Abstract
Meiotic chromosome segregation leads to the production of haploid germ cells. During meiosis I (MI), the paired homologous chromosomes are separated. Meiosis II (MII) segregation leads to the separation of paired sister chromatids. In the budding yeast Saccharomyces cerevisiae, both of these divisions take place in a single nucleus, giving rise to the four-spored ascus. We have modeled the microtubules in 20 MI and 15 MII spindles by using reconstruction from electron micrographs of serially sectioned meiotic cells. Meiotic spindles contain more microtubules than their mitotic counterparts, with the highest number in MI spindles. It is possible to differentiate between MI versus MII spindles based on microtubule numbers and organization. Similar to mitotic spindles, kinetochores in either MI or MII are attached by a single microtubule. The models indicate that the kinetochores of paired homologous chromosomes in MI or sister chromatids in MII are separated at metaphase, similar to mitotic cells. Examination of both MI and MII spindles reveals that anaphase A likely occurs in addition to anaphase B and that these movements are concurrent. This analysis offers a structural basis for considering meiotic segregation in yeast and for the analysis of mutants defective in this process.
INTRODUCTION
Two meiotic divisions reduce the ploidy of diploid cells in half to give rise to germ cells. The reduction in ploidy during meiosis is accomplished by having two rounds of chromosome segregation without an intervening DNA synthesis phase (reviewed in Nasmyth, 2001). The specialized nature of the meiosis I (MI) division results in the segregation of homologous chromosomes. This segregation event reduces the chromosome number, yet ensures that each MI product has a complete set of chromosomes. Meiosis II (MII) segregation is similar to a mitotic segregation, wherein sister chromatids are separated from each other. Despite the significance of meiotic chromosome segregation and despite the importance and specialization of the MI spindle, little fine structure work has been done on meiotic spindles.
Meiosis has been studied in genetically tractable organisms such as the budding yeast Saccharomyces cerevisiae (reviewed in Smith, 2001). Like other fungi, yeast packages all of the meiotic products into spores (reviewed in Byers, 1981). The ability to recover all four meiotic products in a tetrad has been the basis of powerful analysis of meiotic functions. Notably, the genetic analysis of homologous chromosome pairing and recombination has led to significant insights into these processes. Genetic analysis of meiotic chromosome segregation has been more limited, but some important features have been revealed (reviewed in Lee and Amon, 2001; Nasmyth, 2001; Lee and Amon, 2003). As expected, genes required for proper function of the mitotic spindle also may be required for the function of meiotic spindles. An example is genes of the FEAR pathway (SLK19, SPO12, CDC5, ESP1, and CDC14) that in mitosis help control spindle behavior as cells complete mitosis, and in meiosis coordinate the transition from a meiosis I spindle to meiosis II spindles (Stegmeier et al., 2002; Buonomo et al., 2003; Marston et al., 2003). There also are genes whose function is limited to meiosis, such as MAM1, that give a molecular basis to the unusual attachment of homologous chromosomes to the MI spindle (Toth et al., 2000). The MAM1 gene product in complex with the CSM1 and LRS4 gene products enforce the monopolar attachment of paired sister chromatids in MI (Rabitsch et al., 2003). The special nature of the MI spindle function raises a number of questions concerning the structure of these spindles.
Early conventional electron microscopy of yeast meiotic spindles led to a general understanding of meiotic progression from prophase through chromosome segregation to spore formation (reviewed in Byers, 1981). As with mitotic cells, the meiotic spindles consist of microtubules (MTs) nucleated by the nuclear envelope-embedded organelle, the spindle pole body (SPB). SPBs are duplicated during meiotic prophase and the two resultant SPBs separate to form the MI spindle. After anaphase I, the two SPBs of the MI spindle duplicate to form the four SPBs needed to organize the two MII spindles. Interestingly, all of this occurs in a single nucleus that is not partitioned into four discreet nuclei until spore formation after MII. Spore formation is initiated from the SPBs. During MII, the cytoplasmic face of the SPB is modified by the addition of meiosis-specific components that are necessary to begin prospore membrane formation (Neiman, 1998; Knop and Strasser, 2000). The prospore membranes initiated from each of the four SPBs engulf cytoplasm, part of the nucleus, and the SPB with its associated chromosomes. Finally, the spore coats are built on the prospore membranes.
We have examined the microtubule organization in the meiotic spindles in wild-type budding yeast by using three dimensional (3-D) reconstruction from micrographs of serial thin sections of these cells. We have found that the two types of meiotic spindles can be easily distinguished, with MI spindles occupying a larger volume and containing more microtubules than MII spindles. Nonetheless, chromosome segregation in both MI and MII seems to be based on a single microtubule attachment to kinetochores as seen in mitosis. Finally, both MI and MII spindles clearly carry out anaphase B (pole-to-pole lengthening) movements and seem to carry out anaphase A (poleward chromosome movement).
MATERIALS AND METHODS
Strains and Culture
All electron microscopy was done on SK-1 background wild-type yeast strain (S2682/S2683) that was supplied by Nancy Hollingsworth (SUNY, Stony-brook, NY). Cells were cultured for mitotic growth and induced to enter meiosis as described in Straight et al. (2000). Meiotic progression was assayed by morphological characteristics as described in Straight et al. (2000), and cells were harvested for electron microscopy (EM) after 6, 8, 10, and 12 h in sporulation media.
Light Microscopy of Meiotic Spindle Lengths
Fluorescence imaging of meiotic spindles was done with a green fluorescent protein (GFP)-Tub1p–expressing strain of SK1 (strain YUMY4B1). Cells were induced to sporulate synchronously as described in Straight et al. (2000). Live cells were imaged in samples taken at 6, 7, 8, 9, and 10 h from a sporulating culture at 30°C. MI versus MII spindles were identified by one versus two spindles per cell, respectively. Images were captured using a Leica fluorescence microscope with a motorized stage driven by 3I Slidebook (Intelligent Imaging Innovations, Denver, CO) imaging software. Images of a field of cells were captured in three dimensions by using autofluorescence of GFP and a vertical step size of 0.1 μm over 25 planes. End-to-end measurements of spindles (41 MI spindles, 56 MII spindles) were performed using the digital ruler function of the Slidebook software on 3-D compiled images of fluorescent spindles. For long spindles with significant curvature, measurements were determined by summing a series of 0.1-μm lengths from along the curved spindle. An average of three measurements is reported as the actual spindle length (our unpublished data).
Preparation of Cells for Electron Microscopy and Imaging
Cells were prepared for electron microscopy as described in Giddings et al. (2001). In brief, cells were collected from sporulation media by filtration, and the cell paste was rapidly frozen under high pressure in a BAL-TEC HPM-010 high-pressure freezer (Technotrade International, Manchester, NH). The frozen cell pellet was freeze substituted into acetone containing 2% osmium tetroxide and 0.1% uranyl acetate at –80°C and then slowly warmed to room temperature. After substitution and warming, the samples were embedded in Spurr's resin per manufacturer's instructions (Electron Microscopy Sciences, Port Washington, PA).
