Abstract
Seed size and seed weight are major quality attributes and important determinants of yield that have been strongly selected for during crop domestication. Limited information is available about the genetic control and genes associated with seed size and weight in sorghum. This study identified sorghum orthologs of genes with proven effects on seed size and weight in other plant species and searched for evidence of selection during domestication by utilizing resequencing data from a diversity panel. In total, 114 seed size candidate genes were identified in sorghum, 63 of which exhibited signals of purifying selection during domestication. A significant number of these genes also had domestication signatures in maize and rice, consistent with the parallel domestication of seed size in cereals. Seed size candidate genes that exhibited differentially high expression levels in seed were also found more likely to be under selection during domestication, supporting the hypothesis that modification to seed size during domestication preferentially targeted genes for intrinsic seed size rather than genes associated with physiological factors involved in the carbohydrate supply and transport. Our results provide improved understanding of the complex genetic control of seed size and weight and the impact of domestication on these genes.
Keywords: sorghum, seed size, orthologs, comparative genomics, selection signatures, domestication
Introduction
A growing world population and an increase in affluence is driving demand for agricultural products, especially cereals, which supply more than 75% of the calories consumed by humans (Sands et al., 2009). With limited arable land and water resources, particularly in Sub-Saharan Africa where sorghum is a staple food and the population growth rate is amongst the highest in the world, enhancing yield per unit area of cereal crops will be critical to meet this demand. Seed number per unit area and seed size are critical components of seed yield. Although seed number tends to have a bigger influence on yield (Boyles et al., 2016), seed size can make a significant contribution and may offer prospects for further yield improvement (Yang et al., 2009). In addition, it is often a major quality attribute (Lee et al., 2002). Hence, elucidating the genetic basis of seed size and the impact of domestication on seed size genes in sorghum will enhance the understanding of crop domestication and provide new targets for manipulating seed size in breedingpractice.
Seed size is an important fitness trait for flowering plants and plays an important role in adaptation to particular environments. Under natural conditions, greater seed resources stored in larger seeds enable seedlings to grow more rapidly at the seedling stage and increases competitiveness and survival (Manga and Yadav, 1995). However, increased seed number also translates directly into fitness, resulting in selection pressure to produce more (and thus smaller) seeds (Westoby et al., 1992). For cereal crops, the preference of early farmers for large seeded lines for easier harvesting, processing, and planting has resulted in larger seed size being selected during domestication. This selection process has left observable genetic changes, including a reduction of genetic diversity and an increased frequency of favorable seed size alleles in cultivated lines compared to their wild progenitors (Doebley et al., 2006). For example, in rice, the favorable allele of GS3, which encodes a heterotrimeric G-protein subunit that affects seed weight and length, was highly enriched in a set of cultivated accessions of rice (Oryza sativa L.) (34%) compared to a set of wild accessions (4%; Takano-Kai et al., 2009; Botella, 2012). In maize (Zea mays L.), Bt2, which encodes the small subunit of the ADP-glucose pyrophosphorylase involved in starch biosynthesis and seed weight, has shown a 3.9-fold reduction in genetic diversity in cultivated inbred lines compared to their wild teosinte relatives (Whitt et al., 2002). Likewise, selection signatures have also been identified on other seed size genes, including PBF1 (Lang et al., 2014), GS5 (Li et al., 2011), and GIF1 (Wang et al., 2008). These selection signatures provide a “bottom-up” approach to investigate the genetic basis of domesticated traits, which has been successfully implemented in many species for other traits such as prolificacy (Beissinger et al., 2014) and northern leaf blight resistance (Wisser et al., 2008) in maize.
Seed size is a physiologically complex trait. Sorghum seeds are typically tending toward spherical, although considerable phenotypic variation in length, width and density does exist. The potential size of the seed is often associated with cell number, cell size and number of starch granules and is highly correlated with ovary volume at anthesis (Yang et al., 2009). However, measures associated with seed size have not been used consistently in the literature, where individual grain weight is often used as a surrogate for seed size. As key components of carbon demand (sink) during seed filling, seed size and weight are strongly associated with both carbon supply (source) and transport between carbon sources and the seed (path). The potential mass of individual seeds is determined by the rate and duration of seed filling. In sorghum, seed filling rate is highly correlated with ovary volume at anthesis, which in turn is associated with the size of the meristematic dome during early floret development (Yang et al., 2009).
Although seeds with larger potential size tend to have greater seed mass, the extent to which this increased seed mass is actually achieved is strongly determined by assimilate availability for each seed. The amount of assimilate per seed is driven by factors affecting both seed number and assimilate supply. Total seed number per plant is determined by the number of seeds per panicle and the number of panicles per plant (i.e., tillering and branching), which are affected by a range of genetic and environmental factors (Alam et al., 2014). A negative correlation between seed size and seed number has been observed frequently in cereals (Jakobsson and Eriksson, 2000; Acreche and Slafer, 2006; Peltonen-Sainio et al., 2007; Sadras, 2007). Specifically in sorghum this trade-off has been observed by different groups (Heinrich et al., 1983; Yang et al., 2010; Burow et al., 2014). Traits such as number of seeds per panicle and number of tillers per plant are also commonly negatively correlated with seed size (Moles and Westoby, 2004). Contributors of assimilate availability for seed filling, including photosynthesis (Jagadish et al., 2015), have shown positive correlations with seed size. Environmental factors can also exert a strong influence on seed size by affecting assimilate supply (Jenner, 1994; Borrell et al., 2014) and carbon translocation(Zolkevich et al., 1958).
In accordance with this physiological complexity, seed size has been identified as a quantitative trait controlled by multiple genes, many of which have been cloned in model species (Xing and Zhang, 2010; Li et al., 2013; Zuo and Li, 2014). In Arabidopsis, a kinase cascade consisting of HAIKU1, HAIKU2, and MINISEED3 promotes seed development zygotically (Luo et al., 2005; Wang et al., 2010), while TTG2 (Garcia et al., 2005), AP2 (Ohto et al., 2009), and ARF2 (Okushima et al., 2005) are engaged in the maternal control of seed size. In rice, QTLs including GS3 (Mao et al., 2010), GS5 (Li et al., 2011), GW2 (Song et al., 2007), GW5 (Liu et al., 2017), GW8 (Wang S. et al., 2012), and GL7 (Wang Y. et al., 2015) were reported to regulate seed size by controlling cell division, while the influence of SRS3 (Kitagawa et al., 2010), D61 (Morinaka et al., 2006), and SRS5 (Segami et al., 2012) on seed size is related to the regulation of cell size. Additionally, the role of GIF1 in carbon partitioning during early seed-filling, which can impact seed weight, has been identified using functional analysis in rice (Wang et al., 2008). In maize, the Gln-4 gene (Martin et al., 2006) affects seed weight by controlling nitrogen transport to the kernel during seed-filling, whereas Sh2, which encodes the large subunit of ADP-glucose pyrophosphorylase, affects seed weight by regulating starch biosynthesis (Jiang L. et al., 2013). Pleiotropy is common amongst genes affecting seed size. For example, D2 (Hong et al., 2003) and SMG1 (Duan et al., 2014) also have an effect on plant architecture, TH1 (Li X. et al., 2012) affects seed number, and TGW6 (Ishimaru et al., 2013) influences translocation efficiency from source organs. These genes may thus affect seed size via source-sink dynamics.
Sorghum, second only to maize among C4 cereals in terms of the scale of grain production, is known for its adaptation to heat and drought stress, and is a staple for 500 million of the world's poorest people. Despite the great importance of this crop, the genetic basis of seed size in sorghum has been the subject of relatively few studies and little information is available about genetic control of the trait and signatures of domestication. Hence, this study aims to investigate the polymorphism patterns and signatures of domestication of candidate genes associated with seed size and weight by using resequencing data for a diverse group of wild and weedy and landrace genotypes (Mace et al., 2013) in order to enhance understanding of crop domestication and to provide potential targets for manipulating seed size in sorghum breeding.
Materials and methods
Data collection
Genes associated with seed size and weight (hereafter referred as seed size) in three species, maize, rice and Arabidopsis, were identified through a comprehensive literature review (Table S1). Seed length, seed width, and seed density are all potentially associated with seed size; therefore multiple parameters including thousand seed weight, seed length, and seed width, were used as keywords for literature searches. A subset of high confidence genes were identified with evidence of their association with seed size supported by QTL cloning, transgenic experiments, mutant analysis, association signal, and/or near isogenic lines analysis.
Genome assemblies and predicted gene models and protein sequences for Arabidopsis thaliana (TAIR10), Oryza sativa (IRGSP-1.0), Zea mays (AGPv4), and Sorghum bicolor (v3.0) were downloaded from TAIR (https://www.arabidopsis.org); The Rice Annotation Project database (http://rapdb.dna.affrc.go.jp); Gramene (http://www.gramene.org) and Joint Genome institute (http://www.phytozome.net), respectively.
Identification of orthologos genes
Orthologous genes in sorghum were identified by combining synteny-based and the Bidirectional Best Hit (BBH) approaches (Wolf and Koonin, 2012). Genomic syntenic relationships between sorghum and model species were extracted from Plant Genome Duplication Database (http://chibba.agtec.uga.edu/duplication/) and used to search for syntenic orthologs, while a local BLAST strategy was used for the BBH approach to identify pairs of genes in two genomes that are the best BLAST hits (highest score) to one another, using BLASTP.
Expression analysis of seed size candidate genes
The whole genome expression data from the study by Davidson et al. (2012) was used to investigate the differential expression of the 114 candidate genes. The data set compared expression of genes in the seed at two different time points and two different seed tissues in addition to five non-seed tissues (Davidson et al., 2012). The maximum expression value (Fragments Per Kilobase of transcript per Million mapped reads, FPKM) from any of the seed tissue samples was compared to the maximum expression value in any of the non-seed tissues and a fold difference >2 was used to define genes that were differentially highly expressed in the seed.
Population genetics analysis
Gene level population genetics parameters
The sequence data of the seed size genes in sorghum were extracted from the whole genome resequencing data as described in Mace et al. (2013) for 25 sorghum genotypes, representing two groups: (1) wild and weedy genotypes and (2) landraces. A number of summary statistics based on gene level, including the average pairwise genetic diversity within a group, θπ (Nei and Li, 1979) and Tajima's D (Tajima, 1989), were calculated using a BioPerl module and an in-house perl script. FST (Hudson et al., 1992) was calculated to measure population differentiation using another BioPerl module. Reduction of diversity (RoD) during domestication was calculated as fold of decrease of θπ in the landrace group compared to the wild and weedy group.
