SUMMARY
Nuclear pore complexes (NPCs) are multiprotein channels connecting the nucleus with the cytoplasm. NPCs have been shown to have tissue-specific composition, suggesting that their function can be specialized. However, the physiological roles of NPC composition changes and their impacts on cellular processes remain unclear. Here we show that the addition of the Nup210 nucleoporin to NPCs during myoblast differentiation results in assembly of an Mef2C transcriptional complex required for efficient expression of muscle structural genes and microRNAs. We show that this NPC-localized complex is essential for muscle growth, myofiber maturation, and muscle cell survival and that alterations in its activity result in muscle degeneration. Our findings suggest that NPCs regulate the activity of functional gene groups by acting as scaffolds that promote the local assembly of tissue-specific transcription complexes and show how nuclear pore composition changes can be exploited to regulate gene expression at the nuclear periphery.
Graphical Abstract
INTRODUCTION
Nuclear pore complexes (NPCs) are aqueous channels built by multiple copies of ~30 different proteins known as nucleoporins (Raices and D’Angelo, 2012). In addition to regulating nucleocytoplasmic transport, these structures play important roles in gene expression regulation. In budding yeast, several genes have been found to relocate to NPCs when activated, and NPC-tethering has been linked to efficient expression and transcriptional memory (Schneider et al., 2015; Sood and Brickner, 2014). In mammalian cells and Drosophila, NPC components have also been found to play critical roles in gene expression regulation. However, nucleoporin-gene interactions in these organisms have been shown to occur mostly in the nuclear interior (Capelson et al., 2010; Kalverda et al., 2010; Light et al., 2013), and whether nucleoporins/NPCs play a role in gene expression regulation at the nuclear periphery is poorly understood. In fact, the general belief is that the nuclear periphery in metazoans is nearly always associated with gene repression. This is certainly true for lamina-associated domains (LADs) (Kind et al., 2013) and nuclear envelope transmembrane protein (NET)-associated genes (Robson et al., 2016), which are enriched in heterochromatin markers. However, it has long been observed by electron microscopy that NPCs are surrounded by decondensed chromatin (Capelson and Hetzer, 2009; Lemaitre and Bickmore, 2015), suggesting the association of active genes or chromatin regions with NPCs and the potential role of these structures in positively regulating gene expression. Supporting this hypothesis, the association of super-enhancer sequences with NPCs was recently identified (Ibarra et al., 2016).
NPCs have historically been considered structures of ubiquitous composition. Yet the expression of many nucleoporins varies between different cell types and tissues, and mutations in several NPC components result in tissue-specific diseases (Raices and D’Angelo, 2012). These findings indicate that this highly conserved structure can be specialized to play cell-type-specific roles. Supporting this idea, we recently identified that the addition of Nup210, a tissue-specific nucleoporin (Olsson et al., 1999, 2004), to NPCs is required for myogenic and neuronal differentiation in vitro (D’Angelo et al., 2012). Consistent with a transport-independent function, Nup210 does not change nucleo-cytoplasmic transport through NPCs during myogenesis but is required for the expression of key muscle genes (D’Angelo et al., 2012). How Nup210 regulates gene expression and what role this nucleoporin plays in the physiology of skeletal muscle in vivo have not been established.
Using a combination of the zebrafish model organism and a mammalian myoblast differentiation system, we identified that Nup210 regulates skeletal muscle growth, myofiber maturation, and muscle cell survival by modulating the expression of muscle structure and muscle maturation genes. We determined that Nup210 regulates gene expression by assembling a Mef2C-dependent transcription complex at nuclear pores. We also identified that while the association of genes with NPCs does not require Nup210, this nucleoporin is necessary for the efficient recruitment of Mef2C to NPC-tethered target genes. Our findings indicate that NPCs act as hubs for the regulation of functional gene groups that include sarcomeric genes and microRNAs (miRNAs); and that the activity of these genes is regulated by changes in nuclear pore composition. Our findings expose a novel mechanism of transcriptional regulation and support a model for the existence of different gene expression-regulatory compartments at the nuclear periphery, with a repressive lamina-associated environment and an active NPC-associated environment.
RESULTS
Nup210 Is Required for Zebrafish Skeletal Muscle Development
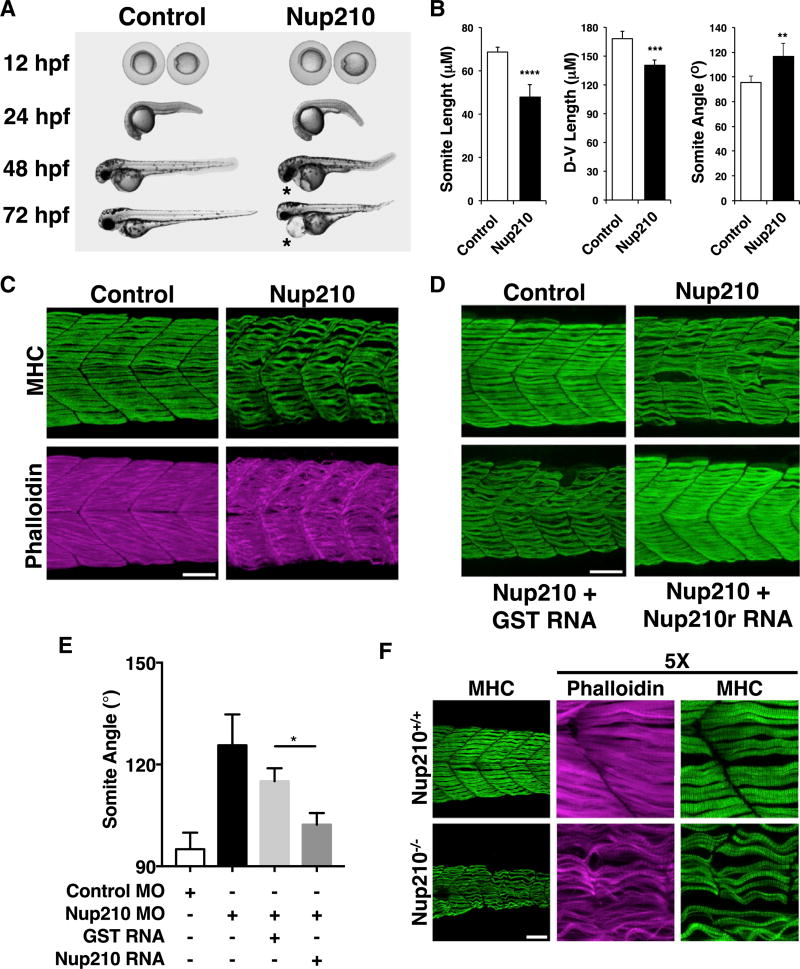
To study the role of Nup210 in the physiology of skeletal muscle we took advantage of the zebrafish model organism, which has emerged as a model to study muscle development and maintenance. We identified that zebrafish has a unique and highly conserved Nup210 ortholog (XM_002667560) coding for a protein with 68% amino acid identity to human Nup210. Consistent with its high degree of conservation, this protein localizes to NPCs when heterologously expressed in mammalian cells (Figure S1A). To evaluate the role of Nup210 in skeletal muscle formation, we first depleted this protein during zebrafish development using two specific morpholino oligonucleotides designed to block Nup210 translation. For this, control or Nup210-specific morpholinos were injected into one-cell zebrafish embryos and animals were analyzed at different times of development. Control animals were injected with morpholinos carrying five sequence mismatches. Although we observed no significant differences in the early stages of embryonic development, at 24 hr post fertilization (hpf) the embryos injected with Nup210-specific morpholinos showed slightly smaller body size and curved tails, and by 48 hpf these animals were characterized by shorter and narrower myotomes with increased myoseptum angles (myotomes in Nup210-depleted animals are U-shaped instead of V-shaped) (Figures 1A and 1B). Nup210-depleted animals showed difficulty in breaking out of the chorion and significantly impaired locomotion in touch-evoked escape assays (data not shown). Downregulation of Nup210 also resulted in the development of cardiac edema by 48–72 hfp, indicating alterations in heart development and/or function (Figure 1A). These phenotypes worsened as animals aged, suggesting alterations in later developmental stages or the deterioration of tissues over time.
Figure 1. Nup210 Is Required for Skeletal Muscle Development.
(A) Zebrafish one-cell embryos injected with Control or Nup210 morpholinos (MO) were imaged at different times of development. Asterisks show cardiac edema.
(B) The length, height (dorsal-ventral length), and angle of somite #20 were quantified in control and Nup210-depleted animals stained with the F59 antibody.
(C) Zebrafish embryos injected with control or Nup210 MOs were stained with F59 and phalloidin at 48 hpf. Images represent the maximal projection of multiple z stacks.
(D) Embryos were co-injected with control or Nup210 morpholinos and GST mRNA, or a Nup210 mRNA resistant to the morpholino (Nup210r) and muscle was analyzed at 48 hpf.
(E) Rescue experiments from (D) were quantified by measuring somite angle.
(F) Muscle structure in Nup210 CRISPR knockouts was analyzed at 48 hpf by staining with F59 and phalloidin.
Bar plots represent mean ± SEM, n ≥ 3 replicates. *p ≤ 0.05, **p ≤ 0.01, ***p<0.001, ****p ≤ 0.0001, two-tailed Student’s t test. Morpholino depletions were performed with n ≥ 50 embryos. Ten to 20 embryos were examined by immunofluorescence and quantified in n ≥ 3 independent experiments. Scale bars, 50 µm. See also Figure S1.
As most of the abnormalities associated with Nup210 depletion were consistent with muscle alterations, we analyzed muscle structure in more detail. Control and Nup210-depleted animals were stained with slow muscle myosin heavy chain antibodies. Fish without Nup210 showed a highly disorganized muscle structure with wavy myofibers, often detached from the myoseptum, and had multiple gaps between them (Figures 1C and S1B; Movies S1 and S2). The presence of wavy skeletal muscle fibers in zebrafish has been linked to alterations in thick and thin filaments (Ferrante et al., 2011; Ha et al., 2013). Phalloidin staining, which labels F-actin, revealed that slow and fast muscle thin filaments were severely disrupted in Nup210 morphants, confirming alterations in myofibril structure (Figure 1C; Movies S1 and S2). All muscle alterations were rescued by co-injection with a mutated Nup210 mRNA not targeted by the morpholinos (Figures 1D and 1E), confirming the specificity of Nup210 muscle phenotypes. To further verify the role of Nup210 in muscle formation, we generated zebrafish Nup210 mutant lines using CRISPR genome-editing technology. Nup210 mutant animals showed all the phenotypes previously identified and even stronger muscle alterations, consistent with a total depletion of Nup210 (Figure 1F).
Nup210 Is Dispensable for Early Embryonic Muscle Development but Essential for Muscle Growth and Myofiber Maturation
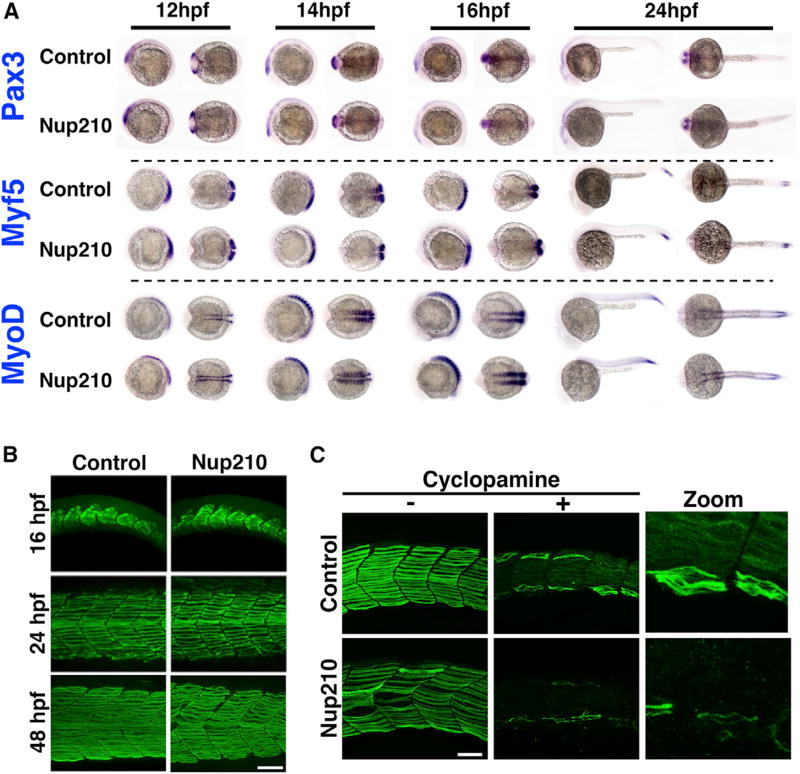
We previously identified that in addition to regulating myoblast differentiation, Nup210 is critical for the differentiation of embryonic stem cells into neuroprogenitors (D’Angelo et al., 2012). This suggests that Nup210 is an important regulator of cell fate. To test whether Nup210 depletion was associated with a reduced number of muscle progenitor cells, we performed whole-mount in situ hybridization against the muscle progenitor markers Pax3, Myf5, and MyoD (Buckingham and Vincent, 2009). In these experiments no differences where observed between control and Nup210-depleted animals (Figure 2A). These observations imply that Nup210 does not regulate the number of embryonic muscle progenitors, and show that myotome formation and the early stages of muscle development are not affected. Consistent with this, when we analyzed the time of onset of muscle defects in these animals we found that the abnormalities in myofiber structure were only detected after 24 hpf (Figure 2B), when early embryonic muscle development ends and muscle growth and maturation begin (Barresi et al., 2001).
Figure 2. Nup210 Depletion Does Not Affect Early Muscle Development.
(A) Whole-mount in situ hybridization against the muscle markers Pax3, Myf5, and MyoD were performed at different times of development in zebrafish embryos injected with control or Nup210 morpholinos.
(B) Slow muscle was stained with F59 at different times of development in control and Nup210-depleted animals and analyzed by confocal microscopy.
(C) Control or Nup210 morphants were incubated with cyclopamine during development to inhibit the Sonic Hedgehog-dependent formation of early embryonic slow muscle. Addition of new muscle fibers during muscle growth was analyzed by F59 staining at 48 hpf.
