Summary
N-glycosylation has a great impact on glycoprotein structure, conformation, stability, solubility, immunogenicity and enzyme activity. Structural characterization of N-glycoproteome has been challenging but can provide insights into the extent of protein folding and surface topology. We describe a highly sensitive proteomics method for large-scale identification and quantification of glycoproteins in Arabidopsis through 15N-metabolic labeling, selective enrichment of glycopeptides, data-dependent MS/MS analysis and automated database searching.
In-house databases of Arabidopsis glycoproteins and glycopeptides containing Asn-X-Ser/Thr/Cys motifs were constructed by reducing 20% and 90% of the public database size, respectively, to enable a rapid analysis of large datasets for comprehensive identification and quantification of glycoproteins and heterogeneous N-glycans in a complex mixture.
Proteome-wide analysis identified c. 100 stress-related N-glycoproteins, of which the endoplasmic reticulum (ER) resident proteins were examined to be up-regulated. Quantitative measurements provided a molecular signature specific to glycoproteins for determining the degree of plant stress at low temperature.
Structural N-glycoproteomics following time-course cold treatments revealed the stress-responsive degradation of high-mannose type N-glycans in ER in response to chilling stress, which may aid in elucidating the cellular mechanisms of protein relocation, transport, trafficking, misfolding and degradation under stress conditions.
Keywords: Arabidopsis, chilling stress, endoplasmic reticulum (ER), glycoproteomics, liquid chromatography-tandem mass spectrometry
Introduction
Degradation of misfolded proteins, through polyubiquitination and degradation in the ubiquitin–proteasome system, is one of the most important plant responses to mitigate the damage caused by accumulation of misfolded proteins under environmental stresses such as cold, heat, drought, salinity, and infections by bacteria and viruses. Proteins fail to fold correctly or function properly as a result of the abnormality of glycan structures or the absence of glycosylation. Glycosylation regulates protein folding, stability, solubility, biogenesis and enzymatic activity, but also prevents proteins from proteolytic degradation. In plants, N-glycosylation of proteins also plays a functional role to regulate endoplasmic reticulum quality control (ERQC), stress tolerance and signal transduction (Ceriotti et al., 1998; von Schaewen et al., 2008; Russell et al., 2009). However, the structural relationship of glycan isoforms with various environmental stresses and developmental biology of plants remains poorly understood. The characterization of N-glycoproteome is of particular importance for understanding the N-glycan distribution, structural topology, cellular location, and molecular adaptation in plant stress responses.
Biosynthesis of plant N-glycans commences with the initial transfer of a dolichol lipid-linked oligosaccharide precursor (i.e., Glc3Man9GlcNAc2) to the asparagine residues localizing at a structural motif Asn-X-Ser/Thr/Cys (to a lesser extent at Cys, X is any amino acid except proline) of nascent proteins in the ER by oligosaccharyltransferase (OST). The terminal glucoses and mannoses are then sequentially trimmed by the ER glucosidases and mannosidases (MNSs) to generate a high-mannose type of N-glycans (Man5–9GlcNAc2) (Supporting Information Fig. S1) (Rayon et al., 1998; Pattison & Amtmann, 2009). ERQC, through binding of N-glycoproteins to the ER chaperones by calnexin (CNX) and calreticulin (CRT), recognizes unfolded or misfolded protein intermediates to be retained in the ER for another cycle refolding. If this fails, the accumulated misfolding proteins are subsequently targeted and translocated for ER-associated degradation (ERAD). Correctly folded glycoproteins are transported to the Golgi, plasma membrane and other subcellular compartments followed by addition of N-acetylglucosamine, fucose and xylose by N-acetylglucosaminyltransferase (GNT1), fucosyltransferase (α1,3 FucT) and xylosyltransferase (β1,2 XylT) to form complex-, hybrid- and paucimannose-type N-glycans in mature proteins (Rayon et al., 1998; Pattison & Amtmann, 2009). As such, N-glycan processing leads to ER-resident glycoproteins having only the high-mannose type N-glycan attachment (e.g. Man8–9GlcNAc2), whereas the glycoproteins localized in the Golgi apparatus and vacuole have hybrid-, complex-type or paucimannosidic N-glycans on different glycosylation sites. Thus, the presence of distinct glycan modifications on a particular protein of interest could be indicative of the protein at different subcellular locations, whereas variable glycan structures often represent the folding state of glycoproteins in the ER and other cellular environments. Relative abundances of N-glycan isoforms of a protein rely heavily on the protein surface accessibility and the intermolecular interaction with the processing enzymes and glycosyltransferases (Rayon et al., 1998).
In Arabidopsis, c. 80% of the gene-encoded proteins are predicted to possess potential N-glycosylation sites based on the Asn-X-Ser/Thr/Cys motif. Previous findings showed that tunicamycin, as a glycan inhibitor, could induce an unfolded protein response (UPR) of the N-glycan-deficient protein substrates, resulting in growth inhibition of the treated plants (Koizumi et al., 1999). To investigate the physiological significance of different types of N-glycans, several plant mutants of triple knockout MNSs (mns 123), N-acetylglucosaminyltransferase (also termed the complex glycan 1, cgl1), GDP-D-mannose-4,6-dehydratase fucose (mur1) and staurosporin-temperature-sensitive 3-like enzymes (stt3a/b), had been utilized to control the biosynthesis of N-glycans in Arabidopsis. The mns123 mutant produced only the high-mannose N-glycan of Man9GlcNAc2 on all glycoproteins, and had a phenotype of short root length and small size (Liebminger et al., 2009; Hüttner et al., 2014). Likewise, the cgl1 mutant accumulated Man5GlcNAc2 but no complex glycans on glycoproteins owing to the lack of GnT-I, and displayed a dwarf phenotype and salt-sensitive root growth in rice (Fanata et al., 2013). A study by Kang et al. (2008) also reported that the plasma membrane protein, KOR1/RSW2, required N-glycans for its proper function in vivo. In the mur 1 mutant, the plant lacked the GDP-fucose synthesis in glycoproteins but retained a normal phenotype assuming the phenomenon was caused by the galactose replacement of fucose at the side chain of N-linked glycan branches (Bonin et al., 1997; van Hengel & Roberts, 2002). The plant, in the absence of the STT3a gene, displayed a salt-sensitive phenotype, and the double knockout mutant of STT3a/b resulted in gametophytic lethality (Koiwa et al., 2003; Pattison & Amtmann, 2009). As a consequence, N-glycosylation functions in regulating plant growth and development through maintaining the proper structural folding of proteins, and the absence of N-glycan enzymes has been associated with the UPR to salt and osmotic stresses (Koiwa et al., 2003; Kang et al., 2008; von Schaewen et al., 2008; Hüttner et al., 2014).
Proteomic methods have been utilized for global characterization of N-glycoproteins, providing valuable information regarding the structure, location and relative abundance of N-glycan isoforms to unravel the secretory pathway related to plant stresses. Such an analysis has been enhanced by the emergence of advanced mass spectrometric technologies with highly improved sensitivity and accuracy, although a full examination of glycoproteome in plants remains a formidable technical challenge owing to the diversity of multiple protein isoforms; the heterogeneity of N-glycan isomeric structures occupying the same site of a protein; the huge variability of glycosylated, nonglycosylated and truncated proteins in dynamic expression; the low intensity of glycopeptide detection; and the complexity of the database search involving the number of protein isoforms and N-glycan structures (Marino et al., 2010; Song et al., 2011). Traditionally, glycoproteins are digested by an endoproteinase and the resulting glycopeptides are captured by a lectin column (e.g. ConA) or hydrophilic interaction liquid chromatography (HILIC). To increase the sensitivity of detection, the negatively charged glycan portion of glycopeptides is often removed by endoglycosidases such as PNGase F,PNGase A or Endo H, to generate completely or partially deglycosylated peptides for mass spectrometric analyses (Song et al., 2013). The technique is feasible to identify N-glycosylation sites of glycopeptides, but misses the structural information of N-glycan isoforms. A full characterization of glycoproteins requires analyses of intact glycopeptides to annotate both peptide sequences and the attached glycans at the side chains. Toward this goal, multistage mass spectrometry (MSn) has been used to identify the complex glycan structure under the low-energy collision-induced dissociation (CID, MS2) or the higher-energy collision-induced dissociation (HCD), followed by MS3 analysis of high-intensity monoglycosylated peptide precursor ions. The method achieved huge success in the analysis of purified glycoproteins with sufficient starting materials; however, the proteomic application is rather limited owing to the low sensitivity of MS3 acquisition. Alternative approaches using either electron-capture dissociation or electron-transfer dissociation for analyzing intact glycopeptides are promising, but the techniques suffer from a low fragmentation efficiency which requires multiply charged precursor ions of glycopeptides to generate high-density fragments for glycopeptide sequencing. Here we describe a highly sensitive proteomics method for high-throughput structural identification of glycoproteins in Arabidopsis through selective enrichment of glycopeptides, data-dependent ultraperformance liquid chromatography (LC)–Orbitrap MS/MS analysis and automated database searching. The method took advantage of the high-sensitivity detection of intact glycopeptides by elucidating the glycan structure with CID and complementary peptide sequencing with HCD in parallel experiments. We also used 15N-metabolic labeling to quantitatively compare the differential expression of glycoproteins during time courses of plant growth under normal and cold-stressed conditions. The study was aimed at establishing a rapid, straightforward method for large-scale structural analyses of glycoproteins in plants, and discovering the functional role of aberrant N-glycans associated with UPR to cold stress.
Materials and Methods
Plant materials
Arabidopsis thaliana (L.) Heynh. seeds (Columbia-0) were surface-sterilized with 0.5% sodium hypochlorite solution and planted on half-strength Murashige and Skoog (½ MS) medium. After low-temperature induction (i.e. vernalization) for 2 d at 4°C, the seedlings were allowed to grow in a chamber at 22°C for 2 wk and subsequently relocated to the growth chamber at 4°C for another 6, 12 and 18 d. As a comparison, the seedlings were also continually grown at 22°C during the same period of time as control samples. The materials were collected, immediately frozen in liquid nitrogen and stored at −80°C until use.
