Abstract
Transcription activator-like effectors (TALEs) are secreted by plant–pathogenic Xanthomonas bacteria into plant cells where they act as transcriptional activators and, hence, are major drivers in reprogramming the plant for the benefit of the pathogen. TALEs possess a highly repetitive DNA-binding domain of typically 34 amino acid (AA) tandem repeats, where AA 12 and 13, termed repeat variable di-residue (RVD), determine target specificity. Different Xanthomonas strains possess different repertoires of TALEs. Here, we study the evolution of TALEs from the level of RVDs determining target specificity down to the level of DNA sequence with focus on rice-pathogenic Xanthomonas oryzae pv. oryzae (Xoo) and Xanthomonas oryzae pv. oryzicola (Xoc) strains. We observe that codon pairs coding for individual RVDs are conserved to a similar degree as the flanking repeat sequence. We find strong indications that TALEs may evolve 1) by base substitutions in codon pairs coding for RVDs, 2) by recombination of N-terminal or C-terminal regions of existing TALEs, or 3) by deletion of individual TALE repeats, and we propose possible mechanisms. We find indications that the reassortment of TALE genes in clusters is mediated by an integron-like mechanism in Xoc. We finally study the effect of the presence/absence and evolutionary modifications of TALEs on transcriptional activation of putative target genes in rice, and find that even single RVD swaps may lead to considerable differences in activation. This correlation allowed a refined prediction of TALE targets, which is the crucial step to decipher their virulence activity.
Keywords: Xanthomonas, transcription activator-like effectors (TALEs), evolution of TALEs, evolutionary mechanisms, plant–pathogen interaction, transcriptional response
Introduction
Xanthomonas bacteria are plant pathogens that infect many crop plants including citrus, tomato, pepper, cabbage and rice, and are responsible for substantial yield losses worldwide. Yield losses for rice are especially harmful, as the food supply of a large part of the world population depends on rice. Rice plants are infected by different strains of Xanthomonas oryzae pv. oryzae (Xoo) and Xanthomonas oryzae pv. oryzicola (Xoc). Different Xanthomonas strains have different repertoires of effector proteins, including transcription activator-like effectors (TALEs). TALEs act as transcription factors in the plant cell. Their DNA-binding domain forms a right-handed supercoil structure wrapping around the DNA and mediating sequence-specific binding to the promoter of plant genes. In detail, the binding domain consists of highly repetitive consecutive repeats of ∼34 highly conserved amino acids (AAs). The AAs on positions 12 and 13 are called repeat variable di-residue (RVD). The TALE target specificity is determined by the sequence of RVDs, where each RVD binds to one nucleotide of the target sequence (Boch et al. 2009; Moscou and Bogdanove 2009).
Xoo and Xoc pathogens typically harbor large TALE repertoires, which differ in number and nature between Asian Xoo (15–26 TALEs), African Xoo (8–10 TALEs), North-American Xoo (no TALEs), and Xoc strains (19–27 TALEs) (Gonzalez et al. 2007; Triplett et al. 2011; Wilkins et al. 2015; Quibod et al. 2016). Several TALEs of different strains are identical on the level of RVD sequences and are, hence, assumed to target the same plant genes, whereas other TALEs show variation in individual RVDs or even in longer, contiguous stretches of their RVD sequence. As TALEs do not only support bacterial infection but may also activate the expression of resistance genes, we expect a permanent selective pressure on TALE genes. Gaining deeper insights into TALE evolution may thus help to understand the evasion from plant resistance, which will be relevant for targeted breeding of resistant plants.
Different aspects of TALE evolution have been in the focus of recent studies. A major driving forces of TALE evolution are modifications in the promoters of the host plant to evade the activation of susceptibility genes or to yield activation of resistance genes. Subsequently, TALEs may be adapted, mostly by modifications of their RVD sequence, such that the activation of susceptibility genes is restored or the activation of resistance genes is avoided (Hutin et al. 2015).
For studying TALE evolution based on the variety of TALEs present in different Xanthomonas strains, we need to define measures of similarity of TALEs and their individual repeats. Such measures may differ in the level of detail from the complete DNA sequence of a TALE to the sequence of RVDs, and in the perspective from evolutionary analyses of nucleotides to functional analyses of target specificity. The tool DisTAL (Pérez-Quintero et al. 2015) compares TALEs on the level of the complete AA sequence of each TALE repeat with the goal of identifying TALEs with common evolutionary origin. FuncTAL instead compares TALEs by the similarity of their binding specificities, which should group TALEs with similar target boxes together (Pérez-Quintero et al. 2015). Previously, we presented AnnoTALE (Grau et al. 2016), a suite of tools for annotating and analyzing TALEs and their putative targets. In AnnoTALE, TALEs of different Xanthomonas strains are grouped into classes based on their RVD sequence, which may yield a reasonable balance between evolutionary relationships and similarities based on target specificity. A similar rationale for grouping TALEs has been used in later studies considering TALE similarity in Xoo (Quibod et al. 2016). On the basis of the AnnoTALE classes, we propose a unified, systematic nomenclature of TALEs. Several parts of this paper are based on these AnnoTALE classes, and we use the unified nomenclature throughout this manuscript.
Recently, the potential of studying TALE evolution in detail has increased dramatically due to a large number of newly sequenced Xoo (Grau et al. 2016; Quibod et al. 2016) and Xoc (Booher et al. 2015; Wilkins et al. 2015) genomes, which are jointly considered in our present study. Such collections of Xanthomonas strains confer the possibility to define sets of TALEs that are conserved across many strains. In the host plant, these TALEs activate similar sets of target genes, and the pattern of presence or absence of TALEs compared with the patterns of gene activation in infected plants may help to distinguish true TALE targets from secondary effects (Wilkins et al. 2015). Some of the target genes seem to be especially important for bacterial growth and have, hence, been termed susceptibility hubs (Hutin et al. 2015), the most prominent gene family being SWEET genes (Streubel et al. 2013). Effects of the presence or absence of TALE genes in different Xoo strains have also been studied with respect to lesion lengths of infected rice plants of near isogenic lines (Quibod et al. 2016).
With multiple sequenced Xoo and Xoc strains available, TALE evolution has also been studied on a macroscopic scale. Larger genomic rearrangements have been identified between different Xoo (Grau et al. 2016) and Xoc (Wilkins et al. 2015) strains, which also affect the location of TALE genes. On the genome, TALE genes are often organized in clusters of multiple TALEs, which have been defined by their flanking genes (Grau et al. 2016). The exact composition of TALEs in a common cluster may vary, and highly similar TALEs may occur in different clusters of different strains (Booher et al. 2015; Grau et al. 2016).
While the impact of alterations of TALEs on gene activation, susceptibility or resistance, or other phenotypic traits has been studied quite extensively, the evolutionary mechanisms leading to these alterations have not been fully resolved yet. Several, consecutive repeats may be deleted from a TALE (Yang et al. 2005; Booher et al. 2015), but the exact mechanism is unknown. In turn, duplication of individual repeats, likely due to slipped strand mispairing, has been described in Ralstonia RipTALEs (Schandry et al. 2016). Finally, putative recombination events between different TALEs and different parts of the same TALE have been observed (Booher et al. 2015).
One potential mechanism promoting TALE duplication and recombination could be transposition. It has been noticed long before elucidation of the TALE code that TALE genes are often flanked by inverted repeats (IRs) (Bonas et al. 1993; Noël et al. 2003). These repeats are believed to confer mobility, for example, as a composite Tn3-like transposon, which might also be the cause of duplications or deletions of TALE repeats (Ferreira et al. 2015).
In this paper, we studied the evolution of TALEs with a focus on rice-pathogenic Xoo and Xoc strains. We first studied the conservation of TALE repeat sequences on the level of individual codons, considering the two codons coding for the RVDs as well as the codons at all flanking positions. We further investigated two alternative hypotheses how novel or altered RVDs could evolve, namely either by base substitutions within the RVD codon-pairs or by modular recombination of TALE repeats. We scrutinized several examples of putative recombinations events or deletions of individual repeats on the DNA level to find indications of the respective evolutionary events. Finally, we compared the TALE repertoires of Xanthomonas strains, investigated the impact of the loss or gain of a TALE on gene expression in the host plant, and assessed the influence of evolutionary modifications of TALE sequences on the expressional response of rice genes to Xoc infection.
Materials and Methods
AnnoTALE
AnnoTALE version 1.2 (Grau et al. 2016) with class builder version 03/09/2017 provides 516 TALE genes of 33 Xanthomonas strains, including TALEs from recently published Xanthomonas strains (Wilkins et al. 2015; Quibod et al. 2016; Jaenicke et al. 2016). All TALEs are listed in supplementary table 2, Supplementary Material online. AnnoTALE is available from http://jstacs.de/index.php/AnnoTALE. (last accessed June 6, 2017)
Codon Usage within Repeats
To calculate the codon usage within TALE repeats, we only considered repeats with standard length of 34 AA and counted the codons used at each AA position of the repeat. We defined synonymous and nonsynonymous substitutions relative to the most frequent codon at each position.
To check whether a reason for the conservation of codon pairs coding for the RVDs might be the probability to yield a nonsynonymous AA substitution by substituting a single nucleotide in a codon pair, we counted for each RVD and all possible codon pairs coding for its AAs, how many synonymous or nonsynonymous substitutions could occur by changing one of the six nucleotides of the codon pair.
We tested the potential selective influence of RNA secondary structures on the codon pair used with RNAalifold of the ViennaRNA Package (Lorenz et al. 2011). For each RVD, we chose the main codon pair and extracted all repeat sequences of standard length containing this codon pair from the AnnoTALE data set. We then aligned these sequences with Clustal Omega (Sievers et al. 2011) and used the alignment as input for RNAalifold to generate the prediction of the secondary RNA structure and to collect the calculated free energy of the structure. To check whether the other codon pairs coding for the same RVD may have different energies, we used the same input repeat sequences and substituted only the codon pair at the RVD by another possible codon pair and compared the resulting free energies.
