Abstract
Genomics data from The Cancer Genome Atlas (TCGA) project has led to the comprehensive molecular characterization of multiple cancer types. The large sample numbers in TCGA offer an excellent opportunity to address questions associated with tumo heterogeneity. Exploration of the data by cancer researchers and clinicians is imperative to unearth novel therapeutic/diagnostic biomarkers. Various computational tools have been developed to aid researchers in carrying out specific TCGA data analyses; however there is need for resources to facilitate the study of gene expression variations and survival associations across tumors. Here, we report UALCAN, an easy to use, interactive web-portal to perform to in-depth analyses of TCGA gene expression data. UALCAN uses TCGA level 3 RNA-seq and clinical data from 31 cancer types. The portal's user-friendly features allow to perform: 1) analyze relative expression of a query gene(s) across tumor and normal samples, as well as in various tumor sub-groups based on individual cancer stages, tumor grade, race, body weight or other clinicopathologic features, 2) estimate the effect of gene expression level and clinicopathologic features on patient survival; and 3) identify the top over- and under-expressed (up and down-regulated) genes in individual cancer types. This resource serves as a platform for in silico validation of target genes and for identifying tumor sub-group specific candidate biomarkers. Thus, UALCAN web-portal could be extremely helpful in accelerating cancer research. UALCAN is publicly available at http://ualcan.path.uab.edu.
Introduction
Recent advances in high throughput technologies such as next-generation sequencing (NGS) and microarrays have enabled basic, translational and clinical cancer researchers to investigate molecular changes in DNA, RNA, and proteins at high throughput scale [1], [2], [3]. Using multiple data platforms (including DNA methylation and copy number, and RNA and protein expression), the Cancer Genome Atlas (TCGA) consortium has generated molecular profiles of over ten thousand samples related to multiple cancer types [4], leading to studies involving the genomic and molecular characterization of individual cancer types [5], [6], [7], [8], [9], [10], [11], [12], [13], [14], [15], [16], [17], [18], [19], [20], [21], [22], [23], [24].
TCGA data provide an opportunity to analyze the associations of various clinicopathologic factors with tumor initiation, progression, and invasion. Publicly available TCGA data (“level 3”, i.e. processed data ready for high-level analyses) can be downloaded via web portals such as Genomic Data Commons (https://gdc.cancer.gov/), cBioPortal [25], [26] and firehose Broad Genome Data Analysis Center (https://gdac.broadinstitute.org/). In addition, various R packages such as CGDS-R (https://cran.r-project.org/web/packages/cgdsr/index.html), TCGA-assembler [27], and RTCGA Toolbox [28] facilitate programmatic access to TCGA data.
The sheer volume of TCGA cancer genomics data, with its availability in different data formats, makes in-depth analyses difficult for clinicians and cancer researchers who lack bioinformatics/programming skills. In order to facilitate basic queries of the data, various analytical tools have been developed. One such tool, cBioPortal, allows users to submit sets of genes for a cancer type of interest. For each gene queried, cBioPortal provides RNA level expression data, mutation events, copy number alterations, protein expression by Reverse Phase Protein Array (RPPA), a survival plot, and a list of co-expressed and mutually expressed genes. Other tools such as miRGator v3.0 [29], TANRIC [30], and ISOexpresso [31] can be used to analyze differential expression of specific biomolecules such as miRNA, lincRNA, and transcript isoforms, respectively. Gene-Drug Interaction for Survival in Cancer (GDISC) web portal [32] aids in estimating the effect of gene-drug interactions on various cancer types using TCGA data. The Cancer Genome Atlas Clinical Explorer (Stanford-TCGA-CE) [33] aids in finding associations between genomic/proteomic features and clinical parameters, hence finding potentially clinically relevant genes. PROGgeneV2 facilitates comprehensive survival analysis of publicly available gene expression data including TCGA [34]. Oncomine [35], [36] provides an interactive platform for gene expression profiling, using TCGA and other published cDNA, Affymetrix, and Illumina microarray data.
While the web resources noted above are highly useful for a multitude of data analyses, there is a need for a tool that allows cancer researchers to perform the following: 1) compare gene expression between specific subsets as defined within each cancer type, e.g. subsets based on pathological stages or tumor grade, patient gender, patient race, patient drinking or smoking history, or molecular subclasses; and 2) examine associations between gene expression and various clinical parameters (e.g. patient's race). As heterogeneity existing within a given cancer type has been recognized as an important factor influencing the patient outcome [37], subgroup analyses can lead to a better understanding of a given disease.
For example, using existing tools, one can readily analyze the expression level of a given gene in primary breast invasive carcinoma (BRCA) as compared to non-cancer (“normal”) samples, but one may also want the ability (not easily facilitated by existing tools) to carry out additional analyses, which might include: 1) surveying the differential expression of a gene in luminal, HER2 positive, or triple negative breast cancer, 2) testing whether post menopause breast cancer patients show higher expression than pre-menopause patients for a given gene, 3) testing whether African American breast cancer patients show higher expression than Caucasian patients for a given gene, 4) testing whether a given gene shows similar expression in patients across age groups, and 5) analyzing the impact of high expression of a given gene on overall patient survival, in either African American or Caucasian patients. In addition, UALCAN provide critical information and graphic ability to make stage, grade, race and other sub status specific expression features from transcriptome sequencing data some of which are unique to this web portal.
To facilitate gene-level queries of TCGA data, we have developed an interactive web resource called UALCAN (http://ualcan.path.uab.edu/index.html). Using TCGA transcriptome and clinical patient data, UALCAN enables researchers to study the expression level of genes, not only to compare primary tumor with normal tissue samples, but also to compare across different tumor subgroups as defined by pathological cancer stage, tumor grade, patient race, and other clinicopathologic features. Furthermore, one can correlate gene expression with patient survival, with patients further stratified using other parameters such as race or smoking status where applicable. The UALCAN data portal also provides quick links to valuable resources like GeneCards (http://www.genecards.org/), TargetScan [38], The Human Protein Atlas [39], and PubMed (https://www.ncbi.nlm.nih.gov/pubmed). The analysis results (box plots, KM-plots, and heatmaps) can be printed directly or downloaded in several formats including PNG (Portable Network Graphics), JPEG (Joint Photographic Experts Group), PDF (Portable Document Format), and SVG (Scalable Vector Graphics).
