Abstract
Gene trap mutagenesis in mouse embryonic stem cells has been widely used for genome-wide studies of mammalian gene function. However, while large numbers of genes can be disrupted, individual mutations may suffer from limitations due to the structure and/or placement of targeting vector. To extend the utility of gene trap mutagenesis, replaceable 3′ [or poly(A)] gene trap vectors were developed that permit sequences inserted in individual entrapment clones to be engineered by Cre-mediated recombination. 3′ traps incorporating different drug resistance genes could be readily exchanged, simply by selecting for the drug-resistance gene of the replacement vector. By substituting different 3′ traps, we show that otherwise identical fusion genes containing a large first exon (804 nt) are not expressed at appreciably lower levels than genes expressing small first exons (384 and 151 nt). Thus, size appears to have less effect on the expression and processing of first exons than has been reported for internal exons. Finally, a retroviral poly(A) trap (consisting of a RNA polymerase II promoter, a neomycin-resistance gene, and 5′-splice site) typically produced mutagenized clones in which vector sequences spliced to the 3′-terminal exons of cellular transcription units, suggesting strong selection for fusion transcripts that evade nonsense-mediated decay. The efficient exchange of poly(A) traps should greatly extend the utility of mutant libraries generated by gene entrapment and provides new strategies to study the rules that govern the expression of exons inserted throughout the genome.
The number and diversity of genes identified by the mammalian genome projects suggest that considerable biology remains to be characterized on a molecular level and have provided the impetus for developing genome-wide strategies to characterize gene functions important in normal and disease processes. Tagged sequence mutagenesis uses gene entrapment vectors to disrupt genes in murine embryonic stem (ES) cells combined with rapid, DNA-sequence-based screens to characterize the genes disrupted in each ES cell (Hicks et al. 1997; Salminen et al. 1998; Stanford et al. 1998; Wiles et al. 2000; Hansen et al. 2003; Stryke et al. 2003; Chen et al. 2004). The resulting libraries of mutant stem cell clones provide large numbers of mutations, characterized at the nucleotide level, that can be transmitted into the mouse germ line (Skarnes et al. 2004).
A variety of gene entrapment vectors have been developed, each with specialized features that address issues of gene targeting and mutagenicity, identification of disrupted genes, and monitoring of disrupted gene expression. However, no single design appears to be best suited for all applications. For example, 5′ gene traps, which rely on splicing of cellular transcripts to activate expression of a drug-resistance gene, disrupt only expressed genes (Skarnes et al. 1992). The U3 retroviral gene traps also target only expressed genes and rely on splicing of cellular transcripts to splice acceptor sites in the flanking cellular DNA (Osipovich et al. 2004). 3′ or poly(A) traps can target nonexpressed genes, but their mutagenic potential varies (Niwa et al. 1993). Vectors with additional features (e.g., to create conditional gene knockouts or to activate expression of cellular genes) are also feasible but often at the expense of other features that would be useful for large-scale mutagenesis. Finally, inserted vector sequences may induce secondary phenotypes unrelated to the disruption of the occupied gene (Al-Shawi et al. 1988; Braun et al. 1990; Kim et al. 1992; Olsen et al. 1996; Pham et al. 1996; Seidl et al. 1999; Huang et al. 2000).
A conceptual approach to these problems involves replaceable gene trap vectors that would allow genes disrupted in individual ES cell clones to be engineered so as to create features not present in the original occupied locus (Araki et al. 1999; Hardouin and Nagy 2000). The replaceable cassettes may contain heterotypic recombination sequences for the Cre, Flp, or ΦC31 DNA site-specific recombinases that do not recombine with each other, but they recombine with other sites of the same type. This allows sequences of the targeting vector to be replaced with DNA introduced into the cells by a process known as recombinase-mediated cassette exchange (RMCE) (Bethke and Sauer 1997; Bouhassira et al. 1997; Seibler et al. 1998; Feng et al. 1999; Soukharev et al. 1999; Araki et al. 2002; Lauth et al. 2002; Belteki et al. 2003). In principle, the replacement cassettes can be used to engineer entrapment loci for several applications such as removal of the entrapment cassette within the vector to restore expression of the occupied cellular gene, creation of an allelic series of mutations, insertion of genes to be expressed from the promoter of the disrupted cellular genes, and modifications resulting in the expression of affinity-tagged fusion proteins for studies of protein-protein interactions. Replaceable gene traps can also be used to identify optimal sequence elements for gene entrapment without the need to construct new vectors.
The present study describes the development and use of replaceable 3′ or poly(A) gene trap vectors. As with previously described 3′ gene traps (Niwa et al. 1993; Yoshida et al. 1995; Salminen et al. 1998; Zambrowicz et al. 1998), gene entrapment involves selection for drug-resistant clones in which the Neo or Zeo sequences splice to downstream exons of cellular genes. We report that most inserts spliced to the 3′-terminal exons of previously characterized transcription units, suggesting the gene-entrapment process selects for the expression of fusion transcripts that evade nonsense-mediated decay (Baker and Parker 2004; Maquat 2004). We also show that 3′ traps flanked by heterotypic loxP cleavage sites (Lee and Saito 1998) and that incorporate different drug-resistance genes can be readily replaced via RMCE simply by selecting for the drug-resistance marker of the replacement vector.
Previous poly(A) traps have used relatively large drug-resistance genes contained within a single 5′-exon to select for inserts capable of splicing to the 3′-ends of cellular genes. This poses a potential problem, since the first exons of cellular genes are typically much smaller, with mean lengths of 151 and 384 nt for untranslated and partially translated exons, respectively (Davuluri et al. 2001). The strong preference for small exons within cellular genes (with the exception of 3′-terminal exons) is thought to result from exon definition, a process by which exons are recognized as functional units by interactions between protein complexes that bind to the ends of the exon (Berget 1995) or between the 5′-CAP and proximal downstream 5′ splice site (Lewis et al. 1996). Increasing the size of internal exons has been shown to result in exon skipping and reduced levels of gene expression (Sterner et al. 1996); however, studies to investigate the effects of first exon size have not been reported. We therefore tested whether 3′ gene traps using smaller 5′-exons might be expressed more efficiently than vectors with large 5′-exons. Otherwise identical fusion genes were generated by RMCE that expressed Neo or Zeo genes with 5′-exons of 804, 375, and 161 nt. However, first exon size did not appear to have a significant effect on the expression of fusion genes generated by gene entrapment.
