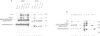Abstract
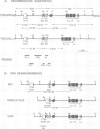
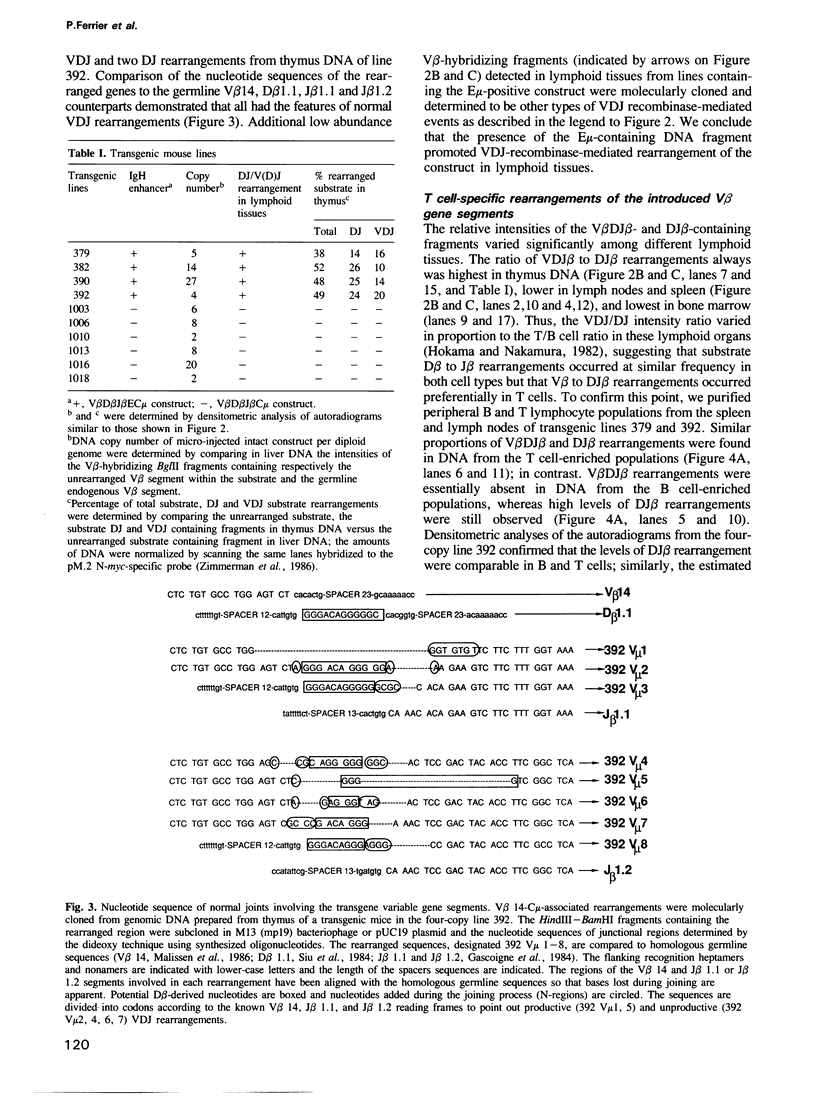
We describe transgenic mice that carry an antigen receptor gene minilocus comprised of germline T cell receptor (TCR) beta variable gene elements (V, D and J) linked to an immunoglobulin (Ig) C mu constant region gene with or without a DNA segment containing the Ig heavy chain transcriptional enhancer (E mu). Transgenic constructs lacking the E mu-containing segment did not undergo detectable rearrangement in any tissue of six independent transgenic lines. In contrast, transgenic constructs containing this DNA segment underwent rearrangement at high frequency in lymphoid tissues, but not other tissues, of four independent lines. Analyses of purified B and T cells, as well as B and T cell lines, from transgenic animals demonstrated that the E mu-containing segment within the construct allowed partial TCR gene assembly (D to J) in both B and T cells. However, complete TCR gene rearrangement within the construct (V to DJ) occurred only in T cells. Therefore, we have demonstrated elements that can control two separate aspects of TCR beta VDJ rearrangement within this construct. One lies within the E mu-containing DNA segment and represents a dominant, cis-acting element that initiates lymphoid cell-specific D beta to J beta rearrangement; various considerations suggest this activity may be related to that of the E mu element. The second element provides T cell-specific control of complete (V beta to DJ beta) variable region gene assembly; it correlates in activity with expression of the unrearranged V beta segment.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aguilera R. J., Akira S., Okazaki K., Sakano H. A pre-B cell nuclear protein that specifically interacts with the immunoglobulin V-J recombination sequences. Cell. 1987 Dec 24;51(6):909–917. doi: 10.1016/0092-8674(87)90578-2. [DOI] [PubMed] [Google Scholar]
- Alt F. W., Baltimore D. Joining of immunoglobulin heavy chain gene segments: implications from a chromosome with evidence of three D-JH fusions. Proc Natl Acad Sci U S A. 1982 Jul;79(13):4118–4122. doi: 10.1073/pnas.79.13.4118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alt F. W., Blackwell T. K., DePinho R. A., Reth M. G., Yancopoulos G. D. Regulation of genome rearrangement events during lymphocyte differentiation. Immunol Rev. 1986 Feb;89:5–30. doi: 10.1111/j.1600-065x.1986.tb01470.x. [DOI] [PubMed] [Google Scholar]
- Alt F. W., Blackwell T. K., Yancopoulos G. D. Development of the primary antibody repertoire. Science. 1987 Nov 20;238(4830):1079–1087. doi: 10.1126/science.3317825. [DOI] [PubMed] [Google Scholar]
- Alt F. W., Rosenberg N., Casanova R. J., Thomas E., Baltimore D. Immunoglobulin heavy-chain expression and class switching in a murine leukaemia cell line. Nature. 1982 Mar 25;296(5855):325–331. doi: 10.1038/296325a0. [DOI] [PubMed] [Google Scholar]