The UALCAN data portal can aid in the identification of candidate biomarkers of specific cancer subclasses, with diagnostic, prognostic or therapeutic implications. It can also be used as a platform for in silico validation of target genes. UALCAN transcriptome data analysis tools help make TCGA data and analysis results more accessible to a larger group of cancer researchers.
Methods
Data Collection
TCGA-Assembler [27], was used to download TCGA level 3 RNA-seq data related to 31 cancer types. It was installed on R 3.2.2 (https://cran.r-project.org/). Using TCGA assembler “rsem.genes.results” files were obtained for ‘Primary Solid Tumor’ and ‘Solid Tissue Normal’ for each cancer. The “rsem.genes.results” file includes gene expression values estimated by RSEM algorithm for 20,502 genes; the “raw_count” column shows the number of unfiltered fragments that are aligned with gene, and the “scaled_estimate” column provides estimation of transcripts generated from the gene. As described by Li and Dewey [40], the “scaled_estimate” was multiplied by 106 to obtain transcripts per million (TPM) expression value using in-house PERL (Practical Extraction and Report Language) program. We used TPM as the measure of expression, as it has been suggested to be more comparable across samples than FPKM (Fragments Per Kilobase of transcript per Million mapped reads) and RPKM (Reads Per Kilobase of transcript per Million mapped reads) [41].
In addition to gene expression data, patient data was obtained for all cancers from Genomic Data commons (GDC) (https://gdc.cancer.gov/) using GDC data transfer tool. The downloaded data included clinical parameters such as age, sex, race, survival status, tumor grade, tumor stage and so on, for each patient in the XML (eXtensible Markup Language) file. A PERL script was written to parse all XML files corresponding to specific cancer and extract them into a tab separated file.
Data Analyses
The gene expression and clinical patient data were downloaded from TCGA and processed to generate three major types of graphical outputs, described as follows:
-
1.
Box and whisker plot showing gene expression level in different cancers and their subtypes/sub-stages.
Level 3 TCGA RNA-seq data corresponding to the primary tumor and normal (if available) samples for each gene is represented as box and whisker plot in every TCGA cancer type. Highcharts (Highsoft AS Highcharts, http://www.highcharts.com/), a javascript library from Highsoft AS, was used to generate the visualization representing interquartile range (IQR) including minimum, 25th percentile, median, 75th percentile and maximum values. Outliers are excluded from the plot. Highcharts also supports exporting visualization plot to an image file.
In addition, primary tumor samples were categorized using clinical patient data and boxplots were generated of the expression level of each gene across various subgroups.
The categories of boxplots are as follows,-
a)Individual cancer stages: based on AJCC (American Joint Committee on Cancer) pathologic tumor stage information, samples were divided into stage I, stage II, stage III and stage IV group.
-
b)Patient race: samples were divided into Caucasian, African-American and Asian groups.
-
c)Patient gender: samples from male and female patients were grouped separately.
-
d)Patient age: samples were also grouped based on the age of the patients. Patients of age 21 to 40, 41 to 60, 61 to 80, and 81 to 100 years were grouped separately.
-
e)Tumor grade: where tumor grade information is available, samples were categorized into grade 1, grade 2, grade 3, and grade 4 groups.
-
f)Body weight: if patient data includes height and weight information, then body mass index (BMI) was calculated using the below mentioned formula (http://www.epic4health.com/bmiformula.html)BMI = (weight in kilograms)/((height in meters) × (height in meters))Using BMI values, patients were categorized into four groups. Patients with BMI ranging from 18 to 24 were classified as “normal weight”, those with BMI ranging from 25 to 29 were classified as “extreme weight”, those with BMI ranging from 30 to 39 were classified as “obese” and patients with BMI equal or above 40 as “extreme obese” (https://www.nhlbi.nih.gov/health/educational/lose_wt/BMI/bmi_tbl.pdf).
-
g)Smoking status: where smoking status information was available, samples were categorized into four groups including smoker, non-smoker, reformed smoker1 (who are current reformed smokers for <= 15 years), and reformed smoker2 (who are current reformed smokers for >15 years).
-
h)Drinking habit: Based on information availability, samples were categorized into groups such as daily drinker, weekly drinker, social drinker, occasional drinker, and non-drinker.
-
i)Menopause status: Patient data corresponding to breast cancer and endometrial carcinoma includes menopause status, with samples categorized as “pre-menopause”, “peri-menopause” and “post-menopause”.
-
j)Molecular signature: In case of prostate adenocarcinoma, 246 primary prostate tumor samples were divided into seven subtypes defined by ERG (ETS transcription factor), ETV1/4 (ETS variant 1/4) and FLI1 (Fli-1 proto-oncogene, ETS transcription factor) gene fusions and SPOP (speckle type BTB/POZ protein), FOXA1 (forkhead box A1) and IDH1 (isocitrate dehydrogenase (NADP(+)) 1, cytosolic) mutations [17]. Similarly, primary breast cancer samples were divided into luminal, HER2 positive, and triple negative subclasses based on estrogen receptor (ER), progesterone receptor (PR), and human epidermal growth factor receptor 2 (HER2) status by immunohistochemistry (IHC). In addition 116 triple negative breast cancer samples were categorized into six TNBC subtypes (such as basal-like1 or BL1, basal-like2 or BL2, immunomodulatory or IM, mesenchymal or M, mesenchymal stem-like or MSL, and luminal androgen receptor or LAR) using TNBCtypes tool [42], [43].TPM values employed for the generation of boxplots were also used to estimate the significance of difference in gene expression levels between groups. The t test was performed using a PERL script with Comprehensive Perl Archive Network (CPAN) module “Statistics::TTest” (http://search.cpan.org/~yunfang/Statistics-TTest-1.1.0/TTest.pm).
-
a)
-
2.
Heatmap showing top differentially expressed genes.
UALCAN also lists genes which show high differential expression between normal and tumor samples in the form of an interactive heatmap. This feature is available to cancer types with normal sample data available, which types include colon adenocarcinoma (COAD), lung adenocarcinoma (LUAD), lung squamous cell carcinoma (LUSC), rectal adenocarcinoma (READ), kidney chromophobe (KICH), kidney renal clear cell carcinoma (KIRC), kidney renal papillary cell carcinoma (KIRP), bladder urothelial carcinoma (BLCA), breast invasive carcinoma (BRCA), prostate adenocarcinoma (PRAD), head and neck squamous cell carcinoma (HNSC), esophageal carcinoma (ESCA), liver hepatocellular carcinoma (LIHC), uterine corpus endometrial carcinoma (UCEC), and thyroid carcinoma (THCA).
