Abstract
Chinese alligator (Alligator sinensis) is an endangered freshwater crocodilian endemic to China, which experienced a severe bottleneck about 30 years ago. In this study, we developed locus-specific primers to investigate the polymorphism of 3 major histocompatibility complex (MHC) loci in 3 Chinese alligator populations, in combination with 6 neutral microsatellite markers as a contrast. We found the genetic trace for the bottleneck effect on the endangered Chinese alligator: the low allelic diversity (2 alleles at each locus), the low nucleotide substitution rate (no more than 0.009) at all sites, the deviation from Hardy-Weinberg Equilibrium/heterozygote deficiency, and the significant Tajima’s D values, indicating the MHC class I and class II loci being at different stages of bottleneck. We also obtained 3 pieces of evidence for balancing selection on this severely bottlenecked reptile: an obvious excess of nonsynonymous substitutions over synonymous at the antigen-binding positions, the mean synonymous substitution rate of MHC exons significantly higher than mean nucleotide substitution rate of introns, and the differentiation coefficient F ST of MHC loci significantly lower than that of microsatellite loci. Consequently, we emphasize that the Chinese alligator holds a pretty low adaptive ability and requires scientific conservation strategies to ensure the long-term population development.
Introduction
The Chinese alligator (Alligator sinensis) is a very ancient species that had once been pushed to the fringe of extinction by massive hunting, territory deprivation and environment deterioration1. In the 1960s, the wild Chinese alligators become very scarce, and the complicated environments of wetlands and swamps made them even harder to be spotted2. The population of Chinese alligator didn’t stop dropping until 1979 when Changxing Yinjiabian Chinese Alligator Nature Reserve (CYCANR) and Anhui Research Center for Chinese Alligator Reproduction (ARCCAR) were founded to conserve this species, starting with 7 and 212 founder alligators respectively. As of today, there are more than 8,000 captive Chinese alligators at both reserve sites, making the ARCCAR and CYCNAR populations not only the last but also the largest captive populations in China. On the other hand, the number of the Chinese alligators in the wild is dropping 20% every year and the current number is predicted to be lower than 1202, 3.
Judging from the population size alone, the Chinese alligators seem to be recovering quite well from the bottleneck event 30 years ago, and previous studies on the major histocompatibility complex (MHC) genes didn’t raise alarms on any of the repercussions of the bottleneck effect. The MHC genes are not only the most functionally polymorphic genes in vertebrates, but also mediate immune responses, thus making them good adaptive markers to assess the evolutionary potential of fighting pathogens4. Previous studies on the Chinese alligator MHC II genes had shown great polymorphism in exon 2, exon 3 and even intron 2 sequences5–8. After the publication of crocodile, alligator and gharial genomes9–11, the structure and polymorphism of the MHC genes in the crocodilians has been massively annotated and studied12, 13. However, all the above-mentioned studies have been using universal primers to investigate the Crocodylia MHC genes. Considering the evidence of MHC gene duplication12, 14, it’s possible that the cross-locus amplification would cause sequences from different loci being regarded as alleles from a single locus, thus distorting the true intra-locus alignment and elevating the rate of non-synonymous (d N) over synonymous (d S) substitutions. Hence, the true level of genetic diversity at single MHC genes of the Crocodylia is still pending for exploration.
In our previous studies on the Chinese alligator, He et al. constructed a bacterial artificial chromosome (BAC) library that contains 6.8-fold genome equivalents15. Ye et al. constructed several contigs containing MHC genes based on the BAC library16. Wan et al. published the Chinese alligator genome as 2.3 Gb in size, and annotated over 22,200 genes16. These works enable us to design locus-specific primers to characterize MHC genes, hoping to investigate whether bottleneck event continues to impact the Chinese alligator population at the single gene level even after an exponential population growth, to unveil the actual polymorphism of a single MHC gene, and to elaborate the evolutionary forces that influence MHC genes.
Results
BLAST results and Genotyping of MHC genes
By BLASTing the BAC-end and known MHC sequences to the A. sinensis genome and predicting potential MHC genes, we found 3 MHC loci with intact coding sequences and gene structures. The 3 MHC loci can be pinpointed to different A. sinensis genome scaffolds: scaffold1303_1 (NCBI accession number: NW_005842753), scaffold364_1 (NCBI accession number: NW_005842546) and scaffold184_1 (NCBI accession number: NW_005842983). The 3 MHC loci were also verified to be from 3 BAC clones that had no overlap sequences, suggesting that they were independent loci. The BAC clones 1327C2 and 20A2 contained 2 MHC I loci and the BAC clone 1085A9 contained 1 MHC II locus; these MHC loci were accordingly named I1327, I20 and Beta1085, respectively.
