Abstract
Methods for exerting and measuring forces on single molecules have revolutionized the study of the physics of biology. However, it is often the case that biological processes involve rotation or torque generation, and these parameters have been more difficult to access experimentally. Recent advances in the single molecule field have led to the development of techniques which add the capability of torque measurement. By combining force, displacement, torque, and rotational data, a more comprehensive description of the mechanics of a biomolecule can be achieved. In this review, we highlight a number of biological processes for which torque plays a key mechanical role. We describe the various techniques that have been developed to directly probe the torque experienced by a single molecule, and detail a variety of measurements made to date using these new technologies. We conclude by discussing a number of open questions and propose systems of study which would be well suited for analysis with torsional measurement techniques.
Keywords: Angular optical trapping, rotor bead tracking, magnetic tweezers, DNA phase transitions, rotary molecular motors
INTRODUCTION
Over the past several decades, techniques for observing and manipulating single biological molecules have opened up new fields of study, allowing researchers in the biological and biophysical sciences to understand how the components of life behave, not just biochemically, but also mechanically. The direct manipulations of physical parameters, such as force, have elucidated the mechanisms of action for a myriad of biological machines and the structures that compose each living cell. While force is certainly a key physical coordinate, it is not the only parameter of importance. Indeed, for a host of cellular machinery, rotation and torque may be even more crucial. To date, these variables have been much harder to observe directly, leading to gaps in our knowledge regarding how key biomolecules function. In recent years, however, researchers have developed a variety of techniques and methodologies which enable direct access to these mechanical parameters, allowing for a more comprehensive understanding of the behavior of certain biological components. In this review, we will detail the major technical advances made to date, which allow for the measurement of rotation and torque in single molecule systems. We will also discuss a variety of results obtained from experiments on biomolecules subjected to torsional strain, and detail the manner in which the direct acquisition of torque data has led to new and important insights. Finally, we propose a range of topics for future study with these new methodologies.
TORQUE IN BIOLOGICAL SYSTEMS
Rotation and the corresponding torque generation are common, though often underappreciated, features of a number of cellular processes. One can broadly identify two kinds of processes which have been of particular interest to researchers: those involving DNA, one of the major chiral molecules of the cell, and its related processing machinery; and those involving the so-called rotary motors, such as F0F1-ATPase and the bacterial flagellar motor. In this section, we will provide an overview of the in vivo relevance of both torque and rotation.
DNA and DNA Based Motors
Since the discovery of the double-helical nature of DNA by Watson and Crick in 1953 (117), researchers have come to realize that the cell must be able to overcome a set of topological challenges in order to perform some of its basic functions. For example, initiation of replication and transcription requires opening of the double helix (57); architectural proteins directionally wrap the DNA (71); torsional stress can be generated as translocases move along the molecule (26, 68). Alteration of the topological state of DNA, which can include structural changes, is called DNA supercoiling. Supercoiling is often characterized by means of the superhelical density, defined as the number of extra turns in the DNA normalized by the total number of superhelical turns in relaxed DNA. Superhelical density is tightly regulated in cells, and is maintained at ~ −0.05 in both prokaryotes and eukaryotes (92). In prokaryotes, approximately half of the supercoiling is unconstrained, while the other half is constrained by various proteins, such as HU, HNS and RNAP (30). Conversely, in eukaryotes, on average all supercoiling is constrained by the nucleosomes (54), although local unconstrained supercoiling can arise (10, 69). The supercoiling balance is maintained by a special class of enzymes – topoisomerases – that are able to relax (and sometimes introduce) supercoiling.
Supercoiling can drastically alter the DNA molecule. Both positive and negative supercoiling can induce the formation of intertwined loop-like structures called plectonemes (111). Negative supercoiling, in particular, has been biochemically shown to disrupt the double-helical structure of DNA, producing a variety of non-B DNA conformations. Strand separation is the most general consequence of underwinding, however specific sequences can produce more exotic structures: cruciforms at palindromic sites, left-handed Z-DNA, and G-quadruplexes within certain GC-rich tracts (55). While initially these structures were considered a mere in vitro curiosity, research in the past decades has demonstrated their existence in vivo and provided examples of important regulatory roles they can play, particularly in transcription.
Transcription initiation requires melting of ~9 bp of the promoter region (13), and is thus modulated, in part, by torsion. The effects of supercoiling on the transcription of individual genes have been extensively documented both in vitro (62) and in vivo (44). Supercoiling can also act as a global regulator of transcription, as has been strikingly illustrated by the studies of circadian gene expression in cyanobacteria (110). Besides the melting of the promoter region, supercoiling can promote formation of non-B DNA structures that can attract regulatory proteins (15).
Torque impacting transcription is only half the story. Transcription can also be a source of torque. During elongation, the RNA polymerase machinery tracks the helical groove of DNA, which requires rotation of the enzyme relative to DNA (Figure 1 top). However, as suggested by Liu and Wang (68), polymerase can become immobilized, requiring DNA to rotate instead and thus generating positive supercoiling in front of, and negative supercoiling behind, the polymerase. The free rotation of polymerase can be prevented by a large viscous drag due to the RNA transcript and associated factors, such as ribosomes in prokaryotes and spliceosomes in eukaryotes, as well as tethering to the cell membrane and other cellular structures (9). This so-called “twin-supercoiled domain” model has been confirmed by numerous in vitro and in vivo experiments (56, 58, 109). Transcription-generated supercoiling can modulate transcription of the same or neighboring genes (23, 56), facilitate formation of RNA-DNA hybrids known as R-loops (1), as well as potentially affect the structure of chromatin (see below).
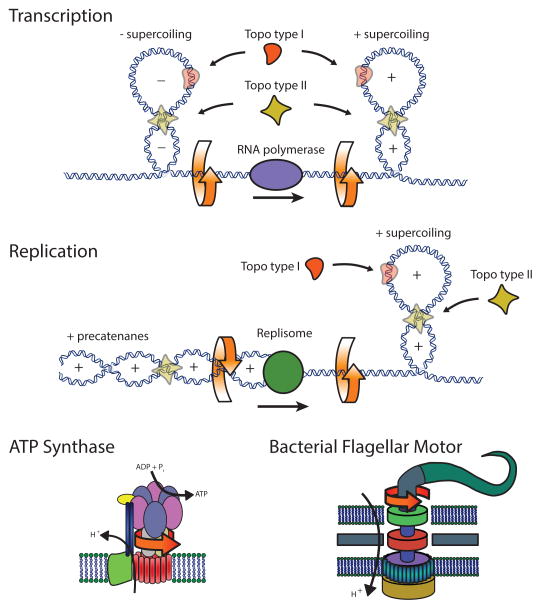
Figure 1. Examples of biological systems involving torque.
(top) Transcription, the process of copying genomic DNA into RNA, involves the rotation of the RNA polymerase enzyme relative to its helical DNA track. Due to the size and typical confinement of the polymerase and associated machinery, the DNA is overtwisted (positive supercoiling) in front of the motor, and undertwisted (negative supercoiling) behind, in accordance with the “twin-supercoiled domain” model.
(middle) During DNA replication, two identical copies are made of a single original DNA molecule. Just as during transcription, positive supercoiling is generated in front of the repliosome complex as it progresses along the DNA track. Behind the replication fork, daughter DNA strands can be twisted and intertwined, forming positive precatenanes. Topoisomerases of various types act during both processes in order to relieve the torsional stress generated along the DNA.
(bottom) Two rotary motors are shown; the F0F1-ATPase, which uses the potential energy from a proton gradient to mechanically rotate and generate ATP from ADP and Pi, and the bacterial flagellar motor, which similarly uses a proton gradient to rotate and provide motility to a swimming bacterium.
Somewhat different topological problems are posed by another important cellular process – replication. Rapid movement of the replication fork, coupled with the unwinding of the parental DNA strands, generates positive supercoiling in the vicinity of the fork (14). If the replication machinery is immobilized, this supercoiling will produce plectonemic loops in front of the fork, in accordance with the “twin-supercoiled domain” model (Figure 1 middle). Alternatively, the replisome may rotate, releasing the torsion upstream, but leading to the intertwining of the daughter strands (generating positive precatenanes) behind the fork (14, 88). Failure to resolve the build-up of torsional stress can lead to fork reversal (31) and genomic instability (89).
In eukaryotes, local generation of torque has additional importance due to its influence on the structure of chromatin. The basic unit of chromatin, the nucleosome, is a protein-DNA complex packaging ~1.7 turns of DNA in a negative supercoil (71). This negative writhe is partially compensated for by the overtwisting of DNA on the nucleosomal surface (71), so that a linking number of −1 is associated with each nucleosome (100). In vitro experiments have observed that nucleosomes preferentially form on negatively supercoiled DNA (25, 87) and are destabilized by positive torsion (61, 86). In the context of the “twin-supercoiled domain” model, this has led to proposals that transcription-dependent supercoiling will destabilize nucleosomes upstream and facilitate their reassembly downstream (25, 86). Torque can also affect higher-order chromatin structures, such as the 30 nm fiber which is believed to have a preferred chirality (97).