A PERL script was written to analyze normalized TCGA level 3 RNA-seq data for each gene. Using CPAN module “Statistics::Descriptive”, mean TPM values of each gene in normal samples and tumor samples were obtained separately. To list top 250 over- and under-expressed genes for each cancer, genes with significantly different TPM values (P < .001) were first selected. Among these genes, only those with median TPM value of 10 or above were retained. Finally, genes were sorted based on the following metric: (mean TPM in tumor samples)/(mean TPM in normal samples).
Using a javascript library from Highcharts, UALCAN provides expression level of these top differentially expressed genes across all normal and tumor samples as a heatmap. Genes can be visualized in sets of 25. Over- and under-expressed genes are shown in separate heatmaps.
-
3.
Kaplan–Meier survival plot.
In addition to gene expression variation across tumor samples, gene-level correlations with patient survival are featured in UALCAN. Available TCGA patient survival data were used for Kaplan–Meier survival analyses and to generate overall survival plots.
Patient clinical data in XML format was parsed with PERL script to obtain, a) patient vital status (Dead/Alive), b) if the patient is alive, then ‘days_to_last_follow_up’ from most recent follow-up, and c) if the patient is dead, then ‘days_to_death’. Overall survival analysis was conducted using only patients with survival data and gene expression data from RNA-seq. For each gene, a tab separated input file was created with columns for TCGA sample id, Time (days_to_death or days_to_last_follow_up), Status (Alive or Dead), and Expression level (High expression or Low/Medium expression).
Samples were categorized into two groups: (1) High expression (with TPM values above upper quartile) and (2) Low/Medium expression (with TPM values below upper quartile).
The Kaplan–Meier survival plot was generated for every gene in each TCGA cancer type, using “survival” package [44] and “survminer” package [45]. The survival curves of samples with high gene expression and low/medium gene expression were compared by log rank test.
In order to assess the combined survival effect of gene expression and clinical parameters such as patient race, gender, BMI, cancer subtypes, tumor grade, etc., we applied multivariate Kaplan–Meier survival analysis [46]. For example, to estimate combined effect of the expression level of a given gene and racial disparity on breast cancer patient survival, the samples were first divided into two groups: samples with high expression of the gene and samples with low/medium expression. Then, within each expression category, patients were further stratified into three subgroups based on race (African American, Caucasian, Asian). R scripts were written to divide all patients into these six categories and to generate Kaplan–Meier plot. The P value obtained from log-rank test was used to indicate statistical significance of survival correlation between groups. Such multivariate survival analyses were performed for all genes within each TCGA cancer type. The plots were also generated in SVG and PDF formats.
Results
Usage of UALCAN
UALCAN is hosted on CentOS server with 72 cores (Intel ® Xeon® CPU E2–2699 v3 @2.30GHz), 98 GB RAM, and 22 TB HDD. The interface was developed using PERL-CGI, while CSS and javascripts were utilized to implement user-friendly features.
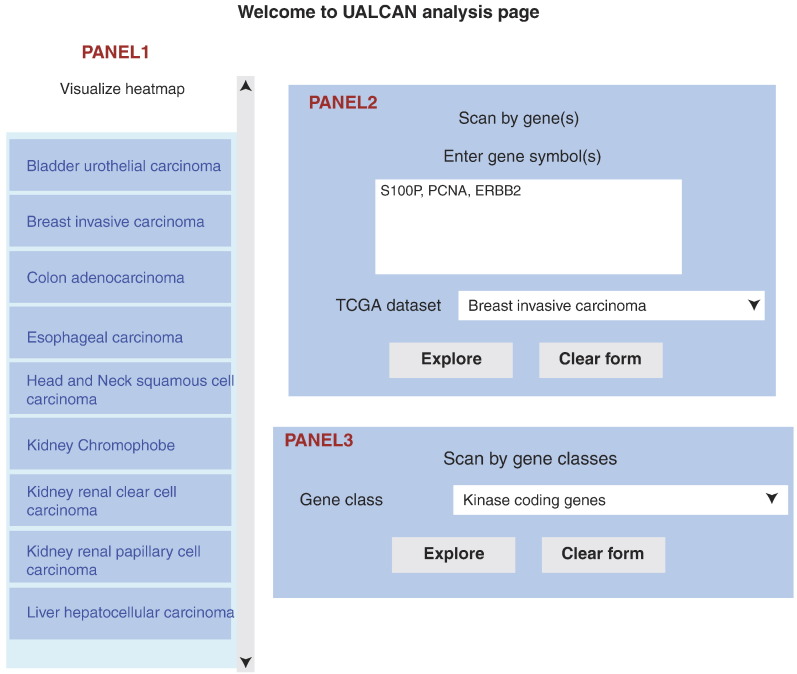
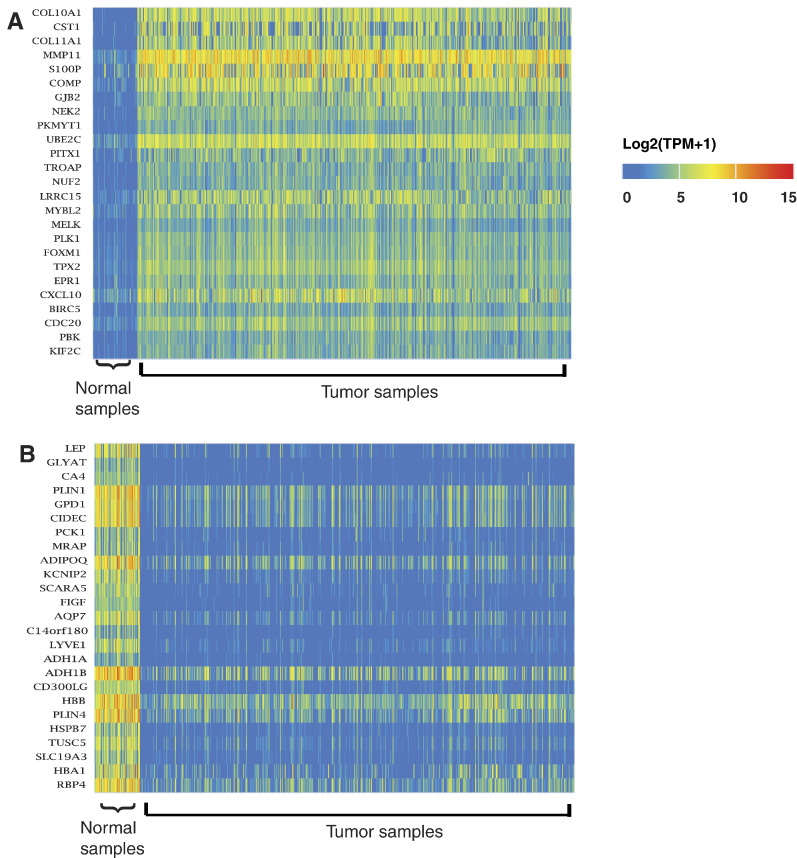
The analysis page of the UALCAN includes three panels (Figure 1). The left side panel on analysis page shows a list of cancer types, which are hyperlinked to a web page showing heatmaps of top differentially expressed genes (Figure 2). The top 250 over- and under-expressed genes are shown separately for those cancer types with data on >10 normal samples. The right side panel includes two options to query UALCAN, listed below:
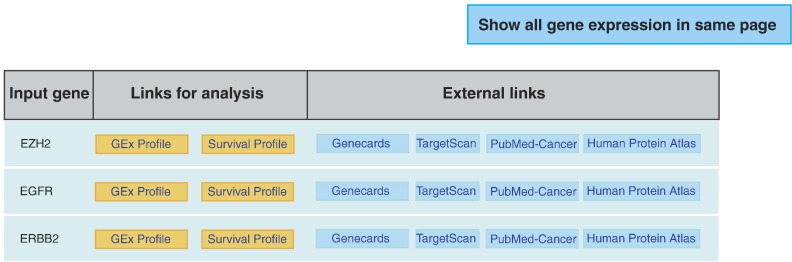
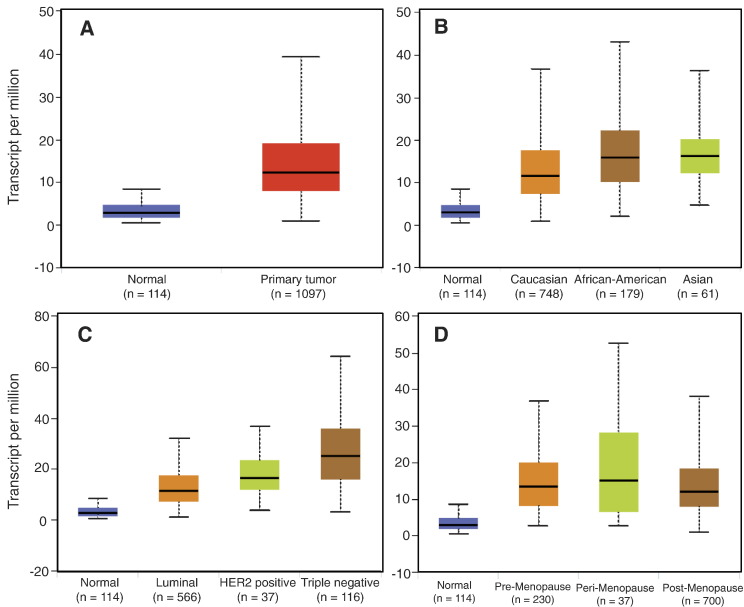
Scan by gene(s): User can paste one or more gene symbols in the text area and choose the cancer type of interest, in order to analyze the expression and survival information of each gene queried. UALCAN lists queried genes with links to gene expression analysis and survival analysis results. In addition, links are also provided to facilitate access of gene related information from external resources (Figure 3). The link to gene expression analysis results provides information about relative expression levels of the gene of interest in normal versus tumor samples and across cancer subgroups, as illustrated in Figure 4. Statistical significance of each comparison performed is provided in tabular form. Similarly, the link to survival analysis results showcases multiple KM-plots showing the association of gene expression levels combined with clinical parameters on patient survival, as illustrated in Figure 5. Log-rank P values show statistical significance of the patterns observed.
Scan by gene classes: The user can choose from a list of precompiled genes sets, to find out which genes of interest show differential expression between tumor and normal samples and which genes are associated with patient survival. The gene-lists were obtained from Uniprot keyword search (http://www.uniprot.org/keywords/), QIAGEN (https://www.qiagen.com/us/resources/), KEGG (obtained using KEGGPATHID2EXTID function of R package KEGG.db [47] and manual curation. For each gene set, the interactive web page shows differential expression and survival associations of each gene across 31 cancer types (Figure 6).
Figure 1.
Snapshot of UALCAN analysis page. The left side panel shows a list of cancer types, each type being hyperlinked to a web page showing the top over- or under-expressed genes in tumor compared to normal samples. The top-right side panel allows the user to query UALCAN by official gene symbol(s) and cancer type of interest, while the bottom-right side panel allows the user to query UALCAN using precompiled gene-sets.
Figure 2.
Heatmaps showing top differentially expressed genes in breast invasive carcinoma (BRCA). (A) The top 25 over-expressed and (B) the top 25 under-expressed genes in BRCA compared to normal samples. Expression level of gene is represented as log2(TPM+ 1). Sample names and associated expression value can be visualized by placing the cursor over the heatmap.
Figure 3.
UALCAN output page listing genes queried, along with links to analyze their expression and survival associations in cancer types of interest. Links to GeneCards, TargetScan, PubMed and Human Protein Atlas are also provided through the interface.
Figure 4.
Box-whisker plots showing the expression of EZH2 in sub groups of breast invasive carcinoma samples (BRCA). (A) Boxplot showing relative expression of EZH2 in normal and BRCA samples. (B) Boxplot showing relative expression of EZH2 in normal, African American, Caucasian and Asian BRCA patients. (C) Boxplot showing relative expression of EZH2 in normal, luminal, HER2 positive and triple negative BRCA patients. (D) Boxplot showing relative expression of EZH2 in normal, pre-menopause, peri-menopause and post-menopause patients.
Figure 5.
Kaplan–Meier plots showing the association of EZH2 expression and other clinical parameters with patient survival. (A) KM plot depicting association of EZH2 expression levels with patient survival. (B) KM plot depicting association of EZH2 expression levels and race with patient survival. (C) KM plot depicting association of EZH2 expression level and BRCA subtype with patient survival. (D) KM plot depicting association of EZH2 expression levels and menopause status with patient survival.
Figure 6.