The single strand conformation polymorphism (SSCP) results of these loci were all di-morphic: 2 alleles at each locus, which were confirmed by the sequencing results. Nonetheless, the gene I1321 is actually mono-morphic at the functional level due to the same amino acid sequence of the 2 alleles (Fig. 1). No more than two sequences were present in each animal, demonstrating no cross-locus amplification in our study, and none of the sequences showed deletions, insertions, or stop codons, showing the accurate genotyping at single functional MHC genes. In total, we obtained 6 nucleotide acid alleles but 5 amino acid alleles from the 3 MHC loci using the locus-specific genotyping primers (Fig. 1), suggesting an extremely low level of adaptive genetic variation in the endangered Chinese alligator.
Figure 1.

Amino acid sequence alignment of 3 MHC loci. Dots represent identical amino acids to the top variant and crosses under the alignment depict putative antigen binding sites.
Allelic distribution, heterozygosity and Hardy-Weinberg Equilibrium test
The sequence analysis exhibits a quite low level of nucleotide diversity (0.0025—0.0128) in all the MHC loci (Table 1). These results were unlike previous MHC studies in A. sinensis 5, 6, likely because of their use of degenerate primers that are capable of amplifying more than one MHC locus. However, our results coincided with results from Wan et al., which revealed low SNP heterozygosity throughout the A. sinensis genome11.
Table 1.
Genetic diversity at the MHC loci.
| Locus | N A | Nucleotide length (bp) | Hd | π | ||||
|---|---|---|---|---|---|---|---|---|
| Exon 2 | Intron 2 | Exon 3 | Exon | Intron | Exon | Intron | ||
| I1327 | 2 | 261 | 15135 | 276 | 0.444 ± 0.064 | 0.467 ± 0.017 | 0.25% | 0.20% |
| I20 | 2 | 267 | 914; 925 | 276 | 0.457 ± 0.055 | 0.356 ± 0.025 | 0.34% | 0.20% |
| Beta1085 | 2 | 282 | 631; 632 | — | 1.000 ± 0.052 | 0.182 ± 0.021 | 0.94% | 0.06% |
Note: N A, the number of alleles; Hd, haplotype diversity; π, nucleotide diversity.
Although varying with loci, the allelic distribution patterns of the same MHC gene are similar among populations (Table 2). The Hardy-Weinberg Equilibrium (HWE) test results show a severe heterozygote deficiency at the Beta1085 locus in ZJ and AH populations. Differently, the Tajima’s D test results are strange (Table 2): below 0 value at MHC class II locus (significant in ZJ population), and above 0 value at both MHC class I loci (significant in ZJ and AH population). The different Tajima’s D values between the class I and II MHC loci might indicate different stages of bottleneck or different selection pressures.
Table 2.
Genetic diversity of MHC in 3 Chinese alligator populations.
| Population | Locus | Allele 01 | Allele 02 | H O | H E | P-value of HWE | Tajima’s D test |
|---|---|---|---|---|---|---|---|
| ZJ | I1327 | 0.73 | 0.27 | 0.469 | 0.396 | 0.396 | 1.711 |
| AH | I1327 | 0.69 | 0.31 | 0.375 | 0.437 | 0.441 | 2.083* |
| USA | I1327 | 0.70 | 0.30 | 0.200 | 0.442 | 0.132 | 1.518 |
| ZJ | I20 | 0.36 | 0.64 | 0.531 | 0.468 | 0.699 | 2.611* |
| AH | I20 | 0.22 | 0.78 | 0.375 | 0.347 | 1.000 | 1.383 |
| USA | I20 | 0.10 | 0.90 | 0.200 | 0.189 | 1.000 | 0.949 |
| ZJ | Beta1085 | 0.98 | 0.02 | 0.031 | 0.092 | 0.015* | −1.953* |
| AH | Beta1085 | 0.91 | 0.09 | 0.125 | 0.231 | 0.008* | −1.524 |
| USA | Beta1085 | 1.00 | 0.00 | — | — | — | — |
Note: the predominant alleles are in bold; * indicates P < 0.05; H O and H E are observed and expected heterozygosities, respectively.
