Abstract
Despite efforts at various levels, racial health disparities still exist in cancer patients. These inequalities in incidence and/or clinical outcome can only be explained by a multitude of factors, with genetic basis being one of them. Several investigations have provided convincing evidence to support epigenetic regulation of cancer-associated genes, which results in the differential transcriptome and proteome, and may be linked to a pre-disposition of individuals of certain race/ethnicity to early or more aggressive cancers. Recent technological advancements and the ability to quickly analyze whole genome have aided in these efforts, and owing to their relatively easy detection, methylation events are much well-characterized, than the acetylation events, across human populations. The early trend of investigating a pre-determined set of genes for differential epigenetic regulation is paving way for more unbiased screening. This review summarizes our current understanding of the epigenetic events that have been tied to the racial differences in cancer incidence and mortality. A better understanding of the epigenetics of racial diversity holds promise for the design and execution of novel strategies targeting the human epigenome for reducing the disparity gaps.
Keywords: Cancer, Racial Disparity, Epigenetics, Methylation, Acetylation
1. Introduction
Health disparities in cancer patients of different racial and ethnic backgrounds have attracted a lot of attention in recent years. Because of the increased awareness and scrutiny, some progress has been made but there is irrefutable evidence to support that racial health disparities still exist in a vast majority of cancer patients [1–4]. When discussing racial cancer health disparities, black patients, most frequently African Americans (AA), are the most common racial group compared to the Caucasian Americans (CA) or white population of European heritage, referred as European Americans. Data on races other than AA and CA, such as Hispanics and Native Americans, is emerging, and points to existence of racial cancer health disparities in these groups as well [5]. Besides racial disparities in cancer incidence and overall clinical outcome, there is incongruence in diagnosis at the initial presentation and the time between the diagnosis and initiation of treatment as well [6, 7]. Even the low participation of minorities in clinical studies is a point of concern [8]. While there are studies that suggest narrowing cancer health disparities between different populations because of concerted efforts of health and local authorities [9], there is still overwhelming data to support existence of racial disparities across almost all human cancers [10–14].
In addition to the non-biological factors (such as socioeconomic, cultural, etc.), biological factors are also believed to play important roles in cancer health disparities [15–18]. Recent data has connected inherent genetic differences, such as those resulting in increased tumor heterogeneity in AA breast cancer patients, with more aggressive tumor biology [19]. AA breast cancer patients have substantially poorer outcomes if they present with a hormone receptor-positive, HER-2 (human epidermal growth factor receptor-2)-negative or the triple negative phenotype [20]. While the genetic basis of disparity is being actively pursued [17, 21], recent findings provide strong suggestion for an epigenetic basis of racial health disparities in cancer, which is discussed in the following sections.
2. Epigenetics in Cancer Health Disparities
Epigenetics is the study of changes in gene expression that are heritable and caused by events other than the change in DNA/gene sequence [22] (Figure 1). Epigenetic events have largely been identified as covalent modifications of either DNA or the histones [23]. However, regulation of gene expression by microRNAs (miRNAs) and other non-coding RNAs (ncRNAs) is also within the broader definition of epigenetics [24]. Covalent modifications include DNA methylation and various modifications of histones viz. acetylation, methylation, phosphorylation, ubiquitination and sumoylation.
Figure 1. Epigenetic events.
Epigenetic events such as methylation and acetylation have profound effect on the transcription of individual genes. These events serve as ‘ON’ / ‘OFF’ switches for transcription of genes and the eventual expression of gene products. While generally tightly regulated in ‘normal’ cells, these epigenetic changes are altered in cancer cells to favor proliferation, anti-apoptotic and pro-metastatic functions. The evidence suggesting a role of epigenetic events in cancer racial disparity is emerging, and more detailed studies are needed to fully understand the complex role of epigenetic events in cancer health disparities.
The research on health disparities has traditionally focused on socioeconomic factors [25]. Socio-economically disadvantaged and low-income populations suffer from disproportionately high incidence and/or mortality rates of cancer [26] as well as other chronic diseases [27, 28]. Social, economic and cultural inequalities, including unsafe living conditions and other stress-inducing conditions, can result in a distinct epigenetic signature with altered epigenetic markers [29–31] and even an accelerated epigenetic aging [32]. Further, lifestyle changes, such as diet and weight loss, have also been reported to influence global DNA methylation levels [33]. The epigenetic changes thus function as a liaison between social/cultural/economic/environmental factors and the genome. Since these inequalities persist for prolonged periods of time, often lifetime, the resulting epigenetic changes keep accumulating and affecting the epigenome in various racial/ethnic groups, resulting in poorer health outcomes, including cancer.
A role of epigenetics in cancer progression, metastasis, angiogenesis and drug resistance is well appreciated, and targeting of epigenome for cancer control is one of the latest areas of focus in cancer research [34, 35]. The information on the role of epigenetics in racial cancer health disparities is scattered, and through the next few sections we will attempt to comprehensively summarize all such information to make a case for the importance of epigenetic events in racial diversity observed in cancer patients.
2.1. DNA Methylation in Cancer Health Disparities
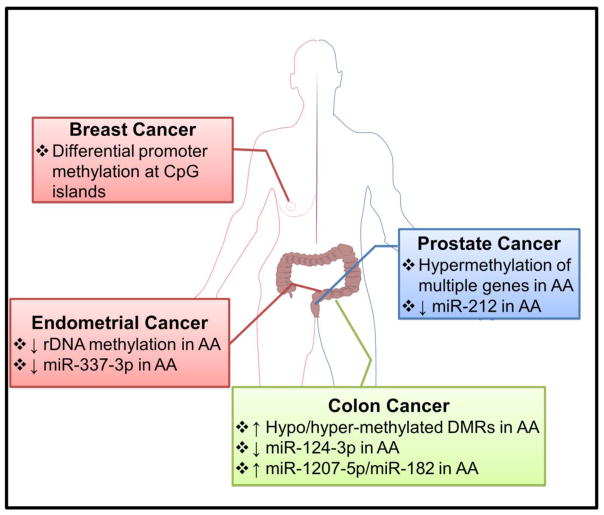
Among all the epigenetic events investigated so far for possible implications in cancer health disparities, methylation is evidently the subject of most interest [36]. Phosphate-linked cytosine and guanine nucleotides, the ‘CpG’ islands, are regions in the DNA with high frequency of CpG repeats. Methylation at CpG islands is an emerging biomarker for cancer diagnosis [37]. There are ~45000 CpG islands per haploid genome in humans [38]. A majority of these (80–90%) are methylated but the rest, found in the promoters of more than half of the genes, remain unmethylated. Methylation status is intricately linked to induction and suppression of genes, wherein reduced methylation switches ‘on’ the expression of genes and increased methylation results in switching ‘off’. Breast, prostate, colorectal and endometrial cancers are the only cancers where a link between DNA methylation and racial cancer health disparities has been suggested (Table 1). A discussion of such reports is presented in the sub-sections below.
Table 1.
Altered epigenetic events, relevant to racial disparity, as reported in cancer patients
| Cancer | Gene(s) | Observation | Reference |
|---|---|---|---|
| Breast | CDH13 | Differential methylation of CDH13 | [56] |
| - | ER-negative tumors are differentially methylated in AA | [43, 56, 57] | |
| DSC2, KCNK4, GSTM1, AXL, DNAJC15, HBII-52, TUSC3 and TES | Differential regulation of gene panel | [63] | |
| Prostate | AR, RARβ2, SPARC, TIMP3 and NKX2-5 | Differential regulation of 5 genes from a panel of 6 genes tested | [76] |
| CD44 | Hypermethylation of CD44 alone in AA, from a panel of 3 genes tested | [93] | |
| CD44 | Hypermethylation of CD44 alone in AA, from a panel of 8 genes tested | [94] | |
| GSTP1 | Hypermethylation of GSTP1 in AA | [92] | |
| TIMP3 and NKX2-5 | Possible differential acetylation | [76] | |
| SNRPN, MST1R and ABCG5 | Increased frequency of differentially mediated genes in AA | [101] | |
| TMS1 | Increased methylation in CA cancer patients, relative to healthy controls | [107] | |
| Colorectal | - | Significantly increased differentially methylated regions. CHL1, NELL1, GDF1, ARHGEF4 and ITGA4 hypermethylated in AA | [110] |
| MGMT | Differential methylation of MGMT in AA patients | [129] | |
| Endometrial | - | Differential ribosomal DNA methylation | [132] |
2.1.1. DNA methylation in Breast Cancer Health Disparity
Breast cancer is the most frequently diagnosed cancer in females in the United States [1, 39]. The data for the year 2013 from Surveillance, Epidemiology and End Results (SEER) database puts breast cancer disparity ratio at 39.14% (28.19 AA deaths Vs. 20.26 CA deaths, per a population size of 100,000). Interestingly, the overall incidence rate of breast cancer among AA women is slightly less than CA women in the US (124.3 vs. 128.1, per a population size of 100,000) [1]. Combined with the mortality rate, this indicates that the breast cancer in AA women is comparatively more aggressive, an idea that is well supported by published literature [17, 19, 40]. Triple-negative breast cancers, the aggressive breast cancer subtype with no targeted therapy, are diagnosed at relatively younger age in AA women [41], and have worse clinical outcomes [42], compared to CA women. A number of studies investigating epigenetic basis of breast cancer disparity have looked at a pre-determined subset of genes, with a focus on the methylation status.
In one of the early studies on the subject [43], AA breast cancer patients were found to harbor higher frequency of methylation in four of five genes studied. The study looked at promoter methylation of 5 select genes viz. HIN-1 (high in normal-1), Twist, Cyclin D2, RAR-β (retinoic acid receptor-beta) and RASSF1A (ras associated domain family 1 isoform A) in 67 AA vs. 44 CA breast tumors. The five genes were selected based on the reports on their hypermethylation in breast cancers, which provided a rationale for this analysis. HIN-1’s selective hypermethylation in breast cancer epithelial cells has been linked to substantial loss of expression [43–45]. Moreover, reintroduction of HIN-1 to breast cancer cells has been demonstrated to reduce the growth of breast cancer cells, suggesting its potent tumor suppressor properties [44]. Further, the hypermethylation-induced inhibition of HIN-1 in majority of AA patients suggests a possible mechanism of methylation-induced disparate cancer health outcomes [43]. Twist is methylated in ~40% primary breast cancers [43, 45], and the study by Mehrotra et al. [43] demonstrated an increased promoter methylation of Twist in the AA cancer patients. In a study by Gort et al. [46], no correlation between Twist promoter methylation and protein/RNA expression was observed even though Twist promoter methylation was observed to be significantly more prevalent in malignant, as compared to healthy tissues. The observations regarding methylation of Twist are intriguing. Twist is a basic helix-loop-helix transcription factor that is considered to play important role in EMT (epithelial-to-mesenchymal transition) and metastasis; thus, correlating with poor overall survival [47, 48]. At the same time, methylation of its promoter has also been reported in breast cancers [49], which is counterintuitive because methylation silences expression. As a possible explanation of how promoter methylation as well as gene product can correlate with cancer progression, it has been suggested that either the proximal part of Twist1 promoter does not influence Twist1 expression or Twist1 methylation is probably an early event that precedes compensatory Twist1 over-expression [46, 50]. Expression of Cyclin D2 is not observed in a majority of breast cancer cells lines [51] and its promoter is methylated in ~50% primary breast cancers [43, 45]. RAR-β is methylated in ~50% invasive breast cancers and was observed to be hypermethylated in 76% of the AA cases vs. only 29% of the CA cases [43, 45]. Restoration of RAR-β expression in breast cancer cell lines has been associated with cell cycle arrest and induction of apoptosis [52]. In other studies, promoter methylation-induced silencing of the RAR-β gene has been reported not only in breast, but cervical cancers as well [53, 54]. Similarly, RASSF1A gene, methylated in 50–60% primary breast cancers, is a potent tumor suppressor, which is found to be hypermethylated in significantly greater number of AA cases than CA cases [43, 45, 55].
