Abstract
High-dose methotrexate (HDMTX) plays an important role in the treatment of acute lymphoblastic leukemia (ALL) although there is great inter-patient variability in the efficacy and toxicity of MTX. The relationship between polymorphisms in genes encoding MTX transporters and MTX response is controversial. In the present study, 322 Chinese children with standard- and intermediate-risk ALL were genotyped for 12 polymorphisms. SLCO1B1 rs10841753 showed a significant association with plasma MTX levels at 48 h (P = 0.017). Patients who had the ABCB1 rs1128503 C allele had longer duration of hospitalization than did those with the TT genotype (P = 0.006). No association was found between oral mucositis and any polymorphism. Long-term outcome was worse in patients with the SLCO1B1 rs4149056 CC genotype than in patients with TT or TC (5-year event-free survival [EFS] 33.3 ± 19.2% vs. 90.5 ± 1.7%, P < 0.001), and was worse in patients with the SCL19A1 rs2838958 AA genotype than in patients with AG or GG (5-year EFS 78.5 ± 4.6% vs. 92.2 ± 1.8%, P = 0.008). Multiple Cox regression analyses revealed associations of minimal residual disease (MRD) at day 33 (hazard ratio 3.458; P = 0.002), MRD at day 78 (hazard ratio 6.330; P = 0.001), SLCO1B1 rs4149056 (hazard ratio 12.242; P < 0.001), and SCL19A1 rs2838958 (hazard ratio 2.324; P = 0.019) with EFS. Our findings show that polymorphisms in genes encoding MTX transporters substantially influence the kinetics and response to HDMTX therapy in childhood ALL.
Keywords: methotrexate, acute lymphoblastic leukemia, polymorphism, SLCO1B1, SLC19A1
INTRODUCTION
Methotrexate (MTX) has been widely used as an anticancer agent in most chemotherapy protocols for childhood acute lymphoblastic leukemia (ALL). Despite its clinical success, MTX may cause serve adverse effects and toxicities [1–3]. MTX pharmacokinetics, efficacy, and toxicity have great inter-patient variability. This diversity can be partly explained by sequence differences in genes encoding MTX membrane transporter proteins [4–6].
MTX enters the cell via the solute carrier family 19 member 1 (SLC19A1) or the solute carrier organic anion transporter 1B1 (SLCO1B1) [7, 8]. It is exported from the cells by ATP binding cassette (ABC) transporters, including ABC subfamily B member 1 (ABCB1), and ABC subfamily G member 2 (ABCG2) [4]. Previous studies suggest that variations in single nucleotide polymorphisms (SNPs) of genes encoding MTX transporters play a great role in ALL prognosis and MTX response [9–15], but the results are inconsistent, which might result from small study cohorts, heterogeneous study populations, and differences in treatment protocols.
Therefore, the main goal of the present study was to investigate the influence of SNPs on the pharmacokinetics and toxicity of high-dose MTX (HDMTX), as well as clinical outcome in ALL patients. A total of 12 SNPs in four genes involved in the MTX transport pathway (SLCO1B1, SLC19A1, ABCB1, and ABCG2) were selected. The association study was carried out in 322 Chinese children with ALL treated according to the Beijing Children's Hospital (BCH)-2003 and Chinese Childhood Leukemia Group (CCLG)-2008 protocols.
RESULTS
Patient and genotyping characteristics
Patient characteristics are shown in Supplementary Table 1. No significant differences were observed between patients included (n = 322) and those not included (n = 142) in this study.
We genotyped 12 SNPs in four genes (SLCO1B1, SLC19A1, ABCB1, and ABCG2). The selected SNPs and allele frequencies are shown in Supplementary Table 2 with an average call rate of 98.23%. ABCB1 rs2032582 was excluded from the following analyses because its allele frequencies deviated from HWE.
SNPs with pharmacokinetics
To explore the impact of SNPs on MTX pharmacokinetics, we tested the association between selected SNPs and MTX plasma level at 48 h from the first dose of HDMTX infusion. Plasma MTX data were available for 317 patients with the range from 0.09 μmol/L to 41.63 μmol/L (median: 0.45 μmol/L). We found a statistically significant association between plasma MTX level and the SLCO1B1 rs10841753. Patients with CC genotypes had a lower MTX plasma level than did patients with the T allele (CT+TT) (P = 0.017) (Table 1 and Supplementary Figure 2A). The MTX plasma concentration at 48 h was higher in patients with the SLCO1B1 rs4149056 CC genotype compared with those with the T allele (TC+TT) (1.1μmol/L vs. 0.45 μmol/L, P = 0.441), and was lower in patients with the SLCO1B1 rs11045879 TT genotype compared with those with the C allele (CC+TC) (0.39 μmol/L vs.0.50 μmol/L, P = 0.287). The trends were consistent with previous studies, but the association did not reach statistical significance (Table 2 and Supplementary Figure 2B and 2C). Both of the patients who died after receiving the first course of HDMTX had a high plasma MTX level at 48 h (13.09 μmol/L and 8.93 μmol/L, respectively). Their genotypes for SLCO1B1 rs10841753, rs4149056, rs11045879, and rs2306283 were TC, TT, TC, TC, and TT, TC, TC, CC, respectively. We also analyzed the association between polymorphisms and MTX plasma levels during administration of the four courses by repeated measures ANOVA; however, none of the associations was significant.
Table 1. BCH-2003 and CCLG-2008 treatment protocol.