- Alt F. W., Rosenberg N., Enea V., Siden E., Baltimore D. Multiple immunoglobulin heavy-chain gene transcripts in Abelson murine leukemia virus-transformed lymphoid cell lines. Mol Cell Biol. 1982 Apr;2(4):386–400. doi: 10.1128/mcb.2.4.386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alt F. W., Yancopoulos G. D., Blackwell T. K., Wood C., Thomas E., Boss M., Coffman R., Rosenberg N., Tonegawa S., Baltimore D. Ordered rearrangement of immunoglobulin heavy chain variable region segments. EMBO J. 1984 Jun;3(6):1209–1219. doi: 10.1002/j.1460-2075.1984.tb01955.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alt F., Rosenberg N., Lewis S., Thomas E., Baltimore D. Organization and reorganization of immunoglobulin genes in A-MULV-transformed cells: rearrangement of heavy but not light chain genes. Cell. 1981 Dec;27(2 Pt 1):381–390. doi: 10.1016/0092-8674(81)90421-9. [DOI] [PubMed] [Google Scholar]
- Atchison M. L., Perry R. P. The role of the kappa enhancer and its binding factor NF-kappa B in the developmental regulation of kappa gene transcription. Cell. 1987 Jan 16;48(1):121–128. doi: 10.1016/0092-8674(87)90362-x. [DOI] [PubMed] [Google Scholar]
- Augereau P., Chambon P. The mouse immunoglobulin heavy-chain enhancer: effect on transcription in vitro and binding of proteins present in HeLa and lymphoid B cell extracts. EMBO J. 1986 Aug;5(8):1791–1797. doi: 10.1002/j.1460-2075.1986.tb04428.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerji J., Olson L., Schaffner W. A lymphocyte-specific cellular enhancer is located downstream of the joining region in immunoglobulin heavy chain genes. Cell. 1983 Jul;33(3):729–740. doi: 10.1016/0092-8674(83)90015-6. [DOI] [PubMed] [Google Scholar]
- Blackwell T. K., Moore M. W., Yancopoulos G. D., Suh H., Lutzker S., Selsing E., Alt F. W. Recombination between immunoglobulin variable region gene segments is enhanced by transcription. Nature. 1986 Dec 11;324(6097):585–589. doi: 10.1038/324585a0. [DOI] [PubMed] [Google Scholar]
- Bucchini D., Reynaud C. A., Ripoche M. A., Grimal H., Jami J., Weill J. C. Rearrangement of a chicken immunoglobulin gene occurs in the lymphoid lineage of transgenic mice. 1987 Mar 26-Apr 1Nature. 326(6111):409–411. doi: 10.1038/326409a0. [DOI] [PubMed] [Google Scholar]
- Coffman R. L. Surface antigen expression and immunoglobulin gene rearrangement during mouse pre-B cell development. Immunol Rev. 1982;69:5–23. doi: 10.1111/j.1600-065x.1983.tb00446.x. [DOI] [PubMed] [Google Scholar]
- Cook W. D., Metcalf D., Nicola N. A., Burgess A. W., Walker F. Malignant transformation of a growth factor-dependent myeloid cell line by Abelson virus without evidence of an autocrine mechanism. Cell. 1985 Jul;41(3):677–683. doi: 10.1016/s0092-8674(85)80048-9. [DOI] [PubMed] [Google Scholar]
- Cook W. D. Thymocyte subsets transformed by Abelson murine leukemia virus. Mol Cell Biol. 1985 Feb;5(2):390–397. doi: 10.1128/mcb.5.2.390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costantini F., Lacy E. Introduction of a rabbit beta-globin gene into the mouse germ line. Nature. 1981 Nov 5;294(5836):92–94. doi: 10.1038/294092a0. [DOI] [PubMed] [Google Scholar]
