Abstract
Keratins 1 (K1) and 10 (K10) are the primary keratins expressed in differentiated epidermis. Mutations in K1/K10 are associated with human skin diseases. We determined the crystal structure of the complex between the distal (2B) helices of K1 and K10 to better understand how human keratin structure correlates with function. The 3.3Å resolution structure confirms many features inferred by previous biochemical analyses but adds new, unexpected insights. It demonstrates a parallel, coiled-coil heterodimer with a predominantly hydrophobic intermolecular interface; this heterodimer formed a higher order complex with a second K1-K10-2B heterodimer via a Cys401K10 disulfide link, although the bond angle is unanticipated. Molecular surface analysis of K1-K10-2B identified several pockets, one adjacent to the disulfide linkage and conserved in K5-K14. The solvent accessible surface area of the K1-K10 structure is 20-25% hydrophobic. The 2B region contains mixed acidic and basic patches proximally (N-terminal) whereas it is largely acidic distally (C-terminal). Mapping of conserved and non-conserved residues between K1-K10 and K5-K14 onto the structure demonstrated the majority of unique residues align along the outer helical ridge. Finally, the structure permitted a fresh analysis of the deleterious effects caused by K1/K10 missense mutations found in patients with phenotypic skin disease.
Keywords: keratin, skin disease, X-ray crystal structure, intermediate filament, protein fibril
INTRODUCTION
Keratins are the most abundant proteins in differentiated keratinocytes. They belong to the larger family of intermediate filament (IF) proteins and form dynamic 10-nm filaments that course from desmosomes to the nuclear membrane. Their function in skin is multifactorial, including cytoskeletal support, intracellular signaling, and scaffolding for protein interactions (Loschke et al., 2015). Important unanswered questions in keratin biology are: why are there so many different keratin proteins and how do KIFs share a common coiled-coil structure yet perform diverse cellular functions? These questions are vital because keratin pairs (54 different functional keratin genes) are expressed in all epithelial tissues where they have varying functions, as evidenced by >25 clinical disorders attributed to keratin mutations. Most disease-causing mutations occur in the well-conserved rod (coiled-coil) domains of keratins and mutations in either K1 or K10 cause the human skin diseases epidermolytic ichthyosis, palmar-plantar keratoderma, and ichthyosis with confetti (Toivola et al., 2015).
KIFs share a basic structural organization, with four helical or coiled-coil segments (denoted 1A, 1B, 2A, 2B) flanked by N- and C-terminal glycine-rich sequences (Fig. 1a). Experimentally determined crystal structures add critical details to biochemical function, validate hypotheses generated from molecular modeling, and reveal unexpected structural information. However, difficulties in crystallizing keratins have limited progress. To overcome this difficulty, a “divide-and-conquer” approach has been utilized to break down other intermediate filaments into manageable, crystallizable segments (Chernyatina et al., 2015, Strelkov et al., 2001). To date, the 2B region of the keratin 5-keratin 14 heterodimer is the only region of a keratin molecule for which a crystal structure has been determined (Lee et al., 2012).
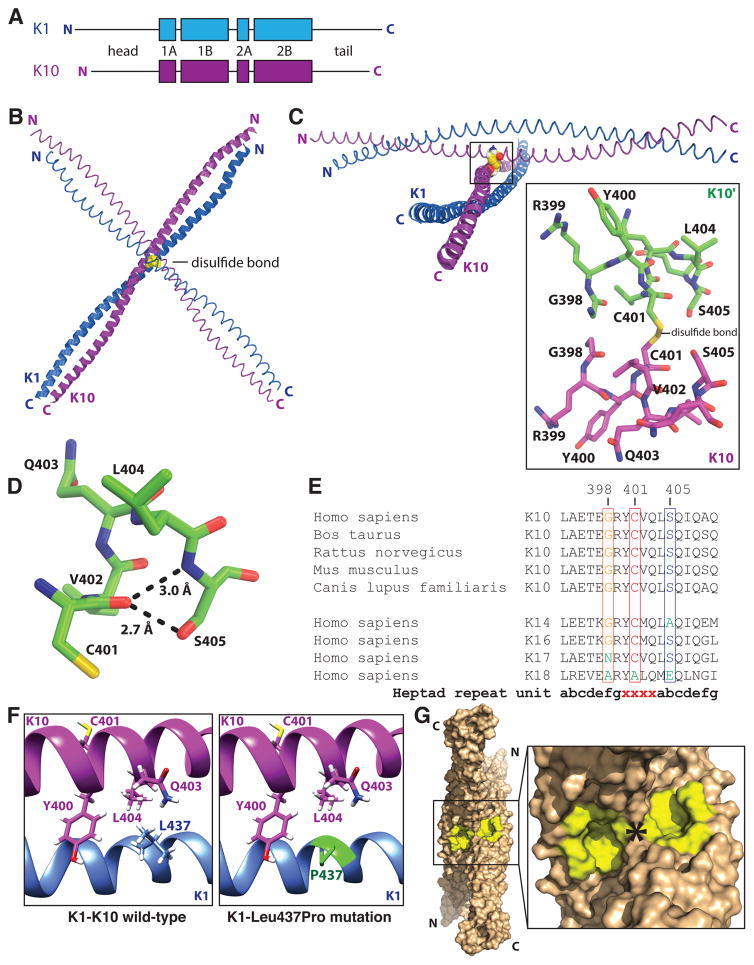
Fig. 1. Disulfide bonding in the keratin 1-keratin 10-2B crystal structure.
(a) K1 and K10 domain organization, which is conserved among KIFs, features four helical segments (1A, 1B, 2A, 2B) flanked by variable head and tail regions. (b) The K1-K10-2B heterodimer formed a trans-dimer, homotypic disulfide bond via Cys401K10. The disulfide bond occurred between K10 molecules in different, adjacent asymmetric units of the crystal lattice. No other cysteine in K1-K10-2B was observed to form disulfide bonds. (c) Rotated global view, compared to panel A, of the disulfide bonded K1-K10 complex; a zoomed in view (boxed) of the residues surrounding the disulfide bond demonstrate Gly398K10 and Ser405K10 are structurally adjacent to Cys401K10. (d) The carbonyl oxygen of Cys401K10 is 2.7 Å from Ser405K10 hydroxyl oxygen and 3.0 Å from Ser405K10 amide nitrogen. (e) Multiple sequence alignment of the K10-2B stutter region (denoted ‘xxxx’) from various species and several type I keratins from Homo sapiens, demonstrating sequence differences in key positions related to disulfide bond formation. (f) Structural consequences of Leu437ProK1 mutation include loss of hydrophobic interactions with Tyr400K10, Gln403K10, and Leu404K10 and loss of hydrogen bonds with Gln403K10 adjacent to Cys401K10; proline may also cause distal helix kinking. (g) Molecular surface representation of the disulfide-linked complex between two K1-K10-2B heterodimers highlighting symmetric concavities (yellow) adjacent to the disulfide bond (*). In vivo, K10 molecules located on the outside of the 10-nm filament will have Cys401K10 exposed and able to form disulfide linkages; it is plausible disulfide-linked K1-K10 influences nucleus behavior and shape in differentiated keratinocytes based on K5-K14 studies (Feng and Coulombe, 2015, Lee et al., 2012).
