Abstract
During fungal spore germination, a resting spore returns to a conventional mode of cell division and resumes vegetative growth, but the requirements for spore germination are incompletely understood. Here, we show that copper is essential for spore germination in Schizosaccharomyces pombe. Germinating spores develop a single germ tube that emerges from the outer spore wall in a process called outgrowth. Under low-copper conditions, the copper transporters Ctr4 and Ctr5 are maximally expressed at the onset of outgrowth. In the case of Ctr6, its expression is broader, taking place before and during outgrowth. Spores lacking Ctr4, Ctr5, and the copper sensor Cuf1 exhibit complete germination arrest at outgrowth. In contrast, ctr6 deletion only partially interferes with formation of outgrowing spores. At outgrowth, Ctr4-GFP and Ctr5-Cherry first co-localize at the spore contour, followed by re-location to a middle peripheral spore region. Subsequently, they move away from the spore body to occupy the periphery of the nascent cell. After breaking of spore dormancy, Ctr6 localizes to the vacuole membranes that are enriched in the spore body relative to the germ tube. Using a copper-binding tracker, results showed that labile copper is preferentially localized to the spore body. Further analysis showed that Ctr4 and Ctr6 are required for copper-dependent activation of the superoxide dismutase 1 (SOD1) during spore germination. This activation is critical because the loss of SOD1 activity blocked spore germination at outgrowth. Taken together, these results indicate that cell-surface copper transporters and SOD1 are required for completion of the spore germination program.
Keywords: copper, metal homeostasis, superoxide dismutase (SOD), yeast, yeast physiology, copper transporters, fission yeast, spore germination
Introduction
Schizosaccharomyces pombe cell proliferation generally occurs in a haploid state through rounds of mitotic cell divisions under nutrient-rich conditions. In contrast, under nutrient-starved conditions such as nitrogen deficiency, haploid cells of the opposite mating types conjugate and form precursor diploid cells that can switch from mitosis to meiosis (1). Once in meiosis, diploid cells undergo one round of DNA replication, followed by two successive rounds of chromosome segregation without an intervening S-phase that generate four haploid sets of chromosomes, termed chromatids. Each set of chromatids is then enclosed in a forespore that by a developmental process results in four mature haploid spores that are enclosed into an ascus (2). After an extended period of time, S. pombe asci are autonomously lysed, releasing spores into the environment. However, under unfavorable conditions, spores remain highly resistant to a variety of environmental stresses due to the presence of a spore-specific structure called the outer spore wall (OSW)4 or the spore coat. The OSW provides a sealed physical barrier between the outside environment and the fungal spore content (3, 4).
Under favorable nutrient and environmental conditions, dormant spores undergo a developmental process called germination in which case each quiescent spore converts itself into a vegetative cell that re-enters the mitotic cell cycle to resume growth and division (5, 6). Spore germination in S. pombe is divided into distinct stages. First, dormancy of the spores is stopped, and their activation is accompanied by loss of spore refractility that can be observed by light microscopy and a decrease in optical absorbance of the spore suspension. Second, spores undergo an isotropic swelling process that results in doubling of their volume. Third, swollen spores enter outgrowth. This stage is characterized by a singular rupture in the OSW that is accompanied by an initial emergence of the germ tube, resulting in the formation of pear-shaped spores. Fourth, the germ tube emerges at one side and grows away from the spore body in an unidirectional manner. Fifth, the duplicated chromosomal material migrates into the cylindrical part, and then the new daughter cell is divided by septation from the mother spore body (5). Previous studies have shown that fungal spore germination is a multistep process whose nutritional requirements differed at the early stage of germination in comparison with subsequent stages such as outgrowth (6–8). In S. pombe, the presence of glucose without any additional nutrients is sufficient for spores to exit dormancy as measured by the loss of spore refractility (9). However, subsequent morphogenesis development fails to occur in glucose-grown spores for which swelling and germ-tube formation are defective and absent, respectively (6). Based on the fact that spore germination implies a profound change in the resting state of the spore, which involves major morphological and metabolic modifications toward its activation, it is expected that essential nutrients are required to satisfy the physiological demand of this multistep developmental process.
Copper is an obligatory micronutrient for aerobic organisms (10). It serves as a catalytic or structural cofactor in a variety of enzymes, including cytochrome c oxidase, copper-zinc superoxide dismutase (SOD1), multicopper ferroxidase, and copper amine oxidase (CAO). These enzymes are essential to fundamental cellular processes such as respiration, superoxide anion detoxification, iron transport, and xenobiotic amine metabolism, respectively (11). In the model organism S. pombe, the copper transport machinery has mostly been studied in dividing cells that grow mitotically. In these cells, copper is taken up by a two-component transporting complex that is composed of the Ctr4 and Ctr5 proteins located at the cell surface (12–16). Studies have shown that Ctr4 and Ctr5 are unable to function independently in copper acquisition. A clear interdependence between Ctr4 and Ctr5 has been established because the secretion of either protein to the plasma membrane requires the concomitant secretion of the partner protein (12, 16). Bimolecular fluorescence complementation experiments have shown that the assembly of a functional heteromeric Ctr4-Ctr5 complex at the cell surface requires the combination of two Ctr4 molecules and one Ctr5 molecule (14). ctr4Δ, ctr5Δ, or ctr4Δ ctr5Δ mutants exhibit phenotypes associated with copper deficiency. These mutants are characterized by their inability to take up radioactive 64Cu and their inability to grow in low-copper medium. In addition, the mutant cells display alterations in copper-dependent enzyme activities, including SOD1, copper amine oxidase 1 (Cao1), and cytochrome c oxidase (15, 17). As is the case for most members of the Ctr family in fungal species, ctr4+ and ctr5+ genes are regulated at the level of transcription as a function of copper availability. They are induced under conditions of copper starvation and repressed in response to high-copper concentrations. The transcription factor Cuf1 is required to activate ctr4+ and ctr5+ gene expression (18–20). Cuf1 associates with ctr4+ and ctr5+ promoters in copper-starved cells in vivo. In contrast, high concentrations of copper inhibit the binding of Cuf1 to chromatin (15).
Based on the fact that meiosis requires copper to successfully undertake and complete its differentiation program, expression and localization of the Ctr4 and Ctr5 proteins have been investigated in meiotic and sporulating cells (21, 22). When the cells switch from mitosis to meiosis, expression and localization profiles of Ctr4 and Ctr5 show that the two proteins are still synchronously co-expressed and co-localized at the cell surface of zygotes but only during the early steps of the meiotic program (22). After meiotic divisions, the transcript levels of ctr4+ and ctr5+ are extinguished with the concomitant disappearance of their encoded proteins. At this stage, the meiosis-specific copper transporter Mfc1 is expressed and subsequently appears at the forespore membrane of ascospores where it serves to transport copper for accurate and timely meiotic differentiation under low-copper conditions (21).
In S. pombe, Ctr6 is a third member of the Ctr family that localizes at the membrane of vacuoles in cells proliferating in mitosis under copper-limiting conditions (23). Ctr6 is an integral membrane protein that assembles as a homotrimer (23). A deletion of the ctr6+ gene (ctr6Δ) results in a reduction of SOD1 activity (22, 23). Similarly, inactivation of ctr4+ (ctr4Δ) also causes a decrease of SOD1 activity but to a greater extent. When both ctr6+ and ctr4+ genes are inactivated, the double mutant (ctr6Δ ctr4Δ) fails to display measurable SOD1 activity, revealing a functional contribution of Ctr6 and Ctr4 in providing copper to at least one cytosolic copper-dependent enzyme under low-copper conditions (22, 23). On the basis of studies on Ctr6 and its ortholog Ctr2 in Saccharomyces cerevisiae (24, 25), Ctr6 has been predicted to function as a vacuolar membrane copper transporter that mobilizes stored copper from the vacuole to the cytosol, thereby participating with a pathway by which copper could be distributed within cells from the organelle according to copper needs. In the case of cells that undergo meiotic differentiation, Ctr6 localizes at the membrane of vacuoles during the first stages of meiosis (22). After meiotic divisions, Ctr6 undergoes an intracellular re-location to co-localize with the forespore membrane at late anaphase II (22). Although the relocation of Ctr6 to the nascent forespore membrane is still unclear, one possibility may be a potential role in transporting stored copper from the prespore to the cytosol where meiotic copper-dependent enzymes are present such as SOD1. As opposed to the expression of ctr4+ and ctr5+ that is exclusively dependent on the presence of Cuf1, ctr6+ expression is broader throughout the entire meiotic process and relies on two distinct regulators, Cuf1 and Mei4 (22).
In this study, we have determined that S. pombe spore outgrowth is a copper-dependent developmental process. During spore germination, ctr4+ and ctr5+ genes are primarily co-expressed in response to low concentrations of copper through Cuf1, with peaks of expression at the end of isotropic swelling and at the onset of outgrowth. In the case of ctr6+, its expression slightly increases in a time-dependent manner from the exit dormancy to outgrowth. Although ctr6+ expression is more robust under low-copper conditions in the presence of Cuf1, a low and constitutive level of expression is observed under basal and copper-replete conditions that is Cuf1-independent. Spores lacking Ctr4, Ctr5, or Cuf1 exhibit a germination arrest at the onset of outgrowth. In the case of ctr6Δ mutant spores, production of developing spores and newborn vegetative cells is reduced in comparison to wild-type spores. During the formation of the polar cap, Ctr4 and Ctr5 co-localize at the periphery of the spore and subsequently at the plasma membrane of the new daughter cell. In the case of Ctr6, it localizes on the membrane of vacuoles. Using the fluorescent copper-binding tracker CNIR4, labile copper is preferentially detected in the spore body, whereas the germ tube exhibits less CNIR4-copper complex-associated fluorescence. Furthermore, results showed the critical importance of copper transport to a specific copper-dependent target because removal of SOD1 leads to a spore developmental arrest at the onset of outgrowth. Taken together, our findings highlight the essential requirement of copper and copper transport proteins for the developmental process of spore germination and outgrowth under low-copper conditions.
Results
Spore germination and outgrowth in S. pombe
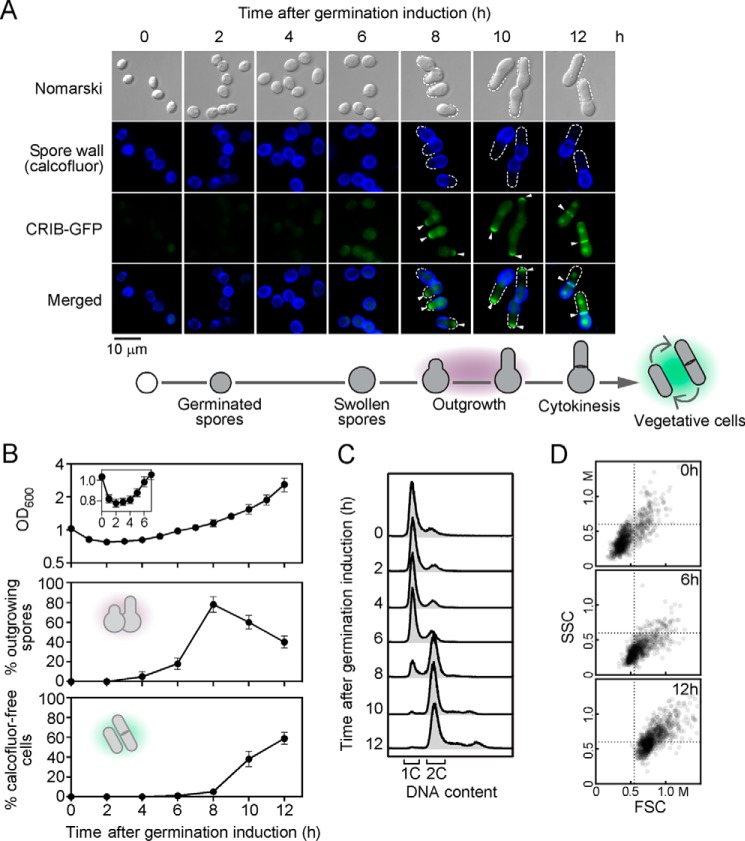
A non-dividing haploid spore represents a resting state in fission yeast. Haploid spores are more resistant to different environmental stresses than vegetative cells that proliferate in mitosis (3, 4). Purified spores are highly refractile when they are observed by phase-contrast microscopy (5, 6). Herein, we used differential interference contrast (DIC or Nomarski) microscopy as a complementary approach (Fig. 1A). Although DIC microscopy gives lower image brightness than standard bright-field microscopy, it provides far more highly defined images of the spore morphology. To monitor morphological changes, we therefore used DIC microscopy and spores harboring a CRIB-GFP allele (26, 27). In S. pombe, the GTP-bound form of Cdc42 binds and activates a group of proteins that possess the CRIB (Cdc42/Rac-interactive binding) domain (27). By itself, the CRIB domain fused to GFP associates with the GTP-bound form of Cdc42 that localizes at new growing cell end(s). CRIB-GFP is therefore used as a marker for spore outgrowth dynamics. Dormant purified spores were first incubated in the presence of calcofluor (10 μg/ml) in glucose-free media. After a 30-min incubation period, dormant spores were washed and synchronously induced to enter germination in glucose-containing and calcofluor-free media in the presence of CuSO4 (50 μm). After initiation of spore germination, we observed that the optical density (OD600) of the spore suspension decreased (Fig. 1B). This observation was consistent with previous reports in which results have shown that light-refractile spores became dark after 2–3 h of induction of germination (5, 6). During the bright- to dark-phase transition, the OD600 of the spore suspension decreased in parallel with darkening (6). Spores then increased their size for ∼4–6 h (Fig. 1A). After 7–8 h of spore germination, a protrusion (also called germ tube) began to emerge at one side of each enlarged spore (Fig. 1A). At this stage and thereafter, an outgrowing spore adopted a bottle-like shape and the germ tube grew away from the spore body to eventually produce a calcofluor-free daughter cell (Fig. 1, A and B). DNA content was determined by flow cytometry (FACS) analysis to assess whether ungerminated spores were restricted to single genome content (1C DNA). At the zero-time point and over a time period of 6 h after induction of germination, spores primarily exhibited 1C DNA content (Fig. 1C). Results showed that spores duplicated their genome (2C DNA) when they entered outgrowth (8-h time point, Fig. 1). At the end of outgrowth, developing spores solely exhibited 2C DNA content (10- and 12-h time points). In addition, forward light scatter (FSC) and side-scatter (SSC) analysis were performed after spores had been synchronously induced into germination. Results showed that at the zero-time point, the FSC/SSC cytogram exhibited the lowest values, which was consistent with a population of single spores that possessed small sizes (FSC) and very low internal granularity (SSC) (Fig. 1D). After 6 h of induction of germination, swollen spores increased their dimensions, which resulted in slightly higher FSC values with a global distribution of points moving toward the right (Fig. 1D). At the 12-h time point, the germ tube has emerged and expended, causing an increase in size of individual cell. This increase in size correlated well with the highest FSC values, whereas further differentiation of cells growing away from the spore body produced higher SSC values, presumably due to formation of organelles that were detected as granularity signals (Fig. 1D).
