Abstract
RalGDS is one of the Ras effectors and functions as a guanine nucleotide exchange factor for the small G-protein, Ral, which regulates membrane trafficking and cytoskeletal remodeling. The translocation of RalGDS from the cytoplasm to the plasma membrane is required for Ral activation. In this study, to understand the mechanism of Ras–Ral signaling we performed a single-molecule fluorescence analysis of RalGDS and its functional domains (RBD and REMCDC) on the plasma membranes of living HeLa cells. Increased molecular density of RalGDS and RBD, but not REMCDC, was observed on the plasma membrane after EGF stimulation of the cells to induce Ras activation, suggesting that the translocation of RalGDS involves an interaction between the GTP-bound active form of Ras and the RBD of RalGDS. Whereas the RBD played an important role in increasing the association rate constant between RalGDS and the plasma membrane, the REMCDC domain affected the dissociation rate constant from the membrane, which decreased after Ras activation or the hyperexpression of Ral. The Y64 residue of Ras and clusters of RalGDS molecules were involved in this reduction. From these findings, we infer that Ras activation not merely increases the cell-surface density of RalGDS, but actively stimulates the RalGDS–Ral interaction through a structural change in RalGDS and/or the accumulation of Ral, as well as the GTP–Ras/RalGDS clusters, to induce the full activation of Ral.
Keywords: small G-protein, fluorescence microscopy, molecular clustering, protein translocation
Ras is a small G-protein that regulates the cell-signaling pathways involved in important cellular functions, including proliferation, differentiation, apoptosis, adhesion, and migration [1,2]. To achieve this regulation, Ras anchors to the inner leaflet of the plasma membrane through posttranscriptional lipid modifications in its C-terminal region [2,3]. On the cell surface, GTPase-activating proteins (GAPs) accelerate the conversion of the GTP-bound active form of Ras (GTP–Ras) to the GDP-bound inactive form (GDP–Ras), and guanine nucleotide exchange factors (GEFs) stimulate the exchange of the GDP molecule bound to Ras for a GTP molecule in the cytoplasm. Thus, Ras acts as a binary molecular switch on the plasma membrane [1–4]. Various extra-cellular signaling molecules, including epidermal growth factor (EGF), activate the GEFs of Ras, and GDP–Ras is then converted to GTP–Ras on the plasma membrane [5]. During the conversion to its active form, structural changes occur in the switch I (residues 30–40) and switch II regions (residues 60–76) of the Ras molecule [6], and these changes strengthen the ability of Ras to binds to its downstream effector molecules, such as RAF kinase, PI3 kinase, and RalGDS [7,8]. These effector molecules are consequently translocated to the plasma membrane from the cytoplasm in response to Ras activation [9–11]. Previous studies have demonstrated that the localization of these effector molecules on the plasma membrane is required for the activation of their further downstream substrates [12–14]. However, how information about Ras activation is transduced to the effectors and how the effectors induce the activation of the downstream molecules on living cell surfaces are not fully understood.
To understand the signal transduction mechanism from Ras to its effectors and downstream molecules, we investigated the interactions of RalGDS with the plasma membranes of cells stimulated with EGF. RalGDS is one of the Ras effectors, with GEF activity for the small G-protein Ral [13–15]. It is thought that RalGDS mediates the signal transduction from Ras to Ral on the plasma membrane, thereby regulating Ral-dependent intracellular events, such as exocytosis, endocytosis, and actin reorganization [14,16,17]. RalGDS has three distinct domains: a Ras exchange motif (REM) domain in the N-terminal region of the protein, a CDC25 homology (CDC25) domain in the central region, and a C-terminal Ras-binding domain (RBD) (Fig. 1A) [14]. The REM domain regulates the GEF activity of the CDC25 domain for Ral and contributes to maintaining the conformation of the CDC25 domain [14]. As with the RBD of RAF kinases, the RBD of RalGDS has higher affinity for GTP–Ras than for GDP–Ras, therefore RalGDS is translocated to the plasma membrane from the cytoplasm depending on the activation status of Ras [10,13]. It has been reported that a RalGDS–CAAX fusion protein, which displayed forced membrane anchoring, activated Ral in the absence of Ras activation, suggesting that the excessive localization of RalGDS at the plasma membrane is sufficient for the activation of Ral [13]. However, it has also been reported that although constitutively active Ras (RasG12V) on the plasma membrane activated Ral in the presence of RalGDS–CAAX lacking the RBD of RalGDS (RalGDSΔRBD–CAAX), the substitution of the tyrosine at residue 64 (Tyr-64, Y64) in the switch II region of RasG12V with tryptophan (Trp, RasG12V,Y64W) abolishes the efficient activation of Ral in the presence of RalGDSΔRBD–CAAX [18]. The latter result suggests that the Y64 residue of Ras and the domains of RalGDS other than the RBD play important roles in the translocation and/or activation of RalGDS. Thus, the mechanism by which Ras activates RalGDS to transmit a signal to Ral remains unclear.
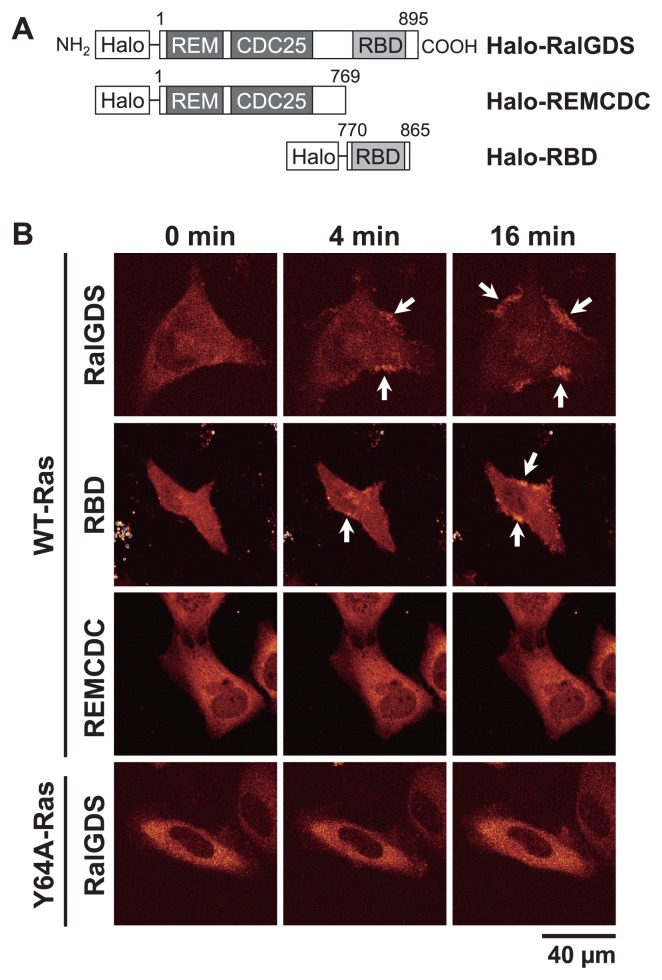
Figure 1.