Blocks of embedded cells were serially sectioned at a thickness of 50 nm, and the sections were stained with 2% uranyl acetate in 70% methanol and lead citrate. Sections were viewed on a Philips CM10 electron microscope (Philips, Eindoven, The Netherlands), and images were collected with a Gatan BioScan digital camera by using the Digital Micrograph software package (Gatan, Pleasanton, CA). Grids mounted in a rotating specimen holder were tilted to angles up to 50° by using a goniometer stage to bring the microtubules into near cross section. Tilt angles were recorded and used for corrections in the construction of models.
3-D Reconstruction and Modeling
The procedure for constructing three-dimensional models of meiotic spindles from the serial micrographs of spindle cross sections was similar to what has been described previously for mitotic spindles (Winey et al., 1995) and for other microtubule arrays (reviewed in McDonald et al., 1996). The centers of microtubule cross sections are marked in each section, and these data points are connected from section to section to map the trajectory and length of given microtubules, resulting in a rough model of the spindles. This modeling and all analyses of the models were done with the IMOD software package (http://bio3d.colorado.edu/imod).
The models of meiotic spindles were processed differently from previous studies because the images were collected and aligned in a slightly different way. In this study, tilting sections during microscopy was not sufficient to bring the majority of microtubules into perfect cross section in many models. Therefore, a more sophisticated adjustment was needed for section tilt when measuring the lengths of microtubules. Furthermore, images were aligned using cellular features in addition to the microtubules as fiducial marks to preserve the true geometry of the spindle. If only the microtubules were used for alignment, then the spindle would seem to be oriented perpendicular to the z-axis even though its microtubules were not. Because of these factors, the following steps were used to transform models for viewing and analysis.
First, the section-to-section alignment of the models was refined by using the positions of the microtubules to derive linear transformations between successive pairs of sections. Regions at the ends of the spindle with few microtubules (typically fewer than 10) were excluded. Only translational alignments were solved for the longest models with few microtubules. These transformations were analyzed to produce transformations that would adjust for local deviations from the trends in the alignment. Applying the latter transformations to the model effectively smoothed the microtubules locally without significantly affecting the overall geometry of the spindle model or the distances between microtubules. A new script in IMOD, Self-align, was used to accomplish this task.
Some of the spindles show significant curvature, so measuring SPB-to-SPB distance does not report an accurate spindle length. A length marker object was added to each model, consisting of a single contour starting among the microtubules near the middle of one SPB, staying near the middle of the spindle, and ending among the microtubules near the middle of the other SPB. This marker was used in the program Mtlengths (below) to determine the spindle lengths that we report.
The program Mtlengths was used to measure the length of each microtubule and of the spindle length marker. This program was modified to take information about the tilt of each section and the location of the tilt axis in each image and to determine what the coordinates of each segment of each microtubule would have been if the sections had been viewed without tilting. These adjusted coordinates were use to compute the true length of each microtubule. There was a potential ambiguity in this step because the sign of the tilt angles was not recorded during microscopy. To resolve this ambiguity, the program computed lengths with both positive and negative angles and reported what fraction of microtubule segments were longer with positive angles. Under the assumption that the tilting was done in the direction that would reduce the apparent obliqueness of the microtubules by as much as possible, the polarity of the angles was chosen so as to maximize the computed lengths. The Mtlengths program also produced a set of transformations that were used to transform the entire model to the coordinates that it would have had if the sections had been imaged without tilting. These transformed models are even more obliquely oriented than the original model.
Finally, a new program, Resamplemod, was used to rotate the model produced in the previous step so that the spindle length marker was parallel to the z-axis, and then resample the microtubule trajectories at the original section thickness. The resampled models are the final models used for display and analysis. Microtubule lengths in these models matched those computed from the original model by the program Mtlengths.
RESULTS
Meiotic Spindle Reconstruction
Meiotic yeast cells of the SK1 genetic background, capable of synchronous sporulation, were prepared for electron microscopy by using high-pressure freezing and freeze substitution (HPF/FS) as described in Giddings et al. (2001). We have previously shown that this technology yields outstanding results in the preservation of mitotic yeast cells. Similar results are obtained with meiotic cells (Figure 1; Straight et al., 2000). Figure 1 shows representative images of meiotic cells prepared by HPF/FS (see Materials and Methods), including images of meiotic spindles, both MI and MII, and images of microtubules in cross section, such as those used to build spindle models.
Figure 1.
Electron micrographs of HPF/FS prepared meiotic yeast cells. MI (A and B) and MII (C and D) spindles are shown in serial longitudinal sections. Arrows indicate SPBs (A and C), and the enhanced outer plaque on the SPB of a MII spindle is indicated in C. Images of spindle cross sections (E and F) show typical microtubule cross sections (E, arrow) and show more oblique microtubule profiles (F, arrow) that had to be traced by hand. Bar, 0.2 μm.
The microtubule arrays in 35 wild-type meiotic spindles have been reconstructed from electron micrographs of spindle cross sections of serially sectioned yeast cells by using the IMOD software package as described previously for mitotic spindles in various cell types (McDonald et al., 1996; see Materials and Methods). Movies of selected micrographs for each of the 35 meiotic spindles in the data set with model points marking the microtubule cross sections are available with the supplemental data. Morphological criteria were used first to demonstrate that a given reconstructed spindle was indeed in a meiotic cell and then to determine whether the spindle was a MI or a MII spindle. Entire serially sectioned nuclei from each cell were surveyed for two characteristics to determine the meiotic stage of the cell. First, cells with a very large vacuole adjacent to the nucleus were presumed to be in meiosis, and a cell with two spindles was definitely meiotic. Both meiotic divisions in yeast occur in the same nucleus, so finding two spindles is indicative not only of meiosis but is also of MII. The distinction between MI and MII was made by determining the number of spindles in meiotic cells (one spindle for MI and two spindles for MII) and was confirmed by seeing the modified outer (cytoplasmic) plaque of the SPB on MII spindles (Figure 1).
