Abstract
Within the endoplasmic reticulum (ER), mannoses and glucoses, donated from dolichol-phosphate-mannose and -glucose, are transferred to N-glycan and GPI-anchor precursors, and serine/threonine residues in many proteins. Glycosyltransferases that mediate these reactions are ER-resident multitransmembrane proteins with common characteristics, forming a superfamily of >10 enzymes. Here, we report an essential component of glycosylphosphatidylinositol-mannosyltransferase I (GPI-MT-I), which transfers the first of the four mannoses in the GPI-anchor precursors. We isolated a Chinese hamster ovary (CHO) cell mutant defective in GPI-MT-I but not its catalytic component PIG-M. The mutant gene, termed phosphatidylinositolglycan-class X (PIG-X), encoded a 252-amino acid ER-resident type I transmembrane protein with a large lumenal domain. PIG-X and PIG-M formed a complex, and PIG-M expression was <10% in the absence of PIG-X, indicating that PIG-X stabilizes PIG-M. We found that Saccharomyces cerevisiae Pbn1p/YCL052Cp, which was previously reported to be involved in autoprocessing of proproteinase B, is the functional homologue of PIG-X; Pbn1p is critical for Gpi14p/YJR013Wp function, the yeast homologue of PIG-M. This is the first report of an essential subcomponent of glycosyltransferases using dolichol-phosphate-monosaccharide.
INTRODUCTION
Posttranslational modifications of proteins by various glycans are initiated in the endoplasmic reticulum (ER) and completed in the Golgi apparatus. Glycosyltransferases in the ER are smaller in number than those in the Golgi, but they are relatively well conserved evolutionarily in a variety of eukaryotic cells and play basic biological functions. Among them, dolichol-phosphate (Dol-P)-monosaccharide using enzymes form a major and distinct group of glycosyltransferases functioning in the lumenal side of the ER (Oriol et al., 2002; Liu and Mushegian, 2003). Dol-P-mannose (Man) is essential for the biosynthesis of glycosylphosphatidylinositol (GPI)-anchors (Kinoshita and Inoue, 2000), O-mannosylation (Manya et al., 2004), and C-mannosylation (Doucey et al., 1998), and both Dol-P-Man and Dol-P-glucose (Glc) are necessary for correct assembly of N-glycan precursor lipid-linked oligosaccharides (Burda and Aebi, 1999).
GPI-anchors that act as membrane anchors for various plasma membrane proteins have a core structure consisting of a phosphatidylinositol (PI), a glucosamine (GlcN), three Mans, and a phosphoethanolamine (EtNP) (Kinoshita and Inoue, 2000). The first two precursors of GPI are synthesized on the cytoplasmic face of the ER by transfer of an α-linked N-acetylglucosamine (GlcNAc) to the inositol residue of PI, generating GlcNAc-PI, followed by deN-acetylation generating GlcN-PI (Nakamura et al., 1997; Watanabe et al., 1998, 2000). It is thought that GlcN-PI is then flipped to the lumenal side of the ER by an unidentified mechanism. Inositol acylation catalyzed by PIG-W generating GlcN-acyl-PI is probably the first reaction to occur once GlcN-PI enters the ER lumen (Murakami et al., 2003). Next, three (or four) Man residues and EtNP are sequentially added, leading to maturation of GPI. The first Man in the core structure is usually modified with an EtNP side chain transferred by PIG-N (Hong et al., 1999). Mature GPIs are attached to the carboxyl termini of the precursor proteins by a GPI-transamidase complex (Ohishi et al., 2001; Hong et al., 2003). The acylchain on inositol is then removed by PGAP1 before leaving the ER in COPII secretory vesicles (Tanaka et al., 2004).
All Man residues in GPI are donated from Dol-P-Man and linked through three different glycosidic bonds by the specific mannosyltransferases (MTs). The first α1→4-linked Man is transferred by Dol-P-Man: GlcN-acyl-PI α1,4 MT (GPI-MT-I); the second α1→6-linked Man is transferred by Dol-P-Man: Man-GlcN-acyl-PI α1,6 MT (GPI-MT-II); and the third and fourth α1→2-linked Mans are transferred by Dol-P-Man: Man2-GlcN-acyl-PI α1,2 MT (GPI-MT-III) and Dol-P-Man: Man3-GlcN-acyl-PI α1,2 MT (GPI-MT-IV), respectively. The genes for GPI-MT-I, -II, -III, and -IV have been cloned and termed PIG-M (GPI14 in Saccharomyces cerevisiae) (Maeda et al., 2001), PIG-V (GPI18 in S. cerevisiae) (Kang et al., 2005), PIG-B (GPI10 in S. cerevisiae) (Takahashi et al., 1996), and SMP3 (Grimme et al., 2001; Taron et al., 2004), respectively. These MTs, as well as the other Dol-P-Man/Glc using glycosyltransferases involved in N-glycan precursor synthesis (ALG3, ALG9, and ALG12 MTs and ALG6, ALG8, and ALG10 glucosyltransferases), show some structural similarity to each other, namely, they have multiple transmembrane domains and the fairly large first ER lumenal domain (Oriol et al., 2002). Among these, PIG-M has a functionally important DXD motif, a typical characteristic of many glycosyltransferases, within the first ER lumenal domain (Maeda et al., 2001). Other enzymes also contain acidic residue(s), if not identical to the DXD motif, important for enzyme activity in the corresponding domain. Thus, they are considered to be catalytic polypeptides. It has been unclear, however, whether they represent complete enzymes or they have partner subcomponents and how these glycosyltransferases are regulated.
In the protozoan parasite Trypanosoma brucei, the GPI synthetic pathway is basically similar to those of mammals and yeast, although there are several differences. One of the striking differences resides in the step involving GPI-MT-I. In trypanosomes, the first Man is added to GlcN-PI lacking inositol acylation by trypanosomal GPI-MT-I (TbGPI14) (Doerrler et al., 1996; Smith et al., 1996), which shows 30% amino acid identity with human PIG-M (Maeda et al., 2001). Therefore, this step is a promising target for antiprotozoan parasite drugs. In fact, a substrate analog GlcN-(2-O-hexadecyl)-PI acts as a specific inhibitor for trypanosomal GPI-MT-I (Ferguson et al., 1999). For developing more effective drugs and understanding inhibitory mechanisms, further characterization of GPI-MT-I from both mammals and protozoans is necessary.
While screening GPI biosynthesis mutants in CHO cells, we found a new mutant cell line that was defective in GPI-MT-I activity but was not restored by transfection with PIG-M cDNA. In the present article, we describe molecular cloning and characterization of a novel component of the GPI-MT-I enzyme complex. This is the first report of an essential subcomponent of Dol-P-Man/Glc using glycosyltransferases involved in the biosyntheses of lipid-linked oligosaccharides in the ER.
MATERIALS AND METHODS
Cell Culture
CHO K1 cells were cultured in Ham's F-12 medium containing 10% fetal calf serum (FCS). The transfectants and mutants derived from CHO K1 cells were cultured in the same medium supplemented with suitable antibiotics. CD59/decay accelerating factor (DAF)-negative human B-cell lymphoma Ramos 517-17 cells were cultured in RPMI 1640 medium containing 10% FCS (Maeda et al., 2001).
Establishment of a Novel Class Mutant in CHO Cells
The parental CHO cell line F21, which stably expressed the GPI-anchored protein markers CD59 and DAF and 12 previously known proteins involved in GPI biosynthesis, was established by three cycles of cDNA transfection and clone selection. First, wild-type CHO K1 cells were transfected with pME vectors containing cDNAs of CD59/DAF/neor, FLAG-PIG-L, GST-DPM2, GST-PIG-A, and SL15-FLAG, and then transfectants were selected with 600 μg/ml G418 (Nacalai Tesque, Kyoto, Japan). In the second cycle, cDNAs of PIG-O/hygr, PIG-U-3FLAG, GnTI-FLAG, FLAG-PIG-F, and FLAG-GAA1 were transfected and transfectants were selected with 600 μg/ml G418 and 400 μg/ml hygromycin B (Wako Pure Chemicals, Osaka, Japan). In the third cycle, cDNAs of PIG-N-GST, GST-PIG-V, GST-DPM1, GST-PIG-B, and bsdr were transfected, and the clone F21 was selected with 600 μg/ml G418, 400 μg/ml hygromycin B, and 50 μg/ml blasticidin S (Invivogen, San Diego, CA).
