Summary
In the past 2 decades, progress in molecular analyses of the plant immune system has revealed key elements of a complex response network. Current paradigms depict the interaction of pathogen‐secreted molecules with host target molecules leading to the activation of multiple plant response pathways. Further research will be required to fully understand how these responses are integrated in space and time, and exploit this knowledge in agriculture. In this review, we highlight systems biology as a promising approach to reveal properties of molecular plant–pathogen interactions and predict the outcome of such interactions. We first illustrate a few key concepts in plant immunity with a network and systems biology perspective. Next, we present some basic principles of systems biology and show how they allow integrating multiomics data and predict cell phenotypes. We identify challenges for systems biology of plant–pathogen interactions, including the reconstruction of multiscale mechanistic models and the connection of host and pathogen models. Finally, we outline studies on resistance durability through the robustness of immune system networks, the identification of trade‐offs between immunity and growth and in silico plant–pathogen co‐evolution as exciting perspectives in the field. We conclude that the development of sophisticated models of plant diseases incorporating plant, pathogen and climate properties represent a major challenge for agriculture in the future.
Keywords: plant–pathogen interactions, systems biology, immunity, network, genome‐scale metabolic network, robustness, trade‐off, modeling
Significance Statement
Plant–pathogen interactions involve a complex interplay of molecular events occurring in the plant and the microbe. In this review we argue that systems biology approaches are particularly well suited to tackle the complexity of these interactions and present an overview of current approaches, challenges and applications of systems biology in plant pathology.
Plant Immunity 101, With a Hint of Networks
Plant immunity is an elaborate, multilayered system involving several lines of defense. In order to get access to nutrients from the plant and complete its lifecycle, a successful pathogen has to pass first the plant passive defense mechanisms. These include structural barriers such as the cuticle, the cell wall, and constitutively produced anti‐microbial compounds (Hückelhoven, 2007; Miedes et al., 2014). In addition to these passive mechanisms, plants possess a two layered actively induced immune system. The first layer of the immune response is termed pathogen‐associated molecular‐pattern (PAMP)‐triggered immunity (PTI). PAMPs are broadly conserved microbial molecules such as bacterial flagellin or fungal chitin, that are perceived by plant surface‐exposed receptors called pattern recognition receptors (Medzhitov and Janeway, 1997; Macho and Zipfel, 2014). A well characterized example is the recognition of the flg22 peptide from bacterial flagellin by the plant FLS2 receptor (Zipfel et al., 2004). This mechanism can be depicted as the simplest network possible, containing two nodes, the flg22 and FLS2 molecules, connected by one edge representing the interaction between them (Figure 1a). The output of this system corresponds to the phenotype observed upon the interaction of these two molecules: PTI. In many cases, PTI is sufficient to fend off pathogen attacks and keep plants healthy. The second layer of plant defense, called effector‐triggered immunity (ETI), is mediated by intracellular resistance (R) proteins that recognize molecules injected by pathogens into plant cells designated effectors (Jones and Dangl, 2006; Dodds and Rathjen, 2010). By contrast to PTI, which confers resistance against a broad group of microorganisms, ETI is specific to isolates of microorganisms producing a given effector, and leads to a complete resistance response often accompanied by a rapid programmed cell death reaction called the hypersensitive response (HR; Coll et al., 2011). In its simplest form, ETI can result from the interaction of a pathogen effector, such as Ralstonia solanacearum (R. solanacearum) PopP2, with a plant R‐protein such as Arabidopsis thaliana RRS1‐R (Deslandes et al., 2003) (Figure 1b). In this particular example, PopP2 was shown to modify RRS1‐R by acetylation (Le Roux et al., 2015). This modification is not reciprocal and can be depicted as a directional interaction.
Figure 1.
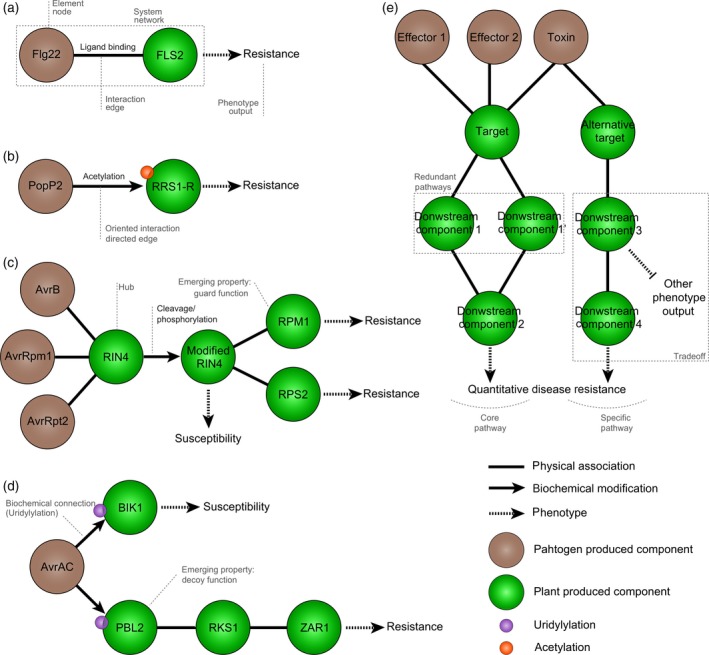
Progression of network‐like representations of molecular plant–pathogen interactions.
(a) The direct perception of a pathogen PAMP (such as flg22) by a plant receptor (FLS2) can be seen as the simplest form of network with two connected nodes.
(b) Similarly, the direct interaction of a pathogen effector (such as Ralstonia solanacearum PopP2) with its plant target (such as the Arabidopsis thaliana R‐protein RRS1‐R) is a simple two‐node network. PopP2 modifies RRS1‐R by acetylation. This modification is not reciprocal, the interaction is oriented.
(c) In the guard model, modification by effectors (such as AvrB, AvrRpm1 and AvrRpt2) of a plant target (such as RIN4) is monitored by plant resistance proteins (RPM1 and RPS2), which are not directly interacting with effectors. In this example, RIN4 in connected to multiple nodes and can be designated as a hub.
(d) In the decoy model, a pathogen effector (such as AvrAC) interacts with an operative target (BIK1) to promote plant susceptibility, but can also interact with a decoy target (PBL2), guarded by a R‐protein complex (RKS1 and ZAR1) leading to plant resistance.
(e) Quantitative disease resistance likely involves a complex network integrating several input signals (Effector 1, Effector 2, toxin,…) perceived simultaneously and signaling through common (‘core’) or specific pathways to initiate plant defense responses. Pathogen molecules are shown as brown circles and plant molecules as green circles.
Research over the past decades has shown however that direct interaction between effector and R‐protein is rather the exception than the norm, leading to more complex models and networks. Instead, the detection of effector proteins usually depends on the detection of their activity within the plant cell. For this, plant R‐proteins monitor the status of other plant proteins (called ‘guardees’), which are the direct targets of pathogen effector proteins. This model is referred to as the ‘guard’ hypothesis (Dangl and Jones, 2001). A typical guard system is composed of the AvrB effector of the pathogenic bacterium Pseudomonas syringae (P. syringae) which directly interacts with the plant RIN4 protein (the guardee) to modify it. The modified RIN4 protein is then recognized by the plant R‐protein RPM1 (Figure 1c) (Jones and Dangl, 2006). In the absence of modified RIN4 protein, RPM1 is not able to detect the AvrB effector and mount ETI. Its function as a ‘guard’ therefore only makes sense in the context of a gene network containing RIN4. This exemplifies so‐called emergent properties of a network, properties that emerge from the connections between multiple elements of a network (see Glossary). The RIN4 example is further complicated by the fact that RIN4 can be modified by several bacterial effectors, AvrB, AvrRmp1 and AvrRps2, and is guarded by two R‐proteins, RPM1 and RPS2 (Figure 1c). RIN4 forms a node with a high number of connections, often designated as ‘hubs’.
In some situations, the modification of a guardee protein by pathogen effectors may be detrimental to the plant. This reasoning leads to the decoy model in which the guardee is a mimic of a plant protein modified by pathogen effectors to promote susceptibility (van der Hoorn and Kamoun, 2008). For example the effector protein AvrAC from Xanthomonas campestris (X. campestris) targets BIK1, a central component of PTI signaling in A. thaliana to promote susceptibility. But instead of guarding BIK1 directly, the plant evolved a related protein (PBL2) which is ‘mistakenly’ modified as well by AvrAC and guarded by a preformed complex of RKS1 and the R‐protein ZAR1 (Figure 1d) (Wang et al., 2015). Here, the decoy function of PBL2 can only by understood in connection with BIK1, RKS1 and ZAR1 function, providing another example of an emergent property of plant immune response networks. Recent work has revealed that some R‐proteins include an integrated decoy domain, combining guard and decoy functions (Le Roux et al., 2015; Kroj et al., 2016; Sarris et al., 2016).
Another form of plant immune response extensively observed in crops and natural plant populations confers partial resistance to pathogens and is usually referred to as quantitative disease resistance (QDR) (Kover and Cheverud, 2007; Poland et al., 2009; Roux et al., 2014). QDR can be considered as the result of interplay between multiple molecular events, including the activity of multiple pathogen effectors and toxins on plant targets and the activation of multiple plant response pathways (Figure 1e) (Roux et al., 2014). However, there is still very limited information about the molecular mechanisms underlying QDR. QDR genes have been identified only in few cases and encode diverse molecular functions underlying durable and broad‐spectrum resistance (Fu et al., 2009; Fukuoka et al., 2009; Krattinger et al., 2009). Interestingly, one of them is RKS1, an atypical kinase also found to play a key role in ETI by interacting with the R‐protein ZAR1 (Huard‐Chauveau et al., 2013; Wang et al., 2015) revealing interplay between different forms of resistance. Overall, thousands of plant and pathogen genes are differently expressed during pathogen infection (Tao et al., 2003; Windram et al., 2012; Lewis et al., 2015), and so far the function of many of these gene products is unknown. Furthermore, under natural conditions, plants are exposed to constantly changing conditions, surrounded by a world of pathogenic and non‐pathogenic microorganisms (the microbiota) and adjusting their physiology to fluctuating environmental conditions (abiotic stress) (Agler et al., 2016; Müller et al., 2016). On the microbial side of the interaction, cell activity needs to be adapted not only to overcome plant defense mechanisms, but also to enable pathogen to feed from resources provided by the plant host, cope with competing microbes and adapt to changes in the environment. This notably implies the manipulation of host cell functions, the conversion of host molecules into easily assimilated compounds and their transport (Chen et al., 2010). Hence, the metabolism of plants and pathogens are interconnected and need to be seen as one unit.
We argue in this review that systems biology approaches are particularly well suited to tackle the complexity of plant–pathogen interactions. In contrast with more classical reductionist approaches, in which a limited number of cell components are studied, systems biology is the attempt to understand biology as the structure and dynamics of cellular and organism functions altogether (Kitano, 2002). The advancements in high‐throughput methods and new approaches in data analysis has made possible the integration of multiomic data from genomic, proteomic and metabolomic sources with the goal to model biological networks and predict the effect of perturbations on these networks. This review aims at providing an overview of current approaches, challenges and applications of systems biology to describe plant–pathogen interactions at the network scale, with the goal to model and predict the outcome of plant–pathogen interactions.
Basic Principles of Systems Biology
Systems biology aims at understanding the properties of living organisms emerging at the network level (or system level), also called emergent properties (Kitano, 2002). Emergent properties are phenomena that cannot be associated to a single component of a system (a unique gene or molecule for biological systems), but rather arise from the interaction between multiple components. As a methodology, systems biology aims at integrating observations on multiple components of the system (cell, organs, or populations) by using mathematical models. As a scientific field, it requires the development of tools in order to: (i) collect qualitative and quantitative data on many elements of the system, at the whole genome scale when feasible, and for different types of cell components such as the genes, RNAs, proteins or metabolites, (ii) the reconstruction of models which are formal descriptions of the components of the system and the interactions between them. These models should be convertible in mathematical format, and (iii) computational algorithms should be able to calculate in a reasonable computational time the behavior of such complex systems, based on the experimental data collected and the model canvas (Sauer et al., 2007).