Figure 1.
Changes in morphology during spore germination and outgrowth. A, purified spores containing a CRIB-GFP allele were synchronously induced to initiate and proceed through germination in the presence of copper (50 μm). When expressed in germinating spores, CRIB-GFP was used as a marker to highlight the new cell tip during outgrowth and the site of cell division (septum) during cytokinesis (center bottom). Calcofluor (blue) was used as a marker to indicate the spore wall and the mother spore side, especially during outgrowth (center top). New cell side of outgrowing cells is indicated with white dashed lines, and white arrowheads show examples of CRIB-GFP cellular locations. The merged images are shown in the far bottom panels. Nomarski optics (far top) was used to examine spore and cell morphologies. Schematic representations of morphological changes of spores during germination and outgrowth are shown below the microscopy panels. Results of microscopy are representative of five independent experiments. B, total growth was measured at OD600 as a function of time after induction of germination. Inset indicates measurements at OD600 for the first 7 h of spore germination (top). The percentage of spores undergoing outgrowth (middle) and the percentage of calcofluor-free vegetative cells (bottom) were determined as a function of time after induction of germination. Values represent the average ± S.D. of three independent experiments. C, FACS analysis of purified spores that contained one nucleus with a single complete genome (1C DNA). Following induction of germination, outgrowing spores duplicated their genome (2C DNA) as shown by FACS analysis. D, dot plot of FSC versus SSC in which each dot represents a single spore or a calcofluor-free cell during early (0 h), middle (6 h), and late (12 h) phases, respectively, of the germination program. FSC is proportional to spore size, and SSC is proportional to spore granularity.
Copper deficiency blocks the outgrowth of developing spores
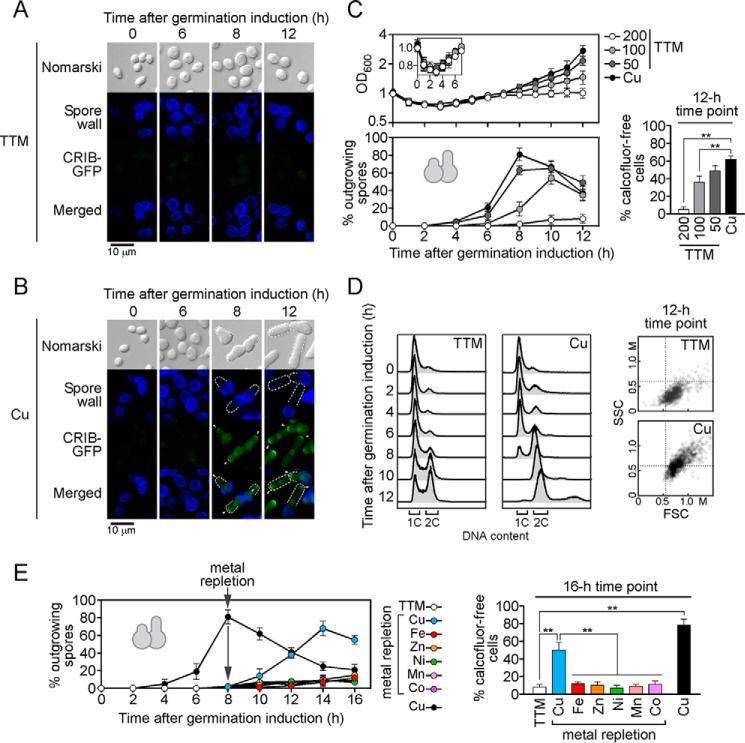
Although yeast cells require copper as a redox cofactor when they are committed to the aerobic mitotic cell cycle or to the meiotic program, little is known about the role of copper in spore germination. To investigate whether insufficient concentrations of copper would perturb spore germination, purified dormant spores harboring a CRIB-GFP allele were incubated in the presence of calcofluor to stain spore walls. Spores were then washed and synchronously induced to enter into germination in the presence of the copper chelator TTM (200 μm) or copper (50 μm). Spores that had been treated with TTM proceeded through the initial phases of germination, including breaking of spore dormancy until they reached isotropic swelling. At this stage, they stopped their differentiation and exhibited a germination arrest (Fig. 2A). A clear phenotype of the TTM-mediated block was the lack of germ-tube formation and therefore absence of outgrowth. Copper-insufficient spores failed to reach the development stage where CRIB-GFP could be localized at one side of an enlarged spore, which represents the site of cell tip emergence. In contrast, spores that had been treated with copper proceeded through the entire germination program and formed bottle-like shaped spores with an outgrowing tip where CRIB-GFP was observed (Fig. 2B). After their entry in germination, OD600 of spores was assessed under copper-replete conditions (50 μm) or in the presence of increasing concentrations of TTM (50, 100, and 200 μm). After initiation of spore germination, light-refractile spores became dark within 2–3 h. During the bright- to dark-phase transition, the OD600 of the spore suspension decreased in parallel with darkening (Fig. 2C). After 3 h of germination, spores then increased their size through a process called isotropic swelling. At this stage (3-h time point) and until the 6-h time point, the OD600 of the spore suspension progressively increased (Fig. 2C). During the first 6 h of observations, spectrophotometric absorbance values (OD600) were similar, irrespective of the presence of TTM or copper (Fig. 2C). After 8 h of germination, at which point there was formation and emergence of a germ tube in copper-replete spores, the presence of TTM inhibited outgrowth of spores as a function of the increase in TTM concentration (Fig. 2C). In the presence of 200 μm TTM, outgrowing spores were absent, whereas 22.3 and 76.5% less outgrowing spores were detected in medium containing 50 and 100 μm TTM, respectively, as compared with that of copper-replete spores (Fig. 2C, bottom left panel). After 12 h of germination, outgrowing spores led to production of vegetative cells that were calcofluor-free, which represent 62% of the total population (outgrowing spores plus new vegetative daughter cells) in the presence of copper (Fig. 2C). Under conditions of copper starvation, production of calcofluor-free cells decreased by 21, 42, and 92% in medium containing 50, 100, and 200 μm TTM, respectively, as compared with that of calcofluor-free cells in medium containing copper (Fig. 2C, bottom right panel). Spores that underwent germination in the presence of 50 μm copper and 200 μm TTM were analyzed by FACS. At the time of outgrowth (8-h time point), the majority of copper-treated spores exhibited a 2C DNA content, whereas most of the TTM-treated spores had a 1C DNA content and were blocked at the stage of isotropic swelling (Fig. 2D). Consistently, the FSC/SSC values for these TTM-treated spores were much lower than those of copper-treated spores after 12 h of germination (Fig. 2D), especially with respect to spore/cell internal granularity (SSC). We investigated whether copper-insufficient spores could be relieved of isotropic swelling-like arrest by transferring the spores to a copper-replete medium. Spores that displayed a germination block were washed and resuspended in a medium containing copper (50 μm), iron (100 μm), zinc (100 μm), nickel (100 μm), manganese (100 μm), or cobalt (100 μm). Results showed that removal of the copper chelator TTM, followed by the addition of exogenous copper, triggered a rescue of spore germination. We noticed that a delay of ∼6 h occurred when copper-insufficient spores were rescued by exogenous copper as compared with control spores for which copper was available during the germination program (Fig. 2E). Although a delayed rescue was observed, supplementation with copper restored spore germination, including outgrowth and generation of novel calcofluor-free cells. Under copper-supplement conditions, pre-treated TTM spores that had been rescued produced calcofluor-free cells with a percentage of 50% with respect to the total population (calcofluor-positive and calcofluor-free cells) as compared with control spores in which case copper was available (78% among the total population: calcofluor-positive and calcofluor-free cells) (Fig. 2E, right panel, 16-h time point). Among the metal ions tested (copper, iron, zinc, nickel, manganese, and cobalt), only copper was able to rescue spore germination caused by copper deficiency (Fig. 2E). Taken together, the results showed that copper is required for spore germination. The evidence is based on the observation that copper deficiency leads to germination arrest at the onset of outgrowth and that arrest of spore germination could be rescue by the addition of copper.
Figure 2.
Copper-insufficient germinating spores undergo arrest at outgrowth. Wild-type spores harboring a CRIB-GFP allele were synchronously induced into germination in the presence of TTM (200 μm) (A) or copper (50 μm) (B). Shown are three representative stages of spore germination and outgrowth that occurred after 6, 8, and 12 h of induction. The spore wall was stained with calcofluor (center top). The daughter cell tip marker CRIB-GFP is shown in green (also indicated with white arrowheads) (center bottom). Spore morphology was examined by Nomarski optics (top). Merged images of calcofluor and CRIB-GFP are shown at the bottom of each panel. Examples of new daughter cell ends are indicated with white dashed lines. C, total growth was measured at OD600 in the presence of copper (50 μm) or three different concentrations of TTM (50, 100, and 200 μm). Inset indicates OD600 values up to 7 h post-induction of germination (top). The percentage of spores undergoing outgrowth (bottom) and the percentage of calcofluor-free cells after 12 h of induction of germination were determined under the indicated conditions of incubation (far right). D, shown is FACS analysis of spores that underwent synchronous germination in the presence of TTM (200 μm) (left) or copper (50 μm) (right). Fluorescence intensities corresponding to 1C and 2C DNA content are indicated. At the 12-h time point, dot plots of FSC versus SSC that correspond to spore size (FSC) and spore granularity (SSC) under low (TTM) and copper (50 μm) conditions. E, aliquots of spores used in C (blocked at outgrowth) were incubated in the presence of exogenous CuSO4 (Cu, 50 μm) (blue), FeSO4 (Fe, 100 μm) (red), ZnSO4 (Zn, 100 μm) (orange), NiCl2 (Ni, 100 μm) (green), MnCl2 (Mn, 100 μm) (pink), or CoCl2 (Co, 100 μm) (violet). The graph (left) depicts the germination profiles of spores. A minimum of 200 spores or calcofluor-free cells was examined every 2 h and under each one of the conditions. The graph on the far right represents the percentage of calcofluor-free cells after 16 h of incubation that included copper supplementation (at the 8-h time point) following a TTM block. Results are shown as the averages ± S.D. of a minimum of three independent experiments. ** corresponds to p < 0.001 (paired Student's t test).
Temporal expression profiles of ctr4+, ctr5+, ctr6+, and cuf1+ transcripts during spore germination and outgrowth
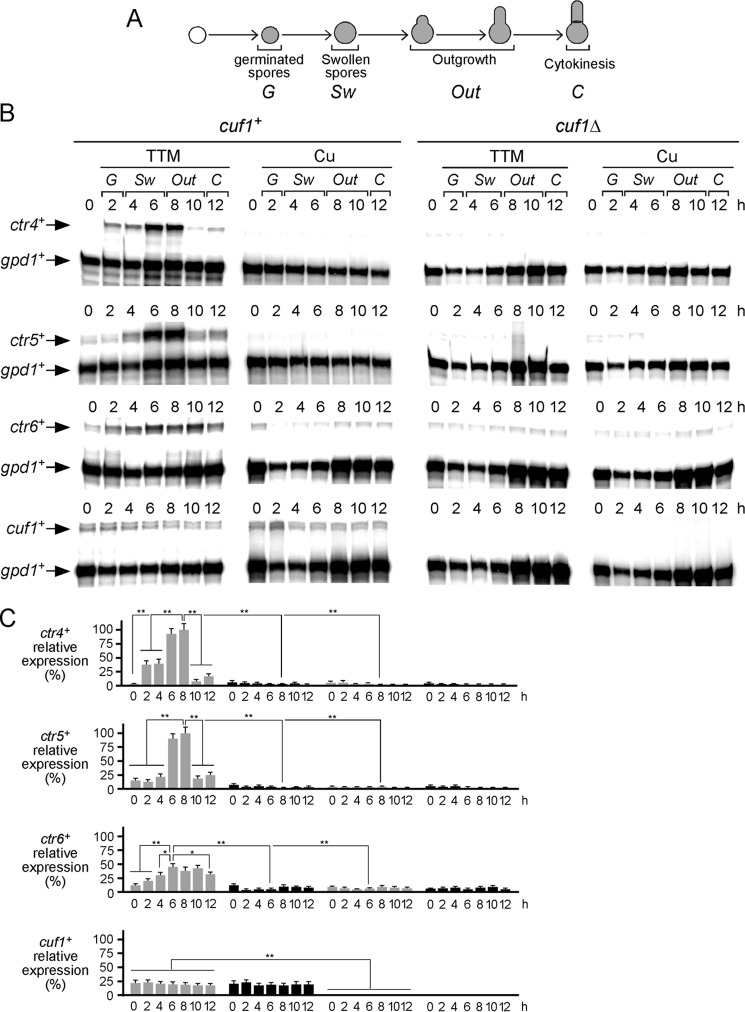
In response to copper starvation conditions, proliferating cells that grow mitotically exhibit transcriptional induction of the copper transport genes, and their activation requires the copper-dependent transcription factor Cuf1 (20). In contrast, ctr4+, ctr5+, and ctr6+ transcripts are repressed in cuf1+ cells in response to copper. Taking into account the fact that copper was required for morphological changes of spores during outgrowth, we examined ctr4+, ctr5+, and ctr6+ transcription profiles during spore germination as a function of time and copper availability. Purified cuf1+ and cuf1Δ spores were synchronously induced into germination and treated with TTM (50 μm) or CuSO4 (50 μm). Aliquots of cultures were taken after induction of germination, and steady-state levels of ctr4+, ctr5+, ctr6+, and cuf1+ mRNA were analyzed by RNase protection assays. Results showed that ctr4+ and ctr5+ mRNA levels were primarily detected in cuf1+ spores treated with TTM (Fig. 3). Under this low-copper condition, ctr4+ and ctr5+ transcript levels were relatively low within the first 4 h compared with their transcript levels detected 6 and 8 h after induction of germination (3–33-fold lower for ctr4+ and 7–8-fold lower for ctr5+). Maximal levels of ctr4+ and ctr5+ transcription were observed 6 and 8 h after induction of germination. This was followed by a reduction of ctr4+ (6–3-fold) and ctr5+ (4–5-fold) mRNA levels within 10–12 h (compared with levels observed after 6 and 8 h of induction of germination). As controls, ctr4+ and ctr5+ mRNAs were virtually absent in wild-type spores treated with copper (Fig. 3). Similarly, RNA samples prepared from cuf1Δ mutant spores showed loss of copper starvation-dependent induction of ctr4+ and ctr5+ gene expression, indicating that the copper-dependent regulation of ctr4+ and ctr5+ mRNAs required Cuf1 during germination and outgrowth.