Results
Replaceable 3′ gene trap vectors
Replaceable 3′ gene trap vectors incorporating neomycin (Neo) or zeocin (Zeo) resistance genes were constructed to assess the effects of 5′-exon size on the expression of fusion transcripts generated by gene entrapment and to compare the relative strengths of the Pol II and PGK promoters. LNPAT1 (Fig. 1) consists of the gene trap elements from the previously described RET vector (Ishida and Leder 1999) (with the longer Pol II promoter sequence [Matsuda et al. 2004]) placed between heterotypic recognition sites (loxP and lox5171) for the Cre DNA site-specific recombinase (Lee and Saito 1998). An RNA instability region from the human GM-CSF gene is intended to suppress the expression of unspliced transcripts, providing further selection for cells that express properly spliced fusion transcripts. Coding sequences for enhanced green fluorescent protein (EGFP) preceded by an internal ribosome entry site (IRES) provide a reporter gene to monitor expression of disrupted cellular genes. Transcripts from occupied cellular genes that splice to the EGFP sequence terminate at downstream polyadenylation signals, thus disrupting their expression. An HSV Tk gene provides a means to select for the loss of vector sequences, enriching for Cre-mediated replacement events.
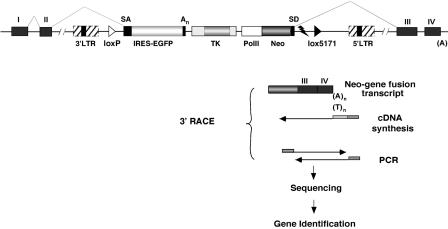
Figure 1.
Tagged sequence mutagenesis with the LNPAT1 gene trap vector. An LNPAT1 provirus is shown integrated after the second exon (black) of a hypothetical gene. The Neo-resistance gene (dark gray shading), expressed from the RNA polymerase II (PolII) promoter, splices to downstream exons (III and IV). Transcripts of the disrupted gene, containing exons I and II, splice to coding sequences for the enhanced green fluorescent protein (EGFP) preceded by the ECMV internal ribosome entry site (IRES) in cells where the occupied gene is expressed. Cellular sequences appended to the 3′-end of virus-cell fusion transcripts are amplified by 3′-RACE and sequenced. The resulting sequence tags identify genes disrupted by the provirus and are subcloned for later use, for example, to prepare DNA microarrays. The resulting library of mutant ES cell clones, cryopreserved in liquid nitrogen, provides a source of mutant alleles that may be transmitted into the mouse germ line. A Herpes virus thymidine kinase gene (TK) and RNA destabilization element (flash symbol) are also present. Heterospecific loxP sites (loxP and loxP5171) allow vector sequences to be replaced by Cre-mediated cassette exchange (see text for details).
A set of plasmid-based 3′ gene traps was also constructed as shown in Figure 2. PolIINeo contains the same entrapment cassette as LNPAT1. NeoPTC is similar to PolNeo, but the drug-resistance gene is expressed from the phosphoglycerate kinase (PGK) promoter (Adra et al. 1987), and the poly(A) trap cassette is flanked by loxP and lox5171 sites (Fig. 2B). ZeoPTC and IntZeoPTC are identical to NeoPTC except the neomycin-resistance gene was replaced with zeocin and intron-containing zeocin-resistance genes, respectively (Fig. 2C,D). The Neo and Zeo drug-resistance genes in NeoPTC and ZeoPTC are thus expressed from 5′-exons of 804 and 375 bp, respectively. A 140-bp synthetic intron splits the Zeo coding sequence in IntZeoPTC into two exons of 161 and 214 bp.
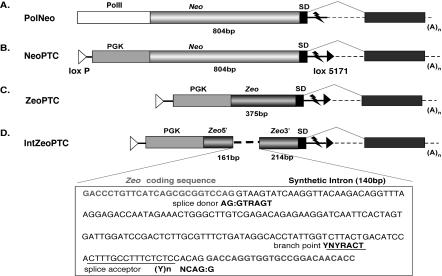
Figure 2.
Replaceable 3′ gene entrapment cassettes. (A,B) neomycin- and (C,D) zeocin-resistance genes ending at a 3′-splice site (SD) and RNA destabilization sequence (flash symbol) were cloned downstream of the (A) RNA polymerase II (Pol II) or (B,C,D) phosphoglycerol kinase (PGK) promoters. IntZeoPTC contains a synthetic intron inserted within the Zeocin-resistance gene (D). After gene entrapment, vector sequences splice to downstream exons of cellular genes (black boxes). The sizes of 5′-exons containing Neo and Zeo sequences is indicated in base pairs (bp). Heterospecific loxP sites (loxP and lox5171) allow vector sequences to be replaced by Cre-mediated cassette exchange.
Tagged sequence mutagenesis with LNPAT1
LNPAT1 was introduced into the Phoenix retrovirus packaging line (Grignani et al. 1998) and cell supernatants were used to infect ES cells at a low multiplicity of infection. The ability of LNPAT1 to work as a 3′ gene trap was confirmed by Northern blot and 3′-RACE analysis of RNAs isolated from G418-resistant ES clones. Transcripts that hybridized to a Neo probe varied in size as expected for fusion transcripts generated by splicing to different cellular genes (data not shown). Cellular sequences appended to Neo fusion transcripts were amplified by 3′-RACE, and then cloned and sequenced (Fig. 1). The sequences were compared with the nonredundant, EST, and mouse genome databases by using the BLASTN program (Altschul et al. 1990). Search results with bit scores higher than 100 were considered significant, and were used to locate 3′-RACE products on the annotated mouse genome sequence. In most cases, this provided information about the identity and structure of the occupied genes, including the locations of exons and coding sequences.