In vivo 15N-isotope metabolic labeling of proteins in Arabidopsis
Metabolic labeling of proteins in Arabidopsis was performed according to the protocol reported previously (Skirycz et al., 2011). The 1/2 MS media were prepared to contain 14N and 15N isotopes. In the 15N-incorporated medium, the heavy labeling reagents of 15NH415NO3 and K15NO3 (Sigma) were used to replace those containing 14N-isotope. Arabidopsis seedlings were grown at 22°C under long-day conditions (16 : 8 h, light : dark) on 15N- or 14N-isotope-containing medium, respectively. After 2 wk, the seedlings grown on the 14N medium were transferred to a growth chamber at 4°C for 6, 12 and 18 d. As such, 15N-labeled plants at 22°C were used for control experiments (i.e. forward labeling), or the time-course cold-treated plants at 4°C were also metabolically labeled by 15N-isotopes in parallel experiments (i.e. reciprocal labeling) (Kline et al., 2010; Qing et al., 2016). The labeled and unlabeled seedlings were harvested separately, and a mixture with the same amount of 14N- and 15N-isotope-labeled tissues (w/w, 1:1) at the two different temperatures was utilized for relative quantification of proteins.
Protein extraction
Arabidopsis seedlings were ground under liquid nitrogen using a chilled mortar and pestle, and 3 g of the materials were suspended in a lysis buffer containing 4% sodium dodecyl sulfate (SDS), 100 mM Tris-HCl (pH 7.6) and 100 mM dithiothreitol (DTT). Following ultrasonication for 10 min and boiling for 3 min to reduce disulfide-binding linkage of proteins, the crude protein extract was harvested as supernatants after centrifugation at 2935 g for 20 min. Proteins were precipitated by 10% trichloroacetic acid-aceton at −20°C overnight, and subsequently washed with cold acetone three times to remove lipids and excess trichloroacetic acid.
Protein digestion using filter-aided sample preparation method
The protein pellet was dried and then solubilized in 500 μl of 8 M urea containing 50 mM iodoacetamide to block the free cysteines of proteins, and finally transferred into a spin column filter (10 kDa MWCO; Pall Life Sciences, Ann Arbor, MI, USA). After sequential washes with 8 M urea and buffer exchanges of 25 mM ammonium bicarbonate (NH4HCO3), the alkylated proteins were digested by sequencing-grade trypsin (Promega) in 25 mM NH4HCO3 at an enzyme: substrate ratio of 1: 200 (w/w) at 37°C in a shaking incubator for 18 h. The tryptic peptides were collected after centrifugation at 13 523 g, and freeze-dried using a refrigerated CentriVap concentrator (Labconco, Kansas, MO, USA).
Glycopeptide enrichment
Glycopeptides were enriched using an HILIC microcolumn that was prepacked with FLASH HILIC resins (20–45 μm, 100 Å; Bonna-Agela Technologies, Wilmington, DE, USA). The column was preconditioned with 3 ml of 0.1% trifluoroacetic acid (TFA) and equilibrated by 3 ml of 0.1% TFA/80% acetonitrile (ACN). The peptide digest was dissolved in 1 ml of 0.1% TFA/80% ACN, and then loaded onto the column. Nonglycosylated peptides were washed away with 3 ml of 0.1% TFA/80% ACN. The glycopeptides were eluted using 1.5 ml of 0.1% TFA/40% ACN, and subsequently dried using a refrigerated CentriVap concentrator.
LC-MS/MS analysis of intact glycopeptides
Samples were reconstituted in 10 μl of 0.1% formic acid (FA) and analyzed by online nanoAcquity ultraperformance LC (Waters, Milford, MA, USA) coupled with an Orbitrap Fusion Tribrid mass spectrometer (Thermo Fisher Scientific, Watham, MA, USA). Nanospary was controlled by a PicoView Nanospray Source (PV550; New Objective, Woburn, MA, USA) at a spray voltage of 1.9 kV. The peptides were trapped by a 2G-V/MT Trap symmetry C18 column (5 μm particles, 180 μm ID × 20 mm length) at the flow rate of 5 μl min−1 for 3 min, and separated on a BEH130 C18 analytical column (1.7 μm particles, 100 μm ID × 250 mm length) at 250 nl min−1. Peptides were eluted using mobile phases consisting of solvent A (0.1% FA) and solvent B (ACN/0.1% FA) through a linear gradient from 10% to 30%, and then 85% of solvent B at a duration of 60 or 120 min. Data-dependent MS/MS acquisition was performed following a full MS survey scan by Orbitrap at a resolution of 60 000 over the m/z range of 350–1800, and MS/MS measurements of the top 20 most intense precursor ions. The target values of automatic gain controls (AGC) were set up to 200 000 for Orbitrap MS and 10 000 for ion-trap MS/MS detection. The fragmentations of the selected multiply charged ions were achieved using helium gas and argon at a normalized collision energy of 35% for CID and HCD, respectively. Dynamic exclusion was enabled for 60 s.
Identification of glycoproteins and N-glycan structure by database search
An in-house N-glycoprotein database was constructed by refining amino acid sequences of the Arabidopsis proteins or peptides containing the Asn-X-Ser/Thr/Cyr motif (X, not proline) from The Arabidopsis Information Resource (TAIR10; downloaded on 6 January 2014; 35 386 sequences) or the National Center for Biotechnology Information (NCBI) nonredundant database (downloaded on 6 January 2014; 226 526 sequences) to exclude non N-glycosylated proteins and redundant peptides, where the generated glycopeptide sequences containing two missed trypsin cleavage sites away from the N-glycosylation motif in each putative glycoprotein were collected (Fig. S2). The MS raw data from the LC-MS/MS analyses of the biological duplicate and three technical repeats were separately converted to MGF files using the Proteome Discoverer 1.4 software (Thermo Fisher Scientific Inc.) and then searched against the in-house databases using Mascot Daemon 2.4 (Matrix Science, London, UK). The search parameter for tryptic digestion was restricted to a maximum of two missed cleavages of proteins. Cysteine carbamidomethylation was designated as a fixed modification. Deamidation of Asn or Gln, oxidation of Met, and a glycan library incorporating 161 common plant N-glycans (Fig. S3) were considered as variable modifications. Mass tolerances were set up to 20 ppm for the Orbitrap-MS ions, and 0.8 Da for ion-trap MS/MS fragment ions. Peptide assignments were filtered by an ion score cutoff of 20, and the significance threshold was adjusted to 0.001 to achieve a false discovery rate (FDR) of < 1%. The retrieved glycopeptide sequences and N-glycan structures were validated by manual inspection of the CID and HCD fragments, as well as the accurate masses that calculated using the predicted sequences. Gene ontology (GO) annotation and functional categorization were conducted using the website tool at https://www.arabidopsis.org/tools/bulk/go/index.jsp.
Protein quantitation and validation by Western blots
Mascot Distiller software (v.2.5.1.0, Matrix Sciences, London, UK) was utilized for the accurate quantification of proteins by 15N-metabolic labeling. The MS data were centroided at a peak half-width of 0.02 and 200 points Da−1. The maximum ion charge state was set to 6, and the Sum method was used as the scan group aggregation at the precursor mass range of 700–16 000 Da. MS/MS processing of the ion-trap data was centroided at a peak half-width of 0.2 and 20 points Da−1, and regridded with the same value. The MS peak picking was accomplished with 500 iterations, and filtered through a correlation threshold of 0.6, minimum signal-to-noise ratio of 2, and m/z range from 50 to 100 000. MS peak profile was determined at a minimum width of 0.002 Da and maximum peak width of 1 Da.
The identified peptides from the MS/MS search were then imported into Mascot Distiller to generate a quantification report of light: heavy (L: H) ratios. The output of L: H ratios was thus limited to the 15N-labeled peptides that were matched with the confident sequence identification and the identical charge state. Precursor ion protocol was used for peptide quantification, and the ratios were calculated using the peak areas of extracted ion chromatograms (XICs) based on the trapezium integration method. To improve the accuracy of the results, the impurity correction of 15N-labeled reagents (98% K15NO3 and 99% 15NH415NO3) was incorporated into the quantification method. The protein ratios were manually examined on the abundances of 14N- and 15N-containing peptides based on the valid sequence identification by Mascot search and calculation by the integrated XIC peak area or the intensity. Reliable quantification of individual proteins was achieved by the mean ± SD value of triplicate experiments on either the glycopeptides or minimum two nonglycopeptides free of methionine (/oxidized methionine).
The relative ratio of protein abundances was validated by western blots. The Arabidopsis proteins were extracted by a buffer containing 4% SDS, 100 mM DTT, and 100 mM Tris-HCl (pH 7.6), and then centrifuged for 20 min at 4°C. Protein concentrations were determined by Bradford assay (Bio-Rad); equal amounts of samples from the control and the cold-treated materials were separated in parallel by 7% SDS-polyacrylamide gel electrophoresis (PAGE), and transferred onto a polyvinylidene flouride membrane on a transfer apparatus at 250 mA for 1.5 h. The membrane was subsequently blocked with 5% nonfat milk and incubated with primary antibodies against maize-CRTs (Pagny et al., 2000), protein disulfide isomerase-like protein (PDI; Rose Biotechnology, MA, USA), and actin (CWBIO, Beijing, China) as described previously (Hong et al., 2008).
The in-house databases of the modified glycoproteins and glycopeptides are publicly accessible at the website of the Shanghai Center for Plant Stress Biology, Chinese Academy of Sciences (http://www.psc.ac.cn/facility.asp#4).