In order to begin building a structure for the keratin 1-keratin 10 complex and compare structures of keratin pairs to each other, we report here the x-ray crystal structure of the complex between the distal (2B) helical segments of keratin 1 and keratin 10. While the structure confirms many features inferred by previous biochemical analyses, it adds new, unexpected insights: most importantly, K1-K10-2B is not identical to K5-K14-2B (despite a common coiled-coil fold) as it demonstrates significant differences in surface shape and chemistry. As predicted (Steinert and Parry, 1993), parallel, in-register, coiled-coil K1-K10 heterodimers form a trans-dimer disulfide bond via Cys401K10, although the angle of the bond is unanticipated. Finally, the crystal structure of the K1-K10-2B region permitted a fresh computational modeling analysis of the deleterious effects caused by K1 and K10 missense mutations found in patients with phenotypic skin disease.
RESULTS
Overall Keratin Structure
We determined the X-ray crystal structure of the human keratin 1-keratin 10 2B complex; it is ~3.5 Å resolution based on I/s cutoff of 2, with weaker reflections to 3.3 Å used for refinement (Supplementary Table S1 and Fig. S1) (Bunick, 2015). The structure demonstrates individual keratin 1 (K1) and keratin 10 (K10) α-helices intertwined in a parallel coiled-coil over the length of the 2B sequence, confirming early predictions (Crick, 1952). The ordered structure spans K1 residues 386–489 and K10 residues 347–456; however, the quality of electron density decreased between K10-2B N-terminus residues 347–356 due to motion of the helical end (Supplementary Fig. S2). This motion is a result of the high solvent content of the crystal (~79%), and stems from a disulfide linkage of heterodimers at K10 residue Cys401which led to a looser protein packing arrangement in the crystal.
Molecular Surface Analysis
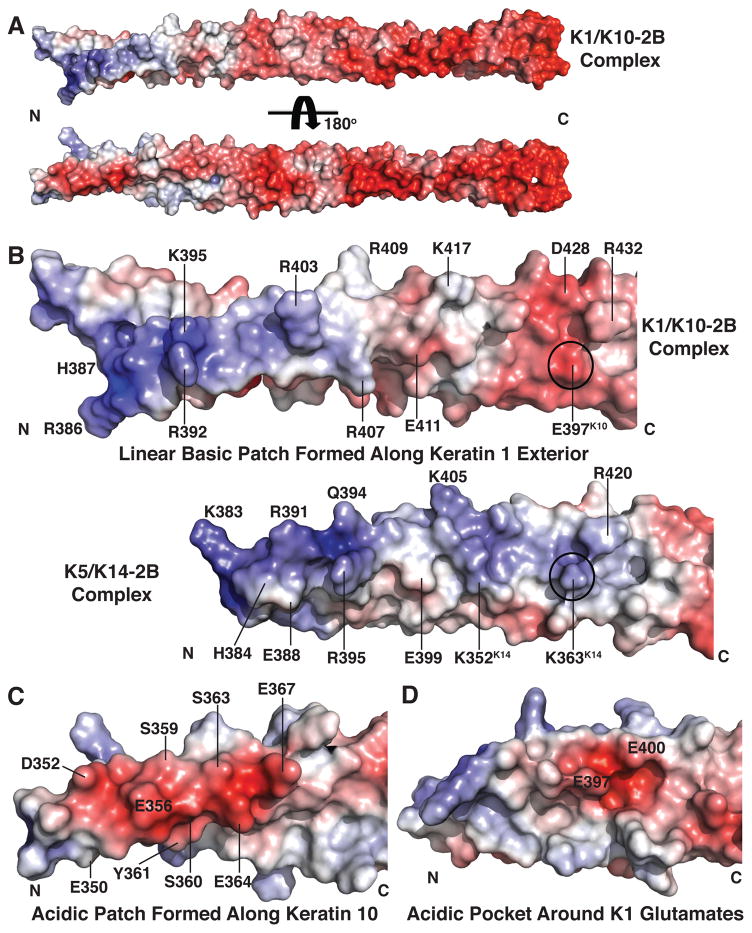
To evaluate the surface chemistry and shape of the K1-K10-2B complex, a detailed analysis on its molecular surface characteristics was performed. The electrostatic surface potential of K1-K10-2B complex demonstrates a polarization of charge: the distal two-thirds of K1-K10-2B is predominantly acidic, whereas the proximal one-third of the molecule contains a single linear basic patch along the K1 protein in addition to two distinct acidic patches (Fig. 2). The N-terminal basic patch is composed of nine K1 residues (Fig. 2b, R386-K417; Supplementary Table S2) that create a linear ridge of surface-exposed positive charge. One large acidic patch is formed at the N-terminal aspect of the 2B region by five K10 residues (Glu350, Asp352, Glu356, Glu364, Glu367) (Fig. 2a, c). This acidic patch contrasts with the surrounding positive charge. A second small acidic patch exists along the sidewall of a pocket created by Glu397K1 and Glu400K1 (Fig. 2d).
Fig. 2. Electrostatic surface properties of K1-K10-2B.
(a) K1-K10-2B heterodimer depicted in two orientations, related by 180°, demonstrating the 2B helix has polarization of charge: a mixed acidic and basic character at the N-terminus and predominantly acidic character at the C-terminus. (b) The N-terminal surface of K1-K10-2B is more acidic than the basal keratin complex K5-K14-2B because it contains a glutamate (Glu397K10) rather than lysine (Lys363K14) at homologous positions (circle). The K1-K10 and K5-K14 molecules are structurally aligned in the panel; there are 27 additional N-terminal residues observed in the K1-K10 structure. (c) Acidic patch formed along the N-terminal surface of K10-2B. (d) Acidic pocket formed between Glu397K1 and Glu400K1.