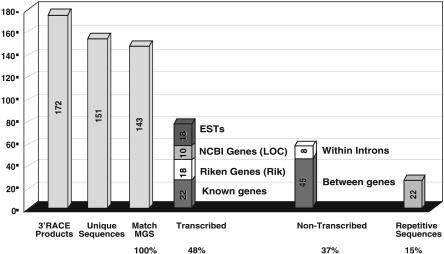
Fusion transcripts were successfully cloned from 171 of 288 (62%) ES cell clones and verified by DNA sequence analysis. Then 20 sequences were derived from sister clones from the same culture dish and were discarded. The cellular sequences in another eight RACE products were too short to produce high scoring matches. Of the RACE products from the 143 remaining clones, 68 (48%) matched transcribed sequences, for example, exons (Fig. 3). Of these, 22 matched previously characterized genes, 28 matched Riken cDNA and NCBI predicted genes (18 and 10, respectively), and 18 matched unannotated EST contigs. Interestingly, 90% of the fusion transcripts that spliced to exons of characterized transcription units spliced to the last exon of the gene. Of the sequence tags, 15% matched repetitive sequences and were therefore difficult to place on the mouse genome, and 37% of the RACE products matched sequences located either within introns or between genes. These matches were located either in introns of known genes or between the annotated genes. These RACE products could represent splicing to sequences capable of functioning as cryptic 3′-exons, to nonannotated, alternative exons or to sequences from novel, previously uncharacterized genes.
Figure 3.
Tagged-sequences mutagenesis with the LNPAT1 3′ gene trap. Here 172 vector-fusion transcripts cloned by 3′-RACE corresponded to genes disrupted in ES cells by the LNPAT1 vector as determined by their DNA sequence (confirmed 3′-RACE products). Of these, 151 provided unique sequence tags (i.e., they were not derived from sister clones from the same culture dish), and 143 matched the mouse genome sequence (MGS). Based on the mouse genome sequence annotation, 68 (48%) of the RACE sequences matched transcribed sequences (i.e., exons), corresponding to 22 previously characterized genes, 18 Riken cDNAs, 10 transcription units identified by gene prediction software (NCBI LOC genes), and 18 EST contigs not included among the Entrez gene annotations (ESTs). Of the remaining 53 sequence tags, eight matched intron sequences within annotated genes, 45 matched genomic sequences not contained within annotated transcription units and therefore may be between genes, and 22 contained repetitive sequences and could not be placed on the genome sequence.
Gene entrapment with plasmid-based 3′ gene traps
Gene entrapment by the PolIINeo, NeoPTC, ZeoPTC, and IntZeoPTC vectors was tested following electroporation into both NIH3T3 and ES cells, and all appeared to function as efficient poly(A) traps. The plasmids generated drug-resistant clones with similar frequencies (0.3-2/μg plasmid DNA); Neo and Zeo transcripts expressed in drug-resistant clones varied in size based on Northern blot analysis, as expected for vector-gene fusion transcripts containing different amounts of appended cell-derived RNA (Supplemental Fig. S1); and fusion transcripts cloned by 3′-RACE all had the expected structures (data not shown). IntZeoPTC fusion transcripts also appeared to be properly spliced (Supplemental Fig. S2), suggesting that the artificial intron was efficiently processed. Zeo-containing fusion transcripts appeared to be expressed at higher levels on average than Neo fusion transcripts, and were more efficiently cloned by 3′-RACE (100% of the Zeo-resistant clones [12 clones tested] as compared to 70% of the Neo-resistant clones). While it is not clear whether these differences reflect higher expression levels of Zeo-fusion transcripts or other factors such as more efficient amplification by Zeo-specific primers, the Zeo-based poly(A) gene traps appear to function at least as efficiently as Neo-based entrapment vectors.
Cre-mediated cassette exchange
We hypothesized that it might be possible to exchange 3′ traps incorporating different drug-resistance genes, simply by selecting for the drug-resistance gene of the replacement vector. To test this hypothesis, several replacement experiments were performed as summarized in Table 1. Entrapment clones were cotransfected with a Cre-expression plasmid (CAGGS-Cre) together with the replacement vector, and potential replacement clones were selected in media containing appropriate antibiotic. The selected clones were analyzed by Southern blot hybridization to determine the frequency of correct gene replacement events (Fig. 4; Supplemental Figs. S3 and S4). Proper cassette exchange was also demonstrated by 3′-RACE, which confirmed that the different entrapment cassettes spliced to the same downstream exons (data not shown). In all cases, cassette exchange involving the selection of one poly(A) trap for another was quite efficient, with 12%-93% of the selected clones having the desired replacement (Table 1). However, in studies involving replacement of the LNPAT1 vector, the selection against the Tk gene with gangcyclovir provided no practical benefit (data not shown), even when previously effective conditions were used (Chang et al. 1993).
Table 1.
Efficient exchange of 3′ gene trap vectors
| Gene trap vector | Disrupted gene | Replacement vector | Clone with correct gene replacement (%) |
|---|---|---|---|
| LNPAT1 | 4930562A09Rik | ZeoPTC | 12 |
| ZeoPTC | 4930562A09Rik | NeoPTC | 30 |
| NeoPTC | LOC228098 | ZeoPTC | 89 |
| NeoPTC | LOC228098 | IntZeoPTC | 93 |
| IntZeoPTC | 4933407O12Rik | NeoPTC | 83 |
Gene trap vectors disrupting the indicated genes were replaced by Cremediated cassette exchange. Clones isolated after selecting for the resistance gene carried by the replacement vector were analyzed by Southern blot hybridization. The percentage of clones in which the entrapment vectors were properly replaced is indicated for each experiment.
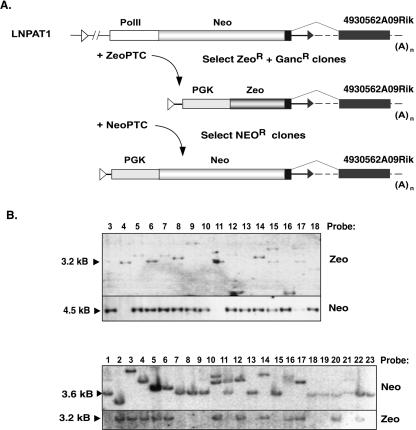
Figure 4.
Cre-mediated cassette exchange at the 4930562A09Rik locus. (A) Sequences of the LNPAT1 provirus inserted in the 4930562A09Rik locus were replaced by the ZeoPTC gene trap cassette, which was then replaced by NeoPTC. (B) Clones containing the proper gene replacements were identified by Southern blot analysis. The replacement of LNAPT1 by ZeoPTC (upper panel) was accompanied by the loss of the 4.5-kb Neo-hybridizing SacI fragment and the presence of a 3.2-kb Zeo-hybridizing fragment (lanes 4,11). SacI sites are located in each LTR and in the Tk gene. The replacement of ZeoPTC by NeoPTC (lower panel) was accompanied by the loss of the 3.2-kb Zeo-hybridizing fragment and the presence of a 3.6-kb Neo-hybridizing fragment (clones 7, 8, 9, 13, 15, 18, 19, 21, and 23).