Snapshot of UALCAN output page showing differential expression status and survival impact across 31 cancer types of genes involved in P53 signaling pathway. In the interface, a pop up text appears on placing the cursor over the buttons showing summary, while the user can access expression and survival information for a given gene by clicking the corresponding button.
UALCAN can facilitate cancer researchers in performing multiple types of analyses. Some of the analysis examples given below help illustrate the utility of this resource.
-
Example analysis 1: Identify the top overexpressed genes in liver hepatocellular carcinoma (LIHC) and examine gene expression differences among Asian, Caucasian and African-American patients.
The left panel in the UALCAN analysis page shows a list of TCGA cancer types. On clicking “Liver hepatocellular carcinoma”, the user is directed to a web page showing a heatmap of the top 25 genes overexpressed in liver hepatocellular carcinoma samples (n = 371) as compared to normal samples (n = 32). On the left side of the interactive heatmap (generated using HighCharts javascript), gene names are listed. Expression information about each gene can be obtained by clicking on the gene name. Glypican 3 (GPC3), Lipocalin 2 (LCN2), Secreted phosphoprotein 1 (SPP1), and ubiquitin conjugating enzyme E2 C (UBE2C) are listed as top four over-expressed genes. Careful observation reveals that expression profile of LCN2 and SPP1 in liver hepatocellular carcinoma show no significant change across patients of different race, GPC3 shows significantly higher expression in Asian patients compared to Caucasian patients, and UBE2C shows significantly higher expression in Asian patients compared to both African American and Caucasian patients. The schematic representation of this analysis is provided in Supplementary Figure 1.
-
Example analysis 2: Stage specific gene expression analysis of genes highly over-expressed in bladder urothelial carcinoma.
Genes that show stage specific expression may represent potential therapeutic biomarkers. Using the UALCAN heatmap feature, the top 25 genes higher in bladder urothelial carcinoma versus normal samples are obtained. Matrix metallopeptidase 11 (MMP11), cyclin dependent kinase inhibitor 2A (CDKN2A), and cystatin E/M (CST6), representing the top three genes over-expressed in cancer, are subsequently analyzed for stage specific expression. CST6 and MMP11 show higher expression in stage 2 to stage 4 samples compared to normal (P < .05). However, the higher expression also observed in stages 3 and 4 compared to stages 1 and 2 (P < .05) indicate steady increases in both CST6 and MMP11 expression from less aggressive to more aggressive stages of bladder cancer. In addition, CDKN2A also shows higher expression in stage 2 to stage 4 compared to normal (P < .05), and uniform expression patterns across stages 2, 3, and 4 suggest that the gene might be considered as a potential stage 2 bladder cancer biomarker (Supplementary Figure 2).
-
Example analysis 3: Pan-cancer gene expression analysis of the P53 signaling pathway.
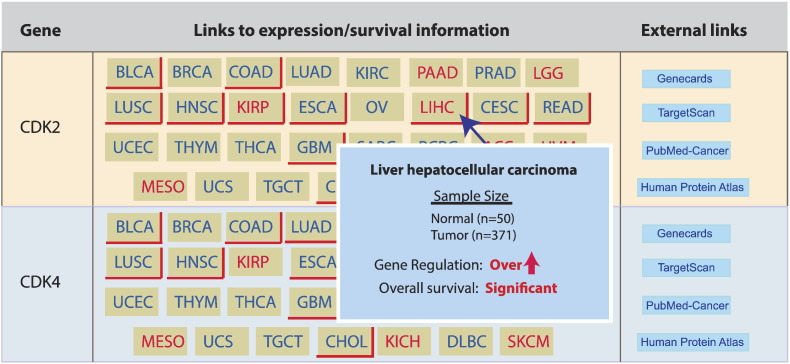
An important feature of UALCAN is the facility to query for genes falling under a specific class. Precompiled lists of genes are available for query, corresponding to pathways most commonly affected in cancer—e.g. P53 signaling, cell cycle, apoptosis and hedgehog signaling—or corresponding to specific molecular classes—e.g. kinases, ubiquitinases, and histone methyltransferases. On scanning UALCAN by the gene class “P53 signaling pathway genes”, the resulting output page provides an overview of differential expression and survival associations involving each of 68 associated genes across 31 TCGA cancer types. The output includes a table of color coded buttons. Buttons with red shadow indicate genes over-expressed in tumor samples compared to normal, while buttons with green shadow indicate genes under-expressed. Similarly, buttons with red text denote genes significantly correlated with patient survival. As shown in Supplementary Figure 3, the output page will readily show, for example, that Cyclin dependent kinase 2 (CDK2) (involved in P53 signaling pathway) is over-expressed in bladder urothelial carcinoma (BLCA), breast invasive carcinoma (BRCA), colon adenocarcinoma (COAD), lung adenocarcinoma (LUAD), kidney renal clear cell carcinoma (KIRC), lung squamous cell carcinoma (LUSC), head and neck squamous cell carcinoma (HNSC), esophageal carcinoma (ESCA), cervical squamous cell carcinoma (CESC), rectal adenocarcinoma (READ), uterine corpus endometrial carcinoma (UCEC), glioblastoma multiforme (GBM), and cholangiocarcinoma (CHOL), as well as under-expressed in kidney chromophobe (KICH). CDK2 is also both over-expressed and associated with patient overall survival in kidney renal papillary cell carcinoma (KIRP) and liver hepatocellular carcinoma (LIHC).
-
Example analysis 4: Understanding the effect of cyclin dependent kinase inhibitor 1A (CDKN1A) expression and racial disparity on overall survival in head and neck squamous cell carcinoma (HNSC).
One of the unique features of UALCAN is that it aids in investigating gene expression patterns in conjunction with clinical parameters, such as patient race, on overall patient survival. To examine the survival association of CDKN1A expression in HNSC, one can use the “scan by gene” option in UALCAN. The output page of the survival analysis provides a set of Kaplan–Meier (KM) plots from both univariate analysis (considering only CDKN1A expression) and multivariate analysis (considering clinical parameters along with CDKN1A expression). The KM plot depicting the effect of high and low/medium CDKN1A expression on overall survival of African American, Caucasian, and Asian patients shows a cumulative significance of 0.039. The user can further focus the analysis on only the African American and Caucasian patients, by selecting the “visualize individual plots” option. On careful observation of the KM plots, one can observe that high expression of CDKN1A significantly (P = .023) correlates with overall survival in Caucasian HNSC patients. This analysis is schematically represented in Supplementary Figure 4.