The repercussions of bottleneck effects can also be observed at the microsatellite loci (Table 3), where allelic diversity is very low (2.5 alleles per locus) in all 3 populations. Despite the higher H O than H E at several loci, this phenomenon should be attributed to the selection of microsatellite markers with high-resolution power in the study of Ma17.
Table 3.
Genetic diversity of microsatellite markers in 3 Chinese alligator populations.
| Locus | ZJ (n = 32) | AH (n = 32) | USA (n = 10) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| N A | H O | H E | N A | H O | H E | N A | H O | H E | |
| CXA-142 | 2 | 0.844* | 0.503 | 2 | 0.536 | 0.399 | 2 | 0.800 | 0.505 |
| CXA-41 | 2 | 0.688* | 0.476 | 2 | 0.393 | 0.431 | 2 | 0.111 | 0.111 |
| CXA-6 | 2 | 0.500 | 0.411 | 2 | 0.615 | 0.507 | 2 | 0.700 | 0.479 |
| CXA-9 | 3 | 0.250* | 0.405 | 3 | 0.654 | 0.578 | 2 | 1.000* | 0.526 |
| CXA-34 | 3 | 0.500* | 0.608 | 3 | 0.786* | 0.590 | 3 | 0.900* | 0.679 |
| CXA-43 | 3 | 0.531* | 0.587 | 4 | 0.536 | 0.543 | 2 | 0.700 | 0.521 |
| Mean | 2.5 | 0.552 | 0.498 | 2.667 | 0.587 | 0.508 | 2.167 | 0.702 | 0.470 |
| SD | 0.548 | 0.200 | 0.086 | 0.816 | 0.132 | 0.078 | 0.408 | 0.312 | 0.190 |
Note: N A, number of alleles; H O, observed heterozygosity; H E, expected heterozytosity; * shows significant deviation from HWE (P < 0.05).
Calculation ofdNanddSsubstitutions
The d N/d S ratio can provide useful information on the degree of selective pressure acting on a protein-coding gene18. The d N/d S > 1 implies positive selection; d N/d S < 1 implies purifying (negative) selection; and d N/d S = 1 indicates neutral (i.e. no) selection. As we can see in Table 4, the Chinese alligator MHC exons exhibit extremely low d N and d S values due to the low allelic and nucleotide diversity, and the Z-tests show no significant P value at any locus. The following trends, however, can be observed: at the MHC class I loci, the d N/d S ratios all exceed 1 at antigen binding sites (ABS), showing the sign of positive selection. Nonetheless, at the MHC class II locus, there is no nucleotide substitution at ABS positions (Fig. 1), thus indicating purifying selection. When the d N/d S values were compared between the ABS and non-ABS sites, the MHC class I loci produced a larger ratio at ABSs than at non-ABSs while the MHC class II locus showed d S at non-ABSs in spite of zero d N changes (Table 4), indicating balancing selection at play in the Chinese alligator.
Table 4.
The d N/d S ratio of 3 MHC loci in 3 Chinese alligator populations.
| Locus | Position | N | ZJ | AH | USA | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| d N | d S | d N/d S | d N | d S | d N/d S | d N | d S | d N/d S | |||
| I1327 | ABS | 56 | 0.008 ± 0.005 | 0.005 ± 0.006 | 1.60 | 0.008 ± 0.006 | 0.006 ± 0.006 | 1.33 | 0.009 ± 0.006 | 0.006 ± 0.006 | 1.50 |
| non-ABS | 125 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| All | 181 | 0.002 ± 0.002 | 0.002 ± 0.002 | 1.00 | 0.003 ± 0.002 | 0.002 ± 0.002 | 1.50 | 0.003 ± 0.002 | 0.002 ± 0.002 | 1.50 | |
| ABS | 56 | 0.007 ± 0.005 | 0 | ∞ | 0.005 ± 0.004 | 0 | ∞ | 0.003 ± 0.002 | 0 | ∞ | |
| I20 | non-ABS | 125 | 0.002 ± 0.002 | 0.005 ± 0.006 | 0.40 | 0.001 ± 0.001 | 0.004 ± 0.004 | 0.25 | 0.001 ± 0.001 | 0.002 ± 0.002 | 0.50 |
| All | 181 | 0.003 ± 0.002 | 0.004 ± 0.004 | 0.75 | 0.003 ± 0.001 | 0.003 ± 0.003 | 1.00 | 0.001 ± 0.001 | 0.002 ± 0.001 | 0.50 | |
| ABS | 49 | 0 | 0 | 0 | 0 | 0 | 0 | — | — | — | |
| Beta1085 | non-ABS | 139 | 0 | 0.001 ± 0.001 | 0.00 | 0.002 ± 0.002 | 0.005 ± 0.004 | 0.40 | — | — | — |
| All | 188 | 0 | 0.001 ± 0.001 | 0.00 | 0.001 ± 0.001 | 0.004 ± 0.003 | 0.25 | — | — | — | |
Note: the d N/d S ratios are calculated separately for antigen-binding site (ABS), non-ABS and all sites. Codon numbers (N) of the sites are shown. In the USA population, Beta1085 locus has only one allele, thus no d N and d S values are calculated.