Fusion gene expression by Pol II and PGK promoters in replacement clones
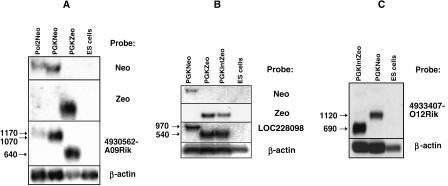
By placing different promoter-marker-splice donor cassettes in the same locus, we were able to compare the effects of different sequences on the expression and processing of otherwise identical fusion genes. A PolIINeo poly(A) trap (LNPAT1) inserted in the 4930562A09Rik gene was replaced first by ZeoPTC, and then the ZeoPTC cassette was replaced by NeoPTC (Table 1; Fig. 4). The resulting clones were used to compare the relative strengths of Pol II and PGK promoters expressing Neo-4930562A09Rik fusion transcripts and to compare the relative levels of expression by PGKZeo and PGKNeo poly(A) traps that splice to the same downstream exon (Fig. 5A). The steady-state levels of Neo-4930562A09Rik fusion transcripts expressed from the PGKNeo poly(A) trap were approximately four times higher than those expressed from the PolIINeo poly(A) trap (normalized to β-actin), providing a measure of the relative strengths of the PGK and Pol II promoters. Moreover, Zeo fusion transcripts were expressed at approximately twofold higher levels than Neo fusion transcripts. This suggests that poly(A) traps incorporating the Zeo 5′-exon may be expressed at higher levels than otherwise identical Neo-based vectors.
Figure 5.
Fusion gene expression in replacement clones. RNA from cell clones expressing poly(A) traps inserted in the (A) 4930562A09Rik,(B) LOC228098, and (C) 4933407O12Rik genes were analyzed by Northern blot hybridization, probing with gene-specific, Neo-specific, and Zeo-specific probes. The clones in A were obtained by replacing an LNPAT1 poly(A) trap (Pol2Neo) inserted in the 4930562A09Rik gene with ZeoPTC (PGKZeo), and then by replacing the ZeoPTC cassette with NeoPTC (PGKNeo). The clones in B were obtained by replacing a NeoPTC poly(A) trap in the LOC228098 gene with either ZeoPTC (PGKZeo) or IntZeoPTC (PGKIntZeo). The clones in C were obtained by replacing a IntZeoPTC (PGKIntZeo) poly(A) trap in the 4933407O12Rik gene with NeoPTC (PGKNeo). All clones expressed fusion transcripts of the expected size as indicated on the left. None of the targeted genes appeared to be expressed in the parental stem cells (ES), confirming the ability of poly(A) traps to disrupt nonexpressed genes.
First exon size does not affect levels of fusion gene expression
We considered the possibility that Zeo-based poly(A) traps might be expressed more efficiently than the Neo-based vectors because the drug-resistance gene is expressed as a smaller first exon, 375 versus 804 nt. We therefore compared the expression of other Neo and Zeo fusion genes generated by replacing a NeoPTC vector (Fig. 2B) inserted in the LOC228098 gene with the ZeoPTC and IntZeoPTC poly(A) traps. The Zeo sequence in ZeoPTC is incorporated into a single, 375-nt 5′-exon, while in IntZeoPTC, it is split by an artificial intron into exons of 161 and 214 nt (Fig. 2C,D). Steady-state levels of fusion transcripts expressed in the original NeoPTC-LOC228098 clone and in the ZeoPTC-LOC228098 and IntZeoPTC-LOC228098 replacement clones were analyzed by Northern blot analysis (Fig. 5B). As before, the levels of Zeo fusion transcripts were higher (1.3×) than Neo transcripts when normalized to a β-actin control. However, no differences were observed in the levels of ZeoPTC and IntZeoPCT transcripts.
Although the expression of fusion genes generated with Neo-based poly(A) traps increased when the targeting vector was replaced with a zeocin-resistance gene, the magnitude of the effect was relatively modest, 1.3- to 2-fold. A potential problem is that gene entrapment with Neo-based vectors may select for specific types of inserts able to express the large, Neo-containing first exon, thus circumventing the potential benefit of a smaller first exon. Precedence for this possibility has been observed with internal exons, where the effect of exon size is significantly reduced when the exon is surrounded by small introns (Sterner et al. 1996).
To explore this issue, we first compared the distances between Zeo- and Neo-poly(A) traps and the downstream exons to which they splice. Genomic DNA from entrapment clones was amplified by nested PCR, using primers to the drug-resistance genes and downstream exons as identified by 3′-RACE (data not shown). While the experiments provided information on only a limited number of inserts, the data suggest that the average intervening sequence downstream of Neo- and Zeo-based poly(A) traps is similar in size.
We next replaced an IntZeoPTC vector, which originally disrupted the 4933407O12Rik gene, with NeoPTC. This fusion gene was selected because IntZeoPTC splices over 2 kb to the last exon of 4933407O12Rik. If smaller 5′-exons are expressed more efficiently, then fusion genes disrupted by IntZeoPTC might be expressed at significantly reduced levels following replacement with NeoPTC. However, Neo fusion transcripts were expressed at only half the level of Zeo fusion transcripts (Fig. 5C).
Stability of Neo and Zeo fusion transcripts
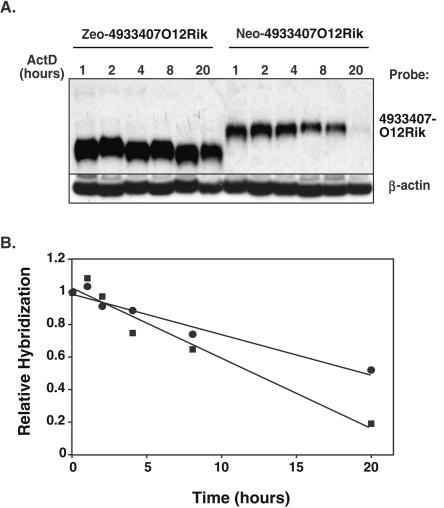
We next tested whether differences in the levels of Neo and Zeo fusion transcripts might result from differences in RNA stability. Cells expressing either IntZeoPTC-4933407O12Rik or NeoPTC-4933407O12Rik fusion genes were switched to media containing 50 μg/mL actinomycin D, and the levels of 4933407O12Rik fusion transcripts were monitored at various times thereafter (Fig. 6). The levels of NeoPTC-4933407O12Rik transcripts declined at about twice the rate of IntZeoPTC-4933407O12Rik transcripts, suggesting that differences in the expression of otherwise identical Zeo and Neo fusion transcripts are due to differences in RNA stability. Consequently, first exon size appears to have no significant effect on the expression of fusion genes generated by gene entrapment.