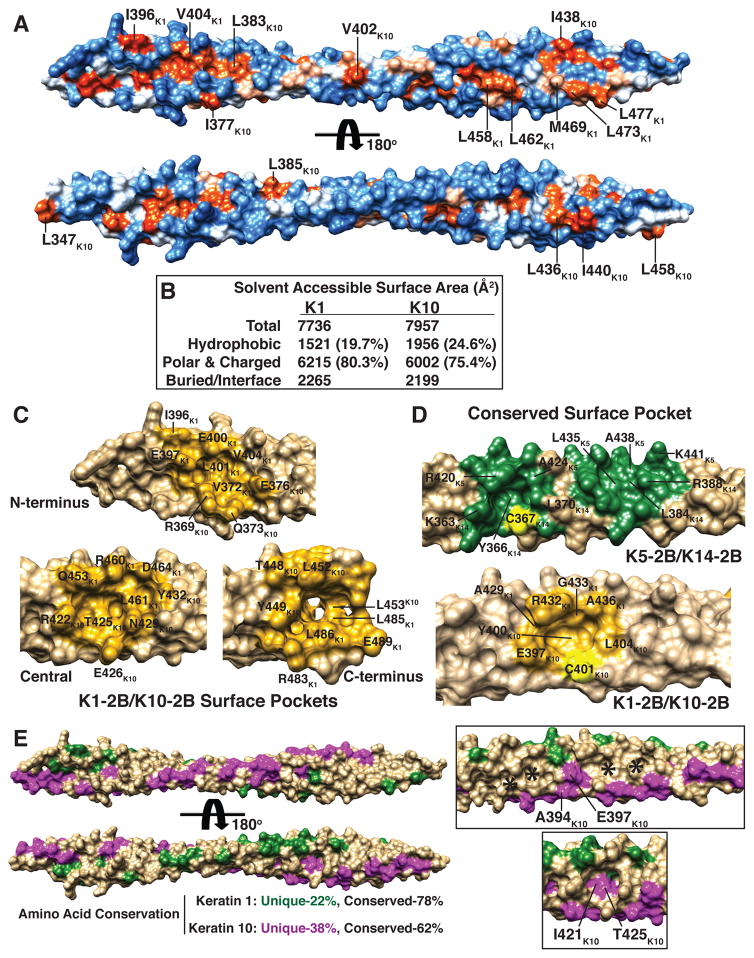
The K1-K10-2B molecular surface contains concentrated regions of both acidic and basic potential (Fig. 2) as well as hydrophobic residues that can be buried (helical interface) or surface-exposed (Fig. 3a, b). Approximately 20% of the K1 and 25% of the K10 solvent accessible surface area (SASA) in the K1-K10-2B structure comes from hydrophobic residues not buried in the dimer interface. This suggests exposed hydrophobic residues have a role in protein-protein interactions and/or KIF formation (such as tetramer stabilization). This is consistent with work showing a four-residue “hydrophobic stripe” in the 1B coil domain of mouse K16 is critical for heterotetramer stability (Bernot et al., 2005).
Fig. 3. Molecular surface properties of K1-K10-2B.
(a) K1-K10-2B heterodimer depicted in two orientations, related by 180°, demonstrating it contains several solvent accessible and surface exposed hydrophobic residues. Non-hydrophobic residues are colored blue. Hydrophobic residues are colored light (less) to dark (more) orange, based on degree of hydrophobicity. (b) Nature of the solvent accessible surface area of the K1-K10-2B heterodimer. (c, d) Concavities/pockets in the molecular surface of K1-K10-2B (gold) and K5-K14-2B (green). Both Cys367K14 and Cys401K10 (cysteines colored yellow) have a pocket adjacent to it; the chemical nature of the pockets is mostly conserved. One pocket wall is formed by Lys363 in K14 (basic) but is formed by Glu397 in K10 (acidic). (e) Residues in K1 (green) and K10 (magenta) that are unique (non-conserved) compared to K5 and K14, respectively, are mapped onto the K1-K10-2B structure, demonstrating alignment mostly along outer helical ridges. Conserved residues align mostly along the helical interface (right upper box), with some regions appearing groove-like (*). Some pockets, such as the ‘Central’ pocket in panel C, have conserved residues (Ile421K10 and Thr425K10) lining the floor.
To identify unique cavities in the surface shape of K1-K10-2B, its structure along with the K5-K14-2B structure were analyzed using MetaPocket (Zhang et al., 2011). Three pockets identified in the K1-K10-2B structure (N-terminal, central, and C-terminal) show deep concavities lined by a mix of hydrophobic, charged and polar residues (Fig. 3c).
To correlate primary sequence conservation between human keratins with molecular surface properties, the conserved (identical) and non-conserved (not identical) residues for K1 and K10 were mapped onto the crystal structure (Fig. 3e). While only 22% of the K1 sequence and 38% of the K10 sequence are unique compared to K5-K14 (Supplementary Fig. S3), the majority of these unique residues are spread along the outer helical ridge of the molecule. This key distribution pattern enables keratins to share a common coiled-coil motif (driven by the high sequence conservation) but generate maximally diverse surface chemistry by strategically placing its small percentage of unique residues on the outer helical ridge. For example, the floor of the “central” pocket (Fig. 3c) is comprised of residues unique to K10 (I421K10 and T425K10) (Fig. 3e). Conserved residues commonly align along the helical interface, at times demonstrating surface grooves (Fig. 3e).
Disulfide-bonding in the K1-K10 Structure
The K1-K10-2B structure demonstrated disulfide-linked heterodimers; Cys401K10 links two K10 molecules from separate heterodimers. This observation was supported by light scattering analysis of the purified K1-K10-2B complex (prior to crystallization) which identified a small fraction of a tetramer species (Supplementary Fig. S4). Further analysis of the K1-K10-2B solution using a non-reducing, denaturing gel showed a keratin dimer that could be abrogated by the addition of TCEP, a disulfide reducing agent, suggesting a disulfide linkage can form between heterodimers to create a tetramer (Supplementary Fig. S4d). This is consistent with the observed disulfide bond in the crystal structure.
Residues surrounding Cys401K10 contribute to a packing interface at the disulfide linkage; two residues, Gly398K10 and Ser405K10, contribute to the packing interface by virtue of their small side chain size. They enable K10-2B helices to pack tightly at Cys401K10. For example, the hydroxyl side chain of Ser405K10 is in close proximity (~2.7 Å) to the carbonyl oxygen of Cys401K10, enabling hydrogen bond formation at the disulfide bond site (Fig. 1b–d). Furthermore, the sequence position occupied by Gly398K10 is one helical turn proximal to the disulfide bond site and Gly398K10 from adjacent K10 molecules pack against each other (Fig. 1c). This glycine is conserved in K14, but not in K17, which contains asparagine; modeling of Gly398Asn mutation in the K1-K10-2B structure introduces unfavorable steric clashes (Supplementary Figure S5). Thus, keratins with residues containing bulkier side chains at this position (or any position in the packing interface) may have altered capacity to form disulfide linkages, altered stability of the disulfide bond, or altered bond angle due to the need to relieve steric clash. To further investigate this concept, an analysis of the change in free folding energy of the disulfide-linked K1-K10-2B structure for all possible single-site mutations in K10 was performed (Supplementary Figures S6–7) (Dehouck et al., 2009, Dehouck et al., 2011). Of all regions of K10, the region surrounding Cys401K10 is least likely to have improved stability by alteration of amino acid sequence; there are very few substitutions that make this disulfide-linked complex more stable. Adjacent to Cys401K10 there exists, unexpectedly, a well-defined pocket; two pockets from linked K1-K10 heterodimers align symmetrically around the disulfide bond (Fig. 1g).