Figure 3.
Expression profiles of ctr4+, ctr5+, ctr6+, and cuf1+ mRNAs during germination and outgrowth. A, shown is a schematic representation of different steps during germination and outgrowth. B, wild-type (cuf1+) and cuf1Δ mutant spores were synchronously induced to undergo germination in the presence of TTM (50 μm) or copper (Cu, 50 μm). Total RNA was isolated from culture aliquots taken at the indicated time points. Following RNA isolation, ctr4+, ctr5+, ctr6+, and cuf1+ steady-state mRNA levels were analyzed by RNase protection assays. The zero-time point (0 h) refers to the onset of germination induction. C, graphs represent quantification of the results of three independent RNase protection assays, including experiments shown in B. Values are represented as averages ± S.D. The asterisks correspond to p < 0.001 (**) and p < 0.05 (*) (paired Student's t test).
Although transcript levels of ctr6+ were detected under all conditions tested, the presence of copper or disruption of Cuf1 (cuf1Δ) resulted in reduced ctr6+ mRNA levels in comparison with those recorded in the case of wild-type (cuf1+) spores incubated under low-copper conditions (Fig. 3). Under this latter condition, ctr6+ mRNA levels progressively increased between 2 and 10 h after induction of germination. This was followed by a slight reduction of ctr6+ mRNA levels within 12 h. Under copper-replete conditions, results showed that steady-state levels of ctr6+ transcripts remained low compared with levels observed in the case of spores that had been exposed to TTM (Fig. 3). Under both conditions (TTM and CuSO4), disruption of Cuf1 (cuf1Δ) resulted in decreased ctr6+ transcript levels in comparison with those observed in copper-starved cuf1+ control spores (Fig. 3). In the absence of Cuf1 (cuf1Δ), although a low level of ctr6+ mRNA was still observed, its expression was not altered by the presence of TTM or copper (Fig. 3).
Steady-state levels of cuf1+ transcripts were constitutively present within the first 12 h of induction of germination. Furthermore, cuf1+ mRNA levels were expressed to a similar extent under copper-starved and copper-replete conditions (Fig. 3). As a control, the cuf1+ mRNA was absent in cuf1Δ spores as determined by RNase protection assays (Fig. 3). Taken together, results showed that ctr4+ and ctr5+ mRNA levels are induced in response to copper starvation in a Cuf1-dependent manner, exhibiting maximal expression during isotropic swelling and outgrowing stages of germination. Furthermore, results indicated that expression of the ctr6+ transcript requires Cuf1 for its maximal induction under copper-limiting conditions during spore germination.
Copper transport proteins Ctr4, Ctr5, and to a lesser extent Ctr6 are required for spore outgrowth under conditions of copper starvation
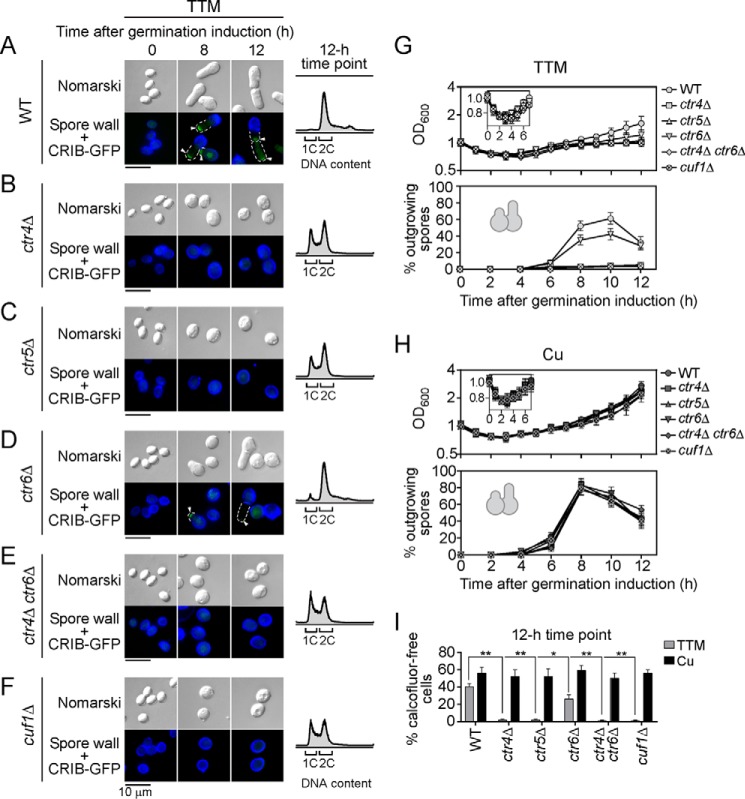
Taking into account the fact that copper ions were required during the spore germination program (Fig. 2), we hypothesized that Ctr4, Ctr5, and Ctr6 could also play important roles in spore germination under low-copper conditions. To test this hypothesis, ctr4Δ, ctr5Δ, ctr6Δ, ctr4Δ ctr6Δ, and cuf1Δ spores were used, and results were compared with wild-type control spores (Fig. 4, A–F). Purified spores were treated with TTM (50 μm) or CuSO4 (50 μm) immediately after induction of germination. In the case of wild-type spores, loss of spore refractility occurred within 2 h, isotropic swelling between 4 and 6 h, and formation of an outgrowing tip between 8 and 10 h following induction of germination, irrespective of the copper status (Figs. 1 and 4, G and H). In the case of ctr4Δ, ctr5Δ, ctr4Δ ctr6Δ, and cuf1Δ mutant spores, the progression of germination primarily stopped at the onset of outgrowth, although loss of spore refractility and isotropic swelling steps were still observed under low-copper conditions (Fig. 4, B, C, and E–G). Expression of the CRIB-GFP reporter produced a fluorescence signal that was visible at the tips of outgrowing wild-type spores after 8 h of induction of germination (Fig. 4A, WT). In contrast, there was an absence of CRIB-GFP signal in ctr4Δ, ctr5Δ, ctr4Δ ctr6Δ, and cuf1Δ mutant spores after 8 and 12 h of induction of germination in the presence of TTM (Fig. 4, B, C, E, and F). Furthermore, observations of ctr4Δ, ctr5Δ, ctr4Δ ctr6Δ, and cuf1Δ mutant spores using Nomarski optics showed that these spores stopped their morphogenesis progression and exhibited an arrest at the isotropic swelling stage (as compared with wild-type spores) (Fig. 4, B, C, and E–G). After 12 h of induction of germination, DNA content of ctr4Δ, ctr5Δ, ctr4Δ ctr6Δ, and cuf1Δ mutant spores was determined by FACS analysis (Fig. 4, B, C, E, and F). Results showed that 40–55% of mutant spores had 1C DNA content, whereas another 45–60% had a 2C DNA content. As a control at the 12-h time point, wild-type spores had 2C DNA content (Fig. 4A). Under copper-limiting conditions, these mutant spores failed to enter outgrowth (>98%) (Fig. 4G) and did not produce newborn calcofluor-free cells (Fig. 4I). The germ-tube formation defect of these mutants could be corrected by addition of CuSO4 (50 μm) to the growth medium (Fig. 4H). Indeed, under copper-replete conditions, ctr4Δ, ctr5Δ, ctr4Δ ctr6Δ, and cuf1Δ mutant spores entered outgrowth with formation of bottle-like-shaped outgrowing spores in a manner comparable with that found in wild-type spores (Fig. 4H). In the case of ctr6Δ mutant spores, the outgrowth defect was present but to a lesser extent than ctr4/5Δ and cuf1Δ mutants (Fig. 4, D and G). Deletion of ctr6 lowered the number of outgrowing spores by 17% (8-h time point), 19% (10-h time point), and 4% (12-h time point) as compared with wild-type spores (Fig. 4, D and G). Inactivation of ctr6 also reduced by 14% the proportion of resulting calcofluor-free cells that were generated and growing away from spore bodies after 12 h of induction of germination (Fig. 4I). As opposed to ctr4/5Δ and cuf1Δ mutants, a large proportion of ctr6Δ mutant spores had 2C DNA content after 12 h of induction of germination (Fig. 4D). Similarly to ctr4/5Δ and cuf1Δ mutants, the addition of exogenous copper to the growth medium rescued the outgrowth defect phenotype of ctr6Δ mutant spores (Fig. 4H). In summary, deletion of ctr4Δ, ctr5Δ, and cuf1Δ stopped the progression of spore germination at outgrowth, whereas deletion of ctr6Δ only partially interfered with the formation of outgrowing spores.
Figure 4.
Effect of the absence of Ctr4, Ctr5, Ctr6, or Cuf1 on the emergence of the germ tube at one side of spores under low-copper conditions. Wild-type (WT) spores and spores harboring deletions of the ctr4+ (ctr4Δ), ctr5+ (ctr5Δ), ctr6+ (ctr6Δ), ctr4+ ctr6+ (ctr4Δ ctr6Δ), and cuf1+ (cuf1Δ) genes were synchronously induced to undergo germination under copper-depleted (50 μm TTM) and copper-replete (50 μm CuSO4) conditions. A–F, representative microscopic images revealed defective outgrowth of ctr4Δ, ctr5Δ, ctr4Δ ctr6Δ, and cuf1Δ spores compared with wild-type (WT) spores under copper starvation conditions (TTM). In the case of ctr6Δ spores, two populations were observed: spores with and without an outgrowing tip. Although wild-type and all mutant spores possess an integrated CRIB-GFP allele, its green fluorescent product was only detected in wild-type and some ctr6Δ germinated spores (A and D). CRIB-GFP (white arrowheads) was used as a marker of the new cell side tip of an outgrowing spore. Calcofluor (blue) was used to stain the wall of the spore body. Morphology of spores was examined by Nomarski optics. At the 12-h time point, FACS analysis of wild-type and mutant spores is shown next to microscopic images (far right). G–H, in the case of each indicated strain, graphs depict growth profiles of spores after induction of germination under copper-starved (G) and copper-replete (H) conditions. I, shown is the percentage of calcofluor-free cells that were produced after 12 h of induction of germination under conditions of low and high levels of copper. A minimum of 200 spores and germinated cells were examined under each condition. Data are represented as the averages ± S.D. of three independent experiments. The asterisks correspond to p < 0.001 (**) and p < 0.05 (*) (paired Student's t test).
Ctr4-GFP and Ctr5-Cherry first co-localize at the spore contour at the onset of outgrowth and then move away from the spore body to occupy the periphery of the nascent cell
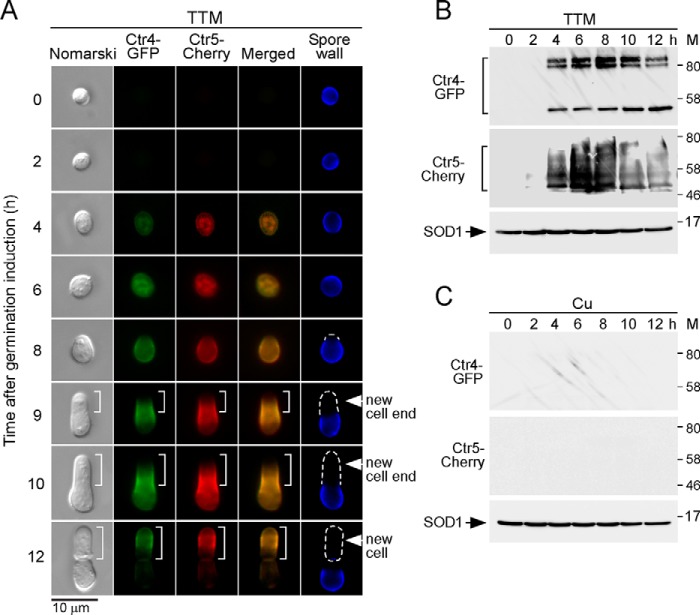
Exogenous copper is transported across the plasma membrane by the Ctr4-Ctr5 heteromeric complex in dividing cells that grow mitotically (12, 16, 21, 28). We next sought to determine the location of Ctr4 and Ctr5 during spore germination because steady-state levels of ctr4+ and ctr5+ transcripts were poorly expressed in early stages of spore germination. However, Ctr4 and Ctr5 were expressed at higher levels at the end of isotropic swelling and at the onset of outgrowth. The experiments were set by integrating functional ctr4+-GFP and ctr5+-Cherry alleles into ctr4Δ ctr5Δ cells. After production and purification of spores, they were synchronously induced to undergo germination in the presence of TTM (50 μm) or CuSO4 (50 μm). Under low copper conditions, Ctr4-GFP and Ctr5-Cherry fluorescent proteins were first co-detected after 4–6 h of induction of germination (Fig. 5A). Their initial intracellular detection revealed the appearance of cytoplasmic vesicular structures within the spore (Fig. 5A). At the onset of outgrowth (8-h time point), Ctr4-GFP and Ctr5-Cherry fluorescent proteins co-localized at the spore contour, except for a short peripheral region that may correspond to the polar cap where there is a local rupture of the OSW (Fig. 5A). After 9 h of induction of germination, green and red fluorescence signals associated with Ctr4-GFP and Ctr5-Cherry, respectively, were primarily seen in a middle peripheral region of the outgrowing elongated spore (Fig. 5A). Ctr4-GFP and Ctr5-Cherry fluorescent signals progressively displayed local shifting toward the germ projection that emerged at one side of the enlarged spore (10-h time point). At 12 h, Ctr4-GFP and Ctr5-Cherry fluorescent signals were mainly restricted to the periphery of the new daughter cells (Fig. 5A). Consistent with the clear interdependence between Ctr4 and Ctr5, Ctr4-GFP- and Ctr5-Cherry-associated fluorescent signals co-localized during spore germination.