F21 cells (3 × 107) were mutagenized with 1.2 μg/ml N-methyl-N′-nitro-N-nitrosoguanidine (NMMG; Nacalai Tesque) for 2 d and cultured in fresh medium for 7 more days. The cells were then treated with 1 nM proaerolysin (Protox Biotech, Victoria, British Columbia, Canada) for 2 d and/or 1 nM Clostridium septicum α-toxin for 2 d. Surviving cells were isolated by limiting dilution.
Wild-type and mutant CHO cells used in this study are listed in Table 1.
Table 1.
CHO cell lines used in this study
| Cell line | Characteristics | Reference |
|---|---|---|
| CHO K1 | Wild type | |
| CHO F21 | CHO K1 transfected with CD59, DAF, and 12 PIG-genes described in Materials and Methods | This study |
| CHO2.46 | PIG-X—defective mutant derived from CHO F21 | This study |
| CHO PA16.1 | PIG-U—defective mutant | Hong et al., 2003 |
| CHO2.38 | Dol-P-Man synthase-defective mutant derived from CHO F21 | This study |
| CHO Lec35 | MPDU1-defective mutant | Camp et al., 1993 |
| CHO1.46 | PIG-L—defective mutant derived from CHO F21 | This study |
| CHO10.14 | PIG-W—defective mutant | Murakami et al., 2003 |
Flow Cytometric Analysis
Cells were stained for CD59 and DAF with anti-CD59 (5H8) plus fluorescein isothiocyanate (FITC)-conjugated anti-mouse IgG (ICN/Cappel, Aurora, OH) and biotinylated anti-DAF (IA10) plus phycoerythrin (PE)-conjugated streptavidin (Biomeda, Foster City, CA). To evaluate the biosynthesis of N-glycan, cells were cultured with or without 10 μg/ml swainsonine (Wako Pure Chemicals) for 4 d, and then stained with 25 μg/ml FITC-conjugated PHA-P lectin (Sigma-Aldrich, St. Louis, MO). Stained cells were analyzed by a FACScan (BD Biosciences, San Jose, CA).
Expression Cloning
The cloning of phosphatidylinositolglycan-class X (PIG-X) cDNA was carried out by the expression cloning method described previously (Inoue et al., 1993) with some modifications. The rat C6 glioma cDNA library constructed in the pME-Py vector bearing the polyoma virus origin of replication, and pcDNA-PyT(ori-) for the expression of the polyoma large T, were cotransfected into the mutant CHO2.46 (see Results for isolation of CHO2.46 cells) by transfection using Lipofectamine 2000 (Invitrogen). After 2 d of cultivation, cells were stained with biotinylated anti-CD59 and PE-conjugated streptavidin, and CD59-positive cells were collected with a cell sorter (FACS-Vantage; BD Biosciences). The plasmids in the sorted cells were rescued in Escherichia coli. After two cycles of transfection, cell sorting, and plasmid rescue, each plasmid clone was tested by transfection into CHO2.46 cells.
Transfection of Cells
Cells were transfected by either electroporation or lipofection. CHO cells (0.4 × 107) suspended in 0.4 ml of culture medium were electroporated at 260 V and 960 μF with 10 μg of DNA by using a Gene Pulser (Bio-Rad, Hercules, CA). Ramos 517-17 cells (0.4 × 107) suspended in 0.4 ml of HEPES-buffered saline were electroporated under similar conditions. For lipofection, CHO cells were transfected using LipofectAMINE 2000 according to the instruction manual.
Plasmids
To construct pME/PIG-X-3FLAG and pME/puror/PIG-X-3FLAG, we amplified full-length PIG-X cDNA by using pME/PIG-X (clone 63B) as a template and the forward SRα primer 5′-TGACCCTGCTTGCTCAACTCTACG and the specific reverse primer with an MluI site 5′-TTACGCGTTAGGGAAAAATGGCCATATTTGAAAAC (R1), digested with XhoI and MluI, and cloned into XhoI- and MluI-cut pME//3FLAG and pME/puror//3FLAG. To construct pME/PBN1–3FLAG, we amplified full-length PBN1 by using genomic DNA from S. cerevisiae as a template and primers 5′-AACTCGAGCATGGTGACAAGACATAGAGTGACTG (forward) and 5′-TTACGCGTTTCCCGTTTTACTGATCTTTTCTTCTTG (reverse), and cloned it in place of the PIG-X cDNA into pME/PIG-X-3FLAG. For pME/GST-GPI14, the full-length GPI14 gene, amplified using 5′-AACTCGAGCATGACTGGCGAAGAATGGGGCTTG (forward) and 5′-TTACGCGTGTTGTTCTTTTTGTTGGAAACTGTGG (reverse), was cloned in place of the PIG-M cDNA into PME/GST-PIG-M. Various 5′ deletion mutants of PIG-X were amplified with the R1 primer and each of the following forward primers: 5′-AACTCGAGGATGGTATCAGAGAGTTTTAATCTAG for A1, 5′-AACTCGAGCACTTGTTCTGAAATTATTTTG for A2, 5′-AACTCGAGCATGTGTTCTGAAATTATTTTG for A2(ATG) 5′-AACTCGAGTGGGCTGGTCGGCAGGCTCGCC for A3, 5′-AACTCGAGCGTCCTGGCTGCCAGCGCCCTT for A4, and 5′-AACTCGAGCGCTCTGCGTGCTCAGGTTCCTC for A5. Site-directed mutagenesis was carried out using a QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA).
Affinity Precipitation and Western Blotting
Cells were lysed in a buffer containing 1.0% Nonidet P-40, 50 mM Tris-HCl, pH 7.7, 5.0 mM EDTA, 150 mM NaCl, and Complete protease inhibitor cocktail (Roche Diagnostics, Indianapolis, IN) (TEN buffer), and cell debris was removed by ultracentrifugation (100,000 × g, 1 h). Anti-FLAG M2 agarose (Sigma-Aldrich) or glutathione-Sepharose 4B (Amersham Biosciences, Piscataway, NJ) beads were added to the supernatant and rotated at 4°C for 2 h. The beads were collected and washed with TEN buffer twice, and then absorbed proteins were eluted with reducing SDS sample buffer. An aliquot was subjected to 10–20% gradient SDS-PAGE gel and electroblotted onto a polyvinylidene difluoride membrane. The blot was treated with the primary antibody, either anti-FLAG BioM2 (Sigma-Aldrich) or anti-GST (Amersham Biosciences), and then with horseradish peroxidase-conjugated protein G (Bio-Rad). Detection was carried out using Western lightning chemiluminescence reagents (PerkinElmer Life and Analytical Sciences, Boston, MA).
Metabolic Labeling of GPI Intermediates and Dolichol-linked Oligosaccharides
For labeling with Man, CHO cells (106) were preincubated in medium containing 100 μg/ml Glc and 10 μg/ml tunicamycin (Sigma-Aldrich) for 1 h, and then incubated in the same medium containing 40 μCi/ml d-[2-3H(N-)]Man (American Radiolabeled Chemicals, St. Louis, MO) for 45 min. To test the effect of YW3548/BE49385A, cells were cultured in medium containing 10 μM BE49385A (a gift from Banyu Pharmaceutical, Tokyo, Japan), followed by labeling with Man. For labeling with inositol, cells (106) were cultured in inositol-free DMEM supplemented with 10% dialyzed FCS and 40 μCi/ml myo-[2-3H(N)]inositol (American Radiolabeled Chemicals) for 1 d. Lipids were extracted from cells with chloroform:methanol [2:1 (vol/vol)] and then partitioned into water-saturated 1-butanol. The extracts were applied onto a silica gel thin layer chromatography (TLC) plate (Merck, Whitehouse Station, NJ), and developed with chloroform:methanol:water (10:10:3) for analyses of mannolipids, and with chloroform:methanol:1 N NH4OH (10:10:3) for analyses of the first three products of GPI-anchor. The radiolabeled lipids were detected by a Fuji BAS1500 image analyzer (Fuji Film, Tokyo, Japan).