Morpholino depletions were performed with n ≥ 50 embryos. Ten to 20 embryos were examined by immunofluorescence and quantified in n ≥ 3 independent experiments. Scale bars, 50 µm. See also Figure S1.
During zebrafish embryogenesis there are two waves of myogenesis that lead to the formation of slow muscle fibers (Figure S1C) (Barresi et al., 2001). Throughout myotome formation muscle fibers originate from muscle precursors derived from adaxial cells. But when segmentation is complete (at ~24 hpf) there is a switch in muscle formation, and muscle now grows by two mechanisms: (1) the growth/maturation of pre-existing myofibers, and (2) the addition of new myofibers to the ventral and dorsal sides of the myotome. The new muscle fibers that are added during this period originate from different muscle precursors, which are Pax7+ cells (Seger et al., 2011), and its formation is under a genetic regulation different from that of early embryonic muscle development. While muscle formation from adaxial-derived cell precursors is dependent on Sonic Hedgehog signaling, muscle growth from Pax7+ progenitors is not (Barresi et al., 2001) (Figure S1C). Thus, inhibition of Sonic Hedgehog signaling blocks the formation of early embryonic muscle fibers without affecting muscle growth after segmentation, and allows studying the addition of new muscle fibers independent of early embryonic muscle development (Barresi et al., 2001). To test whether Nup210 plays a role in muscle growth after segmentation, we injected zebrafish embryos with control or Nup210 morpholinos in the presence or absence of the Sonic Hedgehog inhibitor cyclopamine, and analyzed the addition of new muscle fibers to the myotome at 48 hpf (Figure S1D). As shown in Figure 2C, cyclopamine inhibited the formation of early embryonic muscle fibers in control animals but had no effect on the later addition of new myofibers. In contrast, depletion of Nup210 strongly impaired the addition of the new muscle fibers that result from the second wave of myogenesis. Consistent with an inhibition in the addition of new myofibers during muscle growth, the number of myofibers per myotome was significantly reduced in Nup210-depleted animals (Figure S1E). These findings demonstrate that Nup210 is essential for muscle growth after segmentation, and explain why Nup210-depleted animals have narrower myotomes (Figure 1B).
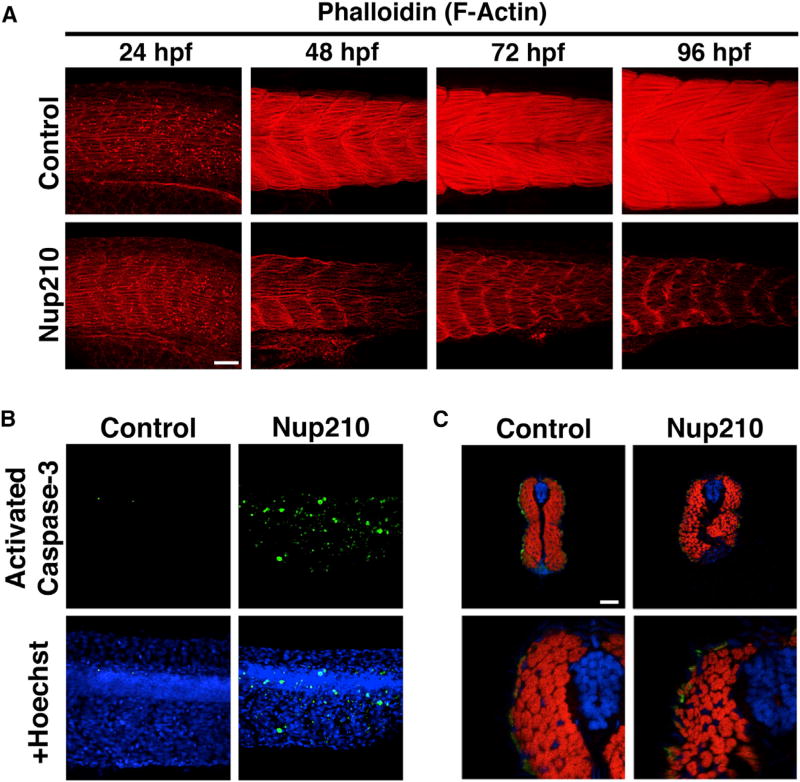
Nup210 Loss Inhibits Myofibrillogenesis and Results in Muscle Degeneration
Our results revealed that Nup210 is not required for the initial formation of myotomes and muscle fibers during zebrafish embryonic development but it is critical for the formation of new fibers during muscle growth. This likely contributes to the abnormal muscle structure that develops in older animals, but does not fully explain the alterations observed at 48 hpf. By this time Nup210-depleted animals show abnormal and missing myofibers in the central part of the myotome, which contains fibers that originate during the first myogenic wave (Figure S2A). This suggests that despite being dispensable for the formation of these fibers, Nup210 might be necessary for their maturation and growth, and its absence might lead to muscle degeneration. To test this, we analyzed muscle structure in Nup210-depleted animals at sequential times of development by staining thin filaments with phalloidin. Consistent with our previous observations, no significant differences in muscle organization between control and Nup210-depleted animals were detected at 24 hpf, but a significant deterioration of the muscle structure was observed as animals grew older in the absence of this nucleoporin (Figure 3A). Nup210-depleted animals failed to properly assemble their thin filament structures, indicating an inhibition of myofibrillogenesis and sarcomere maturation. These animals also showed a progressive accumulation of actin next to the myoseptum (Figure 3A). The aberrant accumulation of actin in this region has also been observed in zebrafish depleted of the transcription factors Mef2C and Mef2D, which have critical roles in sarcomere assembly, myofibrillogenesis, and myofiber maturation (Hinits and Hughes, 2007; Li et al., 2002). Consistent with a degenerating muscle structure, depletion of Nup210 led to a strong increase in apoptosis within the myotomes (Figure 3B). Cross-section examination of animals at 96 hpf showed that Nup210 depletion results in a highly disorganized muscle tissue with fewer myofibers having significant gaps between them (Figure 3C). Nup210-depleted animals also showed a greater number of small muscle fibers, likely due to the inhibition of myofiber maturation and growth (Figure 3C). Smaller fibers are also observed after muscle injury and in dystrophic muscle, and represent new myofibers that repair the damaged tissue (McNally and Pytel, 2007). Thus, these small myofibers could also represent regeneration that results from the death of pre-existing muscle cells in the degenerating muscle. Altogether, these data indicate that Nup210 is important for muscle growth and plays a critical role in the maturation and maintenance of differentiated muscle cells.
Figure 3. Nup210 Depletion Inhibits Myofibrillogenesis and Results in Muscle Cell Death.
(A) Thin filament structure in control and Nup210 morphants was analyzed by phalloidin staining at different times of development.
(B) Tail muscle was stained for apoptotic death with an anti-activated caspase-3 antibody in control and Nup210-depleted animals.
(C) Cross-sections of zebrafish animals at 96 hpf were stained with the slow muscle marker F59 (green), phalloidin (red), and DAPI (blue) (n = 6–10 embryos).
Representative images of n ≥ 3 independent experiments. n = 10–20 embryos unless otherwise specified. Scale bars, 50 µm.
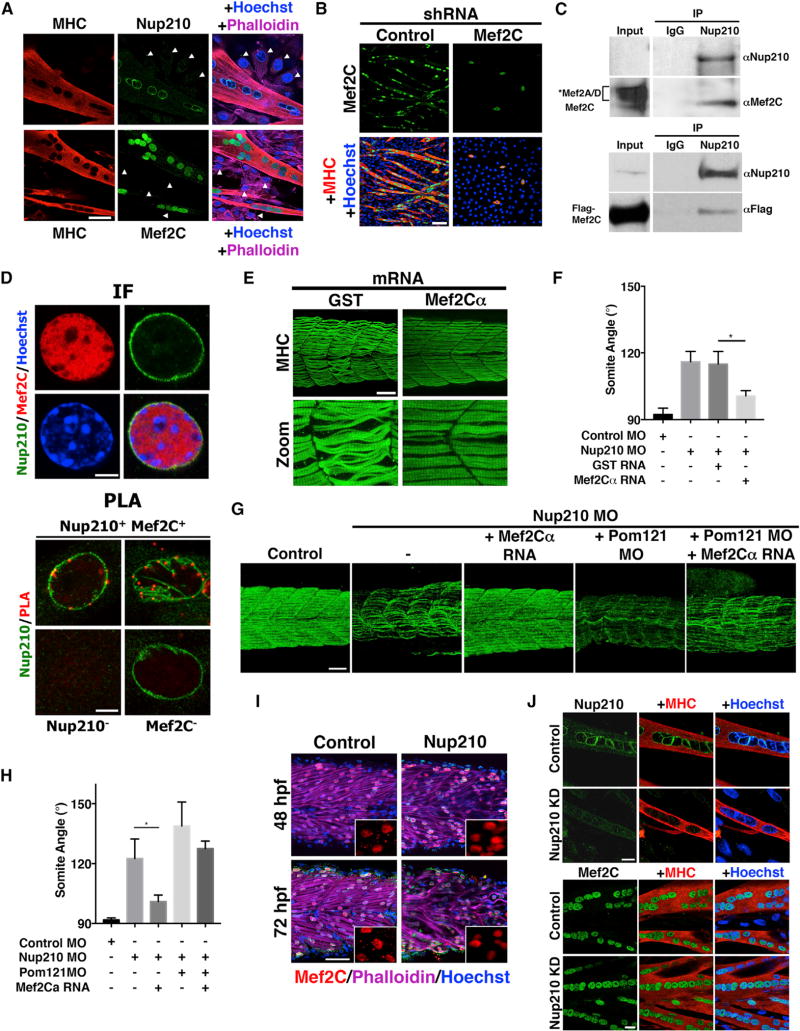
Nup210 Associates with Trip6 to Regulate Muscle Physiology
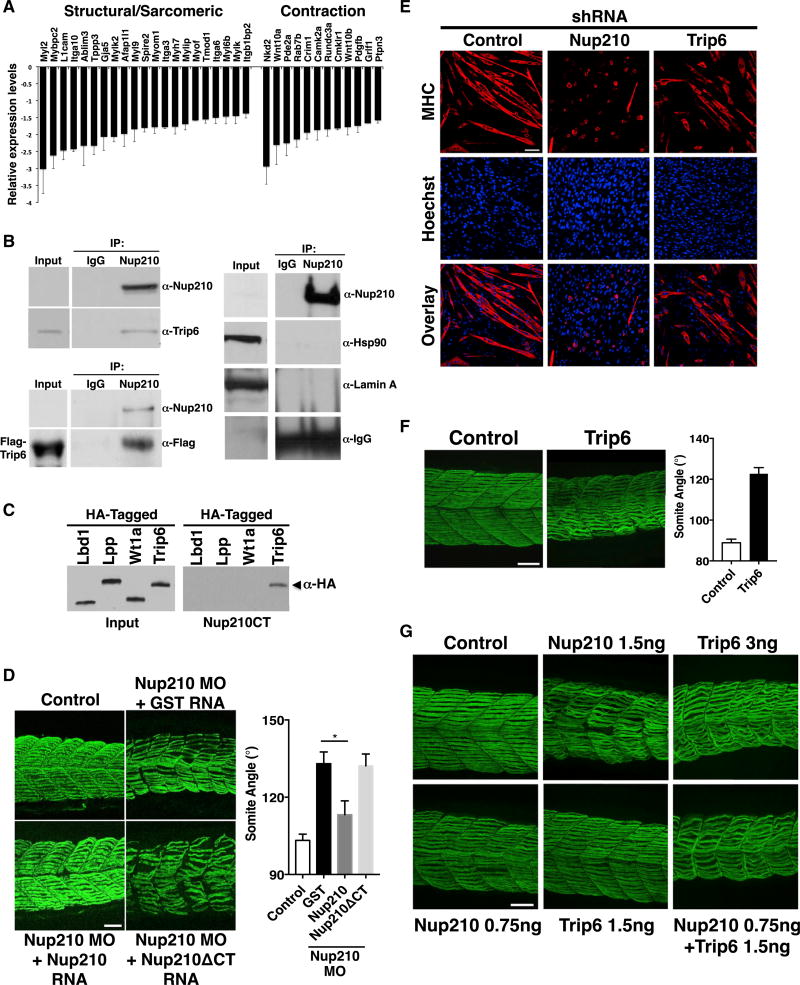
We previously identified that Nup210 regulates myoblast differentiation by modulating the expression of key differentiation genes (D’Angelo et al., 2012). Consistent with its role in myofibrillogenesis, depletion of Nup210 in differentiated muscle cells also results in the downregulation of several sarcomeric, contraction, and muscle structural genes (Figure 4A). How Nup210 addition to the NPC results in changes in gene expression is unknown. Interestingly, Nup210 was previously shown to interact with the LIM-domain protein Trip6 (Yi et al., 2002). Trip6 is an adaptor protein that shuttles between focal adhesions and the nucleus, and acts as a transcriptional co-regulator (Lin and Lin, 2011). Although the function of Trip6 in the regulation of muscle physiology has not been investigated, this protein is known to interact with factors that play key roles in myogenesis and muscle development, such as Mef2C (Kemler et al., 2016) and the LIMdomain proteins FHL3 and ILF3 (Cottle et al., 2007; Shi et al., 2005). Trip6 functions and interaction partners suggest that Nup210 modulation of gene expression might involve this transcriptional co-regulator. As a first approach to investigate this hypothesis, we confirmed the interaction between Nup210 and Trip6 using co-immunoprecipitation approaches. In these experiments, Nup210-specific antibodies but not control antibodies were able to immunoprecipitate endogenous as well as FLAG-tagged Trip6 from protein extracts of myotubes (Figure 4B). The lack of interaction of Nup210 with Lamin A or Hsp90 further confirmed the specificity of Nup210-Trip6 association. Similarly, we found that the zebrafish homolog of Trip6, but not other LIM-domain proteins, interact with the C-terminal/nuclear domain of Nup210 (Figure 4C). Notably, deletion of this domain from Nup210 inhibits the ability of this protein to rescue the muscle phenotypes of Nup210-depleted fish, confirming that the Nup210/Trip6 association is required for its muscle function in vivo (Figure 4D).