Results
High-throughput analysis of N-glycosylated proteins in Arabidopsis
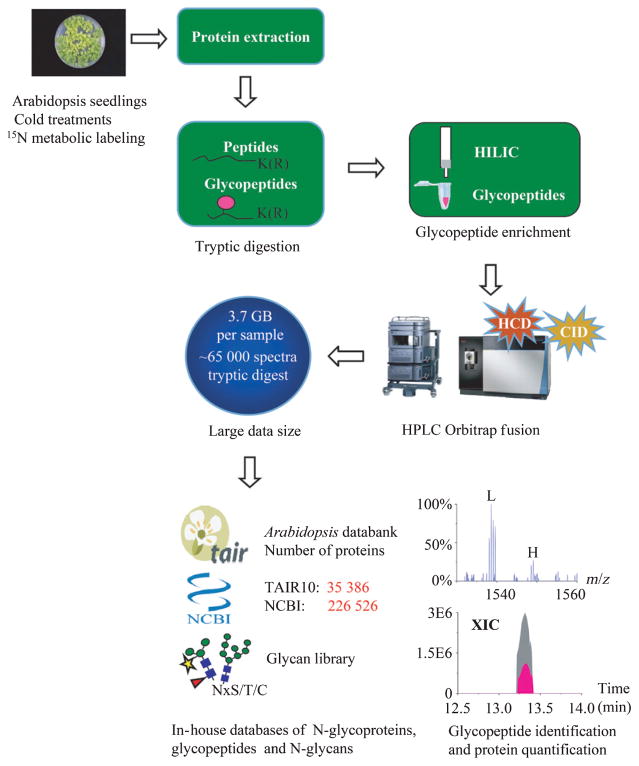
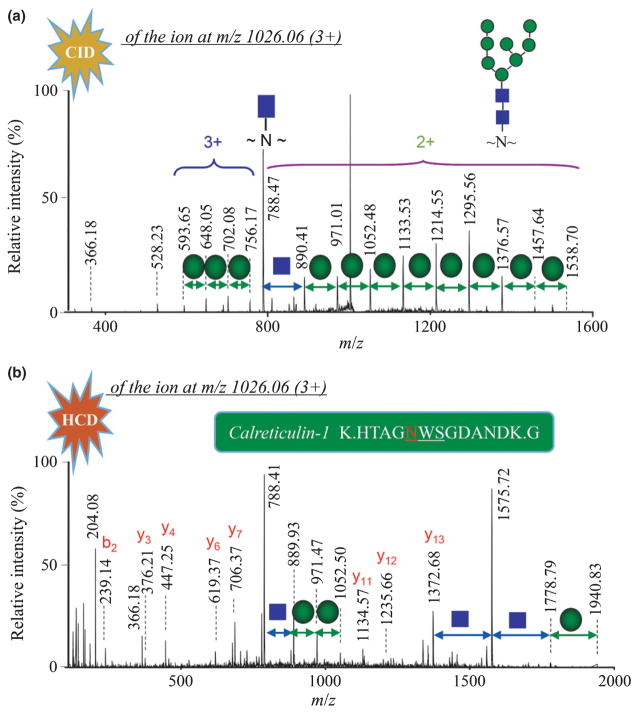
N-glycans attached to asparagine residues of a protein are usually localized at the sequence motif of Asn-X-Ser/Thr/Cys. However, the presence of such a motif is often not sufficient to generate N-glycosylation at a given site in nature, which also heavily depends on the flanking structural surface topology and the close interaction with the processing enzymes and glycosyltransferases. Examination of the whole protein sequences in TAIR10 and NCBI databases indicated that c. 80% of them possess potential N-glycosylation sites. To annotate the putative glycoproteins, we established a proteomic approach for automated analysis of naturally existing N-glycosylated proteome in Arabidopsis. Fig. 1 shows the overall workflow of the experimental procedure used for glycoprotein identification and quantification. Proteins were cleaved by trypsin and the resulting glycopeptides were purified using a HILIC column for unbiased extraction of N-glycosylated species, followed by LC-MS/MS analysis. Owing to a huge data size acquired by highly frequent MS/MS scans of a sample on the Orbitrap Fusion (e.g. 3.7 GB size per 2 h run, 65 000 spectra), the routine database searching was always time-consuming. To rapidly identify heterogeneous N-glycan isoforms, we constructed two in-house databases of N-glycoproteins and N-glycopeptides of Arabidopsis consisting of the N-glycosylation motif. The first refinement of glycoprotein sequences reduced c. 20% of the database size of proteins in TAIR10 and NCBI (Fig. S4a). The second assortment of glycopeptides allowed two missed cleavage sites of trypsin away from the N-glycosylation motif, which dramatically reduced 90.7% and 90.4% of the public database size over the total number of peptides in TAIR10 and NCBI databases, respectively (Figs S2, S4b). In addition, a plant N-glycan library collected from previous reports was also incorporated as variable modifications, which covered 161 known glycan isoforms (Fig. S3; Table S1). Automated database search of LC-MS/MS datasets combined with high-mass accuracy Orbitrap mass spectrometry thus provided a rapid method for large-scale identification of glycoproteins in a complex mixture. The retrieved N-glycan structure was verified manually by CID spectra of the precursor ions based on sequential neutral losses of monosaccharides, and the characteristic oxonium ions of m/z 204.1 [HexNAc+], m/z 366.2 [HexHexNAc+] and m/z 528.2 [HexHexHexNAc+] at the low mass range (Fig. 2a), whereas the peptide sequence was validated by a series of C-terminal yn fragments and N-terminal bn fragments derived from the HCD spectra of the corresponding precursor ions (Fig. 2b).
Fig. 1.
Experimental workflow of glycopeptide identification and protein quantification in Arabidopsis. Arabidopsis seedlings of Col-0 were treated with cold at 4 and 22°C, followed by protein extraction, enzymatic digestion, affinity purification of glycopeptides, and Orbitrap LC-MS/MS analysis. Data were analyzed by Mascot, searching against in-house databases of glycoproteins, glycopeptides and a plant glycan library. Quantification of proteins and glycopeptides was achieved by the relative intensity or the extracted ion chromatogram (XIC) peak area of 14N-labeled peptides and 15N-labeled counterparts. HILIC, hydrophilic interaction liquid chromatography.
Fig. 2.
Identification of the intact calreticulin-1 glycopeptide at m/z 2016.06 (3+) using LC-MS/MS. The N-glycan structure was defined by the sequential losses of monosaccharides and the characteristic oxonium ions of m/z 366.2 (HexHexNAc+) and m/z 528.2 (HexHexHexNAc+) in the low-energy collision-induced dissociation (CID) mass spectrum (a). MASCOT database search identified the peptide sequence matching a series of C-terminal yn fragments (doubly and triply charged ions) in the higher-energy collision-induced dissociation (HCD) mass spectrum (b).
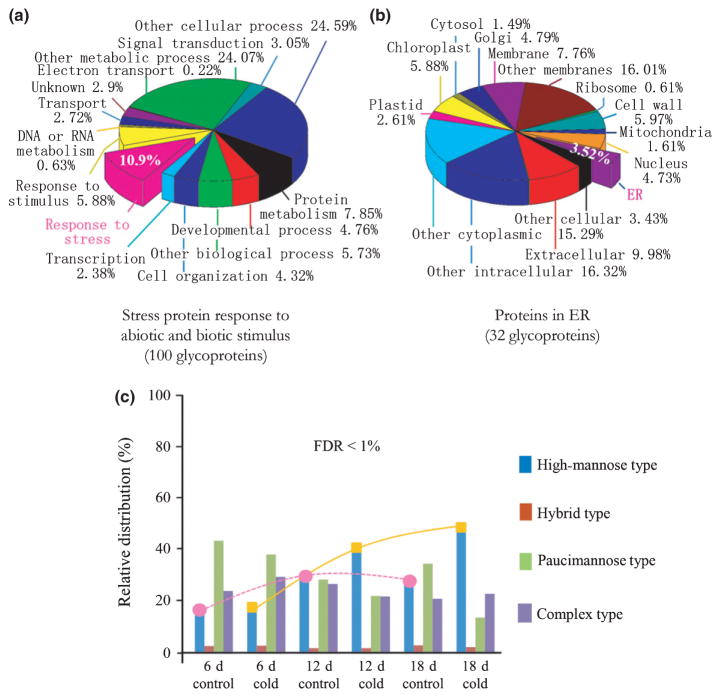
Measurements of a tryptic digest of glycoproteins from wild-type Arabidopsis reliably identified 2000 glycopeptides resulting from a 2 h gradient LC-MS/MS run. Parallel analyses were conducted on three sets of Arabidopsis seedlings grown at two different temperature conditions (22 and 4°C) for periods of 6, 12 and 18 d (Fig. S5). We identified 912 glycoproteins by Mascot database search at the FDR < 1%, c. 90% of which were shared in each biological sample. A total of 100 glycoproteins were found to be related to abiotic and biotic stresses based on the GO functional annotation (Fig. 3a). Some proteins have been reported to be involved in cold stress responses. The GO cellular categorization revealed a wide range of glycoprotein locations, especially in ER, plasma membrane and cell walls (Fig. 3b). The identified N-glycans of high-mannose, hybrid, paucimannosidic and complex-type are summarized in Fig. 3(c). Compared with those variations of N-glycans (labeled with a dashed pink line in Fig. 3c) isolated from the plants grown under the normal condition as a control, the relative distribution of high-mannose N-glycans (labeled with a yellow line in Fig. 3c) dramatically increased with the time period of cold treatments.
Fig. 3.
Structural function, cellular localization and relative distribution of N-glycans of Arabidopsis N-glycoproteome obtained by LC-MS/MS. (a) Pie chart shows the functional categorization of 912 glycoproteins by gene ontology (GO) annotations in which 100 glycoproteins are stress-related. (b) Cellular localization indicated 32 stress-related glycoproteins in endoplasmic reticulum (ER). (c) The N-glycan distribution of glycoproteins determined that the cold-treated Arabidopsis (at 4°C) overall yielded more high-mannose N-glycans than those of plants grown at the normal growth condition (at 22°C). To help view the relative distribution change of N-glycans, the dashed pink line and the solid yellow line show the tendency of increasing percentage of high-mannose N-glycans at the time periods of 6, 12 and 18 d for the control and cold-treated plants, respectively. FDR, false discovery rate.