For analysis of N-glycan precursors, cells were metabolically labeled with [3H]Man in the absence of tunicamycin. Dolichol-linked N-glycan precursors were extracted as described previously (Chantret et al., 2003) and hydrolyzed with 0.1 M HCl/80% tetrahydrofuran at 65°C for 30 min to release oligosaccharides. After neutralization, oligosaccharides were analyzed by TLC by using 1-propanol:acetic acid:water (3:3:2).
Microsomal Assay
To assay Dol-P-Man synthase and GPI-MT-I activities, microsomes were prepared from cells preincubated for 2 h with 5 μg/ml tunicamycin by hypotonic lysis and disruption with a Teflon homogenizer. The microsomes were incubated at 37°C for 60 min with or without 40 μg/ml GlcN-PI(C8) (a gift from Dr. M. A. Lehrman, Texas Southwestern Medical Center, Dallas, TX) in 100 mM HEPES, pH 7.3, containing 20 μCi/ml GDP-[2-3H]Man (American Radiolabeled Chemicals), 20 μM palmitoyl-CoA, 50 mM KCl, 10 mM MgCl2, 10 mM MnCl2, 2 mM 5′AMP, 2 μg/ml tunicamycin, 4 μg/ml leupeptin, and 0.2 mM N-tosyl-l-lysine chloromethyl ketone. Lipid extraction and TLC analysis were carried out as described above.
Cell Viability Assay
The cell viability assay was carried out using 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) (Sigma-Aldrich) as described previously (Hong et al., 2002).
RESULTS
Isolation of a New Class of Mutant Cells Defective in GPI-Anchor Biosynthesis
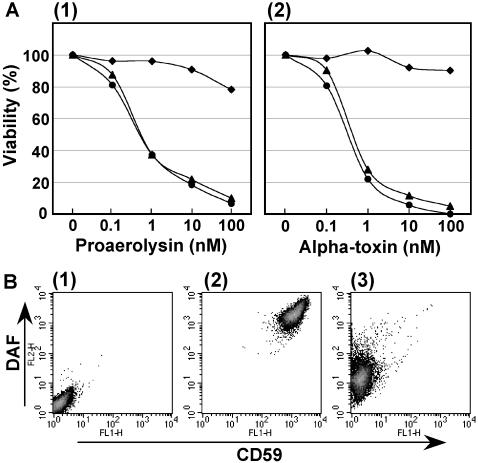
To prevent the generation of mutants with mutations in known genes, we first established the parental cell line, CHO F21, which was stably transfected with the 12 previously identified genes involved in GPI biosynthesis. Then, we mutagenized CHO F21 cells with N-methyl-N′-nitro-N-nitrosoguanidine and selected mutants resistant to the bacterial toxins Aeromonas hydrophila proaerolysin (Buckley, 1999) and/or C. septicum α-toxin (Tweten and Sellman, 1999), both of which bind to GPI-anchors, form pores on the target cell membrane, and kill the cells. We obtained a mutant, CHO2.46, that survived treatment with 1 nM proaerolysin and 1 nM α-toxin. The viabilities of CHO2.46 cells at various concentrations of proaerolysin and α-toxin were measured using MTT (Figure 1A). The binding of proaerolysin to GPI-anchored proteins requires both GPI-anchors and complex-type N-glycans (Hong et al., 2002), and thus some N-glycan biosynthetic mutants, such as N-acetylglucosaminyltransferase I-defective cells, are resistant to proaerolysin. On the other hand, the α-toxin does not require N-glycans for binding. The parental CHO F21 cells and wild-type CHO K1 cells were highly sensitive to both toxins, whereas the mutant CHO2.46 cells were resistant to as much as 100 nM of either toxin, suggesting that the mutation occurred in the GPI-anchor biosynthetic pathway rather than in the N-glycan pathway. The expression of CD59 and DAF, human GPI-anchored proteins introduced as markers, on the surface of CHO2.46 cells as assessed by fluorescence-activated cell sorter (FACS) decreased to <1% of that on the parental CHO F21 cells (Figure 1B). Expression of these marker proteins was not restored by transfection of any known GPI-anchor biosynthetic genes; therefore, CHO2.46 cells seemed to represent a new class of GPI mutant, termed class X.
Figure 1.
CHO2.46 cells are defective in the surface expression of GPI-anchored proteins. (A) Cell viabilities against two microbial toxins, proaerolysin (1) and C. septicum α-toxin (2), assessed by MTT assay. Circles, wild-type CHO K1; triangles, parental CHO F21; diamonds, mutant CHO2.46. (B) FACS analysis showing the cell surface expression of GPI-anchored proteins CD59, and DAF. 1, CHO K1 cells, which do not express CD59 and DAF, used as a negative control; 2, parental CHO F21; and 3, mutant CHO2.46.
CHO2.46 Cells Accumulate GlcN-acyl-PI
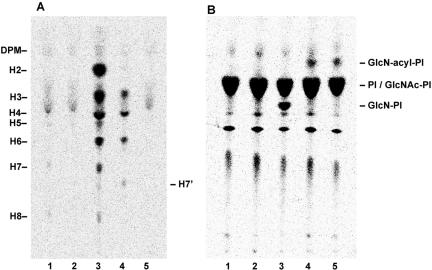
To determine the step at which CHO2.46 cells were defective in GPI-anchor biosynthesis, we metabolically labeled the cells with [3H]Man and then extracted glycolipids containing [3H]Man and analyzed them by TLC. The PIG-U–defective mutant CHO PA16.1 cells, used as a reference, accumulated various GPI intermediates, including the mature forms because PIG-U is an essential component of GPI-transamidase, which is involved in transferring the mature GPI to the precursor proteins (Figure 2A, lane 3) (Hong et al., 2003). In contrast, CHO2.46 cells did not accumulate any Man-containing intermediates (lane 2). The chromatogram of CHO2.46 was similar to that of the parental CHO F21, except that trace amounts of the mature forms H7 and H8 were seen in the latter (lane 1). This result suggested either that CHO2.46 is defective in a GPI biosynthetic step earlier than the first Man addition or that it synthesizes GPI but is defective in a step post-GPI-attachment to proteins. To test the latter possibility, we labeled the cells with [3H]Man in the presence of YW3548/BE49385A (Sutterlin et al., 1997), a specific inhibitor of PIG-N that transfers the EtNP side chain to the first Man (Hong et al., 1999). If Man-containing GPIs were synthesized in CHO2.46, the drug should cause accumulation of intermediates because the action of PIG-N is necessary for efficient downstream reactions. The parental CHO F21 cells accumulated the GPI intermediates H3 and H4 as well as H7′ lacking EtNP on the first Man (lane 4). Because CHO2.46 did not accumulate any GPI intermediates even in the presence of YW3548/BE49385A (lane 5), we concluded that CHO2.46 was defective at a step earlier than the first Man addition.
Figure 2.
CHO2.46 cells accumulate GlcN-acyl-PI. (A) Cells were metabolically labeled with [3H]Man in the absence (lanes 1, 2, and 3) or presence (lanes 4 and 5) of YW3548/BE49385A, and Man-containing lipids were analyzed by TLC. Lane 1, parental CHO F21; lane 2, mutant CHO2.46; lane 3, PIG-U–defective mutant CHO PA16.1; lane 4, CHO F21; and lane 5, CHO2.46. DPM, Dol-P-Man; H2-H8, Man containing GPI species (Hong et al., 2003); H7′, GPI species that accumulated in the presence of YW3548/BE49385A (Hong et al., 1999). (B) Cells were metabolically labeled with myo-[3H]inositol. Lane 1, CHO F21; lane 2, PIG-L–defective mutant CHO1.46; lane 3, PIG-W–defective mutant CHO10.14; lane 4, CHO2.46; lane 5, Dol-P-Man synthase-defective mutant CHO2.38.
Next, to analyze early steps in the pathway, we metabolically labeled CHO2.46 cells with myo-[3H]inositol. The mutant accumulated GlcN-acyl-PI, the third product of GPI-biosynthesis (Figure 2B, lane 4), as did the Dol-P-Man synthase-defective mutant CHO2.38 cells (lane 5). Therefore, CHO2.46 is defective in either Dol-P-Man synthesis, general usage of Dol-P-Man, or the first Man transfer to GlcN-acyl-PI. We tested the restoration of CD59 and DAF expression by transfection of cDNAs for DPM1, DPM2, and DPM3, the components of Dol-P-Man synthase (Maeda et al., 2000); MPDU1, an essential protein for Dol-P-Man usage in the lumenal side of the ER (Ware and Lehrman, 1996); and PIG-M, the catalytic component of GPI-MT-I (Maeda et al., 2001), but none of them restored CD59 and DAF expression (our unpublished data).
