantly, we showed that the dependence on mitochondrial respiration after oncogenic MAPK inhibition sensitized melanoma cells to HKL-induced apoptosis and decreased clonogenic survival (Fig. 5, B–E).
Melanomas have complex genetic and mutational expression profiles (41), which consequently limits the clinical utility of MAPK inhibitors through intrinsic and acquired mechanisms of resistance. Intrinsic resistance refers to melanoma cells that, in addition to BRAFV600E, contain additional genetic alterations, mutations, and/or amplifications (i.e. loss of PTEN, amplification of cylin D1) that override anti-proliferative effect of MAPK inhibition (42). Conversely, acquired resistance to MAPK inhibition frequently occurs within 6–8 months after treatment and is usually due to reactivation of the MAPK pathway (7–11). As single agent therapies, inhibition of MAPK signaling induces apoptosis in only a small fraction of cells, whereas the remainder either enter cell cycle arrest to stabilize disease or do not demonstrate a clinically response (i.e. intrinsically resistant). The subpopulation of cancer cells that display intrinsic resistance to MAPK inhibitors tend to have high rates of mitochondrial biogenesis and are metabolically dependent on mitochondrial respiration (43). The information provided here suggests that this subpopulation of melanoma cells may be sensitive to HKL-induced mitochondrial dysfunction and apoptosis. Consequently, a window of opportunity for alternative therapeutic intervention before reactivation of MAPK signaling and subsequent treatment relapse is often lost. Combined targeting of mitochondrial respiration and oncogenic MAPK signaling, such as HKL and PLX-4032, as presented here may prevent the selection of resistant tumor cells and improve overall patient survival. It would be interesting to further evaluate HKL and MAPK inhibitors using in vivo models of melanoma to establish if simultaneous inhibition of oncogenic signaling and mitochondrial respiration promotes durable responses and ultimately cancer cell apoptosis.
A recent report suggested that HKL negatively affected mitochondrial respiration in non-small cell lung cancer; however, the mechanism was not characterized (24). Our study reveals that HKL directly targets complexes I, II, and V of the ETC (Fig. 2, A, B, and E–I), which causes a state of energetic crisis and, ultimately, induction of cell death (Fig. 4D). We discovered that complex I activity was inhibited by both direct administration of HKL as well as loss of ND1 expression after longer treatment (Fig. 2, C and D). The observation that HKL is able to rapidly decrease complex I activity indicates that HKL may influence complex activity, stability, or assembly. The complex I enzymatic assay used in this study measures the rate of NADH dehydrogenase activity, which is not dependent on ubiquinone to shuttle electrons between complexes I-III. Inhibitors that bind at or near ubiquinone sites (i.e. rotenone) are ineffective at inhibiting complex I activity in this assay. In this context, our data suggest that HKL does not decrease respiration by affecting the transfer of electrons between complexes but most likely directly inhibits NADH dehydrogenase activity. Similarly, we found that HKL diminished the activity of complex II and V, possibly indicating congruent modes of HKL action between ETC complexes. Alternatively, inhibition of multiple ETC complexes by HKL suggests that this drug may also influence mitochondrial ultrastructure or supercomplex assembly (44).
The underlying notion that inhibiting ETC function is an effective anti-cancer strategy is supported by use of metformin in cancer patients (45). Metformin, a type II diabetic drug and complex I inhibitor, has been associated with a decreased risk of cancer development while also possessing anti-cancer properties in vitro and in vivo (45). However, metformin requires organic cation transporters that are only expressed in a few tissues, such as the liver and kidney (46). Likewise, not all cancer cells express organic cation transporters, indicating a potentially low therapeutic index for metformin in certain cancer patients (46). Other ETC inhibitors, such as rotenone (complex I), antimycin A (complex III), and oligomycin (complex V), are either highly toxic or have poor pharmacokinetic properties. HKL is a non-toxic, small molecular weight biphenolic compound capable of inhibiting mitochondrial respiration without the requirement for specific transporters. Moreover, HKL is derived from the bark of Magnolia grandiflora, which has been used in traditional Chinese medicine for centuries without any appreciable toxicity (47, 48), indicating that HKL displays two selective advantages over current ETC inhibitors. First, HKL simultaneously targets multiple ETC complexes, and second, it is well tolerated by normal cells (48). Although there are no known mitochondrial respiratory inhibitors that specifically act within neoplastic cells, it appears the activity of HKL in normal and cancer cells fits within a general class of phenolic compounds, including curcumin and resveratrol. In cancer cells, these compounds inhibit complex V activity, resulting in decreased ATP production, mitochondrial membrane potential, increased ROS generation, and apoptosis (49). Whether or not other phenolic compounds share similar properties to HKL, such as inhibiting ETC complex I and II in cancer cells, remains unknown. Conversely, in normal cells phenolic compounds have cytoprotective effects. For example, both HKL and resveratrol prevent cardiac hypertrophy by enhancing mitochondrial respiration via a Sirt3-dependent mechanism (25, 50), suggesting the phenolic class of small molecules has distinct effects in non-neoplastic and cancer cells. This may highlight a role for mitochondrial ultrastructure and shape mediating the tissue and cellular responses to HKL.