The ‘omics, collecting data at the global scale
Systems biology has emerged as a broadly used methodology with the development of the so‐called ‘omics techniques and the spectacular progress in techniques for the characterization of the main components of the cell during the last two decades: DNA and RNA sequencing for genes, and mass spectrometry (MS) for proteins and metabolites (Metzker, 2010; Altelaar et al., 2013) (Figure 2). Plant and filamentous pathogen genomes can represent several hundreds of megabases and contain numerous repetitive regions, which make genome assembly challenging. The recent development of Single‐Molecule, Real‐Time Sequencing technology (Eid et al., 2009), which sequences DNA molecules longer than 10 000 base pairs with high accuracy, is expected to fill the gap in whole eukaryotic genome sequencing and assembly approaches (Faino et al., 2015; Zapata et al., 2016). This technique has also been emerging as a means to characterize DNA methylation patterns at the whole genome scale (Beaulaurier et al., 2015). Concerning transcriptomics, RNA sequencing (RNA‐seq) technology (Wang et al., 2009) opens new perspectives in measuring the transcriptional reprogramming of any organism, or plant–pathogen pairs at once (Kawahara et al., 2012). Indeed, no prior knowledge on the genomic sequence is required. Thus, exhaustive measurements are theoretically reachable even for non‐sequenced organisms. In addition, structural information on transcripts is available, like alternative splicing of eukaryotes transcripts. Proteomics achieved high level of completeness with the development of fast mass spectrometers performing peptide sequencing within complex samples. Quantitative assessment of a proteome in a sample can reach around 55% completeness (Schmidt et al., 2016), meaning that 55% of the theoretical proteome corresponding to the entire predicted gene set in the genome can be measured. Nevertheless, the true proteome of the sample may be lower than the theoretical proteome since many genes may not have been expressed in conditions tested. Slightly lower amount of proteome completeness are reported in planta compared to ex planta samples. For instance 3168 proteins on the 10 952 predicted genes of Zymoseptoria tritic, 29%, were measured within in planta samples, compared with the 5731 proteins measured, 52%, for the in vitro samples (Yang and Yin, 2016). Methods are now available to characterize protein modifications, like phosphorylation, and obtain relative or absolute protein quantification (Larance and Lamond, 2015). Metabolomics, the analysis of small (<1000 g mol−1) molecules, deals with compounds of very diverse chemical properties (lipids, sugars, organic acids, etc.) requiring diverse techniques for extraction and analysis. Liquid chromatography and gas chromatography are often used to separate complex samples and are coupled with MS or nuclear magnetic resonance for identification and quantification (Heuberger et al., 2014). Classically, several hundreds of metabolites can be unambiguously identified using untargeted metabolomics on in planta samples (Camañes et al., 2015). However this number is still one decade lower than the potential metabolome expected from known metabolic networks. Stable isotopes, like 13C or 15N, can also be used to facilitate metabolite identification and quantification, or gain insight on metabolic activities (Freund and Hegeman, 2016). In addition, isotopic enrichment of molecules and their mapping on metabolic networks (Szyperski, 1995) allows the quantification of fluxes of atoms through the cell. This so‐called fluxomics analysis represents the observation of the system in action, and can be used to reveal the cell physiology which results from the integration of transcriptional, translational and metabolic regulation (Sauer, 2004).
Figure 2.
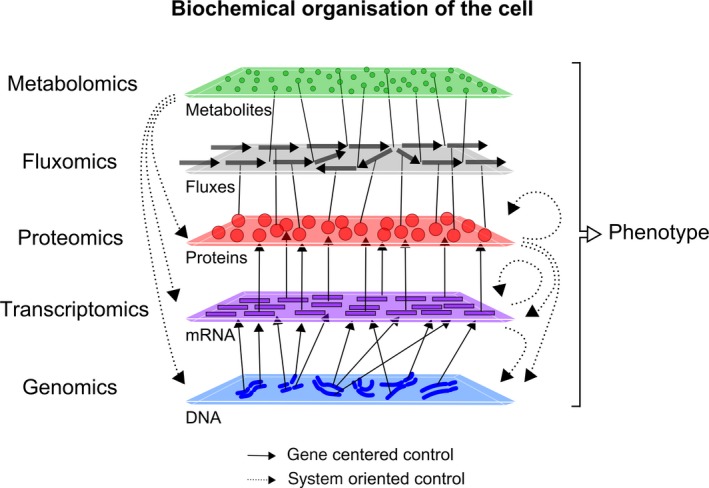
The biochemical nature of the cell components drives cell organization and thus methods used to analyze them.
The cell organization links genes to transcripts to proteins to fluxes of metabolites and backward regulation of the gene expression via biochemical relationships.
Model reconstruction: making sense of the ‘omics
Numerous studies begin with ‘omics experiments providing a large‐scale description of cellular components. Future development of new methodologies will seek to improve the completeness of such system‐scale measurements. If further development of methods will be valuable to shed light on not yet unmeasured components, this is however generally not sufficient to provide new insights into biological processes and their impact on life of the cell or the organism. A classical approach is to follow up with functional analyses focused on cellular components showing the most extreme behavior, such as genes dramatically up‐regulated (Raffaele et al., 2008). However the function of a single gene may be masked in such reverse genetics approaches by redundant or compensatory genes. For instance, single mutants in the Arabidopsis transcription factors TGA2, TGA5 and TGA6 showed wild‐type phenotype whereas a tga6‐1 tga2‐1 tga5‐1 triple mutant was strongly impaired in resistance, revealing redundant roles for these genes (Zhang et al., 2003). This situation should be frequent considering that biological systems, and the plant immune system in particular, are tightly interconnected networks of molecules and macromolecules of various nature with many emergent redundant modules (Pritchard and Birch, 2011) (Figure 2). Furthermore, functional redundancy can emerge at various levels, like gene, enzymatic complex or pathway redundancy. In other words, two independent pathways, not sharing any component, may have the same output and thus compensate each other depending on the genetic perturbation or the environmental conditions (Tsuda et al., 2009). Therefore, a number of biological functions cannot be understood based only on the systematic characterization of cellular components, but also require knowledge on the nature of the connections between components. This nature includes connection between components of a same type (such as protein–protein interactions, post‐translational modifications or catalytic activity for proteins) or between cellular components of a different nature (such as protein–DNA interactions) (Figure 2). Then, the causality between the state of one component and the phenotype can be studied from a network perspective. This effort of integration of the different layers of cellular components and interactions in a formal way is called ‘model reconstruction.’
Models, what kind for what
A model containing molecular components and biochemical interactions is called a mechanistic model. Different kinds of mechanistic models are used in systems biology to analyze cell functions (Figure 3). In this review we will discuss the three main classes used in systems biology, according to the way the interactions between components are described: (i) constraint‐based models, (ii) logical models, and (iii) kinetic models (Bordbar et al., 2014; Le Novère, 2015). Alternative modeling approaches with numerous variants and hybrids have been reviewed elsewhere (Bordbar et al., 2014; Le Novère, 2015).
Figure 3.
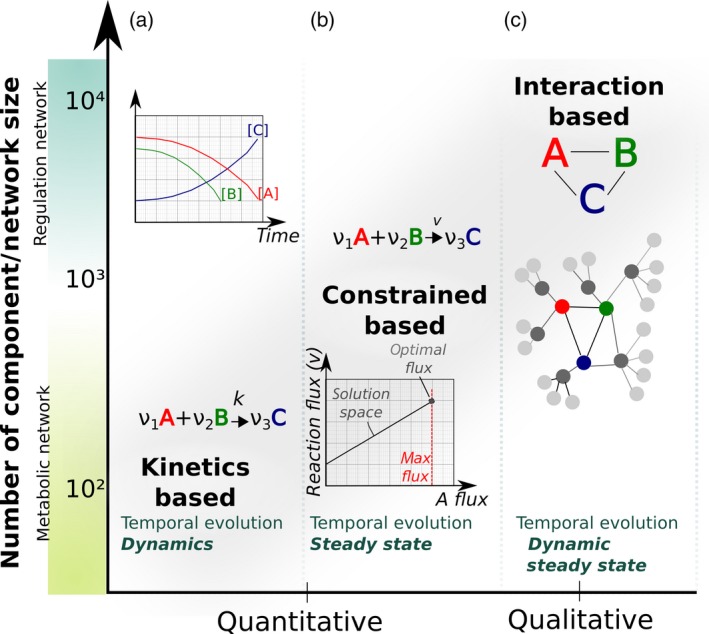
Main type of modeling approaches and their usage.
The three main classes are the kinetic modeling, the constraint‐based modeling and the logical modeling. Usage of one or the other depends of the nature of the network studied (metabolic network, regulatory network), the amount of information available (kinetic parameters) and the temporal response of the system to be investigated (dynamic or steady states).
Constraint‐based models are historically used to study networks of metabolic reactions. They were developed in the frame of metabolic engineering with the aim to optimize metabolic pathways to increase the production of chemicals of interest or biomass (Schatschneider et al., 2013). They are generally successful in predicting phenotypes such as growth rate, based on knowledge of environmental conditions. These models assume constant variation for all properties of all components of the network (called ‘steady states’ for the network; see Glossary), and allow applying a strong constraint of equilibrium between fluxes around each metabolite. As a result, all possible combinations of states for all components of the system (the ‘space of solutions’) can be determined by linear programming for more than 1000 reactions, within a few seconds. The main type of constraint‐based model analysis is called flux balance analysis (Orth et al., 2010) and allows the study of optimality principle in a biological system, that is to say determine the combination of solutions achieving a maximal output (see Glossary) (Poolman et al., 2013).
Logical modeling is based on logical rules of interactions between network entities and is suitable for large‐scale networks. It describes the changes in the state of a given cellular component depending on the states of one or multiple others components. This approach is particularly adapted to model regulatory events. For instance the activation of signaling through the FLS2 receptor will depend on the presence of its flg22 ligand in the extracellular space (Figure 1a). The states of the model are simulated recursively over time, revealing feedback loops within regulatory networks and finding attractors, i.e. recurrent set of network states toward which the regulatory cascades evolved (Figure 3).
Kinetic modeling is the most refined modeling approach to describe the dynamics of living systems. It is based on ordinary differential equations (ODEs) describing the kinetics of changes in the concentration or states of cellular components, based on laws governing chemical equilibrium, such as mass action law (for binding of a ligand to a receptor) and enzyme kinetics (Michaelis–Menten equation). Thus, the systems can be studied far from equilibrium and dynamically through time. However, they require prior knowledge on numerous kinetic parameters which is often not available, and thus the scaling‐up at genome‐scale models remains challenging (Smallbone et al., 2010).
The different steps in model reconstruction
Model reconstruction consists in: (i) defining the purpose of the model and therefore the type of model to be reconstructed (Figure 3), (ii) collecting information on the biological system, by collecting omics data, performing genome annotation, collecting physiological data or measuring kinetics parameters of biochemical reactions, (iii) converting and assembling the raw information in Systems Biology Markup Language (Hucka et al., 2003; Chaouiya et al., 2013), a standardized format suitable for further conversion in mathematical format, (iv) curation of the model by checking in detail the confidence of each component properties and interactions, (v) converting the model in a mathematical format, (vi) running simulations to correct bugs and unveil missing components, and (vii) validating the model by comparing predictions with experimental data (Figure 4). In an ideal process, the data used for the validation should not have been used for building the model. The most time‐consuming part is the curation step which requires in‐depth investigations through databases like KEGG (Kanehisa and Goto, 2000) and BIOCYC (Caspi et al., 2012) for metabolic networks, TCDB (Saier et al., 2006) for transporters, BRENDA (Chang et al., 2015) for reaction kinetics parameters, STRING (Szklarczyk et al., 2015) for regulatory networks, and checking the validity of properties assigned to components of the system, notably by performing BLAST and extensive literature searches. A protocol for generating a high‐quality metabolic network reconstruction has been proposed by Thiele and Palsson (2010).
Figure 4.