CHO2.46 Cells Are Defective in GPI-MT-I But Not in Dol-P-Man Synthesis or Usage
We examined Dol-P-Man synthesis in CHO2.46 cells by in vitro microsomal assay. The microsomes prepared from CHO2.46 synthesized Dol-P-[3H]Man from endogenous Dol-P and exogenously added GDP-[3H]Man (Figure 3A, lane 2), like those from the parental CHO F21 cells (lane 1), indicating that Dol-P-Man synthesis is normal in CHO2.46.
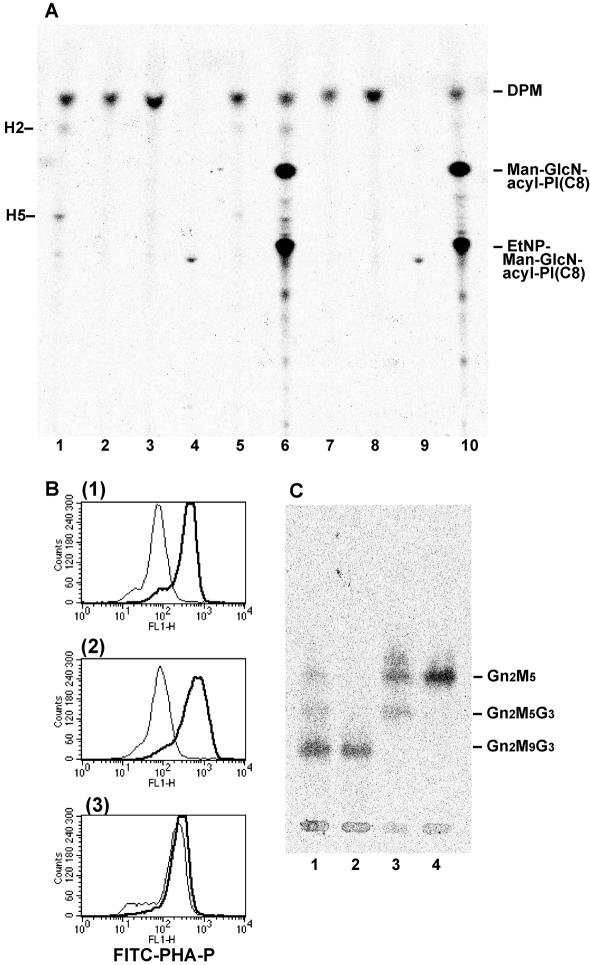
Figure 3.
Dol-P-Man synthesis and usage are normal in CHO2.46 cells, but GPI-MT-I is defective. (A) Microsomes from various cells were incubated with GDP-[3H]Man and palmitoyl-CoA in the absence (lanes 1–5) or presence (lanes 6–10) of GlcN-PI(C8). Man-containing lipids were analyzed by TLC. Lanes 1 and 6, parental CHO F21; lanes 2 and 7, mutant CHO2.46; lanes 3 and 8, PIG-M–defective Ramos 517-17; lanes 4 and 9, Dol-P-Man synthase-defective CHO2.38; and lanes 5 and 10, MPDU1-defective CHO Lec35. DPM, Dol-P-Man; H2, Man-GlcN-acyl-PI; H5, EtNP-Man-GlcN-acyl-PI; Man-GlcN-acyl-PI(C8), H2 analogue generated from GlcN-PI(C8); and EtNP-Man-GlcN-acyl-PI(C8), H5 analogue generated from GlcN-PI(C8). (B) Effect of swainsonine on the expression of complex-type N-glycans. Cells were cultured in the absence (thick lines) or presence (thin lines) of swainsonine for 3 d, stained with FITC-conjugated PHA-P lectin, and analyzed by FACS. 1, CHO F21; 2, CHO2.46; and 3, CHO Lec35. (C) Oligosaccharides from N-glycan precursors were analyzed by TLC. Lane 1, CHO F21; lane 2, CHO2.46; lane 3, CHO2.38; and lane 4, CHO Lec35.
Next, to assess GPI-MT-I activity, we added GlcN-PI(C8) (glucosaminyl-di-octanoyl PI) (Doerrler et al., 1996), a synthetic acceptor substrate for GPI-inositol acyltransferase (PIG-W), to the microsomal assay system. GlcN-acyl-PI(C8) generated from GlcN-PI(C8) by endogenous PIG-W, in turn, acts as a substrate for GPI-MT-I in this system. As expected, the microsomes from the parental CHO F21 cells synthesized two products, Man-GlcN-acyl-PI(C8) and EtNP-Man-GlcN-acyl-PI(C8) (lane 6) (Hong et al., 1999). In contrast, microsomes from CHO2.46 synthesized neither (lane 7), like those from Ramos 517-17 cells, a mutant of PIG-M (lane 8). These results strongly suggest that CHO2.46 is defective in GPI-MT-I.
These results also suggest that the Dol-P-Man usage on the lumenal side of the ER is not affected in CHO2.46 cells. The microsomes from CHO Lec35, which is defective in the usage of Dol-P-Man due to a mutation in MPDU1, synthesized Man-GlcN-acyl-PI(C8) and EtNP-Man-GlcN-acyl-PI(C8) (lane 10). This was known to occur likely due to the destruction of the membrane structure (Camp et al., 1993).
To further confirm that Dol-P-Man usage in the lumenal side of the ER was normal, we tested the effect of swainsonine, an inhibitor of Golgi α1,2-mannosidase II, on N-glycan biosynthesis. It is well known that swainsonine blocks the processing of high-mannose-type N-glycan to complex-type. After 3 d of culturing in the presence of swainsonine, complex-type N-glycans on the cell surface were stained with FITC-conjugated PHA-P lectin and analyzed by FACS. PHA-P staining of CHO F21 cells was lower in the presence of swainsonine (Figure 3B, 1, thin vs. bold lines). On the other hand, PHA-P staining of Dol-P-Man usage-defective cells, such as CHO Lec35, were not affected by swainsonine (3), because those cells did not have Dol-P-Man-derived Man residues, which would be removed by swainsonine-sensitive Golgi α1,2-mannnosidase II. The staining of swainsonine-treated CHO2.46 cells was significantly lower than that of nontreated CHO 2.46 cells (2), like wild-type cells (1), indicating that CHO2.46 cells was normal in Dol-P-Man general usage. We next analyzed oligosaccharides released from N-glycan precursors by acidic hydrolysis. CHO2.46 cells synthesized GlcNAc2Man9Glc3, the mature N-glycan precursor (Figure 3C, lane 2), as in the parental CHO F21 (lane 1). From these results, we concluded that the mutant CHO2.46 was impaired at the GPI-MT-I step.