- Davis M. M., Bjorkman P. J. T-cell antigen receptor genes and T-cell recognition. Nature. 1988 Aug 4;334(6181):395–402. doi: 10.1038/334395a0. [DOI] [PubMed] [Google Scholar]
- Desiderio S. V., Wolff K. R. Rearrangement of exogenous immunoglobulin VH and DJH gene segments after retroviral transduction into immature lymphoid cell lines. J Exp Med. 1988 Feb 1;167(2):372–388. doi: 10.1084/jem.167.2.372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diamond D. J., Nelson F. B., Reinherz E. L. Lineage-specific expression of a T cell receptor variable gene promoter controlled by upstream sequences. J Exp Med. 1989 Apr 1;169(4):1213–1231. doi: 10.1084/jem.169.4.1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Early P., Huang H., Davis M., Calame K., Hood L. An immunoglobulin heavy chain variable region gene is generated from three segments of DNA: VH, D and JH. Cell. 1980 Apr;19(4):981–992. doi: 10.1016/0092-8674(80)90089-6. [DOI] [PubMed] [Google Scholar]
- Ephrussi A., Church G. M., Tonegawa S., Gilbert W. B lineage--specific interactions of an immunoglobulin enhancer with cellular factors in vivo. Science. 1985 Jan 11;227(4683):134–140. doi: 10.1126/science.3917574. [DOI] [PubMed] [Google Scholar]
- Ferrier P., Covey L. R., Suh H., Winoto A., Hood L., Alt F. W. T cell receptor DJ but not VDJ rearrangement within a recombination substrate introduced into a pre-B cell line. Int Immunol. 1989;1(1):66–74. doi: 10.1093/intimm/1.1.66. [DOI] [PubMed] [Google Scholar]
- Forster A., Hobart M., Hengartner H., Rabbitts T. H. An immunoglobulin heavy-chain gene is altered in two T-cell clones. Nature. 1980 Aug 28;286(5776):897–899. doi: 10.1038/286897a0. [DOI] [PubMed] [Google Scholar]
- Gascoigne N. R., Chien Y., Becker D. M., Kavaler J., Davis M. M. Genomic organization and sequence of T-cell receptor beta-chain constant- and joining-region genes. Nature. 1984 Aug 2;310(5976):387–391. doi: 10.1038/310387a0. [DOI] [PubMed] [Google Scholar]
- Gerster T., Matthias P., Thali M., Jiricny J., Schaffner W. Cell type-specificity elements of the immunoglobulin heavy chain gene enhancer. EMBO J. 1987 May;6(5):1323–1330. doi: 10.1002/j.1460-2075.1987.tb02371.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerster T., Picard D., Schaffner W. During B-cell differentiation enhancer activity and transcription rate of immunoglobulin heavy chain genes are high before mRNA accumulation. Cell. 1986 Apr 11;45(1):45–52. doi: 10.1016/0092-8674(86)90536-2. [DOI] [PubMed] [Google Scholar]
- Gillies S. D., Morrison S. L., Oi V. T., Tonegawa S. A tissue-specific transcription enhancer element is located in the major intron of a rearranged immunoglobulin heavy chain gene. Cell. 1983 Jul;33(3):717–728. doi: 10.1016/0092-8674(83)90014-4. [DOI] [PubMed] [Google Scholar]