Identifying selection signatures at the SNP level
CDS of the seed size genes across 25 resequenced genotypes was used to generate population statistics for every SNP using the R package PopGenome (Pfeifer et al., 2014). Specifically, a 1-bp window size with a 1-bp step size was used to define the slide window. θπ (Nei and Li, 1979), Fst (Hudson et al., 1992), and Tajima's D (Tajima, 1989) for each SNP within the CDS were calculated using diversity.stats, F_ST.stats, and neutrality.stats commands. Functional information was estimated by get.codons. RoD in the pairwise ancestor/descendant population comparison was calculated as fold of decrease of θπ in landrace compared to wild and weedy. To identify SNPs under purifying selection the following criteria were used: (1) RoD in the pairwise ancestor/descendant population comparison should be greater than the average RoD based on 159 neutral loci; (2) FST should be positive; (3) Tajima's D should be negative.
mlHKA test
A set of 63 seed size candidate genes under purifying selection were used as input, together with three random selections of 36 genes from 159 neutral genes, for the mlHKA (Wright and Charlesworth, 2004) test for validation purposes. The mlHKA program was run under a neutral model, where numselectedloci = 0, and then under a selection model, where numselectedloci >0. The number of cycles of the Markov chain was set to be 100,000. For each random selection of 36 neutral genes, three random numbers of seed were set to be 10, 20, and 30, respectively. This means 3 × 3 = 9 times of run were performed. Significance was assessed by the mean log likelihood ratio test statistic, where twice the difference in log likelihood between the models is approximately chi-squared distributed with df equal to the difference in the number of parameters.
Haplotype analysis of genes under selection
Haplotype analysis was performed using R package pegas (Population and Evolutionary Genetics Analysis System; Paradis, 2010) and ape package (Paradis et al., 2004) for genes under selection. Functions haplotype, haploFreq and haploNet were called to generate haplotype maps. In addition to landrace and wild & weedy, accessions from improved lines, Guinea margaritiferum race and S. propinquum were used in haplotype analyses (Table S2).
Results
Seed size candidate genes in sorghum
Based on a comprehensive literature survey, 129 genes associated with seed size were identified in three well-studied model species, including 65 genes in rice, 21 in maize and 43 in Arabidopsis (Table S1). By using BBH method and the known syntenic relationship from the Plant Genome Duplication Database to infer orthologs (assembly v3.0), a total of 111 genes were identified in sorghum (Table 1). From the set of 65 seed size-related genes identified in rice, 55 orthologs were identified in sorghum using the BBH method and 47 using the syntenic relationship method. Of these, 30 orthologs were identified by both methods, resulting in a total of 72 unique orthologs identified in sorghum (Figure 1). Additionally, a total of 23 orthologs were identified in sorghum based on the 21 seed size-related genes from maize, including 20 BBH orthologs and 12 syntenic orthologs with 9 orthologs identified by both methods. Finally, 25 sorghum orthologs were identified based on the analysis of the 43 selected seed size-related genes from Arabidopsis (Figure 1). Amongst all putative sorghum orthologs, 9 were in common across a minimum of two species, leading to 111 unique orthologs in sorghum identified as seed size candidate genes (Figure 1). Four seed size candidate genes in sorghum from Zhang et al. (2015) with one overlapped with the 111 seed size orthologs were also taken into consideration, resulting in a final list of 114 seed size candidate genes.
Table 1.
Seed size candidate genes identified in sorghum including details of the identification approach, the original study describing the gene's function and presence of supporting selection.
| Gene IDa | Approachb | Original genec | Selection signatured | Referencese |
|---|---|---|---|---|
| Sobic.001G016200 | BBH | Nuf2 family protein | Yes | Huang et al., 2012b |
| Sobic.001G056700 | Synteny | O2 | Yes | Hartings et al., 1989 |
| Sobic.001G107100 | BBH | SRS5 | Yes | Segami et al., 2012 |
| Sobic.001G113200 | BBH | AHK4 | Yes | Riefler et al., 2006 |
| Sobic.001G154900 | Both | GL3.1/qGL3 | Yes | Qi et al., 2012; Zhang et al., 2012 |
| Sobic.001G170800 | Both | Transport protein | Yes | Huang et al., 2012b |
| Sobic.001G172400 | BBH | BRD1 | Yes | Mori et al., 2002 |
| Sobic.001G184900 | Both | Expressed protein | Yes | Huang et al., 2012b |
| Sobic.001G254100 | Synteny | PGL1 | No | Heang and Sassa, 2012a |
| Sobic.001G254200 | Synteny | OsFBK12 | Yes | Chen et al., 2013 |
| Sobic.001G285000 | BBH | IKU1 | No | Wang et al., 2010 |
| Sobic.001G335800 | Synteny | qGW7/GL7 | Yes | Wang S. et al., 2015; Wang Y. et al., 2015 |
| Sobic.001G336200 | BBH/BBH | KLU/ Grain Length3.2 | No | Adamski et al., 2009; Xu et al., 2015 |
| Sobic.001G341700 | BBH/Both | GS3/ZmGS3 | Yes | Li et al., 2010b; Mao et al., 2010 |
| Sobic.001G382400 | BBH | FER | No | Yu et al., 2014 |
| Sobic.001G445900 | BBH/BBH | CYP90B2/CYP90B1 | No | Wu et al., 2008 |
| Sobic.001G448700 | Both | TUD1 | No | Hu et al., 2013 |
| Sobic.001G468400 | Both | Prol1.1 | No | Wills et al., 2013 |
| Sobic.001G482600 | BBH | TIFY 11b | No | Hakata et al., 2012 |
| Sobic.001G484200 | BBH | RGA1/D1 | No | Ashikari et al., 1999 |
| Sobic.001G485400 | Both | BG1 | No | Liu L. et al., 2015 |
| Sobic.001G488400 | Synteny | PGL1 | No | Heang and Sassa, 2012a |
| Sobic.001G488500 | BBH | OsFBK12 | No | Chen et al., 2013 |
| Sobic.002G021200 | BBH | DDM1 | Yes | Xiao et al., 2006 |
| Sobic.002G022600 | BBH | ANT | No | Mizukami and Fischer, 2000 |
| Sobic.002G054800 | Both | O2 | Yes | Hartings et al., 1989 |
| Sobic.002G056000 | BBH | MET1 | Yes | Xiao et al., 2006 |
| Sobic.002G116000 | BBH | GbssIIa | No | Jiang L. et al., 2013 |
| Sobic.002G216600 | Both/Synteny | DEP1/AGG3 | No | Huang et al., 2009; Li S. et al., 2012 |
| Sobic.002G226500 | Both | SG1 | Yes | Nakagawa et al., 2012 |
| Sobic.002G257900 | Synteny | GW8 | Yes | Wang S. et al., 2012 |
| Sobic.002G272700 | BBH | EOD3/CYP78A6 | Yes | Fang et al., 2012 |
| Sobic.002G308400 | BBH | MYB transcription factor | Yes | Huang et al., 2012b |
| Sobic.002G309600 | BBH | UPF1 | Yes | Yoine et al., 2006 |
| Sobic.002G311000 | BBH | Receptor-like kinase | Yes | Huang et al., 2012b |
| Sobic.002G312200 | Both | GLW7 | No | Si et al., 2016 |
| Sobic.002G367300 | Both | qGW7/GL7 | Yes | Wang S. et al., 2015; Wang Y. et al., 2015 |
| Sobic.002G367600 | BBH | BG2 | No | Xu et al., 2015 |
| Sobic.002G374400 | Both | DEP2 | Yes | Li F. et al., 2010 |
| Sobic.003G014500 | BBH | MHZ7 | No | Ma et al., 2013 |
| Sobic.003G030600 | BBH | D2 | No | Hong et al., 2003 |
| Sobic.003G035400 | Synteny | GW5/qSW5 | No | Liu et al., 2017 |
| Sobic.003G140000 | Synteny | OsSAMS1 | Yes | Chen et al., 2013 |
| Sobic.003G230500 | BBH | Sh2 | Yes | Jiang L. et al., 2013 |
| Sobic.003G277900 | BBH/BBH | D61/ BRI1 | Yes | Morinaka et al., 2006; Jiang W. et al., 2013 |
| Sobic.003G292600 | BBH | AHP4 | No | Hutchison et al., 2006 |
| Sobic.003G358400 | BBH | DET2 | No | Jiang W. et al., 2013 |
| Sobic.003G380900 | Synteny | SERF1 | No | Schmidt et al., 2013 |
| Sobic.003G406600 | Synteny | GW8 | Yes | Wang S. et al., 2012 |
| Sobic.003G407300 | BBH | AHK3 | No | Riefler et al., 2006 |
| Sobic.003G444100 | Both | OsCCS52B | Yes | Su'udi et al., 2012 |
| Sobic.004G065400 | Synteny | GW6 | No | Song et al., 2015 |