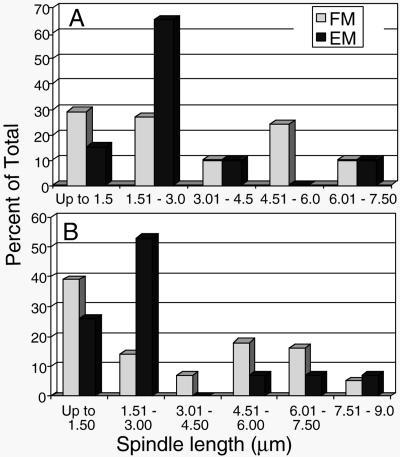
Yeast meiotic spindles have not been well described at either the light microscopic or electron microscopic level. We were interested to know whether our data set of 35 spindles (20 from MI and 15 from MII; Table 1) is representative of the total population of meiotic spindles. We have determined spindle lengths by imaging autofluorescent spindles in live meiotic SK1 cells that were expressing GFP-Tub1p (Straight et al., 2000). Cells containing a single spindle were considered MI, and cells with two spindles were MII. Histograms showing the length distributions of MI and MII spindles from this analysis (Figure 2) are compared with the length distribution of the spindles reconstructed from electron micrographs. As expected, the data set of spindles reconstructed from electron micrographs does not contain examples of all possible lengths, but it contains examples across the length distribution. Multiple reconstructions represent the predominant size classes identified by light microscopy. This abundance of cells in given spindle length classes may reflect pauses in spindle development and elongation suggestive of a cell cycle transition point. For instance, in mitotic cells there is a pause in progression at ∼1.5–2.0 μm in length during the metaphase to anaphase transition (Winey et al., 1995). The high frequency of shorter MI and MII spindles (<3.0 μm) and the dip in the distribution of spindle lengths at 3.0–4.5 μm are suggestive that an important transition, likely metaphase to anaphase, is occurring at these spindle lengths. Such spindles are well represented in the data sets collected by electron microscopy, whereas extremely short or very long spindles are underrepresented. Similar to our analysis of mitotic spindles (Winey et al., 1995), these underrepresented spindles are hard to find and are difficult to model when found (O'Toole et al., 1999). Nonetheless, we believe that the large data set of reconstructed meiotic spindles is generally representative with a number of models of spindles at the metaphase-to-anaphase transition.
Table 1.
Spindle model lengths and microtubule composition
| No. of microtubules:
|
|||||
|---|---|---|---|---|---|
| Model name | Length (μm) | Total | SPB1a | SPB2b | Cont.c |
| Meiosis I | |||||
| MSI-1 | 1.03 | 45 | 28 | 17 | |
| MSI-2 | 1.54 | 57 | 32 | 25 | |
| MSI-3 | 1.59 | 73 | 47 | 26 | |
| MSI-4 | 1.61 | 44 | 21 | 23 | |
| MSI-5 | 1.78 | 62 | 28 | 34 | |
| MSI-6 | 1.79 | 47 | 22 | 24 | 1 |
| MSI-7 | 1.83 | 42 | 22 | 16 | 4 |
| MSI-8 | 1.81 | 47 | 22 | 25 | |
| MSI-9 | 1.98 | 57 | 31 | 26 | |
| MSI-10 | 1.99 | 66 | 31 | 33 | 2 |
| MSI-11 | 2.01 | 50 | 25 | 25 | |
| MSI-12 | 2.02 | 68 | 29 | 39 | |
| MSI-13 | 2.02 | 56 | 27 | 28 | 1 |
| MSI-14 | 2.10 | 66 | 39 | 25 | 2 |
| MSI-15 | 2.24 | 68 | 34 | 32 | 2 |
| MSI-16 | 2.66 | 62 | 28 | 32 | 2 |
| MSI-17 | 3.01 | 47 | 26 | 21 | |
| MSI-18 | 3.14 | 63 | 33 | 28 | 2 |
| MSI-19 | 6.03 | 23 | 14 | 9 | |
| MSI-20 | 6.49 | 26 | 14 | 12 | |
| Meiosis II | |||||
| MSII-1 | 1.15 | 54 | 27 | 26 | 1 |
| MSII-2 | 1.47 | 53 | 26 | 26 | 1 |
| MSII-3 | 1.48 | 45 | 26 | 19 | |
| MSII-4 | 1.50 | 42 | 24 | 18 | |
| MSII-5 | 1.56 | 48 | 22 | 23 | 3 |
| MSII-6 | 1.56 | 52 | 25 | 26 | 1 |
| MSII-7 | 1.62 | 40 | 22 | 17 | 1 |
| MSII-8 | 1.71 | 57 | 28 | 25 | 4 |
| MSII-9 | 1.78 | 40 | 15 | 22 | 3 |
| MSII-10 | 1.92 | 47 | 18 | 24 | 5 |
| MSII-11 | 1.93 | 47 | 23 | 22 | 2 |
| MSII-12 | 1.99 | 47 | 24 | 23 | |
| MSII-13 | 5.03 | 14 | 7 | 7 | |
| MSII-14 | 7.08 | 7 | 4 | 3 | |
| MSII-15 | 7.57 | 10 | 6 | 4 | |
Left SPB in models, red MTs in models and model points
Right SPB in models, green MTs in models and model points
Continuous MTs, blue MTs in models and model points
Figure 2.
Meiotic spindle length distribution from fluorescent microscopy (FM) images of cells with GFP-tagged Tub1p compared with those from 3-D reconstruction from EMs. The spindle lengths were determined as described in Materials and Methods and are reported in 1.5-μm bins. The spindle lengths are reported separately for MI (A) and MII (B) and are reported as percent of total number of spindles. The number of spindles in the data set includes 41 MI spindles and 56 MII spindles identified by immunofluorescence microscopy, and 20 MI spindles and 15 MII spindles identified by electron microscopy (see Table 1).
Meiotic Spindle Models
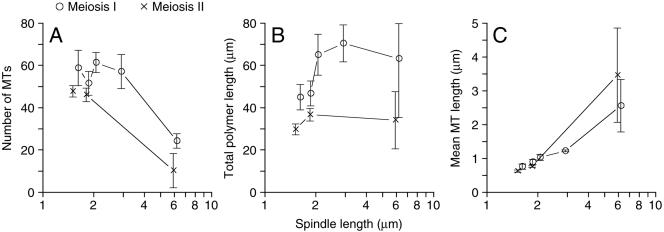
Table 1 lists some basic parameters derived from the 35 wild-type meiotic spindle models, including microtubule numbers and spindle lengths. Microtubules are reported as being from one SPB or the other SPB, with some instances of “continuous” microtubules. Microtubules are assigned to one SPB or the other by tracking the individual microtubule in serial sections until it ends at or within one section (∼40 nm) of a SPB. The SPB proximal end of the microtubule is considered to be the minus-end such that the end in the nucleoplasm will be the plus-end. All microtubules can be tracked to a SPB, but a few microtubules have both ends close enough to each of the two SPBs that the polarity cannot be determined, and these microtubules are called continuous. These microtubules are expected to have a normal plus- and minus-end, and they are found in MI and MII spindles (Table 1), as well as mitotic spindles (Winey et al., 1995). Generally, MI spindles seem to have more microtubules than MII spindles (Figure 3A). MI spindles (up to ∼3.14 μm) have an average of 28 ± 6 microtubules per SPB, more than enough to contact each of the 16 paired homologous chromosomes that are moved to each SPB during MI. MII spindles (<3 μm) have an average of 23 ± 3 microtubules per SPB, again more than enough microtubules to contact the 16 sister chromatids that are moved to each SPB during MII. The greater number of microtubules in MI spindles is reflected in the observation that MI spindles have more total tubulin polymer (Figure 3B). Interestingly, the higher level of total microtubule polymer in MI spindles is caused entirely by the increased microtubule number because the mean microtubule lengths in MI and MII spindles are very similar (Figure 3C).
Figure 3.