- Goodhardt M., Cavelier P., Akimenko M. A., Lutfalla G., Babinet C., Rougeon F. Rearrangement and expression of rabbit immunoglobulin kappa light chain gene in transgenic mice. Proc Natl Acad Sci U S A. 1987 Jun;84(12):4229–4233. doi: 10.1073/pnas.84.12.4229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gough N. M., Bernard O. Sequences of the joining region genes for immunoglobulin heavy chains and their role in generation of antibody diversity. Proc Natl Acad Sci U S A. 1981 Jan;78(1):509–513. doi: 10.1073/pnas.78.1.509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosschedl R., Baltimore D. Cell-type specificity of immunoglobulin gene expression is regulated by at least three DNA sequence elements. Cell. 1985 Jul;41(3):885–897. doi: 10.1016/s0092-8674(85)80069-6. [DOI] [PubMed] [Google Scholar]
- Grosschedl R., Weaver D., Baltimore D., Costantini F. Introduction of a mu immunoglobulin gene into the mouse germ line: specific expression in lymphoid cells and synthesis of functional antibody. Cell. 1984 Oct;38(3):647–658. doi: 10.1016/0092-8674(84)90259-9. [DOI] [PubMed] [Google Scholar]
- Grosveld F. G., Lund T., Murray E. J., Mellor A. L., Dahl H. H., Flavell R. A. The construction of cosmid libraries which can be used to transform eukaryotic cells. Nucleic Acids Res. 1982 Nov 11;10(21):6715–6732. doi: 10.1093/nar/10.21.6715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto Y. T cell receptor beta gene has two downstream DNase I hypersensitive regions. Possible mechanisms of tissue- and stage-specific gene regulation. J Exp Med. 1989 Jun 1;169(6):2097–2107. doi: 10.1084/jem.169.6.2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz H. R., LeBlanc P. A., Russell S. W. Two classes of mouse mast cells delineated by monoclonal antibodies. Proc Natl Acad Sci U S A. 1983 Oct;80(19):5916–5918. doi: 10.1073/pnas.80.19.5916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemp D. J., Harris A. W., Cory S., Adams J. M. Expression of the immunoglobulin C mu gene in mouse T and B lymphoid and myeloid cell lines. Proc Natl Acad Sci U S A. 1980 May;77(5):2876–2880. doi: 10.1073/pnas.77.5.2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiledjian M., Su L. K., Kadesch T. Identification and characterization of two functional domains within the murine heavy-chain enhancer. Mol Cell Biol. 1988 Jan;8(1):145–152. doi: 10.1128/mcb.8.1.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kincade P. W. Experimental models for understanding B lymphocyte formation. Adv Immunol. 1987;41:181–267. doi: 10.1016/s0065-2776(08)60032-2. [DOI] [PubMed] [Google Scholar]
- Krimpenfort P., de Jong R., Uematsu Y., Dembic Z., Ryser S., von Boehmer H., Steinmetz M., Berns A. Transcription of T cell receptor beta-chain genes is controlled by a downstream regulatory element. EMBO J. 1988 Mar;7(3):745–750. doi: 10.1002/j.1460-2075.1988.tb02871.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kronenberg M., Siu G., Hood L. E., Shastri N. The molecular genetics of the T-cell antigen receptor and T-cell antigen recognition. Annu Rev Immunol. 1986;4:529–591. doi: 10.1146/annurev.iy.04.040186.002525. [DOI] [PubMed] [Google Scholar]
- Kurosawa Y., von Boehmer H., Haas W., Sakano H., Trauneker A., Tonegawa S. Identification of D segments of immunoglobulin heavy-chain genes and their rearrangement in T lymphocytes. Nature. 1981 Apr 16;290(5807):565–570. doi: 10.1038/290565a0. [DOI] [PubMed] [Google Scholar]
