Abstract
Background
Implications of different adiposity measures on cardiovascular disease aetiology remain unclear. In this paper we quantify and contrast causal associations of central adiposity (waist:hip ratio adjusted for BMI (WHRadjBMI)) and general adiposity (body mass index (BMI)) with cardiometabolic disease.
Methods
97 independent single nucleotide polymorphisms (SNPs) for BMI and 49 SNPs for WHRadjBMI were used to conduct Mendelian randomization analyses in 14 prospective studies supplemented with CHD data from CARDIoGRAMplusC4D (combined total 66,842 cases), stroke from METASTROKE (12,389 ischaemic stroke cases), type 2 diabetes (T2D) from DIAGRAM (34,840 cases), and lipids from GLGC (213,500 participants) consortia. Primary outcomes were CHD, T2D, and major stroke subtypes; secondary analyses included 18 cardiometabolic traits.
Results
Each one standard deviation (SD) higher WHRadjBMI (1SD~0.08 units) associated with a 48% excess risk of CHD (odds ratio [OR] for CHD: 1.48; 95%CI: 1.28-1.71), similar to findings for BMI (1SD~4.6kg/m2; OR for CHD: 1.36; 95%CI: 1.22-1.52). Only WHRadjBMI increased risk of ischaemic stroke (OR 1.32; 95%CI 1.03-1.70). For T2D, both measures had large effects: OR 1.82 (95%CI 1.38-2.42) and OR 1.98 (95%CI 1.41-2.78) per 1SD higher WHRadjBMI and BMI respectively. Both WHRadjBMI and BMI were associated with higher left ventricular hypertrophy, glycaemic traits, interleukin-6, and circulating lipids. WHRadjBMI was also associated with higher carotid intima-media thickness (39%; 95%CI: 9%-77% per 1SD).
Conclusions
Both general and central adiposity have causal effects on CHD and T2D. Central adiposity may have a stronger effect on stroke risk. Future estimates of the burden of adiposity on health should include measures of central and general adiposity.
Keywords: body mass index, Mendelian randomization
Observational studies have identified associations between adiposity and the risk of developing incident coronary heart disease (CHD), stroke and type 2 diabetes mellitus (T2D)1, 2. Many observational studies report consistent results with different measures of adiposity; for example the Emerging Risk Factors Collaboration found similar associations with both general adiposity measured via body mass index (BMI) and central adiposity measured via waist to hip ratio (WHR) for CHD and ischaemic stroke1. The association of different adiposity measures with T2D has also been found to be similar2.
However, other studies have suggested that central adiposity, measured as either WHR or waist circumference (WC), may have stronger associations with cardiovascular disease. For example, INTERHEART found a stronger association for WHR with myocardial infarction (MI) than BMI, and the association of WHR with MI persisted after adjustment for BMI3. The Million Women Study found that WC increased CHD risk within BMI categories (and vice versa) again suggesting each is independently associated with CHD4. Furthermore, INTERSTROKE found WHR to be more strongly associated with stroke risk than BMI5. While these studies have attempted to separate the independent effects of general and central adiposity, this remains challenging in observational studies due to the high degree of correlation between adiposity measures. Another problem is that adiposity measures may differ in their reproducibility; for example BMI is less affected by regression dilution bias – a bias to the null resulting from measurement error - than WHR 6. In addition, all measures of adiposity suffer from confounding due to underlying ill-health at low or sub-clinical levels, because many chronic conditions lead to weight loss7–9. Consequently it is very difficult, if not impossible, to quantify the true independent effects of different measures of adiposity in observational studies alone.
Whilst Mendelian randomization (MR) studies minimise bias from traditional sources such as confounding, regression dilution bias and reverse causation, they may be susceptible to bias from pleiotropy (association of genetic variants with more than one variable). Pleiotropy can be vertical due to multiple downstream effects that follow the SNP effect on the exposure of interest, but this does not compromise MR assumptions. Alternatively, pleiotropy can be horizontal, whereby the SNP or instrument affects pathways other than those of the exposure of interest and could therefore invalidate the MR assumption that the SNP only affects the outcome through the exposure of interest, potentially leading to biased causal estimates. With multi-SNP instruments, there is a chance that pleiotropic effects might become balanced such that causal inference regarding the exposure is possible. In this study we perform MR analyses of BMI and WHR together with recently developed methods that are robust to horizontal pleiotropy under additional assumptions (Supplemental Figure 1). We therefore employ MR-Egger regression to provide a test for unbalanced pleiotropy and a causal estimate of exposure on outcome in its presence10, 11. In addition we use the weighted median estimator which can give valid estimates even in the presence of horizontal pleiotropy provided at least 50 per cent of the information in the analysis comes from variants that are valid instruments, and has the advantage of retaining greater precision in the estimates compared to MR-Egger12.
This manuscript represents the most comprehensive assessment of the causal role of adiposity on CHD, stroke and T2D to date. It contrasts the causal effects of central adiposity (waist:hip ratio adjusted for BMI (WHRadjBMI) from general adiposity (BMI) on multiple cardiovascular outcomes: new CHD events from 14 prospective studies/ RCTs in addition to data publicly available from the CARDIOGRAMplusC4D13 increasing CHD cases to 66,842, multiple stroke subtypes using data from METASTROKE14 and T2D from DIAGRAM15. We present the largest number of cardiometabolic traits ever examined in a MR analysis of adiposity including lipids from the Global Lipids Genetic Consortium (GLGC; 213,500 participants)16 and many novel intermediate disease end points, including electrocardiogram (ECG) measures of left ventricular hypertrophy, carotid intima media thickness (CIMT) as a measure of sub-clinical atherosclerosis, as well as markers of renal and lung disease. We build distinct multi-SNP genetic instruments for each adiposity measure using the most comprehensive repertoire available from recent genome-wide association (GWA) studies17, 18, with 97 SNPs for BMI and 49 SNPs for WHRadjBMI, thereby more than doubling the phenotypic variance explained in some earlier MR studies19–23.
Methods
Study selection and inclusion of participants
We include individual participant data from 10 studies in the University College London – London School of Hygiene and Tropical Medicine – Edinburgh - Bristol (UCLEB) consortium (see Supplemental Table 1 for study details). We include summary data from a further four studies (Anglo-Scandinavian Cardiac Outcomes Trial (ASCOT), Health and Retirement Study (HRS), Netherlands Epidemiology of Obesity (NEO) and Prospective Study of Pravastatin in the Elderly at Risk (PROSPER)), and summary data from four consortia (CARDIoGRAMplusC4D, METASTROKE, DIAGRAM, Global Lipids Genetics Consortium (GLGC)) (see Appendix). All participating studies received approval from local institutional review boards or ethics committees. All participants gave informed consent.
Clinical Outcomes
Supplemental Table 2 provides details of CHD ascertainment and number of events by study. In UCLEB studies the primary outcome was combined prevalent or incident CHD defined as fatal or non-fatal myocardial infarction, or a coronary revascularisation procedure, but excluding angina. In the majority of studies events were validated (e.g. hospital episode statistics, clinical/laboratory measurements, review of primary care medical records). CARDIoGRAMplusC4D used standard criteria for defining cases of CAD and myocardial infarction with some studies including angiography-confirmed stenosis and stable or unstable angina13. METASTROKE define stroke as a typical clinical syndrome with radiological confirmation; subtyping was done with the Trial of Org 10172 in Acute Stroke Treatment (TOAST) classification system14. We include all ischaemic stroke, three sub-types of ischaemic stroke (large-vessel disease, small-vessel disease and cardioembolic stroke) and haemorrhagic stroke. T2D definitions follow DIAGRAM24.