In summary, there are a multitude of cellular processes that generate DNA supercoiling. In order to stringently control the level of supercoiling cells employ an array of special enzymes called topoisomerases. Type I topoisomerases transiently break one strand of DNA, and type II topoisomerases cleave and rejoin both DNA strands (54). Both type I and type II enzymes relax supercoils, and type II enzymes unknot and decatenate double-stranded DNA (Figure 1, top and middle). Bacterial gyrase, a unique member of the type II family, is also able to introduce negative supercoils (38). While there is a partial redundancy between different topoisomerases (5), they do appear to have preferred functions. Eukaryotic Topoisomerase II (type II) has been shown to relax positive supercoils in nucleosomal DNA faster than Topoisomerase I (type I) (95), while bacterial Topoisomerase I is essential for relieving transcription-generated negative stress (34).
Rotary Protein Motors
Rotary protein motors perform their function by rotating one group of subunits relative to the rest of the enzyme (82). Several motors of this kind have been discovered, including the bacterial flagellar motor and the two motors comprising F0F1-ATPase. The unique architecture of these motors has been extensively studied, providing numerous insights into enzymatic mechanochemistry (82).
F0F1-ATPase (also called ATP synthase) provides the cell with its crucial energy source, ATP, by synthesizing it from ADP and inorganic phosphate (50). The enzyme consists of two rotary motors, the soluble F1 and the transmembrane F0, coupled by a central rotary shaft (Figure 1 bottom). The soluble motor is driven by ATP hydrolysis and rotates the shaft clockwise, while the transmembrane motor is powered by the ion-motive force and rotates the shaft counterclockwise (20). In most cases, the F0 motor is more powerful and drives the F1 motor in the synthesis direction.
The bacterial flagellar motor is the driving force (and torque) behind bacterial flagella, the long filaments that enable bacteria to propel themselves (Figure 1 bottom). The rotary principle of propulsion was demonstrated in the 1970s (7), and incidentally, this was also the first observation of the action of a single molecular motor. This 11 MDa, 45 nm diameter machine can rotate at up to 1700 Hz and generate up to a remarkable 4500 pN·nm of torque (72, 103, 104). Like the F1-ATPase motor, the bacterial flagellar motor is a transmembrane protein driven by the ion-motive force. However, due to its large size and complexity, the exact principle of operation of the motor remains elusive (103).
While the importance of rotation has been well established for the enzymes described above, the list is not exhaustive. Recently, the molecular motors myosin and kinesin have been observed to partially rotate their cargo (42, 53). It has also been suggested that certain members of the AAA family, such as helicases and DNA packaging proteins, are capable of generating rotation (82). The abundance of torsion-generating enzymes underscores the importance of torque for cellular functions.
METHODS OF TORQUE DETECTION
Despite the importance of torque as a key mechanical regulator in a significant number of biological processes, understanding how torsional stress regulates cellular functions has proven to be experimentally challenging. For example, the most commonly employed method to study regulation by DNA supercoiling has been one or two-dimensional gel electrophoresis. However, this biochemical method does not measure torque directly, and also has limited ability to discern dynamical behavior and population heterogeneities. As Cozzarelli and colleagues pointed out, a reason that “twist and torque changes have been underappreciated is that until recently they were not directly measurable” (26).
Single molecule techniques developed in the past two decades have proven to be powerful approaches for the investigation of the response of biological systems to torsional stress. Individual DNA molecules can now be twisted, and molecular motors, acting on these DNA molecules, can be monitored under physiologically relevant conditions. Initial efforts focused on generating and measuring rotation of a torsionally constrained biological molecule. More recent efforts have resulted in the ability to precisely control the torque exerted on a molecule as well as directly measure the torque generated by the molecule, permitting more quantitative measurements of rotational motions and torsional properties in biology. This review is not intended to be exhaustive in the coverage of all methods of torque measurement. Instead, we will focus on methods that have demonstrated impact and applications at the single molecule level. We will highlight four major categories of torque measurement techniques, in a roughly chronological fashion of their development: 1) electrorotation, 2) viscous drag of a rotating body, 3) optical trapping, and 4) magnetic tweezers.
Electrorotation
Direct application of an external torque on a single molecule was first demonstrated by the method of electrorotation, which applies a constant torque to a polarizable and conductive particle by a rotating electric field (8, 115). Briefly, a MHz rotating electric field is created by applying ac voltage to radially oriented metallic probes. The induced dipole of a particle in the field rotates at the same frequency, but lags or leads the electric field due to the conductance and/or dielectric loss of the particle. This phase delay produces a constant torque on the particle, which depends on the field intensity, rotation rate, and properties of the particle and the solution. It is important to note this relation is complex, and rotation of the particle in the opposite sense of field rotation is possible (49). Studies of the bacterial flagella motor were enhanced by applying electrorotation to the tethered bacterial cells (Figure 2a) (8, 115). This assay is not limited to the rotation of cells; it can also be used to rotate typical dielectric probe particles (108). The combination of optical trapping and electrorotation has been demonstrated and allows for high bandwidth detection of rotation by back focal plane interferometry (93).
Figure 2. Experimental configurations for single molecule torque measurements.
(a) Electrorotation of a single bacterium with its flagella bound to the surface (8).
(b) Rotation of the F1-ATPase can be observed by monitoring the rotational orientation of a fluorescent actin filament (78).
(c) The rotor bead tracking assay utilizes the rotation of a small bead to monitor and/or generate torque. Force and externally imposed twist can be controlled at the DNA ends by microsphere handles (19).
(d) The angular optical trap angularly orients a quartz cylinder with the linear polarization of the input trapping laser beam. The polarization state of the transmitted beam, as measured by the transmitted light intensities in the right and left-handed circular polarizations, directly determines the applied torque on a dsDNA molecule and the angular orientation of the DNA (29, 59).
(e) A magnetic tweezers setup by Celedon et al. (21) consists of a magnetic bead and nanorod torque arm to angularly orient one end of a dsDNA tether; twist is introduced by rotating the microscope coverglass.
(f) Magnetic torque tweezers incorporate a supplementary side magnet to add a small horizontal perturbation to the vertical magnetic field created by a cylindrical magnet, producing a low-stiffness angular trap to orient a magnetic bead (65).
(g) Soft magnetic tweezers use a six pole electromagnet to create a rapidly rotating horizontal magnetic field of varying intensity in order to produce either a constant torque or a magnetic angular trap of tunable stiffness (77).
(h) Electromagnetic torque tweezers use two pairs of Helmholtz coils for dynamic control of the horizontal magnetic field (47).
Viscous Drag of a Rotating Body
The earliest reported measurements of torque on single biological molecules were made by exploiting the drag torque experienced by a body rotating through solution (6, 8). The visual observation of a rotating probe can give sufficient estimates of torque generation, either by estimating the viscous drag coefficient or calibration of the viscous drag. The exerted torque can only be varied via the probe geometry and the viscosity of the solution. Thus the main limitation of this method is that torque and rotational velocity are never fully decoupled. It is therefore difficult to exert a user-defined constant torque, which would be advantageous when probing important parameters such as a motor’s stalling torque.
The viscous drag torque method was first used to study single rotary motors. The torque generated by a single bacterial flagellar motor was estimated based on the viscous torque on the bacterium (70). Subsequently, the torque generated by a single F1-ATPase motor was estimated using an actin filament as the rotational probe coupled to the motor (Figure 2b) (78). The viscous drag torque can be calculated for the rotating body as the product of its viscous drag coefficient and rotation rate.
Recent developments in the use of the viscous drag method for DNA based torque measurements have greatly improved the precision and control of this methodology. Bryant et al. (19) developed a clever rotor bead tracking (RBT) method that utilized the rotation of a small bead attached at the side of a DNA tether in order to both apply and directly measure torque (Figure 2c). The rotation of wound DNA, or the action of DNA binding proteins, is observed by visually tracking the spatial position of the rotor bead as it revolves about the long axis of the DNA strand. Torque is determined by measuring the angular velocity of the rotor bead and multiplying it by the viscous drag factor for a sphere rotating about an axis on its edge. The accuracy of the method is enhanced by the calibration of the viscous drag factor for each rotor bead. In order to maintain DNA as a linear rotation axis, the DNA needs to be extended by optical traps, conventional magnetic tweezers, or micropipettes, while passively observing rotation and extension change (39, 40). RBT can also be used to exert a controlled torque by applying twist with one of the force probes, such that the rotor bead rotates at a constant speed while the force is independently controlled by the force probe (80).
Optical Trapping
Since its introduction to biology by Ashkin et al. (3), optical trapping has proven to be an invaluable tool for single molecule research, permitting the dynamics of motor proteins and their substrates to be examined mechanically, one molecule at a time. Systems that have been investigated under force include, but are not limited to, RNA polymerase (113), DNA polymerase (119), helicase (48), the ribosome (118), nucleosomes (16), DNA (102), RNA (67), and viral motors (101). However, until rather recently, measurements were limited to forces and displacements.
In order to use optical trapping to investigate rotational motions, a trapped particle needs to be rotated by the trapping beam. Conventional optical traps employ a Gaussian laser beam and an optically isotropic microsphere that cannot be rotated by a trap with either a linear or circular polarization. Therefore, many of the early demonstrations to rotate the trapping particle relied on breaking the rotational symmetry of the particle and/or the input trapping beam (12, 35, 37, 79). In a seminal work by Friese et al. (35), a calcite particle, which is optically birefringent, was rotated with both linear and circularly polarized light. This work provided the inspiration for a new instrument for single molecule studies which is described below.