In the study by Mehrotra et al. [43], only the ER (estrogen receptor)-negative/PR (progesterone receptor)-negative breast tumors differed in the methylation patterns in AA vs. CA samples. Additionally, age of the patients below 50 was also an important determining factor in differential methylation of genes, suggesting that inherent epigenetic differences could possibly be linked to documented earlier onset of the breast cancer in most AA women, as compared to CA women. Similarly, the study by Wang and co-workers [56] also found the differential methylation status in AA vs. CA patients in a subset of patients of age <50 with ER-negative breast tumors. The finding that the ER-negative breast tumors are more differentially methylated than ER-positive breast tumors in AA, relative to CA patients, was confirmed by a genome-wide methylation study [57].
In another study that compared DNA methylation in 32 AA vs. 33 CA breast cancer patients, CpG islands were evaluated for the genes CDKN2A, RASSF1A, RARβ2, ESR1 (estrogen receptor-1), LINE1 (long interspersed nuclear element-1), CDH13 (cadherin-13), HIN1 and SFRP1 (secreted frizzled related protein-1) [56]. CDKN2A is a known tumor suppressor encoding for the protein p16. ESR1 codes for estrogen receptor 1 which is important for hormone binding and activation of transcription. Mutations in ESR1 have been implicated in resistance to endocrine therapy [58] and metastasis [59] in breast cancer patients. LINE1 is a global methylation marker and LINE1 repeats make up roughly 17% of human genome and, therefore, its promoter methylation is widely used as a surrogate marker for global DNA methylation [60]. Other six genes studied are known growth suppressor genes, whose methylation and consequential suppression of expression is relevant to cancer progression. The one gene that stood out in this study was CDH13, which exhibited significantly differential methylation status in AA patients, compared to the CA patients. More importantly, CDH13 methylation was particularly increased in triple negative breast cancers. Other studies have also demonstrated CDH13 to be frequently silenced in breast tumors because of its promoter methylation [61] and associated its promoter methylation with increased breast cancer risk [62]. More recently [63], 216 AA breast tumors were compared with 301 non-AA breast tumors for differential DNA methylation at the CpG islands of select cancer-related genes. The genes that were observed to be particularly differentially methylated were DSC2 (desmocollin-2), KCNK4 (potassium channel subfamily K member 4), GSTM1 (glutathione S-transferase mu-1), AXL, DNAJC15 (DnaJ heat shock protein family member C15), HBII-52 (also known as SNORD115@ small nucleolar RNA, C/D box 115 cluster), TUSC3 (tumor suppressor candidate 3) and TES (testin). Methylation-induced silencing of a number of these genes is well documented in different human cancers. For instance, DSC2’s [64] and TES’s [65] methylation-induced silencing has been reported in breast cancer. There has been an interest in methylation of AXL because of the observation that it is an imprinted gene whose methylation-influenced expression can be maternally transmitted [66]. As a consequence of this ‘heritability’ of AXL methylation, it has been shown that fetal exposure of children, predominantly girls, to maternal smoking can induce methylation of AXL [67]. Methylation of GSTM1 [68] and TUSC3 [69] has been linked to poor disease-free survival of acute myeloid leukemia and ovarian cancer patients, respectively. Benevolenskaya and colleagues [70], analyzed 75 paraffin-embedded breast samples from 21 CA, 31 AA, and 23 Hispanic patients, and reported hypermethylation of promoters of genes FZD9 (fizzled-9), MME (membrane metalloendopeptidase), BCAP31 (B-cell receptor-associated protein-31), HDAC9 (histone deacetylase-9), PAX6 (paired box-6), SCGB3A1 (secretoglobin family 3A member 1), PDGFRA (platelet-derived growth factor receptor alpha), IGFBP3 (insulin like growth factor binding protein-3), and PTGS2 (prostaglandin-endoperoxide synthase 2), and they all correlated with receptor (ER and PR)-positive disease.
2.1.2. DNA methylation in Prostate Cancer Health Disparity
Prostate cancer is the most common non-cutaneous malignancy in men in the United States [39]. According to the data from US SEER database for year 2013, prostate cancer had a disparity ratio of 118.52% (39.05 AA deaths Vs. 17.87 CA deaths, per a population size of 100,000). There is disparity in both the incidence and mortality rates of prostate cancer, with the numbers being disproportionately higher for AA men; nearly two-thirds higher incidence and more than double mortality [1, 71–75]. In a study that investigated molecular basis of racial disparities in prostate cancer, a set of six genes (GSTP1, glutathione S-transferase pi-1; AR, androgen receptor; RARβ2; SPARC, secreted protein acidic and rich in cysteine; TIMP3, tissue inhibitor of metalloproteinase 3 and NKX2-5, NK2 homeobox-5) was chosen based on the reports on their hypermethylation in patient samples [76]. AR signaling, central to prostate cancer progression [77], is known to be epigenetically regulated [78]. The tumor suppressor RARβ2, similar to its hypermethylation in breast cancer [54, 79], cervical cancer [53] and leukemia [80], is significantly more hypermethylated in prostate cancer patients, and correlates with increased tumor risk [81]. Role of SPARC has been investigated in multiple cancer models [82] and its function largely depends on whether it is produced by cancer cells or the stromal cells in the tumor microenvironment [82]. It is frequently down-regulated in cancer cells [83] and suppresses prostate cancer cells’ proliferation and migration [84]. It is silenced through promoter methylation in metastatic and aggressive prostate cancer cells, with its promoter hypermethylation inversely correlating with prostate cancer patients’ disease-free survival [85]. Hypermthylation-induced silencing of SPARC has also been reported in pancreatic cancer [86], ovarian cancer [87] hepatocellular carcinoma [88] and laryngeal and hypopharyngeal carcinomas [89]. Low TIMP3 expression in many cancers, including prostate cancer, has been demonstrated to facilitate metastatic potential of tumor cells, due to an increase in the activity of matrix metalloproteinase activity in the tumor microenvironment [90]. Similarly, the gene product of NKX2-5 has been proposed as a tumor suppressor in prostate cancer, while its precise biological activity is yet to be determined [91]. Hypermethylation of all of these genes in prostate cancer samples was observed, relative to matched normal samples [76]. However, only five of them, with the exception of GSTP1, were found to be relevant to racial cancer health disparity, exhibiting increased methylation in AA prostate cancers, compared to CA prostate cancers. There was interest in GSTP1 hypermethylation because of an earlier observation where GSTP1 hypermethylation was proposed as a sensitive biomarker in AA prostate cancer patients, compared to CA prostate cancer patients [92]. However, some other studies [93, 94] also failed to find a utility of GSTP1 in racial disparity in prostate cancer patients. One study looked at hypermethylation of GSTP1, CD44 and E-cadherin [93] and, while it failed to find differential hypermethylation of GSTP1 in AA patients, it did find such correlation for CD44, with its 1.7 folds hypermethylation in AA prostate cancer patients, compared to the CA counterparts. Another study by the same group looked at an even larger set of genes viz. GSTP1, RASSF1A, RARβ2, CD44, EDNRB (endothelin receptor type B), E-cadherin, Annexin-2 and Caveolin-1, and again found a possible differential CD44 hypermethylation in AA prostate cancer patients [94]. CD44 is a critical marker of prostate cancer stem cells [95] and its differential expression, through epigenetic regulation, can influence metastasis as well as resistance to therapy. Interestingly, loss of CD44 in primary prostate cancer has also been linked to unfavorable clinical behavior [96]. In addition to prostate cancer [97], hypermethylation-induced regulation of CD44 has been reported in other cancers, including breast [98], colorectal [99] and neuroblastoma [100].
In the study discussed above [76] that investigated the methylation of a six gene panel in prostate cancer patients, the effect of demethylating agent, 5′aza-dC, was also compared with that of histone deacetylase inhibitor, TSA (trichostatin A). First, the findings from methylation part of the study were validated. With a hypothesis that the demethylating agent reduces methylation, resulting in re-expression of genes, it was no surprise to find such effect on almost all genes in prostate cell line pNT1a. However, an effect of TSA was also observed on a few genes namely, TIMP3 and NKX2-5, suggesting that the expression of these two genes is controlled by acetylation as well. While the earlier study [76] employed pyrosequencing to quantitate methylation, a latter study [101] by the same researchers used genome-wide methylation analysis of prostate cancer tissues from AA vs. CA patients. A clear discrepancy in frequency of differentially methylated genes was observed with samples from AA patients exhibiting greater rate of differentially methylated genes. 3303 probe sets showed more than 1.5 fold changes in methylation in AA patients, while only 1075 probe sets showed more than 1.5 fold changes in the CA patients. Of these differentially methylated probes, only 330 probe sets were common to both; 2973 probe sets were unique to AA patients and 745 were unique to CA patients. The top 25 differentially methylated genes in AA vs CA patients were further subjected to pyrosequencing in 7 prostate cancer cell lines representing the two different racial origins. The three genes that stood out with possible differential methylation status in AA vs CA were SNRPN (small nuclear ribonucleoprotein polypeptide N), MST1R (macrophage stimulating 1 receptor) and ABCG5 (ATP binding cassette subfamily G member 5). SNRPN is down-regulated in cancer samples [102], and higher methylation of SNRPN’s promoter CpG islands in prostate cancer tissues, relative to normal tissues, has been confirmed by others as well [103]. The SNRPN encoded protein has a role in the processing of pre-mRNA and possibly governs alternative splicing events in tissue-specific manner [104]. MST1R’s promoter methylation leading to decreased expression has also been reported in cancers other than prostate [105]. Further, stromal cell expression of MST1R within the prostate tumor microenvironment has been observed to drive tumor growth [106]. Knock-down of genes in prostate cancer cell lines of CA origin, to mimick the effect of hypermethylation in AA tissues, resulted in significant inhibition of proliferation and invasion [101].
In a study that looked at the tumor suppressor, TMS1 (target of methylation-induced silencing-1, also known as ASC, apoptosis-associated speck-like protein containing a CARD), an evidence for its differential racial methylation was seen [107]. Prevalence wise, no difference in methylation of this gene was observed in AA (66.7%) vs. CA (62.2%) prostate cancer patients. However, when these numbers were compared to those in healthy individuals, it was observed that TMS1 methylation was more prevalent in CA prostate cancer patients than in the CA healthy controls. AA populations, on the other hand, had similar prevalence in healthy individuals as well as prostate cancer patients. Based on all the available evidence on differentially-methylated genes in AA vs. CA prostate cancer patients, it appears that there are some valid leads that need to be investigated further.
2.1.3. DNA methylation in Colorectal Cancer Disparity
Similar to breast and prostate cancers discussed above, colorectal cancers also have racially disparate mortality rates. SEER database 2013 suggests a mortality disparity ratio of 34.65% among US females and 46.97% among US males, per a population size of 100,000. Colorectal cancer in AAs is suggested to be more aggressive, with 28% higher incidence and 60% increased mortality associated with advance stage colorectal cancers, as compared to CA [108, 109]. There is also evidence supporting epigenetic basis of colorectal cancer disparity [110]. In this study performed on DNA and RNA samples from 6 AA and 7 CA patients, a large number of differentially-methylated regions (DMRs) were identified. More than 27K CpG sites were detected in 1688 differentially-methylated regions in the samples from AA patients, while only 764 methylated CpG sites were detected in 113 DMRs in the samples from CA patients. This is a very significant difference in the status of DMRs in AA patients, compared to CA patients. In this study, hypomethylation vs. hypermethylation in AA vs. CA colorectal cancer patients was also analyzed. 100 DMRs were found to be hypomethylated, while 1588 DMRs were hypermethylated in AA patients whereas only 4 DMRs were hypomethylated and 109 DMRs hypermethylated in CA patients. When extended to differentially methylated genes, the study revealed 23 hypermethylated genes in AA samples and 29 hypermethylated genes in CA samples. Further, 4 genes were observed to be hypomethylated in AA and 1 gene hypomethylated in CA samples. Genes CCDC178 and FLI1 were hypermethylated in both AA and CA tumor samples. However, differential methylation status of these two genes between AA and CA specimens remains unclear as this particular analysis made comparisons of methylation of AA/CA tumor specimens with their respective matched normal tissues. Still, this suggests that these two genes, in general, might be involved in cancer progression. Indeed, FLI1 (Friend leukemia integration 1) has recently been demonstrated to be hypermethylated and, consequently, down-regulated in gastric cancers, relative to non-metaplastic mucosa [111]. FLI1 is relatively abundant in adult hematopoietic tissues, compared to non-hematopoietic tissues, and its activation has been linked to malignant transformation [112]. FLI1 plays an important role in normal development and homeostasis, but its aberrant expression can result in cancer onset [113] as well as metastasis [114].