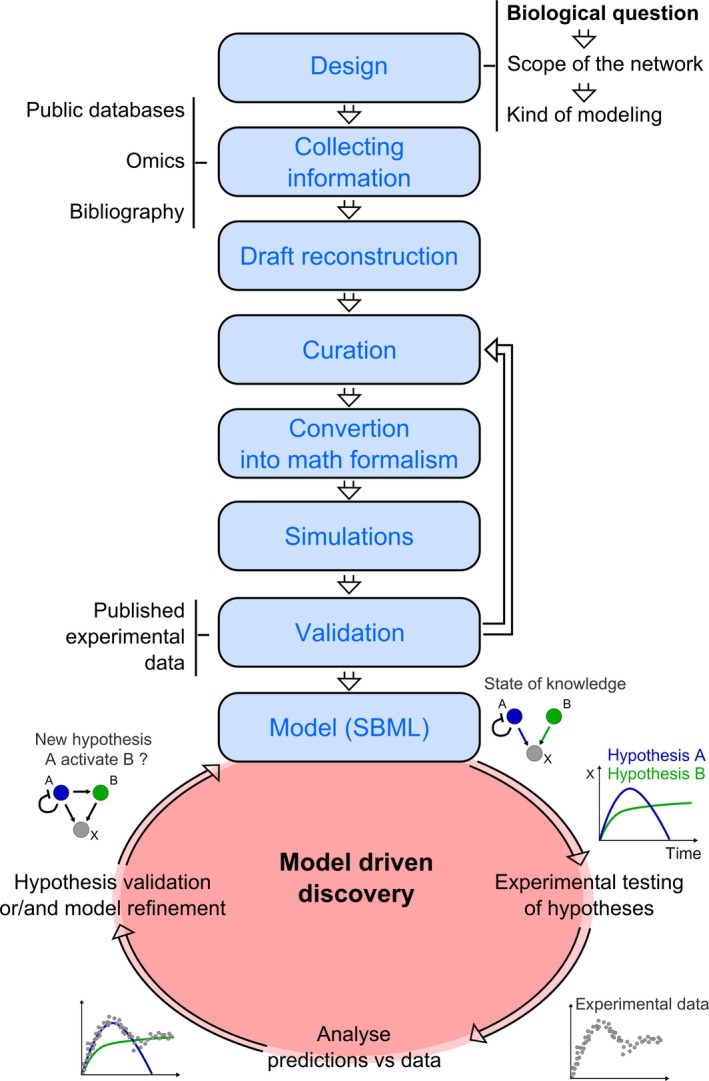
Reconstruction process of mechanistic models of living organisms.
The model can be used to generate hypothesis on the function of a biological system and design experiments to validate this hypothesis. Hence, discovery may be accelerated thanks to focused experiments. The new knowledge obtained can be integrated in the model in an iterative process. In the toy model depicted activation of X by A (hypothesis A) or B (hypothesis B) in a given condition is investigated.
Interactions between regulatory components are difficult to predict from genome annotation because homologous regulatory proteins are easily recycled in different signaling pathways across organisms. Therefore reconstruction of regulatory networks often requires the collection of specific biochemical data on the interactions within the studied organism. Methods for inferring automatically or semi‐automatically regulatory networks from ‘omics data, called network inference, have been used from time course gene expression data (Penfold and Wild, 2011). This process, based on statistical algorithms, determines the probability of a dependency between two or more components investigated, for instance how the expression of a gene is dependent on the activity of a transcription factor previously expressed. This approach successfully predicted a role for the expression of A. thaliana TGA3 transcription factor in controlling Botrytis cinerea colonization (Windram et al., 2012). It also allowed to identify the A. thaliana transcription factors XND1 and FBH3 as putative targets of effectors from P. syringae (Lewis et al., 2015). By providing candidate interactions (direct or indirect), this method should accelerate the discovery of interactions (Sato et al., 2010) and expand the number of reconstructed regulatory networks.
Predictive capacity of models
Systems biology approaches are often useful for their particularly high predictive capacity (Varma and Palsson, 1994). The predictive aspects of systems biology approaches are provided in the one hand by considering physical laws which are universal (Zwieniecki and Dumais, 2011; Barbacci et al., 2015) to govern interactions between components, and in the other hand, by considering the specific biochemical components of each organism. Indeed, the interaction rules have been deduced under reasonable assumptions (steady‐state, isobars and isothermal environments) from general laws of thermodynamics (conservation laws, second principle of thermodynamics, see Biophysical perspectives below). Hence, interactions in models remain valid under known hypothesis and allow the extrapolation of models behavior in conditions not yet tested experimentally. Concerning the consideration of the specific biochemical components of organisms, this consideration allows to predict phenotypes on the basis of known specific features of organisms like genomes. One well established predictive capacity of models is related to nutritional needs for the biomass growth of living organisms (Varma and Palsson, 1994). Indeed, the nature and the amount of matter and energy present in the environment fully constrain growth of organisms. In one side, the amount represents a maximal growth capacity that cannot be exceeded to respect the conservation law of matter and energy. In another side, the specific metabolic capacities of organisms to collect these nutrients are determined by their transporters and enzymes repertoires, and confine their habitat into specific niches (Zhu et al., 2016). In the context of plant–pathogen interactions, growth of both partners can be limited by the amount of matter and energy they can extract from their local environment.
Modeling, an iterative process of knowledge accumulation and validation
Mechanistic models are simplified mathematical representations of the knowledge collected so far on a living organism. Therefore, many yet undiscovered features of the organism may be missing in reconstructed models. Hence, the model performance in predicting observed phenotypes must be assessed rigorously to validate the model (Thiele and Palsson, 2010). When a high accuracy between the model predictions and the experimental data is achieved, the model can be used to explore phenotypes predicted in conditions not yet tested experimentally. This is expected to reveal novel key players in a phenotype of interest. Then, the model can be used to test multiple hypotheses by adding putative components or interactions followed by the design of an experimental setup to validate these hypotheses. This draws a virtuous iterative process between the experimental and the in silico work, enriching the model and the knowledge at each cycle (Figure 4).
Challenges for Plant–Pathogen Systems
Modeling the plant immune system
The knowledge collected so far depicts the plant immune system as a tightly regulated temporal and quantitative system (Schwessinger and Zipfel, 2008). For instance PTI and ETI appear as tightly tuned responses via feedback loop(s) ensuring a transient and appropriate level of response depending on the invader, and compatible with maintaining plant cell viability. Our global understanding of the immune system functioning is mainly based on expository models, i.e. drawn diagrams dedicated to clarify and summarize data and concepts (Pritchard and Birch, 2014). In addition, many components of the immune systems and relationships between them remain to be discovered. In this context, inference of the immune signaling network of A. thaliana upon challenge with the bacterial pathogen P. syringae expressing the effector protein AvrRpt2 constituted an important step (Sato et al., 2010). An integrated regulatory network comprised of 22 components including most of the genetically defined regulators of immunity in Arabidopsis, was inferred based on mRNA profiles for 571 immune response genes of Arabidopsis mutants with defects in immune regulatory genes. Existence of signaling inhibitions in the network suggests that only part of the signaling network is usually used and this organization balances the trade‐off between a high robustness of the plant immune signaling and minimizing negative impacts of the immune response on plant fitness. Moreover, Naseem and co‐workers conducted pioneer research on mathematical modeling of the immune system of A. thaliana by reconstructing a hormonal interaction network merged with the immune network (Naseem et al., 2012). Interaction between components was described using logical‐modeling approach. Then, they converted the logical rules into ODEs to simulate the dynamic of the systems. Among others, the model predicted that cytokinin does not influence early events of PTI during infection by P. syringae and the prediction was experimentally validated. Antagonism between cytokinin and auxin signaling in plant immunity was also predicted (Naseem and Dandekar, 2012; Naseem et al., 2012). This pioneer modeling study should be extended by adding kinetics information and by including recent data. In addition, integration of pathogen network pieces (PAMPs, and effectors) to simulate the presence or absence of the pathogen is clearly a strategy to pursue for a better integration of the pathogen model into the game.
Integrating pathogen and host systems
Until now, pathogens and plants have rarely been studied together as an interacting system, in the sense of systems biology. Systems biology studies on plant cell wall architecture and on pathogen enzymes degrading the plant cell wall are for instance usually conducted separately. So far, to our knowledge, there is no such study coupling genome‐scale models of a phytopathogen with that of a plant, like the analysis of the model for the human pathogen Mycobacterium tuberculosis in combination with the macrophage model to reveal nutritional coupling (Bordbar et al., 2010). This may appear paradoxical for studies aiming at understanding interaction between organisms. The detection of plant cell wall degrading enzymes (PCWDE) in the genome of pathogens is for instance often associated with the ability to feed on dead plant cells, although PCWDE may exclusively be used to overcome physical barriers to plant tissue colonization (Kraepiel and Barny, 2016) (see Biophysical perspectives). The integration of plant and pathogen metabolic networks would be required to determine the exact role of PCWDE in pathogenic lifestyles. More generally, pathogens produce molecules that evolved to function in plant cell systems (effectors sensu lato), and reciprocally. Connecting the models of the two partners is therefore critical to capture fully the dynamics of plant–pathogen interactions.
Data and model integration across scales: from the cell to the environment
Plant–pathogen interactions can be characterized from cell to organism level and depend strongly on abiotic factors. The span of temporal and spatial scales involved in the interaction challenges multiscale data integration and bottom‐up modeling approaches (Figure 5) (Cunniffe et al., 2015). At early steps of the infection process, microbial pathogens initiate primarily cell to cell molecular combat with their hosts which may lead later to systemic invasion and/or systemic host responses (Gjetting et al., 2004; Asai et al., 2014). At the single‐cell level, biological systems are prone to stochasticity, the existence of random noise in the state of network components (see Glossary) (McAdams and Arkin, 1997). Stochasticity, in addition to microenvironment fluctuation, may lead to heterogeneity within cell populations, and to variability in the final output of the interaction (Ackermann, 2015). Besides, the response of the system over time may depend on the initial states for the pathogen and the plant cell. For instance, pathogen success can be impaired by resistance priming due to previous infection events or other signals (Conrath et al., 2015). Current models are often built at the cellular level, and are well adapted to analyze localized plant–pathogen interactions. However, computing kinetics of such complex interaction networks, notably considering stochasticity, remains challenging. Besides, investigations of single‐cell dynamics are mostly carried out via microscopy using reporter genes (Asai et al., 2014), while single‐cell ‘omics data are emerging but remain scarce (Lin et al., 2014; Schiefelbein, 2015). Further efforts will be needed to establish libraries of models for various cell types with time resolution in the minute range.
Figure 5.
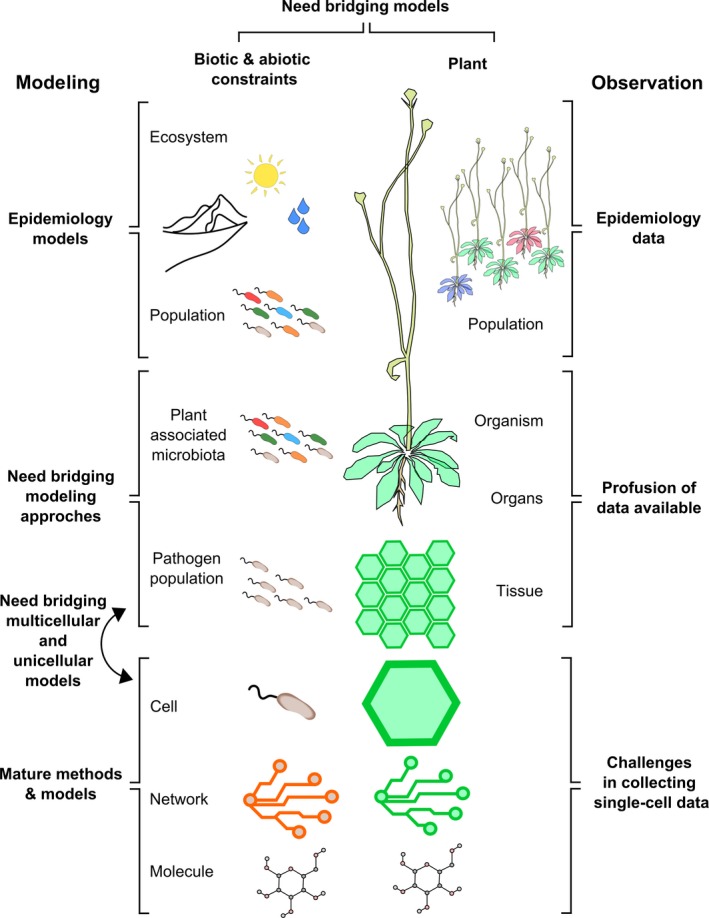
Scale of model and data integration from molecule to ecosystem.
