Abstract
Ongoing elimination efforts have altered the global distribution of Onchocerca volvulus, the agent of river blindness, and further population restructuring is expected as efforts continue. Therefore, a better understanding of population-genetic processes and their effect on biogeography is needed to support elimination goals. We describe O. volvulus genome variation in 27 isolates from early 1990s (before widespread mass treatment) from four distinct locales: Ecuador, Uganda, the West African forest, and the West African savanna. We observed genetic substructuring between Ecuador and West Africa and between the West African forest and savanna bioclimes, with evidence of unidirectional gene flow from savanna to forest strains. We identified forest:savanna-discriminatory genomic regions and report a set of ancestry informative loci that can be used to differentiate between forest, savanna and admixed isolates, which has not previously been possible. We observed mito-nuclear discordance possibly stemming from incomplete lineage sorting. The catalog of the nuclear, mitochondrial and endosymbiont DNA variants generated in this study will support future basic and translational onchocerciasis research, with particular relevance for ongoing control programs and boost efforts to characterize drug, vaccine, and diagnostic targets.
Keywords: river blindness, onchocerciasis, genetic nuclear variations, mitochondrial variations, Wolbachia variations, forest isolates, savanna isolates, admixed isolates
Onchocerca volvulus, the agent of river blindness or onchocerciasis, infects an estimated 37 million people in tropical Africa and in isolated foci in Yemen and Latin America1, causing debilitating eye and skin disease in more than five million among them2. O. volvulus is transmitted by blackflies (Simulium) that breed in fast-flowing, oxygen-rich rivers and streams. The significant human health and socioeconomic burden of the disease inspired massive control efforts that have been underway since the 1970’s (reviewed by Cupp et al.3), reducing or eliminating the disease in many areas4–6, with plans in place to gear-up for world-wide elimination by 20257.
These elimination efforts have already caused considerable changes in the global distribution of onchocerciasis. Transmission has been interrupted in most of Latin America8 and many areas of West Africa9. Further population restructuring is expected as intervention continues, and other factors (e.g., global warming, deforestation, political unrest, etc.) trigger migrations of parasites, vectors, and human hosts. Thus, a thorough understanding of O. volvulus population structure will be crucial to achieve elimination goals. For example, if parasites are found in a given region following repeated rounds of MDA, it would be informative to know whether they represent a sentinel population that has evaded treatment or parasites reintroduced from other areas. And if resistance should arise10, an understanding of gene flow and population connectivity would give a sense of how readily key alleles might spread.
In the past, studies of O. volvulus population genetics focused on identification of strains associated with severe eye disease. In West Africa, higher rates of blindness were reported in the savannas compared to nearby rainforest regions11,12. Divergent morphology13,14, tropism within the host15, vector preferences16, biochemical variations17,18, and endosymbiont populations19 supported the notion that the savanna and forest populations represented discrete strains distinguishable using the O-150 repeat region sequence20. Thus, savanna populations were intensely targeted and mostly eliminated from their distribution zones3,5,6. However, populations outside West Africa (e.g., Uganda21, Northern Sudan22, etc.) did not adhere to this orderly classification scheme, and studies based on other markers (e.g. nuclear ITS rDNA sequence23, antigenic proteins, and mitochondrial RFLP profiles24) indicated a lack of population differentiation, in conflict with the two-strain hypothesis.
Given the opposing results of historical studies based on limited genetic markers, we undertook an O. volvulus population genomics survey, assessing global diversity in the nuclear (Ov), mitochondrial (mtDNA), and Wolbachia (wOv) genomes of 27 isolates from West Africa, Uganda and Ecuador. All three genomes indicated a degree of population structure, with the nuclear genome providing phylogenetic signals consistent with gene flow and genetic admixture among discrete West African forest and savanna populations. To better understand local adaptation, genomic regions of particularly high divergence were identified, which could serve as markers for tracking O. volvulus populations. A set of Ancestry Informative Markers (AIMs) was obtained that can be used to estimate the relative proportion of forest and savanna ancestry in O. volvulus isolates. These AIMs and the catalog of all variants detected in this study will support future basic and translational onchocerciasis research.
Results
Global patterns of genome variation
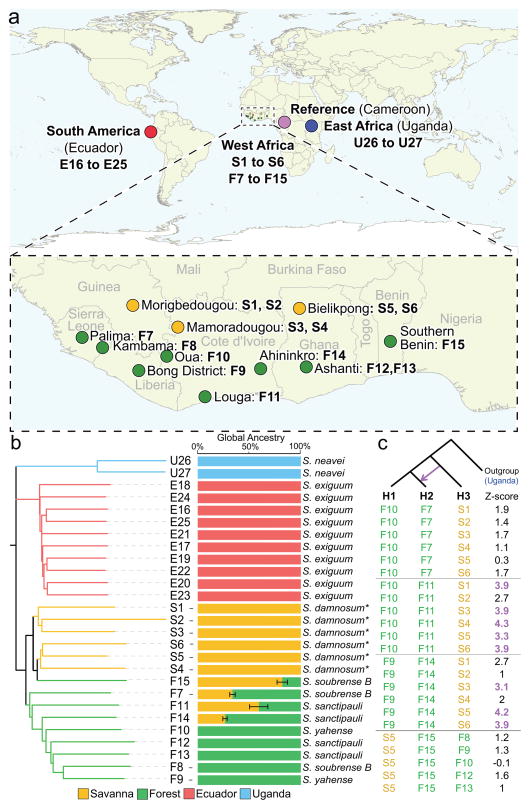
A total of 27 West African (savanna S1–S6, forest F7–F15), Ecuadorian (E16–E25) and Ugandan (U26–U27) O. volvulus isolates were subjected to analysis (Fig. 1a), after removing 5 isolates with low sequence depth (Supplementary Table 1 and Supplementary Fig. 1). All DNA samples were collected in the early 1990’s during the course of previously described studies20,21,25,26. The West African isolates were classified as “forest” or “savanna” by Southern blot based on the O-150 repeat20,21. Our genomic DNA was derived from worm tissues (dissected from a nodule) representing one or more potentially polyandrous females full of embryos in various stages of intrauterine development (Supplementary Table 1). The contribution from embryonic DNA was estimated to be very high in most samples (Supplementary Fig. 2). Paired-end sequence reads were generated (median 59M reads per sample; Supplementary Table 1) and aligned to reference nuclear O. volvulus (Ov), mitochondrial O. volvulus (mtDNA), and Wolbachia endosymbiont (wOv) genome assemblies27, resulting in an average 98.9%, 99.7% and 99.9% of the genome covered by mapped reads and average 66-, 4593- and 216-fold depth of mapped read coverage, respectively (Table 1). Variant calling resulted in a final set of 1.3M SNPs in Ov, 167 SNPs in the mtDNA and 771 SNPs detected in the wOv genomes (Table 1, Supplementary Table 2). Density of segregating sites was similar in Ov (13.6 SNPs/kb) and mtDNA (11.6 SNPs/kb) but an order of magnitude lower in wOv (0.8 SNPs/kb). Genome-wide mean synonymous and nonsynonymous diversity (πS and πN) in Ov were estimated to be 4.0 × 10−3 and 1.0 × 10−3, respectively. The resulting πN/πS ratio of ~0.25 suggests pervasive purifying selection acting on nonsynonymous variants, while the πN/πS ratios were overall significantly higher (median πN/πS =0.43, P = 3.2 × 10−22; Supplementary Fig. 3) for genes specific to Onchocercidae (n = 1274)27 suggesting that these genes evolve under relaxed purifying selection compared to the rest of the coding genome.
Figure 1.
The geographical and genomic relationship between O. volvulus isolates. (a) Twenty- seven individual isolates were collected from Ecuador, Uganda and 11 sites in West Africa (3 savanna and 8 forest sites). (b) Clustering of isolates based on single nucleotide polymorphisms in the three nuclear autosomes, and admixture analysis was used to assess patterns of global ancestry among samples. Error bars represent standard errors estimated using bootstrapping. Local Simulium vector species are presented for each isolate. * or S. sirbanum (c) Gene flow between allopatric populations (Patterson’s D test). Isolate F15 was treated as a savanna sample, and showed no significant gene flow from any forest isolate. Numbers colored in purple were significant according to the test (z-score > 3).
Table 1.