Figure 5.
Colocalization of Ctr4-GFP and Ctr5-Cherry during spore germination and outgrowth. A, ctr4Δ ctr5Δ spores harboring functional ctr4+-GFP and ctr5+-Cherry alleles were synchronously induced to undergo germination. Once induced, spores were allowed to germinate in the presence of TTM (50 μm). Fluorescence signals of Ctr4-GFP and Ctr5-Cherry were observed at different stages of germination (center left). Merged images of Ctr4-GFP and Ctr5-Cherry are shown in the center right panels. Nomarski optics (far left) were used to examine spore or cell morphology. Calcofluor staining was performed to visualize the spore wall (far right). White brackets, dashed lines, and arrowheads indicate the formation of a new cell tip that subsequently produced a new cell. B and C, at the indicated time points, lysates from aliquots of copper-starved or copper-replete spores were analyzed by immunoblotting using anti-GFP, anti-Cherry, and anti-SOD1 antibodies. Positions of the molecular weight standards (M) are indicated to the right in B and C.
To further detect the presence of Ctr4-GFP and Ctr5-Cherry proteins, aliquots of TTM-treated spore cultures were taken at various time points after induction of germination, and steady-state levels of Ctr4-GFP and Ctr5-Cherry were analyzed by Western blottings. Results showed that steady-state protein levels of Ctr4-GFP followed those of Ctr5-Cherry as they were present during the same stages of spore germination (Fig. 5B). The two approaches (fluorescence microscopy and immunoblotting) indicated the absence of both proteins during the first 2 h of spore germination, whereas their presence was detected between 4 and 12 h (Fig. 5, A and B). Because transcription of the ctr4+ and ctr5+ genes was repressed under copper-replete conditions (Fig. 3), results showed that Ctr4-GFP and Ctr5-Cherry steady-state levels (expressed under their native promoters) were undetectable by immunoblot assays in copper-treated spores, irrespective of the indicated time point after induction of germination (Fig. 5C). Taken together, the results revealed that at the end of spore germination, the plasma membrane of a new daughter cell is the preferred location of Ctr4 and Ctr5 where their presence is highly enriched as observed by fluorescence microscopy.
Ctr6 localizes in the vacuole membranes of swollen spores and spores that undergo outgrowth
In the case of Ctr6, it is known that it localizes to the membrane of vacuoles in cells proliferating in mitosis under copper-limiting conditions (22, 23). A similar localization was observed when a functional Ctr6-HA4 fusion protein was expressed in ctr6Δ cells grown in malt medium after 1 day of incubation in the presence of TTM (50 μm) (Fig. 6A). As reported previously (22), Ctr6-HA4 underwent an intracellular re-location and co-localized with the forespore membrane after 2 and 3 days (malt medium) that corresponded to late meiosis (Fig. 6A). Once spores were formed and released from the ascus, Ctr6-HA4-associated fluorescence progressively disappeared after 4 days in malt medium (Fig. 6A). Spores were then purified and cultured under conditions to induce synchronous germination. Under low-copper conditions, the Ctr6-HA4 fluorescent signal was barely detectable during the first 2 h of germination, which corresponded to breaking of spore dormancy. In contrast, during isotropic swelling, Ctr6-HA4-associated fluorescence progressively appeared in the vacuolar membranes of spores, especially after 6 h of induction of germination (Fig. 6B). Swollen spores were also incubated in the presence of the vacuole-staining dye FM4-64 (Fig. 6C). Results showed that Ctr6-HA4 and FM4-64 exhibited similar subcellular location patterns within the spores, especially after 6 h of induction of germination (Fig. 6, B and C). In the case of outgrowing spores, results showed that vacuoles were enriched in the spore body, which was the portion-stained with calcofluor (Fig. 6C). Consistently, results showed that Ctr6-HA4-associated fluorescence was also enriched in the spore body of outgrowing spores after 10–12 h of induction of germination (Fig. 6B). Aliquots of single-spore cultures were analyzed at various time points after induction of germination in the presence of TTM (50 μm) or CuSO4 (50 μm). Immunoblotting analysis showed that Ctr6-HA4 was detected at each time point under copper-starved conditions (Fig. 6D). Steady-state levels of Ctr6-HA4 increased after 6, 8, and 10 h of induction of germination and were expressed to a lesser degree after 12 h (Fig. 6D). Immunoblotting analysis showed that protein extracts from copper-treated swelling and outgrowing spores were negative for the presence of Ctr6-HA4 (Fig. 6E). Taken together, the results revealed that Ctr6-HA4 localizes in the vacuole membranes of spores that had undergone germination and that it is enriched in the spore body following production of a newborn daughter cell.
Figure 6.
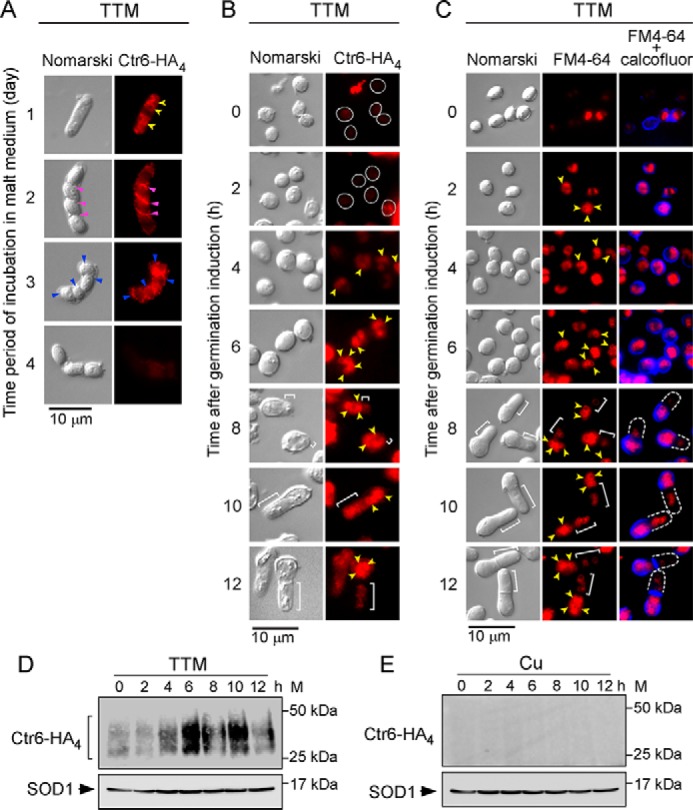
Subcellular localization of Ctr6-HA4 during spore germination and outgrowth. A, h90 ctr6Δ strain expressing a ctr6+-HA4 allele was incubated on ME media under low-copper conditions (50 μm TTM). Four representative stages of the sporulation program that occurred within 4 days are shown. Cells, asci, and spores were analyzed by indirect immunofluorescence microscopy to assess subcellular localization of Ctr6-HA4. Yellow arrowheads indicate examples of vacuole membranes in which Ctr6-HA4 was found (day 1). During the forespore membrane formation (violet arrowheads) (day 2), Ctr6-HA4 is known to become a resident of the spore membrane (blue arrowheads) (day 3). Ctr6-HA4-associated fluorescence disappeared from the surface of spores that had been released from asci after 4 days of meiotic induction. B, purified ctr6Δ mutant spores expressing Ctr6-HA4 were induced into germination in the presence of TTM (50 μm) or copper (50 μm) for the indicated time points. Fixed cells were observed by indirect immunofluorescence (Ctr6-HA4; right) and Normarski optics (left). White circles indicate cells observed within 2 h after induction of germination. After the 4-h time point, Ctr6-HA4-associated fluorescent signal was more prominent and was detected in the vacuole membranes (yellow arrowheads). White brackets indicate examples of new cell tips of outgrowing spores. C, FM4-64 (middle) staining was visualized by fluorescence microscopy as a marker of vacuolar membranes (yellow arrowheads). Double staining (FM4-64 plus calcofluor) was performed to identify spore walls (blue). After the 8-h time point, calcofluor-associated fluorescent signal (blue) was found at the bottom part of the bottle-like shape of spores that had undergone outgrowth. D and E, at the indicated time points, total extracts from aliquots of spores used in B under low and high (not shown by fluorescence microscopy due to the absence of fluorescent signal) copper conditions were analyzed by immunoblotting using anti-HA and anti-SOD1 antibodies. Positions of the molecular weight standards (M) are indicated to the right in D and E.
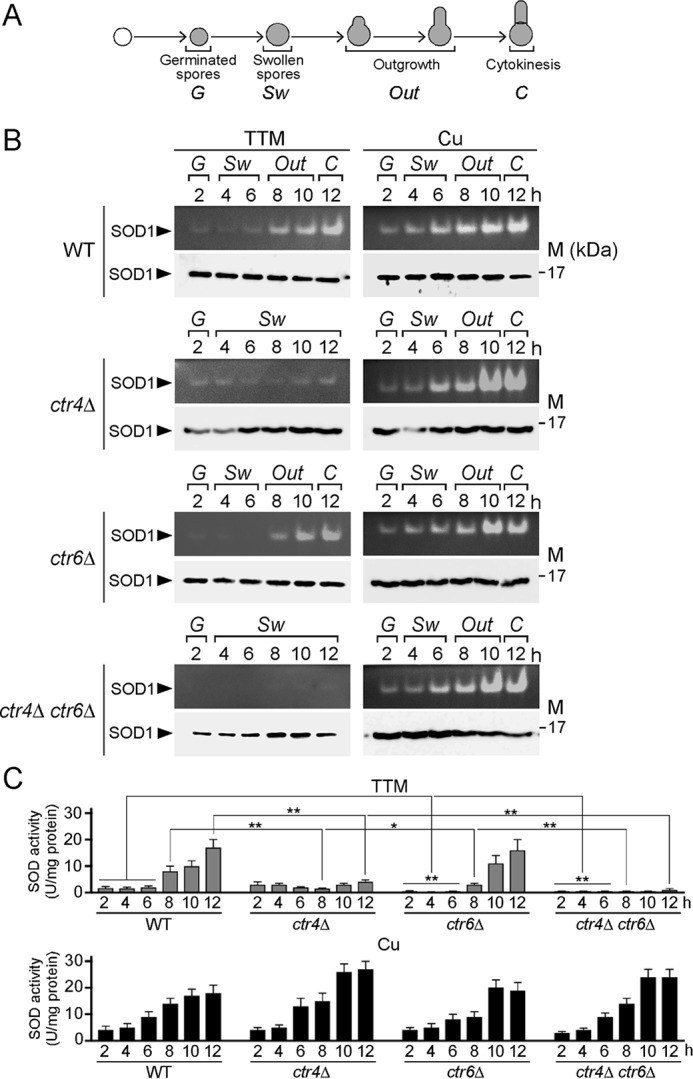
Ctr4 and Ctr6 are required for the production of a fully active SOD1 during spore germination and outgrowth
Previous studies have shown that Ctr4 and Ctr6 transporters are required to produce a fully active copper-dependent SOD1 enzyme in cells undergoing vegetative growth or meiosis for production of spores (22, 23). To determine whether Ctr4 or Ctr6 was needed to provide copper to SOD1 during spore germination, purified h90 wild type (ctr4+ ctr6+), ctr4Δ, ctr6Δ, and ctr4Δ ctr6Δ spores were synchronously induced to undergo germination in the presence of TTM (50 μm) or CuSO4 (50 μm). SOD activity was assayed from spore lysates using a standard in-gel assay with nitro blue tetrazolium staining (22). In the case of wild-type spores, results showed that levels of SOD1 activity increased as a function of germination time to reach maximal activity within 12 h under both copper-depleted and copper-replete conditions (Fig. 7). During the first 6 h of induction of germination (isotropic swelling), copper-starved ctr4Δ mutant spores exhibited a weak SOD1 activity comparable with that of wild-type spores (Fig. 7). At 8 h and later time points, results showed that SOD1 activity of ctr4Δ spores was 5.3-fold (8 h), 3.3-fold (10 h), and 4.3-fold (12 h) lower than that of wild-type spores (Fig. 7). In the case of copper-starved ctr6Δ spores, SOD1 activity was 3.5-, 5.3-, 6.7-, and 2.7-fold lower than that of wild-type spores after 2, 4, 6, and 8 h of induction of germination, respectively (Fig. 7). At 10- and 12-h time points, ctr6Δ spores exhibited SOD1 activity levels comparable with those of wild-type spores. In the case of ctr4Δ ctr6Δ double mutant spores, results showed an absence of measurable SOD1 activity under low-copper conditions, highlighting the fact that both Ctr4 and Ctr6 were required to provide copper to SOD1 during spore germination and outgrowth (Fig. 7). To assess expression of SOD1 in wild-type and mutant spores, whole-spore extracts were analyzed by Western blottings at the indicated time points during germination and outgrowth (Fig. 7). Results showed that SOD1 was present in wild-type and mutant spores, revealing that the decrease or lack of activity was not due to the absence of SOD1 expression. As shown in previous studies and reported here, SOD1 activity could be rescued by addition of exogenous CuSO4 concentrations (50 μm) to the spores (Fig. 7). This copper-remedial phenotype was likely due to the fact that copper ions were assimilated by way of a putative low-affinity copper transport system, thereby circumventing requirements for high-affinity Ctr copper transporters. Taken together, the results revealed a distinct requirement of Ctr4 and Ctr6 for activation of SOD1 under copper starvation during spore germination. Although Ctr6 plays an important role during early stages of germination such as isotropic swelling, Ctr4 is needed to ensure maximal SOD1 activity during outgrowth and cytokinesis.