Cloning of PIG-X That Normalizes the Defective Phenotype of CHO2.46 Mutant Cells
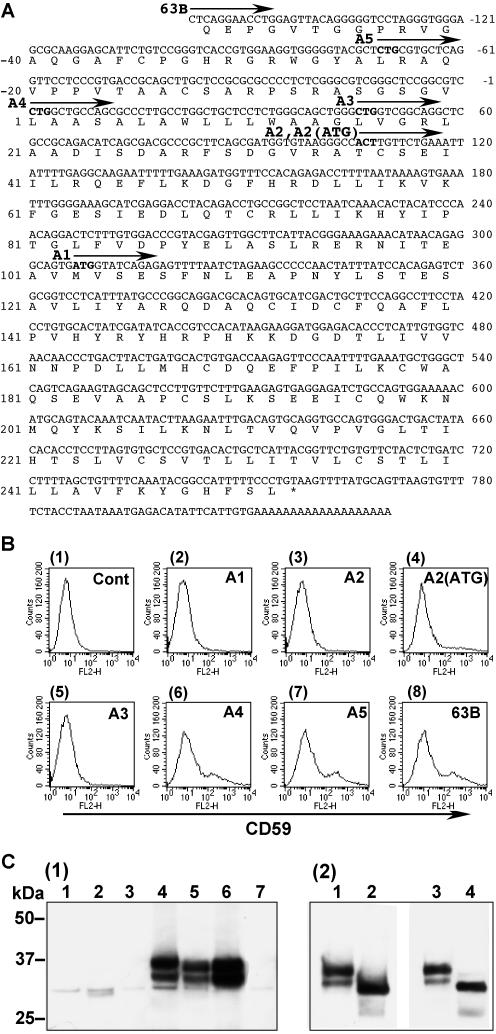
To clone the gene responsible for CHO2.46 mutant cells, we used an expression cloning strategy. We transfected CHO2.46 cells with a rat cDNA library, sorted out CD59 positive cells and rescued the plasmids from the collected cells. After two cycles of transfection and sorting, we tested each rescued plasmid and finally obtained six clones that were able to restore the surface CD59 expression on CHO2.46. These clones shared the same nucleotide sequence but had 5′-upstream regions of different lengths. Clone 63B contained the longest cDNA fragment consisting of 988 base pairs (Figure 4A; accession no. AB177393). Clone 11G was 40 base pairs shorter (starting at G-118) and clones 6D, 7F and 37F were 44 base pairs shorter (starting at G-114). Because this cDNA encoded a previously uncharacterized protein, we termed it PIG-X. A search of the genome databases uncovered human (BC022542) and mouse (AK008583) PIG-X homologues. The human PIG-X gene consists of 6 exons spanning 25 kb in the chromosome 3q29.
Figure 4.
Characterization of PIG-X. (A) Nucleotide and deduced amino acid sequences of the rat PIG-X clone 63B. The arrows indicate the starting positions of various 5′ deletion mutants. Candidate initiation codons are in boldface type. Amino acid positions are on the left and nucleotide positions are on the right. (B) Restoration of the surface expression of GPI-anchored protein CD59. CHO2.46 cells were transiently transfected with each construct shown in A and analyzed by FACS. 1, control empty plasmid; 2, deletion construct A1; 3, A2; 4, A2(ATG), the first ACT codon in A2 was replaced with ATG; 5, A3; 6, A4; 7, A5; and 8, the full-length clone 63B. (C) Western blotting of PIG-X with a FLAG-tag at the C terminus. 1, CHO K1 cells were transfected with various mutant constructs of FLAG-tagged PIG-X followed by immunoprecipitation and Western blotting by using anti-FLAG antibody. Lane 1, A2; lane 2, A2(ATG); lane 3, A3; lane 4, A4; lane 5, clone 63B; lane 6, 63B(CTG→ATG); and lane 7, 63B(CTG→CTT). 2, affinity-purified FLAG-tagged PIG-X was treated with N-glycan releasing enzymes. Lanes 1 and 3, buffer-treated controls; lane 2, peptide-N-glycanase treated; and lane 4, endoglycosidase H treated.
The PIG-X Gene Uses an Unusual CTG Initiation Codon
In the nucleotide sequence of the rat PIG-X (clone 63B), the first candidate initiation codon ATG was found at nucleotides 307–309 and was also conserved in the human and mouse PIG-X sequences. We constructed a deletion mutant PIG-X (A1) lacking the region 5′ of this ATG codon (Figure 4A). When this construct was transfected into CHO2.46 cells CD59 expression was not recovered (Figure 4B, 2), indicating that the initiation codon must exist further upstream. GTG and CTG are alternative initiation codons in some mammalian genes (Touriol et al., 2003). However, Kozak's rule is still important for efficient translation, especially the placement of an A or G at the –3 position. We next constructed various 5′ deletion mutant cDNAs, starting at the candidate initiation codons (Figure 4A), and analyzed their abilities to restore CD59 expression on CHO 2.46 cells. A3 did not show any activity, whereas A4, as well as A5, had activities similar to the full-length clone 63B (Figure 4B, 5–8). Thus, the CTG codon at nucleotides 1–3 could be an initiation codon. Because Thr-36 in rat PIG-X is replaced with Met in human PIG-X, we constructed deletion mutants A2 and A2(ATG), which start at Thr-36 and Met-36, respectively. Notably, CD59 expression was weakly restored by A2(ATG), but not by A2 (3 and 4).
The protein expression levels and the molecular sizes of various deletion mutants of PIG-X with C-terminal triple FLAG-tags were analyzed by Western blotting by using anti-FLAG antibody. The active cDNA construct A4 produced two proteins of 33 and 35 kDa (Figure 4C, 1, lane 4). The full-length construct (clone 63B) produced the same bands (lane 5) as A4, indicating that the CTG at position 1–3 acted as the major initiation codon. In contrast to these active constructs, inactive A2 and A3 produced only trace amounts of proteins of much smaller size (lanes 1 and 3). Similarly, weakly active A2(ATG) did not produce detectable 33- and 35-kDa bands, indicating that translation initiation at that position is inefficient (lane 2).
To further confirm that position 1–3 is the major initiation site, point mutations were introduced. When CTG (Leu) was replaced with ATG (Met) in the clone 63B, the protein expression level was approximately twofold higher than in the clone 63B (Figure 4C, 1, lane 5 vs. 6). On the other hand, when CTG (Leu) was replaced with CTT (Leu), the PIG-X expression was almost completely lost (lane 7).
Characterization of PIG-X Protein
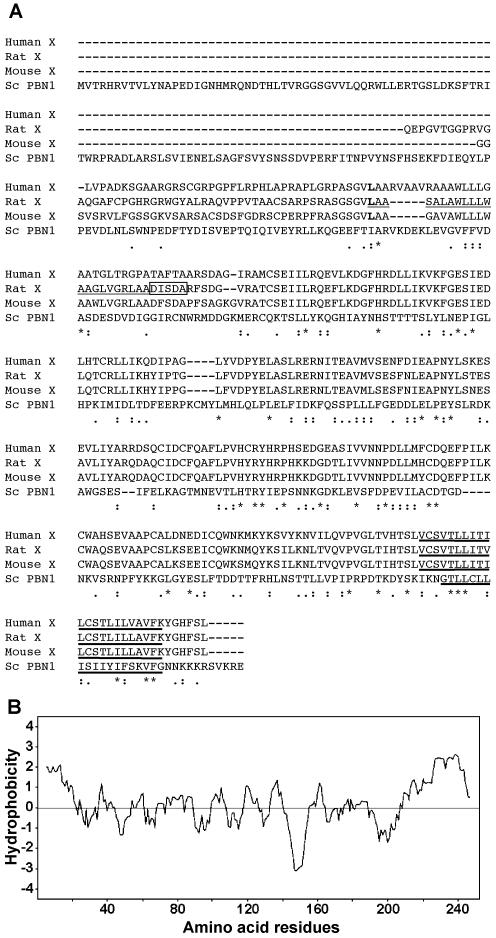
The above-mentioned results indicated that the main initiation codon of rat PIG-X gene is CTG at positions 1–3 and the open reading frame (ORF) consists of 756 base pairs, encoding a 252-amino acid polypeptide. The amino acid sequences of PIG-X from rat, mouse, and human were aligned (Figure 5A). The N-terminal amino acid sequence (Leu-1∼) as well as the nucleotide sequence around the initiation codon of rat PIG-X are well conserved in those of human and mouse, suggesting that this CTG (Leu) may be an initiation codon in the human and mouse PIG-X genes. Within these ORFs, human and mouse PIG-X showed 77 and 93% amino acid identities, respectively, with rat PIG-X. Putative homologues also were found in Arabidopsis thaliana (AB022221; BAA97219.1), Anopheles gambiae (EAA15002), Caenorabditis elegans (T25787), Schizosaccharomyces pombe (AL035075; SPCC1919.02), and S. cerevisiae (X59720; YCL052C), although the similarities are not high (<20% identity within the overlapping region).
Figure 5.
(A) Alignment of amino acid sequences of human, rat, and mouse PIG-X, and S. cerevisiae Pbn1p. Asterisks, amino acids conserved in all four proteins; single dots, positions in which similar residues are conserved; double dots, positions in which highly similar residues are conserved; leucine residues (L) at the translation initiation site are in boldface type. N-terminal sequence of mature rat PIG-X are boxed. Signal peptide in rat PIG-X and putative transmembrane domains are underlined and bold-underlined, respectively. (B) Hydropathy plot of rat PIG-X according to the Kyte–Doolittle method (Kyte and Doolittle, 1982).