Translocation of RalGDS to the plasma membranes of living HeLa cells after EGF stimulation. (A) Schematic structures of RalGDS constructs. The HaloTag was fused to the N-termini of the constructs. Numbers represent the positions of amino acid residues in RalGDS. (B) Halo–RalGDS, Halo–RBD, or Halo–REMCDC was coexpressed with WT-Ras or Y64A-Ras. After serum starvation (0 min), EGF was added to a final concentration of 100 ng/mL. The fluorescent images were taken at 0, 4, and 16 min at 25°C with a CLS microscope. Arrows show the accumulation of RalGDS on the plasma membrane.
In this study, to understand how RalGDS mediates the signal transduction from Ras to Ral on the plasma membrane after cell stimulation, we analyzed the translocation dynamics and kinetics of the RalGDS molecule in its interaction with the plasma membrane using single-molecule imaging in living cells. We successfully estimated the relative association rate constant and dissociation rate constants of RalGDS and its functional domains with the plasma membrane components, and developed a model of the regulation of the RalGDS–Ral interaction by GTP–Ras at the molecular level.
Methods
Construction of plasmids
pCMV–Ras (human H-Ras) and pCMV–Ral (human RalA) were obtained from Takara Bio Inc. (Japan) and RIKEN BRC (Japan), respectively. pmEGFP-C2 vector with a monomeric mutation (A206K) was constructed as described previously [19]. To construct the pmEGFP–Ral transfer vector, the fragment of pCMV–Ral encoding Ral was subcloned into the EcoRI–SalI sites of pmEGFP-C2. The Halo transfer vector (pHalo-C2) was constructed by exchanging mEGFP in pmEGFP-C2 for Halo, as described previously [20]. To produce pHalo–RalGDS or pHalo–RBD, the cDNA fragment encoding full-length Rattus RalGDS (kindly provided by Akira Kikuchi, Hiroshima University) or encoding human RBD (kindly provided by Yoshiyuki Arai, Osaka University) was subcloned into the EcoRI–SalI sites of the pHalo-C2 transfer vector. Halo–REMCDC was produced by introducing a stop codon at the position of the Ser-769 residue of Halo–RalGDS (Fig. 1A). The cDNA of Y64A-Ras was constructed by the direct point mutation of pCMV–Ras using the QuikChange Lightning Site-Directed Mutagenesis Kit (Agilent Technologies, USA) and PrimeSTAR HS DNA Polymerase (Takara Bio Inc., Japan).
Preparation of HeLa cells
HeLa cells were transfected with the expression vectors using a method described previously [20]. After transfection, the cells were cultured in Dulbecco’s modified Eagle’s medium (Wako Pure Chemical Industries, Japan) supplemented with 10% fetal bovine serum (FBS; Hyclone, USA) at 37°C under 5% CO2 for about 20 h. For serum starvation, the cells were then cultured in minimal essential medium (MEM; Nissui, Japan) in the presence of 1% bovine serum albumin (BSA) without FBS at 37°C under 5% CO2 for about 24 h. Immediately before the observations of the cells, the HaloTag moiety on the RalGDS constructs in the cells was labeled with tetramethylrhodamine (TMR), as described previously [20]. Briefly, the cells were incubated with 1 nM (for total internal reflection fluorescence [TIRF] microscopy) or 100 nM (for confocal laser scanning microscopy) HaloTag TMR ligand (Promega, Japan) in culture medium at 37°C under 5% CO2 for 15 min. The cells were then washed repeatedly with Hank’s balanced salt solution and MEM. Before microscopic observation, the medium was replaced with MEM containing 5 mM HEPES (pH 7.4) and 1% BSA. Under the microscope, the cells were stimulated with EGF (Sigma-Aldrich, Japan) at a final concentration of 100 ng/mL. For fixation, the cells were incubated with phosphate-buffered saline (PBS) containing 4% paraformaldehyde and 0.2% glutaraldehyde at 25°C for 30 min, and then washed three times in MEM containing 1% BSA.
Fluorescence microscopy
The localization of RalGDS in living HeLa cells was observed with a confocal laser scanning (CLS) microscope (TCS SP2; Leica, Germany) equipped with a 63×, NA 1.20 objective lens (HCX PL Apo; Leica), as described previously [21]. The TMR ligand conjugated to the Halo protein tagging RalGDS and its domains was excited at a wavelength of 543 nm and the fluorescence images were acquired at an emission wavelength of 560–650 nm. The TIRF microscopic observations were made as previously described, with some modifications [19]. Single molecules of Halo-tagged RalGDS were observed on the plasma membrane with an in-house TIRF microscope based on an inverted fluorescence microscope (TE 2000; Nikon, Japan) equipped with a 60×, NA 1.49 objective lens (PlanApo; Nikon, Japan). A 559 nm wavelength laser (NTT Electronics, Japan) was used for TMR excitation and the fluorescent images were acquired with an EM-CCD camera (ImagEM; Hamamatsu Photonics, Japan) at a frame rate of 32.8 fps. All fluorescence microscopic observations were made at 25°C.
Kinetic analysis
The single-molecule detection and tracking of RalGDS on the plasma membrane were performed with the G-Count software (G-Angstrom, Japan). The statistical and kinetic analyses were performed as described previously [19,20]. Briefly, the cumulative distribution of the dwell times of RalGDS on the plasma membrane was fitted to the following equation:
Here, k1 and k2 are the dissociation rate constants for RalGDS from the plasma membrane components. These values are apparent ones and include the effect of photobleaching. Because a proportion of RalGDS molecules seems to form oligomers as well as the usual monomers, it is difficult to exactly correct for photobleaching. However, the photobleaching rate constant of TMR conjugated to RalGDS (0.07 s−1; Supplementary Fig. S1) was significantly smaller than the apparent values of k1 (4–6 s−1) and k2 (0.5–1 s−1). The fraction sizes (ratios) of the dissociation rate constants were determined as A1 and A2 (A1+A2=1).