Evolution by Point Mutations
We considered all AnnoTALE classes with aligned RVD sequences to identify RVD swaps within TALEs of the same class. We counted one RVD swap, if the codon pair coding for a RVDs at one position showed a synonymous or nonsynonymous substitution on the DNA level, regardless of the corresponding number of base substitutions in the codon pair. Hence, we additionally determined the number of observed RVD swaps given a specific number of base substitutions that lead to the RVD swap.
We divided the data set of repeats with 34 AAs into two disjoint data sets. The first contained all repeats not involved in RVD swaps and the second contained all repeats showing an RVD swap within AnnoTALE classes at this position. We analyzed the flanking sequences of repeats not involved in RVD swaps for each repeat type and generated a DiffLogo (Nettling et al. 2015) for each pair of repeat types to check whether the flanking sequences show differences depending on the repeat type.
We additionally calculated the mutual information of each flanking position of a repeat and the RVD codon pairs using the sequences of all repeats that are not involved in RVD swaps. The mutual information Ij between the RVD codon pairs and the nucleotides at flanking position is defined as:
| (1) |
where is the joint probability of codon pair r and nucleotide aj (estimated as relative frequencies) and P(r) and are the corresponding marginal probabilities.
To distinguish between a pair of repeat types, we trained a binary classifier using the flanking sequences of the corresponding RVD types. To yield a clean training data set, we restricted the training set to only repeats that are not involved in RVD swaps within the corresponding AnnoTALE classes. We generated such classifiers only for repeat types with at least 50 input sequences. We did not consider 33 AA repeats with RVD N*, as these do not fit the schema of individual base substitutions. We weighted the sequences in each training data set to account for the different abundances of RVDs, that is, we assigned each sequence a weight of a constant divided by the number of sequences in the set. We implemented the classifiers within the Java framework Jstacs (Grau et al. 2012). The flanking sequences of the different repeat types were modeled by position weight matrices (PWMs) for each class. For training the parameters of the PWMs in each pairwise classifier, we used the discriminative maximum conditional likelihood principle. Classification performance was assessed in a stratified 5-fold cross-validation on the training data. After training, we classified flanking sequences of repeats, which show an RVD swap within AnnoTALE classes. For each flanking sequence, we assumed the class with the largest a posteriori probability.
Evolution by Recombination
We searched for perfect matches of RVD stretches of at least five RVDs between members of different TALE classes. Some of the classes contain completely identical (on the RVD level) TALEs, which were collapsed into one query sequence to avoid combinatorial explosion of matches. The matches reported are maximal in that sense that an elongation of the RVD sequence at either side would lead to a mismatch or extend beyond the first or last repeat, respectively. We explicitly excluded cases, where the RVD sequence of one TALE could be explained by a deletion of individual TALEs (see below). For each of the matches found, we noted the distance of the match from the N-terminus and C-terminus (in terms of repeats), and its length.
For assessing the enrichment of duplicated RVD sequences between different classes at the terminal regions, we considered two null models. In the first model, columns of RVDs in the class alignments may be permuted such that the structure of (terminal) gaps is not affected. In the second model, the length w of the matching sequences between TALEs from any two classes stays identical, but its position within the TALE is drawn from a uniform distribution in . Under both models, we create 1,000 random instances and plot inverse empirical cumulative frequencies for the original values and those from the null model.
We further studied a selection of examples on the level of DNA sequences to gain further evidence of a common evolutionary origin of such duplicated RVD subsequences. To this end, we selected specific cases from the previous step, extracted the corresponding DNA sequences, aligned those as induced by the alignment of duplicated RVD subsequences. We additionally aligned the N-terminus and C-terminus of the corresponding sequences. For regions that already differed in the alignment of RVD sequences, because these repeats likely did not participate in the potential duplication, we considered four additional repeats in the alignment of DNA sequences, but at most to the first or last repeat, respectively.
For the examples considered, several TALEs of one of the classes shared the duplicated RVD sequence. Hence, we repeated this procedure for all those TALEs: Six TALEs in class TalBC (TalBC3, TalBC9, TalBC10, TalBC1, TalBC4, TalBC6) and six TALEs in class TalBB (TalBB6, TalBB3, TalBB1, TalBB2, TalBB7, TalBB5) were compared with TalCS1; all nine TALEs in class TalAC and four TALEs in class TalAS (TalAS7, TalAS3, TalAS4, TalAS8) with identical trailing RVD stretch were compared with TalAS2.
Evolution of TALE Clusters
We extract the genomic sequence of all Xoo and Xoc strains around prominent TALEs (TalAG in Xoo and TalBF, TalBB in Xoc) such that all TALE genes in a cluster and flanking genes are covered. In this region, we visualize annotated TALE genes according to AnnoTALE (Grau et al. 2016) and other genes from the corresponding Genbank annotation (accessions listed in supplementary table 2, Supplementary Material online). In these regions, we further scan for IRs using Inverted Repeats Finder (Warburton et al. 2004) with parameters 2 5 8 80 10 30 200 100000 -h -d -i2 -t4 10000 -t5 100000 -t7 100000 to allow for longer DNA stretches between the IR left and right half. We obtain alignments of spacer regions using Kalign (Lassmann and Sonnhammer 2005) available from http://www.ebi.ac.uk/Tools/msa/kalign/ (last accessed June 6, 2017). We predict secondary structures of putative hairpin structures by Mfold (Zuker 2003) using DNA parameters available from http://unafold.rna.albany.edu/?q=mfold/DNA-Folding-Form (last accessed June 6, 2017).
Evolution by Deletion or Duplication of Individual Repeats
For evaluating potential deletions or duplications of TALE repeats, we searched for pairs of TALEs that 1) share common RVD subsequences at the N-terminus and C-terminus of the repeat array, and 2) do not have an identical length (which would rather be an indication of RVD swaps). For these, we identified the repeats that might need to be deleted and performed a gapless alignment of the corresponding TALE DNA sequences. We used one of the TALEs as a reference (the singleton TALE in all cases except TalCM, where we used TalCM1 and TalCM5 independently) and counted the number of mismatches to this reference in each column of the alignment.
Finally, we created a novel tool “TALE Repeat Differences” in AnnoTALE that compares the repeats of two TALEs on the level of their DNA or AA sequence and visualizes pairwise repeat differences. To this end, the tool extracts the sequence of each TALE repeat and compares those sequences in a global, pairwise alignment using the Levenshtein distance (i.e., costs of 0 for a match and 1 for a mismatch or an insertion or deletion). Those pairwise distances are then visualized as a matrix of colored squares, where a distance of zero is represented by white color and larger distances are drawn on a scale from yellow (low) to red (high).
TALE Presence in Xoo and Xoc Strains
We extracted all AnnoTALE classes and the corresponding member TALEs from the AnnoTALE class builder (version 03/09/2017). In AnnoTALE, each TALE already carries the information about its strain of origin. Hence, it was only necessary to create a matrix of all AnnoTALE classes by all Xoo and Xoc strains, and mark those entries that are represented by the class and strain origin of at least one TALE. This procedure is automated in an additional tool in AnnoTALE called “TALE Class Presence”, which may be used to repeat the analysis for updated versions of the class builder and/or other sets of Xanthomonas strains.
Impact of Evolutionary Events on Target Gene Activation
For analyzing the impact of evolutionary events on the activation of target genes in rice, we downloaded raw RNA-seq data for ten Xoc strains (Wilkins et al. 2015) available at NCBI Gene Expression Omnibus under accession GSE67588 in FastQ format. As reference transcripts for the expression analysis, we downloaded the transcripts of the MSU7 annotation (Kawahara et al. 2013) available from http://rice.plantbiology.msu.edu/pub/data/Eukaryotic_Projects/o_sativa/annotation_dbs/ (last accessed June 6, 2017). We quantified transcript expression using kallisto (Bray et al. 2016) (v0.43.0) with parameters kallisto quant --single -b 10 -t 8 -l 200 -s 40 -i all.cdna.idx -o ¡out¿ ¡fastq¿. For plotting expression values on log scale, we added a constant of 1 to each expression value.
We then determined differentially expressed genes using the R-package sleuth (Pimentel et al. 2016) (v0.28.1) and aggregated differential expression on the level of genes using the parameter target_mapping of the sleuth function sleuth_prep(), and recorded the log2-fold change and Benjamini–Hochberg-corrected P-value as returned by sleuth for each rice gene and each of the ten Xoc strains compared with the control mock experiment.
For the following analyses, we considered rice genes as putative targets of a TALE that fulfilled the following criteria for at least one of the ten Xoc strains. First, the gene needed to have an expression value (kallisto normalized TPM) of at least 35 to account for large fluctuations in the expression values of lowly expressed genes and due to the typically strong activation by TALEs. Second, genes needed to pass a significance level of and show a log2-fold change of at least 2 to consider genes significantly and substantially upregulated. Third, we limited the analysis to genes that have a target box among the top 100 predictions of TALgetter (Grau et al. 2013) in their promoter (defined as −300bp to 200bp relative to the annotated transcription start) in rice (MSU7) for at least one TALE of the corresponding strain. Finally, the observed effects on gene expression should be traceable to one specific TALE class. Hence, we additionally filtered candidate genes for those that were predicted targets of exactly one and not multiple, alternative TALE classes. Raw TALgetter prediction scores were normalized by subtracting the log-likelihood according to the uniform distribution and dividing the result by the length of the target site.
