Abstract
Targeted delivery of drug-encapsulated nanoparticles is a promising new approach to safe and effective therapeutics for cancer. Here we investigate the pharmacokinetics and biodistribution of a prostate-specific membrane antigen (PSMA)-targeted nanoparticle based on a poly(lactic acid)–polyethylene glycol copolymer by utilizing single photon emission computed tomography (SPECT) and fluorescence imaging of a low-molecular-weight, PSMA-targeting moiety attached to the surface and oriented toward the outside environment. Tissue biodistribution of the radioactive, PSMA-targeted nanoparticles in mice containing PSMA(+) PC3 PIP and PSMA(−) PC3 flu (control) tumors demonstrated similar accumulation compared to the untargeted particles within all tissues except for the tumor and liver by 96 h postinjection. For PSMA(+) PC3 PIP tumor, the targeted nanoparticle demonstrated retention of 6.58% injected dose (ID)/g at 48 h and remained nearly at that level out to 96 h, whereas the untargeted nanoparticle showed a 48 h retention of 8.17% ID/g followed by a significant clearance to 2.37% ID/g at 96 h (P < 0.02). On the other hand, for control tumor, both targeted and untargeted particles displayed similar 48 h retentions and rates of clearance over 96 h. Ex vivo microscopic analysis with near-infrared versions of the nanoparticles indicated retention within PSMA(+) tumor epithelial cells as well as tumor-associated macrophages for targeted particles and primarily macrophage-associated uptake for the untargeted particles. Retention in control tumor was primarily associated with tumor vasculature and macrophages. The data demonstrate the utility of radioimaging to assess nanoparticle biodistribution and suggest that active targeting has a modest positive effect on tumor localization of PSMA-targeted PLA–PEG nanoparticles that have been derivatized for imaging.
Graphical abstract
INTRODUCTION
The key contribution of nanoparticle-based therapeutics to cancer lies in the ability to increase the concentration of therapeutic payload where it is needed, at the tumor site.1–7 Since the appearance of DOXIL, which was designed to mitigate the side effects of systemically administered doxorubicin, a number of micro- and nanoparticle-based anticancer therapeutics have emerged.8–12 One such agent, BIND-014 (ClinicalTrials.gov identifier NCT01300533), is a poly(lactic acid)– polyethyene glycol (PLA–PEG) copolymer-based particle that contains and ultimately releases docetaxel while targeting the prostate-specific membrane antigen (PSMA) by virtue of surface expression of a low-molecular-weight targeting ligand that binds with high affinity to PSMA.13,14 Often debated is the utility of actively targeting such particles, such as with an affinity agent, rather than merely allowing them to accumulate passively at tumor sites through the enhanced permeability and retention (EPR) effect or more recently described super enhanced EPR (SUPR) effects.15,16 It has been proposed that active targeting would increase the concentration of the nanoagent to the target site over that achievable by EPR, particularly at small metastatic sites (<100 mm3) that are poorly vascularized and do not induce EPR.15 However, there are a number of factors beyond the placement of affinity agents on the surface of nanoparticles that determine the effect of active targeting on the efficacy of a nanoencapsulated drug, such as the concentration of the targeting ligand or access of the particle to the target, which may be hampered by improper orientation of the targeting units or insufficient steric freedom.17,18 In addition to facilitating the binding of particles to target cells or tissues, functionalizing the surface of a particle with affinity agents may alter its pharmacokinetics, ability to penetrate barriers, or its immunogenicity.19 Studies performed in vitro may provide hints as to whether targeting will improve target cell localization or uptake of the payload within the particle, but they cannot predict the ultimate performance of nanoparticles in vivo.
One way to ascertain the effects of active targeting on particle biodistribution is to perform pharmacokinetic studies with radiolabeled or fluorescent particles and extract organs of interest for quantification of particle uptake. A less invasive procedure would involve imaging. Other benefits of imaging include the ability to be performed repeatedly (and quantitatively) in the same subject and the ability to translate to the clinic, which is currently easiest with radionuclide-based techniques due to their high sensitivity of detection and limited attenuation of signal through tissue. The generation of particles that can be imaged as well as used for therapy (theranostics) is a burgeoning field. By using multimodal in vivo imaging techniques, Bartlett et al. demonstrated that theranostic untargeted and transferrin (Tf)-targeted polymeric nanoparticles containing siRNA have the same whole body distribution (and kinetics) and accumulation in tumor, but the targeted particles led to more pronounced inhibition of gene expression within cancer cells.20 The same investigators and others concluded that targeting does not increase total accumulation of nanoparticles in tumor as compared with untargeted controls but rather increases targeted particle localization within cancer cells.18,21,22 Therefore, the real value of targeting drug-loaded nanoparticles may lie in their ability to be internalized upon receptor-mediated endocytosis. Cell internalization is potentially important because many FDA-approved nanoparticle-based medications, such as ABRAXANE (paclitaxel) and DOXIL (doxorubicin), contain drugs that act on intracellular targets, and it is expected to be critical for agents that do not otherwise enter cells such as many oligonucleotide therapeutics. Commonly used targeting ligands, such as Tf, epidermal growth factor (EGF), and antibodies, already benefit from receptor-mediated endocytosis. Even ligands that do not naturally internalize often undergo internalization when displayed in multivalent fashion on the nanoparticle surface. Relatively few studies have attempted to address how the pharmacokinetics and biodistribution of nanoparticles are affected by attachment of targeting ligands, yet such studies are important for designing more effective nanoparticle carriers.
Leveraging urea-based inhibitors of PSMA that we have developed for imaging and localization of PLGA and other nanoparticles,23–26 we have synthesized and tested a PSMA-targeted PLA–PEG particle that is radiolabeled with 111In (half-life, 2.8 days, γ − 245 keV (94% abundance), 171 keV (90% abundance), 23 keV (69% abundance)) and could be followed through quantitative tissue biodistribution and semiquantitative single photon emission computed tomography (SPECT)-computed tomography (CT) ex vivo and in vivo, respectively, in experimental models of prostate cancer. We found that targeting provides a small but measurable increase in particle localization to target-expressing tumor. Moreover, in vivo near-infrared (NIR) fluorescence imaging studies using IRDye680RD-labeled, PSMA-targeted nanoparticles (TNPs) and untargeted nanoparticles (UNPs) followed by ex vivo tissue microscopy revealed a marked difference of nanoparticle distribution within tumor tissues; the TNPs and UNPs differed with respect to their distribution within malignant cells versus noncancer cells such as PSMA(+) tumor epithelium versus vascular endothelium and leukocytes. While TNPs and UNPs both accumulate within tumors to a greater extent than within tissues other than those of the reticuloendothelial system, the UNPs do so mainly through association with macrophages rather than malignant cells.
EXPERIMENTAL SECTION
Solvents and chemicals obtained from commercial sources were of at least analytical grade and were used without further purification. A detailed description of the synthesis and characterization of the nanoparticles employed has been published elsewhere.13 Size of the nanoparticles is approximately 100 nm.13 Carrier-free [111In]InCl3 was purchased from MDS Nordion (Ottawa, ON, Canada). DOTA-mono-NHS-tris (t-Bu ester) was purchased from Macrocyclics (Dallas, TX). NH2–PEG4-alkyne was purchased from Click Chemistry Tools (Scottsdale, AZ). IRDye680RD alkyne was purchased from LI-COR (Lincoln, NE) and was stored in the dark as a dry powder under nitrogen, at −80 °C, until use. PLA–PEG nanoparticles functionalized with 10% of the terminal PEG end groups as azide moieties were supplied by BIND Therapeutics (Cambridge, MA). A Phenomenex C18 150 × 3.9 mm2 column was used for the purification of 111In-DOTA-PEG-alkyne by reverse phase high performance liquid chromatography (HPLC) (see Supporting Information for HPLC chromatograms). HPLC purification of [111In]DOTA-PEG-alkyne was performed on a Varian Prostar System (Palo Alto, CA), equipped with a model 490 UV absorbance detector and a Bioscan NaI scintillation detector connected to a Bioscan Flow-count system (Bioscan, Washington, DC). HPLC was performed according to the following methods. Solvent A (0.1% trifluoroacetic acid (TFA) in water) and solvent B (0.1% TFA in acetonitrile) at a flow rate 1 mL/min. Zeba Spin 7K MWCO size exclusion spin columns (Thermo Scientific, Rockford, IL) were used for nanoparticle purification.