The study on DMRs in colorectal cancer [110] also identified 108 down-regulated genes and 34 up-regulated genes in AA samples, when compared directly with CA patient samples. Among the top down-regulated genes in AA patient samples, compared to CA samples, the most significantly differentially expressed gene, RPL13 (ribosomal protein L13) has actually been reported as an oncogene in gastrointestinal cancers [115]; its knockdown causes cell cycle arrest and increases sensitivity to DNA damaging agents. The next gene in the list, HMGCS2 (3-hydroxy-3-methylglutaryl-CoA synthase) is involved in cell differentiation and there is conflicting evidence in the literature regarding its role in tumorigenesis. It has been positively correlated with tumor progression, resulting in poor prognosis [116] but has also been observed to be down-regulated in several intestinal cancers in a myc-dependent way [117]. Other top down-regulated genes in AA samples with a reported oncogenic function include CES2 (carboxylesterase 2; overexpressed in pancreatic cancer and suggested as a determinant of response to FOLFIRINOX therapy [118]), KRT19 (keratin 19: influences invasiveness of breast cancers [119] and radioresistance and cancer stem cell phenotype in colorectal cancers [120]) and RPS2 (ribosomal protein S2: over-expressed in prostate cancer tissues and cell lines [121]). Further, TFF3 (trefoil factor 3) is expressed in normal breast lubules and ducts, benign lesions as well as in invasive carcinomas [122]. Given the comparatively more aggressive disease in AA, this apparent disconnect and the lack of enough information on individual genes is indicative of the enormous work that needs to be done to fully understand the genetic and epigenetic basis of cancer health disparities. On a positive note, the top differentially up-regulated genes in AA samples, compared to CA samples, have also been reported as tumor-promoting genes. These include THBS2 (thrombospondin-2; associated with poor disease-free and overall survival of colorectal cancer patients [123]), PCA3 (prostate cancer antigen-3: over-expressed in prostate cancer cells [124] and a suggested prostate cancer prognostic marker [125]), CYP1B1 (cytochrome P450 1B1; overexpressed in tumor tissues [126]) and BCAT1 (branched chain amino-acid transaminase 1; over-expressed in ovarian tumors [127], possibly through a mechanism involving hypomethylation of the gene [128]) In a study that did not specially focus on cancer health disparities, a differential-methylation of MGMT (O-6-methylguanine-DNA methyltransferase) in AA colorectal patients, particularly the elderly population, was noted [129]. Silencing of MGMT has been shown to precede and associate with the appearance of G-to-A point mutations in the KRAS gene during colorectal tumorigenesis [130]. These early reports on epigenetic basis of racial disparity in colorectal cancer patients have provided encouraging preliminary data, however, more such studies are urgently needed.
2.1.4. DNA methylation in Endometrial Cancer Disparity
For endometrial cancer, the racially disparate mortality ratio is 92.68 % (7.9 AA deaths Vs. 4.1 CA deaths, per population size of 100,000, according to US SEER database). Similar to breast cancer discussed above, while the death rate is higher in AA women, incidence rate is higher in CA women by 9.8% [1]. Endometrial cancers are diagnosed at later stage, and with higher grade disease, in AA women [131]. In addition, the incidence rate of aggressive endometrial cancer subtypes is particularly high in AA women [13]. In endometrial cancer, epigenetic changes in DNA region that codes for ribosomal RNA (referred as ribosomal DNA or rDNA) have been tied with racial cancer health disparity [132]. A comparison of methylation of rDNAs from 176 CA and 39 AA patients-derived tumors revealed that the tumors with high levels of rDNA methylation were the ones with relatively favorable prognosis. Conversely, the tumors with low levels of rDNA methylation correlated with poor prognosis and the AA patients seemed to have relatively higher percentage of such low rDNA methylations (46% vs. 22% in CA). The role of rDNA methylation in endometrial tumorigenesis remains unclear. It has been suggested that rDNA methylation might be a surrogate for overall aberrant methylation of key genes [132]. No specific genes have yet been identified that are differentially methylated between AA and CA endometrial cancers. The AA women have seen an annual percent change of 2.5% in the incidence of endometrial cancer and the 5-year survival rate of AA endometrial cancer patients is significantly less than that of CA patients [13]. While the factors responsible for this disparity are under investigation, the role of epigenetic events, such as the differential rates of rDNA methylation, needs further evaluation.
2.2. Histone modifications in Cancer Health Disparities
As discussed so far, DNA methylation has been shown to be different to some extent in AA vs CA populations (Figure 2). Another important epigenetic event is the modifications of histones, with acetylation and methylation of histones as two well-studied histone modifications. In contrast to DNA methylation, there has been little progress on our understanding of histone modifications in the context of cancer racial health disparities. This is best exemplified by a very recent report that looked at 59,089 men of African and European ancestries and did not find any significant differences in the histone acetylation [133]. The study specifically looked at H3k27 acetylation as a biomarker. The one positive indication for a role of histone modification in cancer racial disparity came from the study in prostate cancer model where an effect of TSA was seen on TIMP3 and NKX2-5 [76]. TSA is an inhibitor of histone deacetylases and thus influences the acetylation of histones. The effect of TSA was seen when it was used to treat prostate cancer cell lines. This is an indirect evidence, which needs to be corroborated directly in patient samples. The lack of convincing evidence in support of histone acetylation, and even histone methylation, is not necessarily a verdict on the relevance of these histone modifications in cancer racial disparity. We need to be cognizant that DNA methylation is relatively easy to detect and thus the reports on DNA methylation are much more forthcoming. This will surely change with time, and with the advances in technological capabilities.
Figure 2. Differential epigenetic events that underline racial cancer health disparities.
Differences in methylation patterns have been proposed as underlying causes for disparate breast, prostate, colorectal and endometrial cancers in AA vs. CA populations. There is also evidence for differential expression of individual microRNAs across racial groups. AA, African American; DMRs, Differentially Methylated Regions; rDNA, ribosomal DNA.
2.3. miRNAs and ncRNAs in Cancer Health Disparities
Gene regulation involving miRNAs and ncRNAs is yet another important epigenetic event, and a role of miRNAs and ncRNAs in cancer health disparities has started emerging (Table 2). Interestingly, expression of miRNAs may, in turn, also be regulated by epigenetic events. In this section, a discussion on the differential expression of miRNAs/ncRNAs, and the resulting functional consequence, in racially disparate cancers is provided.
Table 2.
microRNAs in racial cancer health disparities
| miRNA (s) | Observation | Cancer | Reference |
|---|---|---|---|
| miR-9 | Hypermethylated in AA | Colorectal | [110] |
| miR-26a | Up-regulated in AA | Prostate | [143] |
| miR-31 | Up-regulated in CA | Thyroid | [154] |
| miR-34 | Hypermethylated in CA | Colorectal | [110] |
| miR-124 | Hypermethylated in AA | Colorectal | [110] |
| miR-137 | Hypermethylated in AA | Colorectal | [110] |
| miR-152 | Hypermethylated | Prostate | [144] |
| miR-182 | Up-regulated in AA | Colorectal | [153] |
| miR-212 | Down-regulated in AA | Prostate | [135] |
| miR-221 | Up-regulated in CA | Thyroid | [154] |
| miR-337 | Down-regulated in CA | Endometrial | [157] |
| miR-548 | Hypermethylated in AA | Colorectal | [110] |
| miR-663 | Hypermethylated in AA | Colorectal | [110] |
| miR-1207 | Increased expression in AA | Colorectal | [151] |
| miR-1279 | Up-regulated in AA, compared to CA | Colorectal | [110] |
| miR-2682 | Hypermethylated in AA | Colorectal | [110] |
| miR-6130 | Hypermethylated in AA | Colorectal | [110] |
2.3.1. miRNAs in Prostate Cancer Health Disparity
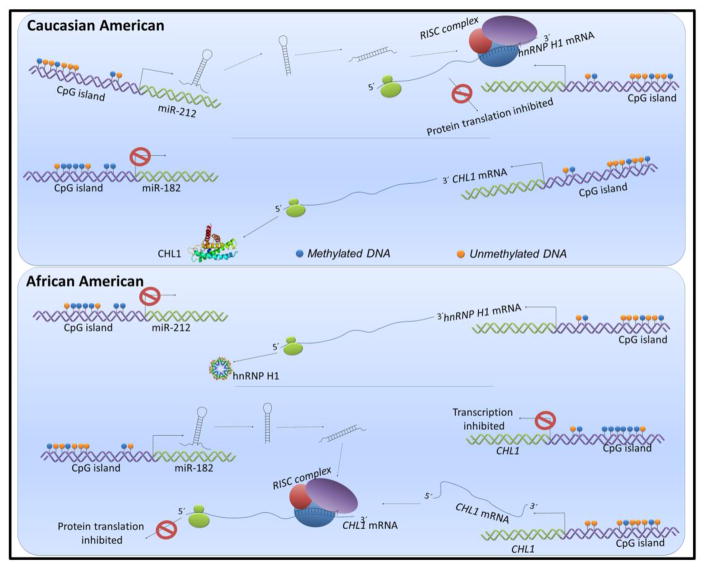
As mentioned above, prostate cancer is one of the most racially disparate cancers, with disproportionally higher mortality of AA patients, as compared to CA patients. Apart from genetic and epigenetic events, the role of miRNAs in prostate cancer health disparities is being appreciated [134]. In prostate cancer patients, miR-212, a negative regulator of splicing factor heterogeneous nuclear ribonucleoprotein H1 (hnRNP H1), was found to be down-regulated in AA patients, compared to CA patients [135]. Such down-regulation of miR-212 correlated with aberrant expression of hnRNP H1, as well as the androgen receptor, resulting in castration-resistance. In a more recent article that focused on TMPRSS2 (transmembrane protease, serine-2) translocations, a panel of 18 differentially altered miRNAs was identified [136]. These miRNAs were associated with DNA CpG methylation, and the resulting aggressive disease in TMPRSS2 fusion-negative prostate cancers. Among these, miR-125b, miR-17, miR-29 and miR-200b have been studied in prostate cancer [136]. miR-125b has been demonstrated to facilitate the development of castration-resistant prostate cancer by targeting the repressor of AR signaling, NCOR2/SMRT (nuclear receptor corepressor 2/silencing mediator of retinoic acid and thyroid hormone receptor) [137]. Similarly, in prostate cancer, miR-17 suppresses the levels of PTEN [138] and miR-29 suppresses the metastatic cascade at multiple steps [139]. miR-29 also regulates the expression of anti-apoptotic and matrix molecules after up-regulation by MBP-1 (c-myc promoter binding protein) in prostate cancer [140]. Williams et al. [141] demonstrated that miR-200b inhibits spontaneous metastasis in an orthotopic mouse model through reversal of EMT with increased E-cadherin, a phenomenon similar to one reported for this miRNA earlier in breast cancer [142]. In another study, miR-26a was found to be significantly overexpressed in AA prostate cancer cells, as compared to CA-derived cell lines [143]. While the precise mechanism of action of miR-26a in prostate cancer has not been delineated, it was observed to modulate apoptosis and cell survival, partly through the inactivation of caspase 3/7 [143]. Theodore et al. [144], observed decreased levels of miR-152 in aggressive prostate cancer cell lines, concomitant with increased promoter methylation, which could be reversed by the treatment of 5-aza-2-deoxycytidine. Clinically, a statistically significant down-regulation of miR-152 was found in about 50% of the AA cancer tissue samples, compared to 35% of the CA samples. A reciprocal inhibitory loop of miR-152-DNMT1 (DNA methyltransferase-1) has been deduced to be the underlying mechanism of action of miR-152 in prostate [144], ovarian [145] and breast cancer [146], underlying the epigenetic connection of this miRNA.