| Stratification Criteria | Treatment Block | |||||||
|---|---|---|---|---|---|---|---|---|
| Remission induction |
Early intensification |
Consolidation | Delay Intensification I |
Maintenance I |
Delay intensification II | Maintenance II |
||
| BCH-2003 | ||||||||
| SR | 1 ≤ age < 6 WBC < 20 × 109/L Good prednisone response No T-cell and not mature B-cell No t(9;22) or MLL rearrangements Bone marrow morphology was M1 at day 33 |
VDLP (DNR×2) |
CAM | HDMTX 3g/m2×4 |
VDLD+CAM | 6-MP+MTX/VD+IT | HDMTX | 6-MP+MTX/VD+IT |
| IR | Good prednisone response No t(9;22) Bone marrow morphology was M1 at day 33 Any one of : Age ≥ 6 or < 1 WBC ≥ 20 × 109/L T-cell MLL rearrangement |
VDLP (DNR×4) |
CAM×2 | HDMTX 5g/m2×4 |
VDLD+CAM | 6-MP+MTX/VD+IT | VDLA+VM26+ HD-Ara-c |
6-MP+MTX/VD+IT |
| CCLG-2008 | 1 ≤ age < 10 WBC < 50 × 109/L Good prednisone response No T-cell and not mature B-cell |
|||||||
| SR | No t(9;22), t(1;19) or MLL rearrangements Bone marrow morphology was M1/M2 at day 15 and M1 at day33 No CNSL MRD < 10–4 at day 33 |
VDLP (DNR×2) |
CAM | HDMTX 2g/m2×4 |
VDLD+CAM | / | / | 6-MP+MTX/VD+IT |
| IR | Good prednisone response No t(9;22) or MLL rearrangements Bone marrow morphology at day 15 was M1/M2 with IR protocol or M3 with SR protocol MRD < 10–2 at day 33 and < 10–3 at day 78 Any one of : Age ≥ 10 or < 1 WBC ≥ 50 × 109/L T-cell CNSL with no other high risk factor |
VDLP (DNR×4) |
CAM×2 | HDMTX 5g/m2×4 |
VDLD+CAM | 6-MP+MTX | VDLD+CAM | 6-MP+MTX/VD+IT |
WBC: white blood cell; SR: standard risk; IR: Intermediate-risk. MRD: minimal residual disease. CNSL: central nervous system leukemia.
Table 2. Association of selected SNPs with plasma levels of MTX at 48 h from the first MTX infusion.
| SNPs | Median (Min-Max) (μmol/L) | P |
|---|---|---|
| SLCO1B1 rs11045879 | 0.287 | |
| CC +TC | 0.50 (0.09–41.63) | |
| TT | 0.39 (0.12–19.88) | |
| SLCO1B1 rs4149056 | 0.441 | |
| TC+TT | 0.45 (0.09–41.63) | |
| CC | 1.1. (0.15–7.53) | |
| SLCO1B1 rs2306283 | 0.294 | |
| CC +TC | 0.45 (0.09–41.63) | |
| TT | 0.37 (0.12–3.91) | |
| SLCO1B1 rs10841753 | 0.017 | |
| TC+TT | 0.47 (0.09–41.63) | |
| CC | 0.32 (0.14–5.50) | |
| SLC19A1 rs1051266 | 0.384 | |
| AA +AG | 0.41 (0.09–34.05) | |
| GG | 0.59 (0.14–41.63) | |
| SLC19A1 rs3788200 | 0.491 | |
| AG+GG | 0.45 (0.09–41.63) | |
| AA | 0.47 (0.11–34.05) | |
| SLC19A1 rs1131596 | 0.219 | |
| TC+CC | 0.41 (0.09–34.05) | |
| TT | 0.6 (0.14–41.63) | |
| SLC19A1 rs2838958 | 0.259 | |
| AG+GG | 0.41 (0.09–34.05) | |
| AA | 0.59 (0.16–41.63) | |
| ABCB1 rs1128503 | 0.149 | |
| TC+CC | 0.47 (0.09–19.88) | |
| TT | 0.39 (0.12–41.63) | |
| ABCB1 rs1045642 | 0.340 | |
| TC+TT | 0.41 (0.12–41.63) | |
| CC | 0.46 (0.09–10.88) | |
| ABCG2 rs2231137 | 0.688 | |
| AG+GG | 0.45 (0.09–41.63) | |
| AA | 0.46 (0.15–10.29) |
SNP: single nucleotide polymorphism.
SNPs with MTX-induced toxicity and duration of hospitalization for HDMTX treatment
Oral mucositis is a common side effect attributed to the use of HDMTX. To investigate genetic determinants for oral mucositis after HDMTX, information regarding oral mucositis during the consolidation therapy period was available for 317 patients. Table 3 shows the risk evaluation for the development of grade 3–4 oral mucositis, calculated by univariate analysis. A total of 38 patients developed grade 3–4 oral mucositis (12.0%). No polymorphism was found to be associated with oral mucositis, although it occurred more frequently in patients with the ABCB1 rs1128503C allele (CC and TC) than in those with the TT genotype (OR 2.009, 95% CI 0.987–4.090, P = 0.054). With regard to SLCO1B1 rs4149056, patients with the C allele had a lower risk of developing mucositis. None of the six patients with the CC genotypes developed oral mucositis. The frequency was 8.47% for patients with TC, and 13.25% for those with TT; however, the differences among genotypes were not statistically significant (Table 3 and Supplementary Figure 3).
Table 3. Logistic regression analysis of association between selected SNPs and oral mucositis (grade 3–4).