| Sobic.004G075600 | Both | Zinc finger protein | Yes | Huang et al., 2012b |
| Sobic.004G085100 | Both | Bt1 | Yes | Shannon et al., 1998 |
| Sobic.004G093900 | BBH | CKX2 | Yes | Li et al., 2013 |
| Sobic.004G107300 | Both/Both/Both | GW2/ZmGW2-4/ZmGW2-5 | No | Song et al., 2007; Li et al., 2010a |
| Sobic.004G133600 | BBH/original | ZmSWEET4c/NA | No | Sosso et al., 2015; Zhang et al., 2015 |
| Sobic.004G163700 | BBH | SbeIIb | No | Jiang L. et al., 2013 |
| Sobic.004G176000 | Both | SDG725 | Yes | Sui et al., 2012 |
| Sobic.004G214100 | Both | BC14 | No | Zhang et al., 2011 |
| Sobic.004G237000 | Both | PGL2 | No | Heang and Sassa, 2012b |
| Sobic.004G245000 | Synteny | AHK4 | Yes | Riefler et al., 2006 |
| Sobic.004G247000 | Both | Gln-4 | No | Martin et al., 2006 |
| Sobic.004G269900 | Synteny | GS2/GL2 | Yes | Che et al., 2015; Hu et al., 2015 |
| Sobic.004G307800 | Both | SGL1 | Yes | Nakagawa et al., 2012 |
| Sobic.004G317300 | BBH | O1 | Yes | Wang G. et al., 2012 |
| Sobic.004G323600 | Both | SMG1 | No | Duan et al., 2014 |
| Sobic.004G330200 | BBH | TGW6 | Yes | Ishimaru et al., 2013 |
| Sobic.004G338400 | BBH | TH1 | No | Li X. et al., 2012 |
| Sobic.005G001500 | Both | PBF1 | Yes | Lang et al., 2014 |
| Sobic.005G132000 | BBH | ARF2 | Yes | Okushima et al., 2005 |
| Sobic.006G059900 | Both | ZmIPT2 | Yes | Weng et al., 2013 |
| Sobic.006G080500 | BBH | RGE1 | Yes | Kondou et al., 2008 |
| Sobic.006G114600 | BBH/BBH | D11/CYP724B3 | Yes | Tanabe et al., 2005; Wu et al., 2008 |
| Sobic.006G203400 | Synteny | GS2/GL2 | Yes | Che et al., 2015; Hu et al., 2015 |
| Sobic.006G239000 | Both | FLO2 | Yes | She et al., 2010 |
| Sobic.006G240700 | BBH | AP2 | No | Ohto et al., 2009 |
| Sobic.006G268800 | Original | NA | Yes | Zhang et al., 2015 |
| Sobic.007G032400 | Both/BBH | OsFIE2/FIE | No | Luo et al., 2000; Na et al., 2012 |
| Sobic.007G101500 | BBH | Bt2 | Yes | Jiang L. et al., 2013 |
| Sobic.007G149200 | Synteny/BBH | DEP1/AGG3 | Yes | Huang et al., 2009; Li S. et al., 2012 |
| Sobic.007G156800 | Synteny | SGL1 | No | Nakagawa et al., 2012 |
| Sobic.007G166600 | Original | NA | No | Zhang et al., 2015 |
| Sobic.007G193500 | Both | GW8 | No | Wang S. et al., 2012 |
| Sobic.008G001700 | Synteny | PBF1 | Yes | Lang et al., 2014 |
| Sobic.008G100400 | BBH | SMK1 | Yes | Li et al., 2014 |
| Sobic.008G152800 | BBH | CBL3 | No | Eckert et al., 2014 |
| Sobic.008G173900 | Both | OsPPKL3 | Yes | Zhang et al., 2012 |
| Sobic.008G193300 | BBH | OsSUT2 | No | Eom et al., 2011 |
| Sobic.009G024600 | Both | RSR1 | No | Fu and Xue, 2010 |
| Sobic.009G033600 | Both | OsSAMS1 | Yes | Chen et al., 2013 |
| Sobic.009G036400 | BBH | APG | No | Heang and Sassa, 2012b |
| Sobic.009G040700 | Both | OsPPKL2 | Yes | Zhang et al., 2012 |
| Sobic.009G049400 | Both | SRS3 | Yes | Kitagawa et al., 2010 |
| Sobic.009G053600 | BBH | GS5 | Yes | Li et al., 2011 |
| Sobic.009G070000 | Both | GW5/qSW5 | Yes | Liu et al., 2017 |
| Sobic.009G141500 | Synteny | SERF1 | No | Schmidt et al., 2013 |
| Sobic.010G022600 | BBH | Wx1 | Yes | Shure et al., 1983 |
| Sobic.010G047400 | Both | HGW | Yes | Li J. et al., 2012 |
| Sobic.010G064600 | BBH | DA1 | No | Li et al., 2008 |
| Sobic.010G064800 | BBH | CKI1 | Yes | Deng et al., 2010 |
| Sobic.010G069600 | Synteny | SMG1 | Yes | Duan et al., 2014 |
| Sobic.010G072300 | Both | Sh1 | No | Jiang L. et al., 2013 |
| Sobic.010G091700 | Synteny | PGL2 | No | Heang and Sassa, 2012b |
| Sobic.010G110100 | BBH | A transcription factor | Yes | Huang et al., 2012b |
| Sobic.010G111200 | BBH | GASR7 | No | Huang et al., 2012b |
| Sobic.010G144900 | Original | NA | No | Zhang et al., 2015 |
| Sobic.010G184100 | Synteny | Bt1 | No | Shannon et al., 1998 |
| Sobic.010G210100 | Both | GW6 | Yes | Song et al., 2015 |
| Sobic.010G228100 | Both | DEP3 | Yes | Qiao et al., 2011 |
| Sobic.010G273900 | BBH | SbeI | No | Jiang L. et al., 2013 |
| Sobic.010G277300 | BBH | BRD2 | No | Hong et al., 2005 |
| Sobic.K041100 | BBH | GIF1 | Yes | Wang et al., 2008 |
| Sobic.K041200 | BBH | Mn1 | Yes | Miller and Chourey, 1992 |
Based on sorghum genome assembly 3.0.
Bioinformatics approach used to identify seed size candidate genes.
Gene name from the original publication in either maize, rice, or Arabidopsis.
Selection signature based on SNP level analysis.
Publication documenting the genes associated with seed size.
Figure 1.
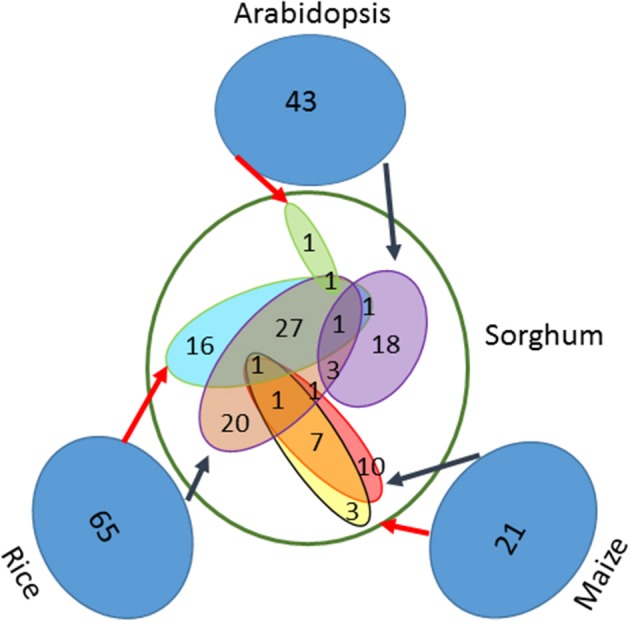
One hundred eleven orthologs of seed size genes identified in sorghum. Both the BBH method and the known syntenic relationships were used to identify orthologs of previously identified seed size genes in Arabidopsis (43), maize (21), and rice (65). The black arrows indicate BBH-identified orthologs, while the red arrows indicate syntenic orthologs.
The 114 identified seed size candidate genes were unevenly distributed across the 10 sorghum chromosomes, ranging from 23 genes located on chromosome 1 to only 2 genes located on chromosome 5. Whole genome expression data from the study by Davidson et al. (2012) was used to investigate the differential expression of the 114 candidate genes. A total of 22 genes exhibited differentially high levels of expression in the seed (Table S3).
Genetic diversity in seed size genes in sorghum
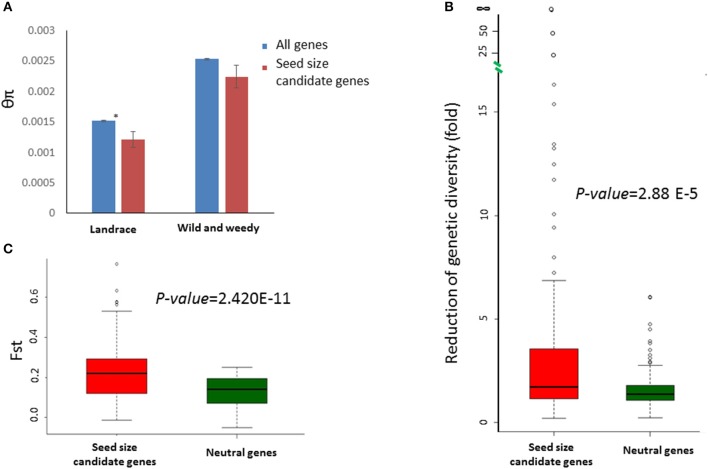
Sequence data for all 114 candidate genes was extracted from a previously described set of wild and weedy genotypes and landraces (Table S2; Mace et al., 2013). Overall, the selected genes exhibited a wide range of variation in sequence diversity in both genotype groups (the wild and weedy genotype group and the landraces group), with diversity measures (θπ) varying from 0.0085 (Sobic.002G311000) to 0 (Sobic.003G380900) in the wild and weedy genotypes, and from 0.0070 (Sobic.004G317300) to 0 (Sobic.003G035400, Sobic.003G380900, Sobic.004G065400, and Sobic.006G059900) in the landraces (Table S4). The SERF1 (a negative regulator of seed filling in rice) ortholog, Sobic.003G380900, was invariant in all the genotypes included in the current study. The sequence diversity observed in the seed size candidate genes in the wild and weedy genotypes was not significantly different to the genome-wide averages. However, the seed size candidate genes in the landraces were significantly less diverse than the genome-wide averages (p = 0.026, t-test) (Figure 2A) and were significantly less diverse in comparison to the wild and weedy genotypes (p = 3.68E-11, paired t-test). The RoD in the seed size candidates between the two genotype groups during domestication was greater when compared to 159 neutral genes identified in a previous study (Mace et al., 2013; Table S5, Figure 2B). The degree of population differentiation, measured by the fixation index FST, based on the seed size candidate genes was significantly higher between the landrace and wild and weedy genotypes (Figure 2C) in contrast to the neutral genes.
Figure 2.
Sequence variation identified in the seed size candidate genes in sorghum. (A) A comparison of sequence diversity (θπ) between the seed size candidate genes (red) and genome-wide averages (blue) in both the landrace and wild and weedy groups. Error bars indicate the standard error; * indicates a significant difference (p < 0.05, t-test) between the groups. (B), Box-plots showing the distributions of sequence diversity reduction fold) of 114 seed size candidate genes (red) and 159 neutral genes (blue) during domestication. The p-value was calculated based on a t-test. (C), Box-plots showing the distributions of FST between the landrace and wild and weedy genotype groups for 114 seed size candidate genes (red) and 159 (blue) neutral genes. The p-value was calculated based on a t-test.