The mature rat PIG-X with a C-terminal triple FLAG-tag was expressed in CHO K1 and purified with anti-FLAG beads, and then the N-terminal was sequenced by Edman degradation. The peptide sequence was found to be DISDA (corresponding to amino acids 23–27), indicating that the first 22 amino acid residues were removed by a signal peptidase and that the N-terminal portion of the mature PIG-X was inserted into the lumenal side of the ER. Using TMpred software (Hofmann and Stoffel, 1993), PIG-X was predicted to have one transmembrane region near its C terminus (Figure 5, A and B).
There are two N-glycosylation motifs in a large N-terminal hydrophilic region (97N-I-T and 209N-L-T). To test whether these N-glycosylation sites are used, we treated C-terminal triple FLAG-tagged PIG-X expressed in CHO K1 cells with peptide-N-glycanase and analyzed the products by Western blotting by using anti-FLAG antibody. The two major bands (35 and 33 kDa) seen in the absence of peptide-N-glycanase treatment disappeared, and a smaller single band (31 kDa) occurred (Figure 4C, 2, lanes 1 and 2). This indicates that one of the two N-glycosylation sites was fully glycosylated and that the other was partially glycosylated. Because the large N-terminal region was N-glycosylated, it must be oriented to the lumenal side of the ER, consistent with the presence of an N-terminal signal peptide. These two N-glycans also were released by a treatment with endoglycosidase H, indicating that they were high-mannose-type N-glycans (lanes 3 and 4). The calculated molecular mass of mature PIG-X (without signal peptide) is 26,215 and coincides with the mass estimated by SDS-PAGE (28 kDa, without FLAG-tag and N-glycans).
The intracellular localization of PIG-X was analyzed by confocal microscopy. PIG-X colocalized with the ER marker BiP (our unpublished data), indicating that PIG-X is expressed in the ER. This result was consistent with the endoglycosidase H sensitivity of N-glycans on PIG-X. In summary, PIG-X is an ER-resident type-I transmembrane protein.
PIG-X Associates with and Stabilizes PIG-M
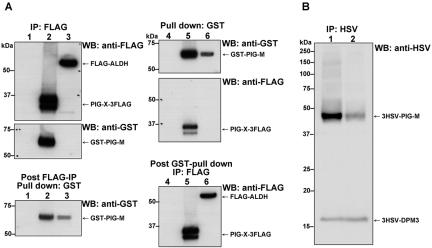
Because both PIG-M-defective Ramos 517-17 cells and PIG-X-defective CHO2.46 cells lacked GPI-MT-I, we predicted that GPI-MT-I might be a complex consisting of PIG-M and PIG-X. To test this, we analyzed the binding of PIG-X and PIG-M with coprecipitation experiments by using differentially tagged PIG-X and PIG-M. We transiently cotransfected CHO K1 cells with pME/PIG-X-3FLAG and pME/GST-PIG-M. After 2.5 d of culturing, we lysed the cells with 1.0% Nonidet P-40, immunoprecipitated PIG-X with anti-FLAG beads, and assessed the coprecipitated PIG-M by Western blotting by using anti-glutathione S-transferase (GST) (Figure 6A). More than 80% of expressed GST-PIG-M was coprecipitated with FLAG-PIG-X, whereas only ∼20% of GST-PIG-M remained in the post-anti-FLAG immunoprecipitation fraction (lane 2, middle vs. bottom). On the contrary, when FLAG-ALDH (aldehyde dehydrogenase), a control ER protein, and GST-PIG-M were coexpressed, no significant binding was detected (lane 3). Similarly, when GST-PIG-M was captured by glutathione-beads, ∼30% of PIG-X-3FLAG was coprecipitated (lane 5), whereas FLAG-ALDH was not coprecipitated at all (lane 6). These results clearly indicate that PIG-X is specifically associated with PIG-M. Moreover, we noticed that the expression level of PIG-M was ∼10-fold higher when PIG-X was coexpressed, suggesting that PIG-X stabilizes PIG-M. To further test this, we stably transfected tagged PIG-M into PIG-X–defective CHO2.46 cells and then transfected PIG-X cDNA or an empty vector. The expression of tagged PIG-M was very low in the absence of PIG-X (Figure 6B, lane 2), whereas it was expressed 10 times more efficiently with PIG-X (lane 1), indicating that PIG-X stabilized PIG-M.
Figure 6.
PIG-X associates with and stabilizes PIG-M. (A) CHO K1 cells were cotransfected with two cDNAs, followed by affinity precipitation with either anti-FLAG- or glutathione-beads. From the supernatant of affinity precipitation, unbound proteins were recovered by using other affinity beads as indicated. The corresponding amount of samples were applied onto SDS-PAGE and detected by Western blotting with either anti-FLAG or anti-GST antibodies. Lanes 1 and 4, empty plasmid only; lanes 2 and 5, FLAG-tagged PIG-X and GST-tagged PIG-M; lanes 3 and 6, FLAG-tagged ALDH and GST-tagged PIG-M. (B) CHO2.46 cells were first cotransfected with HSV-tagged-PIG-M and -DPM3 (as a control). The transfectant cells were further transfected transiently with PIG-X (lane 1) or an empty vector (lane 2). Expression levels of PIG-M and DPM3 were assessed by immunoprecipitation and Western blotting with anti-HSV.
Cotransfection of S. cerevisiae PBN1 and S. cerevisiae GPI14 into the Mutant CHO2.46 Normalized Its Defective Phenotype
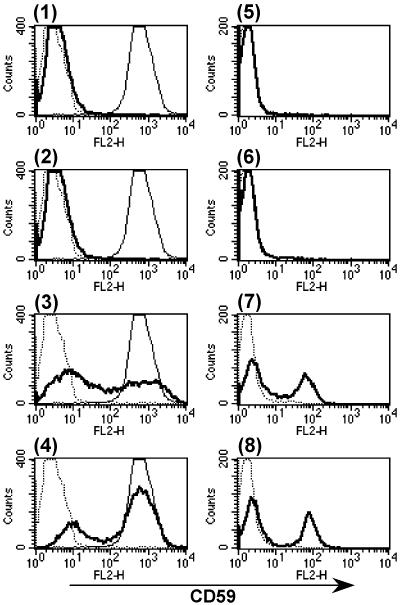
We found that the ORF YCL052C of S. cerevisiae encodes a PIG-X homologue, which was reported as PBN1 (proteinase B negative 1) involved in autoprocessing of proproteinase B in the ER (Naik and Jones, 1998). Pbn1p is much larger in size than rat PIG-X (416- vs. 252-amino acid residues), and the C-terminal part of Pbn1p had only 16% amino acid identity with PIG-X (Figure 5A). The predicted membrane topology of the C-terminal part of Pbn1p was similar to that of PIG-X. To test whether Pbn1p was a functional homologue of PIG-X, we transiently transfected CHO2.46 cells with pME/PBN1–3FLAG; however, cell surface expression of CD59 was not restored (Figure 7, 1). Then, we speculated that Pbn1p might not be able to associate with endogenous hamster PIG-M resulting in a lack of an active enzyme complex. To test this hypothesis, we cotransfected CHO2.46 cells with both pME/PBN1–3FLAG and pME/GST-GPI14, the S. cerevisiae homologue (YJR013W) of PIG-M having ∼35% amino acid sequence identity. Although the transfection of GPI14 alone did not restore the surface expression of CD59 (2), the combination with PBN1 did complement expression (3) to a level comparable with that restored by rat PIG-X (4). Moreover, the combination of PBN1 and GPI14 restored the surface expression of CD59 on human PIG-M mutant Ramos 517-17 cells (7). Transfection of PBN1 (5) or GPI14 (6) alone did not work. These results clearly indicate that Pbn1p is the functional homologue of mammalian PIG-X and that the association of PIG-X and PIG-M is not interchangeable between mammals and yeasts.
Figure 7.