| SNP (n) | Oral mucositis n (%) | OR (95% CI) | P |
|---|---|---|---|
| SLCO1B1 rs11045879 | |||
| CC (52) | 6 (11.54) | Reference | |
| TC (136) | 18 (13.24) | 1.177 (0.558–2.480) | 0.669 |
| TT (122) | 14 (11.46) | 1.006 (0.364–2.781) | 0.990 |
| TC+TT vs. CC | 0.921 (0.364–2.330) | 0.862 | |
| SLCO1B1 rs4149056 | |||
| CC (6) | 0 | Reference | |
| TC (59) | 5 (8.47) | 0.606 (0.226–1.626) | 0.320 |
| TT (249) | 33 (13.25) | 0 | 0.999 |
| TC+TT vs. CC | 0 | 0.999 | |
| SLCO1B1 rs2306283 | |||
| CC (170) | 23 (13.53) | Reference | |
| TC (115) | 13 (11.30) | 1.643 (0.361–7.478) | 0.521 |
| TT (23) | 2 (8.70) | 1.338 (0.281–6.375) | 0.715 |
| TC+TT vs. CC | 1.283 (0.641–2.566) | 0.481 | |
| SLCO1B1 rs10841753 | |||
| CC (33) | 4 (12.12) | Reference | |
| T C (120) | 17 (14.17) | 1.379 (0.672–2.828) | 0.381 |
| TT (159) | 17 (10.69) | 1.152 (0.361–3.676) | 0.811 |
| TC+TT vs. CC | 0.994 (0.329–3.001) | 0.991 | |
| SLC19A1 rs1051266 | |||
| AA (73) | 7 (9.59) | Reference | |
| A G(142) | 20 (14.08) | 1.252 (0.570–2.749) | 0.576 |
| GG (95) | 11 (11.58) | 0.810 (0.298–2.204) | 0.680 |
| AG+GG vs. AA | 0.705 (0.297–1.675) | 0.428 | |
| SLC19A1 rs3788200 | |||
| AA (73) | 7 (9.59) | Reference | |
| A G(143) | 20 (13.99) | 1.271 (0.579–2.789) | 0.549 |
| GG (97) | 11 (11.34) | 0.829 (0.305–2.255) | 0.714 |
| AG+GG vs. AA | 0.715 (0.301–1.699) | 0.448 | |
| SLC19A1 rs1131596 | |||
| TT (101) | 11 (10.89) | Reference | |
| T C(144) | 20 (13.89) | 1.117 (0.411–3.039) | 0.828 |
| CC (71) | 7 (9.86) | 1.475 (0.592–3.671) | 0.404 |
| TC+CC vs. TT | 0.851 (0.404–1.792) | 0.671 | |
| SLC19A1 rs2838958 | |||
| AA (89) | 10 (11.24) | Reference | |
| AG (145) | 19 (13.10) | 0.970 (0.373–2.526) | 0.951 |
| GG (78) | 9 (11.54) | 1.156 (0.496–2.693) | 0.737 |
| AG+GG vs. AA | 0.882 (0.409–1.900) | 0.748 | |
| ABCB1 rs1128503 | |||
| TT (153) | 13 (8.50) | Reference | |
| T C(121) | 18 (14.88) | 1.882 (0.882–4.014) | 0.102 |
| CC (38) | 7 (18.42) | 2.432 (0.897–6.596) | 0.081 |
| TC+CC vs. TT | 2.009 (0.987–4.090) | 0.054 | |
| ABCB1 rs1045642 | |||
| CC (118) | 14 (11.86) | Reference | |
| T C(148) | 18 (12.16) | 0.965 (0.348–2.676) | 0.945 |
| TT (49) | 6 (12.24) | 0.992 (0.370–2.660) | 0.988 |
| TC+TT vs. CC | 0.970 (0.481–1.959) | 0.933 | |
| ABCG2 rs2231137 | |||
| AA (30) | 5 (16.67) | Reference | |
| AG (123) | 13 (10.57) | 0.821 (0.391–1.724) | 0.603 |
| GG (159) | 20 (12.58) | 1.390 (0.478–4.406) | 0.546 |
| AG+GG vs. AA | 1.509 (0.54104.212) | 0.432 |
SNP: single nucleotide polymorphism; OR: odds ratio; CI: confidential interval.
To study the impact of SNPs on duration of hospitalization for HDMTX treatment, we analyzed the association between genotypes and average length of hospitalization for the four courses of HDMTX treatment. Table 4 shows that patients with the ABCB1 rs1128503 C allele had prolonged duration of hospitalization compared with those with the TT genotype (mean 5.92 vs. 5.51 days, P = 0.006).
Table 4. Association of selected SNPs with duration of hospitalization for HDMTX treatment.
| SNPs | Mean ± SD (days) | P |
|---|---|---|
| SLCO1B1 rs11045879 | 0.355 | |
| CC +TC | 5.77 ± 1.39 | |
| TT | 5.62 ± 1.21 | |
| SLCO1B1 rs4149056 | 0.634 | |
| TC+TT | 5.72 ± 1.33 | |
| CC | 5.46 ± 0.62 | |
| SLCO1B1 rs2306283 | 0.345 | |
| CC +TC | 5.73 ± 1.35 | |
| TT | 5.46 ± 1.04 | |
| SLCO1B1 rs10841753 | 0.988 | |
| TC+TT | 5.72 ± 1.34 | |
| CC | 5.72 ± 1.35 | |
| SLC19A1 rs1051266 | 0.544 | |
| AA +AG | 5.69 ± 1.20 | |
| GG | 5.79 ± 1.59 | |
| SLC19A1 rs3788200 | 0.339 | |
| AG+GG | 5.75 ± 1.44 | |
| AA | 5.58 ± 0.85 | |
| SLC19A1 rs1131596 | 0.522 | |
| TC+CC | 5.68 ± 1.20 | |
| TT | 5.79 ± 1.58 | |
| SLC19A1 rs2838958 | 0. 906 | |
| AG+GG | 5.70 ± 1.19 | |
| AA | 5.72 ± 1.62 | |
| ABCB1 rs1128503 | 0.006 | |
| TC+CC | 5.92 ± 1.37 | |
| TT | 5.51 ± 1.28 | |
| ABCB1 rs1045642 | 0.112 | |
| TC+TT | 5.63 ± 1.33 | |
| CC | 5.88 ± 1.34 | |
| ABCG2 rs2231137 | 0.094 | |
| AG+GG | 5.67 ± 1.29 | |
| AA | 6.18 ± 1.59 |
SNP: single nucleotide polymorphism.
Haplotypes with pharmacokinetics and MTX-induced toxicity
To test the association between haplotypes and MTX plasma levels/toxicity, we first determined the linkage disequilibrium (LD) block structure for each gene. Supplementary Figure 4 shows that SLCO1B1 rs10841753, rs2306283, rs4149056, and SLC19A1 rs3788200, rs1051266, rs1131596 were in LD, respectively. None of the haplotypes was associated with MTX level or MTX-induced oral mucositis (Supplementary Tables 3 and 4).
SNPs with treatment outcome
To define the influence of selected SNPs on treatment outcomes of ALL patients, we did both univariate and multivariate analyses. Univariate analyses showed that SLCO1B1 rs4149056 and SLC19A1 rs2838958 were significantly associated with the risk of events (P < 0.001 and 0.008, respectively). Compared with patients with the rs4149056 T allele (TC+TT), patients with the CC genotype had a poorer outcome (5-year EFS 33.3 ± 19.2% vs. 90.5 ± 1.7; Figure 1A). Among the six patients with the rs4149056 CC genotype, three patients relapsed and one died of infection. The SLC19A1 rs2838958 AA genotype was associated with poorer outcome than was the G allele (AG+GG) (5-year EFS 78.5 ± 4.6% vs 92.2 ± 1.8%; Figure 1B). In our study, the SLC19A1 rs1051266 A allele (AA+AG) was associated with better outcome than was the GG genotype (5-year EFS 91.9 ± 1.9% vs. 82.1 ± 4.1%; Supplementary Figure 5), but this association was not statistically significant (P = 0.08).
Figure 1. Polymorphisms and event-free survival (EFS).