Chemistry and Radiochemistry
2,2′,2″-(10-(2-Oxo-6,9,12,15-tetraoxa-3-azaoctadec-17-yn-1-yl)-1,4,7,10-tetraazacyclododecane-1,4,7-triyl)triacetic acid, (DOTA-PEG-Alkyne)
To a solution of DOTA-mono-NHS-tris (t-Bu ester) (106 mg, 0.13 mmol in 1 mL DMSO) was added NH2–PEG4-alkyne (34.4 mg, 0.157 mmol in 0.5 mL DMSO) followed by DIEA (227 μL, 1.3 mmol), and the resulting solution was stirred at room temperature for 4 h. The solution was the diluted with 6 mL of water and purified by HPLC and lyophilized. ESI-MS: 876 [M + H]+. The obtained colorless solid was then treated with 4 mL of ice-cold TFA/CH2Cl2 (1/1) followed by warming to room temperature while stirring for 4 h. After solvent evaporation, the residue was further purified by HPLC (Supporting Information for spectral data). HR-ESI-MS: 618.2913 [M + H]+.
Radiolabeling of Nanoparticles Was Performed Following a Two-Step Procedure
Step 1: Synthesis of 2,2′,2″-(10-(2-Oxo-6,9,12,15-tetraoxa-3-azaoctadec-17-yn-1-yl)-1,4,7,10-tetraazacyclododecane-1,4,7-triyl)triacetic acid 111Indium(III), 111In-DOTA-PEG-Alkyne
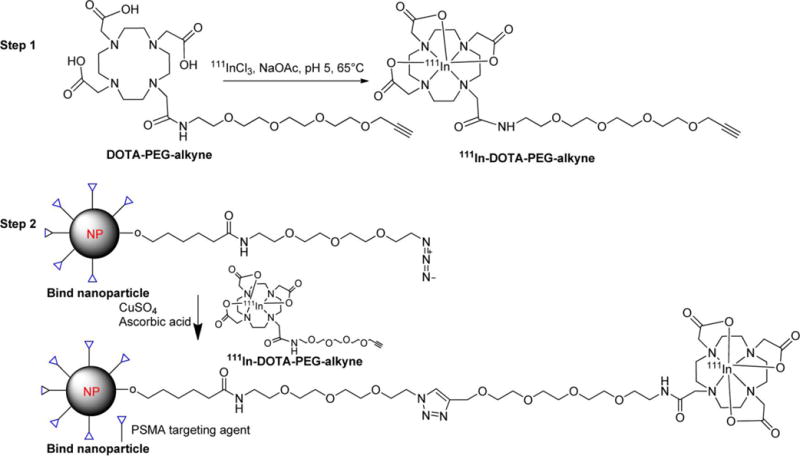
To a solution of 111InCl3 (1998 MBq (54 mCi) in 30 μL 0.1 M HCl) in a 0.5 mL polypropylene vial was added 40 μL of 0.2 M NaOAc and DOTA-PEG-alkyne (100 μg, 162 nmol in 50 μL of 0.2 mM NaOAc) with the reaction mixture incubated at 65 °C for 30 min. Completion of radiolabeling was monitored by injecting aliquots of 1–2 μL onto the HPLC. After 30 min, the radiolabeled solution was diluted with 600 μL of water followed by purification by HPLC. The radiolabeled product, 111In-DOTA-PEG-alkyne, was obtained in ∼50–70% radiochemical yield with radiochemical purity >95% as measured by HPLC and instant thin layer chromatography (ITLC) (Gelman, Ann Arbor, MI) in 10 mM EDTA. The Rf values for free In(III) and for the product were 0.9 and 0.0, respectively. A gradient HPLC method with a flow rate of 1 mL/min, starting at 92/8 water/acetonitrile (0.1% TFA in each solvent) to 90/10 water/acetonitrile at 30 min, was used.
Step 2: Synthesis of Radiolabeled Nanoparticles 111In-TNP (PSMA-targeted) and 111In-UNP (Untargeted Nanoparticle)
All reagents and buffer solutions were purged under nitrogen thoroughly before the click reaction was attempted. 111In-DOTA-PEG-alkyne, 316.4 MBq (8.55 mCi) was dissolved in 10 μL of 10× PBS buffer before the mixture was transferred to a 0.5 mL polypropylene vial. To that solution was added 80 μL of azide-functionalized nanoparticles, either targeted with a urea-bearing moiety (ACUPA) for the PSMA-targeted nanoparticle (TNP) (84.5 mg/mL, ACUPA 2.5%,13 10% azide) or not (untargeted nanoparticle, UNP, control), (83.0 mg/mL, 10% azide), followed by 5 μL of ascorbic acid (100 mM). The mixture was purged with nitrogen briefly (1 min) and finally 2 μL of CuSO4 (100 mM) was added, after which the lid of the vial was closed and the contents were mixed gently for 2 min at room temperature. The vial was then incubated at 4 °C for 24 h. Zeba Spin 7K MWCO size exclusion spin columns were pre-equilibrated with 0.9% saline and used for purification. The nanoparticle solutions were purified four times to remove any unreacted [111In]DOTA-PEG-alkyne. Yield ∼40%. Specific radioactivity ∼1923 MBq/nmol (52 mCi/nmol) or 20 MBq/mg (0.55 mCi/mg). Ratio of 111In/particle ∼0.6–1.1. An example of radiolabeling yield and calculation of specific radioactivity for the particles are shown in the Supporting Information (Table S1).
Synthesis of IRDye680RD-Labeled Nanoparticles
IRDye680RD-alkyne (50 μg, 53 nmol) was dissolved in 20 μL of 10× PBS and was added to a 0.5 mL Eppendorf vial containing 160 μL (13.4 mg) of azide-functionalized nanoparticles, either TNP or UNP, followed by 5 μL of ascorbic acid (100 mM). The mixture was purged with nitrogen briefly (1 min) and finally 2 μL of CuSO4 (100 mM) was added, after which the lid of the vial was closed and the contents of the vial were mixed gently for 2 min at room temperature. The vial was then incubated at 4 °C for 24 h. Zeba Spin 7K MWCO size exclusion spin columns were pre-equilibrated with 0.9% saline and used for purification. The nanoparticle solutions were purified four times to remove any unreacted IRDye680RD-alkyne. Yield ∼60% (determined from the fluorescence intensity measurement associated with the particles at 680 nm).
Animal Studies and Generation of Experimental Models
All animal studies were conducted in compliance with guidelines of the Johns Hopkins Animal Care and Use Committee. Mice were prepared as described previously,27 except that athymic nude mice averaging 25–30 g were used instead of nonobese diabetic severe-combined immunodeficiency (NOD SCID) mice. PSMA(+) PC-3 PIP cells were selected for PSMA expression by using 20 μg/mL of puromycin in the culture media prior to harvesting cells and subcutaneous implantation. The percent of cells expressing PSMA was assessed using fluorescence activated cell sorting (FACS) for membrane-bound PSMA expression (Supplementary Figure 5) and was observed to be ≥99+%.
Ex Vivo and in Vivo Testing of the 111In-Labeled PLA–PEG Nanoparticles
Acute Blood Measurements
PSMA(+) PC-3 PIP and PSMA(−) PC-3 flu dual xenograft bearing mice were injected with either 111In-TNP or 111In-UNP. Three mice per group were injected intravenously with 3.26 ± 0.26 MBq (88 ± 7 μCi) of TNP or 3.59 ± 0.33 MBq (97 ± 9 μCi) of UNP, respectively, which correspond to 0.16–0.18 mg (8–9 mg/kg) of particles. The mice underwent a 45 min uptake time prior to sacrifice by cervical dislocation followed by cardiac puncture for blood collection (500 μL) into cold lithium heparin-containing microtainers (Becton Dickenson, Franklin Lakes, NJ). The packed cells were separated from plasma via centrifugation at 1500g at 4 °C and were each assessed for radioactivity using a 1282 Compugamma CS Universal gamma counter (LKB Wallac, Mt. Waverly Vic., AUS) with results expressed as percentage injected dose per gram (%ID/g) of cells or volume of plasma ± standard deviation (SD).