An additional finding from our study is that HKL causes mitochondrial fusion after a decrease in phosphorylated DRP1 Ser-616 levels (Fig. 3, A and D). Interestingly, this loss of function did not occur through inhibition of oncogenic MAPK signaling but rather through loss of CDK1 activity (Fig. 3, H–I). Similarly, we found CDK2 is a HKL target resulting in G1 cell cycle arrest (Fig. 4B). Previous studies found that HKL induces a similar cell cycle arrest phenotype, albeit through targeting the G1 regulators CDK4 and CDK6 (51–53), indicating that HKL may act as a pan-CDK inhibitor. The recent resurgence in developing targeted therapies against cell cycle regulators, such as CDKs, suggests this approach to be a viable anti-cancer strategy. However, given the level of redundancy across the CDK family (with the exception of CDK1), developing specific cell cycle inhibitors has thus far been problematic, whereas pan-CDK inhibitors are highly toxic (54). The ability of HKL to target multiple CDKs with a high therapeutic index suggests that it may overcome many of the issues surrounding previous CDK inhibitors. Taken together, this study illuminates how HKL affects mitochondrial respiration, mainly through the inhibition of ETC activity, and provides insights into the therapeutic rationale of duel inhibition of oncogenic signaling and mitochondrial function in melanoma.
Experimental procedures
Cell lines and reagents
All cell culture and transfection reagents were obtained from ThermoFisher Scientific, and standard reagents were from Sigma or Fisher. A375, SK-MEL-28, and MeWo lines were obtained from American Type Cell Culture (ATCC) and were maintained in Dulbecco's modified Eagle's medium (DMEM) containing 10% fetal bovine serum, 2 mm l-glutamine, and 1% antibiotics. Drugs were obtained from: ABT-737 (Abbott), PLX-4032/GSK-1120212 (Selleck Chemicals), zVAD-fmk (Calbiochem), Hoechst-33342 (Anaspec), and cycloheximide (CHX; Sigma). Honokiol was kindly provided by the laboratory of Jack Arbiser (Emory University School of Medicine, Atlanta, GA). Antibodies are presented in supplemental Table 1.
Seahorse analyses
For mitochondrial respiration experiments, cells were seeded in XFe96 plates (Agilent) using the following plating densities, A375 (100,000 cells/well), SKMEL-28 (80,000 cells/well), MeWo (100,000 cells/well), and treated with DMSO or 40 μm HKL for 8 h. OCR were measured using the XFe96 Extracellular Flux Analyzer and the XF Cell Mito stress test kit (Agilent) according to the manufacturer's instructions. Briefly, media containing treatments were aspirated, and cells were washed once with unbuffered XF Mito stress media (DMEM supplemented with 100 mm glucose, 200 mm glutamine, and 100 mm pyruvate, pH 7.4). Cells in XF Mito stress media were incubated at 37 °C in the absence of CO2 for 1 h. Baseline OCR measurements were determined before administration of oligomycin (1 μm), FCCP (1 μm), and a combination of rotenone and antimycin A (0.5 μm). Cells were stained with methylene blue and destained, and the absorbance was measured at 668 nm using a plate reader (Synergy H1 Hybrid multi-mode micro-plate reader, Biotek). OCR measurements were normalized against the cell densities, and all data analysis was conducted with the Seahorse Wave software version 2.2.0.276 (Agilent). For cell permeabilization experiments, cells were seeded in XFe96 plates and treated with DMSO or 40 μm HKL for 8 h. Media were removed, and cells were washed in 1× mitochondrial assay buffer (MAS: 220 mm mannitol, 70 mm sucrose, 10 mm KH2PO4, 5 mm MgCl2, 2 mm HEPES, 1 mm EGTA, 0.2% fatty acid free BSA). Cells in 1× MAS buffer containing 1 nm plasma membrane permeabilizer (Agilent) were incubated at 37 °C in the absence of CO2 for 30 min. Baseline OCR measurements were recorded before administration of complex metabolites and/or inhibitors as indicated. OCR measurements were normalized to total cell number as indicated above.