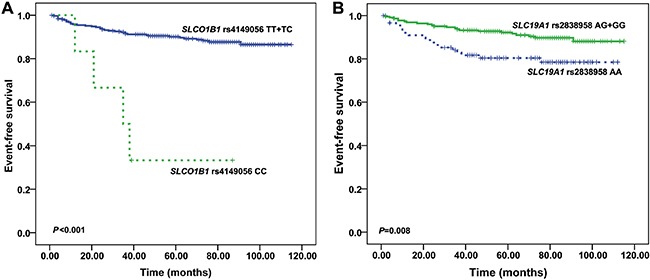
(A) The Kaplan-Meier survival curve is shown to illustrate the difference between SLCO1B1 rs4149056 CC variant and TT/TC variants (log-rank P < 0.001). (B) The Kaplan-Meier survival curve is shown to illustrate the difference between SLC19A1 rs2838958 AA variant and GG/AG variants (log-rank P = 0.008).
In a multivariate Cox regression analysis that included immunophenotype, WBC, TEL/AML1, MRD class (day 33 and day 78), and SLCO1B1 rs4149056 and SLC19A1 rs2838958 genotypes as independent variables, SLCO1B1 rs4149056 and SLC19A1 rs2838958 genotypes and MRD class were shown to be independent prognostic factors (Table 5). The adjusted hazard ratios were 12.242 (95% CI = 3.937–38.059; P < 0.001) for patients with the SLCO1B1 rs4149056 CC genotype vs those with the T allele (TC+TT). The SLC19A1 rs2838958 AA genotype still had a poorer prognosis than did the G allele (AG+GG) (hazard ratio, 2.324, 95% CI 1.147–4.710; P = 0.019). In addition, the presence of MRD at day 33 and day 78 also had significant effect on outcome (hazard ratio, 3.458, 95% CI 1.598–7.483; P = 0.002; hazard ratio, 6.330, 95% CI 2.181–18.369; P = 0.001). Other clinical features, such as WBC, TEL-AML1, and immunophenotype were not independent prognostic factors.
Table 5. Multivariate Cox regression analysis for EFS.
| HR | 95% CI | P | |
|---|---|---|---|
| Immunophenotype | |||
| B-ALL | 1 | Reference | 0.870 |
| T-ALL | 1.094 | 0.371–3.230 | |
| WBC | |||
| < 50 × 109 | 1 | Reference | 0.059 |
| ≥ 50 × 109 | 2.385 | 0.968–5.877 | |
| TEL-AML1 | |||
| Negative | 1 | Reference | 0.935 |
| Positive | 0.959 | 0.344–2.668 | |
| MRD at day 33 | |||
| < 10–4 | 1 | Reference | 0.002 |
| ≥ 10–4 | 3.458 | 1.598–7.483 | |
| MRD at day 78 | |||
| < 10–3 | 1 | Reference | 0.001 |
| ≥ 10–3 | 6.330 | 2.181–18.369 | |
| SLCO1B1 rs4149056 | |||
| TT+TC | 1 | Reference | < 0.001 |
| CC | 12.242 | 3.937–38.059 | |
| SLC19A1 rs2838958 | |||
| AG+GG | 1 | Reference | 0.019 |
| AA | 2.324 | 1.147–4.710 |
EFS: event-free survival; WBC: white blood cell; MRD: minimal residual disease; HR: hazard ratio; CI: confidence interval.
DISCUSSION
HDMTX plays an important role in the improvement of cure rates in ALL. There is great inter-individual variability in MTX response [4, 5, 16, 17]. Several SNPs in genes involved in the MTX transporter pathway have been implicated in the kinetics and effects of MTX in previous studies [9–15], but some results are ambiguous. In the present study, we evaluated the correlation of 12 polymorphisms in four genes involved in the MTX transport pathway with pharmacokinetics and toxicity of HDMTX as well as outcomes in 322 Chinese ALL patients.
SLCO1B1 rs10841753 showed a significant association with the MTX plasma level at 48 h from the first dose of HDMTX infusion in our study. Lower plasma MTX concentrations were found in patients with the rs10841753 CC genotype than in patients with TC or TT genotype. The SLCO1B1 gene encodes an organic anion transporter (OATP1B1) that is located on the sinusoidal membrane of human hepatocytes, and mediates disposition of many medications [18–20]. Previous studies reported that SLCO1B1 can transport MTX in vitro [21, 22], and in vivo [23]. Although many SNPs have been identified in the SLCO1B1 gene, only a few are known to have functional significance [24]. The common SLCO1B1 rs4149056 variant results in decreased transport in vitro [8], whereas the SLCO1B1 rs2306283 variant increases the transport activity for MTX in vitro [22]. The genome-wide study by Trevino et al. demonstrated that SLCO1B1 rs11045879, rs4149056, and eight additional SNPs including rs10841753 are strongly associated with MTX clearance in childhood ALL [25]. The functional rs2306283 in SLCO1B1 was recognized by Ramsey et al. as a predictor of MTX clearance in ALL patients [22]. The association between SLCO1B1 rs4149056 and MTX clearance was confirmed in subsequent studies [5, 14, 15, 26]. Li et al. replicated the association between SLCO1B1 rs11045879 and MTX plasma levels in 280 Chinese pediatric B–ALL patients [27]; however, other studies show conflicting results. Lopez-Lopez et al. failed to show a statistically significant association between rs11045879 and MTX plasma concentrations in patients with ALL (corrected P = 0.08) [12]. In our study, we could not validate the influence of SLCO1B1 rs4149056, rs11045879, and rs2306283 on MTX pharmacokinetics. Our results showed that lower MTX plasma concentration at 48 h was found in patients with the rs4149056 T allele (TC+TT) than in patients with the CC genotype (0.45 μmol/L vs. 1.1 μmol/L), in patients with the rs11045879 TT genotype compared with the CC or TC genotype (0.39μmol/L vs.0.50μmol/L), and in patients with the rs2306283 TT genotype compared with the CC or TC genotype (0.37 μmol/L vs. 0.45 μmol/L), which was consistent with previous results. However, the association did not reach statistical significance (P = 0.441, 0.287, 0.294, respectively). Trevino et al. reported that SLCO1B1 genetic variation accounted for only 9.3% of the MTX inter-patient variability, compared with 17.9% for the treatment regimen [25]. Thus, the differences between our protocol and other protocols, including MTX dosage, the methodology of leucovorin rescue, and concomitant hydration and alkalinization may be the possible reasons for the inconsistent results. SLCO1B1 rs10841753 is an SNP that is located in the intron of the gene. It is unknown how this variant affects the function of SLCO1B1, perhaps by effects on transcription and post-transcriptional processing [25]. We replicated the association between MTX plasma level with rs10841753, confirming that SLCO1B1 was a predictor for MTX pharmacokinetics in our cohort, and that different polymorphisms can affect the functions of SLCO1B1 and MTX clearance.