Cardiometabolic traits
For analysis of individual participant data studies, data on sex, age, measured standing height, weight, waist circumference and hip circumference were used to derive BMI and WHRadjBMI traits. WHRadjBMI was calculated by generating the predicted residuals from the linear regression of WHR on BMI. Biomarkers included in analyses were grouped into the following categories; Lipids (triglycerides, HDL-C and LDL-C), inflammation (IL-6), lung function (ratio of FEV1 to FVC), metabolic (glucose, insulin and albumin), renal (creatinine, estimated glomerular filtration rate (EGFR), MDRD) and systolic blood pressure. The following electrocardiogram (ECG) measures of left ventricular hypertrophy were recorded: QRS voltage sum, QRS voltage sum product, Cornell product and Sokolow-Lyon index as well as PR interval (see Supplemental Method 1 for definitions). Cardiometabolic traits that were not normally distributed were transformed to the natural logarithmic scale. For comparability across biomarkers, measurements were z-score standardised. Self-reports of current smoking status (ever/ never) and alcohol consumption (drinker/ non-drinker) were considered to be potential confounders of adiposity-cardiovascular disease (CVD) associations.
Genotyping
Supplemental Table 1 details genotyping by study. Genotyping in all UCLEB studies was conducted with the Metabochip array (except a subset of ELSA study that used a GWAS array)25. The remaining studies used GWAS arrays (HRS, PROSPER) or Exome Chip (NEO). Individuals were excluded from the analyses on the basis of gender mismatch, excessive or minimal heterozygosity, relatedness or individual missingness (>3%). Individuals of non-European ancestry were removed to minimise confounding by population structure. SNPs with a low call rate or evidence of departure from Hardy–Weinberg equilibrium were excluded from analyses (see Supplemental Table 1 for thresholds employed in different studies).
Statistical Analyses
Observational Analyses
In individual participant data studies adiposity (BMI or WHRadjBMI) was z-score standardised and linear or logistic regression models were fitted for each cardiometabolic trait or disease outcome. Observational models were adjusted for age and sex. Fixed-effect meta-analyses were employed to derive combined observational estimates across studies. We calculated I2 statistics to quantify heterogeneity between studies and derived P-values from Cochran’s Q test26.
Genetic Analyses
SNP selection and construction of the genetic instruments
Selection of SNPs for the genetic instruments was based on analyses from the Genetic Investigation of ANthropometric Traits (GIANT) consortium, which included 339,224 individuals from 125 separate studies for BMI17 and 224,459 individuals from 101 studies for WHRadjBMI18. These studies identified 97 independent SNPs for BMI and 49 independent SNPs for WHRadjBMI at GWAS significance. We found no overlap between the BMI SNPs and WHRadjBMI SNPs. In studies where the SNP identified by GIANT was not available in the Metabochip array, we used proxy SNPs in linkage disequilibrium (R2>0.8) with the specified SNP. Details of proxy SNPs used by platform (Metabochip/ GWAS) are given in Supplemental Tables 3 and 4.
Genetic association analyses in individual participant data
We performed a within study genetic association analysis with adiposity (standardised BMI and WHRadjBMI) as a continuous trait using an additive model. We used linear or logistic regression models to estimate the additive effect of each SNP on cardiometabolic traits and outcomes. We used logistic regression to test the association of each SNP with smoking and alcohol consumption as potential confounders of the adiposity-CVD association.
Instrumental variable analyses in summary data
We conducted three tests for the causal estimation of each adiposity measure on cardiometabolic outcomes: 1) Inverse-variance weighted method (IVW), 2) MR-Egger and 3) Weighted median. In the absence of horizontal pleiotropy, we would expect all three tests to give consistent results. All IV estimates in summary data were calculated using the mrrobust package (available from https://github.com/remlapmot/mrrobust) in Stata version 1427, 28. The proportion of variance in adiposity explained by the genetic instruments in summary data was calculated using the grs.summary function from the gtx package in R29, 30. A threshold of statistical significance of P<0.025 (0.05/2=0.025) was used to reflect testing for two different adiposity traits (BMI and WHRadjBMI).
1) IVW instrumental variable analyses
To combine data across studies with summary level data we pooled the association of each SNP on risk of each CVD outcome/ cardiometabolic trait using fixed effects meta-analysis. To provide external weights for the SNP-adiposity associations, the effect of each SNP on adiposity (BMI; WHRadjBMI) in GIANT was pooled with that in all other contributing studies, excluding studies that had already contributed to GIANT (1958BC, EAS, HRS, NSHD, PROSPER, Whitehall II). To quantify heterogeneity in the SNP effects across studies we calculated I2 and derived P-values from Cochrane’s Q tests. All P-values were two-sided. Inverse-variance weighted meta-analysis (IVW) was used to provide a combined estimate of the causal estimates (SNP-outcome/ SNP-adiposity) from each SNP. IVW is equivalent to a two-stage least squares or allele score analysis using individual-level data, and is hence referred to here as “conventional MR”31. However, it can lead to over-rejection of the null, particularly when there is heterogeneity between the causal estimates from different genetic variants.
2) MR-Egger instrumental variable analyses
To account for potential horizontal pleiotropy in the multi-SNP adiposity instruments, we re-estimated the instrumental variable associations using MR-Egger regression10, 11. MR-Egger tests for presence of, and accounts for, unbalanced pleiotropy by introducing a parameter for this bias10. Specifically, linear regression of the instrument-outcome effects is performed on the instrument-exposure effects, with the slope representing the causal effect estimate and the intercept the net bias due to horizontal pleiotropy. An additional assumption is required that the individual SNP effects on the exposure are independent of their pleiotropic effects on the outcome (termed the ‘InSIDE assumption’)12.
3) Weighted median estimate instrumental variable analyses
Finally, we applied a complementary approach termed the weighted median estimator which can give valid estimates even in the presence of horizontal pleiotropy provided at least half of the weighted variance is valid12.
Power calculations
Power to detect causal estimates was calculated based on the proportion of variance of the exposure explained by the instruments (R2), the total number of individuals in the analysis, and the number of cases and controls using the online tool http://cnsgenomics.com/shiny/mRnd/32. Power estimates are provided in (Supplemental Table 5).
Results
Studies and participants
Full descriptive details of the included studies are given in Supplemental Table 1. Data from 14 prospective studies and randomised trials and four consortia were included with 66,842 CHD cases (3,716 from UCLEB/ other non-consortia studies), 12,389 ischaemic stroke cases and 34,840 T2D cases. The number of individuals included in the analyses of cardiometabolic traits ranged from 6,625 to 213,556. The mean age in individual participant data studies was 63.5 years, the mean BMI 27.4 kg/m2 (SD 4.6) and the mean WHR 0.89 (SD 0.13) (Supplemental Tables 1 & 6). Distribution of binary traits by study are given in Supplemental Table 7.
Instrument validation
We identified Metabochip proxies for 13 BMI SNPs and 7 WHRadjBMI SNPs; the median R2 was 0.965 & 0.913 respectively (Supplemental Tables 3 and 4). The proportion of variance of BMI explained by the BMI genetic instrument was 1.7% while the WHRadjBMI instrument explained 0.7% WHRadjBMI variance. The associations of individual SNPs with adiposity are shown in Supplemental Tables 8 and 9.