- Lenardo M., Pierce J. W., Baltimore D. Protein-binding sites in Ig gene enhancers determine transcriptional activity and inducibility. Science. 1987 Jun 19;236(4808):1573–1577. doi: 10.1126/science.3109035. [DOI] [PubMed] [Google Scholar]
- Lennon G. G., Perry R. P. C mu-containing transcripts initiate heterogeneously within the IgH enhancer region and contain a novel 5'-nontranslatable exon. Nature. 1985 Dec 5;318(6045):475–478. doi: 10.1038/318475a0. [DOI] [PubMed] [Google Scholar]
- Lieber M. R., Hesse J. E., Mizuuchi K., Gellert M. Developmental stage specificity of the lymphoid V(D)J recombination activity. Genes Dev. 1987 Oct;1(8):751–761. doi: 10.1101/gad.1.8.751. [DOI] [PubMed] [Google Scholar]
- Malissen M., McCoy C., Blanc D., Trucy J., Devaux C., Schmitt-Verhulst A. M., Fitch F., Hood L., Malissen B. Direct evidence for chromosomal inversion during T-cell receptor beta-gene rearrangements. Nature. 1986 Jan 2;319(6048):28–33. doi: 10.1038/319028a0. [DOI] [PubMed] [Google Scholar]
- Malynn B. A., Wortis H. H. Role of antigen-specific B cells in the induction of SRBC-specific T cell proliferation. J Immunol. 1984 May;132(5):2253–2258. [PubMed] [Google Scholar]
- Marrack P., Kappler J. The T cell receptor. Science. 1987 Nov 20;238(4830):1073–1079. doi: 10.1126/science.3317824. [DOI] [PubMed] [Google Scholar]
- McDougall S., Peterson C. L., Calame K. A transcriptional enhancer 3' of C beta 2 in the T cell receptor beta locus. Science. 1988 Jul 8;241(4862):205–208. doi: 10.1126/science.2968651. [DOI] [PubMed] [Google Scholar]
- Mercola M., Wang X. F., Olsen J., Calame K. Transcriptional enhancer elements in the mouse immunoglobulin heavy chain locus. Science. 1983 Aug 12;221(4611):663–665. doi: 10.1126/science.6306772. [DOI] [PubMed] [Google Scholar]
- Meyer K. B., Neuberger M. S. The immunoglobulin kappa locus contains a second, stronger B-cell-specific enhancer which is located downstream of the constant region. EMBO J. 1989 Jul;8(7):1959–1964. doi: 10.1002/j.1460-2075.1989.tb03601.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelsen B., Hellman L., Sen R. The NF-kappa B-binding site mediates phorbol ester-inducible transcription in nonlymphoid cells. Mol Cell Biol. 1988 Aug;8(8):3526–3531. doi: 10.1128/mcb.8.8.3526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuberger M. S. Expression and regulation of immunoglobulin heavy chain gene transfected into lymphoid cells. EMBO J. 1983;2(8):1373–1378. doi: 10.1002/j.1460-2075.1983.tb01594.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okazaki K., Davis D. D., Sakano H. T cell receptor beta gene sequences in the circular DNA of thymocyte nuclei: direct evidence for intramolecular DNA deletion in V-D-J joining. Cell. 1987 May 22;49(4):477–485. doi: 10.1016/0092-8674(87)90450-8. [DOI] [PubMed] [Google Scholar]
- Parslow T. G., Granner D. K. Chromatin changes accompany immunoglobulin kappa gene activation: a potential control region within the gene. Nature. 1982 Sep 30;299(5882):449–451. doi: 10.1038/299449a0. [DOI] [PubMed] [Google Scholar]