Furthermore, the extent of RoD varied among the seed size candidate genes. Two genes, Sobic.006G059900 (ZmIPT2 ortholog) and Sobic.003G035400 (GW5 ortholog), were invariant in the landrace genotypes, despite having high levels of sequence diversity in the wild and weedy genotypes. The signature of significantly reduced sequence diversity in the landrace group, in comparison to the wild and weedy group, was also observed in four other genes, with RoD ranging from 15- to 58-fold: Sobic.003G030600 (58-fold decrease), Sobic.003G277900 (25-fold decrease), Sobic.007G149200 (20-fold decrease), and Sobic.003G230500 (15-fold decrease). A contrasting signature of increased sequence diversity in the landraces was observed for 16 seed size candidate genes, including Sobic.004G237000, a syntenic ortholog of PGL2, with θπ of 0.0048 in the landrace genotypes in comparison to just 0.0021 in the wild and weedy genotypes. In addition to reduced sequence diversity in the landraces, a more skewed allele frequency, as determined through a negative Tajima's D value, was observed in the majority of cases.
Signatures of selection in seed size candidate genes
Based on the genome-wide thresholds for the gene-level rankings described in Mace et al., (2013), 6 seed size candidate genes were identified with signatures of purifying selection during sorghum domestication (Table S6). Previous studies (Whitt et al., 2002; Brugiere et al., 2008; He et al., 2011; Hufford et al., 2012; Jiao et al., 2012; Xu et al., 2012; Luo et al., 2013; Weng et al., 2013; Wills et al., 2013; Lang et al., 2014; Zuo and Li, 2014; Sosso et al., 2015; Si et al., 2016) revealed purifying selection signals in 7 maize and 9 rice seed size genes included in this study (Table S1). Twenty one orthologs were identified in sorghum from 15 of the 16 genes under selection in either maize or rice, however, only one of them, Sobic.006G059900 (ZmIPT2 ortholog), was identified with signatures of purifying selection in sorghum based on the gene-level rankings (Table S6).
To investigate the domestication signature in the 114 sorghum seed size candidate genes at a higher resolution, signatures of purifying selection at the SNP level were analyzed. In total, 2,317 SNPs were identified in the CDS of all 114 candidate genes, consisting of 1,202 synonymous SNPs and 1,115 non-synonymous SNPs. In addition to sequence diversity (θπ) metrics, FST, Tajima's D, and RoD during domestication were calculated for each SNP. Based on the specified criteria regarding these metrics (see methods), 283 SNPs from 63 genes were identified with signatures of purifying selection, including Sobic.003G406600 (GW8 ortholog), Sobic.008G100400 (SMK1 ortholog), and Sobic.009G053600 (GS5 ortholog). Out of the 63 genes under selection, 42 contained non-synonymous SNPs under selection (Table S7). The selection signatures identified at the SNP level included 5 out of 6 genes under selection at the gene level.
To validate whether the 63 selection candidates displayed patterns of genetic variation consistent with purifying selection, the mlHKA test was employed. A model of directional selection best explained the patterns of polymorphism observed relative to 159 neutral loci (mean log likelihood ratio test statistic = 661, P < 7.49E-94 for all comparisons, Table S8). Additionally, out of 22 seed size candidates exhibiting differentially high levels of expression in the seed, 17 (77%) were under selection. The percentage is significantly higher than the remaining 92 seed size genes not exhibiting differentially higher levels of expression in the seed, where only 46 genes (50%) in this group were found to be under selection (χ2 = 6.546, p-value < 0.05), indicating seed size genes highly expressed in the seed are more likely to be targeted during domestication.
Parallel domestication of seed size in cereals
Seed size genes under selection across species were also identified. Among 15 seed size genes under selection in maize or rice, 12 were also found to be under selection in sorghum based on the SNP level CDS analysis in this study. A broader investigation of parallel domestication selection signals across syntenic orthologs of all the 114 seed size candidate genes in maize (Hufford et al., 2012; Jiao et al., 2012) and rice (He et al., 2011; Huang et al., 2012a; Xu et al., 2012) identified 30 seed size candidate genes in sorghum that have orthologs under selection in maize and/or rice (Table S6). Among these 30 sorghum genes, only one gene was under selection based on the gene level analysis, but 21 genes were identified as being under selection based on the SNP level CDS analysis (Table S6, Figure 3), with 4 of the 9 remaining genes having paralogs under purifying selection in sorghum. The sorghum seed size candidate genes under selection in multiple cereals included Sobic.009G070000 (GW5 ortholog), Sobic.003G406600 (the of GW8 ortholog), Sobic.007G101500 (Bt2 ortholog), Sobic.K041100 (GIF1 ortholog), and Sobic.005G001500 (PBF1 ortholog).
Figure 3.
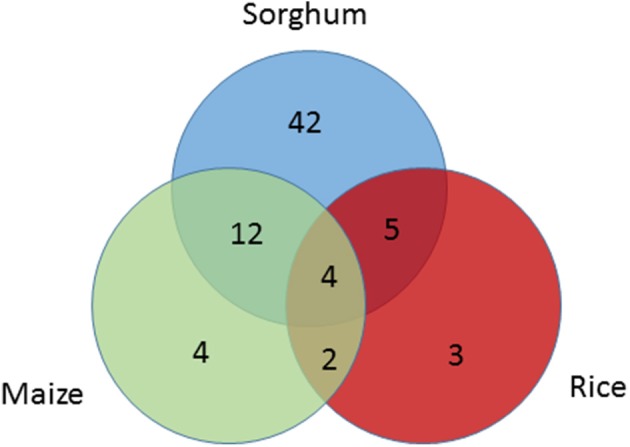
Venn-diagram showing the number seed size genes under selection across species; sorghum (blue), maize (green), and rice (red). Seed size candidate genes under selection in sorghum were identified based on SNP analysis in sorghum, while selection signals on their orthologs in maize and rice were extracted from previous studies.
Discussion
Seed size is a typical domestication syndrome trait, with cultivated cereal crops having larger seeds in comparison to their wild progenitors (Doebley et al., 2006). During domestication, large seeded genotypes were selected for their contribution to increased grain yield, but perhaps more importantly also for their positive effect on the quality of end-use products. Utilising the power of whole genome sequencing of diverse sorghum germplasm at the SNP level, combined with comparative genomic analysis of well researched cereal crops such as rice and maize, we identified 114 seed size candidate genes in sorghum. Signatures of domestication were identified in over half (63) of these genes through SNP level analysis of the CDS regions, with a high degree of concordance of seed size candidate genes under selection across species observed. Additionally, a group of seed size candidate genes that exhibited differentially high levels of expression in the seed were found to be more likely under selection during domestication. These results provide new insights into the genetic control of seed size in sorghum and the domestication of the seed size trait in cereal crops. Candidate genes included in this study provide a useful entry point into investigating the genetic factors controlling seed size. An understanding of genetic diversity and evolutionary pressures on these seed size candidate genes in sorghum provides potential targets for manipulating seed size via marker-assist selection or genome editing. In particular, intrinsic seed size genes may prove more amenable to relatively simple interventions in comparison to genes which effect seed size indirectly, for example via grain number.
Seed size candidate genes under selection are more likely to be intrinsic seed size genes rather than pleiotropic seed size genes
Of the 111 orthologs identified in sorghum based on seed size genes from maize, rice, and Arabidopsis, only 9 orthologous genes were identified as being associated with seed size in more than one species (Figure 1). This limited overlap suggests that the sample of seed size genes identified to date in each species is incomplete and/or that the genetic factors influencing seed size vary among species. This is likely to be due to the complexity of the genetic control of seed size, which is controlled by factors involved in intrinsic seed size, such as cell number, cell size, structure and composition, and by physiological factors involved in the carbohydrate supply-demand balance and transport.
Given the differences in plant architecture and physiology across the four species, it seems likely that genes under selection in sorghum that have also been identified as seed size genes in more than one species, either affect intrinsic seed size or directly affect seed number through an effect on panicle architecture, rather than affecting seed size via carbohydrate supply or indirectly affecting seed number. Both situations occurred in this study, as Sobic.001G341700, the ortholog of GS3 and ZmGS3 directly influences cell number in the seed, whereas Sobic.002G216600, the ortholog of DEP1 and AGG3, changes panicle branching and therefore seed number (Huang et al., 2009; Mao et al., 2010; Chakravorty et al., 2011; Li S. et al., 2012).
Of the 63 seed size candidate genes identified as being under selection in sorghum, 21 were identified as being under selection in at least one of the other species (Table S6). Genes that exhibited differentially high levels of expression in the seed are more likely to be associated with intrinsic variation for seed size. Our data shows that these genes were much more likely to be under selection during domestication. This provides support for the hypothesis that the modification to seed size during domestication preferentially targeted genes for intrinsic seed size rather than genes that indirectly impact on seed size.
Base pair level analysis provides a high resolution approach to study domestication signatures on seed size genes
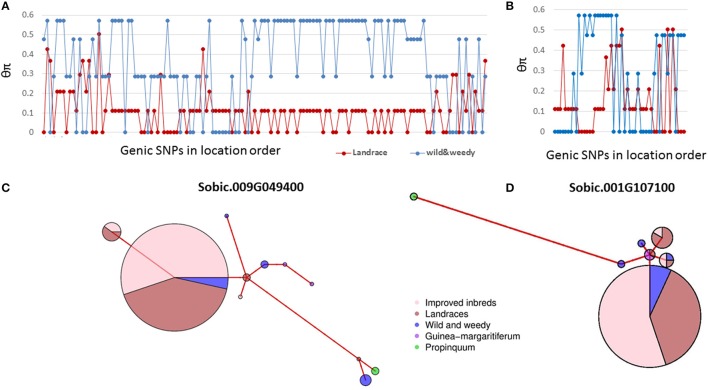
Domestication has shaped sorghum into a productive crop from a wild grass. Previous studies in sorghum have identified thousands of genes underpinning sorghum domestication based on whole genome analyses (Mace et al., 2013; Morris et al., 2013). This study detected selection signals in 63 seed size candidate genes in sorghum identified from cross species analyses based on individual nucleotide level analyses. The nucleotide level analyses provide greater resolution to study domestication signatures than whole gene level rankings. In general, when genes are under strong purifying selection, the gene level analysis may provide sufficient power to identify the signature of selection. For example, in Sobic.009G049400, the ortholog of SRS3 conferring a round seed phenotype in rice (Kitagawa et al., 2010), 44% of the SNPs were identified with signatures of purifying selection (Figure 4A). The majority of the remaining SNPs in this gene also exhibited the same trend of sequence diversity patterns, resulting in this gene being identified as under purifying selection at both the gene and nucleotide levels (Figure 4C, Mace et al., 2013). However, during domestication, contrasting selections can be imposed on different mutant loci of the same gene (particularly genes with pleiotropic effects) at different times, which results in a gene with chimeric positive and purifying selection signals (Purugganan and Fuller, 2009; Campbell et al., 2016). This situation was observed in this study, where 11 SNPs in the SRS5 ortholog, Sobic.001G107100, clustering within 50 bp of each other, were identified with signatures of purifying selection (Figure 4B). However, the gene was not identified as being under selection based on the gene level analysis due to the heterogeneous sequence diversity patterns observed across the entire gene length (Figure 4D). In such cases, analyzing each mutant locus separately provides increased resolution to identify the selection signature in comparison to gene level analysis in which contrasting selection signals within the same gene may cancel each other out.