Results
Codon Usage within Repeats
In this section, we reconsider the conservation of TALE repeat sequences, with a focus on the codon pair coding for the RVD, on the level of their DNA sequence. We first studied the conservation of AAs and codons in TALE repeats based on all 34 AA repeats present in the AnnoTALE database containing TALEs from 20 Xoo, 10 Xoc, and 3 X. translucens strains. A complete list of the Xanthomonas strains and TALEs studied is given in supplementary table 2, Supplementary Material online.
As expected, we found that the AAs at most positions of all these TALE repeats are highly conserved. Notable exceptions were the two positions (12 and 13) coding for the RVDs, which originally lead to the term “repeat variable di-residue”. Furthermore, positions 2, 3, 4, 11, 22, and 32 showed a substantial amount of nonsynonymous substitutions compared with the most frequent codon (and AA) at these positions (see supplementary table 1, Supplementary Material online). Notably, most positions were also highly conserved on the codon level with low numbers not only of nonsynonymous but also of synonymous mutations.
The RVDs are commonly perceived as the most variable AAs of TALE repeats, which is also represented in supplementary table 1, Supplementary Material online. As the codons at most flanking positions are highly conserved, we studied if this is also the case for individual RVD types. The codon usage within different RVDs is shown in table 1. For all of the RVD types considered, only one to three codon pairs coding for an RVD occurred in known TALEs even though the number of theoretically possible codon pairs is substantially larger. For most RVDs, one of the possible codon pairs was used almost exclusively in naturally occurring TALEs. For instance, the RVDs of type HD were encoded by two different codon pairs in known TALEs, with 2,511 occurrences of the main codon pair (CACGAT) and only three of the other (CATGAT). Theoretically, HD could be encoded by two more codon pairs (CACGAC and CATGAC), which, however, were present in none of the TALEs. The only exceptions from this strong conservation were RVDs NS and ND, where two codon pairs (AATAGT, AATAGC, and AACGAT, AATGAC, respectively) occurred with higher frequencies. Hence, we conclude that RVDs are conserved on the codon level to a similar degree as the flanking positions when considering individual RVD types, for example, only HD repeats, although the two RVD codons are highly variable considering all TALE repeats. In general, this observation would comply to an evolutionary model, where the generation of new TALEs and their RVD sequences is the result of recombinations of complete repeats leaving their sequence widely untouched. We will reconsider this possibility in the remainder of this manuscript.
Table 1.
Codon Pair (CP) Usage in RVDs of Known TALEs Compared with the Theoretical Possible Number of Codon Pairs for All RVDs with At Least 50 Examples in the Data Set. Columns “CP I” to “CP III” List the Frequency of the Individual Codon Pairs Present in the Data
| RVD | Used CPs | Possible CPs | CP I | CP II | CP III | |||
|---|---|---|---|---|---|---|---|---|
| HD | 2 | 4 | CACGAT: | 2511 | CATGAT: | 3 | ||
| NN | 3 | 4 | AATAAC: | 1874 | AATAAT: | 15 | AACAAC: | 1 |
| NI | 2 | 6 | AATATT: | 1422 | AATATA: | 13 | ||
| NG | 2 | 8 | AATGGC: | 1128 | AATGGG: | 1 | ||
| HG | 1 | 8 | CATGGC: | 540 | ||||
| NS | 2 | 12 | AATAGT: | 486 | AATAGC: | 195 | ||
| ND | 2 | 4 | AACGAT: | 103 | AATGAC: | 26 | ||
| HA | 1 | 12 | CACGCT: | 57 | ||||
| HH | 1 | 4 | CATCAC: | 63 | ||||
Although the general observation of strong conservation not only of flanking but also of the codons coding for a specific RVD is compelling, its evolutionary background was not instantly clear. Hence, we considered several potential explanations of the observed conservation. A first hypothesis was that the reason might be the stability of RVDs against single base substitutions, namely that substituting a single nucleotide in a codon pair is more (or less) likely to yield a nonsynonymous AA substitution. For this reason, using specific codons might be favorable compared with others. However, in almost all cases, the different possible codon pairs were equal in this regard.
A second reason might be that different codon pairs coding for the same RVD form different RNA secondary structures (possibly influencing translation or its rate). Considering predictions of secondary structures of the different codon pairs of common repeats in RNAalifold (Lorenz et al. 2011), we found that the predicted free energy is nearly identical, however.
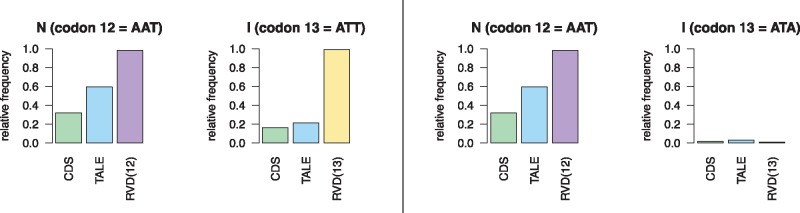
A third idea could be that Xanthomonas tRNAs for the codons of common RVDs are especially frequent (or rare), which could also influence translation rates. Hence, we compared the relative frequency of codons coding for the AAs at position 12 and 13 with 1) their relative frequency within the TALE, and 2) within all coding sequences of the Xanthomonas strains studied, where the latter should be an approximate proxy of tRNA frequency. Exemplarily, figure 1 shows the relative histogram for the two NI codon-pairs. The two codon-pairs coding for NI occurring in RVDs were AATATT and AATATA. Both codons of the first codon pair occurred with higher frequency at RVD positions compared with TALE sequences in general and compared with all coding sequences, whereas the second codon (ATA) of the second codon pair was rare in all three sets. However, no general pattern emerged when also considering codon pairs of other RVDs (see supplementary figs. 1–3, Supplementary Material online).
Fig. 1.
—Relative frequency of the codons of the two codon pairs coding for RVD NI counted at RVD positions only, in complete TALE sequences, and in all coding sequences of Xanthomonas genomes. In case of the codon pair AATATT (left), both codons of the pair occur with higher frequency in RVD positions than in the remainder of TALE sequences and than in all coding sequences. In case of the codon pair AATATA (right), the codon at position 13 occurs with exceptionally low frequency in all regions considered.
Evolutionary Mechanisms in TALEs
In this section, we set out to investigate whether point mutations or recombination events lead to differences in RVDs between TALEs of potential common evolutionary origin.
Our further analyses were based on the current AnnoTALE (Grau et al. 2016) data set, with 516 TALEs from 33 Xanthomonas strains (see supplementary table 2, Supplementary Material online). AnnoTALE is an application for annotating TALEs in Xanthomonas genomes, for analyzing their terminal and repeat sequences, for clustering TALEs by similarity of their RVD sequence, naming TALEs according to a unified system, and for predicting putative TALE target genes. In AnnoTALE, a group of TALEs with similar RVD sequences forms a class. The similarity of RVD sequences is determined from a global alignment, strongly disfavoring internal gaps. AnnoTALE groups the 516 currently represented TALEs into 100 different classes. The number of TALEs in these classes is shown in supplementary figure 4, Supplementary Material online. The majority of AnnoTALE classes comprises between 2 and 20 TALEs. As an example, class TalAD is shown in supplementary figure 5, Supplementary Material online, which contains two subgroups of highly similar TALEs from different Xoo and Xoc strains. Notably, only two positions (7 and 10) show different RVDs between the two subgroups, although these stem from different pathovars.
While highly similar RVD sequences of TALEs, which are the basis of AnnoTALE classes, might also be the result of convergent evolution for a common target gene in the host plant, we considered it more likely that such TALEs evolved from a (fairly recent) common ancestor TALE. For this reason, the present study was based on the assumption that TALEs of a common AnnoTALE class are not only highly similar on the RVD level but also share a common evolutionary origin.
For the following considerations, it is important to distinguish modifications on the level of RVDs from those on the level of individual AAs or DNA bases. To make this distinction obvious, we refer to modifications on the level of RVDs as RVD swaps, where we ignore the specific AAs of RVDs, that is, the RVD swap of “HD” for “NG” is considered to be on a par with the RVD swap of “HD” for “ND”. In contrast, we refer to modifications on the level of individual AAs or bases as substitutions.
Evolution by Point Mutations
Here, we investigated RVD swaps observed in AnnoTALE classes for signs of point mutations as opposed to recombination events. To this end, we considered all members of the individual AnnoTALE classes. For each class, we compared the aligned RVDs on the DNA level and found 202 RVD swaps in total. In our data, synonymous substitutions occurred only for two RVDs, NN, and NS, although, in general, further synonymous substitutions for other RVDs would be possible. The remaining substitutions on the DNA level were nonsynonymous and lead to a modification of the RVD. The histogram in supplementary figure 5, Supplementary Material online shows that most frequently (93 cases, 46% of the observed RVD swaps), only one nucleotide is substituted between the codon pairs of aligned RVDs in a common AnnoTALE class. Substitution of two nucleotides occurred in 22% and of three nucleotides in 11% of the cases, while frequency increased to 20% for four nucleotide substitutions. The remaining 1% was substitutions in five of the six nucleotides of the RVD. One reason for the increasing frequency of four-nucleotide substitutions might be the frequency of HD repeats in the data set (2,514 cases), where most RVD swaps for other frequent RVDs like NN, NI, or NG (4,454 cases in total) lead to four base substitutions in the codon pair.
In contrast to the general assumption of evolution by recombination of repeats within a TALE or between different TALEs, the high frequency of single nucleotide substitutions complies to the assumption that different RVD types evolved through individual point mutations on the DNA level. For this reason, we scrutinized TALE repeats for further indications of evolution by point mutations.