Sequencing coverage and SNP statistics
| Statistic | Nucleusa | Mitochondria | Wolbachiab | ||
|---|---|---|---|---|---|
| Sample coverage statistics | Average depth (Fold) | All samples | 66.5 | 4593.4 | 216.1 |
| Savanna | 26.3 | 2068.6 | 81.3 | ||
| Forest | 51.2 | 3589.5 | 310.7 | ||
| West Africa | 41.3 | 2981.1 | 218.9 | ||
| Ecuador | 101.0 | 5877.2 | 57.5 | ||
| Uganda | 83.0 | 10266.8 | 987.4 | ||
|
| |||||
| Average callable loci | All samples | 95.6% | 88.7% | 98.9% | |
| Savanna | 92.5% | 88.4% | 99.1% | ||
| Forest | 95.6% | 88.6% | 99.2% | ||
| West Africa | 94.3% | 88.5% | 99.1% | ||
| Ecuador | 97.3% | 88.9% | 98.7% | ||
| Uganda | 96.8% | 88.9% | 99.2% | ||
|
| |||||
| SNP statistics | Overview | Variants | 1,308,938 | 167 | 771 |
| Variants rate (per kb) | 13.6 | 11.6 | 0.8 | ||
|
| |||||
| SNP counts | Intergenic | 677,763 (51.8%) | 29 (17.4%) | 336 (43.6%) | |
| Genic | 631,175 (48.2%) | 138 (82.6%) | 435 (56.4%) | ||
| Synonymous | 49,065 (3.7%) | 80 (47.9%) | 203 (26.3%) | ||
| Nonsynonymous | 62,761 (4.8%) | 38 (22.8%) | 232 (30.1%) | ||
| UTR | 1,935 (0.1%) | 26 (15.6%) | 0 (0.0%) | ||
| Intronic | 518,454 (39.6%) | 0 (0.0%) | 0 (0.0%) | ||
|
| |||||
| Number of genes containing SNP | Synonymous | 9,160 | 12 | 159 | |
| Nonsynonymous | 9,800 | 11 | 183 | ||
| UTR | 667 | 7 | 0 | ||
| Intronic | 11,053 | 0 | 0 | ||
For nuclear genome, the counts are based on the longest isoforms
For Wolbachia genome, the counts are based on refseq genecalls after filtering the questionable regions (see Methods)
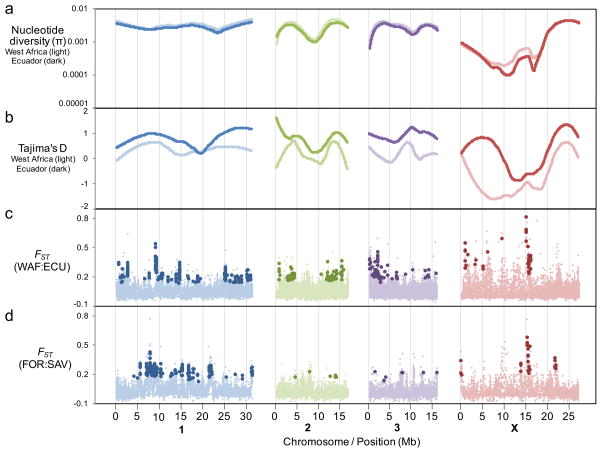
The ancestral karyotype of filarial nematodes is thought to be 5A+X0, the same as Caenorhabditis elegans. O. volvulus was reduced to 3A+XY, likely through chromosomal fusion28, represented by the four large assembled contigs27. The decreased depth of read coverage of one of the four chromosomes suggests that it corresponds to the X chromosome, consistent with findings of Cotton et al.27 (Supplementary Fig. 2a). When nucleotide diversity (π) was plotted across the length of the four chromosomes, chromosome X shows a reduced sequence diversity (Fig. 2a), consistent with the expected lower mutation rate and the smaller population size of the X chromosome relative to the autosomes. Chromosomes 2 and 3, which correspond to C. elegans chromosomes III and II, respectively27, show a decrease in sequence diversity near the chromosome centers. These patterns of sequence diversity are likely to have been shaped by intrachromosomal variations in local recombination rates and/or gene density which influence the effectiveness of linked selection (e.g., background selection)29,30. Local reduction in Tajima’s D (representing a skew in the site frequency spectrum toward an excess of low-frequency variants) near these chromosome centers is in line with the action of linked selection (Fig. 2b). Because the impact of evolutionary processes (mutation, selection, and genetic drift, etc.) differs substantially between sex chromosomes and autosomes31 (Fig. 2a and 2b), separate analyses were performed on the X chromosome and the autosomes.
Figure 2.
Sequence variation in the nuclear genome of O. volvulus. (a) Local polynomial regression lines of best fit of π based on West Africa (light lines) and Ecuador (dark lines) samples (per 50kb window on each chromosome). (b) Local polynomial regression lines of best fit of Tajima’s D based on West Africa (light lines) and Ecuador (dark lines) samples (per 50kb window on each chromosome). (c and d) The average fixation index (FST) of sliding windows across the chromosomes was plotted to compare West African and Ecuadorian populations. The “outlier” windows with high inter-population variation are indicated with lager, darker markers (n=293 autosomal and n=40 X chromosomal regions for West Africa:Ecuador, and n=194 autosomal and n=28 X chromosomal regions for forest:savanna).
Population structure and differentiation inferred from nuclear genome
Clustering based on autosomal allele-sharing distances indicated a clear separation between Ugandan, West African, and Ecuadorian isolates (Fig. 1b), although a mtDNA based phylogeny shows that, as expected, this diversity represents a much more homogenous population when compared to inter-species differences (Supplementary Fig. 4). While mean nucleotide diversity (π) was comparable across populations (Supplementary Fig. 5), genetic variance was partitioned between the populations (Weir and Cockerham weighted FST: 0.08 between West Africa and Ecuador, WAF:ECU; 0.04 between West African forest and savanna, FOR:SAV). Historical records suggest that the Ecuadorian population was founded by a single shipwrecked slaving vessel32,33. A more positive value of Tajima’s D in Ecuador as compared to West Africa (Fig. 2b and Supplementary Fig. 5) is consistent with a recent population bottleneck (likely associated with the colonization of a new habitat in South America) during which nucleotide diversity is reduced proportionally less than segregating sites, thereby departing from mutation-drift equilibrium.
A genome-wide sliding window analysis of FST was used to identify regions that may have experienced diversifying selection for WAF:ECU (Fig. 2c) and FOR:SAV comparisons (Fig. 2d; Supplementary Table 3 and Supplementary Table 4). In the WAF:ECU comparison, we identified 293 and 40 windows with high (top 1%) sequence divergence on autosomal and X chromosomes, respectively (Fig. 2c). These contained 271 genes (Supplementary Table 5) that were significantly enriched for nine gene ontology (GO) terms including vesicle trafficking (actin cytoskeleton reorganization) and G-protein mediated signaling (Table 2).
Table 2.
Gene Ontology term enrichment among genes in FST outlier regions
| Comparison | Gene Ontology Term Description | FUNC Enrichment Significance (P value) | ||
|---|---|---|---|---|
| Root Term | Term Name | Term ID | ||
| West Africa vs Ecuador (WAF:ECU) | Biological Process | G-protein coupled receptor signaling | GO:0007186 | 5.9E-03 |
| Molecular Function | methyltransferase activity | GO:0008168 | 3.4E-02 | |
| Molecular Function | cytoskeletal protein binding | GO:0008092 | 3.4E-02 | |
| Molecular Function | potassium channel activity | GO:0005267 | 3.6E-02 | |
| Biological Process | potassium ion transport | GO:0006813 | 3.9E-02 | |
| Biological Process | actin cytoskeleton organization | GO:0030036 | 6.8E-03 | |
| Molecular Function | G-protein coupled receptor activity | GO:0004930 | 1.2E-02 | |
| Molecular Function | endopeptidase inhibitor activity | GO:0004866 | 1.9E-02 | |
| Molecular Function | motor activity | GO:0003774 | 4.8E-02 | |
|
| ||||
| Forest vs savanna (FOR:SAV) | Molecular Function | endopeptidase activity | GO:0004175 | 4.9E-02 |
| Molecular Function | GTPase activity | GO:0003924 | 4.4E-02 | |
| Cellular Component | voltage-gated potassium channel complex | GO:0008076 | 7.2E-03 | |
| Molecular Function | voltage-gated potassium channel activity | GO:0005249 | 1.7E-02 | |
| Molecular Function | pyridoxal phosphate binding | GO:0030170 | 2.8E-02 | |
West African forest and savanna isolates were differentiated by variations in the Ov genome, largely in line with the two-strain hypothesis. As one exception, F15 (which had a forest type O-150 sequence) clusters with savanna isolates, contains 82% savanna-derived ancestry (Fig. 1b), and does not show significant gene flow from forest populations (measured by Patterson’s D statistic) (Fig. 1c and Supplementary Table 6). Despite the substantial substructuring observed, we found evidence for gene flow from savanna to forest populations. Of the three forest-clustering isolates that showed evidence of mixed ancestry (F7, F11, and F14; Fig. 1b), statistically significant gene flow from savanna populations was detected in F11 and F14 (Fig. 1c and Supplementary Table 6). This pattern is consistent with the observation that savanna flies have been reported to migrate longer distances compared to the forest species34, and deforestation has led to extension of the savanna species into formerly forested areas35. The genetic admixture observed in F7, on the other hand, could be a reflection of a sustained level of low genetic isolation due to the ability of the local vector species, Simulium soubrense B, to efficiently support the development of parasites of both the forest and the savanna origin36. Given the evidence for genetic admixture and inter-population gene flow, our data suggest that a simple dichotomy between forest and savanna parasites is an inadequate description of the parasite variations in West Africa. In addition, there was a lack of correlation between genetic distance and geographic distance among the non-admixed samples within each population (Supplementary Fig. 6), suggesting an overall high level of intra-population gene flow, in part, as a result of host migration.
In the FOR:SAV comparison of FST analysis (in which F15 is grouped with savanna samples), 194 and 28 outlier windows were identified from the autosomal and X chromosomes, respectively (Fig. 2d, and Supplementary Table 4); these contained 300 genes (Supplementary Table 5) that were enriched for five different GO terms, including endopeptidase activity, GTPase activity and two terms related to voltage-gated potassium channels (Table 2). These 300 genes showed significantly higher transcript abundance than other genes in the microfilarial stage (the infective stage for the vector), but not in any other stages (Supplementary Fig. 7).
