Abstract
We developed an automated approach for generating quantitative image analysis metrics (imaging biomarkers) that are then analysed with a set of 13 machine learning algorithms to generate an overall risk score that is called a Q‐score. These methods were applied to a set of 120 “difficult” dermoscopy images of dysplastic nevi and melanomas that were subsequently excised/classified. This approach yielded 98% sensitivity and 36% specificity for melanoma detection, approaching sensitivity/specificity of expert lesion evaluation. Importantly, we found strong spectral dependence of many imaging biomarkers in blue or red colour channels, suggesting the need to optimize spectral evaluation of pigmented lesions.
Keywords: dermoscopy, imaging biomarkers, machine learning, machine vision, melanoma, pigmented lesion, screening, skin optics
Abbreviations
- MC
multicolour
- MIB
melanoma imaging biomarker
- ROC
receiver‐operator characteristic
- SC
single colour
1. Background
Clinical melanoma evaluation depends largely on recognizing abnormal pigmentation patterns that distinguish benign or atypical nevi from cancerous growths. Detection through screening saves lives but remains challenging visually, as the degree of disorder that exists between these different pigmented growths is highly variable and, at times, can be quite subtle and difficult to determine. This level of diagnostic difficulty is reflected across the general practice of dermatology in which melanomas are confirmed by histopathology in only 3‐25%1 or a mean of 10%2 of excised suspicious lesions. However, visual examination of pigmented lesions by expert dermatologists using dermoscopy and following criteria such as the Menzies (or CASH3) method has yielded diagnostic accuracy as high as 98% sensitivity and 68% specificity in some studies. At present, there is an ongoing effort across the medical and scientific communities to improve melanoma diagnosis by developing standardized evaluation criteria of pigmented lesions that can be performed by more medical care givers (beyond expert dermoscopists) and, if possible, might even be performed by automated analysis systems that use image processing and artificial intelligence algorithms.4, 5 However, current methods yield specificity and sensitivity outcomes that are far inferior to expert dermoscopy evaluations. Furthermore, proprietary computational algorithms use “black box” image feature extraction and diagnostic algorithms that do not help healthcare providers to identify possible new lesion features that may be useful in clinical evaluation. Thus, there is a need for transparent methods that extract discrete diagnostic image features and combine them in screening algorithms.
2. Questions Asked
We sought to determine whether automated image analysis of pigmented lesions could generate useful (and potentially novel) melanoma imaging biomarkers (MIBs) to assess risk. Our digital, analytical framework for dermoscopy interpretation was used to answer two key questions: how sensitive and specific is the diagnostic, and do MIBs exhibit any spectral dependence whose exploitation may improve diagnosis?
3. Experimental Design
In our study (Figure S1), 120 dermoscopy images (60 melanomas, 60 atypical nevi) were analysed by a series of computer programs (all detailed in supplement) that determined the border of the lesion, its centre and from that point a radius was projected along which different image features were calculated over a 360° sweep (similar to a clock sweep). Figure 1A,B, with further detail in Figure S2, exemplifies this approach by plotting lesion brightness vs a sweep in angle θ, using blue channel colour information and graphically yielding MIB B12 (B=from the blue colour channel, metric #12). Other programs evaluated symmetry and organization of pigmentation patterns, networks and substructures across red, green and blue (RGB) colour channels. Programs also determined the number of colours present in lesions (Figure 1C) and the pigmented network pattern (Figure 1D). This set of programs was run on each pigmented lesion to generate 50 quantitative metrics. The 33/50 metrics that had a significant difference (P<.05) in values between atypical nevi vs melanomas became the MIB set. MIBs, evaluated in particular colour channels, became the basis of machine learning classification algorithms to construct an overall quantitative score (Q‐score) between zero and one, in which a higher number indicate a higher probability of a lesion being a melanoma (Figure S3).
Figure 1.