Figure 6.
Stability of Neo- and Zeo-4933407O12Rik fusion transcripts. (A) Northern blot analysis of RNA decay in actinomycin D-treated cells. RNAs were extracted from actinomycin D-treated cells expressing Neo-4933407O12Rik and Zeo-4933407O12Rik fusion transcripts, and the Northern blots were hybridized to a 4933407O12Rik-specific probe. RNAs were also probed with a β-actin probe to assess relative levels of RNA in each sample. (B) Kinetics of fusion transcript turnover. Relative levels of Neo-4933407O12Rik (squares) and Zeo-4933407O12Rik (circles) fusion transcripts in A were measured by PhosphorImager densitometry and plotted as a function of time. Zeo-4933407O12Rik fusion transcripts were twice as stable as the otherwise identical Neo fusion transcripts.
Discussion
In the present study, replaceable 3′ [or poly(A)] gene trap vectors were developed and used to study factors that influence the expression of 5′-exons. We show that 3′ traps incorporating different drug-resistance genes can be readily exchanged simply by selecting for the drug-resistance marker of the replacement vector. By substituting different 3′ traps, we show that fusion genes containing a relatively large first exon (804 nt) are not expressed at appreciably lower levels than genes expressing small first exons (384 and 151 nt). Moreover, vector sequences in most entrapment clones spliced to 3′-terminal exons of previously characterized genes. This suggests that gene entrapment by the LNPAT1 vector strongly selects for fusion transcripts capable of evading nonsense-mediated decay.
Cassette exchange involving the selection of one poly(A) trap for another was remarkably efficient, with 10%-90% of the selected clones having the desired replacement. The background of improperly targeted clones presumably results from Cre-independent integration at sites in the genome capable of providing functional 3′-exons as required for expression of the incoming poly(A) trap. Since ∼10% of randomly integrated poly(A) traps are expressed, we estimate that RMCE was responsible for the integration of 1%-10% of transfected replacement vectors in the genome.
LNPAT1 preferentially targeted genes by inserting into the last intron and expressed fusion transcripts that spliced to a single downstream exon. This is in contrast to the 5′ gene traps (i.e., a promoterless 3′-exon) that are preferentially expressed by inserts positioned near the 5′-ends of cellular genes. These results are consistent with a mechanism involving selection for fusion transcripts that do not activate nonsense-mediated decay (NMD). NMD is triggered by termination codons positioned more than 50 nt upstream from a splice junction, thus providing a mechanism to eliminate transcripts that might express deleterious truncated proteins resulting from nonsense mutations (Baker and Parker 2004; Maquat 2004). The 3′ gene traps used in the present study can theoretically splice to single downstream exons without engaging NMD, because the termination codons for the Neo and Zeo drug-resistance genes are positioned <50 nt upstream of the splice donor site. However, fusion transcripts incorporating multiple downstream exons are expected to activate NMD. The apparent selection against clones expressing multiply spliced fusion transcripts implies that NMD can be activated within a wide variety of cellular genes. Moreover, since only modest levels of Neo expression are sufficient for drug resistance, the predicted degradation of multiply spliced Neo transcripts appears to be relatively efficient. Of the 50 fusion transcripts that involved well-annotated genes, only four contained multiple cellular exons. It will be interesting to determine if the appended sequences contain elements capable of suppressing NMD.
Although LNPAT1 preferentially targets the last intron of cellular genes, the provirus is generally positioned in the middle of the occupied transcription unit, because of the relatively large size of 3′-terminal exons. For example, the average gene disrupted by LNPAT1 encoded 1268 nt of exon sequence of which an average of 596 nt of exon sequence was located downstream of the provirus. These results are similar to those that have been described for the related RET poly(A) trap vector (Matsuda et al. 2004), but are at odds with studies involving VICTR3 and VICTR20 in which 44% of the integration events were reported to be near the 5′-ends of cellular genes (Zambrowicz et al. 1998). Additional experiments will be required to identify factors that may influence the expression of fusion transcripts with multiple downstream exons.
The present study raises important issues with regard to the use of poly(A) entrapment vectors for large-scale mutagenesis in murine ES cells. First, the ability of poly(A) traps to disrupt cellular gene expression relies on splicing between cellular transcripts and a splice acceptor-reporter gene cassette located at the 5′-end of the provirus. While the vectors are mutagenic in that they block the expression of cellular exons downstream of the provirus, vectors inserting downstream of most or all proteincoding sequences may not disrupt cellular gene expression. Our results indicate that the mutagenic potential of poly(A) entrapment vectors may be enhanced by modifications that enable 3′ fusion transcripts to evade nonsense mediated decay or that destabilize 5′ fusion transcripts.
Second, our results illustrate the use of RCME to engineer entrapment loci, thereby creating features not present in the original targeting vector. Potential applications include removal of the entrapment cassette within the vector sequences to restore expression of the occupied cellular gene, creation of an allelic series of mutations, insertion of genes to be expressed from the promoter of the disrupted cellular genes, and modifications resulting in the expression of affinity-tagged fusion proteins for studies of protein-protein interactions. In short, post-entrapment genome engineering can greatly extend the utility of libraries of ES cell clones generated by gene entrapment.
Finally, the present study illustrates how replaceable gene traps can be used to optimize features of vectors important for tagged sequence mutagenesis (e.g., that determine mutagenicity or levels of fusion gene expression). Sequence elements can be tested side by side in otherwise identical fusion genes. For retrovirus-based vectors, the process eliminates the time-consuming step of preparing new viruses for each construct.
Methods
Construction of poly(A) trap vectors
The LNPAT1 vector was made by PCR-mediated subcloning of a part of the Moloney mouse leukemia virus 3′-LTR (XbaIU3-RU5) sequence from the pBabe vector (Morgenstern and Land 1990) and the following sequences from the pRET vector (Ishida and Leder 1999): splice acceptor (Bcl2 intron2/exon3)-IRES-EGFP-pA (bovine growth hormone gene), HSV thymidine kinase cassette (promoter-gene-polyA site), RNA polymerase II promoter-neomycinphospho-transpherase gene-splice donor (Hprt gene exon 8/intron 8)-mRNA instability signal (human GM-CSF), and 5′-retrovirus sequences. Wild-type LoxP site and mutant Lox5171 (Lee and Saito 1998) were made by oligonucleotide annealing and cloned in between the retrovirus backbone and gene trapping sequences.