2.3.2. miRNAs and ncRNAs in Colorectal Cancer Health Disparity
Colorectal cancer is a relatively better studied malignancy in terms of miRNA regulation of cancer health disparity. For instance, when extended to differential epigenetic regulation of miRNAs, the study on cancer health disparity in colorectal cancer, discussed above [110], found a small set of differentially-hypermethylated miRNAs, with no miRNA being hypomethylated. A total of 7 miRNAs (miR-9, miR-124, miR-137, miR-548, miR-663, miR-6130 and miR-2682) were hypermethylated in AA patients’ tumors, relative to their adjacent normal tissue, while only miR-34 was hypermethylated in CA patients. Hypermethylation of the promoter region of these miRNAs leads to a decrease in the expression of these miRNAs [147–150]. Almost all these miRNAs, hypermethylated in AA tumors, are known tumor suppressors, with no reports available for the function of miR-6130 and miR-2682. Interestingly, miR-34 has been demonstrated as a potential regulator of metastasis in several cancers, but was observed to be the only miRNA hypermethylated in the CA patients. Also, miR-1279 was identified as the only miRNA dysregulated between AA and CA patient samples [110]. This miRNA was up-regulated in AA samples, however, the functional relevance of miR-1279 is not known. Another study with colorectal cancer patients (53 AA and 47 CA), not only found an evidence of miRNAs, but also of cancer stem cells and long noncoding RNA PVT1, the host for miR-1207-5p, in cancer racial disparity. AA patients had significantly high levels of miR-1207-5p, which was mechanistically linked to increased ‘stemness’, as evidenced by increase in various markers of cancer stem cells and EMT [151]. This increase in cancer stem cells can possibly explain the relatively aggressive colorectal cancers in AA patients.
Identification of differential epigenetic regulation of miRNAs is only the first step in determining a role of miRNAs in cancer health disparity. Since a single miRNA can affect the expression of a number of target genes, epigenetic regulation of just one miRNA can potentially affect the expression and function of many key genes. This underlines the impact of miRNAs. At the same time, it presents a challenge to the researchers because the identification of exact targets, among the plethora of putative targets, is very important to understand the mechanistic details as well as to exploit this knowledge for possible intervention and therapy. In the colorectal study [110], RNA sequencing was done to list the differentially-expressed genes. When the tumor samples from AA and CA patients were compared, 34 genes were found to be up-regulated, while 108 genes were down-regulated in AA samples, compared to the CA samples. The researchers compared the two different sub-sets: the list of epigenetically differentially-expressed miRNAs, and the differentially-expressed genes. This was done to possibly locate differentially expressed miRNA(s) and the target gene(s). This led to the realization that in this particular study, hypermethylation of miR-124-3p correlated with the up-regulation of two of its target genes, POLR2B (RNA polymerase II subunit B) and CYP1B1 (cytochrome P450 family 1 subfamily B member 1). Interestingly, miR-124-3p was differentially hypermethylated in AA patients and consequently these two target genes were up-regulated only in the AA patients, and not in CA patients. This observation calls for further scrutiny of POLR2B and CYP1B1 in racial disparity among colorectal AA vs. CA patients.
As mentioned above, methylation of genes is an epigenetic regulation, which controls their eventual expression. Wang et al. [110] also observed CHL1 (cell adhesion molecule L1 like) to be hypermethylated in colorectal tumors from AA patients. Such hypermethylation should result in suppression of this gene. As cross-referenced by the authors of this study, there is evidence for down-regulated CHL1 in up to 48% of colorectal cancers [152]. This suggests that CHL1, which is down-regulated in a large number of colorectal patients, is particularly hypermethylated in AA patients. Whether or not such down-regulation of CHL1 is significantly more prevalent in AA patients, is something that needs to be investigated further. Interestingly, miR-182, a miRNA that targets CHL1, was previously reported by the same group to be differentially up-regulated in AA patients [153], relative to CA patients. While this provides a complete loop that the up-regulated miR-182 in AA patients can possibly be linked to the down-regulated CHL1 in AA colorectal patients, the observed hypermethylation of CHL1 needs to be explored further. The study that identified miR-182 to be up-regulated in AA patients, also identified FOX01 (forkhead box O1) and FOXO3A (forkhead box O3) as its two gene targets that themselves were differentially-expressed in AA patients [153].
2.3.3. miRNAs in Health Disparity among other Cancers
In a study involving thyroid cancer patients [154], miRNA-array profiling identified a panel of miRNAs that was differentially-regulated in formalin-fixed paraffin-embedded tissue specimens from AA vs. CA patients. Two miRNAs (miR-221 and miR-31) were found to be of particular interest with respect to racial disparity. miR-221 was up-regulated in 92% of CA tumors, but only in 40% of AA tumors. miR-31 was up-regulated in all CA tumors, but only in 75% AA tumors. miR-31 and miR-221/222 have been associated with induction of chemoresistance in cancers [155, 156]. In an array-based determination of differentially-expressed miRNAs [157], miR-337-3p was found to be frequently down-regulated in CA endometrial tumors, compared to AA endometrial tumors. A comparison of samples from 9 patients each led to the identification of this differentially-expressed miRNA and it was further validated in an independent set of 24 AA and 23 CA patients. miR-337-3p inhibits tumor progression through the repression of matrix metalloproteinase 14 [158, 159].
The emerging knowledge on epigenetic regulation of cancer racial disparity is complex (Figure 3). As discussed above, miR-182 is up-regulated in AA colorectal cancer patients [153], compared to CA patients, and its target CHL1 is suppressed by hypermethylation in AA patients as well [152]. Thus, CHL1 seems to be regulated at multiple levels in AA patients; through its own methylation and through miR-182. miR-212, on the other hand, is down-regulated in AA prostate cancer patients [135], compared to CA patients, resulting in attenuated repression of its target hnRNP H1. Interestingly, both miR-182 and miR-212 have been reported to be regulated by methylation [160, 161], albeit in different cancer models, and not in the context of cancer racial disparity. Clearly, the information on complex epigenetic regulation, particularly the one involving miRNAs, is fragmented and warrants further thorough investigations.
Figure 3. Implications of differential miRNA expression on racial cancer health disparities.
Expression of miR-182 and miR-212 has been shown to be influenced by methylation. In AA prostate cancer patients, lower miR-212 expression results in abrogation of suppression of its target hnRNP H1. Conversely, a higher miR-182 expression in AA colorectal cancer patients leads to suppression of its target CHL1 which can, additionally, itself be regulated through differential methylation of its own CpG islands. This model describes the complex interplay of multiple epigenetic events, namely, methylation and regulation through miRNAs, in making an impact on the racially disparate cancer outcomes.
3. Race-associated epigenetic changes in healthy individuals
Some individuals might be pre-disposed to developing cancer, and there might be a role of epigenetic changes in this predisposition. Hypermethylation of ADAMTS14 (a disintegrin and metalloproteinase with thrombospondin motifs 14) was observed in the normal colon mucosa of AA colorectal cancer patients [129]. Although preliminary, this observation lands credibility to the hypothesis that differential methylation of individual or a set of genes could predispose individuals to the risk of developing cancer. Since the analysis in this study was limited to ‘healthy’ regions of colon in colorectal cancer patients, it is difficult to ascertain whether or not such hypermethylation events are actual determinants of cancer disposition, or they are just the correlated epigenetic events that associate with tumorigenesis in the larger vicinity.
Cancer racial disparity in methylation patterns in healthy individuals has been a subject of quite a few investigations [162]. This has resulted in some preliminary evidence, often indirect, supporting a differential methylation status of specific genes across ethnic populations. Examples include hypermethylated NKX2 and TIMP3 in healthy prostates of AA individuals [76], lower levels of colorectal methylated ERα and SFRP1 in healthy AA individuals [163] and hypermethylated p16(INK4) in healthy breasts of CA women [164]. In a study that looked at racial differences in DNA methylation in healthy AA vs. CA women [165], a number of distinct hypermethylated CpG sites were observed – 282 in AA and 203 in CA. Interestingly, there was a noticeable difference in the genome regions that were hypermethylated. Whereas CpG hypermethylations in AAs were more common in intergenic regions, the hypermethylations in CA were more prominent in promoter regions. It thus appears that not only is there a differential hypermethylation in AA vs CA populations, there also seem to be differential regions within the genome that are differentially methylated.
A number of epigenetic differences among races can possibly be attributed to true genetic variations, as suggested by evaluation of cytosine modifications in individuals of African ancestry (Yoruba people from Nigeria) vs. those of Caucasian of European ancestry [166]. This study found approximately 13% differentially-methylated CpG sites between the two distinct populations. These observations confirmed a preceding work [167] that reported 13.7% differentially methylated autosomal CpG sites in AA vs. CA newborns. Interestingly, the earlier work [167] also found a number of cancer pathway genes, particularly those related to pancreatic, prostate, bladder and meningioma, among those with differentially-methylated CpGs. No further data is available on the topic, but it will be very interesting to see if the differential-methylations in ‘normal’ tissue(s) correlate with a progression to tumorigenesis. This will open up new avenues for the use of epigenetic biomarkers in prediction of human cancers.
4. Epigenetic differences among other ethnic groups
Not just the CA vs. AA populations, there seem to be epigenetic differences in other populations and ethnic groups as well. For example, it has been suggested that the Chinese bladder cancer patients may have distinct methylated genes, compared to the bladder cancer patients from US [168]. This study compared its own results with two previous ones [169, 170]. One of the earlier studies [169] reported methylation of 5′ regions of multiple apoptosis-associated genes, such as DAPK (death associated protein kinase-1), BCL2, TERT (telomerase reverse transcriptase), RASSF1A and TNFRSF25 (tumor necrosis factor receptor superfamily member 25) in the urine sediments of bladder cancer patients. The other study [170] observed promoter methylation in at least one of the four genes (CDKN2A, cyclin dependent kinase inhibitor 2A; ARF, ADP ribosylation factor; MGMT and GSTP1) of 175 bladder cancer patients. The study in Chinese patients found the predictive power of a set of 11 genes, methylation of which could confirm the existing diagnosis of 87% bladder cancers studied, with a sensitivity of 91.7%. While a number of genes from the earlier US studies were investigated, only BCL2 and RASSF1A managed to make it to the 11-gene list in Chinese patients. It was pointed out that the methylation markers in Chinese bladder cancer patients might be very distinct from the patients in the US.
The discrepancy in the validity of epigenetic events in Chinese vs. Western populations was further confirmed by evaluating the DNA methylation markers for the detection of bladder cancer, where it was found that only hypermethylation of SFRP1 was critical for detection of bladder cancer in Chinese population, as opposed to several other dysregulated methylations observed in other studies conducted on bladder cancer patients from western countries [171]. Looks like the observations from bladder cancer patients are true for lung cancer patients as well because the same research team found a set of genes methylated in Chinese non-small cell lung cancer patients that was exclusively different from the one previously recommended in the studies in western populations [172].
Not just the comparison of Chinese populations with Western populations, there has also been an effort to look at the differential epigenetic markers in ethnic populations within China. In one such study [173], differential methylation of TFPI2 (tissue factor pathway inhibitor 2) was evaluated in Uygur vs. Han ethnicity cervical cancer patients. The incidence of cervical cancer is disproportionately high in Uygur women. However, no difference in TFPI2 methylation was observed between the two ethnicities. A correlation between increased methylation and HPV16 infection was, however, observed in Uygur women, which was absent in Han women. With the known importance of HPV in the etiology of cervical cancer, this comes across as preliminary evidence supporting the epigenetic basis of ethnic disparity, with a possibility of such disparity across races as well.
Currently additional reports on epigenetic differences between other racial/ethnic groups are not available; however, significant differences between the incidence of cancer and/or cancer-associated mortalities between ethnic populations in Asia and Europe Vs. those in US have been reported. These differences have been largely attributed to diet and life-style. For example, the consumption of soy in the East and the utilization of olive oil in the Mediterranean region have been argued as the basis of reduced prostate cancer-associated deaths in these populations [174, 175]. Genistein, a major constituent of soy with significant anticancer properties [176, 177], has been reported to enhance the expression of tumor suppressor genes in an epigenetic mechanism [178]. Similarly, an up-regulation of CB1 tumor suppressor to inhibit colon cancer cells via epigenetic mechanisms by extra-virgin olive oil has also been reported [179]. The consumption of tobacco and tobacco-related products in India is identified as the underlying cause of oral cancer, which contributes to ~26% of the global burden of oral cancer [180]. Tobacco constituents have been identified to cause epigenetic changes in a number of cancers including oral cancer [181]. Thus, these studies provide further in-direct suggestions to potential differences in epigenetic make-up of different racial/ethnic backgrounds, which need to be further investigated.