A prior study based on the rDNA ITS2 reported a lack of significant genetic differentiation between the West African forest and savanna isolates suggesting a substantial gene flow across the geographical distribution23. In our study, despite evidence of savanna to forest gene flow in some isolates, the four populations were readily differentiated by variations in the nuclear genome, supporting the notion of a geographically and bioclimatically structured population.
Geographically distinct parasite populations are transmitted by different species within the Simulium genus. For example, geographic isolation has resulted in clear biochemical differences and an inability of American parasites to develop in African blackflies37. O. volvulus is transmitted by a number of species of the S. damnosum s.l. complex throughout Africa, while members of the S. neavei complex are also prevalent vectors in Central and East Africa. In the Americas, S. ochraceum s.l. and S. exiguum s.l. complexes transmit onchocerciasis. These species have varying vectorial capacity, and cross-infection experiments demonstrated strong adaptation of parasites to sympatric vectors, giving rise to the concept of Onchocerca-Simulium complexes38. In line with previous hypotheses proposed by various authors, it seems reasonable to postulate that divergent selection in response to different vector species is one of the key driving forces of local adaption in O. volvulus, and this may play an important role in generating population structure. For example, GPCR pathway-related proteins, which play a role in nematode chemosensation39, were significantly enriched (Table 2; P = 0.001) in regions of high genomic divergence between the Ecuadorian and West African isolates. Chemosensation may be important in facilitating migration of microfilariae to the bite site, a process that depends on the worm’s detection of and attraction towards components of the fly’s saliva40. Crossover experiments on American and West African parasite-vector complexes demonstrated that microfilarial chemotropism towards simuliids is a locally adaptive trait that occurs only in sympatric parasite-vector combinations37. In addition, the over-expression in the microfilarial stage for genes with higher divergence between the forest and savanna populations suggests selective pressures related to the parasite infection of the vector.
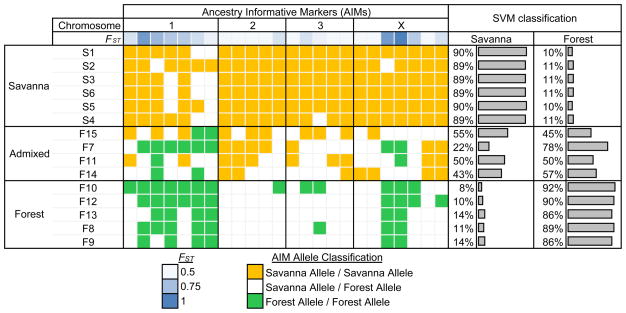
We leveraged our catalog of variants to obtain a set of Ancestry Informative Markers (AIMs) that may be used to assign individual isolates more reliably to the forest and savanna populations, based on selected criteria (high FST values, being separated from each other by a distance of at least 3MB, and belonging to the four chromosomes; See online methods, Supplementary Fig. 8a to 8e), compared to the O-150 marker. As expected, FST values for FOR:SAV partition were significantly higher than for sets with randomly assigned samples, especially for loci with highest FST values (Supplementary Fig. 8f and 8g). This supports the use of FST as an informative metric for FOR:SAV discrimination. An SVM based classification approach41 using the selected 24 markers shows the admixed nature of F11, F14 and F15 samples (forest classification probability of 50%, 57% and 45% respectively; Fig. 3, Supplementary Table 7). F7 had 78% probability of classification as forest, which is much higher than other admixed samples, but is less compared to non-admixed samples (minimum and median 86% and 89% respectively, for O-150 classification). A jackknife approach (i.e., training on all non-admixed samples except 1, then testing the classification of remaining samples) was also used to guard against potential overfitting of our data. All the non-admixed samples were again correctly classified, even when they were not part of the training set. A complete set of loci with high FST (> 0.5) is provided in Supplementary Table 8, to guide the selection of an alternative set of markers. In general, a larger set of markers is likely to increase the accuracy of classification, although improvement in the generalizability of AIMs may require ascertainment of markers based on additional sampling of West African isolates.
Figure 3.
Ancestry-informative markers (AIMs) that exhibit substantially different allele frequencies between the West African forest and savanna populations were used to assign individual isolates to source populations and identify genetic admixture. These 24 markers were selected based on informativeness (i.e., high FST values) and genome-wide distribution (Supplementary Table 7). Prediction of the individuals’ population membership based on Support Vector Machine (SVM) shows that the AIMs provide the discriminatory power to assign individuals to the correct origin and identify admixture.
Population structure and differentiation inferred from maternally inherited genomes
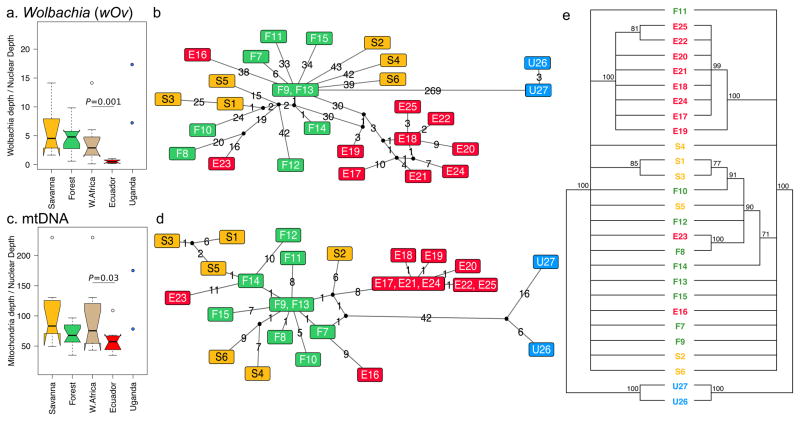
Mapping reads derived from whole-worm genomic DNA to the three reference genomes resulted in high depth of coverage of the mtDNA and wOv genomes compared to the nuclear genome (Table 1). The copy-number ratios of mtDNA or wOv to nuclear DNA were variable from sample to sample (which, for wOv, may have phenotypic effects, for example in worm fecundity42) but both were significantly lower in Ecuador compared to African samples (Fig. 4a and 4c; Supplementary Table 1). This may suggest a potential mechanistic link between the two cytoplasmic genomes. However, because Wolbachia level varies considerably during filarial life-cycle and adult maturation43, we are unable to exclude the age of the worms as being a possible confounding factor in our analysis.
Figure 4.
Haplotype characterization and relative abundance of Wolbachia and mitochondria in O. volvulus. (a) The relative abundance of Wolbachia genomes across isolate groups was calculated as the depth of read coverage across the bacterial genome divided by the depth of coverage of the worm nuclear genome. Relative Wolbachia abundance was significantly lower in Ecuador compared to West Africa. Statistical comparisons of depth ratios were performed using a two-tailed T-test with unequal variance, using the log values of the depth ratios to achieve a parametric distribution (normal distribution confirmed using a Shapiro Wilk test, W = 0.953) (b) Median-joining network of the most abundant (representative) Wolbachia haplotype in each sample. Numbers on edges indicate the number of mutations separating two representative haplotypes, and black nodes represent ancestor sequences inferred by the median-joining algorithm. (c) The relative abundance of mtDNA genomes. Statistical test same as in panel (a) (Shapiro Wilk W for mtDNA data = 0.959). (d) Median-joining network of the most abundant (representative) mtDNA haplotype in each sample. (e) Tanglegram showing concordance of the mitochondrial (left) and wOv (right) maximum-likelihood phylogenetic trees. All nodes with a bootstrap support below 70% were collapsed to polytomy.
Multiple wOv and mtDNA haplotypes were identified from every isolate (median 5 for both) (Supplementary Fig. 9a and 9b). In all sample pairs, between-sample diversity was far greater than within-sample diversity (AMOVA, P = 0.001 based on a 1000 replicate permutation), justifying the use of a single representative haplotype for each sample in inter-sample comparison (Supplementary Fig. 9c and 9d).
Median-joining networks based on the representative mtDNA and wOv haplotypes (Fig. 4b and 4d) indicate that, as with the Ov genome, the Ugandan and Ecuadorian samples mostly cluster away from the West African isolates and form distinct clades. However, discordant patterns between mtDNA and Ov were also evident in samples E16 and E23, which group with other Ecuadorian samples based on nuclear variants but separate from other Ecuadorian samples based on mtDNA and wOv variants. The wOv and mtDNA haplotype networks have a star-like topology, branching out primarily from isolates F9 and F13 (which have identical representative haplotypes) with a degree of connection of 11 and 9 for both of these samples in mtDNA and wOv (respectively). This indicates that, isolates F9 and F13 likely represent ancestral haplotypes that gave rise to multiple descendant haplotypes. The peripheral location of the savanna isolates in the network suggests that the savanna population was derived from the forest population. Interestingly, Ecuadorian haplotypes were invariably connected to the forest haplotypes, suggesting that the founding population in Ecuador was derived from the West African forest population. A previous analysis based on the O-150 marker indicated that isolates from Guatemala and Brazil are closely related to the West African savanna isolates44. This likely suggests an overall heterogeneous genetic ancestry among the American parasites, reflecting the regional variations in the origins of the founding migrants. The representative haplotypes of the maternally inherited genomes were also used to systematically compare the mtDNA and wOv phylogenies and they were found to be significantly congruent (P = 5 × 10−4; maximum likelihood phylogenies shown in Fig. 4e). This result, also supported by tests based directly on inter-sample haplotype distances (P = 1 × 10−4), is consistent with (1) strict maternal co-inheritance of the two genomes and (2) minimal (if any) inter-genome recombination.