Mutations in human skin disease alter intermediate filament organization and structure
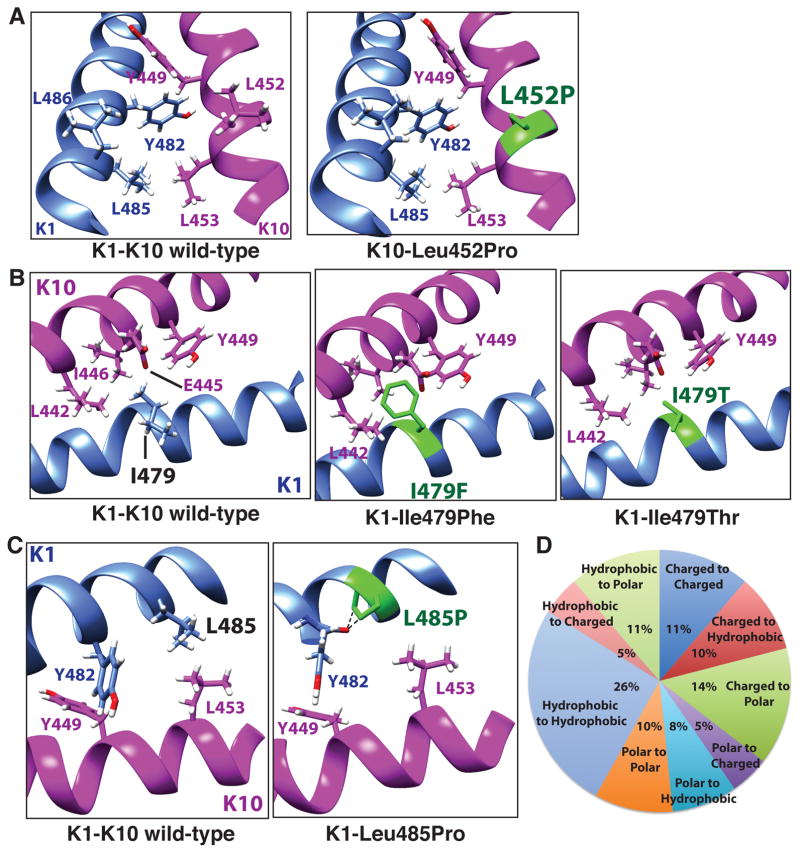
Mutations identified in epithelial keratins that are associated with human skin disease exist mostly in helix IA or helix 2B, so called mutational hot spots. For K1 and K10, several human skin diseases (epidermolytic ichthyosis, cyclic ichthyosis with epidermolytic hyperkeratosis, and epidermolytic and non-epidermolytic palmoplantar keratosis) are associated with missense, nonsense, insertion, and deletion mutations of helix 2B (Supplementary Table S3). Of the ~30 missense mutations in helix 2B of K1 and K10, most reside in the C-terminal aspect of helix 2B which is thought to be critical for KIF nucleation (Supplementary Fig. S8) (Wu et al., 2000). The consequences of these mutations on the structural chemistry of the keratin heterodimer vary; for example, Leu452ProK10 eliminates a surface-exposed hydrophobic residue (Fig. 4a), Ile479PheK1 creates multiple steric clashes from the Phe aromatic ring (Fig. 4b), Ile479ThrK1 eliminates stabilizing hydrophobic interactions from the isoleucine and adds hydrogen bonding from the threonine side chain, and Leu485ProK1 eliminates hydrophobic interactions (with L453K10) and creates a steric clash between the proline and the carbonyl oxygen of Tyr482K1 that drives kinking of the K1-2B helix (Fig. 4c). Single missense mutations are predicted to cause local disturbances to the coiled-coil structure, and not completely disrupt heterodimer assembly based on modeling of K5-K14 helix 1A mutations (Smith et al., 2004). Furthermore, mutant K1 and K10 molecules did not eliminate filament formation, but caused aberrant filament networks (Syder et al., 1994).
Fig. 4. Structural consequences of K1-K10-2B mutations.
(a) Leu452ProK10 mutation associated with epidermolytic ichthyosis eliminates a surface-exposed leucine and generates proline ring clashes leading to helix kinking. (b) Ile479PheK1, associated with epidermolytic ichthyosis, generates steric clashes from the phenylalanine aromatic ring; Ile479ThrK1, associated with epidermolytic ichthyosis and epidermolytic palmoplantar keratosis, fails to provide the same stabilizing hydrophobic interactions or hydrogen bonds that wild-type isoleucine provides. (c) Leu485ProK1 mutation associated with cyclic ichthyosis with epidermolytic hyperkeratosis eliminates hydrophobic interactions (with L453K10) and generates clashes between proline and the carbonyl oxygen of Tyr482K1 that drives kinking of the K1-2B helix. (d) All human keratin mutations (for K1, K5, K6a, K6b, K6c, K10, K14, K16, K17) documented in the Human Intermediate Filament Database were analyzed for the type of chemical change occurring and results for K1/K10 are summarized. The most common type of change was a hydrophobic residue to a hydrophobic residue (26%), and second most common was charged residue to a polar residue (14%). Definitions of residues were: acidic (D, E), basic (R, K), polar (Q, N, H, S, T, Y, C, G) and hydrophobic (A, I, L, F, V, P, M, W). Data for K5-K14 and pachyonychia congenita keratins are summarized in Figure S14.
A mutation of Leu437K1 to proline is associated with the human skin disease diffuse palmoplantar keratosis (DPPK) (Liu et al., 2009). The K1-K10 structure demonstrates Leu437K1 is involved in key hydrophobic and hydrogen bond interactions with three K10 residues, Tyr400 K10, Gln403 K10, and Leu404 K10 (Fig. 1f). These three K10 residues along with Leu437K1 form the helical interface at the location of the surface-exposed Cys401K10. Structurally, Leu437ProK1 eliminates major stabilizing contacts at the site of disulfide bond formation. This is a less damaging mutation than those causing more generalized skin disease; therefore, DPPK due to Leu437ProK1 may require the more severe frictional forces experienced by the palms and soles. Additional biochemical alterations to the structure of K1 and K10 caused by missense mutations are detailed in Supplementary Table S4 and Figures S9–S13.