Figure 7.
Effect of ctr4Δ, ctr6Δ, and ctr4Δ ctr6Δ deletions on SOD1 activity during germination and outgrowth. A, shown is a schematic representation of different steps during germination and outgrowth. B, wild-type (WT), ctr4Δ, ctr6Δ, and ctr4Δ ctr6Δ spores were synchronously induced to undergo germination under copper-starved (50 μm TTM) and copper-replete (50 μm CuSO4) conditions. At the indicated time points, total extracts from aliquots of cultures were analyzed for SOD1 activity using an in-gel assay (upper panels). Protein extracts were analyzed for steady-state protein levels of SOD1 by immunoblotting using an anti-SOD1 antibody (lower panels). M, position of the molecular weight reference band (17 kDa) is indicated to the right in B. C, spectrophotometric determination of SOD activity was also performed from these wild-type and mutant spores using a cytochrome c/xanthine oxidase method. Values are represented as the averages ± S.D. of three independent experiments. The asterisks correspond to p < 0.001 (**) and p < 0.05 (*) (paired Student's t test).
Labile copper pools primarily remain in the spore body during outgrowth
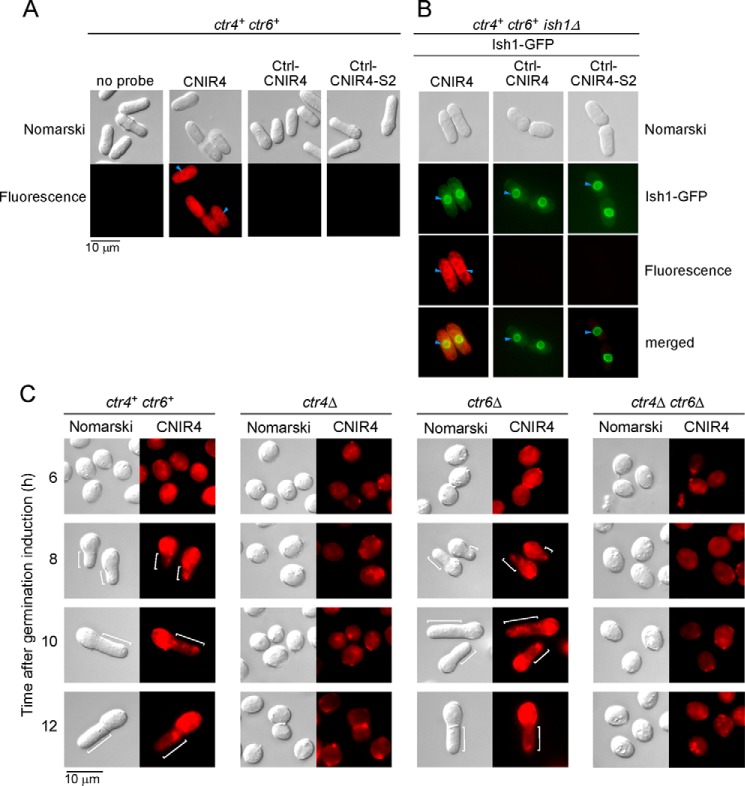
The physiological contribution of copper and cuproproteins to spore germination led us to investigate the status of labile copper pools at different stages of the germination process. Wild-type ctr4+ ctr6+ and ctr4Δ, ctr6Δ, and ctr4Δ ctr6Δ mutant spores were used in these experiments. We utilized the fluorescent copper-binding probe CNIR4 to track exchangeable copper pools in living cells or spores. CNIR4 is a second-generation silicon rhodamine analog of the silicon rhodol CSR1 (29). As chemical control analogs, Ctrl-CNIR4 and Ctrl-CNIR4-S2 were used under the same conditions as CNIR4. These control probes, in which metal-interacting sulfur atoms have been partially or completely replaced by isosteric carbon atoms, do not bind copper but have the same dye scaffold as CNIR4 and thus can be used to help disentangle metal- and dye-dependent signals (29, 30). We first tested CNIR4 and its control analogs using ctr4+ ctr6+ cells that were proliferating in mitosis. Cells were cultured to mid-logarithmic phase and then grown in the presence of CuSO4 (5 μm) for 1 h. The treatment ensured intracellular copper sequestration by the cells. After washings, cells were incubated in low copper (∼0.16 nm CuSO4; a concentration that is 100 times lower than standard culture conditions)-containing Edinburgh minimal medium in the presence of 5 μm CNIR4, Ctrl-CNIR4, and Ctrl-CNIR4-S2 or were left untreated (no probe) for 30 min. Results showed that the cells displayed fluorescent CNIR4-copper complexes that were distributed throughout the cells with a preferred location to the yeast secretory pathway, including the endoplasmic reticulum (Fig. 8A). As expected, there was an absence of fluorescence in the case of Ctrl-CNIR4 and Ctrl-CNIR4-S2 analogs (Fig. 8A). ctr4+ ctr6+ ish1Δ + ish1+-Cherry cells were grown under similar conditions in the absence of CNIR4, Ctrl-CNIR4, or Ctrl-CNIR4-S2 (Fig. 8B, top center). Cellular location of the Ish1-GFP fluorescent signal was detected as a nuclear envelope/endoplasmic reticulum resident marker (31). In the presence of CNIR4, results showed that a fraction of CNIR4–copper-associated fluorescence appeared in a common perinuclear region where Ish1-GFP was detected (Fig. 8B).
Figure 8.
Intracellular copper distribution during spore germination and outgrowth. A and B, live-cell copper imaging with the fluorescent copper-binding probe CNIR4 (5 μm) in ctr4+ ctr6+ cells (A) or ctr4+ ctr6+ ish1Δ cells expressing Ish1-GFP (B) as a marker of the nuclear envelope/endoplasmic reticulum secretory network. Cells proliferating in mitosis were first incubated in the presence of copper (5 μm) and then transferred and analyzed in a low-copper-containing media. Ctrl-CNIR4 and Ctrl-CNIR4-S2 are control analogs that cannot bind to copper due to the fact that metal-interacting sulfur atoms had been replaced by isosteric carbon atoms. Nomarski microscopy was used to examine cell morphology (upper panels). Fluorescent CNIR4-copper complexes were detected with a transmission window at 640 nm (red) (fluorescence). The merged images are shown at bottom right (B). C, live-cell copper imaging with CNIR4 (5 μm) after 6, 8, 10, and 12 h of induction of germination in ctr4+ ctr6+, ctr4Δ, ctr6Δ, and ctr4Δ ctr6Δ cells. White brackets indicate the new cell tips during outgrowth.
To assess copper distribution during spore germination, wild-type ctr4+ ctr6+ and ctr4Δ, ctr6Δ, and ctr4Δ ctr6Δ mutant strains were plated on ME media that contained CuSO4 (5 μm). After sporulation and purification, spores from each strain were synchronized to initiate and proceed through the germination program. At the indicated time points, spores were harvested and resuspended in the presence of CNIR4 (5 μm) for 30 min and then analyzed by direct fluorescence microscopy. After 6 h of germination, CNIR4-copper complexes generated a fluorescent signal that was dispersed within the spores, irrespective of the presence or absence of Ctr4, Ctr6, or both. In the case of ctr4+ ctr6+ and ctr6Δ spores undergoing outgrowth, CNIR4-copper-associated fluorescence was primarily observed within the old spore body after 8, 10, and 12 h of germination induction (Fig. 8C). In contrast, the CNIR4-copper fluorescent signal was weak at the new germ projection end in comparison with that of the spore body. In the case of ctr4Δ and ctr4Δ ctr6Δ spores, there were no changes in the pattern of fluorescence signal of CNIR4-copper complexes after 8, 10, and 12 h of induction of germination in comparison with that observed after 6 h, because spore outgrowth was blocked in these mutants (Fig. 8C). Taken together, these results suggested the existence of a mechanism whereby labile copper pools are preferentially retained in the spore body during outgrowth.
Cuproprotein SOD1 is required for spore outgrowth
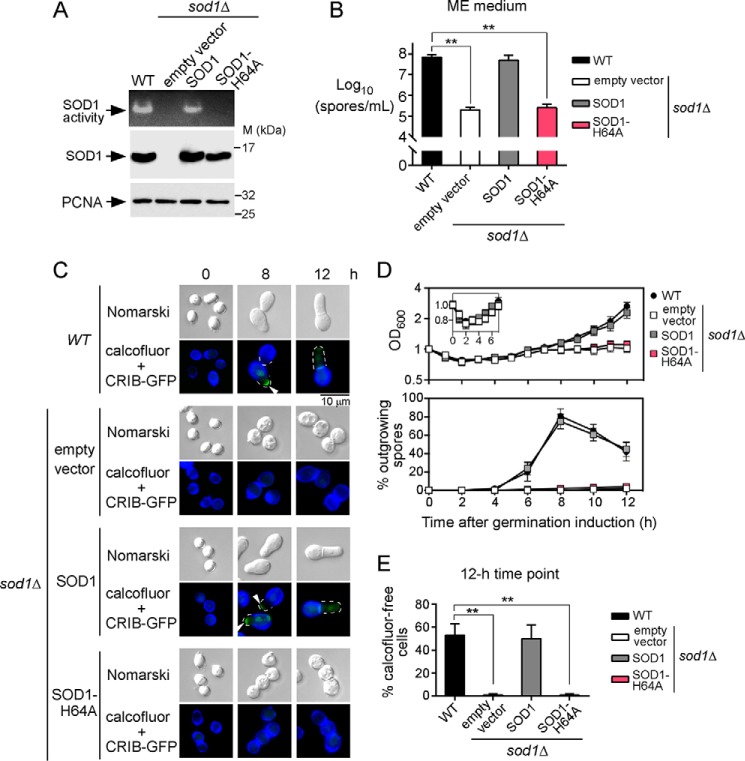
Based on the fact that copper and copper transporters were required for spore outgrowth, we hypothesized that a copper-dependent enzyme may be necessary to ensure spore outgrowth during germination. Given that previous studies had suggested that the loss of copper-dependent SOD1 activity negatively affected fungal spore germination (32–34), we tested the outcome of the absence of SOD1 on the germination and outgrowth of spores. We used sod1Δ cells carrying an integrated empty vector or expressing either a re-integrated wild-type sod1+ or sod1H64A mutant allele and compared the behavior to sod1+ control cells. When these strains were proliferating in mitosis, wild-type (sod1+) cells exhibited high levels of SOD1 activity (Fig. 9A). In contrast, there was no detectable SOD1 activity in sod1Δ null cells harboring an empty plasmid or expressing the sod1H64A mutant allele that was expected to alter both structural and catalytic properties of the enzyme (35, 36) (Fig. 9A). In sod1Δ mutant strain, the loss of SOD1 activity was rescued to ∼50% of the activity of control parental strain by returning a wild-type copy of the sod1+ gene expressed from an integrated plasmid (Fig. 9A). To ascertain that SOD1 and its H64A mutant version were expressed in sod1Δ cells, total protein extracts from transformed cells were analyzed by Western blottings. Results showed that SOD1 and the H64A mutant were produced in sod1Δ cells, indicating that the absence of SOD1 activity in H64A mutant cells was not due to the lack of SOD1-H64A expression (Fig. 9A).
Figure 9.
Active SOD1 is required to undergo spore outgrowth. A, cell extracts were prepared from mitotically proliferating strains and analyzed for SOD1 activity using an in-gel assay. A sod1Δ mutant strain was transformed with an empty vector or a plasmid encoding the sod1+ or sod1+H64A mutant allele. The wild-type (WT) strain was used as a control. Protein extracts prepared from these strains were analyzed for steady-state levels of SOD1 by immunoblotting using anti-SOD1 and anti-proliferating cell nuclear antigen (PCNA) antibodies. B, production of spores was determined from the same strains from A after incubation on ME medium for 4 days. C, h90 sod1Δ mutant cells containing a CRIB-GFP allele were transformed with an empty vector and plasmid pJKsod1+ or plasmid pJKsod1H64A. After sporulation, dormant spores were purified and synchronously induced to germinate. Wild-type (WT) spores were used as a germination control. Three representative stages of the germination and outgrowth that occurred at the zero-time point and after 8 and 12 h of induction are shown. Calcofluor (blue) was used as a marker to visualize the spore wall. Nomarski optics (top) was used to monitor spore morphology. D, total growth was measured at OD600 as a function of time after induction of germination. Inset indicates measurements at OD600 for the first 7 h of spore germination. The percentage of spores undergoing outgrowth were determined as a function of time after induction of germination. E, in the case of each indicated strain, the graph represents the percentage of calcofluor-free cells (new vegetative cells) that were produced after 12 h of induction of germination. A minimum of 200 spores and germinated cells was examined in the case of each condition. Data are represented as the averages ± S.D. of three independent experiments. The asterisks correspond to p < 0.001 (**) (paired Student's t test).