To estimate the cluster size distribution of RalGDS, the fluorescence intensities of the TMR-labeled Halo–RalGDS particles in the living cells were compared with the photobleaching step sizes in fixed cells, as described previously [20]. The cells that expressed TMR-labeled Halo–RalGDS were fixed, as described above, to prevent the dissociation of the Halo–RalGDS molecules from the plasma membrane. The fluorescence intensities of the Halo–RalGDS particles were then measured immediately before the final photobleaching on the plasma membrane to determine the fluorescence intensity of the single Halo–RalGDS molecules. The distribution of the fluorescence intensities was fitted to the following Gaussian equation:
Here, μ and σ are the mean and standard deviation of the single-molecule fluorescence intensities, respectively. The distribution of the fluorescence intensities of the Halo–RalGDS particles measured in living HeLa cells was fitted to the sum of the N Gaussian function:
Here, n is the oligomer size of RalGDS and Wn is the fraction of n-mer. N was estimated using the Akaike information criterion (AIC) [22] (Supplementary Fig. S4 and Table S1). Values of μ and σ determined from the photobleaching step-size distribution were used in the equation.
To estimate the relative expression level of the RalGDS molecules in the cytoplasm, fluorescence intensities of Halo-RalGDS were measured in the epi-illumination mode in the same microscope for TIRF observation. Thickness of cells before and after stimulation was 5.6±0.4 μm and 5.4±0.3 μm (n=15) for cells with EGF stimulation, and 5.6±0.3 μm and 5.3±0.2 μm (n=15) for cells with vehicle stimulation, respectively. Differences among the values of thickness are statistically insignificant (on t-test).
Western blotting
The western blotting analysis of RalGDS was performed as described previously [19], with some modifications. Briefly, the transfected cells were harvested in Laemmli SDS sample buffer. The proteins in the cell lysates were separated on 8% or 12.5% polyacrylamide gel and transferred onto PVDF membrane (Millipore, USA). The membrane was incubated with the following primary antibodies to detect each protein: anti-Halo (Promega, Japan), anti-pan-Ras (Abcam, Japan), anti-actin (Sigma-Aldrich, Japan), anti-RalGDS (Abcam, Japan), and anti-RalA (Cell Signaling Technology, USA). After the membrane was washed three times with PBS, it was incubated with anti-mouse IgG secondary antibody (Cell Signaling Technology, USA) or anti-rabbit IgG secondary antibody peroxidase (Cell Signaling Technology, USA), both conjugated with horseradish peroxidase. Antibody binding was visualized with the ECL Prime Western Blotting Detection Reagent Kit (GE Healthcare, Japan).
Results and Discussion
Translocation of RalGDS to the plasma membrane from the cytoplasm in living HeLa cells after EGF stimulation
We first observed the translocation dynamics of the RalGDS constructs (full-length RalGDS, RBD, and REMCDC; Fig. 1A) to the plasma membranes of living HeLa cells after EGF stimulation, using CLS microscopy (Fig. 1B). To visualize the cells, a HaloTag was fused to the N-termini of the RalGDS constructs (Fig. 1A). The apparent molecular masses of the Halo-tagged proteins, determined from their mobility on SDS-PAGE, were 133, 119, and 46 kDa for Halo–RalGDS, Halo–REMCDC, and Halo–RBD, respectively (Supplementary Fig. S2A). These values agree with the calculated molecular weights expected for these constructs. To enhance the interaction between Ras and RalGDS, the Ras protein was coexpressed with RalGDS molecules. Expression of endogenous RalGDS molecules was not detectable in our experimental condition, and the expression levels of the exogenous Ras molecules were considerably higher than those of the endogenous molecules (Supplementary Fig. S2A). Therefore, the transfected cells allowed us to analyze the interaction between RalGDS and Ras on the plasma membrane.
After the proteins were expressed in the cells, the HaloTag moiety of the fusion proteins was labeled with TMR. As expected, full-length Halo–RalGDS was diffusely distributed in the cytoplasm of the serum-starved (quiescent) cells, and was translocated to the plasma membrane after EGF stimulation (Fig. 1B). The membrane localization of RalGDS was sustained more than 16 min after cell stimulation. These translocation dynamics of RalGDS are similar to the previously reported time course of Ras activation on the plasma membrane [23–25]. Similar EGF-dependent translocation was also observed for Halo–RBD, but not for Halo–REMCDC (Fig. 1B), suggesting that the RBD, but not the REMCDC domain, mainly causes the Ras-activity-dependent translocation of RalGDS to the membrane. The colocalization of Halo–RalGDS with GFP-tagged Ras was observed on the plasma membrane of the cells after EGF stimulation (Supplementary Fig. S3).
Linnemann et al. [18] suggested that the REMCDC domain of RalGDS interacts with the switch II region of Ras during the signal transduction from Ras to Ral. Therefore, we examined the localization of RalGDS when it was coexpressed with Y64A-Ras (Tyr 64 in the switch II region of Ras was substituted with Ala) (Fig. 1B). A western blotting analysis showed that the expression levels of exogenous Y64A-Ras in the transfected cells were much higher than that of the endogenous Ras protein (Supplementary Fig. S2A). The translocation of RalGDS to the plasma membrane after EGF stimulation was significantly attenuated by its coexpression with Y64A-Ras, therefore the Y64 residue of Ras is required for the specific membrane translocation of RalGDS. The RBD of RalGDS recognizes the switch I region of Ras [26]. The effect of the Y64A mutation in Ras suggests that REMCDC inhibits the association between RBD and Ras, and that the interaction between the switch II region in GTP–Ras and the REMCDC releases this inhibition.
Single-molecule imaging of RalGDS on the plasma membrane of living HeLa cells
We next observed individual RalGDS molecules on the plasma membrane under a TIRF microscope to analyze the interaction of RalGDS with the plasma membrane components, including Ras and Ral. Fluorescent particles were observed on the basal plasma membrane of the HeLa cells expressing Halo–RalGDS labeled with TMR, and these particles appeared and disappeared on a subsecond-to-second time scale (Fig. 2A, B, Supplementary Fig. S4A, and Supplementary Movie S1). In contrast, few fluorescent particles were observed in cells that did not express Halo–RalGDS under the same TMR staining conditions (Supplementary Fig. S4B), indicating that the particles attached to the plasma membrane were Halo–RalGDS molecules. The detection of the transient appearance of single particles on the plasma membrane allows us to analyze the interaction kinetics between cytoplasmic proteins and membrane components [27,28]. The fraction of monomeric Halo–RalGDS among the particles was estimated to be about 40% (Supplementary Table S1), based on the fluorescence intensity distribution (Supplementary Fig. S4C) and the score of AIC (Supplementary Fig. S4D), suggesting that a proportion of RalGDS molecules forms oligomers.