Figure 4. Nup210 Interacts with and Regulates Muscle Development through Trip6.
(A) The expression levels of several sarcomeric and muscle contraction genes in Nup210-depleted C2C12 myotubes was extracted from D’Angelo et al. (2012).
(B) Nup210 was immunoprecipitated (IP) from wild-type or FLAG-Trip6-expressing myotubes and the presence of Trip6 was determined using anti-Trip6 or anti-FLAG antibodies. Nup210 is expressed at low levels and is not detected in the input. Lamin A and Hsp90 were analyzed in Nup210 immunoprecipitates to determine the specificity of Trip6 binding (right panel). IgG, immunoglobulin G.
(C) The association of zebrafish Nup210 C-terminal domain fused to GST with different in vitro translated HA-tagged LIM-domain proteins was analyzed using pull-down assays.
(D) Embryos were co-injected with control or Nup210 morpholinos and mRNA for GST, full-length Nup210, or Nup210ΔCT, and muscle structure was analyzed 48 hpf with the F59 antibody. Myotome angle was used to quantify muscle alterations (n = 10–20 embryos, performed in triplicate).
(E) C2C12 cells were infected with lentiviruses carrying control, Nup210, or Trip6 shRNAs, selected and induced to differentiate. Differentiated myotubes were stained for myosin heavy chain (MHC) at 72 hr post differentiation. Nuclei were stained with Hoechst.
(F) Zebrafish embryos were injected with control or Trip6 morpholinos and slow muscle was stained with F59 at 48 hpf (left panel). Muscle alterations were quantified by measuring myotome angle (right panel). n = 10–20 embryos, performed in triplicate.
(G) Control, Nup210, or Trip6 morpholinos (top panel) and subphenotypic concentrations of Nup210, Trip6, or both morpholinos (bottom panel) were injected into one-cell zebrafish embryos and slow muscle was stained at 48 hpf. Bar plots represent mean ± SEM, n ≥ 3 replicates. *p ≤ 0.05. Scale bar, 50 µm. See also Figure S2.
Supporting a role for Trip6 in muscle physiology, downregulation of this LIM-domain protein in C2C12 cells strongly inhibited myogenic differentiation (Figure 4E). Furthermore, depletion of Trip6 during zebrafish development, either by injection of specific morpholino oligonucleotides or by CRISPR-dependent gene mutation, resulted in muscle alterations that resemble Nup210 mutants (Figures 4F and S2A). More importantly, we found that while co-injection of Nup210 and Trip6 morpholinos at phenotypic concentrations did not show an additive effect on muscle alterations (data not shown), co-injection of subphenotypic concentrations, which independently have no effect on muscle structure, resulted in strong myofiber defects (Figure 4G). This additive effect confirms a genetic interaction between Nup210 and Trip6, and indicates that these factors are part of the same pathway. These findings identify a role for Trip6 in skeletal muscle formation and development, and support the hypothesis that Nup210 regulates muscle physiology through its interaction with this LIM-domain protein.
Mef2C Is a Key Player in Nup210/Trip6 Regulation of Muscle Physiology
Trip6 does not possess transcriptional activity on its own, but acts as a co-regulator of gene expression through its interaction with different transcription factors and nuclear hormone receptors, such as v-Rel, NF-κF, AP-1, and glucocorticoid receptor (Lin and Lin, 2011). Recently, Trip6 was identified to interact with the transcription factor Mef2C (Kemler et al., 2016), a critical regulator of skeletal and cardiac muscle development (Potthoff and Olson, 2007). Similar to Nup210, Mef2C is dispensable for early embryonic muscle development but essential for myofiber growth, maturation, and survival (Hinits and Hughes, 2007; Hinits et al., 2012; Lin et al., 1997; Potthoff et al., 2007). Mef2C expression is also induced during C2C12 myoblast differentiation, and its downregulation with short hairpin RNAs (shRNAs) completely inhibits myotube formation (Figures 5A and 5B). These findings suggest that Nup210/Trip6 might regulate muscle gene expression and muscle physiology by modulating the activity of Mef2C. To investigate whether Nup210 forms a complex with Mef2C, we differentiated C2C12 myoblasts into myotubes to induce Nup210 expression and subjected them to immunoprecipitation assays. In myotube extracts, Nup210-specific antibodies, but not control antibodies, were able to precipitate Mef2C (Figure 5C). Notably, other non-Mef2C proteins also recognized by the pan-Mef2 antibody used for these experiments, likely Mef2A or Mef2D (Figure S2B), show virtually no signal in the immunoprecipitation experiment, suggesting that Nup210 has a greater specificity for Mef2C within the Mef2 family (Figure 5C). We further confirmed the interaction between Nup210 and Mef2C by expressing FLAG-tagged Mef2C in C2C12 myotubes and performing Nup210 co-immunoprecipitations (Figure 5C). Since Nup210 is a transmembrane nucleoporin, we could expect that its association with Mef2C will result in its localization to the nuclear periphery. Yet in post-mitotic myotubes we found Mef2C evenly distributed within the nuclear space, with no clear enrichment at the nuclear periphery (Figure 5D). This is not surprising, as Mef2C is an essential muscle transcription factor with target genes all over the genome. But when Nup210-Mef2C interaction was analyzed by proximity ligation assays (PLA), which allow the visualization of protein-protein interactions within cells, we confirmed that a subset of Mef2C is associated with Nup210 at the nuclear periphery (Figure 5D). Interestingly, we found a significantly lower number of Nup210-Mef2C interaction foci compared with Nup210-NPC foci (using the mAb4141 antibody) (Figure S2C). This could suggest a functional heterogeneity of NPCs within the cell, but could also indicate that Nup210-Mef2C interactions are dynamic or temporally regulated, or that the accessibility of antibodies to Nup210 and/or Mef2C in the complex is limited, preventing the detection of more interactions. In fact, even though we have previously observed that Nup210 is present in most, if not all, NPCs of muscle cells (D’Angelo et al., 2012), Nup210-mAb414 PLA does not label every NPC at the nuclear envelope (Figure S2C).
Figure 5. Nup210 Interacts with and Recruits Mef2C to NPCs.
(A) Nup210 and Mef2C immunofluorescence in differentiated C2C12 cells (day 4). MHC was used as a marker for differentiated myotubes. Nuclei were stained with Hoechst. Arrowheads show quiescent myoblasts with non-detectable Nup210 or Mef2C.
(B) C2C12 cells were infected with lentiviruses carrying control and Mef2C shRNAs, induced to differentiate and stained at 72 hr after differentiation with MHC.
(C) Nup210 was immunoprecipitated from wild-type or FLAG-Mef2C expressing C2C12 myotubes, and the presence of Mef2C was determined with a pan-MEF2 antibody (top panel) or an anti-FLAG (bottom panel) antibody. Mef2C-specific band was confirmed by shRNA depletion (Figure S2B). Asterisk shows the potential Mef2A and Mef2D bands recognized by the pan-MEF2 antibody as described by the manufacturer. The specificity of these bands was not confirmed in this study. Nup210 is expressed at low levels in myotubes and not detected in the input.
(D) Differentiated C2C12 cells were analyzed by immunofluorescence against Nup210 and Mef2C (IF), and proximity ligation assays (PLA). PLA shows the interaction of Mef2C with Nup210 at the nuclear periphery (red dots). Representative images (n ≥ 3). Scale bars, 5 µm.
(E) Zebrafish embryos were co-injected with Nup210 morpholinos plus GST or Mef2Cα mRNAs and slow muscle was stained at 48 hpf.
(F) Rescue experiments from (E) were quantified by measuring somite angle (n = 10–20 embryos, performed in triplicate).
(G) Zebrafish embryos were injected with control or Nup210 morpholinos alone or in combination with Mef2Cα RNA, Pom121 MO, or Pom121MO+ Mef2Cα RNA. Slow muscle was analyzed 48 hpf.
(H) Quantification of myotome angle from (G) (n = 10–20 embryos, performed in triplicate).
(I) Control or Nup210 morpholino-depleted embryos were stained at 48 hpf for Mef2C (red dots) and F-actin (magenta). Nuclei were stained with Hoechst (blue).
(J) The localization of Mef2C in control or Nup210-depleted myotubes was analyzed by immunofluorescence. Top panel shows the downregulation of Nup210 in shRNA-depleted myotubes. Scale bar, 20 µm.
Bar plots represent mean ± SEM, n ≥ 3 replicates. *p ≤ 0.05. Scale bars represent 50 µm unless otherwise specified. See also Figure S2.
If Nup210 works by modulating Mef2C function, we reasoned that increasing the activity of this transcription factor during zebrafish development could be sufficient to rescue the muscle abnormalities of Nup210 depletion. Zebrafish has two Mef2C genes, Mef2Cα and Mef2Cβ (Hinits et al., 2012). Depletion of Mef2Cα, but not Mef2C, during zebrafish development led to muscle alterations similar to Nup210-depleted animals (Figure S2D and data not shown), consistent with its critical role in regulating myofibrillogenesis and fiber growth (Yogev et al., 2013). To investigate whether Mef2C could rescue the muscle defects of Nup210 depletion, we co-injected zebrafish embryos with Nup210 morpholinos and glutathione S-transferase (GST) or Mef2Cα mRNAs and analyzed them for muscle abnormalities. We determined that overexpression of Mef2Cα, but not control GST, significantly rescued the muscle defects of Nup210 morphants (Figures 5E and 5F). If Nup210 is required to recruit Mef2C to the nuclear periphery to regulate muscle gene expression, why the overexpression of soluble Mef2C can rescue its muscle defects is puzzling. One possibility is that Nup210 is required to increase the local concentration of Mef2C at the nuclear periphery and its ectopic expression is sufficient to reach the necessary levels for gene regulation. It is also possible that another nuclear pore complex component might compensate for Nup210 loss. There are two additional transmembrane components of the NPC, Pom121 and NDC1. Notably, Pom121 has also been shown to interact with Trip6. Even though we found that depletion of Pom121 does not lead to muscle alterations in zebrafish (Figures S2E and S2F), we identified that its codepletion with Nup210 results in greater muscle defects and strongly blocks the rescue by Mef2Cα RNA (Figures 5G and 5H). These findings suggest that Pom121 might act as an additional anchor for Trip6/Mef2C that is not required for muscle function when Nup210 is expressed, but which might be sufficient to partially compensate Nup210 loss when Mef2C levels are high enough. The lack of Mef2C rescue due to the elimination this potential secondary anchor site at NPCs supports the idea that its localization to nuclear pores is required for Nup210 muscle function. This is further supported by our findings showing that a Nup210 mutant lacking the nucleoplasmic domain is unable to rescue the muscle defects of Nup210-depleted animals (Figure 4D). Altogether, these results confirm that Nup210 and Mef2C form a complex at NPCs in differentiated muscle cells and support the hypothesis that Nup210 regulates muscle physiology through Mef2C. Consistent with a transport-independent function of Nup210 in regulating Mef2C activity, Nup210-depleted fish and C2C12 myotubes showed no alterations in the levels or nuclear accumulation of this transcription factor (Figures 5I and 5J).
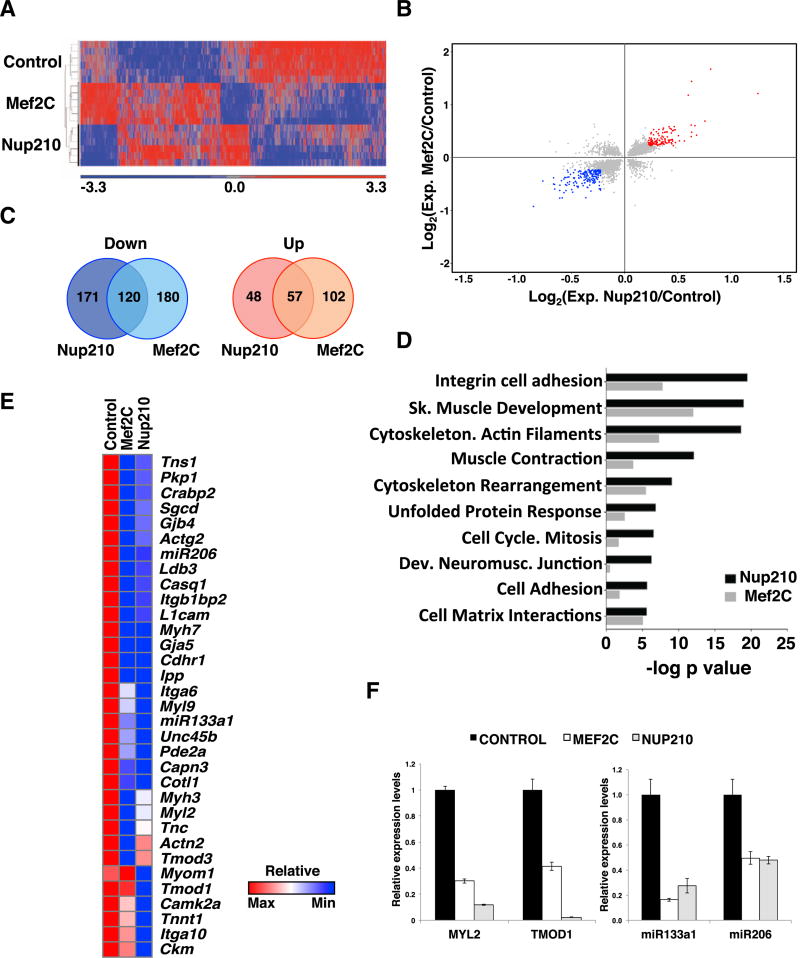
Nup210 and Mef2C Co-regulate Muscle Structural, Sarcomeric, and Cell Adhesion Genes
Our findings suggest that Nup210 regulates muscle gene expression by modulating the activity of Mef2C, but which genes are coregulated by this complex is unknown. Like Nup210, depletion of Mef2C in vivo leads to a deterioration of differentiated myofibers (Hinits and Hughes, 2007; Potthoff et al., 2007). The degeneration of muscle fibers in Mef2C-depleted animals is a consequence of alterations in the assembly of the sarcomeric structures that result from the misexpression of muscle structural genes (Hinits and Hughes, 2007; Potthoff et al., 2007). As shown in Figure 4A, we found that Nup210 also regulates many structural and sarcomeric genes in muscle cells. To identify genes that are co-regulated by both proteins, we independently depleted Nup210 or Mef2C from differentiated C2C12 myotubes and analyzed whole-genome expression using microarrays. For this, C2C12 cells were infected with lentiviruses carrying specific shRNAs 36 hr after the induction of differentiation. Since it takes ~48–72 hr post infection (hpi) to have a significant knockdown of Nup210 or Mef2C, no differences in cell differentiation are observed during this period and this protocol results in multinucleated myotubes lacking Nup210 or Mef2C (Figures S3A and S3B) (D’Angelo et al., 2012). Because Nup210 depletion from post-mitotic myotubes results in cell death starting at ~96–120 hpi (D’Angelo et al., 2012), we restricted our analysis to 48 hpi. This ensured that no alterations in gene activity are an indirect consequence of the initiation of apoptotic cell death, but also resulted in smaller fold changes in gene expression (Figures S3B–S3D). We identified many common genes that showed altered expression in Nup210- and Mef2C-depleted myotubes (Figure 6A and Table S1). Considering a 1.25-fold change, 291 genes were downregulated in Nup210 knockdowns and 300 were downregulated in Mef2C depletion, while 120 were shared by both treatments (Figures 6B and 6C; Table S1). On the other hand, Nup210 depletion resulted in 105 upregulated genes and Mef2C knockdown in 159, 57 of which were upregulated in both treatments. These findings indicate that ~45% of Nup210-regulated genes are also modulated by Mef2C. Meta-core process analysis of Nup210/Mef2C co-regulated genes showed highest enrichment in cell adhesion, muscle development and contraction, and cytoskeleton (Figure 6D). Consistent with our identified role of Nup210 in the maintenance of sarcomere integrity and myofibril maturation; and confirming previous findings for Mef2C, both treatments showed significant downregulation of many cytoskeletal, cell adhesion, and muscle contraction genes (Figure 6E). When we further confirmed the gene expression alterations by real-time PCR, a much stronger decrease in gene expression levels was detected for these and other genes (Figures 6F and S4), indicating that the fold changes observed in the microarrays considerably underestimate the downregulation induced by Nup210 or Mef2C depletion. Interestingly, among the most downregulated genes we found the miRNAs miR-133a-1 and miR-206 (Figures 6E and 6F). These miRNAs, known as myomiRs, play critical roles in myogenesis and skeletal muscle homeostasis by modulating the activity of many target mRNAs. Altogether, these findings indicate that Nup210 and Mef2C regulate the expression of a common set of muscle genes and might indirectly regulate part of their target genes by modulating the levels of muscle-specific miRNAs.