A recent advance in optical trapping techniques, namely the angular optical trap (AOT), also termed the optical torque wrench (OTW), has enabled direct torque and rotation detection of individual biological molecules (Figure 2d) (29, 59). There are three core features of this instrument.
First, the trapping particle is a nanofabricated quartz cylinder, which has its extraordinary optical axis perpendicular to its cylinder axis and one of its ends chemically derivatized for attachment to a biological molecule of interest (29). Quartz has positive optical anisotropy, with a single axis more polarizable than the other two, so that a quartz particle is angularly confined by a linearly polarized light in two of its three Euler angles. (In contrast, a calcite particle (35) can only be confined in one of its three Euler angles.) The remaining Euler angle of the quartz cylinder is confined by the shape anisotropy intrinsic to an elongated cylinder. When a quartz cylinder is trapped by a linearly polarized laser, its cylinder axis aligns with the direction of light propagation so that the cylinder can be rotated about its axis by rotation of the laser polarization. Attaching a biological molecule specifically to one end of the cylinder allows the application of force to the molecule along the laser propagation direction, permitting independent control of force and torque. Nanofabrication techniques allow for the mass production of cylinders of uniform size, shape, and optical properties, as well as specific chemical derivatization of only one end of each cylinder. The cylinders may be fabricated using optical lithography (29) or, for more selective localization of a molecule’s attachment point, by electron-beam lithography (45).
The second feature of an AOT is a rapid and flexible control of the input linear polarization of the trapping laser beam, so that the instrument may function in different modes of operation (59). The use of a pair of acousto-optic modulators (AOMs) provides continuous and rapid control (~ 100 kHz) of the input polarization (59). Alternatively, an electro-optic modulator (EOM) may also be used to rotate the input polarization (41). In the rotation mode, the particle is rotated by simple rotation of the polarization, with the particle’s optical axis closely tracking the electric field of the laser beam (35, 59). In the active torque wrench mode, a constant torque on the trapped particle is maintained via active feedback on the input polarization angle (59). This mode is best suited for applications at high torques. In the passive torque wrench mode, a constant optical torque is achieved by rotating the polarization at a rate much faster than the particle is able to respond, resulting in a minute constant torque exerted on the particle (46). This technique establishes a clear relationship between the rapid polarization rotation rate and the value of the torque acting on the particle, thereby allowing for an easily controllable torque. This passive torque wrench operates as if the angular optical trap has zero torsional stiffness, and as the torque approaches zero the particle can freely rotate in the trap. This mode is optimal for zero and low torque applications.
Finally, torque detection in an AOT is based on the change in the ellipticity of the trapping beam after it interacts with the trapping particle, a method independently demonstrated by two different groups (12, 59). In an AOT, a quartz cylinder is trapped such that its extraordinary axis, which is more polarizable than the other two axes, is aligned with the input beam’s linear polarization. If the cylinder is rotated away from this stable trapping orientation, there will be a restoring torque, which arises due to misalignment of the particle’s polarization and the electric field. Direct torque measurements are subsequently made by measuring the change in the angular momentum of the transmitted beam downstream of the trapped particle, which is accomplished by splitting the beam into its left- and right-circular components and determining their differential intensities (12, 59).
In an AOT, the same trapping beam is used for both torque and angle detection of the trapped particle, without the need for a secondary detection beam or imaging method. Such a detection method is exceedingly direct, relying solely on conservation of angular momentum, and thus distinguishes AOT from other methods described in this review. During a typical experiment, force, displacement, torque, and angle of the cylinder are simultaneously measured at kilohertz frequencies, making this method well suited for the study of fast events.
AOT takes advantage of a combination of optical and shape anisotropy to angularly orient a trapped particle. Particle orientation may also be achieved solely via shape anisotropy with particles made as disks (81), long rods (12), or more complex objects (37). In particular, Oroszi et al. (81) trapped a disk shaped particle with linearly polarized light and measured the torsional stiffness of DNA by imaging the particle’s angular deflection. In addition, although AOT employs a linearly polarized trapping beam to rotate a particle, rotation may also be achieved via circular and elliptical (35, 36) polarizations, laser beams carrying both spin and orbital momentums (84, 85), and asymmetrical trapping beams (79). Future studies may reveal whether these types of variations will further lend themselves to direct torque measurements in single molecule experiments.
Magnetic Tweezers
Magnetic tweezers are the most well-known technique used to apply twist to a single biological molecule. In an elegant demonstration by Strick et al. (105), single DNA molecules were supercoiled, via rotation of a magnetic bead, using a pair of permanent magnets oriented transverse to the DNA molecule. This technique is relatively simple to employ, and a major advantage is the ability to monitor the behavior of many molecules simultaneously, a feature which is typically lacking in many other rotation methods. Even though torque was not directly measured, this technique has proven to be a powerful tool to study torsional properties of DNA (105), chromatin (4), RNA polymerase (91), and topoisomerases (107).
Conventional magnetic tweezers used for rotational experiments tightly confine a magnetic bead’s angular orientation, resulting in a stiff angular trap (52). Measuring torque in this scenario would require the detection of a minute angular deviation between the applied field and the magnetic bead, well below the typical resolution of an optical microscopy based method. Torque detection with magnetic tweezers thus requires dramatic reduction in the torsional stiffness about the axis parallel to the applied force. There has been a recent surge of magnetic tweezers based devices suitable for making such torque measurements.
A solution is to orient the magnets axially instead of transversely, dramatically reducing the horizontal component of the magnetic force. Indeed Harada et al. (43) used this approach to track DNA rotation generated by E. coli RNA polymerase as visualized by attaching small fluorescent beads to the magnetic bead. Although direct torque measurement was not obtained in this study, it demonstrated that torsional stiffness can indeed be greatly reduced by orienting the magnets axially.
The first realization of direct torque measurements using magnetic tweezers with axially oriented magnets was made by Celedon et al. (21, 22). Their assay involved an axially oriented cylindrical magnet to apply force on a magnetic bead coupled to a nanorod torque arm (Figure 2e). A small force on the nanorod kept the probe aligned horizontally, while rotation was applied mechanically by moving the sample stage. The torsional stiffness of the probe was made sufficiently low to allow optical microscopy measurements of the angular deviations, and the method was shown to be capable of resolving single pN·nm scale torques.
Lifpert et al. (65) developed magnetic torque tweezers (MTT), a simpler configuration that does not require nanofabricated handles. MTT utilizes a cylindrical magnet to produce an axial magnetic field and a side-located magnet for a small horizontal field to orient the magnetic bead, and rotation is achieved by rotating the magnets (Figure 2f). Kauert et al. (51) showed that small field asymmetries generated in the main magnets oriented axially can also be sufficient to orient the bead for torque measurements. Lipfert et al. (66) further demonstrated that when the magnetic bead is located in the exact center of the field of a cylindrical magnet, the bead will rotate freely about the axis of force application, and referred to this approach as freely orbiting magnetic tweezers (FOMT).
More recent efforts for torque detection with magnetic tweezers have focused on the use of electromagnets to provide more precise control of the magnetic field. Mosconi et al. (77) developed the soft magnetic tweezers (SMT) apparatus that used electromagnets to rapidly rotate the field in such a way as to simultaneously apply and measure an arbitrary torque on a magnetic bead (Figure 2g). Janssen et al. (47) replaced the side magnet of the MTT with two pairs of Helmholtz coils to achieve full control of the transverse magnetic field. This instrument, named electromagnetic torque tweezers (eMTT), combines the features of MTT and FOMT and allows independent control of the vertical force and torsional stiffness (Figure 2h).
Although magnetic tweezers for torque measurement come in different configurations, they share the same torque measurement principle. Torque is determined by observing the angular orientation of the magnetic particle relative to the applied magnetic field with image tracking techniques and multiplying by a calibrated angular trap stiffness to produce physical torque units (21, 64, 65, 77).
Comparison of Different Techniques
Methods of torque measurement described above each have their advantages and disadvantages. Electrorotation has been the method of choice to exert a user-defined constant torque in single-molecule experiments (though other techniques in principle also possess this capability). However, electrorotation has not been adapted to incorporate force control, and associated heating can be severe (116). On the other hand, rotor bead tracking, angular optical trapping, and magnetic tweezers based techniques are all suited for simultaneous torque and force measurements and manipulation. Torque resolution, one of the critical parameters in investigating minute biological torques, is limited by the viscous drag coefficient of the probe particle, which scales as the cube of the probe’s dimension (18). A smaller probe, however, limits the amount of force that can be exerted. This limitation is circumvented in the RBT assay, which decouples force and torque probes. One of the prerequisites to probe fast dynamics of the biological systems is a high acquisition rate. Because the detection of the linear and angular parameters in an AOT is performed by directly monitoring the transmitted laser beam with photodiodes, acquisition rates in the kilohertz range can be achieved. In comparison, RBT and magnetic tweezers rely on video-based imaging, generally limiting acquisition rates to, at most, several hundred Hz. Methods employing magnetic tweezers to exert force, however, don’t suffer from potential laser induced damage and heating. On the other hand, AOT offers flexible control of both force and torque, enabling rapid switching between different modes of operation (46). In summary, it is clear that all of these methods offer benefits and challenges to a user, and the needs of the experimental system of study should dictate which choice is to be preferred.
TORQUE MEASUREMENTS ON BIOLOGICAL SYSTEMS
To date, there have been a number of reported measurements of biological torques. These can be parsed into two broad categories: those performed on DNA and DNA based systems, and those performed on rotary molecular motors. In this section, we detail the major findings made with the techniques described above.