Interestingly, no discernible geographic pattern was detected in the distribution of West African mtDNA clades, in contrast to the bioclimatic separation observed in the Ov genomes (Fig. 1b). Discordance in the nuclear versus wOv and mtDNA phylogenies could be due to incomplete lineage sorting of mtDNA and the maintenance of ancestral polymorphisms in maternally inherited genomes that are not subject to recombination. If the effective population size of the nuclear genome is small (relative to that of the mitochondria), the process of stochastic lineage-sorting, where ancient polymorphisms are lost over time, progresses more rapidly for the nuclear DNA45, which is not unlikely in a strongly polygamous species, such as O. volvulus, where some males have much higher reproductive success than others, resulting in a small diploid autosomal effective size46. The observed lower FOR:SAV population differentiation in the mtDNA and wOv genomic sequences possibly explains results of the previous PCR-RFLP analysis of the mtDNA that indicated an absence of population substructuring in West Africa24. Overall our analyses help resolve the seemingly conflicting models of population structure put forward in previous studies.
A number of hypotheses have been proposed to explain the higher prevalence of onchocercal blindness in the savanna as compared to the forest areas in West Africa (e.g., differences in transmission patterns, vector biting habits, host response and immunity, environmental factors, etc.)37. Population structure in O. volvulus that mirrors the epidemiological patterns of blindness has been suggested as evidence supporting the hypothesis that intrinsic differences in the pathogenicity of parasites determine the observed clinical variation20. However, not all parasitological and entomological data conform neatly to this simple two-strain model, suggesting a greater degree of parasite heterogeneity in West Africa and/or multifactorial causes for pathology47,48. Our analysis of the forest and savanna isolates do not show any significant difference in Wolbachia levels (a suggested factor19), or an easily identifiable molecular/functional differentiator directly related to pathogenesis. Determining the genetic variants associated with severe blindness in O. volvulus will be challenging given the phenotypic heterogeneity (i.e., not all savanna infections lead to blindness) and the strong population stratification that would result in many false positive associations.
Discussion
As an initial step towards a comprehensive population genomic survey of O. volvulus, we used 27 historical isolates collected from West Africa, Ecuador, and Uganda to assess (1) baseline genetic variation present in populations with little or no exposure to the anthelmintic ivermectin and (2) variation among nearly extinct strains with documented phenotypic differences. Variations in the genomic sequences reveal biogeographical structuring in O. volvulus and candidate loci underlying local adaptation in line with the hypothesis that divergent selection in response to different vector species plays an important role in population differentiation in O. volvulus.
Our data indicate that an isolate may be derived, in large part, from savanna ancestry while maintaining a forest-type O-150 sequence, highlighting the limitations of single-locus studies and a necessity for using multi-locus genotype data to reconstruct robust phylogenetic relationships and reliably infer the ancestry of individual isolates. We have identified and demonstrated the usefulness of a set of AIMs to more accurately classify the origin of O. volvulus isolates (including samples with mixed ancestry). These may be further optimized in the future (using the databases generated) to provide the desired level of discriminatory power to suit specific application needs (e.g, epidemiological monitoring of parasite dispersal).
Examination of additional isolates from under-sampled geographical regions (including Central and East Africa) and areas covered by MDA programs will be required for a more complete and robust understanding of the phylogeography of O. volvulus and the dynamics of population restructuring in the face of on-going control efforts. Nevertheless, the catalog of all variants detected in this study provides a solid foundation for future basic and translational onchocerciasis research.
Methods
Parasite material, DNA isolation and sequencing
De-identified O. volvulus specimens were collected from forest and savanna regions of West Africa, Uganda and Ecuador by nodulectomy and DNA was isolated from worms or nodules during the course of studies that took place in the early 1990’s (Supplementary Table 1 and Fig. 1); approval of sample collections and study designs by relevant institutions were described previously20,21,25,26. West African samples were genotyped by Southern blot using forest (pFS-1) and savanna specific (pSS-1BT) probes20,21.
The yield and integrity of genomic DNA was verified by a PicoGreen assay (Invitrogen) and an 0.8% Agarose gel, respectively. For each whole genome shotgun library, 100–250ng of genomic DNA was fragmented to the desired size in 10mM Tris-HCl, pH 8.5 using the Covaris LE220. Library preparation methods (either KAPA Low Throughput Library Preparation Kit [Kapa Biosystems, Woburn, MA] or Lucigen) and the specific Illumina sequencing technologies employed for each sample are described in Supplementary Table 9. Small insert Illumina libraries were mostly prepared utilizing the KAPA Low Throughput Library Preparation Kit (Kapa Biosystems, Woburn, MA) according to the manufacturer’s recommendations with a few exceptions: Each sample was ligated with 5.0μL of a 1–2.5μM stock of Dual Indexed Adapters (index sequences are the same on both ends of the adaptors) for a final adaptor concentration of 100–250nM. Libraries were cycle-optimized to prevent over-amplification. One 50μL PCR reaction included 2μL of ligated DNA (~1ng), KAPA HotStart PCR Master Mix, and a final concentration of 500nM from each forward reverse primer pair. After cycles 8, 10, 12, and 14 the program was halted and a 5μL aliquot collected. Each cycle amplification product was evaluated on a 2.2% agarose Flash Gel (Lonza, Switzerland) and the proper cycle number determined. Eight PCR reactions were amplified at the determined cycle number to enrich for proper adaptor ligated fragments. Each library was fractionated on the LabChip XT using the DNA 750 chip (Perkin Elmer, Hopkinton, MA) or Blue Pippin (Sage Science, Inc, Beverly, MA) collecting one of three unique fractions when possible: 375bp, 475bp, and 675bp following the manufacturer’s recommendations. The final concentration of each library was verified through qPCR utilizing the KAPA Library Quantification Kit - Illumina/LightCycler® 480 kit (Kapa Biosystems) according to the manufacturer’s protocol in order to produce cluster counts appropriate for the Illumina sequencing platform according to the manufacturer’s protocol (Illumina, SanDiego, CA).
Genomic read processing and SNP analysis
Reads were trimmed to remove relevant barcodes and adapters and filtered to eliminate low-confidence sequences (reads shorter than 60bp or containing ambiguous base calls) using Flexbar49, and mapped to a combined reference database containing the human (hs38), Ov (WormBase WS245), mtDNA (WormBase WS245) and wOv (RefSeq NZ_HG810405.1) genomes using BWA-MEM (version bwa0.7.5a, default parameters50). The resulting alignments were split into three separate files corresponding to the O. volvulus nuclear genome, mtDNA, and the genome of wOv.
For each individual alignment file, duplicate reads were marked for removal using Picard tools (version 1.92, http://broadinstitute.github.io/picard/). Reads were realigned in the region of indels using the Genome Analysis Toolkit (GATK, v.3.3.051), and variants were determined using the HaplotypeCaller tool of GATK package. Using 32 O. volvulus isolates (Supplementary Table 1), a multi-sample variant calling was performed, which borrows information between samples to detect variants with greater sensitivity and accuracy. High confidence SNPs were identified using GATK’s VariantFiltration with the following parameters: DP (maximum depth) > median depth+(median absolute deviation×1.4826)×2; QD (variant confidence divided by the unfiltered depth of non-reference samples) < 2.0; FS (Phred-scaled p-value using non-parametric Fisher’s Exact Test to detect strand bias in the reads) > 60.0; MQ (Root Mean Square of the mapping quality of the reads across all samples) < 40.0; MQRankSum (non-parametric Mann-Whitney Rank Sum Test for mapping qualities) < −12.5; ReadPosRankSum (non-parametric Mann-Whitney Rank Sum Test for the distance from the end of the read for reads with the alternate allele) < −8.0. Indels were similarly filtered using the following criteria: DP (maximum depth) > median depth+(median absolute deviation×1.4826)×2; QD < 2.0; FS > 200.0; ReadPosRankSum < −20.0. High confidence variants were classified according to their context in genomic features using SnpEff (v 3.552). GATK’s CallableLoci tool was used under default settings to determine the areas of the genome that are callable (based on coverage and mapping quality), and the results were expressed as proportions relative to the total ungapped genome length.
Genetic relationships among isolates were examined by building a neighbor-joining tree53 based on pairwise identity-by-state (IBS) distance computed using autosomal nuclear SNPs in PLINK (v1.90b2n)54. Wright’s F-statistics, Nei’s nucleotide diversity (π), and Tajima’s D55 were estimated using VCFtools (v0.1.12b)56. To estimate nucleotide diversity separately for the nonsynonymous and synonymous sites (πN and πS, Supplementary Fig. 4), nonsynonymous or synonymous average pairwise differences were divided by the number of nonsynonymous or synonymous sites, respectively. The number of nonsynonymous or synonymous sites was determined using KaKs_Calculator 2.057. The statistical significance of the increase in πN/πS ratios among genes specific to Onchocercidae27 (i.e., O. volvulus orthologs present in Wuchereria bancrofti, Brugia malayi, Dirofilaria immitis, Loa loa or Onchocerca ochengi, but absent from Caenorhabditis elegans, Trichuris muris and Ascaris suum) relative to the rest of the coding genome was assessed using the non-parametric Wilcoxon Rank Sum test after log-transformation. The genetic ancestry of potentially admixed individuals was inferred based on autosomal nuclear SNPs using ADMIXTURE (v1.23)47. To distinguish gene flow from incomplete lineage sorting, four-taxon ABBA-BABA test of introgression58 was performed using ANGSD (v0.901)59. The statistical significance of differences in population allele frequencies was determined using non-parametric Fisher’s exact test, and highly differentiated SNPs (requiring both a ≤ 0.01 P value and a minimum frequency difference of 40%) were identified.