Mendelian randomization analysis of adiposity with cardiometabolic traits
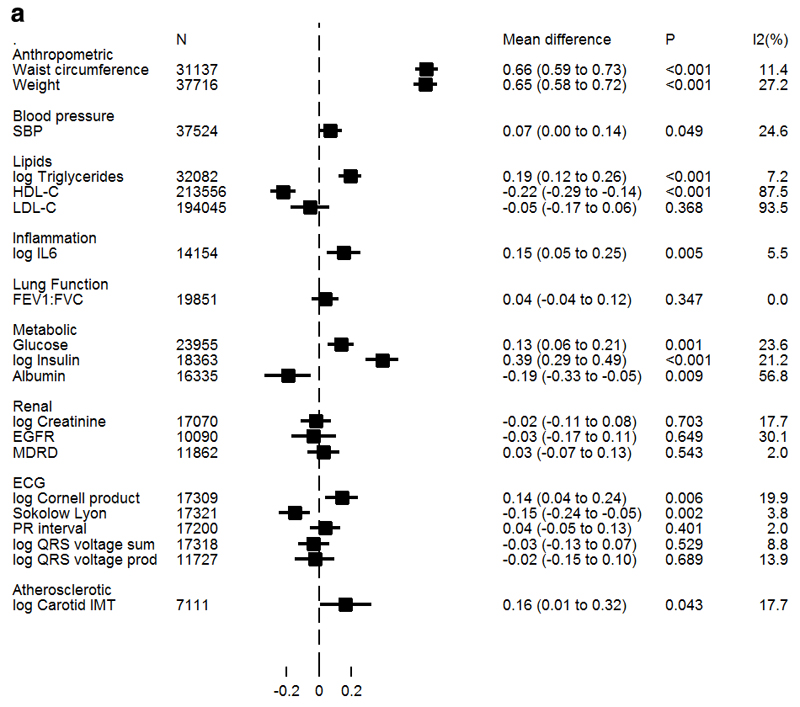
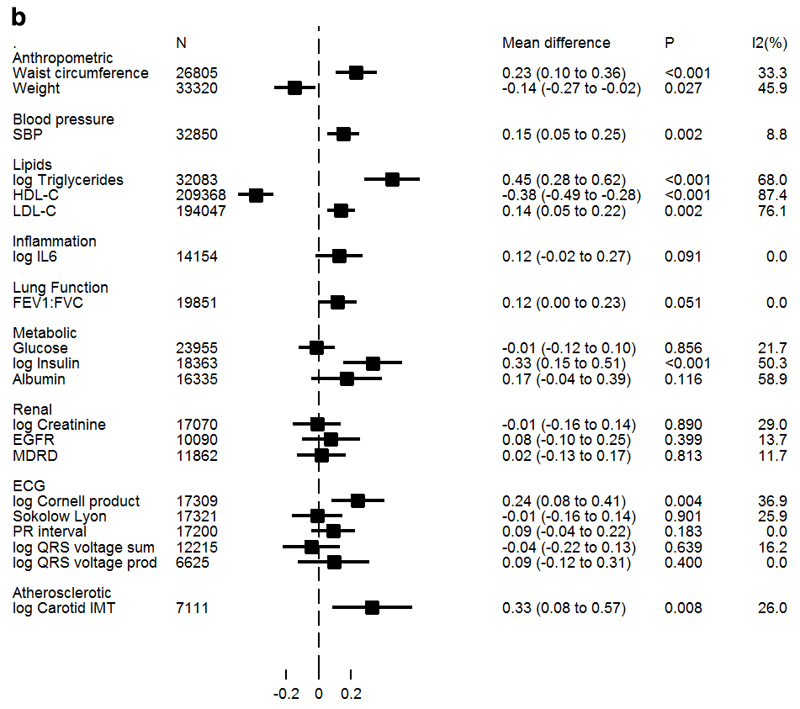
Figure 1a/b presents estimates of associations between BMI and WHRadjBMI with cardiometabolic traits from IV analyses. Both genetically instrumented adiposity measures were found to be causally associated with increased insulin and triglycerides. In addition, BMI was causally associated with higher IL-6, with a directionally consistent result identified for WHRadjBMI. Both adiposity measures were also causally associated with decreased levels of HDL-C. However, only WHRadjBMI was associated with increased LDL-C, and the association with SBP was also stronger. BMI was inversely associated with albumin, while WHRadjBMI was not; but heterogeneity across studies was moderately high (I2=57%).
Figure 1a. Association of BMI with continuous biomarkers derived from Mendelian randomization analysis.
Values represent standardized mean differences of each trait per SD increase in BMI derived from conventional (IVW) Mendelian randomization analysis. Non-normally distributed variables were natural ln transformed; therefore mean differences displayed on the log scale may be anti-logged and interpreted as percentage differences in SD of trait per SD in BMI. Log triglycerides from individual participant data studies only; GLGC triglycerides in Supplemental Table 10.
Figure 1b. Association of WHRadjBMI with continuous biomarkers derived from Mendelian randomization analysis.
Values represent standardized mean differences of each trait per SD increase in WHRadjBMI derived from conventional (IVW) Mendelian randomization analysis. Non-normally distributed variables were natural log transformed; therefore mean differences displayed on the log scale may be anti-logged and interpreted as percentage difference in SD of trait per SD in WHRadjBMI. Log triglycerides from individual participant data studies only; GLGC triglycerides in Supplemental Table 11.
There was evidence for a causal association with some of the ECG measures that index left ventricular hypertrophy with both adiposity measures associated with higher log Cornell Product; in addition BMI, but not WHRadjBMI associated with lower Sokolow-Lyon index. There was no suggestion for a causal association of either measure of adiposity and PR interval.
Both WHRadjBMI and, to a weaker extent, BMI were causally associated with higher CIMT (39%, 95%CI: 9%, 77% and 18%, 95% CI: 1%, 38% higher per SD in WHRadjBMI and BMI, respectively). WHRadjBMI had a weak association with lung function (FEV1:FVC) at 0.12 units per SD (95%CI 0.01, 0.24), but the P-value does not meet the threshold which takes into account testing for multiple measures of adiposity. There was no suggestion of a causal association of either adiposity measure with any of the measures of renal function.
With MR-Egger regression there was no convincing evidence for directional pleiotropy in any of the associations of adiposity traits with continuous cardiometabolic traits (Supplemental Tables 10 and 11).
Supplemental Figures 2a/b illustrate the consistency of observational and IV estimates for associations between adiposity and cardiometabolic traits (Supplemental Tables 12 and 13).