Comparison of nucleotide substitution between exon and intron
All 3 Chinese alligator populations shared the same allele sequences in all 3 MHC loci, and the only differences were the allelic frequencies at each locus. Thus, we computed nucleotide substitution on the basis of these unique allele sequences and without consideration of allelic frequency and population identity. We calculated the mean d S in exons and the mean number of nucleotide substitutions per site (d) in introns of the MHC class I and II genes, and plotted them against the nucleotide position (Fig. 2). The results show a much higher substitution rate in the exon 2 and 3 region than in the intron 2 at all MHC loci, which means the introns are younger than exons, suggesting the exons being constantly maintained by balancing selection for ages.
Figure 2.
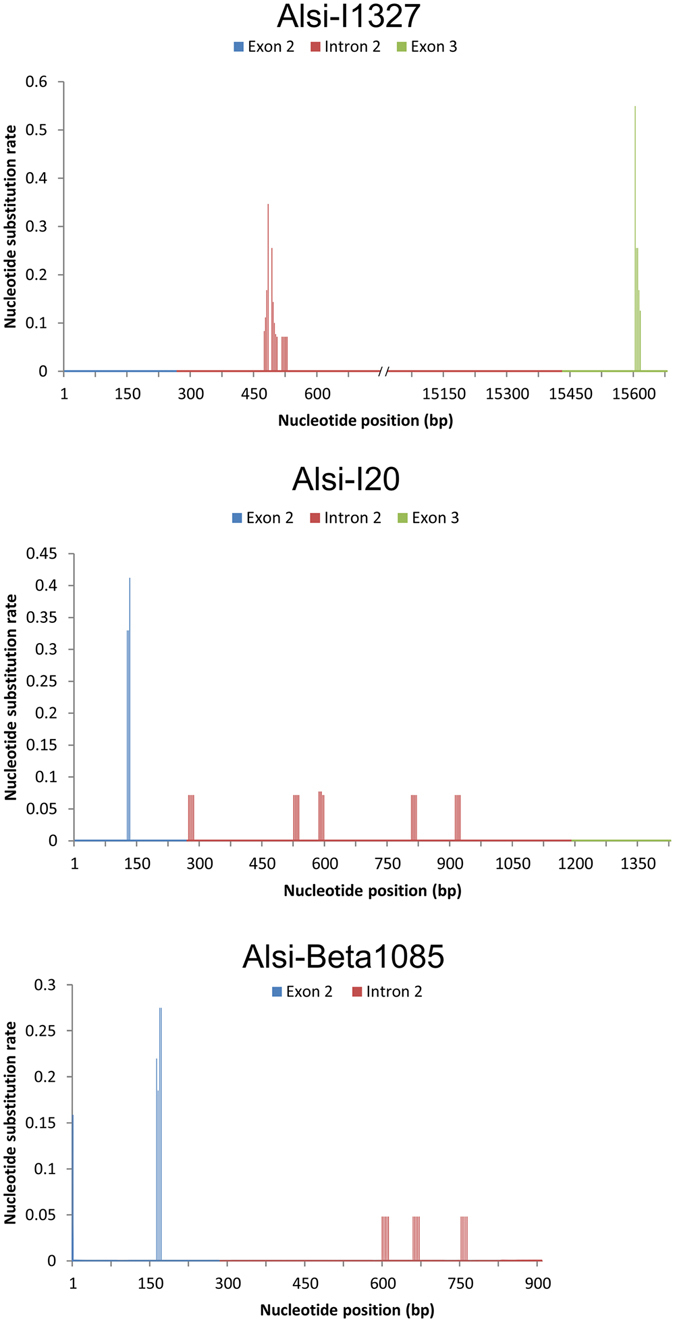
A plot of d S in exon 2 (and exon 3 at class I genes) versus d in intron2.