Biodistribution
PSMA(+) PC-3 PIP and PSMA(−) PC-3 flu dual xenograft bearing mice were injected intravenously with 5.55 ± 0.12 MBq (150 ± 3.24 μCi) of either 111In-TNP or 111In-UNP (0.28 ± 0.006 mg; 14 mg/kg) in physiological saline when the tumors reached 5–8 mm in diameter. Mice were sacrificed by cervical dislocation after 48, 72, and 96 h postinjection with tissues rapidly removed, weighed, and counted using a gamma counter. All tissue counts were compared with two diluted standard doses, which allowed the values to be expressed as %ID/g ± SD, n = 5 per group.
In Vivo SPECT-CT Imaging
Two mice each bearing PSMA(+) PC-3 PIP and PSMA(−) PC-3 flu dual xenografts were injected intravenously with 31.45 ± 0.37 MBq (0.87 ± 0.01 mCi) of either 111In-TNP or 111In-UNP (79 mg/kg or 1.57 ± 0.02 mg) or in 0.9% saline. At 48, 72, and 96 h postinjection, the mice were anesthetized using 3% isoflurane in oxygen (2 L/min) and were placed in mixed pairs (TNP and UNP) upon the scanner bed and scanned with SPECT (X-SPECT, Gamma Medica-Ideas, Inc., Northridge, CA). The mice were scanned using dual medium energy pinhole collimators (0.5 mm), over 64 projections at 55 s/projection. The SPECT data were reconstructed and coregistered with CT using the manufacturer’s software and displayed using AMIDE (www.amide.sourceforge.net). CT scans were acquired over 512 projections and were reconstructed using the manufacturer’s software.
Ex Vivo Fluorescence Imaging and Microscopy
Two mice each of which bore PSMA(+) PC-3 PIP and PSMA(−) PC-3 flu subcutaneous xenografts in opposite flanks, were injected with ∼79 mg/kg (1.57 ± 0.02 mg) of fluorescently labeled TNP or UNP. Mice were sacrificed 72 h later. Mice were then imaged using a LI-COR Pearl Impulse Near-IR fluorescence imager (Lincoln, NE) using the 680 nm excitation/710 nm emission filter set. All four mice were scanned, and the images were normalized for exposure time. Xenografts were rapidly harvested and frozen over dry ice before being cryosectioned to 20 μm thickness onto charged glass slides (VWR, Radnor, PA). Some slides were scanned using a LI-COR Biosciences Odyssey fluorescence scanner using the 680 nm/710 nm excitation and emission filter set. Some slides were probed with mouse-anti-PSMA GCP05 (Abcam; ab66912, 1:83; Cambridge, MA), rat-anti-CD31-PE (ab25644, 1:67), and rat-anti-CD68 (ab53444, 1:83) as indicated for 1 h in 10% FBS in PBS at room temperature. The slides were washed twice with PBS for 5 min each prior to incubation with goat-antimouse-FITC (ab6785, 1:250) or donkey-antimouse-PE (ab7003, 1:250) or sheep-antirat-FITC (ab6848–1, 1:250) as indicated for 30 min at room temperature. Slides were then incubated with Hoechst 33342 (Sigma, St. Louis, MO; 1:1000) for 1.5 min before being washed twice with PBS. Slides were mounted in Dako Cytomation (Carpinteria, CA) aqueous mounting media with a glass coverslip and viewed 30 min later using a Nikon 80i upright epifluorescence microscope equipped with a Nikon DS-Qi1Mc darkfield CCD camera and excited by a Nikon Intensilight C-HGFI lamp (Melville, NY). All images were recorded and processed using Nikon Imaging Software Elements.
PSMA(+) PC-3 PIP and PSMA(−) PC-3 flu cells were cultured at 37 °C in 5% CO2 in humidified air in an eight-well chamber slide (Fisher Scientific, Waltham, MA) to 90% confluence using RPMI 1640 media supplemented with 10% fetal bovine serum (Sigma, St. Louis, MO) with penicillin–streptomycin (Corning, Tewksbury, MA), 1000 IU/mL, 1000 μg/mL). Media for the PSMA(+) PC-3 PIP cells also contained 20 μg/mL of puromycin (Sigma). Four-hundred micrograms of either TNP-IRDye680RD or UNP-IRDye680RD in a final volume of 510 μL in culture media was applied to live cells, and the cells were returned to the incubator for 20 min for particle binding and uptake. After 20 min, the culture media was removed, the cells were washed twice briefly with PBS, and then the cells were fixed for 30 min at room temperature with formalin. The cells were then washed twice with PBS followed by addition of 10% FBS in PBS containing anti-PSMA antibody (Abcam, 1:67, #ab66912) and antitubulin antibody (Novus Biologicals, Littleton, CO), 1:67, #NB600–506PE), which incubated at room temperature for 1 h. Cells were then washed twice with PBS, and then secondary antibody was applied (Abcam (Cambridge, MA), 1:250, #ab97022) for 30 min. Hoechst 33342 (Sigma, 1:1000) was applied for 1.5 min followed by two washes with PBS. Slides were then mounted and visualized as described above.
In vitro FACS Analysis
PSMA(+) PC3-PIP or PSMA(−) PC3-flu cells were detached using enzyme-free cell dissociation buffer (Gibco), and the harvested cells were washed twice with flow cytometry buffer (1× PBS with 2 mM EDTA and 0.5% FBS) and stained with antihuman PSMA antibody conjugated with phycoerythrin (PE) (catalog # 341503, clone LNI-17, Bioegend) according to the manufacturer’s protocol. PSMA expression was analyzed on a FACS Calibur flow cytometer (Becton Dickinson). Data analysis was carried out using FlowJo (Ashland, OR) Tree Star software.
RESULTS
Synthesis
To ensure the stability of the PLA–PEG particles,13 we performed the radiolabeling of those nanoparticles at 4 °C. Since efficient radiolabeling of DOTA with 111In requires temperatures of at least 40–45 °C, we developed the two-step radiolabeling method shown in Scheme 1. Copper-assisted click chemistry proved efficient to radiolabel the azide-functionalized nanoparticles at 4 °C. The first step involved radiolabeling of DOTA-PEG-alkyne with 111InCl3 to produce 111In-DOTA-PEG-alkyne. For the second step, a copper-assisted click reaction was undertaken between the azide-functionalized version of the nanoparticles, either PSMA-targeted TNP or untargeted UNP, and alkyne-function-alized 111In-DOTA-PEG-alkyne from step 1. Both steps proceeded smoothly with particles produced in moderate yields and sufficient specific radioactivity for biodistribution and imaging.
Scheme 1.
Two-Step Radiolabeling Method for the PSMA-Targeted Nanoparticles
Acute Blood Measurements
We evaluated the distribution of both 111In-TNP and 111In-UNP within the blood of tumor-bearing mice at an early time point following injection (45 min) to determine whether the particles were soluble in plasma or were rapidly associated with phagocytic or other blood cells. Tumor-bearing mice were injected with equal mass and radioactivity doses of either 111In-TNP or 111In-UNP. For the TNP, 35.0 ± 5.2% ID/g of the particles was associated with the plasma fraction, while 4.7 ± 1.2% ID/g of the particles was associated with the cellular fraction. For the UNP, 50.2 ± 3.1% ID/g of the particles was associated with the plasma fraction and 3.8 ± 0.2% ID/g with the cellular fraction. These results indicate that the bulk of the particles remain in the plasma and are not associated with blood cells.