Introduction to DNA Mechanics
DNA has long been of great interest to biophysicists, who have sought to detail its mechanical behavior utilizing the terms of polymer physics. For example, the elastic behavior of B-DNA under tension has been successfully described by the well-known modified Marko-Siggia model using two parameters, the bending and stretching moduli (74, 114). In contrast to tension, DNA response to torsion can be described by a simple harmonic potential , where L is the DNA contour length, θ is the added twist, and C is the torsional modulus, a measure of the torsional stiffness of the molecule. While early biochemical experiments were able to estimate the torsional modulus (28), direct measurements have only recently become possible due to the experimental advances detailed above. By measuring the change in torque Δτ as additional twist Δθ is added to the molecule, an effective torsional modulus can be experimentally measured as . Interestingly, Ceff has been observed to increase with force and approach C at ~ 3 pN (65, 76); this behavior has been explained by taking into account the writhe fluctuations of the DNA molecule (75). While a harmonic approximation describes the torsional response of B-DNA rather well, sensitive experiments have detected nonzero coupling between twist and tension (39, 63, 99), leading to a more precise formulation of the torsional energy . Here, z is the DNA extension and g is the so called twist-stretch coupling. Surprisingly, this coupling is negative for forces up to 30 pN, signifying that DNA overwinds when stretched (39).
One should note that the above discussion relates solely to B-DNA, a very important, but by no means unique, state of DNA. The last two decades of biophysical experiments have witnessed a plethora of DNA structures that can be formed under the impact of torsion and tension. For example, under moderate tension and torque DNA can absorb additional twist by buckling, forming intertwined supercoiled loops (also called plectonemes). Such supercoiled DNA (scB-DNA) can exist at low forces under both positive and negative torque (105). Undertwisting DNA at somewhat higher forces will torsionally break the base-paired interactions leading to a state known as ‘L-DNA’, which differs in important ways from thermally melted DNA (19, 98). Substantial overtwisting of DNA will create an exotic state termed ‘P-DNA’, which has a smaller helical repeat than standard B-DNA and unpaired bases extruding to the exterior of the molecule (2, 19).
Direct torque measurements are crucial for the identification of these DNA structural transitions, which display a phase transition behavior so that the phase co-existence state at a set force is characterized by a constant torque (73). It is therefore convenient to introduce a force-torque phase diagram (Figure 3). Below, we will describe key studies that have characterized the torsional properties of various DNA states.
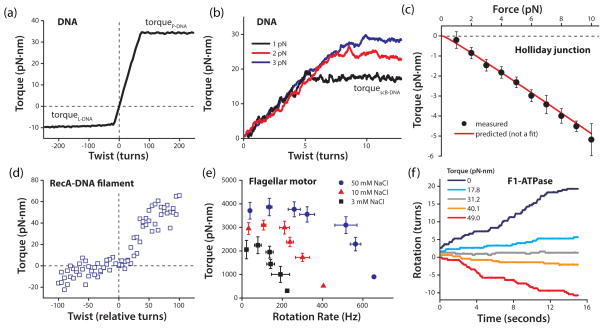
Figure 3.
DNA force-torque phase diagram. Phase transitions between specific states of DNA are represented by solid black lines (19, 73, 96). Red points indicate torque values measured during phase transitions using an angular optical trap (29, 33, 98, 99). Adapted from Sheinin et al. (98).
B-DNA Torsional Modulus
Traditional magnetic tweezers methods, despite lacking direct torque readout during the early years of their use in single molecule assays, have nonetheless been used to estimate the torsional stiffness of DNA by considering the difference in work required to stretch molecules at different supercoiling densities (106). Using this approach, the B-DNA torsional modulus was determined to be approximately 85 nm·kBT. A similar methodology was later employed by Mosconi et al., wherein the integration of a molecule’s extension change, with respect to force, allowed for the deduction of torque applied within the DNA molecule (76). This method allowed for the estimation of the effective torsional modulus across a range of forces.
The first direct measurement of the torsional modulus of DNA was performed by using the rotary bead method (19). A series of measurements were performed at high forces (> 15 pN), and the torsional modulus was found to be in the range of 100 to 110 nm·kBT (Figure 4a). Shortly thereafter, Oroszi et al. employed a linearly polarized optical trap in conjunction with a polystyrene particle possessing shape anisotropy to determine the torsional modulus of DNA, reporting a value of 75 nm·kBT at very low forces and predicting a saturation value of about 100 n·kBT (81).
Figure 4. Torque measurements on single biological molecules.
(a) Averaged torque trace obtained using rotor bead tracking during under- and over-winding of 14.8 kb DNA molecules held at 15 and 45 pN of force. Torque plateaus correspond to transitions to P-DNA (positive twist) or melted L-DNA (negative twist). The torsional modulus was extracted from the slope of the linear region. Adapted from Bryant et al. (19).
(b) Individual torque traces obtained with an angular optical trap as a single 2.2 kb DNA molecule was overwound under tension. The torsional modulus was extracted from the slopes of the linear regions. Torque plateaued as the DNA buckled to form plectonemes (scB-DNA). Adapted from Forth et al. (33).
(c) The torque-force relationship during the migration of a fully homologous Holliday junction, as determined by an angular optical trap. Shown are the mean torque values as a function of force (points) and theoretical prediction (line; not a fit). Adapted from Forth et al. (32).
(d) Torsional response of a RecA filament at 3.5 pN, obtained with magnetic torque tweezers. The wide spread in torque values reflects the dynamic nature of the RecA filament (Lipfert, personal comm.). Adapted from Lipfert et al. (65).
(e) Torque-speed relationship measured for the Na+-driven flagellar motor of Vibrio alginolyticus by monitoring rotation of a bead attached to the flagellum. Adapted from Sowa et al. (104).
(f) Rotational trajectories of a single F1-ATPase motor, under external torque, in the presence of ATP, ADP and Pi. At the stalling torque (~31 pN·nm), the motor still exhibited bidirectional stepwise fluctuations. Adapted from Toyabe et al. (108).
Using an angular optical trap, the torsional modulus of DNA at moderate forces, ranging from 1 to 3.5 pN, was directly measured for DNA molecules of differing lengths, and was found to be 90 n·kBT, in good agreement with previous measurements (Figure 4b) (33). Force dependence of the torsional modulus has been further explored using magnetic torque tweezers (65). As the tension on the DNA was increased from 0.25 to 6 pN, the torsional modulus was found to increase from 40 to 100 n·kBT. Interestingly, the force-dependence of the torsional modulus was shown to deviate from the previously developed theoretical model (75).
Plectonemic DNA
Forth et al. (33) made the first direct measurement of the critical buckling torque during the B-DNA to scB-DNA transition using an angular optical trap (Figure 4b). The buckling torque was found to depend on force, in accordance with a refined DNA mechanical model (73). This analysis also allowed extraction of the plectonemic torsional rigidity, which was found to be 26 nm·kBT, in good agreement with previous bulk experiments (94). Further analysis of the torque measurements in the pre- and post-buckling states revealed an overshoot of torque of ~ 3 pN·nm at the buckling transition; such an overshoot was predicted based on an elastic rod theory (27). Subsequently Celedon et al. (21) utilized a special magnetic tweezers apparatus to determine the buckling torques at low forces down to 0.3 pN, further validating the Marko model (73).
Forth et al. (33) also discovered that the buckling transition takes place abruptly and is highly dynamic. At the buckling transition, the DNA extension hops rapidly between two distinctive states: an extended pre-buckled state and a plectonemic post-buckled state. Furthermore, the initial plectonemic loop absorbed approximately twice as much extension as each subsequent turn. Interestingly, such an abrupt transition was absent in previous magnetic tweezers measurements, where instead a smooth and gradual transition was observed (105). The angular trapping method allowed for the detection of this abrupt transition, due to higher bandwidth, increased spatial resolution, and the use of shorter DNA tethers. More recent experiments, using an upgraded magnetic tweezers setup, provided systematic measurements of the buckling transition under different DNA lengths and salt conditions (17).
Underwound DNA
Under moderate forces (> 0.6 pN) negative torque is able to induce DNA strand separation to melt DNA without undergoing buckling transition. Bryant et al. (19) first directly measured the melting torque using the rotary bead assay and found it to be ~ 10 pN·nm (Figure 4a). This value was further supported with measurements by Sheinin et al. (98) using an angular optical trap. Torsionally melted DNA was long thought to be equivalent to a thermally melted DNA ‘bubble’. Interestingly, Bryant et al. (19) and Sheinin et al. (98) discovered that melted DNA actually exists in a unique left-handed configuration, termed L-DNA, with mechanical properties distinct from both B-DNA and Z-DNA, and inconsistent with the predicted mechanical behavior of thermally melted parallel strands (2). Sheinin et al. (98) also revealed that under low force, different DNA sequences exhibited drastically different behavior when underwound, and the underwinding process was not reversible under the experimental time scale. These results suggest that the transition from B-DNA to L-DNA at low force occurs along a complex pathway, with multiple secondary structures being formed while off equilibrium. Subsequent to this discovery, Oberstrass et al. (80) studied the torsional behavior of a range of DNA sequences using a feedback enhanced rotary bead method. The L-DNA state was also observed, and was found to possess mechanical parameters similar to those measured previously (98). Additionally, it was observed that GC-rich tracts form Z-DNA under moderate negative torsional stress of ~−3 pN·nm, while tracts of mismatched DNA behave similarly to a B-DNA-like helical structure during underwinding.