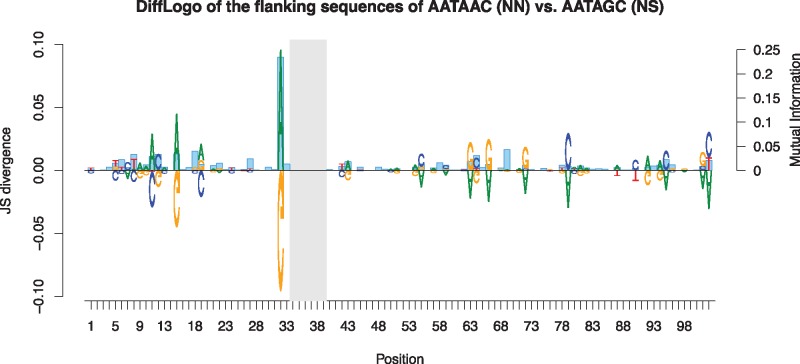
Although the flanking sequences of RVDs are highly conserved on the DNA level, we also observed (cf. see supplementary table 1, Supplementary Material online) several variable positions besides the RVDs. In figure 2, we present a DiffLogo (Nettling et al. 2015) of the flanking sequences of all NN and NS repeats that are not involved in RVD swaps in their corresponding class, excluding RVD codons. We found differences between NN and NS repeats at several positions, indicating that nucleotides at some of the flanking positions show dependencies on the RVD type. Such differences were also present for other combinations of RVD types, although with different positional patterns (see supplementary figs. 6–8, Supplementary Material online). Hence, we computed mutual information values between the nucleotide distribution at individual positions of the flanking region of repeats and the codon pair at the RVD positions for all RVDs. The positions showing large mutual information values widely overlapped with those positions for which we also observed differences between NN and NS repeats (fig. 2). We found considerable dependencies between the RVDs and the nucleotide at position 32 but also those at positions 6, 8, 11, 12, 15, 18, 27, 64, 69, 95, and 102. Hence, we concluded that the sequence at flanking regions of TALE repeats comprises substantial information about the RVD it surrounds. This also suggests that recombination of TALE repeats is the predominant path of TALE evolution.
Fig. 2.
—DiffLogo of the flanking sequences of the two RVD types NN (AATAAC, top) and NS (AATAGC, bottom). For some positions, differences between NN and NS repeats are clearly visible. As similar differences are also present for other pairs of RVDs, we obtain an overall assessment of the dependencies between RVD codon pairs and flanking sequence by computing the mutual information between the codon pairs coding for all the RVDs and the nucleotides of the corresponding flanking regions (blue bars). Several positions with larger mutual information values coincide with positions that show differences between NN and NS repeats, whereas further positions obtain large mutual information values due to dependencies on other RVD types.
This finding lead to the idea of training pairwise binary classifiers that may distinguish RVD types by their flanking regions alone. Trained classifiers may be applied to repeats which show an RVD swap in the alignment of AnnoTALE classes to answer the question whether the RVD swaps between class members have more likely derived from point mutations or from recombination events.
If RVD swaps are the result of a point mutation, the original flanking sequence should be strongly preserved and still be present in the aligned repeats of this class, regardless of their current RVDs. Hence, all repeats at this position should obtain the same classification label based on the flanking sequence.
If RVD swaps are the result of a recombination event, instead, where entire repeats are replaced, the flanking sequence should still resemble the typical flanking sequence of the respective RVD type. Hence, all repeats at this position should obtain a classification label corresponding to their RVD type.
We chose this schema involving pairwise classifiers instead of direct sequence comparisons to yield stable and consistent criteria and results across the AnnoTALE classes studied.
Initially, we trained a binary classifier for distinguishing NN (AATAAC) and NS (AATAGC) repeats by means of the flanking sequences of these two RVD types. We assessed classification accuracy in a 5-fold cross validation on the training data, which resulted in a classification rate of 0.898. The sensitivity for NN was 0.89, which means that 89% of the NN repeats have been classified correctly and only 11% have been misclassified as NS. The sensitivity of NS was 0.97, which means that 97% of the NS repeats have been classified correctly and only 3% have been misclassified as NN. These initial results motivated us to train binary classifiers for all combinations of RVDs with at least 50 training examples in the data set. The sensitivities of the different binary classifiers in 5-fold cross validations are shown in supplementary table 3, Supplementary Material online. The classifiers yielded sensitivities between 0.71 and 0.97, where the lowest sensitivity was obtained for the classifier distinguishing HD and NI.
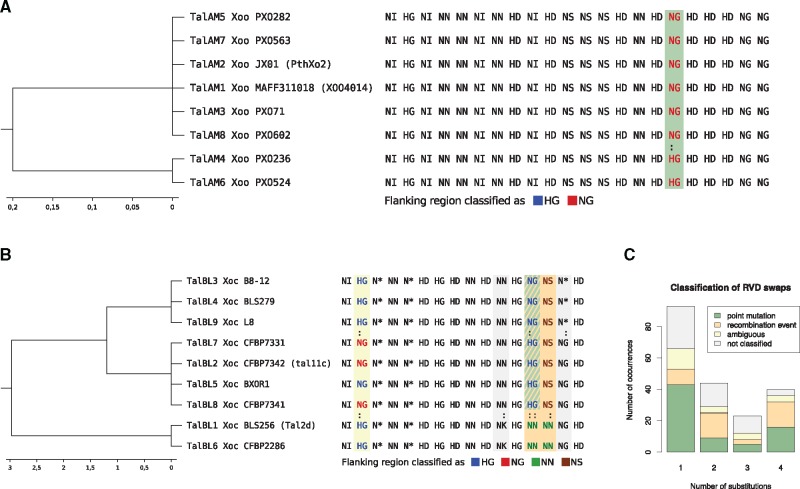
After training and validation, we applied the resulting classifiers to the flanking regions of RVDs showing apparent RVD swaps within AnnoTALE classes. An example for classification of repeats involved in RVD swaps in class TalAM is shown in figure 3A. Class TalAM consists of TALEs from eight different Xanthomonas strains. The corresponding RVD sequences are nearly identical with only one RVD swap between HG and NG at position 17 of the alignment. We used the trained NG-HG-classifier to classify the flanking sequences of each TALE at this position. In this case, the classifier identified all flanking sequences as NG repeats. This indicates that the HG-type repeats at this position (TalAM4 and TalAM6) evolved by point mutations from the corresponding NG-repeat. Figure 3B shows the more complex example of class TalBL with several RVD swaps at alignment positions 2, 11, 13, 14, and 15. We only had classifiers distinguishing between the RVDs at position 2, 13, and 14, as the tree-nucleotide deletion in N* repeats does not fit the schema of point mutations and the training sets for other RVDs did not contain enough examples. The NG-HG-classifier identified each flanking region of HG repeats on position 2 as HG. The flanking regions of the NG repeat were classified as NG in three of four cases. Hence, the classification result did not give a clear indication whether the RVD swap arose from a point mutation, a recombination event, or two separate events at this position. The RVD swap at position 12 between NG and HG repeats showed a much clearer result. The flanking regions of the NG and HG repeats were classified by the NG-HG-classifier as HG, which suggests that the NG repeats arose from the corresponding HG repeats by point mutations. The second RVD swap at this position between HG and NN repeats classified by the HG-NN classifier showed a different result. All flanking sequences matched their respective RVD. The same holds true for the NG-NN-classifier applied to the NG and NN repeats. This suggests that the NN repeats at this position emerged rather from a recombination event. The classification of the NN and NS repeats at position 14 indicated a recombination event, too. In summary, these results indicated that the last four repeats of TalBL1 and TalBL6 evolved rather through recombination events, although the last two repeats match their counterparts in the remaining class members.
Fig. 3.
—Classification of RVD swaps by flanking sequence indicating point mutations or recombination. (A) Example for classification of flanking sequences in class TalAM. RVD swaps between different TALEs are marked with a colon. Common RVDs at swap positions are colored depending on the classification result of their flanking sequence by the corresponding binary classifier. In this case, all repeats at column 17 of the alignment are classified as NG repeats based on their flanking sequence, which indicates a point mutation in the RVD codon pair as indicated by the green shading of that column. (B) Example for classification of flanking sequences in class BL. RVD swaps between different TALEs are marked with a colon. Common RVDs at swap positions are colored depending on the classification result of their flanking sequence by the corresponding binary classifier. Columns with RVD swaps are shaded according to the colors used in C. (C) Results of classification for TALE repeats involved in RVD swaps in all AnnoTALE classes with four different outcomes: (a) all repeats are classified as the same RVD, which suggests point mutation within the RVD (green), (b) all flanking sequences are classified like the RVD of the respective RVD, indicating a recombination event (orange), (c) the classification is ambiguous (yellow). (d) No classifier trained for the combination of these two RVDs and repeats have not been classified (grey).
A summary of all classification results of all RVD swaps in the AnnoTALE classes is shown in figure 3C. If all repeats at the position of an RVD swap were consistently classified by their flanking sequence as repeats of the same type, we counted a point mutation. If the classification based on the flanking sequence is consistent with the actual RVDs, we counted a recombination event. We counted cases where neither of the previous consistent outcomes is obtained as ambiguous. Finally, RVD swaps for which either of the two RVDs did not have at least 50 training examples were not classified.
In cases where we observed only one base substitution in the codon pair coding for an RVD, the majority of classification results indicated a point mutation. For two base substitution, we found the opposite result with more classification results indicating recombination events than point mutations. For larger numbers of base substitutions, both alternatives occurred with comparable frequencies. To get a more detailed view on putative base substitutions, we visualize classification results for a subset of RVD combinations, covering the majority of classified cases, as a network in supplementary figure 9, Supplementary Material online. We found that most putative substitution events could be traced down to the specific RVD pairs NG/HG and NI/NS.
In summary, our findings indicate that one way how TALEs evolve is by single point mutations in RVD codon pairs.