An overview of all SNP classifications per genome is available in Table 1, nonsynonymous SNPs and their detailed annotation and allele frequencies are available in Supplementary Table 2, and SNP counts summarized per gene are available in Supplementary Table 5 (along with detailed functional annotation of every gene).
Genomic FST sliding window analysis
Using the GenWin (Spline Based Window Boundaries for Genomic Analyses) package in R60, average FST and Wstat values were calculated for dynamic sliding windows across the four chromosomes (94.3% of the total assembly) to compare Ecuador versus West Africa and forest versus savanna populations (Fig. 2a and Table 2). Analyzing the putative X chromosome independently from the other chromosomes, the top 1% of identified windows with highest Wstat value were classified as FST outlier windows, representing genomic regions of high genetic differentiation. FUNC61 (which considers the hierarchical structure of GO) was used to determine significant functional enrichment among gene sets identified within the FST outlier windows, with a P ≤ 0.05 significance threshold (after FDR population correction), and only considering GO terms represented by at least 5 genes in the background set. For each comparison, only genes overlapping the windows being tested were used as a background set. Average expression levels (log RPKM) for genes within FST outlier regions, and other genes surveyed in the genome scan, per stage and per FST comparison were calculated based on existing transcriptomic datasets (Supplementary Fig. 6). Significance values were tested using a two-tailed T-test with unequal variance, since this logged genome-wide expression data is parametric (normally distributed; 0.966, Shaprio Wilk test). In accordance with the genome publication27, figures present the six contigs merged into four (OM1a and OM1b are combined in order and OM5 is appended to the end of OM2), but statistical analysis utilized the six contigs defined by the WS245 genome release.
Identifying a set of Ancestry Informative Markers (AIMs) for forest and savanna strains
The variant loci were sorted according to FST values and then top N loci (excluding those on the small contigs that could not be placed on a chromosome) were selected as markers such that no locus is within distance D of an already selected marker. Different values of N and D were explored (Supplementary Fig. 8a–e) and a final set of 24 AIMs was arrived at by keeping only the loci on the 4 chromosomes with D=3Mb (Fig. 3). The number 24 was chosen to facilitate convenient multiplexing on commercial arrays, but a complete set of loci ranked by FST is provided in Supplementary Table 8. The 11 samples that were deemed not to be admixed by ADMIXTURE were used to train an SVM (libsvm v. 3.2141) using the individual sample genotypes at the AIM loci as inputs and the sample strains as output. This model was then used to classify all 15 samples. SVM classification accuracy was also ascertained by using a jackknife approach for non-admixed samples. For this, each of the 11 non-admixed samples was classified after training the SVM using the other 10 non-admixed samples.
Estimating the abundance of Wolbachia
The average depth of read coverage across each sample was calculated for the O. volvulus nuclear genome, mtDNA, and wOv (Table 1). Relative depths for wOv and mtDNA were calculated by dividing by the nuclear genomic depth. P values on box plot figures (Fig. 4a and 4c) were calculated according to a T-test, after confirming a normal distribution for the data using a Shapiro Wilk test for normality (W ≥ 0.95).
Filtering of data for mtDNA and wOv haplotype generation
To generate a set of likely mtDNA and wOv haplotype sequences, a high confidence set of aligned reads was first obtained for each sample by including only reads that are mapped in proper pairs with mapping quality ≥ 60. Since mtDNA had a much higher depth as compared to wOv (median depth 3135 vs 84), we used a downsampled set of mapped reads to generate a comparable set of haplotypes (downsampled to a depth of ~100X). This was also done for 4 wOv samples with particularly high sequencing depth (F9, F15, U27, U27, depth > 500). Manual inspection of sequence alignments revealed consistent coverage of the mtDNA but discrete regions of unusually high read pile-up in the wOv genome; this was most striking in Ecuadorian worms with low wOv to nuclear genome ratios. We hypothesized that it could be due to aberrant mapping of reads related to nuclear Wolbachia transfers (nuwts)62. To ensure highest confidence, these regions were filtered prior to analysis (Supplementary Fig. 10a and 10b). They were identified based on high depth of mapping (median + 4.5 median absolute deviation) or high SNP rate (median + 4.5 median absolute deviation) in any of the samples. Regions thus identified were joined if separated by < 500 bases. This resulted in exclusion of 41,899 bases (4.4% of the wOv genome) from downstream analyses.
Within- and between-sample mtDNA and wOv diversity comparison
The haplotypes were constructed using EVORhA63, a frequency based haplotype reconstruction method, with 4 wOv samples excluded because of low depth of coverage (F7, E17, E21 and E24; average depth <30). For quality filtering, any reads with >1 base with quality <23 were discarded, as previously described64. The inter-haplotype distances were calculated using the K80 model65; within-sample and across-sample K80 distance comparisons were done using the non-parametric Kolmogorov-Smirnoff (KS) test66. The AMOVA phi statistic was used as a more robust method to compare intra- and inter-sample haplotype diversity as it implicitly considers both the sequence distance and the frequency of haplotypes67. The input alignment for AMOVA was generated using MAFFT68. The R package “ade4” was used to run AMOVA and a permutation test with 999 repetitions for all sample pairs to compare within-sample and between-sample pair diversity.
Analysis of representative haplotypes for mtDNA and wOv
A single representative haplotype for each sample was generated using the GATK based SNPs. The genotype at every SNP locus, if called homozygous by HaplotypeCaller, was accepted. The heterozygous calls were resolved using the read counts supporting the alleles. Median-joining phylogenetic networks69 were generated for the mitochondrial and Wolbachia haplotypes using PopART (http://popart.otago.ac.nz) and visualized using Cytoscape70. Maximum Likelihood genealogies were generated using RAxML version 8.2.371, using the two Uganda samples as outgroups. A general time reversible (GTR) model of nucleotide substitution with Γ rate heterogeneity was used, and all model parameters were estimated by RAxML. A full ML search was done for the best-scoring tree and a rapid bootstrap analysis72 with 1000 alternative runs estimated the branch support values. Congruence between ML trees for mitochondrial and Wolbachia sequences was assessed using ParaFit73, a method to evaluate coevolution between parasites and hosts that has previously been used for ascertaining phylogenetic tree congruence74. The ParaFit comparison was done both a) using the patristic distances corresponding to the ML trees and b) directly using the K80 distances without using the ML tree reported by RAxML.
Supplementary Material
Supplementary Figure 1 (a) The average depth of O. volvulus nuclear genome and Wolbachia genome per sample. Red lines indicate the 6X threshold required to include samples in downstream analysis. (b) The genetic distance to the reference sequence vs. the average depth of coverage of the nuclear genome for each sample. To minimize bias stemming from a systematic variation in false negative variant call rates due to undersampling, low-depth samples were excluded from downstream analysis.
Supplementary Figure 2 (a) Median depth of coverage per 50kb region across the four chromosomes. Adult female worms were used in this study. Depth of coverage for the X chromosome (excluding pseudoautosomal region) is expected to be lower than the autosomal chromosomes because male embryonic cells are hemizygous for the X chromosome. (b) The ratio of average X chromosomal depth (excluding the pseudoautosomal region) to the average depth across all autosomal chromosomes, plotted against the average depth of the autosomal chromosomes.
Supplementary Figure 3 Onchocercidae-specific genes27 have higher piN/piS values as compared to other genes in the genome. Overall, most of the values are <1 (i.e. log values on x-axis are <0) in both cases, suggesting purifying selection, but this seems relaxed for the Onchocercidae-specific genes (more negative values on the x-axis indicate higher purifying selection).
Supplementary Figure 4 Maximum Likelihood phylogeny based on mitochondrial sequences from O. volvulus isolates, along with 3 O. ochengi isolates. The inter-sample distances for O. volvulus are much smaller than their distance with O. ochengi isolates.
Supplementary Figure 5 Genome-wide mean Tajima’s D and nonsynonymous, synonymous and non-coding sequence diversity (π) across the chromosomes in various sample sets.
Supplementary Figure 6 Genome-wide identity-by-state (IBS) estimates vs. geographic distance for all pairs of non-admixed West African samples. The lack of positive correlation between genetic and geographic distance indicates that in West Africa the spatial patterns of genetic diversity are not structured by geographic distance (isolation by distance) and there is an overall high level of intra-population gene flow. Geographic distance was calculated using a locally generated perl script, which calculated distance given the latitude and longitude coordinates of the two points (considering the curvature of the earth).
Supplementary Figure 7 Average transcript abundance levels for genes within FST outlier regions, and other genes surveyed in the sliding window analysis, per life-cycle stage and per FST comparison. Values represent average log-RPKM normalized to the average log-RPKM for all genes in the sample. Error bars represent standard error of the mean. Significance values were tested using a two-tailed T-test with unequal variance (normal distribution confirmed according to Shapiro-Wilk test, P = 0.956).