Besides Z-DNA, a number of other sequence-specific non-B DNA structures can form under negative torsion (83). One notable example is a DNA cruciform, or Holliday junction, which is favored at palindromic or near-palindromic sequences. Forth et al. (32) directly measured the torque generated during Holliday junction migration for both fully homologous and single-base heterologous sequences using an angular optical trap. The minute torques observed during smooth migration, on the order of 1 pN·nm, were found to depend on force in a predictable manner (Figure 4c). However, it took approximately 7 pN·nm (consistent with the magnitude of torque needed to melt DNA) to migrate through just a single base mismatch.
Overwound DNA and Twist-Stretch Coupling
While the existence of highly overwound P-DNA was first inferred based on extension measurements using magnetic tweezers (2) and micropippetes (60), crucial additional confirmation was provided by rotary bead experiments (19). Under high force, B-DNA transitions to P-DNA at ~35 pN·nm (Figure 4a), and the helical pitch of P-DNA was found to be ~2.7 bp/turn. Deufel et al. (29) measured a similar torque value for the transition from B-DNA to supercoiled P-DNA (scP-DNA), which is P-DNA shortened by secondary structure formation (2).
Conventional wisdom predicts that DNA should shorten when overwound under tension. Surprisingly, Gore et al. (39) discovered that DNA lengthens instead, achieving a maximum extension before beginning to shorten again. These findings yielded a negative twist-stretch coupling coefficient of −22kBT. It was thought that the extension peak corresponded to the location of a phase transition between B and scP-DNA (63). Sheinin et al. (99) further investigated the twist-stretch coupling using an angular optical trap and measured a value of −21kBT, in agreement with Gore et al. (39). However, examination of the concurrently measured torque signal showed that the phase transition did not occur at the peak of the extension, but instead at a higher degree of supercoiling. This result underscores the importance of direct experimental access to all possible variables.
DNA with Bound Proteins/Small Molecules
In addition to measuring the torsional properties of naked DNA, several studies have been performed on protein-bound DNA filaments and DNA intercalated with small molecules. Celedon et al. (21) directly measured the torque required to twist nucleosomal arrays. Although a wide experimental variability in both extension and torque behavior existed among molecules due to variations in nucleosome occupancy, it was still unambiguously shown that chromatin has a much softer torsional stiffness than that of naked DNA. Subsequent work by the same group also found that ethidium bromide intercalation of DNA leads to a torsional softening of the molecule, but the bending stiffness remains largely unchanged (22). In contrast, magnetic torque tweezers experiments showed (65) that RecA filaments were approximately twice as torsionally stiff as bare DNA, with a torsional modulus of ~175 nm·kBT (Figure 4d).
Motor Proteins
Torque measurements have also been performed on several molecular motors. The E. coli flagellar motor has been extensively studied with a variety of techniques, including viscous drag on cells (70) or beads (24, 90), optical trapping (11), and electrorotation (8). While a range of torque values has been obtained, a recent work has determined the maximum torque to be ~1300 pN·nm (90). In comparison, the flagellar motor of Vibrio alginolyticus was shown to generate torques of up to 4000 pN·nm during its rotation (104). The flagellar motor was also observed to display a nonlinear torque-speed relationship (8, 24, 104), as the torque remains nearly constant for speeds up to several hundred Hz, and subsequently decreases quickly (Figure 4e). More experimental and theoretical work is still required to further understand the mechanochemical cycle of the flagellar motor (103).
The significantly smaller rotary motor F1-ATPase has also been studied in single molecule detail. By attaching a fluorescent actin filament to a surface immobilized motor and observing the resulting rotation rate, it has been estimated that a single rotary unit can generate torques of up to ~ 40 pN·nm (78). This finding, together with the discovery that the rotary shaft of F1-ATPase makes 120° steps (120), allowed Kinosita and coworkers to conclude that nearly 100% of the energy from ATP hydrolysis is expended to perform work against the viscous drag. This makes F1-ATPase a highly efficient molecular machine (120). One should note that the non-conservative nature of the viscous drag force complicates the interpretation of these results in relationship to the true thermodynamic efficiency (112). To overcome this limitation, recent work by Toyabe et al (108) employed the electrorotation method to investigate the rotational behavior of the F1-ATPase under a constant externally imposed torque (Figure 4f). The motor was found to stall at a torque of 31 pN·nm, which corresponded to a thermodynamic efficiency of ~80% (108).
Similarly, RNA polymerase has been shown to be capable of generating torques of at least 5 pN·nm, determined by visualizing the rotation of a DNA-bound bead (43). However, the upper limit of torque generation has not, to date, been directly reported. Such experiments for RNAP and other motor proteins could be repeated using one of the more precise and controllable methodologies outlined above, yielding a more refined insight into the operation of motors that twist.
CONCLUSIONS AND FUTURE DIRECTIONS
It is clear that the ability to directly measure the minute torques relevant to biological structures will be increasingly important moving forward. Applying these innovative techniques to study the torsional properties of protein-bound DNA, as well as the enzymes which process DNA during transcription, replication, and packaging will be paramount to our understanding of their operation. Determining the behavior of chromatin under twist will give insight into the role that torque has on chromatin assembly and stability in vivo. Investigation of the AAA family of DNA processing machines, such as helicases and phage packaging motors, should resolve the long-standing issue of their torque-generating potential.
While DNA has been studied extensively due to the relative ease of adapting the biological substrate to direct torque measurement techniques, a wide class of other biomolecules will most certainly be examined as well. The stepping behavior of microtubule-based molecular motors such as kinesins and dyneins can be modulated by force, and it is believed that such motors must be also capable of generating and withstanding torsional loads during cargo transport. The direct measurement of stall torques and motor behavior under constant torque will further our understanding of how these motors behave in vivo. Similarly, studying the kinetics under torsional load of processive myosins (such as Myosin V) as they walk along their actin substrates should lead to insights into their mechanics.
Single molecule methods have proven to be incredibly effective at elucidating mechanisms of action that traditional ensemble methods cannot probe. Direct manipulation of the mechanical properties of molecules has proven to be a rich and vibrant pursuit, leading to a greater understanding of our biological world. With the addition of direct torque measurement capabilities, we can move even further forward in our efforts to understand the physical principles underlying biological systems and the role that mechanics plays in regulating the building blocks of life.
Acknowledgments
We would like to acknowledge Drs. Z. Bryant, S. Toyabe, J. Lipfert, and A. Ishijima for sharing data for figure reproduction, and Drs. K Kinosita, H. Berg, M. Yoshida, and F. Oberstrass for helpful communications. We thank Dr. S. Fellman for critical comments on the manuscript. We wish to acknowledge postdoctoral support to S. F. from National Institutes of Health NRSA Fellowship (F32GM099380), graduate traineeship support to J.I. from Cornell’s Molecular Biophysics Training Grant (T32GM008267), and support to M.D.W. from National Institutes of Health grant (GM059849) and National Science Foundation grant (MCB-0820293).