Comparison of differentiation degree between the MHC and microsatellite loci
Since the MHC genes are constantly maintained by balancing selection, even in different populations the MHC allelic frequencies will be pretty similar due to the influence of balancing selection. The microsatellite loci, however, will take more diverted evolutionary paths under the pressure of genetic drift. We tested pairwise F ST of the 3 Chinese alligator populations at both MHC and microsatellite loci, and found significant population differentiation among 3 populations at the microsatellite loci (P < 0.05) with no differentiation at the MHC loci (Table 5). This contrast indicates that even though severely damaged by the bottleneck effect, the Chinese alligator MHC loci are still influenced by balancing selection, i.e. MHC genes maintain more similar alleles than neutral markers do.
Table 5.
Pairwise F ST test among 3 Chinese alligator populations.
| ZJ | AH | USA | |
|---|---|---|---|
| ZJ | — | 0.01449 | 0.04720 |
| AH | 0.08424* | — | 0.00670 |
| USA | 0.08990* | 0.06212* | — |
Note: the numbers on the left side below the diagonal and on the right side above the diagonal are the F st values of microsatellite and MHC loci, respectively. * indicates P < 0.05.
Discussion
Normally, when a population encounters bottleneck events, rare alleles are more likely to be lost than the common alleles, and positive Tajima’s D values are expected; when population expands, the segregating sites accumulate at the rare frequencies, thus leading to negative Tajima’s D values19. In the Chinese alligator’s case, the bottleneck effect 30 years ago was too severe for the population to attain ample rare alleles with low frequencies, as evidently shown by the di-morphism MHC loci in this study. Therefore, the positive Tajima’s D value at the MHC class I loci should be a normal case for the Chinese alligator population. The negative Tajima’s D value at the MHC class II locus, however, could indicate the ongoing population expansion or purifying selection, whose trace can also be found in the sequence alignments (Fig. 1) and d N, d S results (Table 4); the MHC class I loci have non-synonymous nucleotide substitutions at and next to the ABS sites whereas the MHC class II locus has no nucleotide substitution at all at any ABS sites.
As our abovementioned results have shown, even after 30 years, the bottleneck effect still influences the MHC genes of the Chinese alligator. That’s why conventional methods – such as d N/d S values – function poorly in detecting balancing selection. The significant d N/d S values in previous studies5, 7 are more likely caused by their usage of universal primers, as Miller and Lambert pointed out, the usage of universal primers could produce a lower than expected d S value, leading an elevated d N/d S ratio and thus giving “anomalous” results20.
Therefore, we need to compare the theoretically balancing-selection-influenced sites to those non-balancing-selection-influenced sites to reveal the presence of balancing selection. The comparison between exons and introns would be an effective tool. Hughes and Yeager21, 22 discovered that in most genes, the mean number of nucleotide substitutions per site (d) in introns and the mean d S in exons are usually about equal – for their mutations are both selectively neutral to a gene – except when it comes to the MHC genes, the mean d S in exons is always much higher than the mean d in introns. They explained that exons in the MHC genes are very ancient because they have been maintained by balancing selection for a very long time, while the mutations in introns are controlled by recombination and genetic drift. When an ancient intron polymorphism is lost due to recombination or drift, it will be selectively neutral to the MHC gene. As time goes by, introns in the MHC genes will become evolutionarily younger on average than are the exons constantly maintained by balancing selection. The much older exons will consequently possess a much higher d S than that of the younger introns, hence mean d S > mean d in MHC genes22. Our findings in this study support Hughes and Yeager’s theory.
The environmental factors could be another cause of the similar MHC genes among populations. Both AH and ZJ populations are intensively managed as captive populations, and their habitats are modified to natural wetlands surrounded by farming countryside. The isolated habitats may provide an environment with low level exposure to pathogens, and cause the MHC genes lack motivations to change; similar situations happen to many other species20, 23–25. Even if the Chinese alligator populations are sustainable at present, they would still be susceptible to new pathogens when reintroduced to the wild in the future. While the balancing selection is working on the MHC genes, proper conservation strategy should be devised to protect this endangered species.
Materials and Methods
Animal experiment ethics statement
All experiments were carried out in accordance with the guidelines issued by the Ethical Committee of Laboratory Animal of Zhejiang University, and all experimental protocols were approved by the Ethical Committee of Laboratory Animal of Zhejiang University.