Figure 6. Nup210 and Mef2C Regulate a Common Set of Muscle Genes.
(A) Whole-genome gene expression was analyzed by microarray in C2C12 myotubes infected with lentiviruses carrying control, Nup210, or Mef2C shRNAs. Heatmap shows the gene expression patterns of Nup210- and Mef2C-depleted myotubes (2,410 common genes, p ≤ 0.05).
(B) Scatterplots of log2 gene expression changes in Nup210- and Mef2C-depleted myotubes (p ≤ 0.05). Genes changing ≥ 1.25-fold for Nup210 and Mef2C are shown in blue (down) or red (up). Gene expression values are mean changes of microarray sextuplicate samples.
(C) Venn diagram of down- and upregulated genes in Nup210- and Mef2C-depleted myotubes relative to scramble control (p ≤ 0.05, ≥ 1.25-fold change).
(D) Process enrichment for Nup210 and Mef2C co-regulated genes was analyzed using Metacore software.
(E) Heatmap for gene expression changes of sarcomeric, cell adhesion, and muscle contraction genes in Nup210- and Mef2C-depleted myotubes.
(F) The expression levels of Myl2, Tmod1, miR-133a-1, and miR-206 in Nup210 and Mef2C-depleted myotubes were analyzed by qPCR. mRNA levels for each gene were normalized to Hprt1 expression and are presented relative to scramble control-treated cells. Data represent mean ± SEM, n ≥ 3 replicates.
See also Figures S3 and S4.
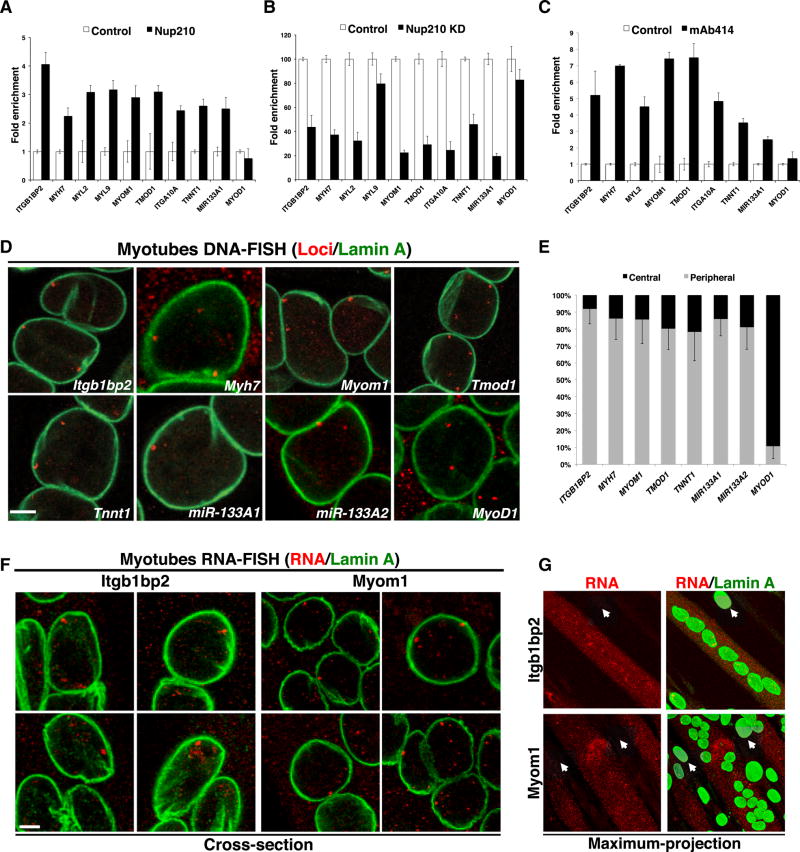
The Nup210/Mef2C Complex Regulates Genes at NPCs
Our data suggest that Nup210 regulates muscle gene expression by recruiting Mef2C to the nuclear envelope. Although many of the genes that are altered by depletion of these proteins will be indirectly regulated, we expect that some, if not most, of the genes that are directly controlled by the Nup210/ Mef2C complex will (1) be differentially induced during myoblast differentiation, when the complex is assembled, (2) be affected by the depletion of either protein in differentiated muscle cells, and (3) have putative Mef2C binding sites in their regulatory sequences, as they should be directly bound by this transcription factor. Because of the critical role of Nup210/Mef2C in the regulation of muscle structural genes and the fact that some sarcomeric genes have been found to associate with NPCs during cardiomyocyte hypertrophic growth (Kehat et al., 2011), we decided to test our prediction on the sarcomeric, cytoskeletal, and muscle contraction genes we found deregulated in Nup210 and Mef2C knockdowns (Figure 6E). To identify genes potentially associated with NPCs within this group, we first analyzed their expression during myoblast differentiation and scanned their upstream regulatory regions for potential Mef2C binding sites. We identified several candidate genes for direct regulation by Nup210/Mef2C that were induced during myoblast differentiation and show putative Mef2C binding sequences (Figures S5 and S6; Table S2). Notably, miR-133a-1 fits the criteria for genes potentially regulated by Nup210/Mef2C directly. This miRNA is upregulated during myogenesis (Chen et al., 2006) and downregulated in Nup210- and Mef2C-depleted myotubes (Figures 6E and 6F), and has been shown to be directly regulated by this Mef2C binding (Granjon et al., 2009; Liu et al., 2007). Ten candidate genes present in different chromosomes and with different number of putative Mef2C binding sites were selected to test their association with Nup210. Itga10 and Itgb1bp2 code for an integrin and an integrin-binding chaperone, respectively; Myl2, Myl9, Myh7, Tnnt1, Tmod1, and Myom1 genes code for structural proteins of the sarcomere; Tmod1 and Myom1 have been previously identified as Mef2C targets (Potthoff et al., 2007). As already mentioned, miR-133a-1 is a muscle-specific miRNA and has one confirmed Mef2C binding site (Liu et al., 2007) plus three additional ones predicted in our studies. MyoD gene was chosen as potential negative control because it relocates from the nuclear lamina to the nuclear interior during myogenesis (Yao et al., 2011) and should not be associated with NPCs in differentiated muscle cells. To establish whether these genes associate with Nup210 in differentiated C2C12 muscle cells, we performed chromatin immunoprecipitation (ChIP) assays using an anti-Nup210 antibody. As shown in Figure7A, all the candidate genes analyzed, but not the MyoD gene, were enriched in immunoprecipitates of Nup210 but not control antibodies. To control for the specificity of association with the Nup210 antibody, we repeated our analysis in multinucleated cells depleted of this nucleoporin. For this, we generated an inducible Nup210 knockdown C2C12 cell line by infecting myoblasts with a lentivirus carrying a tetracycline-regulated Nup210-specific shRNA. In these experiments C2C12 myoblasts were induced to differentiate, and Nup210 was downregulated with doxycycline in myotubes prior to ChIP assays. Nup210 downregulation was confirmed by real-time PCR and western blots (Figures S7A and S7B). We also confirmed that Nup210 depletion resulted in the downregulation of Nup210-target genes (Figure S7B). ChIP assays in this cell line confirmed that all candidate genes, except MylF9, were specifically immunoprecipitated by the Nup210 antibody (Figure 7B).
Figure 7. Nup210/Mef2C-Regulated Genes Associate with NPCs.
(A) Chromatin immunoprecipitations (ChIPs) were performed in differentiated C2C12 myotubes using a Nup210-specific antibody and analyzed by real-time PCR against the specified genes.
(B) Nup210 ChIPs were performed in control or Nup210-depleted C2C12 myotubes using a Nup210-specific antibody and analyzed as described in (A).
(C) ChIPs were performed in differentiated C2C12 myotubes using mAb414 antibody and analyzed as described in (A).
(D) DNA fluorescence in situ hybridization (DNA-FISH) was performed in differentiated C2C12 myotubes for several Nup210-binding genes. Representative images show gene loci (red) and Lamin A (green) as marker of the nuclear periphery. Note that C2C12 cells are tetraploid (four copies for each gene, Figure S8B). The specificity of probes used for FISH was determined using mouse chromosome spreads (Figure S8C). n ≥ 4.
(E) The percentage of nuclei per myotube that show one or more loci associated with the nuclear periphery was quantified from 3D nuclear reconstructions as described in Figure S8D. n ≥ 3, 100–150 nuclei per experiment.
(F) RNA-FISH using probes specific for Itbg1bp2 and Myom1 was performed on differentiated C2C12 cells counterstained with Lamin A antibody (green). Representative images reveal that the nuclear envelope-associated loci are actively transcribing (red). n ≥ 3.
(G) Representative images show the maximum projection of differentiated C2C12 cells stained with Lamin A (green) and the Itgb1bp2 and Myom1 RNA-FISH probes (red). RNA is detected in myotubes but not in quiescent myoblasts (white arrows).
Bar plots represent mean ± SEM, n ≥ 3 replicates. Scale bars, 5 µm. See also Figures S5–S8.
Nup210 is a transmembrane nucleoporin that at endogenous levels localizes exclusively to NPCs (D’Angelo et al., 2012). However, a recent report showed that during myogenesis ectopic expression of an Nup210 fragment that fails to localize to NPCs can partially rescue differentiation (Gomez-Cavazos and Hetzer, 2015). This suggests that Nup210 can potentially function independently of NPCs. As a first approach to determine whether Nup210-associated genes localize to NPCs, we performed additional ChIP assays using the NPC antibody mAb414, which recognizes four nucleoporins. It is important to note that some mAb414-recognized nucleoporins have been shown to localize to the nuclear interior (Capelson et al., 2010; Jacinto et al., 2015; Kalverda et al., 2010). Thus, association with these nucleoporins does not fully probe NPC localization. However, as Nup210 localizes to the nuclear envelope, the NPC is most likely the place where these nucleoporins co-localize. As shown in Figure 7C, we found that all Nup210-bound genes were also associated with mAb414, indicating that the function of this nucleoporin in the transcriptional regulation of Mef2C target genes likely takes place at nuclear pores. To confirm this hypothesis, we analyzed the intranuclear positioning of several Nup210-associated genes by fluorescence in situ hybridization (FISH). We included Itgb1bp2, Myh7, Myom1, Tnnt1, Tmod1, and miR-133a-1 that we found associated with Nup210 by ChIP and also the miR-133a-2 gene, which we had not analyzed before but also fits the criteria for genes potentially regulated at NPCs. Again, Myod1 was used as negative control for association. As shown in Figures 7D, 7E, and S8A–S8D, we found that all the Nup210-associated genes tested showed one or more loci localized to the nuclear periphery in >80%–90% of the nuclei examined. It is important to note that the quantification includes nuclei where not all of the four loci of the tetraploid C2C12 myotubes were detected, and thus the percentage of nuclei with nuclear envelope association could be slightly higher. In contrast, we detected <10% of Myod1 loci associated with the nuclear envelope. Lack of nuclear envelope association was also observed for the nucleoporin genes Nup160 and Nup62 (Figure S8A), whose expression is not regulated by Nup210 or Mef2C (Table S1). Using RNA-FISH assays we established that the peripheral loci of Itgb1bp2 and Myom1 are active in post-mitotic myotubes but not in quiescent myoblasts (Figures 7F and 7G). In agreement with our previous findings, we also determined that depletion of Nup210 reduces the activity of Myom1 gene but not of Gapdh (Figure S8E). Depletion of Nup210 reduced the transcription of the Myom1 gene but was not associated with an aberrant accumulation of Myom1 or Gapdh mRNA inside the nucleus, indicating that Nup210 phenotypes do not result from alterations in global mRNA export.