Mitochondrial membrane potential (ΔϕM) and mitochondrial ROS measurements
Cells seeded (60,000 cells/well) in 48-well plates were treated with either DMSO or increasing doses of HKL for 8 h or with 50 μm FCCP for 2 h at 37 °C. Either 100 nm TMRE or 5 μm MitoSOX (ThermoFisher Scientific) was added, incubated at 37 °C for 30 min, trypsinized, and analyzed on a Canto II flow cytometer (BD Biosciences).
Cell cycle assays
Cells were seeded (100,000 cells/well) in 12-well plates and treated as indicated for 24 h. Cells were harvested by trypsinization, fixed in ice-cold 70% ethanol with gentle vortexing, and incubated at 4 °C overnight. The following day the cells were pelleted, washed twice with ice-cold 1× PBS, resuspended in 1× PBS containing 60 μg/ml propidium iodine and 50 μg/ml RNase (Qiagen), and incubated at room temperature for 30 min. Cells were analyzed on a Canto II flow cytometer (BD Biosciences).
Heavy membrane isolations
Cells were seeded in 15-cm dishes and treated with either DMSO or 40 μm HKL for 24 h. Cells were harvested by trypsinization and centrifuged at 1000 × g for 10 min. Heavy membrane fractions (i.e. mitochondria) were isolated as previously described (27). Briefly, cell pellets were washed once in 1× PBS and resuspended in mitochondrial isolation buffer (MIB: 200 mm mannitol, 68 mm sucrose, 10 mm HEPES-KOH, pH 7.4, 10 mm KCl, 1 mm EDTA, 1 mm EGTA, 0.1% BSA) supplemented with protease inhibitors (ThermoFisher Scientific). Cells were incubated on ice for 20 min and homogenized using a 2-ml Potter-Elvehjem Dounce. To ensure proper clearance of unlysed cells and nuclei, the homogenate was subjected to 2 rounds of centrifugation at 800 × g for 10 min at 4 °C. The resulting supernatant was centrifuged for 10 min at 8000 × g at 4 °C, supernatants were aspirated, and the pellet was collected as the heavy membrane fraction. Pellets were resuspended in 100 μl of MIB and quantified by absorbance at 520 nm. An absorbance reading of 0.25 equates to ∼20 mg/ml. Samples were diluted to 100 μg and subjected to SDS-PAGE and Western blot analyses.
Complex I and II enzymatic ELISA
Complex I and II enzymatic activity measurements were performed using enzyme microplate kits (Abcam) and heavy membrane isolates from wild-type BALB/C mouse livers and A375 and SKMEL-28 cells. Briefly, A375 and SKMEL-28 cells were treated with either DMSO or 40 μm HKL for 24 h. Heavy membranes were isolated as described above. Mouse liver mitochondrial isolations were performed as described (27). Total protein concentrations were quantified using the Pierce BCA protein assay kit (ThermoFisher Scientific), and between 10 and 100 μg of mitochondrial proteins were used in each assay. Complex I and II activity assays were performed according to the manufacturer's instructions, and kinetic measurements were recorded by absorbance readings at 450 nm and 600 nm, respectively, every 30 s for 60 min.
Live cell imaging
Cells were seeded (40,000 cells/6-cm plate) for 24 h before the indicated treatments. Mitochondria and nuclei were labeled with 100 nm Mito Tracker Green (ThermoFisher Scientific) and 4 μm Hoechst 33342 (ThermoFisher Scientific) for 30 min at 37 °C, respectively. All imaging was performed on a Zeiss Imager.Z1 equipped with an N-Achroplan 40×/0.75 water immersion lens and an AxioCAM MRm digital camera. Images were captured using AxioVision 4.8 and Zeiss Zen software (Zeiss).
Immunofluorescence
SKMEL-28 cells were seeded (10,000/chamber) on 1.8 cm2 NuncTM Lab-TekTM chamber slides (ThermoFisher Scientific) for 24 h before the indicated treatments. Cells were washed with 1× PBS, fixed in 4% formaldehyde, and incubated in blocking buffer (1× PBS supplemented with 5% normal goat serum and 0.3% TritonTM X-100) for 60 min at room temperature. Primary antibodies for DRP1 Ser-616 (1:3200) and HSP60 (1:300) were diluted in 1× PBS supplemented with 1% BSA and 0.3% TritonTM X-100 and incubated overnight at 4 °C. Cells were washed 3× for 5 min each with PBS, and the secondary antibodies Alexa Fluor® 488 (1:1000, Cell Signaling) and Texas Red (1:200, Cell Signaling) were diluted in the above primary antibody buffer and incubated for 90 min at room temperature in the dark. Cells were washed 3× for 5 min each in PBS and then mounted with ProLong® Gold Antifade Reagent containing DAPI, cured overnight at room temperature, and imaged with a Zeiss Imager.Z1 at 40× magnification and an AxioCAM MRm digital camera; images were captured using AxioVision 4.8 software.