At the multicellular scale and at a time frame of hours, the challenges concern modeling approaches rather than data collection. Indeed, to date most transcriptomics data have been collected at tissue or cell‐type scale using dissection methods like laser microdissection (Nelson et al., 2008; Gautam and Sarkar, 2015). Also, metabolomic analyses of intercellular fluid (like xylem) or whole organs were performed (Abeysekara et al., 2016). However, extraction methods remain limited in their ability to distinguish between intracellular and intercellular metabolites. One promising method is imaging MS. Ryffel et al. (2016) used this technique to reveal the spatial heterogeneity of substrates available at the surface of leaves upon challenge with P. syringae. Although some properties of plant and pathogen organisms can only be studied at the multi‐cellular level, such as intercellular communication and cooperation, the spatial dimension is rarely considered in mechanistic models. One key phenomenon of plant resistance appearing at this scale is the spread and restriction of the HR (Pogány et al., 2009). Modeling the spatial dimension of the plant tissue may be crucial to gain insight in how cell death during HR is restricted only to the infection site. Modeling plants at the organism level has been achieved recently through a digital multiscale model of Arabidopsis (Chew et al., 2014). This model is a hybrid, bridging multiple model modules and can depict the contribution of one individual organ on the whole plant for different environmental conditions. Such bottom‐up modeling requires the coupling between cellular and multicellular oriented methods. The latter was mostly developed by the biophysics scientific community (cf. next subsection). The next step will be to bridge biophysics multicellular models with cellular genome‐scale models (Figure 5).
At an even larger‐scale, ecosystems are well known to influence plant disease epidemics, through the presence of beneficial or competing organisms in the microbiota associated with the plant (Wei et al., 2015; Müller et al., 2016) or by changes in nutrient availability, humidity and temperature notably (Hacquard et al., 2016). For instance, Wei and co‐workers manipulated the diversity of bacterial species present in the soil to demonstrate that rhizosphere bacteria competing specifically with the soil pathogen R. solanacearum for plant nutrients prevented tomato infection and disease onset (Wei et al., 2015). This experimental evidence calls for investigating the function of ecosystems associated with plants. Systems biology may allow in the future the connection of well established epidemiology models at the population level with the molecular level understanding of plant–pathogen interactions.
Biophysical perspectives
The finest description of biochemical network behavior is provided by kinetics‐based models (Figure 3). The state of the system can be described by kinetics of chemical reactions driven by chemical potentials of biomolecules. Such a framework provides a quantitative description of Gibbs free energy which corresponds only to the chemical part of the whole internal energy of compounds (see Glossary) (Callen, 1985). Interaction between chemical energy and other forms of energy (mainly thermal and mechanical) are often not regarded in kinetics‐based models. A finer description of kinetics of reactions, especially in fluctuating temperature conditions, would require consequently the extension of classical framework to other forms of energy. At a higher scale, taking into account other forms of energy is indispensable. Indeed, plant–pathogen interactions are also physical in nature as host successful colonization requires penetration and movement through the host tissue (Abramovitch and Martin, 2004; Money, 2008; Sanati Nezhad and Geitmann, 2013). Thus, the sole chemical description of the system is likely to be insufficient to explain plant–pathogen interactions at the higher scale. To address this limitation, the description of how molecular networks and molecules interfere to provide geometrical and mechanical properties through scales must be considered. At the cellular and molecular scale, the colonization of plant tissues by filamentous pathogens requires the fine regulation of water quantity, mechanical properties and shape of the apical cell (Goriely and Tabor, 2008; Money, 2008). On the plant side, mechanical properties determine the intensity of mechanical signals perceived during interaction. These signals are involved in many regulation and feedback loops (Coutand and Moulia, 2000; Moulia et al., 2015) and may prime QDR (Mbengue et al., 2016). Hence, to have a better understanding of molecular events governing plant–pathogen interactions, a description of how mechanical properties are generated by the assembly of polysaccharides and proteins is crucial. Genome‐scale metabolic models can reveal functional links between genes and cell wall polysaccharides, but modeling the emergence of mechanical properties of a cell from knowledge on polysaccharide regulation remains challenging (Somerville et al., 2004).
These themes have enjoyed a long history of research in biophysics (Ali et al., 2014) and many models have been developed to understand the links between molecules, physical properties and growth (Bartnicki‐Garcia et al., 1995; Barbacci et al., 2013; Ali and Traas, 2016) or shape (Dumais et al., 2006; Goriely and Tabor, 2008). Frameworks allowing the integration of mechanical regulation loops in biological processes to design systems biology approaches have also been proposed (Moulia and Fournier, 2009; Moulia et al., 2011, 2015). Feedback of mechanical forces on growth regulation has also been emphasized for key mechanisms such as growth (Hamant et al., 2008; Moulia et al., 2015), proprioception (Hamant and Moulia, 2016) or cell division (Louveaux et al., 2016), illustrating how bottom‐up approaches can integrate a physical framework and knowledge on molecular components of the cell (Figure 5). This integration should provide powerful tools to investigate the link between genetic regulation and plant function through multiscale mechanisms.
The pursuit of metabolic model reconstruction for pathogens and plants
Enrichment of the plant–pathogen field with studies using systems biology approaches will go hand in hand with the release of freely available models for pathogens and plants. Although the metabolic reconstruction for microorganisms should be easier than for plants due to the smaller size of their genomes, models exist only for X. campestris (Schatschneider et al., 2013) and R. solanacearum (Peyraud et al., 2016) (Figure 6), and the R. solanacearum model is the only genome‐scale model. The first plant genome‐scale metabolic reconstruction was performed by Poolman et al., (2009) on A. thaliana. This model was dedicated to study heterotrophic cell suspension culture, whereas the later AraGEM model was used to simulate C3 photosynthesis (de Oliveira Dal'Molin et al., 2010). The same authors built C4GEM, a model‐frame dedicated to study C4 photosynthesis (Dal'Molin et al., 2010). Several crops such as rapeseed (Pilalis et al., 2011), barley (Grafahrend‐Belau et al., 2009), maize (Saha et al., 2011), rice (Poolman et al., 2013), millet (de Oliveira Dal'Molin et al., 2016) and tomato (Yuan et al., 2016) were reconstructed, with either complete genome‐scale models or specific metabolic pathways only. Considering the few pathogen models reconstructed so far and the remarkable diversity of plant–pathogen lifestyles and infection strategies, systems biology should be a fertile ground for future advances in plant–pathogen interaction studies.
Figure 6.
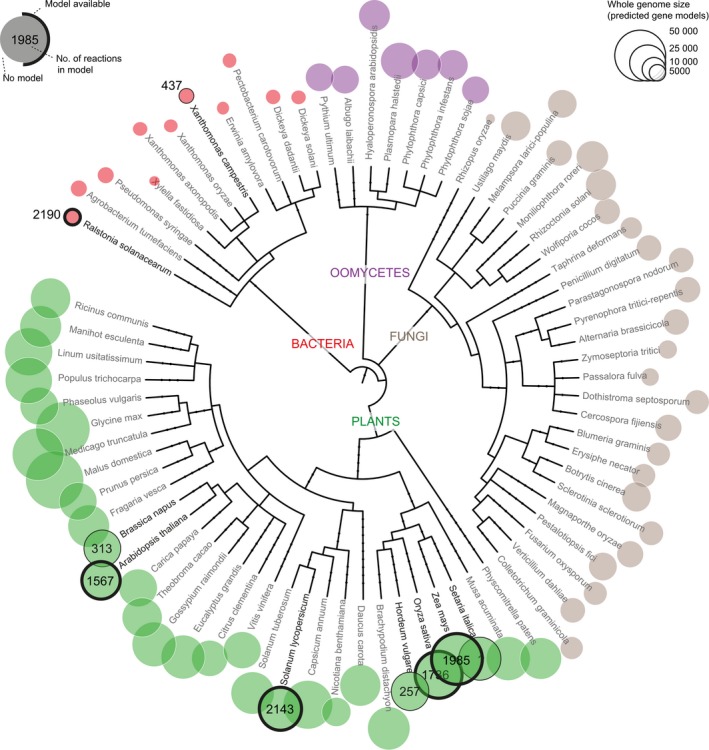
Reconstructed models of plants and plant‐associated microbes across the tree of life.
The phylogenetic tree was generated using PhyloT with a selection of fully sequenced plants (green) and associated microbes, including fungi (brown), oomycetes (purple) and bacteria (red). Circles at the tip of branches are scaled to whole genome sizes (given as the number of predicted genes). Genomes for which a reconstructed model exists are shown with a black line and the number of reactions in the model is indicated.
The Promises of Systems Biology to Plant Pathology: What Can We Do With This?
Robustness of the plant immune system and the virulence effector interactome, a network paradigm
The organization of biological pathways in networks underpins several emergent properties that individual signaling pathways do not have, such as robustness, the ability of a system to maintain its function(s) despite external or internal perturbations. The plant immune system and the arsenal of virulence factors used by pathogens can be considered as robust biological networks (Figure 7a). Microbial pathogens typically possess a battery of effectors aimed at disabling immunity after being translocated into plant cells: the bacterial pathogen R. solanacearum injects up to 75 Type III effectors into plant cells (Peeters et al., 2013), and the genome of some filamentous plant pathogens encodes hundreds of effector proteins (Haas et al., 2009; Pedersen et al., 2012). With few exceptions, the deletion of a single (or even multiple) effector gene(s) has no or only a minor impact on microbial virulence (Schirawski et al., 2010; Cunnac et al., 2011; Büttner, 2016), supporting the view that effector networks exhibit robustness (Figure 7b). Several pathogen effectors can have at least partly redundant functions, such as Phytophthora infestans EPIC1, EPIC2B and AVRblb2 that suppress the activity of tomato C14 protease at different levels (Kaschani et al., 2010; Bozkurt et al., 2011) (Figure 7c). Such robustness might be evolutionarily favorable for pathogens by facilitating evolvability, allowing frequent gains and losses of effector genes to avoid recognition by the plant immune system, without compromising global virulence (Yoshida et al., 2016). The loss of complete dispensable plasmids and chromosomes has also been associated with virulence in some pathogen species (Jackson et al., 2011; Raffaele and Kamoun, 2012). Reciprocally, the plant immune system generally ensures an invariant output (immune response) in the presence of considerable noise (e.g. non‐self recognition) (Figure 7b). In the arms race with pathogens, the plant immune network has evolved to detect invasion in spite of selection acting on microbes to escape recognition (Cook et al., 2015). Such robustness can be achieved through the deployment of partly redundant receptors at the cell surface, such as the rice OsCEBiP, OsLYP4 and OsLYP6 that all detect fungal chitin (Kaku et al., 2006; Liu et al., 2012) (Figure 7d). These extracellular receptors lack a cytoplasmic transduction domain and require interaction with OsCERK1 acting as a hub in chitin signaling (Gust, 2015, for review). Redundancy in the plant immune system is also notable for plant decoy proteins that detect the molecular activity of pathogen effectors and likely arose from gene duplication of high‐value guarded targets or convergent evolution (Figure 1d) (Cesari et al., 2014). Establishment of the Arabidopsis protein–protein interactome (Arabidopsis Interactome Mapping Consortium, 2011)and subsequent mapping of the interactome network between pathogen effectors and Arabidopsis proteins revealed that hubs of the plant immune system are targeted by numerous pathogen effectors (Mukhtar et al., 2011; Weßling et al., 2014).
Figure 7.