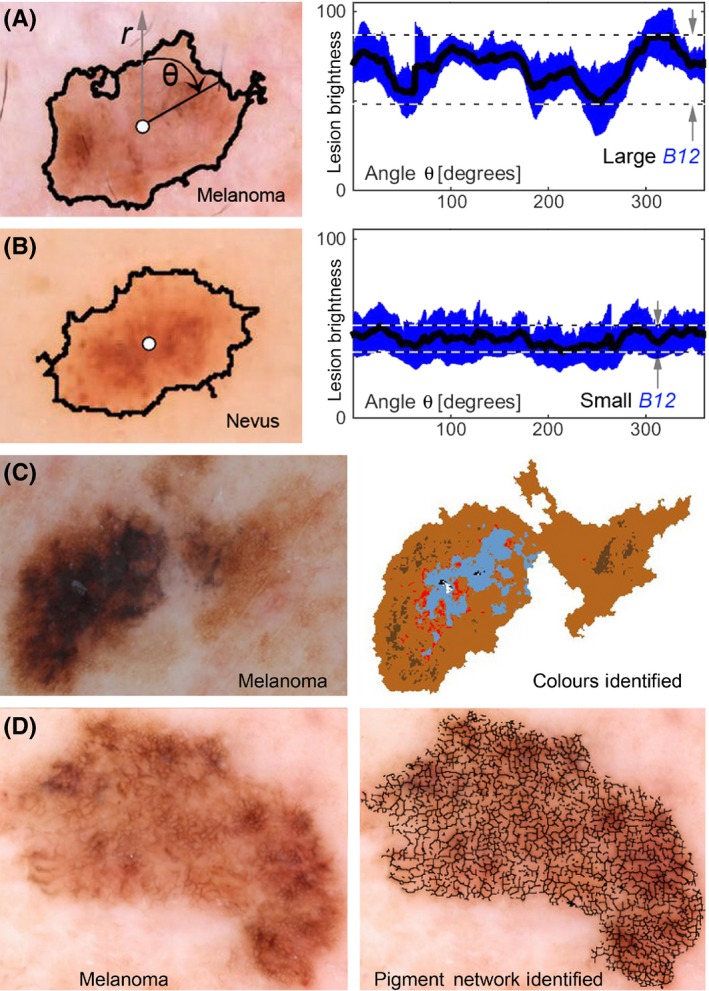
Example melanoma imaging biomarkers. (A) and (B) show a melanoma and a nevus, respectively, where lesion centre (white circle) and peripheral border (black line) between lesion and normal skin are illustrated. The mean lesion brightness along the sweeping arm as a function of angle θ is plotted (black line) to the right with the standard deviation shown in blue. The melanoma imaging biomarker (MIB) B12 is graphically shown to be the brightness range over an angular sweep of the mean lesion pixel brightness. The range is divided by the mean to achieve the final B12 MIB. The images shown are of a melanoma that yields a large B12 value and a nevus that yields a small B12 value. A melanoma with multiple colours (C) is shown in colour map illustrating MIB MC1. A melanoma with an atypical reticular pigmented network (D) is shown with an overlay of the pigmented network branches. Each black line segment terminates on each end in either a branch point or an endpoint. Statistical analysis of these branches yielded MIBs B8, B11, B15, R3, R7 and R8
4. Results
The single most significant MIB (P<10−6) was number of colours in a lesion (MC1, Figure 2). Individual MIBs that evaluate features, such as the lesion brightness, border, diameter and symmetry, were often highly significant only in the red or blue colour channels, while green‐channel MIBs were not as important. A visual example of the selection of a representative MIB in a particular colour channel to maximize its diagnostic power is shown in Figure S4. The most significant MIBs for the red and blue channels, respectively, were MIB R1: variation in the sharpness of lesion demarcation (Figure S5, Equation S24) and MIB B1: angular lesional brightness variation (Figure 1A,B, Equation S10). Figure S5 shows all MIBs on all data, and Figure S6 shows raw MIB values for the most significant six MIBs on representative normal and abnormal lesions.
Figure 2.