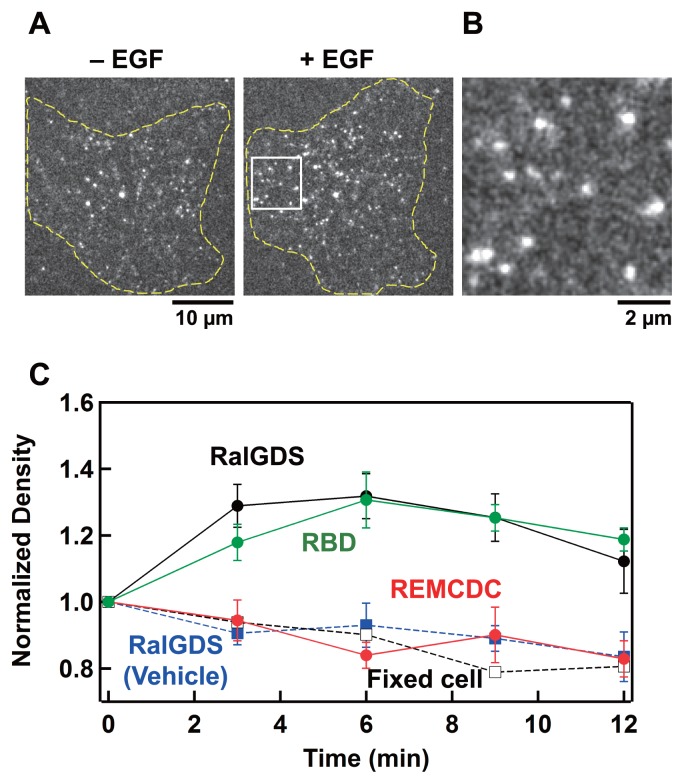
Figure 2.
TIRF microscopic observation of individual RalGDS molecules on the plasma membrane in living HeLa cells coexpressing WT-Ras. (A) TIRF images of Halo–RalGDS particles on the basal plasma membrane before (left) and 3 min after (right) EGF stimulation (final 100 ng/mL) at 25°C. (B) Magnified view of the boxed area in (A). (C) Normalized densities of Halo–RalGDS (black symbols), Halo–RBD (green symbols), and Halo–REMCDC (red symbols) particles on the basal plasma membrane before (0 min) and after (3, 6, 9, and 12 min) EGF stimulation. The density of Halo–RalGDS was also measured in serum-starved fixed cells (open squires) and in serum-starved living cells (closed blue squires) after the addition of control buffer (MEM containing 1% BSA). Values were normalized to those before EGF stimulation. The mean values for 6 cells are plotted along with the SE.
After the cells were stimulated with EGF, the observed density of the Halo–RalGDS particles on the plasma membrane increased by about 30%, peaking at 6 min, compared with that in the quiescent state (0 min) (Fig. 2C and Supplementary Movie S2). These dynamics are consistent with those observed with CLS microscopy (Fig. 1B). It is important to emphasize that for the Halo–RalGDS particles associated with the cell surface without EGF stimulation, and even after cell stimulation, the turnover of individual RalGDS particles (Supplementary Fig. S4A, and Supplementary Movies S1 and S2) was much faster than the dynamics of RalGDS translocation observed in ensemble (Fig. 2C) using CLS microscopy (Fig. 1B). Thus, the membrane accumulation of RalGDS after cell stimulation involves a temporal shift in the dynamic molecular interaction kinetics. However, the EGF-dependent monomer–oligomer transition of RalGDS is unlikely to be involved in its translocation because the distribution of the fluorescence intensity of the particles in the EGF-stimulated cells was indistinguishable from that in the quiescent cells (Supplementary Fig. S4C and Supplementary Table S1). The density of the RBD molecules on the plasma membrane also increased after EGF stimulation, whereas the density of the REMCDC molecules did not change significantly (Fig. 2C). The slight reduction in the density of REMCDC may have been caused by the photobleaching of TMR. A similar reduction was observed after the addition of control buffer and in the cells fixed before observation (Fig. 2C). The observed membrane translocation dynamics of the RBD and REMCDC domain in the single molecules are also consistent with those observed with CLS microscopy (Fig. 1B).
Association kinetics of RalGDS with the plasma membrane components in living cells
During its transient stay on the plasma membrane, RalGDS receives signals from Ras and transduces them to Ral. The association and dissociation kinetics of RalGDS provide information on the mechanism of signal transduction. The extremely sensitive detection possible with single-molecule imaging enabled the reaction kinetics of REMCDC, in addition to those of full-length RalGDS and RBD, to be determined, despite its low affinity for the plasma membrane. To examine the association rate constants of the three constructs for the plasma membrane when moving from the cytoplasm, we measured the appearance frequency of fluorescent particles per unit time and unit area on the basal plasma membrane, which is proportional to the first-order association rate constant. To obtain the relative value for the second-order association rate constant, the frequency was normalized to the relative expression level of the molecules in the cytoplasm (Fig. 3). Although the relative (second-order) association rate constant of REMCDC did not change after EGF stimulation, the rate constants of RalGDS and RBD both increased by about 50% after EGF stimulation (Fig. 3). This suggests that the RBD, but not the REMCDC domain, plays an important role in determining the association rate constant between RalGDS and Ras anchored to the plasma membrane. The association rate constants of RalGDS coexpressed with Y64A-Ras did not differ from those expressed with wild-type (WT)-Ras, either before or after cell stimulation (Fig. 3), as expected from the weak association between REMCDC and the plasma membrane (Figs 1B and 2). These results suggest that at least a proportion of the increases in the molecular densities of RalGDS and RBD on the plasma membrane were caused by increases in the association rate constants for the membrane components, including GTP–Ras. A conformational change in the switch I region of Ras, which is the principal association site for RBD, must cause this increase in the association rate constant. The association rate constant of RBD was significantly greater than that of RalGDS, supporting the suggestion that the REMCDC domain in the full-length inactive conformation of RalGDS inhibits the Ras–RBD interaction (Fig. 1).
Figure 3.