General description of the microtubules in MI (○) and MII (×) spindles derived from the 35 meiotic spindles (see Table 1) as described in Materials and Methods. (A) The total number of microtubules versus spindle length. (B) The microtubule total polymer versus spindle length. (C) Mean microtubule length versus spindle length. Spindles of similar length were grouped together. Error bars represent 74% confidence interval.
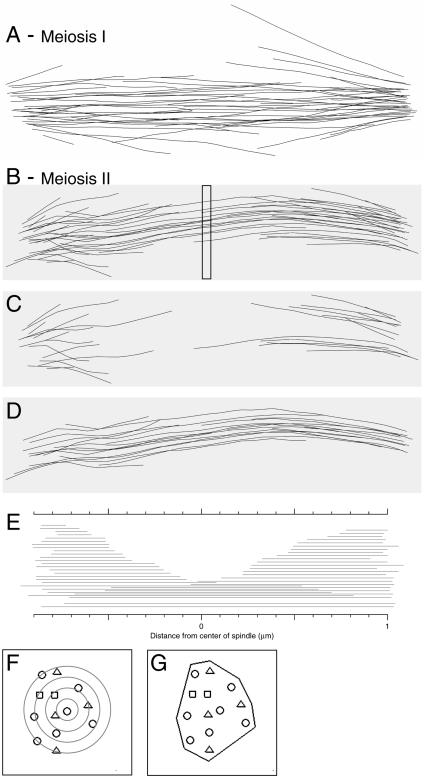
Representative spindle models are shown in Figure 4, and all 35 models are available with the supplemental data. Figure 4 includes different views of a spindle model, including models of how the microtubules of a MI and a MII spindle are organized in space (Figure 4, A and B, respectively). The models can be rotated around the spindle axis for examination from various angles, and movies of each model are available in the supplemental data. Also shown is a deconstruction of the MII spindle, as described below, to show putative kinetochore microtubules (Figure 4C) and the core bundle or central spindle microtubules (Figure 4D). There also is a graph of microtubule overlap (Figure 4E) that shows the microtubules from each SPB arranged by length from one SPB or the other (microtubule overlap graphs for all models are with the supplemental data). A schematic of the model points in a spindle cross section is shown to indicate how neighbor density analysis (NDA) (Figure 4F) and spindle volume (Figure 4G) are determined, both of which are discussed below. Overall, the IMOD software package (see Materials and Methods) has the tools for the analysis of microtubule arrays that make it possible to quantitatively distinguish between MI and MII spindles, as well as distinguish between different microtubule classes within a spindle.
Figure 4.
Representative meiotic spindle models (models MSI-9 and MSII-11; see Table 1). (A) The 3-D model of the microtubules in the MI spindle MSI-9. The supplemental data includes this image in a movie where the microtubules are color coded, allowing one to roll the model around the spindle axis. The supplemental data contains similar movies for all of the models and movies of parts of the EM data sets used to derive the models. (B) The 3-D model of the microtubules in the MII spindle MSII-11. The position of a spindle cross section (for 1 serial section of 50 nm) shown in F and G is indicated by a box (∼40 sections made up the spindle). This spindle model is deconstructed into putative kinetochore microtubules (C) and the core spindle microtubules (D) as determined in Figure 6 (also see text). (E) A microtubule overlap graph showing the length of each microtubule from one SPB or the other with two microtubules at the bottom whose polarity cannot be determined (“continuous microtubules”; see text). The microtubule overlap graphs for all spindle models are in the supplemental data. (F and G) Representation of the positions of microtubules in a single cross-section of spindle MSII-11 (position shown in B) where the polarity of the microtubules is indicated by a circle for microtubules from one SPB, a triangle for microtubules from the other SPB, and squares for microtubules that are continuous. (F) Neighbor density analysis used in Figure 7 is done by drawing concentric circles (gray) of user-defined radii (20 nm) around each microtubule and recording the number of microtubules in each ring created by the circles. (G) The outermost microtubules are used to define a polygon whose area is the cross sectional area for this section of the spindle. These areas are multiplied by section thickness to determine a volume, and the volumes are summed for all of the sections yielding the spindle volume (see Figure 5).
Distinguishing MI from MII Spindles
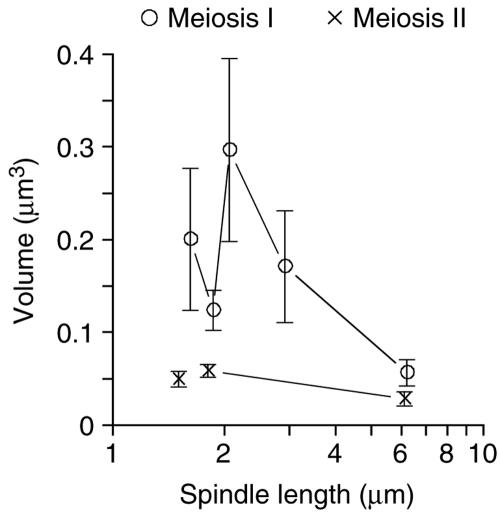
As discussed above, MI is a special type of chromosome segregation that separates paired homologous chromosomes instead of separating paired sister chromatids as in MII or in mitosis. For this reason, we thought that the organization of microtubules in the MI spindle would be unique and would be informative about how these spindles function. The MI spindles have more microtubules than MII spindles, but the difference is not significant enough to distinguish between the two spindle types. However, we observed that MI spindles seem to have several microtubules with trajectories that are at a greater angle from the spindle axis than observed in other yeast spindles (compare Figure 4, A and B). This gives the MI spindles the appearance that they occupy a larger volume than MII spindles. To develop statistics to describe these aspects of spindle structure, we measured the angle between each microtubule and the spindle axis. The distribution of these angles made it clear that MI spindles contain many more splayed out microtubules with trajectories away from the spindle axis (our unpublished data). This difference is apparent when the volume of the spindle is calculated by determining the cross-sectional area for each section of spindle (Figure 4G), converting the area to a volume by incorporating the section thickness and then summing the section volumes for the entire spindle (Figure 5). The MI spindles occupy significantly more volume at spindle lengths of 2–3 μm than do MII spindles of the same length.
Figure 5.
Spindle volumes distinguish MI (○) from MII (×) spindles. Spindle volumes were determined as described in Figure 4 and volumes of spindles of similar length were grouped together. Average spindle volumes are plotted versus spindle length. Error bars represent 74% confidence interval.