Supplementary Figure 8 (a–e) Classification of samples as forest and savanna using different sets of AIMs from the 4 chromosomes. The min, median, mean and max probabilities (in red and green colors) represent the classification probabilities for the 11 non-admixed samples. The 4 admixed samples are shown separately in blue. Distance (X-axis) is the minimum allowable distance between any two markers. Note that the size of the chromosomes puts a constraint on how large the distance threshold can get for a given number of markers. In general, the non-admixed samples are classified correctly with high probability and F7 can be distinguished as an admixed sample using only relatively high number of markers. The classification SVM was trained using non-admixed samples, before testing all the samples using the trained model. (f) Cumulative distribution of FST values shows that forest:savanna FST values tend to be higher than randomly chosen partition of samples, thereby supporting a small set of informative markers that can be used for classification. (g) Distribution of FST values among the loci with top-100 highest FST values. Black is forest:savanna and colored lines represent the 10 randomly chosen partitions of samples.
Supplementary Figure 9: Estimated frequencies of EVORhA based haplotypes for (a) mitochondria and (b) Wolbachia. These sum of frequencies for a sample’s haplotypes ranges from ~0.6 to >2. This is partially because these are means of the distribution of frequency estimates of the best-fitting deconvolution of the data. In some cases, EVORhA splits a single distribution into two overlapping ones resulting in two nearly identical haplotypes with slightly different frequencies (e.g., mitochondria from F11, E16, E20 and E21). (c) Mutation distances and statistical comparison of within- and between-sample distances for mitochondria (left) and Wolbachia (right). For all samples, except 1 for mitochondria and 2 for Wolbachia, the within-sample haplotype distances are significantly smaller than between-sample distances (P-values plotted in top panel, sample mutation distances in bottom panel). The red bar represents a significance level of 0.05. The blue bar represents the mean between-sample mutation distance. This relationship is more rigorously verified using AMOVA (see text).
Supplementary Figure 10. (a) Reference and Alternate allele depth per variable position, per sample, in Wolbachia, before filtering. A clean set has clusters along a line (x+y=c), e.g. S3, F8, F15, and U27. Many of the Ecuador samples show large clusters along a vertical, showing signs of aberrant read mapping potentially due to nuwts. (b) Reference and Alternate allele depth per variable position, per sample, in Wolbachia, after filtering. Most of the sample with clusters along a vertical in panel a are now cleaned up. (c) Reference and Alternate allele depth per variable position, per sample, in mitochondria. The data does not show signs of aberrant read mapping.
Classifications, sources and read mapping data for each of the samples utilized
SnpEff annotations and allele frequency data per SNP position
Summary of results from the FST sliding window analysis
FST genome scan sliding window positions, summary statistics, and overlapping genes
Detailed functional annotation and SNP annotation per gene
ABBA-BABA analysis results for gene flow
Ancestry Informative Markers for distinguishing savanna from forest O. volvulus isolates
Variants ranked by FST (FOR:SAV comparison, minimum value 0.5)
Sources, dates, and processing procedures for each of the whole genome shotgun libraries in each of the samples
Acknowledgments
The sequencing work was funded by NIH-NHGRI (U54HG003079) and the genetic variation analysis was funded by NIH-NIAID (R01AI081803) and the Bill & Melinda Gates Foundation (OPP GH 1083853). The study was also funded, in part, by the Division of Intramural Research, National Institute of Allergy and Infectious Diseases. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. The findings and conclusions contained within are those of the authors and do not necessarily reflect positions or policies of the Bill & Melinda Gates Foundation. We thank the faculty and staff of the McDonnell Genome Institute, who contributed to this study and we thank the physicians and field workers in the endemic countries for their extensive help in collecting the parasite material. The unpublished RNAseq data used in this study were produced by the parasite genomics group at the Wellcome Trust Sanger Institute in collaboration with the laboratories of Drs. Thomas B. Nutman and Sara Lustigman.
Footnotes
Authors contributions. These authors contributed equally to this work: Y.C., R.T.; Conceived and planned the project: M.M., P.U.F., G.J.W; Led the project, analysis and manuscript preparation: M.M.; Provided material: T.R.U., C.T.N., T.B.N., P.F.U. Sequence data production, annotation, and submission: K.H.P., P.O., J.M.; Genome-based variant studies: Y.C., R.T., B.A.R., S.N.M; Drafted, edited and wrote the manuscript: M.M., R.T., Y.C., S.N.M., B.A.R.
Competing Interests. The authors declare no competing financial interest.
PDF files
Supplementary Figures 1 to 10 and Supplementary Table 3 are provided as a single PDF file (file size 3.5MB).
Excel files
Supplementary Tables 1, 2, and Supplementary Tables 4 to 8 are provided as individual excel-format spreadsheets (total file size 26.3MB)
URLs. Picard tools, http://broadinstitute.github.io/picard/; PopART, http://popart.otago.ac.nz; Variants and a browseable genome is available at Nematode.net, http://nematode.net; The reference O. volvulus genome used was from Cotton et al27 and is available in Wormbase, http://parasite.wormbase.org/Onchocerca_volvulus_prjeb513/Info/Index/. The unpublished RNAseq data used in this study is available from the sequence data archives under BioProject PRJEB2965: http://www.ncbi.nlm.nih.gov/bioproject/?term=PRJEB2965.
Accession codes. The whole-genome sequence of the 32 O. volvulus isolates have been deposited in SRA under BioProject ID PRJNA289926 and accession number SRP066374, and under the individual accessions SRX1437566, SRX1437699, SRX1437754, SRX1437813, SRX1437815, SRX1437816, SRX1437859, SRX1437919, SRX1438064, SRX1438065 - SRX1438067, SRX1438077, SRX1438078, SRX1438123, SRX1438124, SRX1438126, SRX1438129, SRX1438132, SRX1438135, SRX1438157, SRX1438163, SRX1438186 - SRX1438195.
References
- 1.Dunn C, et al. The Contributions of Onchocerciasis Control and Elimination Programs toward the Achievement of the Millennium Development Goals. PLoS Negl Trop Dis. 2015;9:e0003703. doi: 10.1371/journal.pntd.0003703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Crump A, Morel CM, Omura S. The onchocerciasis chronicle: from the beginning to the end? Trends Parasitol. 2012;28:280–8. doi: 10.1016/j.pt.2012.04.005. [DOI] [PubMed] [Google Scholar]
- 3.Cupp EW, Sauerbrey M, Richards F. Elimination of human onchocerciasis: history of progress and current feasibility using ivermectin (Mectizan((R))) monotherapy. Acta Trop. 2011;120(Suppl 1):S100–8. doi: 10.1016/j.actatropica.2010.08.009. [DOI] [PubMed] [Google Scholar]
- 4.Coffeng LE, et al. African programme for onchocerciasis control 1995–2015: updated health impact estimates based on new disability weights. PLoS Negl Trop Dis. 2014;8:e2759. doi: 10.1371/journal.pntd.0002759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Progress towards eliminating onchocerciasis in the WHO Region of the Americas: verification by WHO of elimination of transmission in Colombia. Wkly Epidemiol Rec. 2013;88:381–5. [PubMed] [Google Scholar]
- 6.Elimination of onchocerciasis in the WHO Region of the Americas: Ecuador’s progress towards verification of elimination. Wkly Epidemiol Rec. 2014;89:401–5. [PubMed] [Google Scholar]
- 7.Mackenzie CD, Homeida MM, Hopkins AD, Lawrence JC. Elimination of onchocerciasis from Africa: possible? Trends Parasitol. 2012;28:16–22. doi: 10.1016/j.pt.2011.10.003. [DOI] [PubMed] [Google Scholar]
- 8.Centers for Disease, C. & Prevention. Progress toward elimination of onchocerciasis in the Americas - 1993–2012. MMWR Morb Mortal Wkly Rep. 2013;62:405–8. [PMC free article] [PubMed] [Google Scholar]
- 9.WHO. African Programme for Onchocerciasis Control: progress report 2013–2014. Weekly Epidemiological Record. 2014;89:545–560. [Google Scholar]
- 10.Osei-Atweneboana MY, Eng JK, Boakye DA, Gyapong JO, Prichard RK. Prevalence and intensity of Onchocerca volvulus infection and efficacy of ivermectin in endemic communities in Ghana: a two-phase epidemiological study. Lancet. 2007;369:2021–9. doi: 10.1016/S0140-6736(07)60942-8. [DOI] [PubMed] [Google Scholar]
- 11.Dadzie KY, Remme J, Baker RH, Rolland A, Thylefors B. Ocular onchocerciasis and intensity of infection in the community. III. West African rainforest foci of the vector Simulium sanctipauli. Trop Med Parasitol. 1990;41:376–82. [PubMed] [Google Scholar]
- 12.Remme J, Dadzie KY, Rolland A, Thylefors B. Ocular onchocerciasis and intensity of infection in the community. I. West African savanna. Trop Med Parasitol. 1989;40:340–7. [PubMed] [Google Scholar]
- 13.Botto C, Escalante A, Arango M, Yarzabal L. Morphological differences between Venezuelan and African microfilariae of Onchocerca volvulus. J Helminthol. 1988;62:345–51. doi: 10.1017/s0022149x00011755. [DOI] [PubMed] [Google Scholar]