Evolution by Recombination
As an alternative mode of TALE evolution, we considered putative recombination events between repeat arrays of different TALEs. To this end, we initially switched to a view of TALEs as sequences of their RVDs and searched for contiguous, identical RVD stretches in TALEs stemming from different AnnoTALE classes. TALEs in a class that are completely identical on the RVD level were collapsed to prevent combinatorial overrepresentation of such cases. In addition, we excluded cases where one TALE could be derived by deleting individual RVDs from another TALE, as this case is considered separately below.
We found a multitude of RVD stretches apparently duplicated between different classes. We also observed that a surprisingly large number of such duplicate RVD stretches started at the first repeat or extended to the last repeat of the repeat array. To assess the significance of this finding, we considered two null models (for details, see Methods). The first model assumes that the composition of RVDs in each TALE remains constant, but the order of these RVDs is random. We permuted all TALEs in a common AnnoTALE class consistently to preserve alignment similarities. Under this model, we found (see supplementary fig. 10A, Supplementary Material online) that duplicates of length 5 and above occurred with higher frequency in natural TALEs than in the permuted ones. Hence, we conclude that such duplicated RVD stretches occur more likely in natural TALEs than expected by chance. This finding seems reasonable under the assumption that TALEs have a common evolutionary origin and parts of the repeat array have either been preserved during evolution or have been exchanged between TALEs by recombination. The increase in frequency compared with the null model was greater in case of either terminal duplication. However, this might also be an effect of the length of the duplicates as longer duplicates have a greater chance than short ones to extend to the first or last repeat.
To further assess the latter observation, we considered a second null model, where the length of duplicates stayed constant, but their position within the repeat array was drawn uniformly from all possible positions (see supplementary fig. 10B, Supplementary Material online). Under this null model, the distribution of all duplicate lengths remains unchanged by conception. However, for the terminal duplicates, we found substantially larger frequencies than expected from the null model, whereas for central duplicates the observed frequencies were even lower than expected by chance. Hence, we conclude that duplicated RVD stretches are enriched at the N-terminal and C-terminal end of the repeat array, whereas they appear to be depleted in the central region. Evolutionary, this is compatible with a model, where one recombination breakpoint occurs within the repeat regions of two TALEs and the second recombination breakpoint occurs either within the N-terminal or C-terminal region or outside of the TALE gene. A list of putative recombination events is given in supplementary table 4, Supplementary Material online, while we discuss selected examples in the following.
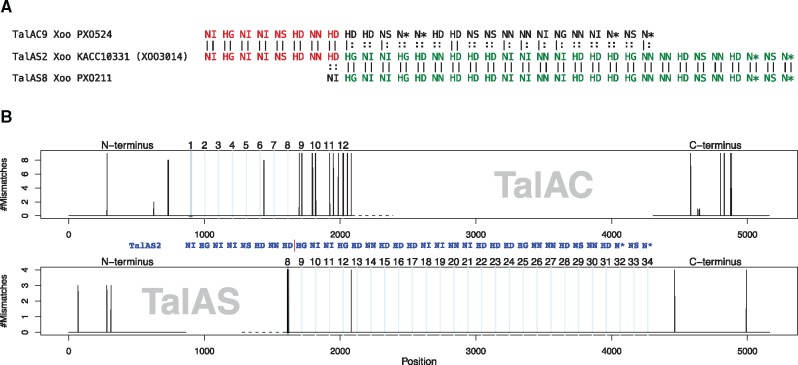
We compared specific examples of putative recombination events on the DNA level to assess the similarity of the corresponding subsequences. In figure 4, TalAS2 differs in its first eight RVDs from the other members of class TalAS, including TalAS8. This RVD sequence, “NI-HG-NI-NI-NS-HD-NN-HD”, also occurred in all class members of class TalAC. To clarify if the beginning of TalAS2 might have evolved by recombination, we compared the DNA sequence of TalAS2 with all members of class TalAC and four members of class TalAS with identical trailing RVD sequence. We found that the DNA sequence of TalAS2 is almost identical to all members of class TalAC in the N-terminus and the first eight repeats. Starting from repeat 9, the DNA sequences of TalAS and the TalAC members differed substantially. However, in this region TalAS2 was highly similar to the other members of class TalAS. Hence, we consider it highly probable that a recombination event has occurred between members of the classes TalAC and TalAS to yield the specific TALE TalAS2.
Fig. 4.
—(A) Alignment of TalAS2 with TalAS8 and TalAC9 as a representative members of their class. The first eight RVDs of TalAS2 match those of TalAC9 (red), whereas the remaining RVDs of TalAS2 are identical to those of other, shorter class TalAS members (green). (B) Visualization of the alignment of the DNA sequence of TalAS2 to those of nine members of class TalAC and four members of class TalAS. The top panel shows the number of mismatches between TalAS2 and members of class TalAC in the N-terminus, the first 12 repeats, and the C-terminus with 1bp resolution. The bottom panel shows the corresponding alignment of the N-terminus, repeats 8–34 and the C-terminus to shorter members of class TalAS. Powder-blue lines indicate position of RVDs.
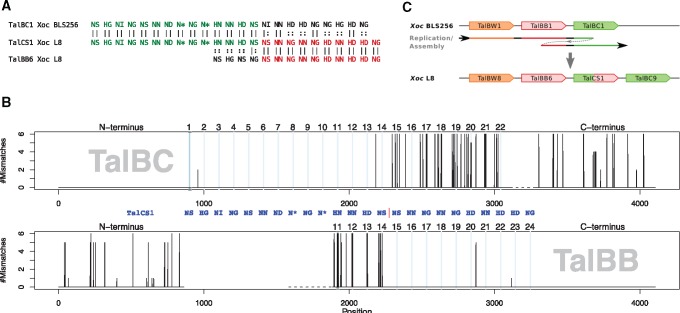
Another example where a recombination might have occurred is shown in figure 5. In this case, TalCS1 might be the result of a recombination of TALEs from two other classes. The first 14 RVDs of TalCS1 are identical to the beginning of six members of class TalBC, whereas the directly following 10 RVDs are identical to the end of six members of class BB. By comparing TalCS1 to the six members of class TalBC on DNA level, we found that the N-terminus and the first 14 repeats are highly similar. Two members show a different nucleotide in repeat 2 and all members have a different nucleotide in repeat 14, which might be the result of a point mutation in TalCS1 after the recombination event. The remaining sequence of TalCS1 and the members of TalBC show substantial differences. By comparing TalCS1 to the six members of class TalBB on DNA level, in turn, we find a very similar region beginning with repeat 15 and ending with an identical C-terminus. Five of the six members of TalBB have a different nucleotide at repeat 20 and one has a different nucleotide at repeat 23. One possibility is that the apparent sequence of TalCS1 is the result of a misassembly of the Xoc L8 genome, but the remaining differences in some of the repeats render this relatively unlikely.
Fig. 5.
—(A) Alignment of TalCS1 with TalBC1 as a representative member of shorter members of class TalBC and TalBB6 as a representative member of that class. The first 14 RVDs of TalCS1 match those of TalBC1, whereas the remaining RVDs of TalCS1 are identical to those of TalBB6. (B) Visualization of the alignment of the DNA sequence of TalCS1 to those of six members of class TalBC and six members of class TalBB. The top panel shows the number of mismatches between TalCS1 and members of class TalBC in the N-terminus, the first 22 repeats, and the C-terminus with 1bp resolution. The bottom panel shows the corresponding alignment of the N-terminus, repeats 11–24 and the C-terminus to members of class TalBB. Powder-blue lines indicate position of RVDs. (C) Putative model of the emergence of TalCS1.
An alternative explanation of the relationship between TalCS1 and class TalBB could be a deletion/insertion of repeats 5–14 in TalCS1, as four of the TalBB members (TalBB3, TalBB1, TalBB2, and TalBB7) also start with the RVD sequence “NS-HG-NI-NG”. However, aligning TalCS1 with deletion of repeats 5–14 to those members of class TalBB (see supplementary fig. 11, Supplementary Material online), we found a substantially larger number of substitutions in the N-terminal region and those first four repeats than in the alignment to members of class TalBC (cf. fig. 5). Hence, we consider it more likely that, indeed, TalCS1 evolved as a combination of TalBC and TalBB TALEs.
An intriguing possibility (fig. 5C), how TalCS1 could have emerged is a tandem duplication of the region containing the end of TalBB and the beginning of TalBC, for instance during replication.
Evolution of TALE Clusters
It has been proposed that TALE genes can also be part of a composite Tn3-related transposon (Ferreira et al. 2015). Matching IRs were found flanking to some TALE genes (Bonas et al. 1993; Noël et al. 2003; Ferreira et al. 2015), but TALE genes in Xoo are typically organized in clusters (Salzberg et al. 2008; Grau et al. 2016) of conserved genomic context, which suggests less mobility than for a typical transposon.
Here, we consider cluster II defined by flanking “tetratricopeptide repeat domain” and “RND superfamily” genes (Grau et al. 2016) present in all Asian Xoo strains studied (see supplementary fig. 12, Supplementary Material online). This cluster contains TalAS2 from Xoo KACC10331 (see above) as well as several other TALEs (TalDV1/2, TalBX1/2, TalAG1) that may be the result of recombination events. The spacer regions between TALEs in this cluster contain the known IRs (see supplementary fig. 12, Supplementary Material online, orange boxes) and also occurs in other TALE clusters of Xoo strains but not in those of Xoc strains.
In Xoc a different mechanism seems to be in place. Here, TALE genes are found more widely dispersed in the genome and TALE genes within some of the clusters are separated by very short (∼132bp) and highly conserved spacer regions. For instance, this applies to the cluster containing TalCS1 (see supplementary fig. 15, Supplementary Material online) and for the cluster containing TALEs from class TalBF in all Xoc strains studied (see supplementary fig. 13, Supplementary Material online). Notably, we also find such short spacer sequences in one cluster of the Xoo strain AXO1947 but not in any of the other Xoo strains studied.