Functional Classes of Microtubules
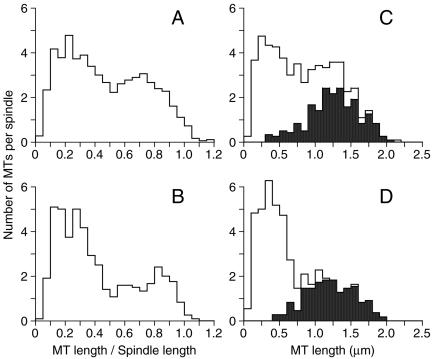
Beyond identifying microtubule organization in spindles, it is critical to identify the distinct classes of microtubules to understand their function, to decipher mutant phenotypes, and to interpret immunolocalization of components. We are interested in two functional classes: kinetochore microtubules that attach to the chromosomes and nonkinetochore microtubules that form the central spindle. No structures that could be identified as kinetochores were observed to be associated with MI or MII spindles, so we needed to identify putative kinetochore microtubules via a different method. In the case of mitotic spindles, yeast kinetochore microtubules are among the shortest microtubules in the spindle and show little or no association with microtubules from the other SPB (Winey et al., 1995). Interestingly, if microtubule lengths as a percentage of spindle length are plotted for the aggregate MI spindles up to 3.01 μm and the MII spindles up to 1.99 μm in length (Figure 6, A and B, respectively), two populations of microtubules are observed. One population has lengths of approximately one-half of the spindle length or less, and the other consists of microtubules longer than one-half the spindle length. If the distribution of shorter (50% of spindle length or shorter) versus longer microtubules is examined in individual spindles, the surprising observation is that the spindles contain very nearly 32 shorter microtubules in both MI and MII spindles. This number coincides perfectly with the number of kinetochores that need to be attached to the spindle if one microtubule attaches to each kinetochore in meiosis, as is observed in mitosis.
Figure 6.
Putative kinetochore microtubules seem to be the shortest microtubules and to have the least amount of pairing between microtubules in the spindle models. Histograms of the number of microtubules per spindle sorted by the ratio of microtubule length to spindle length for MI spindles (A) and MII (B) spindles normalizes microtubule lengths across all of the spindle models and reveals two classes of microtubules, where the relatively short microtubules (≤0.5 of spindle length) are thought to be kinetochore microtubules (see text). Histograms of the number of microtubules per spindle sorted by microtubule lengths for MI spindles (C) and MII spindles (D). The shaded areas indicate microtubules with significant pairing (lengths of ≥0.2 μm in MI spindles and of >0.3 μm in MII spindles) with microtubules from the opposing SPB. Here again, the shorter microtubules without significant pairing (unshaded) seem to be kinetochore microtubules and number very near 32 per spindle (1 per kinetochore; see text).
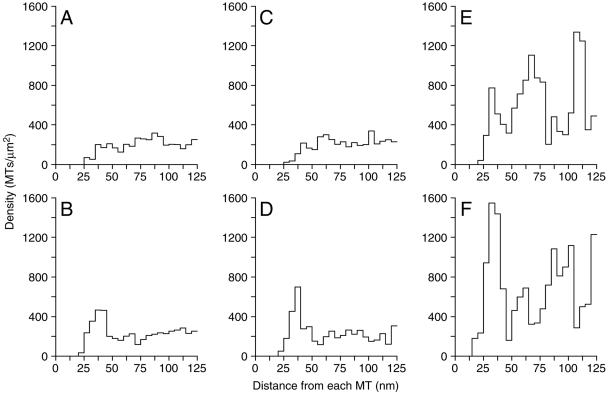
A second method to distinguish functional classes of microtubules is to identify those microtubules that form the central spindle by virtue of their interaction with microtubules from the other SPB. The initial part of this analysis was done by NDA (Figure 4F), wherein the density of microtubules in the vicinity of a given test microtubule is determined by finding the number of microtubules in concentric rings of increasing radius around the test microtubule. The analysis is usually limited to the spindle midzone where the concentration of the central spindle microtubules is high and the density of putative kinetochore microtubules is low. Such an analysis of the central spindle regions of the MII spindles showed a significant preferred distance between antiparallel microtubules. The MII spindles were examined in three different size classes, and the NDA was determined for both the parallel and antiparallel microtubules in the spindle midzone (Figure 7). In all size classes, a strong preference of ∼40 nm was detected between the antiparallel microtubules, particularly at longer spindle lengths (Figure 7, D and F). For the longest MII spindles, a preferred packing distance of ∼35 nm was detected among parallel microtubules (those microtubules from the same SPB; Figure 7, A, C, and E). This interaction is less significant than the interaction with antiparallel microtubules that yields a peak of ∼70 nm between parallel microtubules with an antiparallel microtubule between them. This finding suggests that the interdigitated microtubules of the central spindle in MII anaphase spindles are packed with a fixed distance between them.
Figure 7.
Antiparallel microtubules in the middle of MII spindles show a preferred packing distance. Neighbor density analysis (see Figure 4F) of spindle midzone microtubules was examined in three size classes of MII spindles (1.47–1.62 μm, A and B; 1.7–2.0, C and D; >2.0 μm, E and F) and plotted as histograms showing the density of microtubules (x-axis) at given distances from the reference microtubule (y-axis, the reference microtubule is the one in the center in the Figure 4F schematic). The data for each microtubule in the models being used as the reference microtubule is summed. No significant preferred packing was detected between microtubules from the same SPB (parallel) in the shorter spindles (A and C), but a peak of ∼35 nm was seen in the longest spindles (E). A preferred distance of ∼40 nm is observed between microtubules from opposite SPBs (antiparallel) at all spindle lengths (B, D, and F). This distance of 40 nm is particularly apparent in longer spindle lengths (D and F). Such organization is not apparent in MI spindles (see text; our unpublished data).
Similar analysis of the central spindle microtubules of the spindle midzone of the MI spindles revealed a similar preferred packing distance between the interdigitated antiparallel microtubules for some of the longer spindles (our unpublished data). In keeping with the generally disorganized look of the MI spindles, the degree of MI spindle organiaztion was more variable than in MII spindles, particularly in the shorter MI spindles. Further analysis of these interactions aimed at determining whether the packing had a particular organization (e.g., square or hexagonal) was inconclusive. Nonetheless, the preferred packing distance can be used for determining the length over which the interacting microtubules pair.
The apparent preferred spacing between antiparallel microtubules suggested the possibility that one microtubule is paired over a significant interval with an antiparallel partner. We determined the distance over which the two microtubules are close to each other, which is called the pairing distance (Winey et al., 1995). The pairing distances between all potential antiparallel combinations of microtubules were examined in both MI and MII spindles of lengths that likely correspond to metaphase. The microtubule lengths are plotted in a histogram, and microtubules with significant antiparallel interactions are shaded black, whereas microtubules lacking antiparallel interactions are unshaded (MI, Figure 6C, pairing ≥0.2 μm; MII, Figure 6D, pairing >0.3 μm). This analysis shows that nearly all of the longer microtubules in MII spindles have significant pairing interactions with microtubules from the other SPB, consistent with the idea that these microtubules make up the central spindle and are unlikely to interact with the kinetochores. As predicted by the above-mentioned analysis of relative microtubule length, the shorter microtubules do not have significant antiparallel interactions (Figure 6D, unshaded) and are potential kinetochore microtubules. Each MII spindle model contains a sufficient number of kinetochore microtubules to allow for one microtubule per kinetochore. The pairing analysis of MI spindles was less effective at determining which microtubules might be the kinetochore microtubules. The paired microtubules in the MI spindles are the longer microtubules (Figure 6C), but there are many longer microtubules that do not show significant pairing interactions leaving open the possibility that they are kinetochore microtubules. If this is so, the number of potential kinetochore microtubules (short and long unpaired microtubules) identified in the shorter MI spindles exceeds the number of kinetochores. However, these excess microtubules show no sign of bundling in a parallel orientation that might indicate kinetochore fibers of greater than one microtubule in MI spindles.