LITERATURE CITED
- 1.Aguilera A, García-Muse T. R loops: from transcription byproducts to threats to genome stability. Mol Cell. 2012;46:115–24. doi: 10.1016/j.molcel.2012.04.009. [DOI] [PubMed] [Google Scholar]
- 2.Allemand JF, Bensimon D, Lavery R, Croquette V. Stretched and overwound DNA forms a Pauling-like structure with exposed bases. Proc Natl Acad Sci U S A. 1998;95:14152–7. doi: 10.1073/pnas.95.24.14152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ashkin A, Dziedzic JM, Yamane T. Optical trapping and manipulation of single cells using infrared-laser beams. Nature. 1987;330:769–71. doi: 10.1038/330769a0. [DOI] [PubMed] [Google Scholar]
- 4.Bancaud A, Conde e Silva N, Barbi M, Wagner G, Allemand JF, et al. Structural plasticity of single chromatin fibers revealed by torsional manipulation. Nat Struct Mol Biol. 2006;13:444–50. doi: 10.1038/nsmb1087. [DOI] [PubMed] [Google Scholar]
- 5.Baranello L, Levens D, Gupta A, Kouzine F. The importance of being supercoiled: How DNA mechanics regulate dynamic processes. Biochim Biophys Acta. 2012 doi: 10.1016/j.bbagrm.2011.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berg HC. The rotary motor of bacterial flagella. Annual Review of Biochemistry. 2003;72:19–54. doi: 10.1146/annurev.biochem.72.121801.161737. [DOI] [PubMed] [Google Scholar]
- 7.Berg HC, Anderson RA. Bacteria swim by rotating their flagellar filaments. Nature. 1973;245:380–2. doi: 10.1038/245380a0. [DOI] [PubMed] [Google Scholar]
- 8.Berg HC, Turner L. Torque generated by the flagellar motor of Escherischia coli. Biophysical Journal. 1993;65:2201–16. doi: 10.1016/S0006-3495(93)81278-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bermejo R, Lai MS, Foiani M. Preventing replication stress to maintain genome stability: resolving conflicts between replication and transcription. Mol Cell. 2012;45:710–8. doi: 10.1016/j.molcel.2012.03.001. [DOI] [PubMed] [Google Scholar]
- 10.Bermúdez I, García-Martínez J, Pérez-Ortín JE, Roca J. A method for genome-wide analysis of DNA helical tension by means of psoralen-DNA photobinding. Nucleic Acids Res. 2010;38:e182. doi: 10.1093/nar/gkq687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Berry RM, Berg HC. Absence of a barrier to backwards rotation of the bacterial flagellar motor demonstrated with optical tweezers. Proceedings of the National Academy of Sciences of the United States of America. 1997;94:14433–37. doi: 10.1073/pnas.94.26.14433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bishop AI, Nieminen TA, Heckenberg NR, Rubinsztein-Dunlop H. Optical application and measurement of torque on microparticles of isotropic nonabsorbing material. Physical Review A. 2003;68:8. [Google Scholar]
- 13.Borukhov S, Nudler E. RNA polymerase holoenzyme: structure, function and biological implications. Curr Opin Microbiol. 2003;6:93–100. doi: 10.1016/s1369-5274(03)00036-5. [DOI] [PubMed] [Google Scholar]
- 14.Branzei D, Foiani M. Maintaining genome stability at the replication fork. Nat Rev Mol Cell Biol. 2010;11:208–19. doi: 10.1038/nrm2852. [DOI] [PubMed] [Google Scholar]
- 15.Brooks TA, Hurley LH. The role of supercoiling in transcriptional control of MYC and its importance in molecular therapeutics. Nat Rev Cancer. 2009;9:849–61. doi: 10.1038/nrc2733. [DOI] [PubMed] [Google Scholar]
- 16.Brower-Toland BD, Smith CL, Yeh RC, Lis JT, Peterson CL, Wang MD. Mechanical disruption of individual nucleosomes reveals a reversible multistage release of DNA. Proceedings of the National Academy of Sciences of the United States of America. 2002:99. doi: 10.1073/pnas.022638399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brutzer H, Luzzietti N, Klaue D, Seidel R. Energetics at the DNA Supercoiling Transition. Biophysical Journal. 2010;98:1267–76. doi: 10.1016/j.bpj.2009.12.4292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bryant Z, Oberstrass FC, Basu A. Recent developments in single-molecule DNA mechanics. Curr Opin Struct Biol. 2012;22:304–12. doi: 10.1016/j.sbi.2012.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bryant Z, Stone MD, Gore J, Smith SB, Cozzarelli NR, Bustamante C. Structural transitions and elasticity from torque measurements on DNA. Nature. 2003;424:338–41. doi: 10.1038/nature01810. [DOI] [PubMed] [Google Scholar]
- 20.Capaldi RA, Aggeler R. Mechanism of the F(1)F(0)-type ATP synthase, a biological rotary motor. Trends Biochem Sci. 2002;27:154–60. doi: 10.1016/s0968-0004(01)02051-5. [DOI] [PubMed] [Google Scholar]
- 21.Celedon A, Nodelman IM, Wildt B, Dewan R, Searson P, et al. Magnetic Tweezers Measurement of Single Molecule Torque. Nano Letters. 2009;9:1720–25. doi: 10.1021/nl900631w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Celedon A, Wirtz D, Sun S. Torsional Mechanics of DNA Are Regulated by Small-Molecule Intercalation. Journal of Physical Chemistry B. 2010;114:16929–35. doi: 10.1021/jp107541q. [DOI] [PubMed] [Google Scholar]
- 23.Chen D, Bowater R, Dorman CJ, Lilley DM. Activity of a plasmid-borne leu-500 promoter depends on the transcription and translation of an adjacent gene. Proc Natl Acad Sci U S A. 1992;89:8784–8. doi: 10.1073/pnas.89.18.8784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen X, Berg HC. Torque-speed relationship of the flagellar rotary motor of Escherichia coli. Biophys J. 2000;78:1036–41. doi: 10.1016/S0006-3495(00)76662-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Clark DJ, Felsenfeld G. Formation of nucleosomes on positively supercoiled DNA. EMBO J. 1991;10:387–95. doi: 10.1002/j.1460-2075.1991.tb07960.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cozzarelli NR, Cost GJ, Nollmann M, Viard T, Stray JE. Giant proteins that move DNA: bullies of the genomic playground. Nature Reviews Molecular Cell Biology. 2006:7. doi: 10.1038/nrm1982. [DOI] [PubMed] [Google Scholar]
- 27.Daniels BC, Forth S, Sheinin MY, Wang MD, Sethna JP. Discontinuities at the DNA supercoiling transition. Physical Review E. 2009;80:4. doi: 10.1103/PhysRevE.80.040901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Depew DE, Wang JC. Conformational fluctuations of DNA helix. Proceedings of the National Academy of Sciences of the United States of America. 1975;72:4275–9. doi: 10.1073/pnas.72.11.4275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Deufel C, Forth S, Simmons CR, Dejgosha S, Wang MD. Nanofabricated quartz cylinders for angular trapping: DNA supercoiling torque detection. Nature Methods. 2007;4:223–25. doi: 10.1038/nmeth1013. [DOI] [PubMed] [Google Scholar]
- 30.Drlica K. Control of bacterial DNA supercoiling. Mol Microbiol. 1992;6:425–33. doi: 10.1111/j.1365-2958.1992.tb01486.x. [DOI] [PubMed] [Google Scholar]
- 31.Fierro-Fernández M, Hernández P, Krimer DB, Stasiak A, Schvartzman JB. Topological locking restrains replication fork reversal. Proc Natl Acad Sci U S A. 2007;104:1500–5. doi: 10.1073/pnas.0609204104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Forth S, Deufel C, Patel SS, Wang MD. Direct measurements of torque during Holliday junction migration. Biophys J. 2011;101:L5–7. doi: 10.1016/j.bpj.2011.05.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Forth S, Deufel C, Sheinin MY, Daniels B, Sethna JP, Wang MD. Abrupt buckling transition observed during the plectoneme formation of individual DNA molecules. Physical Review Letters. 2008;100:4. doi: 10.1103/PhysRevLett.100.148301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.French SL, Sikes ML, Hontz RD, Osheim YN, Lambert TE, et al. Distinguishing the roles of Topoisomerases I and II in relief of transcription-induced torsional stress in yeast rRNA genes. Mol Cell Biol. 2011;31:482–94. doi: 10.1128/MCB.00589-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Friese MEJ, Nieminen TA, Heckenberg NR, Rubinsztein-Dunlop H. Optical alignment and spinning of laser-trapped microscopic particles. Nature. 1998;394:348–50. [Google Scholar]
- 36.Funk M, Parkin SJ, Stilgoe AB, Nieminen TA, Heckenberg NR, Rubinsztein-Dunlop H. Constant power optical tweezers with controllable torque. Optics Letters. 2009;34:139–41. doi: 10.1364/ol.34.000139. [DOI] [PubMed] [Google Scholar]
- 37.Galajda P, Ormos P. Orientation of flat particles in optical tweezers by linearly polarized light. Optics Express. 2003;11:446–51. doi: 10.1364/oe.11.000446. [DOI] [PubMed] [Google Scholar]
- 38.Gellert M, Mizuuchi K, O’Dea MH, Nash HA. DNA gyrase: an enzyme that introduces superhelical turns into DNA. Proc Natl Acad Sci U S A. 1976;73:3872–6. doi: 10.1073/pnas.73.11.3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gore J, Bryant Z, Noellmann M, Le MU, Cozzarelli NR, Bustamante C. DNA overwinds when stretched. Nature. 2006;442:836–39. doi: 10.1038/nature04974. [DOI] [PubMed] [Google Scholar]