Because altered MTX pharmacokinetics may affect adverse drug reactions and treatment efficacy, MTX toxicity and survival were also investigated in this study. Oral mucositis is a common side effect attributed to the use of HDMTX. One previous study found that SLCO1B1 rs4149056 was associated with the risk of mucositis [15]; however, this association was not replicated in two other studies [14, 25]. In our study, patients with the SLCO1B1 rs4149056 C allele had a lower risk of developing mucositis. None of the six patients with the CC genotype developed oral mucositis, but the differences among genotypes were not statistically significant. Several factors, including differences in early symptomatic treatment for toxicity, toxicity criteria, treatment protocols, and sample sizes of groups may be responsible for the inconsistent results. Compared with patients with the TT genotype, those with the ABCB1 rs1128503C allele had a higher risk of developing oral mucositis after receiving HDMTX (P = 0.054)), and had a longer duration of hospitalization for the HDMTX treatment (P = 0.006). Zgheib et al. found that rs1128503 was associated with neutropenia [28]. It is possible that the ABCB1 rs1128503C allele is associated with several different manifestations of drug toxicity, which may lead to prolongation of the treatment period. A larger study should be performed to confirm the association.
We next analyzed the prognostic roles of selected polymorphisms in childhood ALL. Our results showed that SLCO1B1 rs4149056 and SLC19A1 rs2838958 were independent prognostic factors in our study. Zhang et al. found that rs4149056 was not associated with the risk of relapse in 136 Chinese pediatric ALL patients. But interestingly, their results showed that patients with the C allele (CC+TC) were given higher leucovorin doses than were patients with the TT genotype [15]. In addition, Skaby et al. reported that higher leucovorin doses during HDMTX treatment may raise the risk for relapse [29]. Based on these data, it seemed that the rs4149056 C allele should be associated with higher risk for relapse. We found that patients with the rs4149056 CC genotype had poorer outcome than did those with the TC or TT genotype (5-year EFS 33.3 ± 19.2%% vs. 90.5 ± 1.7; Figure 1A), which were theoretically reasonable. The sample size in the study by Zhang et al. was relatively small; there were only two patients with the rs4149056 CC alleles, which might obscure the prognostic role of rs4149056. Furthermore, the median follow-up time in the present study was longer than the follow-up time in the study by Zhang et al (75 months vs. 60 months), which is important because relapses tend to occur late in many low-risk ALL patients [15]. Thus, the correlation between rs4149056 and EFS should be further validated.
It is noteworthy that we also found that SLC19A1 rs2838958 was an independent prognostic factor in our study. SLC19A1 encodes the reduced folate carrier (RFC) which delivers folate into cells [10]. SLC19A1 rs2838958 is a tag SNP of SLC19A1 in the CHB population and is located in the intron of the gene. Univariate analysis showed that SLC19A1 rs2838958 was significantly associated with the risk of adverse events (P = 0.008). The SLC19A1 rs2838958 AA genotype was associated with poorer outcome than was the G allele (AG+GG) (5-year EFS 78.5 ± 4.6% vs 92.2 ± 1.8%; Figure 1B). In a multivariate Cox regression analysis, the SLC19A1 rs2838958 AA genotype still was associated with a poorer prognosis than was the G allele (AG+GG) (hazard ratio, 2.324, 95% CI 1.147–4.710; P = 0.019). As far as we know, this is the first study to correlate this polymorphism with outcome in pediatric ALL; however, the exact mechanism of rs2838958 is unknown. The rs1051266 (80G>A, His27Arg) was the most studied SNP in the gene SLC19A1. In previous studies, there were conflicting results on the prognostic significance of SLC19A1 rs1051266 [9, 10, 14, 30]. Several studies have previously investigated the effect of rs1051266 on outcome in childhood ALL, but the results were inconsistent. Rocha et al. and Radtke et al. could not demonstrate any significant effect on cure rates in 246 and 499 children with ALL, respectively [14, 30], although Laverdiere et al., in a study of 204 patients, found a reduced rate of relapse for patients with the G-allele, and Gregers et al., in a study of 500 patients, found a decreased EFS rate in patients who had the G-allele [9, 10]. Our data showed that patients with the SLC19A1 rs1051266 GG genotype had a poorer outcome than did patients with AA or AG genotypes (5-year EFS 82.1 ± 4.1%; vs. 91.9 ± 1.9%, Supplementary Figure 5), but this association was not statistically significant (P = 0.08).
This study has some limitations. It is a retrospective study with a modest sample size and limited SNPs in the MTX transport pathway. In addition, oral mucositis was the only manifestation of MTX toxicity that we studied; we did not include other common adverse drug reactions. Further study and replication in an independent cohort are needed to confirm our recent results.
In conclusion, our results show that polymorphisms in the MTX transport pathway are associated with MTX plasma levels and outcome in childhood ALL, which may help to guide and optimize MTX treatment in patients with childhood ALL.
MATERIALS AND METHODS
Patients and treatment
Between October 1, 2006 and August 1, 2010, 553 Chinese children were newly diagnosed with acute lymphoblastic leukemia in Beijing Children's Hospital. Of these, patients with standard-risk ALL (SR) and intermediate-risk (IR) were included in this study if a sufficient sample of good quality DNA was available. A total of 322 patients (aged 1.0–15.0 years; median: 4.0 years) were eligible according to these criteria (Supplementary Figure 1). The BCH-2003 protocol was followed in 104 patients between October 2006 and March 2008, and the CCLG-2008 protocol was followed in 218 patients after April 2008. Of these, 117 patients were classified as having SR ALL, and 205 as having IR ALL. Details of the stratification and treatment regimens of the two protocols have been published previously [16] and are outlined in Table 1. At a median follow-up time of 75 months (range, 1.0–115.0), 278 patients were in first remission, 33 patients had relapsed, five had died of severe infection (one patient before the first HDMTX), two had died of severe HDMTX toxicity, and four patients had been lost follow-up before the first HDMTX. The BCH-2003 and CCLG-2008 protocols were approved by the Beijing Children's Hospital Institutional Ethics Committee. Informed consent was obtained from all guardians.