SDS-PAGE analyses
To generate whole cell protein lysates, cells were treated were indicated, trypsinized, pelleted, and washed in 1× PBS. Cell pellets were resuspended in radioimmune precipitation assay buffer (1× PBS containing 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulfate) supplemented with protease inhibitors (HALT; Pierce). Cells were incubated on ice for 20 min and centrifuged for 10 min at 21,000 × g at 4 °C. Quantification of lysates was performed using the Pierce BCA protein assay kit (ThermoFisher Scientific). Proteins (30–50 μg/lane) were subjected to SDS-PAGE before transferring to nitrocellulose by standard Western blot protocols and blocked in 5% milk/TBST (TBS-Tween). Primary antibodies (1:1000 in blocking buffer) were added to membranes and incubated overnight at 4 °C. Blots were washed 3× for 10 min each with TBST, and the secondary antibody (1:2000 in blocking buffer) was incubated at room temperature for 30 min followed by 3× for 10 min TBST wash steps. Protein bands were detected by standard enhanced chemiluminescence detection and exposure to film.
Quantitative real-time PCR
Total cellular RNA extractions and first strand cDNA synthesis were performed as described (13). Gene expression was analyzed using SYBR Green detection (FastStart Universal SYBR Green Master Mix, Roche Applied Science) and Applied Biosciences ViiA 7 Real-Time PCR system (ThermoFisher Scientific). Relative gene expression was determined using the comparative CT method and normalized to 18S. Primer sequences for all genes investigated are located in supplemental Table 2.
RNA interference and stable clone generation
The human pSUPER-shDRP1 plasmid was kindly provided by Dr. David Kashatus (University of Virginia, School of Medicine, Charlottesville, VA), and scrambled shRNA construct was kindly provided by the laboratory of Dr. R. Premkumar Reddy (Icahn School of Medicine at Mount Sinai). HEK-293T cells were used to produce lentiviral particles for the generation of stable scramble shRNA and shDRP1 cell lines. Virus supernatants were harvested at 48 and 72 h, pooled, and 0.45-μm filtered. SKMEL-28 stable clones were generated using puromycin (0.5 mg/ml).
Apoptosis and clonogenic survival assays
For annexin V cell death assays, cells were seeded for 24 h and treated as indicated. Floating and attached cells were harvested, stained with annexin V-FITC in binding buffer (10 mm HEPES, pH 7.4, 150 mm NaCl, 5 mm KCl, 1 mm MgCl2, 1.8 mm CaCl2), and analyzed by flow cytometry as described (13). For IncuCyte ZOOM (Essen Bioscience) experiments, cells were plated in 96-well plates and treated where indicated with annexin V labeled with AlexaFluor 594 NHS ester (ThermoFisher Scientific) as described (55). Briefly, experiments were conducted for 24–48 h with data collection every 2 h using a 10× objective lens. Data were processed and analyzed using the IncuCyte ZOOM software package (Version 6.2.9200.0). Processing definitions are presented in supplemental Table 3. Clonogenic survival studies were performed by seeding cells in 24-well plates and treated every 2 days as described until control-treated cells reach confluency. Colonies were stained with methylene blue and imaged. Destaining buffer (20% methanol in 5% acetic acid) was added to each well and measured for absorbance at 668 nm using a microplate reader (Biotek).
Statistical analyses
Statistical analyses were performed as indicated in the figure legends. Student's t tests were performed in Microsoft Excel to determine statistical differences between two groups. Significance was set at p < 0.05.
Author contributions
A. P. T. and J. E. C. were involved in the conception and design of experiments. A. P. T., P. L., J. D. G., M. N. S., and J. E. C. were involved in data acquisition, analysis, and interpretation. A. P. T. and J. E. C. prepared the figures and wrote the manuscript. All authors were involved in critically reviewed the results and approved the final version of the manuscript.
Supplementary Material
Acknowledgments
We thank everyone in the J. E. C. laboratory for assistance and support. We also thank Dr. Robert Fisher and members of his laboratory (Icahn School of Medicine at Mount Sinai) for critical reagents and assistance. The Tisch Center Institute was supported by the Cancer Center Support Grant p30 CA196521 from the National Institutes of Health.