Biodistribution and in Vivo SPECT-CT Imaging
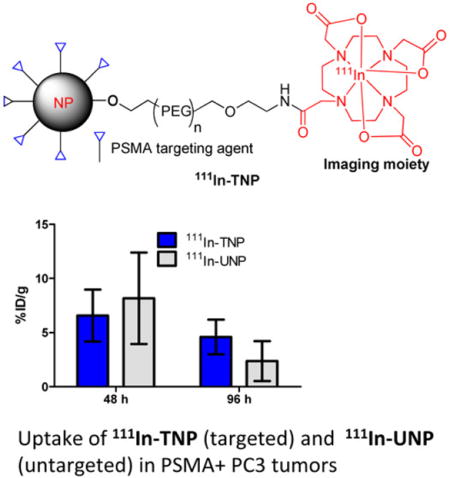
The pharmacokinetics and biodistribution of systemically injected, 111In-TNP and 111In-UNP in tumor-bearing mice were evaluated at 48, 72, and 96 h postinjection. Table 1 displays the values obtained at all time points, expressed as %ID/g of tissue ± SD (n = 4). At 48 h, the 111In-UNP achieved a nonsignificant but slightly higher average retention across all tissues including both tumor types relative to the 111In-TNP (8.2 ± 4.2 vs 6.6 ± 2.4% ID/g in PSMA(+) PC-3 PIP and 6.3 ± 4.5 versus 5.0 ± 1.3% ID/g in PSMA(−) PC-3 flu). Blood circulation also remained higher for 111In-UNP over 111In-TNP at this time (4.8 ± 1.1 vs 2.3 ± 0.5% ID/g). By 72 h postinjection, the 111In-TNP exhibited almost equal retention in several tissues (liver, spleen) and slightly higher retention in PSMA(+) PC-3 PIP as well as PSMA(−) PC-3 flu tumors (5.0 ± 3.1 vs 3.5 ± 1.2% ID/g in PSMA(+) PC-3 PIP; 2.5 ± 1.7 vs 1.7 ± 1.1% ID/g in PSMA(−) PC-3 flu). By 96 h postinjection, 111In-TNP exhibited slightly higher average retention over 111In-UNP in the liver and PSMA(+) PC-3 PIP tumors (4.6 ± 1.6 vs 2.4 ± 1.9% ID/g) and PSMA(−) PC-3 flu tumors (4.0 ± 0.3 vs 2.6 ± 0.2% ID/g). Indeed, the clearance of 111In-UNP from PSMA(+) PC-3 tumor uptake was significantly faster (P < 0.016) during 48 h-to-96 h compared to 111In-TNP in PSMA(+) PC-3 tumor (P < 0.176). The highest retention across all time points occurred in the spleen for both nanoparticles with UNP having the highest retention in spleen at 48 h postinjection (46.3 ± 22.6 vs 23.6 ± 3.2% ID/g) and lowest at 96 h (12.8 ± 8.2 vs 6.0 ± 1.8% ID/g).
Table 1.
Tissue Biodistribution of 111In-TNP and 111In-UNP in Tumor-Bearing Athymic Nude Micea
| tissue (%ID/g) | 48 h TNP | 48 h UNP | 72 h TNP | 72 h UNP | 96 h TNP | 96 h UNP |
|---|---|---|---|---|---|---|
| blood | 2.29 ± 0.47 | 4.77 ± 1.07 | 0.30 ± 0.20 | 0.27 ± 0.12 | 0.03 ± 0.01 | 0.03 ± 0.00 |
| heart | 1.64 ± 0.26 | 3.12 ± 0.91 | 0.74 ± 0.33 | 0.69 ± 0.28 | 0.89 ± 0.22 | 0.62 ± 0.47 |
| liver | 19.49 ± 4.32 | 33.90 ± 4.65 | 19.55 ± 3.35 | 12.29 ± 3.94 | 18.15 ± 2.21 | 10.01 ± 6.86 |
| lung | 2.01 ± 0.56 | 4.90 ± 1.49 | 0.54 ± 0.19 | 0.68 ± 0.32 | 0.91 ± 0.17 | 0.21 ± 0.67 |
| stomach | 2.29 ± 0.43 | 3.33 ± 0.79 | 1.15 ± 0.62 | 1.13 ± 0.55 | 1.39 ± 0.09 | 1.19 ± 0.52 |
| pancreas | 0.58 ± 0.14 | 1.51 ± 0.54 | 0.71 ± 0.59 | 0.49 ± 0.25 | 0.50 ± 0.26 | 0.55 ± 0.56 |
| spleen | 23.64 ± 3.16 | 46.33 ± 22.60 | 9.93 ± 3.19 | 11.39 ± 5.85 | 6.04 ± 1.78 | 12.78 ± 8.24 |
| kidney | 3.89 ± 0.34 | 8.01 ± 5.09 | 2.21 ± 0.69 | 1.90 ± 0.77 | 2.92 ± 0.60 | 2.46 ± 1.38 |
| white fat | 0.37 ± 0.13 | 1.11 ± 0.29 | 0.27 ± 0.11 | 0.41 ± 0.19 | 0.39 ± 0.10 | 1.23 ± 1.19 |
| sm. intestine | 8.49 ± 2.75 | 17.81 ± 5.78 | 4.43 ± 2.79 | 3.84 ± 2.19 | 5.45 ± 1.92 | 4.57 ± 1.39 |
| lg. intestine | 4.23 ± 1.93 | 3.60 ± 1.51 | 1.18 ± 0.52 | 1.70 ± 1.44 | 1.50 ± 0.30 | 2.49 ± 1.92 |
| muscle | 1.04 ± 0.38 | 1.97 ± 1.22 | 0.45 ± 0.14 | 0.63 ± 0.35 | 1.00 ± 0.80 | 0.79 ± 0.42 |
| PC-3 PIP | 6.58 ± 2.40 | 8.17 ± 4.23 | 4.96 ± 3.11 | 3.49 ± 1.21 | 4.59 ± 1.60 | 2.37 ± 1.85 |
| PC-3 flu | 4.94 ± 1.26 | 6.27 ± 4.48 | 2.77 ± 1.72 | 2.53 ± 1.10 | 3.95 ± 0.26 | 2.61 ± 0.20 |
Values shown represent mean ± SD (n = 4).
Four mice with similarly paired tumors, as described for the biodistribution, were injected with ∼31 MBq (838 μCi) of either 111In-TNP or 111In-UNP and were serially imaged. Of note, the injected dose of particles in SPECT imaging studies was five-times higher than the dose used in the biodistribution studies. The mice were imaged by SPECT-CT at 48, 72, and 96 h postinjection. The mice were scanned together as a pair side-by-side of those receiving 111In-TNP or 111In-UNP for comparison. Representative images are shown in Figure 1. The mice were staggered slightly in position to align their tumors. Tumor uptake by TNP was higher across all three scan times relative to the UNP. The SPECT-CT images suggested that nanoparticle uptake in both PSMA(+) PC-3 PIP and PSMA(−) PC-3 flu tumors was largely confined to the outer rim, which is well perfused and densely populated by macrophages.28,29 As noted in the biodistribution studies, both PSMA(+) PC-3 PIP and PSMA(−) PC-3 flu sequester TNP and the vascularity of the tumor may be a positive factor in this uptake. Low tumor uptake by UNP in Figure 1 demonstrated, in this particular pair of mice, the heterogeneous nature of particle uptake, which is also reflected in the Table 1 tumor SD values.
Figure 1.
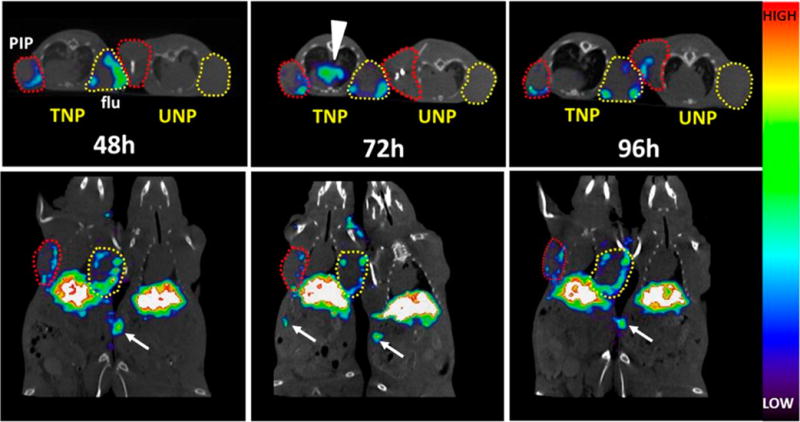
Longitudinal in vivo SPECT-CT of 111In-TNP and 111In-UNP in xenograft-bearing mice. Male athymic nude mice bearing both a PC-3 PIP (PSMA(+), red circles) and PC-3 flu (PSMA(−), yellow circles) xenograft were injected with ∼31 MBq of labeled TNP or UNP and scanned side-by-side at the indicated times. Images are scaled to the same maximum. TNP uptake at 48 h shows tumor uptake in both the PSMA(+) PC-3 PIP and PSMA(−) PC-3 flu xenografts, while both tumors in the UNP mouse remain comparatively low in uptake. After 72 h, TNP is still present along the edges of both PSMA(+) PC-3 PIP and PSMA(−) PC-3 flu xenografts and mediastinal uptake is apparent, possibly within lymphatic tissue (arrowhead). UNP uptake at 72 h remains comparatively low. By 96 h, TNP uptake is still present at the edges of both tumor types, and some UNP uptake is apparent within the PSMA(+) PC-3 PIP tumor (upper panel). Distal spleen uptake is visible within the slices and is depicted by white arrows.