The PolNeo vector was made by subcloning into the pBlue-Script vector (Stratagene) of a BamH1/HindIII fragment containing a Pol2 promoter-neo-splice donor-mRNA instability signal cassette from the pRET vector (Ishida and Leder 1999) (a gift from Philip Leder). The NeoPTC vector was made by, first, replacing the Pol2 promoter in PolNeo vector for the PGK promoter derived by PCR from PGKPuro vector (a gift from Peter Laird, Univ. of Southern California, Los Angeles). Subsequently, the wild-type LoxP site and mutant Lox5171 (Lee and Saito 1998) were made by annealing of oligonucleotides and subcloned at both sides of a gene trap cassette in the NeoPTC vector. The ZeoPTC vector was made by replacement of Neo sequences in the NeoPTC vector for Zeo sequences derived by PCR from the pcDNA3.1/Zeo vector (Invitrogen). A synthetic intron was introduced into the IntZeo-PTC vector by PCR-mediated cloning using primers 1IntZeo (5′-GGATCCTTCTCTGTCTCGACAAGCCCAGTTTCTATTGGTCTCCTTAAACCTGTCTTGTAACCTTGATACTTACCTGGACCGCGCTGATGAACAGGGTC-3′) and 2IntZeo (5′-GGATCCGACTCTTGCGTTTCTGATAGGCACCTATTGGTCTTACTGACATCCACTTTGCCTTTCTCTCCACAGGACCAGGTGGTGC-3′).
Cell culture, gene transfer, and virus production
Mouse embryonic stem cells D3 (Doetschman et al. 1985) were prepared and used as described previously (Hicks et al. 1997). Phoenix Eco cells (Grignani et al. 1998) and NIH3T3 cells were cultured in DMEM with 10% of fetal bovine serum. Retrovirus producer lines were prepared by transfecting packaging cells with virus plasmid constructs by coprecipitation with calcium phosphate and selected in 2 mg/mL G418 (Sigma) for 7 d. Virus production by individual clones was measured as NeoR colony-forming units after infection on NIH3T3 cells (Ausubel et al. 1999). Supernatants from producer lines with titers of ∼3 × 103 CFU/mL were used to infect ES cells as described (Hicks et al. 1997).
Plasmid poly(A) entrapment vectors were linearized and introduced into ES cells by electroporation. In all, 107 cells were suspended with DNA in 0.5 mL of PBS in a 0.4-cm electroporation cuvette (BioRad) and then subjected to electric pulse at 400 V, 25 μF in a BioRad Gene Pulser. After 24 h, cells were fed with medium supplemented either with 400 μg/mL of G418 (Sigma) or with 100 μg/mL of Zeocin (Invitrogen). For Cre-mediated cassette exchange experiments, circular replacement vectors were cotransfected with Cre-expression plasmid pCAGGSCre (Araki et al. 1995) at a 1:2 ratio.
3′-RACE
Disrupted genes were identified using 3′-RACE. RNA was extracted with Trizol (Invitrogen) reagent as specified by the manufacturer. cDNA was synthesized using the 3′-RACE kit (Invitrogen) as described in the manufacturer's protocol. The reverse-transcription reaction was held in a 20-μL volume using ∼5 μg of total RNA and AP primer (5′-GGCCACGCGTCGACTAGTACTTTTTTTTTTTTTTTTT-3′). cDNA from Neo-resistant clones was then amplified by two rounds of PCR in a 50-μL reaction using Taq polymerase (Perkin Elmer), first with 2 μL of the above cDNA mix with NeoExt primer (5′-ACCGCTTCCTCGTGCTTTAC-3′) and AUAP primer (5′-GGCCACGCGTCGACTAGTAC-3′), and second with 1 μL of the first PCR mix with NeoInt primer (5′-TCGCCTTCTTGACGAGTTCT-3′) and AUAP primer. cDNA from Zeocin-resistant clones was amplified with ZeoExt (5′-GACCGAGATCGGCGAGCAGCCGTG-3′) and ZeoInt (5′-CGTGCACTTCGTGGCCGAGGAGCA-3′) primers. RT-PCR for IntZeo clones was performed in the same conditions using the 3′-Zeo (5′-CGGGATCCTCAGTCCTGCTCCTCGGCCACGAAGTG 3′) primer for RT and the 3′-Zeo and 5′Zeo (5′-CGCTCGAGATGGCCAAGTTGACCAGTGCCGTTCC-3′) primers for one round of PCR reactions.
Actinomycin D chase experiments and Northern blot analysis
ES cells were grown to subconfluency on 10-cm dishes, and fresh medium was added 12 h prior to blocking of transcription with actinomycin D (50 μg/mL). Actinomycin D was added to the media, and cells were harvested after 0, 1, 2, 4, 8, and 20 h of incubation. RNA was isolated and analyzed by Northern blot hybridization (Ausubel et al. 1999). For detection of the gene-specific fusion transcripts (4930562A09Rik, LOC228098, and 4933407O12Rik), we used the probes derived from cloned 3′-RACE products.
Acknowledgments
We thank Phil Leder and Yasumasa Ishida for providing modified RET vectors, Kimi Araki for the gift of the pCAGGS-Cre plasmid, and Tracy Moore-Jarrett and Abudi Nashabi for technical assistance. This work was supported by Public Health Service Grants (R01NS043952, R01RR13166, and P01HL68744 to H.E.R.). Additional support was provided by Cancer Center Support grant P30CA68485 to the Vanderbilt-Ingram Cancer Center.
Footnotes
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.3258105.
[Supplemental material is available online at www.genome.org. Sequence tags (3′-RACE products) of genes disrupted by the LNPAT1 vector have been deposited in dbGSS (GenBank accession nos. CL943680-CL943829). The following individuals kindly provided reagents, samples, or unpublished information as indicated in the paper: P. Leder, Y. Ishida, and K. Araki.]