This work was supported by National Institutes of Health Grants CA157740 (to J. E. C.), CA206005 (to J. E. C.), and AR4790 (to J. L. A). This work was also supported by the JJR Foundation, the William A. Spivak Fund, the Fridolin Charitable Trust, the Rabinowitch-Davis Foundation, an American Cancer Society Research Scholar Award, a Leukemia & Lymphoma Society Career Development Award, and an Irma T. Hirschl/Monique Weill-Caulier Trust Research Award. This work was also supported in part by two research grants (5FY1174 and 1FY13416) from the March of Dimes Foundation and the Developmental Research Pilot Project Program within the Department of Oncological Sciences at the Icahn School of Medicine at Mount Sinai. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains supplemental Figs. S1–S4 and Tables S1–S3.
- DRP1
- dynamin-related protein 1
- HKL
- honokiol
- ETC
- electron transport chain
- OCR
- oxygen consumption rate(s)
- ROS
- reactive oxygen species
- ND
- NADH dehydrogenase
- CDK
- cyclin-dependent kinase
- FCCP
- carbonyl cyanide-4-(trifluoromethoxy)phenylhydrazone
- TMRE
- tetramethylrhodamine ethyl ester
- MFN
- mitofusin
- PLX
- PLX-4032
- PP1
- protein phosphatase 1
- zVAD-fmk
- benzyloxycarbonyl-Val-Ala-Asp-fluoromethyl ketone
- CHX
- cycloheximide.
References
- 1. Burotto M., Chiou V. L., Lee J. M., and Kohn E. C. (2014) The MAPK pathway across different malignancies: a new perspective. Cancer 120, 3446–3456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Flaherty K. T., Puzanov I., Kim K. B., Ribas A., McArthur G. A., Sosman J. A., O'Dwyer P. J., Lee R. J., Grippo J. F., Nolop K., and Chapman P. B. (2010) Inhibition of mutated, activated BRAF in metastatic melanoma. N. Engl. J. Med. 363, 809–819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Joseph E. W., Pratilas C. A., Poulikakos P. I., Tadi M., Wang W., Taylor B. S., Halilovic E., Persaud Y., Xing F., Viale A., Tsai J., Chapman P. B., Bollag G., Solit D. B., and Rosen N. (2010) The RAF inhibitor PLX4032 inhibits ERK signaling and tumor cell proliferation in a V600E BRAF-selective manner. Proc. Natl. Acad. Sci. U.S.A. 107, 14903–14908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Solit D. B., Garraway L. A., Pratilas C. A., Sawai A., Getz G., Basso A., Ye Q., Lobo J. M., She Y., Osman I., Golub T. R., Sebolt-Leopold J., Sellers W. R., and Rosen N. (2006) BRAF mutation predicts sensitivity to MEK inhibition. Nature 439, 358–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Infante J. R., Fecher L. A., Falchook G. S., Nallapareddy S., Gordon M. S., Becerra C., DeMarini D. J., Cox D. S., Xu Y., Morris S. R., Peddareddigari V. G., Le N. T., Hart L., Bendell J. C., Eckhardt G., et al. (2012) Safety, pharmacokinetic, pharmacodynamic, and efficacy data for the oral MEK inhibitor trametinib: a phase 1 dose-escalation trial. Lancet Oncol. 13, 773–781 [DOI] [PubMed] [Google Scholar]
- 6. Flaherty K. T., Robert C., Hersey P., Nathan P., Garbe C., Milhem M., Demidov L. V., Hassel J. C., Rutkowski P., Mohr P., Dummer R., Trefzer U., Larkin J. M., Utikal J., Dreno B., et al. (2012) Improved survival with MEK inhibition in BRAF-mutated melanoma. N. Engl. J. Med. 367, 107–114 [DOI] [PubMed] [Google Scholar]
- 7. Larkin J., Ascierto P. A., Dréno B., Atkinson V., Liszkay G., Maio M., Mandalà M., Demidov L., Stroyakovskiy D., Thomas L., de la Cruz-Merino L., Dutriaux C., Garbe C., Sovak M. A., Chang, et al. (2014) Combined vemurafenib and cobimetinib in BRAF-mutated melanoma. N. Engl. J. Med. 371, 1867–1876 [DOI] [PubMed] [Google Scholar]
- 8. Long G. V., Stroyakovskiy D., Gogas H., Levchenko E., de Braud F., Larkin J., Garbe C., Jouary T., Hauschild A., Grob J. J., Chiarion Sileni V., Lebbe C., Mandalà M., Millward M., Arance A., Bondarenko I., et al. (2014) Combined BRAF and MEK inhibition versus BRAF inhibition alone in melanoma. N. Engl. J. Med. 371, 1877–1888 [DOI] [PubMed] [Google Scholar]