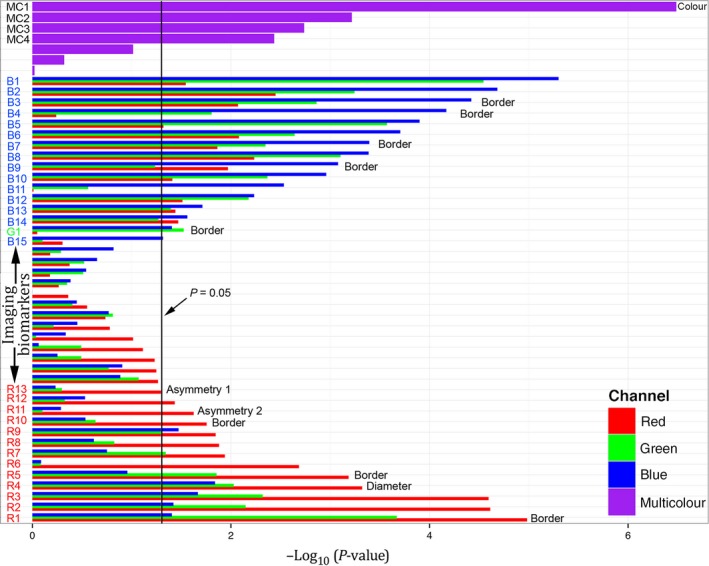
The length of the horizontal bar, for each image feature extracted, is negative the base 10 logarithm of the P‐value, where the P‐value is the standard statistical significance metric, calculated using univariate, two‐tailed, unpaired t‐tests (for continuous variables) and Fisher's exact test (for categorical variables). For single colour channel metrics, three adjoined bars, colour‐coated red, green and blue show the importance when evaluated in the respective colour channels of the image. The melanoma imaging biomarkers (MIBs) with statistical significance for melanoma discrimination (P<.05, vertical black line) are labelled on the vertical axis describing the colour channel they were used in: B1‐B14 from the blue channel, G1 from the green channel and R1‐R13 from the red channel. MC1‐MC4 denote MIBs that used multiple colour channel information. The text to the right of the bars indicates MIBs that contain information based on the dermoscopic ABCD criteria. The most significant MIB was the number of colours identified in the lesion while the diameter of the lesion had intermediate significance and the asymmetry of the lesion silhouette (Asymmetry 1, illustrated in Figure S10) had borderline significance. The lesion border features (see Figure S4) pertain to the edge demarcation.
The MIBs became inputs for a series of 13 machine learning/classification algorithms (Figure S3, Table S2), which individually computed probabilities of melanoma diagnosis. An example of classification using the C5.0 (decision tree) approach is illustrated in Figure S8. The Q‐score was calculated as the median value of melanoma probability across all 13 machine learning approaches. As shown in Figure S9, most lesions with a high Q‐score were diagnosed as melanomas and most lesions with a low Q‐score were diagnosed as nevi. This classification approach achieved 98% sensitivity for melanoma detection and there was 36% specificity for predicting a melanoma diagnosis as illustrated in the receiver‐operator curve of Figure S3. Examples of accurate and inaccurate Q‐scores are presented in Figure S10.
5. Conclusions
This method independently determined that the number of colours present in a lesion is important for melanoma evaluation and is thus aligned with conventional dermoscopic evaluation criteria.6 We also determined that MIBs have spectral dependence in RGB channels and that clear visual differences exist in defined colour channels (Figure S7). Another important feature of this approach is computational transparency, as the derivation of each quantitative biomarker and full description of our statistical analysis are disclosed in supplemental information to this letter. Overall, these results raise a question about whether spectrally dependent MIBs might be further enhanced by extended spectral imaging and analysis.
At high sensitivity, the Q‐score achieved significantly higher specificity than today's estimated standard of 10% in practice, albeit in our study with small sample size and artificially high prevalence. As this method does not depend on expert evaluators, it has the potential to improve upon diagnosis and classification of pigmented lesions widely. Overall, our sensitivity/specificity is similar to an electrical impedance spectroscopy device recently described.7 For expert evaluators, our method has identified some new lesion characteristics, for example border demarcation features, that might improve visual evaluations. Widespread application, if validated in larger clinical trials, could decrease unnecessary biopsies and increase life‐saving early detection events.
Conflict of Interest
All authors have no conflict of interest to report.
Author Contribution
D.G. conceived, designed and implemented the image analysis framework to quantify the MIBs and led the writing of the manuscript. The statistics team (Co‐Authors J.C.R. and M.S.F.) was blind to the image analysis framework and the resulting image processing algorithm but received its output MIBs. D.S.G. was blind to the gold standard diagnoses until the image analysis framework was frozen, and the final MIB set, used in this manuscript, was transferred to the statistics team. The statistics team received the gold standard histopathological diagnoses from S.Y. and defined the machine learning framework upon which the classifier was built, conducting all statistical data analysis and modelling. S.Y. curated the data. J.L.D. and A.M. were the physicians who saw the patients, performed the biopsies and acquired the image data that we analysed. M.S.F. supervised experimental design, data analysis and participated in results interpretation. J.A.C. and N.G. assisted in writing the manuscript and providing clinical context to optimize translational impact of the study. F.H. completed the literature review of the competing technologies outside the peer‐reviewed literature such as the Apps listed and Watson. J.G.K. initiated this project, participated in the experimental design, definition of the MIBs and interpretation of the results. All authors participated in writing of the manuscript or reviewed the manuscript.