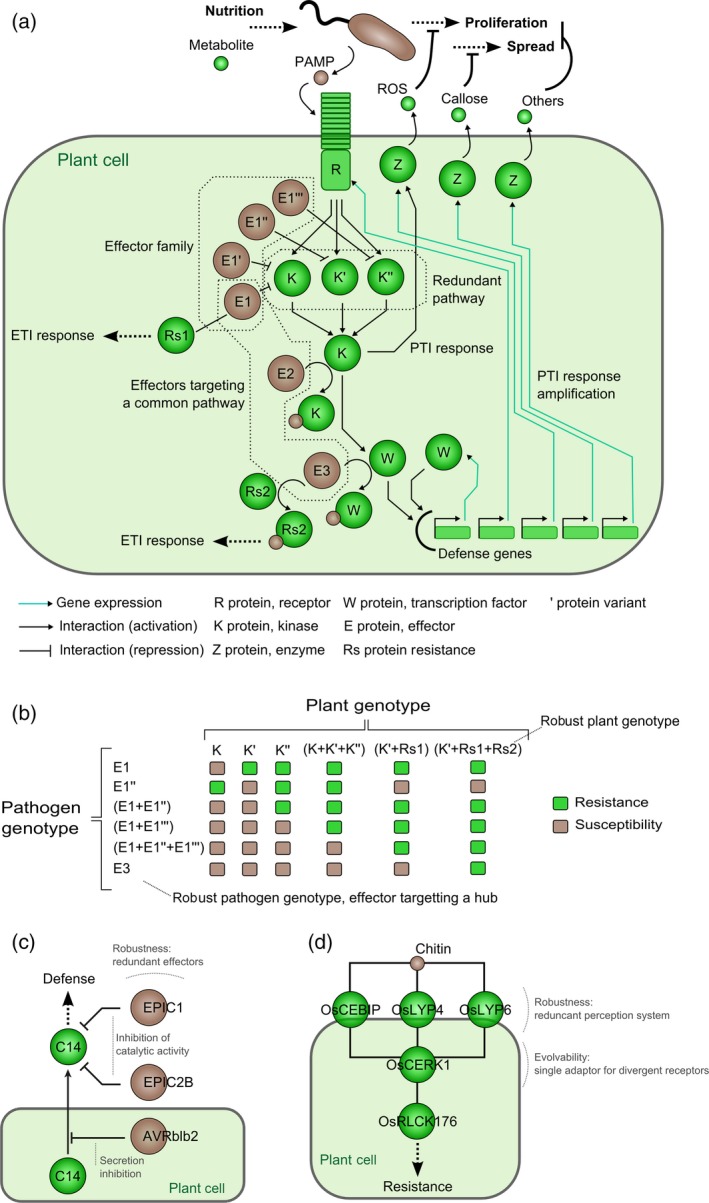
Robustness of plant–pathogen interaction network.
(a) The interaction between plant and pathogen involved a complex network. Here is depicted a sensing pathway recognizing the presence of the pathogen by PRR or R‐protein and the clearance of pathogen infection via PTI or ETI activation. The redundancy of pathogen effectors or plant components may provide robustness to each system.
(b) Predicted behavior of the previous toy model (a) depending of the pathogen and the plant genotypes. None of the outputs (resistance or susceptibility) can be deduced from knowledge on only one single component illustrating that plant resistance is an emergent property of the complex network of interaction between plant and pathogen.
(c) Example of robustness provided by redundancy in Phytophthora infestans effectors (EPIC1, EPIC2B and AVRblb2) that suppress the activity of tomato C14 protease.
(d) Example of robustness provided by redundancy in rice receptor (OsCEBiP, OsLYP4 and OsLYP6) that all detect fungal chitin.
Remarkably, the output of the interaction between different plant and pathogen genotypes shown in Figure 7(b) and deduced from the simple theoretical network shown in Figure 7(a) cannot be predicted based on knowledge limited to a single plant or pathogen component, highlighting the value of systems biology approaches in plant–pathogen interactions. These approaches are powerful to understand how remodeling of the plant network conditions the system's response towards disease susceptibility or resistance, and how different sets of effector molecules influence this remodeling. This knowledge should allow in the future better prediction of durability (i.e. robustness) of the plant resistance phenotype, especially if modeling studies also integrate the impact of environmental factors on interactions and their evolution (see below).
Studying trade‐offs between complex traits
Systems biology approaches, such as flux balance analysis, are powerful to predict the energetic cost of specific biochemical pathway(s), function(s) or even complex traits. Energetic costs are essential drivers for resource allocation to various biological traits in organisms. In turn, resource allocation is classically associated to the concept of trade‐off, by which gaining benefit for a trait then involves losing benefit for another trait (see Glossary). This resource allocation dilemma is paramount when resources are limited; however, some growth/defense trade‐offs are not always governed by metabolic competition and can be a consequence of hormone‐linked transcriptional network rewiring (Campos et al., 2016). There is a substantial cost of resistance to pathogens for plants which results in a trade‐off between growth and immunity (Todesco et al., 2010; Chae et al., 2016). Therefore, maintenance of R and other defense‐associated genes in plant populations can negatively impact growth and development (Tian et al., 2003; Todesco et al., 2010). Several plant hormones, including auxins, abscisic acid, ethylene, cytokinins, and gibberellins, that have been thoroughly described to regulate multiple aspects of plant growth, have recently emerged as key regulators of plant immunity (Denancé et al., 2013). Mechanistic studies revealed for example a hub transcriptional factor involved in brassinosteroid signaling and immunity (Lozano‐Durán and Zipfel, 2015). Although representing undeniable progress at the mechanistic level, these studies remain fragmentary and need to be interconnected to evaluate the impact of these cellular trade‐offs on complex phenotypes.
Virulence is a costly trait for pathogens, involving the secretion of a broad set of macromolecules, so that a trade‐off between virulence and growth exists in phytopathogens (Thrall and Burdon, 2003; Meyer et al., 2010). Such a trade‐off was recently illustrated using the genome‐scale model of R. solanacearum (Mansfield et al., 2012). This model encompasses a metabolic network module and a macromolecule module dedicated to the secretion of virulence factors (Peyraud et al., 2016). Simulations using this reconstructed model suggested that the massive production of virulence factors, such as exopolysaccharides, significantly impairs bacterial growth. These predictions were experimentally confirmed using bacterial mutants and by identifying PhcA, the major regulatory player shown to govern this trade‐off. Flux balance analyses also revealed that a drastic reduction of growth is observed in the presence of substrates that cannot be efficiently used to support growth in addition to virulence. Interestingly, substrates with high usage capacity were found to be the most abundant metabolites present in apoplastic and xylem fluids of the host plant tomato (Zuluaga et al., 2013). This example illustrates how the growth–virulence trade‐off constraints metabolic adaptation of pathogens towards a specific host plant for a broad host range pathogen like R. solanacearum. Systems biology approaches allow quantification of the costs and constraints based on multicellular and multi–organs models (Martins Conde et al., 2016) and therefore hold immense potential in understanding the trade‐offs between complex traits linked to agricultural productivity.
Replaying the tape of evolution
Predicting adaptive trajectories is crucial to understand the constraints that lead to the emergence of phenotypes observed in nature and anticipating the impact of external constraints on biological systems. Because genetic components are interconnected and do not evolve independently, the evolution of genes should be considered in the context of the systems in which they function. Evolutionary systems biology is an integrative approach aiming at generating mechanistic and evolutionary understanding of genotype‐phenotype relationships at multiple levels (Soyer and O'Malley, 2013). In plant–pathogen interaction systems, network components encoded by pathogen genes have typically co‐evolved with the plant network to alter its function, and vice‐versa (Dong et al., 2014). Determining how evolutionary forces shape the structure of gene networks in the context of plant–pathogen interactions is therefore particularly relevant. To this end, mechanistic models can be combined with in silico evolution to replay the tape of evolution. To date, this approach has mostly been implemented on virtual host‐pathogen systems, making generic assumptions about the architecture of immune systems. This allowed showing that co‐evolution with parasites is expected to promote gene redundancy and robust network architectures in hosts (Salathé and Soyer, 2008). A similar approach showed that evasion from host immune receptors by pathogens shifts the optimal strategy towards constitutive over inducible defense in hosts. Evasion from downstream defense pathways however did not alter optimal host defense strategy (Kamiya et al., 2016). Accounting for realistic mechanistic models, such as by coupling flux balance analysis and in silico evolution, managed to reconcile simulations with the results of long‐term experimental evolution in E. coli (Großkopf et al., 2016). Hence, the likelihood of pathogen emergence may be predicted by generating in silico pathogens and comparing their predicted performance on different hosts. For instance, horizontal gene transfer, a well known phenomena allowing quick acquisition of new traits (metabolic enzymes or effectors) (Richards et al., 2011; Danchin et al., 2016; Yin et al., 2016) could be modeled to predict risk of pathogen emergence. These developments would greatly enrich our understanding of plant–pathogen co‐evolution at the molecular level.
Conclusion
The recent extension of international trading, movement of people and global changes is associated with an increase of emerging disease outbreaks (Fisher et al., 2012; Bebber and Gurr, 2015). In this context, an in‐depth knowledge on how pathogens and their hosts adapt to the environment is a prerequisite for further developments of rapid pathogen identification, treatment and surveillance strategies. For agriculture, a major challenge is to develop sophisticated models of plant diseases that incorporate more subtle climate predictions. Several studies have underlined that one of the most important predictors of the magnitude of climate change effects is the adaptive potential of plant and pathogen populations (Bebber, 2015). However, including adaptation parameters in models for disease predictions remains challenging due to the lack of knowledge for predicting the pathogen and host fitness in varying environments. Systems biology may provide a framework to fill this gap by predicting pathogen and host fitness and their evolution from the profusion of molecular knowledge collected in the past decades.
Glossary
Biological network: a group of multiples biological entities connected to each other via biochemical interactions.
Biochemical interaction: any kind of biochemical relationship between biomolecules interacting physically, e.g. substrate of an enzyme, co‐factor binding, gene transcription, etc.
Omics: technologies measuring the entire set, or at least a large part, of a single kind of biomolecules (RNA, proteins, etc.). For instance proteomics provide experimental measurement of cellular proteins. The entire set of proteins expressed in the cell is called the proteome.
Mechanistic model: mathematical representation of a living organism based on chemical and physical laws. It contains information on a set of molecular components and nature of the biochemical interactions between them. An alternative, the statistical model, describes the relationships between system variables without constraints on their chemical and physical properties, but rather based on correlation or probability of dependencies.
Emergent property: property of a living system arising at the network level, that is to say a property that cannot be associated to particular elements of the system (genes or molecules), but rather results from the way elements of the system are organized and interact.
Attractor: An attractor is a recurrent set of network states toward which the network evolves.
Functional redundancy: identical functions provided by two different biological entities. Classically, the two entities can complement each other in at least one condition.
Robustness: emerging property of a system resulting in an invariant phenotype upon genetic or environmental perturbations.
Evolvability: ability of a system to generate novel phenotypes with adaptive value upon environmental changes
Cellular noise: variation in quantity of biomolecules, like gene expression, within the cell which is not genetically determined but instead results from random.
Guard: R‐Protein monitoring the biochemical status of host proteins, which are targeted by pathogen effectors.
Decoy: Host protein which evolved to mimic pathogen effector targets, and which is guarded by R‐proteins.
Hub proteins: proteins which are highly connected in a protein–protein interaction network, being in limited number and interacting with many other proteins.
Steady‐state: stable conditions in which molecule concentration and rate of conversion are stable, i.e. do not change in time, like during exponential growth of microorganisms.
Optimality principle: maximal performance of the system toward one or multiple objective(s) under limitation of the environmental or structural constraints of the system.
Stochasticity: uncertainty in biological systems behavior due to cellular noise.
Trade‐off: Constrain on the range of phenotypes open to organisms: it occurs when a beneficial change in one trait is linked to a detrimental change in another trait, e.g. expressing resistance genes may be detrimental to plant growth and yield.
Gibbs free energy: also coined free enthalpy. Mathematically, It is a Legendre's transformation of the internal energy. It corresponds to the maximum reversible work that may be performed by a system under the assumptions of constant pressure and temperature.
Conflict of Interest
The authors declare no conflict of interest.
Acknowledgements
We are grateful to the Plant Health & Environment Division of INRA for stimulating discussions and support. We apologize to researchers whose work could not be cited due to space limitations. Authors would like to thank Dr Olivier Ali (Virtual Plants INRIA, France) for rich discussions about physical perspectives. U.D. is funded by the ANR project RIPOSTE (ANR‐14‐CE19‐0024). R.P and S.R. are funded by grants from the European Research Council (ERC‐StG‐336808) and Marie Slodowska‐Curie Actions of the European Commission (CIG‐334036). RP received a EMBO grant (Long‐Term Fellowship ALTF 1627‐2011 and Marie Curie Actions EMBOCOFUND2010, GA‐2010‐267146). This work was supported by the French Laboratory of Excellence project ‘TULIP’ (ANR‐10‐LABX‐41).