- 9. Karoulia Z., Wu Y., Ahmed T. A., Xin Q., Bollard J., Krepler C., Wu X., Zhang C., Bollag G., Herlyn M., Fagin J. A., Lujambio A., Gavathiotis E., and Poulikakos P. I. (2016) An integrated model of RAF inhibitor action predicts inhibitor activity against oncogenic BRAF signaling. Cancer cell 30, 485–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lito P., Pratilas C. A., Joseph E. W., Tadi M., Halilovic E., Zubrowski M., Huang A., Wong W. L., Callahan M. K., Merghoub T., Wolchok J. D., de Stanchina E., Chandarlapaty S., Poulikakos P. I., Fagin J. A., and Rosen N. (2012) Relief of profound feedback inhibition of mitogenic signaling by RAF inhibitors attenuates their activity in BRAFV600E melanomas. Cancer Cell 22, 668–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Poulikakos P. I., Persaud Y., Janakiraman M., Kong X., Ng C., Moriceau G., Shi H., Atefi M., Titz B., Gabay M. T., Salton M., Dahlman K. B., Tadi M., Wargo J. A., Flaherty K. T., et al. (2011) RAF inhibitor resistance is mediated by dimerization of aberrantly spliced BRAF(V600E). Nature 480, 387–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Abildgaard C., and Guldberg P. (2015) Molecular drivers of cellular metabolic reprogramming in melanoma. Trends Mol. Med. 21, 164–171 [DOI] [PubMed] [Google Scholar]
- 13. Serasinghe M. N., Wieder S. Y., Renault T. T., Elkholi R., Asciolla J. J., Yao J. L., Jabado O., Hoehn K., Kageyama Y., Sesaki H., and Chipuk J. E. (2015) Mitochondrial division is requisite to RAS-induced transformation and targeted by oncogenic MAPK pathway inhibitors. Mol. Cell 57, 521–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wakabayashi J., Zhang Z., Wakabayashi N., Tamura Y., Fukaya M., Kensler T. W., Iijima M., and Sesaki H. (2009) The dynamin-related GTPase Drp1 is required for embryonic and brain development in mice. J. Cell Biol. 186, 805–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kashatus J. A., Nascimento A., Myers L. J., Sher A., Byrne F. L., Hoehn K. L., Counter C. M., and Kashatus D. F. (2015) Erk2 phosphorylation of Drp1 promotes mitochondrial fission and MAPK-driven tumor growth. Mol. Cell 57, 537–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Haq R., Shoag J., Andreu-Perez P., Yokoyama S., Edelman H., Rowe G. C., Frederick D. T., Hurley A. D., Nellore A., Kung A. L., Wargo J. A., Song J. S., Fisher D. E., Arany Z., and Widlund H. R. (2013) Oncogenic BRAF regulates oxidative metabolism via PGC1α and MITF. Cancer Cell 23, 302–315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Trotta A. P., and Chipuk J. E. (2017) Mitochondrial dynamics as regulators of cancer biology. Cell. Mol. Life Sci. 74, 1999–2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wieder S. Y., Serasinghe M. N., Sung J. C., Choi D. C., Birge M. B., Yao J. L., Bernstein E., Celebi J. T., and Chipuk J. E. (2015) Activation of the mitochondrial fragmentation protein DRP1 correlates with BRAF(V600E) melanoma. J. Investig. Dermatol. 135, 2544–2547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Fried L. E., and Arbiser J. L. (2009) Honokiol, a multifunctional antiangiogenic and antitumor agent. Antioxid. Redox Signal. 11, 1139–1148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Prasad R., and Katiyar S. K. (2016) Honokiol, an active compound of magnolia plant, inhibits growth, and progression of cancers of different organs. Adv. Exp. Med. Biol. 928, 245–265 [DOI] [PubMed] [Google Scholar]
- 21. Xie L., Jiang F., Zhang X., Alitongbieke G., Shi X., Meng M., Xu Y., Ren A., Wang J., Cai L., Zhou Y., Xu Y., Su Y., Liu J., Zeng Z., et al. (2016) Honokiol sensitizes breast cancer cells to TNF-α induction of apoptosis by inhibiting Nur77 expression. Br. J. Pharmacol. 173, 344–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhou B., Li H., Xing C., Ye H., Feng J., Wu J., Lu Z., Fang J., and Gao S. (2017) Honokiol induces proteasomal degradation of AML1-ETO oncoprotein via increasing ubiquitin conjugase UbcH8 expression in leukemia. Biochem. Pharmacol. 128, 12–25 [DOI] [PubMed] [Google Scholar]