- 40.Gore J, Bryant Z, Stone MD, Nollmann MN, Cozzarelli NR, Bustamante C. Mechanochemical analysis of DNA gyrase using rotor bead tracking. Nature. 2006;439:100–04. doi: 10.1038/nature04319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gutierrez-Medina B, Andreasson JOL, Greenleaf WJ, LaPorta A, Block SM. An optical apparatus for rotation and trapping. Methods in Enzymology, Vol 475: Single Molecule Tools, Pt B: Super-Resolution, Particle Tracking, Multiparameter, and Force Based Methods. 2010;475:377–404. doi: 10.1016/S0076-6879(10)75015-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gutiérrez-Medina B, Fehr AN, Block SM. Direct measurements of kinesin torsional properties reveal flexible domains and occasional stalk reversals during stepping. Proc Natl Acad Sci U S A. 2009;106:17007–12. doi: 10.1073/pnas.0907133106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Harada Y, Ohara O, Takatsuki A, Itoh H, Shimamoto N, Kinosita K. Direct observation of DNA rotation during transcription by Escherichia coli RNA polymerase. Nature. 2001;409:113–15. doi: 10.1038/35051126. [DOI] [PubMed] [Google Scholar]
- 44.Hatfield GW, Benham CJ. DNA topology-mediated control of global gene expression in Escherichia coli. Annu Rev Genet. 2002;36:175–203. doi: 10.1146/annurev.genet.36.032902.111815. [DOI] [PubMed] [Google Scholar]
- 45.Huang ZX, Pedaci F, van Oene M, Wiggin MJ, Dekker NH. Electron Beam Fabrication of Birefringent Microcylinders. Acs Nano. 2011;5:1418–27. doi: 10.1021/nn1034108. [DOI] [PubMed] [Google Scholar]
- 46.Inman J, Forth S, Wang MD. Passive torque wrench and angular position detection using a single-beam optical trap. Optics Letters. 2010;35:2949–51. doi: 10.1364/OL.35.002949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Janssen XJA, Lipfert J, Jager T, Daudey R, Beekman J, Dekker NH. Electromagnetic Torque Tweezers: A Versatile Approach for Measurement of Single-Molecule Twist and Torque. Nano Letters. 2012;12:3634–39. doi: 10.1021/nl301330h. [DOI] [PubMed] [Google Scholar]
- 48.Johnson DS, Bai L, Smith BY, Patel SS, Wang MD. Single-molecule studies reveal dynamics of DNA unwinding by the ring-shaped T7 helicase. Cell. 2007:129. doi: 10.1016/j.cell.2007.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jones TB. Basic theory of dielectrophoresis and electrorotation. Ieee Engineering in Medicine and Biology Magazine. 2003:22. doi: 10.1109/memb.2003.1304999. [DOI] [PubMed] [Google Scholar]
- 50.Junge W, Sielaff H, Engelbrecht S. Torque generation and elastic power transmission in the rotary F(O)F(1)-ATPase. Nature. 2009;459:364–70. doi: 10.1038/nature08145. [DOI] [PubMed] [Google Scholar]
- 51.Kauert DJ, Kurth T, Liedl T, Seidel R. Direct Mechanical Measurements Reveal the Material Properties of Three-Dimensional DNA Origami. Nano Letters. 2011;11:5558–63. doi: 10.1021/nl203503s. [DOI] [PubMed] [Google Scholar]
- 52.Klaue D, Seidel R. Torsional Stiffness of Single Superparamagnetic Microspheres in an External Magnetic Field. Physical Review Letters. 2009;102:4. doi: 10.1103/PhysRevLett.102.028302. [DOI] [PubMed] [Google Scholar]
- 53.Komori Y, Iwane AH, Yanagida T. Myosin-V makes two brownian 90 degrees rotations per 36-nm step. Nat Struct Mol Biol. 2007;14:968–73. doi: 10.1038/nsmb1298. [DOI] [PubMed] [Google Scholar]
- 54.Koster DA, Crut A, Shuman S, Bjornsti MA, Dekker NH. Cellular strategies for regulating DNA supercoiling: a single-molecule perspective. Cell. 2010;142:519–30. doi: 10.1016/j.cell.2010.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kouzine F, Levens D. Supercoil-driven DNA structures regulate genetic transactions. Front Biosci. 2007;12:4409–23. doi: 10.2741/2398. [DOI] [PubMed] [Google Scholar]
- 56.Kouzine F, Sanford S, Elisha-Feil Z, Levens D. The functional response of upstream DNA to dynamic supercoiling in vivo. Nat Struct Mol Biol. 2008;15:146–54. doi: 10.1038/nsmb.1372. [DOI] [PubMed] [Google Scholar]
- 57.Kowalski D, Eddy MJ. The DNA unwinding element: a novel, cis-acting component that facilitates opening of the Escherichia coli replication origin. EMBO J. 1989;8:4335–44. doi: 10.1002/j.1460-2075.1989.tb08620.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Krasilnikov AS, Podtelezhnikov A, Vologodskii A, Mirkin SM. Large-scale effects of transcriptional DNA supercoiling in vivo. J Mol Biol. 1999;292:1149–60. doi: 10.1006/jmbi.1999.3117. [DOI] [PubMed] [Google Scholar]
- 59.La Porta A, Wang MD. Optical torque wrench: Angular trapping, rotation, and torque detection of quartz microparticles. Physical Review Letters. 2004;92:4. doi: 10.1103/PhysRevLett.92.190801. [DOI] [PubMed] [Google Scholar]
- 60.Leger J, Romano G, Sarkar A, Robert J, Bourdieu L, et al. Structural transitions of a twisted and stretched DNA molecule. Physical Review Letters. 1999;83:1066–69. [Google Scholar]
- 61.Levchenko V, Jackson B, Jackson V. Histone release during transcription: displacement of the two H2A-H2B dimers in the nucleosome is dependent on different levels of transcription-induced positive stress. Biochemistry. 2005;44:5357–72. doi: 10.1021/bi047786o. [DOI] [PubMed] [Google Scholar]
- 62.Lim HM, Lewis DE, Lee HJ, Liu M, Adhya S. Effect of varying the supercoiling of DNA on transcription and its regulation. Biochemistry. 2003;42:10718–25. doi: 10.1021/bi030110t. [DOI] [PubMed] [Google Scholar]
- 63.Lionnet T, Joubaud S, Lavery R, Bensimon D, Croquette V. Wringing out DNA. Physical Review Letters. 2006;96:4. doi: 10.1103/PhysRevLett.96.178102. [DOI] [PubMed] [Google Scholar]
- 64.Lipfert J, Kerssemakers JJW, Rojer M, Dekker NH. A method to track rotational motion for use in single-molecule biophysics. Review of Scientific Instruments. 2011:82. doi: 10.1063/1.3650461. [DOI] [PubMed] [Google Scholar]
- 65.Lipfert J, Kerssemakers JWJ, Jager T, Dekker NH. Magnetic torque tweezers: measuring torsional stiffness in DNA and RecA-DNA filaments. Nature Methods. 2010;7:977–U54. doi: 10.1038/nmeth.1520. [DOI] [PubMed] [Google Scholar]
- 66.Lipfert J, Wiggin M, Kerssemakers JWJ, Pedaci F, Dekker NH. Freely orbiting magnetic tweezers to directly monitor changes in the twist of nucleic acids. Nature Communications. 2011;2:9. doi: 10.1038/ncomms1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Liphardt J, Onoa B, Smith SB, Tinoco I, Bustamante C. Reversible unfolding of single RNA molecules by mechanical force. Science. 2001:292. doi: 10.1126/science.1058498. [DOI] [PubMed] [Google Scholar]
- 68.Liu LF, Wang JC. Supercoiling of the DNA template during transcription. Proc Natl Acad Sci U S A. 1987;84:7024–7. doi: 10.1073/pnas.84.20.7024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ljungman M, Hanawalt PC. Localized torsional tension in the DNA of human cells. Proc Natl Acad Sci U S A. 1992;89:6055–9. doi: 10.1073/pnas.89.13.6055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lowe G, Meister M, Berg HC. Rapid rotation of flagellar bundles in swimming bacteria. Nature. 1987;325:637–40. [Google Scholar]
- 71.Luger K, Mäder AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–60. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 72.Magariyama Y, Sugiyama S, Muramoto K, Maekawa Y, Kawagishi I, et al. Very fast flagellar rotation. Nature. 1994;371:752. doi: 10.1038/371752b0. [DOI] [PubMed] [Google Scholar]
- 73.Marko JF. Torque and dynamics of linking number relaxation in stretched supercoiled DNA. Physical Review E. 2007;76:13. doi: 10.1103/PhysRevE.76.021926. [DOI] [PubMed] [Google Scholar]
- 74.Marko JF, Siggia ED. Stretching DNA. Macromolecules. 1995;28:8759–70. [Google Scholar]
- 75.Moroz JD, Nelson P. Torsional directed walks, entropic elasticity, and DNA twist stiffness. Proceedings of the National Academy of Sciences of the United States of America. 1997;94:14418–22. doi: 10.1073/pnas.94.26.14418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mosconi F, Allemand JF, Bensimon D, Croquette V. Measurement of the Torque on a Single Stretched and Twisted DNA Using Magnetic Tweezers. Physical Review Letters. 2009;102:4. doi: 10.1103/PhysRevLett.102.078301. [DOI] [PubMed] [Google Scholar]
- 77.Mosconi F, Allemand JF, Croquette V. Soft magnetic tweezers: A proof of principle. Review of Scientific Instruments. 2011;82:12. doi: 10.1063/1.3531959. [DOI] [PubMed] [Google Scholar]
- 78.Noji H, Yasuda R, Yoshida M, Kinosita K. Direct observation of the rotation of F-1-ATPase. Nature. 1997;386:299–302. doi: 10.1038/386299a0. [DOI] [PubMed] [Google Scholar]
- 79.O’Neil AT, Padgett MJ. Rotational control within optical tweezers by use of a rotating aperture. Optics Letters. 2002;27:743–45. doi: 10.1364/ol.27.000743. [DOI] [PubMed] [Google Scholar]