N-glycan structure and functional annotation of glycoproteins
Glycoproteome analysis revealed that many glycoproteins are encoded by genes involved in plant defense responses to abiotic stresses, including temperature and salts (Table S2). These included CRT1b, PDIL1-1, thioglucoside glucohydrolases, UDP-glucose-glycoprotein glucosyltransferase (UGGT), germin-like proteins, beta-glucosidase 35 and STT3a/b. The N-glycosylation sites of these proteins were all occupied by exclusively oligomannosidic glycoforms of Man8–9GlcNAc2, which support the cellular location as ER-resident proteins. By contrast, the presence of only complex N-glycans, instead of mannose-type, in the receptor-like protein kinases (e.g. RLK, AT5G16590) implies the continuous N-glycan processing of the glycoproteins to ensure its proper folding and subsequent transfer from ER to Golgi apparatus.
Glycosylphosphatidylinositol (GPI)-anchor functions in the attachment of proteins to the surface of the membrane. LC-MS/MS analysis reliably identified two distinct glycopeptides of ICSIHSSNLTSSSCPVINVDEFESTVDTAK and NKPCTND-TGSATVPGTEPQFANYPQILAK with the different types of N-glycoforms Man6–8GlcNAc2 and (GlcNAc)0–2Man3(Xyl)(Fuc) (GlcNAc)2, corresponding to the tryptic fragments of two uncharacterized GPI-anchored membrane proteins with the accession numbers of gi|30387511 and gi|18397452, respectively (data not shown). Considering that the GPI-anchored protein precursor normally contains an N-terminal signal peptide and a C-terminal tail of GPI attachment signal sequence (CAAX), the encoding large protein (gi|30387511, 433 aa) from the specific gene AT1G61900 possessed exclusively high-mannose N-glycans of Man6–8GlcNAc2, suggesting that the precursor protein might lose GPI signal at the C-terminus, causing the extensive accumulation mainly in ER (Rivier et al., 2010). In comparison, the existence of complex-type (GlcNAc)0–2Man3(Xyl)(Fuc)(GlcNAc)2 in the small GPI membrane glycoprotein (gi|18397452, 200 aa), derived from the gene AT3G06035, indicated that the glycoprotein sequence has an N-terminal signal peptide that is part of residues 1–106, as seen in the NCBI database (gi|24417290), which was likely cleaved, resulting in a mature protein transported to the cell surface membrane. As such, our results suggest possible subcellular location-specific GPI-anchored membrane proteins in Arabidopsis.
Two distinct glycosylation sites of GDSL esterase/lipase isoenzymes (AT1G54000, AT1G54010 and AT3G14210), a known cold stress-responsive protein, were identified by MS/MS analyses as the complex N-glycans of (GlcNAc)0–1Man3(Xyl)(Fuc)(GlcNAc)2 at varying sequence regions of Asn105 and Asn146 and the high mannose-type of Man3–9GlcNAc2 at the highly conserved residues of Asn288 and Asn290, respectively (Fig. S6). Structure modeling of GDSL esterase/lipase (AT1G54000; gi| 332194912) using the Phyre2 server indicated that the two glycosylation sites of Asn146 and Asn288 are all localized at the structural loop regions (Kelley et al., 2015), and Asn288 is positioned nearby the cluster of four α-helices (Fig. S7). The observation of trimmed high-mannose N-glycans suggests that the Asn288-glycosylation might function in regulating the protein conformation, which is sensitive to cold stress. As reported previously (Zielinska et al., 2010; Song et al., 2013), the identification of heterogeneous N-glycan isoforms by mass spectrometry thus offers a method to determine the surface topology and different subcellular locations of the same protein or different enzyme isoforms.
ER-associated N-glycan degradation in the early stage of cold treatments
Among the identified 912 glycoproteins, c. 100 proteins are stress-related and 3.5% of these (i.e. 32 glycoproteins) are localized in the ER (Table S3). The N-glycan analyses showed that most ER glycoproteins of Arabidopsis grown at 22°C have merely the predominate high-mannose N-glycans of Man8–9GlcNAc2. To examine the possible effect on the glycan structure and glycoprotein misfolding caused by cold stress, we conducted a thorough inspection on N-glycan modifications of the individual ER-resident proteins (Fig. S8), except for CNXs, which are known as nonglycosylated type I integral membrane proteins (Miernyk, 1999).
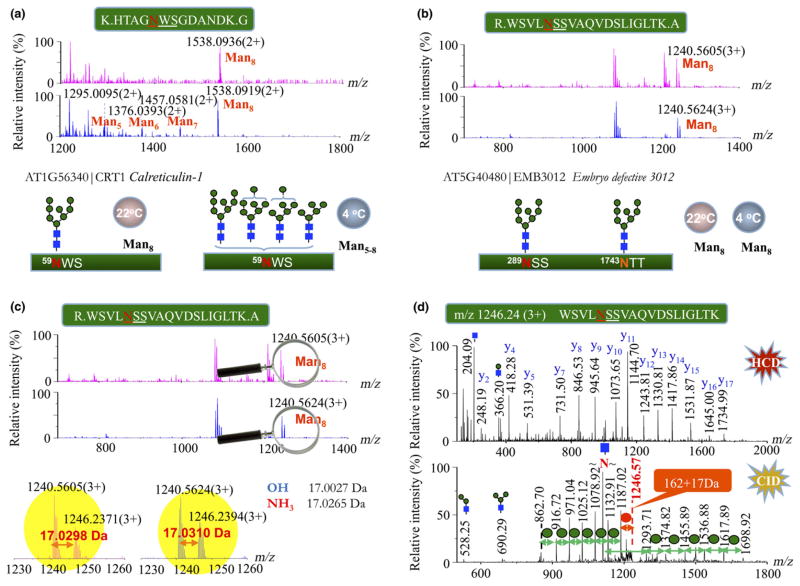
As shown in Fig. 4(a), a predominate glycopeptide at m/z 1538.0936 (2+) was identified as a Man8GlcNAc2 attached to the residue Asn59 at peptide 55–67 of CRT1 (AT1G56340) of plants grown under normal conditions, whereas this unique glycopeptide was detected to have additional trimming by MNSs into heterogeneous Man5–8GlcNAc2 during plant growth at the cold temperature of 4°C between 6 and 18 d (Fig. 4a). Similar phenomena occurred in the other ER stress-related glycoproteins, such as OST1B and UGGT; the N-glycans of these glycoproteins retained the stable glycan iso-forms of Man8–9GlcNAc2 under the normal growth condition, but degraded under the cold-stressed environment (Figs S9a, b). It is worth noting that PDIL1-1, a protein folding assistant, originally possessed a series of high-mannose N-glycans of Man5–8GlcNAc2 at 22°C, and the intensities of these trimmed N-glycans were all enhanced by the low temperature (Fig. S9c). We thus concluded that these cold-regulated ER glycoproteins all have the N-glycan degradation resulting from chilling stress.
Fig. 4.
Discriminating the N-glycan profiles of endoplasmic reticulum (ER)-resident glycoproteins in plants under normal and cold-treated conditions for 18 d. (a) The mass spectra show the distinguishable high-mannose type N-glycans of the stress-responsive calreticulin-1 under two different conditions of plant growth: Man8GlcNAc2 (upper panel, 22°C) vs Man5-8GlcNAc2 (lower panel, 4°C). (b) Similar N-glycan profiles of Man8GlcNAc2 in the spectra were observed at the two glycosylation sites of Asn289 and Asn1743 of EMB3012 in plants grown under both conditions. The expanded m/z range of 1230–1260 from (b) showed the exact mass difference of 17.0299 ± 0.0001 Da between two peptide ions (c) at m/z 1240.56 (3+) and m/z 1246.24 (3+), which is close to that calculated from ammonium. (d) MS/MS spectra of the peptide precursor ion at m/z 1246.24 identified the same peptide sequence as the peptide of m/z 1240.56 at the higher-energy collision-induced dissociation (HCD, upper panel), and the additional modification of 17 Da was localized at the terminal mannose by the low-energy collision-induced dissociation (CID, lower panel) according to the initial neutral loss of 179 Da at m/z 1187.02 ([M + 3H-59.55]3+), that is, the 17 Da adduct of mannose from the precursor ion.
By contrast, identical MS profiles of glycopeptides were observed on the nonregulated proteins isolated from plants grown at the two different temperatures (22 and 4°C), As shown in Fig. 4(b), the spectra of a glycopeptide of embryo defective 3012 (EMB3012) exhibited a pair of nascent peaks after expanding m/z scale, corresponding to an N-glycan of Man8GlcNAc2 attached to the peptide 285–304 at m/z 1240.5605 (3+) and the other component having a mass increase of 17 Da at m/z 1246.2371 (3+). Accurate mass measurements of the triply charged ions showed a difference of 17.0299 ± 0.0001 Da (Fig. 4c), matching one molecule of ammonia (calculated mass: 17.0265 Da). Based on a set of similar C-terminal y fragment ions resulting from the peptide bond cleavages in the HCD condition, MS/MS analysis of the larger peptide ion at m/z 1246.24 (3+) identified the same peptide sequence as the peptide of m/z 1240.56 (3+). The 17 Da increment at the larger peptide was localized at the terminal mannose of Man8GlcNAc2 by the preferential fragmentation of low-energy CID at relatively weak glycosidic bonds, which resulted in the initial neutral loss of 179 Da at m/z 1187.02 ([M + 3H-59.55]3+), that is, the 17 Da adduct of mannose from the precursor ion (Fig. 4d). Similar results were also obtained for a pair of tryptic glycopeptides of the transmembrane CLPTM1 family protein (Fig. S10). Accurate mass analysis on the EMB3012 glycopeptide pairs in a protein mixture of unlabeled and 15N-labeled Arabidopsis seedlings determined that the mass difference of 17.0280 Da (i.e. 14NH3 adduct) between the 14N-coded glycopeptides had shifted to 18.0210 Da between the 15N-coded glycopeptide counterparts, corresponding to one molecule of 15N-labeled ammonia (theoretical mass 18.0236 Da) (Fig. S11). The reproducible results in biological triplets indicated that the modification of 17 Da on the terminal mannose of N-glycans was caused by in vivo ammonolysis of plants. Structural glycoproteomics therefore provided evidence for N-glycan ammonolysis of nonregulated glycoproteins in Arabidopsis, which might protect the N-glycan from degradation under chilling stress.