Together, these analyses of the spindle models indicate that kinetochores are attached to MI and MII spindles via a single microtubule, as is seen in mitotic spindles (Winey et al., 1995). In MII spindles, the kinetochore microtubules can be easily distinguished from core spindle microtubules based on pairing interactions. The microtubules in MI spindles are more difficult to separate into functional classes. However, the longer, presumptive anaphase I, spindles do show signs of an organized core spindle. MI spindles contain more than enough unpaired microtubules to have one microtubule for each paired set of sister chromatids with extra, unpaired microtubules, but not enough microtubules to have a microtubule for each chromatid of the paired homologous chromosomes.
Preanaphase Spindles versus Anaphase Spindles
The identification of spindle microtubules of different functional classes made it possible to explore how these classes of microtubules change as meiotic segregation progresses. We were unable to sample very short spindles to analyze spindle assembly, but we have sampled longer spindles making it possible to address questions about progression from metaphase into anaphase A and B. The analysis of MI and MII spindle models reveals that both meiotic stages feature anaphase B, the separation of the SPBs, wherein the spindles triple or more in length (Table 1 and Figure 2). As the spindles lengthen, the number of interdigitated central spindle microtubules decline by 35–40% in both MI and MII spindles, suggesting that some of the microtubules recruited to form the central spindle are stable, whereas other central spindle microtubules are lost as the spindles elongate. The stable microtubules must also elongate at the plus-end to maintain the zone of overlap of the antiparallel microtubules. For the four longest spindles, these microtubules grow to a mean length that is 57–63% of the length of these spindles. These characteristics of meiotic spindles are commonly found in mitotic spindles as well (Winey et al., 1995).
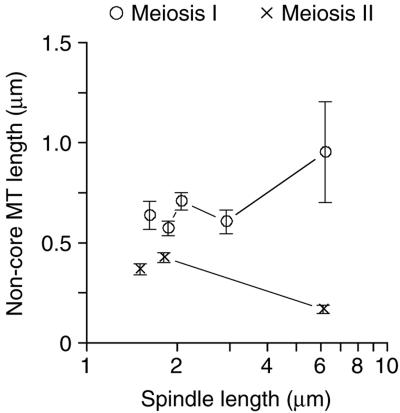
Anaphase A, movement of the chromosomes toward the SPBs, is not so obvious as anaphase B in the models of meiotic spindles. This movement would be inferred from observing the shortening of the putative kinetochore microtubules. Plotting the average lengths of the microtubules that do not show antiparallel interactions (identified in Figure 6, C and D; see above) versus spindle length (Figure 8) does show that the putative kinetochore microtubules get shorter in MII spindles, indicating that anaphase A occurs in MII, concurrent with anaphase B (increasing pole-to-pole distance as seen by spindle length). Furthermore, the kinetochore microtubule length in the longer spindles is artificially high because we did not correct for kinetochore microtubules of “zero” length that we cannot detect in serial thin sections (50 nm or shorter in length), but they are expected to be there based on electron tomographic analysis of mitotic spindles and spindle pole bodies (O'Toole et al., 1999). We also expect that these spindles contain very short microtubules because only one short microtubule (<1 μm) was detected in the two longest MII spindles. These data indicates that anaphase A occurs in meiosis II.
Figure 8.
Apparent kinetochore microtubules diminish in length as MII proceeds indicating anaphase A movements. Plotting the mean length of noncore spindle microtubules in different spindle length classes versus spindle length shows that these microtubules shorten during MII (×), but not during MI (○). This analysis can be biased by a population of stable noncore microtubules, or by the loss (below detection) of microtubules from the spindle models (see text). Error bars represent 74% confidence interval.
The plot of putative kinetochore microtubule length versus spindle length does not show a clear MI anaphase A (Figure 8). However, these spindles also are expected to contain fewer putative kinetochore microtubules if anaphase A occurs because some of the microtubules would become too short to detect as discussed above. Examination of the number of nonpaired microtubules in the MI spindles clearly indicates that anaphase A occurs. MI spindles up to ∼3.0 μm in length contain ∼35 nonpaired microtubules, whereas the two longest MI spindles (∼6.0 μm) average 10 nonpaired microtubules of which only six microtubules in each spindle are <1.0 μm in length. The significant loss (80%) of short, nonpaired microtubules indicates that these microtubules, probably attached to kinetochores, do shrink during the course of meiosis I, hence indicating that anaphase A does occur in these spindles. Like MII and mitotic spindles, it seems that anaphase A and B are concurrent in meiosis I.
DISCUSSION
We report fine structure description and analysis of the microtubule arrays in 35 meiotic spindles in a wild-type yeast strain. The identification of meiotic cells was based on known cytological markers and distinguishing between MI and MII spindles in wild-type cells was straight forward because all yeast meiotic segregation occurs in a single nucleus. Although our data set has representatives for most of the spindle length classes that can be detected by light microscopy, it is lacking in very short spindles. The more complete analysis of mitotic spindles in yeast has included reconstruction from serial thin sections (Winey et al., 1995), stereopair analysis of entire spindles in thick sections (Peterson and Ris, 1976), and electron tomography (O'Toole et al., 1999). The analysis presented here based on reconstruction from serial thin sections is the only high-resolution, 3-D structural information available on yeast meiotic spindles.
The analysis of microtubule numbers and spatial organization in wild-type MI and MII spindles revealed features of these spindles that are similar with each other and with mitotic spindles. First, spindle attachment to kinetochores is via a single microtubule in all three spindle types. This inference is made on the basis that no bundling of presumptive kinetochore microtubules is detected in either MI or MII spindles, as observed previously in mitotic spindles (Winey et al., 1995). This notion is supported by the fact that most half spindles (microtubules from one SPB) in the data set contain enough microtubules to have one per kinetochore (16) plus some extra microtubules to form the central spindle of overlapping antiparallel microtubules, but not 32 microtubules (2 per kinetochore) plus extra for the central spindle. Second, analysis of microtubule lengths and interactions with microtubules from the opposing SPB allows for the identification of functionally distinct microtubules—the shorter microtubules that presumably bind kinetochores and the longer microtubules that interdigitate with microtubules from the opposing SPB to form the central spindle. Although the distinction is less clear in the MI spindles, both MI and MII spindles have these two classes of microtubules that were previously reported for mitotic spindles (Winey et al., 1995).