Figure 4.
Genetic diversity pattern of two genes under selection. (A), The sequence diversity of SNPs within Sobic.009G049400; (B), The sequence diversity of SNPs within Sobic.001G107100; (C), The haplotype network of Sobic.009G049400; (D), The haplotype network of Sobic.001G107100. Group classification of sorghum accessions used as detailed in Table S2. Colour-coding as follows; improved inbred lines (pink), landraces (red), wild and weedy genotypes (blue), S. propinquum (green), and guinea margaritiferums (purple). The size of the circles in the haplotype networks is proportionate to the number of accessions with that haplotype. The branch length represents the genetic distance between two haplotypes.
Common seed size genes under selection across cereals supports parallel domestication of seed size in grass cereals
During crop domestication, human demands have led to a similar suite of traits being changed across a wide range of crops, a phenomenon known as convergent domestication (Lenser and Theißen, 2013). However, whether the same genetic basis underlies parallel changes in different species is still under debate. Early QTL mapping studies found close correspondence of QTLs for seed size, shattering, and flowering time across cereal crops (Paterson et al., 1995), with subsequent detailed QTL analyses identifying high levels of concordance in flowering time QTLs across sorghum and maize (Mace et al., 2013). Recently, Sh1, a major QTL controlling shattering, and HD1, a major locus conferring flowering time, have been reported to be under parallel selection in multiple cereals (Lin et al., 2012; Liu H. et al., 2015). In this study, among 15 seed size genes previously identified to be under selection in rice or maize, 12 were shown to have orthologs in sorghum under selection during domestication. Genes under parallel selection have been found to be major effect loci of seed size explaining a large proportion of the phenotypic variation (Lenser and Theißen, 2013). The significant overlap of selection signatures on seed size genes in cereals provides support for the role of parallel domestication.
Conclusions
Seed size and weight are physiologically complex traits controlled by many loci, some of which have been selected during the domestication of cereals. In this study, we have collated a large number of genes controlling seed size and weight across three extensively studied plant model species and identified their sorghum orthologs using comparative genomics analyses. We demonstrated that has domestication in sorghum left signatures of selection genetic signatures on multiple seed size candidate genes. For a number of the seed size genes we found signatures of selection that were common across sorghum, maize and rice, consistent with parallel domestication of the seed size trait. We also found that seed size candidate genes that exhibited differentially high levels of expression in the seed were more likely to be under selection during domestication. Our work sheds light on the processes involved in cereal domestication and provides potential targets for breeding to increase seed size and potentially yield.
Author contributions
DJ, EM, and IG conceived and designed the experiments: YT, AC, EM, DJ, and XZ collected data; YT, ST, BC, EV, JB, DJ, and EM analyzed data; YT and EM wrote the manuscript. EV, JB, IG, and DJ revised the manuscript. All authors read and approved the final manuscript.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We acknowledge access to background IP from Grains Research and Development Corporation and support from the University of Queensland and the Department of Agriculture and Fisheries Queensland.
Glossary
Abbreviations
- BBH
bidirectional best hit
- RoD
reduction of diversity.
Footnotes
Funding. This work was supported by the Australian Research Council (ARC) Discovery project DP14010250, and Technology Innovation Program (CXZZ20150330171810060) and Basic Research Program (NO. JCYJ20150831201123287) from Shenzhen Municipal Government.
Supplementary material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2017.01237/full#supplementary-material
References
- Acreche M. M., Slafer G. A. (2006). Grain weight response to increases in number of grains in wheat in a Mediterranean area. Field Crops Res. 98, 52–59. 10.1016/j.fcr.2005.12.005 [DOI] [Google Scholar]
- Adamski N. M., Anastasiou E., Eriksson S., O'Neill C. M., Lenhard M. (2009). Local maternal control of seed size by KLUH/CYP78A5-dependent growth signaling. Proc. Natl. Acad. Sci. U.S.A. 106, 20115–20120. 10.1073/pnas.0907024106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alam M. M., Mace E. S., van Oosterom E. J., Cruickshank A., Hunt C. H., Hammer G. L., et al. (2014). QTL analysis in multiple sorghum populations facilitates the dissection of the genetic and physiological control of tillering. Theor. Appl. Genet. 127, 2253–2266. 10.1007/s00122-014-2377-9 [DOI] [PubMed] [Google Scholar]
- Ashikari M., Wu J., Yano M., Sasaki T., Yoshimura A. (1999). Rice gibberellin-insensitive dwarf mutant gene Dwarf 1 encodes the α-subunit of GTP-binding protein. Proc. Natl. Acad. Sci. U.S.A. 96, 10284–10289. 10.1073/pnas.96.18.10284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beissinger T. M., Hirsch C. N., Vaillancourt B., Deshpande S., Barry K., Buell C. R., et al. (2014). A genome-wide scan for evidence of selection in a maize population under long-term artificial selection for ear number. Genetics 196, 829–840. 10.1534/genetics.113.160655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borrell A. K., van Oosterom E. J., Mullet J. E., George-Jaeggli B., Jordan D. R., Klein P. E., et al. (2014). Stay-green alleles individually enhance grain yield in sorghum under drought by modifying canopy development and water uptake patterns. New Phytol. 203, 817–830. 10.1111/nph.12869 [DOI] [PubMed] [Google Scholar]
- Botella J. R. (2012). Can heterotrimeric G proteins help to feed the world? Trends Plant Sci. 17, 563–568. 10.1016/j.tplants.2012.06.002 [DOI] [PubMed] [Google Scholar]
- Boyles R. E., Cooper E. A., Myers M. T., Brenton Z., Rauh B. L., Morris G. P., et al. (2016). Genome-wide association studies of grain yield components in diverse sorghum germplasm. Plant Genome. 9, 1–17. 10.3835/plantgenome2015.09.0091 [DOI] [PubMed] [Google Scholar]
- Brugiere N., Humbert S., Rizzo N., Bohn J., Habben J. E. (2008). A member of the maize isopentenyl transferase gene family, Zea mays isopentenyl transferase 2 (ZmIPT2), encodes a cytokinin biosynthetic enzyme expressed during kernel development. Plant Mol. Biol. 67, 215–229. 10.1007/s11103-008-9312-x [DOI] [PubMed] [Google Scholar]
- Burow G., Xin Z., Hayes C., Burke J. (2014). Characterization of a multiseeded () mutant of sorghum for increasing grain yield. Crop Sci. 54, 2030–2037. 10.2135/cropsci2013.08.0566 [DOI] [Google Scholar]
- Campbell B. C., Gilding E. K., Mace E. S., Tai S., Tao Y., Prentis P. J., et al. (2016). Domestication and the storage starch biosynthesis pathway: signatures of selection from a whole sorghum genome sequencing strategy. Plant Biotechnol. J. 14, 2240–2253 10.1111/pbi.12578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakravorty D., Trusov Y., Zhang W., Acharya B. R., Sheahan M. B., McCurdy D. W., et al. (2011). An atypical heterotrimeric G-protein γ-subunit is involved in guard cell K+-channel regulation and morphological development in Arabidopsis thaliana. Plant J. 67, 840–851 10.1111/j.1365-313X.2011.04638.x [DOI] [PubMed] [Google Scholar]
- Che R., Tong H., Shi B., Liu Y., Fang S., Liu D., et al. (2015). Control of grain size and rice yield by GL2-mediated brassinosteroid responses. Nat. Plants 2:15195. 10.1038/nplants.2015.195 [DOI] [PubMed] [Google Scholar]
- Chen Y., Xu Y., Luo W., Li W., Chen N., Zhang D., et al. (2013). The F-box protein OsFBK12 targets OsSAMS1 for degradation and affects pleiotropic phenotypes, including leaf senescence, in rice. Plant Physiol. 163, 1673–1685. 10.1104/pp.113.224527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson R. M., Gowda M., Moghe G., Lin H., Vaillancourt B., Shiu S. H., et al. (2012). Comparative transcriptomics of three Poaceae species reveals patterns of gene expression evolution. Plant J. 71, 492–502. 10.1111/j.1365-313x.2012.05005.x [DOI] [PubMed] [Google Scholar]
- Deng Y., Dong H., Mu J., Ren B., Zheng B., Ji Z., et al. (2010). Arabidopsis histidine kinase CKI1 acts upstream of histidine phosphotransfer proteins to regulate female gametophyte development and vegetative growth. Plant Cell 22, 1232–1248. 10.1105/tpc.108.065128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doebley J. F., Gaut B. S., Smith B. D. (2006). The molecular genetics of crop domestication. Cell 127, 1309–1321. 10.1016/j.cell.2006.12.006 [DOI] [PubMed] [Google Scholar]
- Duan P., Rao Y., Zeng D., Yang Y., Xu R., Zhang B., et al. (2014). SMALL GRAIN 1, which encodes a mitogen-activated protein kinase kinase 4, influences grain size in rice. Plant J. 77, 547–557. 10.1111/tpj.12405 [DOI] [PubMed] [Google Scholar]
- Eckert C., Offenborn J. N., Heinz T., Armarego-Marriott T., Schültke S., Zhang C., et al. (2014). The vacuolar calcium sensors CBL2 and CBL3 affect seed size and embryonic development in Arabidopsis thaliana. Plant J. 78, 146–156. 10.1111/tpj.12456 [DOI] [PubMed] [Google Scholar]
- Eom J. S., Cho J. I., Reinders A., Lee S. W., Yoo Y., Tuan P. Q., et al. (2011). Impaired function of the tonoplast-localized sucrose transporter in rice, OsSUT2, limits the transport of vacuolar reserve sucrose and affects plant growth. Plant Physiol. 157, 109–119. 10.1104/pp.111.176982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang W., Wang Z., Cui R., Li J., Li Y. (2012). Maternal control of seed size by EOD3/CYP78A6 in Arabidopsis thaliana. Plant J. 70, 929–939. 10.1111/j.1365-313X.2012.04907.x [DOI] [PubMed] [Google Scholar]