Chilling stress induces ER glycoprotein up-regulation
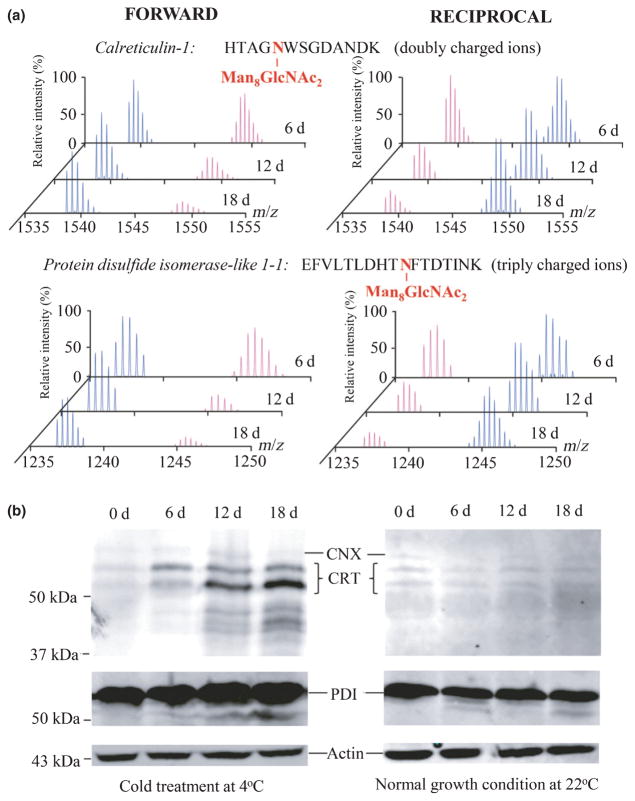
To further investigate the effect on protein expression by cold-induced ER stress, we used a quantitative proteomics strategy by means of the forward and reciprocal 15N-isotope metabolic labeling approaches for accurate measurements of protein abundances at the plant growth stages of 6, 12 and 18 d. Time-course qualification of glycoproteins was performed by LC-MS/MS analyses at the levels of both glycopeptides and nonglycopeptides. The protein abundances of Arabidopsis seedlings were compared between two plant growth environments of 22°C (heavy 15N-isotope labeled proteins) and 4°C (light 14N-isotope labeled proteins) at the same period of time, and vice versa in the reciprocal ones (Fig. 5a). Initial analysis confirmed the similar L: H ratio of relative intensities of 14N- and 15N-labeled glycopeptides to that of the integrated XIC peak areas (data not shown). Using the N-linked glycopeptides of Man8–9GlcNAc2 as references, the abundance of ER-resident glycoproteins after cold treatments for 18 d was determined to be twofold higher than that from plants grown at the normal condition. The quantitative results from LC-MS/MS analyses on the L: H ratios (more than twofold) of 14N- and 15N-isotope-labeled glycopeptide pairs are summarized in Table 1 (Table S4), in which the ER stress-responsive glycoproteins all showed a significant stepwise up-regulation during the time periods of cold treatments.
Fig. 5.
Validation of up-regulated endoplasmic reticulum (ER)-resident glycoproteins in Arabidopsis under chilling stress. (a) Accurate protein quantification was performed by the forward and reciprocal 15N-metabolic labeling and LC-MS/MS analyses as shown of two representative proteins of calreticulin-1 (CRT1) and the protein disulfide isomerase-like 1-1 (PDI). In forward experiments (‘FORWARD’), the 14N-isotope was labeled on plants grown at 4°C (peaks in blue) whereas the 15N-isotope was labeled on plants grown at 22°C (peaks in pink), and in the reciprocal experiments (‘RECIPROCAL’) the reverse was the case. (b) Total protein extracts from the cold-treated Arabidopsis at 4°C and the plants grown at 22°C for 0, 6, 12 and 18 d were separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) followed by western blots using antibodies specific to actin, calnexin (CNX), CRT and PDI.
Table 1.
Quantitative measurements of the endoplasmic reticulum (ER)-resident glycoproteins using 15N-metabolic labeling glycopeptides
| Protein (gene no.) | Peptide sequence | N-Glycan modification | m/z (L : H) (charge state) | Forward L : H (6 d) | Reciprocal L : H (6 d) | Forward L : H (12 d) | Reciprocal L : H (12 d) | Forward L : H (18 d) | Reciprocal L : H (18 d) |
|---|---|---|---|---|---|---|---|---|---|
| PDIL1-1 (AT1G21750) | K.EFVLTLDHTN39FTDTINK.H | Man8GlcNAc2 | 1237.5332 (3+); 1245.5095 (3+) | 1.13 ± 0.03 | 0.85 ± 0.02 | 2.31 ± 0.21 | 0.42 ± 0.03 | 4.12 ± 0.3 | 0.24 ± 0.02 |
| PDIL1-4 (AT5G60640) | K.IGPGVYN213LTTLDDAEK.V | Man7GlcNAc2 | 1623.7006 (20+); 1633.6709 (2+) | 1.11 ± 0.04 | 0.89 ± 0.02 | 1.65 ± 0.04 | 0.61 ± 0.04 | 2.52 ± 0.12 | 0.41 ± 0.02 |
| CRT1b (AT1G56340) | K.HTAGN59WSGDANDK.G | Man8GlcNAc2 | 1538.0887 (2+); 1548.5575 (2+) | 1.11 ± 0.05 | 0.86 ± 0.03 | 2.27 ± 0.03 | 0.44 ± 0.005 | 3.98 ± 0.30 | 0.25 ± 0.02 |
| UGGT (AT1G71220) | K.LN373ESIKDEILSNQR.M | Man8GlcNAc2 | 1121.1561 (3+); 1128.8000 (3+) | 1.10 ± 0.08 | 0.87 ± 0.03 | 2.40 ± 0.03 | 0.43 ± 0.02 | 4.54 ± 0.44 | 0.22 ± 0.01 |
| HSP70 (AT4G16660) | R.N518LSAPIK.A | Man8GlcNAc2 | 1223.0177 (2+); 1228.5014 (2+) | 1.42 ± 0.09 | 0.71 ± 0.07 | 6.63 ± 0.45 | 0.16 ± 0.01 | 10.33 ± 0.24 | 0.10 ± 0.02 |
| STT3a (AT5G19690) | R.TVIVDN604NTWNNTHIATVGTAMSSPEK.A | Man8GlcNAc2 | 1501.6515 (3+); 1513.6159 (3+) | 1.21 ± 0.06 | 0.81 ± 0.04 | 2.13 ± 0.12 | 0.46 ± 0.01 | 3.54 ± 0.18 | 0.28 ± 0.01 |
| R.TVIVDN604NTWNNTHIATVGTAMSSPEK.A | Man9GlcNAc2 | 1555.6691 (3+); 1567.6336 (3+) | 1.17 ± 0.11 | 0.85 ± 0.06 | 1.66 ± 0.06 | 0.59 ± 0.03 | 3.49 ± 0.04 | 0.28 ± 0.01 | |
| STT3b (AT1G34130) | R.TVIVDN581NTWNNTHIATVGR.A | Man8GlcNAc2 | 1276.5587 (3+); 1286.8614 (3+) | 1.05 ± 0.02 | 0.92 ± 0.03 | 1.90 ± 0.09 | 0.52 ± 0.02 | 3.38 ± 0.26 | 0.29 ± 0.02 |
| R.TVIVDN581NTWNNTHIATVGR.A | Man9GlcNAc2 | 1330.5763 (3+); 1340.8789 (3+) | 1.06 ± 0.04 | 0.91 ± 0.005 | 1.40 ± 0.07 | 0.68 ± 0.04 | 2.86 ± 0.15 | 0.35 ± 0.02 | |
| AQI (AT4G38220) | R.N317MSFEFK.Q | Man8GlcNAc2 | 1302.9987 (2+); 1308.4824 (2+) | 1.11 ± 0.05 | 0.89 ± 0.05 | 1.50 ± 0.02 | 0.66 ± 0.008 | 2.14 ± 0.05 | 0.44 ± 0.04 |
| R.N317MSFEFK.Q | Man9GlcNAc2 | 1384.0251 (2+); 1389.5088 (2+) | 1.06 ± 0.04 | 0.96 ± 0.009 | 1.55 ± 0.05 | 0.62 ± 0.01 | 2.88 ± 0.16 | 0.37 ± 0.03 | |
| P-value | < 0.01 | 0.01 | < 0.01 |
LC-MS/MS analysis was performed in biological duplicate samples using ‘forward’ and ‘reciprocal’ isotope labeling approaches, and mean ± SD light : heavy (L : H) ratios were calculated based on manual inspection of the isotope peak intensity of the 14N-labeled (L) and 15N-labeled (H) peptide ion pairs. Student’s t-test was used to compare data from two groups. In forward experiments, the 14N-isotope was labeled on plants grown at 4°C whereas the 15N-isotope was labeled on plants grown at 22°C, and in the reciprocal experiments the reverse was the case. A P-value < 0.05 was considered to be statistically significant. The underlined residues are shown the glycosylation sites localizing at the structural motif Asn-X-Ser/Thr.
A similar comparison was performed on the nonglycosylated peptides using LC-MS/MS analyses, and the L : H ratios of the proteins containing an ER retention [K/R/H]DEL motif were selectively examined as shown in Table 2 (Table S5). The relative abundances of nonglycosylated peptides in the stress-responsive proteins showed up-regulated L: H ratios in the forward 15N-isotope metabolic labeling and further verified by the reciprocal experiments, which were consistent with the quantitative results of glycopeptides combined from the multiple N-glycan distributions. As expected, the normal- and chilling stress-grown plants showed no differences in the overall expression levels of actin, rubisco activase, ribulose bisphosphate carboxylase and other housekeeping proteins as measured by the L: H ratios at c. 1. By contrast, an increase of four- to sixfold in the protein abundances was determined in the amounts of PDIL1-1 (4.43), PDIL1-2 (5.39) and PDIL1-4 (4.52). In terms of the chaperone-like activity of protein disulfide isomerases (Cai et al., 1994), the finding suggests that a large amount of PDI isoforms are required to help the refolding of unfolded or misfolded proteins caused by chilling stress.