Minimal residual disease (MRD) analysis
MRD was measured using RQ-PCR-based quantification of IgH gene rearrangements, as previously reported [31–33]. In this study, 297 samples were examined for MRD at day 33, and 278 samples were examined at day 78. Patients with an MRD <10–4 at day 33 were considered to have a good response to early treatment. An MRD> 10–2 at day 33 or an MRD >10–3 at day 78 represented a poor response.
Candidate gene and polymorphism selection
The selection of candidate genes and SNPs was based on the results of genome-wide association studies, candidate gene association studies, and tag SNPs for CHB (Han Chinese in Beijing) population selected from the HapMap database (http://hapmap.ncbi.nlm.nih.gov/). A total of 12 SNPs in four genes encoding MTX membrane transporter proteins were selected. Detailed information regarding the selected genes and SNPs is shown in Supplementary Table 1. The SNP inclusion criteria were minor allele frequency (MAF)>10%, and in that the gene was demonstrably in Hardy-Weinberg equilibrium (HWE) in the population.
DNA extraction and genotyping
Genomic DNA was extracted using a Genomic DNA Isolation Kit (U-gene, Anhui, China) from bone marrow specimens at diagnosis according to the manufacturer's instructions. Genotyping was performed on the Sequenom MassARRAY (San Diego, CA, USA) platform in OEbiotech Corporation in Shanghai, China. SNPs with genotyping call rate less than 95% were excluded.
HDMTX
Patients received four courses of MTX every 2 weeks during the consolidation phase of chemotherapy (day 8, 22, 36, 50). MTX was administered as follows: 10% of the total dose was administered by intravenous infusion over 0.5 hours, and the remaining 90% over 23.5 hours (MTX doses for each treatment arms are shown in Table 1). Leucovorin rescue was initiated with a 15 mg/m2 dose at 42 h after the start of the HDMTX infusion, and given twice more, at 48 and 54 h after the infusion. A high plasma MTX level after 48 h (> 0.25 μmol/L) was defined as an indication for prolonged rescue. Monitoring of MTX concentration in plasma was carried out every day until the level was below 0.2 umol/L.
Blood samples of 2 ml were collected at 48 h after the continuous MTX infusion over 24 h. The MTX plasma levels were evaluated by fluorescence polarization immunoassay according to the manufacturer's instructions (TDx Abbott Laboratories, Chicago, IL). Before receiving HDMTX therapy, one patient died from infection and four patients were lost to follow-up. Thus, plasma MTX data at 48 h was available for 317 patients. After receiving the first course of HDMTX, one patient died from Stevens-Johnson syndrome and one from severe gastrointestinal mucositis and infection.
Toxicity
Oral mucositis is a common manifestation of toxicity that has been attributed specifically to HDMTX [14]. In this study, oral mucositis was graded according to the National Cancer Institute (NCI) Common Terminology Criteria for Adverse Events version 4.0 scoring system (http://evs.nci.nih.gov/ftp1/CTCAE/CTCAE_4.03_2010-06-14_QuickReference_5x7.pdf). The most severe mucositis (Grade 3–4) observed in each patient during the consolidation therapy period was recorded objectively from electronic patient files (blinded to genotypes and MTX levels).
Statistical analysis
The allele frequencies of all polymorphisms were tested for HWE using χ2. An SNP was excluded from the statistical analysis if the frequencies of the wild type and variant alleles deviated from the HWE requirement. The association between MTX concentrations (first course of HDMTX) and polymorphisms was evaluated by the Mann-Whitney U test. The association between MTX concentrations of the four courses of HDMTX and polymorphisms was evaluated by repeated measures ANOVA. The differences in duration of hospitalization between genotypes were compared using Student's t test. Toxicity was represented by the value 1 or 0, indicating whether an adverse event did or did not occur during the MTX course. Statistical associations between the susceptibility to toxicity and the polymorphisms were analyzed using logistic regression. Haploview v.4.2 was used to determine the haplotype block and to infer haplotype frequencies between individuals with and without toxicity. The duration of event-free survival (EFS) was defined as the time from diagnosis to the date of relapse or death for any reasons (whichever came first), or the last contact with patients in continuous hematologic remission. EFS probabilities were estimated using the Kaplan-Meier method. Differences between groups were compared using the log-rank test. Multivariate Cox regression models adjust for potential confounding variables were used to analyze the prognostic significance of candidate polymorphisms. Two-sided P-values < 0.05 were considered statistically significant. SPSS 16.0 software (SPSS Inc, Chicago, IL, USA) was used for statistical analysis.
SUPPLEMENTARY MATERIALS FIGURES AND TABLES
Acknowledgments
This study was supported by in part by Beijing Nova Project (No.Z151100000315093), National Natural Science Foundation Project (No.81300434, 81300432, 81170504), Beijing Municipal Natural Science Foundation (No.7152054), Beijing Health Qualified Personnel Program (No. 2011-3-049), National Science & Technology Major Project of the 12th 5-Year Plan (No. 2011ZX09302007-01), Beijing Municipal Administration of Hospitals Clinical Medicine Development of Special Grant (No.ZY201404), Beijing Municipal Administration of Hospitals Deng Feng Program (No.DFL20151101), and the Capital Health and Development of Special Grant (No.2016-1-2091). We gratefully thank all staff of the Hematology Oncology Centre of Beijing Children's Hospital, Capital Medical University, for their help in sampling.
Authors’ contributions
SGL and ZGL designed the study. SGL performed the SNP genotyping, reviewed medical records, analyzed the data, and wrote the article. CG collected the medical data and participated in the statistical analysis. CG, XXZ, LC, and WJL performed DNA extraction and MRD analysis. ZGL, XXZ, ZXY, and ZPC participated in the statistical analysis. HYZ, RDZ, MYW, and YYZ recruited and treated the patients. HYZ and RDZ reviewed medical records. JXW gave advice on the data analysis, evaluated the results, and modified the article. HYZ, ZGL, and JXW reviewed the final manuscript. All authors have read and approved the final manuscript.
CONFLICTS OF INTEREST
None.