Four mice bearing subcutaneous xenografts as above were injected with ∼79 mg/kg of fluorescently labeled TNP (n = 2) or UNP (n = 2) to ascertain whether a pattern of localization existed for each nanoparticle type within each tumor type. Mice were first imaged in vivo at 24, 48, and 72 h postinjection (Supporting Figures S1 and S6). At 72 h postinjection, the mice were sacrificed and imaged ex vivo with the skin reflected back (Figure 2A). Near-infrared fluorescence (NIRF) imaging revealed similar gross tumor uptake among TNP and UNP for both tumor types, although heterogeneous tumor uptake was seen among different mice (Supporting Figure S1). Those results support the biodistribution and SPECT-CT findings and may reflect the heterogeneous stromal composition among individual tumors, including vascularity and phagocyte content. Because of the heterogeneous nature of tumor uptake among TNP and UNP in both radiolabeled and fluorescently labeled formats, we then undertook ex vivo fluorescence imaging of whole tumor sections, augmented with additional immunostaining. Whole-section fluorescence imaging (Figure 2B) showed similar uptake in all sections for both TNP and UNP, demonstrating uptake in both the tumor rim and focal uptake in the center. Epifluorescence micrographs (Figure 3) showed delivery of TNP (Figure 3A, red) to CD68+ tumor-associated macrophages (green) at the edges of tumor sections for both PSMA(+) PC-3 PIP and PSMA(−) PC-3 flu.
Figure 2.
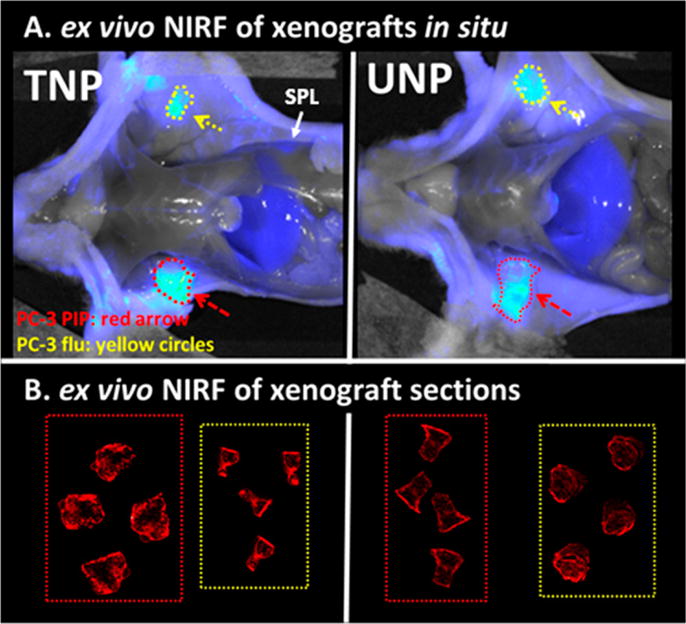
Ex vivo NIRF imaging of TNP and UNP at 72 h postinjection. Panel A depicts NIRF imaging of male athymic nude mice after sacrifice, each bearing a PSMA(+) PC-3 PIP xenograft (red dotted circles, arrow) and PSMA(−) PC-3 flu xenograft (yellow circles, arrow) 72 h after IV injection with fluorescent TNP or UNP. Both images are normalized to exposure time and show similarly luminescent tumors. Panel B shows high resolution NIRF images of 20 μm sections of the tumors in panel A. Tumor types in dotted boxes are labeled as in panel A. TNP distribution in PSMA(+) PC-3 PIP tumors is slightly more intense and is distributed throughout the tumor as compared with particle uptake in all of the other tumor sections. PSMA(−) PC-3 flu sections is also distributed throughout the tumor. All tumor section measurements in panel B were acquired simultaneously and are scaled to the same maximum.
Figure 3.
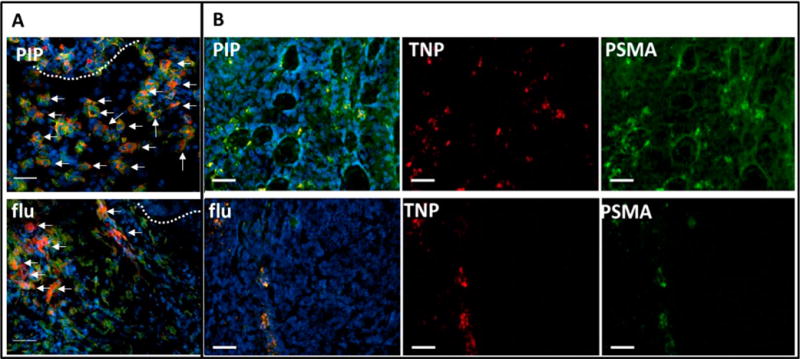
Epifluorescence micrographs of PSMA(+) PC-3 PIP (top panel) and PSMA(−) PC-3 flu (bottom panel) frozen sections containing ex vivo TNPs after 72 h of uptake. (A) Tumor rim (dotted line) of indicated xenografts showing TNP (red) accumulation (arrows) within abundant CD68+ macrophages (green). (B) Tumor parenchyma of indicated sections showing TNP (red) colocalization with anti-PSMA antibody staining (green) in PSMA(+) PC-3 PIP (top row) and within a linear pattern in PSMA(−) PC-3 flu (bottom row). Tumor sections were probed with anti-PSMA antibody (green) to define PSMA(+) tissues and Hoechst 33342 (blue) to reveal nuclei. These micrographs demonstrate delivery of TNPs to PSMA(+) tumor parenchyma (B, top row), and to PSMA(+) cells in a linear orientation, possibly within a vessel (B, bottom row). Colocalized TNP with PSMA is yellow (B, PIP and flu). Tumor sections are 20 μm thick, scale bar = 50 μm.
Figure 3B depicts TNP distribution (red) within the tumor parenchyma colocalized with PSMA(+) PC-3 PIP tumor epithelium (green). Linear colocalization between TNP and anti-PSMA antibody suggests tumor vascular binding in the PC-3 flu section. Slides costained with anti-CD68 antibody to delineate macrophages frequently show nanoparticle localization to these phagocytic cells (Figure 3A, 4) for both TNP and UNP. Intracellular delivery of TNPs and colocalization with internalized PSMA at the tubulin-rich centrosome were confirmed in cultured PSMA(+) PC-3 PIP cells (Figure 5), consistent with our prior report using a fluorescent PSMA-targeted small molecule.30 TNPs incubated with PSMA(−) PC-3 flu and UNP incubated with both PSMA(+) PC-3 PIP and PSMA(−) PC-3 flu cells exhibited a small amount of background uptake and internalization as well (Supporting Figures S2–S4).
Figure 4.