Each strain was cultured on ME medium for 4 days, and spore production was monitored. We observed that during the procedure of spore isolation and purification, sod1Δ mutant cells that harbored an empty vector or that expressed the sod1H64A allele exhibited dramatic decreased levels of spore production in comparison with wild-type control cells or sod1Δ cells expressing an integrated sod1+ gene (Fig. 9B). Results showed that the two strains that lacked a functional SOD1 produced almost 2 orders of magnitude less spores, which corresponded to 99.7% (empty vector) and 99.6% (SOD1H64A) less spores than those of cells expressing SOD1. However, there was a sufficient number of spores to allow their isolation and purification from each strain. Purified spores containing the CRIB-GFP allele were induced to synchronous germination and were examined at the indicated time points (Fig. 9C). Results showed that after 8 h of induction of germination, a germ tube emerged in spores expressing active SOD1. In contrast, there was no emergence and elongation of a germ tube in spores that were defective in SOD1 activity. The CRIB-GFP reporter was observed at the outgrowing tips of wild-type sod1+ spores, whereas CRIB-GFP did not show formation of outgrowing polar caps in sod1Δ or sod1H64A mutants (Fig. 9C). Results further showed that the breaking of dormancy occurred in both wild-type (sod1+) and mutant (sod1Δ or sod1H64A) spores because we detected the loss of refractility (phase-contrast microscope) and a decrease in optical absorbance of spore suspensions after 2 h of induction of germination (Fig. 9D, inset). In contrast, spore outgrowth and production of new calcofluor-free cells were exclusively seen in spores that were competent for SOD1 activity (Fig. 9, D and E). Taken together, these results indicated that deletion or inactivation of SOD1 leads to a defect in spore outgrowth, blocking the production of new daughter cells from the mother spore.
Discussion
Spores are thought to be infectious particles for many pathogenic organisms, including bacteria, fungi, and protozoa (37, 38). Spores are highly resistant to harsh conditions under which vegetative cells lose their viability. Spores are adapted to resist environmental stress and for efficient dispersal through airflow or fluids (39). When conditions are favorable, spores undergo a specialized program called germination. This process requires copper because copper-insufficient spores proceeded through the initial phases of germination until they reached outgrowth and then they stopped their differentiation (Fig. 2). The observation that copper can be a limiting factor for complete and normal progression of fungal spore germination suggests that copper acquisition may depend on specialized transport proteins to deliver copper to critical copper-dependent enzymes essential to germination.
This situation is reminiscent of that found in the fungal plant pathogen Colletotrichum gloeosporioides in which case copper is necessary for pathogenic spore germination (32). In C. gloeosporioides, CgCtr2 is a protein of the Ctr family that localizes to the membrane of vacuoles. CgCtr2 is involved in intracellular delivery of copper to copper-requiring cytosolic enzymes, including SOD1 (32). Cgctr2 mutant cells consistently display reduced SOD1 activity and reduced spore germination rates (32). In this study, results showed that under low-copper conditions, spores lacking the vacuolar membrane copper transporter Ctr6 (ctr6Δ) displayed less SOD1 activity compared with that of wild-type spores, especially during the first 8 h after induction of germination. Furthermore, the percentage of calcofluor-free cells that were produced by ctr6Δ mother spores was 35% lower than that of wild-type spores. This analogy between the two yeast models reinforces the notion that spore germination is a developmental process that requires mobilization of intravacuolar stores of copper.
An additional intriguing observation is the fact that results showed a preferential retention of vacuoles and Ctr6 in the spore body during emergence and elongation of the germ tube. The fact that vacuoles are known to play a role as a copper-storage compartment (24, 25, 40–42) may favor a reserve of copper within the mother spore. Consistent with this possibility, experiments using the fluorescent CNIR4 copper probe revealed a preferential accumulation of labile copper pools in the spore body as compared with the germ tube. Before losing its OSW, a mother spore could produce three consecutive and distinct germ tubes that subsequently results in three novel daughter cells (5). Because of these three rounds of outgrowth, it is presumed that the mother spore contains high levels of micronutrients such as copper to fulfill the demand of three consecutive formations of germ tubes. Concerning the preferential retention of vacuoles in the spore body, one possible explanation is that autophagy would be more active in this area where spore wall un-coating occurs to resume mitotic cell growth. The process of spore development may implicate delivery of cellular materials to the vacuole for degradation. In contrast, a weak autophagic activity is predicted during the formation of a germ tube in which de novo protein and organelle biogenesis occurs to produce a novel daughter cell.
Consistent with an asymmetric copper distribution between mother spore and germ tube, results showed that Ctr4 and Ctr5 were preferentially expressed at the germ tube periphery in comparison with that of the spore body after 12 h of induction of germination. Given the fact that labile copper pools are lower in the germ projection, the asymmetric enrichment of Ctr4 and Ctr5 at the cell surface of the germ tube could compensate and contribute to mediate copper uptake in this new cellular portion. Furthermore, the preferential germ tube localization of Ctr4 and Ctr5 may contribute to save labile copper ions that are present in the spore body. Elucidating the molecular nature of this asymmetric Ctr4 and Ctr5 distribution between the spore body and a germ tube is an interesting challenge for future studies.
Analyses of expression profiles of ctr4+ and ctr5+ have shown that these two genes are co-expressed in response to low-copper conditions during the mitotic cell cycle and during the early stages of meiosis (16, 22). In both cell-cycle programs, expression of ctr4+ and ctr5+ requires the copper-responsive transcription factor Cuf1 (16, 20, 22). In the case of spores undergoing a transition from cellular dormancy to proliferation, expression profiles of ctr4+ and ctr5+ showed that the two genes were still co-expressed, reaching a maximum 6–8 h after induction of germination. At this middle germination phase, spores entered outgrowth that was followed by an extension process of a single germ tube that hatches out the OSW. As observed in the case of mitotic and meiotic cells, expression of ctr4+ and ctr5+ during spore germination was copper starvation- and Cuf1-dependent. Although copper-regulated gene expression profiling during spore germination has not been characterized in other fungal species, global gene expression patterns in S. cerevisiae has revealed that a response to nutrients other than glucose occurs during the second phase of germination (8). This second phase is known to occur after breaking of spore dormancy and is accompanied by loss of spore refractility. Therefore, based on the general transcription program of spore germination in S. cerevisiae (8, 43), the highest expression levels of ctr4+ and ctr5+ appear to be consistent with a timely response that occurs after the first phase of germination, especially in the context of the onset and promotion of spore outgrowth.
In the case of cuf1+, its expression pattern differed from that of ctr4+ and ctr5+. The cuf1+ gene exhibited a constitutive transcription that was independent of developmental stage or copper availability during spore germination. The transcription of ctr6+ was detected in all stages of spore germination and was slightly increased after 6, 8, and 10 h of induction of germination, which corresponds to the period where ctr4+ and ctr5+ mRNA levels were highly expressed (except for the 10-h time point). Deletion of cuf1+ (cuf1Δ) resulted in a significant decrease of ctr6+ mRNA levels under low-copper conditions. However, its expression was not completely abolished and remained detectable throughout the spore germination process under both low- and high-copper conditions. On the basis of these observations, ctr6+ expression is predicted to require at least an additional transcriptional regulator to ensure the continuous presence of Ctr6 during the developmental program. This observation was reminiscent of cells undergoing meiosis in which case ctr6+ expression relies on Cuf1 and the meiosis-specific transcription factor Mei4. Under conditions whereby Cuf1 is inactive, Mei4 ensures the expression of Ctr6 during the meiotic divisions and the process of spore formation, especially under basal and copper-replete conditions (22). Because of the fact that Mei4 is a meiosis-specific regulator that is required for expression of several middle-phase genes (44), it is unlikely that Mei4 co-regulates the timely expression of ctr6+ during spore germination. In S. cerevisiae, spore germination is driven by a gene expression program during which several genes are induced or repressed as a function of the developmental path (8, 43). Although such a transcriptional regulatory network is still awaiting characterization in fission yeast, it is possible that one (or more) transcriptional regulator involved in the control of germination is also required for the expression of the ctr6+ gene.
Previous studies have shown that S. pombe cells lacking Ctr4 and Ctr6 (ctr4Δ ctr6Δ) are devoid of measurable SOD1 activity under copper-limiting conditions. This phenotype has been observed in cells proliferating in mitosis as well as in meiotic and sporulating cells (22, 23). When ctr4Δ ctr6Δ double mutant spores were induced to undergo germination under low-copper conditions, SOD1 activity was undetectable. Furthermore, ctr4Δ ctr6Δ spores exhibited a developmental block at the onset of outgrowth. Based on these phenotypic observations, we sought to further examine whether sod1Δ mutant spores or spores expressing a catalytically inactive mutant form of SOD1 (SOD1H64A) underwent a block of spore germination. Results showed an arrest of spore germination at the onset of outgrowth, suggesting a role for SOD1 in the formation or maintenance of the germ tube. In fungal species, spore outgrowth involves the establishment of a polarized growth axis that is visualized by a tip extension. After hatching out the OSW, a polar cap serves as a platform to foster local membrane addition and cell-wall remodeling that are needed for polar growth. In filamentous fungi, it is generally accepted that polarized growth by tip extension must exhibit apical dominance whereby the growing tip is dominant, suppressing the formation of other tips in its vicinity. One contributing factor of the enforcement of apical dominance is the localized accumulation of reactive oxygen species (ROS) in the apical region of polarized cells. According to previous studies, ROS are produced by NADPH oxidase-like proteins or related flavoproteins (45, 46). In Aspergillus nidulans, ROS accumulation is detected using nitro blue tetrazolium (NBT). When NBT is reduced by superoxide ions, it forms a readily detectable blue-purple formazan precipitate. In the case of A. nidulans spores that undergo germination and formation of a germ tube, NBT stains primarily the tip of the germ tube 12 h after germination induction (45). Accumulation of discrete superoxide ions at the tip of the germ tube suggests the presence of active SOD1 to protect the sequential development of the germ tube against oxidative insults, including membrane damage due to lipid peroxidation. Assuming that an analogous situation occurs to establish a polarized growth axis for formation of germ tubes in S. pombe, this possibility could largely explain the reason why SOD1 activity is required to ensure protection against ROS accumulation at the growing tips of germ tubes during apical dominance.
Experimental procedures
Strains, media, sporulation, and spore purification
Genotypes of S. pombe strains used in this study are described in Table 1. Standard yeast genetic methods were used for growth, mating, and sporulation of cells (1). Untransformed strains were cultured on yeast extract medium (YES) that was supplemented with 225 mg/liter adenine, histidine, leucine, uracil, and lysine. Cells transformed with gene-swap knock-out or knock-in cassettes were selected on YES medium supplemented with the geneticin antibiotic (G418, 200 μg/ml) (Sigma). When plasmid integration was required, S. pombe h90 cells were cultured in synthetic dextrose (SD) minimal medium containing bacto-yeast nitrogen base (0.67%), dextrose (2%), and 225 mg/liter adenine, uracil, and lysine. Leucine was omitted in SD medium to allow selection and maintenance of the integrated DNA fragment into the yeast genome (47).
Table 1.
S. pombe strains used in this study
| Strain | Genotype | Source or ref. |
|---|---|---|
| FY12806 | h90 ura4-Δ18 ade6-M210 leu1-32 | 57 |
| SPY09 | h90 ura4-Δ18 ade6-M210 leu1-32 ctr4Δ::KANr | This study |
| SPY10 | h90 ura4-Δ18 ade6-M210 leu1-32 ctr5Δ::KANr | This study |
| SPY11 | h90 ura4-Δ18 ade6-M210 leu1-32 ctr6Δ::KANr | This study |
| SPY12 | h90 ura4-Δ18 ade6-M210 leu1-32 ctr6Δ::loxP ctr4Δ::KANr | This study |
| SPY13 | h90 ura4-Δ18 ade6-M210 leu1-32 cuf1Δ::KANr | This study |
| SPY14 | h90 ura4-Δ18 ade6-M210 leu1-32 ctr4-GFP:loxP ctr5-mCherry:KANr | This study |
| SPY15 | h90 ura4-Δ18 ade6-M210 leu1-32 ctr6-HA4:KANr | This study |
| SPY17 | h90 ura4-Δ18 ade6-M210 leu1-32 sod1Δ::KANr | This study |
| SPY18 | h90 ura4-Δ18 ade6-M210 leu1-32 ish1Δ::KANr | This study |
| SPY19 | h90 ura4-Δ18 ade6-M210 leu1-32 ish1Δ::KANr ish1+-GFP:leu1+ | This study |
| FY435 | h+ his7–366 leu1-32 ura4-Δ18 ade6-M210 | 53 |
| VNY10 | h+ his7–366 leu1-32 ura4-Δ18 ade6-M210 ish1Δ::KANr | This study |
| VNY11 | h+ his7–366 leu1-32 ura4-Δ18 ade6-M210 ish1Δ::KANr ish1+-GFP:leu1+ | This study |
To obtain spores, homothallic h90 cells were freshly pre-cultured in YES until the mid-logarithmic phase. Cells were then washed and plated on malt extract (ME) medium for 4 days. Sporulation efficiency was assessed by light microscopy examination. Although S. pombe asci break down by themselves, preparations containing ascospores, spores, vegetative cells, and cellular debris were digested by glusulase (5%) for 1 h. The glusulase-dependent step ensured killing of the remaining vegetative cells. The digested mixture was washed, and spores were purified by Percoll gradient centrifugation as described (48, 49). After centrifugation at 15,000 × g at 4 °C for 1 h, the top three layers (50, 60, and 70% Percoll in 2.5 m sucrose and 0.5% Triton X-100) consisting of vegetative cells and debris were removed and discarded. The remaining layer (80% Percoll) containing spores was washed three times with 0.5% Triton X-100. Spores were then suspended in water and stored at 4 °C. To synchronize spores for their entry into germination, they were adjusted to a titer of 1 × 107 spores/ml in glucose-free YE medium. At this point, glucose was added to induce germination, and spore differentiation was monitored by optical density measurements at 600 nm and microscopic examination of changes in spore morphology.