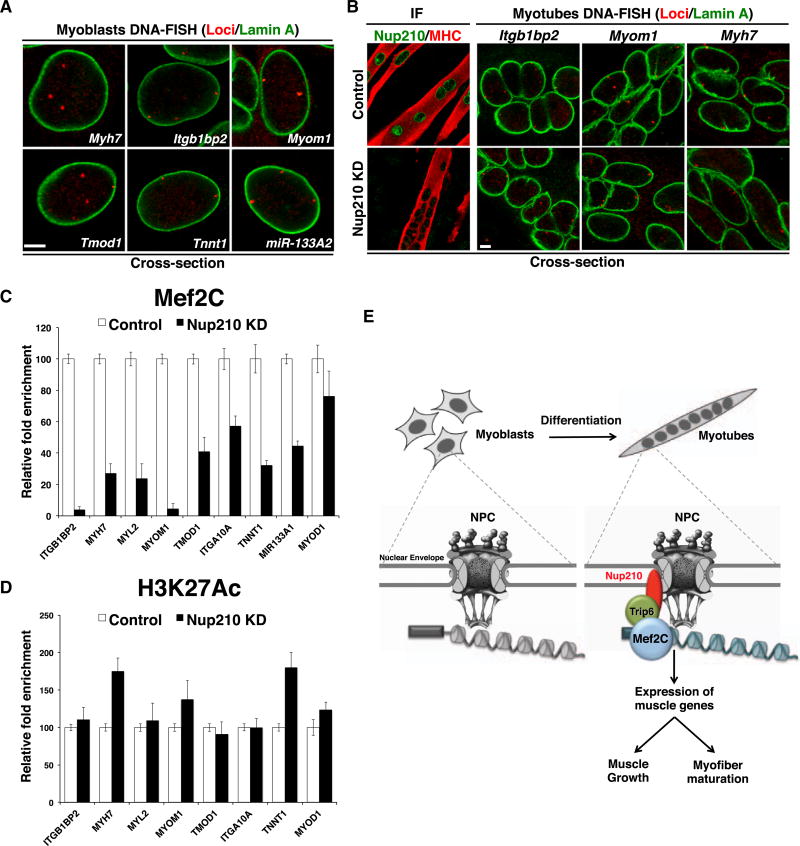
Nup210 Is Required for the Recruitment of Mef2C to NPC-Associated Genes, but Not for Gene Localization to the Nuclear Periphery
In yeast, the association of several genes with NPCs is mediated by transcription factor binding (Randise-Hinchliff et al., 2016). This raises the possibility that the interaction of Nup210 with Mef2C might be required for gene localization to the nuclear periphery. However, we observed that with the exception of Myh7, all Nup210-associated genes analyzed already showed peripheral localization in quiescent myoblasts that do not express Nup210 (Figure 8A). To further confirm that Nup210 is dispensable for gene localization to the nuclear periphery, we performed DNA-FISH for Itgb1bp2, Myom1, and Myh7 in control or Nup210-depleted myotubes. We found that in the absence of Nup210 these genes still localized to the nuclear envelope (Figure 8B). Our data suggest that Nup210 is dispensable for gene-NPC association but is required for the assembly of a localized Mef2C-dependent transcription machinery at nuclear pores and, thus, for the recruitment Mef2C. Consistent with this model, ChIP assays performed with a specific Mef2C antibody show decreased recruitment of the transcription factor to Nup210-bound genes in myotubes depleted of this nucleoporin. This is particularly clear for Itgb1bp2 and Myom1, which show the most prominent nuclear envelope association (Figure 8C). Notably, we observed no changes in the levels of the active chromatin marker H3K27Ac in the promoter of these genes, indicating that the effect on Mef2C recruitment is specific and not due to major changes in chromatin accessibility (Figure 8D). Altogether, our findings support a model in which Nup210 regulates the expression of muscle genes during myogenesis by assembling an Mef2C-dependent transcription complex at NPCs (Figure 8E).
Figure 8. Nup210 Is Required for the Recruitment of Mef2C to Its Target Genes but Is Dispensable for Their Association with NPCs.
(A) DNA-FISH for Nup210 target genes was performed on quiescent myoblasts. Gene loci (red), Lamin A (green). Representative images of four independent experiments.
(B) DNA-FISH was performed on control C2C12 myotubes or myotubes depleted of Nup210. Left panel shows that Nup210 is efficiently downregulated by the specific shRNA in myotubes. Right panel: gene loci (red), Lamin A (green). Representative images of two independent experiments.
(C) ChIPs were performed in control or Nup210-depleted differentiated C2C12 myotubes using a Mef2C-specific antibody and analyzed by real-time PCR against the specified genes. Values represent the relative fold enrichment of knockdown cells to control cells.
(D) ChIPs were performed in control or Nup210-depleted differentiated C2C12 myotubes using an H3K27Ac-specific antibody by real-time PCR against the specified genes. Values represent the relative fold enrichment of knockdown cells to control cells.
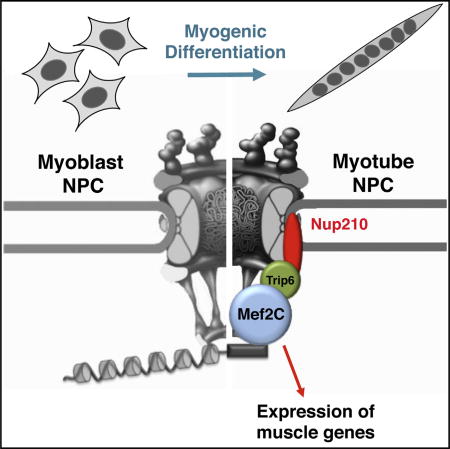
(E) Schematic model of Nup210 regulation of muscle gene expression during myoblast differentiation. Undifferentiated myoblasts do not express Nup210. When cells are induced to differentiate, Nup210 is added to NPCs and recruits Mef2C through Trip6. The assembled complex regulates the expression of local muscle genes involved in sarcomere assembly, myofiber maturation, and muscle growth.
Bar plots represent mean ± SEM, n ≥ 3 replicates. Scale bars, 5 µm. See also Figure S8.
DISCUSSION
In this work we identified that the tissue-specific nucleoporin Nup210 (D’Angelo et al., 2012; Olsson et al., 1999, 2004) is essential for skeletal muscle growth and maintenance, and that its depletion results in muscle fiber degeneration. Our results reveal a key role for this nucleoporin in the maintenance of sarcomere integrity and in the maturation of differentiated myofibers, and indicate that Nup210 regulates these processes by assembling an NPC-localized Mef2C-dependent transcriptional complex required for the expression of muscle structural genes. Our work indicates that mammalian NPCs can act as scaffolds for the organization of gene neighborhoods and that the assembly of these localized transcription sites can be regulated by modifying the composition of these structures.
In yeast, it is well established that NPCs play an important role in gene expression regulation. Several genes have been shown to associate with NPCs upon activation, and the tethering of these genes to the nuclear periphery is important for their efficient expression and for transcriptional memory (Sood and Brickner, 2014). In metazoans, the role of NPCs in gene expression regulation at the nuclear periphery is less clear. Although a few studies have shown the association of genes with NPCs (Kalverda et al., 2010; Kehat et al., 2011; Liang et al., 2013), the physiological relevance of these associations and whether NPCs play any role in modulating gene activity is unknown. In fact, the current view of gene expression regulation by nuclear pore complex components in metazoans is that it mostly occurs within the nuclear interior (Capelson et al., 2010; Kalverda et al., 2010; Liang et al., 2013). Our findings show that the NPC structure plays a critical role in regulating the activity of associated muscle structural genes, and indicate that the function of these structures in regulating active gene expression at the nuclear periphery is conserved in mammals. Moreover, work in yeast has revealed that the tethering of genes to NPCs is mostly performed by transcription factors (Brickner et al., 2012; Randise-Hinchliff et al., 2016). In this organism, the association of transcription factors with NPCs is not only important for gene tethering but is also responsible for gene clustering at the nuclear periphery (Brickner et al., 2012; Randise-Hinchliff et al., 2016). Our findings that Nup210 recruits Mef2C to NPCs to locally regulate a subset of its target genes further support the conservation of NPC function and of the molecular mechanisms of gene expression regulation between yeast and mammals, although in this case Nup210 is required for gene regulation but not gene tethering.
Interestingly, a recent study by the Tjian laboratory showed that the differential subnuclear distribution of general transcription factors regulates the occupancy level of the MyoD gene promoter during myogenesis and suggests that the local compartmentalized availability of transcription factors in the nucleus could be an important mechanism of gene expression regulation during development (Yao et al., 2011). Our findings align with this idea by showing that the NPC-localized activity of the key muscle transcription factor Mef2C is important for myogenesis and muscle development. Further supporting this model, recent findings showed the association of super-enhancers with nuclear pore complexes and identified that super-enhancer tethering to NPCs is important for the expression of cell identity genes (Ibarra et al., 2016). Altogether these findings raise the exciting possibility that NPCs might help to organize different transcriptional machineries at the nuclear periphery.
Our data show that Nup210 is required for the correct expression of sarcomeric and muscle structural genes at NPCs. Similarly, it was previously reported that during cardiac hypertrophic growth the association of several genes, including sarcomeric and calcium-handling genes, with NPCs is important for their efficient transcription and for sarcomeric organization (Kehat et al., 2011). Interestingly, in cardiomyocytes the association of these genes with NPCs is negatively regulated by HDAC4, which is a negative regulator of Mef2C activity. These findings together with ours suggest that the interplay between these factors at NPCs is critical for muscle gene expression and indicate that NPCs are important hubs for the regulation of muscle structural genes. Our results provide an explanation of how Nup210 regulates myofiber growth and maturation by regulating the activity of muscle structural genes through Mef2C. Further work will be required to identify other factors that might work with Nup210 to regulate early myogenic differentiation and cell death.
STAR*METHODS
Detailed methods are provided in the online version of this paper and include the following:
CONTACT FOR REAGENT AND RESOURCE SHARING
-
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Cells Lines
Zebrafish
-
METHOD DETAILS
Morpholino Injections
Whole-Mount In Situ Hybridization (WISH)
Cyclopamine Treatment
Cell Culture
shRNA Production, Infection and Selection
C2C12 Inducible Cell Lines
Immunofluorescence
RNA Extraction and qPCR
Microarrays
Immunoprecipitations and Western Blots
GST Pull Down Assays
Proximity Ligation Assays (PLA)
Chromatin Immunoprecipitations (CHIPs)
DNA Fluorescent In Situ Hybridizations (FISH)
RNA Fluorescent In Situ Hybridization (FISH)
QUANTIFICATION AND STATISTICAL ANALYSIS
DATA AND SOFTWARE AVAILABILITY
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse Monoclonal anti-MYH1A Myosin heavy chain (all fast isoforms) | Developmental Studies Hybridoma Bank, Iowa | F59 |
| Rabbit Polyclonal Anti-MEF2C | Sparrow Biosciences | Catalog # SBS-002 |
| Purified Rabbit Anti-Caspase 3 (Clone C92-605) | BD Biosciences | Catalog # 559565 |
| Purified Rabbit Polyclonal Anti-NUP210 | Bethyl Laboratories | Catalog # A301-795A |
| Rabbit Monoclonal Anti-MEF2C (D80C1) | Cell Signaling Technology | Catalog # 5030 |
| Mouse Monoclonal Anti-Myosin Heavy Chain (MYH1E) | Developmental Studies Hybridoma Bank, Iowa | MF20 |
| Rabbit Monoclonal Anti-Paxillin antibody (Y113) | Abcam | Catalog # ab32084 |
| Rabbit Polyclonal Anti-Trip6 | Dr. Mary Beckerle, University of Utah | N/A |
| Rabbit Polyclonal Anti MEF2 (C21) | Santa Cruz | Catalog # sc-313 |
| Mouse Monoclonal Anti-Trip6 | Proteintech | Catalog # 60205-1-Ig |
| Monoclonal Mouse Anti-FLAG M2 clone | Sigma Aldrich | SKU # F1804 |
| Purified Mouse Monoclonal Anti-HA.11 (clone 16B12) | Covance | Catalog # MMS-101P |
| Rabbit Polyclonal Anti-GFP (IF) | Abcam | Catalog # ab290 |
| A Rabbit Polyclonal Anti-GFP (WB) | Abcam | Catalog # ab6556 |
| Purified anti-Nuclear Pore Complex Proteins Antibody mAB414 | BioLegened | Catalog # 902902 |
| Mouse Monoclonal Anti-HSP90 | R&D Systems | Catalog # MAB3286 |
| Rabbit Polyclonal Anti Lamin A (C-Terminal) | Sigma-Aldrich | Catalog # L1293 |
| Mouse Monoclonal Anti Lamin A/C (Alexa Fluor 488 Conjugate) | Cell Signaling Technology | Catalog # 8617 |
| Purified Goat Polyclonal anti-Mef2C (E-17) X Chip concentration | Santa Cruz | Catalog # sc-13266X |
| Mouse Monoclonal Anti Histone H3K27ac | Active Motif | Catalog # 39685 |
| Rabbit Control IgG | EMD Millipore | Catalog # 12-370 |
| Mouse Control IgG | EMD Millipore | Catalog # 12-371 |
| Bacterial and Virus Strains | ||
| BL21 | New England BioLabs | Catalog # C2530H |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Phalloidin Alexa Fluor 647 | Molecular Probes | Catalog # A22287 |
| N-Phenylthiourea | Sigma-Aldrich | SKU # P7629 |
| Pronase | Roche-Sigma-Aldrich | SKU # 10165921001 |
| Paraformaldehyde (PFA) | Electron Microscopy Sciences | Catalog # 50980487 |
| Cyclopamine | Fisher Scientific | N/A |