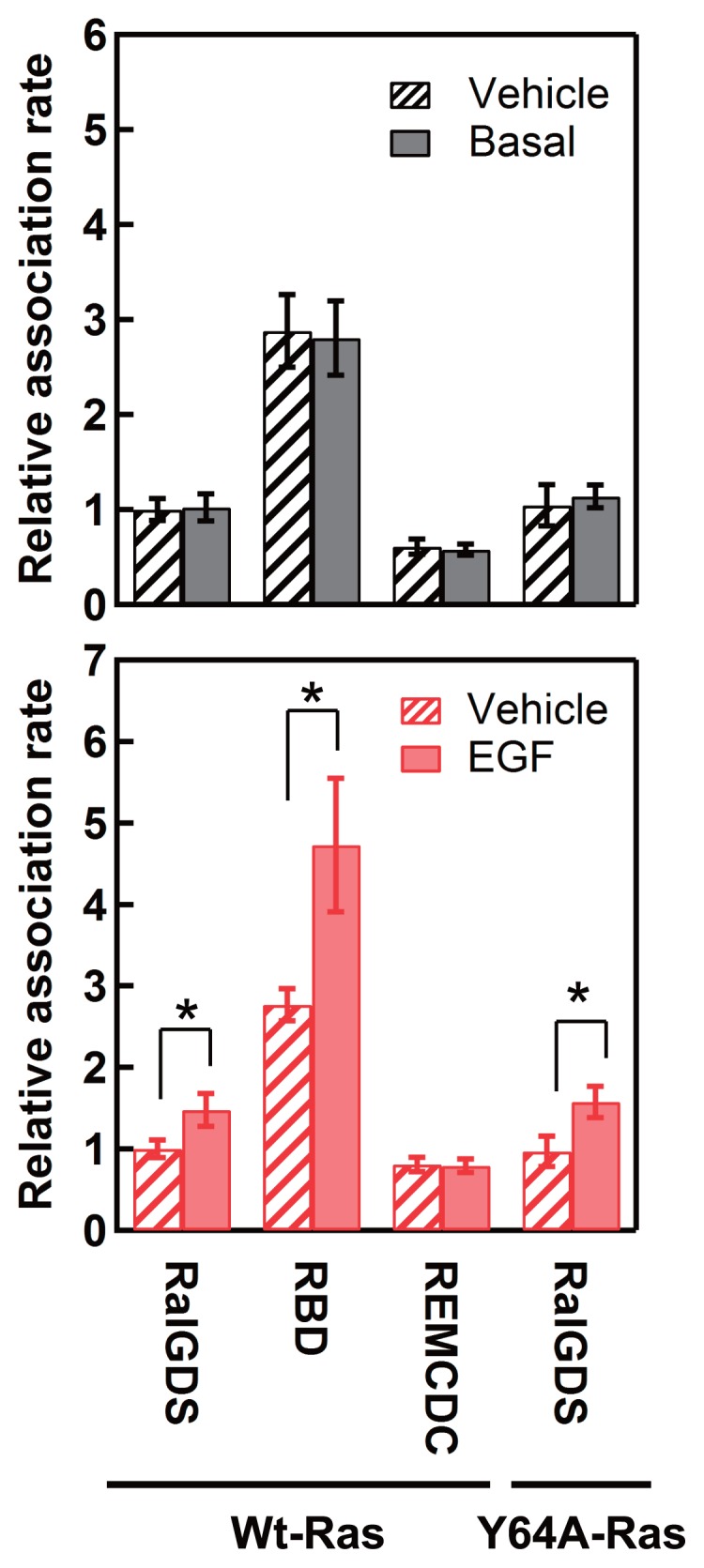
Relative association rate constants of RalGDS with plasma membrane components. Solid bars represent the relative association rate constants for the RalGDS constructs before (upper graph) and 3 min after (lower graph) EGF stimulation. Hatched bars represent relative association rate constants for the RalGDS constructs before (upper graph) and after (lower graph) the addition of MEM containing 1% BSA. The mean values for 5–13 cells are plotted along with the SE. Asterisks denote statistical significance (p<0.05 using the t test).
Dissociation kinetics of RalGDS with the plasma membrane components
We next measured the dwell times of individual RalGDS particles on the basal plasma membrane. The distribution of the dwell times fitted two-component exponential decay for all the molecules and under all the conditions examined (Fig. 4A). The values of the dissociation rate constants and the fractions of each component were calculated (Fig. 4B, C, and Supplementary Table S2). A highly plausible interpretation of the two-component kinetics is that there are at least two independent types of binding state (or binding site) on the plasma membrane. Fractions of the two components were independent of the fluorescence intensity of the particles (Supplementary Fig. S5). It is possible that both binding states are involved in the EGF-stimulated translocation of RalGDS, because the fraction sizes (ratios) of the two components before and after EGF stimulation were indistinguishable (Fig. 4C), even though the molecular densities of RalGDS and RBD on the plasma membrane increased after EGF stimulation (Fig. 2). Compared with the dissociation rate constants of RalGDS before EGF stimulation, the rate constants for both the fast (k1) and slow (k2) dissociation components decreased after stimulation (Fig. 4B). The values of both dissociation rate constants (k1 and k2) for RBD in the stimulated cells were indistinguishable from those in quiescent cells, and the values of k2 were similar to that for RalGDS before EGF stimulation (Fig. 4B). The dissociation rate constants for REMCDC were also independent of EGF stimulation, but in contrast to RBD and the values of k2 were similar to that of RalGDS after EGF stimulation (Fig. 4B).
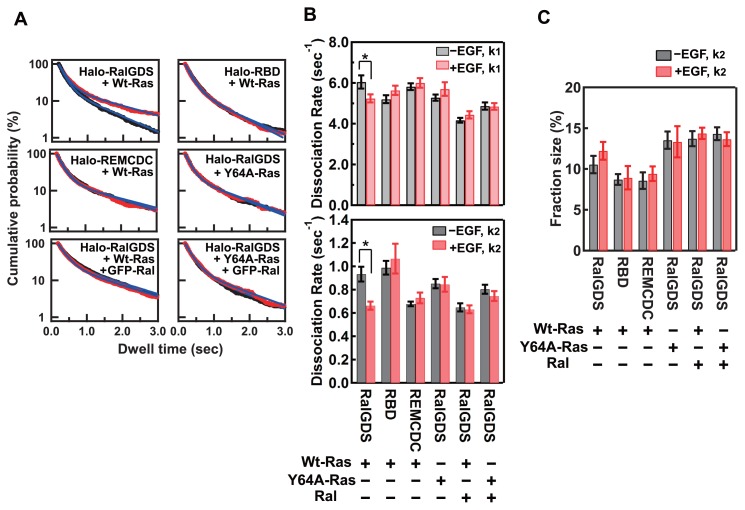
Figure 4.
Dissociation kinetics for RalGDS from the plasma membrane components. (A) Dwell time distributions of Halo–RalGDS constructs coexpressed with small G-proteins on the basal plasma membrane before (black) and 3 min after (red) EGF stimulation. The blue lines are the best-fit curves of the two-component exponential function described in the “METHODS”. (B) Dissociation rate constants for the fast (k1, upper graph) and slow (k2, lower graph) dissociation components of RalGDS before (black bars) and 3 min after (red bars) EGF stimulation. The mean values for 5–8 cells are plotted along with the SE. Asterisks denote statistical significance (p<0.05 using the t test). (C) Fraction sizes for the slow dissociation component of RalGDS. The mean values for 5–8 cells are shown along with the SE.