The identification of the distinct classes of microtubules with their putative functions allows us to begin to model the events of meiotic segregation. As with mitosis in budding yeast, a clear metaphase configuration for the microtubules in MI or MII is not evident. An expectation of metaphase could be a well defined central spindle with kinetochore microtubules of relatively equal length that nearly meet at the spindle midzone, separated only by the paired chromosomes to which they are attached. This is not observed in mitotic cells because of the dynamic “breathing” of centromeric regions of chromosomes that have a bipolar attachment (reviewed in Winey and O'Toole, 2001). We predicted separation of attached kinetochores in mitotic cells based on electron microscopic data similar to what is presented here (Winey et al., 1995). Therefore, the failure to observe metaphase in MI or MII leads us to conclude that centromere breathing is likely to occur in both MI and MII as it does in mitosis. Shonn et al. (2003) have reported that the centromeres of homologous chromosomes do separate on meiotic metaphase spindles by using GFP-tagged centromeres. Finally, analysis of plus-end-to-plus-end distances for presumptive kinetochore microtubules did not yield a satisfying answer for what the extent of kinetochore separation might be (our unpublished data).
Beyond metaphase, both anaphase A and B are detected during MI and MII. Anaphase A is difficult to discern in MI spindles because the function, if any, of the excess, unpaired microtubules is unknown (see below). An alternative hypothesis is that anaphase A does not occur in MI and that the loss of microtubules in these spindles as they elongate is from loss of the excess microtubules. However, the longest MI spindles have too few kinetochore microtubules (<16/half spindle), suggesting that some of the kinetochore microtubules have shortened enough as to be undetectable (O'Toole et al., 1999), therefore constituting anaphase A movement. As in mitosis, anaphase A and B are concurrent in MI and MII. Also similar to mitosis, anaphase B involves increased organization of the core bundle of microtubules along with increased length of those microtubules. Therefore, microtubule shortening of the kinetochore microtubules and of some of the core bundle microtubules, and microtubule lengthening in the central spindle are occurring simultaneously in MI and MII spindles, similar to mitotic spindles. There is little information on how this is achieved, but in mitotic cells it is likely the result of differential plus-end behavior because minus-end flux at the spindle pole body has not been observed in yeast (Maddox et al., 2000). Finally, we were able to observe the unique situation of having two spindles in a single nucleus, as is the case in MII (reviewed in Byers, 1981). We found that the microtubules from one MII spindle do not interact with the microtubules from the other MII spindle, and this is likely achieved by significant separation of the spindles in space.
A major impetus for this study was the supposition that the unique segregation of paired homologous chromosomes by the MI spindle might be reflected in the microtubule organization of these spindles. It is clear from our work that the MI spindle is uniquely organized and very different from MII or mitotic spindles. However, the odd, somewhat random-looking organization of microtubules in the MI spindles raises more questions than it answers about how this spindle functions. Two prominent questions emerge upon inspection of the MI models. The first has to do with the nature of the attachment of the paired homologous chromosomes. If attachment is via a single kinetochore microtubule, as we assert above, how is that accomplished? Each pair of sister chromatids would have two potential kinetochores, such that there would be four kinetochores on the paired homologous chromosomes in a MI spindle, yet only two of the kinetochores capture microtubules. If this is the case, perhaps the “monopolin” complex (Toth et al., 2000; Rabitsch et al., 2003) contributes to silencing one of the two sister kinetochores, which predicts that mutants in MAM1 or the other components would exhibit increased kinetochore attachment, including the erroneous bipolar attachment of sisters. Alternatively, the sister chromatids in the paired homologous chromosomes could have a shared single kinetochore, as observed in Drosophila (Goldstein, 1981), possibly created by the action of the monopolin complex (Toth et al., 2000; Rabitsch et al., 2003). Because it is difficult to identify kinetochore microtubules in MI and because we cannot recognize kinetochore structures by EM in MI spindles, our analysis cannot resolve the issue.
The second question arising from the models of MI spindles concerns the function, if any, of the additional microtubules in these spindles. mRNA levels for the tubulin genes transcripts do increase during meiosis (Chu et al., 1998), suggesting that the tubulin pools increase to accommodate the large amount of polymer in MI spindles and to accommodate the two MII spindles. The excess microtubules in the MI spindles are additional in the sense that there are more microtubules than necessary to contact all of the kinetochores and to have a few extra for the formation of the central spindle based on the apparent organizational principles of MII and mitotic spindles. There is no evidence for MI kinetochore fibers of more than one microtubule, which would require more microtubules in the spindles. The excess microtubules may reflect a “search and capture” mechanism that is needed for MI chromosome attachments. Mitotic cells seem to have kinetochore attachments outside of mitosis because nuclear microtubules are always present (reviewed in Winey and O'Toole, 2001), and it is plausible that kinetochore attachments are maintained during the transition from MI to MII, and during MII. However, the paired homologous chromosomes in meiotic prophase may not be attached requiring capture by microtubules in preparation for MI. This process would be more efficient if more microtubules were “probing” the nucleoplasm for kinetochores to capture, such that the MI spindles end up with the additional microtubules. In keeping with this, centromeres are clustered as cells enter meiosis but are dispersed in prophase (Hayashi et al., 1998; Jin et al., 1998; Trelles-Sticken et al., 1999). So it may indeed be true that microtubules have to search a greater volume to find kinetochores in meiotic prometaphase than in mitosis or MII. This model of MI chromosome attachment that explains the high microtubule numbers also might explain why these spindles are more sensitive to the loss of the spindle assembly checkpoint than MII or mitotic spindles (Shonn et al., 2000, 2003). In the latter cases, the existing attachments have to be disrupted to allow one to observe a requirement for the checkpoint, whereas the MI attachment would be more like that of a vertebrate cell where there is no attachment, and it requires the checkpoint to block mitotic (meiotic, in this case) progression until all of the kinetochores are attached. As such, mutations in the checkpoint would lead to chromosome missegregation in otherwise normal cells as is observed in checkpoint defective (mad1Δ or mad2D) yeast strains (Shonn et al., 2000, 2003).
There is little doubt that much is yet to be learned about the structure, function, and regulation of meiotic spindles. This initial structural analysis of meiotic spindles has revealed a number of features that need further mechanistic explanation. Future analysis can also include other high-resolution imaging tactics, such as electron tomography, to examine stages of meiotic spindle assembly not revealed in this study. Nonetheless, the data presented here provides a structural foundation for yeast meiotic spindle function that will be useful in interpreting mutant phenotypes and in understanding the localization of components of the meiotic apparatus.
Supplementary Material
Acknowledgments
We appreciate the gift of strains from Nancy Hollingsworth. We are grateful to Kim Nasmyth for the communication of results before publication. Discussions with Dean Dawson and Bill Saunders were very valuable, and we thank Dean Dawson for reading the manuscript. We thank the staff of the Boulder Laboratory for 3-D Electron Microscopy of Cells (P.I. J. Richard McIntosh) for help, particularly Eileen O'Toole for training, and they are supported by RR-00592. This work was primarily supported by research grant no. 1-FY00807 from the March of Dimes Birth Defects Foundation (to M. W.). P.D.S. was a National Institutes of Health trainee (GM-07135).