Mendelian randomization analysis of adiposity with cardiometabolic diseases
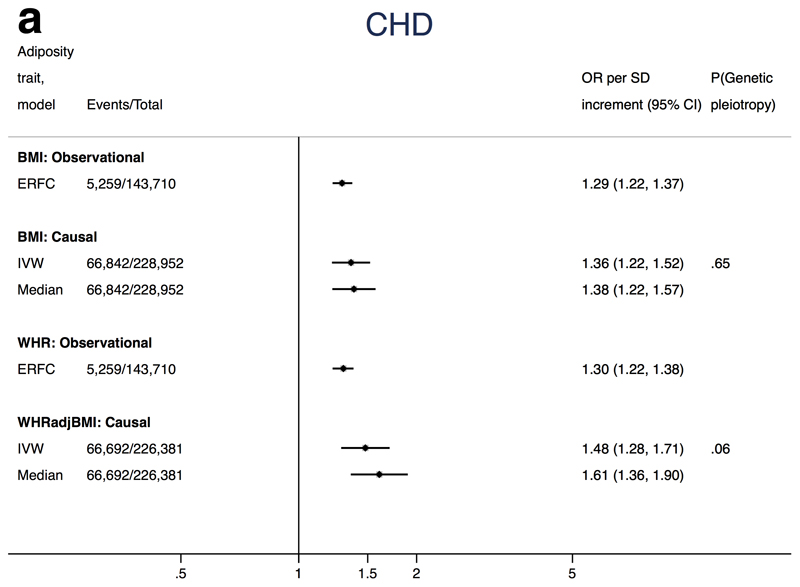
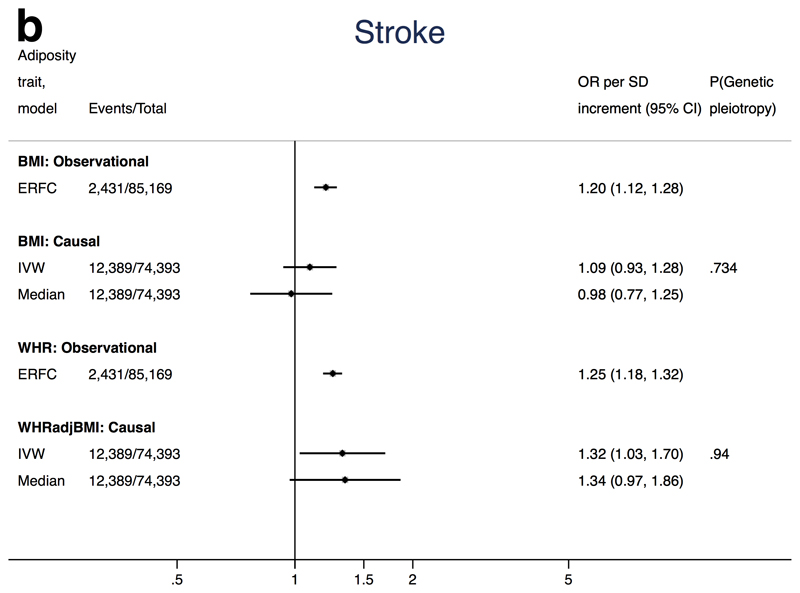
Figures 2a-c show the association of each adiposity measure with CHD, ischaemic stroke and T2D from conventional IVW and weighted median MR analyses. MR-Egger estimates tended to be much more imprecise and are therefore presented separately in Supplemental Table 14 to facilitate interpretation.
Mendelian randomization analysis of adiposity with CHD
The summary causal estimate per 1SD increment in BMI from conventional IVW MR was an OR for CHD of 1.36 (95%CI: 1.22, 1.52) (Figure 2a). MR-Egger regression suggested little evidence for unbalanced pleiotropy in the genetic instrument (intercept P-value=0.65), and both MR-Egger and weighted median estimates were consistent with the IVW estimate (Supplemental Figure 3a). Furthermore, MR estimates were consistent with observational estimates reported by the Emerging Risk Factors Collaboration (Figure 2a)
Figure 2a. Associations of adiposity with risk of CHD from observational and Mendelian randomization analyses.
Association between coronary heart disease and adiposity (BMI and WHRadjBMI) comparing causal odds ratios (OR) per SD of adiposity trait derived from instrumental variable analysis and observational analysis from the Emerging Risk Factors Consortium hazard ratio (HR per SD of BMI or waist:hip adjusted for age, sex and smoking status)1. Causal estimates are derived from Mendelian randomization and include conventional (ratio) approach and weighted median (see Methods for further details). P(genetic pleiotropy) relates to the P-value derived from the intercept of MR-Egger; a small P-value denotes presence of directional pleiotropy.
Similarly, we found an association between WHRadjBMI and CHD using conventional MR (OR 1.48, 95% CI 1.28, 1.71 per SD WHRadjBMI, Figure 2a and Supplemental Figure 3b). The intercept for the MR-Egger test was 0.0134 (95%CI -0.0004, 0.0278; P-value=0.06). The causal estimate from MR-Egger was imprecise (OR 0.89, 95% CI 0.52, 1.53), but the weighted median estimator (which retains more power than MR-Egger) provided a causal effect of 1.61 (95% CI 1.36, 1.90) which was consistent with the IVW result.
Mendelian randomization analysis of adiposity with ischaemic stroke
The causal OR for the association between BMI and ischaemic stroke was 1.09 (95%CI 0.93, 1.28 per SD) (Figure 2b). Results from the MR-Egger analysis were compatible with no unbalanced pleiotropy (intercept P-value=0.73), and the weighted median estimator suggested no causal association (Supplemental Figure 3c). Estimates for association between BMI and stroke sub-types were imprecise and 95% confidence intervals all included the null (Table 1). Thus, while all IV estimates for BMI and stroke include the Emerging Risk Factors Collaboration estimate (Figure 2b), lack of precision hinders any clear causal evidence for an association between BMI and ischaemic stroke.
Figure 2b. Associations of adiposity with risk of ischaemic stroke from observational and Mendelian randomization analyses.
Association between ischaemic stroke and adiposity (BMI and WHRadjBMI) comparing causal odds ratios (OR) per SD of adiposity trait derived from instrumental variable analysis and observational analysis from the Emerging Risk Factors Consortium (HR of ischaemic stroke per SD of BMI or waist:hip adjusted for age, sex and smoking status)1. Causal estimates are derived from Mendelian randomization and include conventional (ratio) approach and weighted median (see Methods for further details). P(genetic pleiotropy) relates to the P-value derived from the intercept of MR-Egger; a small P-value denotes presence of directional pleiotropy.
Table 1.
Mendelian randomization estimates for the association of adiposity and stroke sub-types
| IVW | Weighted median | |||||||
|---|---|---|---|---|---|---|---|---|
| OR | LCI | UCI | I2 | P(Genetic pleiotropy) | OR | LCI | UCI | |
| BMI | ||||||||
| All ischaemic stroke | 1.09 | (0.93, | 1.28) | 20% | 0.734 | 0.98 | (0.77, | 1.25) |
| - Cardioembolic | 1.18 | (0.89, | 1.55) | 0% | 0.507 | 1.40 | (0.87, | 2.24) |
| - Large vessel disease | 1.14 | (0.82, | 1.59) | 19% | 0.625 | 1.12 | (0.65, | 1.91) |
| - Small vessel disease | 0.93 | (0.64, | 1.35) | 30% | 0.270 | 1.15 | (0.67, | 1.97) |
| Haemorrhagic stroke | 1.51 | (0.73, | 3.13) | 0% | 0.435 | 1.28 | (0.37, | 4.40) |
| WHRadjBMI | ||||||||
| All ischaemic stroke | 1.32 | (1.03, | 1.70) | 38% | 0.936 | 1.34 | (0.96, | 1.87) |
| - Cardioembolic | 1.24 | (0.84, | 1.83) | 0% | 0.588 | 1.32 | (0.73, | 2.38) |
| - Large vessel disease | 1.37 | (0.90, | 2.09) | 0% | 0.470 | 0.87 | (0.48, | 1.58) |
| - Small vessel disease | 1.57 | (0.98, | 2.51) | 13% | 0.861 | 1.71 | (0.89, | 3.29) |
| Haemorrhagic stroke | 1.89 | (0.69, | 5.18) | 0% | 0.430 | 1.73 | (0.42, | 7.06) |
IVW: inverse variance weighted (also termed ‘conventional’ MR) and weighted median. P(genetic pleiotropy) relates to the P-value derived from the intercept of MR-Egger; a small P-value denotes presence of directional pleiotropy.
Results do, however, provide some evidence for a causal association of WHRadjBMI with ischaemic stroke (OR 1.32, 95%CI 1.03, 1.70 per SD in WHRadjBMI) (Figure 2b). MR-Egger regression was consistent with no unbalanced pleiotropy (intercept P-value=0.94), and the weighted median estimator was very close to the IVW estimate (causal OR 1.34, 95%CI 0.97, 1.86 per SD increase in WHRadjBMI) (Supplemental Figure 3d). Limited evidence was found for a causal association with stroke sub-types; all point estimates were consistently above one but precision was poor and 95% confidence intervals included the null (Table 1).