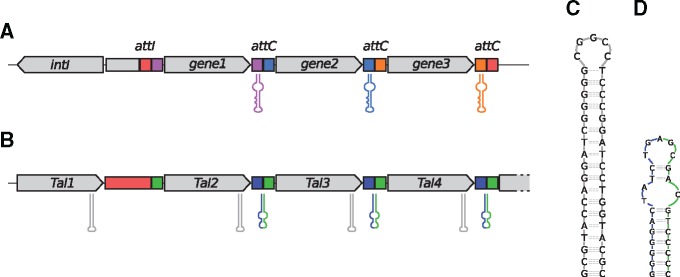
These short spacer regions do not contain the known IRs, although TALE genes appear to be also actively reshuffled in Xoc. The cluster containing TALEs from class TalBF is composed of different, apparently sequentially added TALE genes. This arrangement is reminiscent of integrons where individual cassettes are inserted in a sequential order into a genomic locus and separated by conserved sequences that may form hairpin structures (Nivina et al. 2016).
The general organization of the short spacers, which may be divided into two subregions (blue and green), is also somewhat similar to that of attI and attC regions in integrons. Specifically, we always find almost identical subsequences downstream (blue) and upstream (green) of TALE genes, where the latter also constitutes the 3′ end of the longer spacer region between the first two TALEs (fig. 6, cf. see supplementary figs. 13 and 14, Supplementary Alignment 1, Supplementary Material online).
Fig. 6.
—An integron-like mechanism may be responsible for the reassortment of TALE genes. (A) Organization of gene clusters formed by integrons, including the hairpin structures at attC sites, adapted from Nivina et al. (2016). (B) Schematic arrangement of TALE genes in clusters with short spacer regions in Xoc. Putative hairpin structures may be found at the C-terminal region of TALE genes (grey) as well as in the spacer regions (green/blue). Colors chosen in analogy to supplementary fig. 13, Supplementary Material online. (C) Secondary structure of the C-terminal hairpin as predicted by Mfold. (D) Secondary structure of the hairpin in the spacer region.
Notably, we also find palindromic sequences and, hence, putative hairpin structures at the C-terminal region of TALE genes and within the short spacer regions (fig. 6C and D). The border between the two subregions of the spacer is located in the terminal loop of this hairpin, which might indicate that this hairpin is indeed the breakpoint of TALE insertion/excision within the cluster.
Evolution by Deletion or Duplication of Individual Repeats
As a final process how TALEs may evolve, we considered the deletion or duplication of repeats from the repeat array. Deletions of multiple TALE repeats have been described before (Yang et al. 2005; Booher et al. 2015) and attributed to recombination events. Recently, duplication of single repeats from Ralstonia RipTALEs has been described and slipped strand mispairing has been proposed as the underlying mechanism (Schandry et al. 2016). Here, we focus on the deletion or duplication of individual repeats.
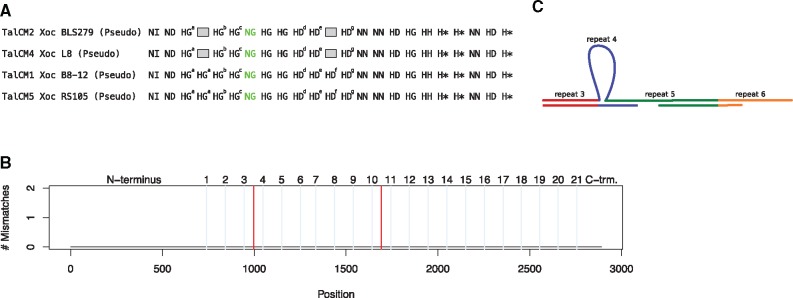
The first example we consider is TalCM1/TalCM5 aligned with TalCM2 and TalCM4 of the same class (fig. 7). In this case, the TALEs aligned without a single substitution if we removed repeats 4 and 12 from TalCM1 and TalCM5. Notably, this includes aberrant, short repeats (Richter et al. 2014) at position 7, which aligned as well. The absence of substitutions is even more remarkable, as these genes have an incomplete C-terminus and were, for this reason, classified as pseudo-genes by AnnoTALE. The strong conservation despite deletion events may be due to the function of these TALEs as interfering TALEs that play a role in suppression of plant resistance (Read et al. 2016; Ji et al. 2016).
Fig. 7.
—(A) The RVD sequence of TalCM2/TalCM4 is identical to TalCM1/TalCM5 with repeats 4 (HG) and 12 (HD) deleted. Small superscript letters at RVDs indicate unique repeat sequences. Aberrant, short repeats highlighted in green. (B) The DNA sequence of TalCM1/TalCM5 with repeats 4 and 12 deleted (red lines) is completely identical to that of TalCM2/TalCM4. Powder-blue vertical lines indicate position of RVDs in repeats. (C) Model of the deletion of a repeat during replication. In single-stranded DNA, repeat 4 loops out and repeat 5 binds to the already synthesized fragment on the lagging strand.
Because repeats 3 and 4 of TalCM1/TalCM5 are identical on the DNA level (see supplementary fig. 16, Supplementary Material online), the missing repeat 4 in TalCM2 and TalCM4 compared with TalCM1/TalCM5 could either be explained by a deletion of repeat 4 from TalCM1/TalCM5 or, conversely, by a duplication of repeat 3 of TalCM2/TalCM4. Both explanations appear to be equally valid. Regarding the missing repeat 12, however, we found differences between the DNA sequences of all HD repeats from repeat 10 to 13 in TalCM1/TalCM5, which renders the duplication of one of these HD repeats less likely. In contrast, this observation would still be compatible with a deletion of repeat 12 from TalCM1/TalCM5, yielding TalCM2/TalCM4.
We found several further examples of possible deletions of single repeats in the evolution of TALEs. TalDS1 may have its origin in members of class TalAC by a deletion of repeat 7 as we found only four positions with substitutions in the N-terminal and C-terminal regions in an alignment of the remaining repeats (see supplementary fig. 17, Supplementary Material online). Similarly, we found substitutions only at three positions between TalAI2 and TalDR1/TalDR2 with repeat 13 deleted (see supplementary fig. 18, Supplementary Material online), substitutions only at one position between TalDT1 and TalAR3/TalAR4/TalAR6 with repeat 9 deleted (see supplementary fig. 19, Supplementary Material online), and no substitutions between TalDU1 and TalAQ5/TalAQ11/TalAQ9 with repeat 13 deleted (see supplementary fig. 20, Supplementary Material online). In all these cases, the missing repeat (from the perspective of the longer TALE(s)) and its neighboring repeats harbor different RVDs. Hence, we consider deletion events more likely than duplication events in all four cases.
Such deletions might happen during replication. Single stranded DNA of the template lagging strand may form loops such that a conserved part of a repeat forms pairs with the corresponding region in the following repeat (cf. fig. 7C).
Impact of Evolutionary Events on Target Gene Activation in Xoc
The relationship between TALEs and their target genes may be established using a combination of experimental expression data and computational predictions of putative target sites in the promoters of these genes (Grau et al. 2013; Cernadas et al. 2014; Pérez-Quintero et al. 2013). While predicted target sites may help to distinguish direct from indirect expressional responses, expression data may help to identify putative false positive predictions of target sites. With the large number of sequenced Xoc genomes associated with expression data of plants infected with these strains, such analyses may be raised to a new level by leveraging the absence/presence pattern of specific TALE classes in the strains studied (Wilkins et al. 2015). TALE repertoires differ substantially between Xoo and Xoc strains, but also among different Xoc strains (see supplementary fig. 21, Supplementary Material online). This should also be reflected by a transcriptional response of the host plant that is congruent with this presence/absence pattern of TALEs.
The present study goes beyond previous approaches in extending this general schema to quantitative comparisons. Evolutionary modifications of different TALEs from the same class could lead to different levels of activation of target genes, or even the loss of functional activation. How well different variants of a TALE match a putative target site may be captured by the corresponding scores from computational target site predictions. In turn, differences in expressional response are reflected by the expression values and fold changes determined from experimental data. Here, we correlated both quantitive measures to yield further evidence of direct TALE targets and to gain insights into the impact of evolutionary events on target gene activation.
To study the impact of TALE evolution on the activation of putative target genes, we considered RNA-seq expression data of rice plants infected with 10 Xoc strains and mock inoculation as control (Wilkins et al. 2015). We analyzed these data using kallisto (Bray et al. 2016) and sleuth (Pimentel et al. 2016) to obtain Benjamini–Hochberg-corrected P-values and log2-fold changes for all annotated rice genes and each Xoc strain compared with the control. In the following, we considered rice genes as putative targets that, for at least one of the Xoc strains, 1) had an expression value (kallisto normalized TPM) of at least 35, 2) passed a significance level of , 3) showed a log2-fold change of at least 2, and 4) had a target box among the top 100 predictions of TALgetter (Grau et al. 2013) in their promoter for at least one TALE of the corresponding strain. A complete list of TALE classes and the corresponding predicted target genes is given in supplementary table 5, Supplementary Material online. For the following analyses, the observed effects on gene expression need to be linked to a specific TALE class. Hence, we additionally filtered candidate genes for those that are predicted targets of exactly one TALE class.