Table 2.
Quantification of the Arabidopsis proteins containing a C-terminal endoplasmic reticulum (ER) retention motif by LC-MS/MS analyses of nonglycopeptides
| Protein (gene no.) | ER motif (H/K/RDEL) | Forward L : H (6 d) | Reciprocal L : H (6 d) | Forward L : H (12 d) | Reciprocal L : H (12 d) | Forward L : H (18 d) | Reciprocal L : H (18 d) |
|---|---|---|---|---|---|---|---|
| BIP1 (AT5G28540) | HDEL | 1.22 ± 0.04 | 0.86 ± 0.04 | 3.32 ± 0.17 | 0.35 ± 0.02 | 5.60 ± 0.28 | 0.25 ± 0.01 |
| BIP2 (AT5G42020) | HDEL | 1.22 ± 0.04 | 0.86 ± 0.04 | 3.32 ± 0.17 | 0.35 ± 0.02 | 4.25 ± 0.05 | 0.25 ± 0.01 |
| CRT1a (AT1G56340) | HDEL | 1.46 ± 0.04 | 1.09 ± 0.06 | 2.50 ± 0.11 | 0.52 ± 0.03 | 5.02 ± 0.26 | 0.32 ± 0.02 |
| CRT1b (AT1G09210) | HDEL | 1.27 ± 0.09 | 0.96 ± 0.01 | 2.86 ± 0.01 | 0.52 ± 0.02 | 4.69 ± 0.22 | 0.38 ± 0.03 |
| PDIL1-1 (AT1G21750) | KDEL | 1.23 ± 0.03 | 0.89 ± 0.02 | 2.55 ± 0.14 | 0.44 ± 0.03 | 4.43 ± 0.25 | 0.21 ± 0.01 |
| PDIL1-2 (AT1G77510) | KDEL | 1.52 ± 0.08 | 0.99 ± 0.07 | 2.24 ± 0.05 | 0.60 ± 0.03 | 5.39 ± 0.13 | 0.22 ± 0.03 |
| PDIL1-4 (AT5G60640) | KDEL | 1.32 ± 0.20 | 0.83 ± 0.13 | 2.67 ± 0.11 | 0.35 ± 0.02 | 4.52 ± 0.17 | 0.16 ± 0.01 |
| BGLU22 (AT1G66280) | KDEL | 0.97 ± 0.02 | 1.60 ± 0.16 | 1.36 ± 0.08 | 0.89 ± 0.04 | 1.40 ± 0.04 | 0.80 ± 0.01 |
| PYK10 (AT3G09260) | KDEL | 1.14 ± 0.04 | 1.74 ± 0.02 | 1.64 ± 0.03 | 1.55 ± 0.04 | 1.37 ± 0.05 | 1.17 ± 0.11 |
| AQI (AT4G38220) | RDEL | 0.86 ± 0.03 | 1.631 ± 0.25 | 1.61 ± 0.11 | 0.77 ± 0.11 | 3.57 ± 0.15 | 0.33 ± 0.01 |
| BGLU21 (AT1G66270) | RDEL | 0.97 ± 0.02 | 1.60 ± 0.16 | 1.36 ± 0.08 | 0.89 ± 0.04 | 1.40 ± 0.04 | 0.80 ± 0.01 |
| P-value | < 0.01 | 0.04 | 0.03 |
The list includes only the ER-resident proteins containing a C-terminal ER-retrieving H/K/RDEL motif. The light : heavy (L : H) values were determined by LC-MS/MS analyses of biological duplicate samples using ‘forward’ and ‘reciprocal’ isotope labeling approaches at mean ± SD, and validated manually on a minimum of two peptides for each protein. Student’s t-test was used to compare data sets from two groups, in which the 14N-isotope was labeled on plants grown at 4°C and the 15N-isotope was labeled on plants grown at 22°C in forward experiments, and the reverse was true in reciprocal experiments. A P-value < 0.05 was considered to be statistically significant.
To ensure the reliability of the quantitative proteomics method, we also used western blotting to verify the expression level of stress-responsive proteins when reliable antibodies were available. A direct comparison of densitometry analyses on the protein bands of plant materials at 0, 6, 12 and 18 d determined the relative abundance change, which was equivalent to that of up-regulated proteins detected by LC-MS/MS quantification (Fig. 5b). The small amount of low-molecular-weight protein bands in Fig. 5(b) was also visualized by western blots to have an increasing abundance with the time of cold treatments of 12 and 18 d for CRT and PDI, but not nonglycosylated CNX and actin, suggesting the existence of degraded products of the glycoproteins. Thus, the data show that LC-MS/MS analysis of glycoproteins using 15N-metabolic labeling provides a reliable and accurate method for comparing the difference in abundance of stress-related glycoproteins between plants grown under normal and stressed conditions.
Discussion
Structural N-glycoproteomics targets proteins in response to chilling stress
Endoplasmic reticulum stress is one of the important responses to mitigate the plant damage caused by adverse environmental conditions, and is activated by unfolded proteins that accumulate in the ER (Howell, 2013). The rapid determination of stress-responsive protein targets and the quantitative assessment on the extent of stress-caused protein misfolding represent a significant bottleneck currently in plant stress proteomics research. Recent advances in mass spectrometry allowed measurements of protein folding, unfolding intermediate and conformational change using hydrogen-deuterium exchange of protein backbone amides and LC-MS/MS (Engen & Wales, 2015); the technology has proved successful but possesses drawbacks such as difficult sample handing, hydrogen/deuterium scrambling, a back-exchange issue and complicated data interpretation from the nonspecific protease cleavage of proteins in the acidic condition (e.g. pepsin) (Hoofnagle et al., 2003). The utility of this method is therefore restricted to purified proteins or isolated protein complexes. As an alternative strategy, we report here a structural glycoproteomics method for large-scale analysis of glycoproteins to provide insights into the folding status of chilling stress-responsive proteins in Arabidopsis.
Structural characterization of the N-glycosylated proteome through analysis of intact glycopeptides by LC-MS/MS has traditionally been difficult as a result of the large protein database and heterogeneous N-glycan structures. The unprecedented LC-MS/MS data acquisition leads to a huge data set from a complicated biological sample. To reduce the complexity of sample analysis and the time-consuming process of database search, we thus established a proteomic method based on selective HILIC enrichment of glycopeptides, and database refinements of glycoproteins or glycopeptides, as well as an N-glycan library of Arabidopsis. Automated LC-MS/MS identification and accurate quantification of glycopeptides and the attached N-glycans were achieved through two highly sensitive data-dependent acquisitions by CID and HCD on the Orbitrap Fusion, and in vivo 15N-metabolic labeling of plant materials. The stress-related protein targets were discriminated from the nonstressed proteins according to the abnormal N-glycan structure together with a significant abundance change of glycoproteins between the plants grown under two different temperature conditions. Using this method, we have identified 912 glycoproteins consisting of 2000 glycopeptides in a 2 h LC-MS/MS run, and resolved the N-glycan isoform structures of cellular location-specific proteins which enabled an in-depth understanding of structural folding and stability of ER chilling stress-responsive glycoproteins.
Chilling stress leads to ER N-glycan degradation and protein up-regulation
As described earlier, mass spectrometry-based glycoproteomic analysis provided a direct comparison on N-glycan components of the same peptide of individual proteins. The results showed that the N-glycan degradation occurred in the ER stress-regulated proteins of plants under cold treatment, but not in the nonregulated glycoproteins (Figs 4b, S10). Further quantitative analysis of MNSs (MNS1, MNS2 and MNS3) revealed the up-regulation of the proteins by increasing abundances over twofold in the chilling stress condition (Fig. S12), and this may be responsible for trimming of N-linked high-mannose of the interacting protein substrates. Protein mislocalization resulting from either the retention of partial MNSs in the ER by trafficking slowdown at low temperature or the escape of ER stress-related proteins to the Golgi could cause the N-glycan cleavage of the proteins by alpha-MNSs. The stable N-glycan forms of nonregulated glycoproteins could be contributing to the possible protection of reversible N-glycan ammonolysis from the N-glycan degradation, in which the modified terminal mannose might not be recognized by MNS for its cleavage (Fig. S13). Mannose trimming of ER stress-regulated proteins from Man8–9GlcNAc2 to Man6–7GlcNAc2 appeared as an ERAD signal for misfolded proteins (Määttänen et al., 2010), and the continued removal of mannose residues to Man4–5GlcNAc2 might result in protein transport and exit from the ER (Fig. S14). Our results thus suggest that ER N-glycan trimming is a consequence of plant responses to environmental stress, which may be caused by mislocalization of stress-responsive proteins. The proper regulation of protein ERQC via UPR is probably an important mechanism for plant adaptation to chilling stress.
Quantitative analyses of glycoproteins at the levels of both glycopeptides and nonglycosylated peptides showed a correlation consistent with the protein abundance changes vs the time of plant cold treatments (Tables 1, 2, S4, S5). The relative abundances of ER stress-responsive proteins were all up-regulated following a stepwise increase after the first 12 d cold treatment and then a dramatic escalation at 18 d. Among the ER glycoproteins examined, the stress-related proteins of PDIs and CRTs were of particular interest, as quantitative differences were detected ranging from four- to sixfold at the top of the list (Fig. 5a; Table 2). The heat shock protein 70 family proteins (BIP1, BIP2, HSP70) had a stable increase in the L : H ratio of one- to threefold after the initial cold treatments of 6 and 12 d followed by a dramatic increase (to fivefold) after 18 d of treatment. Considering the tight interaction of CNX, CRT and UGGT with misfolded glycoproteins, the ER chaperone-binding proteins (BIP1, BIP2, HSP70) and PDI isoforms assist in the formation of intramolecular disulfide bonds and protein refolding to prevent the proteins from misfolding and aggregation (Sung et al., 2001). Our quantification results are highly consistent with previous findings on the up-regulation of PDI and HSPs in Arabidopsis and Abelmschus mosschatus grown at low temperature (Bae et al., 2003; Li et al., 2012). Collective evidence thus suggests that the fold change of ER stress-regulated proteins can be considered a molecular signature for evaluating the degree of plant stress.