| Puromycin Dihydrochloride | GIBCO-Life Technologies | Catalog # A1113803 |
| Polybrene | EMD-Millipore | Catalog # TR-1003-G |
| Doxycycline | Clontech | Catalog # 631311 |
| TRIzol | Ambion- Thermo Fisher Scientific | Catalog # 15596018 |
| iTaq UniverSYBR Green SMX 2500 | BioRad Laboratories | Catalog # 1725124 |
| TaqMan Fast Advanced Master Mix | Thermo Fisher Scientific | Catalog # 444557 |
| Proteinase K | Invitrogen | Catalog # 25530049 |
| Heparin | Sigma-Aldrich | SKU # H3393 |
| NBT/BCIP tablets | Roche-Sigma-Aldrich | SKU # 11697471001 |
| GeneJet transfection reagent (C2C12) | Signagen Laboratories | Catalog # SL100489-C2C12 |
| Complete Mini protease inhibitor cocktail (EDTA Free) | Roche | Catalog # 11836170001 |
| PMSF | Sigma-Aldrich | SKU # 93482 |
| PhosSTOP | Roche-Sigma-Aldrich | SKU # 04906845001 |
| Dynabeads Protein G | Invitrogen- Thermo Fisher Scientific | Catalog # 10003D |
| B-PER Bacterial Protein Extraction Reagent | Thermo Fisher Scientific | Catalog # 78248 |
| Pierce Glutathione Agarose | Thermo Fisher Scientific | Catalog # 16100 |
| Glutathione | Thermo Fisher Scientific | Catalog # 78259 |
| NuPAGE LDS Sample Buffer | Invitrogen- Thermo Fisher Scientific | Catalog # NP007 |
| Novex Sample Reducing Agent | Invitrogen- Thermo Fisher Scientific | Catalog # NP009 |
| Formaldehyde Solution 37% | Thermo Fisher Scientific | Catalog # BP531500 |
| Formamide (deionized) | EMD-Millipore | Catalog # S4117 |
| Dextran Sulfate | Sigma-Aldrich | SKU # D8906 |
| Salmon Sperm DNA | Invitrogen- Thermo Fisher Scientific | Catalog # 15632011 |
| Yeast tRNA | Invitrogen- Thermo Fisher Scientific | Catalog # AM7119 |
| Mouse CotI DNA | Life Technologies | Catalog # 18440016 |
| Critical Commercial Assays | ||
| MAXIscript kit | Invitrogen- Thermo Fisher Scientific | Catalog # AM1312 |
| Gateway Cloning system | Invitrogen- Thermo Fisher Scientific | Catalog # 12535-019 & # 11826-021 |
| mMESSAGE T7 kit | Invitrogen- Thermo Fisher Scientific | Catalog # AM1345 |
| Qiagen RNeasy kit | Qiagen | Catalog # 74104 |
| Quantitect kit | Qiagen | Catalog # 205311 |
| DIG RNA Labeling SP6/T7 Kit | Roche- Sigma-Aldrich | SKU # 11175025910 |
| pGEMT easy cloning system | Promega | Catalog # A1360 |
| NE-PER Nuclear and Cytoplasmic Extraction Reagent | Thermo Fisher Scientific | Catalog # 78833 |
| TNT Quick Coupled Transcription/Translation System | Promega | Catalog # L1170 |
| SuperSignal West Pico Chemiluminescent Substrate | Thermo Fisher Scientific | Catalog # 34087 |
| SuperSignal West Femto Chemiluminescent Substrate | Thermo Fisher Scientific | Catalog # 34095 |
| EZ-Magna ChIP A/G Kit | Millipore | Catalog # 17-10086 |
| Duolink In Situ Red Starter Kit | Sigma-Aldrich | SKU # DUO92101 |
| Biotin-Nick Translation Mix | Sigma-Aldrich | SKU #11745824910 |
| Stellaris Immuno-RNA fish | Biosearch Technologies | Catalogs # SMF-HB1-10, SMF-WA1-60 & SMF-WB1-20 |
| Deposited Data | ||
| Raw and analyzed Microarray data | This paper | GEO: GSE98318 https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=etyjcksytzczrif&acc=GSE98318 |
| Experimental Models: Cell Lines | ||
| Mouse: NIH3T3 | ATCC | CRL-1658 |
| Human: U-2 OS | ATCC | HTB-96 |
| Mouse: C2C12 Cell line | ATCC | CRL-1772 |
| C2C12 cells - inducible Nup210 shRNA | This paper | N/A |
| Experimental Models: Organisms/Strains | ||
| Zebrafish: TL | Zebrafish International Resource Center | N/A |
| Oligonucleotides | ||
| Morpholinos used for Zebrafish experiments, see Table S3 | Gene Tools, LCC | N/A |
| Nup210 Exon II sgRNA target 5’GGTGGCCAGTATCGAGGCTG3’ | http://zifit.partners.org/ZiFiT/ | Gene ID 570945 http://zfin.org/ZDB-GENE-050208-132 |
| TRIP6 Exon I sgRNA target 5’GGTGAAGCGTGGGCCGGTTG3’ (target site on the reverse strand) | http://zifit.partners.org/ZiFiT/ | Gene ID 792697 http://zfin.org/ZDB-GENE-140106-91 |
| Primers used for qPCR, see Table S4 | IDT Oligos | N/A |
| Taqman Gene Expression Assays used for qPCR, see Table S4 | Applied BioSystems - Thermo Fisher Scientific | N/A |
| Primers used for CHIPs, see Table S4 | IDT Oligos | N/A |
| Recombinant DNA | ||
| pCDNA6.2 /nLumio-DEST | Invitrogen - Thermo Fisher Scientific | Catalog # 12489027 |
| pCDNA6.2 nLumio-zfNup210 | This Paper | Gene ID 570945 http://zfin.org/ZDB-GENE-050208-132 |
| pCDNA6.2 nLumio-zfMef2ca | This Paper | Gene ID 30575 http://zfin.org/ZDB-GENE-980526-253 |
| pCDNA6.2 nLumio-zfTrip6 | This Paper | Gene ID 792697 http://zfin.org/ZDB-GENE-140106-91 |
| pCDNA6.2 nLumio-GST | This Paper | N/A |
| pDR274-zfNup210 exon2 | This Paper | Gene ID 570945 http://zfin.org/ZDB-GENE-050208-132 |
| pDR274-zfTrip6 exon1 | This Paper | Gene ID 792697 http://zfin.org/ZDB-GENE-140106-91 |
| pGEMTeasy-myf5 | This Paper | Gene ID 58097 http://zfin.org/ZDB-GENE-000616-6 |
| pGEMTeasy-pax3 | This Paper | Gene ID 30532 http://zfin.org/ZDB-GENE-980526-52 |
| pBSK-myoD | Dr. Daniel Hart, UCSF | N/A |
| pDest15 | Invitrogen - Thermo Fisher Scientific | Catalog # 11802014 |
| pDest15-CTNup210zf | This Paper | Gene ID 30245 http://zfin.org/ZDB-GENE-980526-558 |
| pCDNA6.2 nLumio-HA-zfLDB1 | This Paper | Gene ID 30579 https://zfin.org/ZDB-GENE-990415-138 |
| pCDNA6.2 nLumio-HA-zfWT1A | This Paper | http://zfin.org/ZDB-GENE-980526-558 |
| pCDNA6.2 nLumio-HA-zfTrip6 | This Paper | https://zfin.org/ZDB-GENE-140106-91 |
| pCDNA6.2 nLumio-HA-zfLPP | This Paper | Gene ID 573024 https://zfin.org/ZDB-GENE-040426-918 |
| gp210–EGFP3–CT (Nup210-GFP) | Rabut et al., 2004 | N/A |
| pcDNA4T/O-FLAG-MEF2C | Brian Black, UCSF | N/A |
| pcDNA-FLAG-Trip6 | Addgene | Plasmid # 27254 |
| pLKO-shNup210 | D’Angelo et al., 2012 | N/A |
| Tet-pLKO-puro | Addgene | Plasmid # 21915 |
| ITGB1BP2 BAC | BACPAC resources, CHORI | RP23-370M1 |
| Myh7 BAC | BACPAC resources, CHORI | RP24-263N11 |
| Myom1 BAC | BACPAC resources, CHORI | RP23-34I13 |
| Tmod1 BAC | BACPAC resources, CHORI | RP24-210F14 |
| Tnnt1 BAC | BACPAC resources, CHORI | RP24-388L7 |
| miR133a-1 BAC | BACPAC resources, CHORI | RP24-167F23 |
| miR133a-2 BAC | BACPAC resources, CHORI | RP24-251H23 |
| MyoD BAC | BACPAC resources, CHORI | RP23-209P3 |
| Nup160 BAC | BACPAC resources, CHORI | RP24-171N23 |
| Nup62 BAC | BACPAC resources, CHORI | RP23-403D23 |
| Software and Algorithms | ||
| ZiFiT Targeter Software Package | Sander et al., 2010 | http://zifit.partners.org/ZiFiT/ |
| Partek Genomics Suite Software (version 6.6) | Partek | N/A |
| Gene-E | Broad Institute (broadinstitute.org) | http://www.broadinstitute.org/cancer/software/GENE-E/ |
| BioRad CFX Manager 3.1 Real Time PCR Analysis Software | BioRad Laboratories | N/A |
| Transfac (Biobase Biological Databases) | QIAGEN Bioinformatics | https://portal.biobase-international.com/cgi-bin/portal/login.cgi |
| Stellaris Probe Designer | Biosearch Technologies | N/A |
| LAS X (Leica Suite X Microscope Imaging Software) | Leica | N/A |
STAR*METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for reagents should be directed to and will be fulfilled by the Lead Contact, Maximiliano D’Angelo (mdangelo@sbpdiscovery.org).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Cells Lines
C2C12, U2OS and NIH3T3 cells were obtained from ATCC.
Zebrafish
Wild-type Zebrafish (TL strain) were raised and maintained in an AAALAC-accredited facility accordingly with IACUC regulations at Sanford Burnham Prebys Medical Discovery Institute. Zebrafish gender is not reported because analyzed embryos were between 12-96 hpf. Genomic targets against Nup210 (exon 2) and Trip6 (exon 1) were generated with the ZiFiT Targeter software package at http://zifit.partners.org/ZiFiT/ (Sander et al., 2010). Respective forward and reverse sequences were cloned into BsaI digested pDR274 vector. Positive clones were confirmed by sequencing and linearized with DraI for further use as a template of the guide RNA (sgRNA) synthesis with the MAXIscript kit. Cas9 RNA was a provided by Dr. Daniel Hart. TL embryos at 1-cell stage were co-injected with 100pg of Cas9 RNA and 30 to 100 pg of sgRNA (either Nup210 exon 2 or Trip6 exon 1) as described by Jao (Jao et al., 2013). Fish were fin-clipped at 2 months old for genomic DNA extraction. Deletion mutants at the corresponding targets regions were confirmed by sequencing.
METHOD DETAILS
Morpholino Injections
To prevent pigmentation, egg water containing embryos at stage ~22 hpf was supplemented with 0.225% phenylathiourea (PTU). Media was replaced every 24 h. For dechorionation, embryos were treated for 1 minute with 1% pronase at room temperature. For morpholino injections, 1-cell stage embryos were injected with 0.5 nl of oligo solution containing 0.1% phenol red. The morpholinos employed in this study are described in Table S3 and were used at concentrations ranging from 0.5 to 4 ng. For rescue experiments embryos were co-injected with up to 200 pg of Nup210, Trip6, Mef2Cα, or GST RNA obtained as follows. Zebrafish Nup210, Trip6 and Mef2cα cDNAs were amplified by RT-PCR from total RNA from embryos and cloned into the pCDNA6.2/nLumio-DEST vector using the Gateway cloning system. These constructs were used as template specific RNA synthesis. RNA synthesis was performed with the mMESSAGE Ambion RNA kit and quality was confirmed by agarose gel analysis.
Whole-Mount In Situ Hybridization (WISH)
Whole in situ hybridization was performed as described by Thisse and Thisse (Thisse and Thisse, 2008) with minor modifications. Embryos were fixed 8 h at 4°C with 4% PFA permeabilized with 5 µg/ml of proteinase K for 0.5 minutes (12–16 hpf); 4 minutes (24 hpf); 8 minutes (48 hpf); 20 minutes (54–72 hpf); or 30 minutes for older stage embryos.
For antisense riboprobes, total Zebrafish RNA was used as a template in a RT-PCR for cDNA synthesis. 500bp partial cDNA sequences of Myf5 and Pax3 (an identical region shared among Pax3a and Pax3b paralogues) were cloned into the pGEMT-easy vector, linearized by restriction enzyme digestion and labeled with The DIG RNA Labeling SP6/T7 kit. Riboprobes were purified using BioRad microspin columns and riboprobe quality was confirmed by agarose gel analysis. The pBSK(−)-MyoD construct for Zebrafish MyoD riboprobe synthesis was a kind gift from Dr. Daniel Hart. Probes were used at 1.5 ng/µl in hybridization buffer (50% formamide, 5X SSC, 0.1% Tween 20, 9 mM Citric acid, 50 µg/ml Heparin, 500 µg/ml tRNA). Staining was performed using the NBT/BCIP tablets as indicated by the manufacturer. In situ hybridization images and live embryos movies were taken with a G12 powershot camera (Canon) mounted on a Zeiss Stemi 2000 stereomicroscope. Whole embryo immunofluorescence images were obtained on a Leica TCS SP8 confocal microscope using 1 µm confocal sections (~30–40 sections). Myotubes images optimized for 3 µm z-sections. For Zebrafish fiber quantification and measurements, non-saturated maximal projections were made with the Leica Application Suite X software.
Cyclopamine Treatment
Cyclopamine assays were performed as described by Barresi et al (Barresi et al., 2001) with minor modifications. Briefly, wild-type Zebrafish embryos were injected with control or Nup210 morpholinos and Cyclopamine was added 5.5 h post fertilization at 30 µM concentration. Embryos were stained 48 hpf with the slow muscle marker F59.