- Perez-Mutul J., Macchi M., Wasylyk B. Mutational analysis of the contribution of sequence motifs within the IgH enhancer to tissue specific transcriptional activation. Nucleic Acids Res. 1988 Jul 11;16(13):6085–6096. doi: 10.1093/nar/16.13.6085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson C. L., Calame K. L. Complex protein binding within the mouse immunoglobulin heavy-chain enhancer. Mol Cell Biol. 1987 Dec;7(12):4194–4203. doi: 10.1128/mcb.7.12.4194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson C. L., Orth K., Calame K. L. Binding in vitro of multiple cellular proteins to immunoglobulin heavy-chain enhancer DNA. Mol Cell Biol. 1986 Dec;6(12):4168–4178. doi: 10.1128/mcb.6.12.4168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picard D., Schaffner W. A lymphocyte-specific enhancer in the mouse immunoglobulin kappa gene. Nature. 1984 Jan 5;307(5946):80–82. doi: 10.1038/307080a0. [DOI] [PubMed] [Google Scholar]
- Pierce J. H., Di Fiore P. P., Aaronson S. A., Potter M., Pumphrey J., Scott A., Ihle J. N. Neoplastic transformation of mast cells by Abelson-MuLV: abrogation of IL-3 dependence by a nonautocrine mechanism. Cell. 1985 Jul;41(3):685–693. doi: 10.1016/s0092-8674(85)80049-0. [DOI] [PubMed] [Google Scholar]
- Queen C., Baltimore D. Immunoglobulin gene transcription is activated by downstream sequence elements. Cell. 1983 Jul;33(3):741–748. doi: 10.1016/0092-8674(83)90016-8. [DOI] [PubMed] [Google Scholar]
- Rosenberg N., Baltimore D. A quantitative assay for transformation of bone marrow cells by Abelson murine leukemia virus. J Exp Med. 1976 Jun 1;143(6):1453–1463. doi: 10.1084/jem.143.6.1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakano H., Maki R., Kurosawa Y., Roeder W., Tonegawa S. Two types of somatic recombination are necessary for the generation of complete immunoglobulin heavy-chain genes. Nature. 1980 Aug 14;286(5774):676–683. doi: 10.1038/286676a0. [DOI] [PubMed] [Google Scholar]
- Schatz D. G., Baltimore D. Stable expression of immunoglobulin gene V(D)J recombinase activity by gene transfer into 3T3 fibroblasts. Cell. 1988 Apr 8;53(1):107–115. doi: 10.1016/0092-8674(88)90492-8. [DOI] [PubMed] [Google Scholar]
- Schlissel M. S., Baltimore D. Activation of immunoglobulin kappa gene rearrangement correlates with induction of germline kappa gene transcription. Cell. 1989 Sep 8;58(5):1001–1007. doi: 10.1016/0092-8674(89)90951-3. [DOI] [PubMed] [Google Scholar]
- Sen R., Baltimore D. Inducibility of kappa immunoglobulin enhancer-binding protein Nf-kappa B by a posttranslational mechanism. Cell. 1986 Dec 26;47(6):921–928. doi: 10.1016/0092-8674(86)90807-x. [DOI] [PubMed] [Google Scholar]
- Sen R., Baltimore D. Multiple nuclear factors interact with the immunoglobulin enhancer sequences. Cell. 1986 Aug 29;46(5):705–716. doi: 10.1016/0092-8674(86)90346-6. [DOI] [PubMed] [Google Scholar]
- Siu G., Kronenberg M., Strauss E., Haars R., Mak T. W., Hood L. The structure, rearrangement and expression of D beta gene segments of the murine T-cell antigen receptor. 1984 Sep 27-Oct 3Nature. 311(5984):344–350. doi: 10.1038/311344a0. [DOI] [PubMed] [Google Scholar]