Supporting information
Figure S1 Study design
Figure S2 Coordinate transformation and illustration of MIB derivation using angular sweep analysis
Figure S3 Diagnostic performance results Vs. published techniques
Figure S4 Spectral diagnostic content variation
Figure S5 Fitting for edge demarcation
Figure S6 Imaging biomarkers heat map
Figure S7 Good metrics
Figure S8 Decision tree built with the C5.0 algorithm
Figure S9 Distribution of the melanoma Q‐score
Figure S10 Examples of well‐classified (top two rows) and poorly‐classified (bottom two rows) lesions
Figure S11 Examples of failed border detection
Figure S12 Determination of border asymmetry
Appendix S1 Supplemental methods and discussion
Video S1 Study summary video
Table S1 Diagnostic sensitivity and specificity for melanoma detection by human pattern recognition (*) and by machine‐augmented pattern recognition (**)
Table S2 Classification algorithms applied to melanoma discrimination
Table S3 Mean ratios for intensities between color channel pairs
Table S4 Median estimates for the performance of MIBs according to different channels across all methods
Acknowledgements
This work was supported in part by grant #UL1TR001866 from the National Institutes of Health and in part by the Paul and Irma Milstein Family Foundation and the American Skin Association.
References
- 1. Hansen C., Wilkinson D., Hansen M., Argenziano G., J. Am. Acad. Dermatol. 2009, 61, 599. [DOI] [PubMed] [Google Scholar]
- 2. Salerni G., Teran T., Puig S., Malvehy J., Zalaudek I., Argenziano G., Kittler H., J. Eur. Acad. Dermatol. Venereol. 2013, 27, 805. [DOI] [PubMed] [Google Scholar]
- 3. Henning J. S., Dusza S. W., Wang S. Q., Marghoob A. A., Rabinovitz H. S., Polsky D., Kopf A. W., J. Am. Acad. Dermatol. 2007, 56, 45. [DOI] [PubMed] [Google Scholar]
- 4. Monheit G., Cognetta A. B., Ferris L., Rabinovitz H., Gross K., Martini M., Grichnik J. M., Mihm M., Prieto V. G., Googe P., King R., Toledano A., Kabelev N., Wojton M., Gutkowicz‐Krusin D., Arch. Dermatol. 2011, 147, 188. [DOI] [PubMed] [Google Scholar]
- 5. Doyle‐Lindrud S., Clin. J. Oncol. Nurs. 2015, 19, 31. [DOI] [PubMed] [Google Scholar]
- 6. Marghoob A. A., Malvehy J., Braun R. P., Atlas of Dermoscopy. 2012. [Google Scholar]
- 7. Malvehy J., Hauschild A., Curiel‐Lewandrowski C., Mohr P., Hofmann‐Wellenhof R., Motley R., Berking C., Grossman D., Paoli J., Loquai C., Olah J., Reinhold U., Wenger H., Dirschka T., Davis S., Henderson C., Rabinovitz H., Welzel J., Schadendorf D., Birgersson U., Br. J. Dermatol. 1099, 2014, 171. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 Study design
Figure S2 Coordinate transformation and illustration of MIB derivation using angular sweep analysis
Figure S3 Diagnostic performance results Vs. published techniques
Figure S4 Spectral diagnostic content variation
Figure S5 Fitting for edge demarcation
Figure S6 Imaging biomarkers heat map
Figure S7 Good metrics
Figure S8 Decision tree built with the C5.0 algorithm
Figure S9 Distribution of the melanoma Q‐score
Figure S10 Examples of well‐classified (top two rows) and poorly‐classified (bottom two rows) lesions
Figure S11 Examples of failed border detection
Figure S12 Determination of border asymmetry
Appendix S1 Supplemental methods and discussion
Video S1 Study summary video
Table S1 Diagnostic sensitivity and specificity for melanoma detection by human pattern recognition (*) and by machine‐augmented pattern recognition (**)
Table S2 Classification algorithms applied to melanoma discrimination
Table S3 Mean ratios for intensities between color channel pairs
Table S4 Median estimates for the performance of MIBs according to different channels across all methods