-
Example analysis 5: Exploring class specific expression of the top breast cancer associated genes.
Breast cancer involves various histopathological features known to have treatment implications [48]. Therefore, identification of biomarkers specific to breast cancer subtype can be considered extremely important. In UALCAN, TCGA BRCA tumors can be subdivided into “luminal,” “HER2 positive,” and “TNBC” groups, with the levels of a given gene being shown across these groups. Starting with the expression profile of the top 25 over-expressed genes in breast cancer (as shown in the associated heatmap), one can observe, for example, that both Baculoviral IAP repeat containing 5 (BIRC5) and Ubiquitin conjugating enzyme E2 C (UBE2C) show higher expression in TNBC samples compared to other tumor samples. Using UALCAN, the expression patterns of BIRC5 and UBE2C across molecular subtypes of TNBC can also be explored (Supplementary Figure 5).
Discussion
The molecular profiling data generated by TCGA consortium has great value in increasing our understanding of the underlying molecular mechanisms involved in various cancers, as well as in the identification of novel therapeutic and diagnostic biomarkers [49], [50], [51], [52]. In order to maximize TCGA data as a community resource, it is important to provide web resources that allow cancer researchers and clinicians (regardless of their levels of computational expertise) to access, analyze, visualize, and interpret the data with ease. For example, one of the possible ways to prioritize genes for further study, in terms of their potential oncogenic or tumor suppressor properties, is to identify genes with expression associated with patient survival. The user friendly interface of UALCAN facilitates the identification of survival associations involving any gene of interest, across different cancer types as well as cancer subtypes as defined by various clinicopathologic features. Multiple public resources such as cBioPortal [25], [26], miRGator v 3.0 [29], TANRIC [30], and ISOexpresso [31] aid in the comprehensive analysis of transcriptomic TCGA data. While cBioPortal, for example, is extremely useful in exploring gene-level associations across different cancers involving mutation frequency or gene expression, there remains a need for tools allowing one to examine RNA level expression differences or survival associations across different cancer subsets as defined by clinicopathologic features. In future, we will incorporate additional transcriptome sequencing datasets from various cancers as well as additional utilities like co-expression analysis, long non-coding RNA analysis and microRNA analysis from the available datasets.
We believe that UALCAN can greatly aid cancer biologists and clinicians in the identification of novel diagnostic and therapeutic targets, investigate the gene expression and its disease association in any particular cancer. With its intuitive features, UALCAN will enable researchers across disciplines to easily query for the target or gene of their interest in cancer and make cross-disease associations.
Acknowledgements
The data generated by TCGA Research Network (http://cancergenome.nih.gov/) has been used for UALCAN development. We thank many researchers who tested this web portal and gave us very valuable feedbacks.
Footnotes
This work was supported by funding from The University of Alabama Birmingham and Breast Cancer Research Foundation of Alabama to SV. SV is also supported by NIH/NCI R01CA157845 and R01CA154980. CC is supported by NIH CA125123.
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.neo.2017.05.002.
Appendix A. Supplementary data
Supplementary Figure 1. Race based expression analysis of hepatocellular carcinoma associated genes. (A) Screen shot of the left side panel in UALCAN analysis page, which directs to heatmap of top differentially expressed genes. (B) Heatmap showing top 25 over-expressed genes in liver hepatocellular carcinoma [LIHC]. Each row shows expression level of specific gene across tumor (n=371) and normal(n=50) samples. Gene names on heatmap is linked to gene expression analysis page. Boxplots (C-F) depict expression level of GPC3, LCN2, SPP1 and UBE2C in cacausian, african american and asian LIHC patients.
Supplementary Figure 2. Cancer stage specific expression analysis of bladder urothelial carcinoma associated genes. (A) Heatmap showing top 25 over-expressed genes in bladder urothelial carcinoma [BLCA]. Each row shows expression level of specific gene across tumor (n=408) and normal(n=19) samples. Gene names on heatmap is linked to gene expression analysis page. Boxplots (B-D) depict expression level of CST6, CDKN2A and MMP11 in stage1, stage2, stage3 and stage4 BLCA patients.
Supplementary Figure 3. Pan cancer analysis of P53 signaling pathway genes. (A) Screen shot of the panel in analysis page that facilitates to scan UALCAN with precompiled gene class. (B) Screen shot of the output page providing bird’s eye view on expression and survival profile of each gene across 31 TCGA cancer types. Each button links to gene’s expression profile in different cancers. The styles of the button indicate the expression status (Over-/under-regulation or No change) and overall survival impact (significant/not significant) of gene.
Supplementary Figure 4. Assessment of combined impact of CDKN1A expression and patient’s race on overall survival of head and neck squamous cell carcinoma [HNSC] patients. (A) Screen shot of UALCAN output page providing links to survival and gene expression analysis. (B) Kaplan meier plot showing result of multivariate overall survival analysis considering CDKN1A expression and patient’s race. (C-D) Kaplan meier plot showing influence of CDKN1A expression level on overall survival of Caucasian and African American patients.
Supplementary Figure 5. Breast cancer subtype specific gene expression analysis. (A) Heatmap showing top 25 over-expressed genes in breast invasive carcinoma [BRCA]. (B-C) Boxplots showing expression profile of BIRC5 and UBE2C in BRCA patients with luminal (n=566), HER2 positive (n=37) and TNBC (n=116) types of breast cancer. (D-E) Boxplots showing BIRC5 and UBE2C expression in TNBC sutypes (samples categorized using TNBCtype tool).