References
- Abeysekara, N.S. , Swaminathan, S. , Desai, N. , Guo, L. and Bhattacharyya, M.K. (2016) The plant immunity inducer pipecolic acid accumulates in the xylem sap and leaves of soybean seedlings following Fusarium virguliforme infection. Plant Sci. 243, 105–114. [DOI] [PubMed] [Google Scholar]
- Abramovitch, R.B. and Martin, G.B. (2004) Strategies used by bacterial pathogens to suppress plant defenses. Curr. Opin. Plant Biol. 7, 356–364. [DOI] [PubMed] [Google Scholar]
- Ackermann, M. (2015) A functional perspective on phenotypic heterogeneity in microorganisms. Nat. Rev. Microbiol. 13, 497–508. [DOI] [PubMed] [Google Scholar]
- Agler, M.T. , Ruhe, J. , Kroll, S. , Morhenn, C. , Kim, S.‐T. , Weigel, D. and Kemen, E.M. (2016) Microbial hub taxa link host and abiotic factors to plant microbiome variation. PLoS Biol. 14, e1002352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali, O. and Traas, J. (2016) Force‐driven polymerization and turgor‐induced wall expansion. Trends Plant Sci. 21, 398–409. [DOI] [PubMed] [Google Scholar]
- Ali, O. , Mirabet, V. , Godin, C. and Traas, J. (2014) Physical models of plant development. Annu. Rev. Cell Dev. Biol. 30, 59–78. [DOI] [PubMed] [Google Scholar]
- Altelaar, A.F.M. , Munoz, J. and Heck, A.J.R. (2013) Next‐generation proteomics: towards an integrative view of proteome dynamics. Nat. Rev. Genet. 14, 35–48. [DOI] [PubMed] [Google Scholar]
- Arabidopsis Interactome Mapping Consortium (2011) Evidence for network evolution in an Arabidopsis interactome map. Science, 333, 601–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asai, S. , Rallapalli, G. , Piquerez, S.J.M. et al. (2014) Expression profiling during Arabidopsis/downy mildew interaction reveals a highly‐expressed effector that attenuates responses to salicylic acid. PLoS Pathog. 10, e1004443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbacci, A. , Lahaye, M. and Magnenet, V. (2013) Another brick in the cell wall: biosynthesis dependent growth model. PLoS ONE, 8, e74400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbacci, A. , Magnenet, V. and Lahaye, M. (2015) Thermodynamical journey in plant biology. Front. Plant Sci. 6, 481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartnicki‐Garcia, S. , Bartnicki, D.D. , Gierz, G. , López‐Franco, R. and Bracker, C.E. (1995) Evidence that Spitzenkörper behavior determines the shape of a fungal hypha: a test of the hyphoid model. Exp. Mycol. 19, 153–159. [DOI] [PubMed] [Google Scholar]
- Beaulaurier, J. , Zhang, X.‐S. , Zhu, S. et al. (2015) Single molecule‐level detection and long read‐based phasing of epigenetic variations in bacterial methylomes. Nat. Commun. 6, 7438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bebber, D.P. (2015) Range‐expanding pests and pathogens in a warming world. Annu. Rev. Phytopathol. 53, 335–356. [DOI] [PubMed] [Google Scholar]
- Bebber, D.P. and Gurr, S.J. (2015) Crop‐destroying fungal and oomycete pathogens challenge food security. Fungal Genet. Biol. 74, 62–64. [DOI] [PubMed] [Google Scholar]
- Bordbar, A. , Lewis, N.E. , Schellenberger, J. , Palsson, B.Ø. and Jamshidi, N. (2010) Insight into human alveolar macrophage and M. tuberculosis interactions via metabolic reconstructions. Mol. Syst. Biol. 6, 422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordbar, A. , Monk, J.M. , King, Z.A. and Palsson, B.O. (2014) Constraint‐based models predict metabolic and associated cellular functions. Nat. Rev. Genet. 15, 107–120. [DOI] [PubMed] [Google Scholar]
- Bozkurt, T.O. , Schornack, S. , Win, J. et al. (2011) Phytophthora infestans effector AVRblb2 prevents secretion of a plant immune protease at the haustorial interface. Proc. Natl. Acad. Sci. USA, 108, 20832–20837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Büttner, D. (2016) Behind the lines‐actions of bacterial type III effector proteins in plant cells. FEMS Microbiol. Rev. 40, 894–937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callen, H.B. (1985) Thermodynamics and an Introduction to Thermostatistics, 2nd edn New York: Wiley. [Google Scholar]
- Camañes, G. , Scalschi, L. , Vicedo, B. , González‐Bosch, C. and García‐Agustín, P. (2015) An untargeted global metabolomic analysis reveals the biochemical changes underlying basal resistance and priming in Solanum lycopersicum, and identifies 1‐methyltryptophan as a metabolite involved in plant responses to Botrytis cinerea and Pseudomonas syringae . Plant J. 84, 125–139. [DOI] [PubMed] [Google Scholar]
- Campos, M.L. , Yoshida, Y. , Major, I.T. et al. (2016) Rewiring of jasmonate and phytochrome B signalling uncouples plant growth‐defense tradeoffs. Nat. Commun. 7, 12570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspi, R. , Altman, T. , Dreher, K. et al. (2012) The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of pathway/genome databases. Nucleic Acids Res. 40, D742–D753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cesari, S. , Bernoux, M. , Moncuquet, P. , Kroj, T. and Dodds, P.N. (2014) A novel conserved mechanism for plant NLR protein pairs: the ‘integrated decoy’ hypothesis. Front. Plant Sci. 5, 606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chae, E. , Tran, D.T.N. and Weigel, D. (2016) Cooperation and conflict in the plant immune system. PLoS Pathog. 12, e1005452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, A. , Schomburg, I. , Placzek, S. , Jeske, L. , Ulbrich, M. , Xiao, M. , Sensen, C.W. and Schomburg, D. (2015) BRENDA in 2015: exciting developments in its 25th year of existence. Nucleic Acids Res. 43, D439–D446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaouiya, C. , Bérenguier, D. , Keating, S.M. et al. (2013) SBML qualitative models: a model representation format and infrastructure to foster interactions between qualitative modelling formalisms and tools. BMC Syst. Biol. 7, 135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, L.‐Q. , Hou, B.‐H. , Lalonde, S. et al. (2010) Sugar transporters for intercellular exchange and nutrition of pathogens. Nature, 468, 527–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chew, Y.H. , Wenden, B. , Flis, A. et al. (2014) Multiscale digital Arabidopsis predicts individual organ and whole‐organism growth. Proc. Natl. Acad. Sci. USA, 111, E4127–E4136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coll, N.S. , Epple, P. and Dangl, J.L. (2011) Programmed cell death in the plant immune system. Cell Death Differ. 18, 1247–1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrath, U. , Beckers, G.J.M. , Langenbach, C.J.G. and Jaskiewicz, M.R. (2015) Priming for enhanced defense. Annu. Rev. Phytopathol. 53, 97–119. [DOI] [PubMed] [Google Scholar]
- Cook, D.E. , Mesarich, C.H. and Thomma, B.P.H.J. (2015) Understanding plant immunity as a surveillance system to detect invasion. Annu. Rev. Phytopathol. 53, 541–563. [DOI] [PubMed] [Google Scholar]
- Coutand, C. and Moulia, B. (2000) Biomechanical study of the effect of a controlled bending on tomato stem elongation: local strain sensing and spatial integration of the signal. J. Exp. Bot. 51, 1825–1842. [DOI] [PubMed] [Google Scholar]
- Cunnac, S. , Chakravarthy, S. , Kvitko, B.H. , Russell, A.B. , Martin, G.B. and Collmer, A. (2011) Genetic disassembly and combinatorial reassembly identify a minimal functional repertoire of type III effectors in Pseudomonas syringae . Proc. Natl. Acad. Sci. USA, 108, 2975–2980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunniffe, N.J. , Koskella, B. , Metcalf, C.J.E. , Parnell, S. , Gottwald, T.R. and Gilligan, C.A. (2015) Thirteen challenges in modelling plant diseases. Epidemics, 10, 6–10. [DOI] [PubMed] [Google Scholar]
- Dal'Molin, C.G. , Quek, L.E. , Palfreyman, R.W. , Brumbley, S.M. and Nielsen, L.K. (2010) C4GEM, a genome‐scale metabolic model to study C4 plant metabolism. Plant Physiol. 154, 1871–1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danchin, E.G.J. , Guzeeva, E.A. , Mantelin, S. , Berepiki, A. and Jones, J.T. (2016) Horizontal gene transfer from bacteria has enabled the plant‐parasitic nematode Globodera pallida to feed on host‐derived sucrose. Mol. Biol. Evol. 33, 1571–1579. [DOI] [PubMed] [Google Scholar]
- Dangl, J.L. and Jones, J.D. (2001) Plant pathogens and integrated defence responses to infection. Nature, 411, 826–833. [DOI] [PubMed] [Google Scholar]
- Denancé, N. , Sánchez‐Vallet, A. , Goffner, D. and Molina, A. (2013) Disease resistance or growth: the role of plant hormones in balancing immune responses and fitness costs. Front. Plant Sci. 4 Available at: http://journal.frontiersin.org/article/10.3389/fpls.2013.00155/abstract [Accessed October 3, 2016]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deslandes, L. , Olivier, J. , Peeters, N. , Feng, D.X. , Khounlotham, M. , Boucher, C. , Somssich, I. , Genin, S. and Marco, Y. (2003) Physical interaction between RRS1‐R, a protein conferring resistance to bacterial wilt, and PopP2, a type III effector targeted to the plant nucleus. Proc. Natl. Acad. Sci. USA, 100, 8024–8029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodds, P.N. and Rathjen, J.P. (2010) Plant immunity: towards an integrated view of plant–pathogen interactions. Nat. Rev. Genet. 11, 539–548. [DOI] [PubMed] [Google Scholar]
- Dong, S. , Stam, R. , Cano, L.M. et al. (2014) Effector specialization in a lineage of the Irish potato famine pathogen. Science, 343, 552–555. [DOI] [PubMed] [Google Scholar]
- Dumais, J. , Shaw, S.L. , Steele, C.R. , Long, S.R. and Ray, P.M. (2006) An anisotropic‐viscoplastic model of plant cell morphogenesis by tip growth. Int. J. Dev. Biol. 50, 209–222. [DOI] [PubMed] [Google Scholar]
- Eid, J. , Fehr, A. , Gray, J. et al. (2009) Real‐time DNA sequencing from single polymerase molecules. Science, 323, 133–138. [DOI] [PubMed] [Google Scholar]
- Faino, L. , Seidl, M.F. , Datema, E. , van den Berg, G.C. , Janssen, A. , Wittenberg, A.H. and Thomma, B.P. (2015) Single‐molecule real‐time sequencing combined with optical mapping yields completely finished fungal genome. mBio, 6, e00936‐15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher, M.C. , Henk, D.A. , Briggs, C.J. , Brownstein, J.S. , Madoff, L.C. , McCraw, S.L. and Gurr, S.J. (2012) Emerging fungal threats to animal, plant and ecosystem health. Nature, 484, 186–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freund, D.M. and Hegeman, A.D. (2016) Recent advances in stable isotope‐enabled mass spectrometry‐based plant metabolomics. Curr. Opin. Biotechnol. 43, 41–48. [DOI] [PubMed] [Google Scholar]
- Fu, D. , Uauy, C. , Distelfeld, A. , Blechl, A. , Epstein, L. , Chen, X. , Sela, H. , Fahima, T. and Dubcovsky, J. (2009) A kinase‐START gene confers temperature‐dependent resistance to wheat stripe rust. Science, 323, 1357–1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuoka, S. , Saka, N. , Koga, H. et al. (2009) Loss of function of a proline‐containing protein confers durable disease resistance in rice. Science, 325, 998–1001. [DOI] [PubMed] [Google Scholar]
- Gautam, V. and Sarkar, A.K. (2015) Laser assisted microdissection, an efficient technique to understand tissue specific gene expression patterns and functional genomics in plants. Mol. Biotechnol. 57, 299–308. [DOI] [PubMed] [Google Scholar]
- Gjetting, T. , Carver, T.L.W. , Skøt, L. and Lyngkjaer, M.F. (2004) Differential gene expression in individual papilla‐resistant and powdery mildew‐infected barley epidermal cells. Mol. Plant‐Microbe Interact. 17, 729–738. [DOI] [PubMed] [Google Scholar]
- Goriely, A. and Tabor, M. (2008) Mathematical modeling of hyphal tip growth. Fungal Biol. Rev. 22, 77–83. [Google Scholar]