Cotransfection of S. cerevisiae PBN1 and GPI14 cDNAs restored surface expression of CD59 in the mutant cells. CHO2.46 cells (1–4) and Ramos 517-17 cells (5–8) were transiently transfected with cDNAs and analyzed by FACS. 1 and 5, PBN1 cDNA alone; 2 and 6, GPI14 cDNA alone; 3 and 7, PBN1 and GPI14 cDNAs; 4, rat PIG-X cDNA alone; and 8, human PIG-M cDNA alone. Thick lines, transfected cells stained with anti-CD59; dotted lines, nontransfected cells stained with anti-CD59; thin lines in 1–4, CHO F21 cells stained with anti-CD59.
DISCUSSION
PIG-X Is the Essential Component of GPI-MT-I
We have established a novel class of mutant, CHO2.46, defective in the cell-surface expression of GPI-anchored proteins by using two GPI-anchor–recognizing microbial toxins. Biochemical analyses revealed that CHO2.46 cells were impaired in GPI-MT-I activity as in PIG-M–defective Ramos 517-17 cells (Figures 2 and 3). By expression cloning with these mutant cells, we identified a cDNA, termed PIG-X, that fully restored the defective phenotype after transfection. In the strict sense, however, we are not able to declare that PIG-X is a gene responsible for this mutant, because we have not sequenced PIG-X locus in the genome of Chinese hamster. The translation of PIG-X started at the CUG initiation codon generating an ER-resident type-I transmembrane protein with a large N-terminal region oriented toward the lumenal side of the ER (Figures 4 and 5). PIG-X was specifically coimmunoprecipitated with PIG-M, which had been previously identified as GPI-MT-I (Maeda et al., 2001) (Figure 6A). It is highly likely that PIG-M is the catalytic subunit of GPI-MT-I because it has structural similarity with other Dol-P-Man/Glc using glycosyltransferases and contains a DXD motif essential for its enzyme activity. Thus, PIG-X may not have a catalytic domain but is a critical subunit of GPI-MT-I. We demonstrated that the stable expression of PIG-M is dependent upon PIG-X (Figure 6B). Dol-P-Man/Glc using glycosyltransferases involved in the biosyntheses of lipid-linked oligosaccharides, GPI-anchor, and N-glycan precursor form a superfamily characterized by similar transmembrane topology (Oriol et al., 2002; Liu and Mushegian, 2003). Among these, GPI-MT-I is the first example of a heterodimeric enzyme.
Yeast Pbn1p Is the Functional Homologue of Mammalian PIG-X
We found that S. cerevisiae Pbn1p is the functional homologue of mammalian PIG-X. Although neither PBN1 nor GPI14 alone could restore the defective phenotype of the PIG-X mutant CHO2.46 or PIG-M mutant Ramos 517-17, cotransfection of PBN1 and GPI14 restored both mutant cell lines (Figure 7). Pbn1p was first identified as an ER protein essential for autoprocessing of proproteinase B (Prb1p), a vacuolar protease (Naik and Jones, 1998). Because the pbn1 deletant yeast is inviable despite the nonessential nature of proteinase B for vegetative growth, other important roles of Pbn1p were suggested (Naik and Jones, 1998). Here, we show that Pbn1p is involved in the biosynthesis of GPI, which is essential for growth of yeast. The relationship between the biosynthesis of GPI and the autoprocessing of proteinase B is unknown. Because Pbn1p has an additional 164-amino acid region containing a coiled-coil structure at the N terminus compared with mammalian PIG-X, it may be a dual-functional protein. Another report described that yeast cells carrying multiple copies of both PBN1 and LRE1 (laminarinase resistance 1) showed resistance to the cell wall β1,3-glucan degrading enzyme, although the cells carrying either gene showed no significant alterations (Lai et al., 1997). These observations may suggest that Pbn1p serves as a rate limiting factor for GPI-anchor biosynthesis and interacts with several molecules other than Gpi14p.
GPI-MT-I Is a Promising Target for Antimicrobial Drugs
GPI-biosynthesis is essential for growth of fungi and protozoa; thus, specific inhibitors for the GPI-biosynthetic pathway would be antifungal and antiprotozoan drugs. The complex of yeast Pbn1p and Gpi14p was functional in mammalian cells, although neither alone was sufficient for function (Figure 7). This fact indicates that the correct assembly of these two components is necessary for active GPI-MT-I formation and the intermolecular recognition between these two is highly specific in each organism. A compound inhibitory to the assembly of Pbn1p and Gpi14p in yeast could be a specific antifungal drug.
In the genome of T. brucei, a protozoan parasite that causes sleeping sickness in human, we did not find a homologue of mammalian PIG-X and yeast Pbn1p. It is well known that the substrate specificity of GPI-MT-I in T. brucei is different from that in mammals and yeast, namely, the former prefers GlcN-PI as an acceptor substrate, whereas the latter prefer GlcN-acyl-PI (Smith et al., 1996). It is possible that TbGPI14 requires a counter-subcomponent nonhomologous with mammalian and yeast PIG-X, and those subcomponents including PIG-X define the substrate specificities of GPI-MT-I. If this hypothesis is correct, a trypanosome-specific component could be a target of antiprotozoan drugs.
Acknowledgments
We thank Kohjiro Nakamura for cell sorting, Fumiko Ishii and Keiko Kinoshita for technical assistance, and Dr. Tadashi Suzuki for discussion. This work was supported by grants from the Ministry of Education, Sports, Science, Culture and Technology of Japan. Y. H. was partially supported by a grant from the Ministry of Health and Welfare of the Republic of Korea (01-PJ10-PG6-01GM00-0002).
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E04-09-0802) on January 5, 2005.
Abbreviations used: CHO, Chinese hamster ovary; DAF, decay accelerating factor; Dol-P, dolichol-phosphate; ER, endoplasmic reticulum; EtNP, phosphoethanolamine; FACS, fluorescence-activated cell sorter; GlcN-PI(C8), glucosaminyl-PI with di-octanoyl groups; GPI, glycosylphosphatidylinositol; MT, mannosyltransferase; PI, phosphatidylinositol.
References
- Buckley, J. T. (1999). The channel-forming toxin aerolysin. In: The Comprehensive Sourcebook of Bacterial Protein Toxins, ed. J. E. Alouf and J. H. Freer, London: Academic Press, 362-372.