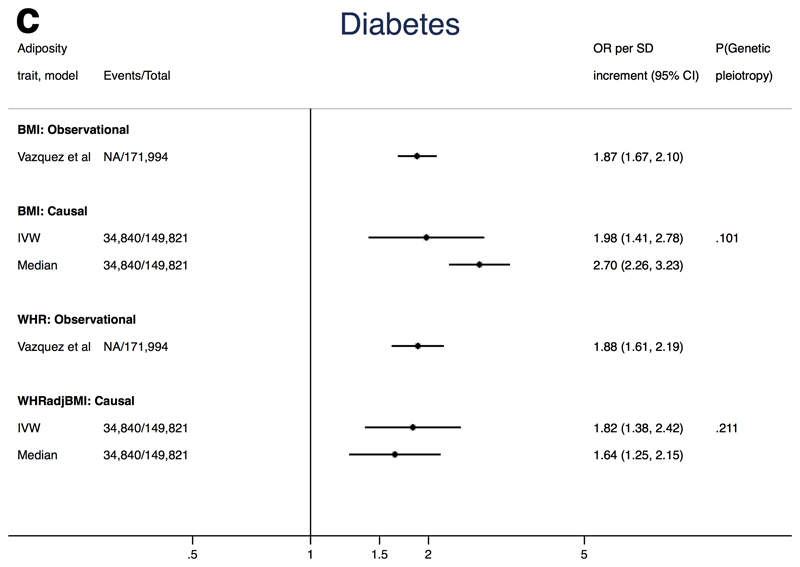
Mendelian randomization analysis of adiposity with T2D
We found a causal OR for T2D of 1.98 (95%CI: 1.41, 2.78) per SD increase in BMI (Figure 2c). Similar but stronger estimates were identified using MR-Egger (OR 3.70, 95% CI 1.63, 8.41; P-value for pleiotropy=0.10) and weighted median estimator (OR 2.70, 95% CI 2.26, 3.23). One BMI SNP (rs7903146) was an outlier (Supplemental Figure 3e) and is a marker for the TCF7L2 gene, a GWAS-identified locus for T2D33. We therefore repeated the T2D analysis excluding rs7903146 (Supplemental Table 15 yielding an IVW OR of 2.25 (95%CI: 1.87, 2.71) per SD increase in BMI, with similar estimates from MR-Egger and weighted median estimators.
Figure 2c. Associations of adiposity with risk of T2D from observational and Mendelian randomization analyses.
Association between T2D and adiposity (BMI and WHRadjBMI) comparing causal odds ratios (OR) per SD of adiposity trait derived from instrumental variable analysis and observational analysis from Vazquez et al., 20072. Causal estimates are derived from Mendelian randomization and include conventional (ratio) approach and weighted median (see Methods for further details). P(genetic pleiotropy) relates to the P-value derived from the intercept of MR-Egger; a small P-value denotes presence of directional pleiotropy.
Likewise, we found a causal relationship between WHRadjBMI and T2D (OR 1.82, 95% CI 1.38, 2.42 per SD increase in WHRadjBMI, Figure 2c). MR-Egger did not provide evidence of unbalanced pleiotropy (P-value for pleiotropy=0.21), and the weighted median estimator result was consistent with the IVW (OR 1.64, 95% CI 1.25, 2.15) (Supplemental Figure 3g).
Multivariate Mendelian randomization
We found some evidence for association of both adiposity instruments with smoking, but not with other major confounders (Supplemental Table 16). To account for this, sensitivity analyses were undertaken for each cardiometabolic disease using multivariate MR including the effect of each SNP used as instrument for BMI and WHRadjBMI on smoking. MR estimates were found to be robust to this adjustment (Supplemental Table 17), with generally consistent point estimates measured with greater imprecision reflecting the reduced power in these analyses. The multivariate MR (adjusted for smoking) for the causal association of WHRadjBMI with ischaemic stroke was 1.27 (95% CI 0.84-1.93) broadly similar to 1.32 (95% CI 1.03-1.70) in the main IVW analysis, but with a wider confidence interval. We also included FEV1:FVC in these sensitivity analyses due to the likely association of this trait with smoking; again adjusted results were very similar to the main IVW results (Supplemental Table 17).
Discussion
We conducted the most comprehensive MR analysis to date comparing the causal role of central and general adiposity in the development of multiple cardiovascular disease outcomes (CHD, multiple stroke sub-types and T2D). Owing to benefits of MR to minimize residual confounding by common lifestyle factors and underlying ill-health, we are able to quantify that one standard deviation increase in genetically instrumented WHRadjBMI (~0.08 units) results in a ~50% increase in risk of CHD independent of BMI. This compares with the ~40% increase in risk of CHD we find per 1SD increase in genetically instrumented BMI (~4.6 kg/m2) which is consistent with the observational effect derived from large prospective population cohorts including the Emerging Risk Factors Collaboration 1 (CHD HR 1.29 [1.22-1.37] per 1SD) and the Prospective Studies Collaboration33 Thus, while observational studies such as the Emerging Risk Factors Collaboration have found risk to be consistent across different measures of adiposity, our results suggest WHRadjBMI may have a stronger effect, although the greater imprecision in the MR estimates should also be considered.
Similarly, while observational studies have found different measures of adiposity to have similar associations with risk of ischaemic stroke1, our result again suggest that WHRadjBMI may be more strongly associated (increased risk ~30% per 1SD). Recent findings from INTERSTROKE also suggest that WHR is a much stronger deleterious risk factor for ischaemic stroke5. Our SBP results follow a similar pattern, with a much stronger association between central adiposity and SBP than general adiposity. This is also the first MR study to suggest potential causal association between central adiposity ischaemic stroke subtypes, and CIMT, a widely used surrogate measure of sub-clinical atherosclerosis.
Previous adiposity MR studies used limited numbers of SNPs, (with weaker genetic instruments), fewer events and generally failed to find evidence for a causal association between BMI and CHD19, 21. However, one MR study using a 3-SNP allele score (FTO, MC4R, TMEM18) reported an OR of 1.52 (95% CI 1.12-2.05 for a 4 kg/m2 increase in BMI20 , and most recently a MR study using a 32-SNP instrument for BMI found similar results for CHD to ours22. We do not however replicate the causal association between BMI and ischaemic stroke reported by the same study (hazard ratio per SD-increase of BMI 1.83; 95% CI 1.05-3.20)22, despite increasing the number of stroke cases tenfold. Furthermore, our results are in line with those for ischaemic stroke from the Emerging Risk Factors Collaboration and INTERSTROKE, including the apparently stronger association we find between central adiposity and stroke relative to general adiposity. Results for the causal association of WHRadjBMI with CHD and T2D are consistent with those from a recent MR analysis34.
We present the largest number of cardiometabolic traits ever examined in a MR analysis of adiposity. The current findings are broadly consistent with earlier MR studies for glucose, triglycerides, HDL-C, SBP, and IL-6, providing further support for a detrimental impact of adiposity on the cardiovascular system19, 21, 23. However, we find no evidence for a causal association between BMI and LDL-C, consistent with some but not all earlier studies21, 23. A recent MR study found a causal effect of BMI and a wide range of lipid metabolites, including all LDL metabolites35, but was conducted in a younger, healthier population (average BMI ~24kg/m2) than is commonly included in MR studies (including the current one) and this could explain the discrepancy with our findings (as observational studies suggests the association of BMI and LDL-C plateaus beyond 27kg/m2) 33. We also report novel positive causal associations of adiposity with the ECG measure log Cornell product (a measure of left ventricular hypertrophy; LVH). The negative association of BMI with Sokolow Lyon (an alternative measure of LVH) was unexpected and may represent a false positive. While both log Cornell product and Sokolow Lyon measure left ventricular hypertrophy, log Cornell product is considered to be the better test for identifying LVH when measured against a gold standard36.