5. Conclusions and future perspectives
Research on racial health disparities in cancer is a rapidly emerging area of emphasis, and, sure enough, epigenetic basis of these disparities is gaining ground. Early detection is key to successful management of human cancers leading to improved overall survival of patients. Health guidelines call for periodical screening of select cancers, such as breast, prostate and colorectal cancers, all with the aim of diagnosing the disease early. It is envisioned that screening for epigenetic changes, such as DNA methylation status, might be a good strategy [182]. This also has potential of simultaneous screening for multiple cancers because there seems to be a few epigenetic events that are observed across different malignancies. The idea is still in its infancy; not yet well-tested, but worth trying.
As discussed in this article, methylation remains one of the better studied epigenetic events in the context of cancer health disparities. It is evident that a number of genes are differentially methylated in AA cancer patients, relative to CA cancer patients. Some of these genes have either already been proposed as biomarkers for specific cancers, or have been shown to be functionally involved in different stages of cancer progression. While epigenetic regulation of these genes can put individuals of any race/ethnicity at higher risk of cancer, AA populations with seemingly higher degree of basal epigenetic changes in these genes, might start at a disadvantage, and are, therefore, much more pre-disposed to cancer onset, and an even aggressive disease. In most of these studies on differential methylation in AA vs. CA cancer patient samples, the selection of genes is based on their previously reported methylation-dependent regulation of expression. While this provides a good rationale for their evaluation in studies focused on differential methylation, such as those on cancer racial disparity, it remains to be seen whether the differential methylation of genes in racially disparate populations, as observed in these studies, correlates with actual differential expression in these populations. This is an aspect that is not adequately addressed in most of the studies. Further, differential expression of these genes and their products needs to be tied to differential disease outcome in AA vs. CA populations, before the fundamental involvement of epigenetics in racial cancer health disparities can be established.
A number of factors can affect epigenetic events. Minority populations such as AAs and Hispanics often live in socio-economically disadvantaged and low income neighborhoods, and, as discussed above, social, cultural and economic inequalities leave a profound imprint on the epigenome through distinct epigenetic changes. The overall stress-full living conditions also contribute to these changes. Therefore, complex interactions among genes, environment, and the diseases, including cancer, require an examination of how epigenetic changes regulate susceptibility to various stressors. To better understand cancer disparities in disease susceptibility, future studies will need to assess the cumulative effect of various social, and environmental stressors on genetic substrates. It can also be envisioned that the populations that have migrated and inhabited a new geographical location for many generations can show distinct epigenetic traits, relative to the populations at the site of origin. This adds a new layer of complexity to the phenomenon that is already poorly understood. At the very least this means that any new observations have to be analyzed with caution. In the US, a lot of effort has traditionally been directed towards understanding the cancer health disparity between AA and CA populations. But slowly and steadily, data from Hispanic and Native American populations is also being made available. This is still not as robust as the one comparing AA vs. CA populations, but the changing ethnic proportions in the US will undoubtedly result in inclusion of other significant minorities in the studies in due course of future. When looked from the global perspective, the task seems even more challenging. Clearly, large collaborative projects with shared specimens, data and resources will be needed to address this. Alternatively, knowledge gained from simple comparisons of ethnic and racial groups in a population will need to be evaluated in entirely different population for possible comparison, overlap or exclusivity. Yet another challenge in the studies involving race is the way race/ethnicity is determined. Most often, designation of race is based on self-reporting. However, it has been suggested that a better approach would be to evaluate ‘genetic admixture’ which takes into account the intermixing of races that has happened over the last several centuries resulting in individuals/populations with admixed background [183]. This concept was tested recently in a pilot study involving endometrial cancer patients [183] and results from larger cohorts as well as other cancers will be interesting to look forward to.
The increasing interest and advancements in epigenetic connection of racial cancer health disparity are made possible by the recent advancements in techniques that have enabled a critical and quick look at epigenetic events throughout the genome. Emerging techniques such as single cell epigenomics make it possible to characterize epigenetic events within a single cell [184]. Hence, it is envisioned that further refining of techniques will make it possible to evaluate all epigenetic changes, especially those that have not yet been explored, for their relevance in cancer racial disparity.
Highlights.
Racial health disparities exist among cancer patients
Epigenetic basis of cancer health disparities is increasingly being recognized
Racially disparate genes are often differentially methylated
Regulation involving microRNAs is important for differential cancer racial outcomes
Acknowledgments
Funding
Cancer Health Disparity Research at USA MCI is supported by NCI, NIH grants CA185490 (APS) and CA204801 (SS).
Footnotes
Conflict of Interest
APS and SS are co-founders and serve on executive management team of Tatva Biosciences LLC, which is involved in the development of tools and models for cancer health disparity research.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.DeSantis CE, et al. Cancer statistics for African Americans, 2016: Progress and opportunities in reducing racial disparities. CA Cancer J Clin. 2016 doi: 10.3322/caac.21340. [DOI] [PubMed] [Google Scholar]
- 2.Hunt BR, Whitman S, Hurlbert MS. Increasing Black:White disparities in breast cancer mortality in the 50 largest cities in the United States. Cancer Epidemiol. 2014;38(2):118–23. doi: 10.1016/j.canep.2013.09.009. [DOI] [PubMed] [Google Scholar]
- 3.Hunt B, Balachandran B. Black:White disparities in lung cancer mortality in the 50 largest cities in the United States. Cancer Epidemiol. 2015;39(6):908–16. doi: 10.1016/j.canep.2015.10.003. [DOI] [PubMed] [Google Scholar]
- 4.DeSantis CE, et al. Breast cancer statistics, 2015: Convergence of incidence rates between black and white women. CA Cancer J Clin. 2016;66(1):31–42. doi: 10.3322/caac.21320. [DOI] [PubMed] [Google Scholar]
- 5.Garza AL, et al. Liver and Other Gastrointestinal Cancers Are Frequent in Mexican Americans. J Racial Ethn Health Disparities. 2016;3(1):1–10. doi: 10.1007/s40615-015-0104-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kinlock BL, et al. Racial Disparity in Time Between First Diagnosis and Initial Treatment of Prostate Cancer. Cancer Control. 2016;23(1):47–51. doi: 10.1177/107327481602300108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dosch AR, et al. Racial and ethnic disparities in the diagnosis of breast cancer: changes in presenting stage in minority populations in Florida during 1981–2009. Breast Cancer Res Treat. 2014;148(2):379–87. doi: 10.1007/s10549-014-3158-5. [DOI] [PubMed] [Google Scholar]
- 8.Scalici J, et al. Minority participation in Gynecologic Oncology Group (GOG) Studies. Gynecol Oncol. 2015;138(2):441–4. doi: 10.1016/j.ygyno.2015.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rust G, et al. Counties eliminating racial disparities in colorectal cancer mortality. Cancer. 2016 doi: 10.1002/cncr.29958. [DOI] [PubMed] [Google Scholar]
- 10.Mahdi H, et al. Racial disparity in survival of patients with uterine serous carcinoma: Changes in clinical characteristics, patterns of care and outcomes over time from 1988 to 2011. Gynecol Oncol. 2016 doi: 10.1016/j.ygyno.2016.03.002. [DOI] [PubMed] [Google Scholar]
- 11.Taioli E, et al. Racial disparities in esophageal cancer survival after surgery. J Surg Oncol. 2016;113(6):659–64. doi: 10.1002/jso.24203. [DOI] [PubMed] [Google Scholar]
- 12.Osazuwa-Peters N, et al. Race and sex disparities in long-term survival of oral and oropharyngeal cancer in the United States. J Cancer Res Clin Oncol. 2016;142(2):521–8. doi: 10.1007/s00432-015-2061-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cote ML, et al. The Growing Burden of Endometrial Cancer: A Major Racial Disparity Affecting Black Women. Cancer Epidemiol Biomarkers Prev. 2015;24(9):1407–15. doi: 10.1158/1055-9965.EPI-15-0316. [DOI] [PubMed] [Google Scholar]
- 14.McCarthy AM, Yang J, Armstrong K. Increasing disparities in breast cancer mortality from 1979 to 2010 for US black women aged 20 to 49 years. Am J Public Health. 2015;105(Suppl 3):S446–8. doi: 10.2105/AJPH.2014.302297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McGinley KF, Tay KJ, Moul JW. Prostate cancer in men of African origin. Nat Rev Urol. 2016;13(2):99–107. doi: 10.1038/nrurol.2015.298. [DOI] [PubMed] [Google Scholar]
- 16.Schwartz K, et al. Racial disparities in overall survival among renal cell carcinoma patients with young age and small tumors. Cancer Med. 2016;5(2):200–8. doi: 10.1002/cam4.578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Daly B, Olopade OI. A perfect storm: How tumor biology, genomics, and health care delivery patterns collide to create a racial survival disparity in breast cancer and proposed interventions for change. CA Cancer J Clin. 2015;65(3):221–38. doi: 10.3322/caac.21271. [DOI] [PubMed] [Google Scholar]
- 18.Daraei P, Moore CE. Racial Disparity Among the Head and Neck Cancer Population. J Cancer Educ. 2015;30(3):546–51. doi: 10.1007/s13187-014-0753-4. [DOI] [PubMed] [Google Scholar]
- 19.Keenan T, et al. Comparison of the Genomic Landscape Between Primary Breast Cancer in African American Versus White Women and the Association of Racial Differences With Tumor Recurrence. J Clin Oncol. 2015;33(31):3621–7. doi: 10.1200/JCO.2015.62.2126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tao L, et al. Breast Cancer Mortality in African-American and Non-Hispanic White Women by Molecular Subtype and Stage at Diagnosis: A Population-Based Study. Cancer Epidemiol Biomarkers Prev. 2015;24(7):1039–45. doi: 10.1158/1055-9965.EPI-15-0243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Deshmukh SK, et al. Resistin and interleukin-6 exhibit racially-disparate expression in breast cancer patients, display molecular association and promote growth and aggressiveness of tumor cells through STAT3 activation. Oncotarget. 2015;6(13):11231–41. doi: 10.18632/oncotarget.3591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ahmad A, et al. Epigenetic regulation of miRNA-cancer stem cells nexus by nutraceuticals. Mol Nutr Food Res. 2014;58(1):79–86. doi: 10.1002/mnfr.201300528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ahmad A. Epigenetics in Personalized Management of Lung Cancer. Adv Exp Med Biol. 2016;890:111–22. doi: 10.1007/978-3-319-24932-2_6. [DOI] [PubMed] [Google Scholar]
- 24.Liz J, Esteller M. lncRNAs and microRNAs with a role in cancer development. Biochim Biophys Acta. 2016;1859(1):169–76. doi: 10.1016/j.bbagrm.2015.06.015. [DOI] [PubMed] [Google Scholar]