Plasmids
S. cerevisiae GIC2 coding region corresponding to amino acid residues 1- 208, denoted the CRIB domain (27), was isolated by PCR and then cloned into the EcoRI and SalI sites of the pJK148 plasmid (50). The resulting construct was named pJKCRIB. The GFP coding sequence was isolated by PCR using primers designed to generate SalI and ApaI sites at the 5′ and 3′ termini, respectively, of the GFP gene. The resulting DNA fragment was digested with SalI and ApaI and then fused in-frame with CRIB into the corresponding sites of pJKCRIB, generating pJKCRIB-GFP. The S. pombe shk1+ promoter up to −636 from the start codon of the shk1+ gene was isolated by PCR and then inserted into pJKCRIB-GFP at the SacI and EcoRI sites. We therefore ensured that the expression of the CRIB-GFP reporter, which localizes to the cell tips and to the site of cell division, was under the control of the shk1+ promoter and was used as described previously (27).
To create h90 strains in which GFP, Cherry, and HA4 coding sequences were integrated at the chromosomal locus of ctr4+, ctr5+, and ctr6+, respectively, a cassette-based gene replacement approach was used. In the case of Ctr4, a ctr4+-GFP DNA fragment containing a sub-region of the ctr4+ locus (starting at +514 downstream of the first base of the translational initiator codon) up to the stop codon of GFP was amplified by PCR from pJKctr4+-GFP plasmid (22). The resulting DNA fragment was inserted into the BamHI and EcoRI sites of pKSloxP-kanMX6-loxP. Similarly, a 320-bp SalI–Asp-718 PCR-amplified DNA segment containing a 3′-UTR region of the ctr4+ locus was inserted downstream of the second loxP sequence using the SalI and Asp-718 sites of pKSloxP-kanMX6-loxP. Once generated, the ctr4+-GFP-loxP-kanMX6-loxP-ctr4–3′UTR cassette was isolated using NheI and Asp-718 digestion and then integrated at the chromosomal locus of ctr4+. In the case of Ctr5, a ctr5+-Cherry DNA fragment containing a sub-region of the ctr5+ locus (starting at +314 downstream of the first base of the translational initiator codon) up to the stop codon of Cherry was amplified by PCR of the pJKctr5+-Cherry plasmid (22). The resulting DNA fragment was inserted into the SpeI and EcoRI sites of pKSloxP-kanMX6-loxP. Likewise, a 336-bp SalI–Asp-718 PCR-amplified DNA segment containing a 3′-UTR region of the ctr5+ locus was inserted downstream of the second loxP sequence using the SalI and Asp-718 sites of pKSloxP-kanMX6-loxP. Once generated, the ctr5+-Cherry-loxP-kanMX6-loxP-ctr5–3′UTR cassette was isolated using SpeI and Asp-718 digestion and then integrated at the chromosomal locus of ctr5+. In the case of Ctr6, a ctr6+-HA4 DNA fragment containing the ctr6+ locus starting at −350 from the translational initiator codon up to the stop codon was amplified by PCR of the pJKctr6+-HA4 plasmid (22). The resulting DNA fragment was inserted into the NotI and EcoRI sites of pKSloxP-kanMX6-loxP. Similarly, a 452-bp SalI–Asp-718 PCR-amplified DNA segment containing a 3′-UTR region of the ctr6+ locus was inserted downstream of the second loxP sequence using the SalI and Asp-718 sites of pKSloxP-kanMX6-loxP. Once generated, the ctr6+-HA4-loxP-kanMX6-loxP-ctr6–3′UTR cassette was isolated using NotI and Asp-718 digestion and then integrated at the chromosomal locus of ctr6+.
To generate the integrative pJKsod1+ plasmid, a 1115-bp EcoRI–XhoI PCR-amplified DNA segment containing the S. pombe sod1+ locus starting at position −650 from the translational start codon up to the stop codon was inserted into the EcoRI and XhoI sites of pJK148. Nucleotide substitutions that gave rise to sod1H64A mutant allele were performed by an overlap extension method as described previously (51).
RNA isolation and analysis
Total RNA was isolated using a standard hot phenol method as described previously (52). Analysis of gene transcript levels was performed using RNase protection assays as described previously (53). To detect gpd1+ mRNA levels, a plasmid pSKgpd1+ was created by inserting a 154-bp BamHI–EcoRI fragment from the gpd1+ gene into the same restriction sites of pBluescript SK (Stratagene, La Jolla, CA). The antisense RNA hybridizes to the region between positions +51 and +205 downstream of the first base of the translational start codon of gpd1+. Plasmids pSKctr4+ (18), pSKctr5+-v1, pSKctr6+-v2 (22), pSKcuf1+-v7 (21), and pSKgpd1+ were used to produce antisense RNA probes, allowing the detection of steady-state levels of ctr4+, ctr5+, ctr6+, cuf1+, and gpd1+ mRNAs, respectively. 32P-Labeled antisense RNA probes were produced using the above-mentioned BamHI-linearized plasmids, [α-32P]UTP, and the T7 RNA polymerase, as described previously (53). The gpd1+ riboprobe was used to detect gpd1+ transcript as an internal control for normalization during quantification of the RNase protection products.
Calcofluor, FM4-64, and CNIR4 labeling of spores and outgrowing cells
To stain the OSW, dormant spores that were found to be refractile under phase-contrast microscopy were incubated in the presence of calcofluor (10 μg/ml) in glucose-free media. After a 30-min incubation, dormant spores were washed and synchronously induced to enter into germination in glucose-containing and calcofluor-free media. At the zero-time point, when spores had just entered germination, and for the subsequent time points, aliquots were retrieved every hour. At each time point, spores were subjected to microscopic analysis using ×1000 magnification and the following filters: 340–380 nm (calcofluor); 465–495 nm (GFP) (when spores contained an integrated CRIB-GFP or ctr4+-GFP allele); and 510–560 nm (Cherry) (when spores harbored an integrated ctr5+-Cherry allele). This microscopic procedure allowed monitoring of the progression of germination of individual spores and simultaneously visualization of fluorescent markers. Calcofluor was used to stain the outer spore wall because it binds to constituents in the spore wall of S. pombe. CRIB-GFP was used as a marker of the new cell end during outgrowth.
Vacuole membrane staining using FM4-64 (Sigma) was performed as described previously (54), except that germinating spores were not resuspended in distilled water prior to microscopic analysis. At the indicated germination phase, aliquots were harvested, washed, and resuspended in germination YES media containing FM4-64 (16 μm) for 30 min at 30 °C. After this step, spores and outgrowing spores were pelleted, washed, and resuspended in fresh germination YES media and then incubated at 30 °C for an additional 30 min before performing fluorescence microscopy.
Wild-type h90 cells were grown mitotically in the presence of CuSO4 (5 μm) to an A600 of 1.0 at 30 °C. This step allowed accumulation of copper within cells. Subsequently, cells were harvested, washed, and resuspended in low copper-containing media (0.16 nm CuSO4, which corresponds to 100 times less copper than basal conditions). Cells were divided into four treatment groups as follows: no addition of copper sensor (control) but addition of the same concentration of DMSO when a chemical probe was added; addition of CNIR4 (5 μm in 1% DMSO); addition of Ctrl-CNIR4 (5 μm in 1% DMSO); and addition of Ctrl-CNIR4-S2 (5 μm in 1% DMSO). After 30 min following each treatment, aliquots were examined by direct fluorescence microscopy. Ctrl-CNIR4 and Ctrl-CNIR4-S2 are control analogs where the metal-interacting sulfur atoms are replaced by isosteric carbons. Ctrl-CNIR4 has no sulfur atoms, and Ctrl-CNIR4-S2 contains two sulfur atoms. These control probes do not respond to copper but have the same dye scaffold as CNIR4 and thus can be used to help disentangle metal- and dye-dependent signals (29, 30). To study intracellular copper distribution during spore germination and outgrowth, wild-type h90 and h90 ctr4Δ, h90 ctr6Δ, and h90 ctr4Δ ctr6Δ mutant strains were plated on ME media that contained CuSO4 (5 μm). After sporulation and purification of spores from each strain, spores were induced to undergo germination in low copper-containing media. At the indicated step of germination, spores or outgrown spores were harvested, washed, and resuspended in the presence of CNIR4 (5 μm in 1% DMSO) for 30 min and then analyzed by direct fluorescence microscopy using ×1000 magnification at 633 nm excitation to match the absorption maximum of the Cu+-bound CNIR4 probe.
Indirect immunofluorescence microscopy
Localization of Ctr6-HA4 in meiotic and sporulating cells was performed as described previously (22). In the case of Ctr6-HA4 localization during germination and outgrowth, purified h90 ctr6Δ mutant spores harboring a ctr6+-HA4 allele were induced to germinate in the presence of TTM (50 μm) for the indicated time points. Germinating spores were fixed and adsorbed on poly-l-lysine-coated (0.1%) multiwall slides as described previously (23). After a 30-min block with TNB (10 mm Tris-HCl, pH 7.5, 150 mm NaCl, 1% BSA, 0.02% sodium azide), spores were incubated with an anti-HA antibody (F-7) (Santa Cruz Biotechnology) diluted 1:250 in TNB. After an 18-h period of incubation at 4 °C, spores were washed with TNB and incubated for 4 h with a goat anti-mouse Alexa Fluor 546-labeled IgG antibody (Invitrogen) diluted 1:250 in TNB. After spores were washed, mounting medium (Invitrogen) was added to each well prior to microscopic examination. Fluorescence and differential interference contrast images of germinating spores and outgrown cells were obtained using a Nikon Eclipse E800 epifluorescent microscope (Nikon, Melville, NY) equipped with a Hamamatsu ORCA-ER digital cooled charge-coupled device camera (Hamamatsu, Bridgewater, NJ). Merged images were obtained using the Simple PCI software version 5.3.0.1102 (Compix, Sewickly, PA). Cell fields shown in this study represent a minimum of five independent experiments.
Flow cytometry analysis
At various time points after spores had entered germination, aliquots of germinating spores were retrieved and fixed with cold ethanol to perform flow cytometry assays as described previously (55). Spores were stained with propidium iodide and analyzed using a CytoFLEX (or FACScan) flow cytometer with an argon laser tuned to 488 nm. The FL3 detector with a 670/690-nm long pass filter was used to collect propidium iodide fluorescence. Results were analyzed using the FlowJo version 10 software (FlowJo LLC).
SOD activity and Western blot assays
At the indicated time points, germinating spores or outgrown cells were harvested and washed with lysis buffer containing 25 mm Tris-HCl, pH 7.5, 150 mm NaCl, 1 mm PMSF, 1 mm DTT, 0.1 mm Na3VO4, and a protease inhibitor mixture (Sigma; P8340). After washings, germinating spores were suspended in lysis buffer, and then disrupted with an equivalent volume of glass beads using a Fastprep instrument (MP Biomedicals). Lysates were centrifuged at 13,000 × g, and then the supernatants that contained soluble proteins were partially purified using Spin-X centrifuge tube filters (Costar-Corning; 8161). Equal concentrations of lysates were analyzed on 10% native polyacrylamide gels as described previously (56), and SOD activity assays were carried out using in-gel nitro blue tetrazolium staining as described previously (20). Spectrophotometric determination of SOD activity was performed using a cytochrome c/xanthine oxidase method (23).
In the case of Ctr4-GFP, Ctr5-Cherry, Ctr6-HA4, and SOD1 detection, germinating spore lysates were centrifuged at 100,000 × g for 30 min at 4 °C. The supernatant that contained soluble proteins was set aside for SOD1 detection, whereas the membrane fraction (pellet) was treated with 1% Triton X-100 for 30 min on ice and then re-fractionated at 100,000 × g. The resulting supernatant fraction contained solubilized membrane proteins. The following antibodies were used for immunodetection of Ctr4-GFP, Ctr5-Cherry, Ctr6-HA4, and SOD1: monoclonal anti-GFP antibody B-2 (Santa Cruz Biotechnology); polyclonal anti-Cherry antibody 16D7 (Thermo Fisher Scientific); monoclonal anti-HA antibody F-7; and polyclonal anti-SOD1 antibody 100 (StressGen), respectively. Following incubation, Western blotting membranes were washed and incubated with the appropriate horseradish peroxidase-conjugated secondary antibodies (Amersham Biosciences), developed with enhanced chemiluminescence (ECL) reagents (Amersham Biosciences), and visualized by chemiluminescence using an ImageQuant LAS 4000 instrument (GE Healthcare) equipped with a Fujifilm High Sensitivity F0.85 43 mm camera.
Author contributions
S. P. designed and performed most of the experiments. V. N. produced several DNA constructs and performed a number of fluorescence microscopy experiments. K. M. R. T. produced and purified the fluorescent-based copper probe and its chemical control analogs. S. P., V. N., and S. L. analyzed data. S. P. and S. L. conceptualized research and wrote the manuscript. All authors reviewed the results and approved the final version of the manuscript.
Acknowledgments
We are indebted to Dr. Gilles Dupuis for critical review of the manuscript and for valuable comments, as well as to Dr. Chris Chang for help in providing copper probes. We gratefully acknowledge Dr. Léonid Volkov for excellent assistance in flow cytometry experiments. The S. pombe strain FY12806 was provided by the Yeast Genetic Resource Center of Japan.
This work was supported by Grant MOP-CP-243929 from the Canadian Institutes of Health Research (to S. L.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- OSW
- outer spore wall
- Cherry
- red fluorescent protein
- CRIB
- Cdc42/Rac interactive-binding
- Ctr
- copper transporter
- Cuf1
- copper factor 1
- DIC
- differential interference contrast
- FSC
- forward-scattered light
- ME
- malt extract
- NBT
- nitro blue tetrazolium
- SOD1
- copper-zinc superoxide dismutase 1
- SSC
- side-scattered light
- TTM
- ammonium tetrathiomolybdate
- YE
- yeast extract
- YES
- yeast extract plus supplements
- ROS
- reactive oxygen species.