Cell Culture
Proliferating myoblasts were maintained in 20% FBS/DMEM. Differentiation into myotubes was induced by shifting to 2% horse serum. Differentiation media was replaced every 48 h. Myoblasts transfections were performed using GeneJet Transfection Reagent. NIH3T3 and U2OS cells were grown in 10%FBS/DMEM with penicillin/streptomycin as recommended by the manufacturer and transfected using Lipofectamine 2000.
shRNA Production, Infection and Selection
To generate stable cell lines, C2C12 were infected with pLKO lentiviral vectors carrying shRNAs targeting Nup210 (TRCN0000101935 and TRCN0000101938); Mef2C (TRCN0000012069 to TRCN0000012070), Trip6 (TRCN0000113515 to TRCN0000113519) or non-target control (GE Healthcare). Lentiviruses were packaged into 293T cells grown in 20% FBS/DMEM. Cells were transfected in 10 cm plates with 5.2 µg of pLKO-shRNA vector and 2.8 µg of packaging mix using 24 µl of Lipofectamine 2000. Media was replaced 12 h after transfection and supernatants were collected at 36 and 60 h. Myoblasts were infected at 30% confluency with virus-containing supernatants supplemented with 6 µg/ml of polybrene. Selection was made 48 hpi with media 5 µg/ml puromycin. Myoblasts were induced to differentiate and efficiency of gene knockdown was confirmed by qPCR and immunofluorescence.
C2C12 Inducible Cell Lines
shRNA targeting Nup210 (TRCN0000101935) was sublcloned in a pLKO inducible lentiviral vector (Tet-pLKO-puro) and used to generate C2C12 lines as described above. Selection was made 48 hpi with media 5 µg/ml puromycin. Inducible cell lines we were isolated and induced to differentiate. 1 µg/ml of doxycicline or vehicle were added 24 h later to ensure that differentiation was not affected. Differentiated myotubes were collected 72 h after doxycicline was added. The efficiency of gene knockdown was confirmed by qPCR, western blot and immunofluorescence.
Immunofluorescence
Zebrafish were euthanized with tricaine prior to fixation. After overnight (ON) incubation in 0.2% Triton X-100/ PBS (0.2% TxPBS), embryos were permeabilized with 2% Triton X-100 1xPBS (T-PBS) at RT with gentle agitation (4 h for 24 hpf; ON for later stages). Blocking was made with 2%BSA/0.2% TxPBS (BB) 1 h at RT. All incubations for Zebrafish immunofluorescence were made for 2 days at 4°C with gentle agitation. Embryos were mounted on 0.5% low melting point agarose for confocal imaging. For immunofluorescence C2C12 myoblasts and myotubes were cultured in 8-well plates. For Nup210 staining, cells were fixed in −20°C methanol for 2 min and then permeabilized in 1 × PBS/1% Triton X-100 for 1 min. For MHC, cells were either fixed in methanol when costained with Nup210 or in 4% PFA for 5 min. For all other antibodies and cells, fixation was done in 4% PFA for 5 min. Fixed cells were blocked using IF buffer (1 × PBS, 10 µg/ml BSA, 0.02% SDS, 0.1% Triton X-100) and incubated with primary antibody in IF buffer for 1 h or at RT or ON at 4°C. Cells were washed in IF buffer and incubated with secondary antibody for an additional hour at RT. Cells were washed in IF buffer and incubated with Hoechst for 5 min before mounting.
RNA Extraction and qPCR
Total RNA from cells or whole Zebrafish embryos was extracted with Trizol and purified with the Qiagen RNeasy kit. cDNA was synthesized from 1 µg of RNA with the Quantitect kit. Real time qPCR experiments were performed using SYBR Green or Taqman gene expression Assays on the BioRad CFX384 Touch real-time PCR detection system. qPCR primers and Taqman Assays are described in Table S4.
Microarrays
Myotube infections were performed with 2×108 TU at 36h after initiation of differentiation as described before (D’Angelo et al., 2012). 48 h post-infection myotubes were resuspended in Trizol reagent and RNA was purified following the manufacturer specifications and after the addition of ethanol 70% the RNA was loaded on an RNeasy purification column and further purified following the kit instructions. RNA quality and concentration were determined using Pico chips on a Bioanalyser 2100 (Agilent Technologies). Before using the RNA for microarray analysis, Nup210 down regulation was confirmed by qPCR (Figure S5B). Two independent microarray studies performed by triplicate on Affymetrix GeneChip 2.0 ST mouse arrays were used to identify genes co-regulated by Nup210 and Mef2C depletion.
Immunoprecipitations and Western Blots
C2C12 cells were harvested and washed with PBS and lysed using the NE-PER Nuclear and Cytoplasmic Extraction Reagent containing protease inhibitor cocktail (Complete Mini EDTA Free), 1mM PMSF and phosphatase inhibitors (PhosSTOP) according to manufacturer’s instructions. Nuclear extracts were diluted with 20 mM HEPES pH 7.6, 10% glycerol, 100 mM KCl, 1.5 mM MgCl2, 0.2 mM EDTA, 0.02% NP40, 0.5 mM DTT, 1mM PMSF and protease inhibitor cocktail to perform immunoprecipitations. Extracts were incubated with Protein A/G Dynabeads for 1h to remove non-specific binding. 2µg of anti Nup210 antibody or control rabbit IgG was added to each clarified nuclear extract. Immunoprecipitations were performed overnight at 4°C on a rotator. The next morning 50µL of Protein A/G Dynabeads were added and samples were incubated on a rotator for 1 h at 4°C. Immunoprecipitations were washed 3 times with 20 mM HEPES pH 7.6, 100 mM KCl, 1.5 mM MgCl2, 0.2 mM EDTA, 0.01% NP40, 0.5 mM DTT for 5 min at 4°C and eluted from beads by incubation with 2X LDS Sample Buffer with Reducing Agent 15 min at 70°C. Ten percent of the input and the whole volume of each immunoprecipitation were analyzed by using Novex 3–8% Tris Acetate Gels (Life Technologies) and blotted to nitrocellulose membranes using an iBlot2. Membranes were stained with Ponceau, washed with TBS-tween, blocked with 5% Milk and blotted with specific antibodies overnight. Western blot was visualized using Thermo Scientific Chemiluminescent Substrates SuperSignal West Pico and SuperSignal West Femto.
GST Pull Down Assays
GST pull-down assays were performed using standard procedures. GST-fused C-Terminal Nup210 was expressed in BL21 bacteria induced with 250 µM IPTG at 18°C, purified using B-PER Bacterial Protein Extraction Reagent and bound to Glutathione Agarose beads. HA-tagged LIM domain proteins were cloned in pCDNA6.2 nLumio and expressed in vitro using TNT Coupled Transcription/Translation System following the manufacturers protocol. Pull downs were performed by incubation 12 µl of de TNT reactions with 8µl of C-Terminal Nup210-GST coupled beads in 150 µl of GST binding buffer containing 50 mM TrisHcl pH7.6, 120 mM NaCl, 0.1% NP40 and protease inhibitors for 2 h at 4°C. Beads were washed 3 times with 500 µl of GST binding buffer and eluted from beads by incubation with 2X LDS Sample Buffer with Reducing Agent 15 min at 70°C and analyzed by SDS-PAGE and Western Blot. For Mef2 coimmunoprecipitations Nup210 and control rabbit IgG were crosslinked to Dynabeads using BS crosslinking reagent according to manufacturer’s protocol.
Proximity Ligation Assays (PLA)
C2C12 myoblasts were co-transfected with FLAG-Mef2C and Nup210-GFP vectors in µ-slide 8 well plates from Ibidi (Cat. # 80826) and induced to differentiate into myotubes as described above. Myotubes were washed once with PBS and fixed in 4% formaldehyde in PBS for 10 min at room temperature ice. Cells were incubated in IF-buffer (1xPBS, 10 mg/ml BSA, 0.02% SDS, 0.1% Triton-X100) for 1 h before adding primary antibodies. Anti-GFP and anti-FLAG, or anti-GFP and mAb4141 were incubated in IF buffer 1 h or ON, washed 3 times with IF buffer and PLA was performed using the Duolink® In Situ Red Starter Kit Mouse/Rabbit according to the manufacturer’s instructions. Briefly, cells were incubated with the mouse/rabbit PLA probes for 60 min at 37°C, washed with wash buffer A twice for 5 min and incubated with the ligation mixture for 30 min at 37°C and washed again. Cells were incubated with the amplification mixture for 100 min at 37°C. After final washes in buffer B, cells were incubated with Hoechst in PBS, and washed 3 times in PBS before imaging. Controls with each individual antibody alone or cells transfected with only one construct were performed following the same protocol.
Chromatin Immunoprecipitations (CHIPs)
ChIPs were performed using the EZ-Magna ChIP™ A/G Kit according to the manufacturer’s recommendations. Briefly, day 4 differentiated myotubes were washed with PBS and fixed with 1% formaldehyde for 10 min at room temperature followed by 5 min quenching with 125 mM glycine. Cells were washed 3–5 times with ice-cold PBS, scrapped and centrifuged at 1,000 × G for 10 min at 4°C. Cell pellets were resuspended in SDS lysis buffer with Protease Inhibitor Cocktail II and chromatin shearing was performed with a Misonix sonicator 3000 until DNA bands were between 300–500 bp.
The chromatin solution was clarified by centrifugation and an aliquot of sheared chromatin was put aside for preparation of input sample. The remaining was immunoprecipitated using mouse monoclonal antibody mAb414 (2 µg), rabbit polyclonal anti-Nup210 antibody (2 µg), anti-Mef2C (5 µg), anti-H3K27Ac (5 µg) or the corresponding isotype and Chip blocked Protein A/G magnetic beads. Protein complexes were eluted and cross-links were reverted following manufacturer’s protocol. Immunoprecipitated DNA fragments were purified using spin columns and analyzed by real time PCR using the primers described in Table S4.
DNA Fluorescent In Situ Hybridizations (FISH)
For immune-DNA FISH, coverslips containing cells were fixed in PBS with 4% PFA for 10 minutes, washed twice with PBS and subject to immunostaining with an anti-Lamin A. For this, fixed cells were blocked using IF buffer (1x PBS, 10 µg/ml BSA, 0.02% SDS, 0.1% Triton X-100) for 1 h at RT, incubated with primary antibody in IF buffer for 1 h at RT, washed 3 times with IF buffer and incubated with secondary antibody in IF buffer for 1 h at RT. After 3 additional washes with IF buffer, cells were washed once with PBS 1x and fixed for 10 min in PBS with 4% PFA. Cells were washed 3 times with PBS 1x at RT, and then placed in ice. After one wash with ice-cold 2xSSC, cells were permeabilized with 0.1M HCl/0.7% Triton for 15 min on ice, washed 3 times in 2xSSC, and then incubated with 2xSSC/50% formamide at 80°C for 30 min. Cells were washed with ice-cold 2xSSC and the coverslips were inverted into 10 µl of the specified probe. Biotinylated probes were prepared by labeling 1 µg of BAC DNA using Biotin-Nick Translation Mix and following the manufacturer instructions. Probes were precipitated at 4°C by adding 0.1 volume of 3M Sodium Acetate pH 5.2, 2.5 volumes of DNA, 20 µl of Cot DNA, 20 µl salmon sperm DNA and 15 µl of yeast tRNA. Probes were resuspended in 100 µl of hybridization buffer (2xSSC/50% formamide/20% dextran sulfate). Cells with probes were hybridized overnight at 42°C in a humidity chamber. Cells were washed at 42°C 3 times for 5 min with 2xSSC/50% formamide and 3 times with 2xSSC and subjected to IF with an anti-biotin antibody as described above. Metaphase spreads were prepared from mouse splenocytes as described by Hesed M Padilla-Nash et al (Padilla-Nash et al., 2006).
RNA Fluorescent In Situ Hybridization (FISH)
Immuno-RNA FISH was performed with Stellaris probes and buffers following the manufacturer’s protocol. Briefly, cells were fixed in PBS with 4% PFA for 10 minutes, washed twice with PBS. After fixation, cells were permeabilized in 70% ethanol for at least 1 h at 4 °C. Cells were subsequently incubated in Buffer A (2xSSC, 10% formamide) at room temperature for 5 min and then, hybridized in a humidified chamber with 125 nM probe in hybridization buffer (2xSSC, 10% formamide, 10% dextran sulfate) in the presence of Alexa488 labeled Lamin A/C antibody. Cells were incubated at 37 °C for 16 h, washed with Buffer A for 30 min at 37 °C once and for an additional 30 minutes in the presence of 6 ng/mL Hoechst. Cells were washed with PBS and imaged. Probes were designed using Stellaris Probe Designer.
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical Analysis of normalized Microarray data was made at the Bioinformatics & Data Management core at SBP using Partek Genomics Suite software. Analysis of gene expression was performed with Partek® Genomics Suite® software (version 6.6; 2016). RMA background correction, quantile normalization, log2 transformation and median polished probeset summarization was performed to generate intensity values. After the initial preprocessing, one-way ANOVA using Method of Elements and Fisher’s Least Significant Difference (LSD) (Tamhane and Dunlop, 2000) was run on the data to identify significantly-changing microarray features and corresponding genes for either Mef2C or Nup210 versus the scramble control. Filters were applied for gene prioritization and Venn diagram overlaps (e.g. significant p-value, p<0.05, and/or absolute fold-change, abs(FC) > 1.25 ). Figure 6F heatmap was rendered with Gene-E software using negative correlation clustering (1 – Pearson). Cell process and pathway analysis were performed with the MetaCore software.
Real time PCR experiments were quantified using the BioRad CFX Manager 3.1 Real Time PCR Analysis Software. For other experiments statistical analysis was done using GraphPad Prism software. Differences between samples were tested using a two-tailed Student’s t-test. Error bars represent SEM unless otherwise indicated. Please refer to figures and figure legends for number of animals or cells used per experiment.
DATA AND SOFTWARE AVAILABILITY
The accession number for the raw and analyzed microarray data reported in this paper is GEO: GSE98318 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=etyjcksytzczrif&acc=GSE98318).
Supplementary Material
Highlights.
Nucleoporin Nup210 is required for myofiber maturation, survival, and muscle growth
Nup210 recruits Mef2C to the nuclear periphery during myogenic differentiation
Nup210/Mef2C regulate the expression of muscle genes and miRNAs at nuclear pores
Nuclear pores act as scaffolds for the assembly of specific transcription complexes
In Brief.
Nup210 is a tissue-specific nuclear pore complex component with a role in myogenic differentiation. Raices, Bukata et al. show in zebrafish and mammalian myoblasts that Nup210 regulates myofiber maturation, growth, and survival by promoting assembly of a Mef2C-dependent transcription complex at nuclear pores to regulate muscle structural and miRNA genes.