References
- 1.Sheehan KM, Calvert VS, Kay EW, Lu Y, Fishman D, Espina V, Aquino J, Speer R, Araujo R, Mills GB. Use of reverse phase protein microarrays and reference standard development for molecular network analysis of metastatic ovarian carcinoma. Mol Cell Proteomics. 2005;4(4):346–355. doi: 10.1074/mcp.T500003-MCP200. [DOI] [PubMed] [Google Scholar]
- 2.Trevino V, Falciani F, Barrera-Saldana HA. DNA microarrays: a powerful genomic tool for biomedical and clinical research. Mol Med. 2007;13(9–10):527–541. doi: 10.2119/2006-00107.Trevino. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Reuter JA, Spacek DV, Snyder MP. High-throughput sequencing technologies. Mol Cell. 2015;58(4):586–597. doi: 10.1016/j.molcel.2015.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tomczak K, Czerwinska P, Wiznerowicz M. The Cancer Genome Atlas (TCGA): an immeasurable source of knowledge. Contemp Oncol (Pozn) 2015;19(1A):A68–A77. doi: 10.5114/wo.2014.47136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cancer Genome Atlas Research N Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455(7216):1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cancer Genome Atlas N Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487(7407):330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cancer Genome Atlas N Comprehensive molecular portraits of human breast tumours. Nature. 2012;490(7418):61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cancer Genome Atlas Research N Comprehensive genomic characterization of squamous cell lung cancers. Nature. 2012;489(7417):519–525. doi: 10.1038/nature11404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cancer Genome Atlas Research N Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med. 2013;368(22):2059–2074. doi: 10.1056/NEJMoa1301689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cancer Genome Atlas Research N, Kandoth C, Schultz N, Cherniack AD, Akbani R, Liu Y, Shen H, Robertson AG, Pashtan I, Shen R. Integrated genomic characterization of endometrial carcinoma. Nature. 2013;497(7447):67–73. doi: 10.1038/nature12113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cancer Genome Atlas Research N Comprehensive molecular characterization of urothelial bladder carcinoma. Nature. 2014;507(7492):315–322. doi: 10.1038/nature12965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cancer Genome Atlas Research N Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014;511(7511):543–550. doi: 10.1038/nature13385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cancer Genome Atlas Research N Comprehensive molecular characterization of gastric adenocarcinoma. Nature. 2014;513(7517):202–209. doi: 10.1038/nature13480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cancer Genome Atlas Research N Integrated genomic characterization of papillary thyroid carcinoma. Cell. 2014;159(3):676–690. doi: 10.1016/j.cell.2014.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cancer Genome Atlas N Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature. 2015;517(7536):576–582. doi: 10.1038/nature14129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cancer Genome Atlas N Genomic Classification of Cutaneous Melanoma. Cell. 2015;161(7):1681–1696. doi: 10.1016/j.cell.2015.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cancer Genome Atlas Research N The Molecular Taxonomy of Primary Prostate Cancer. Cell. 2015;163(4):1011–1025. doi: 10.1016/j.cell.2015.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ciriello G, Gatza ML, Beck AH, Wilkerson MD, Rhie SK, Pastore A, Zhang H, McLellan M, Yau C, Kandoth C. Comprehensive Molecular Portraits of Invasive Lobular Breast Cancer. Cell. 2015;163(2):506–519. doi: 10.1016/j.cell.2015.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cancer Genome Atlas Research N, Linehan WM, Spellman PT, Ricketts CJ, Creighton CJ, Fei SS, Davis C, Wheeler DA, Murray BA, Schmidt L. Comprehensive Molecular Characterization of Papillary Renal-Cell Carcinoma. N Engl J Med. 2016;374(2):135–145. doi: 10.1056/NEJMoa1505917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ceccarelli M, Barthel FP, Malta TM, Sabedot TS, Salama SR, Murray BA, Morozova O, Newton Y, Radenbaugh A, Pagnotta SM. Molecular Profiling Reveals Biologically Discrete Subsets and Pathways of Progression in Diffuse Glioma. Cell. 2016;164(3):550–563. doi: 10.1016/j.cell.2015.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zheng S, Cherniack AD, Dewal N, Moffitt RA, Danilova L, Murray BA, Lerario AM, Else T, Knijnenburg TA, Ciriello G. Comprehensive Pan-Genomic Characterization of Adrenocortical Carcinoma. Cancer Cell. 2016;29(5):723–736. doi: 10.1016/j.ccell.2016.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cancer Genome Atlas Research N Integrated genomic and molecular characterization of cervical cancer. Nature. 2017;543(7645):378–384. doi: 10.1038/nature21386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cancer Genome Atlas Research N, Analysis Working Group: Asan U, Agency BCC, Brigham, Women's H, Broad I, Brown U, Case Western Reserve U, Dana-Farber Cancer I, Duke U, Greater Poland Cancer C Integrated genomic characterization of oesophageal carcinoma. Nature. 2017;541(7636):169–175. doi: 10.1038/nature20805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fishbein L, Leshchiner I, Walter V, Danilova L, Robertson AG, Johnson AR, Lichtenberg TM, Murray BA, Ghayee HK, Else T. Comprehensive Molecular Characterization of Pheochromocytoma and Paraganglioma. Cancer Cell. 2017;31(2):181–193. doi: 10.1016/j.ccell.2017.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2(5):401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6(269):pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhu Y, Qiu P, Ji Y. TCGA-assembler: open-source software for retrieving and processing TCGA data. Nat Methods. 2014;11(6):599–600. doi: 10.1038/nmeth.2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Samur MK. RTCGAToolbox: a new tool for exporting TCGA Firehose data. PLoS One. 2014;9(9):e106397. doi: 10.1371/journal.pone.0106397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cho S, Jang I, Jun Y, Yoon S, Ko M, Kwon Y, Choi I, Chang H, Ryu D, Lee B. MiRGator v3.0: a microRNA portal for deep sequencing, expression profiling and mRNA targeting. Nucleic Acids Res. 2013;41(Database issue):D252–D257. doi: 10.1093/nar/gks1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li J, Han L, Roebuck P, Diao L, Liu L, Yuan Y, Weinstein JN, Liang H. TANRIC: An Interactive Open Platform to Explore the Function of lncRNAs in Cancer. Cancer Res. 2015;75(18):3728–3737. doi: 10.1158/0008-5472.CAN-15-0273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang IS, Son H, Kim S, Kim S. ISOexpresso: a web-based platform for isoform-level expression analysis in human cancer. BMC Genomics. 2016;17(1):631. doi: 10.1186/s12864-016-2852-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Spainhour JC, Lim J, Qiu P. GDISC: a web portal for integrative analysis of gene-drug interaction for survival in cancer. Bioinformatics. 