These results suggest that both the RBD and the REMCDC domain of RalGDS play important roles in determining the dissociation rate constant of the RalGDS molecule from the association sites on the plasma membrane. The REMCDC domain is especially required for the reduction in the rate constant for the slow dissociation component, which mainly causes the extension of the dwell time after EGF stimulation, whereas this function of the REMCDC domain in full-length RalGDS is impeded in cells before stimulation. We investigated the dissociation of RalGDS coexpressed with Y64A-Ras, and observed no reduction in the slow dissociation rate constant after cell stimulation (Fig. 4 and Supplementary Table S2). Therefore, the interaction between Ras and the REMCDC domain in the switch II region of GTP–Ras was confirmed as important for the reduction in the slow dissociation rate constant of full-length RalGDS. The slow dissociation rate constant (k2) of the bright RalGDS particles (mainly oligomers) before and after EGF stimulation was lower than that of the dark RalGDS particles (mainly monomers) (Supplementary Fig. S5B), suggesting that the oligomerization of RalGDS is also involved in determining the dissociation rate constant between RalGDS and the plasma membrane components.
We next measured the dwell times of individual RalGDS molecules on the plasma membranes in cells coexpressing GFP-tagged Ral (GFP–Ral) and WT-Ras (Fig. 4 and Supplementary Table S2). A Western blotting analysis revealed that the expression level of GFP–Ral in the transfected cells was much higher than that of endogenous Ral (Supplementary Fig. S2B). The colocalization of Ral and RalGDS on the plasma membrane was observed after EGF stimulation, and, consistent with previous reports [29,30], Ral localized to the plasma membrane independently of EGF stimulation (Supplementary Fig. S6). We expected that the interaction with Ral would affect the dissociation rate constants of RalGDS from the plasma membrane. Actually, although the coexpression of GFP–Ral did not affect the fractions of the two dissociation components of RalGDS, both dissociation rate constants (k1 and k2) decreased (Fig. 4). In the quiescent cells, the slow dissociation rate constant (k2) was reduced to a similar extent as in the cells after EGF stimulation without the coexpression of GFP–Ral. The co-expression of GFP–Ral with Y64A-Ras instead of WT-Ras resulted in slight reductions in the dissociation rate constants for both of the components. Whereas EGF stimulation had no effect on the dissociation rate constants in cells with a hyperexpression of GFP–Ral (Fig. 4B). The similar values for the slow dissociation rate constants in cells after EGF stimulation (but without GFP–Ral expression) and in cells with GFP–Ral hyperexpression (before and after EGF stimulation) suggest that the reduction in the slow dissociation rate constant is caused by the interaction of RalGDS with Ral. However, the fraction of the slow component did not increase with GFP–Ral expression (Fig. 4C), meaning that Ral does not determine the ratio of the two dissociation components. Ras is a candidate for the factor that determines the component ratio. Then, the reduced slow dissociation may be attributable to the ternary complex formed by Ras, RalGDS, and Ral.
Model of RalGDS interaction with Ras and Ral on the plasma membrane
The results of this study allow us to construct a model of the interaction between RalGDS and the plasma membrane components that causes the signal transduction from Ras to Ral (Fig. 5). In this model, we considered only Ras, RalGDS, and Ral, for simplicity, although other membrane components, including lipids, may be involved in the actual system, performing some of the molecular functions described below. The involvement of RalGDS oligomerization is also not fully described in this model because its details are as yet unknown.
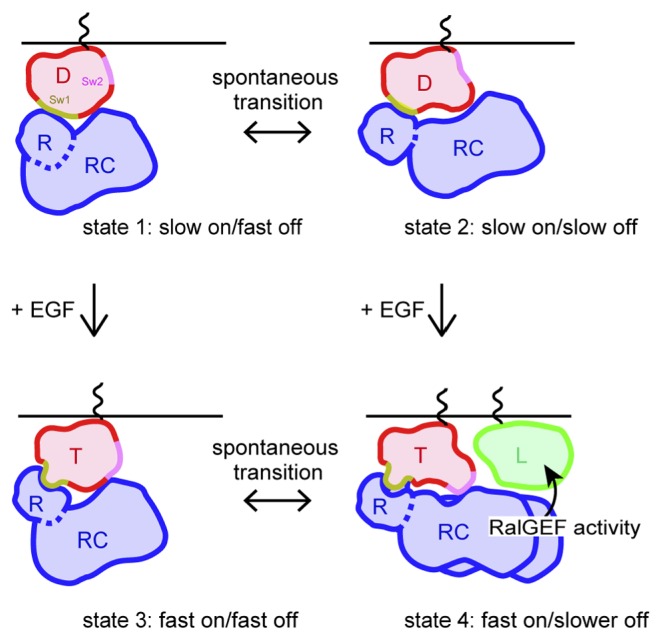
Figure 5.
Model of the interaction of RalGDS and its counterpart proteins on the plasma membrane. In quiescent cells, RalGDS associates with GDP–Ras on the plasma membrane via both of the RBD and REMCDC domain with two different binding states (states 1 and 2). When GDP–Ras becomes GTP–Ras on the plasma membrane after cell stimulation, structural changes in the switch I and switch II regions of Ras occur. These changes increase the association rate constant with the RBD (states 3 and 4) and promote the interaction between the REMCDC domain and Ral on the plasma membrane (state 4). D, GDP–Ras; R, RBD of RalGDS; RC, REMCDC domain of RalGDS; T, GTP–Ras; L, Ral; Sw1, switch I region of Ras; Sw2, switch II region of Ras.
In quiescent cells, RalGDS recognizes GDP–Ras on the plasma membrane using the RBD for its initial recognition, which reflects the association rate constant. The complex formed between GDP–Ras and RalGDS has at least two different states in the dissociation rate constant, and the REMCDC domain of RalGDS is also involved in the interaction in both association states (Figs. 3 and 4). The two-component exponential kinetics suggests that the transition between these two (or more) states is slower than the dissociation of RalGDS. Full-length RalGDS displayed a smaller association rate constant than RBD (Fig. 3). A possible explanation of this phenomenon is that the Ras–RBD interaction is sterically hindered by the REMCDC domain in full-length RalGDS. Previous reports [14,31,32] have suggested that the N-terminal region (containing REM) of RalGDS is phosphorylated by PKC in quiescent cells, in the region of RalGDS that associates with the catalytic CDC25 domain, thereby inhibiting GEF activity. This autoinhibitory conformation of RalGDS might disturb the interaction between the RBD in full-length RalGDS and Ras. However, the structural information available for full-length RalGDS is currently insufficient to confirm this.