- 14.Eichner M, Renz A. Differential length of Onchocerca volvulus infective larvae from the Cameroon rain forest and savanna. Trop Med Parasitol. 1990;41:29–32. [PubMed] [Google Scholar]
- 15.Vuong PN, et al. Forest and savanna onchocerciasis: comparative morphometric histopathology of skin lesions. Trop Med Parasitol. 1988;39:105–10. [PubMed] [Google Scholar]
- 16.Duke BO, Lewis DJ, Moore PJ. Onchocerca-Simulium complexes. I. Transmission of forest and Sudan-savanna strains of Onchocerca volvulus, from Cameroon, by Simulium damnosum from various West African bioclimatic zones. Ann Trop Med Parasitol. 1966;60:318–26. [PubMed] [Google Scholar]
- 17.Omar MS, Prost A, Marshall TF. Histochemical enzyme variation in Onchocerca volvulus microfilariae from rain-forest and Sudan-savanna areas of the Onchocerciasis Control Programme in West Africa. Bull World Health Organ. 1982;60:933–44. [PMC free article] [PubMed] [Google Scholar]
- 18.Flockhart HA, Cibulskis RE, Karam M, Albiez EJ. Onchocerca volvulus: enzyme polymorphism in relation to the differentiation of forest and savannah strains of this parasite. Trans R Soc Trop Med Hyg. 1986;80:285–92. doi: 10.1016/0035-9203(86)90037-4. [DOI] [PubMed] [Google Scholar]
- 19.Higazi TB, et al. Wolbachia endosymbiont levels in severe and mild strains of Onchocerca volvulus. Mol Biochem Parasitol. 2005;141:109–12. doi: 10.1016/j.molbiopara.2005.02.006. [DOI] [PubMed] [Google Scholar]
- 20.Zimmerman PA, et al. Onchocerca volvulus DNA probe classification correlates with epidemiologic patterns of blindness. J Infect Dis. 1992;165:964–8. doi: 10.1093/infdis/165.5.964. [DOI] [PubMed] [Google Scholar]
- 21.Fischer P, Bamuhiiga J, Kilian AH, Buttner DW. Strain differentiation of Onchocerca volvulus from Uganda using DNA probes. Parasitology. 1996;112(Pt 4):401–408. doi: 10.1017/s0031182000066634. [DOI] [PubMed] [Google Scholar]
- 22.Higazi TB, et al. Onchocerca volvulus: genetic diversity of parasite isolates from Sudan. Exp Parasitol. 2001;97:24–34. doi: 10.1006/expr.2000.4589. [DOI] [PubMed] [Google Scholar]
- 23.Morales-Hojas R, Cheke RA, Post RJ. A preliminary analysis of the population genetics and molecular phylogenetics of Onchocerca volvulus (Nematoda: Filarioidea) using nuclear ribosomal second internal transcribed spacer sequences. Mem Inst Oswaldo Cruz. 2007;102:879–82. doi: 10.1590/s0074-02762007005000114. [DOI] [PubMed] [Google Scholar]
- 24.Keddie EM, et al. Onchocerca volvulus: limited heterogeneity in the nuclear and mitochondrial genomes. Exp Parasitol. 1999;93:198–206. doi: 10.1006/expr.1999.4450. [DOI] [PubMed] [Google Scholar]
- 25.Gallin M, et al. Epidemiological studies of onchocerciasis in southern Benin. Trop Med Parasitol. 1993;44:69–74. [PubMed] [Google Scholar]
- 26.Nutman TB, Parredes W, Kubofcik J, Guderian RH. Polymerase chain reaction-based assessment after macrofilaricidal therapy in Onchocerca volvulus infection. J Infect Dis. 1996;173:773–6. doi: 10.1093/infdis/173.3.773. [DOI] [PubMed] [Google Scholar]
- 27.Cotton JA, et al. The genome of Onchocerca volvulus, the agent of River Blindness. Nat Microbiology. 2016 doi: 10.1038/nmicrobiol.2016.216. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Post R. The chromosomes of the Filariae. Filaria J. 2005;4:10. doi: 10.1186/1475-2883-4-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Andersen EC, et al. Chromosome-scale selective sweeps shape Caenorhabditis elegans genomic diversity. Nat Genet. 2012;44:285–90. doi: 10.1038/ng.1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cutter AD. Integrating phylogenetics, phylogeography and population genetics through genomes and evolutionary theory. Mol Phylogenet Evol. 2013;69:1172–85. doi: 10.1016/j.ympev.2013.06.006. [DOI] [PubMed] [Google Scholar]
- 31.Johnson NA, Lachance J. The genetics of sex chromosomes: evolution and implications for hybrid incompatibility. Ann N Y Acad Sci. 2012;1256:E1–22. doi: 10.1111/j.1749-6632.2012.06748.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zimmerman PA, et al. Migration of a novel DQA1* allele (DQA1*0502) from African origin to North and South America. Hum Immunol. 1995;42:233–40. doi: 10.1016/0198-8859(94)00107-2. [DOI] [PubMed] [Google Scholar]
- 33.Pezzi PJP, Trebeschi P. Aporte Hacia la Consolidacion de la Identidad Cultural del Negro Esmeraldeno. Es el Negro Esmeraldeno un Afroamericano? In: Pezzi PJP, Nunez GC, Minda P, editors. Coleccion Antropolgia Aplicada no 10. U.P.S; Quito, Ediciones: 1996. pp. 12–98. [Google Scholar]
- 34.Boakye DA, Back C, Fiasorgbor GK, Sib AP, Coulibaly Y. Sibling species distributions of the Simulium damnosum complex in the west African Onchocerciasis Control Programme area during the decade 1984–93, following intensive larviciding since 1974. Med Vet Entomol. 1998;12:345–58. doi: 10.1046/j.1365-2915.1998.00118.x. [DOI] [PubMed] [Google Scholar]
- 35.Wilson MD, et al. Deforestation and the spatio-temporal distribution of savannah and forest members of the Simulium damnosum complex in southern Ghana and south- western Togo. Trans R Soc Trop Med Hyg. 2002;96:632–9. doi: 10.1016/s0035-9203(02)90335-4. [DOI] [PubMed] [Google Scholar]
- 36.Baker RH, et al. Progress in controlling the reinvasion of windborne vectors into the western area of the Onchocerciasis Control Programme in West Africa. Philos rans R Soc Lond B Biol Sci. 1990;328:731–47. doi: 10.1098/rstb.1990.0141. discussion 747–50. [DOI] [PubMed] [Google Scholar]
- 37.Duke BO. Geographical aspects of onchocerciasis. Ann Soc Belg Med Trop. 1981;61:179–86. [PubMed] [Google Scholar]
- 38.Basanez MG, Churcher TS, Grillet ME. Onchocerca-Simulium interactions and the population and evolutionary biology of Onchocerca volvulus. Adv Parasitol. 2009;68:263–313. doi: 10.1016/S0065-308X(08)00611-8. [DOI] [PubMed] [Google Scholar]
- 39.Thomas JH, Robertson HM. The Caenorhabditis chemoreceptor gene families. BMC Biology. 2008;6:1–17. doi: 10.1186/1741-7007-6-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stallings T, Cupp MS, Cupp EW. Orientation of Onchocerca lienalis stiles (Filarioidea: Onchocercidae) microfilariae to black fly saliva. J Med Entomol. 2002;39:908–14. doi: 10.1603/0022-2585-39.6.908. [DOI] [PubMed] [Google Scholar]
- 41.Chang CC, Lin CJ. LIBSVM: a library for support vector machines. ACM Transactions on Intelligent Systems and Technology (TIST) 2011;2:27. [Google Scholar]
- 42.Taylor MJ, Voronin D, Johnston KL, Ford L. Wolbachia filarial interactions. Cell Microbiol. 2013;15:520–6. doi: 10.1111/cmi.12084. [DOI] [PubMed] [Google Scholar]
- 43.McGarry HF, Egerton GL, Taylor MJ. Population dynamics of Wolbachia bacterial endosymbionts in Brugia malayi. Mol Biochem Parasitol. 2004;135:57–67. doi: 10.1016/j.molbiopara.2004.01.006. [DOI] [PubMed] [Google Scholar]
- 44.Zimmerman PA, Katholi CR, Wooten MC, Lang-Unnasch N, Unnasch TR. Recent evolutionary history of American Onchocerca volvulus, based on analysis of a tandemly repeated DNA sequence family. Mol Biol Evol. 1994;11:384–92. doi: 10.1093/oxfordjournals.molbev.a040114. [DOI] [PubMed] [Google Scholar]
- 45.Toews DP, Brelsford A. The biogeography of mitochondrial and nuclear discordance in animals. Mol Ecol. 2012;21:3907–30. doi: 10.1111/j.1365-294X.2012.05664.x. [DOI] [PubMed] [Google Scholar]
- 46.Ballard JW, Whitlock MC. The incomplete natural history of mitochondria. Mol Ecol. 2004;13:729–44. doi: 10.1046/j.1365-294x.2003.02063.x. [DOI] [PubMed] [Google Scholar]
- 47.Alexander DH, Novembre J, Lange K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009;19:1655–64. doi: 10.1101/gr.094052.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cheke RA, Garms R. Indices of onchocerciasis transmission by different members of the Simulium damnosum complex conflict with the paradigm of forest and savanna parasite strains. Acta Trop. 2013;125:43–52. doi: 10.1016/j.actatropica.2012.09.002. [DOI] [PubMed] [Google Scholar]
- 49.Dodt M, Roehr JT, Ahmed R, Dieterich C. Flexbar flexible barcode and adapter processing for next-generation sequencing platforms. MDPI Biology. 2012;1:895–905. doi: 10.3390/biology1030895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–60. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.McKenna A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cingolani P, et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 2012;6:80–92. doi: 10.4161/fly.19695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–25. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 54.Purcell S, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123:585–95. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Danecek P, et al. The variant call format and VCFtools. Bioinformatics. 2011;27:2156–8. doi: 10.1093/bioinformatics/btr330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang D, Zhang Y, Zhang Z, Zhu J, Yu J. KaKs_Calculator 2.0: a toolkit incorporating gamma-series methods and sliding window strategies. Genomics Proteomics Bioinformatics. 2010;8:77–80. doi: 10.1016/S1672-0229(10)60008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Patterson N, et al. Ancient admixture in human history. Genetics. 2012;192:1065–93. doi: 10.1534/genetics.112.145037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Korneliussen TS, Albrechtsen A, Nielsen R. ANGSD: Analysis of Next Generation Sequencing Data. BMC Bioinformatics. 2014;15:356. doi: 10.1186/s12859-014-0356-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Beissinger TM, Rosa GJ, Kaeppler SM, Gianola D, de Leon N. Defining window-boundaries for genomic analyses using smoothing spline techniques. Genet Sel Evol. 2015;47:30. doi: 10.1186/s12711-015-0105-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Prufer K, et al. FUNC: a package for detecting significant associations between gene sets and ontological annotations. BMC bioinformatics. 2007;8:41. doi: 10.1186/1471-2105-8-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dunning Hotopp JC, et al. Widespread lateral gene transfer from intracellular bacteria to multicellular eukaryotes. Science. 2007;317:1753–6. doi: 10.1126/science.1142490. [DOI] [PubMed] [Google Scholar]