- Grafahrend‐Belau, E. , Schreiber, F. , Koschützki, D. and Junker, B.H. (2009) Flux balance analysis of barley seeds: a computational approach to study systemic properties of central metabolism. Plant Physiol. 149, 585–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Großkopf, T. , Consuegra, J. , Gaffé, J. , Willison, J.C. , Lenski, R.E. , Soyer, O.S. and Schneider, D. (2016) Metabolic modelling in a dynamic evolutionary framework predicts adaptive diversification of bacteria in a long‐term evolution experiment. BMC Evol. Biol. 16, 163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gust, A.A. (2015) Peptidoglycan perception in plants. PLoS Pathog. 11, e1005275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas, B.J. , Kamoun, S. , Zody, M.C. et al. (2009) Genome sequence and analysis of the Irish potato famine pathogen Phytophthora infestans . Nature, 461, 393–398. [DOI] [PubMed] [Google Scholar]
- Hacquard, S. , Kracher, B. , Hiruma, K. et al. (2016) Survival trade‐offs in plant roots during colonization by closely related beneficial and pathogenic fungi. Nat. Commun. 7, 11362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamant, O. and Moulia, B. (2016) How do plants read their own shapes? New Phytol. 212, 333–337. [DOI] [PubMed] [Google Scholar]
- Hamant, O. , Heisler, M.G. , Jönsson, H. et al. (2008) Developmental patterning by mechanical signals in Arabidopsis. Science, 322, 1650–1655. [DOI] [PubMed] [Google Scholar]
- Heuberger, A.L. , Robison, F.M. , Lyons, S.M.A. , Broeckling, C.D. and Prenni, J.E. (2014) Evaluating plant immunity using mass spectrometry‐based metabolomics workflows. Front. Plant Sci. 5, 291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Hoorn, R.A.L. and Kamoun, S. (2008) From Guard to Decoy: a new model for perception of plant pathogen effectors. Plant Cell, 20, 2009–2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huard‐Chauveau, C. , Perchepied, L. , Debieu, M. , Rivas, S. , Kroj, T. , Kars, I. , Bergelson, J. , Roux, F. and Roby, D. (2013) An atypical kinase under balancing selection confers broad‐spectrum disease resistance in Arabidopsis. PLoS Genet. 9, e1003766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hucka, M. , Finney, A. , Sauro, H.M. et al. (2003) The Systems Biology Markup Language (SBML): a medium for representation and exchange of biochemical network models. Bioinformatics, 19, 524–531. [DOI] [PubMed] [Google Scholar]
- Hückelhoven, R. (2007) Cell wall‐associated mechanisms of disease resistance and susceptibility. Annu. Rev. Phytopathol. 45, 101–127. [DOI] [PubMed] [Google Scholar]
- Jackson, R.W. , Vinatzer, B. , Arnold, D.L. , Dorus, S. and Murillo, J. (2011) The influence of the accessory genome on bacterial pathogen evolution. Mob. Genet. Elements, 1, 55–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, J.D.G. and Dangl, J.L. (2006) The plant immune system. Nature, 444, 323–329. [DOI] [PubMed] [Google Scholar]
- Kaku, H. , Nishizawa, Y. , Ishii‐Minami, N. , Akimoto‐Tomiyama, C. , Dohmae, N. , Takio, K. , Minami, E. and Shibuya, N. (2006) Plant cells recognize chitin fragments for defense signaling through a plasma membrane receptor. Proc. Natl. Acad. Sci. USA, 103, 11086–11091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamiya, T. , Oña, L. , Wertheim, B. and van Doorn, G.S. (2016) Coevolutionary feedback elevates constitutive immune defence: a protein network model. BMC Evol. Biol. 16, 92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa, M. and Goto, S. (2000) KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28, 27–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaschani, F. , Shabab, M. , Bozkurt, T. et al. (2010) An effector‐targeted protease contributes to defense against Phytophthora infestans and is under diversifying selection in natural hosts. Plant Physiol. 154, 1794–1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawahara, Y. , Oono, Y. , Kanamori, H. , Matsumoto, T. , Itoh, T. and Minami, E. (2012) Simultaneous RNA‐seq analysis of a mixed transcriptome of rice and blast fungus interaction. PLoS ONE, 7, e49423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitano, H. (2002) Systems biology: a brief overview. Science, 295, 1662–1664. [DOI] [PubMed] [Google Scholar]
- Kover, P.X. and Cheverud, J. (2007) The genetic basis of quantitative variation in susceptibility of Arabidopsis thaliana to Pseudomonas syringae (Pst DC3000): evidence for a new genetic factor of large effect. New Phytol. 174, 172–181. [DOI] [PubMed] [Google Scholar]
- Kraepiel, Y. and Barny, M.‐A. (2016) Gram‐negative phytopathogenic bacteria, all hemibiotrophs after all? Mol. Plant Pathol. 17, 313–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krattinger, S.G. , Lagudah, E.S. , Spielmeyer, W. , Singh, R.P. , Huerta‐Espino, J. , McFadden, H. , Bossolini, E. , Selter, L.L. and Keller, B. (2009) A putative ABC transporter confers durable resistance to multiple fungal pathogens in wheat. Science, 323, 1360–1363. [DOI] [PubMed] [Google Scholar]
- Kroj, T. , Chanclud, E. , Michel‐Romiti, C. , Grand, X. and Morel, J.‐B. (2016) Integration of decoy domains derived from protein targets of pathogen effectors into plant immune receptors is widespread. New Phytol. 210, 618–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larance, M. and Lamond, A.I. (2015) Multidimensional proteomics for cell biology. Nat. Rev. Mol. Cell Biol. 16, 269–280. [DOI] [PubMed] [Google Scholar]
- Le Novère, N. (2015) Quantitative and logic modelling of molecular and gene networks. Nat. Rev. Genet. 16, 146–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Roux, C. , Huet, G. , Jauneau, A. et al. (2015) A receptor pair with an integrated decoy converts pathogen disabling of transcription factors to immunity. Cell, 161, 1074–1088. [DOI] [PubMed] [Google Scholar]
- Lewis, L.A. , Polanski, K. , de Torres‐Zabala, M. et al. (2015) Transcriptional dynamics driving MAMP‐triggered immunity and pathogen effector‐mediated immunosuppression in Arabidopsis leaves following infection with Pseudomonas syringae pv tomato DC3000. Plant Cell, 27, 3038–3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, K. , Limpens, E. , Zhang, Z. et al. (2014) Single nucleus genome sequencing reveals high similarity among nuclei of an endomycorrhizal fungus. PLoS Genet. 10, e1004078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, T. , Liu, Z. , Song, C. et al. (2012) Chitin‐induced dimerization activates a plant immune receptor. Science, 336, 1160–1164. [DOI] [PubMed] [Google Scholar]
- Louveaux, M. , Julien, J.‐D. , Mirabet, V. , Boudaoud, A. and Hamant, O. (2016) Cell division plane orientation based on tensile stress in Arabidopsis thaliana . Proc. Natl. Acad. Sci. USA, 113, E4294–E4303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozano‐Durán, R. and Zipfel, C. (2015) Trade‐off between growth and immunity: role of brassinosteroids. Trends Plant Sci. 20, 12–19. [DOI] [PubMed] [Google Scholar]
- Macho, A.P. and Zipfel, C. (2014) Plant PRRs and the activation of innate immune signaling. Mol. Cell, 54, 263–272. [DOI] [PubMed] [Google Scholar]
- Mansfield, J. , Genin, S. , Magori, S. et al. (2012) Top 10 plant pathogenic bacteria in molecular plant pathology. Mol. Plant Pathol. 13, 614–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins Conde, P. do R. , Sauter, T. and Pfau, T. (2016) Constraint based modeling going multicellular. Front. Mol. Biosci. 3, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mbengue, M. , Navaud, O. , Peyraud, R. , Barascud, M. , Badet, T. , Vincent, R. , Barbacci, A. and Raffaele, S. (2016) Emerging trends in molecular interactions between plants and the broad host range fungal pathogens Botrytis cinerea and Sclerotinia sclerotiorum . Front. Plant Sci. 7, 422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAdams, H.H. and Arkin, A. (1997) Stochastic mechanisms in gene expression. Proc. Natl. Acad. Sci. USA, 94, 814–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medzhitov, R. and Janeway, C.A. (1997) Innate immunity: the virtues of a nonclonal system of recognition. Cell, 91, 295–298. [DOI] [PubMed] [Google Scholar]
- Metzker, M.L. (2010) Sequencing technologies – the next generation. Nat. Rev. Genet. 11, 31–46. [DOI] [PubMed] [Google Scholar]
- Meyer, S.E. , Stewart, T.E. and Clement, S. (2010) The quick and the deadly: growth vs virulence in a seed bank pathogen. New Phytol. 187, 209–216. [DOI] [PubMed] [Google Scholar]
- Miedes, E. , Vanholme, R. , Boerjan, W. and Molina, A. (2014) The role of the secondary cell wall in plant resistance to pathogens. Front. Plant Sci. 5 Available at: http://journal.frontiersin.org/article/10.3389/fpls.2014.00358/abstract [Accessed September 26, 2016]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Money, N.P. (2008) Insights on the mechanics of hyphal growth. Fungal Biol. Rev. 22, 71–76. [Google Scholar]
- Moulia, B. and Fournier, M. (2009) The power and control of gravitropic movements in plants: a biomechanical and systems biology view. J. Exp. Bot. 60, 461–486. [DOI] [PubMed] [Google Scholar]
- Moulia, B. , Der Loughian, C. , Bastien, R. et al. (2011) Integrative mechanobiology of growth and architectural development in changing mechanical environments In Mechanical Integration of Plant Cells and Plants (Wojtaszek P., ed.). Berlin, Heidelberg: Springer Berlin Heidelberg, pp. 269–302. Available at: http://link.springer.com/10.1007/978-3-642-19091-9_11 [Accessed September 27, 2016]. [Google Scholar]
- Moulia, B. , Coutand, C. and Julien, J.‐L. (2015) Mechanosensitive control of plant growth: bearing the load, sensing, transducing, and responding. Front. Plant Sci. 6, 52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhtar, M.S. , Carvunis, A.‐R. , Dreze, M. et al. (2011) Independently evolved virulence effectors converge onto hubs in a plant immune system network. Science, 333, 596–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller, D.B. , Vogel, C. , Bai, Y. and Vorholt, J.A. (2016) The plant microbiota: systems‐level insights and perspectives. Annu. Rev. Genet. 50, 211–234. [DOI] [PubMed] [Google Scholar]
- Naseem, M. and Dandekar, T. (2012) The role of auxin‐cytokinin antagonism in plant–pathogen interactions. PLoS Pathog. 8, e1003026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naseem, M. , Philippi, N. , Hussain, A. , Wangorsch, G. , Ahmed, N. and Dandekar, T. (2012) Integrated systems view on networking by hormones in Arabidopsis immunity reveals multiple crosstalk for cytokinin. Plant Cell, 24, 1793–1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson, T. , Gandotra, N. and Tausta, S.L. (2008) Plant cell types: reporting and sampling with new technologies. Curr. Opin. Plant Biol. 11, 567–573. [DOI] [PubMed] [Google Scholar]
- de Oliveira Dal'Molin, C.G. , Quek, L.‐E. , Palfreyman, R.W. , Brumbley, S.M. and Nielsen, L.K. (2010) AraGEM, a genome‐scale reconstruction of the primary metabolic network in Arabidopsis. Plant Physiol. 152, 579–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Oliveira Dal'Molin, C.G. , Orellana, C. , Gebbie, L. et al. (2016) Metabolic reconstruction of Setaria italica: a systems biology approach for integrating tissue‐specific omics and pathway analysis of bioenergy grasses. Front. Plant Sci. 7, 1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orth, J.D. , Thiele, I. and Palsson, B.Ø. (2010) What is flux balance analysis? Nat. Biotechnol. 28, 245–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen, C. , Ver Loren van Themaat, E. , McGuffin, L.J. et al. (2012) Structure and evolution of barley powdery mildew effector candidates. BMC Genomics, 13, 694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peeters, N. , Carrère, S. , Anisimova, M. , Plener, L. , Cazalé, A.‐C. and Genin, S. (2013) Repertoire, unified nomenclature and evolution of the Type III effector gene set in the Ralstonia solanacearum species complex. BMC Genomics, 14, 859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penfold, C.A. and Wild, D.L. (2011) How to infer gene networks from expression profiles, revisited. Interface Focus, 1, 857–870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peyraud, R. , Cottret, L. , Marmiesse, L. , Gouzy, J. and Genin, S. (2016) A resource allocation trade‐off between virulence and proliferation drives metabolic versatility in the plant pathogen Ralstonia solanacearum . PLoS Pathog. 12, e1005939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilalis, E. , Chatziioannou, A. , Thomasset, B. and Kolisis, F. (2011) An in silico compartmentalized metabolic model of Brassica napus enables the systemic study of regulatory aspects of plant central metabolism. Biotechnol. Bioeng. 108, 1673–1682. [DOI] [PubMed] [Google Scholar]