- Fu F., Xue H. (2010). Coexpression analysis identifies Rice Starch Regulator1, a rice AP2/EREBP family transcription factor, as a novel rice starch biosynthesis regulator. Plant Physiol. 154, 927–938. 10.1104/pp.110.159517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia D., Gerald J. N. F., Berger F. (2005). Maternal control of integument cell elongation and zygotic control of endosperm growth are coordinated to determine seed size in Arabidopsis. Plant Cell, 17, 52–60. 10.1105/tpc.104.027136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakata M., Kuroda M., Ohsumi A., Hirose T., Nakamura H., Muramatsu M., et al. (2012). Overexpression of a rice TIFY gene increases grain size through enhanced accumulation of carbohydrates in the stem. Biosci. Biotechnol. Biochem. 76, 2129–2134. 10.1271/bbb.120545 [DOI] [PubMed] [Google Scholar]
- Hartings H., Maddaloni M., Lazzaroni N., Di Fonzo N., Motto M., Salamini F., et al. (1989). The O2 gene which regulates zein deposition in maize endosperm encodes a protein with structural homologies to transcriptional activators. EMBO J. 8, 2795–2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Z., Zhai W., Wen H., Tang T., Wang Y., Lu X., et al. (2011). Two evolutionary histories in the genome of rice: the roles of domestication genes. PLoS Genet. 7:e1002100. 10.1371/journal.pgen.1002100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heang D., Sassa H. (2012a). An atypical bHLH protein encoded by POSITIVE REGULATOR OF GRAIN LENGTH 2 is involved in controlling grain length and weight of rice through interaction with a typical bHLH protein APG. Breed. Sci. 62, 133–141. 10.1270/jsbbs.62.133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heang D., Sassa H. (2012b). Overexpression of a basic helix–loop–helix gene Antagonist of PGL1 (APG) decreases grain length of rice. Plant Biotechnol. 29, 65–69. 10.5511/plantbiotechnology.12.0117a [DOI] [Google Scholar]
- Heinrich G., Francis C., Eastin J. (1983). Stability of grain sorghum yield components across diverse environments. Crop Sci. 23, 209–212. 10.2135/cropsci1983.0011183X002300020004x [DOI] [Google Scholar]
- Hong Z., Ueguchi-Tanaka M., Fujioka S., Takatsuto S., Yoshida S., Hasegawa Y., et al. (2005). The rice brassinosteroid-deficient dwarf2 mutant, defective in the rice homolog of Arabidopsis DIMINUTO/DWARF1, is rescued by the endogenously accumulated alternative bioactive brassinosteroid, dolichosterone. Plant Cell 17, 2243–2254. 10.1105/tpc.105.030973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong Z., Ueguchi-Tanaka M., Umemura K., Uozu S., Fujioka S., Takatsuto S., et al. (2003). A rice brassinosteroid-deficient mutant, ebisu dwarf (d2), is caused by a loss of function of a new member of cytochrome P450. Plant Cell, 15, 2900–2910. 10.1105/tpc.014712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J., Wang Y., Fang Y., Zeng L., Xu J., Yu H., et al. (2015). A rare allele of GS2 enhances grain size and grain yield in rice. Mol. Plant 8, 1455–1465. 10.1016/j.molp.2015.07.002 [DOI] [PubMed] [Google Scholar]
- Hu X., Qian Q., Xu T., Zhang Y. E., Dong G., Gao T., et al. (2013). The U-box E3 ubiquitin ligase TUD1 functions with a heterotrimeric G α subunit to regulate brassinosteroid-mediated growth in rice. PLoS Genet. 9:e1003391. 10.1371/journal.pgen.1003391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X., Kurata N., Wei X., Wang Z., Wang A., Zhao Q., et al. (2012a). A map of rice genome variation reveals the origin of cultivated rice. Nature 490, 497–501. 10.1038/nature11532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X., Qian Q., Liu Z., Sun H., He S., Luo D., et al. (2009). Natural variation at the DEP1 locus enhances grain yield in rice. Nat. Genet. 41, 494–497. 10.1038/ng.352 [DOI] [PubMed] [Google Scholar]
- Huang X., Zhao Y., Wei X., Li C., Wang A., Zhao Q., et al. (2012b). Genome-wide association study of flowering time and grain yield traits in a worldwide collection of rice germplasm. Nat. Genet. 44, 32–39. 10.1038/ng.1018 [DOI] [PubMed] [Google Scholar]
- Hudson R. R., Slatkin M., Maddison W. P. (1992). Estimation of levels of gene flow from DNA sequence data. Genetics 132, 583–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hufford M. B., Xu X., Van Heerwaarden J., Pyhäjärvi T., Chia J. M., Cartwright R. A., et al. (2012). Comparative population genomics of maize domestication and improvement. Nat. Genet. 44, 808–811. 10.1038/ng.2309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchison C. E., Li J., Argueso C., Gonzalez M., Lee E., Lewis M. W., et al. (2006). The Arabidopsis histidine phosphotransfer proteins are redundant positive regulators of cytokinin signaling. Plant Cell 18, 3073–3087. 10.1105/tpc.106.045674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishimaru K., Hirotsu N., Madoka Y., Murakami N., Hara N., Onodera H., et al. (2013). Loss of function of the IAA-glucose hydrolase gene TGW6 enhances rice grain weight and increases yield. Nat. Genet. 45, 707–711. 10.1038/ng.2612 [DOI] [PubMed] [Google Scholar]
- Jagadish K. S., Kishor P. B. K., Bahuguna R. N., von Wirén N., Sreenivasulu N. (2015). Staying alive or going to die during terminal senescence-an enigma surrounding yield stability. Front. Plant Sci. 6:01070. 10.3389/fpls.2015.01070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakobsson A., Eriksson O. (2000). A comparative study of seed number, seed size, seedling size and recruitment in grassland plants. Oikos 88, 494–502. 10.1034/j.1600-0706.2000.880304.x [DOI] [Google Scholar]
- Jenner C. (1994). Starch synthesis in the kernel of wheat under high temperature conditions. Funct. Plant Biol. 21, 791–806. 10.1071/pp9940791 [DOI] [Google Scholar]
- Jiang L., Yu X., Qi X., Yu Q., Deng S., Bai B., et al. (2013). Multigene engineering of starch biosynthesis in maize endosperm increases the total starch content and the proportion of amylose. Transgenic Res. 22, 1133–1142. 10.1007/s11248-013-9717-4 [DOI] [PubMed] [Google Scholar]
- Jiang W., Huang H., Hu Y., Zhu S., Wang Z., Lin W. (2013). Brassinosteroid regulates seed size and shape in Arabidopsis. Plant Physiol. 162, 1965–1977. 10.1104/pp.113.217703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y., Zhao H., Ren L., Song W., Zeng B., Guo J., et al. (2012). Genome-wide genetic changes during modern breeding of maize. Nat. Genet. 44, 812–815. 10.1038/ng.2312 [DOI] [PubMed] [Google Scholar]
- Kitagawa K., Kurinami S., Oki K., Abe Y., Ando T., Kono I., et al. (2010). A novel kinesin 13 protein regulating rice seed length. Plant Cell Physiol. 51, 1315–1329. 10.1093/pcp/pcq092 [DOI] [PubMed] [Google Scholar]
- Kondou Y., Nakazawa M., Kawashima M., Ichikawa T., Yoshizumi T., Suzuki K., et al. (2008). RETARDED GROWTH OF EMBRYO1, a new basic helix-loop-helix protein, expresses in endosperm to control embryo growth. Plant Physiol. 147, 1924–1935. 10.1104/pp.108.118364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang Z., Wills D. M., Lemmon Z. H., Shannon L. M., Bukowski R., Wu Y., et al. (2014). Defining the role of prolamin-box binding factor1 gene during maize domestication. J. Hered. 105, 576–582. 10.1093/jhered/esu019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee W., Pedersen J. F., Shelton D. (2002). Relationship of sorghum kernel size to physiochemical, milling, pasting, and cooking properties. Food Res. Int. 35, 643–649. 10.1016/S0963-9969(01)00167-3 [DOI] [Google Scholar]
- Lenser T., Theißen G. (2013). Molecular mechanisms involved in convergent crop domestication. Trends Plant Sci. 18, 704–714. 10.1016/j.tplants.2013.08.007 [DOI] [PubMed] [Google Scholar]
- Li F., Liu W., Tang J., Chen J., Tong H., Hu B., et al. (2010). Rice DENSE AND ERECT PANICLE 2 is essential for determining panicle outgrowth and elongation. Cell Res. 20, 838–849. 10.1038/cr.2010.69 [DOI] [PubMed] [Google Scholar]
- Li J., Chu H., Zhang Y., Mou T., Wu C., Zhang Q., et al. (2012). The rice HGW gene encodes a ubiquitin-associated (UBA) domain protein that regulates heading date and grain weight. PLoS ONE 7:e34231. 10.1371/journal.pone.0034231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Nie X., Tan J. L. H., Berger F. (2013). Integration of epigenetic and genetic controls of seed size by cytokinin in Arabidopsis. Proc. Natl Acad. Sci. U.S.A. 110, 15479–15484. 10.1073/pnas.1305175110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q., Li L., Yang X., Warburton M. L., Bai G., Dai J., et al. (2010a). Relationship, evolutionary fate and function of two maize co-orthologs of rice GW2 associated with kernel size and weight. BMC Plant Biol. 10:143. 10.1186/1471-2229-10-143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q., Yang X., Bai G., Warburton M. L., Mahuku G., Gore M., et al. (2010b). Cloning and characterization of a putative GS3 ortholog involved in maize kernel development. Theor. Appl. Genet. 120, 753–763. 10.1007/s00122-009-1196-x [DOI] [PubMed] [Google Scholar]
- Li S., Liu Y., Zheng L., Chen L., Li N., Corke F., et al. (2012). The plant-specific G protein γ subunit AGG3 influences organ size and shape in Arabidopsis thaliana. New Phytol. 194, 690–703. 10.1111/j.1469-8137.2012.04083.x [DOI] [PubMed] [Google Scholar]
- Li X. J., Zhang Y. F., Hou M., Sun F., Shen Y., Xiu Z. H., et al. (2014). Small kernel 1 encodes a pentatricopeptide repeat protein required for mitochondrial nad7 transcript editing and seed development in maize (Zea mays) and rice (Oryza sativa). Plant J. 79, 797–809. 10.1111/tpj.12584 [DOI] [PubMed] [Google Scholar]