- Burda, P., and Aebi, M. (1999). The dolichol pathway of N-linked glycosylation. Biochim. Biophys. Acta 1426, 239-257. [DOI] [PubMed] [Google Scholar]
- Camp, L. A., Chauhan, P., Farrar, J. D., and Lehrman, M. A. (1993). Defective mannosylation of glycosylphosphatidylinositol in Lec35 CHO cells. J. Biol. Chem. 268, 6721-6728. [PubMed] [Google Scholar]
- Chantret, I., et al. (2003). A deficiency in dolichyl-P-glucose: Glc1Man9GlcNAc2-PP-dolichyl α3-glucosyltransferase defines a new subtype of congenital disorders of glycosylation. J. Biol. Chem. 278, 9962-9971. [DOI] [PubMed] [Google Scholar]
- Doerrler, W. T., Ye, J., Falck, J. R., and Lehrman, M. A. (1996). Acylation of glucosaminyl phosphatidylinositol revisited. J. Biol. Chem. 271, 27031-27038. [DOI] [PubMed] [Google Scholar]
- Doucey, M. A., Hess, D., Cacan, R., and Hofsteenge, J. (1998). Protein C-mannosylation is enzyme-catalysed and uses dolichyl-phosphate-mannose as a precursor. Mol. Biol. Cell 9, 291-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson, M. A., Brimacombe, J. S., Brown, J. R., Crossman, A., Dix, A., Field, R. A., Guther, M. L., Milne, K. G., Sharma, D. K., and Smith, T. K. (1999). The GPI biosynthetic pathway as a therapeutic target for African sleeping sickness. Biochim. Biophys. Acta 1455, 327-340. [DOI] [PubMed] [Google Scholar]
- Grimme, S. J., Westfall, B. A., Wiedman, J. M., Taron, C. H., and Orlean, P. (2001). The essential Smp3 protein is required for addition of the side-branching fourth mannose during assembly of yeast glycosylphosphatidylinositols. J. Biol. Chem. 276, 27731-27739. [DOI] [PubMed] [Google Scholar]
- Hofmann, K., and Stoffel, W. (1993). TMbase - a database of membrane spanning proteins segments. Biol. Chem. Hoppe-Seyler 347, 166 [Google Scholar]
- Hong, Y., Maeda, Y., Watanabe, R., Ohishi, K., Mishkind, M., Riezman, H., and Kinoshita, T. (1999). Pig-n, a mammalian homologue of yeast Mcd4p, is involved in transferring phosphoethanolamine to the first mannose of the glycosylphosphatidylinositol. J. Biol. Chem. 274, 35099-35106. [DOI] [PubMed] [Google Scholar]
- Hong, Y., Ohishi, K., Inoue, N., Kang, J. Y., Shime, H., Horiguchi, Y., van der Goot, F. G., Sugimoto, N., and Kinoshita, T. (2002). Requirement of N-glycan on GPI-anchored proteins for efficient binding of aerolysin but not Clostridium septicum α-toxin. EMBO J. 21, 5047-5056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong, Y., Ohishi, K., Kang, J. Y., Tanaka, S., Inoue, N., Nishimura, J., Maeda, Y., and Kinoshita, T. (2003). Human PIG-U and yeast Cdc91p are the fifth subunit of GPI transamidase that attaches GPI-anchors to proteins. Mol. Biol. Cell 14, 1780-1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue, N., Kinoshita, T., Orii, T., and Takeda, J. (1993). Cloning of a human gene, PIG-F, a component of glycosylphosphatidylinositol anchor biosynthesis, by a novel expression cloning strategy. J. Biol. Chem. 268, 6882-6885. [PubMed] [Google Scholar]
- Kang, J. Y., Hong, Y., Ashida, H., Shishioh, N., Murakami, Y., Morita, Y. S., Maeda, Y., and Kinoshita, T. (2005). PIG-V involved in transferring the second mannose in glycosylphosphatidylinositol. J. Biol. Chem. (in press). [DOI] [PubMed]
- Kinoshita, T., and Inoue, N. (2000). Dissecting and manipulating the pathway for glycosylphosphatidylinositol-anchor biosynthesis. Curr. Opin. Chem. Biol. 4, 632-638. [DOI] [PubMed] [Google Scholar]
- Kyte, J., and Doolittle, R. F. (1982). A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 157, 105-132. [DOI] [PubMed] [Google Scholar]
- Lai, M. H., Silverman, S. J., Gaughran, J. P., and Kirsch, D. R. (1997). Multiple copies of PBS2, MHP1 or LRE1 produce glucanase resistance and other cell wall effects in Saccharomyces cerevisiae. Yeast 13, 199-213. [DOI] [PubMed] [Google Scholar]
- Liu, J., and Mushegian, A. (2003). Three monophyletic superfamilies account for the majority of the known glycosyltransferases. Protein Sci. 12, 1418-1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda, Y., Tanaka, S., Hino, J., Kangawa, K., and Kinoshita, T. (2000). Human dolichol-phosphate-mannose synthase consists of three subunits, DPM1, DPM2 and DPM3. EMBO J. 19, 2475-2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda, Y., Watanabe, R., Harris, C. L., Hong, Y., Ohishi, K., Kinoshita, K., and Kinoshita, T. (2001). PIG-M transfers the first mannose to glycosylphosphatidylinositol on the lumenal side of the ER. EMBO J. 20, 250-261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manya, H., Chiba, A., Yoshida, A., Wang, X., Chiba, Y., Jigami, Y., Margolis, R. U., and Endo, T. (2004). Demonstration of mammalian protein O-mannosyltransferase activity: coexpression of POMT1 and POMT2 required for enzymatic activity. Proc. Natl. Acad. Sci. USA 101, 500-505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami, Y., Siripanyapinyo, U., Hong, Y., Kang, J. Y., Ishihara, S., Nakakuma, H., Maeda, Y., and Kinoshita, T. (2003). PIG-W is critical for inositol acylation but not for flipping of glycosylphosphatidylinositol-anchor. Mol. Biol. Cell 14, 4285-4295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naik, R. R., and Jones, E. W. (1998). The PBN1 gene of Saccharomyces cerevisiae: an essential gene that is required for the post-translational processing of the protease B precursor. Genetics 149, 1277-1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura, N., Inoue, N., Watanabe, R., Takahashi, M., Takeda, J., Stevens, V. L., and Kinoshita, T. (1997). Expression cloning of PIG-L, a candidate N-acetylglucosaminyl-phosphatidylinositol deacetylase. J. Biol. Chem. 272, 15834-15840. [DOI] [PubMed] [Google Scholar]
- Ohishi, K., Inoue, N., and Kinoshita, T. (2001). PIG-S and PIG-T, essential for GPI anchor attachment to proteins, form a complex with GAA1 and GPI8. EMBO J. 20, 4088-4098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oriol, R., Martinez-Duncker, I., Chantret, I., Mollicone, R., and Codogno, P. (2002). Common origin and evolution of glycosyltransferases using Dol-P-monosaccharides as donor substrate. Mol. Biol. Evol. 19, 1451-1463. [DOI] [PubMed] [Google Scholar]
- Smith, T. K., Cottaz, S., Brimacombe, J. S., and Ferguson, M. A. (1996). Substrate specificity of the dolichol phosphate mannose:glucosaminyl phosphatidylinositol α1–4-mannosyltransferase of the glycosylphosphatidylinositol biosynthetic pathway of African trypanosomes. J. Biol. Chem. 271, 6476-6482. [DOI] [PubMed] [Google Scholar]
- Sutterlin, C., Horvath, A., Gerold, P., Schwarz, R. T., Wang, Y., Dreyfuss, M., and Riezman, H. (1997). Identification of a species-specific inhibitor of glycosylphosphatidylinositol synthesis. EMBO J. 16, 6374-6383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi, M., Inoue, N., Ohishi, K., Maeda, Y., Nakamura, N., Endo, Y., Fujita, T., Takeda, J., and Kinoshita, T. (1996). PIG-B, a membrane protein of the endoplasmic reticulum with a large lumenal domain, is involved in transferring the third mannose of the GPI anchor. EMBO J. 15, 4254-4261. [PMC free article] [PubMed] [Google Scholar]
- Tanaka, S., Maeda, Y., Tashima, Y., and Kinoshita, T. (2004). Inositol deacylation of glycosylphosphatidylinositol-anchored proteins is mediated by mammalian PGAP1 and yeast Bst1p. J. Biol. Chem. 279, 14256-14263. [DOI] [PubMed] [Google Scholar]
- Taron, B. W., Colussi, P. A., Grimme, J. M., Orlean, P., and Taron, C. H. (2004). Human Smp3p adds a fourth mannose to yeast and human glycosylphosphatidylinositol precursors in vivo. J. Biol. Chem. 279, 36083-36092. [DOI] [PubMed] [Google Scholar]
- Touriol, C., Bornes, S., Bonnal, S., Audigier, S., Prats, H., Prats, A. C., and Vagner, S. (2003). Generation of protein isoform diversity by alternative initiation of translation at non-AUG codons. Biol. Cell 95, 169-178. [DOI] [PubMed] [Google Scholar]
- Tweten, R. K., and Sellman, B. R. (1999). Clostridium septicum pore-forming and lethal α-toxin. In: The Comprehensive Sourcebook of Bacterial Protein Toxins, ed. J. E. Alouf and J. H. Freer, London: Academic Press, 435-442.
- Ware, F. E., and Lehrman, M. A. (1996). Expression cloning of a novel suppressor of the Lec15 and Lec35 glycosylation mutations of CHO cells. J. Biol. Chem. 271, 13935-13938. [DOI] [PubMed] [Google Scholar]
- Watanabe, R., Inoue, N., Westfall, B., Taron, C. H., Orlean, P., Takeda, J., and Kinoshita, T. (1998). The first step of glycosylphosphatidylinositol biosynthesis is mediated by a complex of PIG-A, PIG-H, PIG-C and GPI1. EMBO J. 17, 877-885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe, R., Murakami, Y., Marmor, M. D., Inoue, N., Maeda, Y., Hino, J., Kangawa, K., Julius, M., and Kinoshita, T. (2000). Initial enzyme for glycosylphosphatidylinositol biosynthesis requires PIG-P and is regulated by DPM2. EMBO J. 19, 4402-4411. [DOI] [PMC free article] [PubMed] [Google Scholar]