To investigate the chemical nature of missense mutations occurring in K1 and K10, a detailed analysis was performed on the type of biochemical change resulting from mutation. Surprisingly, the majority of pathologic mutations throughout K1 and K10 involve substitution of a hydrophobic residue with another hydrophobic residue (26%); the second most common pattern was substitution of a charged residue with a polar one (Fig. 4d). Comparing K1/K10 with K5/K14 and with keratins involved in pachyonychia congenita (keratins 6a, 6b, 6c, 16, 17), hydrophobic to hydrophobic residue substitution was also the most common mutation pattern (Supplementary Fig. S14).
Key structural comparisons between K1-K10 2B and K5-K14 2B
Superposition of the K1-K10-2B and K5-K14-2B heterodimer structures has a root-mean-square deviation (RMSD) of 1.74 Å (Suppl. Fig. 15a). There are notable differences in the electrostatic surface potential, exposed hydrophobic surface, and surface shape (pockets) observed in the K1-K10-2B structure compared to K5-K14 (Figs. 2–3). The K1-K10-2B structure contains 27 additional residues at the N-terminal end of the 2B region compared to a K5-K14-2B structure (Lee, Kim, 2012); this enabled more complete visualization of an N-terminal basic patch formed by nine K1 residues (Fig. 2). All of the residues comprising this basic patch are conserved in K5, but not all of them were visible in the K5-K14 structure. A small basic patch created by Lys363K14 and Arg420K5 in the K5-K14 structure is not present in K1-K10 because the analogous sequence position in K10 contains glutamate (Glu397K10) rather than lysine (Fig. 2b). This shifts the overall electrostatic potential of the N-terminal aspect of K1-K10-2B to a more acidic environment than K5-K14-2B.
Similar to the K5-K14-2B structure, K1-K10-2B heterodimers formed a disulfide linked aggregate. The interhelical angle between K10 molecules in the disulfide-linked complex for K1-K10-2B is 77.4°; this is slightly more acute than the interhelical angle (79.4°) between K14 molecules in the disulfide-linked complex of K5-K14-2B (calculated using INTERHLX, K. Yap, Univ. of Toronto). It is unexpected that two independently derived keratin heterodimer crystal structures, using crystals differing in growth conditions and spacegroups, have only 2° difference in the angle relating the disulfide-linked keratin proteins; it implies residues surrounding the cysteine form a critical protein interaction interface. There is a 2.49 Å RMSD when superposing the disulfide-linked K10 from the K1-K10-2B structure and the disulfide-linked K14 from the K5-K14-2B structure (Suppl. Fig. 15b). Chemical differences do exist between K10 and K14 near Cys401K10: serine resides at position 405 in K10 as opposed to alanine in K14 (Fig. 1c–e). Interestingly, one surface pocket is conserved between the K1-K10 and K5-K14 structures and is located adjacent to the cysteine (Cys401K10 or Cys 367K14) involved in disulfide bond formation (Figs. 1g, 3d).
DISCUSSION
Decades of outstanding biochemistry, molecular modeling, sequence analysis and evaluations of pathological mutations have provided great insights into the structure of keratins. Our crystal structure analysis of K1-K10 confirms many of the previous predictions while adding new understandings to epidermal keratin structure and function. The K1-K10 structure enabled a comparison with K5-K14, where identification of key structural differences illustrates the capacity of various keratin pairs to have biochemically distinct surface properties. The K1-K10-2B structure fulfills a major need to determine more atomic resolution structures of human keratins in order to gain better understanding of the biochemical relationships between KIF structure and pathogenesis of human skin diseases.
Important unresolved questions regarding KIF function include: 1) how do keratins generate functional diversity despite sharing a coiled-coil fold, and 2) what are the molecular determinants of higher-order filament formation by heterodimers? Detailed molecular surface analysis of the K1-K10 structure, and its comparison to K5-K14, demonstrated that the structural chemistry at the molecular surface of keratin filaments involves a complex mix of acidic, basic, and hydrophobic patches distributed over various ridges, grooves, and pockets. For example, just one residue change between type I keratins (e.g. Glu397K10 rather than Lys363K14) alters the electrostatic surface potential of the N-terminus of the 2B region (Fig. 2b). We observed one profound mechanism for generating differences in molecular surface properties of keratins: the clustering of nonconserved residues (observed between K1-K10-2B and K5-K14-2B in Fig. 3e) along the outer helical ridges of the coiled-coil. Despite sharing 60–80% of the amino acid sequence, keratins (in this case K1/K10) have evolved so that the majority of the 20–40% unique residues align in a special distribution (outer helical ridges) so that keratins only need a small number of residue differences to create substantial changes in surface shape, charge, and hydrophobicity. How these distinct surface properties within helical regions affect keratin function and packing of KIFs needs investigation.
The K1-K10 heterodimer contains up to 25% hydrophobic SASA. Since K1-K10 filament assembly involves aggregation of numerous heterodimers and oligomeric species (Steinert, 1991), one hypothesis is these solvent-exposed hydrophobic residues help drive filament aggregation, align filaments, and stabilize formed filaments (Lee, Kim, 2012, Woods, 1989). Electrostatic interactions also are expected to affect axial alignment during higher order filament formation (Lee, Kim, 2012, Woods, 1989); K1-K10-2B displays polarization of surface charge similar to K5-K14-2B (Fig. 2a). It is possible that phosphorylation of specific tyrosine, threonine or serine residues could add negative surface charge (Figure S16). Our comprehensive analysis of the chemical and structural nature of K1 and K10 mutations associated with human skin disease revealed the most common type of missense mutation occurring in keratins is mutation of one hydrophobic residue to another hydrophobic residue; while this appears conservative, these mutations are associated with clinical phenotype, suggesting that even small alterations in the chemical structure of surface-exposed hydrophobic residues can impact KIF formation and/or function.
The K5-K14-2B crystal structure enabled the discovery of a trans-dimer, homotypic disulfide bond in keratin 14 (at Cys367) (Lee, Kim, 2012). This led further to identification of a filament cage, formed by disulfide-linked K14, around the nucleus that helped regulate nuclear shape. More recent studies on K14 demonstrated disulfide bonding as an important mechanism for regulating filament elongation, as well as regulating the dynamics of filament organization around pan-cytoplasmic, perinuclear, and peripheral keratin networks (Feng and Coulombe, 2015). It had been surmised that K10 has similar capacity to form a disulfide linkage, based on sequence conservation of Cys401 (analogous to Cys367K14), identification of disulfide bond cross-links in the 2B region (Steinert and Parry, 1993), and characterization of disulfide-bonded K1-K10 oligomers from cultured human epidermal cells (Sun and Green, 1978).