Sampling
The A. sinensis samples (see Supplementary Table S1) came from 3 captive populations, kindly provided by CYCANR (the Zhejiang population, acronym ZJ), ARCCAR (the Anhui population, acronym AH) and the Rockefeller Wildlife Refuge (the USA population, acronym USA). Although there are more than 8,000 Chinese alligators in the ZJ and AH populations, most of them are still juvenile while several hundreds of Chinese alligators are breeding adults. In this study, all sample donors were adult male/female Chinese alligators that were randomly captured, and we believe they can represent the species’ natural range. The blood samples were taken during routine medical examinations. DNA was extracted using a traditional phenol-chloroform method26.
Genome BLAST and primer design
We screened the BAC clones using the universal MHC primers of Ye et al.16 and pinpointed each MHC gene to their corresponding genome scaffolds (requiring >95% in both gene coverage and sequence similarity) by BLASTing the end sequences of the target BACs and the known MHC sequence against the A. sinensis genome11 using default parameters of a local BLAST tool downloaded from NCBI (ftp://ftp.ncbi.nlm.nih.gov/blast/executables/blast+/LATEST/). Then, we used Augustus (http://augustus.gobics.de/) to predict exon positions for each MHC sequence, and manually translated each exon to amino acid sequences in order to search for MHC loci with intact coding sequences and gene structures. All the MHC genes from the BAC and the genome were manually checked for gene structural integrity to rule out pseudogenes. The MHC genes with a complete gene structure were selected for subsequent studies. We gave prefix ‘Alsi’ to all the MHC genes.
We then designed locus-specific primers to amplify the antigen binding domains of the MHC loci (exons 2 and 3 for the MHC class I, and exon 2 for the MHC class II). Intron 2 sequences were also amplified and sequenced to ensure the correctness of genotyping as well as to be a useful comparison material (see Supplementary Table S2). Primers were designed using Primer Premier 5 (http://www.premierbiosoft.com/), and the primers’ binding positions are illustrated in Supplementary Fig. S1. We performed pre-experiments to investigate the polymorphism of each locus using 10 randomly selected blood samples in each population. We chose the polymorphic loci to genotype all the samples in 3 populations. Six microsatellite markers from our former works were also used as a neutral contrast17.
PCR amplification and genotyping
The PCR amplifications were performed in a 10 μl reaction system containing 1 L of template DNA (30–50 ng/µL), 0.5 U rTaq DNA polymerase (TaKaRa), 1 µL of 10 × ExTaq buffer (TaKaRa), 1 µL of 2.5 mmol/L dNTPs, 0.2 µL of each primer (diluted to 10 mol/µL), and 6.5 µL of double-distilled water. The PCR was programmed as follows: initial denaturation at 95 °C for 5 min, followed by 35 cycles of 95 °C for 30 s, 63 °C for 30 s, and extension at 72 °C for 30 s, with a final extension of 72 °C for 5 min.
We adopted Single-Strand Conformation Polymorphism - heteroduplex (SSCP-HD) techniques27 to genotype each individual and screen 3 Chinese alligator populations in order to examine the level of adaptive genetic variation in this endangered reptile. In the SSCP-HD analysis, we added 5 μl of 2X loading buffer (95% formamide, 10 mm NaOH, 0.25% bromophenol blue, 0.25% xylene cyanol) for each 10 μl of PCR product. And the mixture was denatured at 95 °C for 5 min, and swiftly transferred onto ice for cooling down. Then, the PCR products were run in an acrylamide gel consisting of 12% 37.5:1 acrylamide/bisacrylamide with 2.5% crosslinking, and sequences were separated in 0.5 × TBE running buffer at 16 °C by 150 V for 6.5 h on the Decode System (Bio-Rad). Finally, the SSCP gel was fixed in 10% acetic acid for 30 min, washed with dH2O, and achieved silver staining pictures. We repeated the PCR-SSCP process for 3 times at each locus to make sure the banding pattern was stable and consistent, and obtained the allele sequences by sequencing the homozygous individuals with unique banding patterns in BGI, Shanghai. Microsatellite loci were amplified and genotyped as described by Ma17.
Data analyses
We used Lasergene 7 (DNASTAR) for nucleotide sequence editing and Mega 528 for sequence alignment. We used DnaSP29 to calculate the haplotype diversity (Hd), the nucleotide diversity (π), and Tajima’s D values. The non-synonymous (d N) and synonymous (d S) substitution rates of the MHC exons were calculated using Mega 5, and we used K-estimator30 with Kimura-2p method to compute nucleotide substitution rate of intron 2 (d) as well as the d S of exon 2 and exon 3, which were then plotted against nucleotide position using a sliding window size of 15 base pairs and steps of 3 base pairs. We used Kaufman’s study31 as a reference to annotate antigen-binding sites.