This study demonstrates that central obesity (as quantified by WHRadjBMI) has a causal effect on CHD that is independent of BMI. This finding demonstrates the potential of MR approaches for investigating highly correlated adiposity measures that have proved challenging to disentangle in observational studies37. In these analyses we find that WHRadjBMI has a more deleterious lipid profile than BMI, with detrimental associations of greater magnitude with triglycerides and HDL-C and association with LDL-C not found for BMI. The association of WHRadjBMI with CIMT is also of greater magnitude. Conversely, BMI appears to have a greater inflammatory effect than WHRadjBMI, and potentially a stronger effect on the ECG measures that index left ventricular hypertrophy as well as with glucose and T2D. The apparent lack of association of WHRadjBMI with glucose is surprising, but is potentially explained by a negative association of WHRadjBMI SNPs with BMI. Interestingly, a recent paper showed WHRadjBMI to associate with 2-hour fasting glucose suggesting that WHRadjBMI may have differential effects according to how glucose is measured; different mechanisms are likely to regulate fasting and 2-hour glucose34. In keeping with our findings, the discovery GWAS that identified 49 SNPs associated with WHRadjBMI18 found associations of the SNPs with concentrations of HDL-C, TG, LDL-C, adiponectin and fasting insulin. Furthermore, the study identified enrichment of WHRadjBMI SNPs for T2D and CHD.
This study suggests that it is not only the volume of adiposity, but also its location, that is relevant for disease, lending weight to the emerging theory that the deposition of body fat plays important roles that are independent of total fat. For example, at a given BMI, there is considerable inter-individual variation in the amount of visceral fat, which shows associations with disease38. Our results also suggest that efforts to quantify the effect of adiposity on burden of disease should include multiple measures of adiposity to avoid underestimating the true burden of adiposity on health39. As regards specific interventions that focus WHR more than BMI, there is observational evidence that physical activity can modify WHR independent of BMI40. Thus it may be possible to mitigate the effects of WHR through increased population-wide physical activity. In addition, our findings open potentially new avenues of investigation. For example, identifying these causal effects of WHRadjBMI can enable research to focus on the downstream consequences of this trait, and potentially identify traits (such as metabolites)35 that could mediate the relationship between WHRadjBMI and disease which may themselves be amenable to pharmacological modification. Such traits downstream of WHRadjBMI could be unique (and not shared with BMI) raising the possibility of novel opportunities for drug discovery and disease prevention.
Strengths
This study has many strengths. First, independent multi-SNP instruments comparing the effect of central and general adiposity on multiple CVD outcomes; second, the use of powerful genetic instruments for BMI and WHRadjBMI which explained up to twice the phenotypic variation compared with previous MR studies; third, large number of clinical events that provided ample power to detect the associations of adiposity with cardiometabolic diseases fourth, the use of methods to minimise the impact of unbalanced pleiotropy in the genetic instruments that may invalidate findings from conventional MR.
In addition to this being the most comprehensive evaluation of adiposity-related traits with cardiovascular and metabolic risk factors and diseases, our analysis also facilitates their direct comparison, and therefore contrasts the effects of general adiposity with body fat distribution in the same datasets. This provides novel insights, demonstrating that WHRadjBMI is more relevant to the development of subclinical atherosclerosis and stroke compared to BMI, whereas both BMI and WHRadjBMI are important for CHD and diabetes.
Limitations
Limitations include the potential pleiotropic effects of the multi-SNP instruments. However, results suggest little evidence for unbalanced pleiotropy. Re-estimates of the causal associations using MR-Egger regression were broadly consistent with our conventional MR analysis, albeit with a loss of precision and consequently a loss of power, while weighted median estimates (that retains more power than MR-Egger) proved remarkably similar to IVW.
The InSIDE (Instrument Strength Independent of Direct Effect), which is untestable, assumes that the pleiotropic effects of the genetic variants are uncorrelated with the association of the genetic variants with the exposure. Violation of InSIDE would give rise to biased causal estimates from MR-Egger; however each MR approach has different strengths and assumptions, for example, violation of InSIDE does not affect the weighted median MR approach 12. This highlights the importance of using the three MR approaches (IVW, median and MR-Egger) in our study. General concordance of MR estimates derived from these approaches helps reinforce the conclusions that can be drawn. We used a multi-SNP instrument for WHR that had already been adjusted for BMI as part of the GIANT GWAS18. Genetic instruments for phenotypes adjusted for heritable components may show association with the adjusted phenotype through collider bias41, which could violate the InSIDE assumption. Indeed, we found WHRadjBMI SNPs to be associated with BMI beyond what would be expected by chance (Supplemental Table 18). This could lead to biased results; however in the current scenario the bias will tend to be towards the null (and underestimate the true effect) as the WHRadjBMI SNPs are associated negatively with BMI.
We selected cardiometabolic traits a priori on the basis that previous studies have shown them to be observationally and genetically associated with BMI. Therefore, although we test multiple outcomes use of a conventional Bonferroni would over-penalize the interpretation.
Future studies should look to include emerging CVD outcomes such as heart failure and atrial fibrillation, and consider additional potential confounders. In addition, more stroke cases should be added to improve precision in these analyses, in particular for multivariate MR analyses.
Given that our MR analysis on CHD was largely based on summary data, we were unable undertake more detailed investigations of the linear relationship between BMI or WHRadjBMI and risk of CHD and/ or to explore the causal effects of very low levels of BMI or WHR on CHD42. These are important next steps to investigate, given the uncertainty regarding whether the U-shape association of BMI with disease reflects a true causal relationship, or whether it is an artefact from residual confounding and/or underlying ill-health. The recent finding of a J-shaped (rather than U-shaped) association between BMI and mortality in healthy non-smokers reinforces the likely role of artefact this association43. Therefore, application of methods for non-linear MR could help to determine the true optimal level of BMI for health44. However, such analyses would require access to individual participant data in all studies.
Finally, although we identify several downstream biological mechanisms by which general and central adiposity may mediate the effects on risk of CHD, these results should be considered as exploratory and further studies using adequate methodology for mediation analysis should be conducted45, 46, including the analysis of finer resolution for cardio-metabolic traits for example using NMR metabolomics.
Conclusions
Our study supports evidence for a causal role of both central and general adiposity in risk of CHD and T2D, and central adiposity in risk of ischaemic stroke. Furthermore, our results suggest that central adiposity may pose higher risk for stroke and CHD. Efforts to estimate the role of adiposity on cardiovascular disease should consider the potential independent effects of different measures of adiposity.
Supplementary Material
Clinical Perspective.
What is new?
This large-scale genetic analysis presents the most comprehensive causal assessment of adiposity with cardiometabolic diseases to date, including new data for stroke subtypes from METASTROKE and novel cardiometabolic traits including ECG measures and CIMT.
We find that waist:hip ratio adjusted for BMI, a measure of central body fat distribution that aims to be independent of general adiposity, is causally related to higher risks of coronary heart disease, ischaemic stroke and a multitude of cardiometabolic traits.
Our findings also reinforce existing evidence on the causal relevance of general adiposity (BMI) to these diseases and provide more precise estimates.
What are the clinical implications?