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E04-09-0765) on January 5, 2005.
Abbreviations used: 3-D, three-dimensional; EM, electron microscopy; MI, meiosis I; MII, meiosis II; SPB, spindle pole body.
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
References
- Buonomo, S. B., Rabitsch, K. P., Fuchs, J., Gruber, S., Sullivan, M., Uhlmann, F., Petronczki, M., Toth, A., and Nasmyth, K. (2003). Division of the nucleolus and its release of CDC14 during anaphase of meiosis I depends on separase, SPO12, and SLK19. Dev. Cell 4, 727-739. [DOI] [PubMed] [Google Scholar]
- Byers, B. (1981). Cytology of the yeast life cycle. In: Molecular Biology of the Yeast, Saccharomyces. I. Life Cycle and Inheritance, ed. J. N. Strathern, E. W. Jones, and J. R. Broach, Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press, 59-96.
- Chu, S., DeRisi, J., Eisen, M., Mulholland, J., Botstein, D., Brown, P. O., and Herskowitz, I. (1998). The transcriptional program of sporulation in budding yeast. Science 282, 699-705. [DOI] [PubMed] [Google Scholar]
- Giddings, T. H., Jr., O'Toole, E. T., Morphew, M., Mastronarde, D. N., McIntosh, J. R., and Winey, M. (2001). Using rapid freeze and freeze-substitution for the preparation of yeast cells for electron microscopy and three-dimensional analysis. Methods Cell Biol. 67, 27-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein, L. S. (1981). Kinetochore structure and its role in chromosome orientation during the first meiotic division in male D. melanogaster. Cell 25, 591-602. [DOI] [PubMed] [Google Scholar]
- Hayashi, A., Ogawa, H., Kohno, K., Gasser, S. M., and Hiraoka, Y. (1998). Meiotic behaviours of chromosomes and microtubules in budding yeast: relocalization of centromeres and telomeres during meiotic prophase. Genes Cells 3, 587-601. [DOI] [PubMed] [Google Scholar]
- Jin, Q., Trelles-Sticken, E., Scherthan, H., and Loidl, J. (1998). Yeast nuclei display prominent centromere clustering that is reduced in nondividing cells and in meiotic prophase. J. Cell Biol. 141, 21-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knop, M., and Strasser, K. (2000). Role of the spindle pole body of yeast in mediating assembly of the prospore membrane during meiosis. EMBO J. 19, 3657-3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, B., and Amon, A. (2001). Meiosis: how to create a specialized cell cycle. Curr. Opin. Cell Biol. 13, 770-777. [DOI] [PubMed] [Google Scholar]
- Lee, B. H., and Amon, A. (2003). Polo kinase–meiotic cell cycle coordinator. Cell Cycle 2, 400-402. [PubMed] [Google Scholar]
- Maddox, P. S., Bloom, K. S., and Salmon, E. D. (2000). The polarity and dynamics of microtubule assembly in the budding yeast Saccharomyces cerevisiae. Nat. Cell. Biol. 2, 36-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marston, A. L., Lee, B. H., and Amon, A. (2003). The Cdc14 phosphatase and the FEAR network control meiotic spindle disassembly and chromosome segregation. Dev. Cell 4, 711-726. [DOI] [PubMed] [Google Scholar]
- McDonald, K., O'Toole, E. T., Mastronarde, D., Winey, M., and McIntosh, R. (1996). Mapping the three-dimensional organization of microtubules in mitotic spindles of yeast. Trends Cell Biol. 6, 235-239. [DOI] [PubMed] [Google Scholar]
- Nasmyth, K. (2001). Disseminating the genome: joining, resolving, and separating sister chromatids during mitosis and meiosis. Annu. Rev. Genet. 35, 673-745. [DOI] [PubMed] [Google Scholar]
- Neiman, A. (1998). Prospore membrane formation defines a developmentally regulated branch of the secretory pathway in yeast. J. Cell Biol. 140, 29-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Toole, E. T., Winey, M., and McIntosh, J. R. (1999). High-voltage electron tomography of spindle pole bodies and early mitotic spindles in the yeast Saccharomyces cerevisiae. Mol. Biol. Cell 10, 2017-2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson, J., and Ris, H. (1976). Electron-microscopic study of the spindle and chromosome movement in the yeast Saccharomyces cerevisiae. J. Cell Sci. 22, 219-242. [DOI] [PubMed] [Google Scholar]
- Rabitsch, K. P., Petronczki, M., Javerzat, J. P., Genier, S., Chwalla, B., Schleiffer, A., Tanaka, T. U., and Nasmyth, K. (2003). Kinetochore recruitment of two nucleolar proteins is required for homolog segregation in meiosis I. Dev. Cell 4, 535-548. [DOI] [PubMed] [Google Scholar]
- Shonn, M. A., McCarroll, R., and Murray, A. W. (2000). Requirement of the spindle checkpoint for proper chromosome segregation in budding yeast meiosis. Science 289, 300-303. [DOI] [PubMed] [Google Scholar]
- Shonn, M. A., Murray, A. L., and Murray, A. W. (2003). Spindle checkpoint component Mad2 contributes to biorientation of homologous chromosomes. Curr. Biol. 13, 1979-1984. [DOI] [PubMed] [Google Scholar]
- Smith, G. R. (2001). Homologous recombination near and far from DNA breaks: alternative roles and contrasting views. Annu. Rev. Genet. 35, 243-274. [DOI] [PubMed] [Google Scholar]
- Stegmeier, F., Visintin, R., and Amon, A. (2002). Separase, polo kinase, the kinetochore protein Slk19, and Spo12 function in a network that controls Cdc14 localization during early anaphase. Cell 108, 207-220. [DOI] [PubMed] [Google Scholar]
- Straight, P. D., Giddings, T. H., Jr., and Winey, M. (2000). Mps1p regulates meiotic spindle pole body duplication in addition to having novel roles during sporulation. Mol. Biol. Cell 11, 3525-3537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toth, A., Rabitsch, K. P., Galova, M., Schleiffer, A., Buonomo, S. B., and Nasmyth, K. (2000). Functional genomics identifies monopolin. A kinetochore protein required for segregation of homologs during Meiosis I. Cell 103, 1155-1168. [DOI] [PubMed] [Google Scholar]
- Trelles-Sticken, E., Loidl, J., and Scherthan, H. (1999). Bouquet formation in budding yeast: initiation of recombination is not required for meiotic telomere clustering. J. Cell Sci. 112, 651-658. [DOI] [PubMed] [Google Scholar]
- Winey, M., Mamay, C., O'Toole, E. T., Mastronarde, D., Giddings, T., McDonald, K., and McIntosh, J. R. (1995). Three dimensional untrastructural analysis of the Saccharomyces cerevisiae mitotic spindle. J. Cell Biol. 129, 1601-1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winey, M., and O'Toole, E. T. (2001). The spindle cycle in budding yeast. Nat. Cell Biol. 3, E23-27. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.