References
- Adra, C.N., Boer, P.H., and McBurney, M.W. 1987. Cloning and expression of the mouse pgk-1 gene and the nucleotide sequence of its promoter. Gene 60: 65-74. [DOI] [PubMed] [Google Scholar]
- Al-Shawi, R., Burke, J., Jones, C.T., Simons, J.P., and Bishop, J.O. 1988. A Mup promoter-thymidine kinase reporter gene shows relaxed tissue-specific expression and confers male sterility upon transgenic mice. Mol. Cell. Biol. 8: 4821-4828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul, S.F., Gish, W., Miller, W., Myers, E.W., and Lipman, D.J. 1990. Basic local alignment search tool. J. Mol. Biol. 215: 403-410. [DOI] [PubMed] [Google Scholar]
- Araki, K., Araki, M., Miyazaki, J., and Vassalli, P. 1995. Site-specific recombination of a transgene in fertilized eggs by transient expression of Cre recombinase. Proc. Natl. Acad. Sci. 92: 160-164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araki, K., Imaizumi, T., Sekimoto, T., Yoshinobu, K., Yoshimuta, J., Akizuki, M., Miura, K., Araki, M., and Yamamura, K. 1999. Exchangeable gene trap using the Cre/mutated lox system. Cell Mol. Biol. 45: 737-750. [PubMed] [Google Scholar]
- Araki, K., Araki, M., and Yamamura, K. 2002. Site-directed integration of the cre gene mediated by Cre recombinase using a combination of mutant lox sites. Nucleic Acids Res. 30: e103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausubel, F., Brent, R., Kingston, R., Moore, D., Seidman, J., Smith, J., and Struhl, K. 1999. Current protocols in molecular biology. John Wiley, New York.
- Baker, K.E. and Parker, R. 2004. Nonsense-mediated mRNA decay: Terminating erroneous gene expression. Curr. Opin. Cell Biol. 16: 293-299. [DOI] [PubMed] [Google Scholar]
- Belteki, G., Gertsenstein, M., Ow, D.W., and Nagy, A. 2003. Site-specific cassette exchange and germline transmission with mouse ES cells expressing phiC31 integrase. Nat. Biotech. 21: 321-324. [DOI] [PubMed] [Google Scholar]
- Berget, S.M. 1995. Exon recognition in vertebrate splicing. J. Biol. Chem. 270: 2411-2414. [DOI] [PubMed] [Google Scholar]
- Bethke, B. and Sauer, B. 1997. Segmental genomic replacement by Cre-mediated recombination: Genotoxic stress activation of the p53 promoter in single-copy transformants. Nucleic Acids Res. 25: 2828-2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouhassira, E.E., Westerman, K., and Leboulch, P. 1997. Transcriptional behavior of LCR enhancer elements integrated at the same chromosomal locus by recombinase-mediated cassette exchange. Blood 90: 3332-3344. [PubMed] [Google Scholar]
- Braun, R.E., Lo, D., Pinkert, C.A., Widera, G., Flavell, R.A., Palmiter, R.D., and Brinster, R.L. 1990. Infertility in male transgenic mice: Disruption of sperm development by HSV-tk expression in postmeiotic germ cells. Biol. Reprod. 43: 684-693. [DOI] [PubMed] [Google Scholar]
- Chang, W., Hubbard, C., Friedel, C., and Ruley, H.E. 1993. Enrichment of insertional mutants following retrovirus gene trap selection. Virology 193: 737-747. [DOI] [PubMed] [Google Scholar]
- Chen, W.V., Delrow, J., Corrin, P.D., Frazier, J.P., and Soriano, P. 2004. Identification and validation of PDGF transcriptional targets by microarray-coupled gene-trap mutagenesis. Nat. Genet. 36: 304-312. [DOI] [PubMed] [Google Scholar]
- Davuluri, R.V., Grosse, I., and Zhang, M.Q. 2001. Computational identification of promoters and first exons in the human genome. Nat. Genet. 29: 412-417. [DOI] [PubMed] [Google Scholar]
- Doetschman, T.C., Eistetter, H., Katz, M., Schmidt, W., and Kemler, R. 1985. The in vitro development of blastocyst-derived embryonic stem cell lines: Formation of visceral yolk sac, blood islands and myocardium. J. Embryol. Exp. Morphol. 87: 27-45. [PubMed] [Google Scholar]
- Feng, Y.Q., Seibler, J., Alami, R., Eisen, A., Westerman, K.A., Leboulch, P., Fiering, S., and Bouhassira, E.E. 1999. Site-specific chromosomal integration in mammalian cells: Highly efficient CRE recombinase-mediated cassette exchange. J. Mol. Biol. 292: 779-785. [DOI] [PubMed] [Google Scholar]
- Grignani, F., Kinsella, T., Mencarelli, A., Valtieri, M., Riganelli, D., Lanfrancone, L., Peschle, C., Nolan, G.P., and Pelicci, P.G. 1998. High-efficiency gene transfer and selection of human hematopoietic progenitor cells with a hybrid EBV/retroviral vector expressing the green fluorescence protein. Cancer Res. 58: 14-19. [PubMed] [Google Scholar]
- Hansen, J., Floss, T., Van Sloun, P., Fuchtbauer, E.M., Vauti, F., Arnold, H.H., Schnutgen, F., Wurst, W., von Melchner, H., and Ruiz, P. 2003. A large-scale, gene-driven mutagenesis approach for the functional analysis of the mouse genome. Proc. Natl. Acad. Sci. 100: 9918-9922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardouin, N. and Nagy, A. 2000. Gene-trap-based target site for cre-mediated transgenic insertion. Genesis 26: 245-252. [DOI] [PubMed] [Google Scholar]
- Hicks, G.G., Shi, E.G., Li, X.M., Li, C.H., Pawlak, M., and Ruley, H.E. 1997. Functional genomics in mice by tagged sequence mutagenesis. Nat. Genet. 16: 338-344. [DOI] [PubMed] [Google Scholar]
- Huang, W.Y., Aramburu, J., Douglas, P.S., and Izumo, S. 2000. Transgenic expression of green fluorescence protein can cause dilated cardiomyopathy. Nat. Med. 6: 482-483. [DOI] [PubMed] [Google Scholar]
- Ishida, Y. and Leder, P. 1999. RET: A poly A-trap retrovirus vector for reversible disruption and expression monitoring of genes in living cells. Nucleic Acids Res. 27: e35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, C.G., Epner, E.M., Forrester, W.C., and Groudine, M. 1992. Inactivation of the human β-globin gene by targeted insertion into the β-globin locus control region. Genes & Dev. 6: 928-938. [DOI] [PubMed] [Google Scholar]