Acknowledgments
We thank Dr. B. Black and Dr. M. Beckerle for reagents and the M.A.D. laboratory for critical discussion. Research reported in this publication was supported by the NIAMS of the NIH under award number R01AR065083, R01AR065083-S1 and by the award 10SDG2610290 from American Heart Association. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. M.A.D. is a Pew Scholar in the Biomedical Sciences.
Footnotes
Supplemental Information includes eight figures, four tables, and two movies and can be found with this article online at http://dx.doi.org/10.1016/j.devcel.2017.05.007.
AUTHOR CONTRIBUTIONS
Conceptualization, M.A.D., M.R., and L.B.; Methodology, M.A.D., M.R., and L.B.; Investigation, M.R., L.B., S.S., L.S.H., and J.B.; Resources, D.O.H.; Writing, M.A.D. and M.R.; Funding Acquisition, M.A.D.; Project Administration, M.A.D.
References
- Barresi MJ, D’Angelo JA, Hernandez LP, Devoto SH. Distinct mechanisms regulate slow-muscle development. Curr. Biol. 2001;11:1432–1438. doi: 10.1016/s0960-9822(01)00428-6. [DOI] [PubMed] [Google Scholar]
- Brickner DG, Ahmed S, Meldi L, Thompson A, Light W, Young M, Hickman TL, Chu F, Fabre E, Brickner JH. Transcription factor binding to a DNA zip code controls interchromosomal clustering at the nuclear periphery. Dev. Cell. 2012;22:1234–1246. doi: 10.1016/j.devcel.2012.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckingham M, Vincent SD. Distinct and dynamic myogenic populations in the vertebrate embryo. Curr. Opin. Genet. Dev. 2009;19:444–453. doi: 10.1016/j.gde.2009.08.001. [DOI] [PubMed] [Google Scholar]
- Capelson M, Hetzer MW. The role of nuclear pores in gene regulation, development and disease. EMBO Rep. 2009;10:697–705. doi: 10.1038/embor.2009.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capelson M, Liang Y, Schulte R, Mair W, Wagner U, Hetzer MW. Chromatin-bound nuclear pore components regulate gene expression in higher eukaryotes. Cell. 2010;140:372–383. doi: 10.1016/j.cell.2009.12.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JF, Mandel EM, Thomson JM, Wu Q, Callis TE, Hammond SM, Conlon FL, Wang DZ. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat. Genet. 2006;38:228–233. doi: 10.1038/ng1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cottle DL, McGrath MJ, Cowling BS, Coghill ID, Brown S, Mitchell CA. FHL3 binds MyoD and negatively regulates myotube formation. J. Cell Sci. 2007;120:1423–1435. doi: 10.1242/jcs.004739. [DOI] [PubMed] [Google Scholar]
- D’Angelo MA, Gomez-Cavazos JS, Mei A, Lackner DH, Hetzer MW. A change in nuclear pore complex composition regulates cell differentiation. Dev. Cell. 2012;22:446–458. doi: 10.1016/j.devcel.2011.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrante MI, Kiff RM, Goulding DA, Stemple DL. Troponin T is essential for sarcomere assembly in zebrafish skeletal muscle. J. Cell Sci. 2011;124:565–577. doi: 10.1242/jcs.071274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Cavazos JS, Hetzer MW. The nucleoporin gp210/Nup210 controls muscle differentiation by regulating nuclear envelope/ER homeostasis. J. Cell Biol. 2015;208:671–681. doi: 10.1083/jcb.201410047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granjon A, Gustin MP, Rieusset J, Lefai E, Meugnier E, Guller I, Cerutti C, Paultre C, Disse E, Rabasa-Lhoret R, et al. The microRNA signature in response to insulin reveals its implication in the transcriptional action of insulin in human skeletal muscle and the role of a sterol regulatory element-binding protein-1c/myocyte enhancer factor 2C pathway. Diabetes. 2009;58:2555–2564. doi: 10.2337/db09-0165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha K, Buchan JG, Alvarado DM, McCall K, Vydyanath A, Luther PK, Goldsmith MI, Dobbs MB, Gurnett CA. MYBPC1 mutations impair skeletal muscle function in zebrafish models of arthrogryposis. Hum. Mol. Genet. 2013;22:4967–4977. doi: 10.1093/hmg/ddt344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinits Y, Hughes SM. Mef2s are required for thick filament formation in nascent muscle fibres. Development. 2007;134:2511–2519. doi: 10.1242/dev.007088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinits Y, Pan L, Walker C, Dowd J, Moens CB, Hughes SM. Zebrafish Mef2ca and Mef2cb are essential for both first and second heart field cardiomyocyte differentiation. Dev. Biol. 2012;369:199–210. doi: 10.1016/j.ydbio.2012.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibarra A, Benner C, Tyagi S, Cool J, Hetzer MW. Nucleoporin-mediated regulation of cell identity genes. Genes Dev. 2016;30:2253–2258. doi: 10.1101/gad.287417.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacinto FV, Benner C, Hetzer MW. The nucleoporin Nup153 regulates embryonic stem cell pluripotency through gene silencing. Genes Dev. 2015;29:1224–1238. doi: 10.1101/gad.260919.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jao LE, Wente SR, Chen W. Efficient multiplex biallelic zebrafish genome editing using a CRISPR nuclease system. Proc. Natl. Acad. Sci. USA. 2013;110:13904–13909. doi: 10.1073/pnas.1308335110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalverda B, Pickersgill H, Shloma VV, Fornerod M. Nucleoporins directly stimulate expression of developmental and cell-cycle genes inside the nucleoplasm. Cell. 2010;140:360–371. doi: 10.1016/j.cell.2010.01.011. [DOI] [PubMed] [Google Scholar]
- Kehat I, Accornero F, Aronow BJ, Molkentin JD. Modulation of chromatin position and gene expression by HDAC4 interaction with nucleoporins. J. Cell Biol. 2011;193:21–29. doi: 10.1083/jcb.201101046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemler D, Dahley O, Rosswag S, Litfin M, Kassel O. The LIM domain protein nTRIP6 acts as a co-repressor for the transcription factor MEF2C in myoblasts. Sci. Rep. 2016;6:27746. doi: 10.1038/srep27746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kind J, Pagie L, Ortabozkoyun H, Boyle S, de Vries SS, Janssen H, Amendola M, Nolen LD, Bickmore WA, van Steensel B. Single-cell dynamics of genome-nuclear lamina interactions. Cell. 2013;153:178–192. doi: 10.1016/j.cell.2013.02.028. [DOI] [PubMed] [Google Scholar]
- Lemaitre C, Bickmore WA. Chromatin at the nuclear periphery and the regulation of genome functions. Histochem. Cell Biol. 2015;144:111–122. doi: 10.1007/s00418-015-1346-y. [DOI] [PubMed] [Google Scholar]
- Li J, Puceat M, Perez-Terzic C, Mery A, Nakamura K, Michalak M, Krause KH, Jaconi ME. Calreticulin reveals a critical Ca(2+) checkpoint in cardiac myofibrillogenesis. J. Cell Biol. 2002;158:103–113. doi: 10.1083/jcb.200204092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Y, Franks TM, Marchetto MC, Gage FH, Hetzer MW. Dynamic association of NUP98 with the human genome. PLoS Genet. 2013;9:e1003308. doi: 10.1371/journal.pgen.1003308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Light WH, Freaney J, Sood V, Thompson A, D’Urso A, Horvath CM, Brickner JH. A conserved role for human Nup98 in altering chromatin structure and promoting epigenetic transcriptional memory. PLoS Biol. 2013;11:e1001524. doi: 10.1371/journal.pbio.1001524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin VT, Lin FT. TRIP6: an adaptor protein that regulates cell motility, antiapoptotic signaling and transcriptional activity. Cell Signal. 2011;23:1691–1697. doi: 10.1016/j.cellsig.2011.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Q, Schwarz J, Bucana C, Olson EN. Control of mouse cardiac morphogenesis and myogenesis by transcription factor MEF2C. Science. 1997;276:1404–1407. doi: 10.1126/science.276.5317.1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu N, Williams AH, Kim Y, McAnally J, Bezprozvannaya S, Sutherland LB, Richardson JA, Bassel-Duby R, Olson EN. An intragenic MEF2-dependent enhancer directs muscle-specific expression of microRNAs 1 and 133. Proc. Natl. Acad. Sci. USA. 2007;104:20844–20849. doi: 10.1073/pnas.0710558105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNally EM, Pytel P. Muscle diseases: the muscular dystrophies. Annu. Rev. Pathol. 2007;2:87–109. doi: 10.1146/annurev.pathol.2.010506.091936. [DOI] [PubMed] [Google Scholar]
- Olsson M, Ekblom M, Fecker L, Kurkinen M, Ekblom P. cDNA cloning and embryonic expression of mouse nuclear pore membrane glycoprotein 210 mRNA. Kidney Int. 1999;56:827–838. doi: 10.1046/j.1523-1755.1999.00618.x. [DOI] [PubMed] [Google Scholar]
- Olsson M, Scheele S, Ekblom P. Limited expression of nuclear pore membrane glycoprotein 210 in cell lines and tissues suggests cell-type specific nuclear pores in metazoans. Exp. Cell Res. 2004;292:359–370. doi: 10.1016/j.yexcr.2003.09.014. [DOI] [PubMed] [Google Scholar]
- Padilla-Nash HM, Barenboim-Stapleton L, Difilippantonio MJ, Ried T. Spectral karyotyping analysis of human and mouse chromosomes. Nat. Protoc. 2006;1:3129–3142. doi: 10.1038/nprot.2006.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potthoff MJ, Olson EN. MEF2: a central regulator of diverse developmental programs. Development. 2007;134:4131–4140. doi: 10.1242/dev.008367. [DOI] [PubMed] [Google Scholar]
- Potthoff MJ, Arnold MA, McAnally J, Richardson JA, Bassel-Duby R, Olson EN. Regulation of skeletal muscle sarcomere integrity and postnatal muscle function by Mef2c. Mol. Cell. Biol. 2007;27:8143–8151. doi: 10.1128/MCB.01187-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabut G, Doye V, Ellenberg J. Mapping the dynamic organization of the nuclear pore complex inside single living cells. Nat. Cell Biol. 2004;6:1114–1121. doi: 10.1038/ncb1184. [DOI] [PubMed] [Google Scholar]
- Raices M, D’Angelo MA. Nuclear pore complex composition: a new regulator of tissue-specific and developmental functions. Nat. Rev. Mol. Cell Biol. 2012;13:687–699. doi: 10.1038/nrm3461. [DOI] [PubMed] [Google Scholar]
- Randise-Hinchliff C, Coukos R, Sood V, Sumner MC, Zdraljevic S, Meldi Sholl L, Garvey Brickner D, Ahmed S, Watchmaker L, Brickner JH. Strategies to regulate transcription factor-mediated gene positioning and interchromosomal clustering at the nuclear periphery. J. Cell Biol. 2016;212:633–646. doi: 10.1083/jcb.201508068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robson MI, de Las Heras JI, Czapiewski R, Le Thanh P, Booth DG, Kelly DA, Webb S, Kerr AR, Schirmer EC. Tissue-specific gene repositioning by muscle nuclear membrane proteins enhances repression of critical developmental genes during myogenesis. Mol. Cell. 2016;62:834–847. doi: 10.1016/j.molcel.2016.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sander JD, Maeder ML, Reyon D, Voytas DF, Joung JK, Dobbs D. ZiFiT (Zinc Finger Targeter): an updated zinc finger engineering tool. Nucleic Acids Res. 2010;38:W462–W468. doi: 10.1093/nar/gkq319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider M, Hellerschmied D, Schubert T, Amlacher S, Vinayachandran V, Reja R, Pugh BF, Clausen T, Kohler A. The Nuclear pore-associated TREX-2 complex employs mediator to regulate gene expression. Cell. 2015;162:1016–1028. doi: 10.1016/j.cell.2015.07.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seger C, Hargrave M, Wang X, Chai RJ, Elworthy S, Ingham PW. Analysis of Pax7 expressing myogenic cells in zebrafish muscle development, injury, and models of disease. Dev. Dyn. 2011;240:2440–2451. doi: 10.1002/dvdy.22745. [DOI] [PubMed] [Google Scholar]
- Shi L, Zhao G, Qiu D, Godfrey WR, Vogel H, Rando TA, Hu H, Kao PN. NF90 regulates cell cycle exit and terminal myogenic differentiation by direct binding to the 3’-untranslated region of MyoD and p21WAF1/CIP1 mRNAs. J. Biol. Chem. 2005;280:18981–18989. doi: 10.1074/jbc.M411034200. [DOI] [PubMed] [Google Scholar]
- Sood V, Brickner JH. Nuclear pore interactions with the genome. Curr. Opin. Genet. Dev. 2014;25:43–49. doi: 10.1016/j.gde.2013.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamhane A, Dunlop D. Statistics and Data Analysis: From Elementary to Intermediate. Prentice Hall; 2000. pp. 473–474. [Google Scholar]
- Thisse C, Thisse B. High-resolution in situ hybridization to whole-mount zebrafish embryos. Nat. Protoc. 2008;3:59–69. doi: 10.1038/nprot.2007.514. [DOI] [PubMed] [Google Scholar]
- Yao J, Fetter RD, Hu P, Betzig E, Tjian R. Subnuclear segregation of genes and core promoter factors in myogenesis. Genes Dev. 2011;25:569–580. doi: 10.1101/gad.2021411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi J, Kloeker S, Jensen CC, Bockholt S, Honda H, Hirai H, Beckerle MC. Members of the Zyxin family of LIM proteins interact with members of the p130Cas family of signal transducers. J. Biol. Chem. 2002;277:9580–9589. doi: 10.1074/jbc.M106922200. [DOI] [PubMed] [Google Scholar]
- Yogev O, Williams VC, Hinits Y, Hughes SM. eIF4EBP3L acts as a gatekeeper of TORC1 in activity-dependent muscle growth by specifically regulating Mef2ca translational initiation. PLoS Biol. 2013;11:e1001679. doi: 10.1371/journal.pbio.1001679. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.