Both the amount of adiposity and its distribution play important roles in influencing multiple cardiometabolic traits and the development of cardiometabolic diseases.
Furthermore, our findings indicate that body fat distribution has multiple causal roles in disease that are independent of general adiposity.
This suggests that physicians should pay attention to measures of adiposity beyond BMI as measurement of such traits may identify patients at risk of cardiometabolic disease and provides opportunities to the scientific community to identify novel approaches to disease prevention.
Sources of Funding
CED is supported by a University College London Springboard Population Science Fellowship.
JP Casas is supported by the NIHR University College London Hospitals Biomedical Research Centre. Aroon D Hingorani is supported by an NIHR Senior Investigator Award. Work in his laboratory is supported by a British Heart Foundation Grant (RG/10/12/28456). The UCLEB consortium is supported by funding from NIHR, British Heart Foundation (RG/10/12/28456), and Medical Research Council.
Footnotes
Disclosures
None
References
- 1.Wormser D, Kaptoge S, Di Angelantonio E, Wood AM, Pennells L, Thompson A, Sarwar N, Kizer JR, Lawlor DA, Nordestgaard BG, Ridker P, et al. Separate and combined associations of body-mass index and abdominal adiposity with cardiovascular disease: collaborative analysis of 58 prospective studies. Lancet. 2011;377:1085–1095. doi: 10.1016/S0140-6736(11)60105-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vazquez G, Duval S, Jacobs DR, Jr, Silventoinen K. Comparison of body mass index, waist circumference, and waist/hip ratio in predicting incident diabetes: a meta-analysis. Epidemiol Rev. 2007;29:115–128. doi: 10.1093/epirev/mxm008. [DOI] [PubMed] [Google Scholar]
- 3.Yusuf S, Hawken S, Ounpuu S, Bautista L, Franzosi MG, Commerford P, Lang CC, Rumboldt Z, Onen CL, Lisheng L, Tanomsup S, et al. Obesity and the risk of myocardial infarction in 27,000 participants from 52 countries: a case-control study. Lancet. 2005;366:1640–1649. doi: 10.1016/S0140-6736(05)67663-5. [DOI] [PubMed] [Google Scholar]
- 4.Canoy D, Cairns BJ, Balkwill A, Wright FL, Green J, Reeves G, Beral V, Million Women Study C Coronary heart disease incidence in women by waist circumference within categories of body mass index. Eur J Prev Cardiol. 2013;20:759–762. doi: 10.1177/2047487313492631. [DOI] [PubMed] [Google Scholar]
- 5.O'Donnell MJ, Chin SL, Rangarajan S, Xavier D, Liu L, Zhang H, Rao-Melacini P, Zhang X, Pais P, Agapay S, Lopez-Jaramillo P, et al. Global and regional effects of potentially modifiable risk factors associated with acute stroke in 32 countries (INTERSTROKE): a case-control study. Lancet. 2016;388:761–775. doi: 10.1016/S0140-6736(16)30506-2. [DOI] [PubMed] [Google Scholar]
- 6.Yusuf S, Anand S. Body-mass index, abdominal adiposity, and cardiovascular risk. Lancet. 2011;378:226–7. doi: 10.1016/S0140-6736(11)61120-3. author reply 228. [DOI] [PubMed] [Google Scholar]
- 7.Lawlor DA, Harbord RM, Timpson NJ, Lowe GD, Rumley A, Gaunt TR, Baker I, Yarnell JW, Kivimaki M, Kumari M, Norman PE, et al. The association of C-reactive protein and CRP genotype with coronary heart disease: findings from five studies with 4,610 cases amongst 18,637 participants. PLoS One. 2008;3:e3011. doi: 10.1371/journal.pone.0003011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lawlor DA, Hart CL, Hole DJ, Davey Smith G. Reverse causality and confounding and the associations of overweight and obesity with mortality. Obesity (Silver Spring) 2006;14:2294–2304. doi: 10.1038/oby.2006.269. [DOI] [PubMed] [Google Scholar]
- 9.Dale C, Nuesch E, Prieto-Merino D, Choi M, Amuzu A, Ebrahim S, Casas JP, Davey-Smith G. Why do thin people have elevated all-cause mortality? Evidence on confounding and reverse causality in the association of adiposity and COPD from the British Women's Heart and Health Study. PLoS One. 2015;10:e0115446. doi: 10.1371/journal.pone.0115446. eCollection 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bowden J, Davey Smith G, Burgess S. Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Intern J Epidemiol. 2015;44:512–525. doi: 10.1093/ije/dyv080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.White J, Sofat R, Hemani G, Shah T, Engmann J, Dale C, Shah S, Kruger FA, Giambartolomei C, Swerdlow DI, Palmer T, et al. Plasma urate concentration and risk of coronary heart disease: a Mendelian randomisation analysis. Lancet Diabetes Endocrinol. 2016;4:327–336. doi: 10.1016/S2213-8587(15)00386-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bowden J, Davey Smith G, Haycock PC, Burgess S. Consistent Estimation in Mendelian Randomization with Some Invalid Instruments Using a Weighted Median Estimator. Genet Epidemiol. 2016;40:304–314. doi: 10.1002/gepi.21965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Consortium CAD. Deloukas P, Kanoni S, Willenborg C, Farrall M, Assimes TL, Thompson JR, Ingelsson E, Saleheen D, Erdmann J, Goldstein BA, et al. Large-scale association analysis identifies new risk loci for coronary artery disease. Nat Genet. 2013;45:25–33. doi: 10.1038/ng.2480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Traylor M, Farrall M, Holliday EG, Sudlow C, Hopewell JC, Cheng YC, Fornage M, Ikram MA, Malik R, Bevan S, Thorsteinsdottir U, et al. Genetic risk factors for ischaemic stroke and its subtypes (the METASTROKE collaboration): a meta-analysis of genome-wide association studies. Lancet Neurol. 2012;11:951–962. doi: 10.1016/S1474-4422(12)70234-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.DIAGRAM. 2016 http://diagram-consortium.org/index.html.