- 80.Oberstrass FC, Fernandes LE, Bryant Z. Torque measurements reveal sequence-specific cooperative transitions in supercoiled DNA. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:6106–11. doi: 10.1073/pnas.1113532109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Oroszi L, Galajda P, Kirei H, Bottka S, Ormos P. Direct measurement of torque in an optical trap and its application to double-strand DNA. Physical Review Letters. 2006;97:4. doi: 10.1103/PhysRevLett.97.058301. [DOI] [PubMed] [Google Scholar]
- 82.Oster G, Wang H. Rotary protein motors. Trends Cell Biol. 2003;13:114–21. doi: 10.1016/s0962-8924(03)00004-7. [DOI] [PubMed] [Google Scholar]
- 83.Palecek E. Local supercoil-stabilized DNA structures. Crit Rev Biochem Mol Biol. 1991;26:151–226. doi: 10.3109/10409239109081126. [DOI] [PubMed] [Google Scholar]
- 84.Parkin S, Knoner G, Nieminen TA, Heckenberg NR, Rubinsztein-Dunlop H. Measurement of the total optical angular momentum transfer in optical tweezers. Optics Express. 2006;14:6963–70. doi: 10.1364/oe.14.006963. [DOI] [PubMed] [Google Scholar]
- 85.Paterson L, MacDonald MP, Arlt J, Sibbett W, Bryant PE, Dholakia K. Controlled rotation of optically trapped microscopic particles. Science. 2001;292:912–14. doi: 10.1126/science.1058591. [DOI] [PubMed] [Google Scholar]
- 86.Pfaffle P, Gerlach V, Bunzel L, Jackson V. In vitro evidence that transcription-induced stress causes nucleosome dissolution and regeneration. J Biol Chem. 1990;265:16830–40. [PubMed] [Google Scholar]
- 87.Pfaffle P, Jackson V. Studies on rates of nucleosome formation with DNA under stress. J Biol Chem. 1990;265:16821–9. [PubMed] [Google Scholar]
- 88.Postow L, Crisona NJ, Peter BJ, Hardy CD, Cozzarelli NR. Topological challenges to DNA replication: conformations at the fork. Proc Natl Acad Sci U S A. 2001;98:8219–26. doi: 10.1073/pnas.111006998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Postow L, Ullsperger C, Keller RW, Bustamante C, Vologodskii AV, Cozzarelli NR. Positive torsional strain causes the formation of a four-way junction at replication forks. J Biol Chem. 2001;276:2790–6. doi: 10.1074/jbc.M006736200. [DOI] [PubMed] [Google Scholar]
- 90.Reid SW, Leake MC, Chandler JH, Lo CJ, Armitage JP, Berry RM. The maximum number of torque-generating units in the flagellar motor of Escherichia coli is at least 11. Proc Natl Acad Sci U S A. 2006;103:8066–71. doi: 10.1073/pnas.0509932103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Revyakin A, Ebright RH, Strick TR. Promoter unwinding and promoter clearance by RNA polymerase: detection by single-molecule DNA nanomanipulation. Proc Natl Acad Sci U S A. 2004;101:4776–80. doi: 10.1073/pnas.0307241101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Roca J. The torsional state of DNA within the chromosome. Chromosoma. 2011;120:323–34. doi: 10.1007/s00412-011-0324-y. [DOI] [PubMed] [Google Scholar]
- 93.Rowe AD, Leake MC, Morgan H, Berry RM. Rapid rotation of micron and submicron dielectric particles measured using optical tweezers. Journal of Modern Optics. 2003;50:1539–54. [Google Scholar]
- 94.Rybenkov VV, Vologodskii AV, Cozzarelli NR. The effect of ionic conditions on DNA helical repeat, effective diameter and free energy of supercoiling. Nucleic Acids Res. 1997;25:1412–8. doi: 10.1093/nar/25.7.1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Salceda J, Fernández X, Roca J. Topoisomerase II, not topoisomerase I, is the proficient relaxase of nucleosomal DNA. EMBO J. 2006;25:2575–83. doi: 10.1038/sj.emboj.7601142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Sarkar A, Leger JF, Chatenay D, Marko JF. Structural transitions in DNA driven by external force and torque. Physical Review E. 2001;63:10. doi: 10.1103/PhysRevE.63.051903. [DOI] [PubMed] [Google Scholar]
- 97.Scheffer MP, Eltsov M, Frangakis AS. Evidence for short-range helical order in the 30-nm chromatin fibers of erythrocyte nuclei. Proc Natl Acad Sci U S A. 2011;108:16992–7. doi: 10.1073/pnas.1108268108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Sheinin MY, Forth S, Marko JF, Wang MD. Underwound DNA under Tension: Structure, Elasticity, and Sequence-Dependent Behaviors. Physical Review Letters. 2011;107:5. doi: 10.1103/PhysRevLett.107.108102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Sheinin MY, Wang MD. Twist-stretch coupling and phase transition during DNA supercoiling. Physical Chemistry Chemical Physics. 2009;11:4800–03. doi: 10.1039/b901646e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Simpson RT, Thoma F, Brubaker JM. Chromatin reconstituted from tandemly repeated cloned DNA fragments and core histones: a model system for study of higher order structure. Cell. 1985;42:799–808. doi: 10.1016/0092-8674(85)90276-4. [DOI] [PubMed] [Google Scholar]
- 101.Smith DE, Tans SJ, Smith SB, Grimes S, Anderson DL, Bustamante C. The bacteriophage phi 29 portal motor can package DNA against a large internal force. Nature. 2001:413. doi: 10.1038/35099581. [DOI] [PubMed] [Google Scholar]
- 102.Smith SB, Cui YJ, Bustamante C. Overstretching B-DNA: The elastic response of individual double-stranded and single-stranded DNA molecules. Science. 1996:271. doi: 10.1126/science.271.5250.795. [DOI] [PubMed] [Google Scholar]
- 103.Sowa Y, Berry RM. Bacterial flagellar motor. Q Rev Biophys. 2008;41:103–32. doi: 10.1017/S0033583508004691. [DOI] [PubMed] [Google Scholar]
- 104.Sowa Y, Hotta H, Homma M, Ishijima A. Torque-speed relationship of the Na+-driven flagellar motor of Vibrio alginolyticus. J Mol Biol. 2003;327:1043–51. doi: 10.1016/s0022-2836(03)00176-1. [DOI] [PubMed] [Google Scholar]
- 105.Strick TR, Allemand JF, Bensimon D, Bensimon A, Croquette V. The elasticity of a single supercoiled DNA molecule. Science. 1996;271:1835–37. doi: 10.1126/science.271.5257.1835. [DOI] [PubMed] [Google Scholar]
- 106.Strick TR, Bensimon D, Croquette V. Micro-mechanical measurement of the torsional modulus of DNA. Genetica. 1999;106:57–62. doi: 10.1023/a:1003772626927. [DOI] [PubMed] [Google Scholar]
- 107.Strick TR, Croquette V, Bensimon D. Single-molecule analysis of DNA uncoiling by a type II topoisomerase. Nature. 2000;404:901–04. doi: 10.1038/35009144. [DOI] [PubMed] [Google Scholar]
- 108.Toyabe S, Watanabe-Nakayama T, Okamoto T, Kudo S, Muneyuki E. Thermodynamic efficiency and mechanochemical coupling of F1-ATPase. Proc Natl Acad Sci U S A. 2011;108:17951–6. doi: 10.1073/pnas.1106787108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Tsao YP, Wu HY, Liu LF. Transcription-driven supercoiling of DNA: direct biochemical evidence from in vitro studies. Cell. 1989;56:111–8. doi: 10.1016/0092-8674(89)90989-6. [DOI] [PubMed] [Google Scholar]
- 110.Vijayan V, Zuzow R, O’Shea EK. Oscillations in supercoiling drive circadian gene expression in cyanobacteria. Proc Natl Acad Sci U S A. 2009;106:22564–8. doi: 10.1073/pnas.0912673106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Vologodskii AV, Cozzarelli NR. Conformational and thermodynamic properties of supercoiled DNA. Annu Rev Biophys Biomol Struct. 1994;23:609–43. doi: 10.1146/annurev.bb.23.060194.003141. [DOI] [PubMed] [Google Scholar]
- 112.Wang H, Oster G. The Stokes efficiency for molecular motors and its applications. Europhysics Letters. 2002;57:134–40. [Google Scholar]
- 113.Wang MD, Schnitzer MJ, Yin H, Landick R, Gelles J, Block SM. Force and velocity measured for single molecules of RNA polymerase. Science. 1998:282. doi: 10.1126/science.282.5390.902. [DOI] [PubMed] [Google Scholar]
- 114.Wang MD, Yin H, Landick R, Gelles J, Block SM. Stretching DNA with optical tweezers. Biophysical Journal. 1997;72:1335–46. doi: 10.1016/S0006-3495(97)78780-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Washizu M, Kurahashi Y, Iochi H, Kurosawa O, Aizawa S, et al. Dielectrophoretic measurement of bacterial motor characteristics. Ieee Transactions on Industry Applications. 1993:29. [Google Scholar]
- 116.Watanabe-Nakayama T, Toyabe S, Kudo S, Sugiyama S, Yoshida M, Muneyuki E. Effect of external torque on the ATP-driven rotation of F1-ATPase. Biochem Biophys Res Commun. 2008;366:951–7. doi: 10.1016/j.bbrc.2007.12.049. [DOI] [PubMed] [Google Scholar]
- 117.Watson JD, Crick FH. Molecular structure of nucleic acids; a structure for deoxyribose nucleic acid. Nature. 1953;171:737–8. doi: 10.1038/171737a0. [DOI] [PubMed] [Google Scholar]
- 118.Wen J-D, Lancaster L, Hodges C, Zeri A-C, Yoshimura SH, et al. Following translation by single ribosomes one codon at a time. Nature. 2008:452. doi: 10.1038/nature06716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Wuite GJL, Smith SB, Young M, Keller D, Bustamante C. Single-molecule studies of the effect of template tension on T7 DNA polymerase activity. Nature. 2000:404. doi: 10.1038/35003614. [DOI] [PubMed] [Google Scholar]
- 120.Yasuda R, Noji H, Kinosita K, Yoshida M. F1-ATPase is a highly efficient molecular motor that rotates with discrete 120 degree steps. Cell. 1998;93:1117–24. doi: 10.1016/s0092-8674(00)81456-7. [DOI] [PubMed] [Google Scholar]