- Lauth, M., Spreafico, F., Dethleffsen, K., and Meyer, M. 2002. Stable and efficient cassette exchange under non-selectable conditions by combined use of two site-specific recombinases. Nucleic Acids Res. 30: e115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, G. and Saito, I. 1998. Role of nucleotide sequences of loxP spacer region in Cre-mediated recombination. Gene 216: 55-65. [DOI] [PubMed] [Google Scholar]
- Lewis, J.D., Izaurralde, E., Jarmolowski, A., McGuigan, C., and Mattaj, I.W. 1996. A nuclear cap-binding complex facilitates association of U1 snRNP with the cap-proximal 5′ splice site. Genes & Dev. 10: 1683-1698. [DOI] [PubMed] [Google Scholar]
- Maquat, L.E. 2004. Nonsense-mediated mRNA decay: Splicing, translation and mRNP dynamics. Nat. Rev. Mol. Cell. Biol. 5: 89-99. [DOI] [PubMed] [Google Scholar]
- Matsuda, E., Shigeoka, T., Iida, R., Yamanaka, S., Kawaichi, M., and Ishida, Y. 2004. Expression profiling with arrays of randomly disrupted genes in mouse embryonic stem cells leads to in vivo functional analysis. Proc. Natl. Acad. Sci. 101: 4170-4174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgenstern, J.P. and Land, H. 1990. Advanced mammalian gene transfer: High titre retroviral vectors with multiple drug selection markers and a complementary helper-free packaging cell line. Nucleic Acids Res. 18: 3587-3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa, H., Araki, K., Kimura, S., Taniguchi, S., Wakasugi, S., and Yamamura, K. 1993. An efficient gene-trap method using poly A trap vectors and characterization of gene-trap events. J. Biochem. (Tokyo) 113: 343-349. [DOI] [PubMed] [Google Scholar]
- Olsen, E.N., Arnold, H.-H., Rigby, P.W.J., and Wold, B.J. 1996. Know your neighbors: Three phenotypes in null mutants of the myogenic bHLH gene MRF4. Cell 85: 1-4. [DOI] [PubMed] [Google Scholar]
- Osipovich, A.B., White-Grindley, E.K., Hicks, G.G., Roshon, M.J., Shaffer, C., Moore, J.H., and Ruley, H.E. 2004. Activation of cryptic 3′ splice sites within introns of cellular genes following gene entrapment. Nucleic Acids Res. 32: 2912-2924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham, C.T., MacIvor, D.M., Hug, B.A., Heusel, J.W., and Ley, T.J. 1996. Long-range disruption of gene expression by a selectable marker cassette. Proc. Natl. Acad. Sci. 93: 13090-13095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salminen, M., Meyer, B.I., and Gruss, P. 1998. Efficient poly A trap approach allows the capture of genes specifically active in differentiated embryonic stem cells and in mouse embryos. Dev. Dyn. 212: 326-333. [DOI] [PubMed] [Google Scholar]
- Seibler, J., Schubeler, D., Fiering, S., Groudine, M., and Bode, J. 1998. DNA cassette exchange in ES cells mediated by Flp recombinase: An efficient strategy for repeated modification of tagged loci by marker-free constructs. Biochemistry 37: 6229-6234. [DOI] [PubMed] [Google Scholar]
- Seidl, K.J., Manis, J.P., Bottaro, A., Zhang, J., Davidson, L., Kisselgof, A., Oettgen, H., and Alt, F.W. 1999. Position-dependent inhibition of class-switch recombination by PGK-neor cassettes inserted into the immunoglobulin heavy chain constant region locus. Proc. Natl. Acad. Sci. 96: 3000-3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skarnes, W.C., Auerbach, B.A., and Joyner, A.L. 1992. A gene trap approach in mouse embryonic stem cells: The lacZ reported is activated by splicing, reflects endogenous gene expression, and is mutagenic in mice. Genes & Dev. 6: 903-918. [DOI] [PubMed] [Google Scholar]
- Skarnes, W.C., von Melchner, H., Wurst, W., Hicks, G., Nord, A.S., Cox, T., Young, S.G., Ruiz, P., Soriano, P., Tessier-Lavigne, M., et al. 2004. A public gene trap resource for mouse functional genomics. Nat. Genet. 36: 543-544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soukharev, S., Miller, J.L., and Sauer, B. 1999. Segmental genomic replacement in embryonic stem cells by double lox targeting. Nucleic Acids Res. 27: e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanford, W.L., Caruana, G., Vallis, K.A., Inamdar, M., Hidaka, M., Bautch, V.L., and Bernstein, A. 1998. Expression trapping: Identification of novel genes expressed in hematopoietic and endothelial lineages by gene trapping in ES cells. Blood 92: 4622-4631. [PubMed] [Google Scholar]
- Sterner, D.A., Carlo, T., and Berget, S.M. 1996. Architectural limits on split genes. Proc. Natl. Acad. Sci. 93: 15081-15085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stryke, D., Kawamoto, M., Huang, C.C., Johns, S.J., King, L.A., Harper, C.A., Meng, E.C., Lee, R.E., Yee, A., L'Italien, L., et al. 2003. BayGenomics: A resource of insertional mutations in mouse embryonic stem cells. Nucleic Acids Res. 31: 278-281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiles, M.V., Vauti, F., Otte, J., Fuchtbauer, E.M., Ruiz, P., Fuchtbauer, A., Arnold, H.H., Lehrach, H., Metz, T., von Melchner, H., et al. 2000. Establishment of a gene-trap sequence tag library to generate mutant mice from embryonic stem cells. Nat. Genet. 24: 13-14. [DOI] [PubMed] [Google Scholar]
- Yoshida, M., Yagi, T., Furuta, Y., Takayanagi, K., Kominami, R., Takeda, N., Tokunaga, T., Chiba, J., Ikawa, Y., and Aizawa, S. 1995. A new strategy of gene trapping in ES cells using 3′RACE. Transgenic Res. 4: 277-287. [DOI] [PubMed] [Google Scholar]
- Zambrowicz, B.P., Friedrich, G.A., Buxton, E.C., Lilleberg, S.L., Person, C., and Sands, A.T. 1998. Disruption and sequence identification of 2,000 genes in mouse embryonic stem cells. Nature 392: 608-611. [DOI] [PubMed] [Google Scholar]