- Li X., Sun L., Tan L., Liu F., Zhu Z., Fu Y., et al. (2012). TH1, a DUF640 domain-like gene controls lemma and palea development in rice. Plant Mol. Biol. 78, 351–359. 10.1007/s11103-011-9868-8 [DOI] [PubMed] [Google Scholar]
- Li Y., Fan C., Xing Y., Jiang Y., Luo L., Sun L., et al. (2011). Natural variation in GS5 plays an important role in regulating grain size and yield in rice. Nat. Genet. 43, 1266–1269. 10.1038/ng.977 [DOI] [PubMed] [Google Scholar]
- Li Y., Zheng L., Corke F., Smith C., Bevan M. W. (2008). Control of final seed and organ size by the DA1 gene family in Arabidopsis thaliana. Genes Dev. 22, 1331–1336. 10.1101/gad.463608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Z., Li X., Shannon L. M., Yeh C.-T., Wang M. L., Bai G., et al. (2012). Parallel domestication of the Shattering1 genes in cereals. Nat. Genet. 44, 720–724. 10.1038/ng.2281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Liu H., Zhou L., Zhang Z., Zhang X., Wang M., et al. (2015). Parallel domestication of the heading date 1 gene in cereals. Mol. Biol. Evol. 32, 2726–2737. 10.1093/molbev/msv148 [DOI] [PubMed] [Google Scholar]
- Liu J., Chen J., Zheng X., Wu F., Lin Q., Heng Y., et al. (2017). GW5 acts in the brassinosteroid signalling pathway to regulate grain width and weight in rice. Nat. Plants 3:17043. 10.1038/nplants.2017.43 [DOI] [PubMed] [Google Scholar]
- Liu L., Tong H., Xiao Y., Che R., Xu F., Hu B., et al. (2015). Activation of Big Grain1 significantly improves grain size by regulating auxin transport in rice. Proc. Natl. Acad. Sci. U.S.A. 112, 11102–11107. 10.1073/pnas.1512748112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo J., Liu H., Zhou T., Gu B., Huang X., Shangguan Y., et al. (2013). An-1 encodes a basic helix-loop-helix protein that regulates awn development, grain size, and grain number in rice. Plant Cell 25, 3360–3376. 10.1105/tpc.113.113589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo M., Bilodeau P., Dennis E. S., Peacock W. J., Chaudhury A. (2000). Expression and parent-of-origin effects for FIS2, MEA, and FIE in the endosperm and embryo of developing Arabidopsis seeds. Proc. Natl. Acad. Sci. U.S.A. 97, 10637–10642. 10.1073/pnas.170292997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo M., Dennis E. S., Berger F., Peacock W. J., Chaudhury A. (2005). MINISEED3 (MINI3), a WRKY family gene, and HAIKU2 (IKU2), a leucine-rich repeat (LRR) KINASE gene, are regulators of seed size in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 102, 17531–17536. 10.1073/pnas.0508418102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma B., He S., Duan K., Yin C., Chen H., Yang C., et al. (2013). Identification of rice ethylene-response mutants and characterization of MHZ7/OsEIN2 in distinct ethylene response and yield trait regulation. Mol. Plant 6, 1830–1848. 10.1093/mp/sst087 [DOI] [PubMed] [Google Scholar]
- Mace E. S., Tai S., Gilding E. K., Li Y., Prentis P. J., Bian L., et al. (2013). Whole-genome sequencing reveals untapped genetic potential in Africa's indigenous cereal crop sorghum. Nat. Commun. 4:2320. 10.1038/ncomms3320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manga V. K., Yadav O. P. (1995). Effect of seed size on development traits and ability to tolerate drought in pearl millet. J. Arid Environ. 29, 169–172. 10.1016/S0140-1963(05)80087-4 [DOI] [Google Scholar]
- Mao H., Sun S., Yao J., Wang C., Yu S., Xu C., et al. (2010). Linking differential domain functions of the GS3 protein to natural variation of grain size in rice. Proc. Natl. Acad. Sci. U.S.A. 107, 19579–19584. 10.1073/pnas.1014419107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin A., Lee J., Kichey T., Gerentes D., Zivy M., Tatout C., et al. (2006). Two cytosolic glutamine synthetase isoforms of maize are specifically involved in the control of grain production. Plant Cell 18, 3252–3274. 10.1105/tpc.106.042689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller M. E., Chourey P. S. (1992). The maize invertase-deficient miniature-1 seed mutation is associated with aberrant pedicel and endosperm development. Plant Cell 4, 297–305. 10.1105/tpc.4.3.297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizukami Y., Fischer R. L. (2000). Plant organ size control: AINTEGUMENTA regulates growth and cell numbers during organogenesis. Proc. Natl. Acad. Sci. U.S.A. 97, 942–947. 10.1073/pnas.97.2.942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moles A. T., Westoby M. (2004). Seedling survival and seed size: a synthesis of the literature. J. Ecol. 92, 372–383. 10.1111/j.0022-0477.2004.00884.x [DOI] [Google Scholar]
- Mori M., Nomura T., Ooka H., Ishizaka M., Yokota T., Sugimoto K., et al. (2002). Isolation and characterization of a rice dwarf mutant with a defect in brassinosteroid biosynthesis. Plant Physiol. 130, 1152–1161. 10.1104/pp.007179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morinaka Y., Sakamoto T., Inukai Y., Agetsuma M., Kitano H., Ashikari M., et al. (2006). Morphological alteration caused by brassinosteroid insensitivity increases the biomass and grain production of rice. Plant Physiol. 141, 924–931. 10.1104/pp.106.077081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris G. P., Ramu P., Deshpande S. P., Hash C. T., Shah T., Upadhyaya H. D., et al. (2013). Population genomic and genome-wide association studies of agroclimatic traits in sorghum. Proc. Natl. Acad. Sci. U.S.A. 110, 453–458. 10.1073/pnas.1215985110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Na J. K., Seo M. H., Yoon I. S., Lee Y. H., Lee K. O., Kim D. Y. (2012). Involvement of rice Polycomb protein OsFIE2 in plant growth and seed size. Plant Biotechnol. Rep. 6, 339–346. 10.1007/s11816-012-0229-0 [DOI] [Google Scholar]
- Nakagawa H., Tanaka A., Tanabata T., Ohtake M., Fujioka S., Nakamura H., et al. (2012). Short grain1 decreases organ elongation and brassinosteroid response in rice. Plant Physiol. 158, 1208–1219. 10.1104/pp.111.187567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M., Li W. H. (1979). Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc. Natl. Acad. Sci. U.S.A. 76, 5269–5273. 10.1073/pnas.76.10.5269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohto M. A., Floyd S. K., Fischer R. L., Goldberg R. B., Harada J. J. (2009). Effects of APETALA2 on embryo, endosperm, and seed coat development determine seed size in Arabidopsis. Sex. Plant Reprod. 22, 277–289. 10.1007/s00497-009-0116-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okushima Y., Mitina I., Quach H. L., Theologis A. (2005). AUXIN RESPONSE FACTOR 2 (ARF2): a pleiotropic developmental regulator. Plant J. 43, 29–46. 10.1111/j.1365-313X.2005.02426.x [DOI] [PubMed] [Google Scholar]
- Paradis E. (2010). pegas: an R package for population genetics with an integrated–modular approach. Bioinformatics 26, 419–420. 10.1093/bioinformatics/btp696 [DOI] [PubMed] [Google Scholar]
- Paradis E., Claude J., Strimmer K. (2004). APE: analyses of phylogenetics and evolution in R language. Bioinformatics 20, 289–290. 10.1093/bioinformatics/btg412 [DOI] [PubMed] [Google Scholar]
- Paterson A. H., Lin Y. R., Li Z., Schertz K. F. (1995). Convergent domestication of cereal crops by independent mutations at corresponding genetic loci. Science 269, 1714–1718. [DOI] [PubMed] [Google Scholar]
- Peltonen-Sainio P., Kangas A., Salo Y., Jauhiainen L. (2007). Grain number dominates grain weight in temperate cereal yield determination: evidence based on 30 years of multi-location trials. Field Crops Res. 100, 179–188. 10.1016/j.fcr.2006.07.002 [DOI] [Google Scholar]
- Pfeifer B., Wittelsbürger U., Onsins S. E. R., Lercher M. J. (2014). PopGenome: an efficient Swiss army knife for population genomic analyses in R. Mol. Biol. Evol. 31, 1929–1936. 10.1093/molbev/msu136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purugganan M. D., Fuller D. Q. (2009). The nature of selection during plant domestication. Nature 457, 843–848. 10.1038/nature07895 [DOI] [PubMed] [Google Scholar]
- Qi P., Lin Y., Song X., Shen J., Huang W., Shan J., et al. (2012). The novel quantitative trait locus GL3. 1 controls rice grain size and yield by regulating Cyclin-T1; 3. Cell Res. 22, 1666–1680. 10.1038/cr.2012.151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao Y., Piao R., Shi J., Lee S. I., Jiang W., Kim B. K., et al. (2011). Fine mapping and candidate gene analysis of dense and erect panicle 3, DEP3, which confers high grain yield in rice (Oryza sativa L.). Theor. Appl. Genet. 122, 1439–1449. 10.1007/s00122-011-1543-6 [DOI] [PubMed] [Google Scholar]
- Riefler M., Novak O., Strnad M., Schmülling T. (2006). Arabidopsis cytokinin receptor mutants reveal functions in shoot growth, leaf senescence, seed size, germination, root development, and cytokinin metabolism. Plant Cell 18, 40–54. 10.1105/tpc.105.037796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadras V. O. (2007). Evolutionary aspects of the trade-off between seed size and number in crops. Field Crops Res. 100, 125–138. 10.1016/j.fcr.2006.07.004 [DOI] [Google Scholar]