In EGF-stimulated cells, SOS converts GDP–Ras to GTP–Ras, and this conversion induces structural changes in the switch I and switch II regions of Ras [33,34]. These structural changes in Ras enhance the association rate constant between the RBD of RalGDS and Ras (Fig. 3). Subsequently, the REMCDC domain of RalGDS interacts with Ral adjacent to the GTP–Ras molecule, reducing the slow dissociation rate constant. During this interaction, RalGDS exchanges the bound GDP on Ral for GTP to activate Ral. The Y64 residue of GTP–Ras is also involved in this process. The switch II region of Ras, in which Y64 is located, is thought to interact with the REMCDC domain, therefore changes in the interaction between Ras and the REMCDC domain after Ras activation might produce a RalGDS–Ral interaction. Such a structural change in the switch II region may not be necessary for the interaction between RalGDS and Ral under anomalous conditions, because the REMCDC molecule (lacking the RBD) and full-length RalGDS showed a reduced slower dissociation rate constant in the presence of excess Ral (but without Ras stimulation). However, under the normal conditions of cells, a structural change in the Ras–RalGDS (and Ral) complex induced by the activation of Ras is required for a prolonged interaction between RalGDS and Ral (Figs. 4 and 5). This must explain why the mutation at Y64 of Ras impairs the full activation of Ral [18].
Conclusion
To understand the signal transduction mechanism from Ras to Ral mediated by RalGDS, we analyzed the translocation dynamics and kinetics of the RalGDS molecule in its interaction with the plasma membrane using single-molecule fluorescence imaging in living cells. We successfully estimated the relative association rate constant and dissociation rate constants of RalGDS with the plasma membrane components, and developed a model in which the RalGDS–Ral interaction is regulated by GTP–Ras. Of particular interest is the fact that Ras activation regulates RalGDS and therefore Ral activity, not simply increasing the affinity of RalGDS for Ras. The switch I region of Ras seems to determine the association rate constant of RalGDS with the plasma membrane, but appears insufficient to regulate Ral activation. A conformational change in the switch II region of Ras upon activation is also required for its interaction with the REMCDC domain of RalGDS to promote the interaction of RalGDS with Ral. Therefore, multiple regions and domains of both Ras and RalGDS act in concert to regulate the interaction between RalGDS and Ral. Such concerted functions of multiple domains in single molecules seem to be a general feature of the regulation of intracellular signal transduction [20,21,27]. The nonlinear effects of the concerted functions of multiple domains in signal transduction proteins are probably important in increasing the accuracy of cell signaling.
Significance.
Despite the importance of the signal transduction from Ras to Ral in tumorigenesis and inflammation, the mechanism is not fully understood. Several reports have demonstrated that RalGDS translocates from the cytoplasm to the plasma membrane to receive the Ras signal and transduces it to effectors, including Ral. However, it is difficult to measure the kinetics and dynamics of protein interactions in living cells with conventional methods. Single-molecule fluorescence imaging is a powerful tool with which to clarify the mechanisms of protein reactions. We used this technique to determine the mechanism of the Ras–Ral signaling mediated by RalGDS.
Supplementary Information
Acknowledgments
This work was supported, in part, by the Uehara Memorial Foundation (to NU), the Takeda Science Foundation (to NU), and MEXT, Japan (JP16H00788, JP15H02394, JP15KT0087). We thank Hiromi Sato for technical assistance.
Footnotes
Conflicts of Interest
The authors declare that they have no conflicts of interest.
Author Contributions
Conceived and designed the experiments: RY, NU, MM, and YS. Performed the experiments: RY and NU. Analyzed the data: RY and MY. Wrote the paper: NU and YS.
References
- 1.Guin S, Theodorescu D. The RAS-RAL axis in cancer: evidence for mutation-specific selectivity in non-small cell lung cancer. Acta Pharmacol Sin. 2015;36:291–297. doi: 10.1038/aps.2014.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cox AD, Der CJ. Ras history: The saga continues. Small GTPases. 2010;1:2–27. doi: 10.4161/sgtp.1.1.12178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hancock JF. Ras proteins: different signals from different locations. Nat Rev Mol Cell Biol. 2003;4:373–384. doi: 10.1038/nrm1105. [DOI] [PubMed] [Google Scholar]
- 4.Milburn MV, Tong L, deVos AM, Brunger A, Yamaizumi Z, Nishimura S, et al. Molecular switch for signal transduction: structural differences between active and inactive forms of protooncogenic ras proteins. Science. 1990;247:939–945. doi: 10.1126/science.2406906. [DOI] [PubMed] [Google Scholar]
- 5.Mochizuki N, Yamashita S, Kurokawa K, Ohba Y, Nagai T, Miyawaki A, et al. Spatio-temporal images of growth-factor-induced activation of Ras and Rap1. Nature. 2001;411:1065–1068. doi: 10.1038/35082594. [DOI] [PubMed] [Google Scholar]
- 6.Lu S, Jang H, Muratcioglu S, Gursoy A, Keskin O, Nussinov R, et al. Ras Conformational Ensembles, Allostery, and Signaling. Chem Rev. 2016;116:6607–6665. doi: 10.1021/acs.chemrev.5b00542. [DOI] [PubMed] [Google Scholar]
- 7.Mello LV, van Aalten DM, Findlay JB. Comparison of ras-p21 bound to GDP and GTP: differences in protein and ligand dynamics. Protein Eng. 1997;10:381–387. doi: 10.1093/protein/10.4.381. [DOI] [PubMed] [Google Scholar]
- 8.Buhrman G, O’Connor C, Zerbe B, Kearney BM, Napoleon R, Kovrigina EA, et al. Analysis of binding site hot spots on the surface of Ras GTPase. J Mol Biol. 2011;413:773–789. doi: 10.1016/j.jmb.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hibino K, Watanabe TM, Kozuka J, Iwane AH, Okada T, Kataoka T, et al. Single- and multiple-molecule dynamics of the signaling from H-Ras to cRaf-1 visualized on the plasma membrane of living cells. Chemphyschem. 2003;4:748–753. doi: 10.1002/cphc.200300731. [DOI] [PubMed] [Google Scholar]