References
- 1. Sabatinos S. A., and Forsburg S. L. (2010) Molecular genetics of Schizosaccharomyces pombe. Methods Enzymol. 470, 759–795 [DOI] [PubMed] [Google Scholar]
- 2. Shimoda C. (2004) Forespore membrane assembly in yeast: coordinating SPBs and membrane trafficking. J. Cell Sci. 117, 389–396 [DOI] [PubMed] [Google Scholar]
- 3. Fukunishi K., Miyakubi K., Hatanaka M., Otsuru N., Hirata A., Shimoda C., and Nakamura T. (2014) The fission yeast spore is coated by a proteinaceous surface layer comprising mainly Isp3. Mol. Biol. Cell 25, 1549–1559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. García I., Tajadura V., Martín V., Toda T., and Sánchez Y. (2006) Synthesis of α-glucans in fission yeast spores is carried out by three α-glucan synthase paralogues, Mok12p, Mok13p and Mok14p. Mol. Microbiol. 59, 836–853 [DOI] [PubMed] [Google Scholar]
- 5. Bonazzi D., Julien J. D., Romao M., Seddiki R., Piel M., Boudaoud A., and Minc N. (2014) Symmetry breaking in spore germination relies on an interplay between polar cap stability and spore wall mechanics. Dev. Cell 28, 534–546 [DOI] [PubMed] [Google Scholar]
- 6. Hatanaka M., and Shimoda C. (2001) The cyclic AMP/PKA signal pathway is required for initiation of spore germination in Schizosaccharomyces pombe. Yeast 18, 207–217 [DOI] [PubMed] [Google Scholar]
- 7. Herman P. K., and Rine J. (1997) Yeast spore germination: a requirement for Ras protein activity during re-entry into the cell cycle. EMBO J. 16, 6171–6181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Joseph-Strauss D., Zenvirth D., Simchen G., and Barkai N. (2007) Spore germination in Saccharomyces cerevisiae: global gene expression patterns and cell cycle landmarks. Genome Biol. 8, R241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Shimoda C. (1980) Differential effect of glucose and fructose on spore germination in the fission yeast Schizosaccharomyces pombe. Can. J. Microbiol. 26, 741–745 [Google Scholar]
- 10. Kim B. E., Nevitt T., and Thiele D. J. (2008) Mechanisms for copper acquisition, distribution and regulation. Nat. Chem. Biol. 4, 176–185 [DOI] [PubMed] [Google Scholar]
- 11. Festa R. A., and Thiele D. J. (2011) Copper: an essential metal in biology. Curr. Biol. 21, R877–R883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Beaudoin J., Laliberté J., and Labbé S. (2006) Functional dissection of Ctr4 and Ctr5 amino-terminal regions reveals motifs with redundant roles in copper transport. Microbiology 152, 209–222 [DOI] [PubMed] [Google Scholar]
- 13. Beaudoin J., Thiele D. J., Labbé S., and Puig S. (2011) Dissection of the relative contribution of the Schizosaccharomyces pombe Ctr4 and Ctr5 proteins to the copper transport and cell surface delivery functions. Microbiology 157, 1021–1031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ioannoni R., Beaudoin J., Mercier A., and Labbé S. (2010) Copper-dependent trafficking of the Ctr4-Ctr5 copper transporting complex. PLoS One 5, e11964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Labbé S., Beaudoin J., and Ioannoni R. (2013) in Metals in Cells (Culotta V. C., and Scott R. S., eds) pp. 163–174, John Wiley & Sons, Inc., Chichester, UK [Google Scholar]
- 16. Zhou H., and Thiele D. J. (2001) Identification of a novel high affinity copper transport complex in the fission yeast Schizosaccharomyces pombe. J. Biol. Chem. 276, 20529–20535 [DOI] [PubMed] [Google Scholar]
- 17. Peter C., Laliberté J., Beaudoin J., and Labbé S. (2008) Copper distributed by Atx1 is available to copper amine oxidase 1 in Schizosaccharomyces pombe. Eukaryot. Cell 7, 1781–1794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Beaudoin J., and Labbé S. (2001) The fission yeast copper-sensing transcription factor Cuf1 regulates the copper transporter gene expression through an Ace1/Amt1-like recognition sequence. J. Biol. Chem. 276, 15472–15480 [DOI] [PubMed] [Google Scholar]
- 19. Beaudoin J., Mercier A., Langlois R., and Labbé S. (2003) The Schizosaccharomyces pombe Cuf1 is composed of functional modules from two distinct classes of copper metalloregulatory transcription factors. J. Biol. Chem. 278, 14565–14577 [DOI] [PubMed] [Google Scholar]
- 20. Labbé S., Peña M. M., Fernandes A. R., and Thiele D. J. (1999) A copper-sensing transcription factor regulates iron uptake genes in Schizosaccharomyces pombe. J. Biol. Chem. 274, 36252–36260 [DOI] [PubMed] [Google Scholar]
- 21. Beaudoin J., Ioannoni R., López-Maury L., Bähler J., Ait-Mohand S., Guérin B., Dodani S. C., Chang C. J., and Labbé S. (2011) Mfc1 is a novel forespore membrane copper transporter in meiotic and sporulating cells. J. Biol. Chem. 286, 34356–34372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Plante S., Ioannoni R., Beaudoin J., and Labbé S. (2014) Characterization of Schizosaccharomyces pombe copper transporter proteins in meiotic and sporulating cells. J. Biol. Chem. 289, 10168–10181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bellemare D. R., Shaner L., Morano K. A., Beaudoin J., Langlois R., and Labbe S. (2002) Ctr6, a vacuolar membrane copper transporter in Schizosaccharomyces pombe. J. Biol. Chem. 277, 46676–46686 [DOI] [PubMed] [Google Scholar]
- 24. Rees E. M., Lee J., and Thiele D. J. (2004) Mobilization of intracellular copper stores by the ctr2 vacuolar copper transporter. J. Biol. Chem. 279, 54221–54229 [DOI] [PubMed] [Google Scholar]
- 25. Rees E. M., and Thiele D. J. (2007) Identification of a vacuole-associated metalloreductase and its role in Ctr2-mediated intracellular copper mobilization. J. Biol. Chem. 282, 21629–21638 [DOI] [PubMed] [Google Scholar]
- 26. Das M., Wiley D. J., Chen X., Shah K., and Verde F. (2009) The conserved NDR kinase Orb6 controls polarized cell growth by spatial regulation of the small GTPase Cdc42. Curr. Biol. 19, 1314–1319 [DOI] [PubMed] [Google Scholar]
- 27. Tatebe H., Nakano K., Maximo R., and Shiozaki K. (2008) Pom1 DYRK regulates localization of the Rga4 GAP to ensure bipolar activation of Cdc42 in fission yeast. Curr. Biol. 18, 322–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Beaudoin J., Ekici S., Daldal F., Ait-Mohand S., Guérin B., and Labbé S. (2013) Copper transport and regulation in Schizosaccharomyces pombe. Biochem. Soc. Trans. 41, 1679–1686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ackerman C. M., Lee S., and Chang C. J. (2017) Analytical methods for imaging metals in biology: from transition metal metabolism to transition metal signaling. Anal. Chem. 89, 22–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Cotruvo J. A. Jr, Aron A. T., Ramos-Torres K. M., and Chang C. J. (2015) Synthetic fluorescent probes for studying copper in biological systems. Chem. Soc. Rev. 44, 4400–4414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Taricani L., Tejada M. L., and Young P. G. (2002) The fission yeast ES2 homologue, Bis1, interacts with the Ish1 stress-responsive nuclear envelope protein. J. Biol. Chem. 277, 10562–10572 [DOI] [PubMed] [Google Scholar]
- 32. Barhoom S., Kupiec M., Zhao X., Xu J. R., and Sharon A. (2008) Functional characterization of CgCTR2, a putative vacuole copper transporter that is involved in germination and pathogenicity in Colletotrichum gloeosporioides. Eukaryot. Cell 7, 1098–1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lambou K., Lamarre C., Beau R., Dufour N., and Latge J. P. (2010) Functional analysis of the superoxide dismutase family in Aspergillus fumigatus. Mol. Microbiol. 75, 910–923 [DOI] [PubMed] [Google Scholar]
- 34. Yao S. H., Guo Y., Wang Y. Z., Zhang D., Xu L., and Tang W. H. (2016) A cytoplasmic Cu-Zn superoxide dismutase SOD1 contributes to hyphal growth and virulence of Fusarium graminearum. Fungal Genet. Biol. 91, 32–42 [DOI] [PubMed] [Google Scholar]
- 35. Graden J. A., Ellerby L. M., Roe J. A., and Valentine J. S. (1994) Role of the bridging histidyl imidazolate ligand in yeast copper-zinc superoxide dismutase. Characterization of the His63Ala mutant. J. Am. Chem. Soc. 116, 9743–9744 [Google Scholar]
- 36. Strange R. W., Antonyuk S., Hough M. A., Doucette P. A., Rodriguez J. A., Hart P. J., Hayward L. J., Valentine J. S., and Hasnain S. S. (2003) The structure of holo and metal-deficient wild-type human Cu, Zn superoxide dismutase and its relevance to familial amyotrophic lateral sclerosis. J. Mol. Biol. 328, 877–891 [DOI] [PubMed] [Google Scholar]
- 37. Botts M. R., and Hull C. M. (2010) Dueling in the lung: how Cryptococcus spores race the host for survival. Curr. Opin. Microbiol. 13, 437–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Oiartzabal-Arano E., Perez-de-Nanclares-Arregi E., Espeso E. A., and Etxebeste O. (2016) Apical control of conidiation in Aspergillus nidulans. Curr. Genet. 62, 371–377 [DOI] [PubMed] [Google Scholar]
- 39. Geib E., Gressler M., Viediernikova I., Hillmann F., Jacobsen I. D., Nietzsche S., Hertweck C., and Brock M. (2016) A non-canonical melanin biosynthesis pathway protects Aspergillus terreus conidia from environmental stress. Cell Chem. Biol. 23, 587–597 [DOI] [PubMed] [Google Scholar]
- 40. Eide D. J., Bridgham J. T., Zhao Z., and Mattoon J. R. (1993) The vacuolar H(+)-ATPase of Saccharomyces cerevisiae is required for efficient copper detoxification, mitochondrial function, and iron metabolism. Mol. Gen. Genet. 241, 447–456 [DOI] [PubMed] [Google Scholar]
- 41. Portnoy M. E., Schmidt P. J., Rogers R. S., and Culotta V. C. (2001) Metal transporters that contribute copper to metallochaperones in Saccharomyces cerevisiae. Mol. Genet. Genomics 265, 873–882 [DOI] [PubMed] [Google Scholar]
- 42. Szczypka M. S., Zhu Z., Silar P., and Thiele D. J. (1997) Saccharomyces cerevisiae mutants altered in vacuole function are defective in copper detoxification and iron-responsive gene transcription. Yeast 13, 1423–1435 [DOI] [PubMed] [Google Scholar]
- 43. Geijer C., Pirkov I., Vongsangnak W., Ericsson A., Nielsen J., Krantz M., and Hohmann S. (2012) Time course gene expression profiling of yeast spore germination reveals a network of transcription factors orchestrating the global response. BMC Genomics 13, 554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Mata J., Wilbrey A., and Bähler J. (2007) Transcriptional regulatory network for sexual differentiation in fission yeast. Genome Biol. 8, R217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Semighini C. P., and Harris S. D. (2008) Regulation of apical dominance in Aspergillus nidulans hyphae by reactive oxygen species. Genetics 179, 1919–1932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Takemoto D., Tanaka A., and Scott B. (2007) NADPH oxidases in fungi: diverse roles of reactive oxygen species in fungal cellular differentiation. Fungal Genet. Biol. 44, 1065–1076 [DOI] [PubMed] [Google Scholar]
- 47. Matsuyama A., Yabana N., Watanabe Y., and Yamamoto M. (2000) Schizosaccharomyces pombe Ste7p is required for both promotion and withholding of the entry to meiosis. Genetics 155, 539–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kloimwieder A., and Winston F. (2011) A screen for germination mutants in Saccharomyces cerevisiae. G3 1, 143–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Shi L., Li Z., Tachikawa H., Gao X. D., and Nakanishi H. (2014) Use of yeast spores for microencapsulation of enzymes. Appl. Environ. Microbiol. 80, 4502–4510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Keeney J. B., and Boeke J. D. (1994) Efficient targeted integration at leu1–32 and ura4–294 in Schizosaccharomyces pombe. Genetics 136, 849–856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Ho S. N., Hunt H. D., Horton R. M., Pullen J. K., and Pease L. R. (1989) Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77, 51–59 [DOI] [PubMed] [Google Scholar]
- 52. Chen D., Toone W. M., Mata J., Lyne R., Burns G., Kivinen K., Brazma A., Jones N., and Bähler J. (2003) Global transcriptional responses of fission yeast to environmental stress. Mol. Biol. Cell 14, 214–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Mercier A., Watt S., Bähler J., and Labbé S. (2008) Key function for the CCAAT-binding factor Php4 to regulate gene expression in response to iron deficiency in fission yeast. Eukaryot. Cell 7, 493–508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Iwaki T., Giga-Hama Y., and Takegawa K. (2006) A survey of all 11 ABC transporters in fission yeast: two novel ABC transporters are required for red pigment accumulation in a Schizosaccharomyces pombe adenine biosynthetic mutant. Microbiology 152, 2309–2321 [DOI] [PubMed] [Google Scholar]
- 55. Sabatinos S. A., and Forsburg S. L. (2009) Measuring DNA content by flow cytometry in fission yeast. Methods Mol. Biol. 521, 449–461 [DOI] [PubMed] [Google Scholar]
- 56. Laliberté J., Whitson L. J., Beaudoin J., Holloway S. P., Hart P. J., and Labbé S. (2004) The Schizosaccharomyces pombe Pccs protein functions in both copper trafficking and metal detoxification pathways. J. Biol. Chem. 279, 28744–28755 [DOI] [PubMed] [Google Scholar]
- 57. Itadani A., Nakamura T., Hirata A., and Shimoda C. (2010) Schizosaccharomyces pombe calmodulin, Cam1, plays a crucial role in sporulation by recruiting and stabilizing the spindle pole body components responsible for assembly of the forespore membrane. Eukaryot. Cell 9, 1925–1935 [DOI] [PMC free article] [PubMed] [Google Scholar]