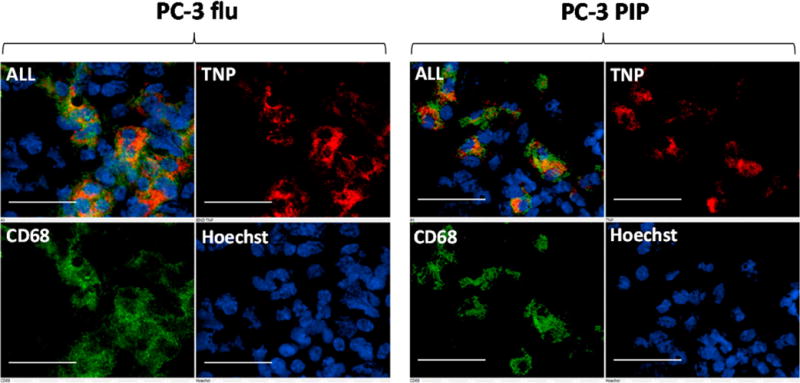
Ex vivo epifluorescence micrographs of PSMA(+) PC-3 PIP and PSMA(−) PC-3 flu frozen sections containing TNPs. Tumor sections were subsequently probed with anti-CD68 antibody to delineate macrophages and Hoechst 33342 to reveal nuclei. TNPs (red) predominantly colocalize with CD68 (green) suggesting substantial phagocytosis of TNPs by macrophages. Tumor sections are 20 μm thick, scale bar = 50 μm.
Figure 5.
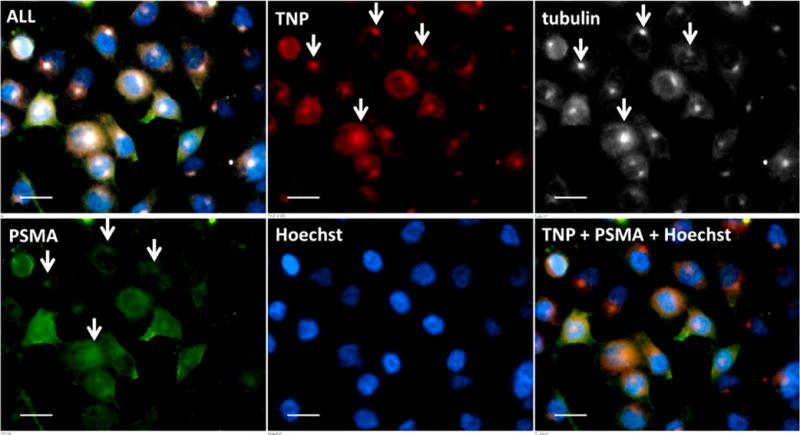
Ex vivo epifluorescence micrograph of cultured PSMA(+) PC-3 PIP cells containing TNPs. This magnification shows the intracellular distribution of TNPs (red) with PSMA (green), tubulin (white), and nuclei (blue). White arrows indicate colocalization of TNPs with endocytosed PSMA at the centrosome. Scale bar = 10 μm.
DISCUSSION
The goal of this study was to use imaging to investigate the effect in vivo of active targeting using a small-molecule PSMA-targeting moiety that has been attached to a PLA–PEG nanoparticle. We did this to assess the feasibility of developing a theranostic agent based on a targeted nanoparticle therapeutic.13 To provide fully quantitative results and corroborate cellular and intracellular location of the nanoparticles, we also performed ex vivo biodistribution and microscopy studies, respectively. The two-step, copper-assisted click chemistry procedure produced radiolabeled nanoparticles under mild conditions to ensure their stability. The nanoparticles were similarly labeled with a NIRF probe using copper-assisted click chemistry.
We evaluated two parameters in this study, namely, targeted versus untargeted nanoparticles (TNP vs UNP) and PSMA(+) and PSMA(−) PC-3 tumor xenografts that were otherwise isogenic. We observed similar but slightly higher overall tumor targeting on SPECT-CT with the TNP in the PSMA(+) PC-3 PIP tumor xenografts as compared to the PSMA(−) control tumors. The biodistribution studies showed similar relative accumulation of TNP and UNP in almost all tissues except for liver and tumor (Table 1), in keeping with the observation that irrespective of the presence of targeting agent, the pharmacokinetics of nanoparticles is dependent on particle size, shape, and surface properties.3 The targeting moiety likely serves to enhance cellular uptake when the target receptor, as in the case of PSMA, is internalized (Figure 4) and recirculated.30,31 Such internalization and recirculation have been posited to provide multiple opportunities for interaction between receptor and targeting ligand. In the biodistribution studies, which were performed to uncover more subtle differences in overall tumor targeting than could be done by imaging, we observed moderately more rapid clearance from the PSMA(+) tumors of the UNP relative to the TNP in vivo. We demonstrated that accumulation of UNP within, or, more correctly at the rim of the tumor, was less epithelium-specific than that for TNP and was dependent on a combination of EPR and, even more importantly, on phagocytosis by tumor-associated macrophages (Figures 3 and 4). Despite the single genetic difference of PSMA expression by PSMA(+) PC-3 PIP but not PSMA(−) PC-3 flu cells, we have observed that subcutaneous PSMA(+) PC-3 PIP tumor xenografts contain approximately twice the number of CD68-expressing macrophages as PSMA(−) PC-3 flu cells as well as approximately 25% more CD31+ vasculature (unpublished results), which may also account for the small positive preference of TNP accumulation within PSMA(+) PC-3 PIP tumors. We have seen nearly 400:1 target to non-target ratios using the same urea-based targeting compounds as imaging agents in the same isogenic PSMA(+) PC-3 PIP versus PSMA(−) PC-3 flu models used here.32 The comparatively modest benefit of targeting we observed with the PSMA-targeted nanoparticles appears to be a consequence of passive targeting associated with the EPR effect together with particle uptake by tumor associated macrophages. These processes occur in the absence of PSMA expression and appear to impact the uptake of TNPs and UNPs to a similar degree. We observed that tumor-associated macrophages had a substantial impact on tumor uptake of NIRF particles as determined by fluorescence microscopy. We also noted that for both TNP and UNP, the nanoparticles were primarily concentrated at the rim of both tumor types (Figures 2 and 3).
Previous studies with other targeted delivery systems have also suggested that targeting moieties do not necessary increase the total tumor accumulation but are instead responsible for increased internalization by the tumor cells.3,10,18,20–22,33–37 The question often arises as to the relevance of studies performed in experimental models, particularly in heterotopic rodent xenografts, to clinical studies. That is important in the case at hand as we and others intend to develop theranostic nanoparticles for a variety of indications, including for dosing PSMA-targeted entities, clinically.13,38–44 Because human tumors are much more heterogeneous, both in terms of blood flow and receptor expression, than carefully controlled animal tumors, there is a sense that the animal models may not be very predictive of the human situation.3 Nevertheless, the modest increase in tumor accumulation, whether due to malignant PSMA-expressing epithelium or to tumor-associated macrophages, that we saw with the TNP is consistent with the reported preliminary benefit of PSMA-targeted nanoparticles in phase I trial13,14 and phase II clinical trials.45
CONCLUSIONS
Surface functionalization of PEG–PLA particles with a urea-based PSMA-targeting moiety had a modest, positive effect on PSMA(+) tumor association relative to EPR. Nanoparticles, which were shown to internalize, were associated not only with tumor epithelium, but also associated macrophages. Radionuclide imaging in the animal studies could not definitively distinguish PSMA(+) from PSMA(−) lesions nor could TNP rather than UNP generate a visually different signal, owing to the contributions of EPR and macrophage uptake to accumulation of the nanoparticles in tumors. Importantly, however, these studies demonstrate the potential utility of imaging of radiolabeled particles for predicting the biodistribution and tumor accumulation in patients of therapeutic TNPs with similar size and surface characteristics. This approach may provide a basis for selection of patients most likely to respond to treatment.
Supplementary Material
Acknowledgments
We thank Gilbert Green for technical assistance and Dr. Samit Chatterjee for the FACS analysis.
Funding
We would like to thank NIH NCI K25 CA148901, CA134675, CA151838, CA184228, CA183031, CA058236, and BIND Therapeutics for financial support.
ABBREVIATIONS
- PLA–PEG
poly(lactic acid)–polyethylene glycol copolymer
- PSMA
prostate-specific membrane antigen
- SPECT
single photon emission computed tomography
- DOTA
1,4,7,10-tetraazacyclododecane-N,N′,N″,N‴-tetraacetic acid
- TNP
targeted nanoparticle
- UNP
untargeted nanoparticle
- SPECT
single-photon emission computed tomography
- NIR
near-infrared
Footnotes
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.biomac.6b01485.