- 25.Fiscella K, et al. Inequality in quality: addressing socioeconomic, racial, and ethnic disparities in health care. JAMA. 2000;283(19):2579–84. doi: 10.1001/jama.283.19.2579. [DOI] [PubMed] [Google Scholar]
- 26.Chornokur G, et al. Disparities at presentation, diagnosis, treatment, and survival in African American men, affected by prostate cancer. Prostate. 2011;71(9):985–97. doi: 10.1002/pros.21314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Puckrein GA, Egan BM, Howard G. Social and Medical Determinants of Cardiometabolic Health: The Big Picture. Ethn Dis. 2015;25(4):521–4. doi: 10.18865/ed.25.4.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Didsbury MS, et al. Socio-economic status and quality of life in children with chronic disease: A systematic review. J Paediatr Child Health. 2016;52(12):1062–1069. doi: 10.1111/jpc.13407. [DOI] [PubMed] [Google Scholar]
- 29.Saban KL, et al. Epigenetics and social context: implications for disparity in cardiovascular disease. Aging Dis. 2014;5(5):346–55. doi: 10.14336/AD.2014.0500346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Janusek LW, et al. Relationship of childhood adversity and neighborhood violence to a proinflammatory phenotype in emerging adult African American men: An epigenetic link. Brain Behav Immun. 2016 doi: 10.1016/j.bbi.2016.10.006. [DOI] [PubMed] [Google Scholar]
- 31.Chen E, et al. The Great Recession and health risks in African American youth. Brain Behav Immun. 2016;53:234–41. doi: 10.1016/j.bbi.2015.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miller GE, et al. Self-control forecasts better psychosocial outcomes but faster epigenetic aging in low-SES youth. Proc Natl Acad Sci U S A. 2015;112(33):10325–30. doi: 10.1073/pnas.1505063112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Delgado-Cruzata L, et al. Dietary modifications, weight loss, and changes in metabolic markers affect global DNA methylation in Hispanic, African American, and Afro-Caribbean breast cancer survivors. J Nutr. 2015;145(4):783–90. doi: 10.3945/jn.114.202853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brien GL, Valerio DG, Armstrong SA. Exploiting the Epigenome to Control Cancer-Promoting Gene-Expression Programs. Cancer Cell. 2016;29(4):464–76. doi: 10.1016/j.ccell.2016.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hamm CA, Costa FF. Epigenomes as therapeutic targets. Pharmacol Ther. 2015;151:72–86. doi: 10.1016/j.pharmthera.2015.03.003. [DOI] [PubMed] [Google Scholar]
- 36.Mohammed SI, Springfield S, Das R. Role of epigenetics in cancer health disparities. Methods Mol Biol. 2012;863:395–410. doi: 10.1007/978-1-61779-612-8_25. [DOI] [PubMed] [Google Scholar]
- 37.Kim Y, Kim DH. CpG island hypermethylation as a biomarker for the early detection of lung cancer. Methods Mol Biol. 2015;1238:141–71. doi: 10.1007/978-1-4939-1804-1_8. [DOI] [PubMed] [Google Scholar]
- 38.Antequera F, Bird A. Number of CpG islands and genes in human and mouse. Proc Natl Acad Sci U S A. 1993;90(24):11995–9. doi: 10.1073/pnas.90.24.11995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66(1):7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 40.Danforth DN., Jr Disparities in breast cancer outcomes between Caucasian and African American women: a model for describing the relationship of biological and nonbiological factors. Breast Cancer Res. 2013;15(3):208. doi: 10.1186/bcr3429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Boyle P. Triple-negative breast cancer: epidemiological considerations and recommendations. Ann Oncol. 2012;23(Suppl 6):vi7–12. doi: 10.1093/annonc/mds187. [DOI] [PubMed] [Google Scholar]
- 42.Dietze EC, et al. Triple-negative breast cancer in African-American women: disparities versus biology. Nat Rev Cancer. 2015;15(4):248–54. doi: 10.1038/nrc3896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mehrotra J, et al. Estrogen receptor/progesterone receptor-negative breast cancers of young African-American women have a higher frequency of methylation of multiple genes than those of Caucasian women. Clin Cancer Res. 2004;10(6):2052–7. doi: 10.1158/1078-0432.ccr-03-0514. [DOI] [PubMed] [Google Scholar]
- 44.Krop IE, et al. HIN-1, a putative cytokine highly expressed in normal but not cancerous mammary epithelial cells. Proc Natl Acad Sci U S A. 2001;98(17):9796–801. doi: 10.1073/pnas.171138398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fackler MJ, et al. DNA methylation of RASSF1A, HIN-1, RAR-beta, Cyclin D2 and Twist in in situ and invasive lobular breast carcinoma. Int J Cancer. 2003;107(6):970–5. doi: 10.1002/ijc.11508. [DOI] [PubMed] [Google Scholar]
- 46.Gort EH, et al. Methylation of the TWIST1 promoter, TWIST1 mRNA levels, and immunohistochemical expression of TWIST1 in breast cancer. Cancer Epidemiol Biomarkers Prev. 2008;17(12):3325–30. doi: 10.1158/1055-9965.EPI-08-0472. [DOI] [PubMed] [Google Scholar]
- 47.Wushou A, et al. Twist-1 up-regulation in carcinoma correlates to poor survival. Int J Mol Sci. 2014;15(12):21621–30. doi: 10.3390/ijms151221621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tang H, et al. AKT-ions with a TWIST between EMT and MET. Oncotarget. 2016 doi: 10.18632/oncotarget.11232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Huang KT, et al. DNA methylation profiling of phyllodes and fibroadenoma tumours of the breast. Breast Cancer Res Treat. 2010;124(2):555–65. doi: 10.1007/s10549-010-0970-4. [DOI] [PubMed] [Google Scholar]
- 50.Khan A, Fu J. Epigenetics of Transcription Factor Twist1 and Cancer. JSM Clin Oncol Res. 2014;2(3):2. [Google Scholar]
- 51.Evron E, et al. Loss of cyclin D2 expression in the majority of breast cancers is associated with promoter hypermethylation. Cancer Res. 2001;61(6):2782–7. [PubMed] [Google Scholar]
- 52.Seewaldt VL, et al. Expression of retinoic acid receptor beta mediates retinoic acid-induced growth arrest and apoptosis in breast cancer cells. Cell Growth Differ. 1995;6(9):1077–88. [PubMed] [Google Scholar]
- 53.Ivanova T, et al. Methylation and silencing of the retinoic acid receptor-beta 2 gene in cervical cancer. BMC Cancer. 2002;2:4. doi: 10.1186/1471-2407-2-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Widschwendter M, et al. Epigenetic downregulation of the retinoic acid receptor-beta2 gene in breast cancer. J Mammary Gland Biol Neoplasia. 2001;6(2):193–201. doi: 10.1023/a:1011360724350. [DOI] [PubMed] [Google Scholar]
- 55.Dammann R, Yang G, Pfeifer GP. Hypermethylation of the cpG island of Ras association domain family 1A (RASSF1A), a putative tumor suppressor gene from the 3p21.3 locus, occurs in a large percentage of human breast cancers. Cancer Res. 2001;61(7):3105–9. [PubMed] [Google Scholar]
- 56.Wang S, et al. Relationship between tumor DNA methylation status and patient characteristics in African-American and European-American women with breast cancer. PLoS One. 2012;7(5):e37928. doi: 10.1371/journal.pone.0037928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ambrosone CB, et al. Genome-wide methylation patterns provide insight into differences in breast tumor biology between American women of African and European ancestry. Oncotarget. 2014;5(1):237–48. doi: 10.18632/oncotarget.1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Toy W, et al. ESR1 ligand-binding domain mutations in hormone-resistant breast cancer. Nat Genet. 2013;45(12):1439–45. doi: 10.1038/ng.2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Angus L, et al. ESR1 mutations: Moving towards guiding treatment decision-making in metastatic breast cancer patients. Cancer Treat Rev. 2016;52:33–40. doi: 10.1016/j.ctrv.2016.11.001. [DOI] [PubMed] [Google Scholar]
- 60.Nusgen N, et al. Inter-locus as well as intra-locus heterogeneity in LINE-1 promoter methylation in common human cancers suggests selective demethylation pressure at specific CpGs. Clin Epigenetics. 2015;7:17. doi: 10.1186/s13148-015-0051-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Toyooka KO, et al. Loss of expression and aberrant methylation of the CDH13 (H-cadherin) gene in breast and lung carcinomas. Cancer Res. 2001;61(11):4556–60. [PubMed] [Google Scholar]
- 62.Yang J, et al. A Systematic Analysis of the Relationship of CDH13 Promoter Methylation and Breast Cancer Risk and Prognosis. PLoS One. 2016;11(5):e0149185. doi: 10.1371/journal.pone.0149185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Conway K, et al. Racial variation in breast tumor promoter methylation in the Carolina Breast Cancer Study. Cancer Epidemiol Biomarkers Prev. 2015;24(6):921–30. doi: 10.1158/1055-9965.EPI-14-1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Oshiro MM, et al. Epigenetic silencing of DSC3 is a common event in human breast cancer. Breast Cancer Res. 2005;7(5):R669–80. doi: 10.1186/bcr1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tobias ES, et al. The TES gene at 7q31.1 is methylated in tumours and encodes a novel growth-suppressing LIM domain protein. Oncogene. 2001;20(22):2844–53. doi: 10.1038/sj.onc.1204433. [DOI] [PubMed] [Google Scholar]
- 66.Choufani S, et al. A novel approach identifies new differentially methylated regions (DMRs) associated with imprinted genes. Genome Res. 2011;21(3):465–76. doi: 10.1101/gr.111922.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Breton CV, Salam MT, Gilliland FD. Heritability and role for the environment in DNA methylation in AXL receptor tyrosine kinase. Epigenetics. 2011;6(7):895–8. doi: 10.4161/epi.6.7.15768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ohgami RS, et al. DNA methylation analysis of ALOX12 and GSTM1 in acute myeloid leukaemia identifies prognostically significant groups. Br J Haematol. 2012;159(2):182–90. doi: 10.1111/bjh.12029. [DOI] [PubMed] [Google Scholar]
- 69.Pils D, et al. Methylation status of TUSC3 is a prognostic factor in ovarian cancer. Cancer. 2013;119(5):946–54. doi: 10.1002/cncr.27850. [DOI] [PubMed] [Google Scholar]
- 70.Benevolenskaya EV, et al. DNA methylation and hormone receptor status in breast cancer. Clin Epigenetics. 2016;8:17. doi: 10.1186/s13148-016-0184-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.O’Keefe EB, Meltzer JP, Bethea TN. Health disparities and cancer: racial disparities in cancer mortality in the United States, 2000–2010. Front Public Health. 2015;3:51. doi: 10.3389/fpubh.2015.00051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cooperberg MR. Re-examining racial disparities in prostate cancer outcomes. J Clin Oncol. 2013;31(24):2979–80. doi: 10.1200/JCO.2013.50.7723. [DOI] [PubMed] [Google Scholar]
- 73.Hoffman RM, et al. Racial and ethnic differences in advanced-stage prostate cancer: the Prostate Cancer Outcomes Study. J Natl Cancer Inst. 2001;93(5):388–95. doi: 10.1093/jnci/93.5.388. [DOI] [PubMed] [Google Scholar]
- 74.Thompson I, et al. Association of African-American ethnic background with survival in men with metastatic prostate cancer. J Natl Cancer Inst. 2001;93(3):219–25. doi: 10.1093/jnci/93.3.219. [DOI] [PubMed] [Google Scholar]
- 75.Wu I, Modlin CS. Disparities in prostate cancer in African American men: what primary care physicians can do. Cleve Clin J Med. 2012;79(5):313–20. doi: 10.3949/ccjm.79a.11001. [DOI] [PubMed] [Google Scholar]
- 76.Kwabi-Addo B, et al. Identification of differentially methylated genes in normal prostate tissues from African American and Caucasian men. Clin Cancer Res. 2010;16(14):3539–47. doi: 10.1158/1078-0432.CCR-09-3342. [DOI] [PubMed] [Google Scholar]
- 77.Coutinho I, et al. Androgen receptor signaling in castration-resistant prostate cancer: a lesson in persistence. Endocr Relat Cancer. 2016;23(12):T179–T197. doi: 10.1530/ERC-16-0422. [DOI] [PubMed] [Google Scholar]
- 78.Nakayama T, et al. Epigenetic regulation of androgen receptor gene expression in human prostate cancers. Lab Invest. 2000;80(12):1789–96. doi: 10.1038/labinvest.3780190. [DOI] [PubMed] [Google Scholar]
- 79.Sirchia SM, et al. Endogenous reactivation of the RARbeta2 tumor suppressor gene epigenetically silenced in breast cancer. Cancer Res. 2002;62(9):2455–61. [PubMed] [Google Scholar]