Disulfide bond formation is critical for regulating K5-K14 keratin filament behavior in vivo (Feng and Coulombe, 2015). It is important that in cultured mouse keratinocytes as little as ~ 5% of K14 is estimated to be in this cross-linked form, representing a subset of KIFs for a unique cellular function (Feng and Coulombe, 2015, Lee, Kim, 2012). Thus, the disulfide cross-linkage between heterodimer pairs likely contributes to higher order arrangement of 10-nm KIFs rather than an essential characteristic of the packing within the 10-nm filament. True mechanisms behind protein organization within 10-nm filaments remain crystallographically elusive. We establish that K1-K10 forms a trans-dimer disulfide bond (Fig. 1 and Supplementary Figs. S17–18), but we have no data that address its quantitative importance or the function of that disulfide in vivo. Structure analysis suggests sequence positions adjacent to the cysteine forming the disulfide bond influence the stereochemistry and spatial parameters of disulfide linkage; they create a particular interaction interface. This interface reflects selection against residues that will not allow for correct protein-protein packing; for example, residues with bulky side chains will not pack similar to Gly398K10 or Ser405K10 at those positions (Supplementary Figs. S5–7). Differences in residues at sequence positions adjacent to Cys401K10 thus may affect the angle with which K1-K10 dimers pack, the type of disulfide bond (the K1-K10 disulfide bond is classified as a left handed spiral), and the stability of the higher order complex (Wong and Hogg, 2010). The concept that subtle sequence differences at select positions may dictate cellular behaviors for different keratins is plausible given the noted differences between K1-K10 and K5-K14, and their stark contrast with K18 (Fig. 1e). K18, which is expressed in liver, does not conserve the glycine at position 398 or the cysteine at position 401 or the serine at position 405. This implies this disulfide linkage is not critical for KIF formation. Therefore, K18 cannot form 2B filament crosslinks analogous to K1/K10 or K5/K14, and this difference emphasizes how small amino acid alterations within helical regions may impact the cellular functions of the various keratins (Zatloukal et al., 2007). To date no skin disease is associated with a mutation to Cys401K10, however, palmoplantar keratoderma is associated with a Leu437ProK1 mutation for which modeling demonstrates structural instability at the disulfide bond site (Fig. 1f).
An intriguing and unanticipated aspect of the K1-K10 structure is the identification of pockets and grooves in the molecular surface (Figs. 1g, 3c–e), raising the question of their functional purpose. One hypothesis is that during KIF formation many of the surface pockets bind with residues from other heterodimers; thus, crystal structures of higher order keratin complexes would be highly beneficial to better understand the structural determinants of filament packing. However, it is also possible that other non-keratin proteins bind to K1 and K10 at these pockets to modify KIF functions in the cell. The unexpected finding of a pocket adjacent to the disulfide bond forming cysteine seems relevant given that it is present in both the K1-K10-2B structure and the K5-K14-2B structure (Fig. 1g). Perhaps it represents an interaction site with sulfhydryl oxidase, a disulfide catalyst which participates in terminal epidermal differentiation (Coppock and Thorpe, 2006, Hashimoto et al., 2000, Matsuba et al., 2002, Saaranen and Ruddock, 2013).
A valuable outcome of this K1-K10-2B crystal structure is that it exposes the biological questions most critical for taking our understanding of KIFs to the next level. This includes identification of protein-binding partners and the functions of surface pockets, the determination of additional structures of keratins, particularly their higher-order assemblies, and how the head and tail domains interact with the keratin helices in the intact molecule. Recent molecular dynamics simulation of an intact K1-K10 heterodimer model proposed ways in which all domains of a keratin (head, helices, linkers, and tail) relate in 3D space; it further emphasized the need for experimental crystal structures to validate the models (Bray et al., 2015). Future studies must define more clearly how point mutations cause disease – at the heterodimer level and in the KIF aggregate. All of these difficult, future experiments are necessary to gain greater mechanistic insight into the correlation between genotype, 3D keratin structure, and human phenotype.
MATERIALS AND METHODS
Keratin Constructs
cDNA for full-length keratin 1 (K1) and keratin 10 (K10) was provided by Dr. Keith Choate (Yale Univ.). The 2B helical regions of K1 (res. 370–489) and K10 (res. 337–456) were PCR-amplified from cDNA using the following primers: K1-2B-For: 5′-GCCATATGGAGTCCTTGTACCAGAGCAAG-3′; K1-2B-Rev: 5′-CAGGACCCTCCTGGAGGGAGAATAGTAGGAATTCGC-3′; K10-2B-For: 5′-GCCATATGGAAGCCTGGTTCAATGAAAAGAGC-3′; K10-2B-Rev: 5′-CGCAGCCTGCTAGAAGGAGAGTAGTAGGAATTCGC-3′. K1-2B was sub-cloned using NdeI and EcoRI into pGS-21A plasmid such that K1-2B did not contain a fusion tag. K10-2B was sub-cloned using NdeI and EcoRI into pET-28A plasmid such that K10-2B expressed an N-terminal 6-histidine tag. DNA sequencing verified plasmid constructs (Keck Biotechnology Resource Laboratory, Yale Univ.).