Reaction scheme to synthesize DOTA-PEG-alkyne; In-DOTA-PEG-alkyne associated HPLC chromatograms; LC–MS spectra; specific activity calculation of nanoparticles; in vivo and ex vivo NIRF imaging in mouse model; FACS analysis of PSMA expression; epifluorescence micrographs in cultured PSMA(+) PC3 PIP and PSMA(−) PC3 PIP cells (PDF)
ORCID
Martin G. Pomper: 0000-0001-6753-3010
Author Contributions
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
Notes
The authors declare no competing financial interest.
References
- 1.Davis ME, Chen ZG, Shin DM. Nanoparticle therapeutics: an emerging treatment modality for cancer. Nat Rev Drug Discovery. 2008;7(9):771–782. doi: 10.1038/nrd2614. [DOI] [PubMed] [Google Scholar]
- 2.Chen F, Ehlerding EB, Cai W. Theranostic nanoparticles. J Nucl Med. 2014;55(12):1919–1922. doi: 10.2967/jnumed.114.146019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Torchilin VP. Multifunctional, stimuli-sensitive nanoparticulate systems for drug delivery. Nat Rev Drug Discovery. 2014;13(11):813–827. doi: 10.1038/nrd4333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Iyer AK, Singh A, Ganta S, Amiji MM. Role of integrated cancer nanomedicine in overcoming drug resistance. Adv Drug Delivery Rev. 2013;65(13–14):1784–1802. doi: 10.1016/j.addr.2013.07.012. [DOI] [PubMed] [Google Scholar]
- 5.Beech JR, Shin SJ, Smith JA, Kelly KA. Mechanisms for targeted delivery of nanoparticles in cancer. Curr Pharm Des. 2013;19(37):6560–6574. doi: 10.2174/1381612811319370002. [DOI] [PubMed] [Google Scholar]
- 6.Kijanka M, Dorresteijn B, Oliveira S, van Bergen en Henegouwen PM. Nanobody-based cancer therapy of solid tumors. Nanomedicine. 2015;10(1):161–174. doi: 10.2217/nnm.14.178. [DOI] [PubMed] [Google Scholar]
- 7.Kaittanis C, Shaffer TM, Thorek DL, Grimm J. Dawn of advanced molecular medicine: nanotechnological advancements in cancer imaging and therapy. Crit Rev Oncog. 2014;19(3–4):143–176. doi: 10.1615/critrevoncog.2014011601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheng Z, Al Zaki A, Hui JZ, Muzykantov VR, Tsourkas A. Multifunctional nanoparticles: cost versus benefit of adding targeting and imaging capabilities. Science. 2012;338(6109):903–910. doi: 10.1126/science.1226338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zamboni WC, Torchilin V, Patri AK, Hrkach J, Stern S, Lee R, Nel A, Panaro NJ, Grodzinski P. Best practices in cancer nanotechnology: perspective from NCI nanotechnology alliance. Clin Cancer Res. 2012;18(12):3229–3241. doi: 10.1158/1078-0432.CCR-11-2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bertrand N, Wu J, Xu X, Kamaly N, Farokhzad OC. Cancer nanotechnology: the impact of passive and active targeting in the era of modern cancer biology. Adv Drug Delivery Rev. 2014;66:2–25. doi: 10.1016/j.addr.2013.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Danhier F, Ansorena E, Silva JM, Coco R, Le Breton A, Preat V. PLGA-based nanoparticles: an overview of biomedical applications. J Controlled Release. 2012;161(2):505–522. doi: 10.1016/j.jconrel.2012.01.043. [DOI] [PubMed] [Google Scholar]
- 12.Delehanty JB, Breger JC, Gemmill KB, Stewart MH, Medintz IL. Controlling the actuation of therapeutic nanomaterials: enabling nanoparticle-mediated drug delivery. Ther Delivery. 2013;4(11):1411–1429. doi: 10.4155/tde.13.110. [DOI] [PubMed] [Google Scholar]
- 13.Hrkach J, Von Hoff D, Ali MM, Andrianova E, Auer J, Campbell T, De Witt D, Figa M, Figueiredo M, Horhota A, Low S, McDonnell K, Peeke E, Retnarajan B, Sabnis A, Schnipper E, Song JJ, Song YH, Summa J, Tompsett D, Troiano G, Van Geen Hoven T, Wright J, LoRusso P, Kantoff PW, Bander NH, Sweeney C, Farokhzad OC, Langer R, Zale S. Preclinical development and clinical translation of a PSMA-targeted docetaxel nanoparticle with a differentiated pharmacological profile. Sci Transl Med. 2012;4(128):128ra39. doi: 10.1126/scitranslmed.3003651. [DOI] [PubMed] [Google Scholar]
- 14.Von Hoff DD, Mita MM, Ramanathan RK, Weiss GJ, Mita AC, LoRusso PM, Burris HA, Hart LL, Low SC, Parsons DM, Zale SE, Summa JM, Youssoufian H, Sachdev JC. Phase I Study of PSMA-Targeted Docetaxel-Containing Nanoparticle BIND-014 in Patients with Advanced Solid Tumors. Clin Cancer Res. 2016;22(13):3157–3163. doi: 10.1158/1078-0432.CCR-15-2548. [DOI] [PubMed] [Google Scholar]
- 15.Jain RK, Stylianopoulos T. Delivering nanomedicine to solid tumors. Nat Rev Clin Oncol. 2010;7(11):653–664. doi: 10.1038/nrclinonc.2010.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nakamura Y, Mochida A, Choyke PL, Kobayashi H. Nanodrug Delivery: Is the Enhanced Permeability and Retention Effect Sufficient for Curing Cancer? Bioconjugate Chem. 2016;27(10):2225–2238. doi: 10.1021/acs.bioconjchem.6b00437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bhattacharyya S, Bhattacharya R, Curley S, McNiven MA, Mukherjee P. Nanoconjugation modulates the trafficking and mechanism of antibody induced receptor endocytosis. Proc Natl Acad Sci U S A. 2010;107(33):14541–14546. doi: 10.1073/pnas.1006507107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Choi CH, Alabi CA, Webster P, Davis ME. Mechanism of active targeting in solid tumors with transferrin-containing gold nanoparticles. Proc Natl Acad Sci U S A. 2010;107(3):1235–1240. doi: 10.1073/pnas.0914140107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang J, Tian S, Petros RA, Napier ME, Desimone JM. The complex role of multivalency in nanoparticles targeting the transferrin receptor for cancer therapies. J Am Chem Soc. 2010;132(32):11306–11313. doi: 10.1021/ja1043177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bartlett DW, Su H, Hildebrandt IJ, Weber WA, Davis ME. Impact of tumor-specific targeting on the biodistribution and efficacy of siRNA nanoparticles measured by multimodality in vivo imaging. Proc Natl Acad Sci U S A. 2007;104(39):15549–15554. doi: 10.1073/pnas.0707461104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kirpotin DB, Drummond DC, Shao Y, Shalaby MR, Hong K, Nielsen UB, Marks JD, Benz CC, Park JW. Antibody targeting of long-circulating lipidic nanoparticles does not increase tumor localization but does increase internalization in animal models. Cancer Res. 2006;66(13):6732–6740. doi: 10.1158/0008-5472.CAN-05-4199. [DOI] [PubMed] [Google Scholar]