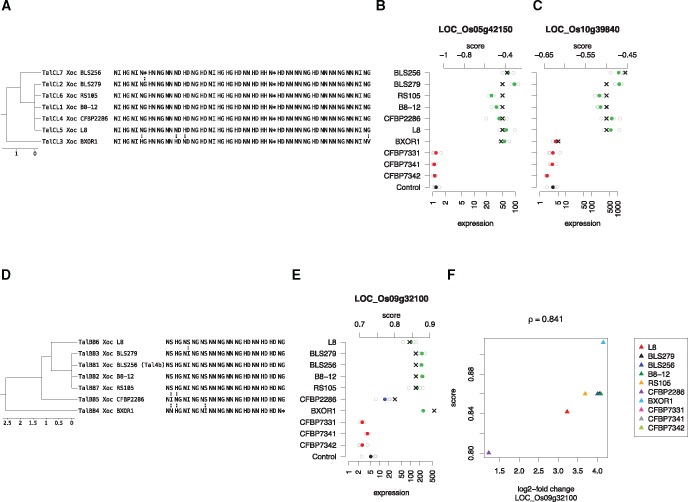
TALEs from class TalCL are present in 7 of the 10 Xoc strains studied. Here, we found two novel putative target genes with an activation pattern compatible with the absence and presence of TALEs (fig. 8A–C). Gene Os05g42150 (probable indole-3-acetic acid-amido synthetase) was clearly activated in those seven strains that posses TALEs from class TalCL, namely BLS256, BSL279, RS105, B8-12, CFBP2286, L8, and BXOR1, although with slightly different expression levels. The corresponding normalized prediction scores varied between -0.38 and -0.44. In contrast, the prediction scores for gene Os10g39840 (glycosyl hydrolase) varied between -0.45 and -0.50 for six of the strains, whereas the prediction score for TalCL3 from BXOR1 was substantially lower (-0.62). In the expression data, we found that this target gene was not activated for BXOR1 but only for the six remaining strains, likely because the target box in the promoter of Os05g42150 (TATAAATACCGCCTTCACCTCGCTCGCTGTC) is less affected by the four RVD swaps in TalCL3 than the target box in the promoter of Os10g39840 (TATAAATGACTCACTCCAACAAGTGAGAGAT). In the latter case, prediction scores were also highly correlated () with the corresponding log2-fold changes (see supplementary fig. 22, Supplementary Material online).
Fig. 8.
—Impact of variation in TALEs on expressional activation. (A) Alignment of TALEs in class TalCL. (B) Expression of LOC_Os05g42150 after infection with different Xoc strains compared with mock control. Filled circles represent mean expression and open circles expression values in individual experiments. Color indicates log2-fold change (red: , blue: > 1 and , green: > 2). The corresponding prediction scores from TALgetter (scale at top) are represented by black crosses. The pattern of expressional activation resembles the presence/absence of TalCL in these strains. (C) Expression of LOC_Os10g39840 after infection with different Xoc strains compared with mock control. For BXOR1, we do not find activation of LOC_Os10g39840 and also a low prediction score for the corresponding target site. (D) Alignment of TALEs in class TalBB. (E) Expression of LOC_Os09g32100 after infection with different Xoc strains compared with mock control in analogy to panel B. (F) Correlation of TALgetter prediction scores for the putative target site in the promoter of LOC_Os09g32100 to the log2-fold changes of this gene after infection with different Xoc strains compared with mock control.
Another example of a clear correlation () between prediction scores and corresponding log2-fold changes is Os04g49194 (naringenin,2-oxoglutarate 3-dioxygenase; Cernadas et al. 2014) for members of class TalBL (see supplementary fig. 23, Supplementary Material online). In this case, class members were present for all studies strains but RS105, for which Os04g49194 showed an expression level comparable to the control case. For the other class members, we found slightly lower expression levels after infection with B8-12, BLS279, and L8, where the corresponding TALEs (TalBL3, TalBL4, and TalBL9) are identical on the RVD level and also yielded slightly lower prediction scores. We also found a clear correlation between prediction scores and expressional response for class TalAX (see supplementary fig. 24, Supplementary Material online) and the novel putative target gene Os02g02190 (transporter, major facilitator family), as well as previously predicted target genes Os02g34970 (no apical meristem protein) and Os07g36430 (expressed protein) (Cernadas et al. 2014).
The TALEs of class TalAV (see supplementary fig. 25, Supplementary Material online) form two groups of TALEs, differing only in the last RVD. These two groups were also represented by the transcriptional response of the novel putative target gene Os02g51110 (aquaporin protein), which suggests that single RVD swaps at the last repeat may have a decisive influence on transcriptional activation. A similar pattern could be found for class TalBN (see supplementary fig. 26, Supplementary Material online) and Os09g21380 (expressed protein), where only TalBN2 from Xoc CFBP7342, which differs from the remaining TALEs in the last RVD, did not result in a clear transcriptional activation. For the long-known target Os07g06970 (OsHEN1) of class TalAK, we found transcriptional activation only in those six Xoc strains harboring TalAK TALEs (see supplementary fig. 27, Supplementary Material online).
Finally, we found several RVD swaps between the members of class TalBB (fig. 8D–F), which resulted in varying prediction scores for the putative target box TGTATAGTATCCCCT in the promoter of Os09g32100 (expressed protein), which has been predicted previously (Cernadas et al. 2014). Again, the prediction scores of the putative target box correlated well with the corresponding log2-fold changes (). In this case, the RVD swap in the last repeat of TalBB4 (Xoc BXOR1) had a slightly positive influence on transcriptional activation, whereas the RVD swap at the first position of TalBB5 (Xoc CFBP2286) lead to a reduced transcriptional response.
Discussion
Plant–pathogenic Xanthomonas bacteria are responsible for substantial yield losses of many important crop plants including rice. TALEs are secreted by the bacteria into plant cells where they act as transcriptional activators and, hence, are major drivers in reprogramming the plant cell transcriptome for the benefit of the pathogen. Understanding the different mechanisms of TALE evolution is pivotal for developing successful strategies to breed or design stably pathogen-resistant crop plants. In this paper, we systematically studied different aspects of TALE evolution. Key findings of this study and their consequences for plant breending are listed in table 2 and discussed in the following.
Table 2.
Summary of the Evolutionary Events Considered in This Study and Their Consequence for Breeding Resistant Plants
| Evolutionary Event | Present in | Consequence for Plant Breeding |
|---|---|---|
| Point mutations | Xoo, Xoc | Resistance by alleles with SNPs may be overcome |
| Recombination of TALE repeats | Xoo, Xoc | Resistance by longer modifications, for example, by genome editing, might be overcome if new site is matching terminal RVD stretches of different TALEs |
| Deletion/duplication of individual TALE repeats | Xoo, Xoc | Resistance by deletions or insertion of single nucleotides in alleles may be overcome |
| Reassortment of TALEs by Tn3-like transposases | Xoo | Transposition may lead to recombination (see above) |
| Reassortment of TALEs by integron-like mechanism | Xoc, (Xoo AXO1947) |
First, we found that the codon pairs coding for specific RVDs were remarkably conserved, while the most obvious explanations—namely codon stability, RNA structure, or general codon frequency—appeared to be unlikely in our analyses. Selective pressure might also stem from differing accuracy of tRNAs with respect to loading with AAs and recognition accuracy of anticodons (Zhang et al. 2015; Bullwinkle and Ibba 2016), but no data on the accuracy aminoacyl tRNA synthetases in Xanthomonas are currently available.
Second, we checked for potential evidence of TALE evolution by base substitutions in RVD codon pairs. Here, we find clear indications of base substitutions, which are especially pronounced for modifications of a single base in the RVD codon pair, where it is 4-fold as likely that a modification of an RVD occurred by a base substitution than by a recombination event. Considering a network of possible RVD conversions by single or double base mutations, we also observe that the majority of RVD modifications by base substitution may be traced to two specific cases, namely, the conversion of NS to NI repeats and the conversion of NG to HG repeats, which both have overlapping target specificities.
Third, we considered the alternative mechanism of TALE evolution by recombination of existing TALEs. Here, we find that duplicated RVD stretches are significantly enriched at the N-terminus and C-terminus of the repeat array. Scrutinizing such cases in alignments on the DNA level, we found base substitutions in the shared repeats and their adjacent N- or C-terminal sequence with surprisingly low frequency. Hence, our results provide strong evidence that these shared parts of the TALEs indeed have a common origin and have been recombined.
Type III effectors can be found as part of transposons and active translocation has been demonstrated for some of those (Landgraf et al. 2006) which increases their capacity for horizontal gene transfer and recombination. One possibility, besides the transposition mechanism proposed before (Ferreira et al. 2015), how this can lead to TALE reassortment is that one DNA strand is cleaved by the action of a transposase or recombinase resulting in the displacement of a ssDNA 3′-OH strand encoding either the N- or C-terminal region of a TALE gene, which can be used for a nucleophile attack at homologous sequences to form holiday junctions which are resolved at some distance, for example, within the repeat region. This mechanism should result in recombination products with partially replaced TALE genes. If the initial DNA cleavage occurs flanking to the TALE gene, it can be expected that predominantly either the N- or C-terminal domains and the repeats immediately bordering these domains are replaced which is exactly what can be observed in Xoo (cf. see supplementary fig. 12, Supplementary Material online).
Considering the organization of TALE clusters in Xoc, we propose that reassortment of TALEs in Xoc is mediated by a different mechanism than in Xoo, and shows similarities to integrons. Specifically, we found putative hairpin structures at the C-terminal region of TALE genes and within short spacer regions between TALE genes, which might be the breakpoint of TALE insertion/excision within the cluster. In general, hairpins at the 3′ end of ORFs might also function as Rho-independent termination structures, but the required 3′ poly-T stretch is not present downstream of these hairpin structures. For this reason, we consider it highly possible that single TALE genes together with parts of the spacer region form gene cassettes that have inserted into a TALE array in Xoc in an analogous fashion to integrons, possibly involving a site-specific recombinase that mediates excision and integration at precise sequences.