Allelic frequency, observed heterozygosity (H O), expected heterozygosity (H E) and HWE, as well as pairwise F ST among 3 populations were calculated in Arlequin 3.532.
Data availability statement
The genomic sequences of MHC genes are collected from the scaffolds 1303_1 (GenBank accession number: NW_005842753), 364_1 (NW_005842546) and 184_1 (NW_005842983) of the Chinese alligator genome (GCA_000455745.1). The new nucleotide acid sequences obtained in this study are available in the supplementary Data S1.
Electronic supplementary material
Acknowledgements
Special thanks go to the CYCANR and the ARCCAR for providing samples. This work was supported by a grant from the National Natural Science Foundation of China (No. 31470439), a special grant from the State Forestry Administration, and the Fundamental Research Funds for the Central Universities of the P. R. China.
Author Contributions
T.Z. and H.Q.Y. conducted the experiments, interpreted data and wrote the manuscript. R.C.Z., L.M.F. and G.H.Z. collected samples and participated in experiments. S.G.F. conceived the project, designed the research, interpreted data and wrote the manuscript. All authors reviewed and approved the manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Teng Zhai and Hai-Qiong Yang contributed equally to this work.
Electronic supplementary material
Supplementary information accompanies this paper at doi:10.1038/s41598-017-05640-2
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Thorbjarnarson, J. & Wang, X. M. The Chinese Alligator: Ecology, Behavior, Conservation and Culuture. (The Johns Hopkins University Press, 2010).
- 2.Wang ZH, Yao H, Ding YZ, John T, Wang XM. Testing reintroduction as a conservation strategy for the critically endangered Chinese alligator: movements and home range of released captive individuals. Chin Sci Bull. 2011;56:2586–2593. doi: 10.1007/s11434-011-4615-8. [DOI] [Google Scholar]
- 3.Zhao L, et al. The sex ration of wild Chinese alligators Alligator sinensis. Current Zoology. 2013;59:725–731. doi: 10.1093/czoolo/59.6.725. [DOI] [Google Scholar]
- 4.Hedrick PW. Pathogen resistance and genetic variation at MHC loci. Evolution. 2002;56:1902–1908. doi: 10.1111/j.0014-3820.2002.tb00116.x. [DOI] [PubMed] [Google Scholar]
- 5.Shi Y, Wu XB, Yan P, Chen BH. Cloning and sequences analysis of the second exon of MHC class IIB genes in Chinese alligator (Alligator sinensis) Zool Res. 2004;25:415–421. [Google Scholar]
- 6.Liu H, Wu XB, Yan P, Jiang ZG. Polymorphism of exon 3 of MHC class IIB gene in Chinese alligator (Alligator sinensis) J Genet Genomics. 2007;34:918–929. doi: 10.1016/S1673-8527(07)60103-9. [DOI] [PubMed] [Google Scholar]
- 7.Nie CP, Zhao J, Li YY, Wu XB. Diversity and selection of MHC class IIb gene exon3 in Chinese alligator. Mol Biol Rep. 2013;40:295–301. doi: 10.1007/s11033-012-2061-6. [DOI] [PubMed] [Google Scholar]
- 8.Nie CP, Li YY, Zhao Z, Wu XB. Extremely high MHC class IIb gene intron 2 variation and population structure in Chinese alligator. J Genet. 2012;91:86–91. [PubMed] [Google Scholar]
- 9.Green RE, et al. Three crocodilian genomes reveal ancestral patterns of evolution among archosaurs. Science. 2014;346:1254449. doi: 10.1126/science.1254449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.St John JA, et al. Sequencing three crocodilian genomes to illuminate the evolution of archosaurs and amniotes. Genome Biol. 2012;13:415. doi: 10.1186/gb-2012-13-1-415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wan QH, et al. Genome analysis and signature discovery for diving and sensory properties of the endangered Chinese alligator. Cell Res. 2013;23:1091–105. doi: 10.1038/cr.2013.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jaratlerdsiri W, Isberg SR, Higgins DP, Gongora J. MHC class I of saltwater crocodiles (Crocodylus porosus):polymorphism and balancing selection. Immunogenetics. 2012;64:825–38. doi: 10.1007/s00251-012-0637-x. [DOI] [PubMed] [Google Scholar]
- 13.Jaratlerdsiri W, Isberg SR, Higgins DP, Miles LG, Gongora J. Selection and trans-species polymorphism of major histocompatibility complex class II genes in the order Crocodylia. PLoS One. 2014;9:e87534. doi: 10.1371/journal.pone.0087534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jaratlerdsiri W, et al. Comparative genome analyses reveal distinct structure in the saltwater crocodile MHC. PLoS ONE. 2014;9:e114631. doi: 10.1371/journal.pone.0114631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.He K, et al. A bacterial artificial chromosome library for the Chinese alligator (Alligator sinensis) Gene. 2012;507:74–78. doi: 10.1016/j.gene.2012.06.035. [DOI] [PubMed] [Google Scholar]
- 16.Ye, Q. Construction of genomic BAC library of Chinese alligator and contigs of MHC class II and OR gene. (Zhejiang University, 2011).