- 80.Di Croce L, et al. Methyltransferase recruitment and DNA hypermethylation of target promoters by an oncogenic transcription factor. Science. 2002;295(5557):1079–82. doi: 10.1126/science.1065173. [DOI] [PubMed] [Google Scholar]
- 81.Gao T, et al. The association of retinoic acid receptor beta2(RARbeta2) methylation status and prostate cancer risk: a systematic review and meta-analysis. PLoS One. 2013;8(5):e62950. doi: 10.1371/journal.pone.0062950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Said N, Theodorescu D. Secreted Protein Acidic and Rich in Cysteine (Sparc) in Cancer. J Carcinogene Mutagene. 2013;4 [Google Scholar]
- 83.Said N, et al. Loss of SPARC in bladder cancer enhances carcinogenesis and progression. J Clin Invest. 2013;123(2):751–66. doi: 10.1172/JCI64782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Shin M, et al. Exogenous SPARC suppresses proliferation and migration of prostate cancer by interacting with integrin beta1. Prostate. 2013;73(11):1159–70. doi: 10.1002/pros.22664. [DOI] [PubMed] [Google Scholar]
- 85.Sharma S, et al. Secreted Protein Acidic and Rich in Cysteine (SPARC) Mediates Metastatic Dormancy of Prostate Cancer in Bone. J Biol Chem. 2016;291(37):19351–63. doi: 10.1074/jbc.M116.737379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sato N, et al. SPARC/osteonectin is a frequent target for aberrant methylation in pancreatic adenocarcinoma and a mediator of tumor-stromal interactions. Oncogene. 2003;22(32):5021–30. doi: 10.1038/sj.onc.1206807. [DOI] [PubMed] [Google Scholar]
- 87.Socha MJ, et al. Aberrant promoter methylation of SPARC in ovarian cancer. Neoplasia. 2009;11(2):126–35. doi: 10.1593/neo.81146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zhang Y, et al. Aberrant methylation of SPARC in human hepatocellular carcinoma and its clinical implication. World J Gastroenterol. 2012;18(17):2043–52. doi: 10.3748/wjg.v18.i17.2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.He Q, et al. Aberrant methylation of secreted protein, acidic and rich in cysteine in human laryngeal and hypopharyngeal carcinoma. Oncol Lett. 2011;2(4):725–729. doi: 10.3892/ol.2011.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Adissu HA, et al. Timp3 loss accelerates tumour invasion and increases prostate inflammation in a mouse model of prostate cancer. Prostate. 2015;75(16):1831–43. doi: 10.1002/pros.23056. [DOI] [PubMed] [Google Scholar]
- 91.Daremipouran M, et al. Abstract 3033: NKX2–5, a potential tumor suppressor gene in prostate cancer. Cancer Research. 2014;71:3033–3033. [Google Scholar]
- 92.Enokida H, et al. Ethnic group-related differences in CpG hypermethylation of the GSTP1 gene promoter among African-American, Caucasian and Asian patients with prostate cancer. Int J Cancer. 2005;116(2):174–81. doi: 10.1002/ijc.21017. [DOI] [PubMed] [Google Scholar]
- 93.Woodson K, et al. Hypermethylation of GSTP1, CD44, and E-cadherin genes in prostate cancer among US Blacks and Whites. Prostate. 2003;55(3):199–205. doi: 10.1002/pros.10236. [DOI] [PubMed] [Google Scholar]
- 94.Woodson K, Hanson J, Tangrea J. A survey of gene-specific methylation in human prostate cancer among black and white men. Cancer Lett. 2004;205(2):181–8. doi: 10.1016/j.canlet.2003.11.027. [DOI] [PubMed] [Google Scholar]
- 95.Bucay N, et al. MicroRNA-383 located in frequently deleted chromosomal locus 8p22 regulates CD44 in prostate cancer. Oncogene. 2016 doi: 10.1038/onc.2016.419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Verkaik NS, et al. Silencing of CD44 expression in prostate cancer by hypermethylation of the CD44 promoter region. Lab Invest. 2000;80(8):1291–8. doi: 10.1038/labinvest.3780137. [DOI] [PubMed] [Google Scholar]
- 97.Lou W, et al. Methylation of the CD44 metastasis suppressor gene in human prostate cancer. Cancer Res. 1999;59(10):2329–31. [PubMed] [Google Scholar]
- 98.Kagara N, et al. Epigenetic regulation of cancer stem cell genes in triple-negative breast cancer. Am J Pathol. 2012;181(1):257–67. doi: 10.1016/j.ajpath.2012.03.019. [DOI] [PubMed] [Google Scholar]
- 99.Stallmach A, et al. Downregulation of CD44v6 in colorectal carcinomas is associated with hypermethylation of the CD44 promoter region. Exp Mol Pathol. 2003;74(3):262–6. doi: 10.1016/s0014-4800(03)00025-x. [DOI] [PubMed] [Google Scholar]
- 100.Yan P, et al. Hypermethylation-mediated regulation of CD44 gene expression in human neuroblastoma. Genes Chromosomes Cancer. 2003;36(2):129–38. doi: 10.1002/gcc.10150. [DOI] [PubMed] [Google Scholar]
- 101.Devaney JM, et al. Genome-wide differentially methylated genes in prostate cancer tissues from African-American and Caucasian men. Epigenetics. 2015;10(4):319–28. doi: 10.1080/15592294.2015.1022019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Feng W, et al. Imprinted tumor suppressor genes ARHI and PEG3 are the most frequently down-regulated in human ovarian cancers by loss of heterozygosity and promoter methylation. Cancer. 2008;112(7):1489–502. doi: 10.1002/cncr.23323. [DOI] [PubMed] [Google Scholar]
- 103.Jacobs DI, et al. Dysregulated methylation at imprinted genes in prostate tumor tissue detected by methylation microarray. BMC Urol. 2013;13:37. doi: 10.1186/1471-2490-13-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Lee MS, et al. Modulation of alternative splicing by expression of small nuclear ribonucleoprotein polypeptide N. FEBS J. 2014;281(23):5194–207. doi: 10.1111/febs.13059. [DOI] [PubMed] [Google Scholar]
- 105.Dai W, et al. Whole-exome sequencing identifies MST1R as a genetic susceptibility gene in nasopharyngeal carcinoma. Proc Natl Acad Sci U S A. 2016;113(12):3317–22. doi: 10.1073/pnas.1523436113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Gurusamy D, et al. Myeloid-specific expression of Ron receptor kinase promotes prostate tumor growth. Cancer Res. 2013;73(6):1752–63. doi: 10.1158/0008-5472.CAN-12-2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Das PM, et al. Methylation mediated silencing of TMS1/ASC gene in prostate cancer. Mol Cancer. 2006;5:28. doi: 10.1186/1476-4598-5-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Robbins AS, Siegel RL, Jemal A. Racial disparities in stage-specific colorectal cancer mortality rates from 1985 to 2008. J Clin Oncol. 2012;30(4):401–5. doi: 10.1200/JCO.2011.37.5527. [DOI] [PubMed] [Google Scholar]
- 109.Jackson CS, et al. Health disparities in colorectal cancer among racial and ethnic minorities in the United States. J Gastrointest Oncol. 2016;7(Suppl 1):S32–43. doi: 10.3978/j.issn.2078-6891.2015.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Wang X, et al. Aberrant DNA Methylation: Implications in Racial Health Disparity. PLoS One. 2016;11(4):e0153125. doi: 10.1371/journal.pone.0153125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Sepulveda JL, et al. High-definition CpG methylation of novel genes in gastric carcinogenesis identified by next-generation sequencing. Mod Pathol. 2016;29(2):182–93. doi: 10.1038/modpathol.2015.144. [DOI] [PubMed] [Google Scholar]
- 112.Truong AH, Ben-David Y. The role of Fli-1 in normal cell function and malignant transformation. Oncogene. 2000;19(55):6482–9. doi: 10.1038/sj.onc.1204042. [DOI] [PubMed] [Google Scholar]
- 113.Li Y, et al. The ets transcription factor Fli-1 in development, cancer and disease. Oncogene. 2015;34(16):2022–31. doi: 10.1038/onc.2014.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Song W, et al. Friend leukemia virus integration 1 activates the Rho GTPase pathway and is associated with metastasis in breast cancer. Oncotarget. 2015;6(27):23764–75. doi: 10.18632/oncotarget.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Kobayashi T, et al. Activation of the ribosomal protein L13 gene in human gastrointestinal cancer. Int J Mol Med. 2006;18(1):161–70. [PubMed] [Google Scholar]
- 116.Lee YE, et al. The prognostic impact of lipid biosynthesis-associated markers, HSD17B2 and HMGCS2, in rectal cancer treated with neoadjuvant concurrent chemoradiotherapy. Tumour Biol. 2015;36(10):7675–83. doi: 10.1007/s13277-015-3503-2. [DOI] [PubMed] [Google Scholar]
- 117.Camarero N, et al. Ketogenic HMGCS2 Is a c-Myc target gene expressed in differentiated cells of human colonic epithelium and down-regulated in colon cancer. Mol Cancer Res. 2006;4(9):645–53. doi: 10.1158/1541-7786.MCR-05-0267. [DOI] [PubMed] [Google Scholar]
- 118.Capello M, et al. Carboxylesterase 2 as a Determinant of Response to Irinotecan and Neoadjuvant FOLFIRINOX Therapy in Pancreatic Ductal Adenocarcinoma. J Natl Cancer Inst. 2015;107(8) doi: 10.1093/jnci/djv132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Saha SK, et al. KRT19 directly interacts with beta-catenin/RAC1 complex to regulate NUMB-dependent NOTCH signaling pathway and breast cancer properties. Oncogene. 2016 doi: 10.1038/onc.2016.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Asfaha S, et al. Krt19(+)/Lgr5(−) Cells Are Radioresistant Cancer-Initiating Stem Cells in the Colon and Intestine. Cell Stem Cell. 2015;16(6):627–38. doi: 10.1016/j.stem.2015.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Wang M, Hu Y, Stearns ME. RPS2: a novel therapeutic target in prostate cancer. J Exp Clin Cancer Res. 2009;28:6. doi: 10.1186/1756-9966-28-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Ahmed AR, et al. TFF3 is a normal breast epithelial protein and is associated with differentiated phenotype in early breast cancer but predisposes to invasion and metastasis in advanced disease. Am J Pathol. 2012;180(3):904–16. doi: 10.1016/j.ajpath.2011.11.022. [DOI] [PubMed] [Google Scholar]
- 123.Wang X, et al. THBS2 is a Potential Prognostic Biomarker in Colorectal Cancer. Sci Rep. 2016;6:33366. doi: 10.1038/srep33366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Marks LS, Bostwick DG. Prostate Cancer Specificity of PCA3 Gene Testing: Examples from Clinical Practice. Rev Urol. 2008;10(3):175–81. [PMC free article] [PubMed] [Google Scholar]
- 125.Merola R, et al. PCA3 in prostate cancer and tumor aggressiveness detection on 407 high-risk patients: a National Cancer Institute experience. J Exp Clin Cancer Res. 2015;34:15. doi: 10.1186/s13046-015-0127-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Gajjar K, Martin-Hirsch PL, Martin FL. CYP1B1 and hormone-induced cancer. Cancer Lett. 2012;324(1):13–30. doi: 10.1016/j.canlet.2012.04.021. [DOI] [PubMed] [Google Scholar]
- 127.Wang ZQ, et al. BCAT1 expression associates with ovarian cancer progression: possible implications in altered disease metabolism. Oncotarget. 2015;6(31):31522–43. doi: 10.18632/oncotarget.5159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Keita M, et al. Global methylation profiling in serous ovarian cancer is indicative for distinct aberrant DNA methylation signatures associated with tumor aggressiveness and disease progression. Gynecol Oncol. 2013;128(2):356–63. doi: 10.1016/j.ygyno.2012.11.036. [DOI] [PubMed] [Google Scholar]
- 129.Alonso S, et al. Methylation of MGMT and ADAMTS14 in normal colon mucosa: biomarkers of a field defect for cancerization preferentially targeting elder African-Americans. Oncotarget. 2015;6(5):3420–31. doi: 10.18632/oncotarget.2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Shen L, et al. MGMT promoter methylation and field defect in sporadic colorectal cancer. J Natl Cancer Inst. 2005;97(18):1330–8. doi: 10.1093/jnci/dji275. [DOI] [PubMed] [Google Scholar]
- 131.Long B, Liu FW, Bristow RE. Disparities in uterine cancer epidemiology, treatment, and survival among African Americans in the United States. Gynecol Oncol. 2013;130(3):652–9. doi: 10.1016/j.ygyno.2013.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Powell MA, et al. Ribosomal DNA methylation in patients with endometrial carcinoma: an independent prognostic marker. Cancer. 2002;94(11):2941–52. doi: 10.1002/cncr.10559. [DOI] [PubMed] [Google Scholar]