2017;33(9):1426–1428. doi: 10.1093/bioinformatics/btw830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee H, Palm J, Grimes SM, Ji HP. The Cancer Genome Atlas Clinical Explorer: a web and mobile interface for identifying clinical-genomic driver associations. Genome Med. 2015;7:112. doi: 10.1186/s13073-015-0226-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Goswami CP, Nakshatri H. PROGgeneV2: enhancements on the existing database. BMC Cancer. 2014;14:970. doi: 10.1186/1471-2407-14-970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D, Barrette T, Pandey A, Chinnaiyan AM. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6(1):1–6. doi: 10.1016/s1476-5586(04)80047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rhodes DR, Kalyana-Sundaram S, Mahavisno V, Varambally R, Yu J, Briggs BB, Barrett TR, Anstet MJ, Kincead-Beal C, Kulkarni P. Oncomine 3.0: genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles. Neoplasia. 2007;9(2):166–180. doi: 10.1593/neo.07112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Allison KH, Sledge GW. Heterogeneity and cancer. Oncology (Williston Park) 2014;28(9):772–778. [PubMed] [Google Scholar]
- 38.Agarwal V, Bell GW, Nam JW, Bartel DP. Predicting effective microRNA target sites in mammalian mRNAs. Elife. 2015;4 doi: 10.7554/eLife.05005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Uhlen M, Fagerberg L, Hallstrom BM, Lindskog C, Oksvold P, Mardinoglu A, Sivertsson Å, Kampf C, Sjöstedt E, Asplund A. Proteomics. Tissue-based map of the human proteome. Science. 2015;347(6220):1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 40.Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li B, Ruotti V, Stewart RM, Thomson JA, Dewey CN. RNA-Seq gene expression estimation with read mapping uncertainty. Bioinformatics. 2010;26(4):493–500. doi: 10.1093/bioinformatics/btp692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen X, Li J, Gray WH, Lehmann BD, Bauer JA, Shyr Y, Pietenpol JA. TNBCtype: A Subtyping Tool for Triple-Negative Breast Cancer. Cancer Informat. 2012;11:147–156. doi: 10.4137/CIN.S9983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lehmann BD, Bauer JA, Chen X, Sanders ME, Chakravarthy AB, Shyr Y, Pietenpol JA. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J Clin Invest. 2011;121(7):2750–2767. doi: 10.1172/JCI45014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Therneau T. 2015. A Package for Survival Analysis in S. R package version 2.38. [Google Scholar]
- 45.Kassambara A, Kosinski M, Biecek P. 2017. survminer: Drawing Survival Curves using 'ggplot2'. R package version 0.3.1. [Google Scholar]
- 46.Bradburn MJ, Clark TG, Love SB, Altman DG. Survival analysis part II: multivariate data analysis--an introduction to concepts and methods. Br J Cancer. 2003;89(3):431–436. doi: 10.1038/sj.bjc.6601119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Carlson M. 2016. KEGG.db: A set of annotation maps for KEGG. R package version 3.1.2. [Google Scholar]
- 48.Blows FM, Driver KE, Schmidt MK, Broeks A, van Leeuwen FE, Wesseling J, Cheang MC, Gelmon K, Nielsen TO, Blomqvist C. Subtyping of breast cancer by immunohistochemistry to investigate a relationship between subtype and short and long term survival: a collaborative analysis of data for 10,159 cases from 12 studies. PLoS Med. 2010;7(5):e1000279. doi: 10.1371/journal.pmed.1000279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ricketts CJ, Hill VK, Linehan WM. Tumor-specific hypermethylation of epigenetic biomarkers, including SFRP1, predicts for poorer survival in patients from the TCGA Kidney Renal Clear Cell Carcinoma (KIRC) project. PLoS One. 2014;9(1):e85621. doi: 10.1371/journal.pone.0085621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang W. TCGA divides gastric cancer into four molecular subtypes: implications for individualized therapeutics. Chin J Cancer. 2014;33(10):469–470. doi: 10.5732/cjc.014.10117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Brodie SA, Li G, Brandes JC. Molecular characteristics of non-small cell lung cancer with reduced CHFR expression in The Cancer Genome Atlas (TCGA) project. Respir Med. 2015;109(1):131–136. doi: 10.1016/j.rmed.2014.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Peters I, Tezval H, Kramer MW, Wolters M, Grunwald V, Kuczyk MA, Serth J. Implications of TCGA Network Data on 2nd Generation Immunotherapy Concepts Based on PD-L1 and PD-1 Target Structures. Aktuelle Urol. 2015;46(6):481–485. doi: 10.1055/s-0041-106169. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. Race based expression analysis of hepatocellular carcinoma associated genes. (A) Screen shot of the left side panel in UALCAN analysis page, which directs to heatmap of top differentially expressed genes. (B) Heatmap showing top 25 over-expressed genes in liver hepatocellular carcinoma [LIHC]. Each row shows expression level of specific gene across tumor (n=371) and normal(n=50) samples. Gene names on heatmap is linked to gene expression analysis page. Boxplots (C-F) depict expression level of GPC3, LCN2, SPP1 and UBE2C in cacausian, african american and asian LIHC patients.
Supplementary Figure 2. Cancer stage specific expression analysis of bladder urothelial carcinoma associated genes. (A) Heatmap showing top 25 over-expressed genes in bladder urothelial carcinoma [BLCA]. Each row shows expression level of specific gene across tumor (n=408) and normal(n=19) samples. Gene names on heatmap is linked to gene expression analysis page. Boxplots (B-D) depict expression level of CST6, CDKN2A and MMP11 in stage1, stage2, stage3 and stage4 BLCA patients.
Supplementary Figure 3. Pan cancer analysis of P53 signaling pathway genes. (A) Screen shot of the panel in analysis page that facilitates to scan UALCAN with precompiled gene class. (B) Screen shot of the output page providing bird’s eye view on expression and survival profile of each gene across 31 TCGA cancer types. Each button links to gene’s expression profile in different cancers. The styles of the button indicate the expression status (Over-/under-regulation or No change) and overall survival impact (significant/not significant) of gene.
Supplementary Figure 4. Assessment of combined impact of CDKN1A expression and patient’s race on overall survival of head and neck squamous cell carcinoma [HNSC] patients. (A) Screen shot of UALCAN output page providing links to survival and gene expression analysis. (B) Kaplan meier plot showing result of multivariate overall survival analysis considering CDKN1A expression and patient’s race. (C-D) Kaplan meier plot showing influence of CDKN1A expression level on overall survival of Caucasian and African American patients.
Supplementary Figure 5. Breast cancer subtype specific gene expression analysis. (A) Heatmap showing top 25 over-expressed genes in breast invasive carcinoma [BRCA]. (B-C) Boxplots showing expression profile of BIRC5 and UBE2C in BRCA patients with luminal (n=566), HER2 positive (n=37) and TNBC (n=116) types of breast cancer. (D-E) Boxplots showing BIRC5 and UBE2C expression in TNBC sutypes (samples categorized using TNBCtype tool).