REFERENCES
- 1.Pui CH, Carroll WL, Meshinchi S, Arceci RJ. Biology, risk stratification, and therapy of pediatric acute leukemias: an update. J Clin Oncol. 2011;29:551–565. doi: 10.1200/JCO.2010.30.7405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liu SG, Li ZG, Cui L, Gao C, Li WJ, Zhao XX. Effects of methylenetetrahydrofolate reductase gene polymorphisms on toxicities during consolidation therapy in pediatric acute lymphoblastic leukemia in a Chinese population. Leuk Lymphoma. 2011;52:1030–1040. doi: 10.3109/10428194.2011.563883. [DOI] [PubMed] [Google Scholar]
- 3.Liu SG, Gao C, Zhang RD, Jiao Y, Cui L, Li WJ, Chen ZP, Wu MY, Zheng HY, Zhao XX, Yue ZX, Li ZG. FPGS rs1544105 polymorphism is associated with treatment outcome in pediatric B-cell precursor acute lymphoblastic leukemia. Cancer Cell Int. 2013;13:107. doi: 10.1186/1475-2867-13-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mikkelsen TS, Thorn CF, Yang JJ, Ulrich CM, French D, Zaza G, Dunnenberger HM, Marsh S, McLeod HL, Giacomini K, Becker ML, Gaedigk R, Leeder JS, et al. PharmGKB summary: methotrexate pathway. Pharmacogenet Genomics. 2011;21:679–686. doi: 10.1097/FPC.0b013e328343dd93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Csordas K, Lautner-Csorba O, Semsei AF, Harnos A, Hegyi M, Erdelyi DJ, Eipel OT, Szalai C, Kovacs GT. Associations of novel genetic variations in the folate-related and ARID5B genes with the pharmacokinetics and toxicity of high-dose methotrexate in paediatric acute lymphoblastic leukaemia. Br J Haematol. 2014;166:410–420. doi: 10.1111/bjh.12886. [DOI] [PubMed] [Google Scholar]
- 6.Moriyama T, Relling MV, Yang JJ. Inherited genetic variation in childhood acute lymphoblastic leukemia. Blood. 2015;125:3988–3995. doi: 10.1182/blood-2014-12-580001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhao R, Diop-Bove N, Visentin M, Goldman ID. Mechanisms of membrane transport of folates into cells and across epithelia. Annu Rev Nutr. 2011;31:177–201. doi: 10.1146/annurev-nutr-072610-145133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Niemi M, Pasanen MK, Neuvonen PJ. Organic anion transporting polypeptide 1B1: a genetically polymorphic transporter of major importance for hepatic drug uptake. Pharmacol Rev. 2011;63:157–181. doi: 10.1124/pr.110.002857. [DOI] [PubMed] [Google Scholar]
- 9.Laverdière C, Chiasson S, Costea I, Moghrabi A, Krajinovic M. Polymorphism G80A in the reduced folate carrier gene and its relationship to methotrexate plasma levels and outcome of childhood acute lymphoblastic leukemia. Blood. 2002;100:3832–3834. doi: 10.1182/blood.V100.10.3832. [DOI] [PubMed] [Google Scholar]
- 10.Gregers J, Christensen IJ, Dalhoff K, Lausen B, Schroeder H, Rosthoej S, Carlsen N, Schmiegelow K, Peterson C. The association of reduced folate carrier 80G>A polymorphism to outcome in childhood acute lymphoblastic leukemia interacts with chromosome 21 copy number. Blood. 2010;115:4671–4677. doi: 10.1182/blood-2010-01-256958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gregers J, Gréen H, Christensen IJ, Dalhoff K, Schroeder H, Carlsen N, Rosthoej S, Lausen B, Schmiegelow K, Peterson C. Polymorphisms in the ABCB1 gene and effect on outcome and toxicity in childhood acute lymphoblastic leukemia. Pharmacogenomics J. 2015;15:372–379. doi: 10.1038/tpj.2014.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lopez-Lopez E, Martin-Guerrero I, Ballesteros J, Piñan MA, Garcia-Miguel P, Navajas A, Garcia-Orad A. Polymorphisms of the SLCO1B1 gene predict methotrexate-related toxicity in childhood acute lymphoblastic leukemia. Pediatr Blood Cancer. 2011;57:612–619. doi: 10.1002/pbc.23074. [DOI] [PubMed] [Google Scholar]
- 13.Lopez-Lopez E, Ballesteros J, Piñan MA, Sanchez de Toledo J, Garcia de Andoin N, Garcia-Miguel P, Navajas A, Garcia-Orad A. Polymorphisms in the methotrexate transport pathway: a new tool for MTX plasma level prediction in pediatric acute lymphoblastic leukemia. Pharmacogenet Genomics. 2013;23:53–61. doi: 10.1097/FPC.0b013e32835c3b24. [DOI] [PubMed] [Google Scholar]
- 14.Radtke S, Zolk O, Renner B, Paulides M, Zimmermann M, Möricke A, Stanulla M, Schrappe M, Langer T. Germline genetic variations in methotrexate candidate genes are associated with pharmacokinetics, toxicity, and outcome in childhood acute lymphoblastic leukemia. Blood. 2013;121:5145–5153. doi: 10.1182/blood-2013-01-480335. [DOI] [PubMed] [Google Scholar]
- 15.Zhang HN, He XL, Wang C, Wang Y, Chen YJ, Li JX, Niu CH, Gao P. Impact of SLCO1B1 521T>C variant on leucovorin rescue and risk of relapse in childhood acute lymphoblastic leukemia treated with high-dose methotrexate. Pediatr Blood Cancer. 2014;61:2203–2207. doi: 10.1002/pbc.25191. [DOI] [PubMed] [Google Scholar]
- 16.Kałużna E, Strauss E, Zając-Spychała O, Gowin E, Świątek-Kościelna B, Nowak J, Fichna M, Mańkowski P, Januszkiewicz-Lewandowska D. Functional variants of gene encoding folate metabolizing enzyme and methotrexate-related toxicity in children with acute lymphoblastic leukemia. Eur J Pharmacol. 2015;769:93–99. doi: 10.1016/j.ejphar.2015.10.058. [DOI] [PubMed] [Google Scholar]
- 17.Xue Y, Rong L, Tong N, Wang M, Zhang Z, Fang Y. CCND1 G870A polymorphism is associated with toxicity of methotrexate in childhood acute lymphoblastic leukemia. Int J Clin Exp Pathol. 2015;8:11594–11600. [PMC free article] [PubMed] [Google Scholar]