- 133.Gusev A, et al. Atlas of prostate cancer heritability in European and African-American men pinpoints tissue-specific regulation. Nat Commun. 2016;7:10979. doi: 10.1038/ncomms10979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Bhardwaj A, et al. Racial disparities in prostate cancer: a molecular perspective. Front Biosci (Landmark Ed) 2017;22:772–782. doi: 10.2741/4515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Yang Y, et al. Dysregulation of miR-212 Promotes Castration Resistance through hnRNPH1-Mediated Regulation of AR and AR-V7: Implications for Racial Disparity of Prostate Cancer. Clin Cancer Res. 2016;22(7):1744–56. doi: 10.1158/1078-0432.CCR-15-1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Yates C, et al. miRNAs as drivers of TMPRSS2-ERG negative prostate tumors in African American men. Front Biosci (Landmark Ed) 2017;22:212–229. doi: 10.2741/4482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Yang X, et al. miR-125b Regulation of Androgen Receptor Signaling Via Modulation of the Receptor Complex Co-Repressor NCOR2. Biores Open Access. 2012;1(2):55–62. doi: 10.1089/biores.2012.9903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Dhar S, et al. Resveratrol and pterostilbene epigenetically restore PTEN expression by targeting oncomiRs of the miR-17 family in prostate cancer. Oncotarget. 2015;6(29):27214–26. doi: 10.18632/oncotarget.4877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Yan B, et al. The role of miR-29b in cancer: regulation, function, and signaling. Onco Targets Ther. 2015;8:539–48. doi: 10.2147/OTT.S75899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Steele R, Mott JL, Ray RB. MBP-1 upregulates miR-29b that represses Mcl-1, collagens, and matrix-metalloproteinase-2 in prostate cancer cells. Genes Cancer. 2010;1(4):381–387. doi: 10.1177/1947601910371978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Williams LV, et al. miR-200b inhibits prostate cancer EMT, growth and metastasis. PLoS One. 2013;8(12):e83991. doi: 10.1371/journal.pone.0083991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Ahmad A, et al. Phosphoglucose isomerase/autocrine motility factor mediates epithelial-mesenchymal transition regulated by miR-200 in breast cancer cells. Cancer Res. 2011;71(9):3400–9. doi: 10.1158/0008-5472.CAN-10-0965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Theodore SC, et al. MiRNA 26a expression in a novel panel of African American prostate cancer cell lines. Ethn Dis. 2010;20(1 Suppl 1):S1-96–100. [PMC free article] [PubMed] [Google Scholar]
- 144.Theodore SC, et al. MicroRNA profiling of novel African American and Caucasian Prostate Cancer cell lines reveals a reciprocal regulatory relationship of miR-152 and DNA methyltranferase 1. Oncotarget. 2014;5(11):3512–25. doi: 10.18632/oncotarget.1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Xiang Y, et al. MiR-152 and miR-185 co-contribute to ovarian cancer cells cisplatin sensitivity by targeting DNMT1 directly: a novel epigenetic therapy independent of decitabine. Oncogene. 2014;33(3):378–86. doi: 10.1038/onc.2012.575. [DOI] [PubMed] [Google Scholar]
- 146.Xu Q, et al. A regulatory circuit of miR-148a/152 and DNMT1 in modulating cell transformation and tumor angiogenesis through IGF-IR and IRS1. J Mol Cell Biol. 2013;5(1):3–13. doi: 10.1093/jmcb/mjs049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Li Y, et al. Epigenetic silencing of miRNA-9 is correlated with promoter-proximal CpG island hypermethylation in gastric cancer in vitro and in vivo. Int J Oncol. 2014;45(6):2576–86. doi: 10.3892/ijo.2014.2667. [DOI] [PubMed] [Google Scholar]
- 148.Wang LQ, et al. Epigenetic inactivation of miR-9 family microRNAs in chronic lymphocytic leukemia--implications on constitutive activation of NFkappaB pathway. Mol Cancer. 2013;12:173. doi: 10.1186/1476-4598-12-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Xu L, et al. Methylation-regulated miR-124–1 suppresses tumorigenesis in hepatocellular carcinoma by targeting CASC3. Oncotarget. 2016;7(18):26027–41. doi: 10.18632/oncotarget.8266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Banno K, et al. Carcinogenic mechanisms of endometrial cancer: involvement of genetics and epigenetics. J Obstet Gynaecol Res. 2014;40(8):1957–67. doi: 10.1111/jog.12442. [DOI] [PubMed] [Google Scholar]
- 151.Farhana L, et al. Role of cancer stem cells in racial disparity in colorectal cancer. Cancer Med. 2016 doi: 10.1002/cam4.690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Senchenko VN, et al. Differential expression of CHL1 gene during development of major human cancers. PLoS One. 2011;6(3):e15612. doi: 10.1371/journal.pone.0015612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Li E, et al. Differential expression of miRNAs in colon cancer between African and Caucasian Americans: implications for cancer racial health disparities. Int J Oncol. 2014;45(2):587–94. doi: 10.3892/ijo.2014.2469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Suresh R, et al. Differential Expression of MicroRNAs in Papillary Thyroid Carcinoma and Their Role in Racial Disparity. J Cancer Sci Ther. 2015;7(5):145–154. doi: 10.4172/1948-5956.1000340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Li M, et al. MiR-31 regulates the cisplatin resistance by targeting Src in gallbladder cancer. Oncotarget. 2016 doi: 10.18632/oncotarget.13067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Karimi G, et al. MiR 221/222 as new players in tamoxifen resistance. Curr Pharm Des. 2016 doi: 10.2174/1381612822666161102100211. [DOI] [PubMed] [Google Scholar]
- 157.Maxwell GL, et al. MicroRNAs in endometrial cancers from black and white patients. Am J Obstet Gynecol. 2015;212(2):191.e1–10. doi: 10.1016/j.ajog.2014.08.028. [DOI] [PubMed] [Google Scholar]
- 158.Zheng L, et al. miRNA-337–3p inhibits gastric cancer progression through repressing myeloid zinc finger 1-facilitated expression of matrix metalloproteinase 14. Oncotarget. 2016;7(26):40314–40328. doi: 10.18632/oncotarget.9739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Xiang X, et al. miRNA-337–3p suppresses neuroblastoma progression by repressing the transcription of matrix metalloproteinase 14. Oncotarget. 2015;6(26):22452–66. doi: 10.18632/oncotarget.4311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Liu S, Howell PM, Riker AI. Up-regulation of miR-182 expression after epigenetic modulation of human melanoma cells. Ann Surg Oncol. 2013;20(5):1745–52. doi: 10.1245/s10434-012-2467-3. [DOI] [PubMed] [Google Scholar]
- 161.Xu L, et al. Down-regulation of miR-212 expression by DNA hypermethylation in human gastric cancer cells. Med Oncol. 2011;28(Suppl 1):S189–96. doi: 10.1007/s12032-010-9691-0. [DOI] [PubMed] [Google Scholar]
- 162.Xia YY, et al. Racial/ethnic disparities in human DNA methylation. Biochim Biophys Acta. 2014;1846(1):258–62. doi: 10.1016/j.bbcan.2014.07.001. [DOI] [PubMed] [Google Scholar]
- 163.Wallace K, et al. Association between folate levels and CpG Island hypermethylation in normal colorectal mucosa. Cancer Prev Res (Phila) 2010;3(12):1552–64. doi: 10.1158/1940-6207.CAPR-10-0047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Dumitrescu RG, et al. Familial and racial determinants of tumour suppressor genes promoter hypermethylation in breast tissues from healthy women. J Cell Mol Med. 2010;14(6b):1468–75. doi: 10.1111/j.1582-4934.2009.00924.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Song MA, et al. Racial differences in genome-wide methylation profiling and gene expression in breast tissues from healthy women. Epigenetics. 2015;10(12):1177–87. doi: 10.1080/15592294.2015.1121362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Moen EL, et al. Genome-wide variation of cytosine modifications between European and African populations and the implications for complex traits. Genetics. 2013;194(4):987–96. doi: 10.1534/genetics.113.151381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Adkins RM, et al. Racial differences in gene-specific DNA methylation levels are present at birth. Birth Defects Res A Clin Mol Teratol. 2011;91(8):728–36. doi: 10.1002/bdra.20770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Yu J, et al. A novel set of DNA methylation markers in urine sediments for sensitive/specific detection of bladder cancer. Clin Cancer Res. 2007;13(24):7296–304. doi: 10.1158/1078-0432.CCR-07-0861. [DOI] [PubMed] [Google Scholar]
- 169.Friedrich MG, et al. Detection of methylated apoptosis-associated genes in urine sediments of bladder cancer patients. Clin Cancer Res. 2004;10(22):7457–65. doi: 10.1158/1078-0432.CCR-04-0930. [DOI] [PubMed] [Google Scholar]
- 170.Hoque MO, et al. Quantitation of promoter methylation of multiple genes in urine DNA and bladder cancer detection. J Natl Cancer Inst. 2006;98(14):996–1004. doi: 10.1093/jnci/djj265. [DOI] [PubMed] [Google Scholar]
- 171.Sun J, et al. Hypermethylated SFRP1, but none of other nine genes “informative” for western countries, is valuable for bladder cancer detection in Mainland China. J Cancer Res Clin Oncol. 2009;135(12):1717–27. doi: 10.1007/s00432-009-0619-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Lin Q, et al. RASSF1A, APC, ESR1, ABCB1 and HOXC9, but not p16INK4A, DAPK1, PTEN and MT1G genes were frequently methylated in the stage I non-small cell lung cancer in China. J Cancer Res Clin Oncol. 2009;135(12):1675–84. doi: 10.1007/s00432-009-0614-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Dong Y, et al. Hypermethylation of TFPI2 correlates with cervical cancer incidence in the Uygur and Han populations of Xinjiang, China. Int J Clin Exp Pathol. 2015;8(2):1844–54. [PMC free article] [PubMed] [Google Scholar]
- 174.Kimura T. East meets West: ethnic differences in prostate cancer epidemiology between East Asians and Caucasians. Chin J Cancer. 2012;31(9):421–9. doi: 10.5732/cjc.011.10324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Lopez-Guarnido O, et al. Mediterranean diet adherence and prostate cancer risk. Nutr Hosp. 2014;31(3):1012–9. doi: 10.3305/nh.2015.31.3.8286. [DOI] [PubMed] [Google Scholar]
- 176.Ullah MF, et al. Soy isoflavone genistein induces cell death in breast cancer cells through mobilization of endogenous copper ions and generation of reactive oxygen species. Mol Nutr Food Res. 2011;55(4):553–9. doi: 10.1002/mnfr.201000329. [DOI] [PubMed] [Google Scholar]
- 177.Ahmad A, et al. Perspectives on the role of isoflavones in prostate cancer. AAPS J. 2013;15(4):991–1000. doi: 10.1208/s12248-013-9507-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Majid S, et al. Genistein induces the p21WAF1/CIP1 and p16INK4a tumor suppressor genes in prostate cancer cells by epigenetic mechanisms involving active chromatin modification. Cancer Res. 2008;68(8):2736–44. doi: 10.1158/0008-5472.CAN-07-2290. [DOI] [PubMed] [Google Scholar]
- 179.Di Francesco A, et al. Extravirgin olive oil up-regulates CB(1) tumor suppressor gene in human colon cancer cells and in rat colon via epigenetic mechanisms. J Nutr Biochem. 2015;26(3):250–8. doi: 10.1016/j.jnutbio.2014.10.013. [DOI] [PubMed] [Google Scholar]
- 180.D’Souza W, Saranath D. Clinical implications of epigenetic regulation in oral cancer. Oral Oncol. 2015;51(12):1061–8. doi: 10.1016/j.oraloncology.2015.09.006. [DOI] [PubMed] [Google Scholar]
- 181.Yang CM, et al. Aberrant DNA hypermethylation-silenced SOX21-AS1 gene expression and its clinical importance in oral cancer. Clin Epigenetics. 2016;8:129. doi: 10.1186/s13148-016-0291-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Guerrero-Preston R. Global epigenetic screening technologies: a novel tool to address cancer health disparities in high-risk population groups. P R Health Sci J. 2008;27(4):350–6. [PubMed] [Google Scholar]
- 183.Rocconi RP, et al. The role of racial genetic admixture with endometrial cancer outcomes: An NRG Oncology/Gynecologic Oncology Group study. Gynecol Oncol. 2016;140(2):264–9. doi: 10.1016/j.ygyno.2015.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Schwartzman O, Tanay A. Single-cell epigenomics: techniques and emerging applications. Nat Rev Genet. 2015;16(12):716–26. doi: 10.1038/nrg3980. [DOI] [PubMed] [Google Scholar]