- 63.Pulido-Tamayo S, et al. Frequency-based haplotype reconstruction from deep sequencing data of bacterial populations. Nucleic Acids Res. 2015 doi: 10.1093/nar/gkv478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li M, et al. Detecting heteroplasmy from high-throughput sequencing of complete human mitochondrial DNA genomes. Am J Hum Genet. 2010;87:237–49. doi: 10.1016/j.ajhg.2010.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16:111–20. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 66.Massey FJ., Jr The Kolmogorov-Smirnov Test for Goodness of Fit. Journal of the American Statistical Association. 1951;46:68–78. [Google Scholar]
- 67.Excoffier L, Smouse PE, Quattro JM. Analysis of Molecular Variance Inferred from Metric Distances among DNA Haplotypes: Application to Human Mitochondrial DNA Restriction Data. Genetics. 1992;131:479–491. doi: 10.1093/genetics/131.2.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30:772–80. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bandelt HJ, Forster P, Röhl A. Median-joining networks for inferring intraspecific phylogenies. Molecular Biology and Evolution. 1999;16:37–48. doi: 10.1093/oxfordjournals.molbev.a026036. [DOI] [PubMed] [Google Scholar]
- 70.Shannon P, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome research. 2003;13:2498–504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Stamatakis A, Hoover P, Rougemont J. A rapid bootstrap algorithm for the RAxML Web servers. Syst Biol. 2008;57:758–71. doi: 10.1080/10635150802429642. [DOI] [PubMed] [Google Scholar]
- 73.Legendre P, Desdevises Y, Bazin E. A statistical test for host-parasite coevolution. Syst Biol. 2002;51:217–34. doi: 10.1080/10635150252899734. [DOI] [PubMed] [Google Scholar]
- 74.Oton EV, Quince C, Nicol GW, Prosser JI, Gubry-Rangin C. Phylogenetic congruence and ecological coherence in terrestrial Thaumarchaeota. The ISME journal. 2015 doi: 10.1038/ismej.2015.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1 (a) The average depth of O. volvulus nuclear genome and Wolbachia genome per sample. Red lines indicate the 6X threshold required to include samples in downstream analysis. (b) The genetic distance to the reference sequence vs. the average depth of coverage of the nuclear genome for each sample. To minimize bias stemming from a systematic variation in false negative variant call rates due to undersampling, low-depth samples were excluded from downstream analysis.
Supplementary Figure 2 (a) Median depth of coverage per 50kb region across the four chromosomes. Adult female worms were used in this study. Depth of coverage for the X chromosome (excluding pseudoautosomal region) is expected to be lower than the autosomal chromosomes because male embryonic cells are hemizygous for the X chromosome. (b) The ratio of average X chromosomal depth (excluding the pseudoautosomal region) to the average depth across all autosomal chromosomes, plotted against the average depth of the autosomal chromosomes.
Supplementary Figure 3 Onchocercidae-specific genes27 have higher piN/piS values as compared to other genes in the genome. Overall, most of the values are <1 (i.e. log values on x-axis are <0) in both cases, suggesting purifying selection, but this seems relaxed for the Onchocercidae-specific genes (more negative values on the x-axis indicate higher purifying selection).
Supplementary Figure 4 Maximum Likelihood phylogeny based on mitochondrial sequences from O. volvulus isolates, along with 3 O. ochengi isolates. The inter-sample distances for O. volvulus are much smaller than their distance with O. ochengi isolates.
Supplementary Figure 5 Genome-wide mean Tajima’s D and nonsynonymous, synonymous and non-coding sequence diversity (π) across the chromosomes in various sample sets.
Supplementary Figure 6 Genome-wide identity-by-state (IBS) estimates vs. geographic distance for all pairs of non-admixed West African samples. The lack of positive correlation between genetic and geographic distance indicates that in West Africa the spatial patterns of genetic diversity are not structured by geographic distance (isolation by distance) and there is an overall high level of intra-population gene flow. Geographic distance was calculated using a locally generated perl script, which calculated distance given the latitude and longitude coordinates of the two points (considering the curvature of the earth).
Supplementary Figure 7 Average transcript abundance levels for genes within FST outlier regions, and other genes surveyed in the sliding window analysis, per life-cycle stage and per FST comparison. Values represent average log-RPKM normalized to the average log-RPKM for all genes in the sample. Error bars represent standard error of the mean. Significance values were tested using a two-tailed T-test with unequal variance (normal distribution confirmed according to Shapiro-Wilk test, P = 0.956).
Supplementary Figure 8 (a–e) Classification of samples as forest and savanna using different sets of AIMs from the 4 chromosomes. The min, median, mean and max probabilities (in red and green colors) represent the classification probabilities for the 11 non-admixed samples. The 4 admixed samples are shown separately in blue. Distance (X-axis) is the minimum allowable distance between any two markers. Note that the size of the chromosomes puts a constraint on how large the distance threshold can get for a given number of markers. In general, the non-admixed samples are classified correctly with high probability and F7 can be distinguished as an admixed sample using only relatively high number of markers. The classification SVM was trained using non-admixed samples, before testing all the samples using the trained model. (f) Cumulative distribution of FST values shows that forest:savanna FST values tend to be higher than randomly chosen partition of samples, thereby supporting a small set of informative markers that can be used for classification. (g) Distribution of FST values among the loci with top-100 highest FST values. Black is forest:savanna and colored lines represent the 10 randomly chosen partitions of samples.
Supplementary Figure 9: Estimated frequencies of EVORhA based haplotypes for (a) mitochondria and (b) Wolbachia. These sum of frequencies for a sample’s haplotypes ranges from ~0.6 to >2. This is partially because these are means of the distribution of frequency estimates of the best-fitting deconvolution of the data. In some cases, EVORhA splits a single distribution into two overlapping ones resulting in two nearly identical haplotypes with slightly different frequencies (e.g., mitochondria from F11, E16, E20 and E21). (c) Mutation distances and statistical comparison of within- and between-sample distances for mitochondria (left) and Wolbachia (right). For all samples, except 1 for mitochondria and 2 for Wolbachia, the within-sample haplotype distances are significantly smaller than between-sample distances (P-values plotted in top panel, sample mutation distances in bottom panel). The red bar represents a significance level of 0.05. The blue bar represents the mean between-sample mutation distance. This relationship is more rigorously verified using AMOVA (see text).
Supplementary Figure 10. (a) Reference and Alternate allele depth per variable position, per sample, in Wolbachia, before filtering. A clean set has clusters along a line (x+y=c), e.g. S3, F8, F15, and U27. Many of the Ecuador samples show large clusters along a vertical, showing signs of aberrant read mapping potentially due to nuwts. (b) Reference and Alternate allele depth per variable position, per sample, in Wolbachia, after filtering. Most of the sample with clusters along a vertical in panel a are now cleaned up. (c) Reference and Alternate allele depth per variable position, per sample, in mitochondria. The data does not show signs of aberrant read mapping.
Classifications, sources and read mapping data for each of the samples utilized
SnpEff annotations and allele frequency data per SNP position
Summary of results from the FST sliding window analysis
FST genome scan sliding window positions, summary statistics, and overlapping genes
Detailed functional annotation and SNP annotation per gene
ABBA-BABA analysis results for gene flow
Ancestry Informative Markers for distinguishing savanna from forest O. volvulus isolates
Variants ranked by FST (FOR:SAV comparison, minimum value 0.5)
Sources, dates, and processing procedures for each of the whole genome shotgun libraries in each of the samples