- Sands D. C., Morris C. E., Dratz E. A., Pilgeram A. L. (2009). Elevating optimal human nutrition to a central goal of plant breeding and production of plant-based foods. Plant Sci. 177, 377–389. 10.1016/j.plantsci.2009.07.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt R., Mieulet D., Hubberten H. M., Obata T., Hoefgen R., Fernie A. R., et al. (2013). SALT-RESPONSIVE ERF1 regulates reactive oxygen species–dependent signaling during the initial response to salt stress in rice. Plant Cell 25, 2115–2131. 10.1105/tpc.113.113068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segami S., Kono I., Ando T., Yano M., Kitano H., Miura K., et al. (2012). Small and round seed 5 gene encodes alpha-tubulin regulating seed cell elongation in rice. Rice 5, 1–10. 10.1186/1939-8433-5-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon J. C., Pien F. M., Cao H., Liu K. (1998). Brittle-1, an adenylate translocator, facilitates transfer of extraplastidial synthesized ADP-glucose into amyloplasts of maize endosperms. Plant Physiol. 117, 1235–1252. 10.1104/pp.117.4.1235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- She K. C., Kusano H., Koizumi K., Yamakawa H., Hakata M., Imamura T., et al. (2010). A novel factor FLOURY ENDOSPERM2 is involved in regulation of rice grain size and starch quality. Plant Cell 22, 3280–3294. 10.1105/tpc.109.070821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shure M., Wessler S., Fedoroff N. (1983). Molecular identification and isolation of the Waxy locus in maize. Cell 35, 225–233. 10.1016/0092-8674(83)90225-8 [DOI] [PubMed] [Google Scholar]
- Si L., Chen J., Huang X., Gong H., Luo J., Hou Q., et al. (2016). OsSPL13 controls grain size in cultivated rice. Nat. Genet. 48, 447–456 10.1038/ng.3518 [DOI] [PubMed] [Google Scholar]
- Song X. J., Kuroha T., Ayano M., Furuta T., Nagai K., Komeda N., et al. (2015). Rare allele of a previously unidentified histone H4 acetyltransferase enhances grain weight, yield, and plant biomass in rice. Proc. Natl. Acad. Sci. U.S.A. 112, 76–81. 10.1073/pnas.1421127112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song X., Huang W., Shi M., Zhu M., Lin H. (2007). A QTL for rice grain width and weight encodes a previously unknown RING-type E3 ubiquitin ligase. Nat. Genet. 39, 623–630. 10.1038/ng2014 [DOI] [PubMed] [Google Scholar]
- Sosso D., Luo D., Li Q. B., Sasse J., Yang J., Gendrot G., et al. (2015). Seed filling in domesticated maize and rice depends on SWEET-mediated hexose transport. Nat. Genet. 47, 1489–1493. 10.1038/ng.3422 [DOI] [PubMed] [Google Scholar]
- Su'udi M., Cha J. Y., Ahn I. P., Kwak Y. S., Woo Y. M., Son D. (2012). Functional characterization of a B-type cell cycle switch 52 in rice (OsCCS52B). Plant Cell Tissue Organ Cult. 111, 101–111. 10.1007/s11240-012-0176-z [DOI] [Google Scholar]
- Sui P., Jin J., Ye S., Mu C., Gao J., Feng H., et al. (2012). H3K36 methylation is critical for brassinosteroid-regulated plant growth and development in rice. Plant J. 70, 340–347. 10.1111/j.1365-313X.2011.04873.x [DOI] [PubMed] [Google Scholar]
- Tajima F. (1989). Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123, 585–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takano-Kai N., Jiang H., Kubo T., Sweeney M., Matsumoto T., Kanamori H., et al. (2009). Evolutionary history of GS3, a gene conferring grain length in rice. Genetics 182, 1323–1334. 10.1534/genetics.109.103002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanabe S., Ashikari M., Fujioka S., Takatsuto S., Yoshida S., Yano M., et al. (2005). A novel cytochrome P450 is implicated in brassinosteroid biosynthesis via the characterization of a rice dwarf mutant, dwarf11, with reduced seed length. Plant Cell 17, 776–790. 10.1105/tpc.104.024950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang A., Garcia D., Zhang H., Feng K., Chaudhury A., Berger F., et al. (2010). The VQ motif protein IKU1 regulates endosperm growth and seed size in Arabidopsis. Plant J. 63, 670–679. 10.1111/j.1365-313X.2010.04271.x [DOI] [PubMed] [Google Scholar]
- Wang E., Wang J., Zhu X., Hao W., Wang L., Li Q., et al. (2008). Control of rice grain-filling and yield by a gene with a potential signature of domestication. Nat. Genet. 40, 1370–1374. 10.1038/ng.220 [DOI] [PubMed] [Google Scholar]
- Wang G., Wang F., Wang G., Wang F., Zhang X., Zhong M., et al. (2012). Opaque1 encodes a myosin XI motor protein that is required for endoplasmic reticulum motility and protein body formation in maize endosperm. Plant Cell 24, 3447–3462. 10.1105/tpc.112.101360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Li S., Liu Q., Wu K., Zhang J., Wang S., et al. (2015). The OsSPL16-GW7 regulatory module determines grain shape and simultaneously improves rice yield and grain quality. Nat. Genet. 47, 949–954. 10.1038/ng.3352 [DOI] [PubMed] [Google Scholar]
- Wang S., Wu K., Yuan Q., Liu X., Liu Z., Lin X., et al. (2012). Control of grain size, shape and quality by OsSPL16 in rice. Nat. Genet. 44, 950–954. 10.1038/ng.2327 [DOI] [PubMed] [Google Scholar]
- Wang Y., Xiong G., Hu J., Jiang L., Yu H., Xu J., et al. (2015). Copy number variation at the GL7 locus contributes to grain size diversity in rice. Nat. Genet. 47, 944–948. 10.1038/ng.3346 [DOI] [PubMed] [Google Scholar]
- Weng J., Li B., Liu C., Yang X., Wang H., Hao Z., et al. (2013). A non-synonymous SNP within the isopentenyl transferase 2 locus is associated with kernel weight in Chinese maize inbreds (Zea mays L.). BMC Plant Biol. 13:98. 10.1186/1471-2229-13-98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westoby M., Jurado E., Leishman M. (1992). Comparative evolutionary ecology of seed size. Trends Ecol. Evol. 7, 368–372. 10.1016/0169-5347(92)90006-W [DOI] [PubMed] [Google Scholar]
- Whitt S. R., Wilson L. M., Tenaillon M. I., Gaut B. S., Buckler E. S. (2002). Genetic diversity and selection in the maize starch pathway. Proc. Natl. Acad. Sci. U.S.A. 99, 12959–12962. 10.1073/pnas.202476999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wills D. M., Whipple C. J., Takuno S., Kursel L. E., Shannon L. M., Ross-Ibarra J., et al. (2013). From many, one: genetic control of prolificacy during maize domestication. PLoS Genet. 9:e1003604. 10.1371/journal.pgen.1003604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisser R. J., Murray S. C., Kolkman J. M., Ceballos H., Nelson R. J. (2008). Selection mapping of loci for quantitative disease resistance in a diverse maize population. Genetics 180, 583–599. 10.1534/genetics.108.090118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf Y. I., Koonin E. V. (2012). A tight link between orthologs and bidirectional best hits in bacterial and archaeal genomes. Genome Bio Evol. 4, 1286–1294. 10.1093/gbe/evs100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright S. I., Charlesworth B. (2004). The HKA test revisited. Genetics 168, 1071–1076. 10.1534/genetics.104.026500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C., Trieu A., Radhakrishnan P., Kwok S. F., Harris S., Zhang K., et al. (2008). Brassinosteroids regulate grain filling in rice. Plant Cell 20, 2130–2145. 10.1105/tpc.107.055087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao W., Brown R. C., Lemmon B. E., Harada J. J., Goldberg R. B., Fischer R. L. (2006). Regulation of seed size by hypomethylation of maternal and paternal genomes. Plant Physiol. 142, 1160–1168. 10.1104/pp.106.088849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing Y., Zhang Q. (2010). Genetic and molecular bases of rice yield. Annu. Rev. Plant Biol. 61, 421–442. 10.1146/annurev-arplant-042809-112209 [DOI] [PubMed] [Google Scholar]
- Xu F., Fang J., Ou S., Gao S., Zhang F., Du L., et al. (2015). Variations in CYP78A13 coding region influence grain size and yield in rice. Plant Cell Environ. 38, 800–811. 10.1111/pce.12452 [DOI] [PubMed] [Google Scholar]
- Xu X., Liu X., Ge S., Jensen J. D., Hu F., Li X., et al. (2012). Resequencing 50 accessions of cultivated and wild rice yields markers for identifying agronomically important genes. Nat. Biotechnol. 30, 105–111. 10.1038/nbt.2050 [DOI] [PubMed] [Google Scholar]
- Yang Z., van Oosterom E. J., Jordan D. R., Hammer G. L. (2009). Pre-anthesis ovary development determines genotypic differences in potential kernel weight in sorghum. J Exp Bot. 60, 1399–1408. 10.1093/jxb/erp019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z., van Oosterom E. J., Jordan D. R., Doherty A., Hammer G. L. (2010). Genetic variation in potential kernel size affects kernel growth and yield of sorghum. Crop Sci. 50, 685–695. 10.2135/cropsci2009.06.0294 [DOI] [Google Scholar]
- Yoine M., Nishii T., Nakamura K. (2006). Arabidopsis UPF1 RNA helicase for nonsense-mediated mRNA decay is involved in seed size control and is essential for growth. Plant Cell Physiol. 47, 572–580. 10.1093/pcp/pcj035 [DOI] [PubMed] [Google Scholar]
- Yu F., Li J., Huang Y., Liu L., Li D., Chen L., et al. (2014). FERONIA receptor kinase controls seed size in Arabidopsis thaliana. Mol. Plant 7, 920–922. 10.1093/mp/ssu010 [DOI] [PubMed] [Google Scholar]
- Zhang B., Liu X., Qian Q., Liu L., Dong G., Xiong G., et al. (2011). Golgi nucleotide sugar transporter modulates cell wall biosynthesis and plant growth in rice. Proc. Natl. Acad. Sci. U.S.A. 108, 5110–5115. 10.1073/pnas.1016144108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D., Li J., Compton R. O., Robertson J., Goff V. H., Epps E., et al. (2015). Comparative genetics of seed size traits in divergent cereal lineages represented by sorghum (Panicoidae) and rice (Oryzoidae). G3: Genes Genom. Genet 5, 1117–1128. 10.1534/genetics.115.177170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Wang J., Huang J., Lan H., Wang C., Yin C., et al. (2012). Rare allele of OsPPKL1 associated with grain length causes extra-large grain and a significant yield increase in rice. Proc. Natl. Acad. Sci. U.S.A. 109, 21534–21539. 10.1073/pnas.1219776110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolkevich V., Prusakova L., Lizandr A. (1958). Translocation of assimilates and respiration of conductive tissues in relation to soil moisture. Fiziologiya Rastenii. 5, 337–344. [Google Scholar]
- Zuo J., Li J. (2014). Molecular genetic dissection of quantitative trait loci regulating rice grain size. Annu. Rev. Genet. 48, 99–118. 10.1146/annurev-genet-120213-092138 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.