We also identified several TALEs that have likely evolved from other TALEs by deletion of individual repeats. It has been proposed previously that deletions of repeats could occur during transposition (Ferreira et al. 2015). However, the TALEs from class TalCM, for instance, are located in the same cluster (also containing TALEs from class TalBF) in different Xoc strains, which renders a transposition event unlikely in this case (cf. see supplementary fig. 13, Supplementary Material online). We rather propose that deletion of repeats may happen during replication if the single stranded DNA of one repeat loops out and conserved parts pair with the corresponding region in the following repeat (cf. fig. 7C). Repetitive genomic DNA sequences are typically highly dynamic involving recombination and deletions. In particular, the repeat region of TALEs is well known to hamper PCR amplification possibly due to mispriming of truncated amplification products or template switching resulting in TALEs with variable numbers of repeats (Hommelsheim et al. 2014). The situation in vivo could be related, for example, during replication. The average length of an Okazaki fragment in Escherichia coli is 1–2kb (Bzymek and Lovett 2001) which should be sufficient to allow looping of a 102 nucleotide single TALE repeat.
Finally, we investigate the impact of evolutionary events on transcriptional activation of putative target genes. In several cases, the observed differences in transcriptional activation correlate well with prediction scores for the corresponding target box. This approach increases the prediction accuracy for target genes, which is pivotal for deciphering the impact of TALEs on virulence. Consequently, our predictions include not only some known target genes of Xoc TALEs (Moscou and Bogdanove 2009; Cernadas et al. 2014; Wilkins et al. 2015) but also several novel target genes as listed in supplementary table 5, Supplementary Material online. In summary, our finding suggest that 1) even single RVD swaps, which might be the result of individual point mutations, may have a decisive influence on transcriptional activation, 2) the direction and strength of this influence depends on the target box and, hence, the effect of a RVD swap cannot be anticipated from the RVD sequence alone, and 3) scores from computational target box predictions may give a valid indication of differential transcriptional responses that can be expected in infection experiments.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
We thank Stefan Posch and Cornelius Schmidtke for valuable discussions. This work was supported by grants from the Deutsche Forschungsgemeinschaft (BO 1496/8-1 to J.B. and GR 4587/1-1 to J.G.) and by the COST action FA1208 “SUSTAIN”.
Literature Cited
- Boch J, et al. 2009. Breaking the code of DNA binding specificity of TAL-type III effectors. Science 326(5959):1509–1512. [DOI] [PubMed] [Google Scholar]
- Bonas U, Conrads-Strauch J, Balbo I.. 1993. Resistance in tomato to Xanthomonas campestris pv. vesicatoria is determined by alleles of the pepper-specific avirulence gene avrBs3. Mol Gen Genet. 238(1):261–269. [DOI] [PubMed] [Google Scholar]
- Booher NJ, et al. 2015. Single molecule real-time sequencing of Xanthomonas oryzae genomes reveals a dynamic structure and complex TAL (transcription activator-like) effector gene relationships. Microb Genom. 1(4). doi:10.1099/mgen.0.000032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray NL, Pimentel H, Melsted P, Pachter L.. 2016. Near-optimal probabilistic RNA-seq quantification. Nat Biotechnol. 34(5):525–527. [DOI] [PubMed] [Google Scholar]
- Bullwinkle TJ, Ibba M.. 2016. Translation quality control is critical for bacterial responses to amino acid stress. Proc Natl Acad Sci U S A. 113(8):2252–2257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bzymek M, Lovett ST.. 2001. Instability of repetitive DNA sequences: the role of replication in multiple mechanisms. Proc Natl Acad Sci U S A. 98(15):8319–8325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cernadas RA, et al. 2014. Code-assisted discovery of TAL effector targets in bacterial leaf streak of rice reveals contrast with bacterial blight and a novel susceptibility gene. PLOS Pathog. 10(4):e1004126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira RM, et al. 2015. A TALE of transposition: Tn3-like transposons play a major role in the spread of pathogenicity determinants of Xanthomonas citri and other Xanthomonads. mBio 6(1):e02505–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez C, et al. 2007. Molecular and pathotypic characterization of new Xanthomonas oryzae strains from West Africa. Mol Plant Microbe Interact. 20(5):534–546. [DOI] [PubMed] [Google Scholar]
- Grau J, et al. 2012. Jstacs: a Java framework for statistical analysis and classification of biological sequences. J Mach Learn Res. 13:(Jun):1967–1971. [Google Scholar]
- Grau J, et al. 2013. Computational predictions provide insights into the biology of TAL effector target sites. PLoS Comput Biol. 9(3):e1002962.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grau J, et al. 2016. AnnoTALE: bioinformatics tools for identification, annotation, and nomenclature of TALEs from Xanthomonas genomic sequences. Sci Rep. 6:21077.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hommelsheim CM, Frantzeskakis L, Huang M, Ülker B.. 2014. PCR amplification of repetitive DNA: a limitation to genome editing technologies and many other applications. Sci Rep. 4:5052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutin M, Pérez-Quintero AL, Lopez C, Szurek B.. 2015. MorTAL Kombat: the story of defense against TAL effectors through loss-of-susceptibility. Front Plant Sci. 6:535.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaenicke S, et al. 2016. Complete genome sequence of the barley pathogen Xanthomonas translucens pv. translucens DSM 18974T (ATCC 19319T). Genome Announc. 4(6):e01334–e01316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Z, et al. 2016. Interfering TAL effectors of Xanthomonas oryzae neutralize R-gene-mediated plant disease resistance. Nat Commun. 7:13435.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawahara Y, et al. 2013. Improvement of the Oryza sativa Nipponbare reference genome using next generation sequence and optical map data. Rice 6(1):4.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landgraf A, Weingart H, Tsiamis G, Boch J.. 2006. Different versions of Pseudomonas syringae pv. tomato DC3000 exist due to the activity of an effector transposon. Mol Plant Pathol. 7(5):355–364. [DOI] [PubMed] [Google Scholar]
- Lassmann T, Sonnhammer EL.. 2005. Kalign – an accurate and fast multiple sequence alignment algorithm. BMC Bioinformatics. 6(1):298.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz R, et al. 2011. ViennaRNA package 2.0. Algorithms Mol Biol. 6(1):26.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moscou MJ, Bogdanove AJ.. 2009. A simple cipher governs DNA recognition by TAL effectors. Science 326(5959):1501.. [DOI] [PubMed] [Google Scholar]
- Nettling M, et al. 2015. DiffLogo: a comparative visualization of sequence motifs. BMC Bioinformatics. 16(1):387.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nivina A, Escudero JA, Vit C, Mazel D, Loot C.. 2016. Efficiency of integron cassette insertion in correct orientation is ensured by the interplay of the three unpaired features of attC recombination sites. Nucleic Acids Res. 44(16):7792–7803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noël L, Thieme F, Gäbler J, Bättner D, Bonas U.. 2003. XopC and XopJ, two novel type III effector proteins from Xanthomonas campestris pv. vesicatoria. J Bacteriol. 185(24):7092–7102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Quintero AL, et al. 2013. An improved method for TAL effectors DNA-binding sites prediction reveals functional convergence in TAL repertoires of Xanthomonas oryzae strains. PLoS ONE. 8(7):1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Quintero AL, et al. 2015. QueTAL: a suite of tools to classify and compare TAL effectors functionally and phylogenetically. Front Plant Sci. 6:545.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pimentel H, Bray NL, Puente S, Melsted P, Pachter L.. 2017. Differential analysis of RNA-seq incorporating quantification uncertainty. Nat Meth. Advance online publication. doi: 10.1038/nmeth.4324. [DOI] [PubMed]
- Quibod IL, et al. 2016. Effector diversification contributes to Xanthomonas oryzae pv. oryzae phenotypic adaptation in a semi-isolated environment. Sci Rep. 6:34137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Read AC, et al. 2016. Suppression of Xo1-mediated disease resistance in rice by a truncated, non-DNA-binding TAL effector of Xanthomonas oryzae. Front Plant Sci. 7:1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter A, et al. 2014. A TAL effector repeat architecture for frameshift binding. Nat Commun. 5:3447.. [DOI] [PubMed] [Google Scholar]
- Salzberg S, et al. 2008. Genome sequence and rapid evolution of the rice pathogen Xanthomonas oryzae pv. oryzae PXO99A. BMC Genomics. 9(1):204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schandry N, de Lange O, Prior P, Lahaye T.. 2016. TALE-like effectors are an ancestral feature of the Ralstonia solanacearum species complex and converge in DNA targeting specificity. Front Plant Sci. 7:1225.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievers F, et al. 2011. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 7:539.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streubel J, et al. 2013. Five phylogenetically close rice SWEET genes confer TAL effector-mediated susceptibility to Xanthomonas oryzae pv. oryzae. New Phytol. 200(3):808–819. [DOI] [PubMed] [Google Scholar]
- Triplett LR, et al. 2011. Genomic analysis of Xanthomonas oryzae isolates from rice grown in the United States reveals substantial divergence from known X. oryzae pathovars. Appl Environ Microbiol. 77(12):3930–3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warburton PE, Giordano J, Cheung F, Gelfand Y, Benson G.. 2004. Inverted repeat structure of the human genome: the X-chromosome contains a preponderance of large, highly homologous inverted repeats that contain testes genes. Genome Res. 14(10a):1861–1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkins K, Booher N, Wang L, Bogdanove A.. 2015. TAL effectors and activation of predicted host targets distinguish Asian from African strains of the rice pathogen Xanthomonas oryzae pv. oryzicola while strict conservation suggests universal importance of five TAL effectors. Front Plant Sci. 6:536.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang B, Sugio A, White FF.. 2005. Avoidance of host recognition by alterations in the repetitive and C-terminal regions of AvrXa7, a type III effector of Xanthomonas oryzae pv. oryzae. Mol Plant Microbe Interact. 18(2):142–149. [DOI] [PubMed] [Google Scholar]
- Zhang J, Ieong KW, Johansson M, Ehrenberg M.. 2015. Accuracy of initial codon selection by aminoacyl-tRNAs on the mRNA-programmed bacterial ribosome. Proc Natl Acad Sci U S A. 112(31):9602–9607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31(13):3406–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.