- 16.Global Lipids Genetics Consortium. Willer CJ, Schmidt EM, Sengupta S, Peloso GM, Gustafsson S, Kanoni S, Ganna A, Chen J, Buchkovich ML, Mora S, et al. Discovery and refinement of loci associated with lipid levels. Nat Genet. 2013;45:1274–1283. doi: 10.1038/ng.2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Locke AE, Kahali B, Berndt SI, Justice AE, Pers TH, Day FR, Powell C, Vedantam S, Buchkovich ML, Yang J, Croteau-Chonka DC, et al. Genetic studies of body mass index yield new insights for obesity biology. Nature. 2015;518:197–206. doi: 10.1038/nature14177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shungin D, Winkler TW, Croteau-Chonka DC, Ferreira T, Locke AE, Magi R, Strawbridge RJ, Pers TH, Fischer K, Justice AE, Workalemahu T, et al. New genetic loci link adipose and insulin biology to body fat distribution. Nature. 2015;518:187–196. doi: 10.1038/nature14132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fall T, Hagg S, Magi R, Ploner A, Fischer K, Horikoshi M, Sarin AP, Thorleifsson G, Ladenvall C, Kals M, Kuningas M, et al. The role of adiposity in cardiometabolic traits: a Mendelian randomization analysis. PLoS medicine. 2013;10:e1001474. doi: 10.1371/journal.pmed.1001474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nordestgaard BG, Palmer TM, Benn M, Zacho J, Tybjaerg-Hansen A, Davey Smith G, Timpson NJ. The effect of elevated body mass index on ischemic heart disease risk: causal estimates from a Mendelian randomisation approach. PLoS medicine. 2012;9:e1001212. doi: 10.1371/journal.pmed.1001212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Holmes MV, Lange LA, Palmer T, Lanktree MB, North KE, Almoguera B, Buxbaum S, Chandrupatla HR, Elbers CC, Guo Y, Hoogeveen RC, et al. Causal effects of body mass index on cardiometabolic traits and events: a Mendelian randomization analysis. Am J Hum Genet. 2014;94:198–208. doi: 10.1016/j.ajhg.2013.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hagg S, Fall T, Ploner A, Magi R, Fischer K, Draisma HH, Kals M, de Vries PS, Dehghan A, Willems SM, Sarin AP, et al. Adiposity as a cause of cardiovascular disease: a Mendelian randomization study. Intern J Epidemiol. 2015;44:578–586. doi: 10.1093/ije/dyv094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fall T, Hagg S, Ploner A, Magi R, Fischer K, Draisma HH, Sarin AP, Benyamin B, Ladenvall C, Akerlund M, Kals M, et al. Age- and sex-specific causal effects of adiposity on cardiovascular risk factors. Diabetes. 2015;64:1841–1852. doi: 10.2337/db14-0988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morris AP, Voight BF, Teslovich TM, Ferreira T, Segre AV, Steinthorsdottir V, Strawbridge RJ, Khan H, Grallert H, Mahajan A, Prokopenko I, et al. Large-scale association analysis provides insights into the genetic architecture and pathophysiology of type 2 diabetes. Nat Genet. 2012;44:981–990. doi: 10.1038/ng.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Voight BF, Kang HM, Ding J, Palmer CD, Sidore C, Chines PS, Burtt NP, Fuchsberger C, Li Y, Erdmann J, Frayling TM, et al. The metabochip, a custom genotyping array for genetic studies of metabolic, cardiovascular, and anthropometric traits. PLoS genetics. 2012;8:e1002793. doi: 10.1371/journal.pgen.1002793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. Bmj. 2003;327:557–560. doi: 10.1136/bmj.327.7414.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.mrrobust [computer program] 2016 https://github.com/remlapmot/mrrobust/blob/master/mrrobust.pkg.
- 28.StataCorp [computer program] College Station; Texas, USA: [Google Scholar]
- 29.The Comprehensive R Archive Network. 2013 gtx [computer program] [Google Scholar]
- 30.R: A language and environment for statistical computing [computer program] Vienna; Austria: 2013. [Google Scholar]
- 31.Burgess S, Dudbridge F, Thompson SG. Combining information on multiple instrumental variables in Mendelian randomization: comparison of allele score and summarized data methods. Stat Med. 2016;35:1880–1906. doi: 10.1002/sim.6835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brion MJ, Shakhbazov K, Visscher PM. Calculating statistical power in Mendelian randomization studies. Intern J Epidemiol. 2013;42:1497–1501. doi: 10.1093/ije/dyt179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Prospective Studies C, Whitlock G, Lewington S, Sherliker P, Clarke R, Emberson J, Halsey J, Qizilbash N, Collins R, Peto R. Body-mass index and cause-specific mortality in 900 000 adults: collaborative analyses of 57 prospective studies. Lancet. 2009;373:1083–1096. doi: 10.1016/S0140-6736(09)60318-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Emdin C, Khera A, Natarajan P, Klarin D, Zekavat S, Hsiao A, Kathiresan S. Genetic Association of Waist-to-Hip Ratio With Cardiometabolic Traits, Type 2 Diabetes, and Coronary Heart Disease. Jama. 2017;317:626–634. doi: 10.1001/jama.2016.21042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wurtz P, Wang Q, Kangas AJ, Richmond RC, Skarp J, Tiainen M, Tynkkynen T, Soininen P, Havulinna AS, Kaakinen M, Viikari JS, et al. Metabolic signatures of adiposity in young adults: Mendelian randomization analysis and effects of weight change. PLoS medicine. 2014;11:e1001765. doi: 10.1371/journal.pmed.1001765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Truong QA, Ptaszek LM, Charipar EM, Taylor C, Fontes JD, Kriegel M, Irlbeck T, Toepker M, Schlett CL, Bamberg F, Blankstein R, et al. Performance of electrocardiographic criteria for left ventricular hypertrophy as compared with cardiac computed tomography: from the Rule Out Myocardial Infarction Using Computer Assisted Tomography trial. J hypertens. 2010;28:1959–1967. doi: 10.1097/HJH.0b013e32833b49cb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pischon T, Boeing H, Hoffmann K, Bergmann M, Schulze MB, Overvad K, van der Schouw YT, Spencer E, Moons KG, Tjonneland A, Halkjaer J, et al. General and abdominal adiposity and risk of death in Europe. N Engl J Med. 2008;359:2105–2120. doi: 10.1056/NEJMoa0801891. [DOI] [PubMed] [Google Scholar]
- 38.Despres JP. Body fat distribution and risk of cardiovascular disease: an update. Circulation. 2012;126:1301–1313. doi: 10.1161/CIRCULATIONAHA.111.067264. [DOI] [PubMed] [Google Scholar]
- 39.Ng M, Fleming T, Robinson M, Thomson B, Graetz N, Margono C, Mullany EC, Biryukov S, Abbafati C, Abera SF, Abraham JP, et al. Global, regional, and national prevalence of overweight and obesity in children and adults during 1980-2013: a systematic analysis for the Global Burden of Disease Study 2013. Lancet. 2014;384:766–781. doi: 10.1016/S0140-6736(14)60460-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Trichopoulou A, Gnardellis C, Lagiou A, Benetou V, Naska A, Trichopoulos D. Physical activity and energy intake selectively predict the waist-to-hip ratio in men but not in women. Am J Clin Nutr. 2001;74:574–578. doi: 10.1093/ajcn/74.5.574. [DOI] [PubMed] [Google Scholar]
- 41.Aschard H, Vilhjalmsson BJ, Joshi AD, Price AL, Kraft P. Adjusting for heritable covariates can bias effect estimates in genome-wide association studies. Am J Hum Genet. 2015;96:329–339. doi: 10.1016/j.ajhg.2014.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Silverwood RJ, Holmes MV, Dale CE, Lawlor DA, Whittaker JC, Smith GD, Leon DA, Palmer T, Keating BJ, Zuccolo L, Casas JP, et al. Testing for non-linear causal effects using a binary genotype in a Mendelian randomization study: application to alcohol and cardiovascular traits. Intern J Epidemiol. 2014;43:1781–1790. doi: 10.1093/ije/dyu187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Aune D, Sen A, Prasad M, Norat T, Janszky I, Tonstad S, Romundstad P, Vatten LJ. BMI and all cause mortality: systematic review and non-linear dose-response meta-analysis of 230 cohort studies with 3.74 million deaths among 30.3 million participants. Bmj. 2016;353:i2156. doi: 10.1136/bmj.i2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Afzal S, Tybjaerg-Hansen A, Jensen GB, Nordestgaard BG. Change in Body Mass Index Associated With Lowest Mortality in Denmark, 1976-2013. Jama. 2016;315:1989–1996. doi: 10.1001/jama.2016.4666. [DOI] [PubMed] [Google Scholar]
- 45.Daniel RM, De Stavola BL, Cousens SN, Vansteelandt S. Causal mediation analysis with multiple mediators. Biometrics. 2015;71:1–14. doi: 10.1111/biom.12248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Burgess S, Daniel RM, Butterworth AS, Thompson SG, Consortium EP-I Network Mendelian randomization: using genetic variants as instrumental variables to investigate mediation in causal pathways. Intern J Epidemiol. 2015;44:484–495. doi: 10.1093/ije/dyu176. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.