Protein Production and Purification
K1-2B and K10-2B were co-expressed in Escherichia coli strain BL21 (DE3) Codon+RIPL (Agilent Technologies) at 37°C in Luria Broth Miller (EMD Millipore). Protein expression induced with 1mM isopropyl-D-thiogalactopyranoside proceeded for 3–4hrs. After pelleting cells by centrifugation at 2500xg, 10min., at 4°C, they were suspended in 50mM Tris-HCl buffer (pH 7.8) containing 0.5M NaCl, 20mM imidazole, 1% Nonidet P-40, 6mM MgCl2, 1mM CaCl2 and 1x EDTA-free protease inhibitor cocktail (Roche Diagnostics). Cells were lysed by sonication on ice, followed by incubation of lysate with ~ 30 units/mL DNase I at 37°C for 15min. The solution was centrifuged at 17500xg, 15min, at 4°C. After decanting the supernatant, the pellet was washed with 50mM Tris-HCl buffer (pH 7.8) containing 0.5M NaCl and 20mM imidazole, and centrifuged again at 17500xg, 15min, at 4°C. Washed pellets were suspended in 8M urea, and agitated at 4°C for 1hr, followed by centrifugation at 17500xg, 10min, at 4°C. The supernatant (20mL) was placed in 3500Da molecular weight cutoff Spectra/Por membrane tubing (Spectrum Laboratories, Inc.) and dialyzed against 1L of 100mM Tris-HCl buffer (pH 7.4) containing 4M urea and 0.2M NaCl for 24hrs at 4°C. The supernatant was subsequently dialyzed against 2L of 100mM Tris-HCl buffer (pH 7.4) containing 0.2M NaCl and 10mM 2-mercaptoethanol for 48hrs at 4°C. Dialyzed sample was centrifuged at 25000xg, 20min, at 4°C, and the supernatant subjected to batch nickel affinity purification using previously described methods (Bunick et al., 2015), with the following modifications: the solution used to equilibrate and wash nickel resin was the final dialysate and 91.2μg (408 units) bovine alpha-thrombin (Haematologic Technologies, Inc.) was used to cleave His-tag. The clarified solution containing the untagged K1-2B–K10-2B heterocomplex was applied to a Superdex75 (26/60) gel filtration column in 100mM Tris-HCl buffer (pH 7.4) containing 0.2M NaCl and 10mM 2-mercaptoethanol. Collected fractions were analyzed by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE), and selected pooled fractions were concentrated in a 3500Da molecular weight cutoff centrifugal filter unit (EMD Millipore).
Crystallization and x-ray data collection
K1–K10-2B heterocomplex at 7.8mg/ml in 100mM Tris-HCl buffer (pH 7.4) containing 0.2M NaCl and 10mM 2-mercaptoethanol was crystallized by sitting drop vapor diffusion at 25°C using a reservoir solution of 100mM Hepes buffer (pH 7.5) containing 2.0M ammonium formate. Crystals were soaked 1–3min in a cryoprotectant solution containing 30% PEG400 in mother liquor prior to flash-freezing in a liquid nitrogen cryostream. A native data set on a single crystal maintained at ~100K was collected using the NSLS X-25 beamline (λ=1.1 Å) at Brookhaven National Laboratory. The crystal belonged to the orthorhombic space group I222 (cell dimensions: a=75.19 Å, b=75.86 Å, c=209.29 A, α=β=γ=90°) with a unit cell solvent content of 79%. Diffraction data was processed using XDS (Kabsch, 2010).
Structure determination, refinement, and analysis
The K1–K10-2B structure was determined by molecular replacement with MOLREP (Vagin and Teplyakov, 2010) using the keratin 5-2B–keratin 14-2B structure (PDB 3TNU) as a search model (Lee, Kim, 2012). The K1–K10-2B structure underwent iterative rounds of model building (Coot) (Emsley and Cowtan, 2004) and refinement (PHENIX) using standard geometric (bond length, bond angle) and secondary structure restraints. The final model of the crystal asymmetric unit contained one K1-2B and one K10-2B molecule in complex. The final Ramachandran statistics were: residues in favorable regions, 98.1%; in allowed regions, 1.43%; in outlier regions, 0.48%. Electrostatics were calculated using PDB2PQR (Dolinsky et al., 2004) and Adaptive Poisson-Boltzmann Software (APBS) (Baker et al., 2001). Structural analyses were performed with Coot, UCSF Chimera (Resource for Biocomputing, Visualization, and Informatics, University of California, San Francisco), and PDBePISA (The European Bioinformatics Institute, European Molecular Biology Laboratory, UK). Figures were prepared using UCSF Chimera or PyMOL Molecular Graphics System (Version 1.5.0.4, Schrödinger, LLC). Atomic coordinates and structure factors have been deposited in the Protein Data Bank under the accession code 4ZRY.
Supplementary Material
Acknowledgments
We thank Professor Thomas A. Steitz (Dept. of Molecular Biophysics and Biochemistry, Yale Univ.) for providing a collegial environment, laboratory space, equipment, and supplies for this project (to C.G.B.). We thank Dr. Jimin Wang for assistance with x-ray data processing and structure determination. This work was supported by the Dermatology Foundation through a Dermatologist Investigator Research Fellowship and a Career Development Award (to C.G.B.), an NIH/NIAMS Dermatology Training Grant to Yale (PI: Richard Edelson) T32 AR007016 (to C.G.B.), and an NIH/NIAMS grant 5K08AR070290-02 to C.G.B. Aspects of this work were presented at the 2015 World Congress of Dermatology Meeting (Vancouver, CA), the 2015 and 2016 Society for Investigative Dermatology Annual Meeting (Atlanta, GA; Scottsdale, AZ), and the 2016 American Academy of Dermatology Annual Meeting (Washington D.C.).
Footnotes
CONFLICT OF INTEREST
The authors state no conflict of interest.
References
- Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc Natl Acad Sci U S A. 2001;98:10037–41. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernot KM, Lee CH, Coulombe PA. A small surface hydrophobic stripe in the coiled-coil domain of type I keratins mediates tetramer stability. J Cell Biol. 2005;168:965–74. doi: 10.1083/jcb.200408116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray DJ, Walsh TR, Noro MG, Notman R. Complete Structure of an Epithelial Keratin Dimer: Implications for Intermediate Filament Assembly. PLoS One. 2015;10:e0132706. doi: 10.1371/journal.pone.0132706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunick CG. X-ray crystal structure of the keratin 1-keratin 10 heterodimer reveals a molecular basis for associated keratinopathies. J Invest Dermatol; Society for Investigative Dermatology 74th Annual Meeting; Atlanta, GA. 2015. pp. S58–S69. [Google Scholar]