Enhancement of plant tolerance to chilling stress
Endoplasmic reticulum stress activates the UPR signaling network and causes cell dysfunction and death (Kim et al., 2008). Investigation of ER stress-induced protein misfolding and aberrant N-glycans could help us to understand the structure and function of stress-responsive glycoproteins, and to develop strategies to improve plant resistance to the ER stress-caused disorders. Based on the glycoproteomics results and the previous reports on the direct involvement of MNSs in targeting misfolding glycoproteins for ERAD to relieve ER stress (Tokunaga et al., 2000; Hosokawa et al., 2001; Termine et al., 2009), we can speculate that there is a general plant chilling stress response pathway, as shown in Fig. 6. ER stress initially triggers the N-glycan trimming of Man8–9GlcNAc2 of stress-related glycoproteins to yield smaller Man5–7GlcNAc2 isoforms at the protein sidechains, resulting in misfolded proteins and subsequent escape from the ER lumen. With a certain amount of misfolded proteins degraded by ubiquitin–proteosome system, ER chaperones and PDI enzymes are up-regulated to maintain the proper function of ERQC and the refolding cycle of misfolded proteins during cold acclimation. With extensive cold treatment, ER stress continues to produce ERQC dysfunction and accumulation of misfolded proteins, leading to subsequent ERAD degradation of the proteins and consequently cell death and damage of plants (Kim et al., 2008).
Fig. 6.
Diagram of the stress-related glycoproteins in the endoplasmic reticulum (ER) and the effects of chilling stress in Arabidopsis. The N-glycan degradation of up-regulated ER glycoproteins caused by extensive chilling stress leads to partial misfolding of the proteins, ER-associated degradation (ERAD) and escape from the ER lumen, which will eventually cause cell death of plants. Thus, a possible strategy to use ER retention of stress-regulated proteins through the retrieval transport from the Golgi apparatus back to the ER by overexpressed ER retention receptors selectively binding the molecules containing a C-terminal motif [K/R/H]DEL or to treat with chemical chaperones is suggested for enhancing plant resistance to the chilling stress. ERQC, endoplasmic reticulum quality control; CRT, calreticulin; CNX, calnexin.
To reduce ER stress and the downstream UPR, the stress-responsive ER-resident glycoproteins were up-regulated to cope with the cold-induced disorder in ER. With the knowledge gained from this study, it may be possible to increase plant tolerance to chilling stress by enhancing ER retrieving signals. Partial escape of ER stress-related proteins resulted in dysfunction of ERQC and the refolding cycle of misfolded proteins during cold acclimation; the overexpression of the ERD2 transmembrane protein, a KDEL receptor, may be helpful to retrieve the escaped ER-resident proteins bearing the C-terminal ER retention sequence motif of [K/R/H]DEL (Table S5) from the Golgi apparatus to the ER. The protein retention could assist in the CNX/CRT chaperone function of the ERQC system. It may also be useful to develop reliable chemical chaperones to complement the refolding role of CRT, HSP70 and PDI isoenzymes to rescue the folding-defective proteins that could eventually alleviate ER-associated chilling stress for plant survival (Molinari, 2007).
Concluding remarks
We have established a highly sensitive proteomics approach for large-scale structural analysis of N-linked glycoproteins and utilized it to determine the chilling stress-related proteins in Arabidopsis. Our results suggest that the ER-associated N-glycan degradation serves as a signature in response to chilling stress, which could aid in elucidating cellular mechanisms of protein relocation, transport, trafficking, misfolding and degradation under stress conditions. The stable 15N-isotope metabolic labeling and LC-MS/MS analysis further provide a quantitative strategy to accurately measure the differential protein expression, and thus the effects of chilling stress on individual glycoproteins. Our observations suggest that the molecular signature of N-glycan degradation and the stoichiometry of stress-regulated proteins could be considered as criteria for evaluating the physiological effect of plants under stress conditions. The findings increase our understanding of the underlying mechanisms of ER-associated N-glycan signaling, low temperature-responsive protein regulation, and the structure–function relationship of chilling stress-related glycoproteins.
Supplementary Material
Fig. S1 Maturation of plant N-glycoproteins.
Fig. S2 Example of glycopeptide collection from Arabidopsis protein sequences to the in-house glycopeptide database.
Fig. S3 Construction of a plant N-glycan library for structural glycoprotein identification.
Fig. S4 Refined databases of Arabidopsis glycoprotein and glycopeptide sequences from the TAIR10 and NCBI.
Fig. S5 Qualitative and quantitative analyses of N-glycoproteins in Arabidopsis in response to chilling stress.
Fig. S6 Identification of N-glycosylation structures in different GDSL esterase/lipase isoforms in Arabidopsis.
Fig. S7 Structure modeling of GDSL esterase/lipase (AT1G54000) using Phyre2 server.
Fig. S8 Endoplasmic reticulum quality control (ERQC)/ER-associated degradation (ERAD) of the ER-resident proteins.
Fig. S9 ER-associated stress-responsive N-glycan degradation to the cold treatments.
Fig. S10 N-linked Man8–9GlcNAc2 of nonregulated ER protein CLPTM1.
Fig. S11 MS spectrum of the tryptic glycopeptide pairs of protein EMB3012 from the mixture of unlabeled and 15N-labeled Arabidopsis seedlings.
Fig. S12 Up-regulation of mannosidases (MNS1, MNS2, MNS3) in the cold treatments in Arabidopsis.
Fig. S13 Proposed reversible ammonolysis of N-glycans at the terminal mannose.
Fig. S14 Overview of the ER stress-related glycoprotein in response to the cold treatment.
Table S1 Delta masses of plant N-glycans
Table S2 Identification of stress-related glycoproteins by LC-MS/MS and database search
Table S3 Summary of the ER-stress related glycoproteins identified by LC-MS/MS analyses
Table S4 Detailed quantitative results of the ER resident glycoproteins in Arabidopsis using 15N-metabolically labeled glycopeptides
Table S5 Detailed quantitative results of ER resident proteins by LC MS/MS analyses of nonglycosylated peptides
Acknowledgments
The research work was supported by the 100 Technical Talents Program of the Chinese Academy of Sciences (to Y-M.S). We also thank Professor Renyi Liu and Dr Xing Fu at the Shanghai Center for Plant Stress Biology, Chinese Academy of Sciences, for their technical support with the construction of the in-house databases of Arabidopsis glycoproteins and glycopeptides.
Footnotes
Author contributions
Y-M.S., J.L. and J-K.Z. planned and designed the research. J.M. and J.S. performed the glycoproteomics experiments. D.W. conducted western blots. J.M., D.W. and J.S. analyzed the data. Y-M.S., J.L. and J-K.Z. wrote the manuscript.