- 22.Davis ME, Zuckerman JE, Choi CH, Seligson D, Tolcher A, Alabi CA, Yen Y, Heidel JD, Ribas A. Evidence of RNAi in humans from systemically administered siRNA via targeted nanoparticles. Nature. 2010;464(7291):1067–1070. doi: 10.1038/nature08956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen Z, Penet MF, Nimmagadda S, Li C, Banerjee SR, Winnard PT, Jr, Artemov D, Glunde K, Pomper MG, Bhujwalla ZM. PSMA-targeted theranostic nanoplex for prostate cancer therapy. ACS Nano. 2012;6(9):7752–7762. doi: 10.1021/nn301725w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Behnam Azad B, Banerjee SR, Pullambhatla M, Lacerda S, Foss CA, Wang Y, Ivkov R, Pomper MG. Evaluation of a PSMA-targeted BNF nanoparticle construct. Nanoscale. 2015;7(10):4432–4442. doi: 10.1039/c4nr06069e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen Z, Penet MF, Krishnamachary B, Banerjee SR, Pomper MG, Bhujwalla ZM. PSMA-specific theranostic nanoplex for combination of TRAIL gene and 5-FC prodrug therapy of prostate cancer. Biomaterials. 2016;80:57–67. doi: 10.1016/j.biomaterials.2015.11.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhu C, Bandekar A, Sempkowski M, Banerjee SR, Pomper MG, Bruchertseifer F, Morgenstern A, Sofou S. Nanoconjugation of PSMA-Targeting Ligands Enhances Perinuclear Localization and Improves Efficacy of Delivered Alpha-Particle Emitters against Tumor Endothelial Analogues. Mol Cancer Ther. 2016;15(1):106–113. doi: 10.1158/1535-7163.MCT-15-0207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Banerjee SR, Pullambhatla M, Byun Y, Nimmagadda S, Foss CA, Green G, Fox JJ, Lupold SE, Mease RC, Pomper MG. Sequential SPECT and Optical Imaging of Experimental Models of Prostate Cancer with a Dual Modality Inhibitor of the Prostate-Specific Membrane Antigen. Angew Chem, Int Ed. 2011;50(39):9167–9170. doi: 10.1002/anie.201102872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bingle L, Brown NJ, Lewis CE. The role of tumour-associated macrophages in tumour progression: implications for new anticancer therapies. J Pathol. 2002;196(3):254–265. doi: 10.1002/path.1027. [DOI] [PubMed] [Google Scholar]
- 29.Rahat MA, Hemmerlein B. Macrophage-tumor cell interactions regulate the function of nitric oxide. Front Physiol. 2013;4:144. doi: 10.3389/fphys.2013.00144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kiess AP, Minn I, Chen Y, Hobbs R, Sgouros G, Mease RC, Pullambhatla M, Shen CJ, Foss CA, Pomper MG. Auger Radiopharmaceutical Therapy Targeting Prostate-Specific Membrane Antigen. J Nucl Med. 2015;56(9):1401–1407. doi: 10.2967/jnumed.115.155929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Banerjee SR, Ngen EJ, Rotz MW, Kakkad S, Lisok A, Pracitto R, Pullambhatla M, Chen Z, Shah T, Artemov D, Meade TJ, Bhujwalla ZM, Pomper MG. Synthesis and Evaluation of GdIII-Based Magnetic Resonance Contrast Agents for Molecular Imaging of Prostate-Specific Membrane Antigen. Angew Chem, Int Ed. 2015;54(37):10778–10782. doi: 10.1002/anie.201503417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen Y, Pullambhatla M, Foss CA, Byun Y, Nimmagadda S, Senthamizhchelvan S, Sgouros G, Mease RC, Pomper MG. 2-(3-{1-Carboxy-5-[(6-[18F]fluoro-pyridine-3-carbonyl)-amino]-pentyl}-ureido)-pen tanedioic acid, [18F]DCFPyL, a PSMA-based PET imaging agent for prostate cancer. Clin Cancer Res. 2011;17(24):7645–7653. doi: 10.1158/1078-0432.CCR-11-1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maeda N, Miyazawa S, Shimizu K, Asai T, Yonezawa S, Kitazawa S, Namba Y, Tsukada H, Oku N. Enhancement of anticancer activity in antineovascular therapy is based on the intratumoral distribution of the active targeting carrier for anticancer drugs. Biol Pharm Bull. 2006;29(9):1936–1940. doi: 10.1248/bpb.29.1936. [DOI] [PubMed] [Google Scholar]
- 34.Kamaly N, Xiao Z, Valencia PM, Radovic-Moreno AF, Farokhzad OC. Targeted polymeric therapeutic nanoparticles: design, development and clinical translation. Chem Soc Rev. 2012;41(7):2971–3010. doi: 10.1039/c2cs15344k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fuchs AV, Tse BW, Pearce AK, Yeh MC, Fletcher L, Huang SS, Heston WD, Whittaker AK, Russell PJ, Thurecht KJ. Evaluation of Polymeric Nanomedicines Targeted to PSMA: Effect of Ligand on Targeting Efficiency. Biomacromolecules. 2015;16(10):3235–3247. doi: 10.1021/acs.biomac.5b00913. [DOI] [PubMed] [Google Scholar]
- 36.Salvati A, Pitek AS, Monopoli MP, Prapainop K, Bombelli FB, Hristov DR, Kelly PM, Aberg C, Mahon E, Dawson KA. Transferrin-functionalized nanoparticles lose their targeting capabilities when a biomolecule corona adsorbs on the surface. Nat Nanotechnol. 2013;8(2):137–143. doi: 10.1038/nnano.2012.237. [DOI] [PubMed] [Google Scholar]
- 37.Sykes EA, Chen J, Zheng G, Chan WCW. Investigating the Impact of Nanoparticle Size on Active and Passive Tumor Targeting Efficiency. ACS Nano. 2014;8(6):5696–5706. doi: 10.1021/nn500299p. [DOI] [PubMed] [Google Scholar]
- 38.Chandran SS, Banerjee SR, Mease RC, Pomper MG, Denmeade SR. Characterization of a targeted nanoparticle functionalized with a urea-based inhibitor of prostate-specific membrane antigen (PSMA) Cancer Biol Ther. 2008;7(6):974–982. doi: 10.4161/cbt.7.6.5968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dhar S, Kolishetti N, Lippard SJ, Farokhzad OC. Targeted delivery of a cisplatin prodrug for safer and more effective prostate cancer therapy in vivo. Proc Natl Acad Sci U S A. 2011;108(5):1850–1855. doi: 10.1073/pnas.1011379108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Abdolahi M, Shahbazi-Gahrouei D, Laurent S, Sermeus C, Firozian F, Allen BJ, Boutry S, Muller RN. Synthesis and in vitro evaluation of MR molecular imaging probes using J591 mAb-conjugated SPIONs for specific detection of prostate cancer. Contrast Media Mol Imaging. 2013;8(2):175–184. doi: 10.1002/cmmi.1514. [DOI] [PubMed] [Google Scholar]
- 41.Kasten BB, Liu T, Nedrow-Byers JR, Benny PD, Berkman CE. Targeting prostate cancer cells with PSMA inhibitor-guided gold nanoparticles. Bioorg Med Chem Lett. 2013;23(2):565–568. doi: 10.1016/j.bmcl.2012.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Valencia PM, Pridgen EM, Perea B, Gadde S, Sweeney C, Kantoff PW, Bander NH, Lippard SJ, Langer R, Karnik R, Farokhzad OC. Synergistic cytotoxicity of irinotecan and cisplatin in dual-drug targeted polymeric nanoparticles. Nanomedicine. 2013;8(5):687–698. doi: 10.2217/nnm.12.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huang B, Otis J, Joice M, Kotlyar A, Thomas TP. PSMA-targeted stably linked “dendrimer-glutamate urea-methotrexate” as a prostate cancer therapeutic. Biomacromolecules. 2014;15(3):915–923. doi: 10.1021/bm401777w. [DOI] [PubMed] [Google Scholar]
- 44.Tse BW, Cowin GJ, Soekmadji C, Jovanovic L, Vasireddy RS, Ling MT, Khatri A, Liu T, Thierry B, Russell PJ. PSMA-targeting iron oxide magnetic nanoparticles enhance MRI of preclinical prostate cancer. Nanomedicine (London, U K) 2015;10(3):375–386. doi: 10.2217/nnm.14.122. [DOI] [PubMed] [Google Scholar]
- 45.Autio KA, Garcia JA, Alva AS, Hart LL, Milowsky MI, Posadas EM, Ryan CJ, Summa JM, Youssoufian H, H I S, R D. A phase 2 study of BIND-014 (PSMA-targeted docetaxel nanoparticle) administered to patients with chemotherapy-naïve metastatic castration-resistant prostate cancer (mCRPC) J Clin Oncol. 2016;34:233. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