- Bunick CG, Presland RB, Lawrence OT, Pearton DJ, Milstone LM, Steitz TA. Crystal Structure of Human Profilaggrin S100 Domain and Identification of Target Proteins Annexin II, Stratifin and hsp27. J Invest Dermatol. 2015;135:1801–9. doi: 10.1038/jid.2015.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernyatina AA, Guzenko D, Strelkov SV. Intermediate filament structure: the bottom-up approach. Curr Opin Cell Biol. 2015;32:65–72. doi: 10.1016/j.ceb.2014.12.007. [DOI] [PubMed] [Google Scholar]
- Coppock DL, Thorpe C. Multidomain flavin-dependent sulfhydryl oxidases. Antioxid Redox Signal. 2006;8:300–11. doi: 10.1089/ars.2006.8.300. [DOI] [PubMed] [Google Scholar]
- Crick FH. Is alpha-keratin a coiled coil? Nature. 1952;170:882–3. doi: 10.1038/170882b0. [DOI] [PubMed] [Google Scholar]
- Dehouck Y, Grosfils A, Folch B, Gilis D, Bogaerts P, Rooman M. Fast and accurate predictions of protein stability changes upon mutations using statistical potentials and neural networks: PoPMuSiC-2.0. Bioinformatics. 2009;25:2537–43. doi: 10.1093/bioinformatics/btp445. [DOI] [PubMed] [Google Scholar]
- Dehouck Y, Kwasigroch JM, Gilis D, Rooman M. PoPMuSiC 2.1: a web server for the estimation of protein stability changes upon mutation and sequence optimality. BMC Bioinformatics. 2011;12:151. doi: 10.1186/1471-2105-12-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolinsky TJ, Nielsen JE, McCammon JA, Baker NA. PDB2PQR: an automated pipeline for the setup of Poisson-Boltzmann electrostatics calculations. Nucleic Acids Res. 2004;32:665–7. doi: 10.1093/nar/gkh381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–32. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Feng X, Coulombe PA. A role for disulfide bonding in keratin intermediate filament organization and dynamics in skin keratinocytes. J Cell Biol. 2015;209:59–72. doi: 10.1083/jcb.201408079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto Y, Suga Y, Matsuba S, Mizoguchi M, Takamori K, Seitz J, et al. Immunohistochemical localization of sulfhydryl oxidase correlates with disulfide crosslinking in the upper epidermis of rat skin. Arch Dermatol Res. 2000;292:570–2. doi: 10.1007/s004030000171. [DOI] [PubMed] [Google Scholar]
- Kabsch W. XDS. Acta Crystallogr D Biol Crystallogr. 2010;66:125–32. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CH, Kim MS, Chung BM, Leahy DJ, Coulombe PA. Structural basis for heteromeric assembly and perinuclear organization of keratin filaments. Nat Struct Mol Biol. 2012;19:707–15. doi: 10.1038/nsmb.2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu XP, Ling J, Xiong H, Shi XL, Sun X, Pan Q, et al. Mutation L437P in the 2B domain of keratin 1 causes diffuse palmoplantar keratoderma in a Chinese pedigree. J Eur Acad Dermatol Venereol. 2009;23:1079–82. doi: 10.1111/j.1468-3083.2009.03175.x. [DOI] [PubMed] [Google Scholar]
- Loschke F, Seltmann K, Bouameur JE, Magin TM. Regulation of keratin network organization. Curr Opin Cell Biol. 2015;32:56–64. doi: 10.1016/j.ceb.2014.12.006. [DOI] [PubMed] [Google Scholar]
- Matsuba S, Suga Y, Ishidoh K, Hashimoto Y, Takamori K, Kominami E, et al. Sulfhydryl oxidase (SOx) from mouse epidermis: molecular cloning, nucleotide sequence, and expression of recombinant protein in the cultured cells. J Dermatol Sci. 2002;30:50–62. doi: 10.1016/s0923-1811(02)00061-0. [DOI] [PubMed] [Google Scholar]
- Saaranen MJ, Ruddock LW. Disulfide bond formation in the cytoplasm. Antioxid Redox Signal. 2013;19:46–53. doi: 10.1089/ars.2012.4868. [DOI] [PubMed] [Google Scholar]
- Smith TA, Steinert PM, Parry DA. Modeling effects of mutations in coiled-coil structures: case study using epidermolysis bullosa simplex mutations in segment 1a of K5/K14 intermediate filaments. Proteins. 2004;55:1043–52. doi: 10.1002/prot.20089. [DOI] [PubMed] [Google Scholar]
- Steinert PM. Analysis of the mechanism of assembly of mouse keratin 1/keratin 10 intermediate filaments in vitro suggests that intermediate filaments are built from multiple oligomeric units rather than a unique tetrameric building block. J Struct Biol. 1991;107:175–88. doi: 10.1016/1047-8477(91)90020-w. [DOI] [PubMed] [Google Scholar]
- Steinert PM, Parry DA. The conserved H1 domain of the type II keratin 1 chain plays an essential role in the alignment of nearest neighbor molecules in mouse and human keratin 1/keratin 10 intermediate filaments at the two- to four-molecule level of structure. J Biol Chem. 1993;268:2878–87. [PubMed] [Google Scholar]
- Strelkov SV, Herrmann H, Geisler N, Lustig A, Ivaninskii S, Zimbelmann R, et al. Divide-and-conquer crystallographic approach towards an atomic structure of intermediate filaments. J Mol Biol. 2001;306:773–81. doi: 10.1006/jmbi.2001.4442. [DOI] [PubMed] [Google Scholar]
- Sun TT, Green H. Keratin filaments of cultured human epidermal cells. Formation of intermolecular disulfide bonds during terminal differentiation. J Biol Chem. 1978;253:2053–60. [PubMed] [Google Scholar]
- Syder AJ, Yu QC, Paller AS, Giudice G, Pearson R, Fuchs E. Genetic mutations in the K1 and K10 genes of patients with epidermolytic hyperkeratosis. Correlation between location and disease severity. J Clin Invest. 1994;93:1533–42. doi: 10.1172/JCI117132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toivola DM, Boor P, Alam C, Strnad P. Keratins in health and disease. Curr Opin Cell Biol. 2015;32:73–81. doi: 10.1016/j.ceb.2014.12.008. [DOI] [PubMed] [Google Scholar]
- Vagin A, Teplyakov A. Molecular replacement with MOLREP. Acta Crystallogr D Biol Crystallogr. 2010;66:22–5. doi: 10.1107/S0907444909042589. [DOI] [PubMed] [Google Scholar]
- Wong JW, Hogg PJ. Analysis of disulfide bonds in protein structures. J Thromb Haemost. 2010 doi: 10.1111/j.1538-7836.2010.03894.x. [DOI] [PubMed] [Google Scholar]
- Woods EF. Aggregation of wool keratin intermediate filament proteins. Int J Biol Macromol. 1989;11:278–84. doi: 10.1016/0141-8130(89)90020-2. [DOI] [PubMed] [Google Scholar]
- Wu KC, Bryan JT, Morasso MI, Jang SI, Lee JH, Yang JM, et al. Coiled-coil trigger motifs in the 1B and 2B rod domain segments are required for the stability of keratin intermediate filaments. Mol Biol Cell. 2000;11:3539–58. doi: 10.1091/mbc.11.10.3539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zatloukal K, French SW, Stumptner C, Strnad P, Harada M, Toivola DM, et al. From Mallory to Mallory-Denk bodies: what, how and why? Exp Cell Res. 2007;313:2033–49. doi: 10.1016/j.yexcr.2007.04.024. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Li Y, Lin B, Schroeder M, Huang B. Identification of cavities on protein surface using multiple computational approaches for drug binding site prediction. Bioinformatics. 2011;27:2083–8. doi: 10.1093/bioinformatics/btr331. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.