- Storb U., Pinkert C., Arp B., Engler P., Gollahon K., Manz J., Brady W., Brinster R. L. Transgenic mice with mu and kappa genes encoding antiphosphorylcholine antibodies. J Exp Med. 1986 Aug 1;164(2):627–641. doi: 10.1084/jem.164.2.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonegawa S. Somatic generation of antibody diversity. Nature. 1983 Apr 14;302(5909):575–581. doi: 10.1038/302575a0. [DOI] [PubMed] [Google Scholar]
- Tsao B. P., Wang X. F., Peterson C. L., Calame K. In vivo functional analysis of in vitro protein binding sites in the immunoglobulin heavy chain enhancer. Nucleic Acids Res. 1988 Apr 25;16(8):3239–3253. doi: 10.1093/nar/16.8.3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uematsu Y., Ryser S., Dembić Z., Borgulya P., Krimpenfort P., Berns A., von Boehmer H., Steinmetz M. In transgenic mice the introduced functional T cell receptor beta gene prevents expression of endogenous beta genes. Cell. 1988 Mar 25;52(6):831–841. doi: 10.1016/0092-8674(88)90425-4. [DOI] [PubMed] [Google Scholar]
- Van Ness B. G., Weigert M., Coleclough C., Mather E. L., Kelley D. E., Perry R. P. Transcription of the unrearranged mouse C kappa locus: sequence of the initiation region and comparison of activity with a rearranged V kappa-C kappa gene. Cell. 1981 Dec;27(3 Pt 2):593–602. doi: 10.1016/0092-8674(81)90401-3. [DOI] [PubMed] [Google Scholar]
- Weinberger J., Baltimore D., Sharp P. A. Distinct factors bind to apparently homologous sequences in the immunoglobulin heavy-chain enhancer. 1986 Aug 28-Sep 3Nature. 322(6082):846–848. doi: 10.1038/322846a0. [DOI] [PubMed] [Google Scholar]
- Winoto A., Baltimore D. A novel, inducible and T cell-specific enhancer located at the 3' end of the T cell receptor alpha locus. EMBO J. 1989 Mar;8(3):729–733. doi: 10.1002/j.1460-2075.1989.tb03432.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wysocki L. J., Sato V. L. "Panning" for lymphocytes: a method for cell selection. Proc Natl Acad Sci U S A. 1978 Jun;75(6):2844–2848. doi: 10.1073/pnas.75.6.2844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yancopoulos G. D., Alt F. W. Developmentally controlled and tissue-specific expression of unrearranged VH gene segments. Cell. 1985 Feb;40(2):271–281. doi: 10.1016/0092-8674(85)90141-2. [DOI] [PubMed] [Google Scholar]
- Yancopoulos G. D., Blackwell T. K., Suh H., Hood L., Alt F. W. Introduced T cell receptor variable region gene segments recombine in pre-B cells: evidence that B and T cells use a common recombinase. Cell. 1986 Jan 31;44(2):251–259. doi: 10.1016/0092-8674(86)90759-2. [DOI] [PubMed] [Google Scholar]
- Zimmerman K. A., Yancopoulos G. D., Collum R. G., Smith R. K., Kohl N. E., Denis K. A., Nau M. M., Witte O. N., Toran-Allerand D., Gee C. E. Differential expression of myc family genes during murine development. 1986 Feb 27-Mar 5Nature. 319(6056):780–783. doi: 10.1038/319780a0. [DOI] [PubMed] [Google Scholar]
- von Boehmer H. The developmental biology of T lymphocytes. Annu Rev Immunol. 1988;6:309–326. doi: 10.1146/annurev.iy.06.040188.001521. [DOI] [PubMed] [Google Scholar]