- 10.Takaya A, Ohba Y, Kurokawa K, Matsuda M. RalA activation at nascent lamellipodia of epidermal growth factor-stimulated Cos7 cells and migrating Madin-Darby canine kidney cells. Mol Biol Cell. 2004;15:2549–2557. doi: 10.1091/mbc.E03-11-0857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Katz M, Amit I, Yarden Y. Regulation of MAPKs by growth factors and receptor tyrosine kinases. Biochim Biophys Acta. 2007;1773:1161–1176. doi: 10.1016/j.bbamcr.2007.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goetz CA, O’Neil JJ, Farrar MA. Membrane localization, oligomerization, and phosphorylation are required for optimal raf activation. J Biol Chem. 2003;278:51184–51189. doi: 10.1074/jbc.M309183200. [DOI] [PubMed] [Google Scholar]
- 13.Matsubara K, Kishida S, Matsuura Y, Kitayama H, Noda M, Kikuchi A. Plasma membrane recruitment of RalGDS is critical for Ras-dependent Ral activation. Oncogene. 1999;18:1303–1312. doi: 10.1038/sj.onc.1202425. [DOI] [PubMed] [Google Scholar]
- 14.Ferro E, Trabalzini L. RalGDS family members couple Ras to Ral signalling and that’s not all. Cell Signal. 2010;22:1804–1810. doi: 10.1016/j.cellsig.2010.05.010. [DOI] [PubMed] [Google Scholar]
- 15.Wolthuis RM, Bos JL. Ras caught in another affair: the exchange factors for Ral. Curr Opin Genet Dev. 1999;9:112–117. doi: 10.1016/s0959-437x(99)80016-1. [DOI] [PubMed] [Google Scholar]
- 16.Bhattacharya M, Anborgh PH, Babwah AV, Dale LB, Dobransky T, Benovic JL, et al. Beta-arrestins regulate a Ral-GDS Ral effector pathway that mediates cytoskeletal reorganization. Nat Cell Biol. 2002;4:547–555. doi: 10.1038/ncb821. [DOI] [PubMed] [Google Scholar]
- 17.Gentry LR, Martin TD, Reiner DJ, Der CJ. Ral small GTPase signaling and oncogenesis: More than just 15minutes of fame. Biochim Biophys Acta. 2014;1843:2976–2988. doi: 10.1016/j.bbamcr.2014.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Linnemann T, Kiel C, Herter P, Herrmann C. The activation of RalGDS can be achieved independently of its Ras binding domain. Implications of an activation mechanism in Ras effector specificity and signal distribution. J Biol Chem. 2002;277:7831–7837. doi: 10.1074/jbc.M110800200. [DOI] [PubMed] [Google Scholar]
- 19.Hibino K, Shibata T, Yanagida T, Sako Y. Activation kinetics of RAF protein in the ternary complex of RAF, RAS-GTP, and kinase on the plasma membrane of living cells: single-molecule imaging analysis. J Biol Chem. 2011;286:36460–36468. doi: 10.1074/jbc.M111.262675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nakamura Y, Hibino K, Yanagida T, Sako Y. Switching of the positive feedback for RAS activation by a concerted function of SOS membrane association domains. Biophys Physicobiol. 2016;13:1–11. doi: 10.2142/biophysico.13.0_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hibino K, Shibata T, Yanagida T, Sako YA. RasGTP-induced conformational change in C-RAF is essential for accurate molecular recognition. Biophys J. 2009;97:1277–1287. doi: 10.1016/j.bpj.2009.05.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Akaike H. Information theory and an extension of the maximum likelihood principle. In: Petrov BN, Caski F, editors. Proceedings of the Second International Symposium on Information Theory. Akademiai Kiado; Budapest: 1973. pp. 267–281. [Google Scholar]
- 23.Murakoshi H, Shibata AC, Nakahata Y, Nabekura J. A dark green fluorescent protein as an acceptor for measurement of Forster resonance energy transfer. Sci Rep. 2015;5:15334. doi: 10.1038/srep15334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Matsunaga-Udagawa R, Fujita Y, Yoshiki S, Terai K, Kamioka Y, Kiyokawa E, et al. The scaffold protein Shoc2/SUR-8 accelerates the interaction of Ras and Raf. J Biol Chem. 2010;285:7818–7826. doi: 10.1074/jbc.M109.053975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Satoh T, Endo M, Nakafuku M, Akiyama T, Yamamoto T, Kaziro Y. Accumulation of p21ras. GTP in response to stimulation with epidermal growth factor and oncogene products with tyrosine kinase activity. Proc Natl Acad Sci USA. 1990;87:7926–7929. doi: 10.1073/pnas.87.20.7926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vetter IR, Linnemann T, Wohlgemuth S, Geyer M, Kalbitzer HR, Herrmann C, et al. Structural and biochemical analysis of Ras-effector signaling via RalGDS. FEBS Lett. 1999;451:175–180. doi: 10.1016/s0014-5793(99)00555-4. [DOI] [PubMed] [Google Scholar]
- 27.Hiroshima M, Saeki Y, Okada-Hatakeyama M, Sako Y. Dynamically varying interactions between heregulin and ErbB proteins detected by single-molecule analysis in living cells. Proc Natl Acad Sci USA. 2012;109:13984–13989. doi: 10.1073/pnas.1200464109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Arata Y, Hiroshima M, Pack CG, Ramanujam R, Motegi F, Nakazato K, et al. Cortical Polarity of the RING Protein PAR-2 Is Maintained by Exchange Rate Kinetics at the Cortical-Cytoplasmic Boundary. Cell Rep. 2016;16:2156–2168. doi: 10.1016/j.celrep.2016.07.047. [DOI] [PubMed] [Google Scholar]
- 29.Gentry LR, Nishimura A, Cox AD, Martin TD, Tsygankov D, Nishida M, et al. Divergent roles of CAAX motif-signaled posttranslational modifications in the regulation and subcellular localization of Ral GTPases. J Biol Chem. 2015;290:22851–22861. doi: 10.1074/jbc.M115.656710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gildea JJ, Harding MA, Seraj MJ, Gulding KM, Theodorescu D. The role of Ral A in epidermal growth factor receptor-regulated cell motility. Cancer Res. 2002;62:982–985. [PubMed] [Google Scholar]
- 31.Tian X, Rusanescu G, Hou W, Schaffhausen B, Feig LA. PDK1 mediates growth factor-induced Ral-GEF activation by a kinase-independent mechanism. EMBO J. 2002;21:1327–1338. doi: 10.1093/emboj/21.6.1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rusanescu G, Gotoh T, Tian X, Feig LA. Regulation of Ras signaling specificity by protein kinase C. Mol Cell Biol. 2001;21:2650–2658. doi: 10.1128/MCB.21.8.2650-2658.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Aronheim A, Engelberg D, Li N, al-Alawi N, Schlessinger J, Karin M. Membrane targeting of the nucleotide exchange factor Sos is sufficient for activating the Ras signaling pathway. Cell. 1994;78:949–961. doi: 10.1016/0092-8674(94)90271-2. [DOI] [PubMed] [Google Scholar]
- 34.Lu S, Jang H, Zhang J, Nussinov R. Inhibitors of Ras-SOS Interactions. Chem Med Chem. 2016;11:814–821. doi: 10.1002/cmdc.201500481. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.