References
- Bae MS, Cho EJ, Choi EY, Park OK. Analysis of the Arabidopsis nuclear proteome and its response to cold stress. Plant Journal. 2003;36:652–663. doi: 10.1046/j.1365-313x.2003.01907.x. [DOI] [PubMed] [Google Scholar]
- Bonin CP, Potter I, Vanzin GF, Reiter WD. The MUR1 gene of Arabidopsis thaliana encodes an isoform of GDP-D-mannose-4,6-dehydratase, catalyzing the first step in the de novo synthesis of GDP-L-fucose. Proceedings of the National Academy of Sciences, USA. 1997;94:2085–2090. doi: 10.1073/pnas.94.5.2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai H, Wang CC, Tsou CL. Chaperone-like activity of protein disulfide isomerase in the refolding of a protein with no disulfide bonds. Journal of Biological Chemistry. 1994;269:24550–24552. [PubMed] [Google Scholar]
- Ceriotti A, Duranti M, Bollini R. Effects of N-glycosylation on the folding of plant proteins. Journal of Experimental Botany. 1998;49:1091–1103. [Google Scholar]
- Engen JR, Wales TE. Analytical aspects of hydrogen exchange mass spectrometry. Annual Review of Analytical Chemistry. 2015;8:127–148. doi: 10.1146/annurev-anchem-062011-143113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanata WI, Lee KH, Son BH, Yoo JY, Harmoko R, Ko KS, Ramasamy NK, Kim KH, Oh DB, Jung HS, et al. N-glycan maturation is crucial for cytokinin-mediated development and cellulose synthesis in Oryza sativa. Plant Journal. 2013;73:966–979. doi: 10.1111/tpj.12087. [DOI] [PubMed] [Google Scholar]
- van Hengel AJ, Roberts K. Fucosylated arabinogalactan-proteins are required for full root cell elongation in Arabidopsis. Plant Journal. 2002;32:105–113. doi: 10.1046/j.1365-313x.2002.01406.x. [DOI] [PubMed] [Google Scholar]
- Hong Z, Jin H, Tzfira T, Li J. Multiple mechanism-mediated retention of a defective brassinosteroid receptor in the endoplasmic reticulum of Arabidopsis. Plant Cell. 2008;20:3418–3429. doi: 10.1105/tpc.108.061879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoofnagle AN, Resing KA, Ahn NG. Protein analysis by hydrogen exchange mass spectrometry. Annual Review of Biophysics and Biomolecular Structure. 2003;32:1–25. doi: 10.1146/annurev.biophys.32.110601.142417. [DOI] [PubMed] [Google Scholar]
- Hosokawa N, Wada I, Hasegawa K, Yorihuzi T, Tremblay LO, Herscovics A, Nagata K. A novel ER alpha-mannosidase-like protein accelerates ER-associated degradation. EMBO Reports. 2001;2:415–422. doi: 10.1093/embo-reports/kve084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell SH. Endoplasmic reticulum response in plants. Annual Review of Plant Biology. 2013;64:477–499. doi: 10.1146/annurev-arplant-050312-120053. [DOI] [PubMed] [Google Scholar]
- Hüttner S, Veit C, Vavra U, Schoberer J, Liebminger E, Maresch D, Grass J, Altmann F, Mach L, Strasser R. Arabidopsis class I α-mannosidases MNS4 and MNS5 are involved in endoplasmic reticulum-associated degradation of misfolded glycoproteins. Plant Cell. 2014;26:1712–1728. doi: 10.1105/tpc.114.123216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang JS, Frank J, Kang CH, Kajiura H, Vikram M, Ueda A, Kim S, Bahk JD, Triplett B, Fujiyama K, et al. Salt tolerance of Arabidopsis thaliana requires maturation of N-glycosylated proteins in the Golgi apparatus. Proceedings of the National Academy of Sciences, USA. 2008;105:5933–5938. doi: 10.1073/pnas.0800237105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJ. The Phyre2 web portal for protein modeling, prediction and analysis. Nature Protocols. 2015;10:845–858. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim I, Xu W, Reed JC. Cell death and endoplasmic reticulum stress: disease relevance and therapeutic opportunities. Nature Reviews Drug Discovery. 2008;7:1013–1030. doi: 10.1038/nrd2755. [DOI] [PubMed] [Google Scholar]
- Kline KG, Barrett-Wilt GA, Sussman MR. In planta changes in protein phosphorylation induced by the plant hormone abscisic acid. Proceedings of the National Academy of Sciences, USA. 2010;107:15986–15991. doi: 10.1073/pnas.1007879107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koiwa H, Li F, McCully MG, Mendoza I, Koizumi N, Manabe Y, Nakagawa Y, Zhu J, Rus A, Pardo JM, et al. The STT3a subunit isoform of the Arabidopsis oligosaccharyltransferase controls adaptive responses to salt/osmotic stress. Plant Cell. 2003;15:2273–2284. doi: 10.1105/tpc.013862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koizumi N, Ujino T, Sano H, Chrispeels MJ. Overexpression of a gene that encodes the first enzyme in the biosynthesis of asparagine-linked glycans makes plants resistant to tunicamycin and obviates the tunicamycin-induced unfolded protein response. Plant Physiology. 1999;121:353–361. doi: 10.1104/pp.121.2.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Wang C, Chen T, Chen P. Quantitative proteomic analysis of cold-responsive proteins in Abelmoschus moschatus. Journal of Animal & Plant Sciences. 2012;14:2006–2023. [Google Scholar]
- Liebminger E, Hüttner S, Vavra U, Fischl R, Schoberer J, Grass J, Blaukopf C, Seifert GJ, Altmann F, Mach L, et al. Class I alpha-mannosidases are required for N-glycan processing and root development in Arabidopsis thaliana. Plant Cell. 2009;21:3850–3867. doi: 10.1105/tpc.109.072363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Määttänen P, Gehring K, Bergeron JJM, Thomas DY. Protein quality control in the ER: the recognition of misfolded proteins. Seminars in Cell & Developmental Biology. 2010;21:500–511. doi: 10.1016/j.semcdb.2010.03.006. [DOI] [PubMed] [Google Scholar]
- Marino K, Bones J, Kattla JJ, Rudd PM. A systematic approach to protein glycosylation analysis: a path through the maze. Nature Chemical Biology. 2010;6:713–723. doi: 10.1038/nchembio.437. [DOI] [PubMed] [Google Scholar]
- Miernyk JA. Protein folding in the plant cell. Plant Physiology. 1999;121:695–703. doi: 10.1104/pp.121.3.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molinari M. N-glycan structure dictates extension of protein folding or onset of disposal. Nature Chemical Biology. 2007;3:313–320. doi: 10.1038/nchembio880. [DOI] [PubMed] [Google Scholar]
- Pagny S, Cabanes-Macheteau M, Gillikin JW, Leborgne-Castel N, Lerouge P, Boston RS, Faye L, Gomord V. Protein recycling from the Golgi apparatus to the endoplasmic reticulum in plants and its minor contribution to calreticulin retention. Plant Cell. 2000;12:739–756. doi: 10.1105/tpc.12.5.739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattison RJ, Amtmann A. N-glycan production in the endoplasmic reticulum of plants. Trends in Plant Science. 2009;14:92–99. doi: 10.1016/j.tplants.2008.11.008. [DOI] [PubMed] [Google Scholar]
- Qing D, Yang Z, Li M, Wong WS, Guo G, Liu S, Guo H, Li N. Quantitative and functional phosphoproteomic analysis reveals that ethylene regulates water transport via the C-terminal phosphorylation of aquaporin PIP2;1 in Arabidopsis. Molecular Plant. 2016;9:158–174. doi: 10.1016/j.molp.2015.10.001. [DOI] [PubMed] [Google Scholar]
- Rayon C, Lerouge P, Faye L. The protein N-glycosylation in plants. Journal of Experimental Botany. 1998;49:1463–1472. [Google Scholar]
- Rivier AS, Castillon GA, Michon L, Fukasawa M, Romanova-Michaelides M, Jaensch N, Hanada K, Watanabe R. Exit of GPI-anchored proteins from the ER differs in yeast and mammalian cells. Traffic. 2010;11:1017–1033. doi: 10.1111/j.1600-0854.2010.01081.x. [DOI] [PubMed] [Google Scholar]
- Russell D, Oldham NJ, Davis BG. Site-selective chemical protein glycosylation protects from autolysis and proteolytic degradation. Carbohydrate Research. 2009;344:1508–1514. doi: 10.1016/j.carres.2009.06.033. [DOI] [PubMed] [Google Scholar]
- von Schaewen A, Frank J, Koiwa H. Role of complex N-glycans in plant stress tolerance. Plant Signaling & Behavior. 2008;3:871–873. doi: 10.4161/psb.3.10.6227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skirycz A, Memmi S, De Bodt S, Maleux K, Obata T, Fernie AR, Devreese B, Inzé D. A reciprocal 15N-labeling proteomic analysis of expanding Arabidopsis leaves subjected to osmotic stress indicates importance of mitochondria in preserving plastid functions. Journal of Proteomics. 2011;10:1018–1029. doi: 10.1021/pr100785n. [DOI] [PubMed] [Google Scholar]
- Song W, Henquet MG, Mentink RA, van Dijk AJ, Cordewener JH, Bosch D, America AH, van der Krol AR. N-glycoproteomics in plants: perspectives and challenges. Journal of Proteomics. 2011;74:1463–1474. doi: 10.1016/j.jprot.2011.05.007. [DOI] [PubMed] [Google Scholar]
- Song W, Mentink RA, Henquet MG, Cordewener JH, van Dijk AD, Bosch D, America AH, van der Krol AR. N-glycan occupancy of Arabidopsis N-glycoproteins. Journal of Proteomics. 2013;93:343–355. doi: 10.1016/j.jprot.2013.07.032. [DOI] [PubMed] [Google Scholar]
- Sung DY, Vierling E, Guy CL. Comprehensive expression profile analysis of the Arabidopsis Hsp70 gene family. Plant Physiology. 2001;126:789–800. doi: 10.1104/pp.126.2.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Termine DJ, Moremen KW, Sifers RN. The mammalian UPR boosts glycoprotein ERAD by suppressing the proteolytic downregulation of ER mannosidase I. Journal of Cell Science. 2009;122:976–984. doi: 10.1242/jcs.037291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokunaga F1, Brostrom C, Koide T, Arvan P. Endoplasmic reticulum (ER)-associated degradation of misfolded N-linked glycoproteins is suppressed upon inhibition of ER mannosidase. Journal of Biological Chemistry. 2000;275:40757–40764. doi: 10.1074/jbc.M001073200. [DOI] [PubMed] [Google Scholar]
- Zielinska DF, Gnad F, Wiśniewski JR, Mann M. Precision mapping of an in vivo N-glycoproteome reveals rigid topological and sequence constraints. Cell. 2010;141:897–907. doi: 10.1016/j.cell.2010.04.012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Maturation of plant N-glycoproteins.
Fig. S2 Example of glycopeptide collection from Arabidopsis protein sequences to the in-house glycopeptide database.
Fig. S3 Construction of a plant N-glycan library for structural glycoprotein identification.
Fig. S4 Refined databases of Arabidopsis glycoprotein and glycopeptide sequences from the TAIR10 and NCBI.
Fig. S5 Qualitative and quantitative analyses of N-glycoproteins in Arabidopsis in response to chilling stress.
Fig. S6 Identification of N-glycosylation structures in different GDSL esterase/lipase isoforms in Arabidopsis.
Fig. S7 Structure modeling of GDSL esterase/lipase (AT1G54000) using Phyre2 server.
Fig. S8 Endoplasmic reticulum quality control (ERQC)/ER-associated degradation (ERAD) of the ER-resident proteins.
Fig. S9 ER-associated stress-responsive N-glycan degradation to the cold treatments.
Fig. S10 N-linked Man8–9GlcNAc2 of nonregulated ER protein CLPTM1.
Fig. S11 MS spectrum of the tryptic glycopeptide pairs of protein EMB3012 from the mixture of unlabeled and 15N-labeled Arabidopsis seedlings.
Fig. S12 Up-regulation of mannosidases (MNS1, MNS2, MNS3) in the cold treatments in Arabidopsis.
Fig. S13 Proposed reversible ammonolysis of N-glycans at the terminal mannose.
Fig. S14 Overview of the ER stress-related glycoprotein in response to the cold treatment.
Table S1 Delta masses of plant N-glycans
Table S2 Identification of stress-related glycoproteins by LC-MS/MS and database search
Table S3 Summary of the ER-stress related glycoproteins identified by LC-MS/MS analyses
Table S4 Detailed quantitative results of the ER resident glycoproteins in Arabidopsis using 15N-metabolically labeled glycopeptides
Table S5 Detailed quantitative results of ER resident proteins by LC MS/MS analyses of nonglycosylated peptides