- Pogány, M. , von Rad, U. , Grün, S. et al. (2009) Dual roles of reactive oxygen species and NADPH oxidase RBOHD in an Arabidopsis‐Alternaria pathosystem. Plant Physiol. 151, 1459–1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poland, J.A. , Balint‐Kurti, P.J. , Wisser, R.J. , Pratt, R.C. and Nelson, R.J. (2009) Shades of gray: the world of quantitative disease resistance. Trends Plant Sci. 14, 21–29. [DOI] [PubMed] [Google Scholar]
- Poolman, M.G. , Miguet, L. , Sweetlove, L.J. and Fell, D.A. (2009) A genome‐scale metabolic model of Arabidopsis and some of its properties. Plant Physiol. 151, 1570–1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poolman, M.G. , Kundu, S. , Shaw, R. and Fell, D.A. (2013) Responses to light intensity in a genome‐scale model of rice metabolism. Plant Physiol. 162, 1060–1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard, L. and Birch, P. (2011) A systems biology perspective on plant‐microbe interactions: biochemical and structural targets of pathogen effectors. Plant Sci. 180, 584–603. [DOI] [PubMed] [Google Scholar]
- Pritchard, L. and Birch, P.R.J. (2014) The zigzag model of plant‐microbe interactions: is it time to move on? Mol. Plant Pathol. 15, 865–870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raffaele, S. and Kamoun, S. (2012) Genome evolution in filamentous plant pathogens: why bigger can be better. Nat. Rev. Microbiol. 10, 417–430. [DOI] [PubMed] [Google Scholar]
- Raffaele, S. , Vailleau, F. , Léger, A. et al. (2008) A MYB transcription factor regulates very‐long‐chain fatty acid biosynthesis for activation of the hypersensitive cell death response in Arabidopsis. Plant Cell, 20, 752–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards, T.A. , Soanes, D.M. , Jones, M.D.M. , Vasieva, O. , Leonard, G. , Paszkiewicz, K. , Foster, P.G. , Hall, N. and Talbot, N.J. (2011) Horizontal gene transfer facilitated the evolution of plant parasitic mechanisms in the oomycetes. Proc. Natl. Acad. Sci. USA, 108, 15258–15263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux, F. , Voisin, D. , Badet, T. , Balagué, C. , Barlet, X. , Huard‐Chauveau, C. , Roby, D. and Raffaele, S. (2014) Resistance to phytopathogens e tutti quanti: placing plant quantitative disease resistance on the map. Mol. Plant Pathol. 15, 427–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryffel, F. , Helfrich, E.J.N. , Kiefer, P. , Peyriga, L. , Portais, J.‐C. , Piel, J. and Vorholt, J.A. (2016) Metabolic footprint of epiphytic bacteria on Arabidopsis thaliana leaves. ISME J. 10, 632–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha, R. , Suthers, P.F. and Maranas, C.D. (2011) Zea mays iRS1563: a comprehensive genome‐scale metabolic reconstruction of maize metabolism. PLoS ONE, 6, e21784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saier, M.H. , Tran, C.V. and Barabote, R.D. (2006) TCDB: the Transporter Classification Database for membrane transport protein analyses and information. Nucleic Acids Res. 34, D181–D186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salathé, M. and Soyer, O.S. (2008) Parasites lead to evolution of robustness against gene loss in host signaling networks. Mol. Syst. Biol. 4, 202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanati Nezhad, A. and Geitmann, A. (2013) The cellular mechanics of an invasive lifestyle. J. Exp. Bot. 64, 4709–4728. [DOI] [PubMed] [Google Scholar]
- Sarris, P.F. , Cevik, V. , Dagdas, G. , Jones, J.D.G. and Krasileva, K.V. (2016) Comparative analysis of plant immune receptor architectures uncovers host proteins likely targeted by pathogens. BMC Biol. 14, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato, M. , Tsuda, K. , Wang, L. , Coller, J. , Watanabe, Y. , Glazebrook, J. and Katagiri, F. (2010) Network modeling reveals prevalent negative regulatory relationships between signaling sectors in Arabidopsis immune signaling. PLoS Pathog. 6, e1001011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer, U. (2004) High‐throughput phenomics: experimental methods for mapping fluxomes. Curr. Opin. Biotechnol. 15, 58–63. [DOI] [PubMed] [Google Scholar]
- Sauer, U. , Heinemann, M. and Zamboni, N. (2007) Genetics. Getting closer to the whole picture. Science, 316, 550–551. [DOI] [PubMed] [Google Scholar]
- Schatschneider, S. , Persicke, M. , Watt, S.A. , Hublik, G. , Pühler, A. , Niehaus, K. and Vorhölter, F.‐J. (2013) Establishment, in silico analysis, and experimental verification of a large‐scale metabolic network of the xanthan producing Xanthomonas campestris pv. campestris strain B100. J. Biotechnol. 167, 123–134. [DOI] [PubMed] [Google Scholar]
- Schiefelbein, J. (2015) Molecular phenotyping of plant single cell‐types enhances forward genetic analyses. Front. Plant Sci. 6, 509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schirawski, J. , Mannhaupt, G. , Münch, K. et al. (2010) Pathogenicity determinants in smut fungi revealed by genome comparison. Science, 330, 1546–1548. [DOI] [PubMed] [Google Scholar]
- Schmidt, A. , Kochanowski, K. , Vedelaar, S. et al. (2016) The quantitative and condition‐dependent Escherichia coli proteome. Nat. Biotechnol. 34, 104–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwessinger, B. and Zipfel, C. (2008) News from the frontline: recent insights into PAMP‐triggered immunity in plants. Curr. Opin. Plant Biol. 11, 389–395. [DOI] [PubMed] [Google Scholar]
- Smallbone, K. , Simeonidis, E. , Swainston, N. and Mendes, P. (2010) Towards a genome‐scale kinetic model of cellular metabolism. BMC Syst. Biol. 4, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somerville, C. , Bauer, S. , Brininstool, G. et al. (2004) Toward a systems approach to understanding plant cell walls. Science, 306, 2206–2211. [DOI] [PubMed] [Google Scholar]
- Soyer, O.S. and O'Malley, M.A. (2013) Evolutionary systems biology: what it is and why it matters. BioEssays, 35, 696–705. [DOI] [PubMed] [Google Scholar]
- Szklarczyk, D. , Franceschini, A. , Wyder, S. et al. (2015) STRING v10: protein–protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 43, D447–D452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szyperski, T. (1995) Biosynthetically directed fractional 13C‐labeling of proteinogenic amino acids. An efficient analytical tool to investigate intermediary metabolism. Eur. J. Biochem. 232, 433–448. [DOI] [PubMed] [Google Scholar]
- Tao, Y. , Xie, Z. , Chen, W. , Glazebrook, J. , Chang, H.‐S. , Han, B. , Zhu, T. , Zou, G. and Katagiri, F. (2003) Quantitative nature of Arabidopsis responses during compatible and incompatible interactions with the bacterial pathogen Pseudomonas syringae . Plant Cell, 15, 317–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiele, I. and Palsson, B.Ø. (2010) A protocol for generating a high‐quality genome‐scale metabolic reconstruction. Nat. Protoc. 5, 93–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thrall, P.H. and Burdon, J.J. (2003) Evolution of virulence in a plant host‐pathogen metapopulation. Science, 299, 1735–1737. [DOI] [PubMed] [Google Scholar]
- Tian, D. , Traw, M.B. , Chen, J.Q. , Kreitman, M. and Bergelson, J. (2003) Fitness costs of R‐gene‐mediated resistance in Arabidopsis thaliana . Nature, 423, 74–77. [DOI] [PubMed] [Google Scholar]
- Todesco, M. , Balasubramanian, S. , Hu, T.T. et al. (2010) Natural allelic variation underlying a major fitness trade‐off in Arabidopsis thaliana . Nature, 465, 632–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuda, K. , Sato, M. , Stoddard, T. , Glazebrook, J. and Katagiri, F. (2009) Network properties of robust immunity in plants. PLoS Genet. 5, e1000772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varma, A. and Palsson, B.O. (1994) Stoichiometric flux balance models quantitatively predict growth and metabolic by‐product secretion in wild‐type Escherichia coli W3110. Appl. Environ. Microbiol. 60, 3724–3731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Z. , Gerstein, M. and Snyder, M. (2009) RNA‐Seq: a revolutionary tool for transcriptomics. Nat. Rev. Genet. 10, 57–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, G. , Roux, B. , Feng, F. et al. (2015) The decoy substrate of a pathogen effector and a pseudokinase specify pathogen‐induced modified‐self recognition and immunity in plants. Cell Host Microbe, 18, 285–295. [DOI] [PubMed] [Google Scholar]
- Wei, Z. , Yang, T. , Friman, V.‐P. , Xu, Y. , Shen, Q. and Jousset, A. (2015) Trophic network architecture of root‐associated bacterial communities determines pathogen invasion and plant health. Nat. Commun. 6, 8413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weßling, R. , Epple, P. , Altmann, S. et al. (2014) Convergent targeting of a common host protein‐network by pathogen effectors from three kingdoms of life. Cell Host Microbe, 16, 364–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Windram, O. , Madhou, P. , McHattie, S. et al. (2012) Arabidopsis defense against Botrytis cinerea: chronology and regulation deciphered by high‐resolution temporal transcriptomic analysis. Plant Cell, 24, 3530–3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, F. and Yin, Q. (2016) Comprehensive proteomic analysis of the wheat pathogenic fungus Zymoseptoria tritici . Proteomics, 16, 98–101. [DOI] [PubMed] [Google Scholar]
- Yin, Z. , Zhu, B. , Feng, H. and Huang, L. (2016) Horizontal gene transfer drives adaptive colonization of apple trees by the fungal pathogen Valsa mali . Sci. Rep. 6, 33129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida, K. , Saunders, D.G.O. , Mitsuoka, C. et al. (2016) Host specialization of the blast fungus Magnaporthe oryzae is associated with dynamic gain and loss of genes linked to transposable elements. BMC Genomics, 17, 370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan, H. , Cheung, C.Y.M. , Poolman, M.G. , Hilbers, P.A.J. and van Riel, N.A.W. (2016) A genome‐scale metabolic network reconstruction of tomato (Solanum lycopersicum L.) and its application to photorespiratory metabolism. Plant J. 85, 289–304. [DOI] [PubMed] [Google Scholar]
- Zapata, L. , Ding, J. , Willing, E.‐M. et al. (2016) Chromosome‐level assembly of Arabidopsis thaliana Ler reveals the extent of translocation and inversion polymorphisms. Proc. Natl. Acad. Sci. USA, 113, E4052–E4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y. , Tessaro, M.J. , Lassner, M. and Li, X. (2003) Knockout analysis of Arabidopsis transcription factors TGA2, TGA5, and TGA6 reveals their redundant and essential roles in systemic acquired resistance. Plant Cell, 15, 2647–2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, B. , Ibrahim, M. , Cui, Z. , Xie, G. , Jin, G. , Kube, M. , Li, B. and Zhou, X. (2016) Multi‐omics analysis of niche specificity provides new insights into ecological adaptation in bacteria. ISME J. 10, 2072–2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipfel, C. , Robatzek, S. , Navarro, L. , Oakeley, E.J. , Jones, J.D.G. , Felix, G. and Boller, T. (2004) Bacterial disease resistance in Arabidopsis through flagellin perception. Nature, 428, 764–767. [DOI] [PubMed] [Google Scholar]
- Zuluaga, A.P. , Puigvert, M. and Valls, M. (2013) Novel plant inputs influencing Ralstonia solanacearum during infection. Front. Microbiol. 4, 349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwieniecki, M.A. and Dumais, J. (2011) Quantifying green life: grand challenges in plant biophysics and modeling. Front. Plant Sci. 2, 31. [DOI] [PMC free article] [PubMed] [Google Scholar]