- 17.Ma, M. Y. The development of high resolving microsatellite markers in Chinese alligator. (Zhejiang University, 2015).
- 18.Hughes AL, Nei M. Nucleotide substitution at major histocompatibility complex class II loci: evidence for overdominant selection. Proc Natl Acad Sci USA. 1989;86:958–962. doi: 10.1073/pnas.86.3.958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123(3):585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miller HC, Lambert DM. Genetic drift outweighs balancing selection in shaping post-bottleneck major histocompatibility complex variation in New Zealand robins (Petroicidae) Mol Ecol. 2004;13:3709–3721. doi: 10.1111/j.1365-294X.2004.02368.x. [DOI] [PubMed] [Google Scholar]
- 21.Hughes AL, Yeager M. Comparative evolutionary rates of introns and exons in murine rodents. J Mol Evol. 1997;45:125–30. doi: 10.1007/PL00006211. [DOI] [PubMed] [Google Scholar]
- 22.Hughes AL, Yeager M. Natural selection at major histocompatibility complex loci of vertebrates. Annu Rev Genet. 1998;32:415–435. doi: 10.1146/annurev.genet.32.1.415. [DOI] [PubMed] [Google Scholar]
- 23.Slade RW. Limited MHC polymorphism in the southern elephant seal: implications for MHC evolution and marine mammal population biology. Proc R Soc London B. 1992;249:163–171. doi: 10.1098/rspb.1992.0099. [DOI] [PubMed] [Google Scholar]
- 24.Ellegren H, Hartman G, Johansson M, Andersson L. Major histocompatibility complex monomorphism and low levels of DNA fingerprinting variability in a reintroduced and rapidly expanding population of beavers. Proc Natl Acad Sci USA. 1993;90:8150–8153. doi: 10.1073/pnas.90.17.8150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hambuch TM, Lacey EA. Enhanced selection for MHC diversity in social tuco-tucos. Evolution. 2002;56:841–845. doi: 10.1111/j.0014-3820.2002.tb01395.x. [DOI] [PubMed] [Google Scholar]
- 26.Sambrook, J., Fritsch, E. & Maniatis, T. Molecular cloning: a laboratory manual 2nd. (Cold Spring Harbor Laboratory Press, 1989).
- 27.Wan QH, et al. A novel HURRAH protocol reveals high numbers of monomorphic MHC class II loci and two asymmetric multi-locus haplotypes in the Père David’s Deer. PLoS ONE. 2011;6:e14518. doi: 10.1371/journal.pone.0014518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tamura K, et al. MEGA5: Molecular evolutionary genetics analysis using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony Methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Librado P, Rozas J. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25:1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- 30.Comeron JM. Bioinformatics. 1999;15:763–764. doi: 10.1093/bioinformatics/15.9.763. [DOI] [PubMed] [Google Scholar]
- 31.Kaufman J, Salomonsen J, Flajnik MF. Evolutionary conservation of MHC class I and class II molecules—different yet the same. Semin Immunol. 1994;6:411–424. doi: 10.1006/smim.1994.1050. [DOI] [PubMed] [Google Scholar]
- 32.Excoffier L, Lischer HEL. Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Res. 2010;10:564–567. doi: 10.1111/j.1755-0998.2010.02847.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The genomic sequences of MHC genes are collected from the scaffolds 1303_1 (GenBank accession number: NW_005842753), 364_1 (NW_005842546) and 184_1 (NW_005842983) of the Chinese alligator genome (GCA_000455745.1). The new nucleotide acid sequences obtained in this study are available in the supplementary Data S1.