- 23. Prasad R., Kappes J. C., and Katiyar S. K. (2016) Inhibition of NADPH oxidase 1 activity and blocking the binding of cytosolic and membrane-bound proteins by honokiol inhibit migratory potential of melanoma cells. Oncotarget 7, 7899–7912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Pan J., Zhang Q., Liu Q., Komas S. M., Kalyanaraman B., Lubet R. A., Wang Y., and You M. (2014) Honokiol inhibits lung tumorigenesis through inhibition of mitochondrial function. Cancer prevention research 7, 1149–1159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Pillai V. B., Samant S., Sundaresan N. R., Raghuraman H., Kim G., Bonner M. Y., Arbiser J. L., Walker D. I., Jones D. P., Gius D., and Gupta M. P. (2015) Honokiol blocks and reverses cardiac hypertrophy in mice by activating mitochondrial Sirt3. Nat. Commun. 6, 6656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Salabei J. K., Gibb A. A., and Hill B. G. (2014) Comprehensive measurement of respiratory activity in permeabilized cells using extracellular flux analysis. Nat. Protoc. 9, 421–438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Renault T. T., Floros K. V., and Chipuk J. E. (2013) BAK/BAX activation and cytochrome c release assays using isolated mitochondria. Methods 61, 146–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wittig I., Carrozzo R., Santorelli F. M., and Schägger H. (2006) Supercomplexes and subcomplexes of mitochondrial oxidative phosphorylation. Biochim. Biophys. Acta 1757, 1066–1072 [DOI] [PubMed] [Google Scholar]
- 29. Dudkina N. V., Eubel H., Keegstra W., Boekema E. J., and Braun H. P. (2005) Structure of a mitochondrial supercomplex formed by respiratory-chain complexes I and III. Proc. Natl. Acad. Sci. U.S.A. 102, 3225–3229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Otera H., Ishihara N., and Mihara K. (2013) New insights into the function and regulation of mitochondrial fission. Biochim. Biophys. Acta 1833, 1256–1268 [DOI] [PubMed] [Google Scholar]
- 31. Viollet B., Guigas B., Sanz Garcia N., Leclerc J., Foretz M., and Andreelli F. (2012) Cellular and molecular mechanisms of metformin: an overview. Clin. Sci. 122, 253–270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lin C. J., Chen T. L., Tseng Y. Y., Wu G. J., Hsieh M. H., Lin Y. W., and Chen R. M. (2016) Honokiol induces autophagic cell death in malignant glioma through reactive oxygen species-mediated regulation of the p53/PI3K/Akt/mTOR signaling pathway. Toxicol. Appl. Pharmacol. 304, 59–69 [DOI] [PubMed] [Google Scholar]
- 33. Zhang Y., Ren X., Shi M., Jiang Z., Wang H., Su Q., Liu Q., Li G., and Jiang G. (2014) Down-regulation of STAT3 and activation of MAPK are involved in the induction of apoptosis by HNK in glioblastoma cell line U87. Oncol. Rep. 32, 2038–2046 [DOI] [PubMed] [Google Scholar]
- 34. Kashatus D. F., Lim K. H., Brady D. C., Pershing N. L., Cox A. D., and Counter C. M. (2011) RALA and RALBP1 regulate mitochondrial fission at mitosis. Nat. Cell Biol. 13, 1108–1115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Taguchi N., Ishihara N., Jofuku A., Oka T., and Mihara K. (2007) Mitotic phosphorylation of dynamin-related GTPase Drp1 participates in mitochondrial fission. J. Biol. Chem. 282, 11521–11529 [DOI] [PubMed] [Google Scholar]
- 36. Kwon Y. G., Lee S. Y., Choi Y., Greengard P., and Nairn A. C. (1997) Cell cycle-dependent phosphorylation of mammalian protein phosphatase 1 by cdc2 kinase. Proc. Natl. Acad. Sci. U.S.A. 94, 2168–2173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Arora S., Bhardwaj A., Srivastava S. K., Singh S., McClellan S., Wang B., and Singh A. P. (2011) Honokiol arrests cell cycle, induces apoptosis, and potentiates the cytotoxic effect of gemcitabine in human pancreatic cancer cells. PloS ONE 6, e21573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Deng J., Qian Y., Geng L., Chen J., Wang X., Xie H., Yan S., Jiang G., Zhou L., and Zheng S. (2008) Involvement of p38 mitogen-activated protein kinase pathway in honokiol-induced apoptosis in a human hepatoma cell line (hepG2). Liver Int. 28, 1458–1464 [DOI] [PubMed] [Google Scholar]