- 18.Abe T, Kakyo M, Tokui T, Nakagomi R, Nishio T, Nakai D, Nomura H, Unno M, Suzuki M, Naitoh T, Matsuno S, Yawo H. Identification of a novel gene family encoding human liver-specific organic anion transporter LST-1. J Biol Chem. 1999;274:17159–17163. doi: 10.1074/jbc.274.24.17159. [DOI] [PubMed] [Google Scholar]
- 19.Nozawa T, Minami H, Sugiura S, Tsuji A, Tamai I. Role of organic anion transporter OATP1B1 (OATP-C) in hepatic uptake of irinotecan and its active metabolite, 7-ethyl-10-hydroxycamptothecin: in vitro evidence and effect of single nucleotide polymorphisms. Drug Metab Dispos. 2005;33:434–439. doi: 10.1124/dmd.104.001909. [DOI] [PubMed] [Google Scholar]
- 20.Weaver YM, Hagenbuch B. Several conserved positively charged amino acids in OATP1B1 are involved in binding or translocation of different substrates. J Membr Biol. 2010;236:279–290. doi: 10.1007/s00232-010-9300-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Trenk D, Hochholzer W, Fromm MF, Zolk O, Valina CM, Stratz C, Neumann FJ. Paraoxonase-1 Q192R polymorphism and antiplatelet effects of clopidogrel in patients undergoing elective coronary stent placement. Circ Cardiovasc Genet. 2011;4:429–436. doi: 10.1161/CIRCGENETICS.111.960112. [DOI] [PubMed] [Google Scholar]
- 22.Ramsey LB, Bruun GH, Yang W, Treviño LR, Vattathil S, Scheet P, Cheng C, Rosner GL, Giacomini KM, Fan Y, Sparreboom A, Mikkelsen TS, Corydon TJ, et al. Rare versus common variants in pharmacogenetics: SLCO1B1 variation and methotrexate disposition. Genome Res. 2012;22:1–8. doi: 10.1101/gr.129668.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van de Steeg E, van der Kruijssen CM, Wagenaar E, Burggraaff JE, Mesman E, Kenworthy KE, Schinkel AH. Methotrexate pharmacokinetics in transgenic mice with liver-specific expression of human organic anion-transporting polypeptide 1B1 (SLCO1B1) Drug Metab Dispos. 2009;37:277–281. doi: 10.1124/dmd.108.024315. [DOI] [PubMed] [Google Scholar]
- 24.Forestier E, Heyman M, Andersen MK, Autio K, Blennow E, Borgström G, Golovleva I, Heim S, Heinonen K, Hovland R, Johannsson JH, Kerndrup G, Nordgren A, et al. Outcome of ETV6/RUNX1-positive childhood acute lymphoblastic leukaemia in the NOPHO-ALL-1992 protocol: frequent late relapses but good overall survival. Br J Haematol. 2008;140:665–672. doi: 10.1111/j.1365-2141.2008.06980.x. [DOI] [PubMed] [Google Scholar]
- 25.Treviño LR, Shimasaki N, Yang W, Panetta JC, Cheng C, Pei D, Chan D, Sparreboom A, Giacomini KM, Pui CH, Evans WE, Relling MV. Germline genetic variation in an organic anion transporter polypeptide associated with methotrexate pharmacokinetics and clinical effects. J Clin Oncol. 2009;27:5972–5978. doi: 10.1200/JCO.2008.20.4156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ramsey LB, Panetta JC, Smith C, Yang W, Fan Y, Winick NJ, Martin PL, Cheng C, Devidas M, Pui CH, Evans WE, Hunger SP, Loh M, Relling MV. Genome-wide study of methotrexate clearance replicates SLCO1B1. Blood. 2013;121:898–904. doi: 10.1182/blood-2012-08-452839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li J, Wang XR, Zhai XW, Wang HS, Qian XW, Miao H, Zhu XH. Association of SLCO1B1 gene polymorphisms with toxicity response of high dose methotrexate chemotherapy in childhood acute lymphoblastic leukemia. Int J Clin Exp Med. 2015;8:6109–6113. [PMC free article] [PubMed] [Google Scholar]
- 28.Zgheib NK, Akra-Ismail M, Aridi C, Mahfouz R, Abboud MR, Solh H, Muwakkit SA. Genetic polymorphisms in candidate genes predict increased toxicity with methotrexate therapy in Lebanese children with acute lymphoblastic leukemia. Pharmacogenet Genomics. 2014(24):387–396. doi: 10.1097/FPC.0000000000000069. [DOI] [PubMed] [Google Scholar]
- 29.Skärby TV, Anderson H, Heldrup J, Kanerva JA, Seidel H, Schmiegelow K, Nordic Society of Paediatric Haematology and Oncology (NOPHO) High leucovorin doses during high-dose methotrexate treatment may reduce the cure rate in childhood acute lymphoblastic leukemia. Leukemia. 2006;20:1955–1962. doi: 10.1038/sj.leu.2404404. [DOI] [PubMed] [Google Scholar]
- 30.Rocha JC, Cheng C, Liu W, Kishi S, Das S, Cook EH, Sandlund JT, Rubnitz J, Ribeiro R, Campana D, Pui CH, Evans WE, Relling MV. Pharmacogenetics of outcome in children with acute lymphoblastic leukemia. Blood. 2005;105:4752–4758. doi: 10.1182/blood-2004-11-4544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gao C, Liu SG, Zhang RD, Li WJ, Zhao XX, Cui L, Wu MY, Zheng HY, Li ZG. NOTCH1 mutations are associated with favourable long-term prognosis in paediatric T-cell acute lymphoblastic leukaemia: a retrospective study of patients treated on BCH-2003 and CCLG-2008 protocol in China. Br J Haematol. 2014;166:221–228. doi: 10.1111/bjh.12866. [DOI] [PubMed] [Google Scholar]
- 32.Cui L, Li Z, Wu M, Li W, Gao C, Deng G. Combined analysis of minimal residual disease at two time points and its value for risk stratification in childhood B-lineage acute lymphoblastic leukemia. Leuk Res. 2010;34:1314–1319. doi: 10.1016/j.leukres.2009.11.031. [DOI] [PubMed] [Google Scholar]
- 33.Gao C, Zhao XX, Li WJ, Cui L, Zhao W, Liu SG, Yue ZX, Jiao Y, Wu MY, Li ZG. Clinical features, early treatment responses, and outcomes of pediatric acute lymphoblastic leukemia in China with or without specific fusion transcripts: a single institutional study of 1,004 patients. Am J Hematol. 2012;87:1022–1027. doi: 10.1002/ajh.23307. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