- 39. Baudy A. R., Dogan T., Flores-Mercado J. E., Hoeflich K. P., Su F., van Bruggen N., and Williams S. P. (2012) FDG-PET is a good biomarker of both early response and acquired resistance in BRAFV600 mutant melanomas treated with vemurafenib and the MEK inhibitor GDC-0973. EJNMMI Res. 2, 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. McArthur G. A., Puzanov I., Amaravadi R., Ribas A., Chapman P., Kim K. B., Sosman J. A., Lee R. J., Nolop K., Flaherty K. T., Callahan J., and Hicks R. J. (2012) Marked, homogeneous, and early [18F]fluorodeoxyglucose-positron emission tomography responses to vemurafenib in BRAF-mutant advanced melanoma. J. Clin. Oncol. 30, 1628–1634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Cancer Genome Atlas Network (2015) Genomic classification of cutaneous melanoma. Cell 161, 1681–1696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Haarberg H. E., and Smalley K. S. (2014) Resistance to Raf inhibition in cancer. Drug Discov. Today Technol. 11, 27–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Zhang G., Frederick D. T., Wu L., Wei Z., Krepler C., Srinivasan S., Chae Y. C., Xu X., Choi H., Dimwamwa E., Ope O., Shannan B., Basu D., Zhang D., Guha M., et al. (2016) Targeting mitochondrial biogenesis to overcome drug resistance to MAPK inhibitors. J. Clin. Investig. 126, 1834–1856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Cogliati S., Frezza C., Soriano M. E., Varanita T., Quintana-Cabrera R., Corrado M., Cipolat S., Costa V., Casarin A., Gomes L. C., Perales-Clemente E., Salviati L., Fernandez-Silva P., Enriquez J. A., and Scorrano L. (2013) Mitochondrial cristae shape determines respiratory chain supercomplexes assembly and respiratory efficiency. Cell 155, 160–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kasznicki J., Sliwinska A., and Drzewoski J. (2014) Metformin in cancer prevention and therapy. Ann. Transl. Med. 2, 57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Weinberg S. E., and Chandel N. S. (2015) Targeting mitochondria metabolism for cancer therapy. Nat. Chem. Biol 11, 9–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Liu Z., Zhang X., Cui W., Zhang X., Li N., Chen J., Wong A. W., and Roberts A. (2007) Evaluation of short-term and subchronic toxicity of magnolia bark extract in rats. Regul. Toxicol. Pharmacol. 49, 160–171 [DOI] [PubMed] [Google Scholar]
- 48. Zhang Q., Li J., Zhang W., An Q., Wen J., Wang A., Jin H., and Chen S. (2015) Acute and sub-chronic toxicity studies of honokiol microemulsion. Regul. Toxicol. Pharmacol. 71, 428–436 [DOI] [PubMed] [Google Scholar]
- 49. Cerella C., Radogna F., Dicato M., and Diederich M. (2013) Natural compounds as regulators of the cancer cell metabolism. Int. J. Cell Biol. 2013, 639401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Chen T., Li J., Liu J., Li N., Wang S., Liu H., Zeng M., Zhang Y., and Bu P. (2015) Activation of SIRT3 by resveratrol ameliorates cardiac fibrosis and improves cardiac function via the TGF-β/Smad3 pathway. Am. J. Physiol. Heart Circ. Physiol. 308, H424–H434 [DOI] [PubMed] [Google Scholar]
- 51. Singh T., Gupta N. A., Xu S., Prasad R., Velu S. E., and Katiyar S. K. (2015) Honokiol inhibits the growth of head and neck squamous cell carcinoma by targeting epidermal growth factor receptor. Oncotarget 6, 21268–21282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Vaid M., Sharma S. D., and Katiyar S. K. (2010) Honokiol, a phytochemical from the magnolia plant, inhibits photocarcinogenesis by targeting UVB-induced inflammatory mediators and cell cycle regulators: development of topical formulation. Carcinogenesis 31, 2004–2011 [DOI] [PubMed] [Google Scholar]
- 53. Hahm E. R., and Singh S. V. (2007) Honokiol causes G0-G1 phase cell cycle arrest in human prostate cancer cells in association with suppression of retinoblastoma protein level/phosphorylation and inhibition of E2F1 transcriptional activity. Mol. Cancer Ther. 6, 2686–2695 [DOI] [PubMed] [Google Scholar]
- 54. Asghar U., Witkiewicz A. K., Turner N. C., and Knudsen E. S. (2015) The history and future of targeting cyclin-dependent kinases in cancer therapy. Nat. Rev. Drug Discov. 14, 130–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Gelles J. D., and Edward Chipuk J. (2016) Robust high-throughput kinetic analysis of apoptosis with real-time high-content live-cell imaging. Cell Death Disease 7, e2493. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.